Abstract
Genetic studies of human evolution require high-quality contiguous ape genome assemblies that are not guided by the human reference. We coupled long-read sequence assembly, full-length cDNA sequencing with a multi-platform scaffolding approach to produce ab initio chimpanzee and orangutan genome assemblies. Comparing these with two long-read de novo human genome assemblies and a gorilla genome assembly, we characterized lineage-specific and shared great ape genetic variation ranging from single base-pair to megabase-sized variants. We identified ~17 thousand fixed human-specific structural variants identifying genic and putative regulatory changes that emerged in humans since divergence from nonhuman apes. Interestingly, these fixed human-specific structural variants are enriched near genes that are downregulated in human compared to chimpanzee cerebral organoids, particularly in cells analogous to radial glial neural progenitors.
Keywords: whole-genome sequence and assembly, single-molecule, real-time (SMRT) sequencing, structural variation, primate genome evolution
INTRODUCTION
Scientists have long been interested in the functional genetic differences that distinguish humans from other ape species (1). Human and chimpanzee protein-encoding changes and structural differences in regulatory DNA or in the copy number of gene families have all been implicated in adaptation (2, 3). Indeed, several potentially high-impact regulatory changes (4, 5) and human-specific genes (6–9) that are important in synapse density, neuronal count, and other morphological differences have been identified. Most of these genetic differences, however, were not initially recognized upon comparison of human and ape genomes because the genetic changes mapped to regions of rapid genomic structural change that were not resolved in draft genome assemblies.
Despite recent efforts to sequence and assemble ape genomes (10–12), our understanding of structural differences, and particularly those specific to the human lineage, remains far from complete. There are two fundamental problems. First, there is considerable heterogeneity in the contiguity of ape genome assemblies. The presence of tens to hundreds of thousands of gaps in ape genomes limits the proportion of the genome that can be compared in a multi-species sequence alignment. Therefore, a large fraction of human-specific insertions and deletions, including those that alter regulatory sequences, are not resolved. Second, the higher quality human genome assemblies have often been used to guide the final stages of nonhuman genome projects, including the order and orientation of sequence contigs and, perhaps more importantly, the annotation of genes. This bias has effectively “humanized” other ape genome assemblies, minimizing potential structural and transcript differences observed between the species. Using long-read, long-range sequence and mapping technologies (13–15), we generated new great ape genome assemblies along with full-length cDNA annotation without guidance from the human genome. We also generated and analyzed an African genome and an effectively haploid human genome complement to distinguish fixed differences in the human ancestral lineage and to further mitigate human genome reference biases.
RESULTS
Genome assembly:
We sequenced two human, one chimpanzee, and one orangutan genome to high depth (>65-fold coverage) using single-molecule, real-time (SMRT, PacBio) sequence data, and we assembled each ab initio using the same underlying assembly algorithm (Table 1) (16). For each species, we generated assemblies ranging from 2.9 to 3.1 Gbp where the majority of the euchromatic DNA mapped to <1000 large contigs (Table 1). We error-corrected sequence contigs with Quiver (17) and Pilon (18), followed by a procedure that reduced the remaining 1-2 bp indels specifically in regions with clustered single-nucleotide variants (SNVs) (16). We next scaffolded the chimpanzee and orangutan genomes without guidance from the human reference genome. In total, 93% (2.79 Gbp; excluding chrY) of the chimpanzee and 92.7% (2.82 Gbp) of the orangutan-assembled bases were incorporated into chromosomal-level scaffolds (Table 1). We confirmed most large-scale chromosomal inversions among the great apes (19), some of which were absent from previous assemblies.
Table 1.
Assembly statistics for the great ape genomes.
| Ape assembly | CHM13_HSAv1e (human) | YRI_HSAv1 (human) | Clint_PTRv1 (chimpanzee) | GSMRT3.2 (gorilla) | Susie_PABv1 (orangutan) |
|---|---|---|---|---|---|
| Estimated deptha | 72 | 116 | 117 | 86.3 | 94.9 |
| Subread length N50 (kbp)b | 16.2 | 13.4 | 17.4 | 18.6 | 16.6 |
| Contig (number) initial/finalc | 1,923/1,916 | 3,645/3,642 | 4,912/5,037 | 15,997 | 5,771/5,813 |
| Assembly size (Gbp) | 2.88 | 2.88 | 2.99 | 3.08 | 3.04 |
| Contig length >3 Mbp, (Gbp) | 2.65 | 2.27 | 2.45 | 2.42 | 2.48 |
| Contig N50 (Mbp) initial/finalc | 29.26 | 6.60 | 12.76/12.42 | 10.02 | 11.27/11.07 |
| Scaffold N50 (Mbp) | 83.02 | ND | 53.1 | ND | 98.47 |
| Longest contig (Mbp)d | 81 | 27 | 80 | 36 | 53 |
| BAC concordance | 97.11% | 97.73% | 99.13% | 96.85% | 96.75% |
| Sequence accuracy (QV) | 36 | 31 | 33-38 | 30-38 | 28-33 |
| Iso-Seq transcripts | 710,974 | ND | 565,691 | 881,801 | 528,145 |
| Contigs in AGP | ND | ND | 685 | 794 | 544 |
| Contigs aligned to GRCh38f [Gbp] | 407 [2.8] | 1,167 [2.8] | 656 [2.8] | 907 [2.8] | 524 [2.8] |
Estimated coverage in raw SMRT subreads based on 3.5 Gbp (gorilla) or 3.2 Gbp (all others) estimated genome size.
N50 subread lengths of raw input data.
Initial contigs/final contigs are the number of contigs before and after resolving chimeras by optical map comparison. These stats do not consider the NCBI minimum contig length filter.
Longest contig without gross assembly error.
Haploid genome assembly derived from a complete hydatidiform mole.
Contigs with less than 95% of sequence aligning to GRCh38, depth-of-coverage greater than two SDs above the mean, or no coverage were excluded.
ND denotes no data.
Sequence accuracy and quality assessment:
Over 96% of our assembled sequence was concordant by length and orientation by different metrics (Table 1) (16). We conservatively estimate that these assemblies have improved contiguity for the chimpanzee and orangutan genomes by 32- and 533-fold, respectively (Fig. 1a-b). Consistent with the gorilla genome (20), the application of long-read sequence data closed most of the genome gaps in earlier assemblies. The extent of the change varied, however, depending on the prior level of finishing. In the case of the chimpanzee, 52% of the remaining 27,797 gaps were closed. We added 6.9 Mbp of new sequence and removed at least 27.2 Mbp of duplicated or extraneous sequence, possibly artifacts of scaffolding and gap filling (21). In the case of orangutan, we added 54.5 Mbp of sequence while removing 4.2 Mbp, closing an estimated 96.8% (305,069/315,124) of the remaining euchromatic gaps. We determined the sequence contigs to be highly accurate at the base-pair level (>99.9%) on the basis of comparisons of each genome to Sanger end-sequence data, completely sequenced clone inserts, and Illumina whole-genome sequencing (WGS) data generated from the same source individuals (Table 1) (16).
