Abstract
Spatial patterns of cell differentiation in developing tissues can be controlled by receptor tyrosine kinase (RTK) signaling gradients, which may form when locally secreted ligands activate uniformly expressed receptors. Graded activation of RTKs can span multiple cell diameters, giving rise to spatiotemporal patterns of signaling through the Extracellular Signal Regulated/Mitogen Activated Protein Kinase (ERK/MAPK), which connects receptor activation to multiple aspects of tissue morphogenesis. This general mechanism has been identified in numerous developmental contexts, from body axis specification in insects to patterning of the mammalian neocortex. We review recent quantitative studies of this mechanism in Drosophila oogenesis, an established genetic model of signaling through the Epidermal Growth Factor Receptor (EGFR), a highly conserved RTK.
Introduction
The Drosophila eggshell is a complex asymmetrical structure that houses the developing embryo, and mediates its interaction with the environment [1]. Some of its most prominent structural features are the micropyle, an anterior tapered tube for sperm entry, and a pair of dorsal respiratory appendages, used for gas exchange during embryogenesis. The establishment of the eggshell polarity is highly interdependent with that of developing embryo. The eggshell polarity can be traced back to two rounds of Epidermal Growth Factor Receptor (EGFR) signaling from the oocyte to the surrounding somatically derived follicle cells (Figure 1a). Both rounds of signaling depend on Gurken (GRK), a TGFa-like ligand, which is secreted from the oocyte and activates EGFR in the follicle cells [2]. GRK secretion is a highly regulated process, reflecting tight localization of the grk mRNA and protein around the oocyte nucleus (Figure 1b).
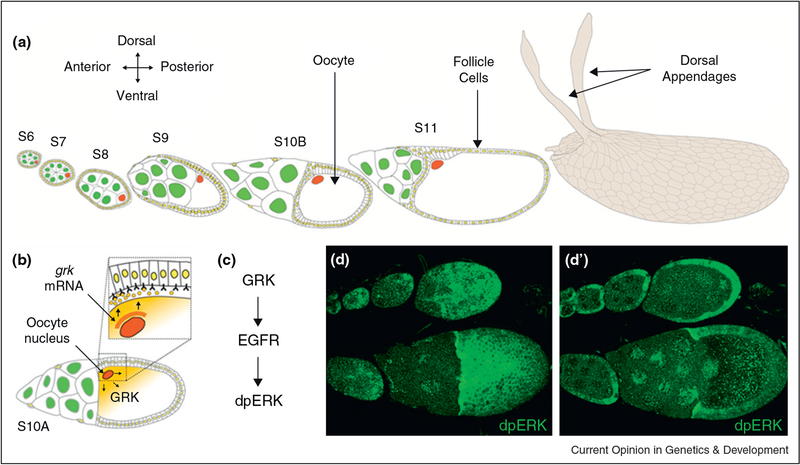
Figure 1.
The body axes of the Drosophila eggshell and the future embryo are established by the asymmetrical localization of the oocyte nucleus, and Gurken (GRK) synthesis, during oogenesis, (a) Schematic of egg chamber development during stages 6–11 of oogenesis (left) and the resulting eggshell (right), (b) grk mRNA accumulates in close proximity of the oocyte nucleus. The protein is locally produced by the oocyte and secreted into the perivitelline space, where it diffuses and binds receptors in the surrounding follicle cells, (c) Activation of EGFR by GRK initiates a protein kinase cascade that leads to double phosphorylation of ERK (dpERK). (d-d’) Dorsal surface and cross section views of egg chambers at different stages of oogenesis. The pattern of ERK activation, as recognized by an antibody specific for the double phosphorylated form of the protein, reflects the distribution pattern of the ligand. During early stages (top), dpERK is activated in a posterior-to-anterior gradient, while at late stages (bottom), dpERK is activated in a dorsal-to-ventral gradient.
The anterior posterior axis of the egg chamber is established early in oogenesis, when the oocyte moves to a posterior position within the nurse cell oocyte complex [3]. The first phase of EGFR activation by GRK occurs at stage 5 of oogenesis, when the oocyte is still very small and only occupies a minor fraction of the egg chamber. At this point, GRK activates EGFR in the follicle cells directly adjacent to the oocyte, instructing them to adopt a posterior fate. In response, these posterior follicle cells send a signal back to the oocyte. While molecular mechanisms of this reverse signal remain to be fully elucidated, its main effect is a polarity reversal of the network of microtubules within the oocyte. Since the oocyte nucleus is associated with the network, it relocates to the anterior of the oocyte, where it is stably anchored at a random position under the plasma membrane. This movement establishes the dorsal ventral asymmetry of the egg chamber, grk mRNA, which is always found in close proximity to the oocyte nucleus, now starts accumulating in the region around the oocyte nucleus.
