We propose that the coral holobiont should be conceptualized as a diverse transient microbial community that is responsive to the surrounding environment and encompasses a simple, redundant, resident microbiome and a small conserved core microbiome. Most importantly, we show that the coral microbiome is comparable to the microbiomes of other organisms studied thus far. Accurately characterizing the coral-microbe interactions provides an important baseline from which the functional roles and the functional niches within which microbes reside can be deciphered.
KEYWORDS: bacteria, coral, holobiont, microbiome, symbiosis
ABSTRACT
Studies of the coral microbiome predominantly characterize the microbial community of the host species as a collective, rather than that of the individual. This ecological perspective on the coral microbiome has led to the conclusion that the coral holobiont is the most diverse microbial biosphere studied thus far. However, investigating the microbiome of the individual, rather than that of the species, highlights common and conserved community attributes which can provide insights into the significance of microbial associations to the host. Here, we show there are consistent characteristics between individuals in the proposed three components of the coral microbiome (i.e., “environmentally responsive community,” “resident or individual microbiome,” and “core microbiome”). We found that the resident microbiome of a photoendosymbiotic coral harbored <3% (∼605 phylotypes) of the 16S rRNA phylotypes associated with all investigated individuals of that species (“species-specific microbiome”) (∼21,654 phylotypes; individuals from Pachyseris speciosa [n = 123], Mycedium elephantotus [n = 95], and Acropora aculeus [n = 91] from 10 reef locations). The remaining bacterial phylotypes (>96%) (environmentally responsive community) of the species-specific microbiome were in fact not found in association with the majority of individuals of the species. Only 0.1% (∼21 phylotypes) of the species-specific microbiome of each species was shared among all individuals of the species (core microbiome), equating to ∼3.4% of the resident microbiome. We found taxonomic redundancy and consistent patterns of composition, structure, and taxonomic breadth across individual microbiomes from the three coral species. Our results demonstrate that the coral microbiome is structured at the individual level.
INTRODUCTION
Deciphering the functional contribution of symbiotic bacteria to host health is imperative to determine the mechanistic basis of coral health, survival, and resilience in a rapidly changing environment. However, to date, the challenge remains to understand which of the thousands of bacteria that are in association with a particular coral host species (“species-specific microbiome”) have a significant contribution to the well-being of individual corals in their natural habitat (1–3). Accurately documenting the taxonomic structure of the coral microbiome has been crucial to this aim, and over the past decade, numerous studies have aimed to define the characteristics of the healthy coral microbiome through the use of amplicon sequencing (2, 4). Importantly though, it is the interactions of the individual with its microbiome (“individual microbiome”) that impact coral health. Distinguishing the individual’s microbiome from that of species-specific microbial consortia is therefore critical to identify the symbiotic microbial roles.
As a holobiont or meta-organism, corals are inhabited by diverse microbial communities including microalgae, bacteria, fungi, archaea, and viruses (5). The endosymbiosis of coral with the zooxanthellae Symbiodinium is, so far, the most studied and best understood biological interaction between coral and any member of the microbial community (6). The coral relies on this symbiotic relationship with the dinoflagellate for up to 95% of its nutrition uptake (7, 8), and when this endosymbiosis is disrupted for long periods of time by environmental stress, the coral can perish (9–11). However, for all other members of the microbial community, the stability and functional contribution of the host-microbe interaction have largely remained unknown. One of the main factors limiting our understanding of these bacterial associations is the variability that is evident in community associations (12, 13), whereby determining conserved functional contributions by specific bacteria has remained elusive. Coral-associated bacterial communities are highly variable, diverse, and abundant, and differentiating the stable, functionally significant interactions within these communities is challenging (4). Variability in the bacterial community is known to occur at different spatial and temporal scales, including within the many microbial niches of a coral colony, at a scale of centimeters (13), and along different biogeographical regions, across a scale of thousands of kilometers (14–16). Thus, corals represent a variable ecosystem and host habitat for bacterial associations.
However, here we argue that the diversity, structure, and potential function of coral bacterial associations are much lower and less complex than has previously been reported (5). We investigated the common and conserved attributes of the healthy coral microbiome using 309 individuals collected from three highly abundant and widespread Indo-Pacific species (17), Acropora aculeus, Mycedium elephantotus, and Pachyseris speciosa. We applied bacterial 16S rRNA gene amplicon sequencing to examine the bacterial associations of each coral specimen, collected from 10 reefs from central and northern parts of the Great Barrier Reef and the Coral Sea, at depth intervals of 10, 20, 40, and 60 to 80 m. We propose that the coral microbiome can be divided into three distinct layers (Fig. 1): (i) the “environmentally responsive community,” which is predominantly a transient community encompassing thousands of distinct 16S rRNA phylotypes, of which very few are associated with a single host individual; (ii) the “resident community” consisting of phylotypes principally from three key bacterial classes, Alpha- and Gammaproteobacteria and Deltaproteobacteria in M. elephantotus or Flavobacteriia in P. speciosa and A. aculeus; and (iii) the “core microbiome” consisting of a few ubiquitous, potentially symbiotic, bacterial phylotypes. The identity of phylotypes composing the coral core microbiome depends on, among other factors, the coral species considered (4).
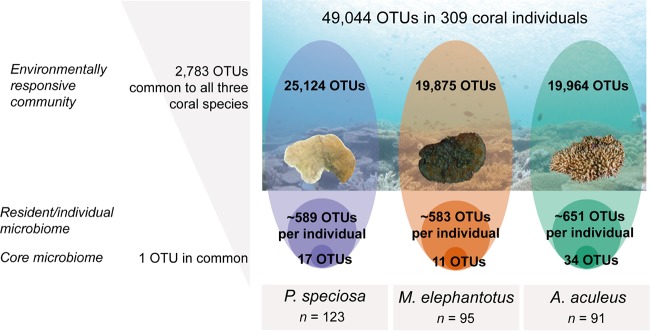
FIG 1.
Coral microbiome conceptualized into three distinct layers. We propose that the microbiome of each coral individual should be categorized into three distinct groups of bacteria with different levels of impact from the environment and host: (i) an environmentally responsive community with thousands of bacterial phylotypes, transient and highly variable across coral individuals; (ii) individual microbiome, ∼500 to 600 OTUs that vary among reefs at the level of OTUs but consistently belong to three major taxonomic classes; and (iii) core microbiome, few bacterial phylotypes, potentially symbiotic. Taxonomic (and potentially functional) redundancy is occurring in the resident community and the core microbiome. The reef picture was provided by Alexander J. Fordyce.
