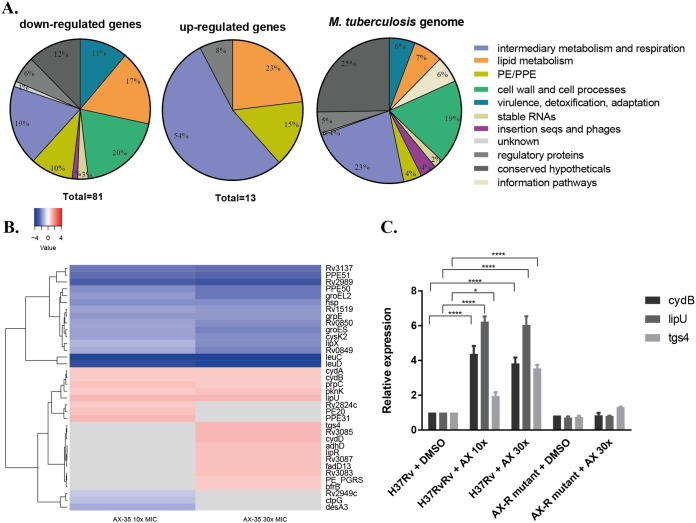
FIG 3.
Transcriptional response of M. tuberculosis to AX-35 treatment by RNA-seq. (A) Global transcriptomic response and involvement of different metabolic responses, based on TubercuList classification (https://mycobrowser.epfl.ch/), after exposure of two independent cultures of M. tuberculosis H37Rv to AX-35 at 10× and 30× MIC for 4 h. (B) Heat map representing top significantly differentially regulated M. tuberculosis genes (Padj ≤ 0.05). The color scale indicates differential regulation as log2 fold change of H37Rv with AX-35 treatment relative to H37Rv with vehicle control, DMSO. Upregulation is indicated in red, downregulation is in blue, and insignificant log2 fold change values for the condition are in gray. Data are from two independent experiments. (C) qPCR validation of genes cydB, lipU, and tgs4 in H37Rv and AX-resistant mutant strains treated with vehicle (DMSO) or AX-35. Data are presented as mean ± SD from two independent cultures. Statistical analysis was performed using two-way ANOVA with Tukey’s multiple-comparison test (*, P < 0.05; ****, P < 0.0001).