Abstract
Amyloids are typically associated with neurodegenerative diseases, but recent research demonstrates that several bacteria utilize functional amyloid fibrils to fortify the biofilm extracellular matrix and thereby resist antibiotic treatments. In Pseudomonas aeruginosa, these fibrils are composed predominantly of FapC, a protein with high-sequence conservation among the genera. Previous studies established FapC as the major amyloid subunit, but its mechanism of fibril formation in P. aeruginosa remained largely unexplored. Here, we examine the FapC sequence in greater detail through a combination of bioinformatics and protein engineering, and we identify specific motifs that are implicated in amyloid formation. Sequence regions of high evolutionary conservation tend to coincide with regions of high amyloid propensity, and mutation of amyloidogenic motifs to a designed, non-amyloidogenic motif suppresses fibril formation in a pH-dependent manner. We establish the particular significance of the third repeat motif in promoting fibril formation and also demonstrate emergence of soluble oligomer species early in the aggregation pathway. The insights reported here expand our understanding of the mechanism of amyloid polymerization in P. aeruginosa, laying the foundation for development of new amyloid inhibitors to combat recalcitrant biofilm infections.
Keywords: protein aggregation, bacterial amyloid, extracellular matrix, protein sequence analysis, peptide microarray
Introduction
Pseudomonas aeruginosa is one of the most prominent causes of health care-associated infections due to its unique arsenal of virulence factors, resistance to a range of antibiotics, and its ability to form biofilms [1–3]. Infections with P. aeruginosa often manifest in the form of ventilator-associated pneumonia, which demonstrates mortality rates as high as 30% in patients with comorbidities [4]. The plastic endotracheal tube readily provides a colonization site for P. aeruginosa, and aerosolization of the mature biofilm by mechanical ventilation or tracheal suctioning can lead to ventilator-associated pneumonia [5]. Mucoid, biofilm-associated P. aeruginosa, is also found in chronic lung infections of patients with cystic fibrosis [6]. When bacteria dwell in biofilms, cells co-associate using a self-produced extracellular matrix (EM). The EM comprises a protective network of polysaccharides, DNA, lipids, and proteins that encases the cells, mediates attachment, and promotes metabolite transport [7]. Proteins in the EM take on a variety of roles, but a growing body of research seeks to understand the role of amyloid fibrils in this complex biological material [8].
Amyloid fibrils are non-branching, rigid protein structures characterized by high β-sheet content and alignment of β-strands perpendicular to the fibril axis [9] (Fig. 1a). Insoluble amyloid deposits form as a result of protein folding dysfunction in diseases like Alzheimer’s and Parkinson’s, but recent evidence suggests that several microbes intentionally produce amyloid fibrils to serve functional roles [10]. Biofilm-associated bacteria, in particular, utilize amyloids as a building material to reinforce the EM and resist dispersion by chemical or mechanical agents [8, 11]. These fibrils may also promote antibiotic resistance and act as a reservoir for small molecules involved in quorum sensing [12, 13]. Amyloid fibrils polymerize in the absence of an energy source, so they serve as a metabolically advantageous molecular scaffold despite the limited resources of the EM environment [14].
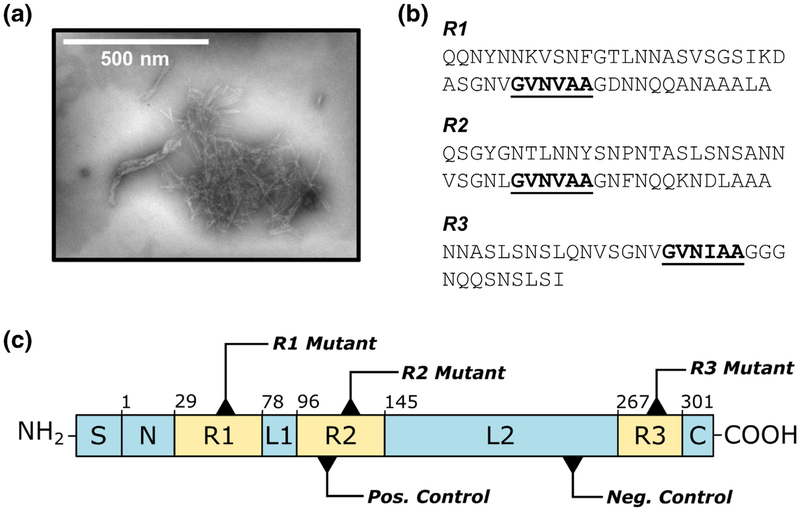
Fig. 1.
The FapC protein from P. aeruginosa. (a) FapC aggregates to form amyloid fibrils. (b) The FapC sequence is characterized by three conserved repeats, R1, R2, and R3, each of which contains a GVNXAA motif indicated by bold/underlined text. (c) Schematic of the PAO1 FapC sequence, including mutation sites for each of the protein variants used in this study. S = signal peptide, N = N-terminal region (residues 1–28), R1–R3 = repeat regions (residues 29–77, 96–144, 267–300); L1–L2 = linker regions (residues 78–95, 145–266); C = C-terminal region (residues 301–316). The first residue of each region is indicated above its corresponding box. The R1, R2, and R3 mutants incorporate GVNXAA→KFDDTK substitution in each of the three repeats, and the TM incorporates this substitution in all three repeats. The negative control (AQGAKD→KFDDTK) consists of a hexapeptide substitution in L2, and the positive control (SANNVS→GVNVAA) inserts a second amyloidogenic hexapeptide in R2.
In P. aeruginosa, the production of functional amyloids is controlled by a single six-gene operon known as (functional amyloid in Pseudomonas) fap [15]. The mature form of the major amyloid subunit, FapC, consists of 316-amino-acid residues without its 24-residue signal peptide, and it includes three imperfect sequence repeats (R1, R2, R3) separated by two “linker” regions (L1, L2) [16] (Fig. 1b, c). The three repeats are highly conserved among pseudomonads and related genera, with 56% ± 25% average residue conservation observed among 65 strains [17]. As such, the current model of FapC fibril formation designates the repeat regions as drivers of amyloid polymerization that ultimately constitute the core of the mature fibril. Analogous to the curli amyloids found in Escherichia coli [18], the fap system also includes a nucleator protein FapB, which demonstrates 38% sequence identity to FapC and is believed to serve as a template for rapid fibril polymerization on the exterior of the cell. Small amounts of FapB are also found in mature fibrils, so it may play an additional role in modulating properties of the fibrils [17]. The remaining proteins encoded by the fap operon serve as outer membrane pores for translocation of amyloid precursors (FapF) [19], chaperones to guide monomers through the periplasm (FapA), or auxiliary regulators and proteases (FapE, FapD) [16]. Wild-type P. aeruginosa PAO1 expresses fap constitutively, with peak promoter activity occurring during the exponential growth phase, but laboratory growth conditions limit the strain’s ability to produce large quantities of functional amyloid. Conversely, overexpression of the fap operon in P. aeruginosa PAO1 leads to highly aggregative phenotypes with five to six times more biofilm than the wild-type strain, and similar effects are observed for overexpression of fap in E. coli [15,16,20].
Despite extensive characterization of Fap proteins under sessile growth conditions, their mechanisms of fibril formation remain largely unexplored. We studied the FapC sequence in greater detail through a combination of bioinformatics and protein engineering. Sequence analysis of the repeat regions predicts that a specific, conserved hexapeptide motif—GVNXAA—is responsible for a significant amyloidogenic effect in each of the three repeats. Mutation of the amyloidogenic motif to a highly soluble, non-amyloidogenic hexapeptide changes aggregation kinetics compared to wild-type FapC in a direction consistent with our predictions. These effects are pH dependent, and we demonstrate the particular significance of the third sequence repeat, R3, in promoting fibril formation. Finally, we highlight a minor role for the FapC disulfide bond in forming small, pre-amyloid oligomers. The insights reported here reveal important mechanistic details of FapC polymerization, paving the way for new strategies to inhibit functional amyloid formation and ultimately provide new therapeutic avenues for biofilm-associated infections.