During later stages of oogenesis, as the volume of the oocyte increases, the asymmetric distribution of grk mRNA results in localized secretion of the GRK protein, which activates the EGFR in the overlying follicle cells and patterns the follicular epithelium by establishing the expression patterns of multiple genes. The two phases of EGFR signaling in the follicle cells can be visualized by the patterns of dual phosphorylated ERK (dpERK, Figure 2d–d’). In both cases, we see graded activation patterns, which decay with distance from the source of GRK production. Here, we focus on the dorsoventral (DV) stage of EGFR activation, reviewing recent work on the distribution of GRK and its transcriptional effects.
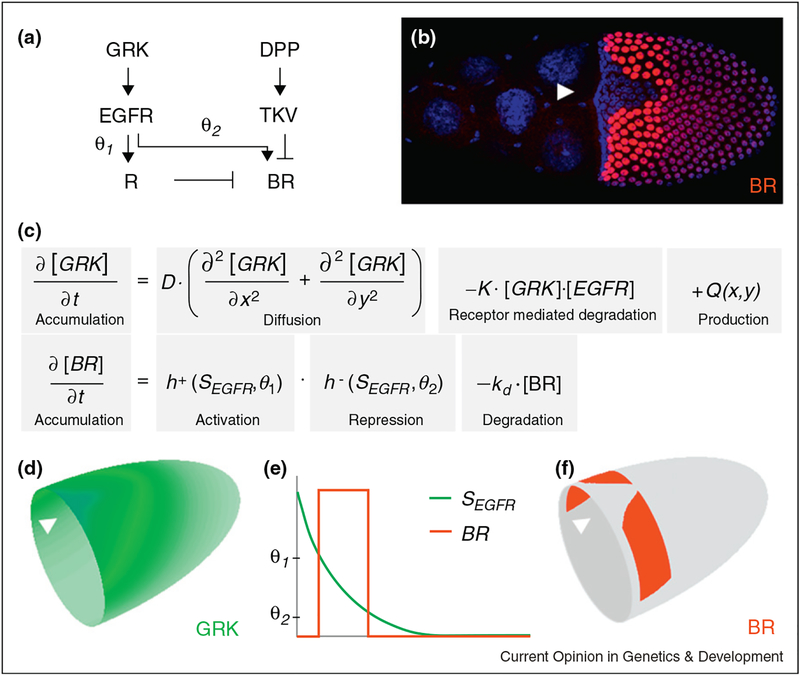
Figure 2.
Model for the regulation of the expression pattern of broad (br). (a, e) The EGFR pathway regulates br in an incoherent feedforward loop, where EGFR activation induces both br (at a low threshold value θ2) and its repressor R (at a higher threshold value θ2). (b) BR (red) is expressed in two groups of follicle cells (nuclei shown in blue) on either side of the dorsal midline, (c–f) Mathematical model for the two-dimensional distribution of GRK and BR expression. Arrow heads mark the position of the dorsal midline.
Quantifying the GRK gradient
Several lines of evidence suggest that GRK acts as a morphogen gradient during the DV patterning of the follicle cells [4,5]. GRK is released from a localized source, and establishes several distinct genes expression boundaries across the follicular epithelium. For example, pipe (pip), which encodes an enzyme essential for the DV patterning of the future embryo and is repressed by EGFR signaling, is expressed in the ventral follicle cells [6–8]. Another target of GRK is broad (br), a transcription factor necessary for the formation of the respiratory eggshell appendages; br is expressed in two dorsal groups of follicle cells, positioned symmetrically on either side of the dorsal midline [9,10]. Finally, sprouty (sty), which encodes an intracellular inhibitor of EGFR signaling, is expressed in a narrow region over the dorsal midline. Together with the border of pip domain, the ventral and dorsal borders of the br pattern and the border of sty expression provide examples of genes controlled by low, intermediate, and high levels of GRK, respectively. In agreement with the morphogen model, these boundaries respond in predictable manner to quantitative manipulations of the GRK gradient [11••,12•].