RESULTS AND DISCUSSION
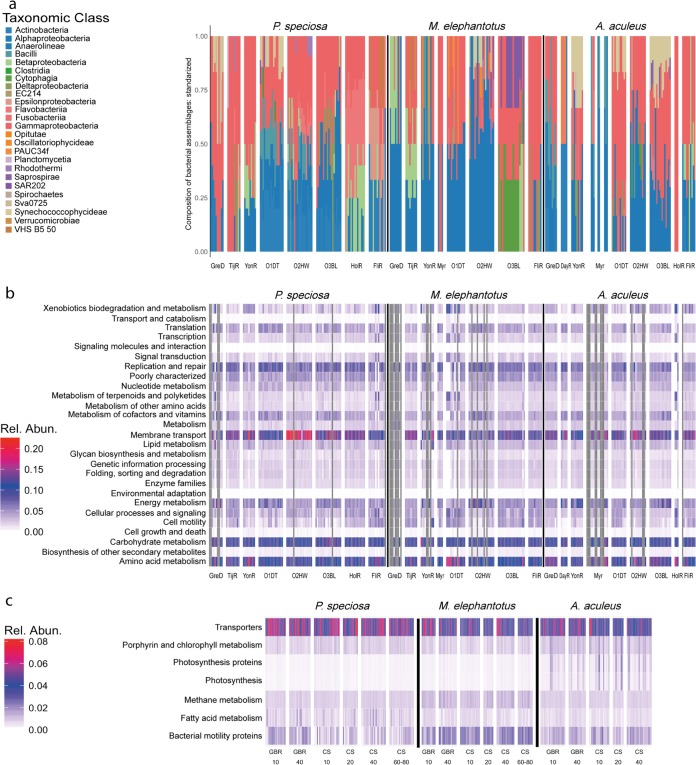
Across ecological systems, the concept of taxonomic and functional redundancy has been employed to characterize and conceptualize healthy and disturbed ecosystem states (18–21). Here, we investigated the microbiomes of 309 coral individuals from three highly abundant and widespread Indo-Pacific species (17), the plating corals Mycedium elephantotus and Pachyseris speciosa and the branching coral Acropora aculeus. The diversity and species-specific patterns of the microbiome observed for these three species were consistent with previous studies (5), in that collectively, corals host highly diverse bacterial interactions which are responsive to the hosts’ reef environment (Fig. 2). We reported more than 79,000 distinct operational taxonomic units (OTUs) generated from 17 million sequences within the collective data set (see Table S1 at https://figshare.com/s/ffadfdf1a1bab8a37088), which resulted in 49,000 distinct OTUs (from ∼6 million sequences) after filtering chloroplasts and unidentified/unassigned (not assigned to Kingdom Bacteria) sequences. From those OTUs, about half were species specific, and half were shared between the three coral species (Fig. 3b; see also Table S1), with each coral species hosting on average ∼20,000 to 25,000 OTUs (19,964 OTUs for M. elephantotus, 25,124 OTUs for P. speciosa, and 19,875 OTUs for A. aculeus). Importantly, we found that each individual coral hosts very few of these species-specific bacterial phylotypes. For example, P. speciosa individuals hosted on average only 589 ± 39 OTUs (n = 123 coral individuals); similarly, on average, M. elephantotus individuals hosted 583 ± 62 OTUs (n = 95 corals) and A. aculeus individuals hosted 651 ± 41 OTUs (n = 91 corals). As such, regardless of the coral species, the reef site in which they reside, or other environmental variables (such as reef depth and nutrient and light availability), an individual coral colony harbors only 2 to 3% of the total number of bacteria that are found in association with the species (species-specific microbiome). We also found that the bacterial associations of individual corals are overwhelmingly constrained to two or three dominant bacterial classes (Fig. 3a and c). These findings are consistent with the characteristics of the microbiome of holobiont models across the phyla that have been studied thus far, including hydra (22, 23), nematode Caenorhabditis elegans (24), plants (25, 26), and humans (20, 27), which similarly identified that the host microbiome is highly structured, habitat specific, and functionally redundant. As such, our findings contrast with previous studies hypothesizing that the coral microbiome is as an exceptionally diverse microbial biosphere compared to other organisms. As it has been widely documented and further supported by this study, corals host thousands of bacteria in species-specific interactions. However, these high numbers of species-specific interactions are due to a highly transient microbiome (Fig. 2), likely reflective of the highly dynamic symbiotic state and open interaction with the surrounding environment. We further suggest that the substantial taxonomic redundancy within an individual coral’s microbiome may reflect functional redundancy of the highly efficient photoendosymbiotic host system, particularly within waste production, utilization, and nutrient cycling.
FIG 2.
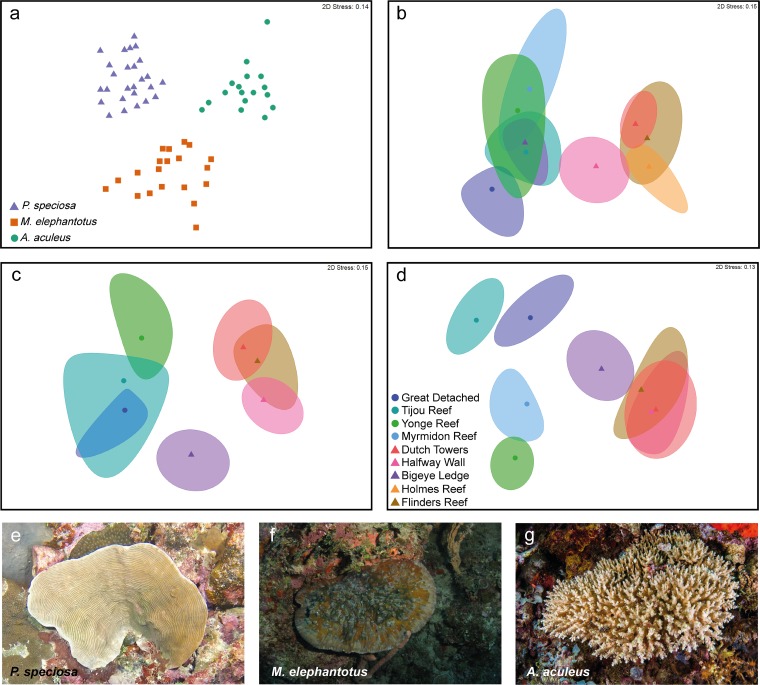
Bacterial community structure differed spatially and between coral species. Nonmetric multidimensional scaling (nMDS) based on relative abundance to illustrate differences between coral species (a) (P < 0.01 by PERMANOVA [see Table S4 at https://figshare.com/s/ffadfdf1a1bab8a37088]) and between reefs for P. speciosa (b and e), M. elephantotus (c and f; excluding Myrmidon reef), and A. aculeus (d and g; excluding Holmes reef). nMDS based on Bray-Curtis dissimilarity of fourth-root-transformed data. (a) Centroids, (b to d) bootstrap area and average for reefs. Circles denote Great Barrier Reef (GBR) reefs, and triangles indicate Coral Sea (CS) reefs. For presence/absence equivalent results, see Fig. S1 at https://figshare.com/s/ffadfdf1a1bab8a37088. The A. aculeus photo was provided by Ed Roberts.
FIG 3.
