Abstract
Roots remain an underexplored frontier in plant genetics despite their well-known influence on plant development, agricultural performance and competition in the wild. Visualizing and measuring root structures and their growth is vastly more difficult than characterizing aboveground parts of the plant and is often simply avoided. The majority of research on maize root systems has focused on their anatomy, physiology, development and soil interaction, but much less is known about the genetics that control quantitative traits. In maize, seven root development genes have been cloned using mutagenesis, but no genes underlying the many root-related quantitative trait loci (QTLs) have been identified. In this review, we discuss whether the maize mutants known to control root development may also influence quantitative aspects of root architecture, including the extent to which they overlap with the most recent maize root trait QTLs. We highlight specific challenges and anticipate the impacts that emerging technologies, especially computational approaches, may have toward the identification of genes controlling root quantitative traits.
Keywords: Genotype to phenotype, Imaging and analysis, Quantitative genetics, Root architecture, Root mutant, Zea mays
Introduction
Plant physiologists have spent many years thoughtfully characterizing roots in fine detail, and creating an intricate vocabulary to describe their observations. Common terminologies have been proposed at various times by the International Society of Root Research (ISRR), the Plant Ontology Consortium and maize researchers (Feix et�al. 2002, Ilic et�al. 2007, Hochholdinger 2009, Zobel and Waisel 2010). Root traits have generally been grouped based on developmental criteria (tissue of origin), anatomy (internal structure at the tissue or cellular level) and topology (branching order or connectedness, i.e. primary, secondary, tertiary, etc.). These groupings have been and continue to be broadly used; however, as technologies advance, new ways to characterize roots arise.
Root system architecture (RSA) has been defined as the growth pattern of roots within, and in response to, their environment, i.e. where, when and what type of roots grow (Fitter 1987). RSA integrates aspects of development and topology, and more recently has been connected to anatomical features (Burton et�al. 2013, Lynch et�al. 2014). Root architecture is measured as a function of increasing root system complexity through plant development, which has been facilitated by technologies that quantify many traits with high throughput. This increased scope has allowed plant geneticists to begin identifying the genetic basis of root traits across a wide spectrum of descriptors.
Zea mays (L.) is one of the most highly produced grain crops in the world and has been the focus of extensive breeding and genetic research over the years. However, maize roots have received far less attention compared with aboveground structures because they are hidden from view and grow in complex patterns that are heavily influenced by the soil environment. Indirect selection for root architecture in response to denser planting is posited as a major factor for increased yield over the past half century in the USA (Hammer et al. 2009), but no genes have been identified or attributed to this phenomenon.
As currently understood, developmentally defined maize root types are controlled by single genes that regulate gross phenotypic differences, often presence or absence (Hochholdinger et�al. 2018). Root architecture and anatomical traits appear instead to be quantitative and controlled by many genes (Price and Tomos 1997, Mano et�al. 2007, Topp et�al. 2013, Burton et�al. 2015, Zurek et�al. 2015). These traits also have a well-documented capacity for phenotypic plasticity (MacMillan et�al. 2006, Rosas et�al. 2013, Meij�n et�al. 2014) that can contribute to crop yield stability across multiple environments (Sandhu et�al. 2016). Quantitative variation for root architecture has been proposed to facilitate trait optimization in different environments by concerted ‘fine-tuning’ of many loci (Gifford et�al. 2013, Rosas et�al. 2013). However, this idea has not been well tested in crop plants, largely because few genes have been identified that control root architecture. Thus, fully realizing the value of root architecture for crop improvement (Lynch 1995, 2013, de Dorlodot et�al. 2007, Hochholdinger and Tuberosa 2009) will require a more thorough understanding of the specific genetic loci involved in quantitative root variation.
Only a small number of genes that control strong developmental phenotypes have been cloned in maize (Hochholdinger et�al. 2018). Additionally, many root quantitative trait locus (QTL) mapping studies and meta-analyses spanning decades of work have been conducted (Hund et�al. 2011), but no causal genes have been reported yet. Amid these data, important questions remain. (i) To what extent could the known major genetic regulators also control quantitative variation, and do they co-align with maize root QTLs? (ii) What are the key experimental factors limiting our current ability to identify the genetic basis of quantitative root traits? (iii) What tools can be leveraged to help survey the diversity of root traits in maize going forward?
Is There a Distinction Between Qualitative and Quantitative Regulators of Root Architecture?
Maize root system development can first be divided into an embryonic root system derived from the seed and the post-embryonic root system derived from the shoot, which eventually dominates the root architecture. The embryonic root system is made up of a primary root and a variable number of seminal roots depending on genotype and environment (Kiesselbach 1949, Sass 1977, Feldman 1994). The post-embryonic root system is comprised of lateral roots and whorls of nodal roots, which are called crown roots if below the soil and brace or prop roots if above the soil. Nodal roots emerge at the base of each node, and so comprise a temporal and developmental axis that increases shootward. Lateral roots initiate from the pericycle of both embryonic and post-embryonic roots, and comprise the first and all subsequent topological orders of branching (Bodner et�al. 2013). Root hairs are single-celled protrusions that initiate from the epidermis of all root types and have been considered both anatomical and developmental structures. The characterization of mutant phenotypes in this way has led to the identification of 12 maize developmental root mutants, which are summarized in Table�1.
Table 1.
Summary of known maize root development mutants and cloned genes
| Root type | Mutagen | Genetic background | Introgressed | Mutant phenotype | Gene name | GeneIDa | Bin | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| Root hair | Mu | Robertson mutator stocks | Q60, Aet/LineC, BMS, NY821, B37 | Root hairs do not elongate | rth1 | roothairless 1 | GRMZM2G099056 | 1.09 | Wen et al. (2005) |
| NA | Root hairs 1/4 length of wild type | rth2 | roothairless 2 | NA | 5.04 | Wen and Schnable (1994) | |||
| NA | Root hairs do not elongate; ‘stocking cap’ phenotype | rth3 | roothairless 3 | GRMZM2G377215 | 1.01 | Hochholdinger et al. (2008) | |||
| EMS | Pioneer inbred | B73, Mo17 | Root hair initiation and elongation | rth5 | roothairless 5 | GRMZM2G426953 | 3.06 | Nestler et al. (2014) | |
| Mu, AcDs | Pioneer inbred, W22 | B73 | Root hairs are arrested after bulge formation | rth6 | roothairless 6 | GRMZM2G436299 | 1.05 | Li et al. (2016) | |
| Embryonic lateral | EMS | B73 | A632 | Lateral root primordia not present on embryonic roots | lrt1 | lateral rootless 1 | NA | 2S | Hochholdinger and Feix (1998) |
| Lateral | Mu | SWS2000 | A632, B73 | Lateral roots 1/4 length of wild type | slr1 | short lateral roots 1 | NA | NA | Hochholdinger et al. (2001) |
| NA | NA | Lateral roots 1/4 length of wild type | slr2 | short lateral roots 2 | NA | NA | |||
| Lateral and seminal | Mu | B73 | F2, F7, Aet, HiIIA | Seminal and lateral root primordia absent on embryonic roots; delayed gravitropic response | rum1 | rootless with undetectable meristem 1 | GRMZM2G037368 | 3.08 | von Behrens et al. (2011), Woll et al. (2005) |
| Seminal and nodal | Enhancer, Mu, AcDs | DK105 | A632, B73, W22, W23 | No nodal or seminal roots | rtcs | rootless concerning seminal and crown roots | GRMZM2G092542 | 1.01 | Taramino et al. (2007) |
| Nodal | Mu | Pioneer inbred | NA | Nodal root length reduced | rtcl | rtcs-like | AC149818.2_FGT009 | 9.07 | Taramino et al. (2007), Xu et al. (2015) |
| Nodal | Unknown | NA | NA | Lateral roots few or absent, reduced crown root number, no aerial brace roots | rt1 | rootless 1 | NA | 3S | Jenkins (1930) |
| No mutant phenotype | NA | Homolog of rum1b | Rul1 | Rum1-like | GRMZM2G163848 | 8.06 | von Behrens et al. (2011), Zhang et al. (2016) | ||
| NA | Blast of AtLRP1; regulated by rum1b | Lrp1 | Lateral root primordia 1 | GRMZM2G077752 | 8.06 | Zhang et al. (2015) | |||
aGeneID is from B73 v3 reference.
bThese genes were identified by homology to rum1 and AtLRP1, and are predicted to have similar phenotypes but no mutants exist to validate these phenotypes.