The gradient-based mechanism of follicle cell patterning by GRK has been elucidated by genetic approaches, but the gradient itself has not been measured directly. At this point, the only quantitative description of the spatial distribution of GRK comes from a computational approach, which is based on a localized production and uniform degradation model of gradient formation [11••]. This model accounts for the localized production of GRK from the oocyte, its diffusion in the extracellular space, and degradation by EGFR-mediated endocytosis. According to this model, the shape of the GRK gradient (i.e. the concentration profile divided by the maximal value at the dorsal most position) depends on a single dimensionless parameter, given by the ratio of the dynamic and geometric lengths scales of the problem.
The geometric length scale is provided by the size of the patterned tissue, whereas the dynamic length scale can be interpreted as the average distance to which the oocyte-derived GRK molecule diffuses before it is internalized by EGFR in the follicle cells. On the basis of reaction-diffusion theory, this distance depends on the diffusivity of secreted GRK, the levels of EGFR expression, and the rates of ligand-receptor binding and internalization. While none of these parameters are known in vivo, what determines the shape of the gradient are not their individual values, but a collective effect, given by the ratio of the dynamic and geometric length scales in the model. Hence, the problem of quantifying the GRK gradient can be reduced to estimating a single parameter in a mathematical model of the source-diffusion-degradation mechanism. Using a genetic system that allowed us to vary one parameter in an otherwise wild type background, precise measurements of the boundary of pipe expression could be used to provide anchor points for the Grk distribution [11••,13]. Using a combination of imaging, genetic, and statistical approaches, we were thus able to provide the first quantitative characterization of the GRK gradient.
On the basis of this framework, the spatial distribution of GRK is graded throughout the entire tissue, which is consistent with notion that GRK directly controls multiple gene expression borders during the DV patterning of the follicle cells. At the same time, a parameter estimation approach relies on a number of assumptions, such as the fact that the gradient is at steady state and that the system operates in a ligand limiting regime, where EGFR is present in excess. While some of these assumptions can be justified a posteriori, a true test of the model will be provided only by direct quantitative analysis of the GRK gradient. Recently, a first step in this direction has been made in a study that used electron microscopy to visualize the spatial distribution of GRK internalized by the follicle cells [14•]. The results of this work are in semi-quantitative agreement with model-based predictions, but more work remains to be done in the future.
Pattern formation by the GRK gradient
Studies of transcriptional interpretation of the GRK gradient are still in early stages. To illustrate the current state of knowledge, we discuss the model proposed for GRK-dependent formation of the two-domain pattern of br, which foreshadows the formation of the dorsal appendages [9,15]. The two symmetrically placed domains of br correspond to the intermediate levels of the GRK gradient. Analyses of br expression in multiple mutants support a cell-autonomous mechanism whereby GRK both induces expression above a critical level of EGFR activation, and represses it at a higher level [16•]. A key element of this regulatory strategy is an incoherent feedforward loop, a highly conserved regulatory network motif, in which a signal induces both a target gene and its repressor, resulting in expression at an intermediate signal level [12•,17] (Figure 2a).
A mathematical model based on a feedforward loop, and the estimated GRK distribution predicts that the expression pattern of br should be shaped like a horseshoe, delimited by the two lines that correspond to two different threshold values of GRK [12•,18]. In contrast, the observed wild-type pattern is well separated from the anterior boundary of the follicular epithelium and split dorsally (Figure 2b). These two differences can be explained by two additional pre-patterning signals, which provide the anterior and posterior limits of the br domains.
The first of these is provided by the anterior-to-posterior activation gradient of the DPP signaling pathway, which represses br in the anterior follicle cells [16•, 19].
The nature of the signal that splits the br pattern is still unclear. One possibility is that it is provided by the earlier, posterior-to-anterior, phase of EGFR signaling, which could define the posterior limit of the competence zone that can later be patterned by the GRK [18]. This possibility is consistent with the changes of br expression induced by loss of Sprouty, which encodes an inhibitor of RTK signaling, and is induced by EGFR activation [20]. Loss of sty increases the level of EGFR signaling and leads to two specific changes in the br pattern: First, the separation between the two patches is increased, which could be explained by an increased expression of a repressor that defines the dorsal limit of br expression. Second, the posterior extent of the two patches is greatly reduced, which could be explained if a higher level of EGFR signaling during the earlier phase of EGFR activation reduces the size of the competence zone for the future DV patterning of the follicle cells.