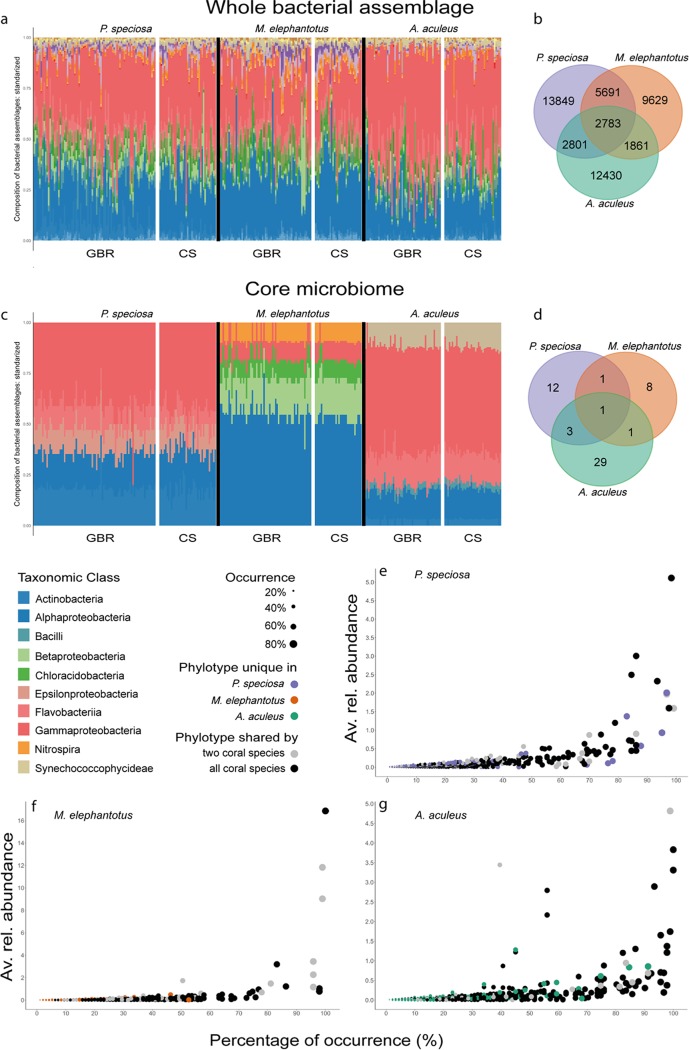
The coral microbiome comprises common and species-specific phylotypes in a stable taxonomic structure across individuals. The taxonomic structure of coral-associated bacteria within individual coral hosts (a) was reflected by that of the core microbiome (c). Alpha- and Gammaproteobacteria dominated the bacterial assemblage composition despite the variability across spatial scales (Fig. 2). Bacterial classes were structurally stable across individuals. Common bacterial phylotypes of the three commonly occurring coral species (species-specific microbiome and resident microbiome [b]) and core microbiome of individual P. speciosa, M. elephantotus, and A. aculeus corals (d). Graphs of the average relative abundance versus percentage of occurrence across coral individuals for P. speciosa (e), M. elephantotus (f), and A. aculeus (g) revealed that highly persistent (≥80%) OTUs are rarely species specific (note the difference in scale in relative abundance). Bacterial assemblage composition was defined by the number of OTUs belonging to the taxonomic level class, standardized by the total per individual. For the extended taxonomic legend, see Fig. S16 at https://figshare.com/s/ffadfdf1a1bab8a37088.
Resident community.
The composition of the bacterial assemblages observed in individual corals, based on the number of OTUs per bacterial class, revealed that Alpha- and Gammaproteobacteria are consistently the dominant taxonomic classes across coral species, geographic regions, and depth (Fig. 3). Here, we report that the numerical dominance of phylotypes within these two bacterial classes, which was reflected in the taxonomic structure, was similar to that which has been previously reported in the numerical abundance of phylotypes within the Alpha- and Gammaproteobacteria (2, 4, 5). Thus, we show that a clear taxonomic redundancy was evident within the individual coral’s microbial assemblage. This pattern of constrained diversity was even more evident when evaluating beta-diversity and the indices of complexity to assess the microbiome.
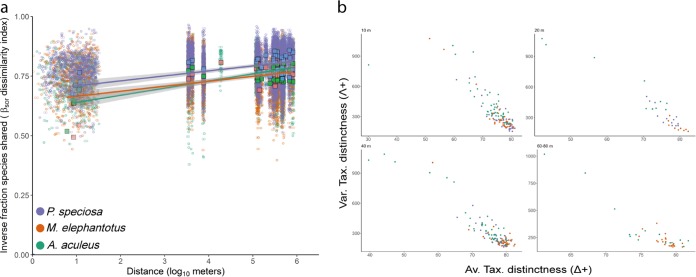
Variations in the composition of bacterial assemblages (beta-diversity [turnover]) (28) showed that the bacterial distance-decay relationship for the three coral species is constant across space or distance, which is a unique feature of the coral microbiome compared to other ecological systems (29–31) (Fig. 4a). This indicates that regardless of community drivers related to the reef environment, the composition of the coral microbiome system is structured at the individual level. The structure and complexity of the assemblage, expressed as taxonomic relatedness (average of taxonomic distinctness [Δ+]) (32) and taxonomic evenness (variation of taxonomic distinctness [Λ+]) (33), provide a measure of the taxonomic spread of communities (34) which has been applied in macroecology (35–37) and more recently utilized in microbial ecology (38, 39). This allows us to assess biodiversity changes on spatial and temporal scales and the response to disturbances. By utilizing this approach in the current study, we found that the taxonomic relatedness is inversely proportional to the taxonomic evenness, thus indicating that the individual microbiome with high taxonomic complexity (high average, Δ+) are more even (low variance, Λ+) and vice versa. This pattern was consistent across all individuals of the three coral species studied, at all depths evaluated, and across all reef locations (Fig. 4b). Interestingly, both M. elephantotus and P. speciosa showed a constrained range in both average and variation of complexity beyond 20 m in reef depth (see Fig. S2 to S6 at https://figshare.com/s/ffadfdf1a1bab8a37088). In contrast, A. aculeus exhibited a broad response ranging across all reef depths. These results suggest that coral growth form, branching versus plating and/or massive forms, may have a substantial influence over the complexity and variation of the coral microbiome, particularly across reef depths.
FIG 4.
Beta-diversity (turnover [a]) and taxonomic breadth (b) of bacterial assemblages. (a) A minimal increase on dissimilarity (i.e., the inverse fraction of species shared) was observed between pairs of individuals across space or distance. Depending on the spatial scales, the composition of the coral microbiome was conserved in 50 to 23% (same reef) and 27 to 18% (distinct reefs). (b) The pattern of high taxonomic relatedness and low taxonomic evenness in the three species regardless of the depth supports the taxonomic structure observed in the coral microbiome. Together, these results suggest that a fraction of the coral microbiome is conserved and taxonomically structured, regardless of the reef environment.
Core microbiome.
A key feature of the individual microbiome in all three corals species studied was the characteristically small group of highly persistent OTUs (core microbiome) (Fig. 3c to g). Each coral species’ core microbiome encompassed phylotypes ranging from rare to highly dominant across reef habitats. In addition, very few bacterial phylotypes were shared between the three coral species. We also found similarities in the taxonomic structure of the core bacteria across species and individuals (core microbiome herein defined as when a bacterial species is present in ≥80% individuals within the study [Fig. 3c and d]). In each coral species, the core microbiome was equivalent in taxonomic complexity and predicted functional capabilities (Fig. 5c; see also Fig. S13 to S15 at https://figshare.com/s/ffadfdf1a1bab8a37088).
FIG 5.
Different bacterial taxonomic structure of representative and highly persistent OTUs (core microbiome) with similar functional capabilities. For each coral species, representative OTUs (a) and core microbiome had a distinct taxonomic structure (Fig. 3c), but equivalent prediction on functional content (b and c). Functional prediction content generated from the relative abundance of KEGG Orthology (KO) genes, normalized and standardized by the total per individual.
The core microbiome of both P. speciosa and A. aculeus was dominated by Gammaproteobacteria phylotypes and included several phylotypes with relative abundance between 0.43 and 5.11% and 0.15 to 4.82%, respectively (Fig. 3e to g; see also Table S2 at https://figshare.com/s/ffadfdf1a1bab8a37088). Overall, the core microbiome of P. speciosa and A. aculeus showed similar taxonomic structure (Fig. 3c), as observed in the bacterial assemblage of the species as a whole (Fig. 3a), with high numbers of phylotypes from Alpha- and Gammaproteobacteria. The P. speciosa core microbiome included phylotypes of the genera Acinetobacter, Cloacibacterium, Corynebacterium, Gluconacetobacter, Mycobacterium, Pseudoalteromonas, Propionibacterium, Pseudomonas, and Rhodobacter and family Hyphomicrobiaceae. The A. aculeus core microbiome comprised OTUs from the genera Alicyclobacillus, Alteromonas, Oleibacter, Prochlorococcus, Propionibacterium, Pseudoalteromonas, Pseudomonas, Synechococcus, and Vibrio and families Endozoicomonadaceae and Flavobacteriaceae. Interestingly, the core microbiome of the coral M. elephantotus was unique compared to the core microbiomes of the other two corals. Most notably, we found that the Alphaproteobacteria are the dominant group within the Mycedium microbiome (Fig. 3a), followed by Betaproteobacteria. Furthermore, in M. elephantotus, the most dominant 11 phylotypes each accounted for a relative abundance between 0.78 and 16.85% (≥80% occurrence in Fig. 3f; see also Table S2 at https://figshare.com/s/ffadfdf1a1bab8a37088). Moreover, M. elephantotus hosted three phylotypes with a high relative abundance (>9%) which together accounted for 37.7% of the relative abundance. These phylotypes belong to the classes Alphaproteobacteria, Betaproteobacteria, and Chloracidobacteria; family Ellin6075, and orders EC94 and Kiloniellales. The family Ellin6075 (class Chloracidobacteria) is unusual in the coral microbiome, but it has been previously found, with variable abundance, in microbialites (40) and soil (41, 42). Members of this family possess a nitrifying capacity (43).
