Summary
Neural stem cells (NSCs) have the ability to exit quiescence and reactivate in response to physiological stimuli. In the Drosophila brain, insulin receptor (InR)/phosphatidylinositol 3-kinase (PI3K)/Akt pathway triggers NSC reactivation. However, intrinsic mechanisms that control the InR/PI3K/Akt pathway during reactivation remain unknown. Here, we have identified heat shock protein 83 (Hsp83/Hsp90), a molecular chaperone, as an intrinsic regulator of NSC reactivation. Hsp83 is both necessary and sufficient for NSC reactivation by promoting the activation of InR pathway in larval brains in the presence of dietary amino acids. Both Hsp83 and its co-chaperone Cdc37 physically associate with InR. Finally, reactivation defects observed in brains depleted of hsp83 were rescued by over-activation of the InR/PI3K/Akt pathway, suggesting that Hsp83 functions upstream of the InR/PI3K/Akt pathway during NSC reactivation. Given the conservation of Hsp83 and the InR pathway, our finding may provide insights into the molecular mechanisms underlying mammalian NSC reactivation.
Keywords: neural stem cells, Drosophila, reactivation, quiescence, chaperone
Graphical Abstract
Highlights
-
•
Hsp83/Hsp90 and its co-chaperone Cdc37 are required for NSC reactivation
-
•
Hsp83 overexpression results in premature NSC reactivation on fed condition
-
•
Hsp83 and Cdc37 physically associate with InR
-
•
Hsp83 and Cdc37 are required for the activation of InR pathway in NSCs
In this article, Huang and Wang show that heat shock protein 83 (Hsp83/Hsp90), a molecular chaperone, is a novel intrinsic regulator of NSC reactivation. Hsp83 is both necessary and sufficient for NSC reactivation in the presence of nutrition. Both Hsp83 and its co-chaperone Cdc37 physically associate with InR and are required for activation of InR pathway during NSC reactivation.
Introduction
The balance between proliferation and quiescence of stem cells is crucial in maintaining tissue homeostasis. Neural stem cells (NSCs) in the brain have the ability to be reactivated from a reversible quiescent state to generate new neurons. In the mammalian adult brain, the majority of adult NSCs are quiescent and are not actively dividing (Doetsch et al., 1999, Morshead et al., 1994). Interestingly, these quiescent NSCs can be reactivated and participate in neurogenesis upon various extrinsic stimuli (Ahn and Joyner, 2005, Daynac et al., 2013, Daynac et al., 2016, Faiz et al., 2015, Kawai et al., 2017, Lugert et al., 2010, Wang et al., 2011). Dysregulation of the balance between proliferation and quiescence of NSCs may contribute to neurodevelopmental disorders such as microcephaly (Baser et al., 2017, Cloetta et al., 2013). Reactivating quiescent NSCs may also provide therapeutic strategies for the treatment of brain injuries or neurodegenerative disorders.
Recently, Drosophila NSCs, neuroblasts, have emerged as an excellent in vivo model for the study of NSC quiescence and reactivation. NSCs exit the cell cycle, shrink in size, and enter quiescence at the end of embryogenesis under the control of the spatial Hox protein, temporal identity factors, and a homeodomain differentiation factor Prospero (Isshiki et al., 2001, Lai and Doe, 2014, Tsuji et al., 2008). At early larval stages, following 24 hr of quiescence, they are reactivated in response to feeding (Britton and Edgar, 1998, Ito and Hotta, 1992, Truman and Bate, 1988). Dietary amino acids present in the food are sensed by the fat body, which functions equivalent to the mammalian liver and adipose tissue (Colombani et al., 2003). The fat body generates mitogens, stimulating blood-brain barrier glial cells to secrete insulin-like peptides (dILPs). dILPs act locally by activating the insulin receptor (InR)/phosphatidylinositol 3-kinase (PI3K)/Akt pathway in underlying NSCs and promoting their reactivation (Britton and Edgar, 1998, Chell and Brand, 2010, Sousa-Nunes et al., 2011). Blood-brain barrier glial cells synchronize NSC reactivation via calcium oscillations through gap junctions (Speder and Brand, 2014), while cortex glial cells remodel to promote new-born neuron survival (Speder and Brand, 2018). Besides InR pathway, the spindle matrix complex intrinsically promotes NSC reactivation (Li et al., 2017), while the Hippo pathway maintains NSC quiescence (Ding et al., 2016, Poon et al., 2016). In mammalian brains, insulin-like growth factor-1 (IGF-1) produced by astroglial cells has a similar role in promoting NSC proliferation in response to brain injury (Mairet-Coello et al., 2009, Yan et al., 2006, Ye et al., 2004). In addition, IGF-1 receptor (IGF-1R) promotes the proliferation of mammalian NSCs (Arsenijevic et al., 2001), and human IGF-1R mutations are associated with microcephaly (Juanes et al., 2015), suggesting that the insulin pathway is likely conserved from flies to humans in promoting NSC proliferation. However, how the insulin pathway is regulated during NSC reactivation is still poorly understood.
Here, we show that a highly conserved chaperone, Hsp83/Hsp90, is required for the activation of the InR/PI3K/Akt pathway during NSC reactivation. Hsp83 functions in a wide variety of biological processes, including cell polarity, DNA transcription, and chromatin remodeling (Andersen et al., 2012, Pratt and Toft, 1997, Sawarkar et al., 2012, Tariq et al., 2009). Unlike Hsp70, eukaryotic Hsp83/Hsp90 does not act in nascent protein folding, rather, it binds to substrate proteins that are in a near native state at a late stage of folding (Young et al., 2001). We show that Hsp83 is necessary and sufficient for NSC reactivation in the presence of dietary amino acids. Its co-chaperones Cdc37 and Hsp70 mediate the association between Hsp83 and its client protein kinases and steroid hormone receptors, respectively (Eckl and Richter, 2013, Young et al., 2001). We show that the activation of the InR/PI3K/Akt pathway is dependent on Hsp83 and Cdc37 function in larval brains. Hsp83 and Cdc37 physically associate with InR, but not Akt. Finally, the hsp83 knockdown phenotype is rescued by over-activation of the InR/PI3K/Akt pathway.
Results
Hsp83 Is Required Intrinsically for NSC Reactivation
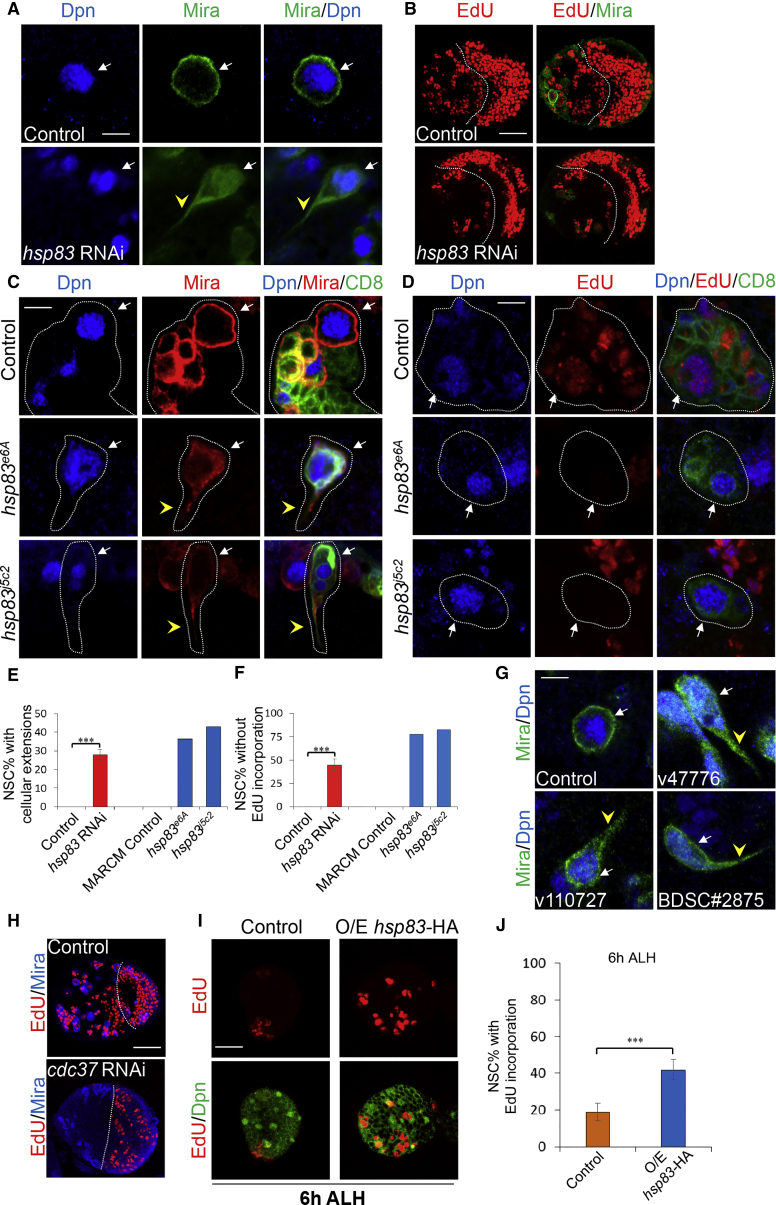