When supplemented by two pre-patterning signals, the feedforward model successfully describes both the wild type pattern of br and its changes in response to variations in the strength and distribution of GRK [18]. In its current form, this model does not explicitly invoke transcription factors controlled by the GRK gradient and approximates their combined effects by simple threshold functions. In the future, this phenomenological description should be made more mechanistic, based on the information about transcription factors involved in br regulation. In particular, the activating effect of GRK on br depends on MAPK-dependent downregulation of Gapicua (Gic), an HMG-box transcriptional repressor involved in a number of RTK signaling events in Drosophila [21,22]. Downregulation of Gic relieves repression of Mirror, an Iroquois transcription factor which acts as an activator of br [23]. At the same time, br repression by high levels of EGFR signaling depends on Pointed, an ETS-family transcription factor [24,25]. Whether any of these effects are direct is currently unknown.
A combinatorial code for two-dimensional patterns
One important question, relevant to all tissue patterning contexts, is the number of geometrically distinct patterns established by inductive signals. Previously, such questions have been addressed only for one-dimensional patterning events. For example, close to 100 gene expression patterns established by the Dorsal morphogen gradient in the early Drosophila embryo can be described as stripes in the dorsal, lateral, and ventral regions [26]. The width of the stripe in each of the classes can vary, but they cannot be deformed into each other (e.g. a ventral stripe cannot be converted to a dorsal stripe). Thus, numerous patterns are assigned to only three geometric groups. Once this is done, one can systematically explore sequence-specific regulation of genes in different spatial classes, their joint dynamics, and response to genetic perturbations.
Our recent transcriptional profiling studies suggest that gene expression patterns in the follicle cells obey a relatively simple combinatorial code that uses only six basic shapes (Figure 3a). The basic shapes are also called ‘primitives’, following the terminology in constructive solid geometry, a field of computer graphics from which we borrowed this idea [27]. Three of the primitives, the dorsal patch (D), the midline cusp (M), and the anterior stripe (A), can be directly related to the EGFR and DPP signaling gradients. In the simplest case, they can be interpreted as their level sets, i.e. lines that correspond to different constant levels of these signals. The uniform pattern (U) can reflect the presence of a uniform activator (yet to be identified). Finally, the roof (R) and floor (F) primitives correspond to the cells that form the roof (top) and floor (bottom) surfaces of the future dorsal appendages. The connection of these two last primitives to the EGFR and DPP signals remains to be fully elucidated, although the R pattern can be explained by the feedforward model discussed above.
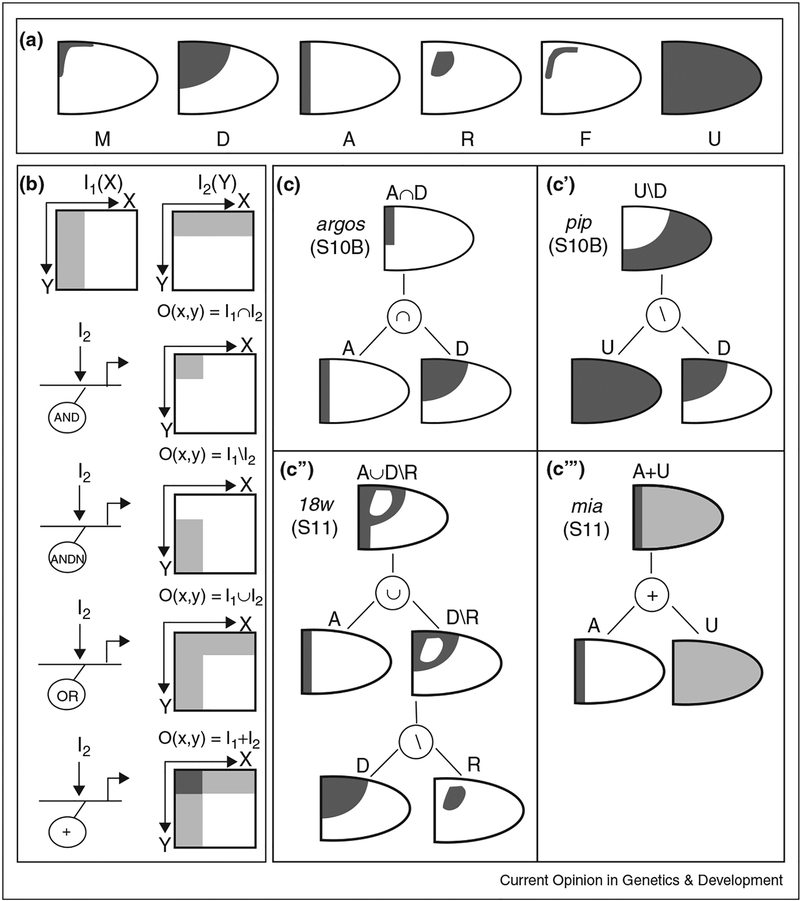
Figure 3.