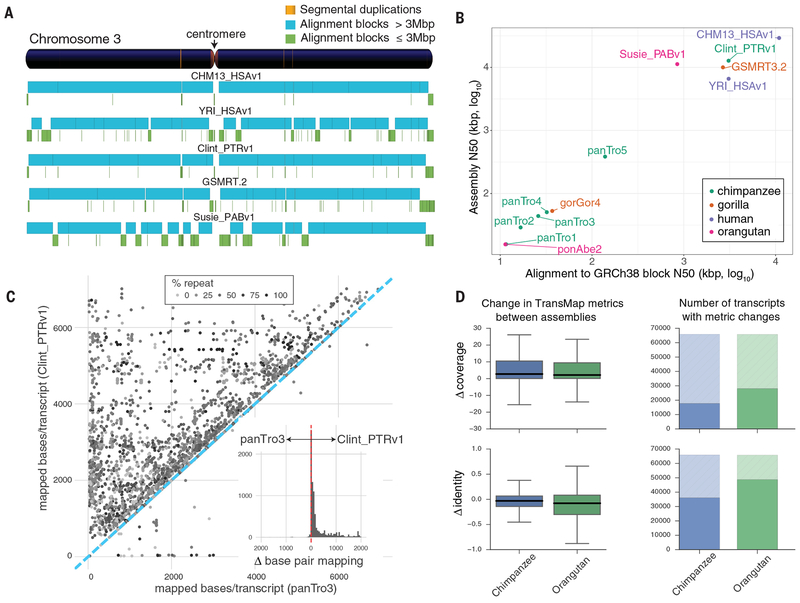
Fig. 1. Assembly and annotation of great ape genomes.
a) Comparison of genome sequence contiguity. Chromosome 3 contiguity is compared among the great ape genome assemblies by alignment to GRCh38. Contigs larger (blue) and smaller (green) than 3 Mbp are compared with the position of segmental duplications (SDs >50 kbp, orange) shown in the reference ideogram. b) Scatterplot of syntenic-alignment block lengths (x-axis) against GRCh38 vs. contig N50 (y-axis) of the great ape assemblies. The SMRT assemblies are Clint_PTRv1, Susie_PABv1, GSMRT3.2, CHM13_HSAv1, and YRI_HSAv1. The previous reference genomes are ponAbe2 (GCF_000001545.3), gorGor4 (GCA_000151905.3), panTro2 (GCF_000001515.2), panTro3 (GCA_000001515.3), panTro4 (GCA_000001515.4), and panTro5 (GCA_000001515.5). c) Full-length assembled transcripts mapped to Clint_PTRv1 and panTro3. Each point denotes the number of bases/transcript matching the two assemblies. Repeat content is indicated by gray shading of the points. While the majority of transcripts map well to both assemblies (Pearson’s correlation = 0.95), the subset of differentially mapped transcripts (12,724; 60% of 21,118) aligns better to Clint_PTRv1 (dots above the blue dashed line). The histogram inset shows the effect, per transcript, with a total of 4.8 Mbp more bases aligned to Clint_PTRv1. d) Comparative Annotation Toolkit (CAT) was used to project transcripts from GRCh38 to Clint_PTRv1, panTro3, Susie_PABv1, and ponAbe2. Alignment coverage and identity were compared for orthologous transcripts found in each assembly pair. The boxplots (left) summarize TransMap differences between the short-read and SMRT assemblies in terms of coverage and identity. The shaded portion of the bar plots (right) represents alignments, which had identical coverage or identity in both assemblies.
Gene annotation:
Nonhuman primate (NHP) genome assemblies have typically relied almost exclusively on the human reference to define gene models (Table S1). To provide a less biased source of gene annotation, we generated long-read transcriptome sequencing data to produce an average of 658,546 full-length non-chimeric (FLNC) transcripts from induced pluripotent stem cells (iPSCs) derived from each of the three nonhuman ape lineages (16). We selected iPSC material to maximize transcript diversity and enrich for early developmental genes. We next annotated the genomes of chimpanzee, gorilla and orangutan, using FLNC transcripts along with short-read RNA-seq to guide gene and novel isoform predictions (22).
The number of genes and most gene models (coding and noncoding, including lncRNA) are consistent among the different ape genomes (Table 2). However, we saw differential mapping of FLNC transcripts that favored the SMRT assemblies, especially in repeat-rich transcripts (Fig. 1c). Concordantly, human transcript models (GENCODE V27) aligned better to SMRT assemblies (Fig. 1d). For chimpanzee, 17,744 human protein-coding transcript models showed an increase of mapping coverage, which averaged 5.6%. This pattern was more pronounced in orangutan where 28,033 of the 91,578 protein-coding transcripts annotations showed an average improvement of 5.7% in mapping coverage. Overall, human protein-coding transcript models mapped to chimpanzee and orangutan SMRT assemblies with 99.1% and 98.8% average coverage, respectively—a 1.5% and 2.5% improvement. This improvement stemmed largely from gap closures, which rescue missing exons and recover more full-length transcripts, including untranslated regions (UTRs).
Table 2.
Great ape gene/transcript annotation summary.
| Clint_PTRv1 | GSMRT3.2 | Susie_PABv1 | |
|---|---|---|---|
| Genes | 55,894 | 55,985 | 55,522 |
| Orthologs in human | 55,594 (95.4%)a | 55,570 (95.4%)a | 54,900 (94.2%)a |
| Isoforms | 192,725 | 192,734 | 190,716 |
| Coding genes | 19,153 | 19,311 | 19,043 |
| Novelb | 300 | 415 | 322 |
| Coding isoforms | 92,610 | 92,713 | 91,578 |
| Transcript predictions with novel splice junctionsc | 2,809 | 2,902 | 2,333 |
| Percent of transcripts with TPM > 0.1 | 66.3 | 67.3 | 50.6 |
| Percent of transcripts supported by Iso-Seq reads | 66.5 | 46.5 | 63.4 |
| Previously unannotated exons identified | 29 | 16 | 16 |
| Putative exons gained in human | 57 | NA | ND |
| Putative exons lost in human | 13 | NA | NA |
Percent of GENCODE V27 represented.
Novel predicted genes based on GENCODE V27 annotation.
Novel splice junctions compared to liftover annotation set from the human reference genome where splice junction is supported by NHP RNA-seq.
ND denotes no data; NA denotes not applicable to this genome.
We identified a small fraction (~1.5%) of putative protein-encoding genes present among NHPs absent in human annotations (GENCODE V27). In addition, a larger fraction (3.1% to 3.8%) of transcripts exhibited RNA-seq or Iso-Seq supported splice junctions present in NHPs but not in human transcripts. Finally, we evaluated the NHP annotations, identifying full exons that affect coding sequence, which have been gained or lost between humans and other great apes (Table S1).
Comparative sequence analyses:
We constructed a five-way genome-wide multiple sequence alignment (MSA) of the ab initio assembled genomes (Table 1) by identifying syntenic (20 kbp) blocks against the human reference genome. In total, 83% of the ape genome was represented in MSAs. This allowed us to identify a comprehensive set of SNVs, indels and structural variants (SVs), calculate divergence, and perform genome-wide phylogenetic analyses (Fig. 2). We observed a modest elevation in SNV divergence compared to previous genome comparisons (Fig. 2a; Table S2) and estimated that 35.6% of the human genome is subject to incomplete lineage sorting among the African apes (Fig. 2b). Human and chimpanzee branch lengths are remarkably similar within coding regions (0.026% difference in branch length); however, we observed a 3.5% slowdown of the human mutation rate in noncoding regions (23, 24) (Fig. 2c). Human and chimpanzee branch lengths were significantly shorter compared to the other apes, consistent with the hominid slowdown hypothesis (25).