To identify regulators of NSC reactivation, we used an NSC-specific driver insc-GAL4 to perform an unbiased RNAi screening in NSCs. Our screen identified an RNAi line (VDRC no. 108568), which knocks down hsp83 expression, exhibiting defective NSC reactivation, with cellular extensions, a hallmark of quiescent NSCs, observed in 27.7% ± 3.0% (Figures 1A and 1E; n = 1,452, 41 brain lobes) of NSCs at 72 hr after larval hatching (ALH) at 29°C. In comparison, all control NSCs were reactivated and lost their cellular extensions at the same stage (Figures 1A and 1E; n = 1,084, 20 brain lobes). This finding suggests that these NSCs failed to be reactivated and remained in a quiescent state upon hsp83 knockdown. Next, in order to examine if NSCs with hsp83 knockdown are able to enter the cell cycle, we carried out 5-ethynyl-2′-deoxyuridine (EdU) incorporation experiments. All control NSCs (Figures 1B and 1F; n = 756, 20 brain lobes) incorporated EdU at 72 hr ALH at 29°C. By contrast, at the same condition, 44.7% ± 6.3% (n = 824, 24 brain lobes) of NSCs failed to incorporate EdU with hsp83 knockdown (Figures 1B and 1F). Next, we examined the phenotype of an amorphic hsp83j5C2 and hypomorphic hsp83e6A in mosaic analysis with a repressible cell marker (MARCM) clones (Lee and Luo, 2001). While all NSCs in wild-type control clones exhibited normal round shapes without any cellular extensions (n = 58), 43.0% (n = 114) of hsp83j5C2 NSC clones and 36.4% (n = 266) of hsp83e6A NSC clones contained NSCs with cellular extensions (Figures 1C and 1E). These observations further show that NSCs remain quiescent after loss of hsp83. To examine the proliferative status of the NSCs, we performed EdU pulse-chase analysis. At 96 hr ALH, all NSCs in wild-type control clones were actively dividing evident by EdU incorporation (n = 101). Unlike the wild-type control clones, 82.8% (n = 82) and 77.9% (n = 87) of NSC clones with hsp83j5C2 and hsp83e6A, respectively, failed to incorporate EdU (Figures 1D and 1F). Furthermore, the NSC quiescence phenotype of hsp83 mutants were fully rescued by introducing a 21.8-kb bacterial artificial chromosome (BAC) CH322-129N17 containing hsp83 (hsp83-BAC). A total of 39.7% (n = 126) and 36.4% (n = 107) NSCs in hsp83j5C2 and hsp83e6A MARCM clones showed cellular extensions (Figure S1A). However, when hsp83-BAC was expressed, none of NSCs in hsp83j5C2 (n = 117) and hsp83e6A (n = 107) MARCM clones exhibited cellular extensions (Figure S1A). Similarly, 78.3% (n = 46) and 70.8% (n = 72) NSCs in hsp83j5C2 and hsp83e6A MARCM clones failed to incorporate with EdU (Figure S1A), while all NSCs from hsp83j5C2 with hsp83-BAC (n = 95) and hsp83e6A with hsp83-BAC (n = 70) were incorporated with EdU (Figure S1A). Taken together, these results indicate that Hsp83 is required for NSC reactivation.
Figure 1.
Hsp83 and Its Co-chaperone Cdc37 Are Required for NSC Reactivation
(A) Larval NSCs of control (UAS-dicer2) and hsp83 RNAi (VDRC no. 108568) induced with insc-Gal4 at 72 hr ALH were stained for Deadpan (Dpn) and Miranda (Mira).
(B) Larval brains of control (UAS-dicer2) and hsp83 RNAi (VDRC no. 108568) induced with insc-Gal4 at 72 hr ALH were labeled with EdU and Mira.
(C) Larval NSC clones of wild-type control, hsp83e6A and hsp83j5C2 at 96 hr ALH were stained for GFP, Dpn, and Mira. NSC lineages were marked with CD8-GFP.
(D) Larval NSC clones of wild-type control, hsp83e6A, and hsp83j5C2 at 96 hr ALH were labeled with GFP, Dpn, and EdU. NSC lineages were marked with CD8-GFP.
(E and F) Quantifications of cellular extensions (E) and EdU incorporation (F) for control, hsp83 RNAi, MARCM control clone, hsp83e6A, and hsp83j5C2 MARCM clones. Data are presented as mean ± SD for RNAi knockdown.
(G) Larval NSCs of control (UAS-dicer2) and cdc37 RNAi (VDRC no. 47776/v47776, v110727 and BDSC no. 28756; induced with insc-Gal4 at 72 hr ALH were stained for Dpn and Mira).
(H) Larval brains of control (UAS-dicer2) and cdc37 RNAi (v47776) induced with insc-Gal4 at 72 hr ALH were labeled with EdU and Mira.
(I and J) Larval brains of control (UAS-dicer2) and UAS-hsp83-HA under the control of insc-Gal4 at 6 hr ALH were labeled with EdU and Dpn. Data are presented in (J) as mean ± SD.
White arrows point to NSCs and yellow arrowheads point to cellular extensions of NSCs (A, C, D, and G). Central brain is to the left of white dotted lines (B and H). Clone outline is indicated by white dotted lines (C and D). Statistical analyses were done comparing two different genotypes using a two-tailed Student's t test (E, F, and J). ∗∗∗p < 0.001. Scale bars, 5 μm in (A), (C), (D), and (G), 30 μm in (B) and (H), and 15 μm in (I).
We carried out a time course experiment and examined larval brains at 24, 48, and 72 hr ALH. At 24 hr ALH, most control NSCs were reactivated, with only 4.9% ± 1.0% (n = 2,174, 37 brain lobes) showing cellular extensions, suggesting that they were still in quiescence (Figures S1C and S1E). With hsp83 knockdown under the insc-Gal4 driver, 15.4% ± 1.3% (n = 1,535, 28 brain lobes) of NSCs possess cellular extensions at 24 hr ALH (Figures S1C and S1E). Loss of cellular extensions of NSCs seemed to precede cell-cycle re-entry. At 24 hr ALH, 19.5% ± 3.8% (n = 1,205, 22 brain lobes) of control NSCs failed to incorporate EdU (Figures S1B and S1D), while 63.1% ± 5.0% (n = 1,193, 24 brain lobes) NSCs with hsp83 knockdown were EdU-negative (Figures S1B and S1D). This observation suggests that NSC reactivation is defective when hsp83 is knocked down at early larval stages. At 48 hr ALH, all control NSCs showed round cell morphology without cellular extension (Figures S1C and S1E; n = 1,625, 30 brain lobes) and nearly all of the NSCs incorporated EdU (Figures S1B and S1D; 98.2% ± 1.4%, n = 704, 21 brain lobes). In comparison, with hsp83 knockdown, 27.2% ± 2.9% (n = 1,254, 20 brain lobes) of NSCs still showed cellular extensions (Figures S1C and S1E), while 48.7% ± 6.2% (n = 526, 13 brain lobes) of NSCs failed to incorporate EdU (Figures S1B and S1D). At 72 hr ALH, all control NSCs had no cellular extensions (Figures S1C and S1E; n = 1,221, 13 brain lobes) and incorporated EdU (Figures S1B and S1D; n = 322, 6 brain lobes). By contrast, with hsp83 knockdown 28.2% ± 2.1% (n = 1,003, 11 brain lobes) of NSCs showed cellular extensions (Figures S1C and S1E) and 49.5% ± 7.4% (n = 290, 5 brain lobes) of NSCs lacked EdU incorporation (Figures S1B and S1D). Taken together, Hsp83 is intrinsically required for NSC reactivation.
Upon hsp83 knockdown at 72 hr ALH, the sizes of NSCs are variable, ranging from 4 to 11 μm, even when they still maintained cellular extensions, a hallmark of quiescent NSCs. At 24 hr ALH, the diameter of control NSCs was 8.7 ± 0.6 μm (n = 121), while NSC diameters were decreased to 6.8 ± 1.3 μm (n = 129) upon hsp83 knockdown under insc-Gal4 (data not shown). At 72 hr ALH, the average NSC diameter upon hsp83 RNAi knockdown is 8.1 ± 1.8 μm (n = 124), still smaller than control NSCs (10.6 ± 0.7 μm; data not shown). Similarly, at 72 hr ALH the NSC diameters of hsp83e6A and hsp83j5c2 were 8.1 ± 1.7 μm (n = 121) and 8.2 ± 1.6 μm (n = 118), respectively. These observations suggest that, although NSC reactivation is dramatically disrupted upon knocking down hsp83, cell growth is incompletely blocked, resulting in enlargement of some of the quiescent NSCs. Thus, cell division and cell growth seemed to be partially uncoupled upon failure of NSC reactivation in NSCs upon hsp83 depletion. Similar uncoupling of cell division and growth were also reported in other mutants defective in NSC reactivation including chromator, which appears to function downstream of the InR signaling pathway (Li et al., 2017).
