Summary
Cellular membrane fluidity is a critical modulator of cell adhesion and migration, prompting us to define the systematic landscape of lineage-specific cellular fluidity throughout differentiation. Here, we have unveiled membrane fluidity landscapes in various lineages ranging from human pluripotency to differentiated progeny: (1) membrane rigidification precedes the exit from pluripotency, (2) membrane composition modulates activin signaling transmission, and (3) signatures are relatively germ layer specific presumably due to unique lipid compositions. By modulating variable lineage-specific fluidity, we developed a label-free “adhesion sorting (AdSort)” method with simple cultural manipulation, effectively eliminating pluripotent stem cells and purifying target population as a result of the over 1,150 of screened conditions combining compounds and matrices. These results underscore the important role of tunable membrane fluidity in influencing stem cell maintenance and differentiation that can be translated into lineage-specific cell purification strategy.
Keywords: pluripotency, membrane fluidity, fluidic modulator, cell adhesion, cell sorting
Graphical Abstract
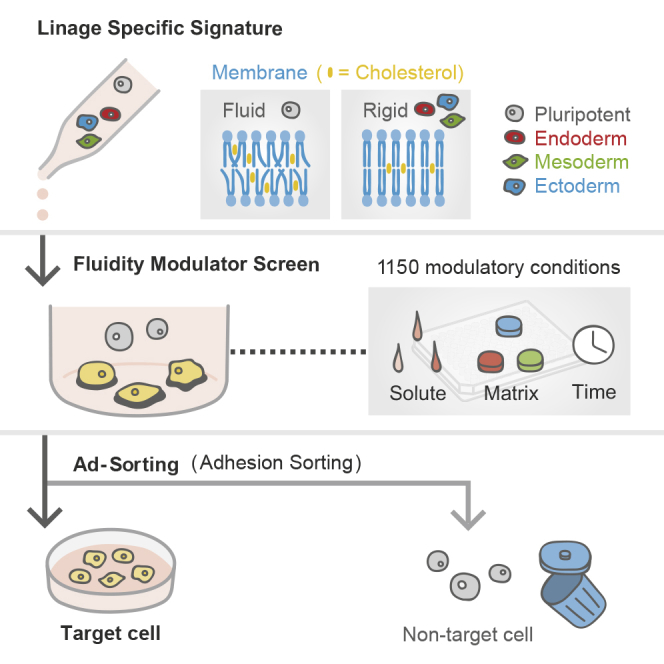
Highlights
-
•
Membrane rigidification precedes the exit from pluripotency
-
•
Germ layer-specific membrane fluidity signature exists
-
•
Identification of polyphenols as a membrane fluidity modulator
-
•
Fluidity-based adhesion sorting purify differentiated progeny from pluripotency
In this article, Takebe and the colleagues unveiled membrane fluidity landscapes in various lineages ranging from human pluripotency to differentiated progeny. By modulating pluripotent fluidity signature, we developed a label-free “adhesion sorting (AdSort)” method with simple cultural manipulation, effectively eliminating pluripotent stem cells as a result of the over 1,150 of screened conditions combining compounds and matrices.
Introduction
Membrane fluidity has been classically considered a key physical property affecting cell adhesion (Juliano and Gagalang, 1979, Santoro and Cunningham, 1981, Schaeffer and Curtis, 1977, Ueda et al., 1976) and communication (Salaita et al., 2010, Zhou et al., 2015). For example, cellular adhesion has been shown to be regulated by the nanocluster formation of adhesion complex-associating ordered lipid rafts (Gaus et al., 2006, Strale et al., 2015); thus, the ease of lateral membrane protein diffusion in optimum membrane fluidity reflects the adhesion characteristics (Eich et al., 2016, Reiss et al., 2011). Both studies indicate that membrane fluidity can be the more generic parameters influencing biological function of cells over adhesion characteristics. Given the important role of lipid membranes (i.e., membrane fluidity) in biological function such as cell adhesion, previous studies have characterized the fluidic properties of human primary neurons (Bonaventura et al., 2014, Noutsi et al., 2016), while others have examined the lipidomic profiles of lysed hepatocyte-like cells (Kiamehr et al., 2017). However, limited numbers of studies have been performed to address its cell-type-specific signatures, especially in the context of stem cell biology; thus, the comprehensive characterization of membrane fluidity in various lineages is essential for gaining key membrane insights into cellular identity and differentiation.
Classical “panning” method is a simplified technique for effective bulk cell purification based on differential adhesion kinetics (Diogo et al., 2012, Lennon and Caplan, 2006, Wysocki and Sato, 1978), waiving antibody-based separation such as fluorescence-activated cell sorting and magnetic-activated cell sorting (MACS). Interestingly, Singh et al. (2013) developed the microfluidic device to eliminate pluripotent stem cell colonies that are sensitive to shear stress and separated from differentiated progeny. Therefore, we hypothesized the cell-type-specific adhesion capacity, if it exists, can be rendered to be an effective and simple panning approach without requiring flow devices or depending on colony morphology for the purification of “early” differentiated progeny from heterogeneous cell mixtures.
Here, we revisited the panning concept by modulating the membrane fluidity of pluripotent membrane and established an adhesion sorting (AdSort) method. Natural polyphenols modulate the pluripotent membrane signature so as to reduce the cell adhesion without any toxic effects relative to its early differentiated progeny. Although the purity and sensitivity of this method is not so comparable with an antibody-based approach, the AdSort method will likely be a complementary separation method for cell manufacturing due to its minimal time, toxicity, and cost.
