Abstract
Micromilling is an underutilized technique for fabricating microfluidic platforms that is well-suited for the diverse needs of the biologic community. This technique, however, produces culture surfaces that are considerably rougher than in commercially available culture platforms and the hydrophilicity of these surfaces can vary considerably depending on the choice of material. In this study, we evaluated the impact of surface topography and hydrophilicity in milled microfluidic devices on the cellular phenotype and function of primary human macrophages. We found that the rough culture surface within micromilled systems affected the phenotype of macrophages cultured in these devices. However, the presence, type, and magnitude of this effect was dependent on the surface hydrophilicity as well as exposure to chemical polarization signals. These findings confirm that while milled microfluidic systems are an effective platform for culture and analysis of primary macrophages, the topography and hydrophilicity of the culture surface within these systems should be considered in the planning and analysis of any macrophage experiments in which phenotype is relevant.
Introduction:
Microfluidic cell culture platforms offer an array of advantages over traditional culture platforms for cell-based, biological research. These platforms utilize small sample volumes, enable efficient use of valuable material such as primary cells and expensive reagents, allow for precise control of the spatio-temporal environment, and offer high assay sensitivity with multiplexed endpoint analysis1. Unfortunately, the high start-up costs and narrow design flexibility of many of the more common microfabrication techniques available, such as injection molding, present major barriers to the wide spread adoption of this technology by the biologic community.
Micromilling is an alternative and underutilized technique for fabricating microfluidic platforms that is well-suited for the diverse needs of the biologic community and is not cost-prohibitive. This technique, which uses mill cutting tools to create microscale culture systems, offers versatility across various materials with short design-to prototype turnaround times and low costs when frequent design iterations are required2. However, micromilling produces culture surfaces with a relatively rough topography and the choice of material for micromilled devices can have dramatic effects on culture surface hydrophilicity. This is in contrast to commercially available cell culture systems, which typically have culture surfaces that are smooth and hydrophilic. As both surface topography and hydrophilicity have been shown to impact cell biology within in vitro cell culture platforms, it is critical to understand the degree to which the topography and hydrophilicity within micromilled systems influence cell biology and whether these factors need to be considered when designing and analyzing experiments.
Macrophages are a cell population whose phenotype and function are particularly sensitive to surface topography and hydrophilicity. These myeloid immune cells have integral roles in immunity, tissue repair, and cancer and can perform a diverse array of functions, including pathogen eradication, matrix remodeling, growth factor secretion, angiogenesis, and immune regulation3–6,4,7–9. These functions are regulated by their phenotype, which can range from a classical or M1 phenotype to an alternative or M2 phenotype. M1 macrophages promote cytotoxic T cell responses and intracellular pathogen destruction through high expression of reactive oxygen intermediates (ROIs) and proinflammatory cytokines7,10. M2 macrophages suppress immune responses and promote tissue repair through high expression of anti-inflammatory cytokines and growth factors10–12.
Environmental stimuli play an important role in regulating macrophage phenotype. These include paracrine factors, such as IL-4 and IL-10, which promote M2 polarization and IFN-g, which promotes an M1 phenotype6,7. Macrophages are also responsive to mechanical stimuli and cell surface interactions with surrounding stroma and surfaces are known regulate their phenotype as well13–15. In vitro, surface topography and hydrophilicity are mechanical and physiochemical factors, respectively, which have been shown to regulate macrophage phenotype 16,17. In this study, we sought to evaluate the effect of surface topography and hydrophilicity within milled microfluidic systems on the phenotype of primary, patient-derived macrophages. We employed acetone treatment of the micromilled surfaces to modulate surface topography and plasma etching to modulate the hydrophilicity of culture surfaces. We evaluated the impact of these surface properties on cell proliferation and gene expression in undifferentiated as well as M1- and M2-polarized macrophages (Figure 1).
Figure 1.