Results
Amyloid-prone segments of FapC coincide with conserved regions
The rapid polymerization of FapC in vitro precludes its structural characterization by traditional means, such as NMR or X-ray crystallography, but a combination of evolutionary sequence analysis and bioinformatics tools helped us to identify potential “hot spots” for aggregation. The three repeat regions of the FapC sequence are highly conserved among pseudomonads, and therefore, we hypothesized that these regions are critical for the macromolecular assembly of amyloid fibrils. A variety of bioinformatics tools has been developed to predict aggregation propensity and solubility of proteins [21], and we applied two of these algorithms to examine the repeat sequences of FapC from P. aeruginosa PAO1 (Fig. 2a, b). Both programs apply a sliding-window approach to analyze query sequences in hexapeptide increments.
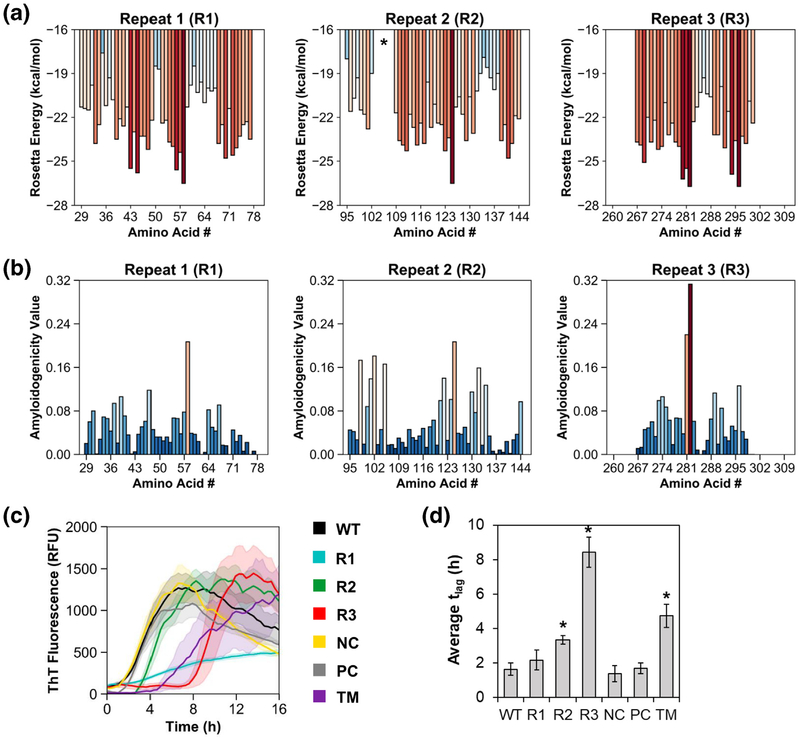
Fig. 2.
Mutation of conserved GVNXAA motifs modulates aggregation kinetics in a direction consistent with computational predictions. ZipperDB (a) and FISH Amyloid (b) algorithms were used to predict amyloidogenic regions within each of the three FapC repeats, R1–R3. Red colored bars indicate high amyloidogenicity, while blue colored bars indicate low amyloidogenicity. R1 spans residues 29–77, R2 spans residues 95–144, and R3 spans residues 267–300. GVNXAA motifs initiate at the following sties: R1 = residue 59; R2 = residue 126; R3 = residue 283. The asterisk in ZipperDB output for R2 indicates no data for residues 104–109 because residue Pro109 is incompatible with the 3D profile method. Amino acid numbering schemes in this figure are based on PAO1 FapC without its signal peptide. (c) Protein variants were expressed and purified, diluted into 10 mM Tris buffer (pH 7.5), and allowed to aggregate with shaking at 37 °C. ThT fluorescence indicates amyloid fibril formation. (d) Lag times define the amount of time elapsed prior to onset of fibril formation in each of the FapC variants. Asterisks indicate significant increase (p < 0.001, two-tailed Student’s t test) in lag time compared to WT. Error bands/bars in panels c and d indicate 1 SD from the mean of at least three replicates.
ZipperDB, which uses structural alignment via the 3D profile method and Rosetta-based scoring to compute aggregation propensities, predicts fibril formation in hexapeptides with Rosetta energies below −23 kcal/mol [22, 23]. In PAO1 FapC, these segments tend to arise in the center of each repeat, with the greatest amyloid propensity occurring in R3 (Fig. 2a). Similarly, FISH Amyloid, a machine learning approach that classifies sequences based on cooccurrence of query residues with those of known amyloidogenic hexapeptides [24], also predicts higher amyloidogenicity in the center of each repeat, with a notable spike in R3 (Fig. 2b). The hexapeptide with the greatest predicted amyloid propensity in each repeat follows the same amino acid motif: GVNXAA, where X = Val in R1 and R2 and X = Ile in R3. Importantly, these amyloidogenic motifs coincide with regions of high-sequence conservation among pseudomonads. We used Clustal Omega [25] to align 31 Pseudomonas FapC sequences originally compiled by Dueholm and coworkers [17], and then we assessed the sequence identity of each hexapeptide motif to its counterpart in the P. aeruginosa PAO1 FapC sequence. Indeed, the R1 hexapeptide site averages 85.5% identity; the R2 site averages 75.6% identity; and the R3 site averages 78.9% identity to PAO1 FapC. Notably, the Gly and Asn residues at positions 1 and 3 are 100% conserved in all repeats across all analyzed sequences (SI Table 1).
Redesigned amyloidogenic segments modulate FapC aggregation kinetics
We sought to further probe the significance of predicted amyloidogenic regions in PAO1 FapC by mutating the conserved hexapeptide motif, GVNXAA, to a hexapeptide with very low amyloid propensity. To select a low-amyloid hexapeptide, candidate sequences were first determined using peptide microar-rays displaying segments from the Pseudomonas sp. UK4 FapC protein, a shorter sequence (226 residues in the mature protein versus 316 for PAO1 FapC) that is more amenable to high-throughput analysis but retains the triple-repeat structure found in P. aeruginosa PAO1 FapC. Solutions of fluorescent-labeled UK4 FapC were applied to cellulose arrays of 384 immobilized peptide spots, each of which displays a 14-residue peptide. One hundred seven of these spots were dedicated to the UK4 FapC sequence. Each spot moves two residues forward in the FapC sequence compared to its predecessor, leading to 12-residue overlaps between successive spots to facilitate high-resolution analysis of protein self-association [26]. Fluorescence measurements indicated low levels of self-association for a charged region around residues 23–56 (numbering based on the first position of the mature protein, highlighted with orange in SI Fig. 1A). Hexapeptide sequences from this region were subsequently analyzed for amyloid propensity in ZipperDB (SI Fig. 1B) and FISH Amyloid, and generic aggregation potential was evaluated using the AGGRESCAN server [27]. The hexapeptide KFDDTK was ultimately selected as the ideal substitution for GVNXAA in PAO1 FapC, with a Rosetta energy of −17.7 kcal/mol and a FISH amyloidogenicity value of 0.025. A total of six mutant proteins were generated (Fig. 1c): GVNXAA→KFDDTK substitution in each of the three repeats (R1, R2, R3); a triple mutant (TM) with substitution in all three repeats (TM); AQGAKD→KFDDTK as a negative control in linker region 1 (NC); and SANNVS→GVNVAA to insert a second amyloidogenic hexapeptide in R2, 9 residues upstream of GVNVAA (PC). All mutant proteins and wild-type (WT) PAO1 FapC were recombinantly expressed in E. coli using pET30a + vectors and a C-terminal His tag for purification.
