The psychiatric genetics field has made notable progress in the past decade, moving from candidate gene approaches whose results were largely unreplicated to statistically robust genome-wide association and sequencing methods. These latter methods have identified replicable genetic risk variants for psychiatric disorders that have provided important insights into underlying biological mechanisms. One barrier to further progress is that well-powered study designs require enormously large sample sizes (i.e., tens to hundreds of thousands of individuals) because the effect sizes of most common genetic variants are extremely small (typically accounting for much less than 1% of the variance). Many research groups have collected case/control samples of a few thousand individuals, a non-trivial investment, and yet these samples are typically too small to permit genetic discovery on their own. Until recently1, this was the case for attention-deficit/hyperactivity disorder (ADHD). For example, a 2010 meta-analysis of genome-wide association studies (GWASs) with nearly a thousand cases and two thousand trios (i.e., mother-father-child) did not uncover any genetic associations with ADHD that met standard statistical correction thresholds2. Given this stalled progress for many psychiatric disorders, an important question is whether existing GWAS samples can be used more efficiently for genetic discovery. One possibility is that an improved understanding of the underlying genetic architecture of psychiatric disorders could lead to more powerful statistical genetic approaches. An emerging insight about the underlying genetic architecture is that there is a surprisingly high degree of genetic overlap across many psychiatric disorders and related traits3. These cross-disorder genetic findings have been remarkably consistent across genetic methods and phenotypes. Approaches which leverage this cross-disorder genetic sharing may boost power for genetic discovery.
A new paper in the Journal by Shadrin et al. illustrates the application of such a cross-disorder approach to ADHD GWAS analysis and identifies some of the first replicable genetic associations with the disorder. The cross-disorder statistical framework, referred to as the conditional/conjunctional false discovery rate (condFDR/conjFDR)4, takes advantage of shared genetic underpinnings for two different phenotypes in order to boost power for genetic discovery. The logic is that if two phenotypes are known to share genetic underpinnings and a genetic variant is found to have effects on both phenotypes, then that genetic variant is more likely to be a true genetic association. This approach boosts power for genetic discovery in one phenotype by leveraging genetic information from a secondary phenotype.
In the current study, the authors apply the condFDR/conjFDR statistical approach to the genetic relationship between ADHD and educational attainment5. The authors used existing GWAS samples for ADHD (2064 trios, 896 cases, 2455 controls)2 and educational attainment (328,917 individuals)6. Individuals with ADHD are 7 times more likely to drop out of high school than their unaffected peers7 and there is evidence that this phenotypic relationship between ADHD and educational attainment is partially due to shared genetic risk factors (genetic correlation = −0.40 in the current study). By applying the condFDR/conjFDR methods, the authors were able to improve power in the existing ADHD GWAS sample and detect five novel associations that were not identified using standard GWAS statistical methods. The associations included variants in or near the genes KDM4A, MEF2C, PINK1, and RUNX1T1, though more research will be needed to conclusively link these variants to implicated genes. Nevertheless, several of these genes have been previously implicated in other neurodevelopmental disorders (i.e., oppositional defiant disorder (ODD), schizophrenia, and intellectual disability), suggesting interesting crossdisorder effects for further study. None of the implicated genes were a priori candidate genes for ADHD. Four of the five variants had the same direction of effect in an independent GWAS of ADHD symptoms, and two of the reported associations reached genome-wide significance in the most recent large-scale GWAS of ADHD that is currently in preprint (20183 cases; 35191 controls)1. As with most genetic discoveries for complex neurobehavioral disorders, these genetic variants only account for a small percent of the variance in ADHD risk, yet identifying replicable associations is the first step in understanding the gene networks and biological pathways that increase risk for ADHD.
These results mark an important milestone for ADHD genetics by reporting some of the first common genetic variant associations using GWAS methods. In addition to this notable achievement, the current paper opens up two complementary lines of inquiry. First, it provides a framework for investigating similar questions with other developmental phenotypes that are also associated with educational attainment. Second, the identification of overlapping genetic effects for ADHD and educational attainment raises mechanistic questions about the causes of this genetic overlap.
One logical next step is to apply the condFDR/conjFDR approach to other developmental phenotypes. The strategy of conditioning on educational attainment is particularly relevant for disorders with developmental cognitive and behavioral symptoms that impact school performance. One illustration of a future research direction concerns the learning disability dyslexia (also known as DSM-5 Specific Learning Disorder with impairment in reading). Dyslexia is strongly associated with educational attainment8 and has been actively studied from a genetic perspective, but sample collections have not yet reached the magnitude needed for genetic discovery despite intensive global collaborative efforts9. This paper by Shadrin et al. provides a dose of optimism about the prospects for genetic discovery in dyslexia using existing samples and this same methodology. This study design also has broad applicability to child and adolescent psychiatry because most early-onset disorders are associated with educational attainment10. The strongest associations are with the externalizing disorders (e.g., ODD, Conduct Disorder, Substance Abuse)10, and so these disorders might be particularly promising targets for this multi-trait GWAS approach.