Combinatorial code for pattern formation during Drosophila oogenesis, (a) Lateral views of the six building blocks (primitives) used to describe two- dimension expression patterns in the follicle cells. The M and D primitives reflect the activation gradient of the EGFR pathway, while the A primitive reflects the activation of the DPP pathway, (b) The primitives are combined by the Boolean and arithmetic operations of intersection (∩), difference (\), union (∪) and addition (+). These operations are realized at the c/s-regulatory level by the combined action of one or more inputs and the AND, AND (AND NOT), OR and + logic gates. I/O refer to the input/output, respectively, (c-c’”) Expression patterns of the genes argos, pip, 18w and mia can be constructed from the primitives and the combinatorial operations.
More complex patterns can be constructed by geometric combinations of primitives (Figure 3b–c”‘). These geometric operations can be interpreted as a consequence of the local algebraic operations at the regulatory regions of genes controlled by inductive signals [28] (Figure 3c). For example, if a gene requires both the EGFR and DPP signals for activation, its expression domain can be defined by the intersection of the level sets of the EGFR and DPP gradients. Geometrically, this would correspond to the intersection of the A and D primitives (Figure 3c). On the other hand, for a gene that can be activated by either one of the two signals, the expression pattern will be given by the union of two level sets (Figure 3c”). As another example, a gene activated by a uniform signal and repressed by a localized signal will be expressed in a pattern that is given by the difference of uniform and localized primitives (Figure 3c’).
This framework allowed us to assign hundreds of experimentally observed patterns to ~30 distinct combinations of a small number of basic shapes [29••]. On the basis of this analysis, we began to explore systems-level dynamics of gene expression patterns. For instance, we found that a group of genes that are expressed in the same pattern at one stage of oogenesis is likely to scatter into different patterns at later stages. Our preliminary results suggest that these dynamics can be explained by a model where gene expression at different stages of oogenesis is controlled by different regulatory modules.
Concluding remarks
Recent studies of EGFR-mediated pattern formation in Drosophila oogenesis provided the first quantitative description of the GRK gradient [11••], began to elucidate the networks that interpret this gradient [12•,18,29••], and established a systems-level view of gene expression patterns in the follicular epithelium [29••,30,31]. These studies led to a number of mechanistic models which should be tested by the identification of m-regulatory modules of br and other transcriptional targets of EGFR signaling in the follicle cells. Future studies of the EGFR signaling in Drosophila oogenesis will be greatly enabled by recently developed new tools for culturing and imaging of live egg chambers, and by a new generation of methods for transgenesis, targeted gene expression and knockdown [18,32,33•,34]. Given the highly conserved nature of EGFR signaling in development, systems-level studies of the GRK gradient will continue to provide insights into general mechanisms of pattern formation by locally activated RTKs (see Figure 4).
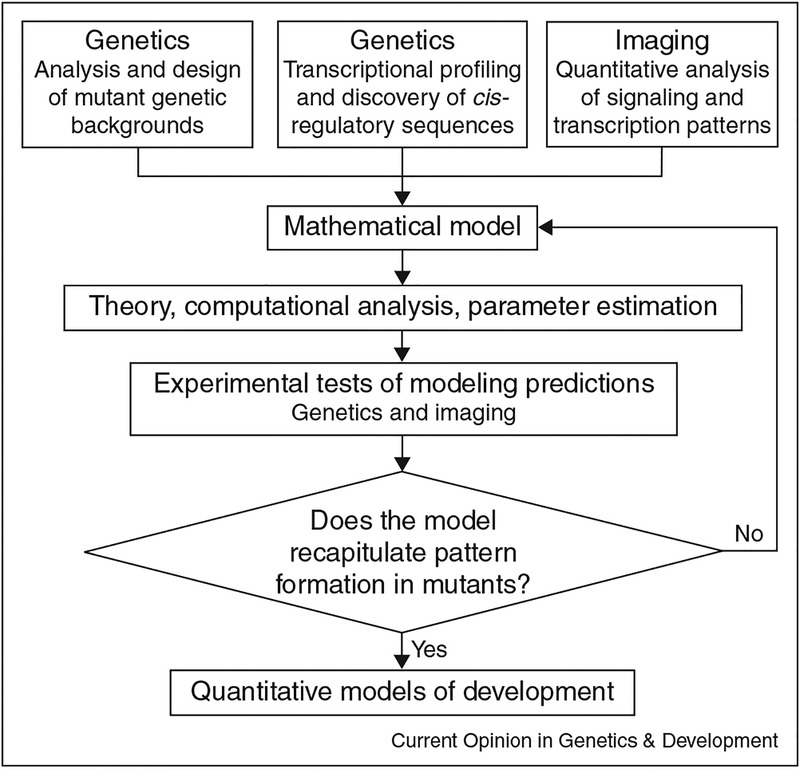
Figure 4.