The aggregation kinetics of PAO1 FapC WT and its associated mutants were compared using a fluorescence assay. Briefly, each protein was desalted from a stock solution containing 8 M urea (which keeps the protein monomeric under storage conditions) into buffer and diluted to a final concentration of 0.2 mg/mL. Samples were incubated with shaking in a plate reader at 37 °C, and thioflavin T (ThT), a compound known to fluoresce in the presence of β-sheet-rich amyloid fibrils [28], enabled monitoring of protein fibril formation in real time. Conversion from random coil to β-sheet secondary structure was verified by circular dichroism spectroscopy (CD) (SI Fig. 2A) and the presence of amyloid fibrils was confirmed by transmission electron microscopy (TEM) (SI Fig. 2B). Fibril conformation was additionally validated by Fourier-transform infrared spectroscopy (FTIR), where recombinant FapC fibrils were compared to Fap fibril extracts from P. aeruginosa. The high content of antiparallel β-sheets in amyloid fibrils leads to a characteristic high frequency band in the 1620- to 1630-cm−1 region [29], and the FapC variants displayed a characteristic band at ~1625 cm−1 (SI Fig. 2C).
As shown in Fig. 2c, FapC WT fibrillated quickly, with essentially no lag time before ThT fluorescence increased. The three GVNXAA→KFDDTK mutants, however, displayed markedly different kinetics compared to the wild-type. Substitutions in R2 and R3 delayed the onset of fibril formation (Fig. 2d), and mutation in R1 substantially slowed the rate of fibril formation, that is, the slope of the transition region from low to high ThT fluorescence. Interestingly, the combined effect of three hexapeptide mutations did not entirely suppress fibril formation in the TM. Rather, the aggregation kinetics of TM reflected a blend of the three single mutants, R1, R2, and R3; the lag time for TM was extended, like R3, but its rate of fibril formation was also somewhat slow, like R1. Note that it was not uncommon to see a decay in ThT signal intensity from the plateau reached after the transition region; this is frequently observed for fibrillating proteins, and while no definitive explanation for its occurrence has been provided, it may be caused by slow isomerization of ThT or bundling of the initially formed fibrils rather than reflecting conformational transitions at the level of the individual molecule. Therefore, we like others chose to disregard it in our analysis of the data [28]. Overall, the most significant changes resulted from GVNIAA→KFDDTK mutation in R3. This mutation induces a remarkable increase in the lag time, tlag, from 1.6 h in the wild-type to 8.4 h in the R3 mutant (Fig. 2d). Analysis of the mutant sequences in ZipperDB supports this observation; mutations in the R3 region are predicted to have a greater energetic impact than mutations in R1 or R2 (SI Table 3). In addition, the negative and positive control mutants NC and PC fibrillated with behavior similar to WT, indicating that the influence of mutations is site-specific. The NC and PC mutants display modest changes in Rosetta energy compared to WT (SI Table 3), but the magnitude of these changes is much smaller than that of the GVNXAA mutants, which serves as a salutary reminder of the importance of context. These observations support our hypothesis that conserved GVNXAA motifs play a key role in amyloid formation in PAO1 FapC.
FapC fibril formation is pH dependent
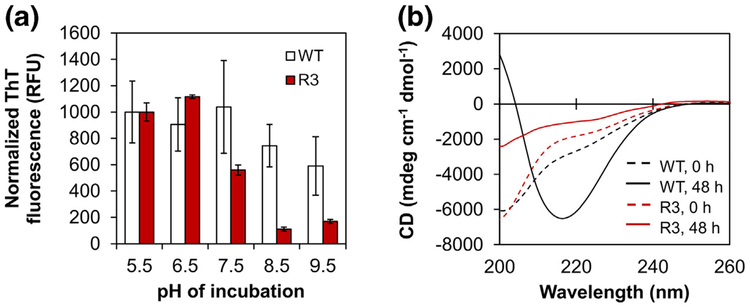
Natural habitats for P. aeruginosa are highly variable, including soil, water, plants, animals, and sewage. The pH of these environments spans a broad range from 4.5 to 9.5, which necessitates adaptation according to extracellular conditions [30]. In the hospital environment, P. aeruginosa encounters both acidic conditions (e.g., the airway of cystic fibrosis patients) and alkaline conditions (e.g., chronic wounds) [31, 32]. We therefore sought to explore the influence of pH on fibril formation of PAO1 FapC in vitro. Desalted FapC stocks were diluted into pH-adjusted buffer and aggregation was monitored in a plate reader using ThT fluorescence as previously described. Three pH values were evaluated to account for three clinical scenarios—pH 6.5 for the cystic fibrosis airway, pH 7.5 for normal physiological conditions, and pH 8.5 for chronic wounds—along with two additional “extreme” pH values at 5.5 and 9.5. ThT becomes inactivated by hydroxylation at pH > 8, so the pH 8.5 and 9.5 samples were incubated without ThT [33]. At the end of the assay (48 h), the pH of these samples was shifted back to 6.5 and then ThT was added to obtain end point fluorescence measurements. In the case of FapC WT, the extent of fibril formation did not change significantly between pH 5.5 and 9.5 (Fig. 3a), and the lag time remained quite short (~1 h) for pH values < 8 (SI Fig. 3A). Like WT, the aggregation kinetics of the R3 mutant were relatively similar to one another at pH 5.5–7.5 (SI Fig. 3B). However, the extent of fibril formation was substantially reduced at pH 8.5 and 9.5 (Fig. 3a). These observations were confirmed by CD spectroscopy, which demonstrated conversion from random coil to β-sheet structure in FapC WT but not the R3 mutant at pH 9.5 (Fig. 3b). These results further support the role of conserved GVNXAA motifs in promoting amyloid formation under a variety of conditions, and they underscore the importance of amino acid charge composition at the mutation site.
Fig. 3.
Fibril formation of FapC depends on pH. (a) FapC WT and FapC R3 mutant were incubated with shaking at 37 °C in 10 mM Tris buffer (pH 5.5, 6.5, 7.5, 8.5, or 9.5). After 48 h, the ThT fluorescence of all samples was measured on a plate reader (high-pH samples were incubated without ThT and adjusted to pH 6.5 prior to measurement). These end point measurements indicated a high degree of fibril formation for WT, regardless of pH, but R3 fibril formation was more pH dependent. Error bars represent the SD from the mean of three replicates. (b) CD spectra for pH 9.5 at t = 0 h (dashed lines) and 48 h (solid lines) indicate secondary structure conversion from random coil to β-sheet in FapC WT (black) but not the R3 mutant (red).
Sequence repeat R3 is essential for robust fibril formation
The reduced aggregation propensity of the R3 mutant encouraged further investigation of the role of R3 in amyloid fibril formation and stability. First, we hypothesized that delayed aggregation of the R3 mutant would enhance its susceptibility to known amyloid inhibitors. Next, we compared the stability of R3 fibrils to their WT counterparts using a novel dissociation approach with formic acid (FA).
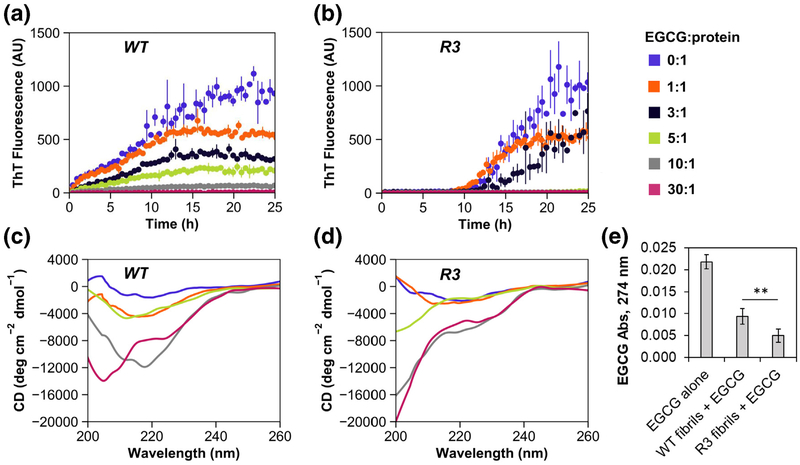
Previous studies have established the utility of epigallotcatechin-3-gallate (EGCG) as an antimicrobial that disrupts biofilm stability, interferes with quorum sensing, and increases susceptibility to antibiotics [34, 35]. EGCG also inhibits PAO1 FapC amyloid formation by stabilizing pre-fibrillar oligomers, and it structurally remodels existing fibrils in vitro through the formation of non-amyloid aggregates [13]. To investigate the effect of EGCG on fibril formation of FapC WT and R3, desalted protein stocks were diluted in Tris buffer with various concentrations of EGCG. Samples were aliquoted into 96-well plates and incubated with shaking in a plate reader at 37 °C, where fibril formation was monitored by ThT fluorescence (Fig. 4a, b). Without EGCG, FapC WT and R3 aggregated in a similar fashion to previous experiments, albeit at a slower rate due to decreased protein concentration and gentler shaking (see Materials and Methods). As expected, the introduction of EGCG substantially inhibited the extent of amyloid formation in both FapC WT and R3, but the R3 mutant responded with greater sensitivity; an EGCG:protein molar ratio of 30:1 was required to fully suppress fibril formation in FapC WT, while a molar ratio of 5:1 was sufficient to elicit the same effect in R3 within the 25-h observation window. Some evidence of aggregation eventually emerged in the 5:1 R3 sample after ~30 h of incubation (SI Fig. 4A).