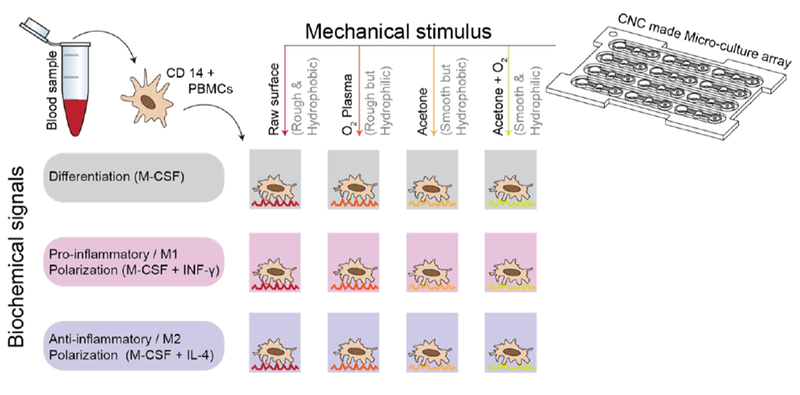
Schematic representation of MDM isolation from patient blood samples followed by culture on untreated and treated micromilled surfaces with varying chemical stimuli.
Materials and Methods:
Device preparation and polishing
Microdevice surfaces were fabricated through standardized rapid prototyping methods. All devices were fabricated from sheets of polystyrene (2 mm, Goodfellow) through micro-CNC milling using a Tormach PCNC 770 mill. Acetone polishing was perfromed by adding 10 μL of acetone directly onto a dry device, allowed to etch for 10 seconds, and then polished by a air hose. The polished device were then washed by water and soked for overnight to remove potential leftover acetone. Plasma treatment was performed shortly before cell seeding through oxygen plasma at 100W for 1 minute. For devices that are acetone polished and oxygen plasma treated, oxygen plasma was performed after the acetone polishing and shortly before cell seeding.
Isolation of Primary Cells
Peripheral blood specimens were collected at the University of Wisconsin with informed written consent under a University of Wisconsin Health Sciences Institutional Review Board (HS-IRB) approved protocol. The HS-IRB complies with the applicable requirements of the Department of Health and Human Services (DHHS) regulations, 45 CFR Part 46; the Food and Drug Administration (FDA regulations, 21 CFR Parts 50, 56, 312, and 812; Veteran’s Administration (VA) Regulations pertaining to the protection of human subjects, 38 CFR Part 16; and the privacy requirements of the Health Insurance Portability and Accountability Act of 1996 implemented by 45 CFR Parts 160 and 164 (Privacy Rule). Blood was drawn into vacutainer tubes (BD Biosciences) with EDTA anticoagulant. Whole blood was diluted 1:1 with Hank’s balanced salt solution (HBSS, Lonza) and 30 mL of diluted blood was underlaid with 10 mL of ficoll-paque PLUS (GE Healthcare) per 50 mL conical tube. The blood was centrifuged for 40 min at 974 g, and resulting buffy coats were washed once with HBSS. Monocytes were enriched from peripheral blood mononuclear cells (PBMCs) using magnetic LS MACS columns (Miltenyi) following incubation with commercially available, pre-conjugated, anti-CD14 magnetic beads (Miltenyi) in a buffer containing 2 mM EDTA (Fisher Scientific) and 0.5% bovine serum albumin (BSA, Sigma-Aldrich) in phosphate buffered saline (PBS, Hyclone).
Cell lines and cell culture
Following isolation, CD14+ cells (isolated from peripheral blood samples collected at the University of Wisconsin as above) were plated in microscale culture devices that had been micromilled followed by: 1) No additional treatment, 2) Acetone treatment, 3) Oxygen plasma treatment, and 4) Acetone and oxygen plasma treatment. Cell from each donor were plated on all 4 surface conditions. In each plate, 10 ul of cells were plated in 12 wells at a concentration of 3×10^6/mL in Corning Cellgro® RPMI 1640 Medium (VWR, USA) containing 10 % FBS, 2% Pen-Strep, 1% glutamine(Life Technologies, USA). Monocyte-derived macrophages (MDMs) were obtained through culture with M-CSF (50ng/mL;TONBO) for 4 days followed by media exchange. Macrophages were then cultured for an additional 3 days in M-CSF (unpolarized macrophages), IFN-g (M1-polarized macrophages), or IL-4 (M2-polarized macrophages).