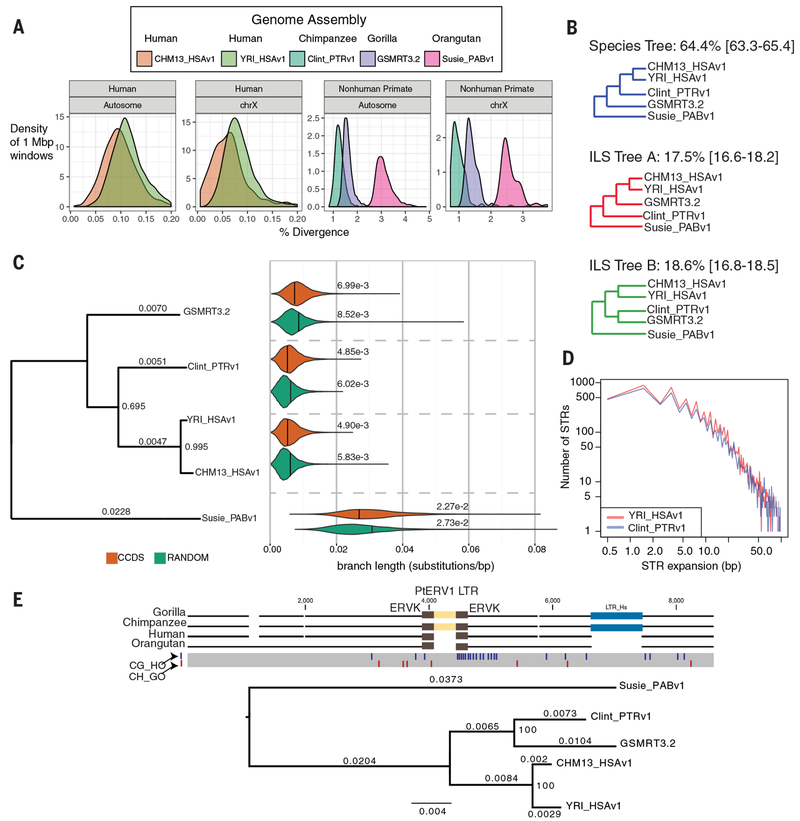
Fig. 2. Ape genetic diversity and lineage sorting.
a) Single-nucleotide variant (SNV) divergence between each primate assembly and GRCh38 was calculated in 1 Mbp non-overlapping windows across all autosomes and chromosome X (excluding X-Y homologous regions). Mean autosomal divergence is 1.27+/−0.20% (human-chimpanzee), 1.61+/−0.21% (human-gorilla) and 3.12+/−0.33% (human-orangutan). The African genome (YRI_HSAv1) shows a 17% increase in SNV diversity. b) Proportion of phylogenetic trees supporting standard species topology and incomplete lineage sorting (ILS). The mean and 95% confidence intervals are based on 100 genome-wide permutations. c) A phylogenetic tree (maximum clade credibility consensus tree) comparing genic regions (~9,000 consensus CDS (CCDS) and 1,000 bp flanking sequence [orange]) to a randomly genome-shuffled set matched to CDS lengths (green). The analysis excludes regions of SDs, SVs and large tandem repeats. Branch lengths (above the lines) and proportion of trees supporting each bifurcation (internal nodes) are shown. Violin plots summarize the distribution and mean divergence (substitutions/bp) for a subset of trees consistent with the species tree. YRI_HSAv1 is the representative human in the violin plots. d) A comparison of the expanded STR sequences (n = 16,138 loci) between human (African) and chimpanzee ab initio genome assemblies shows little to no species bias (0.02 bp). e) A multiple sequence alignment (MSA) of ape genomes (gorilla BAC CH277-16N20, chimpanzee CH251-550G17) identifies an orthologous 379 bp PtERV1 element nested within another LTR and shared between gorilla and chimpanzee. A maximum likelihood phylogenetic tree (GTR+Gamma) built from 12,108 bp supporting ILS. Single-nucleotide polymorphisms that support chimpanzee-gorilla sorting (CG_HO) are shown as blue lines and the red lines show single-nucleotide polymorphisms supporting the species tree (CH_GO). Branch lengths (substitutions per site) are shown above the lineages and internal nodes are labeled with bootstrap support (proportion of replicates supporting split; 1,000 replicates).
Repeat comparisons:
Although the general repeat content of primate genomes has been well established (16), the longest and most complex repetitive regions have been more difficult to assay. Because long-read sequence data resolve most microsatellites and high-copy interspersed repeats (20, 26), we focused on comparative analysis of short tandem repeats (STRs) and endogenous retrovirus elements. Previous studies have suggested differential expansion of STR sequences between humans and other NHPs (27, 28). However, these studies suffer from ascertainment bias due to methodological differences in genome sequencing or STR enrichment, differential access to GC-rich regions, and discovery bias in the human reference genome.
We analyzed each genome independently and, after clustering STRs that mapped within 25 bp, identified a consistent number of STRs per ape genome (344,354–358,622 STR regions; Table S3). Since STRs often map within or adjacent to other classes of repetitive DNA, we restricted our analysis to the subset where orthology and STR lengths were clearly defined (12,694–16,138 STRs; Fig. S28, Table S4). The average length difference between human and chimpanzee STR loci is 0.02 bp with only a slight difference in distributions (p = 0.015 Kolmogorov-Smirnov [KS] test; Table S5; Fig. 2d). Other ape comparisons show a modest increase in overall STR length (e.g., 1.2 average bp increase in gorilla vs. chimpanzee; p = 8.76E-12, KS test). We found no significant difference between human and chimpanzee STR length in coding sequences (n = 2,199, p = 0.28, KS test) or UTRs of genes (n = 2,794, p = 0.16, KS test) although we identified 4,920 loci preferentially expanded in the human lineage (Table S6), including loci associated with genomic instability and disease.
Endogenous retroelements are among the longest retrotransposons within mammalian genomes (up to 10 kbp) and are frequently misassembled because of their copy number and sequence identity. The chimpanzee and gorilla lineages carry an endogenous retrovirus, PtERV1, that is absent in orangutan and human genomes (29, 30). None of the PtERV1 integrations between chimpanzees and gorillas appear orthologous, suggesting independent retroviral integrations in these two lineages (29, 30) or that humans and orangutans contain extrinsic factors that differentially restricted propagation (31). A high-quality map of 540 PtERV1 elements (both full-length and solo long terminal repeat [LTR]) in chimpanzee and gorilla (Table S7) (16) shows that their integration events are nonorthologous (99.8%), biased against genes, and integrated in the antisense orientation (Figs. S30, S31) consistent with the action of purifying selection.
Using the more complete ape genomes, we identified only one chimpanzee–gorilla orthologous PtERV1 element, not present in modern humans, that was lost through incomplete lineage sorting and integrated roughly 4.7 mya (95% HPD: [1.9, 7.2 mya]; Fig. 2e). We named this element the “source PtERV1” as it was present in the common ancestor of all African apes and was likely the progenitor for independent expansions to non-orthologous loci in the chimpanzee and gorilla genomes. The source PtERV1 was likely missed in earlier genomic studies of draft genomes because the locus (sharing orthology with human chromosome 19) (16) is repeat-rich and the integration site is an ancient LTR element.
Structural variation analyses:
We focused on identifying all SVs >50 bp within ape genomes because these are the least well-characterized differences and are more likely to impact gene function than SNVs (32). SVs were identified by mapping each assembly back to the human reference genome, using the two newly assembled human genomes as a control for reference effects and fixed human differences (CHM13_HSAv1 and YRI_HSAv1). We detect 614,186 ape deletions, insertions and inversions with the number of SVs increasing as a function of evolutionary distance from human (Fig. 3, Table 3). We confirmed 92% of 61 events (from 2.7 to 95 kbp) by BAC sequencing (Table S8) (three of the remaining events were polymorphic among the great apes, suggesting a validation rate of >95%). We assigned SVs as shared or lineage-specific and genotyped each at the population level, with a panel of 86 great apes (33) (Fig. 3a). We identified 17,789 fixed human-specific structural variants (fhSVs), including 11,897 fixed human-specific insertions (fhINSs) and 5,892 fixed human-specific deletions (fhDELs) (Fig. 3a, Table S9). Projecting these onto the human genome identifies potential hotspots of structural variation (Fig. 3b).