Results
Membrane Rigidification Prior to Pluripotency Exit of Human iPSCs
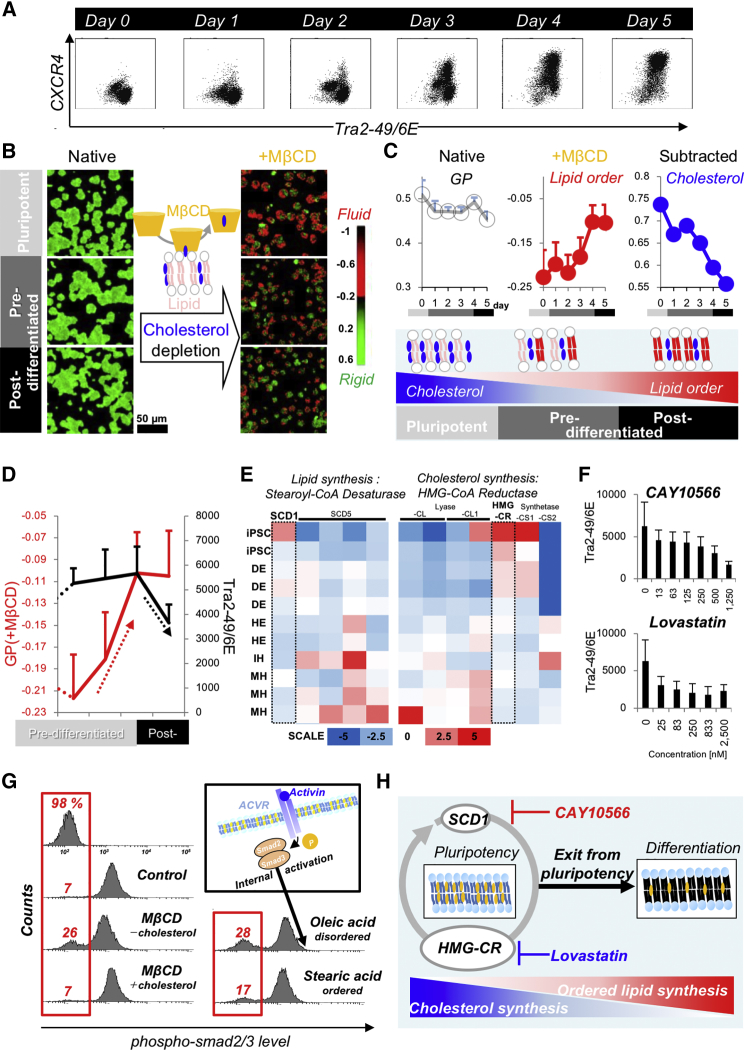
To evaluate the time-progression of membrane fluidity during pluripotency exit and differentiation (Figure 1A), laurdan (Gaus et al., 2003, Owen et al., 2011, Parasassi et al., 1991, Viard et al., 1997) was applied to obtain generalized polarization (GP) as an index of membrane fluidity. The native membrane of pluripotent and pre-/post-differentiated cells showed comparable GP during endodermal induction (Figure 1B, left). In contrast, the cholesterol-depleted membrane obtained by methyl-beta-cyclodextrin (MβCD) (Yamamoto and Ando, 2013, Zidovetzki and Levitan, 2007), treatment revealed a distinguishable fluidic signature for the pluripotent membrane (red in the GP image) from that of the early differentiated progeny (Figures 1B, right and S1A). It should be noted that such abrupt decrease in GP of pluripotent membrane is not influenced by the induced pluripotent stem cell (iPSC) lines (Figures S1E and S1F). The results indicated that the membrane signature of iPSCs (highly cholesterol content) is not influenced by the donor differences. Quantitative fluidic analysis of the cholesterol-depleted membrane of human iPSCs during endodermal induction confirmed the progressive rigidification of lipids (red) and decrease in cholesterol (blue, Figure 1C, upper), resulting in a gradual increase in ordered lipid level (Figure 1C, lower).
Figure 1.
Membrane Rigidification Prior to Pluripotency Exit
(A) Flow cytometry analysis of pluripotent cells during endodermal induction. A differentiation marker (CXCR4, marker of definitive endoderm cells) and a pluripotent marker (Tra2-49/6E) were used.
(B) Representative GP images of pluripotent and pre-/post-differentiation cells in the absence/presence of MβCD. Pre-/post-differentiation cells were obtained at days 3 and 5 during endodermal induction, respectively.
(C) Statistical analysis of the mean GP during endodermal induction in the absence (gray) and presence (red) of MβCD and the calculated cholesterol level (blue). Estimated membrane composition is schematically illustrated in the lower panel.
(D) Differentiation (CXCR4) and pluripotency marker (tra2-49/6E) is co-plotted with GP (+MβCD) during induction. Error bars originate from three independent experiments (n = 3).
(E) Microarray analysis of SCD1/hydroxymethylglutaryl-CoA reductase (HMG-CR) expression in iPSCs during endodermal induction, which was reanalyzed data of our previous study (Takebe et al., 2013).
(F) Concentration dependence of CAY10566 (inhibitor of unsaturated lipid synthesis, SCD1) and lovastatin (inhibitor of cholesterol synthesis, HMG-CR) on pluripotency. Error bars originate from the SD of the histograms of Tra2-49/6E expression.
(G) Phosphorylated-Smad2/3 expression of pluripotent cells in the presence and absence of fluidic modulators.
(H) Estimated role of SCD1 and HMG-CoA reductase in the maintenance of pluripotency.
Error bars show the standard deviation.
Interestingly, such lipid rigidification preceded the exit of pluripotency, based on pluripotency marker analysis by flow cytometry (FCM) (Tra2-49/6E, Figures 1D, S1B, and S1C in detail), indicating a higher specificity to pluripotent state than surface marker profiling. mRNA levels of the other pluripotency marker sets (Oct4/Nanog/Sox2) were analyzed by qPCR during endodermal induction (Figure S1D). Although, of these markers, the SOX2 expression level decreases at almost the same timescale as membrane rigidification, overall membrane fluidity changes precede the downregulation of the other markers. In agreement with this, the timeline of pluripotency exit is similar to endodermal induction of human embryonic stem cells (Vallier et al., 2009) and murine iPSCs (Balasiddaiah et al., 2013), and mesodermal induction of mouse embryonic stem cells (Zhu et al., 2015). To assess the origin of membrane rigidification dynamics, key fluidity determinants, (1) lipid and (2) cholesterol biosynthetic pathways, were assessed. Regarding lipid synthesis, stearoyl-coA desaturase (SCD1) was proposed as a prominent enzyme converted into fluidic unsaturated oleic acid, leading to the proliferation of pluripotent cells (Ben-David et al., 2013). Microarray analysis confirmed the SCD1-specific expression of pluripotent cells (Figure 1E, left). Addition of an inhibitor (CAY10566) at a milder concentration (<500 nM) strongly decreased the pluripotency level (Figure 1F) without affecting viability (Figure S2B, left). In the case of cholesterol synthesis, microarray analysis showed the high expression of hydroxymethylglutaryl- coenzyme A (CoA) reductase (Greenwood et al., 2006) in pluripotent cells (Figure 1E, right). Thus, mild treatment with the inhibitor lovastatin (<833 nM) also accelerated exit from pluripotency (Figure 1F) without affecting viability (Figure S2B, right).