Nucleic Acid Extraction
mRNA isolation was performed using Dynabeads® mRNA Direct Kit (Life Technologies, USA). Cells were lysed within the culture wells using 10 ul of supplied lysis/binding buffer. Lysate was transferred to tubes containing an additional 30ul of lysis/binding buffer. Culture wells were washed with an additional 10 ul of lysis/binding buffer, which was added to lysate. 10ul of washed beads were added to each sample lysate was washed with 200 ul Buffer A x 2 and 200 ul Buffer B x 1.
Quantitative RT-PCR.
The mRNA elution sample containing PMPs was reverse transcribed using a High Capacity cDNA Reverse Transcriptase kit (Life Tech, USA),according to manufacturer’s directions using Bio-Rad C1000 Thermo Cycler (Bio-Rad, USA). The RT reaction (12.5 μL) was then amplified for 10 cycles using TaqMan® PreAmp (Life Tech, USA) according to manufacturer’s directions and diluted 1:3 in 1× TE (10 mM Tris-HCL pH8, 1 mM EDTA). For TaqMan® assays, 5 μL of diluted cDNA template was mixed with 10 μL iTaq® master mix (Bio-Rad, USA), 1 μL TaqMan® Gene Expression Assay (Specified in Table 6, Life Technologies, USA) and 4 μL nuclease free (NF) water. Each reaction was amplified for 45 cycles (denatured at 95 °C for 15 seconds followed by annealing at 60°C for 1 minute) using a CFX Connect® Real-Time PCR System (Biorad, USA). Threshold cycle (Ct) values were reported.
Cell Counting
Macrophages were counted following 7 days of culture on each surface. Dead cells were removed by aspiration of media and remaining adhered macrophages were then stained with Hoechst and imaged on a Nikkon Eclipse TI-E microscope using the 10x objective. Nuclei were identified and the number of cells was quantified by Hoechst thresholding using NIS Elements AR software. Relative cell number was quantified by calculating the ratio of cells on each surface to the total cell number.
Statistical Analysis
All experiments were repeated at least 3 times. Data are reported as means ± s.e.m. Differences among treatment groups were determined by t tests. P ≤ 0.05 was considered significant.
Results:
Acetone and plasma gas treatment can modulate surface topography and hydrophilicity
To evaluate whether the surface topography and hydrophilicity of milled microfluidic systems impacts macrophage phenotype, microscale devices were first fabricated through the micromilling of polystyrene sheets. Devices were then either acetone polished to produce a smooth-hydrophobic surface, treated with oxygen plasma to produce a rough-hydrophilic surface, treated with a combination of acetone polishing and oxygen plasma treatment to produce a smooth-hydrophilic surface, or left untreated, resulting in rough-hydrophobic surface.
Profilometry was used quantify variations in surface topography among each treatment condition. This analysis demonstrated that micromilling produces patterns of ridges and troughs throughout the culture surface, which reflect both the dimensions as well as the motion of the drill bit. Treatment of the micromilled surfaces with acetone polishing resulted in an overall smoothing of this surface with a 55% lowering in the height from peak to trough (Figure 2A). Plasma gas treatment did not result in any appreciable change in surface roughness.
Figure 2.
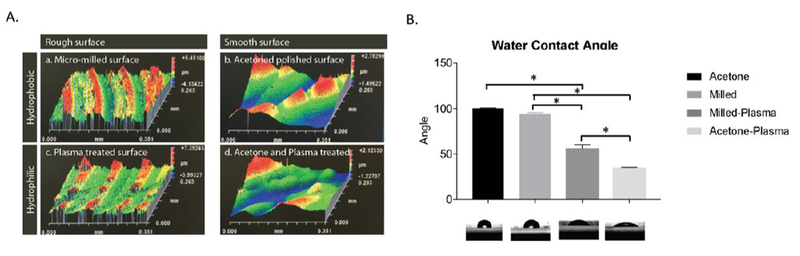
A. Profilometry analysis of the culture surface within micromilled devices after micromilling and after treatment with acetone polishing, plasma etching, and combination of both treatments. B. Measurement of water contact angle on each of the 4 surfaces.