Fig. 4.
FapC R3 is more susceptible to inhibition by the small molecule EGCG. FapC WT (a) and FapC R3 mutant (b) were incubated with shaking in 10 mM Tris buffer (pH 7.5). Various EGCG:protein molar ratios were tested (blue = 0:1, orange = 1:1, black = 3:1, green = 5:1, gray = 10:1, magenta = 30:1). In all cases, EGCG exerted some inhibitory effect on FapC fibril formation, but the effect was much more pronounced for the R3 mutant protein than WT. At the end of the assay, samples were analyzed further by CD. (c) For FapC WT, only the samples with an EGCG:protein molar ratio of 30:1 remained unstructured throughout the 45 h incubation; all other samples converted to β-sheet-rich signals at ~220 nm. (d) For FapC R3, random coil content persisted down to an EGCG:protein ratio of 1:1, indicating much greater susceptibility to inhibition by EGCG. Error bars in panels A and B represent the SD from the mean of three replicates. (e) FapC WT and R3 fibrils were incubated with 15 μM EGCG overnight, and then fibrils were removed from solution by centrifugation. Analysis of the supernatants by UV–Vis spectrometry demonstrated that R3 fibrils bound more EGCG than WT fibrils (p = 0.003, two-tailed Student’s t test). Error bars indicate SD from the mean of three measurements.
Independent tests have shown that EGCG can displace ThT from its fibril binding sites or serve as a quencher, which may bias fluorescence assays [36, 37]. Therefore, we chose to validate the impact of EGCG by structurally evaluating end point samples with CD spectroscopy (Fig. 4c, d). Prefibrillar, soluble FapC is mostly random coil (unstructured), and these types of signals were observed for samples incubated with high concentrations of EGCG. In the case of FapC WT, only the samples with an EGCG:protein molar ratio of 30:1 remained unstructured throughout the 45-h incubation; all other samples converted to β-sheet-rich fibrils with signals at ~220 nm. Conversely, unstructured content persisted in FapC R3 down to an EGCG:protein ratio of 1:1, indicating much greater susceptibility to inhibition by EGCG. For both proteins, weakening of the β-sheet signal at lower EGCG concentrations was attributed to visible fibril precipitation. We also performed a simple spin-down assay to assess the binding of EGCG to WT and R3 fibrils. Fibrils were prepared in 10 mM Tris buffer (pH 7.5), and then washed and resuspended either in buffer alone or buffer +15 μM EGCG. The suspensions were incubated at room temperature overnight and then insoluble fibrils were removed by centrifugation. The supernatants were analyzed by UV–Vis spec-trometry to assess the quantity of unbound EGCG remaining in solution (Fig. 4e). R3 fibrils bound significantly more EGCG than WT fibrils, a result that is in accord with the greater EGCG susceptibility observed for this protein. Finally, to address the possibility of a nonspecific, colloidal inhibition effect [38, 39], we used dynamic light scattering (DLS) and nanoparticle tracking analysis (NTA) to analyze EGCG samples at a range of concentrations. Some particles were observed at a concentration of 90 μM EGCG (i.e., the 30:1 molar ratio used in inhibition experiments), but not in more dilute samples (SI Fig. 4B). Thus, colloidal effects may play a role in inhibiting FapC fibril formation, but only when EGCG is applied at very high concentrations.
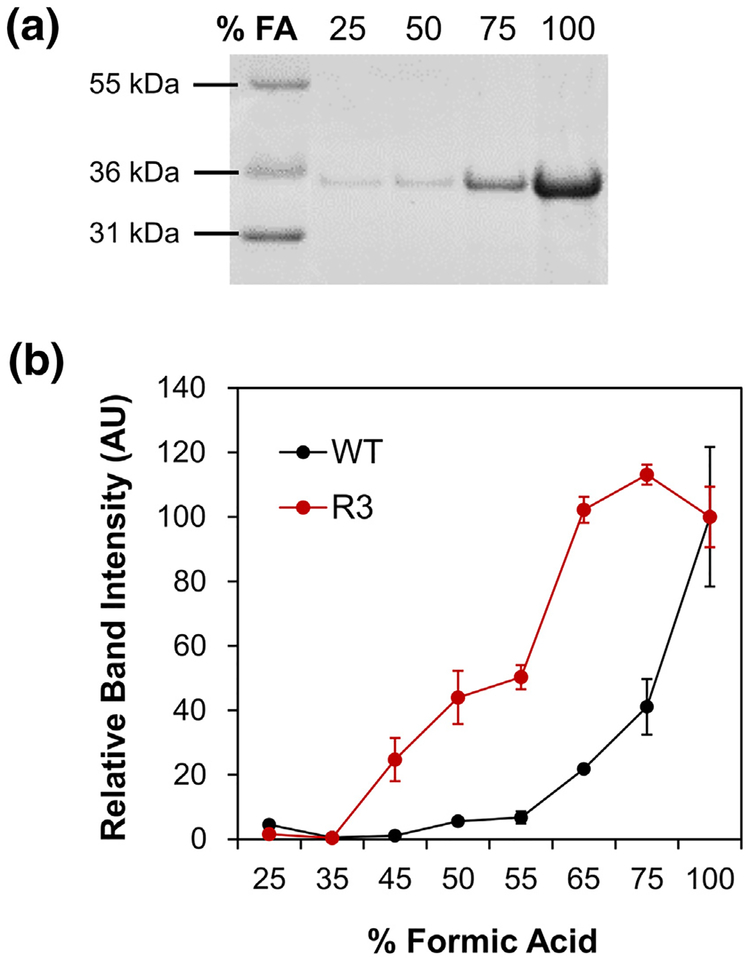
The notable differences in aggregation kinetics and susceptibility to inhibitors between FapC WT and R3 prompted further examination of the fibrils themselves. Treatment of amyloids with concentrated FA is an established technique to solubilize fibrillar material [40], particularly functional amyloids which require much higher concentrations of FA than pathological amyloids to be solubilized [15] (and Line F.B. Christiansen and D.E.O., unpublished results). Note that dissolution of FapC fibrils is not a simple pH effect; the pH drops below 2.0 at low concentrations of FA (<20%), at which stage all side chains are titrated. However, dissolution only occurs at much higher FA concentrations (see below), indicating that FA solubilizatoin must be attributed to its properties as organic solvent. We extended these methods to investigate the stability of FapC fibrils. Samples of FapC WT and R3 were incubated in 96-well plates under the same conditions as the ThT aggregation assays, but with no ThT present. After 48 h, fibril solutions were withdrawn from the plates and subjected to a series of centrifugation and wash steps to remove soluble or loosely adhered protein. The fibrils were then incubated with increasing concentrations of FA (25%–100%) for 10 min prior to freeze drying and resuspension in water. The solubility of each sample was determined by SDS-PAGE, where the intensity of the FapC monomer band corresponded to the fraction of protein dissolved by FA (Fig. 5a). Visible aggregates were present in samples treated with low concentrations of FA, but solutions with 100% FA treatment appeared completely soluble and were used as the reference bands for all other samples. In accordance with its relatively slow polymerization and greater susceptibility to inhibition by EGCG, the R3 mutant fibrils dissolved much more easily and therefore displayed more soluble protein on gels than those from FapC WT (Fig. 5b). The midpoint of dissolution (i.e., the concentration of FA at which 50% of the fibrils have dissolved) was estimated using least-squares regression (SI Fig. 5) at 54% FA for the R3 mutant and 80% FA for FapC WT, demonstrating that WT fibrils are more stable. These results further underscore the importance of the R3 repeat, and the GVNIAA sequence in particular, in reinforcing the intractable structure of FapC fibrils.