Does such a change in membrane composition affect transmission of differentiation stimuli? To test this, the phosphorylation of Smad2/3 activated by activin (Pauklin and Vallier, 2015) was monitored in the presence of lipid-composition modulators (Figure 1G). Cholesterol depletion and fatty acid insertion produced a four times greater silent fraction (17%–28%), with 32 times quicker reaction than activin receptor inhibition (Figures S2E and S2F). This silencing could originate from the restriction of receptor movement under the conditions of optimal physical membrane properties (Salaita et al., 2010). These results indicate that membrane composition to pluripotent state is unique, as shown in previous studies on cancer cell lines (Beloribi-Djefaflia et al., 2016, Edmond et al., 2015, Remorino et al., 2017, Zhao et al., 2016), and the exit from pluripotency significantly alters its composition, which might sensitize collective cells to differentiation stimuli (Figure 1H).
Presence of Lineage-Specific Membrane Fluidity Signatures
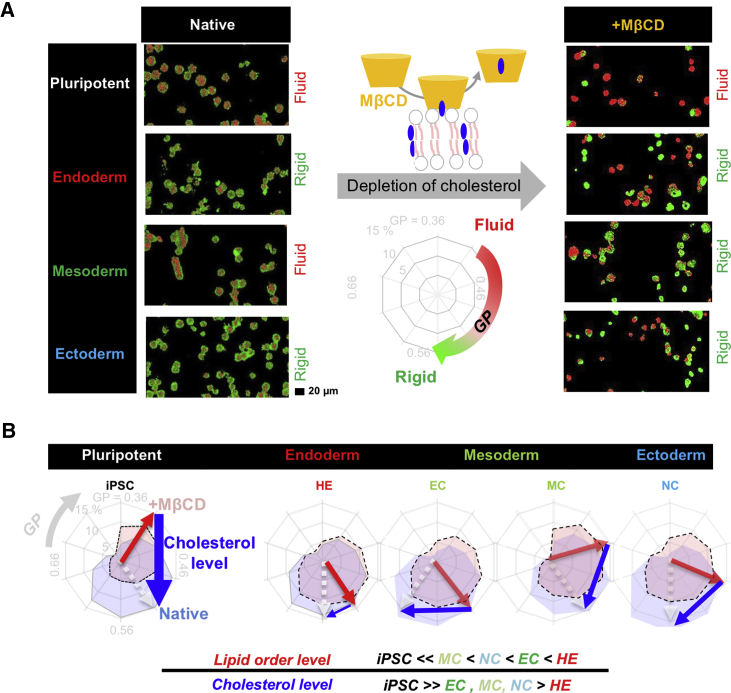
Next, to assess whether the membrane fluidity fingerprint is unique to the other cell states, we systematically evaluated the membrane property of pluripotent cells and the early differentiated progeny, including definitive endoderm (DE) cells, hepatic endoderm (HE) cells, endothelial cells (ECs), mesenchymal cells (MCs), and neural crest (NC) cells, covering all three germ layers (Figure 2A). To assess the lineage specification, gene expression analysis of CXCR4/CER1 (DE), HNF4α (HE), and AFP/ALB (MH), and FCM analysis of CD31/CD144 (EC) and CD166/CD90 (MC) were routinely confirmed as described previously (Takebe et al., 2017). Interestingly, in contrast to iPSCs or mesodermal derivatives, the native membrane of later endoderm/ectoderm cells was clearly recognized as rigidized (Figure 2A, left). Upon cholesterol depletion by MβCD, the mesodermal membrane clearly rigidized, whereas the pluripotent membrane fluidized compared with that of the other early differentiating progeny. Quantitative GP histograms of the cholesterol-depleted (red) or native membrane presented in a radar plot indicated that the pluripotent state also showed lipid disorder (peak position, red arrow) with a high cholesterol content (blue arrow) compared with the endodermal state, and such tendency was preserved for the mesoderm and ectoderm states (Figure 2B, original histograms in Figures S3A and S3B). These results indicated the presence of a distinct physical membrane fingerprint, i.e., a membrane fluidity signature, for pluripotent cells and early differentiated progeny in terms of the (1) native lipid order level (iPSCs ≪ MCs < NC cells < HE cells) and (2) cholesterol level (iPSCs ≫ ECs ∼ MCs ∼ NC cells > HE cells). Thus, the fluidity measurement of native and/or cholesterol-depleted membranes revealed the presence of lineage-specific membrane fluidity signatures, which are temporally regulated throughout cellular differentiation.
Figure 2.
Lineage-Specific Membrane Fluidity Signatures
(A) Representative GP images of pluripotent cell derivatives with different germ layer origins in the absence/presence of MβCD. Early differentiated progeny was induced and characterized with specific markers according to established protocols (Takebe et al., 2017). Pluripotent, iPSC; endoderm, HE; mesoderm, MC; ectoderm, NC.
(B) GP radar plot of iPSCs and derivatives covering all germ layer origins. Dotted arrows show the maximum peaks in the presence (red) and absence (blue) of MβCD. Solid arrows show the differences between the red and blue ends, representing the cholesterol level. Three independent experiments were performed (n = 3, raw histograms are displayed in Figures S3A and S3B).