The three coral species have common and distinct biomechanical, morphological, reproductive, and ecological traits. They are all of colonial morphology attached to the benthos, broadcast spawners, are hermaphrodites, and coincide in their Symbiodinium clade association (C3 and C3h in common) (44). However, they also have distinctive traits that may potentially influence microbial associations. M. elephantotus belongs to a robust major clade with encrusting and laminar growth forms that have large polyps (7.5 to 10 mm). P. speciosa and A. aculeus belong to the complex clade, have small polyps (0.8 to 1 mm and 2.4 to 4.6 mm, respectively) and are variable in growth forms (corymbose and laminar, respectively) (44). While the relationship between some of these traits and the coral microbial community has been explored (morphology [45, 46], phylogeny [45], mode of larval development [47–50], zooxanthellae clade [51]), to date, no direct or conclusive relationships have been drawn.
In the current study, only one phylotype, a member from the family Alteromonadaceae (OTU 806717), was found to be highly persistent in all three depth generalist coral species across the entire 10 to 80 m depth range (Fig. 3d). This bacterial phylotype was not only highly persistent in that it was present in 98.4% of individuals, but it was also present at a high relative abundance within both A. aculeus and P. speciosa individuals. This is the first report of a persistent bacterial phylotype within this group across coral species and a broad range of geographical and depth reef locations. However, members of the family Alteromonadaceae have been previously reported to occur in the core microbiome of other coral species (52) and in early life stages of coral (50, 53) and to be important in chemical defenses against pathogenesis in crustaceans (54). A search comparison of the sequence of this phylotype in the BLAST Nucleotide database (National Center of Biotechnology Information [NCBI] [see Table S3 at https://figshare.com/s/ffadfdf1a1bab8a37088]) showed close affiliation to an uncultured bacterium, a member of the Symbiodinium core microbiome (55), and to Alteromonas sp. and Alteromonas macleodii, which are bacteria found to coexist with cyanobacteria Trichodesmium and Prochlorococcus (56–58). When associated with Prochlorococcus, the opportunistic copiotroph and heterotroph bacteria of the genus Alteromonas degrade algal polysaccharides and remove reactive oxygen species (ROS) (56, 59, 60). The persistence of this specific taxon (OTU 806717) could indicate that it plays a critical functional role for the coral or for the coral-zooxanthellae symbiosis. The potential role of members of the Alteromonadaceae family in association with corals has yet to be explored.
Environmentally responsive community.
Given that the reef environment varies across the depth gradient (61) and abiotic factors within the reef impact nutrient acquisition and cycling within the coral host (62, 63), these are also likely to be drivers of microbiome structure. Therefore, we also aimed to determine whether bacterial associations were reflective of the locations within the reef (depth of sampling) from which the corals were collected. We identified bacteria present within all host corals at various depths for each reef location. All three coral species showed persistent bacterial phylotypes across the sampled depths (see Fig. S7 at https://figshare.com/s/ffadfdf1a1bab8a37088). However, at all depths, the A. aculeus microbiome harbored the greatest diversity and highest number of phylotypes. Interestingly, the family Endozoicomonadaceae was present in the core microbiome of A. aculeus at all sampled depths, but was present in M. elephantotus and P. speciosa corals only occasionally. The family Endozoicomonadaceae was found in some M. elephantotus individuals collected at 10 m (TijR) and 40 m (FliR) and in P. speciosa individuals at 10 m (MyrR, O1DT), 40 m (O3BL), and 60 to 80 m (O2HW). Similarly, phylotypes from the phylum Cyanobacteria, including the genera Synechococcus and Prochlorococcus, were present in all individuals of the coral A. aculeus at all depths and reefs, but only occasionally in M. elephantotus (10 m at O1DT and O3BL, 20 m at O2HW, 40 m GreD and TijR, and 60 to 80 m at O1DT) and P. speciosa (all depths at TijR and YonR, 10 m at O1DT and O3BL, 40 m at MyrR, and 60 to 80 m at O2HW, O3BL, and FliR). This clear structuring of the coral microbiome was evident across the biogeographical and depth ranges of the current study, highlighting that redundancy is a consistent feature of the coral microbiome regardless of the region, site, and depth of the reef.
Significantly, analyses over the different spatial scales and depth gradients of the 309 coral individuals examined in the current study showed that the number of sequences and phylotypes and taxonomic structure of depth generalist corals P. speciosa, M. elephantotus, and A. aculeus were clearly comparable (see Fig. S8 to S12 and Table S4 at https://figshare.com/s/ffadfdf1a1bab8a37088). While we found differences in the microbiome in geographic regions of the three coral species (Great Barrier Reef [GBR] versus the Coral Sea [CS]; P < 0.05 by permutational multivariate analysis of variance [PERMANOVA]), differences between reef localities (within region; P < 0.05 by PERMANOVA; see Tables S5 to S8) and depths were not consistent across species (a posteriori analyses, Fig. 2; see also Tables S9 to S12). For example, we found that at Halfway Wall, Bigeye Ledge (Osprey), and Flinders of the Coral Sea, the bacterial assemblages within these reef localities showed similar assemblage structures at 10 m and 20 m but unique assemblages at 40 m and 60 to 80 m. Likely reflective of changes associated with mesophotic conditions (in this case 40 m and deeper), these results suggest substantial variation between individuals and between environments which have different abiotic drivers (such as light penetration, shading, nutrient availability, and water flow). This is highly likely within the diverse habitats of any coral reef environment whereby corals may be exposed to greater upwelling, flow, or shading depending on local reef factors, all of which are known to affect the physiology of the coral, its nutrient cycling, and its reliance on heterotrophic and autotrophic feeding (61–65).
Taxonomic redundancy.
Finally, representative phylotypes from the transient community in each reef for each coral species were selected as groups of OTUs with a multivariate pattern, reflective of that observed in the species-specific microbiome (BVSTEP algorithm, ρ > 0.95). We then evaluated taxonomic relatedness and conducted functional prediction analysis for each individual coral. We found that taxonomic redundancy is reflected in the functional predictions for the corals' microbiome (Fig. 5a and b). We also found that the core microbiome and representative phylotypes per reef were equivalent in taxonomic complexity (Δ+ and Λ+, Fig. 3c and Fig. 5a; see also Table S13 at https://figshare.com/s/ffadfdf1a1bab8a37088) and in functional predictions (Fig. 5b and c; see also Fig. S13 to S15). This research clearly demonstrates that regardless of the bacterial assemblages at a reef, they are taxonomically, and likely functionally, redundant within the coral host. For example, transporters, porphyrin and chlorophyll metabolism, photosynthesis proteins, photosynthesis, methane metabolism, fatty acid metabolism, and bacterial motility proteins were consistently enriched within the coral-associated bacterial assemblage of individual corals across reef habitats (see level 3 KEGG Orthology (KO) at https://figshare.com/s/ffadfdf1a1bab8a37088). This is likely to indicate that within the nutritionally dynamic coral host, there are relatively few niche microhabitats available (e.g., gut [66], surface mucous layer [67], and skeleton [68]) and that those niches are highly consistent across species and reef environments. The hypothesis of functional redundancy in the coral microbiome has yet to be tested.