Forward genetics approaches have been used to generate mutant alleles of genes involved in maize root anatomy and development. Of the 12 root mutants: two affect seminal root development, three affect shoot-borne root development, four affect lateral root development and five affect root hair development. Only seven of these genes have been cloned (Table�1), and they have been thoroughly reviewed elsewhere (Hochholdinger 2009, Hochholdinger and Tuberosa 2009, Yu et�al. 2016, Hochholdinger et�al. 2018). Mutants have traditionally been screened by eye at the seedling stage (10–14 d after germination) from plants grown in paper towel rolls (Hochholdinger 2009). This screening method allows for rapid scoring of many lines, but is biased toward identifying obvious root phenotypes that affect qualitative aspects of external morphology rather than subtle variation of quantitative phenotypes. Not surprisingly, the genes controlling the most obvious and severe mutants were cloned first. For example, the first cloned mutant was rth1 which has no root hairs visible by eye on the emerging primary root at 3 d after germination (Wen et�al. 2005; Andorf et�al. 2016; images at https://maizegdb.org/data_center/variation?id=61056 (June 29, 2018, date last accessed)). The mutants rth2, slr1 and slr2 have more subtle phenotypes and have not been cloned. Root hairs of the rth2 mutant elongate but are shorter than those of the wild type (Wen and Schnable 1994). Lateral root length of both slr1 and slr2 mutants is reduced compared with the wild type (Hochholdinger et�al. 2001). The mutant lrt1 has a strong effect on the embryonic root system, which has no laterals (Hochholdinger and Feix 1998). However, the nodal root system has apparently normal lateral roots yet is smaller than the wild type, raising the possibility that the lrt1 phenotype may be more quantitative when evaluated at a later growth stage. More robust phenotyping of root mutants, especially throughout development, may aid in gene identification and reveal more quantitative aspects of their phenotype.
Due to the enormous amount of structural, genetic and transcriptional variation known in the maize pan-genome, any mutagenesis study is limited to the variation present in the targeted genotypes (Hirsch et�al. 2014, Jiao et�al. 2017). For example, the ‘B73’ genome only captures an estimated 70% of non-transposon genes present in diverse maize inbreds (Gore et�al. 2009). Additionally, approximately 30% of genes in both ‘B73’ and ‘PH207’ show genotype-specific expression (Hirsch et�al. 2016), suggesting that the genomic environment could also have a significant effect on the phenotypic expression of a mutant allele. While not extensively studied, epistatic modifiers have been observed for half of the mutants when they were crossed or mutated in multiple genetic backgrounds: rum1, rtcs, slr1 and rth2 (Table�1). The epistatic modifiers of rtcs and rum1 are paralogs, known as Rtcl and Rul1, respectively (Taramino et�al. 2007, von Behrens et�al. 2011, Xu et�al. 2015, Zhang et�al. 2016). No mutants have been identified for Rul1 but, interestingly, the rtcl mutant has a less severe phenotype than rtcs, resulting in normal seminal roots and reduced length of crown roots. This observation supports the finding from Schnable and Freeling (2011) that genes from the maize1 subgenome are over-represented in the historic identification of visible mutant phenotypes because of their increased severity. Given that the ancestral duplication of the maize genome led to widespread gene differentiation (Schnable et�al. 2011), it is tantalizing to wonder if the maize2 subgenome paralogs of root developmental genes may generally be more quantitative in nature, and thus more likely to control root system architecture tunability. It will be important to screen diverse maize lines—including inbreds, landrace, and wild teosinte ancestors—if we are to obtain a complete picture of the phenotypic context of a given mutation and any potentially adaptive alleles (Schmidt et�al. 2016, Topp et�al. 2016).
To date, only eight of the 12 known root mutants have been introgressed into at least two genetic backgrounds (Table�1). There has been little further work on quantitative aspects of these phenotypes, with one notable exception: Abdel-Ghani et�al. (2015) directly compared quantitative phenotypic variation across different maize lines to identify associations with functional alleles of root development genes. They queried an association panel of 74 maize inbreds, and reported that allele variants for Rtcl, Rth3, Rum1 and Rul1 contributed to quantitative variation of seedling root traits. However, the lack of genome-wide markers in this experiment made it difficult to understand how these targeted developmental gene associations compare with genome-wide associations with root traits. When Pace et�al. (2015) conducted a genome-wide association study of a different but overlapping set of inbred lines for seedling RSA traits, Rum1 was the only known root gene with significant marker associations nearby. Several other maize root QTLs overlap with or have been found in proximity to known root mutant genes, yet no causal relationship has been identified between genes and the large QTL regions. QTLs for seminal root number, crown root length, brace root whorl number and crown root number overlap with Rul1 (Pestsova et�al. 2016, Salvi et�al. 2016, Gu et�al. 2017, Liu et�al. 2017, Zhang et�al. 2018). Rum1 was under QTLs for seminal root number, lateral root density, lateral root length and crown root number per whorl (Pestsova et�al. 2016, Salvi et�al. 2016, Liu et�al. 2017, Zhang et�al. 2018). Rtcl was under QTLs for lateral root density and brace root number (Pestsova et�al. 2016, Zhang et�al. 2018), and Rtcs was found under a QTL for seminal root number (Salvi et�al. 2016). In total, the evidence suggests that known root mutant genes may plausibly control quantitative variation for maize RSA; however, as most QTL regions contain hundreds of genes, more research is needed to identify bona fide causal relationships.
Current Knowledge and Challenges Towards Gene Identification of Maize Root Architecture QTLs
While the list of QTLs controlling maize root system traits is long, no causative genes underlying any of these QTLs have been reported. In Table�2 we summarize the 19 maize root QTL mapping and genome-wide association studies (GWAS) that have been published since or were not included in the last summary (Hund et�al. 2011). These works were based on 12 biparental populations (19 inbreds and 2 teosintes) and three association populations (396 and 384 inbreds, and 66 landrace F1 doubled haploids).
Table 2.
Summary of 19 maize root trait QTLs and GWAS studies that have been published since or were not included in Hund et al. (2011)
| Trait group | Trait measurement | No. of traits | Population | Pop size | Rep� Repeat | Plants per rep | Time and/ or stage sampled | No. of markers | Map density | Media and treatment | H2 | No. of QTLs | Meta- QTLsa | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nodal root anatomy | Aerenchyma: visual score | 1 | B64�Z. nicaraguensis: F2 | 141 | 1 � 1 | 1 | 28 d V6 | 85 SSRs | 17.2 cM | Granular soil; GH | NA | 6 | N | Mano et al. (2007) |
| Z. nicaraguensis�Mi29: BC2F1 | 214 | 1 � 1 | 1 | 28 d V6 | 94 SSRs | 10.2 cM | Granular soil; GH | NA | 3 | N | Mano and Omori (2008) | |||
| Z. nicaraguensis�Mi29: BC4F1 | 123 | 1 � 1 | 1 | 28 d V6 | 156 SSRs + 38 Indels | NA | Granular soil; GH | NA | 2 | N | Mano and Omori (2009) | |||
| ROL: methylene blue + oxygen electrode | 2 | Z. nicaraguensis�Mi29: BC3F5 | 48 | 4 � 1 | 1 | 25 d | 98 SSRs | NA | Hydroponics | NA | 1 | N | Watanabe et al. (2017) | |
| RootScan: cross-sections | 10 | B73�Mo17 (IBM): RILs | 200 | 3 � 1 | 1;3 | 30 d V6; 56 d V12 | 8,224 GBS-SNPs | 0.7 cM | Potting mix; field | 0.47 to 0.78 | 1 | N | Burton et al. (2015) | |
| Oh43�W64a (OhW): RILs | 200 | 3 � 1 | 1 | 30 d V6 | 5,683 GBS-SNPs | 0.6 cM | Potting mix | 0.47 to 0.78 | 5 | N | ||||
| NY821�H99 (NyH): RILs | 176 | 3 � 1 | 1 | 30 d V6 | 5,320 GBS-SNPs | 0.7 cM | Potting mix | 0.47 to 0.78 | 0 | N | ||||