Fluidity Modulator Screening for Pluripotent Membrane
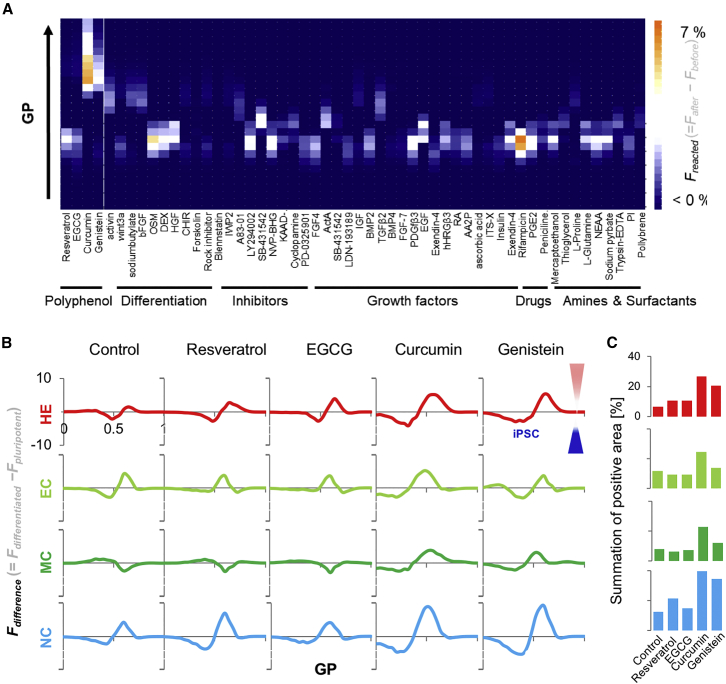
Classical studies have revealed that surfactants fluidize the cell membrane, weakening cell adhesion (Juliano and Gagalang, 1979, Schaeffer and Curtis, 1977, Taraboletti et al., 1989); however, such toxic modification renders the cells unusable (Ulloth et al., 2007). Although MβCD strongly magnifies these potential differences (Figure 1), such a strong depletion of cholesterol reduces cell viability, which is a critical problem for their actual use after characterization. To identify specific fluidic modulators for pluripotent cells without significantly affecting the function and viability of targeted differentiated progeny (Figures S4D–S4F), we screened a self-built small-molecule library combined with an image-based fluidity determination assay and identified polyphenol as a potential augmenter (Figure 3A). The GP (vertical axis) values of the iPSC membrane in the presence of small molecules (horizontal axis) were plotted with z-color strength as Freacted = Fafter − Fbefore to identify effective molecule. F is frequency in the histogram before (Fbefore) and after (Fafter) reaction with small molecules. To further confirm that the identified polyphenols act as fluidic modulators specifically for the pluripotent membrane, Fdifference = Fdifferentiated − Fpluripotent were plotted as GP values in the absence (control) and presence of 100 μM polyphenols (Figure 3B). Stronger modulators (curcumin/genistein) especially enhanced the differences, with 4-fold greater positive area (Figure 3C). These results indicate that fluidity differences between pluripotent cells and early differentiated progeny were successfully augmented by the natural polyphenols.
Figure 3.
Identification of Polyphenols as a Fluidic Modulator for Pluripotent Membrane
(A) Fluidity-based drug screening for iPSC fluidic modulators. The power of Freacted = Fafter−Fbefore is plotted as GP and small molecules.
(B) Fdifference= Fdifferentiated − Fpluripotent is plotted as GP in the absence/presence of polyphenols. Higher Fdifference indicates that the histograms from differentiated cells are dominant.
(C) Summation of positive area in (B).
AdSort Method for Cell Purification
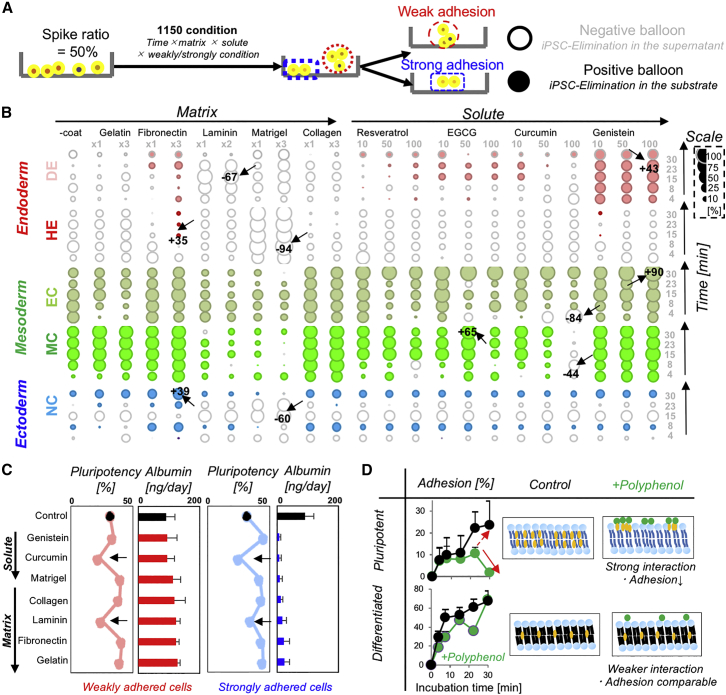
Given that membrane fluidity plays key roles to regulate the subsequent biological function, we further aimed to devise a practical methodology for label-free cell purification by using the cell adhesion characteristics, which are a more specific physical parameters under membrane fluidity. We initially evaluated adhesion differences between two distinct differentiation stages as an elimination ratio with empty (supernatant) and filled (substrate) balloons (Figure 4A). Balloon arrays combining fluidity modulators (i.e., solute), conventional adhesion regulators (i.e., time and matrix), and weakly/strongly adhered conditions were obtained after screening 1,150 different conditions, identifying arrays of specific conditions to separate out specific early progenitors from iPSCs (Figure 4B). Interestingly, cell lineage-specific adhesion strength order was summarized as FHE < FiPSC < FMC. Adhesion strength of differentiated fibroblast has been known to be stronger than those of iPSCs (Yu et al., 2018, i.e., FiPSC < FMC). In contrast, adhesion strength of late stage definitive endodermal cells showed gradual decreases according to the specific integrin subtypes (Ogaki et al., 2016, i.e., FHE < FiPSC). Although both studies support the obtained order of adhesion strength, further studies are necessary to provide the conclusive answer.