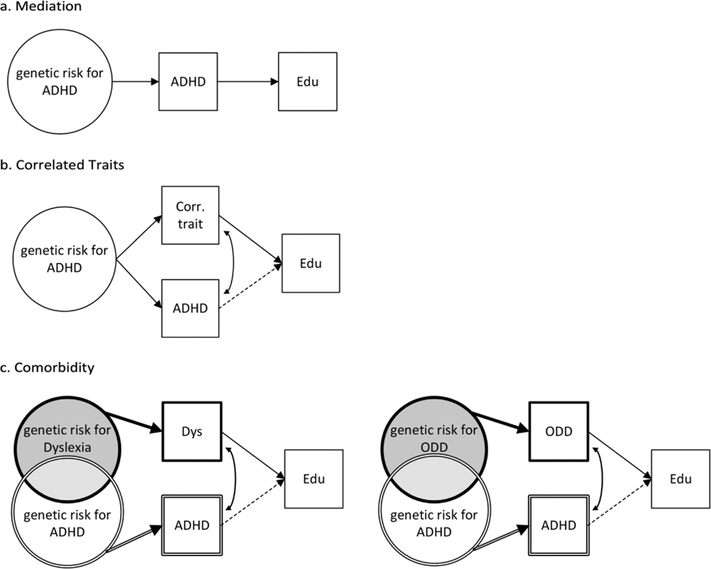
This paper also highlights important mechanistic questions about the nature of the genetic relationship between ADHD and educational attainment, especially since educational attainment is such a multi-determined, distal phenotype. There are at least three, non-exclusive models that could explain a genetic correlation between ADHD and educational attainment: (1) mediation, (2) correlated traits, and (3) comorbidity (see Figure 1)3. These models are intended to provide a broad heuristic to conceptualize underlying shared genetic risk and are not exhaustive with respect to genetic mechanisms (i.e., assortative mating is not included) or levels of analysis (i.e., neural correlates and intermediate phenotypes are not specified). In the mediation model (Figure 1a), genetic risk for ADHD is shared with educational attainment because symptoms that are core to the ADHD diagnosis directly impact school performance (e.g., inattention, impulsivity, executive dysfunction). In the correlated traits model (Figure 1b), genetic risk for ADHD influences multiple traits, in this case ADHD itself and other cognitive and behavioral features that are correlated with ADHD but are not core to the diagnosis (e.g., aggressive behavior, substance use, social skills). These correlated features, alone or in combination with ADHD-specific effects (dotted line Figure 1b), place a child at risk for poor school performance. The last model illustrates the potential confounding influences of comorbidity (Figure 1c). Two of the most common comorbidities of ADHD are dyslexia (25–40% comorbidity)11 and ODD (33–50% comorbidity)12. Similar to ADHD, both dyslexia and ODD are strongly associated with educational attainment8,10 and twin studies further indicate both disorders share genetic risk with ADHD (dyslexia-ADHD genetic correlation ~.7; ODD-ADHD genetic correlation ~.6)13,14. Given these genetic and phenotypic relationships, the comorbidity model illustrates how the genetic association between ADHD and educational attainment might be at least partially driven by comorbid conditions. These comorbidity pathways do not preclude an ADHD-specific effect as well (dotted line Figure 1c). Taken together, each of these models provides a plausible explanation, individually or in combination, for the genetic association between ADHD and educational attainment.
Figure 1.
Potential genetic models explaining shared genetic risk between ADHD and educational attainment. Models are non-exclusive. Circles indicate latent factors representing all of the genetic influences for a disorder, many of which are currently unknown. Squares indicate measured variables. Dotted lines indicated ADHD-specific effects that may be operational but are not required for the model to produce the observed shared genetic risk between ADHD and educational attainment.
ADHD=Attention-deficit/hyperactivity disorder, Edu = educational attainment, Corr. Trait = correlated behavioral or cognitive traits associated with ADHD, Dys = Dyslexia, ODD=Oppositional Defiant disorder.
Distinguishing between these different models will require strategies to address the phenotypic limitations of current GWAS samples. This problem is not specific to ADHD, but more general to psychiatric genetics, and is largely a function of the challenges in harmonizing phenotypes across global collaborative research teams. Nevertheless, distinguishing different models leading to overlapping genetic risk will require more extensive phenotyping in large samples, including theoretically-relevant comorbidities and intermediate traits. Ideally, this phenotypic information would be collected in longitudinal samples that could support causal inferences. Collecting such comprehensive phenotypes in large, longitudinal samples is a daunting prospect, but it is clear that this investment will be necessary to fully specify the genetic pathways underlying shared genetic risk across psychiatric disorders. A few population-based developmental samples have collected genetic information and could provide data to begin addressing these important issues (e.g., ALSPAC, PING, Philadelphia Neurodevelopmental Cohort, Generation R).