| Nodal root | Visual count | 2 | Huangzao 4� CML288: IF2 | 278 | 1 x 1 | 10 | VT + 10 d | 237 SSRs | 12.3 cM | Field | NA | 15 | N | Ku et al. (2012) |
| Huangzao 4�CML288: RILs | 201 | 3 � 1 | 10 | VT + 10 d | 237 SSRs | 8.33 cM | Field | NA | 10 | N | ||||
| Visual count + weight | 8 | Yi17�Yi16: F2 | 276 | 1 � 1 | 1 | R6 | 212 SSRs | 7.35 cM | Field | 0.42 to 0.79 | 44 | Y | Gu et al. (2017) | |
| Yi17�Yi16: F2:3 | 241 | 2 � 3 | 8 | R6 | 212 SSRs | 7.35 cM | Field | 0.40 to 0.66 | 49 | Y | ||||
| Visual count | 9 | Z. parviglumis�W22: BC2S3 | 866 | 1 � 4 | 10 | VT per line | 19,838 GBS-SNPs | 39.6 kbp | Field | NA | 133 | N | Zhang et al. (2018) | |
| RSA | WinRhizo + weight | 7 | NUEC2�NUEC4: DH | 60 | 2 � 1 | 15-30 | 12 d | 754 50k-SNPs | NA | Paper roll | NA | 30 | Y | Pestsova et al. (2016) |
| Root count + weight | 9 | Ye478�Wu312: F8 | 218 | 3 � 3 | 2 | 20 d V5/6 | 184 SSRs | 11.3cM | Hydroponics: HN + LN | 0.36 to 0.69 | 134 | Y | Li et al. (2015) | |
| Count + weight + WinRhizo | 21 | B73�Mo17 (IBM): RILs | 200 | 3 � 1 | 1 | 30 d V6/7 | 8,224 GBS-SNPs | 0.7 cM | Potting mix | 0.10 to 0.60 | 7 | N | Burton et al. (2014) | |
| Oh43�W64a (OhW): RILs | 200 | 3 � 1 | 1 | 30 d V6/7 | 5,683 GBS-SNPs | 0.6 cM | Potting mix | 0.10 to 0.60 | 5 | Y | ||||
| NY821�H99 (NyH): RILs | 176 | 3 � 1 | 1 | 30 d V6/7 | 5,320 GBS-SNPs | 0.7 cM | Potting mix | 0.10 to 0.60 | 3 | Y | ||||
| WinRhizo + weight | 6 | Ye478�Wu312: F8 | 218 | 3 � 2 | 2–3 | 50 d V6; 80 d VT; 120 d R6 | 184 SSRs | 11.3 cM | Field | 0.01 to 0.55 | 36 | N | Cai et al. (2012) | |
| Ye478�Wu312: BC4F3 | 187 | 3 � 2 | 2–3 | 50 d V6; 80 d VT; 120 d R6 | 143 SSRs | 11.3 cM | Field | 0.01 to 0.55 | 36 | N | ||||
| WinRhizo + weight + length | 8 | Ye478�Wu312: F8 | 218 | 2;2;1 | 6; 12; 17 | 12 d; 8 d; 10 d | 184 SSRs | 11.3cM | Hydroponics; paper roll; vermiculite | 0.52 to 0.83 | 46 | Y | Liu et al. (2017) | |
| Ye478�Wu312: BC4F3 | 187 | 2;2;1 | 6; 12; 17 | 12 d; 8 d; 10 d | 143 SSRs | 11.3 cM | 0.52 to 0.83 | 46 | Y | |||||
| Weight + volume | 6 | CIMMYT Asia Panel | 396 | 2 � 2 | 1 | VM | 331,390 GBS-SNPs | 6.2 kbp | Field in tubes | 0.82 to 0.98 | 67 | N | Zaidi et al. (2016) | |
| GiaRoots 3D | 19 | B73�Ki3 NAM RILs | 175 | 3 � 1 | 1 | 4 d, 6 d, 8 d | 1,106 SNPs | 1.3 cM | Hoaglands gel | 0.07 to 0.55 | 102 | Y | Zurek et al. (2015) | |
| ARIA + weight | 24 | 384 Ames Panel | 384 | 3 � 1 | 3 | 14 d | 135,311 GBS-SNPs | 15.1 kbp | Paper roll | 0.12 to 0.49 | 268 | Y | Pace et al. (2015) | |
| Count + length + scans | 10 | B73�Gaspe Flint: BC5F2 | 75 | 2;2 | 9;5 | 7 d; 25 d | 173 SSRs | NA | Paper roll; potting mix | 0.69 to 0.95 | 14 | N | Salvi et al. (2016) | |
| ARIA + weight | 24 | 66 Landrace F1 DH | 300 | 3 � 1 | 1 | 14d | 62,077 GBS-SNPs | 100 Mbp | Paper roll | 0.06 to 0.50 | 39 | N | Sanchez et al. (2018) | |
| RPF | Dynamometer | 1 | Ye478�Wu312: F8 | 218 | 3 � 4 | 4 | VT (silk) + 14 d | 184 SSRs | 11.3 cM | Field | 0.61 | 4 | Y | Liu et al. (2011) |
| Ye478�Wu312: BC4F3 | 187 | 3 � 4 | 4 | VT (silk) + 14 d | 143 SSRs | 11.3 cM | Field | 0.44 | 3 | Y |
RSA, root system architecture; RPF, root pulling force; ROL, radial oxygen loss; ARIA, automated root image analysis.
aQTL overlaps a meta-QTL: Y = yes, N = no.
Currently, the only genes reported to control a root architecture QTL in a crop species were found in rice: deeper rooting 1 (DRO1) (Uga et�al. 2013) and phosphorus starvation tolerance 1 (PSTOL1) (Gamuyao et�al. 2012). Both of these genes underlie stress tolerance traits and were identified in landrace germplasm rather than elite breeding lines, underscoring the importance of querying genetic diversity for root traits. A functional DRO1 gene was found in a drought-tolerant cultivar ‘Kinandang Patong’ from the Philippines, whereas a 1 bp deletion caused a premature stop codon in the widely planted, drought-sensitive rice cultivar ‘IR64’. PSTOL1 was found in the low phosphorus-tolerant aus-type ‘Kasalath’ in a 90 kb indel that is completely absent in the low phosphorus-intolerant ‘Nipponbare’ reference genome. The DRO1 gene enhances root gravitropism, thus generating deeper roots, and PSTOL1 is a protein kinase involved in regulation of early crown root development that promotes root growth under both high and low phosphorus conditions. Both genes have been reported to enhance yield under their respective stress environments, with DRO1 showing no yield penalty under well-watered conditions in some environments (Gamuyao et�al. 2012, Uga et�al. 2013). A study of PSTOL1 orthologs in a sorghum diversity panel has shown a similar role for superior alleles of the gene in grain yield under low phosphorus conditions (Hufnagel et�al. 2014). DRO1 and PSTOL1 are powerful yet isolated examples of the potential for root architecture-based improvement of crops. Decades of work was needed for the identification of these genes in rice. Even though researchers have been studying maize root QTLs for over two decades (Lebreton et�al. 1995), similar success has not been achieved.
Gene identification is hampered by the combination of low root trait heritability and the size of mapping populations that can be adequately measured. Since phenotyping is laborious, practical considerations result in sparse or incomplete measurements of the root system. The strong environmental conditioning of root growth contributes to low heritabilities, especially in the field. Thus, few if any studies have had sufficient power to overcome the Beavis effect (Beavis 1994, Xu 2003), which states that the power to detect QTLs is directly proportional to the size of the mapping population and the heritability of the trait. The numbers of QTLs identified in these studies range from 1 to 268, using on average 227 lines and marker densities of about 10 cM (Table�2). Given these population sizes and root trait heritabilities of ≤50%, at best each would only have the power to detect half the real number of QTLs. As a consequence, the effect size of each QTL is likely to be overestimated, resulting in QTLs that cannot easily be fine-mapped without enormous effort. These are major impediments to identification of genes underlying root quantitative traits in maize.
Meta-QTL analyses have attempted to leverage the many available maize root QTL studies to home in on key loci that are in common across populations and environments (Tuberosa et�al. 2003, Hund et�al. 2011). Hund et�al. (2011) performed a meta-QTL analysis on traits relating to root length in maize from 15 QTL studies that used eight bi-parental mapping populations (15 inbreds) and one association mapping population (74 inbreds). A total of 161 single QTLs from the different studies were condensed into 24 meta-QTLs, with only 16 individual QTLs remaining. Root length traits were grouped based on axile and lateral root type and branching order, and available yield QTLs were overlaid. Seminal root meta-QTLs co-localized with yield QTLs more than any other trait. Six meta-QTLs in bins 1.07, 2.04, 2.08, 3.06, 6.05 and 7.04 were suggested as good candidates for further research and gene identification due to the number of single QTLs combined from different mapping populations and the number of different traits co-localized. For example, a meta-QTL in bin 2.04, located 15 cM from Root-ABA1 (Giuliani et�al. 2005, Landi et�al. 2007), had 10 co-localized QTLs from three populations for length and number of seminal roots, root capacitance, lateral root number and elongation rate, and root pulling force. Notably, the meta-analysis included studies from different developmental times (first/second leaf stage, silking and physiological maturity), in different environments (paper roll, hydroponics, greenhouse and field) and using different measurement techniques (manual measurements, root pulling force, root capacitance and root volume). These sources of variability are likely to limit the genetic signal that could be discerned from the meta-analysis. While comparative approaches have helped refine some of the most promising regions, they have not delivered on gene identification thus far.