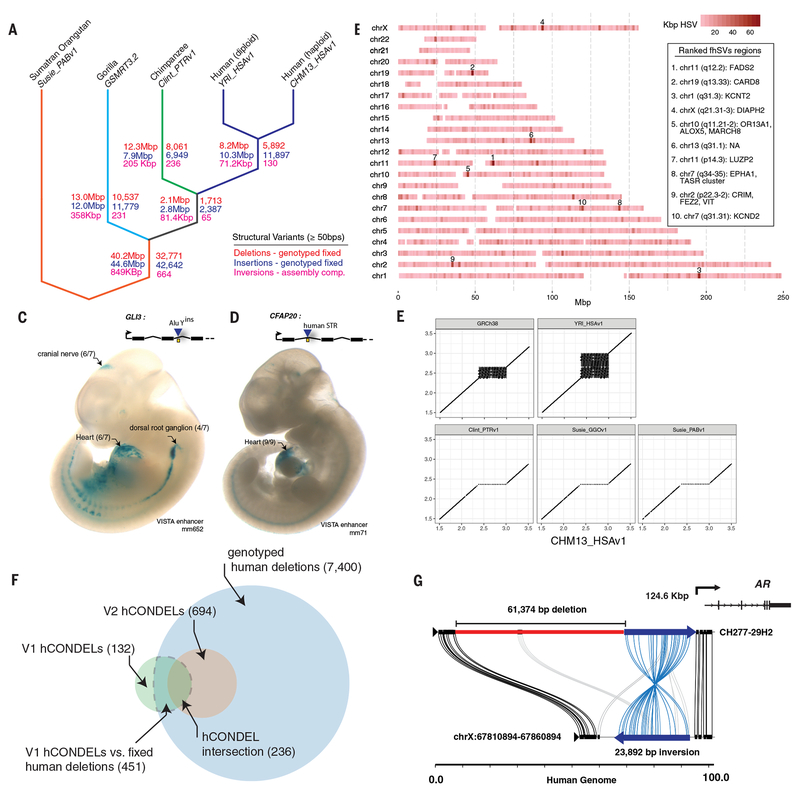
Fig. 3. Fixed structural variation and regulatory mutation.
a) The great ape cladogram with fixed structural variation assigned to lineages on the basis of assembly comparison, genotyping and stratification (except for inversions). The total amount of sequence is shown on the left side of the branches and the number of SVs is shown on the right for deletions (blue), insertions (red) and inversions (magenta). Inversions were assigned to branches on the basis of the comparison of our five assemblies because genotyping was less reliable. The cladogram was rooted against Susie_PABv1, meaning the assignment of SVs to the orangutan or the common ancestor of human, chimpanzee, and gorilla is arbitrary. b) A map of fixed human-specific structural variants (fhSVs). The color denotes number of fhSVs bases (kbp), within a 1 Mbp sliding window (0.5 Mbp step). Each chromosome is labeled on the y-axis. Key regions are annotated with genes. c) The cell specificity for a mouse enhancer element (mm652, represented as a yellow box) that shares orthology in chimpanzee. In human, an AluY element has been inserted directly into the mm652 enhancer. d) A human-specific STR interrupts a mouse heart-specific enhancer shared with chimpanzee (yellow box). The STR is contained within a CFAP20 intron. e) Dotplots of the human-specific STR expansion. The two human assemblies, CHM13_HSAv1 and YRI_HSAv1, show additional STR expansion relative to GRCh38, suggesting the reference is collapsed. f) A comparison of the hCONDEL set reported by McLean et al. (5) (V1) vs. the hCONDELs reported here (V2). The current hCONDELs are from conservation (25 bp MSA windows) between chimpanzee, macaque and mouse. The current hCONDELs are from conservation (25 bp MSA windows) between chimpanzee, macaque and mouse. The dashed gray area shows the overlap between all fixed human deletions and all V1 hCONDELs. g) A Miropeats diagram of the gorilla complex SV (inversion and deletion) upstream of the AR locus; the human reference genome is shown on the bottom.
Table 3. Summary of great ape genome structural variation.
SV events (>50 bp) called against the human reference genome (GRCh38) using smartie-sv.
| CHM13_HSAv1 | YRI_HSAv1 | Clint_PTRv1 | GSMRT3.2 | Susie_PABv1 | |
|---|---|---|---|---|---|
| Deletion count | 9,126 | 11,747 | 63,634 | 73,681 | 136,980 |
| Insertion count | 14,962 | 14,528 | 68,589 | 76,230 | 142,631 |
| Inversion count | 74 | 55 | 446 | 533 | 969 |
| Deletion (Mbp) | 4.76 | 4.85 | 41.88 | 45.48 | 84.76 |
| Insertion (Mbp) | 6.85 | 7.17 | 40.34 | 47.53 | 120.35 |
| Avg. deletion size (bp) | 552 | 413 | 658 | 617 | 618 |
| Avg. insertion size (bp) | 458 | 493 | 588 | 623 | 843 |
| Largest (kbp) [type] | 84 [del] | 124 [ins] | 133 [ins] | 90 [ins] | 123 [ins] |
We annotated fhSVs against chimpanzee and human gene models (Table S10). The Variant Effect Predictor (VEP) annotated the loss of 13 start codons, 16 stop codons, and 61 exonic deletions in the human lineage. By contrast, we estimate that fhSVs disrupt 643 regulatory regions near 479 genes (e.g., Fig. 3c-e). Interestingly, 139 of the fhSVs intersect with regions recently classified as super-enhancers (34). A comparison with a previous analysis of human-conserved deletions (hCONDELs) from earlier versions of the human, chimpanzee and macaque genomes (5) confirms that 77% (451/583) of the hCONDELs intersect the fhDELs, with the remainder corresponding primarily to polymorphic events in the human population (Fig. 3f). We also predicted an additional 694 hCONDELs (Table S11). A comparison of the SMRT gorilla assembly to human identified an hCONDEL sequence previously reported as affecting an androgen receptor enhancer and associated with the loss of penile spines in humans. In gorilla this fhDEL involves a complex SV, including an inversion, that may independently influence AR gene expression in the gorilla lineage (Fig. 3g) (35).
The spectrum of structural variation ranges from simple insertion/deletion events to larger events of increasing complexity (Fig. 4). We identified 46 fhSV deletions that putatively disrupt the orthologous chimpanzee gene, of which only six were previously reported (5). Seven of the 46 fhSV deletions can also be seen in the transcript data (Iso-Seq). The largest novel fhSV deletion is 61,265 bp. It contains the majority of the caspase recruitment domain family member 8 gene (CARD8) and removes 13 exons that are transcribed into full-length cDNA in the chimpanzee (Fig. 4a). We also resolve a 65 kbp human-specific deletion in FADS1 and FADS2, genes involved in fatty acid biosynthesis that have been the target of positive selection (36) and potential dietary changes in human evolution (37, 38). The deletion brings the promoters of FADS1 and FADS2 (major isoform) in closer proximity and shortens the first intron of the other two FADS2 isoforms (Fig. 4b). The fhDEL might alter the relative abundance of the FADS2 isoforms, as supported by quantifying the number of splice-junction-containing reads unique to each isoform (16). The relative abundance of the minor FADS2 isoforms is significantly increased in humans (χ-sq = 165.65, df = 1, p < 2.2e-16). These minor isoforms differ only in their N-terminus, and, of the two, one (NM_001281502.1, designated here “long1”) shows evidence of encoding a signal peptide (39) potentially altering the protein’s subcellular location. Since great ape diets range from herbivorous to omnivorous, genic and structural changes related to diet metabolism may be of particular relevance for the evolution of ape species.