Mushroom body (MB) neuroblasts do not enter quiescence at the end stage of embryogenesis and are capable of dividing in the absence of dietary amino acids throughout larval stages (Ito and Hotta, 1992, Sousa-Nunes et al., 2011, Truman and Bate, 1988). We investigated whether Hsp83 was required for general proliferation of cells, such as MB neuroblasts. At 24 hr ALH, all MB neuroblasts (Dpn+), which were surrounded by Dachshund-positive MB neurons, from both control (n = 224) and hsp83 RNAi (n = 200), were incorporated with EdU (Figure S1F). This result suggests that proliferation of MB neuroblasts is unaffected by hsp83 RNAi knockdown.
Co-chaperone Cdc37 Is Required for NSC Reactivation
Functioning as a chaperone, Hsp83 has two major classes of clients, steroid hormone receptors and protein kinases, and requires co-chaperones heat shock protein 70 (Hsp70) or cell division cycle 37 (Cdc37), respectively (Eckl and Richter, 2013). To investigate whether steroid hormone receptors or protein kinases are the potential clients of Hsp83 during NSC reactivation, we tested whether Hsp70 or Cdc37 plays any role during NSC reactivation. There are seven isoforms of hsp70 in Drosophila and we acquired 14 RNAi stocks for these isoforms (Table S1) and knocked down these genes in NSCs using insc-Gal4. However, knocking down these isoforms of hsp70 or Hsp70/Hsp90 organizing protein (hop), which coordinates the interaction between Hsp70 and Hsp83 by binding to both of them, did not result in any NSC reactivation defect at both 24 and 72 hr ALH (Table S1 and data not shown). By contrast, at 72 hr ALH at 29°C, in cdc37 RNAi knockdown (VDRC no. 47776) brains, 16.8% ± 2.2% (n = 947, 11 brain lobes) of NSCs showed cellular processes (Figure 1G), and only 1.5% ± 1.4% (n = 1,135, 14 brain lobes) of NSCs were incorporated with EdU (Figure 1H). Similarly, in two other cdc37 RNAi lines (VDRC no. 110727 and BDSC no. 28756), 16.1% ± 1.2% (n = 1,352, 18 brain lobes) and 19.7% ± 3.3% (n = 1,812, 28 brain lobes) of NSCs showed cellular extensions, respectively (Figure 1G). These observations suggest that Hsp83 likely functions together with its co-chaperone Cdc37 during NSC reactivation. cdc37e4D is an amorphic allele in which almost the entire coding region of cdc37 is deleted (Cutforth and Rubin, 1994). In wild-type NSC MARCM clones, all NSCs were devoid of cellular extensions at 96 hr ALH (Figures S1G and S1H; n = 50). By contrast, in cdc37e4D MARCM clones, 28.8% (n = 198) of NSCs extend cellular processes (Figures S1G and S1H), suggesting that Cdc37 is required for NSC activation. The process phenotype of cdc37e4D was fully rescued by introducing a 22.1-kb BAC CH322-35F18 containing cdc37 (cdc37-BAC). A total of 25.9% (n = 85) NSCs in cdc37e4D MARCM clones showed cellular extensions, while none of the NSCs (n = 71) from cdc37e4D with cdc37-BAC showed cellular extensions (Figures S1G and S1H). Therefore, we conclude that Cdc37 is required for NSC reactivation.
Overexpression of Hsp83 Leads to Premature NSC Reactivation
To test whether Hsp83 overexpression is sufficient to trigger NSC reactivation, we overexpressed hsp83-hemagglutinin (HA) (Kuo et al., 2013) in NSCs using insc-Gal4 and examined its effect on NSC proliferation at 6 hr ALH. Hsp83-HA is functional, as it largely rescued the NSC quiescence phenotype in hsp83j5C2 and hsp83e6A mutants (data not shown). At 6 hr ALH, most wild-type control NSCs remained in quiescence, as only 18.8% ± 4.8% of NSCs (n = 1,907, 55 brain lobes) incorporated EdU (Figures 1I and 1J). By contrast, upon hsp83 overexpression, 41.9% ± 5.5% (n = 2,211, 62 brain lobes) of NSCs incorporated EdU (Figures 1I and 1J). This result suggests that Hsp83 overexpression leads to premature NSC reactivation. However, in the absence of dietary amino acids, Hsp83 overexpressing in NSCs was unable to promote NSC reactivation (data not shown), suggesting that Hsp83 overexpression effect is nutrition dependent. Taken together, Hsp83 is both necessary and sufficient for NSC reactivation in the presence of dietary amino acids.
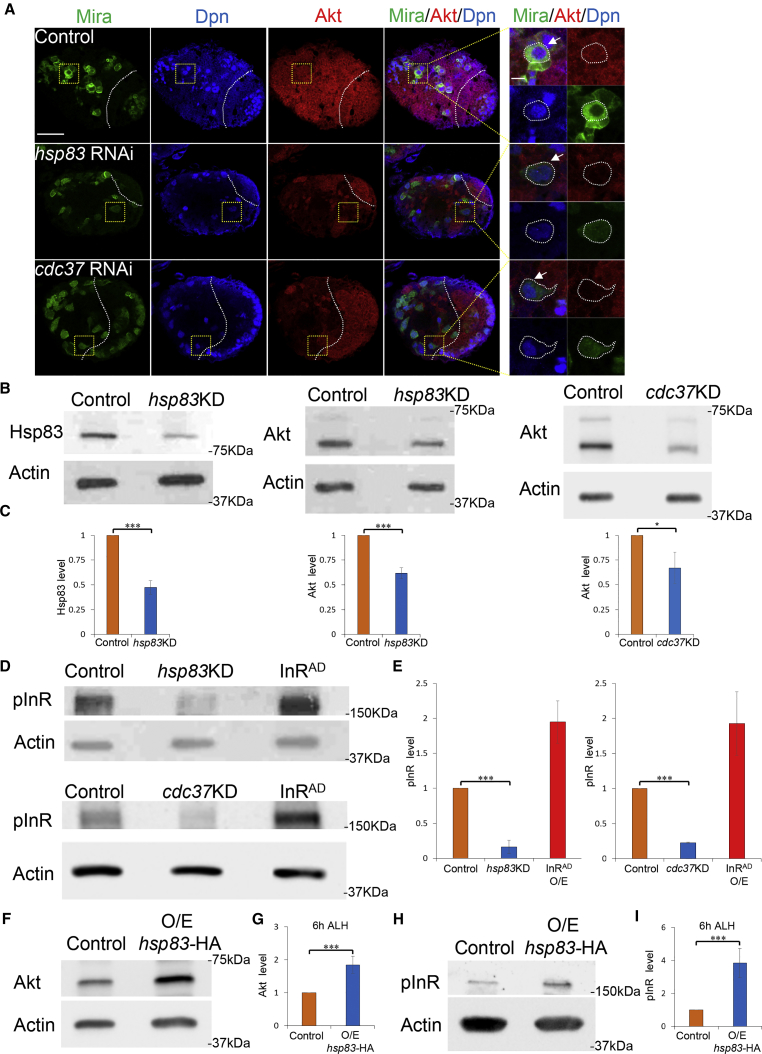
pInR and Akt Levels Are Decreased upon hsp83 Depletion
Activation of the InR/PI3K/Akt pathway promotes NSC reactivation (Chell and Brand, 2010, Sousa-Nunes et al., 2011). We investigated whether the InR/PI3K/Akt pathway is activated in NSCs with hsp83 depleted. To this end, we first analyzed the protein levels of Akt in NSCs, a major component of the InR/PI3K/Akt pathway and a protein kinase that triggers NSC reactivation (Shim et al., 2013). In the central brain of wild-type larvae at 72 hr ALH, Akt protein levels were comparable with its levels in the optic lobe (Figure 2A). However, upon hsp83 knockdown in central brain NSCs by insc-Gal4 at 72 hr ALH, Akt protein levels in the central brain decreased significantly, while its levels in the optic lobe remained similar to control (Figure 2A). Likewise, Akt protein levels in the central brain were reduced significantly upon cdc37 knockdown by RNAi (Figure 2A). These observations suggest that Hsp83 and Cdc37 may be required for the activation of the InR/PI3K/Akt pathway in NSCs. Next, we examined Akt protein levels in whole-brain protein extracts by western blot. hsp83 knockdown in larval brains was efficient, as Hsp83 protein levels in the whole brain were reduced to 47.3% ± 6.8% (n = 3) in hsp83 knockdown samples compared with controls (Figures 2B and 2C). Remarkably, Akt protein levels was only 61.7% ± 5.5% (n = 3) in hsp83 knockdowns compared with wild-type controls (Figures 2B and 2C). Likewise, upon cdc37 knockdown in NSCs Akt level was reduced to 67.1% ± 15.8% (n = 3), compared with wild-type control (Figures 2B and 2C). These results suggest that the InR/PI3K/Akt pathway is likely impaired upon depletion of either hsp83 or cdc37 in NSCs.