Fig. 5.
Fibrils produced by the R3 mutant are less stable than those from FapC WT. FapC WT and R3 were fibrillated under the same conditions as ThT assays, harvested, and incubated with increasing concentrations of FA. The treated samples were then lyophilized and resuspended for analysis by SDS-PAGE; panel a shows an example gel for FapC WT. (b) Bands corresponding to FapC monomers were more pronounced for R3 (red) than WT (black), indicating that R3 mutant fibrils de-fibrillated more easily. Error bars indicate the SD from the mean of two samples.
Small oligomers originate through a conserved disulfide bond
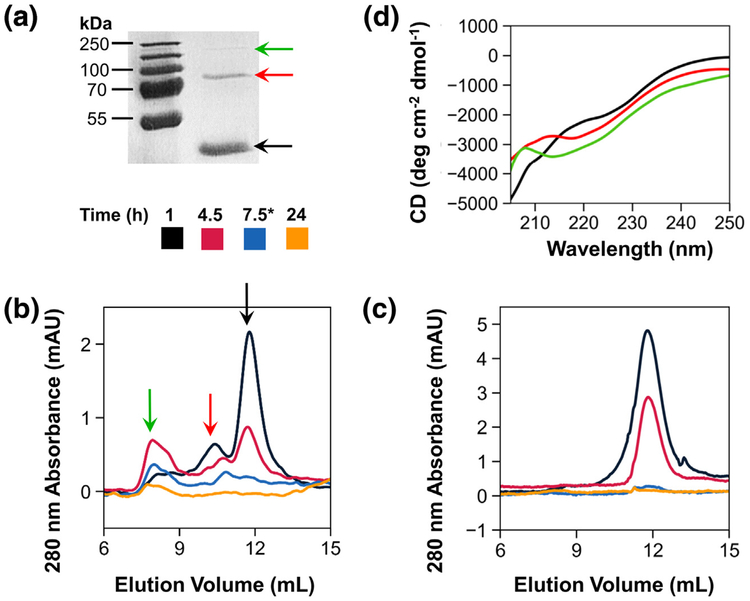
The distinct kinetic behavior observed in the R3 mutant, as well as the other engineered proteins, inspired us to investigate the molecular species formed on the pathway to amyloid formation. First, in FapC, WT samples incubated quiescently for 1 h at room temperature, we noticed the presence of dimers and tetramers on SDS-PAGE gels (Fig. 6a). We hypothesized that two conserved cysteines near the end of the FapC sequence, which comprise a CXXC motif, may form an intermolecular disulfide bridge to initiate dimer formation and subsequent assembly of tetramers and higher-order species. Dimers were proven to arise via the disulfide bond by a simple chemical reduction test, in which reduced (β-mercaptoethanol) FapC WT samples ran as monomers on SDS-PAGE gels, while non-reduced samples retained the dimer and tetramer species. Next, we tracked changes in the populations of soluble species throughout the fibril formation process. FapC WT was incubated in a 96-well plate under the same conditions as Fig. 2c, and samples were periodically withdrawn for analysis by size exclusion chromatography (SEC). Solutions were spun down to remove insoluble material prior to injection, and the resulting chromatograms revealed the presence of distinct dimer and tetramer species in addition to the main monomer peak, particularly at early time points (Fig. 6b). Interestingly, the purified dimer species aggregated on its own, but with slower kinetics than the monomer (SI Fig. 6A). This suggests that some or all of the initial FapC monomer passes through small oligomeric states on the pathway to amyloid formation. However, these results could not establish whether the dimeric and tetrameric states serve as compulsory precursors to FapC amyloid assembly.
Fig. 6.
Small oligomers originate through a conserved disulfide bond. (a) After 1-h incubation at room temperature, FapC WT began to convert from monomeric (black arrow) to dimeric (red arrow) and tetrameric (green arrow) species. To assess the influence of disulfide bonding on small oligomer species, FapC WT (b) and the ΔCys (C304S/C307G) mutant (c) were incubated with shaking at 37 °C, and samples were periodically withdrawn for analysis by SEC. Peaks corresponding to monomeric, dimeric, and tetrameric species elute at approximately 12, 10.5, and 8 mL, respectively, and arrows indicate the same species as in panel A. *The third sample was withdrawn at 7.5 h for WT, but at 12 h for ΔCys. (d) SEC elution fractions were pooled, concentrated, and adjusted to 0.1 mg/mL prior to measurement by CD. Monomers (black), dimers (red), and tetramers (green) all displayed spectra dominated by random-coil signal.
To assess whether dimer formation through the disulfide bond is required for polymerization, we mutated FapC to remove its two cysteine residues (ΔCys, with mutations C304S and C307G). The mutant protein was expressed in E. coli, purified, and desalted in the same manner as FapC WT. Non-reducing SDS-PAGE confirmed that ΔCys lacks the ability to form dimers (SI Fig. 6B). However, ThT assays (SI Fig. 6C) and CD spectra of end point samples (SI Fig. 6D) failed to reveal any substantial differences in the rate, extent, or structure of amyloid formed by ΔCys compared to WT. Furthermore, SEC chromatograms of matched ΔCys samples exhibited only a single peak for the monomeric species, which depleted over time (Fig. 6c); no dimers, tetramers, or other higher-order species were observed. Taken together, these results imply that disulfide bonding is not a prerequisite for amyloid formation in FapC. In addition, FapC WT precursors isolated by fraction collection were dominated by random-coil CD signal in all three species (monomer, dimer, tetramer; Fig. 6d), indicating that most conversion to β-sheet structure occurs in larger oligomers formed later in the aggregation process.
Discussion
Through detailed sequence examination, we identified specific motifs that are implicated in amyloid formation of the P. aeruginosa FapC protein. Our analysis revealed sequence regions with high levels of both evolutionary conservation and amyloid propensity, which facilitated biophysical study of these regions with protein engineering. Mutation of highly conserved segments to corresponding segments with much lower aggregation propensity confirmed the role of evolution in maintaining FapC amyloidogenicity. Modification of just 6-amino-acid residues substantially altered the aggregation kinetics of mutant proteins compared to their wild-type counterparts, and the observed effects were site-specific. The third sequence repeat was shown to be especially sensitive to mutation, underscoring the importance of this conserved region in promoting amyloid assembly. Sequence analysis also identified a highly conserved disulfide motif that facilitates the development of soluble oligomers, but these species are not required for fibril formation.
The repeat mutants developed in this study draw important connections between computational predictions and experimental outcomes for the PAO1 FapC protein. FapC is large compared to most amyloids, and its dynamic nature has eluded structural characterization by traditional means such as NMR or X-ray crystallography. Therefore, we focused only on physicochemical characteristics and known relationships between the PAO1 sequence and its evolutionary relatives. These data were surprisingly rich and, coupled with prediction tools, allowed us to engineer the FapC protein in a way that revealed much about its mechanisms of fibril formation. All three of the site-specific mutants in this study (GVNXAA→KFDDTK) fibrillated with uniquely delayed kinetics, demonstrating that the GVNXAA motif fulfills its predicted role as a highly conserved “driver” of amyloid formation in FapC. Furthermore, the extent of delay varied among the three mutants, with R3 being more profoundly affected than R2 and R1. Analysis in ZipperDB revealed that these discrepancies are likely caused by differences in the energetic impact of each mutation. The wild-type R3 segment has the lowest Rosetta energy (and, therefore, highest amyloid propensity) of the three repeats, so introduction of the KFDDTK mutation in this region causes the greatest reduction in overall amyloid propensity (SI Table 3). Substitution of all three GVNXAA motifs in the TM mutant also delayed the rate and onset of amyloid formation, but the protein was still able to form fibrils. This reflects the complexities of FapC polymerization, while important, the presence of GVNXAA motifs is not the only property required to induce amyloid formation. Our predictions identified several other sequence segments with high amyloid propensity, and it appears that these regions are able to compensate for the loss of GVNXAA in the TM. All of the mutants ultimately formed amyloid fibrils, so the mutations reported here successfully modulated the aggregation kinetics without generating off-pathway aggregates. Thus, mutants derived through this approach could serve as novel tools to study fibril formation in a variety of amyloid proteins, particularly those with little or no structural information. Moreover, mutations to reduce the amyloid propensity of FapC’s third repeat increased its susceptibility to exogenous inhibitors. In theory, compounds designed to target R3, or the GVNIAA hexapeptide site specifically (e.g., peptides or peptide mimetics), could serve as effective inhibitors of fibril formation and biofilm establishment in vivo.