In summary, Shadrin et al. identify some of the first common genetic variants associated with ADHD using GWAS methods. This is an important milestone for ADHD genetics. The strategy for identifying these genetic variants, by leveraging a large-scale genetic study of educational attainment, is also relevant for many early-onset phenotypes, especially learning disabilities and externalizing disorders. The findings also raise important questions about the mechanisms underlying overlapping genetic risk for ADHD and educational attainment. Given the surprising degree of cross-disorder genetic sharing that is being uncovered across psychiatric disorders, understanding the mechanisms underlying shared genetic risk is one of the most important upcoming challenges for psychiatric genetics.
Acknowledgments:
This work was supported by NIH grants P50HD027802 and R15HD086662. Special thanks to colleagues who provided helpful feedback: Laramie Duncan, Ph.D., Stacy Drury, M.D., Ph.D., Christa Hutaff-Lee, Ph.D., Andrew Kirby, Chintan Mehta, Ph.D, Bruce Pennington, Ph.D., Robin Peterson, Ph.D., Erik Willcutt, Ph.D. My thanks to Sarah Pollard, M.A., for administrative help preparing the manuscript.
Footnotes
Disclosure: Dr. McGrath receives book royalties from Guilford Press for the 2nd and upcoming 3rd edition of Diagnosing Learning Disorders: Science into Practice.
References
- 1.Demontis D, Walters RK, Martin J, et al. Discovery of the first genome-wide significant risk loci for ADHD. bioRxiv Prepr. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neale BM, Medland SE, Ripke S, et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49(9):884–897. doi: 10.1016/j.jaac.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14(7):483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreassen OA, Thompson WK, Dale AM. Boosting the power of schizophrenia genetics by leveraging new statistical tools. Schizophr Bull. 2014;40(1):13–17. doi: 10.1093/schbul/sbt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shadrin AA et al. Novel loci associated with Attention-deficit/hyperactivity disorder are revealed by leveraging polygenic overlap with Educational Attainment. J Am Acad Child Adolesc Psychiatry. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okbay A, Beauchamp JP, Fontana MA, et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533(7604):539–542. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barkley RA. Major life activity and health outcomes associated with attention-deficit/hyperactivity disorder. J Clin Psychiatry. 2002;63 Suppl 1:10–15. http://www.ncbi.nlm.nih.gov/pubmed/12562056. [PubMed] [Google Scholar]
- 8.Cortiella C, Horowize SH. The State of Learning Disabilities: Facts, Trends, and Emerging Issues, 3rd Edition New York, NY; 2014. http://www.ncld.org/images/content/files/stateofld2014/2014. State of LD FINAL FOR RELEASE.pdf. [Google Scholar]
- 9.Gialluisi A, Newbury DF, Wilcutt EG, et al. Genome-wide screening for DNA variants associated with reading and language traits. Genes Brain Behav. 2014;13(7):686–701. doi: 10.1111/gbb.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breslau J, Lane M, Sampson N, Kessler RC. Mental disorders and subsequent educational attainment in a US national sample. JPsychiatr Res. 2008;42(9):708–716. doi: 10.1016/j.jpsychires.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willcutt EG, Pennington BF. Comorbidity of reading disability and attention-deficit/hyperactivity disorder: Differences by gender and subtype. J Learn Disabil. 2000;33(2):179–191. doi: 10.1177/002221940003300206. [DOI] [PubMed] [Google Scholar]
- 12.Harvey EA, Breaux RP, Lugo-candelas CI. Early Development of Comorbidity Between Symptoms of Attention-Deficit / Hyperactivity Disorder ( ADHD ) and Oppositional Defiant Disorder ( ODD ). J Abnorm Psychol. 2016;125(2):154–167. doi: 10.1530/ERC-14-0411.Persistent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willcutt EG, Betjemann RS, McGrath LM, et al. Etiology and neuropsychology of comorbidity between RD and ADHD: the case for multiple-deficit models. Cortex. 2010;46(10):1345–1361. doi:S0010-9452(10)00181-4 [pii] 10.1016/j.cortex.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willcutt E Behavioral Genetic Approaches to Understand the Etiology of Comorbidity. Behav Genet Psychopathol. 2014. http://link.springer.com/chapter/10.1007/978-1-4614- 9509-3_8. Accessed January 7, 2015. [Google Scholar]