Figure 2.
Hsp83 and Cdc37 Are Required for the Activation of InR Pathway
(A) Larval brains of control (UAS-dicer2), hsp83 RNAi (v108568), and cdc37 RNAi (v47776) under insc-Gal4 at 72 hr ALH were stained for Mira, Dpn, and Akt. Yellow dotted boxes indicate the region of zoomed-in images. Central brain is to the left of white dotted lines. White arrows point to NSCs and outlines of NSCs are indicated by white dotted lines. Scale bars, 30 μm for whole brain lobe and 5 μm for single cell.
(B and C) Western blot analysis of larval brain extracts of control (UAS-dicer2), hsp83 knockdown (v108568), and cdc37 knockdown (v110727) induced with insc-Gal4 at 72 hr ALH. Blots were probed with anti-Hsp83 antibody or anti-Akt antibody.
(D and E) Western blot analysis of larval brains of control (UAS-dicer2), hsp83 knockdown (v108568), cdc37 knockdown (v110727), and InRAD overexpression (O/E; induced with insc-Gal4) at 72 hr ALH. Blots were probed with anti-pInR antibody.
(F and G) Western blot analysis of whole larvae extracts of control (UAS-dicer2), hsp83-HA overexpression induced with insc-Gal4 at 6 hr ALH. Blots were probed with anti-Akt antibody.
(H and I) Western blot analysis of whole larvae extracts of control (UAS-dicer2), hsp83-HA overexpression induced with insc-Gal4 at 6 hr ALH. Blots were probed with anti-pInR antibody. Loading control, actin (B, D, F, and H).
Statistical analyses were done comparing between two different genotypes using a two-tailed Student's t test (C, E, G, and I). ∗p < 0.05, ∗∗∗p < 0.001. Data are presented as mean ± SD in (C), (E), (G), and (I).
We then examined whether the protein levels of phosphorylated insulin receptor (pInR), the activated form of InR, is altered upon hsp83 or cdc37 depletion in larval brains. After hsp83 knockdown in NSCs under insc-Gal4 at 72 hr ALH, only 16.5% ± 9.7% (n = 3) of pInR remained compared with wild-type controls (Figures 2D and 2E). Similarly, knockdown of cdc37 in NSCs resulted in a dramatic reduction of pInR levels with 23.1% ± 0.6% (n = 3) compared with controls (Figures 2D and 2E). The mRNA levels of InR or akt, were not obviously altered with hsp83 or cdc37 RNAi knockdown (Figures S2A and S2B).
Taken together, we conclude that Hsp83 and Cdc37 are likely required for the activation of the InR/PI3K/Akt pathway.
Hsp83 Overexpression Results in Increase of pInR and Akt Level
Since Hsp83 overexpression triggers premature NSC reactivation, it is worthy to investigate whether Hsp83 overexpression can result in the activation of the InR/PI3K/Akt pathway. Due to insufficient amounts of protein that could be obtained due to the tiny size of larval brains at 6 hr ALH, we examined the protein levels of the whole larvae. Upon hsp83-HA overexpression in NSCs using insc-Gal4, the protein levels of Akt in the whole larvae were increased to 184.6% ± 25.9% (n = 4), compared with controls (Figures 2F and 2G). Similarly, hsp83 overexpression in NSCs resulted in a significant increase of pInR level in whole larvae, with 383.6% ± 88.2% (n = 4) compared with controls (Figures 2H and 2I). These results suggest that overexpression of Hsp83 is sufficient to drive the activation of the InR/PI3K/Akt pathway on the fed condition.
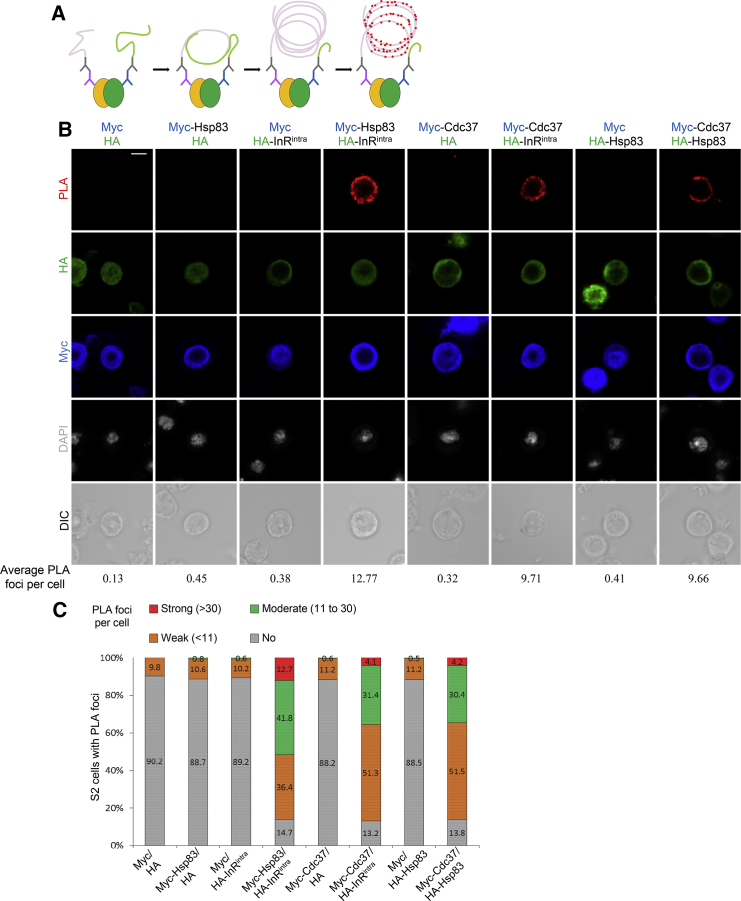
Hsp83, Cdc37, and InR Physically Interact
Given that Hsp83 and Cdc37 are required for the activation of the InR/PI3K/Akt pathway, we explored whether Hsp83 and Cdc37 physically interact with InR or Akt. To test the physical association between Hsp83, Cdc37, and InR, we performed a proximity ligation assay (PLA), a technique enabling high specificity and sensitivity detection of protein-protein interactions (Figure 3A) (Fredriksson et al., 2002). We co-expressed various proteins tagged with Myc or HA in S2 cells and quantified PLA foci that indicated interactions in cells, which co-expressed both Myc and HA-tagged proteins (Figures 3B and 3C). In S2 cells co-expressing both HA and Myc controls, the vast majority of cells (90.2%, n = 1,397) had no PLA signals and the rest of the cells displayed weak PLA fluorescence signal of less than 10 foci (Figures 3B and 3C). On average, these control cells only had 0.13 PLA foci per cell. We failed to generate full-length InR constructs, presumably due to its large size (>6 kb). As the intracellular domain of InR including the kinase domain (InRintra), an active form of InR, was previously used for demonstrating protein-protein interactions for InR (Almudi et al., 2013), we decided to use InRintra in PLA assays. Similarly, in the vast majority of cells co-expressing Myc-Hsp83 with control HA (n = 1,456) and HA-InRintra with control Myc (n = 1,767), no PLA signal was detected and, on average, there were 0.45 and 0.38 PLA foci per cell, respectively (Figures 3B and 3C). By contrast, cells co-expressing Myc-Hsp83 and HA-InRintra displayed 12.77 PLA foci per cell on average; 90.9% of cells exhibited PLA signal, with 41.8% of cells displaying moderate PLA signals (11–30 foci), 36.4% displaying weak PLA signals (≤10 foci), and 12.7% displaying strong PLA signals (>30 foci; Figures 3B and 3C; n = 1784). In a similar PLA assay, we tested whether Hsp83 interacts with Akt. The majority of S2 cells co-expressing Myc-Akt and HA-Hsp83 had no PLA signal (Figures S3A and S3B; 76.1%, n = 1,581), suggesting that Hsp83 did not interact with Akt. These results suggest that Hsp83 physically associated with InRintra, but not Akt.
Figure 3.
Hsp83, Cdc37, and InR Interact in PLA Assays
(A) A schematic representation of the proximity ligation assay performed on S2 cells (refer to Experimental Procedures).
(B) In situ PLA assay among Hsp83, Cdc37, and InRintra in S2 cells. S2 cells transfected with two of the indicated plasmids (Myc, HA, Myc-Hsp83, HA-InRintra, Myc-Cdc37, and HA-Hsp83) were stained for HA, Myc, and DNA and screened for PLA signal. Cell outline was determined by differential interference contrast images.
(C) Graphs showing the percentage of S2 cells with no PLA signal, and weak (≤10 foci), moderate (11–30 foci), and strong (>30 foci) PLA signals for (B).
Scale bars, 4 μm.