When leveraging diverse germplasm for root genetic analysis, controlling for phenology, the timing of developmental events, is a key challenge, yet often ignored. The highest heritability of root crown traits and the greatest number of QTLs were observed after a longer period of growth (at silking), compared with the six leaf stage when phenological differences are not as great (Cai et�al. 2012). Both genotype and environment can lead to differences in the rates of leaf emergence and flowering times, which uncouples calendar time from developmental time in a relative sense. For example, since crown root whorl number is tied directly to leaf formation, a plant with more leaves is likely to have more crown roots if evaluated at the same time. It is then perhaps not surprising that when comparing root traits from tropical teosintes vs. the temperate maize inbred line ‘W22’, many of the QTLs controlling crown root number coincided with flowering time genes (Zhang et�al. 2018). The key flowering time locus vgt1 is linked to a major seminal root number QTL on chromosome 8 (Salvi et�al. 2016), suggesting that phenological effects may control seminal root number. While these results could indicate that flowering time directly regulates root traits, especially in terms of size (lengths, surface areas, volumes and biomass) and numbers (of axile and primary branches), controlling for flowering time, either experimentally or statistically, could reveal important traits that scale proportionally or allometrically. Trait values may be higher in magnitude for larger root systems, but relatively greater for smaller root systems if phenology is included in the calculation. Given that ‘bigger’ is not always ‘better,’ especially in terms of crop cultivation at high plant density (Duvick 2005, Hammer et al. 2009, York et�al. 2015), and trade-offs in root vs. shoot resource allocation (Lynch et�al. 2005, Saengwilai et�al. 2014), it will be important to learn the extent to which there is genetic variation for root allometry. Both the size and relative values of traits should be considered when studying root architecture, especially in diverse germplasm from different latitudes.
Allometric relationships are also important when considering root phenotypic plasticity to the environment. Growth simulations of maize roots computed different optimal lateral root densities in response to varying nitrogen and phosphorus availability, which were corroborated by analysis of excavated root crowns (Postma et�al. 2014). Still, the genetic basis of plasticity remains notably elusive despite the fact that it has been exploited for many investigations of stress tolerance and genotype�environment (G�E) interactions (Trachsel et�al. 2013, Gao and Lynch 2016). In more cases than not, environmental and G�E factors interfere with our ability to understand root genetics. For example, when Li et�al. (2015) repeated their seedling hydroponics experiment three times and mapped QTLs for each repeated experiment, they found only 21 of 114 QTLs in common across repeated experiments. In a much simpler set of controlled environment experiments, Spalding and colleagues showed a profound effect of external environment, developmental time and seed size/composition on Arabidopsis seedling root gravitropism dynamics and their quantitative genetic basis (Durham Brooks et�al. 2010, Moore et�al. 2013). The temporal and subtle environmental dependencies of QTLs identified in these works are striking results that provide context to the current inability to replicate, co-align and move beyond initial root QTL results in maize. Clearly, future work will need to focus on capturing the dynamism of root growth (Kwak et�al. 2014) and environmental interactions, as a function of both real and developmental time.
Acquiring Superior Phenotypes with Higher Throughput for the Investigation of the Genetic Control of Maize Root Architecture
The acquisition of root phenotype data is becoming cheaper, faster and better, largely due to rapid advances in digital imaging and automated analysis (Galkovskyi et�al. 2012, Pierret et�al. 2013, Pace et�al. 2014, Das et�al. 2015, Rell�n-�lvarez et�al. 2015, Symonova et�al. 2015). These exciting technological innovations have been reviewed extensively (Fiorani and Schurr 2013, Paez-Garcia et�al. 2015, Topp et�al. 2016, Morris et�al. 2017), but how can we best exploit this richness of information to understand the genetics of root architecture? The first maize root QTL study was limited to seminal and nodal root number, as well as root pulling force of plants grown in a soil glasshouse (Lebreton et�al. 1995). The first field study of maize root QTLs focused on nodal root numbers, angles and estimated mean diameters of excavated samples (Guingo et�al. 1998). While these measurements are intuitive and (relatively) easily quantified, they are only small samples of an entire maize root system, which can only sufficiently be described considering three dimensions and time. More traits are measured now than ever before; whereas only 2.7 traits per study were averaged prior to 2011, 8.3 traits were averaged since (Table�2), (Hund et�al. 2011). A parallel can be drawn in this progression with the effects that high-throughput genotyping and other ‘omics’ technologies had on our understanding of gene function. Many pathways previously defined as linear by classical mutational analysis are now considered as part of large and complex gene/protein networks governed at a genome-wide scale by systems-level rules (Westerhoff and Palsson 2004). Given that the phenome is vastly more complex than the genome (Houle 2010, Chitwood and Topp 2015), we can now move beyond only historically driven, a priori thinking about what traits are important to measure, and let the wealth of new data define the trait through statistical modeling in our quest to unravel genotype to phenotype relationships in roots (Bodner et�al. 2013, Topp et�al. 2016).
Multivariate statistics, applied mathematics and machine learning (ML) approaches can be particularly powerful when applied to root architecture analysis because they can extract the key relationships among multivariate and multidimensional data (Balduzzi et�al. 2017). There is currently no viable way to capture the 3D/4D shape of a maize root as it grows in the field. Therefore, important trade-offs must be made: minirhizotrons can be used to measure temporal dynamics, but only capture a small fraction of the root system and the topology is lost. Roots can be excavated to varying extents manually or mechanically, but the process is destructive and typically only coarse measurements are taken. Entire growing root systems can be imaged at high resolution using X-ray computed tomography (XRT) or other methods, but these approaches are constrained by the size and resolution of the imaging system, which limits plant and pot sizes as well as growth media. The spectra of phenotype vary in their dimensionality, units, scale, throughput, accuracy and precision, and thus require capable analysis frameworks.
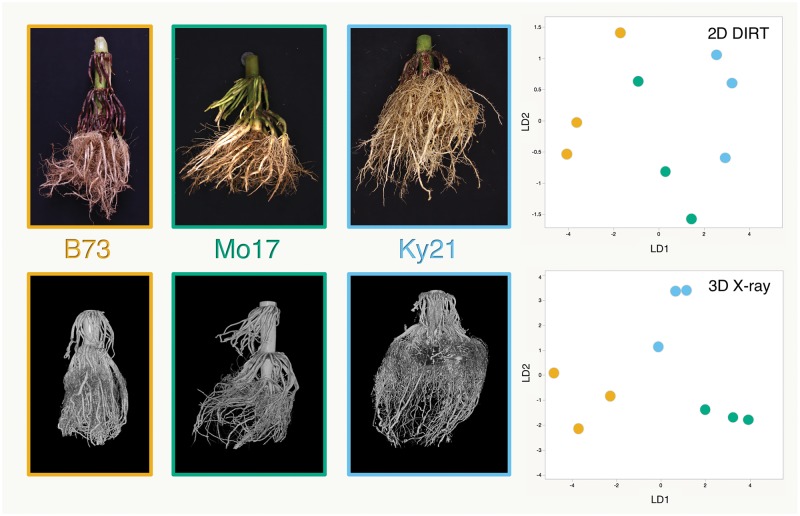
For example, principal component analysis (PCA) and linear discriminant analysis (LDA) are effective statistical methods that can be used to reduce the dimensional space of data to reveal the underlying axes of variation. In Fig.�1 we show an example of how LDA can discriminate the shapes of field-excavated root crowns among three maize inbred lines. The upper left panel shows 2D photographs of each genotype, which were the inputs to the open-access Digital Imaging of Root Traits (DIRT; http://dirt.iplantcollaborative.org/ (July 29, 2018, date last accessed)) software. DIRT calculates dozens of features that describe shape variations in these root images. We applied PCA to reduce the number of traits, and took the first three PCs, which explained >90% of the variation, for LDA. Despite obvious differences in their root structures, LD1 and LD2 do not clearly group each genotype. DIRT has enabled at least an order of magnitude increase in the number of field-grown roots that can be evaluated, along with improved accuracy and precision (Bucksch et�al. 2014, Das et�al. 2015); however, information is lost by representing a complex 3D object into two dimensions. The lower left panel of Fig.�1 shows a 3D reconstruction generated from scanning the same root crowns via XRT. We measured the shapes in 3D using the RSA Gia pipeline (Topp et�al. 2013) and conducted a PCA and LDA. In this analysis, the first two linear discriminants clearly discriminate all three genotypes from one another. This result highlights how multivariate analysis coupled with enhanced phenotypes can improve our ability to make genotype to phenotype associations.
