ABSTRACT
Epithelial cells have characteristic membrane domains. Identification of membrane proteins playing an important role in these membrane domains has progressed and numerous studies have been performed on the functional analysis of these membrane proteins. On the other hand, the precise roles of membrane lipids in the organization of these membrane domains are largely unknown. Historically, the concept of lipid raft arose from the analysis of lipid composition of the apical membrane, and it can be said that epithelial cells are an optimal experimental model for elucidating the functions of lipids. In this review, I discuss the role of lipids in the formation of epithelial polarity and in the formation of cell membrane structures of epithelial cells such as microvilli in the apical domain, cell-cell adhesion apparatus in the lateral domain and cell-matrix adhesion in the basal domain.
KEYWORDS: Tight junction, microvilli, epithelial polarity
Introduction
The cellular membrane is continuous, but the distribution of membrane proteins and lipids constituting the cell membrane is domain-like and heterogeneous. It is possible to classify such cell membrane domains by morphological features of plasma membrane, such as microvilli, tight junctions and caveolae etc., functional roles, existence of specific membrane protein maker, and detergent insolubility. The myriad possible classifications as well as the functional and compositional differences in the membrane domains of epithelial cell have attracted many researchers.
Epithelial cells are polarized and maintain apical and basolateral membranes. In addition to epithelial cells, formation of cell polarity is observed in various biological contexts such as neurons, chemotaxis of neutrophils, asymmetric division of budding yeast, and so on. However, it remains unclear to what extent these various cell polarization phenomena can be explained by a common molecular mechanism. Evolutionally conserved polarity proteins including Par-3/Par-6/aPKC/Cdc42 complex, Crumbs complex, Dlg/Lgl/Scribble complex were identified as key regulators in determining cellular axis in epithelial cells.1 However, among these molecules, only Lgl and Cdc42 have homologs in the budding yeast.2 Considering that diverse lipid molecules are evolutionarily well conserved, it is reasonable to assume that cell membrane lipids play some role in the formation of cell polarity. With regard to the functional importance of lipids in cell polarity, we still have no clear answer for the following questions: Do cell membrane lipid differences contribute to generation of functional differences in cell membranes? If the composition of lipids is important for functional difference of each cell membrane domain, what is a mechanism that makes it possible to maintain restricted distribution of cell membrane lipids by inhibiting the free diffusion of lipids?
In addition to the differentiated apical and basolateral membranes, epithelial cells have morphologically distinct membrane structures with specific functions. For example, in the apical membrane, membrane protrusions called microvilli exist and play essential roles in absorption of nutrients. What role, then, do cell membrane lipids play in the formation of such membrane structures? In this review, I will outline recent findings that address these issues.
Roles of sphingolipids in the formation of apical membrane
Kai Simons and colleagues examined the compositional differences of membrane lipids in the apical and basolateral membranes by taking advantage of the enveloped virus. They extracted lipids from the fowl plague virus that selectively sprouts from the apical membrane and the vesicular somatitis virus that emerges from the basolateral membrane in MDCK cells and compared the lipid composition of these enveloped viruses. This pioneering work revealed that sphingomyelin is enriched, compared to phosphatidylcholine, in the apical membrane but not in the basolateral membrane.3 The fact that sphingolipid-rich domains exist in the apical membrane was later extended to the concept of “lipid raft”.4 Kai Simons and colleagues first described that sphingolipids biosynthesized in the Golgi apparatus form lipid rafts with cholesterol in the trans-Golgi network and then a group of proteins having affinity for lipid rafts such as GPI anchor proteins assemble to form transport vesicles directed to the apical membrane. In fact, GPI-anchored proteins often have saturated fatty acids in their lipid moiety and have affinity to detergent-insoluble fractions.5 Thus, lipid rafts were proposed as a mechanism of polar transport to the apical membranes in epithelial cells.6 In this way, the sphingolipids-enriched membrane domain is assumed to be involved in the establishment of apico-basal polarity.