The hydrophilicity of each surface was quantified through measurement of water contact angle. Plasma etching of both the both the micromilled and acetone treated surfaces resulted in a lowering of water contact angle, indicating an increase in surface hydrophilicity in each of these surfaces (Figure 2B). The combination of acetone polishing and plasma etching produced the surface with the lowest water contact angle.
Surface topography and hydrophilicity regulate attached cell number
MDMs from 3 donors were cultured on each of the micromilled surfaces and the number of MDMs attached to each surface was quantified using fluorescence microscopy. After 7 days of culture, there were significantly more cells attached to the micromilled surface than either of the other 3 surfaces (Figure 3). While acetone treatment reduced the number of attached cells, treatment with plasma etching resulted in the least number of attached MDMs, regardless of surface topography.
Figure 3.
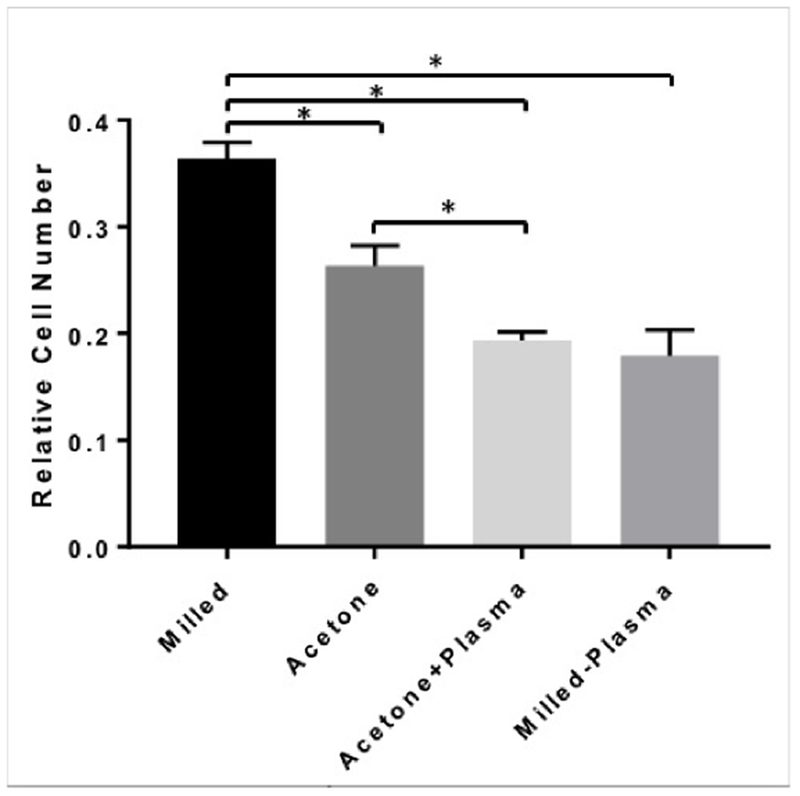
Relative cell number attached to each surface was quantified after 7 days of culture using fluorescence microscopy. *p<0.05.
Surface hydrophilicity regulates gene expression in unpolarized macrophages
MDMs from 3 donors were cultured on each of the micromilled surfaces and evaluated for expression of select M1- and M2-associated genes through quantitative realtime PCR. For MDMs cultured on the non-plasma treated surfaces, variations in surface topography had no effect on expression of either M2 genes (Fig 4A) or M1 genes (Fig 4B). However, when surface hydrophilicity was increased through plasma gas treatment, there were significant changes in both M1 and M2 gene expression. These changes were dependent on the topographical properties of the surface. For macrophages culture on smooth surfaces, an increase in surface hydrophilicity resulted in increased expression of two of the three M2-associated genes evaluated and no significant increase in any of the M1-genes. Conversely, for macrophages cultured on rough surfaces, an increase in hydrophilicity resulted in significant increases in two of the three M1-associated genes and a significant increase in only one of the M2-associated genes.