Flowchart summarizing the cycle of model development, computational analysis, and experimental validation.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
• • of outstanding interest
- 1.Spradling AC: Developmental genetics of oogenesis The Development of Drosophila melanogaster. Plainview: Cold Spring Harbor Laboratory Press; 1993:. 1–70. [Google Scholar]
- 2.Nilson LA, Schupbach T: EGF receptor signaling in Drosophila oogenesis. Curr Top Dev Biol 1999, 44:203–243. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez-Reyes A, St Johnston D: Patterning of the follicle cell epithelium along the anterior-posterior axis during Drosophila oogenesis. Development 1998, 125:2837–2846. [DOI] [PubMed] [Google Scholar]
- 4.Neuman-Silberberg FS, Schupbach T: Dorsoventral axis formation in Drosophila depends on the correct dosage of the gene Gurken. Development 1994, 120:2457–2463. [DOI] [PubMed] [Google Scholar]
- 5.Pai L, Barcelo G, Schupbach T: D-cbl, negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell 2000, 103:51–61. [DOI] [PubMed] [Google Scholar]
- 6.Sen J, Goltz JS, Stevens L, Stein D: Spatially restricted expression of pipe in the Drosophila egg chamber defines embryonic dorsal-ventral polarity. Cell 1998, 95:471–481. [DOI] [PubMed] [Google Scholar]
- 7.Peri F, Technau M, Roth S: Mechanisms of Gurken-dependent pipe regulation and the robustness of dorsoventral patterning in Drosophila. Development 2002, 129:2965–2975. [DOI] [PubMed] [Google Scholar]
- 8.James KE, Dorman JB, Berg CA: Mosaic analyses reveal the function of Drosophila Ras in embryonic dorsoventral patterning and dorsal follicle cell morphogenesis. Development 2002, 129:2209–2222. [DOI] [PubMed] [Google Scholar]
- 9.Deng WM, Bownes M: Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development 1997, 124:4639–4647. [DOI] [PubMed] [Google Scholar]
- 10.Dorman JB, James KE, Fraser SE, Kiehart DP, Berg CA: bullwinkle is required for epithelial morphogenesis during Drosophila oogenesis. Dev Biol 2004, 267:320–341. [DOI] [PubMed] [Google Scholar]
- 11.Goentoro LA, Reeves GT, Kowal CP, Martinelli L, Schupbach T, Shvartsman SY: Quantifying the Gurken morphogen gradient in Drosophila oogenesis. Dev Cell 2006, 11:263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]; • • Combination of modeling, genetic, imaging and computational approaches to estimate the spatial range of a morphogen gradient.
- 12.Lembong J, Yakoby N, Shvartsman SY: Pattern formation by dynamically interacting network motifs. Proc Natl Acad Sci U S A 2009, 106:3213–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Example of dynamical systems analysis of pattern formation by interacting signaling pathways.
- 13.Goentoro LA, Yakoby N, Goodhouse J, Schupbach T, Shvartsman SY: Quantitative analysis of the GAL4/UAS system in Drosophila oogenesis. Genesis 2006, 44:66–74. [DOI] [PubMed] [Google Scholar]
- 14.Chang WL, Liou W, Pen HC, Chou HY, Chang YW, Li WH, Chiang W, Pai LM: The gradient of Gurken, a long-range morphogen, is directly regulated by Cbl-mediated endocytosis. Development 2008, 135:1923–1933. [DOI] [PubMed] [Google Scholar]; • Quantitative analysis of electron microscopy for characterizing the spatial distribution of a morphogen ligand in a developing tissue.