Figure 4.
Purification of Differentiated Progeny from Pluripotency by Fluidity Signature Sorting
(A) Schematic protocols for the cell purification experiment.
(B) Balloon plot of the elimination ratio of iPSCs from differentiated cells.
(C) Quantitative analysis of flow cytometry analysis of cells with high Tra2-49/6E expression (day 1) and albumin secretion at day 18. The definitions are given in the legend to Figure S3C. The error bars originate from three independent experiments.
(D) Representative adhesion kinetics of pluripotent cells and early differentiated progeny (endoderm, HE; mesoderm, EC) in the absence/presence of polyphenols (100 μM genistein). Proposed mechanism for enhancing fluidity and adhesion differences between pluripotent cells and early differentiated progeny. The error bars originate from three independent experiments (n = 3).
Error bars show the standard deviation.
The safety and functionality of separated cells were tested. Most importantly, AdSort using curcumin lowers initial pluripotency levels of HE (50% spiked) under the control (Figure 4C, left dark arrows) according to FCM analysis (Figures S3C–S3E) with the resultant high cell recovery ratio (∼80%, Figure S4C). Moreover, curcumin lowered the coverage ratio of iPSCs (GFP labeled) (Figures S4A and S4B). For functional analysis, equal volumes of iPSCs and iPSC-hepatic endoderm cells were mixed, and albumin secretion of separated hepatic endoderm cells was evaluated after AdSort-based cell separation (Figure 4C, right). For all conditions, the weakly adhered cells secreted higher albumin levels, indicating functional HE cell enrichment in the supernatant. Regarding the other lineages, polyphenol-based separation, in this case genistein, preserves the cells with plug formation capacity or contraction capacity, which are typical characteristics of ECs (Figure S4D) or MCs (Figure S4E), without significantly altering their viability (Figure S4F). Moreover, separated endothelial and MCs under various incubation times facilitated liver organoid formation and supported the enhancement of albumin secretion (Figures S4G and S4H). Hence, these results suggest to confirm the minimal invasiveness of our system. Of note, polyphenols specifically reduced the adhesion of pluripotent cells but not other differentiated progeny, leading to an increased difference in adhesion kinetics (Figure 4D, left). The shape of Fdifference (Figure 3B) resembles that of the cholesterol-depleted membrane (Figure S3B, bottom), suggesting that polyphenols interact with membranes with fluidic lipids and cholesterol confirmed by model membrane experiments (Figure 4D, right) (Hwang et al., 2003, Karewicz et al., 2011, Matsuzaki et al., 2017, Neves et al., 2015, Ogawa et al., 2016, Sun et al., 2009).
Discussion
Membrane fluidity influences stem cell maintenance and differentiation, possibly through the modulation of intra-cellular signaling transmission. For example, the ease of ephrin constriction in fluidic membranes augments internal signaling (Salaita et al., 2010). Here, a stimulated change in the membrane composition transmitted to internal signaling is a comparably short timescale relative to that of conventional phosphorylation inhibitors (Figures S2C–S2F). These results potentially led us to the hypothesis that membrane rigidification can be transmitted to neighboring cells, resulting in the explosive acceleration of a differentiation wave. Salaita et al. (2010) emphasized that intermembrane signaling is initially triggered by the clustering of adhesion ligands in the fluid membrane. Such physical connections among cells with different fluidic membrane potentials can strengthen cell-cell signaling, leading to the “relay” of membrane fluidity signatures. Further studies, such as those using the model membrane system (Salaita et al., 2010), will further delineate the presence of fluidic relays during the stem cell differentiation.
Experimental Procedures
Materials
Deionized water from a Milli-Q device (Millipore, Molsheim, France) was used throughout this study. Unless stated otherwise, all other chemicals were purchased either from Sigma-Aldrich (Tokyo, Japan), Invitrogen (Tokyo, Japan), or Wako (Tokyo, Japan). Pure chemicals (Tokyo, Japan) and were used without further purification.
Cell Culture and Differentiation
All procedures involving the use of human stem cell were approved by ethics commission of Yokohama City University and Tokyo Medical and Dental University.
FfI01, NcGMP1 (ET), and FfI14S04 (M66) human iPSC clones used in this study were kindly provided by CiRA (Kyoto, Japan) and Dr. Xianmin Zeng (XCell, CA, USA). Undifferentiated human iPSCs were maintained on laminin 511 (imatrix-511, nippi)-coated plastic dishes. For germ layer differentiation, we followed slightly modified protocols. DE cells, HE cells, MH, ECs, MCs were obtained based on modified previous protocols (Camp et al., 2017, Takebe et al., 2017), and NC cells were obtained based on previous protocols. To confirm the fluidic signature of iPSCs (high cholesterol content), four cell lines were used (Figure S2). For the demonstration of the AdSort impact on the cell purification, a single-cell line (FFI01) was used combining 1,150 screening conditions. Obtained optimal condition for the purification of cell sources possibly depends on the iPSC lines; however, such dependency can be clarified by using our AdSort method in future studies.