Figure 4.
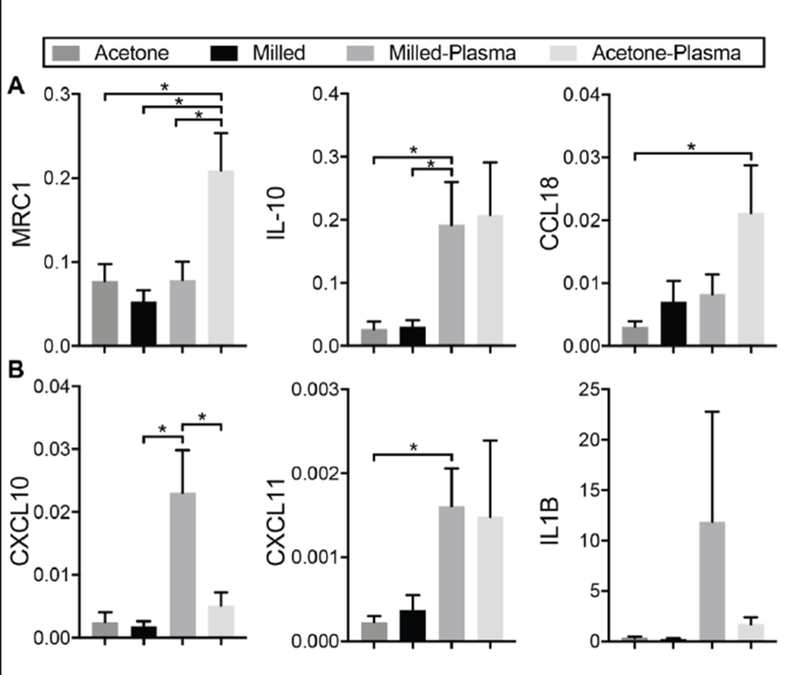
mRNA expression of A. M2-associated genes and B. M1-Associated genes in MDMs cultured on each of the 4 surfaces. Data displayed in normalized relative quantity. *p<0.05.
Surface hydrophilicity regulates gene expression in M1- and M2-polarized macrophages in a polarization-specific manner
Macrophage phenotype is strongly regulated by chemical stimuli, such as cytokines and chemokines, and these are frequently used to polarize and study macrophages in vitro. We therefore evaluated the impact of surface topography and hydrophilicity on macrophage phenotype in the presence of polarizing cytokines. On each of the 4 culture surfaces, macrophages were polarized to either an M1 phenotype with IFN-g or to an M2 phenotype with IL-4. MDMs were then analyzed for expression of M1- and M2-associated genes (Figure 5). In the presence of these IL-4 and IFN-g, surface topography had no significant effect on expression of either M1 or M2 associated genes. However, when macrophages were polarized to an M1 phenotype with IFN-g, culture on surfaces with increased hydrophilicity was associated with an increase in expression of M1 genes with no effect on expression of M2 genes (Figure 5A). This was true on both smooth and rough surfaces. Similarly, for macrophages polarized to an M2 phenotype with IL-4, culture on both of the hydrophilic, plasma-etched surfaces increased expression of M2-associated genes, but did not have a significant effect on M1 genes (Figure 5B).
Figure 5.