We also demonstrated that a second conserved sequence region, a CXXC motif near the C-terminus of FapC, forges a disulfide linkage between two monomers and promotes the formation of early, on-pathway oligomeric species. Despite CXXC conservation among pseudomonads, disulfide formation is not a requirement for amyloid formation in FapC, and prefibrillar oligomers larger than dimeric and tetrameric species are likely involved. It is also possible that the disulfide bond is retained throughout amyloid assembly to serve as an additional molecular reinforcement in mature fibrils. Faint dimer bands were also visible in urea-purified FapC WT stocks (SI Fig. 6B). Thus, the ability to dimerize even under harsh or denaturing conditions indicates that FapC readily forms disulfide contacts. It is also worth noting that faint dimers were observed on SDS-PAGE gels with FA-treated FapC. This suggests that dimers are either retained in the fibrils or re-form from monomers sometime during the treatment with FA. Indeed, in dissolution experiments with FA, fibrils with disulfide bonds intact (FapC WT) were moderately more stable than those without disulfide bonding capability (ΔCys) (SI Fig. 6E). The formation of these bonds may be even more critical in vivo to prevent proteolysis, impart stability, or drive fibril formation [41, 42]. In our evolutionary sequence analyses, only P. putida strains consistently lacked the CXXC motif and, intriguingly, previous attempts to isolate amyloid fibrils from P. putida failed due to fibril instability and inefficient separation from contaminants [16].
The range of pH experienced by bacteria in vivo depends not only on the growth medium but also on a variety of other factors, such as access to fermentable sugars, oxygen availability, and the size of the biofilm. Amyloid fibrils, including those from FapC, are highly resistant to acidic conditions [43], so increased fibril formation at low pH values could help provide a denser mesh of protective protein to shield the biofilm from harsh conditions. The EM of Pseudomonas is highly complex, however, so interactions between FapC and other matrix components—particularly charged macromolecules such as the exopolysaccharides Pel, Psl, and alginate, as well as extracellular DNA [44]—must be considered to fully elucidate the effect of pH on aggregation in vivo. Here, our study of FapC behavior under a variety of pH values was designed to mimic variable biofilm conditions, and we demonstrated that the FapC R3 mutant fibrillates to a greater extent at low pH values (≤ 7.5). Charge state analysis of the R3 repeat (residues 267–300) reveals that mutation of GVNIAA to KFDDTK increases charge sensitivity to pH above and below the isoelectric point (SI Fig. 7). For example, at pH 9.5, the wild-type R3 segment has a predicted net charge of −1, while the mutated R3 segment has a predicted net charge of −1.5. This is in accord with the insertion of charged residues. However, charge state values assume that all residues have pKa values equivalent to those of the isolated residues, so it is possible that the influence of pH and charge becomes more pronounced as FapC associates with itself and neighboring monomers during the course of fibril formation.
Recent advances in prediction tools have enabled accurate prediction of intrinsic aggregation propensity based on specific mutations in mammalian amyloid proteins [45, 46], but here we have extended this approach to the relatively unexplored realm of bacterial amyloids. Taken together, our findings identify segments in the P. aeruginosa FapC sequence that are implicated in amyloid fibril formation. They serve to shed light on the mechanisms of aggregation in these complex proteins, and more importantly, they provide new targets that can be exploited for therapeutics. These insights are highly valuable in the development of antibiotic alternatives for combating biofilm and ultimately reducing the burden of hospital-acquired infections.
Materials and Methods
Sequence analysis
The FASTA sequence for PAO1 FapC (without the 24-residue signal peptide) was imported from the UniProtKB database. The sequence was then analyzed for amyloid “hot spots” in ZipperDB (http://services.mbi.ucla.edu/zipperdb/) with threshold − 23 kcal/mol and FISH Amyloid (http://www.comprec.pwr.wroc.pl/fish/) with threshold 0.19. Predicted hot spots were compared to conserved regions among 31 FapC homologs using sequence alignment in Clustal Omega [25].
Peptide microarrays
Printed peptide libraries were designed to assess the self-recognition of UK4 FapC (SI Methods). Each array contained 384 peptide spots, each 14 residues in length. One hundred seven of these spots were employed to display residues 25–250 in the UK4 FapC sequence (NCBI entry EEP64551.1, excluding the 24-residue signal peptide), advancing stepwise two residues through the FapC sequence with each spot and thus overlapping the previous spot by 12 residues. Recombinant UK4 FapC was expressed in E. coli, purified with Ni-NTA beads, and labeled with AlexaFluor® 546 (A546; Thermo-Fisher, Waltham, MA) according to the manufacturer’s protocols. Prior to protein application, peptide microarrays were blocked overnight at 4 °C with PBS-T containing 3% w/v whey protein. A546-labeled protein was then added to the array at a concentration of 0.05 mg/mL and incubated for 6 h at room temperature on a roller. After incubation, each array was washed with TBS-T for 15 min. The washed arrays were left to dry for approximately 5 min and then the fluorescence of each spot was measured using a Typhoon Trio scanner (GE Life Sciences, Pittsburgh, PA). Spots were quantified using the ImageJ Protein Array Analyzer [47].
Expression and purification of recombinant FapC
A synthetic gene corresponding to the wild-type FapC protein from P. aeruginosa PAO1 was designed and synthesized by GenScript (Piscataway, NJ). The gene was then cloned into the pET30a(+) vector, which added a C-terminal 6× His tag for purification. All FapC mutants were constructed by GenScript using this plasmid as template, with the exception of FapC TM, which was generated in-house using restriction enzyme cloning (synthetic TM gene from Genscript) and FapC ΔCys which was constructed in-house using site-directed mutagenesis (primers in SI Table 2). Mutation in FapC ΔCys (C304S and C307G) and insertion of the synthetic TM gene was verified by Sanger sequencing (Genewiz, South Plainfield, NJ). Plasmids were transformed into E. coli BL21 (DE3) cells, and protein expression was carried out in 2-L baffled shake flasks at 37 °C and 165 rpm (LB medium supplemented with 4 mM MgSO4, 1% v/v glycerol, and 50 μg/mL kanamycin). Cultures grew to an OD600nm of 0.6–0.8 prior to induction with 1 mM IPTG. After 3–4 h of additional growth, cells were harvested by centrifugation at 4000g for 20 min. Cell pellets were re-suspended in 30 mL denaturing buffer (8 M urea, 20 mM sodium phosphate, 500 mM NaCl, pH 7.8) and lysed overnight with stirring at 4 °C. Insoluble material was removed by centrifugation at 14,000g for 30 min and the supernatant was incubated with Ni-NTA beads for ≥4 h at 4 °C. The beads were then washed twice with denaturing buffer, once with denaturing buffer plus 15 mM imidazole, and twice with denaturing buffer plus 30 mM imidazole. Finally, protein was eluted with denaturing buffer plus 300 mM imidazole. Protein purity was assessed by SDS-PAGE (15% acrylamide-bis, Mes–Tris running buffer), and eluents were stored at −20 °C to prevent urea-induced carbamylation. Immediately prior to use, protein eluents were thawed and desalted according to the PD-10 desalting column protocol (GE Life Sciences) into 10 mM Tris (pH 7.5) and kept on ice. Protein concentration was determined by absorption at 280 nm (extinction coefficients calculated with ExPAsy ProtParam [48]). Desalted proteins were of high concentration and purity, so they were not purified further prior to aggregation assays.