Conclusion.
We find that the observed simplicity and structure of the corals’ diverse microbial biosphere, in which there is substantial taxonomic (and potentially functional) redundancy, is consistent across coral species, bioregions, and reef depths. These results thereby suggest that within diverse microbial biospheres, such as coral, simplicity exists despite complex environmental drivers. For the individual coral meta-organisms’ microbiome, this simplicity is on average 605 bacterial phylotypes and is likely to be reflective of coral microhabitats and the processes occurring on them. Conceptualizing the coral microbiome as a system of transient, resident, and core microbiomes is likely to make deciphering the microbial contribution to corals’ symbiotic and dysbiotic states more achievable.
MATERIALS AND METHODS
Fragments of corals Mycedium elephantotus (n = 95) and Acropora aculeus (n = 91) were collected from 10 reefs from northern reefs of the Great Barrier Reef (GBR) and the Coral Sea (CS) during the Caitlin Seaview Survey expeditions. Coral specimens were collected at shallow and intermediate depth (10 to 40 m) using regular open-circuit diving between September and December 2012. At mesophotic depth (60 to 80 m), coral specimens were sampled in November 2013 using a remotely operated vehicle (ROV). Based on the bathymetric distribution of the coral species (61), between three and eight individuals were collected at each depth, from each reef location, and samples were preserved in salt-saturated 20% dimethyl sulfoxide (DMSO) with 0.5 M EDTA and stored at –20°C. A nested hierarchical design was used for collection and data analysis: (i) coral species, (ii) region (fixed factor, two levels: GBR and CS), (iii) reefs (random factor nested in region, 10 levels: for the GBR, Great Detached, Tijou Reef, Day Reef, Yonge Reef, and Myrmidon Reef; for the CS, Flinders Reef, Holmes Reef, and in Osprey Reef Dutch Towers, Halfway Wall [also known as Nautilus Wall], and Bigeye Ledge); and (iv) depth [fixed factor, four levels: 10 m (±3 m), 20 m (± 2 m), 40 m (±3 m), and 60 to 80 m]. Coral specimens were collected under permits supplied by the Great Barrier Reef Marine Park Authority (Townsville, Australia) and Commonwealth Marine Reserves, Department of the Environment (Hobart, Australia). For coordinates and site information of reef localities, see references 69 and 61.
DNA extraction, amplification, and sequencing.
DNA extraction was performed using 0.4 (±0.02) g of each coral fragment using the modified protocol of MoBio PowerPlant pro DNA isolation kit (catalog no. 13400-50; MoBio, Carlsbad, CA) described by S. Sunagawa et al. (45). DNA concentration was estimated in a Qubit fluorometer using the Qubit dsDNA (double-stranded DNA) HS (high-sensitivity) assay kit (Thermo Fisher Scientific, Wilmington, DE). Amplified DNA was stored at −20°C before PCR amplification. Genomic template primers 27F/519R (v1-v3 region) were used to amplify bacterial 16S rRNA gene amplicons for examining bacterial assemblage structure. Gene amplicons were amplified in a single-step, 30-cycle PCR (HotStarTaq plus master mix kit; Qiagen, USA). The conditions for PCR were as follows: (i) 3 min at 94°C; (ii) 28 cycles, with each cycle consisting of 30 s at 94°C, 40 s at 53°C, and 1 min at 72°C; (iii) a final elongation step of 5 min at 72°C. PCR products were checked in 2% agarose gels, and samples were pooled in equal proportions based on molecular weight and DNA concentrations. Pooled samples were purified using calibrated Ampure XP beads. DNA library was prepared following the Illumina TruSeq DNA library protocol. Sequencing was performed by MR DNA (Molecular Research LP; Shallowater, TX, USA) using 300-bp paired ends on an Illumina MiSeq platform following the manufacturer’s guidelines.
Sequence analysis.
To establish common phylotypes among coral species, M. elephantotus and A. aculeus data (newly generated here) were jointly analyzed with P. speciosa sequence data (previously analyzed in reference 69). Sequence data analysis was performed using the open-source software Quantitative Insights Into Microbial Ecology (QIIME, version 1.9) (70). Sequences with ambiguous base calls, with homopolymer runs exceeding 6 bp or below 200 bp, were discarded. Barcodes, primers, and chimeras were removed from sequences prior to analysis (Usearch61 [71] for chimera removal). Operational taxonomic units (OTUs) were defined and taxonomically identified with 97% cluster similarity using the RDP classifier and Greengenes database (version 13_8 [72]).
Statistical analysis, taxonomic redundancy, and core microbiome.
Data mining and statistical and taxonomic redundancy analyses were performed with PRIMER v7 and PERMANOVA+ (73). Bacterial assemblage structure was analyzed by composition and structure. Abundance matrices were normalized using the fourth root transformation and standardized by the total per sample. Fourth root transformation was selected to balance the contribution of rare and highly abundant bacteria (74). Raw abundance OTU tables were also converted to presence/absence data to analyze bacterial composition. For both relative abundance and composition data, significant differences in the factors of the design were identified by permutational multivariate analysis of variance (PERMANOVA) using 9,999 permutations, Bray-Curtis and weighted Unifrac distances (on relative abundance data), and Sorensen and unweighted Unifrac distances (on composition data). All the sequences and OTUs obtained from specimens of a species were considered the species-specific microbiome or the whole microbiome of that coral species. Coral species data were analyzed separately after detection of structural differences between them (i.e., excluding the factor “coral species” from statistical analysis). Pairwise comparisons were used for further exploration of significant differences for any factor. PERMANOVA results were visualized in nonmetric multidimensional scaling (nMDS) (75) plots using 95% bootstrap reefs or centroids (identified in figure legends).
A stepwise selection of species (BVSTEP routine) was used to create subset matrices of selected OTUs reflecting the abundance pattern observed in the normalized relative abundance matrix (76–78). Using Bray-Curtis distance matrices, Spearman rank as the correlation method and Rho (ρ) of >0.95 and Delta Rho of 0.001 as the stop criteria, BVSTEP was run for each reef per coral species. Taxonomic redundancy and functional prediction were evaluated on the resulting subset matrices and the core microbiome for each coral species.
Phylotypes consistently present in ≥80% of the individuals were considered highly persistent core microbiome (79). Core 80% was identified for (i) each coral species (ii) each reef per coral species; and (iii) each depth per reef per coral species using the command compute_core_microbiome.py in QIIME. Core 80% matrices were created as subset matrices selecting core 80% OTUs from the normalized relative abundance matrix. Taxonomic redundancy was evaluated using the indices average [Δ+] (32) and variation [Λ+] (33) of taxonomic distinctness. Venn diagrams for the core 80% and the whole bacterial community were generated using Venn diagram software (Bioinformatics and Evolutionary Genomics [http://bioinformatics.psb.ugent.be/webtools/Venn/]). A search in the BLAST Nucleotide database (National Center of Biotechnology Information [NCBI] [https://blast.ncbi.nlm.nih.gov/]) was conducted for the sequence of the OTU 806717 (family Alteromonadaceae) to identify the closest sequence affiliation. Based on the percentage of identity and E value, the top 20 significant alignments were selected and presented in Table S3 at https://figshare.com/s/ffadfdf1a1bab8a37088.
Predicted functional profiling based on bacterial taxonomy.