Importance of lipid rafts is assumed not only in intracellular transport but also in the regulation of cell signaling. Proteins subject to covalent lipid modification by saturated fatty acids (myristoylated/palmitoylated) also tend to gather in lipid rafts. Therefore, it is thought that lipid rafts enhance the efficiency of signal transduction by promoting the accumulation of signal transduction molecules such as Src and trimeric G proteins that are covalently attached to saturated fatty acids, and increasing the probability of their interaction. Thus, the heterogeneity of the signal transmission intensity in the cell membrane domains, due to the difference in the degree of integration or efficiency of assembly of signaling molecules, may also contribute to axis formation of cell polarity.
Our group addressed this issue by using a sphingomyelin-binding toxin, lysenin. Lysenin is a proteinous toxin derived from earthworms and recognizes the membrane region in which sphingomyelin accumulates at high density in the presence of cholesterol.7 We stained cultured epithelial cells using lysenin to visualize the distribution of sphingomyelin in epithelial cells. Interestingly, lysenin specifically stained the apical membrane of mouse cultured epithelial cells (EpH4 cells), suggesting that sphingomyelin is enriched in the apical membrane. On the other hand, when apical membrane and basolateral membrane were each physically isolated using colloidal silica particles and examined for differences in contents of phosphatidylcholine and sphingomyelin, we found that both apical membrane and basolateral membrane also contained larger amounts of sphingomyelin compared to phosphatidylcholine and that there is no difference in the ratio of sphingomyelin to phosphatidylcholine between these membranes.7
These seemingly contradictory results were later resolved by analyzing the apical membrane domains recognized by lysenin in more detail. Examining the membrane region recognized by lysenin in the apical membrane by immuno-electron microscopy revealed that lysenin selectively recognizes the filamentous-actin-based membrane protrusion of the apical membrane – microvilli (Fig. 1A).8 Thus, sphingomyelin is enriched within a uniquely defined membrane domain of the apical membrane. Intriguingly, when epithelial cells are treated with a sphingomyelin degrading enzyme from the apical side, the establishment of apico-basal polarity was not affected but formation of microvilli was markedly and specifically impaired.8 The accumulation of sphingomyelin in the microvilli was recently confirmed by using nanoscale secondary ion mass spectrometry (NanoSIMS) imaging.9
Figure 1.
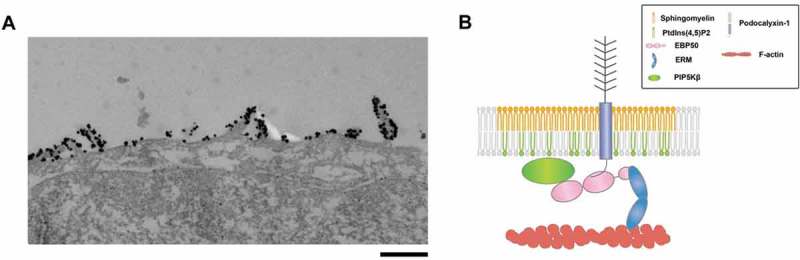
(A) Localization of sphingomyelin clusters was visualized with a sphingomyelin-binding toxin, lysenin. When cultured epithelial cells (EpH4 cells) were stained with green fluorescent protein (GFP)–lysenin, using anti-GFP antibody and the gold-conjugated secondary antibody, immunogold particles were present selectively at the microvilli in the apical membrane, suggesting that microvilli are specialized membrane structures where sphingomyelin clusters localize (Scale bar: 0.5µm). (B) PIP5Kbeta is recruited to the microvilli by its interaction with EBP-50, which in turn binds to podocalyxin-1 associated with sphingomyelin clusters.
Kai Simons and colleagues also conducted a detailed study on the lipid composition of apical membrane by using mass spectrometry.10 They reported that sphingomyelin is enriched in the apical membrane, compared to the whole cell lipids, by physically isolating apical membrane using nitrocellulose membrane. However, since they did not examine sphingomyelin content in the basolateral membrane, further analysis is needed to establish the relative enrichment of sphingomyelin in the epithelial cell plasma membrane.
Further complicating the issue is that enveloped virus choose specific membrane domains of plasma membrane when budding from the cells.11 For example, influenza virus emerges from the tip of microvilli.12 According to our findings, microvilli constitute a unique membrane domain containing higher amount of sphingomyelin in the apical membrane. Therefore, analysis of lipids derived from enveloped virus particles should be evaluated carefully since they may reflect lipid composition in a specialized membrane region, i.e. properties specific to the enveloped virus rather than general to the membrane region of interest.