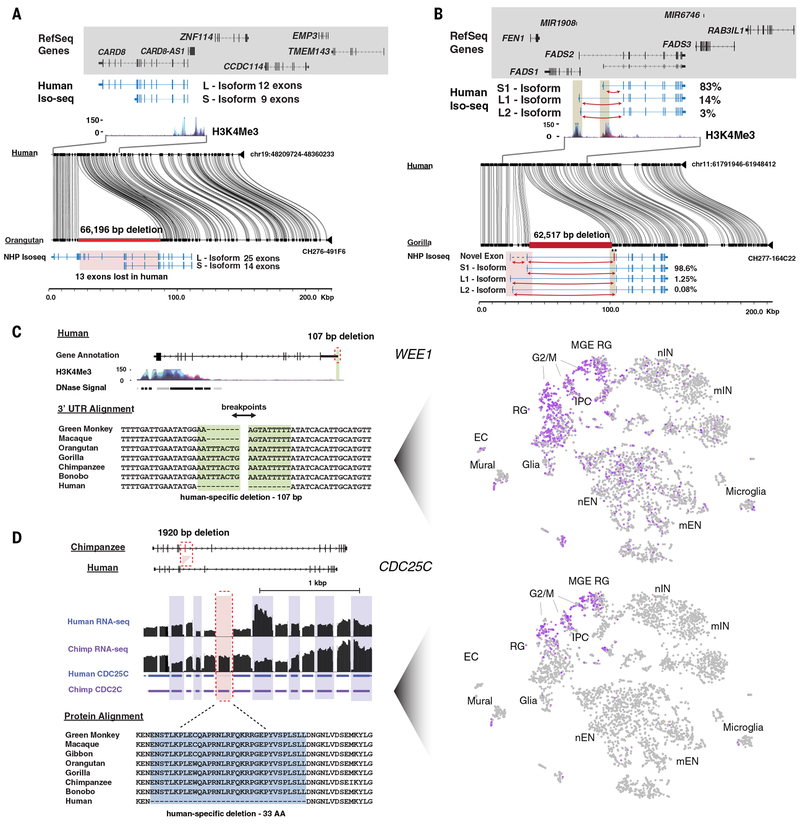
Fig 4. Examples of intragenic human-specific structural variation.
Shown are annotated MSAs between the human reference (GRCh38) and nonhuman primates (NHPs) generated with MAFFT or visualized with Miropeats against sequenced large-insert primate clones. Single-cell gene expression for select genes is highlighted across 4,261 cells developing human telencephalon plotted using t-distributed stochastic neighbor embedding (tSNE) (67). a) A 66.2 kbp intragenic deletion of CARD8 removes 13 putative coding exons in human. Iso-Seq data from chimpanzee and human iPSCs identifies isoforms with and without the deleted exons, respectively. b) A 62.5 kbp intergenic deletion of FADS2 is found in humans, along with an altered isoform ratio: the relative abundance of the long isoforms is increased in humans relative to chimpanzee, as seen in the counts of junction-spanning short reads specific to each isoform. Additionally, a novel, rare (<5%) 75 bp exon is observed in chimpanzee and gorilla but absent in human, likely resulting from a human-specific splice-site mutation. c) A 107 bp deletion in the 3’ UTR of WEE1 reduces AU-rich sequence content in the mRNA. The tSNE plot illustrates that WEE1 is highly expressed in cortical radial glia (RG), intermediate progenitor cells (IPCs), and medial ganglionic eminence progenitors (MGE RG) but shows limited expression in newborn and maturing inhibitory and excitatory neurons (nIN, mIN, nEN, mEN), microglia, endothelial cells (ECs), and glia. d) A 1,920 bp deletion of cell cycle regulator CDC25C removes a 99 bp constitutive exon conserved in mouse, resulting in a 33 amino acid deletion and shorter N-terminal regulatory domain in humans. The tSNE plot illustrates that CDC25C shows restricted expression to telencephalon progenitors in the G2/M cell cycle phase. Human and chimpanzee RNA-seq data were aligned directly to the exonic regions of CDC25C.
We further discovered two fhDELs, in WEE1 (Fig. 4c) and CDC25C (Fig. 4d), two highly conserved cell cycle genes that act as ultrasensitive antagonists during the interphase to mitotic transition, G2/M (40). WEE1 is a serine/threonine protein kinase that delays mitosis by phosphorylating CDK1, while CDC25C is a member of the phosphatase gene family that dephosphorylates CDK1, triggering entry into mitosis. Expression of these genes in radial glia is particularly interesting because additional cell divisions are thought to have played a role in increasing the number of cortical neurons in human evolution (41). These cell cycle regulators displaying different protein sequence or differential expression between chimpanzee and human are, thus, candidates for future investigation to explain neocortical expansion in the human lineage.
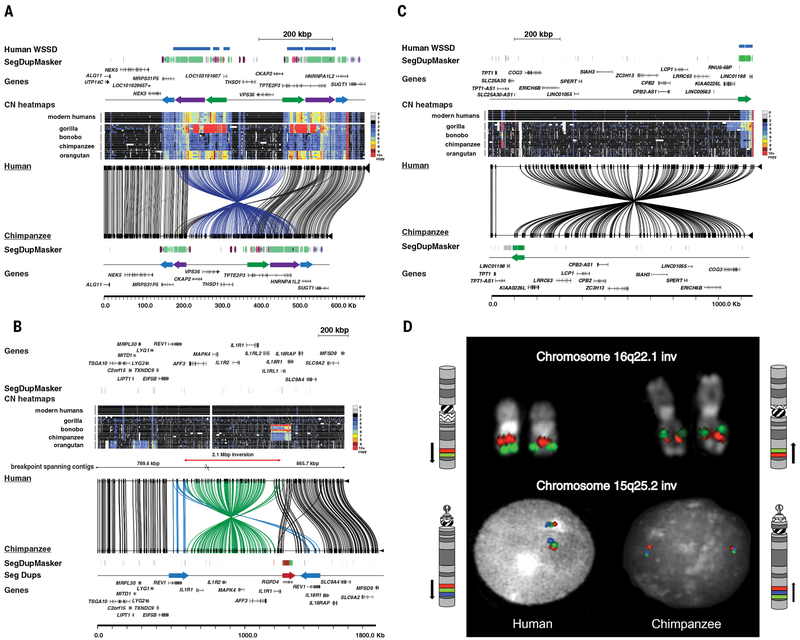
We also identified several larger, subcytogenetic structural differences using optical (Bionano) (42, 43) and BAC end-sequence mapping data that were not detected or sequence-resolved in previous genome assemblies. We validated large inversions and more complex SV events by integrating FISH and large-insert clone sequencing at the breakpoints (Table S12). We identified 29 human–chimpanzee–orangutan inversions (16 chimpanzee, 10 in orangutan and 3 shared between chimpanzee and orangutan) spanning 100 kbp to 5 Mbp in size of which 55% (16/29) have not been previously described (Table S12, Fig. 5) (44–48). More than 93% of inversions are flanked by large complex segmental duplication (SD) blocks, 38% of which show evidence of other structural and copy number variation at the boundaries of the inversion (Fig. 5).
Fig. 5. Complex structural variation.
Large-scale inversions between human and chimpanzee are depicted. The human reference genome sequence (GRCh38) with gene annotation is compared to large-insert clone-based assemblies from the chimpanzee BAC library CH251 using Miropeats. Connecting lines identify homologous regions of high sequence identity. SD organization is depicted as colored arrows as defined by whole-genome shotgun sequence detection (WSSD) and DupMasker. Heatmap indicates copy number (CN) estimated by read-depth from ape genome sequence. a) A ~265 kbp inversion on chromosome 13q14.3 detected by optical mapping in chimpanzee (annotated blue lines). The inverted region is flanked by large ~180 kbp inverted SD blocks that vary with respect to copy number among great apes. b) A 2.7 Mbp inversion on chromosome 2q12-13 detected by BAC end sequencing in chimpanzee (annotated green lines). The inverted region is flanked by duplication blocks containing lineage-specific expansions of the interleukins, an inverted duplication of REV1, and an additional copy of the RGPD4 core duplicon. c) A ~1.1 Mbp inversion at chr13q14.13 identified by optical mapping in chimpanzee encompassing 15 genes. On the telomeric side of the inversion lies a ~60 kbp duplication block that demonstrates lineage-specific duplications in great apes. d) Chromosome inversions, originally detected by optical mapping and BAC end sequencing, confirmed by metaphase analysis and interphase FISH experiments. A human-specific inversion of the chromosome 16q22.1 region was confirmed with orangutan clones CH276-89P20 (red) and CH276-192M7 (green) reported in upper line, and the 15q25.2 inversion was confirmed using chimpanzee clones CH251-321P13 (red), CH251-511D5 (green) and CH251-66E11 (blue) reported in lower line.