The Galaxy web version of Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt [80]) was used to produce a prediction of the metagenomic functional content of the subset matrices of representative phylotypes and the core 80% matrices. Each matrix was normalized by copy number, and the metagenome prediction was produced using KEGG Orthology (KOs) and summarized at levels 2 and 3 of KEGG Pathway. The predicted functional profiles of representative OTUs in P. speciosa individuals from Myrmidon reef were not estimated since the OTUs were not present in the Greengenes database. Differences in KEGG pathways were assessed as indicated previously (see “Statistical analysis, taxonomic redundancy, and core microbiome” above). Graphs were produced with ggplot2 (81) as implemented in R (82).
Beta-diversity (turnover).
Beta-diversity was analyzed using packages betapart (28, 83, 84) and geosphere in R (82). Pair-wise turnover was computed for each presence/absence OTU table using Sorensen dissimilarity index. Distance matrixes (one per coral species) among pairs of samples were calculated using their geographic locations and the Vincenty ellipsoid method. Due to the impossibility of assigning the exact location within the reef, we assigned a random location to each colony sampled in each reef (3 to 33 m from the reef coordinate). The relationship between the fraction of species shared (i.e., turnover) and distance for each pair of samples per coral species was explored in plots produced with ggplot2 (81) in R (82).
Data availability.
Raw sequences are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under project number PRJNA435628, accession numbers SAMN10088816 to SAMN10089001.
ACKNOWLEDGMENTS
This work was made possible by support from the Australian Research Council Centre of Excellence for Coral Reef Studies and the Australian Research Council Discovery Program. A. Hernandez-Agreda was supported by Australia Awards Scholarships (AusAID). Specimen collections were carried out as part of the XL Catlin Seaview Survey, funded by the XL Catlin Group and undertaken by the Global Change Institute at The University of Queensland.
We thank Norbert Englebert, Kyra Hay, David Whillas, and Paul Muir for help with specimen collections. We also thank Danielle Asson-Batzel for discussion on data analysis, Ed Roberts for the A. aculeus photo, and Alexander J. Fordyce for the reef photo. We thank Tessa Hill for editorial assistance.
A.H.-A. wrote the manuscript and managed, interpreted, and analyzed the data. W.L. and C.H.A. helped to write the manuscript and analyzed and interpreted the data. P.B. edited the manuscript and interpreted the data. T.D.A. wrote the manuscript and interpreted the data.
Footnotes
Citation Hernandez-Agreda A, Leggat W, Bongaerts P, Herrera C, Ainsworth TD. 2018. Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. mBio 9:e00812-18. https://doi.org/10.1128/mBio.00812-18.
REFERENCES
- 1.Ainsworth TD, Gates RD. 2016. Corals' microbial sentinels. Science 352:1518–1519. doi: 10.1126/science.aad9957. [DOI] [PubMed] [Google Scholar]
- 2.Bourne DG, Morrow KM, Webster NS. 2016. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol 70:317–340. doi: 10.1146/annurev-micro-102215-095440. [DOI] [PubMed] [Google Scholar]
- 3.Knowlton N, Rohwer F. 2003. Multispecies microbial mutualisms on coral reefs: the host as a habitat. Am Nat 162:S51–S62. doi: 10.1086/378684. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez-Agreda A, Gates RD, Ainsworth TD. 2017. Defining the core microbiome in corals’ microbial soup. Trends Microbiol 25:125–140. doi: 10.1016/j.tim.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Blackall LL, Wilson B, van Oppen MJ. 2015. Coral—the world's most diverse symbiotic ecosystem. Mol Ecol 24:5330–5347. doi: 10.1111/mec.13400. [DOI] [PubMed] [Google Scholar]
- 6.Davy SK, Allemand D, Weis VM. 2012. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev 76:229–261. doi: 10.1128/MMBR.05014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muscatine L, Falkowski P, Porter J, Dubinsky Z. 1984. Fate of photosynthetic fixed carbon in light- and shade-adapted colonies of the symbiotic coral Stylophora pistillata. Proc R Soc Lond B Biol Sci 222:181–202. doi: 10.1098/rspb.1984.0058. [DOI] [Google Scholar]
- 8.Muscatine L, McCloskey R, Marian E. 1981. Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration. Limnol Oceanogr 26:601–611. doi: 10.4319/lo.1981.26.4.0601. [DOI] [Google Scholar]
- 9.Baird A, Marshall P. 2002. Mortality, growth and reproduction in scleractinian corals following bleaching on the Great Barrier Reef. Mar Ecol Prog Ser 237:133–141. doi: 10.3354/meps237133. [DOI] [Google Scholar]
- 10.Eakin CM, Morgan JA, Heron SF, Smith TB, Liu G, Alvarez-Filip L, Baca B, Bartels E, Bastidas C, Bouchon C, Brandt M, Bruckner AW, Bunkley-Williams L, Cameron A, Causey BD, Chiappone M, Christensen TRL, Crabbe MJC, Day O, de la Guardia E, Díaz-Pulido G, DiResta D, Gil-Agudelo DL, Gilliam DS, Ginsburg RN, Gore S, Guzmán HM, Hendee JC, Hernández-Delgado EA, Husain E, Jeffrey CFG, Jones RJ, Jordán-Dahlgren E, Kaufman LS, Kline DI, Kramer PA, Lang JC, Lirman D, Mallela J, Manfrino C, Maréchal J-P, Marks K, Mihaly J, Miller WJ, Mueller EM, Muller EM, Orozco Toro CA, Oxenford HA, Ponce-Taylor D, Quinn N. 2010. Caribbean corals in crisis: record thermal stress, bleaching, and mortality in 2005. PLoS One 5:e13969. doi: 10.1371/journal.pone.0013969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glynn PW. 1984. Widespread coral mortality and the 1982–83 El Niño warming event. Environ Conserv 11:133–146. doi: 10.1017/S0376892900013825. [DOI] [Google Scholar]
- 12.Bourne D, Munn C. 2005. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ Microbiol 7:1162–1174. doi: 10.1111/j.1462-2920.2005.00793.x. [DOI] [PubMed] [Google Scholar]
- 13.Rohwer F, Seguritan V, Azam F, Knowlton N. 2002. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10. doi: 10.3354/meps243001. [DOI] [Google Scholar]
- 14.Li J, Chen Q, Zhang S, Huang H, Yang J, Tian X-P, Long L-J. 2013. Highly heterogeneous bacterial communities associated with the South China Sea Reef corals Porites lutea, Galaxea fascicularis and Acropora millepora. PLoS One 8:e71301. doi: 10.1371/journal.pone.0071301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. 2007. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol 9:2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YY, Ling J, Yang QS, Wen CQ, Yan QY, Sun HY, Van Nostrand JD, Shi Z, Zhou JZ, Dong JD. 2015. The functional gene composition and metabolic potential of coral-associated microbial communities. Sci Rep 5:1–11. doi: 10.1038/srep16191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeVantier L, Turak E. 2017. Species richness and relative abundance of reef-building corals in the Indo-West Pacific. Diversity 9:25. doi: 10.3390/d9030025. [DOI] [Google Scholar]
- 18.Bellwood DR, Hughes TP, Folke C, Nyström M. 2004. Confronting the coral reef crisis. Nature 429:827–833. doi: 10.1038/nature02691. [DOI] [PubMed] [Google Scholar]