Fig. 1.
Comparison of 2D images (top) and 3D X-ray computed tomography (XRT) scans (bottom) of maize root crowns excavated from the field at tasseling. Differences between ‘B73,’ ‘Mo17’ and ‘Ky21’ from the images are visually apparent, showing the narrower, denser habit of ‘B73’ and ‘Ky21’ compared with ‘Mo17’; however, LDA of 2D DIRT traits does not entirely discriminate these genotypes (top right). Additional information gathered from 3D XRT scans and 3D GiaRoot traits group these genotypes in LDA (bottom right). These data are a subset of three excavated root crowns from three maize inbred lines collected at tasseling from a larger experiment conducted at our field site at University of Missouri Genetics Farm, Columbia, MO.
While multivariate approaches are powerful to discern subtle or pleiotropic phenotypes, their biological interpretation is sometimes called into question because the trait space is no longer easily described along a univariate axis. Instead, multivariate models often provide a new descriptor without an intuitive connection to more common traits (Topp et�al. 2013, Kenobi et�al. 2017). However, we argue that data-defined trait descriptors will play a key role in finally realizing the potential of root imaging technologies to identify genes. A 3D QTL analysis of seedling root systems in rice leveraged multivariate analysis of variance (MANOVA) to identify several large effect QTLs that were not identified by the univariate traits alone, and explained more phenotypic variation in the data (Topp et�al. 2013). Composite traits were computed from the relative contributions of each univariate trait to the multivariate, and used to verify the MANOVA-based QTLs. Thus, the very concept of genetic architecture for a complex trait such as root shape is strongly influenced by how we quantify it in the trait space. The impacts of ML on plant science will also be felt in this area. Classification-focused ML methods such as support vector machines and logistic regression have been used recently as multivariate discriminators for root phenotypes (Iyer-Pascuzzi et�al. 2010, Zurek et�al. 2015), and deep learning approaches such as convolutional neural networks are poised to learn salient features of the data that can lead to automated quantification of statistical descriptors (Pound et�al. 2017).
Multivariate methods are a powerful complement to univariate traits, but are still limited to input features that were developed a priori to capture specific aspects of root shape. A much more general statistical approach was used to quantify shoot architectures by their spatial density functions, revealing fundamental similarities in 3D Gaussian density functions across several species and stages of development that were otherwise hidden (Conn et�al. 2017a). The work followed on an evaluation of architectural trade-offs between biomass investment and resource distribution in the same samples using graph theory (Conn et�al. 2017b), which also seems fitting for root architecture studies. Topological data analysis (TDA) methods are another way to provide more comprehensive quantifications of whole plant and root shapes without pre-supposing any specific trait (Li et�al. 2017, Delory et al. 2018). A study evaluating the genetic determinants of tomato root architecture showed that a TDA approach known as persistent homology can be used for QTL analysis. Persistent homology captured nearly all of the loci that the most heritable DIRT traits did, plus nearly two dozen additional QTLs (Li et�al. 2018). These studies demonstrate that the traditional univariate trait methods of quantifying root architecture are simply not adequate to describe all of the phenotypic variation, and therefore have probably limited our ability to discern the underlying genetic relationships in maize and many other species.
Conclusions and Outlooks
Only a small handful of genes are currently known to control maize root development, and none has been shown specifically to influence quantitative traits useful for breeding or understanding plant�environment interactions. While the genotype to phenotype gap is generally a major challenge of current biological research, it is especially wide for root biology because of limitations in phenotyping tools and analysis approaches. Nonetheless, the pace of advances has been increasing so that now higher quality, higher throughput studies can be conducted in many laboratories throughout the world, and excellent open-source platforms for quantifying, describing and modeling root phenotypes are in place (Lobet et�al. 2013, Lobet et�al. 2015, Postma et�al. 2017, Delory et�al. 2018, Schnepf et�al. 2018).
The re-evaluation of maize mutant phenotypes with current tools may be particularly informative to understand how known genes may impact quantitative aspects of root architecture, and to identify the genetic polymorphisms underlying mutants that have not yet been cloned (Table�1). High-throughput phenotyping methods coupled to the current wealth of genotype information across hundreds of maize accessions (Gore et�al. 2009, Hirsch et�al. 2014, Bukowski et�al. 2018) can also be used to scan for common polymorphisms in these genes that may modify their phenotypic expression in more subtle ways than could be previously measured. Similarly, the evaluation of known mutants in many different genetic backgrounds could reveal modifying genes that will help connect root development and architecture at a molecular systems level. It is likely to be this quantitative variation that allows us to advance our understanding of the genetic basis of complex root phenotypes. Dynamic imaging of root growth and the function-valued phenotypes derived from these data will be a key part of increasing the information content of the phenotypes we collect, which is being enabled in 3D and increasingly environmentally realistic scenarios (Fig.�1) (Clark et�al. 2011, Metzner et�al. 2015, Symonova et�al. 2015, Ahmed et�al. 2016, van Dusschoten et�al. 2016, Morris et�al. 2017). However, these tools are still nascent, not widespread, and thus only on the cusp of realizing their potential. It will be important to continue to establish sound and transparent methods for data acquisition and post-processing in order to lower the ‘barriers to entry’ for the many root biologists who wish to incorporate advanced phenotyping into their studies.
Given the crop improvement potential for root traits, a major emphasis will continue to be field studies. Belowground phenotyping capacity will always be less than aboveground (Pauli et�al. 2016), but it is now possible to harvest and evaluate hundreds or thousands of superficial root crown samples for root architecture using computer vision (Fig.�1; Table�2) (Bucksch et�al. 2014, Das et�al. 2015). Minirhizotron and soil coring data provide additional (but sparse) information of roots at depth that can be combined with root crown excavations in high throughput to provide a more complete picture of the true root structure. Such integrated phenotyping approaches could move beyond simple trait comparisons, and directly evaluate QTLs in common from complementary data types, which will be more robust than using only one approach. All things considered, it seems only a matter of time until the combined efforts of laboratory and field work, bolstered by technological and computational advances, will begin to yield a more thorough understanding of the genetics controlling the hidden half of maize.
Funding
This material is based upon work supported by the National Science Foundation [award numbers: IIA-1355406 and IOS-1638507 to C.N.T.].