Interestingly, ~28% (8/29) of these ape–human inversions are also polymorphic among humans (49, 50)—some in regions previously shown to be hotspots of recurrent rearrangement and disease (48, 51). Notably, these regions of genomic instability also associate with expression differences in radial glial and excitatory neurons between the species. For example, among the 18 chimpanzee–human inversions (Table S12), we identified 18 differentially expressed brain genes between chimpanzee and human (10 radial glia, 11 excitatory neurons, 3 common to both sets), of which 78% resided in SD regions. Three of these genes (GLG1, ST3GAL2, and EXOSC6) were significantly upregulated in human and associated with a 5 Mbp human-specific inversion on chr16q22 (Fig. 5d). ST3GAL2 is the main mammalian sialyltransferase for GD1a and GT1b ganglioside biosynthesis in the brain (52).
Radial glial neural progenitor expression differences and human-specific SVs:
Over the course of human evolution, human brain volume has nearly tripled compared to chimpanzees (53), likely due to differential expression of genes during brain development (6, 8, 54). We investigated the association of structural variation with changes in human–chimpanzee brain gene expression using cerebral organoids as a proxy for brain expression differences (55). Importantly, because great ape brain tissue is largely inaccessible, these organoid models provide a realistic window into developmental cell behavior and gene expression differences between human and ape radial glia and other early developmental cell types (56). We processed several single-cell RNA-seq brain datasets from primary human cortex and from human and chimpanzee cortical organoids, focusing on cortical excitatory neurons and radial glia (55–57). Using the new chimpanzee SMRT assembly and genome annotations increases the sensitivity of gene expression analyses—our dataset reveals 2,625 additional chimpanzee genes with expression in the brain relative to previous studies (58). After performing unsupervised clustering, we analyzed 52,875 orthologous genes in 320 primary neurons, 176 human organoid cells, and 210 chimpanzee organoid cells expressing cortical radial glia and excitatory neuron genes.
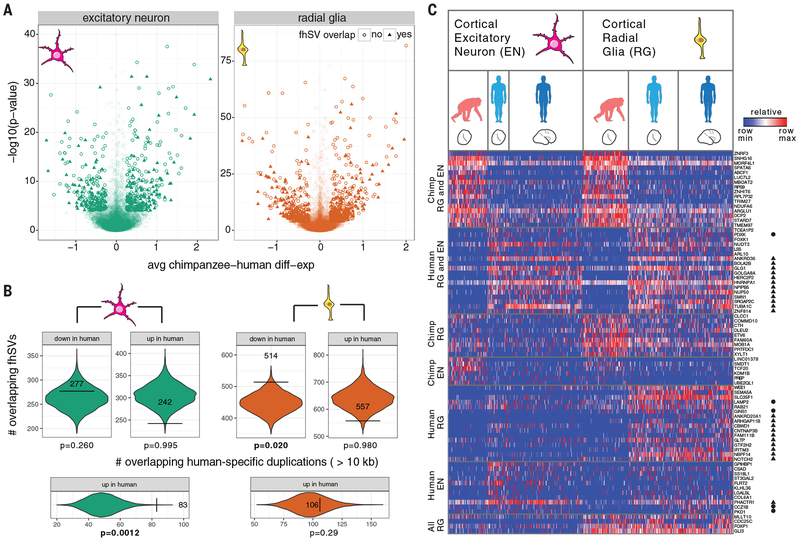
Our analysis identified 383 and 219 genes upregulated in human radial glial and excitatory neurons, respectively, when compared to chimpanzee (Table S13) (16). Conversely, we defined a set of 285 and 165 genes downregulated in human radial glia and excitatory neurons (Fig. 6), respectively; most of these changes have not been identified previously (56, 59). Because SVs are more likely (32) to affect gene expression, we considered fhSV overlap on the basis of VEP annotations (including GRCh38 and Clint_PTRv1 annotation sets), which correlates both coding and noncoding variation to genes (Fig. 6a). Of the differentially expressed genes, 252 radial glia genes (p = 9.78e-8; χ-sq; [252/668]) and 123 excitatory neuron genes (p = 0.27; χ-sq; [123/360]) had annotated fhSVs associated with them. To test if this observation was an artifact of gene size, we shuffled fhSVs and counted the number of fhSVs that mapped within 50 kbp of a differentially expressed gene.
Fig. 6. Structural variation and neural progenitor expression differences between human and chimpanzee.
a) Volcano plots for chimpanzee–human gene expression in excitatory neuron (left) and radial glia (right) organoid single-cell data. Each point represents a gene, with sufficient data to assess significance between human and chimpanzee organoid cells. Genes with fhSVs within 50 kbp are denoted with a triangle. The data points are shaded by significance. b) Spatial permutation test for overlap between fhSVs and differentially expressed genes. Each violin shows the null distribution of human-specific SV overlap (+/−50 kbp of transcript start/end) with genes that are significantly differentially down or upregulated, relative to chimpanzee. The horizontal bars and observed counts are overlaid upon the null distribution. c) Heatmap illustrating the percentile gene expression of differentially expressed genes near fhSVs (rows) across single cells (columns), including genes near the start or end of inversions (circle) and duplicated regions (WSSD) (triangle). Cells include 333 excitatory neurons (97 chimpanzee organoid; 53 human organoid; 183 human primary cells) and 373 radial glia (113 chimpanzee organoid; 123 human organoid; 137 human primary cells) (56, 57). Expression patterns include concerted changes between chimpanzee and human cells across radial glia and excitatory neurons (chimpanzee RG and EN; human RG and EN), cell-type-specific changes (human EN; human RG) and conserved radial glia expression (pan-RG).
Overall, genes downregulated in humans remain enriched for fhSVs, compared to the null distribution, whereas upregulated genes did not show a significant overlap. In particular, genes downregulated in human radial glial neural progenitors showed significant enrichment for structural variation (p = 0.02; 1e4 permutations) (Fig. 6b). Although we observe the same trend in excitatory neurons, the effect did not reach significance. As a control, we repeated the same analysis for genes mapping to human-specific SDs (54), a form of structural variation not accessed in this study. Genes mapping to human SDs were upregulated in radial glial and excitatory neurons when compared to chimpanzee (Fig. 6). This association identifies dozens of putative candidates for functional investigation, including some of the most differentially expressed genes between humans and chimpanzees in neural progenitor cells (Fig. 6, Table S14).
DISCUSSION
Our great ape genome assemblies improved sequence contiguity by orders of magnitude (20, 60), leading to a more comprehensive understanding of the evolution of structural variation. Coupling this effort with full-length cDNA sequencing improved gene annotation, especially for the discovery of new transcripts and isoforms that have recently diverged between closely related species. Because species may be sequenced and assembled using the same platforms and experimental designs, we minimized biases introduced by ascertainment or an uneven sequencing quality between genomes.
These improved genomes yielded a comprehensive view of intermediate-size structural variation among apes. As we focused on SVs that potentially disrupt genes or regulatory sequence, we began to address potential functional effect. Differential gene expression, especially in cortical radial glia, has been hypothesized to be a critical effector of brain size and a likely selective target of human brain evolution (61). Nearly 41% of the genes downregulated in human when compared to chimpanzee radial glial analogs from cerebral organoids associate with an fhSV and most often as a deletion or a retroposon insertion. These findings are consistent with the “less-is-more” hypothesis (62), which argues that the loss of functional elements underlies critical aspects of human evolution. In contrast, human-specific gene duplications associate with upregulated expression in both neural progenitors and excitatory neurons although the effect is stronger for the latter. This finding is consistent with recent studies evidencing that human-specific SDs contribute to cortical differences between humans and chimpanzees (6–8). It is intriguing that the repeat-rich nature of ape genomes and, in particular, the expansion of SDs in the common ancestral lineage of the African ape lineage (63) may have made great ape genomes particularly prone to both deletion and duplication, accelerating the rate of structural changes and large-effect mutations during the evolution of these species.