- 19.Carmona CP, Azcarate FM, de Bello F, Ollero HS, Leps J, Peco B. 2012. Taxonomical and functional diversity turnover in Mediterranean grasslands: interactions between grazing, habitat type and rainfall. J Appl Ecol 49:1084–1093. doi: 10.1111/j.1365-2664.2012.02193.x. [DOI] [Google Scholar]
- 20.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moretti M, de Bello F, Roberts SPM, Potts SG. 2009. Taxonomical vs. functional responses of bee communities to fire in two contrasting climatic regions. J Anim Ecol 78:98–108. doi: 10.1111/j.1365-2656.2008.01462.x. [DOI] [PubMed] [Google Scholar]
- 22.Augustin R, Fraune S, Franzenburg S, Bosch TCG. 2012. Where simplicity meets complexity: Hydra, a model for host–microbe interactions. Adv Exp Med Biol 710:71–81. doi: 10.1007/978-1-4419-5638-5_8. [DOI] [PubMed] [Google Scholar]
- 23.Bosch TCG. 2012. Understanding complex host-microbe interactions in Hydra. Gut Microbes 3:345–351. doi: 10.4161/gmic.20660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F, Berg M, Dierking K, Félix M-A, Shapira M, Samuel BS, Schulenburg H. 2017. Caenorhabditis elegans as a model for microbiome research. Front Microbiol 8:485. doi: 10.3389/fmicb.2017.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 26.Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V. 2015. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baselga A. 2010. Partitioning the turnover and nestedness components of beta diversity. Global Ecol Biogeogr 19:134–143. doi: 10.1111/j.1466-8238.2009.00490.x. [DOI] [Google Scholar]
- 29.Condit R, Pitman N, Leigh EG, Chave J, Terborgh J, Foster RB, Núñez P, Aguilar S, Valencia R, Villa G, Muller-Landau HC, Losos E, Hubbell SP. 2002. Beta-diversity in tropical forest trees. Science 295:666–669. doi: 10.1126/science.1066854. [DOI] [PubMed] [Google Scholar]
- 30.Green JL, Holmes AJ, Westoby M, Oliver I, Briscoe D, Dangerfield M, Gillings M, Beattie AJ. 2004. Spatial scaling of microbial eukaryote diversity. Nature 432:747–750. doi: 10.1038/nature03034. [DOI] [PubMed] [Google Scholar]
- 31.Bates ST, Clemente JC, Flores GE, Walters WA, Parfrey LW, Knight R, Fierer N. 2013. Global biogeography of highly diverse protistan communities in soil. ISME J 7:652–659. doi: 10.1038/ismej.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clarke KR, Warwick RM. 1998. A taxonomic distinctness index and its statistical properties. J Appl Ecol 35:523–531. doi: 10.1046/j.1365-2664.1998.3540523.x. [DOI] [Google Scholar]
- 33.Clarke KR, Warwick RM. 2001. A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar Ecol Prog Ser 216:265–278. doi: 10.3354/meps216265. [DOI] [Google Scholar]
- 34.Gibson R, Barnes M, Atkinson R. 2001. Practical measures of marine biodiversity based on relatedness of species. Oceanogr Mar Biol 39:207–231. [Google Scholar]
- 35.Alahuhta J, Toivanen M, Hjort J, Ecke F, Johnson LB, Sass L, Heino J. 2017. Species richness and taxonomic distinctness of lake macrophytes along environmental gradients in two continents. Freshw Biol 62:1194–1206. doi: 10.1111/fwb.12936. [DOI] [Google Scholar]
- 36.McClanahan TR, Ateweberhan M, Graham NAJ, Wilson SK, Sebastián CR, Guillaume MMM, Bruggemann JH. 2007. Western Indian Ocean coral communities bleaching responses and susceptibility to extinction. Mar Ecol Prog Ser 337:1–13. doi: 10.3354/meps337001. [DOI] [Google Scholar]
- 37.Thrush SF, Hewitt JE, Norkko A, Nicholls PE, Funnell GA, Ellis JI. 2003. Habitat change in estuaries: predicting broad-scale responses of intertidal macrofauna to sediment mud content. Mar Ecol Prog Ser 263:101–112. doi: 10.3354/meps263101. [DOI] [Google Scholar]
- 38.Moss JA, Nocker A, Lepo JE, Snyder RA. 2006. Stability and change in estuarine biofilm bacterial community diversity. Appl Environ Microbiol 72:5679–5688. doi: 10.1128/AEM.02773-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu H, Jiang Y, Al-Rasheid KAS, Al-Farraj SA, Song W. 2011. Application of an indicator based on taxonomic relatedness of ciliated protozoan assemblages for marine environmental assessment. Environ Sci Pollut Res 18:1213–1221. doi: 10.1007/s11356-011-0476-6. [DOI] [PubMed] [Google Scholar]
- 40.Valdespino-Castillo PM, Hu P, Merino-Ibarra M, López-Gómez LM, Cerqueda-García D, Zayas G-D, Pi-Puig T, Lestayo JA, Holman H-Y, Falcón LI. 2018. Exploring biogeochemistry and microbial diversity of extant microbialites in Mexico and Cuba. Front Microbiol 9:510. doi: 10.3389/fmicb.2018.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin C, Mueth N, Hulbert S, Schlatter D, Paulitz TC, Schroeder K, Prescott A, Dhingra A. 2017. Bacterial communities on wheat grown under long-term conventional tillage and no-till in the Pacific Northwest of the United States. Phytobiomes 1:83–90. doi: 10.1094/PBIOMES-09-16-0008-R. [DOI] [Google Scholar]
- 42.Ye J, Zhang R, Nielsen S, Joseph SD, Huang D, Thomas T. 2016. A combination of biochar–mineral complexes and compost improves soil bacterial processes, soil quality, and plant properties. Front Microbiol 7:372. doi: 10.3389/fmicb.2016.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chuang H-P, Imachi H, Tandukar M, Kawakami S, Harada H, Ohashi A. 2007. Microbial community that catalyzes partial nitrification at low oxygen atmosphere as revealed by 16S rRNA and amoA genes. J Biosci Bioeng 104:525–528. doi: 10.1263/jbb.104.525. [DOI] [PubMed] [Google Scholar]
- 44.Madin JS, Anderson KD, Andreasen MH, Bridge TCL, Cairns SD, Connolly SR, Darling ES, Diaz M, Falster DS, Franklin EC, Gates RD, Hoogenboom MO, Huang D, Keith SA, Kosnik MA, Kuo C-Y, Lough JM, Lovelock CE, Luiz O, Martinelli J, Mizerek T, Pandolfi JM, Pochon X, Pratchett MS, Putnam HM, Roberts TE, Stat M, Wallace CC, Widman E, Baird AH. 2016. The Coral Trait Database, a curated database of trait information for coral species from the global oceans. Sci Data 3:160017. doi: 10.1038/sdata.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sunagawa S, Woodley CM, Medina M. 2010. Threatened corals provide underexplored microbial habitats. PLoS One 5:e9554. doi: 10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang J, Yu K, Wang Y, Huang X, Huang W, Qin Z, Pan Z, Yao Q, Wang W, Wu Z. 2017. Distinct bacterial communities associated with massive and branching scleractinian corals and potential linkages to coral susceptibility to thermal or cold stress. Front Microbiol 8:979. doi: 10.3389/fmicb.2017.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Apprill A, Marlow HQ, Martindale MQ, Rappe MS. 2009. The onset of microbial associations in the coral Pocillopora meandrina. ISME J 3:685–699. doi: 10.1038/ismej.2009.3. [DOI] [PubMed] [Google Scholar]
- 48.Leite DCA, Leão P, Garrido AG, Lins U, Santos HF, Pires DO, Castro CB, van Elsas JD, Zilberberg C, Rosado AS, Peixoto RS. 2017. Broadcast spawning coral Mussismilia hispida can vertically transfer its associated bacterial core. Front Microbiol 8:176. doi: 10.3389/fmicb.2017.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lema KA, Bourne DG, Willis BL. 2014. Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora. Mol Ecol 23:4682–4695. doi: 10.1111/mec.12899. [DOI] [PubMed] [Google Scholar]