Similarly, in controls expressing Myc-Cdc37 and HA, we observed that the majority of cells (88.2%) had no PLA signal (Figures 3B and 3C; n = 1,522). However, co-expressing Myc-Cdc37 and HA-InRintra resulted in 86.8% S2 cells showing PLA fluorescence; 51.3% of cells displayed weak PLA signals, 31.4% of cells showed moderate PLA signals, and 4.1% of cells showed strong PLA signals (Figures 3B and 3C; n = 1,144). In controls co-expressing HA-Hsp83 and Myc, 88.5% of S2 cells did not have any PLA signal (Figures 3B and 3C; n = 1,069). By contrast, in cells co-expressing Myc-Cdc37 and HA-Hsp83, 51.5%, 30.4%, and 4.2% of these cells displayed weak, moderate, and strong PLA signals, respectively (Figures 3B and 3C; n = 1,271).
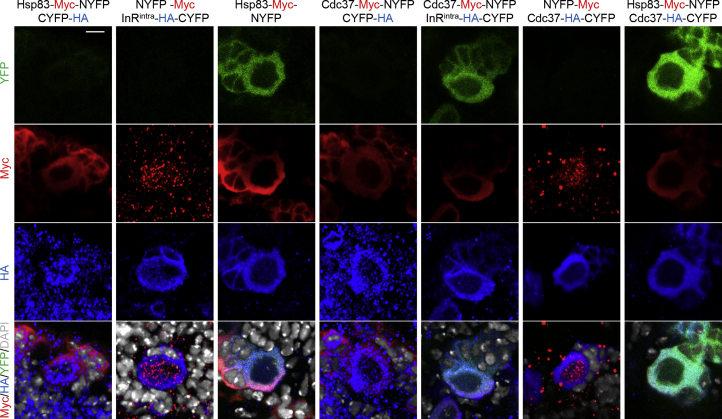
To further validate the interactions between Hsp83, Cdc37, and InR, we employed a protein-protein interaction assay named biomolecular fluorescence complementation (BiFC), which is able to detect transient or weak interactions due to the irreversibility of the BiFC complex formation (Gohl et al., 2010, Shyu and Hu, 2008). We generated chimeric proteins InRintra-HA-CYFP (InRintra with C-terminal YFP tagged with HA) and Hsp83-Myc-NYFP (N-terminal YFP tagged with Myc was fused to the Hsp83). As expected, when transfecting either of these two chimeric constructs into S2 cells with their respective controls, Myc-NYFP and HA-CYFP, no YFP signal was detected in cells (Figure S4B). By contrast, strong YFP was detected when cells were expressing both Hsp83-myc-NYFP and InRintra-HA-CYFP, suggesting that Hsp83 and InRintra physically associate (Figure S4B). To further test the specificity of this interaction, we generated a C-terminal truncation of Hsp83 (Figure S4A; Hsp83C: amino acids 538–717), which presumably abolishes the dimerization of Hsp83 and its subsequent ability to form a pocket-like structure to bind to its client proteins. We tested the ability of this Hsp83 truncation to associate with InRintra. There was no YFP signal upon co-expression of Hsp83C-myc-NYFP with InRintra-HA-CYFP in S2 cells (Figure S4B), suggesting that the association between Hsp83 and InRintra was abolished with the N-terminal Hsp83 deletion. In addition, Hsp83 associated with its co-chaperone Cdc37, but not with an unrelated control WAVE in the BiFC assay (Figure S4B). Cdc37 also physically interacts with InRintra in the BiFC assay, as S2 cells expressing both InRintra-HA-CYFP and Cdc37-myc-NYFP, but not the controls, displayed YFP signal (Figure S4B).
Next, to examine their physical association in an in vivo BiFC assay, we generated transgenic flies expressing Hsp83-myc-NYFP, InRintra-HA-CYFP, Cdc37-myc-NYFP, and Cdc37-HA-CYFP in NSCs under the insc-Gal4 driver. At 72 hr ALH at 29°C, overexpressed Hsp83, Cdc37, and InRintra in NSCs were observed by their respective epitope tags. As expected, there was no detectable YFP when Hsp83-myc-NYFP, Cdc37-myc-NYFP, Cdc37-HA-CYFP Cdc37, or InRintra-HA-CYFP were co-expressed with their corresponding control half-YFP proteins (Figure 4). By contrast, when Hsp83-myc-NYFP and InRintra-HA-CYFP were co-expressed, strong YFP was detected in NSCs (Figure 4), suggesting that Hsp83 and InRintra physically associate in NSCs. Likewise, Cdc37 interacted with both InRintra and Hsp83 in NSCs in the BiFC assay (Figure 4). Taken together, Hsp83, Cdc37, and InRintra physically associate with one another both in vitro and in vivo in BiFC assays.
Figure 4.
Hsp83, Cdc37, and InR Physically Associate in BiFC Assays
UAS-Hsp83-Myc-NYFP and UAS-InRintra-HA-CYFP were co-expressed in larval NSCs by insc-Gal4 at 72 hr ALH, stained for Myc, HA, and DNA and screened for YFP fluorescence. Negative controls were UAS-Hsp83-Myc-NYFP with UAS-CYFP-HA and UAS-NYFP-Myc with UAS-InRintra-HA-CYFP. Other transgenes used in the assay were UAS-Cdc37-Myc-NYFP and UAS-Cdc37-HA-CYFP. Scale bars, 4 μm.
Hsp83 Functions Upstream of the InR/PI3K/Akt Pathway in NSC Reactivation
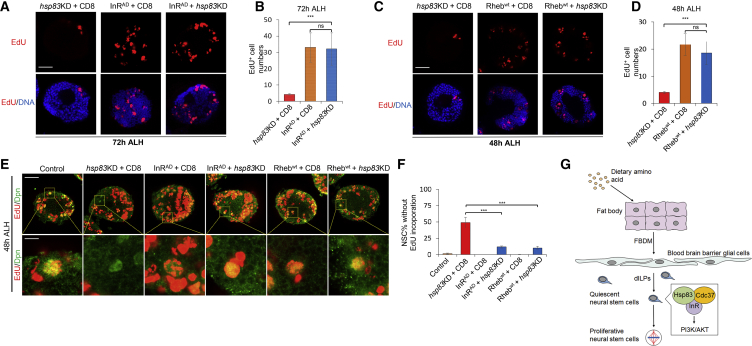
Next, we ascertained whether Hsp83 functions upstream or downstream of the InR/PI3K/Akt pathway during NSC reactivation. Over-activation of the InR/PI3K/Akt pathway by expressing various active components of this pathway overrides the requirement of dietary amino acids for NSC reactivation (Chell and Brand, 2010, Sousa-Nunes et al., 2011). We explored whether depletion of hsp83 was able to abrogate this effect of InR/PI3K/Akt pathway over-activation. Overexpression of an active form of InR (i.e., InRAD, BDSC no. 8440) under the insc-Gal4 drove NSC reactivation in the absence of dietary amino acid (sucrose-only food), as there were 32.8 ± 13.9 (n = 22) EdU+ cells per brain hemisphere at 72 hr ALH (Figures 5A and 5B). Similarly, at the same growth condition, when hsp83 was depleted in InRAD-overexpressing brains, there were 32.5 ± 14.6 (n = 25) EdU+ cells per brain hemisphere (Figures 5A and 5B). Likewise, at 24 hr ALH in sucrose-only food, in InRAD-overexpressing brains with hsp83 RNAi knockdown, there were 17.1 ± 6.6 (n = 20) EdU+ cells per hemisphere (Figures S5A and S5B), similar to InRAD overexpression alone (Figures S5A and S5B; 21.3 ± 5.0 EdU+ cells per hemisphere, n = 32). These observations suggest that over-activation of InR was epistatic to hsp83 depletion in NSCs. Therefore, Hsp83 likely functions upstream of InR during NSC reactivation.
Figure 5.
Hsp83 Functions Upstream of the InR/PI3K/Akt Pathway during NSC Reactivation
(A and B) Larval brains of hsp83 knockdown (v108568) with UAS-CD8-GFP, InRAD overexpression (BDSC no. 8440) with UAS-CD8-GFP, and InRAD overexpression with hsp83 knockdown (v108568) under insc-Gal4 at 72 hr ALH in sucrose-only food were labeled with EdU and DNA. hsp83 knockdown, 4.3 ± 0.5 (n = 16) EdU+ cells per brain hemisphere at 72 hr ALH.
(C and D) Larval brains of hsp83 knockdown (v108568) with UAS-CD8-GFP, Rhebwt overexpression (BDSC no. 9689) with UAS-CD8-GFP, and Rhebwt overexpression with hsp83 knockdown (v108568) under insc-Gal4 at 48 hr ALH in sucrose-only food were labeled with EdU and DNA. hsp83 knockdown, 4.0 ± 0.4 (n = 20) EdU+ cells per brain hemisphere at 48 hr ALH.
(E and F) Larval brains of control (UAS-dicer2), hsp83 knockdown (v108568) with UAS-CD8-GFP, InRAD overexpression (BDSC no. 8440) with UAS-CD8-GFP, InRAD overexpression with hsp83 knockdown (VDRC no. 108568), Rhebwt overexpression (BDSC no. 9689) with UAS-CD8-GFP, and Rhebwt overexpression with hsp83 knockdown (v108568) induced with insc-Gal4 at 48 hr ALH in normal food were labeled with EdU and Dpn. Yellow dotted boxes indicate the region of zoomed-in images.