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- DIRT
Digital Imaging of Root Traits
- DRO1
Deeper Rooting 1
- G�E
genotype�environment
- GWAS
genome-wide association study
- LDA
linear discriminant analysis
- Lrp1
Lateral root primordia 1
- lrt1
lateral rootless 1
- MANOVA
multivariate analysis of variance
- ML
machine learning
- PCA
principal component analysis
- PSTOL1
Phosphorus-Starvation Tolerance 1
- QTL
quantitative trait locus
- rth
roothairless
- RSA
root system architecture
- rtcl
rtcs-like
- rtcs
rootless concerning seminal and crown roots
- Rul1
Rum1-like
- rum1
rootless with undetectable meristem 1
- slr
short lateral roots
- TDA
topological data analysis
- XRT
X-ray computed tomography
References
- Abdel-Ghani A.H., Kumar B., Pace J., Jansen C., Gonzalez-Portilla P.J., Reyes-Matamoros J., et al. (2015) Association analysis of genes involved in maize (Zea mays L.) root development with seedling and agronomic traits under contrasting nitrogen levels. Plant Mol. Biol. 88: 133–147. [DOI] [PubMed] [Google Scholar]
- Ahmed S., Klassen T.N., Keyes S., Daly M., Jones D.L., Mavrogordato M., et al. (2016) Imaging the interaction of roots and phosphate fertiliser granules using 4D X-ray tomography. Plant Soil 401: 125–134. [Google Scholar]
- Andorf C.M., Cannon E.K., Portwood J.L. 2nd, Gardiner J.M., Harper L.C., Schaeffer M.L., et al. (2016) MaizeGDB update: new tools, data and interface for the maize model organism database. Nucleic Acids Res. 44: D1195–D1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balduzzi M., Binder B.M., Bucksch A., Chang C., Hong L., Iyer-Pascuzzi A.S., et al. (2017) Reshaping plant biology: qualitative and quantitative descriptors for plant morphology. Front Plant Sci. 8: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis W.D. (1994) The power and deceit of QTL experiments: lessons from comparative QTL studies InProceedings of the Forty-Ninth Annual Corn and Sorghum Industry Research Conference. pp. 250–266. ASTA, Washington. [Google Scholar]
- Bodner G., Leitner D., Nakhforoosh A., Sobotik M., Moder K., Kaul H.-P. (2013) A statistical approach to root system classification. Front. Plant Sci. 4: 292.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucksch A., Burridge J., York L.M., Das A., Nord E., Weitz J.S., et al. (2014) Image-based high-throughput field phenotyping of crop roots. Plant Physiol. 166: 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowski R., Guo X., Lu Y., Zou C., He B., Rong Z., et al. (2018) Construction of the third generation Zea mays haplotype map. Gigascience 7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A.L., Brown K.M., Lynch J.P. (2013) Phenotypic diversity of root anatomical and architectural traits in Zea species. Crop Sci. 53: 1042–1055. [Google Scholar]
- Burton A.L., Johnson J., Foerster J., Hanlon M.T., Kaeppler S.M., Lynch J.P., et al. (2015) QTL mapping and phenotypic variation of root anatomical traits in maize (Zea mays L.). Theor. Appl. Genet. 128: 93–106. [DOI] [PubMed] [Google Scholar]
- Burton A.L., Johnson J.M., Foerster J.M., Hirsch C.N., Buell C.R., Hanlon M.T., et al. (2014) QTL mapping and phenotypic variation for root architectural traits in maize (Zea mays L.). Theor. Appl. Genet. 127: 2293–2311. [DOI] [PubMed] [Google Scholar]
- Cai H., Chen F., Mi G., Zhang F., Maurer H.P., Liu W., et al. (2012) Mapping QTLs for root system architecture of maize (Zea mays L.) in the field at different developmental stages. Theor. Appl. Genet. 125: 1313–1324. [DOI] [PubMed] [Google Scholar]
- Chitwood D.H., Topp C.N. (2015) Revealing plant cryptotypes: defining meaningful phenotypes among infinite traits. Curr. Opin. Plant Biol. 24: 54–60. [DOI] [PubMed] [Google Scholar]
- Clark R.T., MacCurdy R.B., Jung J.K., Shaff J.E., McCouch S.R., Aneshansley D.J., et al. (2011) Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 156: 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn A., Pedmale U.V., Chory J., Navlakha S. (2017b) High-resolution laser scanning reveals plant architectures that reflect universal network design principles. Cell Syst. 5: 53–62. [DOI] [PubMed] [Google Scholar]
- Conn A., Pedmale U.V., Chory J., Stevens C.F., Navlakha S. (2017a) A statistical description of plant shoot architecture. Curr. Biol. 27: 2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Schneider H., Burridge J., Ascanio A.K.M., Wojciechowski T., Topp C.N., et al. (2015) Digital imaging of root traits (DIRT): a high-throughput computing and collaboration platform for field-based root phenomics. Plant Methods 11: 51.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dorlodot S., Forster B., Pag�s L., Price A., Tuberosa R., Draye X. (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 12: 474–481. [DOI] [PubMed] [Google Scholar]
- Delory B.M., Li M., Topp C.N., Lobet G. (2018) archiDART v3.0: a new data analysis pipeline allowing the topological analysis of plant root systems. F1000Res. 7: 22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham Brooks T.L., Miller N.D., Spalding E.P. (2010) Plasticity of Arabidopsis root gravitropism throughout a multidimensional condition space quantified by automated image analysis. Plant Physiol. 152: 206–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick D.N. (2005) The contribution of breeding to yield advances in maize (Zea mays L.). Adv. Agron. 86: 83–145. [Google Scholar]
- Feix G., Hochholdinger F., Park W.J. (2002) Maize root system and genetic analysis of its formation InPlant Roots: The Hidden Half, 3rd edn Edited by Waisel Y., Eshel A., Beeckman T., Kafkafi U. pp. 239–248. Marcel Dekker, New York. [Google Scholar]
- Feldman L. (1994) The maize root InThe Maize Handbook. Edited by Greeling M., Walbot V. pp. 29–37. Springer, New York. [Google Scholar]
- Fiorani F., Schurr U. (2013) Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 64: 267–291. [DOI] [PubMed] [Google Scholar]
- Fitter A.H. (1987) An architectural approach to the comparative ecology of plant root systems. New Phytol. 106: 61–77. [Google Scholar]
- Galkovskyi T., Mileyko Y., Bucksch A., Moore B., Symonova O., Price C.A., et al. (2012) GiA Roots: software for the high throughput analysis of plant root system architecture. BMC Plant Biol. 12: 116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamuyao R., Chin J.H., Pariasca-Tanaka J., Pesaresi P., Catausan S., Dalid C., et al. (2012) The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488: 535–539. [DOI] [PubMed] [Google Scholar]
- Gao Y., Lynch J.P. (2016) Reduced crown root number improves water acquisition under water deficit stress in maize (Zea mays L.). J. Exp. Bot. 67: 4545–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford M.L., Banta J.A., Katari M.S., Hulsmans J., Chen L., Ristova D., et al. (2013) Plasticity regulators modulate specific root traits in discrete nitrogen environments. PLoS Genet. 9: e1003760.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani S., Sanguineti M.C., Tuberosa R., Bellotti M., Salvi S., Landi P. (2005) Root-ABA1, a major constitutive QTL, affects maize root architecture and leaf ABA concentration at different water regimes. J. Exp. Bot. 56: 3061–3070. [DOI] [PubMed] [Google Scholar]
- Gore M.A., Chia J.-M., Elshire R.J., Sun Q., Ersoz E.S., Hurwitz B.L., et al. (2009) A first-generation haplotype map of maize. Science 326: 1115–1117. [DOI] [PubMed] [Google Scholar]
- Gu D., Mei X., Yu T., Sun N., Xu D., Liu C., et al. (2017) QTL identification for brace-root traits of maize in different generations and environments. Crop Sci. 57: 13–21. [Google Scholar]
- Guingo E., H�bert Y., Charcosset A. (1998) Genetic analysis of root traits in maize. Agronomie 18: 225–235. [Google Scholar]
- Hammer G.L., Dong Z., McLean G., Doherty A., Messina C., Schussler J., et al. (2009) Can changes in canopy and/or root system architecture explain historical maize yield trends in the US corn belt? Crop Sci. 49: 299–312. [Google Scholar]
- Hirsch C.N., Foerster J.M., Johnson J.M., Sekhon R.S., Muttoni G., Vaillancourt B., et al. (2014) Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26: 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C.N., Hirsch C.D., Brohammer A.B., Bowman M.J., Soifer I., Barad O., et al. (2016) Draft assembly of elite inbred line PH207 provides insights into genomic and transcriptome diversity in maize. Plant Cell 28: 2700–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F. (2009) The maize root system: morphology, anatomy, and genetics InHandbook of Maize: Its Biology. Edited by Bennetzen J.L., Hake S.C. pp. 145–160. Springer, New York. [Google Scholar]
- Hochholdinger F., Feix G. (1998) Early post-embryonic root formation is specifically affected in the maize mutant lrt1. Plant J. 16: 247–255. [DOI] [PubMed] [Google Scholar]
- Hochholdinger F., Park W.J., Feix G.H. (2001) Cooperative action of SLR1 and SLR2 is required for lateral root-specific cell elongation in maize. Plant Physiol. 125: 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F., Tuberosa R. (2009) Genetic and genomic dissection of maize root development and architecture. Curr. Opin. Plant Biol. 12: 172–177. [DOI] [PubMed] [Google Scholar]