Amyloid formation assays
The buffer in all aggregation assays was 10 mM Tris (pH 7.5) plus 20 μM ThT. As necessary, pH adjustments were made using 6 M HCl or 6 M NaOH, and all buffers were filtered through a 0.2-μm membrane before use. In the case of inhibition experiments, EGCG (Cayman Chemical, Ann Arbor, MI) was freshly prepared as a 20-mg/mL stock and added to the buffer to a final concentration of 3, 9, 15, 30, or 90 μM. To start the assay, desalted protein stocks were added to black-walled 96-well plates and diluted with appropriate buffer such that the final concentration of protein was 0.2 mg/mL (except for EGCG inhibition assays, where it was 0.1 mg/mL) and the final volume was 100 μL per well. A sterile 3-mm borosilicate glass bead was added to each well to facilitate mixing and the plate was covered with an adhesive seal. ThT fluorescence was monitored in a Tecan (Männedorf, Switzerland) plate reader at 37 °C with excitation at 448 nm and emission at 485 nm. For each measurement, 60 s of orbital shaking at 180 rpm was applied before a 5-s settling time, and then 10 single reads were averaged for each well. In the case of EGCG and ΔCys experiments (Fig. 4a, b; SI Fig. 6C), settling time was extended to 5 min. All experiments consisted of at least three replicates per condition, and plots show data points from every 0.5–0.7 h for clarity. Samples for CD, FTIR, and SEC were treated identically and added to the same plate to ensure matched conditions. Values for the lag time, tlag, and rate of maximum fibril formation were calculated according to the methods of Morris et al. [49].
Spectroscopy
Samples for CD were diluted fresh from desalted protein stocks (0 h samples) or removed from micro-titer plates (end point, fibrillated samples). SEC elution fractions were pooled, concentrated with Amicon® Ultra centrifugal filter units (Sigma-Aldrich), and adjusted to 0.1 mg/mL prior to measurement. CD spectra were recorded on a J-810 spectropolarimeter (Jasco Spectroscopic Co. Ltd., Tokyo, Japan) using a 1-mm quartz cuvette and the following settings: wavelength of 260–200 nm, 0.5-nm data pitch, 1-s integration time, 2-nm bandwidth, 25 °C, and 4–6 accumulations. Only data points with detector voltages below 600 V were used. All spectra were corrected by a blank spectrum of relevant buffer and smoothed with a Savitsky–Golay filter at window size 25–65 and polynomial order 2. FTIR spectroscopy was performed on a Tensor 27 FTIR spectrophotometer (Bruker, Billerica, MA) with a deuterated tri-glycine sulfate mid-infrared detector and a Golden Gate single reflection diamond attenuated total reflectance cell (Specac, Kent, UK). Fibrillated samples were dried on the attenuated total reflectance crystal with a stream of nitrogen and spectra were collected from 4000 to 600 cm−1. Spectra were baseline corrected and atmospheric compensation was performed with OPUS 5.5 software (Bruker). Signals in the amide I region (~1550–1700 cm−1) were normalized and identified by second derivative analysis in Python [50]. For FTIR analysis of fibrils from P. aeruginosa, fibrils were collected from a fap overexpressing strain as previously described [16] (SI Table 2).
DLS and NTA
EGCG stock was prepared by dissolving lyophilized stock powder in PBS to a concentration of 20 mg/mL. This stock was then diluted in 10 mM Tris (pH 7.5) to a concentration of 1, 3, 9, 30, or 90 μM and immediately used for analysis. DLS measurements were performed in triplicate on a Zetasizer Nano Series instrument (Malvern, Salisbury, UK) using 1 mL for each sample. NTA measurements were performed on a NanoSight NS300 (Malvern) at 23 °C, camera level 11, and a detection threshold of 3.
SEC
Samples for SEC were FapC WT and ΔCys, fibrillated under identical conditions and in the same plate as the corresponding ThT assays. At each time point, three well volumes (300 μL total) per condition were removed from the plate and transferred to a 1.5-mL Eppendorf tube. Tubes were flash-frozen for storage. Immediately prior to SEC analysis, each sample was thawed on ice and then centrifuged at >12,000 rpm for at least 10 min to remove insoluble material. Supernatant (250 μL) was loaded onto a Superdex 200 Increase 10/300 GL column (GE Life Sciences) equilibrated in 10 mM Tris (pH 7.5). Fractions of 0.75 mL were collected at a flow rate of 0.75 mL/min, and chromatography traces were collected with a UV cell at 280 nm (ÄKTA Pure system; GE Life Sciences).
FA treatment
FapC WT, R3, and ΔCys were incubated at 0.2 mg/mL in Tris buffer, under the same conditions as the ThT aggregation assays but with 200 μL of solution per well. After 48 h, fibril solutions were removed from wells and transferred to Eppendorf tubes. Fibrils were isolated by centrifugation at 20,000g for 10 min and the concentration of FapC remaining in the supernatant was measured by NanoDrop (Thermo-Fisher). Fibrils were then washed twice in filtered water, resuspended to a concentration of 1 mg/mL, and divided among a series of new Eppendorf tubes. The tubes were centrifuged again and each fibril pellet was resuspended in different concentrations of FA in water (range = 25%–100% FA) such that the final concentration of protein was 120.5 mg/mL. Samples were incubated for 10 min, frozen, and lyophilized overnight. For SDS-PAGE analysis, each dried protein film was resuspended in water and mixed with non-reducing sample buffer. Samples were heated for 5 min at 95° and then loaded into lanes of 15% acrylamide–bis gels. Gels ran at 100 V for 90 min in Mops–Tris running buffer, stained for 30 min in Coomassie Brilliant Blue R-250, and destained overnight prior to imaging on an Azure c300 imaging system (Azure Biosystems, Dublin, CA). Relative band intensities were determined using ImageJ software [51]. Two gels were run for each condition to ensure reproducibility.
Accession numbers
UniProtKB accession nos. Q9I2F0 (PAO1 FapC) and C4IN70 (UK4 FapC).
Supplementary Material
Acknowledgments
We extend thanks to Dr. Morten Simonsen Dueholm (Aalborg University) and Line F. B. Christiansen (Aarhus University) for helpful discussions, and to Ivan Vulovic and Shijie Cao (University of Washington) for assistance with equipment. This research was supported by the Danish Council for Independent Research (6111–00241B to D.E.O. and 7093–00001B to D.E.O. and A.B.) and the National Science Foundation (GROW Fellowship, ID 2015176941 to A.B.).
Abbreviations used:
- CD
dichroism spectroscopy
- DLS
dynamic light scattering
- EGCG
epigallotcatechin-3-gallate
- EM
extracellular matrix
- FA
formic acid
- fap
functional amyloid in Pseudomonas
- FTIR
Fourier-transform infrared spectroscopy
- NTA
nanoparticle tracking analysis
- SEC
size exclusion chromatography
- TEM
transmission electron microscopy
- ThT
thioflavin T
- TM
triple mutant
- VAP
ventilator associated pneumonia
- WT
wildtype
Appendix A. Supplementary Data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2018.06.043.