Data are presented in (B), (D), and (F) as mean ± SD. Statistical analyses were done by pairwise comparison between two different genotypes using a two-tailed Student's t test (B, D, and F). ns, not significant; ∗∗∗p < 0.001. Scale bars, 15 μm in (A) and (C), 30 μm for whole brain lobe, and 5 μm for single cell in (E).
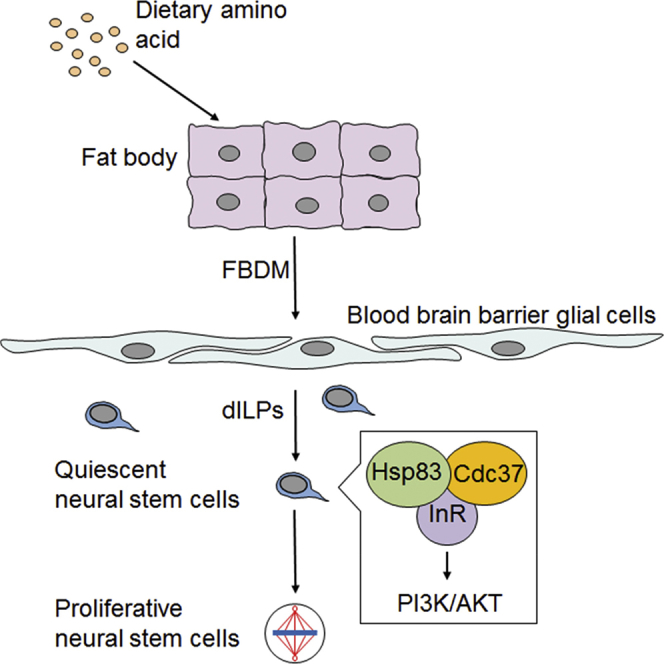
(G) A working model.
Rheb is a GTPase and a downstream effector of the InR/PI3K/Akt pathway in NSC reactivation (Shim et al., 2013). In sucrose-only food at 24 hr ALH, there were 15.2 ± 2.8 (n = 42) EdU+ cells per brain hemisphere upon Rhebwt overexpression (wild-type form, BDSC no. 9689) under the insc-Gal4 (Figures S5B and S5D). Under the same conditions, 13.8 ± 3.4 EdU+ cells (n = 48) were observed in larval brains overexpressing Rhebwt with hsp83 RNAi knockdown (Figures S5B and S5D). Likewise, at 48 hr ALH on sucrose-only food, the number of EdU+ cells were similar in Rhebwt-overexpressing brains with (Figures 5B and 5D; 21.6 ± 4.0, n = 26) and without hsp83 depletion (Figures 5B and 5D; 18.5 ± 4.2, n = 27).
Under fed conditions, InRAD or Rhebwt overexpression also significantly rescued NSC quiescence phenotype caused by hsp83 knockdown. At 48 hr ALH, vast majority of control NSCs were reactivated, except for 1.62% ± 0.8% NSCs without EdU incorporation (Figures 5E and 5F; n = 2410, 24 brain lobes). Similarly, almost all NSCs with InRAD or Rhebwt overexpression were reactivated as well, with none (Figures 5E and 5F; n = 2,412, 24 brain lobes) or 0.3% ± 0.4% (Figures 5E and 5F; n = 1,757, 18 brain lobes) NSCs without EdU incorporation. Upon hsp83 RNAi knockdown in NSCs, 48.9% ± 8.2% (Figures 5E and 5F; n = 1,357, 17 brain lobes) lacked EdU incorporation. By contrast, at the same time point, in NSCs overexpressing InRAD or Rhebwt concomitant with hsp83 RNAi knockdown, the percentages of EdU− NSCs were decreased significantly to 11.7% ± 1.5% (Figures 5E and 5F; n = 2,392, 26 brain lobes) and 10.4% ± 2.3% (Figures 5E and 5F; n = 1,827, 22 brain lobes).
Taken together, overexpression of InRAD or Rhebwt significantly rescued NSC reactivation defects observed upon hsp83 RNAi knockdown.
Amino Acids Deprivation Causes Significant Downregulation of hsp83 mRNA and Inactivation of InR/PI3K/Akt Pathway
To explore whether the expression of InR, hsp83, and cdc37 are nutrition dependent, we assessed the RNA level of InR, hsp83, and cdc37 in larvae upon amino acid deprivation. At 24 hr ALH in sucrose-only food, the mRNA levels of hsp83 were dramatically reduced to 13.4% ± 3.2% and 10.4% ± 3.1% measured by two different primer pairs (n = 4 each), compared with control larvae raised under fed conditions at the same time point (Figure S5E). RNA level of cdc37 remains unchanged on the starvation condition (Figure S5E). This suggests that the expression of hsp83, but not cdc37, is dependent on the presence of dietary amino acids. Surprisingly, the mRNA level of InR and 4E-binding protein (4E-BP) on sucrose-only food was increased significantly to at least 5-fold (Figure S5E; n = 4). This result is in line with the previous finding that increased transcription of InR and 4E-BP was correlated with inactivation of InR pathway and growth inhibition when nutrients are limited due to a feedback control mechanism for the InR pathway (Puig et al., 2003). Since the upregulation of mRNA levels of InR in the entire larvae upon amino acid deprivation was not observed in larval brains upon knocking down of hsp83 alone, this effect likely involves additional factors that are altered by depletion of dietary amino acids or due to different experimental conditions. Therefore, amino acid deprivation causes dramatic downregulation of hsp83 and inactivation of InR/PI3K/Akt pathway.
Discussion
How the InR/PI3K/Akt pathway is regulated during NSC reactivation is poorly understood. Here we show that molecular chaperone Hsp83/Hsp90, together with its co-chaperone Cdc37, play a role in the reactivation of Drosophila NSCs. Mechanistically, Hsp83 and Cdc37 physically associate with InR and are important for the activation of the InR/PI3K/Akt pathway in NSCs. Therefore, we demonstrate that Hsp83 serves as an intrinsic factor within NSCs that is necessary for the activation of the InR/PI3K/Akt pathway and, in turn, reactivation of NSCs. Our evidence suggests that Hsp83 and Cdc37 regulate the protein folding and activation of InR in the nervous system.
The role of Hsp83 in NSC reactivation at early larval stages is distinct from its known role in centrosomes or NSC polarity. Drosophila Hsp83 is a core centrosomal component required for proper mitotic spindle formation and chromosome segregation (Lange et al., 2000). In Drosophila larval CNS, Hsp83 and co-chaperone Sgt1 are required for the stabilization of Polo and centrosome organization in NSCs (Martins et al., 2009). Hsp83 and Sgt1 are also required for the establishment of NSC polarity via the LKB1/AMPK pathway in third-instar larvae (Andersen et al., 2012). However, sgt1 RNAi (BDSC no. 34605) in NSCs did not display any phenotypes during NSC reactivation (data not shown), suggesting that Hsp83 interacts with different co-chaperones to control NSC reactivation and cortical polarity at different developmental stages. Consistent with this notion, we found that Cdc37, but not other co-chaperones of Hsp83, is required for NSC reactivation. We found that the proliferation of MB NSCs were unaffected by hsp83 knockdown, Therefore, Hsp83 promotes NSC reactivation rather than general cell proliferation. Consistent with our observations, there is no significant difference in proliferation between hsp83 mutant and wild-type eye imaginal discs in the proliferating zone (Bandura et al., 2013). Interestingly, in pupal eyes that undergo terminal differentiation, Hsp83 is required for cell-cycle exit by activating the anaphase-promoting complex/cyclosome (Bandura et al., 2013). We found cytokinesis defects in cdc37-depleted NSCs, but not in NSCs depleted of Hsp83 (data not shown). This observation is consistent with a known role of Cdc37 in cell division and cytokinesis in Drosophila (Lange et al., 2002).