Roles of phosphatidylinositol in the formation of membrane domains
Phosphatidylinositol is one of the most well-characterized phospholipid species among membrane lipids. Phosphatidylinositol can take a total of eight different phosphorylation states depending on the phosphorylation of the hydroxyl groups at the 3, 4, 5 position of the inositol ring. Among them, PtdIns(4,5)P2 and PtdIns(3,4,5)P3 are present mainly in the plasma membrane. PtdIns(4,5)P2 is involved in the regulation of actin cytoskeleton and in various phenomena such as endocytosis, cell movement and cell division.13
Pleckstrin homology domains (PH domains) of particular proteins have binding affinities to PtdIns(4,5)P2 or PtdIns(3,4,5)P3, which allows subcellular distribution of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 to be visualized by expressing PH domains fused with fluorescent proteins. In migrating cellular slime molds or neutrophils, PtdIns(3, 4, 5)P3 3 is distributed at the leading edge whereas PtdIns(4,5)P2 is accumulated at the rear end. This asymmetric distribution of phosphoinositide species is reported to be involved in the establishment of front-rear polarity.14
Keith Mostov and colleagues examined the distribution of these lipids in MDCK II cells and reported that PtdIns(4,5)P2 exists mainly in apical membrane whereas PtdIns(3,4,5)P3 distributed in basolateral membrane.15 Furthermore, when PtdIns(4,5)P2 is ectopically supplied to the basolateral membrane, apical marker membrane proteins are re-localized to the basolateral membrane.15 Thus, the asymmetric distribution of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 contributes to asymmetric distribution of membrane proteins in epithelial cells. Polarity proteins regulate the apical localization of the lipid metabolizing enzyme that dephosphorylates PtdIns(3,4,5)P3, PTEN, to drive enrichment of PtdIns(4,5)P2 in the apical membrane.15 However, it remains to be elucidated how the asymmetric distribution of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 regulates localization of membrane proteins in epithelial cells. Recently, it was reported that PtdIns(3,4,5)P3 of the lateral membrane is essential for recruiting β II-spectrin, an essential component of submembranous cytoskeletal networks, to the lateral membrane and is important for the formation of the lateral membrane.16
On the other hand, we showed that PtdIns(4,5)P2 is not distributed uniformly in apical membrane, but rather that PtdIns(4,5)P2 is concentrated in microvilli.8 That is, in the microvilli, sphingomyelin is highly accumulated in the outer layer of the cell membrane, and PtdIns(4,5)P2 is abundantly present on the inner layer side. So, how is such asymmetric distribution of phospholipids achieved? Our group searched for membrane proteins that co-localized with sphingomyelin in the microvilli and identified Podocalyxin-1. Podocalyxin-1 is a single-pass membrane protein that binds to a scaffold protein present in microvilli, EBP-50.
Interestingly, we found that Podocalyxin-1 has an intrinsic affinity to sphingomyelin-enriched membrane domain, irrespective of binding to EBP50. In addition, we found that EBP 50 directly binds to the type I phosphatidylinositol phosphate kinase isoform-beta (PIP5Kbeta) which is responsible for the production of PtdIns(4,5)P2 and forms a tripartite complex of Podocalyxin-1-EBP50-PIPK5Kbeta. Therefore, clustering of Podocalyxin-1-EBP50-PIPK5Kbeta occurs in the sphingomyelin enriched membrane domains, which results in the local accumulation of PtdIns(4,5)P2. We proposed that the local accumulation of PtdIns(4,5)P2 mediated by this protein complex promotes the activation of ERM family proteins and the formation of microvilli (Fig. 1B).8 Thus, we revealed that the two apically-enriched membrane lipids – sphingomyelin and PtdIns(4,5)P2 – are accumulated in the microvilli of the apical membrane and involved in the formation of microvilli.