Despite this more comprehensive assessment of structural variation, not all SV types have been fully resolved among the great apes. In particular, we are still missing many larger, more complex events, including inversions and SDs that have differentially evolved between the lineages. For example, we recovered only one of five ape inversions identified by comparative BAC-based sequencing of a 2 Mbp region of chromosome 16p11.2 (64), although optical mapping techniques did identify four of the events. In this case, all inversions are flanked by large blocks of SDs (>200 kbp) that cannot be currently assembled by long-read WGS. We predict that such large, multi-megabase-pair inversions represent a common uncharacterized source of human–ape genetic variation that has been underestimated. Long-range sequencing and mapping technologies, such as Strand-seq (49), BAC-based sequencing (64), optical mapping (Table S12) and longer-read sequencing (65) will be necessary to sequence-resolve such large, more complex SVs.
MATERIALS AND METHODS
We sequenced and assembled four genomes (chimpanzee [Clint], Sumatran orangutan [Susie], CHM13 [human], and YRI19240 [human]) using long-read PacBio RS II sequencing chemistry and the Falcon genome assembler. Sequence contigs were error-corrected using Quiver (17), Pilon (18), and a FreeBayes-based (66) indel correction pipeline. A chromosomal-level AGP was generated using optical maps (Bionano Genomics Saphyr platform) for scaffold building and bicolor FISH of ~700 large-insert clones. The comparative annotation toolkit (CAT) (22) was used to annotate all of the great ape genomes using the human GENCODE V27 as reference with a combination of RNA-seq obtained from SRA as well as Iso-Seq data specifically from NHP iPSCs. STRs were defined using RepeatMasker v4.0.1 and Tandem Repeats multiple sequence Finder v4.07b. Syntenic regions and MSAs were constructed with MUSCLE (v3.8.31); phylogenetic analyses were performed using a general time-reversible model (“GTR+GAMMA”) under a maximum likelihood RAxML (8.2.3) framework; phylogenetic trees were generated using DendroP. A BLASR-based computational pipeline, smartie-sv, was developed to align, compare, and call insertions, deletions, and inversions (https://github.com/zeeev/smartie-sv). Insertions and deletions were genotyped against a panel of 45 ape genomes using SVTyper (paired-end) and WSSD (read depth). FISH and BAC clone sequencing was used to estimate sequence accuracy and validate the breakpoints of complex rearrangements. We compared SV locations with genes showing differential expression during human and chimpanzee cortical development using single-cell gene expression data from cerebral organoid models and from primary cortex.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Lee and A. Lewis for technical assistance and QC in generating sequencing data and G.I. Saunders from EBI for help with submitting our structural variation data to the EBI database. The authors thank C. Dunn and J. Chin for assistance with troubleshooting newer releases of the Falcon assembler, T. Brown as well as M. Lynn Gage for assistance in editing this manuscript. We thank L.W. Hillier for her insight on the chimpanzee assembly. Z.N.K. would like to thank E.E.E. and E. Kronenberg for their steady guidance. Funding: This work was supported, in part, by grants from the U.S. National Institutes of Health (NIH grants HG002385 to E.E.E.; HG007635 to R.K.W. and E.E.E.; HG003079 to R.K.W.; HG007990 to D.H., M.D. and B.P.; HG006283 to J.S.; HG009081 to S.K.D. and E.E.E.; HG007234 to D.H., B.P., J.A. and M.D.; HG008742 to B.P.; and HG009478 to M.L.D.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also partially supported by RBFR103CE3 by MIUR (MInistero dell’Università e Ricerca – Italy, to M.V.) and GC1R-06673-A by CIRM (California Institute for Regenerative Medicine, to I.T.F. and D.H.). S.C. was supported by a National Health and Medical Research Council (NHMRC) CJ Martin Biomedical Fellowship (#1073726). The work carried out by NCBI was supported by the Intramural Research Program of the NIH, National Library of Medicine. E.E.E., J.S. and D.H. are investigators of the Howard Hughes Medical Institute.
Footnotes
Competing interests: E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc.; A.R.H., A.W.C.P., J.L., E.T.L. and H.C. are employees of Bionano Genomics, Inc.; J.G.U. is an employee of Pacific Biosciences, Inc.
Data and materials availability: A full list of accessions and datatypes can be found in the supplemental section “data deposition and accessions.”
SUPPLEMENTARY MATERIALS
Science supplement link
Materials and Methods
Figs. S1 to S53
Tables S1 to S58
REFERENCES AND NOTES
- 1.Varki A, Geschwind DH, Eichler EE, Explaining human uniqueness: genome interactions with environment, behaviour and culture. Nat. Rev. Genet 9, 749–763 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King MC, Wilson AC, Evolution at two levels in humans and chimpanzees. Science. 188, 107–116 (1975). [DOI] [PubMed] [Google Scholar]
- 3.Fortna A et al. , Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2, E207 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd JL et al. , Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr. Biol 25, 772–779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLean CY et al. , Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 471, 216–219 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dennis MY et al. , Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 149, 912–922 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charrier C et al. , Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 149, 923–935 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Florio M et al. , Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 347, 1465–1470 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Ju X-C et al. , The hominoid-specific gene TBC1D3 promotes generation of basal neural progenitors and induces cortical folding in mice. Elife. 5, 206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scally A et al. , Insights into hominid evolution from the gorilla genome sequence. Nature. 483, 169–175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Locke DP et al. , Comparative and demographic analysis of orang-utan genomes. Nature. 469, 529–533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chimpanzee Sequencing and Analysis Consortium, Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 437, 69–87 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Lam ET et al. , Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol 30, 771–776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton JN et al. , Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol 31, 1119–1125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eid J et al. , Real-time DNA sequencing from single polymerase molecules. Science. 323, 133–138 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Materials and methods are part of the online supplementary materials.