References
- [1].Gellatly SL, Hancock REW, Pseudomonas aeruginosa: new insights into pathogenesis and host defenses, Pathog. Dis 67 (2013) 159–173. [DOI] [PubMed] [Google Scholar]
- [2].Weiner LM, et al. , Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014, Infect. Control Hosp. Epidemiol 37 (2016) 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bjarnsholt T, et al. , The in vivo biofilm, Trends Microbiol 21 (2013) 466–474. [DOI] [PubMed] [Google Scholar]
- [4].Williams BJ, Dehnbostel J, Blackwell TS, Pseudomonas aeruginosa: host defence in lung diseases, Respirology 15 (2010) 1037–1056. [DOI] [PubMed] [Google Scholar]
- [5].Boisvert A-A, Cheng MP, Sheppard DC, Nguyen D, Microbial biofilms in pulmonary and critical care diseases, Ann. Am. Thorac. Soc 13 (2016) 1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bjarnsholt T, et al. , Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients, Pediatr. Pulmonol 44 (2009) 547–558. [DOI] [PubMed] [Google Scholar]
- [7].Flemming H-C, Wingender J, The biofilm matrix, Nat. Rev. Microbiol 8 (2010) 623–633. [DOI] [PubMed] [Google Scholar]
- [8].Taglialegna A, Lasa I, Valle J, Amyloid structures as biofilm matrix scaffolds, J. Bacteriol 198 (2016) 2579–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sipe JD, et al. , Nomenclature 2014: amyloid fibril proteins and clinical classification of the amyloidosis, Amyloid 21 (2014) 221–224. [DOI] [PubMed] [Google Scholar]
- [10].Bleem A, Daggett V, Structural and functional diversity among amyloid proteins: agents of disease, building blocks of biology, and implications for molecular engineering, Biotechnol. Bioeng 114 (2017) 7–20. [DOI] [PubMed] [Google Scholar]
- [11].Depas WH, Chapman MR, Microbial manipulation of the amyloid fold, Res. Microbiol 163 (2012) 592–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Seviour T, et al. , Functional amyloids keep quorum-sensing molecules in check, J. Biol. Chem 290 (2015) 6457–6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Stenvang M, et al. , Epigallocatechin gallate remodels over-expressed functional amyloids in Pseudomonas aeruginosa and increases biofilm susceptibility to antibiotic treatment, J. Biol. Chem 291 (2016) 26540–26553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chapman MR, et al. , Role of Escherichia coli curli operons in directing amyloid fiber formation, Science 295 (2002) 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dueholm MS, et al. , Functional amyloid in Pseudomonas, Mol. Microbiol 77 (2010) 1009–1020. [DOI] [PubMed] [Google Scholar]
- [16].Dueholm MS, et al. , Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation, Microbiol. Open 2 (2013) 365–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dueholm MS, Otzen D, Nielsen PH, Evolutionary insight into the functional amyloids of the pseudomonads, PLoS One 8 (2013), e76630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Barnhart MM, Chapman MR, Curli biogenesis and function, Annu. Rev. Microbiol 60 (2006) 131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rouse SL, et al. , A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis, Nat. Commun 8 (2017) 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Herbst F-A, et al. , Major proteomic changes associated with amyloid-induced biofilm formation in Pseudomonas aeruginosa PAO1, J. Proteome Res 14 (2015) 72–81. [DOI] [PubMed] [Google Scholar]
- [21].Trainor K, Broom A, Meiering EM, Exploring the relationships between protein sequence, structure and solubility, Curr. Opin. Struct. Biol 42 (2017) 136–146. [DOI] [PubMed] [Google Scholar]
- [22].Thompson MJ, et al. , The 3D profile method for identifying fibril-forming segments of proteins, Proc. Natl. Acad. Sci 103 (2006) 4074–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Goldschmidt L, Teng PK, Riek R, Eisenberg D, Identifying the amylome, proteins capable of forming amyloid-like fibrils, Proc. Natl. Acad. Sci 107 (2010) 3487–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gasior P, Kotulska M, FISH Amyloid—a new method for finding amyloidogenic segments in proteins based on site specific co-occurence of aminoacids, BMC Bioinf 15 (2014) 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sievers F, et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega, Mol. Syst. Biol 7 (2011) 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hilpert K, Winkler DFH, Hancock REW, Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion, Nat. Protoc 2 (2007) 1333–1349. [DOI] [PubMed] [Google Scholar]
- [27].Conchillo-Solé O, et al. , AGGRESCAN: a server for the prediction and evaluation of ‘hot spots’ of aggregation in polypeptides, BMC Bioinf. 8 (2007) 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gade Malmos K, et al. , ThT 101: a primer on the use of thioflavin T to investigate amyloid formation, Amyloid 24 (2017) 1–16. [DOI] [PubMed] [Google Scholar]
- [29].Sarroukh R, Goormaghtigh E, Ruysschaert J-M, Raussens V, ATR-FTIR: a ‘rejuvenated’ tool to investigate amyloid proteins, Biochim. Biophys. Acta Biomembr. 1828 (2013) 2328–2338. [DOI] [PubMed] [Google Scholar]
- [30].Klein S, et al. , Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol, Mol. Microbiol 71 (2009) 551–565. [DOI] [PubMed] [Google Scholar]
- [31].Jones EM, Cochrane CA, Percival SL, The effect of pH on the extracellular matrix and biofilms, Adv. Wound Care 4 (2015) 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pezzulo AA, et al. , Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung, Nature 487 (2012) 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Foderá V, et al. , Thioflavin T hydroxylation at basic pH and its effect on amyloid fibril detection, J. Phys. Chem. B 112 (2008) 15174–15181. [DOI] [PubMed] [Google Scholar]
- [34].Steinmann J, Buer J, Pietschmann T, Steinmann E, Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea: anti-infective effects of EGCG, Br. J. Pharmacol 168 (2013) 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang L, Liu Y, Sternberg C, Molin S, Evaluation of enoyl-acyl carrier protein reductase inhibitors as Pseudomonas aeruginosa quorum-quenching reagents, Molecules 15 (2010) 780–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hudson SA, Ecroyd H, Kee TW, Carver JA, The thioflavin T fluorescence assay for amyloid fibril detection can be biased by the presence of exogenous compounds: exogenous compounds can bias thioflavin T assays, FEBS J 276 (2009) 5960–5972. [DOI] [PubMed] [Google Scholar]
- [37].Palhano FL, Lee J, Grimster NP, Kelly JW, Toward the molecular mechanism(s) by which EGCG treatment remodels mature amyloid fibrils, J. Am. Chem. Soc 135 (2013) 7503–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lendel C, et al. , On the mechanism of nonspecific inhibitors of protein aggregation: dissecting the interactions of α-synuclein with Congo red and lacmoid, Biochemistry 48 (2009) 8322–8334. [DOI] [PubMed] [Google Scholar]
- [39].Feng BY, et al. , Small-molecule aggregates inhibit amyloid polymerization, Nat. Chem. Biol 4 (2008) 197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kaplan B, Shtrasburg S, Pras M, Micropurification techniques in the analysis of amyloid proteins, J. Clin. Pathol 56 (2003) 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Heras B, et al. , DSB proteins and bacterial pathogenicity, Nat. Rev. Microbiol 7 (2009) 215–225. [DOI] [PubMed] [Google Scholar]
- [42].Arts IS, et al. , Dissecting the machinery that introduces disulfide bonds in Pseudomonas aeruginosa, MBio 4 (2013) e00912–e00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Otzen D, Functional amyloid: turning swords into plowshares, Prion 4 (2010) 256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jennings LK, et al. , Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix, Proc. Natl. Acad. Sci 112 (2015) 11353–11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Paul KR, et al. , Effects of mutations on the aggregation propensity of the human prion-like protein hnRNPA2B1, Mol. Cell. Biol 37 (2017), e00652–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hee JS, Mitchell SM, Liu X, Leonhardt RM, Melanosomal formation of PMEL core amyloid is driven by aromatic residues, Sci. Rep 7 (2017), 44064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Carpentier G, Protein Array analyzer for ImageJ, ImageJ User and Developer Conference, 2010. [Google Scholar]
- [48].Gasteiger E, et al. , Protein identification and analysis tools on the ExPASy server, The Proteomics Protocols Handbook, Humana Press; Inc. 2005, pp. 571–607. [Google Scholar]
- [49].Morris RJ, et al. , Mechanistic and environmental control of the prevalence and lifetime of amyloid oligomers, Nat. Commun 4 (1891) (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jones E, Oliphant E, Peterson P, SciPy: Open Source Scientific Tools for Python, 2001.
- [51].Schneider CA, Rasband WS, Eliceiri KW, NIH image to ImageJ: 25 years of image analysis, Nat. Methods 9 (2012) 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.