Diffusion barrier of cell membrane
As described above, the epithelial cell apical membrane and the basolateral membrane have different membrane protein and lipid distribution. Given that membrane proteins and lipids freely diffuse in the plane of cell membranes, some barrier that restricts free diffusion of membrane proteins and lipids are required at the boundary between the apical membrane and basolateral membrane to maintain epithelial polarity. As such a molecular mechanism, it is assumed that one of the cell adhesion apparatus, tight junction, which localizes to the boundary between apical membrane and basolateral membrane, is essential for the compartmentalization and maintenance of apical and basolateral membrane domains.17 To examine whether tight junction functions as such a fence, we established cultured epithelial cells lacking tight junctions by knocking out the main scaffolding proteins of tight junction, ZO-1 and ZO-2.
The epithelial polarization was severely delayed in ZO-1 and ZO-2 deficient cells. However, adherens junctions were formed when cultured for a long time. Tight junctions are never formed because ZO-1 and ZO-2 are essential structural components of tight junctions.18 In ZO-1 ZO-2 double knockout cells, the cell adhesion molecules of tight junction, claudins, did not concentrate at the intercellular region and tight junctions were not formed between these cells when observed by the freeze fracture electron microscope. To our surprise – and contrary to the initial expectations – the epithelial cells that lack tight junction showed normal distribution of membrane proteins of apical membrane and basolateral membrane.19 Furthermore, even in the absence of tight junction, free diffusion of lipid molecules is inhibited between apical membrane and basolateral membrane, suggesting the existence of molecular mechanisms to prevent free diffusion of lipid molecules other than tight junction.7
In recent years, the existence of diffusion barrier against membrane proteins has been shown at the root of the primary cilia of apical membrane.20 At the base of primary cilia, ring of septin, a cytoskeletal protein, exists. Therefore, septin-based diffusion barrier may also work in the epithelial cells lacking tight junctions. Of note, it was recently reported that septins are localized at cell-cell junction in endothelial cells.21 At present, how septin ring works as diffusion barrier against free diffusion of membrane proteins and lipids remains unknown.20,22,23
Cell adhesion apparatus in the basolateral domains of epithelial cells
Finally, in this section, I introduce the findings about the roles of lipids in the formation of cell membrane structures involved in cell adhesion. In the basolatelral membrane domains, epithelial cells form membrane structures involved in the cell-cell or cell-matrix adhesion. Among cell adhesion structures, the roles of membrane lipids have been more intensively studied in the cell-matrix adhesion. The cell adhesion receptor responsible for the cell-matrix adhesion is integrin. First, cholesterol-rich membrane domain was reported to be involved in the clustering of integrins, which regfulates their activation.24 Cholesterol depletion significantly affected the formation of integrin-mediated cell adhesion.25 In parallel, activated integrins preferentially localize to cholesterol-rich membrane domains.26 Interestingly, cell detachment triggered internalization of plasma membrane cholesterol and lipid raft markers such as GPI anchored protein.27,28 Adhesion of cells to the matrix via integrins also facilitates the transport of cholesterol-enriched vesicles to the plasma membrane.28 Thus, integrin mediated cell-adhesion reciprocally alters the composition of the plasma membrane.
In addition to cholesterol, PtdIns(4,5)P2 is implicated in integrin-mediated signaling. PtdIns(4,5)P2 is an anionic lipid and can bind to the polybasic sequences present at the juxtamembrane domain of integrins, which help integrin cluster formation by neutralizing the charges.29 An integrin-associated actin-binding protein, Talin, was reported to bind to one isoform of PIP5Kgamma, PIPKIγ661.30 Therefore, Talin binding to integrins may stimulate the local production of PtdIns(4,5)P2 and the accumulation of PtdIns(4,5)P2 binding adhesion proteins such as vinculin, resulting in the formation of integrin clusters.