Hsp90 plays a key role in signal transduction and appears to bind to its substrates in a near native state poised for activation by binding of ligand or other factors (Young et al., 2001). Since Hsp83 overexpression is sufficient to drive the activation of InR/PI3K/Akt pathway and trigger premature NSC reactivation, Hsp83 likely plays an active role in promoting NSC reactivation by binding to InR at a late stage of folding poised for activation by dILP binding. Furthermore, in the absence of dietary amino acids, the expression of hsp83 is downregulated, likely partially contributing to the inactivation of the InR pathway (Figure S5E). We propose that InR is a target of Hsp83 and Cdc37 during NSC reactivation. The physical association among Hsp83, Cdc37, and InR was strongly supported by PLA assays, and both in vitro and in vivo BiFC. Although tandem affinity purification-mass spectrometry in Drosophila S2 cells implied an interaction between Hsp83 and InR (Friedman et al., 2011), we failed to detect a consistent interaction between Hsp83 and InRintra in S2 cells in co-immunoprecipitation experiments, probably due to the transient nature of this interaction. In addition, our genetic interaction experiments indicate that Hsp83 activates the InR/PI3K/Akt pathway to promote NSC reactivation. Taken together, InR is likely a client of Hsp83 in Drosophila NSCs. Consistent with our findings, in human fibroblasts, Hsp90 co-immunoprecipitated with intracellular InR β subunit (Takata et al., 1997). Furthermore, Hsp90 facilitates the maturation of the InR precursor in the ER and, in turn, is required for cell surface expression of InR in both bovine adrenal medullary chromaffin cells and human kidney HEK293 cells (Ramos et al., 2007, Saitoh et al., 2002). Therefore, the interaction between the Hsp90 chaperone family and InR may be conserved from Drosophila to humans. In mammals, the expression level of Hsp90 in the brain is the highest among all tissues (Barrott and Haystead, 2013). Although mammalian Hsp90 proteins are heavily implicated in neurodegenerative diseases (Lackie et al., 2017, Luo et al., 2010), their function in brain development is not well understood. Hsp90/Cdc37 stabilize the intracellular domain of Ryk, a Wnt receptor required for neurogenesis (Lyu et al., 2009). Furthermore, Hsp90 stabilizes hypoxia-inducible factor-1, which promotes NSC proliferation under hypoxia (Xiong et al., 2009). It remains to be determined whether the interaction between mammalian Hsp90 and InR is conserved during mammalian NSC development.
Experimental Procedures
Fly Stocks and Genetics
The fly strains used in this study were: UAS-CYFP-HA, UAS-NYFP-Myc, UAS-Hsp83-Myc-NYFP, UAS-InRintra-HA-CYFP, UAS-Cdc37-Myc-NYFP, UAS-Cdc37-HA-CYFP, UAS-Hsp83-HA (T. Wang), hsp83-BAC, and cdc37-BAC. The following stocks were obtained from Bloomington Drosophila Stock Center (BDSC): RNAi for Hop and various isoforms of Hsp70 listed in Table S1, cdc37 RNAi (BDSC no. 28756), cdc37e4D (BDSC no. 5693), UAS-InR.A1325D (InRAD, BDSC no. 8440), UAS-Rhebwt (BDSC no. 9689), and hsp83e6A (BDSC no. 36576). RNAi lines including hsp83 RNAi (VDRC no. 108568) and cdc37 RNAi (VDRC nos. 47776 and 110727) were obtained from Vienna Drosophila Resource Center (VDRC). Hsp83j5C2 (DGRC no. 111379) was obtained from Kyoto Drosophila Genetic Resource Center. All experiments were carried out at 25°C, except for RNAi knockdown or overexpression at 29°C.
Immunochemistry
Larval brains were dissected in PBS and fixed for 22 min in 0.3% PBS-Triton (PBT) with 4% electron microscopy (EM)-grade formaldehyde (methanol free). Fixed brains were processed for immunostaining as described previously (Koe et al., 2014). Further details and primary antibodies used can be found in Supplemental Experimental Procedures.
MARCM Analysis
To generate MARCM clones, late first-instar larvae were heat shocked for 2 hr at 37°C, and heat shocked for a second time 10–16 hr later after recovering at 25°C. Larvae were dissected at the third instar-larval stage and for the tissue processed by immunochemistry.
EdU Pulse-Chase Analysis
Larvae were fed with standard food supplemented with 0.2 mM EdU from Click-iT EdU Alexa Fluor 555 Imaging Kit (Invitrogen) for 4 hr prior to dissection. The dissected larval brains were then fixed with 4% EM-grade formaldehyde (in 0.3% PBT) for 22 min. The brains were then processed for as described previously (Li et al., 2017).
Author Contributions
Conceptualization, H.W.; Methodology, H.J.; Writing – Original Draft, Review & Editing, H.W. and H.J.; Funding Acquisition, H.W.; Resources, H.W. and H.J.; Supervision, H.W.
Acknowledgments
We thank H. Stocker, T. Wang, S. Bodgan, F. Matsuzaki, C. Doe, J. Knoblich, A.H. Brand, T. Lee, W. Chia, X. Yang, H. Steller, F. Yu, the Bloomington Drosophila Stock Center, Vienna Drosophila Resource Center, Drosophila Genomics Resource Center, and the Developmental Studies Hybridoma Bank for fly stocks and antibodies. We thank X. Wei for isolation of hsp83 RNAi from the RNAi screen. This work is supported by Singapore National Medical Research Council (NMRC/CBRG/0082/2015).
Published: September 20, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.08.014.
Supplemental Information
References
- Ahn S., Joyner A.L. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- Almudi I., Poernbacher I., Hafen E., Stocker H. The Lnk/SH2B adaptor provides a fail-safe mechanism to establish the Insulin receptor-Chico interaction. Cell Commun. Signal. 2013;11:26. doi: 10.1186/1478-811X-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen R.O., Turnbull D.W., Johnson E.A., Doe C.Q. Sgt1 acts via an LKB1/AMPK pathway to establish cortical polarity in larval neuroblasts. Dev. Biol. 2012;363:258–265. doi: 10.1016/j.ydbio.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenijevic Y., Weiss S., Schneider B., Aebischer P. Insulin-like growth factor-I is necessary for neural stem cell proliferation and demonstrates distinct actions of epidermal growth factor and fibroblast growth factor-2. J. Neurosci. 2001;21:7194–7202. doi: 10.1523/JNEUROSCI.21-18-07194.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandura J.L., Jiang H., Nickerson D.W., Edgar B.A. The molecular chaperone Hsp90 is required for cell cycle exit in Drosophila melanogaster. PLoS Genet. 2013;9:e1003835. doi: 10.1371/journal.pgen.1003835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrott J.J., Haystead T.A. Hsp90, an unlikely ally in the war on cancer. FEBS J. 2013;280:1381–1396. doi: 10.1111/febs.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baser A., Skabkin M., Martin-Villalba A. Neural stem cell activation and the role of protein synthesis. Brain Plast. 2017;3:27–41. doi: 10.3233/BPL-160038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton J.S., Edgar B.A. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- Chell J.M., Brand A.H. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell. 2010;143:1161–1173. doi: 10.1016/j.cell.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloetta D., Thomanetz V., Baranek C., Lustenberger R.M., Lin S., Oliveri F., Atanasoski S., Ruegg M.A. Inactivation of mTORC1 in the developing brain causes microcephaly and affects gliogenesis. J. Neurosci. 2013;33:7799–7810. doi: 10.1523/JNEUROSCI.3294-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J., Raisin S., Pantalacci S., Radimerski T., Montagne J., Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- Cutforth T., Rubin G.M. Mutations in Hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell. 1994;77:1027–1036. doi: 10.1016/0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]
- Daynac M., Chicheportiche A., Pineda J.R., Gauthier L.R., Boussin F.D., Mouthon M.A. Quiescent neural stem cells exit dormancy upon alteration of GABAAR signaling following radiation damage. Stem Cell Res. 2013;11:516–528. doi: 10.1016/j.scr.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Daynac M., Tirou L., Faure H., Mouthon M.A., Gauthier L.R., Hahn H., Boussin F.D., Ruat M. Hedgehog controls quiescence and activation of neural stem cells in the adult ventricular-subventricular zone. Stem Cell Rep. 2016;7:735–748. doi: 10.1016/j.stemcr.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., Weynans K., Bossing T., Barros C.S., Berger C. The Hippo signalling pathway maintains quiescence in Drosophila neural stem cells. Nat. Commun. 2016;7:10510. doi: 10.1038/ncomms10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F., Garcia-Verdugo J.M., Alvarez-Buylla A. Regeneration of a germinal layer in the adult mammalian brain. Proc. Natl. Acad. Sci. USA. 1999;96:11619–11624. doi: 10.1073/pnas.96.20.11619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckl J.M., Richter K. Functions of the Hsp90 chaperone system: lifting client proteins to new heights. Int. J. Biochem. Mol. Biol. 2013;4:157–165. [PMC free article] [PubMed] [Google Scholar]