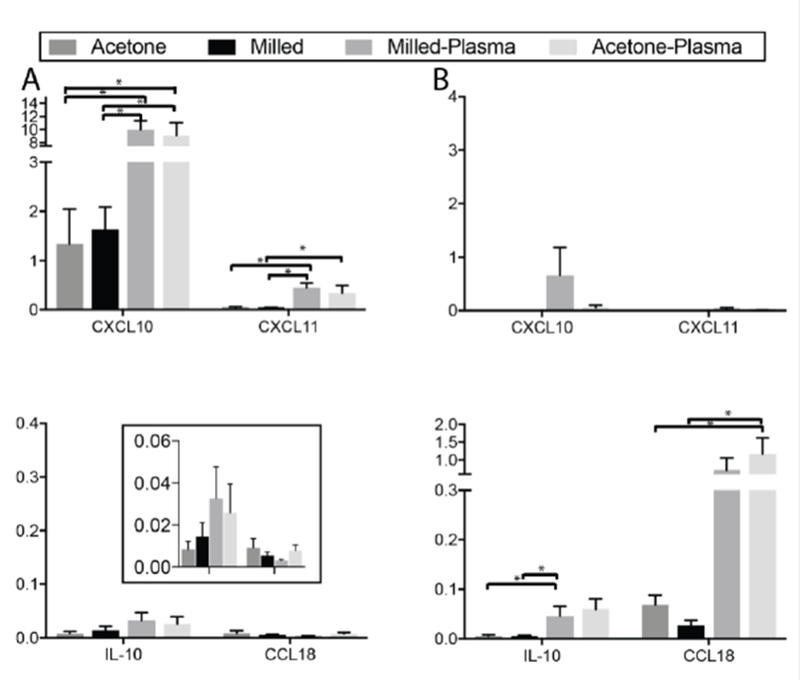
mRNA expression of M1- (top) and M2-associated genes (bottom) in MDMs polarized with A. IFN-g and B. IL-4. *p<0.05.
Discussion:
Micromilling offers numerous advantages for the fabrication of microfluidic devices for cell-based assays, including versatility across various materials, short design-to prototype turnaround times and low costs when frequent design iterations are required. This technique, however, also produces culture surfaces that are rougher than commercial tissue culture devices and can vary considerably in hydrophilicity depending on choice of material. In this study, we evaluated the impact of surface topography and hydrophilicity of milled microfluidic devices on cellular phenotype and function. We utilized macrophages as a model cell type that can be differentially regulated by such platform characteristics and may have be a significant confounding variable to understand the biologic function of different cell populations.
On untreated polystyrene surfaces, the rough surface topography generated by the micromilling process had no effect on macrophage expression of any of the M1 or M2 genes evaluated in this study. Yet, when the hydrophilicity of the polystyrene surfaces was increased through plasma etching, macrophages cultured on the rough micromilled surface expressed higher levels of M1 genes and lower levels of M2 genes than macrophages cultured on surfaces that had been smoothed by acetone polishing. This data demonstrates that while the rough, micromilled surface can promote an M1 phenotype in primary macrophages, the presence of this effect is dependent on surface hydrophilicity.
Prior studies evaluating surface topography and hydrophilicity on macrophage phenotype have demonstrated mixed findings with respect to the effect of these surface properties on macrophage phenotype16–19. While some studies have found that rough surfaces upregulate secretion of pro-inflammatory cytokines, others have found no effect or even and anti-inflammatory effect of surface roughness19,20. Similarly, the impact of surface hydrophilicity has also ranged from pro-inflammatory to anti-inflammatory in the various studies that have evaluated the effect of this physiochemical factor16,18. Our observation that the impacts of topography, hydrophilicity, and chemical polarization are interdependent may explain, at least in part, why these studies have generated discordant conclusions. Additional studies will be needed to validate these findings on additional surfaces, such as titanium, which is frequently utilized for biologic implants and where macrophage-surface interactions may play an important role in clinical outcomes.
Chemical stimuli, such as cytokines, chemokines, and growth factors play an integral role in macrophage polarization and are often utilized to regulate and evaluate macrophage phenotype in vitro. We therefore evaluated how variations in surface roughness and topography affected primary human macrophage phenotype in the presence IL-4 and IFN-g, which promote M2 and M1 polarization respectively. When these cytokines were added to macrophage cultures, we found that variations in surface topography had no effect on M1 and M2 gene expression regardless of surface hydrophilicity/hydrophobicity. Increases in surface hydrophiliciy, however, was associated with dramatic increases expression M1-genes when MDMs were cultured with IFN-g and M2-genes MDMs were exposed to IL-4. This data suggests that hydrophilic surfaces augment the polarizing effect of chemical stimuli in MDMs. While we did not evaluate the mechanism driving this effect, prior studies have demonstrated that increased surface hydrophilicity leads to alterations in the adsorption of a range of biomaterials. Interactions between macrophage surface receptors, such as integrins, and these biomaterials may ultimately regulate the effect of chemical stimuli16. This concept of synergistic chemical-mechanical polarization was demonstrated in a study by Joshi et all, which found that activation of surface integrin receptors regulated M2-polarization by MCSF in macrophages21.