- Hochholdinger F., Wen T.-J., Zimmermann R., Chimot-Marolle P., da Costa e Silva O., Bruce W., et al. (2008) The maize (Zea mays L.) roothairless3 gene encodes a putative GPI-anchored, monocot-specific, COBRA-like protein that significantly affects grain yield. Plant J. 54: 888–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F., Woll K., Sauer M., Feix G. (2005) Functional genomic tools in support of the genetic analysis of root development in maize (Zea mays L. ). Maydica 50: 437–442. [Google Scholar]
- Hochholdinger F., Yu P., Marcon C. (2018) Genetic control of root system development in maize. Trends Plant Sci. 23: 79–88. [DOI] [PubMed] [Google Scholar]
- Houle D. (2010) Colloquium papers: numbering the hairs on our heads: the shared challenge and promise of phenomics. Proc. Natl. Acad. Sci. USA 107: 1793–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel B., de Sousa S.M., Assis L., Guimaraes C.T., Leiser W., Azevedo G.C., et al. (2014) Duplicate and conquer: multiple homologs of PHOSPHORUS-STARVATION TOLERANCE1 enhance phosphorus acquisition and sorghum performance on low-phosphorus soils. Plant Physiol. 166: 659–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hund A., Reimer R., Messmer R. (2011) A consensus map of QTLs controlling the root length of maize. Plant Soil 344: 143–158. [Google Scholar]
- Ilic K., Kellogg E.A., Jaiswal P., Zapata F., Stevens P.F., Vincent L.P., et al. (2007) The plant structure ontology, a unified vocabulary of anatomy and morphology of a flowering plant. Plant Physiol. 143: 587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Pascuzzi A.S., Symonova O., Mileyko Y., Hao Y., Belcher H., Harer J., et al. (2010) Imaging and analysis platform for automatic phenotyping and trait ranking of plant root systems. Plant Physiol. 152: 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins M.T. (1930) Heritable characters of maize: XXXIV—rootless. J. Hered. 21: 79–80. [Google Scholar]
- Jiao Y., Peluso P., Shi J., Liang T., Stitzer M.C., Wang B., et al. (2017) Improved maize reference genome with single-molecule technologies. Nature 546: 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenobi K., Atkinson J.A., Wells D.M., Gaju O., De Silva J.G., Foulkes M.J., et al. (2017) Linear discriminant analysis reveals differences in root architecture in wheat seedlings related to nitrogen uptake efficiency. J. Exp. Bot. 68: 4969–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesselbach T.A. (1949) The Structure and Reproduction of Corn Agricultural Experimental Station Research Bulletin No. 161. University of Nebraska College of Agriculture [Google Scholar]
- Ku L.X., Sun Z.H., Wang C.L., Zhang J., Zhao R.F., Liu H.Y., et al. (2012) QTL mapping and epistasis analysis of brace root traits in maize. Mol. Breed. 30: 697–708. [Google Scholar]
- Kwak I.-Y., Moore C.R., Spalding E.P., Broman K.W. (2014) A simple regression-based method to map quantitative trait loci underlying function-valued phenotypes. Genetics 197: 1409–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi P., Sanguineti M.C., Liu C., Li Y., Wang T.Y., Giuliani S., et al. (2007) Root-ABA1 QTL affects root lodging, grain yield, and other agronomic traits in maize grown under well-watered and water-stressed conditions. J. Exp. Bot. 58: 319–326. [DOI] [PubMed] [Google Scholar]
- Lebreton C., Lazić-Jančić V., Steed A., Pekić S., Quarrie S.A. (1995) Identification of QTL for drought responses in maize and their use in testing causal relationships between traits. J. Exp. Bot. 46: 853–865. [Google Scholar]
- Li L., Hey S., Liu S., Liu Q., McNinch C., Hu H.-C., et al. (2016) Characterization of maize roothairless6 which encodes a D-type cellulose synthase and controls the switch from bulge formation to tip growth. Sci. Rep. 6: 34395.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Duncan K., Topp C.N., Chitwood D.H. (2017) Persistent homology and the branching topologies of plants. Amer. J. Bot. 104: 349–353. [DOI] [PubMed] [Google Scholar]
- Li M., Frank M., Coneva V., Mio W., Chitwood D.H., Topp C.N. (2018) The persistent homology mathematical framework provides enhanced genotype-to-phenotype associations for plant morphology. Plant Physiol. bioRxiv. doi:10.1101/104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Chen F., Cai H., Liu J., Pan Q., Liu Z., et al. (2015) A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. J. Exp. Bot. 66: 3175–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cai H., Chu Q., Chen X., Chen F., Yuan L., et al. (2011) Genetic analysis of vertical root pulling resistance (VRPR) in maize using two genetic populations. Mol. Breed. 28: 463–474. [Google Scholar]
- Liu Z., Gao K., Shan S., Gu R., Wang Z., Craft E.J., et al. (2017) Comparative analysis of root traits and the associated QTLs for maize seedlings grown in paper roll, hydroponics and vermiculite culture system. Front. Plant Sci. 8: 436.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet G., Draye X., P�rilleux C. (2013) An online database for plant image analysis software tools. Plant Methods 9: 38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobet G., Pound M.P., Diener J., Pradal C., Draye X., Godin C., et al. (2015) Root system markup language: toward a unified root architecture description language. Plant Physiol. 167: 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. (1995) Root architecture and plant productivity. Plant Physiol. 109: 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J.P. (2013) Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 112: 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J.P., Chimungu J.G., Brown K.M. (2014) Root anatomical phenes associated with water acquisition from drying soil: targets for crop improvement. J. Exp. Bot. 65: 6155–6166. [DOI] [PubMed] [Google Scholar]
- Lynch J.P., Ho M.D., Phosphorus L. (2005) Rhizoeconomics: carbon costs of phosphorus acquisition. Plant Soil 269: 45–56. [Google Scholar]
- MacMillan K., Emrich K., Piepho H.-P., Mullins C.E., Price A.H. (2006) Assessing the importance of genotype � environment interaction for root traits in rice using a mapping population II: conventional QTL analysis. Theor. Appl. Genet. 113: 953–964. [DOI] [PubMed] [Google Scholar]
- Mano Y., Omori F. (2008) Verification of QTL controlling root aerenchyma formation in a maize � teosinte ‘Zea nicaraguensis’ advanced backcross population. Breed. Sci. 58: 217–223. [Google Scholar]
- Mano Y., Omori F. (2009) High-density linkage map around the root aerenchyma locus Qaer1.06 in the backcross populations of maize Mi29 � teosinte ‘Zea nicaraguensis’. Breed. Sci. 59: 427–433. [Google Scholar]
- Mano Y., Omori F., Takamizo T., Kindiger B., McK Bird R., Loaisiga C.H., et al. (2007) QTL mapping of root aerenchyma formation in seedlings of a maize � rare teosinte ‘Zea nicaraguensis’ cross. Plant Soil 295: 103–113. [Google Scholar]
- Meij�n M., Satbhai S.B., Tsuchimatsu T., Busch W. (2014) Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat. Genet. 46: 77–81. [DOI] [PubMed] [Google Scholar]
- Metzner R., Eggert A., van Dusschoten D., Pflugfelder D., Gerth S., Schurr U., et al. (2015) Direct comparison of MRI and X-ray CT technologies for 3D imaging of root systems in soil: potential and challenges for root trait quantification. Plant Methods 11: 17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C.R., Johnson L.S., Kwak I.-Y., Livny M., Broman K.W., Spalding E.P. (2013) High-throughput computer vision introduces the time axis to a quantitative trait map of a plant growth response. Genetics 195: 1077–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris E.C., Griffiths M., Golebiowska A., Mairhofer S., Burr-Hersey J., Goh T., et al. (2017) Shaping 3D root system architecture. Curr. Biol. 27: R919–R930. [DOI] [PubMed] [Google Scholar]
- Nestler J., Liu S., Wen T.-J., Paschold A., Marcon C., Tang H.M., et al. (2014) Roothairless5, which functions in maize (Zea mays L.) root hair initiation and elongation encodes a monocot-specific NADPH oxidase. Plant J. 79: 729–740. [DOI] [PubMed] [Google Scholar]
- Pace J., Gardner C., Romay C., Ganapathysubramanian B., L�bberstedt T. (2015) Genome-wide association analysis of seedling root development in maize (Zea mays L.). BMC Genomics 16: 47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace J., Lee N., Naik H.S., Ganapathysubramanian B., L�bberstedt T. (2014) Analysis of maize (Zea mays L.) seedling roots with the high-throughput image analysis tool ARIA (Automatic Root Image Analysis). PLoS One 9: e108255.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez-Garcia A., Motes C.M., Scheible W.-R., Chen R., Blancaflor E.B., Monteros M.J. (2015) Root traits and phenotyping strategies for plant improvement. Plants (Basel) 4: 334–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli D., Chapman S.C., Bart R., Topp C.N., Lawrence-Dill C.J., Poland J., et al. (2016) The quest for understanding phenotypic variation via integrated approaches in the field environment. Plant Physiol. 172: 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestsova E., Lichtblau D., Wever C., Presterl T., Bolduan T., Ouzunova M., et al. (2016) QTL mapping of seedling root traits associated with nitrogen and water use efficiency in maize. Euphytica 209: 585–602. [Google Scholar]