On the other hand, in lateral membranes, epithelial cells adhere to each other via cadherin-mediated adherens junction and claudin-mediated tight junction. Of note, cholesterol and PtdIns(4,5)P2 are also involved in the formation of these apical junctions. Similar to integrins, E-cadherin, claudin and occludin are concentrated in the detergents-insoluble fractions.31–33 Furthermore, when the amount of cholesterol at the plasma membrane was decreased, occludin and claudin could not localize to tight junction and remained internalized within the cell.33,34 In addition, claudins are known to undergo multiple posttranslational modifications such as palmitoylation, suggesting that tight junction is a membrane domain with lipid-raft like properties.35,36 Recently we demonstrated that cholesterol is accumulated at the apical junctions in epithelial cells.33 Interestingly, loss of adherens junction led to the reduction of cholesterol at the apical junction, which promotes endocytosis of claudin and inhibits tight junction formation.33
Several reports clearly indicate that PI(4,5)P2 is involved in the formation of adherens junction. E-cadherin binds to two distinct splice variants of PIP5Kgamma, PIPKIγ635 and PIPKIγ661.37 N-cadherin adhesions was shown to lead to local PI(4,5)P2 production, resulting in the actin assembly by uncapping gelsolin from the filament barbed ends.38
In addition to these membrane lipids, certain membrane lipid species may be involved in the formation of tight junction because tight junction is morphologically unique membrane domain of epithelial cells. Ultrathin electron microscopic analysis revealed that tight junctions form focal contacts between plasma membranes of neighboring cells; the focal contacts are called as “kissing points”. On freeze-fracture electron microscopy, tight junctions appear as a set of continuous, anastomosing strands (Fig. 2A). The unique ultrastructural features of tight junctions stimulated the discussion about the molecular models of tight junctions. Historically, two models of tight junction were proposed as the molecular models of tight junction (Fig. 2B). On the one hand, the “membrane protein model” proposed that tight junction is constructed by the interaction of membrane proteins like other cell adhesion apparatus. On the other hand, the “lipid model” proposed that tight junction is a non-bilayer membrane structure and the outer leaflet of plasma membrane is fused (hemi-fusion) at the membrane contacts.39 This membrane structure is called the inverted micelle structure. The formation of inverted micelle structure requires lipid molecules having a conical shape with a small polar head group and bulky fatty acid chains. Tight junction-like strands can be observed in the multi-lamellar membrane vesicles made from specific lipid mixture such as DOPE/DOPC/Cholesterol = 3:1:2 by freeze fracture microscopy.40 Thus, tight junction-like strands can be reconstituted solely by membrane lipids that assume inverted micellar configurations. Based on this finding, tight junction was thought to be local membranous subphases in the lipid bi-layer. In either theory, it was necessary to prove that a special membrane protein or specific lipid molecule was actually present in tight junctions in vivo and functional in the formation of tight junctions. At this point, it is widely accepted that claudin is a bona fide membrane protein of tight junction.41 However, the possibility that claudin forms a supramolecular complex with specific lipid species to form the tight junction is not completely excluded as discussed in recent reviews. 42,43
Figure 2.
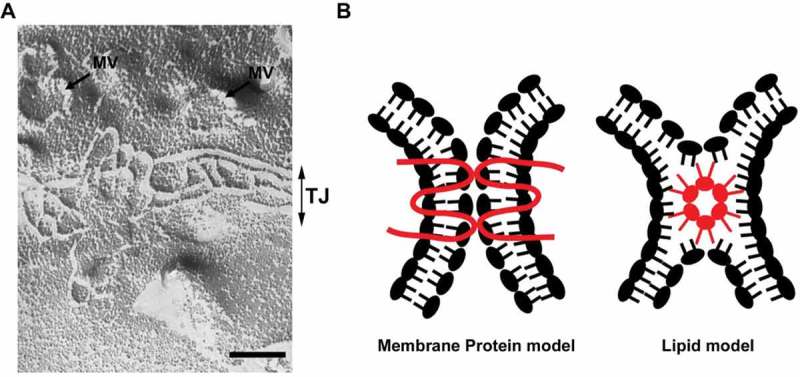
(A) A freeze fracture image showing tight junction strands in epithelial cells. MV: microvilli, TJ: tight junction. Scale bar, 200 nm. (B) Two models of tight junction. It is widely accepted that tight junctions are formed by the membrane protein, claudin (membrane protein model). The possibility that claudins or other tight junction membrane proteins help assembly and stabilization of a lipid-based strand structure is not completely denied (lipid model).