In addition to macrophage phenotype, surface topography and hydrophilicity also impacted macrophage cell number. We found that rougher and more hydrophobic surfaces were associated with higher macrophage cell numbers following seven days of culture. This was likely due to surface effects on macrophage attachment and proliferation, both of which are known to be impacted by culture surface features22–24. Whether the addition of other immune cell populations to these cultures would impact macrophage attachment and/or proliferation on these surfaces is another interesting, which warrants further investigation. In vivo, immune cells often work in concert to facilitate attachment and proliferation. Milled microfluidic culture devices provide a useful platform for investigating the impact of co-culture on immune cell attachment and proliferation on varying surface topographies and hydrophilicities. Additional experiments investigating this important question are in process.
Conclusions:
Macrophage phenotype has important implications in their physiologic roles and is a key focus of macrophage-directed research. Our results demonstrate that the surface topography generated by the micromilling process as well as the hydrophilicity of the culture surface impact the phenotype of primary human macrophages cultured within micromilled systems. Furthermore, we found that the presence and magnitude of these effects are dependent on the presence of chemical polarization stimuli. These findings confirm that while milled microfluidic systems are an effective platform for culture and analysis of primary macrophages, the topography and hydrophilicity of the culture surface within these systems should be considered in the planning and analysis of any macrophage experiments in which phenotype is relevant. We also demonstrated in this study that both surface topography and hydrophilicity can be easily manipulated with post-fabrication modifications, including acetone polishing, which effectively reduced the surface roughness generated by the micromilling process and plasma gas treatment, which increased the hydrophilicity of the relatively hydrophobic material, polystyrene.
Acknowledgements:
We gratefully acknowledge the contributions of Jennifer Schehr to this study and Charlotte Stahlfeld, Anupama Sing, and Rory Bade for their technical assistance. We would like to thank the University of Wisconsin Carbone Cancer Center (UWCCC) Microtechnology Core for use of its facility to complete this research and its director, Dr. Jose A. Jimenez-Torres, for facilitation and helpful discussions, and undergraduate researcher Benjamin Horman for help in microdevice fabrication and preparation. We would also like to thank the patients and their families for their generous contributions to this study.
This work was supported by a Biology of Aging and Age Related Diseases T32 Training Grant (2T32AG000213–34), NIH R01 (CA185251), University of Wisconsin State Economic Engagement & Development (SEED) Research Program, and by the Prostate Cancer Foundation/Movember Challenge Award to Dr. Joshua Lang.