Fluidity Measurements
For the quantitative evaluation of cell membrane fluidity, we basically followed well-established protocols (Gaus et al., 2003, Owen et al., 2011, Parasassi et al., 1991). In brief, dimethyl-6-dodecanoyl-2-naphthylamine (laurdan, AdipoGen Life Science, CA, USA) was selected as a fluidity probe and dissolved into a DMSO solution (9 mM final). The final concentration of the probe was constant (33 μM final) in RPMI 1640 medium (+10 mM MβCD) and StemFit AK02N culture medium. Cells were incubated in the media for 30 min at 37°C and then observed by confocal microscopy (TCS-SP8, Leica microsystems, Tokyo, Japan) with a 37°C incubation chamber (Tokken, Tokyo, Japan). A 405-nm laser diode was utilized as a light source, and the fluorescence intensity at λ = 406–460 nm (I406−460) or λ = 470–530 nm (I470−530) was obtained by the spectrum analyzer inside the confocal unit. To evaluate the cell membrane fluidity quantitatively, GP values were defined:
| (Equation 1) |
| (Equation 2) |
Here, GPref = 0.207, the convention in the literature, was defined as a reference GP value3 such that GP values for model membranes with areas of liquid-ordered and -disordered phases separated at around GP = 0. GPmeas was calculated by measuring laurdan in DMSO (33 μM) using the same microscopic setup, as follows:
| (Equation 3) |
Spike Measurements
To obtain the elimination ratio of Figure 4B, GFP-labeled iPSCs were spiked with differentiated cells at a ratio of 1:1 (3 × 105 in total, 300 μL in final volume). The suspension was seeded on 48-well plates (Falcon) and incubated at 37°C to examine various factors, including time, solute, and matrix. To coat the matrix protein on the well surface, the wells were pre-incubated for 30 min in the concentration used for the induction of differentiation: ×1 gelatin (0.09 mg/cm2), ×1 fibronectin (0.91 μg/cm2), ×1 laminin (0.24 μg/cm2), ×1 Matrigel (7.14 μg/cm2), and ×1 collagen (0.91 μg/cm2). The solute stock solution was dissolved in ethanol (120 mM final). The iPSC elimination ratio of the mixtures was calculated using Equations 4 and 5:
| (Equation 4) |
| (Equation 5) |
Here, contamination ratiobase is 50% due to the 1:1 mixture of the green iPSCs and differentiated cells. Thus, the strength of the elimination ratio ranged from −100 (supernatant elimination) to approximately +100% (adhesion elimination), which are the limits for eliminating the iPSCs from the suspension. It should be noted that the phase and fluorescence images of the wells were captured by an automated microscope (Keyence, USA), and the number of cells was counted by the Keyence counting program. For further chemical fixation methods (Benoit et al., 1988), live imaging dyes (Lulevich et al., 2009) were not used thus avoiding significant effects on viable cell adhesion.
For further differentiation of separated hepatic endoderm, which was spiked with iPSCs in Figure 4C, strongly adhered cells were incubated in the differentiation medium (Hepatocyte Culture Media BulletKit, HCM, Lonza, Tokyo, Japan) with 1 μM rock inhibitor and laminin (1× concentration). Weakly adhered cells in the supernatant were quenched by adding 300 μL of the culturing differentiation medium. All the culture media were exchanged on days 1, 4, and 7, and the albumin concentration of supernatant was measured on day 10. The iPSC coverage ratio in Figures S4A and S4B was calculated according to Equation 6:
| (Equation 6) |
It should be noted that the culture conditions at the day 0 (immediately after the separation) were set to favor iPSCs: (1) rock inhibitor (1 μM) suppressed iPSC apoptosis after separation (Watanabe et al., 2007) and (2) the presence of laminin 511 facilitated iPSC adhesion (Miyazaki et al., 2012).
Author Contributions
T.M. conceived the study, designed and performed the experiments, collected and analyzed the data, and wrote the manuscript. S.M., T.K., E.Y., and S.O. performed the experiments with the technical guidance and expertise of T.M. and T.T. S.O. and H.Y.Y. reviewed the manuscript. H.T. provided advice regarding the research strategy. T.T. conceived the study, designed the experiments, obtained funding, and wrote and reviewed the manuscript.
Acknowledgments
We thank Asuka Kodaka for illustration materials. We also thank Tomoko Hisai and Nahoko Hijikata, Hiroko Nozawa, Yukie Nafune, and Noriko Mizuguchi, Hiroaki Ayabe, Masaru Koido, Shodai Togo, Mami Watanabe and all other members for technical assistance. The present work was supported by grants from the Japan Society for the Promotion of Science (16J11759 to T.M., 16KT0073 to T.T. and H.Y.Y., and 15H05351 to H.Y.Y.), the Japan Science and Technology Agency (PRESTO to T.T.), the Naito Foundation (to H.Y.Y.), and the AMED Research Center Network for the Realization of Regenerative Medicine to H.T., and also to T.T. and H.Y.Y. under grant number JP18bm0704025. T.T. is a New York Stem Cell Foundation Robertson Investigator.
Published: September 6, 2018
Footnotes
Supplemental Information includes four figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.08.010.