Recently, our group conducted a lipid analysis of the membrane fraction of tight junctions and found that sphingomyelin containing long fatty acid tail and cholesterol are enriched in the tight junction fraction.33 Therefore, tight junction is a clearly distinct membrane domain of epithelial cells with a unique lipid composition. In future studies, it will be necessary to elucidate how the distinct lipid species of tight junctions contribute to the formation of tight junctions.
Future direction
Examples of cell polarity are found in various cell types of diverse organisms. However, the mechanism by which cell membrane lipids plays a role in cell polarity as well as the mechanism that maintains the asymmetric distribution of cell membrane lipids is unknown in many cases. Furthermore, there are thousands of lipid species constituting the cell membrane. Although analytical methods for lipid composition using mass spectrometry have been established, the methodology for analyzing the lipid composition of specific cell membrane structures remains to be improved. In particular, tools to visualize the subcellular distribution of membrane lipids are currently limited. Therefore, elucidating the function of membrane lipids in the establishment of epithelial polarity and the associated formation of specific membrane structures awaits further development in this field.
Although analysis of the phenotype caused by knockout of lipid metabolizing enzymes may partly help our understanding about roles of downstream lipids, this approach will not provide an accurate picture of how lipids works in the cells. This is due to the fact that lipid species are globally and indiscriminately lost from the whole cell in such a scenario. For example, even if specific membrane structure is lost when a lipid metabolizing enzyme is knocked out, we cannot tell whether this lipid species is directly necessary for the formation of the structure itself or if its contribution is indirect, such as through the intracellular transport of membrane proteins.
In this regard, reconstituting specific cell membrane structures or polarity formation in vitro using giant liposomes with defined lipid composition will be important in future studies to clarify whether or not a specific lipid species is essential for the formation of specific membrane structure or establishment of polarity. Currently, it is technically difficult to reconstitute the cell membrane structure using complex constituents in liposomes, but this will be essential to advance our understanding of the regulatory roles of cell membrane lipids in cell polarity formation.
Disclosure statement
No potential conflict of interest was reported by the author.
Funding Statement
This work was supported by JSPS KAKENHI (JP16H04786 (J.I.), JP17H06012 (J.I.), JP16K14729 (J.I.), AMED-PRIME (15664862), grants from MSD Life Science Foundation and Takeda Science foundation. The author declares no competing financial interests.
Acknowledgments
I thank all members of the Ikenouchi laboratory (Department of Biology, Faculty of Sciences, Kyushu University) for helpful discussions. J. I. is grateful to Kenji Matsuzawa for helpful discussion in preparing this manuscript. J.I. also thank Leika Ikenouchi for encouragement throughout this work.
References
- 1.Chen J, Zhang M.. The Par3/Par6/aPKC complex and epithelial cell polarity. Exp Cell Res. 2013;319:1357–1364. doi: 10.1016/j.yexcr.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 2.Bi E, Park HO. Cell polarization and cytokinesis in budding yeast. Genetics. 2012;191:347–387. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Meer G, Simons K. Viruses budding from either the apical or the basolateral plasma membrane domain of MDCK cells have unique phospholipid compositions. EMBO J. 1982;1:847–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita T, Fujita M. Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling. J Lipid Res. 2016;57:6–24. doi: 10.1194/jlr.R063313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuck S, Simons K. Polarized sorting in epithelial cells: raft clustering and the biogenesis of the apical membrane. J Cell Sci. 2004;117:5955–5964. doi: 10.1242/jcs.01596. [DOI] [PubMed] [Google Scholar]
- 7.Ikenouchi J, Suzuki M, Umeda K, Ikeda K, Taguchi R, Kobayashi T, et al. Lipid polarity is maintained in absence of tight junctions. J Biol Chem. 2012;287:9525–9533. doi: 10.1074/jbc.M111.327064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikenouchi J, Hirata M, Yonemura S, Umeda M. Sphingomyelin clustering is essential for the formation of microvilli. J Cell Sci. 2013;126:3585–3592. doi: 10.1242/jcs.122325. [DOI] [PubMed] [Google Scholar]