Footnotes
Conflicts of Interest:
Jiaquan Yu and David Beebe hold equity in Stacks to the Future LLC, which licensed some of the technology used in this study
References
- 1.Sackmann EK, Fulton AL, Beebe DJ. The present and future role of microfluidics in biomedical research. Nature 2014;507:181–9. [DOI] [PubMed] [Google Scholar]
- 2.Guckenberger DJ, de Groot TE, Wan AM, Beebe DJ, Young EW. Micromilling: a method for ultra-rapid prototyping of plastic microfluidic devices. Lab on a chip 2015;15:2364–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt T, Carmeliet P. Blood-vessel formation: Bridges that guide and unite. Nature 2010;465:697–9. [DOI] [PubMed] [Google Scholar]
- 4.Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nature reviews Cancer 2008;8:618–31. [DOI] [PubMed] [Google Scholar]
- 5.Bottazzi B, Doni A, Garlanda C, Mantovani A. An integrated view of humoral innate immunity: pentraxins as a paradigm. Annual review of immunology 2010;28:157–83. [DOI] [PubMed] [Google Scholar]
- 6.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nature immunology 2010;11:889–96. [DOI] [PubMed] [Google Scholar]
- 7.Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014;6:1670–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannoni E, Bianchini F, Masieri L, et al. Reciprocal activation of prostate cancer cells and cancer-associated fibroblasts stimulates epithelial-mesenchymal transition and cancer stemness. Cancer research 2010;70:6945–56. [DOI] [PubMed] [Google Scholar]
- 9.Park KY, Li G, Platt MO. Monocyte-derived macrophage assisted breast cancer cell invasion as a personalized, predictive metric to score metastatic risk. Scientific reports 2015;5:13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verreck FA, de Boer T, Langenberg DM, et al. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proceedings of the National Academy of Sciences of the United States of America 2004;101:4560–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffelt SB, Hughes R, Lewis CE. Tumor-associated macrophages: effectors of angiogenesis and tumor progression. Biochimica et biophysica acta 2009;1796:11–8. [DOI] [PubMed] [Google Scholar]
- 12.Filardy AA, Pires DR, Nunes MP, et al. Proinflammatory clearance of apoptotic neutrophils induces an IL-12(low)IL-10(high) regulatory phenotype in macrophages. Journal of immunology (Baltimore, Md : 1950) 2010;185:2044–50. [DOI] [PubMed] [Google Scholar]
- 13.Schaefer L, Babelova A, Kiss E, et al. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. The Journal of clinical investigation 2005;115:2223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Midwood K, Sacre S, Piccinini AM, et al. Tenascin-C is an endogenous activator of Toll-like receptor 4 that is essential for maintaining inflammation in arthritic joint disease. Nature medicine 2009;15:774–80. [DOI] [PubMed] [Google Scholar]
- 15.Kim S, Takahashi H, Lin WW, et al. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature 2009;457:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rostam HM, Singh S, Salazar F, et al. The impact of surface chemistry modification on macrophage polarisation. Immunobiology 2016;221:1237–46. [DOI] [PubMed] [Google Scholar]
- 17.McWhorter FY, Davis CT, Liu WF. Physical and mechanical regulation of macrophage phenotype and function. Cellular and molecular life sciences : CMLS 2015;72:1303–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alfarsi MA, Hamlet SM, Ivanovski S. Titanium surface hydrophilicity modulates the human macrophage inflammatory cytokine response. Journal of biomedical materials research Part A 2014;102:60–7. [DOI] [PubMed] [Google Scholar]
- 19.Tan KS, Qian L, Rosado R, Flood PM, Cooper LF. The role of titanium surface topography on J774A.1 macrophage inflammatory cytokines and nitric oxide production. Biomaterials 2006;27:5170–7. [DOI] [PubMed] [Google Scholar]
- 20.Lee S, Choi J, Shin S, et al. Analysis on migration and activation of live macrophages on transparent flat and nanostructured titanium. Acta Biomater 2011;7:2337–44. [DOI] [PubMed] [Google Scholar]
- 21.Joshi S, Singh AR, Zulcic M, et al. Rac2 controls tumor growth, metastasis and M1-M2 macrophage differentiation in vivo. PloS one 2014;9:e95893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rostam HM, Singh S, Vrana NE, Alexander MR, Ghaemmaghami AM. Impact of surface chemistry and topography on the function of antigen presenting cells. Biomater Sci 2015;3:424–41. [DOI] [PubMed] [Google Scholar]
- 23.Xue L, Greisler HP. Biomaterials in the development and future of vascular grafts. J Vasc Surg 2003;37:472–80. [DOI] [PubMed] [Google Scholar]
- 24.Bartneck M, Heffels KH, Pan Y, Bovi M, Zwadlo-Klarwasser G, Groll J. Inducing healing-like human primary macrophage phenotypes by 3D hydrogel coated nanofibres. Biomaterials 2012;33:4136–46. [DOI] [PubMed] [Google Scholar]