- Faiz M., Sachewsky N., Gascon S., Bang K.W., Morshead C.M., Nagy A. Adult neural stem cells from the subventricular zone give rise to reactive astrocytes in the cortex after stroke. Cell Stem Cell. 2015;17:624–634. doi: 10.1016/j.stem.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gustafsdottir S.M., Ostman A., Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- Friedman A.A., Tucker G., Singh R., Yan D., Vinayagam A., Hu Y., Binari R., Hong P., Sun X., Porto M. Proteomic and functional genomic landscape of receptor tyrosine kinase and ras to extracellular signal-regulated kinase signaling. Sci. Signal. 2011;4:rs10. doi: 10.1126/scisignal.2002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl C., Banovic D., Grevelhorster A., Bogdan S. WAVE forms hetero- and homo-oligomeric complexes at integrin junctions in Drosophila visualized by bimolecular fluorescence complementation. J. Biol. Chem. 2010;285:40171–40179. doi: 10.1074/jbc.M110.139337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T., Pearson B., Holbrook S., Doe C.Q. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Ito K., Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev. Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- Juanes M., Guercio G., Marino R., Berensztein E., Warman D.M., Ciaccio M., Gil S., Bailez M., Rivarola M.A., Belgorosky A. Three novel IGF1R mutations in microcephalic patients with prenatal and postnatal growth impairment. Clin. Endocrinol. (Oxf) 2015;82:704–711. doi: 10.1111/cen.12555. [DOI] [PubMed] [Google Scholar]
- Kawai H., Kawaguchi D., Kuebrich B.D., Kitamoto T., Yamaguchi M., Gotoh Y., Furutachi S. Area-specific regulation of quiescent neural stem cells by Notch3 in the adult mouse subependymal zone. J. Neurosci. 2017;37:11867–11880. doi: 10.1523/JNEUROSCI.0001-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koe C.T., Li S., Rossi F., Wong J.J., Wang Y., Zhang Z., Chen K., Aw S.S., Richardson H.E., Robson P. The Brm-HDAC3-Erm repressor complex suppresses dedifferentiation in Drosophila type II neuroblast lineages. Elife. 2014;3:e01906. doi: 10.7554/eLife.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo Y., Ren S., Lao U., Edgar B.A., Wang T. Suppression of polyglutamine protein toxicity by co-expression of a heat-shock protein 40 and a heat-shock protein 110. Cell Death Dis. 2013;4:e833. doi: 10.1038/cddis.2013.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackie R.E., Maciejewski A., Ostapchenko V.G., Marques-Lopes J., Choy W.Y., Duennwald M.L., Prado V.F., Prado M.A.M. The Hsp70/Hsp90 chaperone machinery in neurodegenerative diseases. Front. Neurosci. 2017;11:254. doi: 10.3389/fnins.2017.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S.L., Doe C.Q. Transient nuclear Prospero induces neural progenitor quiescence. Elife. 2014;3 doi: 10.7554/eLife.03363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B.M., Bachi A., Wilm M., Gonzalez C. Hsp90 is a core centrosomal component and is required at different stages of the centrosome cycle in Drosophila and vertebrates. EMBO J. 2000;19:1252–1262. doi: 10.1093/emboj/19.6.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange B.M., Rebollo E., Herold A., Gonzalez C. Cdc37 is essential for chromosome segregation and cytokinesis in higher eukaryotes. EMBO J. 2002;21:5364–5374. doi: 10.1093/emboj/cdf531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Li S., Koe C.T., Tay S.T., Tan A.L.K., Zhang S., Zhang Y., Tan P., Sung W.K., Wang H. An intrinsic mechanism controls reactivation of neural stem cells by spindle matrix proteins. Nat. Commun. 2017;8:122. doi: 10.1038/s41467-017-00172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugert S., Basak O., Knuckles P., Haussler U., Fabel K., Gotz M., Haas C.A., Kempermann G., Taylor V., Giachino C. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Luo W., Sun W., Taldone T., Rodina A., Chiosis G. Heat shock protein 90 in neurodegenerative diseases. Mol. Neurodegener. 2010;5:24. doi: 10.1186/1750-1326-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu J., Wesselschmidt R.L., Lu W. Cdc37 regulates Ryk signaling by stabilizing the cleaved Ryk intracellular domain. J. Biol. Chem. 2009;284:12940–12948. doi: 10.1074/jbc.M900207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairet-Coello G., Tury A., DiCicco-Bloom E. Insulin-like growth factor-1 promotes G(1)/S cell cycle progression through bidirectional regulation of cyclins and cyclin-dependent kinase inhibitors via the phosphatidylinositol 3-kinase/Akt pathway in developing rat cerebral cortex. J. Neurosci. 2009;29:775–788. doi: 10.1523/JNEUROSCI.1700-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins T., Maia A.F., Steffensen S., Sunkel C.E. Sgt1, a co-chaperone of Hsp90 stabilizes Polo and is required for centrosome organization. EMBO J. 2009;28:234–247. doi: 10.1038/emboj.2008.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morshead C.M., Reynolds B.A., Craig C.G., McBurney M.W., Staines W.A., Morassutti D., Weiss S., van der Kooy D. Neural stem cells in the adult mammalian forebrain: a relatively quiescent subpopulation of subependymal cells. Neuron. 1994;13:1071–1082. doi: 10.1016/0896-6273(94)90046-9. [DOI] [PubMed] [Google Scholar]
- Poon C.L., Mitchell K.A., Kondo S., Cheng L.Y., Harvey K.F. The hippo pathway regulates neuroblasts and brain size in Drosophila melanogaster. Curr. Biol. 2016;26:1034–1042. doi: 10.1016/j.cub.2016.02.009. [DOI] [PubMed] [Google Scholar]
- Pratt W.B., Toft D.O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr. Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- Puig O., Marr M.T., Ruhf M.L., Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 2003;17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos R.R., Swanson A.J., Bass J. Calreticulin and Hsp90 stabilize the human insulin receptor and promote its mobility in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2007;104:10470–10475. doi: 10.1073/pnas.0701114104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Yanagita T., Shiraishi S., Yokoo H., Kobayashi H., Minami S., Onitsuka T., Wada A. Down-regulation of cell surface insulin receptor and insulin receptor substrate-1 phosphorylation by inhibitor of 90-kDa heat-shock protein family: endoplasmic reticulum retention of monomeric insulin receptor precursor with calnexin in adrenal chromaffin cells. Mol. Pharmacol. 2002;62:847–855. doi: 10.1124/mol.62.4.847. [DOI] [PubMed] [Google Scholar]
- Sawarkar R., Sievers C., Paro R. Hsp90 globally targets paused RNA polymerase to regulate gene expression in response to environmental stimuli. Cell. 2012;149:807–818. doi: 10.1016/j.cell.2012.02.061. [DOI] [PubMed] [Google Scholar]
- Shim J., Gururaja-Rao S., Banerjee U. Nutritional regulation of stem and progenitor cells in Drosophila. Development. 2013;140:4647–4656. doi: 10.1242/dev.079087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu Y.J., Hu C.D. Fluorescence complementation: an emerging tool for biological research. Trends Biotechnol. 2008;26:622–630. doi: 10.1016/j.tibtech.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R., Yee L.L., Gould A.P. Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature. 2011;471:508–512. doi: 10.1038/nature09867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speder P., Brand A.H. Gap junction proteins in the blood-brain barrier control nutrient-dependent reactivation of Drosophila neural stem cells. Dev. Cell. 2014;30:309–321. doi: 10.1016/j.devcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speder P., Brand A.H. Systemic and local cues drive neural stem cell niche remodelling during neurogenesis in Drosophila. Elife. 2018;7 doi: 10.7554/eLife.30413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata Y., Imamura T., Iwata M., Usui I., Haruta T., Nandachi N., Ishiki M., Sasaoka T., Kobayashi M. Functional importance of heat shock protein 90 associated with insulin receptor on insulin-stimulated mitogenesis. Biochem. Biophys. Res. Commun. 1997;237:345–347. doi: 10.1006/bbrc.1997.7116. [DOI] [PubMed] [Google Scholar]
- Tariq M., Nussbaumer U., Chen Y., Beisel C., Paro R. Trithorax requires Hsp90 for maintenance of active chromatin at sites of gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:1157–1162. doi: 10.1073/pnas.0809669106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman J.W., Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev. Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Tsuji T., Hasegawa E., Isshiki T. Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Development. 2008;135:3859–3869. doi: 10.1242/dev.025189. [DOI] [PubMed] [Google Scholar]
- Wang Y.Z., Plane J.M., Jiang P., Zhou C.J., Deng W. Concise review: quiescent and active states of endogenous adult neural stem cells: identification and characterization. Stem Cells. 2011;29:907–912. doi: 10.1002/stem.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Zhao T., Huang X., Liu Z.H., Zhao H., Li M.M., Wu L.Y., Shu H.B., Zhu L.L., Fan M. Heat shock protein 90 is involved in regulation of hypoxia-driven proliferation of embryonic neural stem/progenitor cells. Cell Stress Chaperones. 2009;14:183–192. doi: 10.1007/s12192-008-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.P., Sailor K.A., Vemuganti R., Dempsey R.J. Insulin-like growth factor-1 is an endogenous mediator of focal ischemia-induced neural progenitor proliferation. Eur. J. Neurosci. 2006;24:45–54. doi: 10.1111/j.1460-9568.2006.04872.x. [DOI] [PubMed] [Google Scholar]
- Ye P., Popken G.J., Kemper A., McCarthy K., Popko B., D'Ercole A.J. Astrocyte-specific overexpression of insulin-like growth factor-I promotes brain overgrowth and glial fibrillary acidic protein expression. J. Neurosci. Res. 2004;78:472–484. doi: 10.1002/jnr.20288. [DOI] [PubMed] [Google Scholar]
- Young J.C., Moarefi I., Hartl F.U. Hsp90: a specialized but essential protein-folding tool. J. Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.