- Pierret A., Gonkhamdee S., Jourdan C., Maeght J.-L. (2013) IJ_Rhizo: an open-source software to measure scanned images of root samples. Plant Soil 373: 531–539. [Google Scholar]
- Postma J.A., Dathe A., Lynch J.P. (2014) The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiol. 166: 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma J.A., Kuppe C., Owen M.R., Mellor N., Griffiths M., Bennett M.J., et al. (2017) OpenSimRoot: widening the scope and application of root architectural models. New Phytol. 215: 1274–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pound M.P., Atkinson J.A., Townsend A.J., Wilson M.H., Griffiths M., Jackson A.S., et al. (2017) Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. Gigascience 6: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.H., Tomos A.D. (1997) Genetic dissection of root growth in rice (Oryza sativa L.). II: mapping quantitative trait loci using molecular markers. Theor. Appl. Genet. 95: 143–152. [Google Scholar]
- Rell�n-�lvarez R., Lobet G., Lindner H., Pradier P.-L., Sebastian J., Yee M.-C., et al. (2015) GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas U., Cibrian-Jaramillo A., Ristova D., Banta J.A., Gifford M.L., Fan A.H., et al. (2013) Integration of responses within and across Arabidopsis natural accessions uncovers loci controlling root systems architecture. Proc. Natl. Acad. Sci. USA 110: 15133–15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saengwilai P., Tian X., Lynch J.P. (2014) Low crown root number enhances nitrogen acquisition from low-nitrogen soils in maize. Plant Physiol. 166: 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi S., Giuliani S., Ricciolini C., Carraro N., Maccaferri M., Presterl T., et al. (2016) Two major quantitative trait loci controlling the number of seminal roots in maize co-map with the root developmental genes rtcs and rum1. J. Exp. Bot. 67: 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez D.L., Liu S., Ibrahim R., Blanco M., L�bberstedt T. (2018) Genome-wide association studies of doubled haploid exotic introgression lines for root system architecture traits in maize (Zea mays L.). Plant Sci. 268: 30–38. [DOI] [PubMed] [Google Scholar]
- Sandhu N., Raman K.A., Torres R.O., Audebert A., Dardou A., Kumar A., et al. (2016) Rice root architectural plasticity traits and genetic regions for adaptability to variable cultivation and stress conditions. Plant Physiol. 171: 2562–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass J.E. (1977) Morphology InCorn and Corn Improvement. Edited by Sprague G.F., Eberhart S.A. pp. 89–110. American Society of Agronomy, Madison, WI. [Google Scholar]
- Schmidt J.E., Bowles T.M., Gaudin A.C.M. (2016) Using ancient traits to convert soil health into crop yield: impact of selection on maize root and rhizosphere function. Front. Plant Sci. 7: 373.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Freeling M. (2011) Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS One 6: e17855.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Springer N.M., Freeling M. (2011) Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf A., Leitner D., Landl M., Lobet G., Mai T.H., Morandage S., et al. (2018) CRootBox: a structural–functional modelling framework for root systems. Ann. Bot. 121: 1033–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonova O., Topp C.N., Edelsbrunner H. (2015) DynamicRoots: a software platform for the reconstruction and analysis of growing plant roots. PLoS One 10: e0127657.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramino G., Sauer M., Stauffer J.L., Multani D., Niu X., Sakai H., et al. (2007) The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J. 50: 649–659. [DOI] [PubMed] [Google Scholar]
- Topp C.N., Bray A.L., Ellis N.A., Liu Z. (2016) How can we harness quantitative genetic variation in crop root systems for agricultural improvement? J. Integr. Plant Biol. 58: 213–225. [DOI] [PubMed] [Google Scholar]
- Topp C.N., Iyer-Pascuzzi A.S., Anderson J.T., Lee C.-R., Zurek P.R., Symonova O., et al. (2013) 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc. Natl. Acad. Sci. USA 110: E1695–E1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel S., Kaeppler S.M., Brown K.M., Lynch J.P. (2013) Maize root growth angles become steeper under low N conditions. Field Crops Res. 140: 18–31. [Google Scholar]
- Tuberosa R., Salvi S., Sanguineti M.C., Maccaferri M., Giuliani S., Landi P. (2003) Searching for quantitative trait loci controlling root traits in maize: a critical appraisal. Plant Soil 255: 35–54. [Google Scholar]
- Uga Y., Sugimoto K., Ogawa S., Rane J., Ishitani M., Hara N., et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45: 1097–1102. [DOI] [PubMed] [Google Scholar]
- van Dusschoten D., Metzner R., Kochs J., Postma J.A., Pflugfelder D., B�hler J., et al. (2016) Quantitative 3D analysis of plant roots growing in soil using magnetic resonance imaging. Plant Physiol. 170: 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Behrens I., Komatsu M., Zhang Y., Berendzen K.W., Niu X., Sakai H., et al. (2011) Rootless with undetectable meristem 1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize: AUX/IAA regulation of maize root development. Plant J. 66: 341–353. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Takahashi H., Sato S., Nishiuchi S., Omori F., Malik A.I., et al. (2017) A major locus involved in the formation of the radial oxygen loss barrier in adventitious roots of teosinte Zea nicaraguensis is located on the short-arm of chromosome 3. Plant Cell Environ. 40: 304–316. [DOI] [PubMed] [Google Scholar]
- Wen T.-J., Hochholdinger F., Sauer M., Bruce W., Schnable P.S. (2005) The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol. 138: 1637–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T.-J., Schnable P.S. (1994) Analyses of mutants of three genes that influence root hair development in Zea mays (Gramineae) suggest that root hairs are dispensable. Amer. J. Bot. 81: 833–842. [Google Scholar]
- Westerhoff H.V., Palsson B.O. (2004) The evolution of molecular biology into systems biology. Nat. Biotechnol. 22: 1249–1252. [DOI] [PubMed] [Google Scholar]
- Woll K., Borsuk L.A., Stransky H., Nettleton D., Schnable P.S., Hochholdinger F. (2005) Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol. 139: 1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. (2003) Theoretical basis of the Beavis effect. Genetics 165: 2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Tai H., Saleem M., Ludwig Y., Majer C., Berendzen K.W., et al. (2015) Cooperative action of the paralogous maize lateral organ boundaries (LOB) domain proteins RTCS and RTCL in shoot-borne root formation. New Phytol. 207: 1123–1133. [DOI] [PubMed] [Google Scholar]
- York L.M., Galindo-Casta�eda T., Schussler J.R., Lynch J.P. (2015) Evolution of US maize (Zea mays L.) root architectural and anatomical phenes over the past 100 years corresponds to increased tolerance of nitrogen stress. J. Exp. Bot. 66: 2347–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Gutjahr C., Li C., Hochholdinger F. (2016) Genetic control of lateral root formation in cereals. Trends Plant Sci. 21: 951–961. [DOI] [PubMed] [Google Scholar]
- Zaidi P.H., Seetharam K., Krishna G., Krishnamurthy L., Gajanan S., Babu R., et al. (2016) Genomic regions associated with root traits under drought stress in tropical maize (Zea mays L.). PLoS One 11: e0164340.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Marcon C., Tai H., von Behrens I., Ludwig Y., Hey S., et al. (2016) Conserved and unique features of the homeologous maize Aux/IAA proteins ROOTLESS WITH UNDETECTABLE MERISTEM 1 and RUM1-like 1. J. Exp. Bot. 67: 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., von Behrens I., Zimmermann R., Ludwig Y., Hey S., Hochholdinger F. (2015) LATERAL ROOT PRIMORDIA 1 of maize acts as a transcriptional activator in auxin signalling downstream of the Aux/IAA gene rootless with undetectable meristem 1. J. Exp. Bot. 66: 3855–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang X., Lin Z., Wang J., Xu M., Lai J., et al. (2018) The genetic architecture of nodal root number in maize. Plant J. 93: 1032–1044. [DOI] [PubMed] [Google Scholar]
- Zobel R.W., Waisel Y. (2010) A plant root system architectural taxonomy: a framework for root nomenclature. Plant Biosyst. 144: 507–512. [Google Scholar]
- Zurek P.R., Topp C.N., Benfey P.N. (2015) Quantitative trait locus mapping reveals regions of the maize genome controlling root system architecture. Plant Physiol. 167: 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]