- 17.Chin C-S et al. , Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Walker BJ et al. , Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yunis JJ, Prakash O, The origin of man: a chromosomal pictorial legacy. Science. 215, 1525–1530 (1982). [DOI] [PubMed] [Google Scholar]
- 20.Gordon D et al. , Long-read sequence assembly of the gorilla genome. Science. 352, aae0344–aae0344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuderna LFK et al. , A 3-way hybrid approach to generate a new high-quality chimpanzee reference genome (Pan_tro_3.0). Gigascience. 6, 1–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiddes IT et al. , Comparative Annotation Toolkit (CAT) - simultaneous clade and personal genome annotation. bioRxiv, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elango N, Thomas JW, NISC Comparative Sequencing Program, S. V. Yi, Variable molecular clocks in hominoids. Proc. Natl. Acad. Sci. U.S.A 103, 1370–1375 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moorjani P, Amorim CEG, Arndt PF, Przeworski M, Variation in the molecular clock of primates. Proc. Natl. Acad. Sci. U.S.A 113, 10607–10612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li WH, Tanimura M, Sharp PM, An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J. Mol. Evol 25, 330–342 (1987). [DOI] [PubMed] [Google Scholar]
- 26.Bickhart DM et al. , Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet 49, 643–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubinsztein DC et al. , Microsatellite evolution--evidence for directionality and variation in rate between species. Nat. Genet 10, 337–343 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Webster MT, Smith NGC, Ellegren H, Microsatellite evolution inferred from human-chimpanzee genomic sequence alignments. Proc. Natl. Acad. Sci. U.S.A 99, 8748–8753 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yohn CT et al. , Lineage-specific expansions of retroviral insertions within the genomes of African great apes but not humans and orangutans. PLoS Biol. 3, e110 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polavarapu N, Bowen NJ, McDonald JF, Identification, characterization and comparative genomics of chimpanzee endogenous retroviruses. Genome Biol. 7, R51 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaiser SM, Malik HS, Emerman M, Restriction of an extinct retrovirus by the human TRIM5alpha antiviral protein. Science. 316, 1756–1758 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Sudmant PH et al. , An integrated map of structural variation in 2,504 human genomes. Nature. 526, 75–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prado-Martinez J et al. , Great ape genetic diversity and population history. Nature. 499, 471–475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez-Rico YA et al. , Comparative analyses of super-enhancers reveal conserved elements in vertebrate genomes. Genome Res. 27, 259–268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reno PL et al. , A penile spine/vibrissa enhancer sequence is missing in modern and extinct humans but is retained in multiple primates with penile spines and sensory vibrissae. PLoS ONE. 8, e84258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ameur A et al. , Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am. J. Hum. Genet 90, 809–820 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye K, Gao F, Wang D, Bar-Yosef O, Keinan A, Dietary adaptation of FADS genes in Europe varied across time and geography. Nat Ecol Evol. 1, 167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buckley MT et al. , Selection in Europeans on Fatty Acid Desaturases Associated with Dietary Changes. Mol. Biol. Evol 34, 1307–1318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen TN, Brunak S, von Heijne G, Nielsen H, SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Trunnell NB, Poon AC, Kim SY, Ferrell JE Jr., Ultrasensitivity in the Regulation of Cdc25C by Cdk1. Molecular Cell. 41, 263–274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakic P, A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 18, 383–388 (1995). [DOI] [PubMed] [Google Scholar]
- 42.Pendleton M et al. , Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods 12, 780–786 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mak ACY et al. , Genome-Wide Structural Variation Detection by Genome Mapping on Nanochannel Arrays. Genetics. 202, 351–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feuk L et al. , Discovery of human inversion polymorphisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 1, e56 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman TL et al. , A genome-wide survey of structural variation between human and chimpanzee. Genome Res. 15, 1344–1356 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szamalek JM et al. , Polymorphic micro-inversions contribute to the genomic variability of humans and chimpanzees. Hum. Genet 119, 103–112 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Cardone MF et al. , Hominoid chromosomal rearrangements on 17q map to complex regions of segmental duplication. Genome Biol. 9, R28 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zody MC et al. , Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet 40, 1076–1083 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanders AD et al. , Characterizing polymorphic inversions in human genomes by single-cell sequencing. Genome Res. 26, 1575–1587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaisson MJP et al. , Multi-platform discovery of haplotype-resolved structural variation in human genomes, 1–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coe BP et al. , Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet 46, 1063–1071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sturgill ER et al. , Biosynthesis of the major brain gangliosides GD1a and GT1b. Glycobiology. 22, 1289–1301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herculano-Houzel S, The human brain in numbers: a linearly scaled-up primate brain. Front Hum Neurosci. 3, 31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dennis MY et al. , The evolution and population diversity of human-specific segmental duplications. Nat Ecol Evol. 1, 69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Camp JG et al. , Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. U.S.A 112, 15672–15677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mora-Bermúdez F et al. , Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. Elife. 5, 166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pollen AA et al. , Molecular identity of human outer radial glia during cortical development. Cell. 163, 55–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He Z et al. , Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat. Neurosci. 20, 886–895 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Marchetto MCN et al. , Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 503, 525–529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korlach J et al. , De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. Gigascience. 6, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rakic P, A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 18, 383–388 (1995). [DOI] [PubMed] [Google Scholar]
- 62.Olson MV, When less is more: gene loss as an engine of evolutionary change. Am. J. Hum. Genet 64, 18–23 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marques-Bonet T et al. , A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 457, 877–881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nuttle X et al. , Emergence of a Homo sapiens-specific gene family and chromosome 16p11.2 CNV susceptibility. Nature. 536, 205–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jain M, Olsen HE, Paten B, Akeson M, The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 17, 239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garrison E, Scholar GMG, Haplotype-based variant detection from short-read sequencing. arXiv Prepr. arXiv1207 3907 2012; 9. [Google Scholar]
- 67.Nowakowski TJ et al. , Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 358, 1318–1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Durand NC et al. , Juicebox Provides a Visualization System for Hi-C Contact Maps with Unlimited Zoom. Cell Syst. 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koren S et al. , Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lichter P et al. , High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 247, 64–69 (1990). [DOI] [PubMed] [Google Scholar]
- 71.Paten B et al. , Cactus: Algorithms for genome multiple sequence alignment. Genome Res. 21, 1512–1528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.König S, Romoth LW, Gerischer L, Stanke M, Simultaneous gene finding in multiple genomes. Bioinformatics. 32, 3388–3395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shekhar K et al. , Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 166, 1308–1323.e30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaisson MJ, Tesler G, Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics. 13, 238 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khelik K, Lagesen K, Sandve GK, Rognes T, Nederbragt AJ, NucDiff: in-depth characterization and annotation of differences between two sets of DNA sequences. BMC Bioinformatics. 18, 338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stanyon R et al. , Primate chromosome evolution: ancestral karyotypes, marker order and neocentromeres. Chromosome Res. 16, 17–39 (2008). [DOI] [PubMed] [Google Scholar]
- 77.Chaisson MJP, Wilson RK, Eichler EE, Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet 16, 627–640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huddleston J et al. , Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 27, 677–685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weissensteiner MH et al. , Combination of short-read, long-read, and optical mapping assemblies reveals large-scale tandem repeat arrays with population genetic implications. Genome Res. 27, 697–708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiao W-B et al. , Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res. 27, 778–786 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ventura M et al. , Gorilla genome structural variation reveals evolutionary parallelisms with chimpanzee. Genome Res. 21, 1640–1649 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ventura M et al. , Evolutionary formation of new centromeres in macaque. Science. 316, 243–246 (2007). [DOI] [PubMed] [Google Scholar]
- 83.Ramani V et al. , Mapping 3D genome architecture through in situ DNase Hi-C. Nat Protoc. 11, 2104–2121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lek M et al. , Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Danecek P et al. , The variant call format and VCFtools. Bioinformatics. 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Braastad CD, Hovhannisyan H, van Wijnen AJ, Stein JL, Stein GS, Functional characterization of a human histone gene cluster duplication. Gene. 342, 35–40 (2004). [DOI] [PubMed] [Google Scholar]
- 87.Hormozdiari F et al. , Rates and patterns of great ape retrotransposition. Proc. Natl. Acad. Sci. U.S.A 110, 13457–13462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ramsay L et al. , Conserved expression of transposon-derived non-coding transcripts in primate stem cells. BMC Genomics. 18, 214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chan Y-S et al. , Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 13, 663–675 (2013). [DOI] [PubMed] [Google Scholar]
- 90.Renaud G, Stenzel U, Maricic T, Wiebe V, Kelso J, deML: robust demultiplexing of Illumina sequences using a likelihood-based approach. Bioinformatics. 31, 770–772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hartley SW, Mullikin JC, QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics. 16, 224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gordon SP et al. , Widespread Polycistronic Transcripts in Fungi Revealed by Single-Molecule mRNA Sequencing. PLoS ONE. 10, e0132628 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 96.Consortium GTEx, The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harrow J et al. , GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stanke M, Diekhans M, Baertsch R, Haussler D, Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 24, 637–644 (2008). [DOI] [PubMed] [Google Scholar]
- 99.Stanke M et al. , AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–9 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stanke M, Schöffmann O, Morgenstern B, Waack S, Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 7, 62 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mallick S et al. , The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sudmant PH et al. , Global diversity, population stratification, and selection of human copy-number variation. Science. 349, aab3761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.George RD et al. , Trans genomic capture and sequencing of primate exomes reveals new targets of positive selection. Genome Res. 21, 1686–1694 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mohajeri K et al. , Interchromosomal core duplicons drive both evolutionary instability and disease susceptibility of the Chromosome 8p23.1 region. Genome Res. 26, 1453–1467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiang Z et al. , Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet 39, 1361–1368 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.