Supplemental Information
References
- Balasiddaiah A., Moreno D., Guembe L., Prieto J., Aldabe R. Hepatic differentiation of mouse iPS cells and analysis of liver engraftment potential of multistage iPS progeny. J. Physiol. Biochem. 2013;69:835–845. doi: 10.1007/s13105-013-0260-9. [DOI] [PubMed] [Google Scholar]
- Beloribi-Djefaflia S., Vasseur S., Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U., Gan Q.F., Golan-Lev T., Arora P., Yanuka O., Oren Y.S., Leikin-Frenkel A., Graf M., Garippa R., Boehringer M. Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen. Cell Stem Cell. 2013;12:167–179. doi: 10.1016/j.stem.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Benoit J., Cormier M., Wepierre J. Comparative effects of four surfactants on growth, contraction and adhesion of cultured human fibroblasts. Cell Biol. Toxicol. 1988;4:111–122. doi: 10.1007/BF00141290. [DOI] [PubMed] [Google Scholar]
- Bonaventura G., Barcellona M.L., Golfetto O., Nourse J.L., Flanagan L.A., Gratton E. Laurdan monitors different lipids content in eukaryotic membrane during embryonic neural development. Cell Biochem. Biophys. 2014;70:785–794. doi: 10.1007/s12013-014-9982-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp J.G., Sekine K., Gerber T., Loeffler-Wirth H., Binder H., Gac M., Kanton S., Kageyama J., Damm G., Seehofer D. Multilineage communication regulates human liver bud development from pluripotency. Nature. 2017;546:533–538. doi: 10.1038/nature22796. [DOI] [PubMed] [Google Scholar]
- Diogo M.M., da Silva C.L., Cabral J.M. Separation technologies for stem cell bioprocessing. Biotechnol. Bioeng. 2012;109:2699–2709. doi: 10.1002/bit.24706. [DOI] [PubMed] [Google Scholar]
- Edmond V., Dufour F., Poiroux G., Shoji K., Malleter M., Fouque A., Tauzin S., Rimokh R., Sergent O., Penna A. Downregulation of ceramide synthase-6 during epithelial-to-mesenchymal transition reduces plasma membrane fluidity and cancer cell motility. Oncogene. 2015;34:996–1005. doi: 10.1038/onc.2014.55. [DOI] [PubMed] [Google Scholar]
- Eich C., Manzo C., De Keijzer S., Bakker G.-J., Reinieren-Beeren I., García-Parajo M.F., Cambi A. Changes in membrane sphingolipid composition modulate dynamics and adhesion of integrin nanoclusters. Sci. Rep. 2016;6:20693. doi: 10.1038/srep20693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaus K., Gratton E., Kable E.P., Jones A.S., Gelissen I., Kritharides L., Jessup W. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc. Natl. Acad. Sci. USA. 2003;100:15554–15559. doi: 10.1073/pnas.2534386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaus K., Le Lay S., Balasubramanian N., Schwartz M.A. Integrin-mediated adhesion regulates membrane order. J. Cell Biol. 2006;174:725–734. doi: 10.1083/jcb.200603034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood J., Steinman L., Zamvil S.S. Statin therapy and autoimmune disease: from protein prenylation to immunomodulation. Nat. Rev. Immunol. 2006;6:358–370. doi: 10.1038/nri1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang T.-C., Koeppe R.E., Andersen O.S. Genistein can modulate channel function by a phosphorylation-independent mechanism: importance of hydrophobic mismatch and bilayer mechanics. Biochemistry. 2003;42:13646–13658. doi: 10.1021/bi034887y. [DOI] [PubMed] [Google Scholar]
- Juliano R., Gagalang E. The effect of membrane-fluidizing agents on the adhesion of CHO cells. J. Cell Physiol. 1979;98:483–489. doi: 10.1002/jcp.1040980307. [DOI] [PubMed] [Google Scholar]
- Karewicz A., Bielska D., Gzyl-Malcher B., Kepczynski M., Lach R., Nowakowska M. Interaction of curcumin with lipid monolayers and liposomal bilayers. Colloids Surf. B Biointerfaces. 2011;88:231–239. doi: 10.1016/j.colsurfb.2011.06.037. [DOI] [PubMed] [Google Scholar]
- Kiamehr M., Viiri L.E., Vihervaara T., Koistinen K.M., Hilvo M., Ekroos K., Kakela R., Aalto-Setala K. Lipidomic profiling of patient-specific iPSC-derived hepatocyte-like cells. Dis. Model Mech. 2017;10:1141–1153. doi: 10.1242/dmm.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon D.P., Caplan A.I. Isolation of human marrow-derived mesenchymal stem cells. Exp. Hematol. 2006;34:1604–1605. doi: 10.1016/j.exphem.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Lulevich V., Shih Y.P., Lo S.H., Liu G.Y. Cell tracing dyes significantly change single cell mechanics. J. Phys. Chem. B. 2009;113:6511–6519. doi: 10.1021/jp8103358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki T., Ito H., Chevyreva V., Makky A., Kaufmann S., Okano K., Kobayashi N., Suganuma M., Nakabayashi S., Yoshikawa H.Y. Adsorption of galloyl catechin aggregates significantly modulates membrane mechanics in the absence of biochemical cues. Phys. Chem. Chem. Phys. 2017;19:19937–19947. doi: 10.1039/c7cp02771k. [DOI] [PubMed] [Google Scholar]
- Miyazaki T., Futaki S., Suemori H., Taniguchi Y., Yamada M., Kawasaki M., Hayashi M., Kumagai H., Nakatsuji N., Sekiguchi K. Laminin E8 fragments support efficient adhesion and expansion of dissociated human pluripotent stem cells. Nat. Commun. 2012;3:1236. doi: 10.1038/ncomms2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves A.R., Nunes C., Reis S. New insights on the biophysical interaction of resveratrol with biomembrane models: relevance for its biological effects. J. Phys. Chem. B. 2015;119:11664–11672. doi: 10.1021/acs.jpcb.5b05419. [DOI] [PubMed] [Google Scholar]
- Noutsi P., Gratton E., Chaieb S. Assessment of membrane fluidity fluctuations during cellular development reveals time and cell type specificity. PLoS One. 2016;11:e0158313. doi: 10.1371/journal.pone.0158313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogaki S., Omori H., Morooka M., Shiraki N., Ishida S., Kume S. Late stage definitive endodermal differentiation can be defined by Daf1 expression. BMC Dev. Biol. 2016;16:19. doi: 10.1186/s12861-016-0120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K., Hirose S., Nagaoka S., Yanase E. Interaction between tea polyphenols and bile acid inhibits micellar cholesterol solubility. J. Agric. Food Chem. 2016;64:204–209. doi: 10.1021/acs.jafc.5b05088. [DOI] [PubMed] [Google Scholar]
- Owen D.M., Rentero C., Magenau A., Abu-Siniyeh A., Gaus K. Quantitative imaging of membrane lipid order in cells and organisms. Nat. Protoc. 2011;7:24–35. doi: 10.1038/nprot.2011.419. [DOI] [PubMed] [Google Scholar]