- 15.Berg CA: Tube formation in Drosophila egg chambers. Tissue Eng Part A 2008, 14:1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yakoby N, Lembong J, Schupbach T, Shvartsman SY: Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development 2008, 135:343–351. [DOI] [PubMed] [Google Scholar]; • Genetic characterization of signaling and pattern formation networks.
- 17.Alon U: Network motifs: theory and experimental approaches. Nat Rev Genet 2007, 8:450–461. [DOI] [PubMed] [Google Scholar]
- 18.Zartman JJ, Cheung LS, Niepielko M, Bonini C, Haley B, Yakoby N, Shvartsman SY: Pattern formation by a moving morphogen source. Phys Biol 2011, 8:045003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lembong J, Yakoby N, Shvartsman SY: Spatial regulation of BMP signaling by patterned receptor expression. Tissue Eng Part A 2008, 14:1469–1477. [DOI] [PubMed] [Google Scholar]
- 20.Zartman JJ, Kanodia JS, Cheung LS, Shvartsman SY: Feedback control of the EGFR signaling gradient: superposition of domain splitting events in Drosophila oogenesis. Development 2009, 136:2903–2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Astigarraga S, Grossman R, Diaz-Delfin J, Caelles C, Paroush Z, Jimenez G: A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J 2007, 26:668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimenez G, Guichet A, Ephrussi A, Casanova J: Relief of gene repression by Torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev 2000, 14:224–231. [PMC free article] [PubMed] [Google Scholar]
- 23.Atkey MR, Lachance JF, Walczak M, Rebello T, Nilson LA: Capicua regulates follicle cell fate in the Drosophila ovary through repression of mirror. Development 2006, 133:2115–2123. [DOI] [PubMed] [Google Scholar]
- 24.Morimoto AM, Jordan KC, Tietze K, Britton JS, O’Neill EM, Ruohola-Baker H: Pointed, an ETS domain transcription factor, negatively regulates the EGF receptor pathway in Drosophila oogenesis. Development 1996, 122:3745–3754. [DOI] [PubMed] [Google Scholar]
- 25.Boisclair Lachance JF, Fregoso Lomas M, Eleiche A, Bouchard Kerr P, Nilson LA: Graded Egfr activity patterns the Drosophila eggshell independently of autocrine feedback. Development 2009, 136:2893–2902. [DOI] [PubMed] [Google Scholar]
- 26.Hong JW, Hendrix DA, Papatsenko D, Levine MS: How the Dorsal gradient works: insights from postgenome technologies. Proc Natl Acad Sci U S A 2008, 105:20072–20076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foley JD, van Dam A, Feiner SK, Hughes JF: Computer Graphics: Principles and Practice. Addison-Wesley; 1992. [Google Scholar]
- 28.Davidson EH: The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Academic Press; 2006. [Google Scholar]
- 29.Yakoby N, Bristow CA, Gong D, Schafer X, Lembong J, Zartman JJ, Halfon MS, Schupbach T, Shvartsman SY: A combinatorial code for pattern formation in Drosophila oogenesis. Dev Cell 2008, 15:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]; • • Large scale analysis of two-dimensional pattern formation development.
- 30.Zartman JJ, Kanodia JS, Yakoby N, Schafer X, Watson C, Schlichting K, Dahmann C, Shvartsman S: Expression patterns of cadherin genes in Drosophila oogenesis. Gene Expr Patterns 2009, 9:31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zartman JJ, Yakoby N, Bristow CA, Zhou X, Schlichting K, Dahmann C, Shvartsman SY: Cad74A is regulated by BR and is required for robust dorsal appendage formation in Drosophila oogenesis. Dev Biol 2008, 322:289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venken KJ, He Y, Hoskins RA, Bellen HJ: P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 2006, 314:1747–1751. [DOI] [PubMed] [Google Scholar]
- 33.Haley B, Hendrix D, Trang V, Levine M: A simplified miRNA- based gene silencing method for Drosophila melanogaster. Dev Biol 2008, 321:482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A versatile experimental technique for miRNA-based gene silencing.
- 34.Prasad M, Jang ACC, Starz-Gaiano M, Melani M, Montell DJ: A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat Protocols 2007, 2:2467–2473. [DOI] [PubMed] [Google Scholar]