- 50.Sharp KH, Distel D, Paul VJ. 2012. Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J 6:790–801. doi: 10.1038/ismej.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pantos O, Bongaerts P, Dennis PG, Tyson GW, Hoegh-Guldberg O. 2015. Habitat-specific environmental conditions primarily control the microbiomes of the coral Seriatopora hystrix. ISME J 9:1916–1927. doi: 10.1038/ismej.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leite DC, Salles JF, Calderon EN, Elsas JD, Peixoto RS. 2018. Specific plasmid patterns and high rates of bacterial co‐occurrence within the coral holobiont. Ecol Evol 8:1818–1832. doi: 10.1002/ece3.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ceh J, van Keulen M, Bourne DG. 2013. Intergenerational transfer of specific bacteria in corals and possible implications for offspring fitness. Microb Ecol 65:227–231. doi: 10.1007/s00248-012-0105-z. [DOI] [PubMed] [Google Scholar]
- 54.Gil-Turnes MS, Hay ME, Fenical W. 1989. Symbiotic marine bacteria chemically defend crustacean embryos from a pathogenic fungus. Science 246:116–118. doi: 10.1126/science.2781297. [DOI] [PubMed] [Google Scholar]
- 55.Lawson CA, Raina J-B, Kahlke T, Seymour JR, Suggett DJ. 2018. Defining the core microbiome of the symbiotic dinoflagellate, Symbiodinium. Environ Microbiol Rep 10:7–11. doi: 10.1111/1758-2229.12599. [DOI] [PubMed] [Google Scholar]
- 56.Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. 2011. Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean's surface. PLoS One 6:e16805. doi: 10.1371/annotation/468e1735-605a-40a7-a253-884a117f0e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. 2008. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol 74:4530–4534. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou S, López-Pérez M, Pfreundt U, Belkin N, Stüber K, Huettel B, Reinhardt R, Berman-Frank I, Rodriguez-Valera F, Hess WR. 2018. Benefit from decline: the primary transcriptome of Alteromonas macleodii str. Te101 during Trichodesmium demise. ISME J 12:981–996. doi: 10.1038/s41396-017-0034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.López-Pérez M, Gonzaga A, Martin-Cuadrado A-B, Onyshchenko O, Ghavidel A, Ghai R, Rodriguez-Valera F. 2012. Genomes of surface isolates of Alteromonas macleodii: the life of a widespread marine opportunistic copiotroph. Sci Rep 2:696. doi: 10.1038/srep00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neumann AM, Balmonte JP, Berger M, Giebel H-A, Arnosti C, Voget S, Simon M, Brinkhoff T, Wietz M. 2015. Different utilization of alginate and other algal polysaccharides by marine Alteromonas macleodii ecotypes. Environ Microbiol 17:3857–3868. doi: 10.1111/1462-2920.12862. [DOI] [PubMed] [Google Scholar]
- 61.Englebert N, Bongaerts P, Muir PR, Hay KB, Pichon M, Hoegh-Guldberg O. 2017. Lower mesophotic coral communities (60-125 m depth) of the northern Great Barrier Reef and Coral Sea. PLoS One 12:e0170336. doi: 10.1371/journal.pone.0170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Einbinder S, Mass T, Brokovich E, Dubinsky Z, Erez J, Tchernov D. 2009. Changes in morphology and diet of the coral Stylophora pistillata along a depth gradient. Mar Ecol Prog Ser 381:167–174. doi: 10.3354/meps07908. [DOI] [Google Scholar]
- 63.Lesser MP, Slattery M, Stat M, Ojimi M, Gates RD, Grottoli A. 2010. Photoacclimatization by the coral Montastraea cavernosa in the mesophotic zone: light, food, and genetics. Ecology 91:990–1003. doi: 10.1890/09-0313.1. [DOI] [PubMed] [Google Scholar]
- 64.Bongaerts P, Frade PR, Hay KB, Englebert N, Latijnhouwers KRW, Bak RPM, Vermeij MJA, Hoegh-Guldberg O. 2015. Deep down on a Caribbean reef: lower mesophotic depths harbor a specialized coral-endosymbiont community. Sci Rep 5:7652. doi: 10.1038/srep07652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kahng SE, Garcia-Sais JR, Spalding HL, Brokovich E, Wagner D, Weil E, Hinderstein L, Toonen RJ. 2010. Community ecology of mesophotic coral reef ecosystems. Coral Reefs 29:255–275. doi: 10.1007/s00338-010-0593-6. [DOI] [Google Scholar]
- 66.Ainsworth TD, Fine M, Blackall LL, Hoegh-Guldberg O. 2006. Fluorescence in situ hybridization and spectral imaging of coral-associated bacterial communities. Appl Environ Microbiol 72:3016–3020. doi: 10.1128/AEM.72.4.3016-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ritchie KB. 2006. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322:1–14. doi: 10.3354/meps322001. [DOI] [Google Scholar]
- 68.Yang SH, Lee STM, Huang CR, Tseng CH, Chiang PW, Chen CP, Chen HJ, Tang SL. 2016. Prevalence of potential nitrogen-fixing, green sulfur bacteria in the skeleton of reef-building coral Isopora palifera. Limnol Oceanogr 61:1078–1086. doi: 10.1002/lno.10277. [DOI] [Google Scholar]
- 69.Hernandez-Agreda A, Leggat W, Bongaerts P, Ainsworth TD. 2016. The microbial signature provides insight into the mechanistic basis of coral success across reef habitats. mBio 7:e00560-16. doi: 10.1128/mBio.00560-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anderson M, Gorley RN, Clarke RK. 2008. Permanova+ for Primer: guide to software and statistical methods. Primer-E Limited, Plymouth, United Kingdom. [Google Scholar]
- 74.Clarke KR, Gorley RN, Warwick RM. 2014. Change in marine communities: an approach to statistical analysis and interpretation, 3rd ed. PRIMER-E, Plymouth, United Kingdom. [Google Scholar]
- 75.Clarke KR. 1993. Non‐parametric multivariate analyses of changes in community structure. Aust J Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 76.Clarke K, Warwick R. 1998. Quantifying structural redundancy in ecological communities. Oecologia 113:278–289. doi: 10.1007/s004420050379. [DOI] [PubMed] [Google Scholar]
- 77.Clarke KR, Somerfield PJ, Airoldi L, Warwick RM. 2006. Exploring interactions by second-stage community analyses. J Exp Mar Biol Ecol 338:179–192. doi: 10.1016/j.jembe.2006.06.019. [DOI] [Google Scholar]
- 78.Somerfield PJ, Clarke KR. 1995. Taxonomic levels, in marine community studies, revisited. Mar Ecol Prog Ser 127:113–119. doi: 10.3354/meps127113. [DOI] [Google Scholar]
- 79.Ainsworth TD, Krause L, Bridge T, Torda G, Raina JB, Zakrzewski M, Gates RD, Padilla-Gamino JL, Spalding HL, Smith C, Woolsey ES, Bourne DG, Bongaerts P, Hoegh-Guldberg O, Leggat W. 2015. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J 9:2261–2274. doi: 10.1038/ismej.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wickham H. 2016. ggplot2: elegant graphics for data analysis, 2nd ed Springer, New York, NY. [Google Scholar]
- 82.R Core Team. 2013. R v. 3.0.2: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 83.Baselga A, Orme CDL. 2012. Betapart: an R package for the study of beta diversity. Methods Ecol Evol 3:808–812. doi: 10.1111/j.2041-210X.2012.00224.x. [DOI] [Google Scholar]
- 84.Baselga A, Orme D, Villeger S, De Bortoli J, Leprieur F. Partitioning beta diversity into turnover and nestedness components. Package ‘betapart’, version 1. 2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw sequences are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under project number PRJNA435628, accession numbers SAMN10088816 to SAMN10089001.