- Parasassi T., De Stasio G., Ravagnan G., Rusch R., Gratton E. Quantitation of lipid phases in phospholipid vesicles by the generalized polarization of Laurdan fluorescence. Biophys. J. 1991;60:179–189. doi: 10.1016/S0006-3495(91)82041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauklin S., Vallier L. Activin/Nodal signalling in stem cells. Development. 2015;142:607–619. doi: 10.1242/dev.091769. [DOI] [PubMed] [Google Scholar]
- Reiss K., Cornelsen I., Husmann M., Gimpl G., Bhakdi S. Unsaturated fatty acids drive disintegrin and metalloproteinase (ADAM)-dependent cell adhesion, proliferation, and migration by modulating membrane fluidity. J. Biol. Chem. 2011;286:26931–26942. doi: 10.1074/jbc.M111.243485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remorino A., De Beco S., Cayrac F., Di Federico F., Cornilleau G., Gautreau A., Parrini M.C., Masson J.B., Dahan M., Coppey M. Gradients of Rac1 nanoclusters support spatial patterns of Rac1 signaling. Cell Rep. 2017;21:1922–1935. doi: 10.1016/j.celrep.2017.10.069. [DOI] [PubMed] [Google Scholar]
- Salaita K., Nair P.M., Petit R.S., Neve R.M., Das D., Gray J.W., Groves J.T. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science. 2010;327:1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro S.A., Cunningham L.W. Platelet-collagen adhesion—membrane fluidity and the development of high affinity adhesion through multiple interacting sites. Coll. Relat. Res. 1981;1:517–526. doi: 10.1016/s0174-173x(81)80033-7. [DOI] [PubMed] [Google Scholar]
- Schaeffer B.E., Curtis A. Effects on cell adhesion and membrane fluidity of changes in plasmalemmal lipids in mouse L929 cells. J. Cell. Sci. 1977;26:47–55. doi: 10.1242/jcs.26.1.47. [DOI] [PubMed] [Google Scholar]
- Singh A., Suri S., Lee T., Chilton J.M., Cooke M.T., Chen W., Fu J., Stice S.L., Lu H., McDevitt T.C. Adhesion strength-based, label-free isolation of human pluripotent stem cells. Nat. Methods. 2013;10:438–444. doi: 10.1038/nmeth.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strale P.-O., Duchesne L., Peyret G., Montel L., Nguyen T., Png E., Tampé R., Troyanovsky S., Hénon S., Ladoux B. The formation of ordered nanoclusters controls cadherin anchoring to actin and cell-cell contact fluidity. J. Cell Biol. 2015;210:333–346. doi: 10.1083/jcb.201410111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Hung W.C., Chen F.Y., Lee C.C., Huang H.W. Interaction of tea catechin (-)-epigallocatechin gallate with lipid bilayers. Biophys. J. 2009;96:1026–1035. doi: 10.1016/j.bpj.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.R., Ueno Y., Zheng Y.W., Koike N. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- Takebe T., Sekine K., Kimura M., Yoshizawa E., Ayano S., Koido M., Funayama S., Nakanishi N., Hisai T., Kobayashi T. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 2017;21:2661–2670. doi: 10.1016/j.celrep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Taraboletti G., Perin L., Bottazzi B., Mantovani A., Giavazzi R., Salmona M. Membrane fluidity affects tumor-cell motility, invasion and lung-colonizing potential. Int. J. Cancer. 1989;44:707–713. doi: 10.1002/ijc.2910440426. [DOI] [PubMed] [Google Scholar]
- Ueda M.J., Ito T., Okada T., Ohnishi S.-I. A correlation between membrane fluidity and the critical temperature for cell adhesion. J. Cell Biol. 1976;71:670–674. doi: 10.1083/jcb.71.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloth J.E., Almaguel F.G., Padilla A., Bu L., Liu J.W., De Leon M. Characterization of methyl-beta-cyclodextrin toxicity in NGF-differentiated PC12 cell death. Neurotoxicology. 2007;28:613–621. doi: 10.1016/j.neuro.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L., Touboul T., Chng Z., Brimpari M., Hannan N., Millan E., Smithers L.E., Trotter M., Rugg-Gunn P., Weber A. Early cell fate decisions of human embryonic stem cells and mouse epiblast stem cells are controlled by the same signalling pathways. PLoS One. 2009;4:e6082. doi: 10.1371/journal.pone.0006082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viard M., Gallay J., Vincent M., Meyer O., Robert B., Paternostre M. Laurdan solvatochromism: solvent dielectric relaxation and intramolecular excited-state reaction. Biophys. J. 1997;73:2221–2234. doi: 10.1016/S0006-3495(97)78253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S., Nishikawa S., Muguruma K. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Wysocki L., Sato V. “Panning” for lymphocytes: a method for cell selection. Proc. Natl. Acad. Sci. USA. 1978;75:2844–2848. doi: 10.1073/pnas.75.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Ando J. Endothelial cell and model membranes respond to shear stress by rapidly decreasing the order of their lipid phases. J. Cell Sci. 2013;126:1227–1234. doi: 10.1242/jcs.119628. [DOI] [PubMed] [Google Scholar]
- Yu L., Li J., Hong J., Takashima Y., Fujimoto N., Nakajima M., Yamamoto A., Dong X., Dang Y., Hou Y. Low cell-matrix adhesion reveals two subtypes of human pluripotent stem cells. Stem Cell Reports. 2018;11:142–156. doi: 10.1016/j.stemcr.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Prijic S., Urban B.C., Tisza M.J., Zuo Y., Li L., Tan Z., Chen X., Mani S.A., Chang J.T. Candidate antimetastasis drugs suppress the metastatic capacity of breast cancer cells by reducing membrane fluidity. Cancer Res. 2016;76:2037–2049. doi: 10.1158/0008-5472.CAN-15-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Mao H., Joddar B., Umeki N., Sako Y., Wada K., Nishioka C., Takahashi E., Wang Y., Ito Y. The significance of membrane fluidity of feeder cell-derived substrates for maintenance of iPS cell stemness. Sci. Rep. 2015;5:11386. doi: 10.1038/srep11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Yao X., Liang Y., Liang D., Song L., Jing N., Li J., Wang G. Mediator Med23 deficiency enhances neural differentiation of murine embryonic stem cells through modulating BMP signaling. Development. 2015;142:465–476. doi: 10.1242/dev.112946. [DOI] [PubMed] [Google Scholar]
- Zidovetzki R., Levitan I. Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochim. Biophys. Acta. 2007;1768:1311–1324. doi: 10.1016/j.bbamem.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.