- 9.He C, Hu X, Jung RS, Weston TA, Sandoval NP, Tontonoz P, et al. High-resolution imaging and quantification of plasma membrane cholesterol by NanoSIMS. Proc Natl Acad Sci U S A. 2017;114:2000–2005. doi: 10.1073/pnas.1621432114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerl MJ, Sampaio JL, Urban S, Kalvodova L, Verbavatz JM, Binnington B, et al. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J Cell Biol. 2012;196:213–221. doi: 10.1083/jcb.201108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nayak DP, Hui EK. The role of lipid microdomains in virus biology. Subcell Biochem. 2004;37:443–491. [DOI] [PubMed] [Google Scholar]
- 12.Kolesnikova L, Heck S, Matrosovich T, Klenk HD, Becker S, Matrosovich M. Influenza virus budding from the tips of cellular microvilli in differentiated human airway epithelial cells. J Gen Virol. 2013;94:971–976. doi: 10.1099/vir.0.049239-0. [DOI] [PubMed] [Google Scholar]
- 13.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 14.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. [DOI] [PubMed] [Google Scholar]
- 15.Martin-Belmonte F, Gassama A, Datta A, Yu W, Rescher U, Gerke V, et al. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell. 2007;128:383–397. doi: 10.1016/j.cell.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He M, Abdi KM, Bennett V. Ankyrin-G palmitoylation and betaII-spectrin binding to phosphoinositide lipids drive lateral membrane assembly. J Cell Biol. 2014;206:273–288. doi: 10.1083/jcb.201401016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trimble WS, Grinstein S. Barriers to the free diffusion of proteins and lipids in the plasma membrane. J Cell Biol. 2015;208:259–271. doi: 10.1083/jcb.201410071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikenouchi J, Umeda K, Tsukita S, Furuse M. Requirement of ZO-1 for the formation of belt-like adherens junctions during epithelial cell polarization. J Cell Biol. 2007;176:779–786. doi: 10.1083/jcb.200612080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, et al. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126:741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 20.Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, et al. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329:436–439. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, Cooper JA. Septins regulate junctional integrity of endothelial monolayers. Mol Biol Cell. 2018;163:265-272. doi: 10.1093/jb/mvx077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palander O, El-Zeiry M, Trimble WS. Uncovering the roles of septins in cilia. Front Cell Dev Biol. 2017;5:36. doi: 10.3389/fcell.2017.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krapf D. Compartmentalization of the plasma membrane. Curr Opin Cell Biol. 2018;53:15–21. doi: 10.1016/j.ceb.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Van Zanten TS, Cambi A, Koopman M, Joosten B, Figdor CG, Garcia-Parajo MF. Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc Natl Acad Sci U S A. 2009;106:18557–18562. doi: 10.1073/pnas.0905217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leitinger B, Hogg N. The involvement of lipid rafts in the regulation of integrin function. J Cell Sci. 2002;115:963–972. [DOI] [PubMed] [Google Scholar]
- 26.Decker L. ffrench-Constant C. Lipid rafts and integrin activation regulate oligodendrocyte survival. J Neurosci. 2004;24:3816–3825. doi: 10.1523/JNEUROSCI.5725-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Del Pozo MA, Alderson NB, Kiosses WB, Chiang HH, Anderson RG, Schwartz MA. Integrins regulate Rac targeting by internalization of membrane domains. Science. 2004;303:839–842. doi: 10.1126/science.1092571. [DOI] [PubMed] [Google Scholar]
- 28.Norambuena A, Schwartz MA. Effects of integrin-mediated cell adhesion on plasma membrane lipid raft components and signaling. Mol Biol Cell. 2011;22:3456–3464. doi: 10.1091/mbc.E11-04-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore DT, Nygren P, Jo H, Boesze-Battaglia K, Bennett JS, DeGrado WF. Affinity of talin-1 for the beta3-integrin cytosolic domain is modulated by its phospholipid bilayer environment. Proc Natl Acad Sci U S A. 2012;109:793–798. doi: 10.1073/pnas.1117220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling K, Doughman RL, Firestone AJ, Bunce MW, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- 31.Hinck L, Nathke IS, Papkoff J, Nelson WJ. Dynamics of cadherin/catenin complex formation: novel protein interactions and pathways of complex assembly. J Cell Biol. 1994;125:1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nusrat A, Parkos CA, Verkade P, Foley CS, Liang TW, Innis-Whitehouse W, et al. Tight junctions are membrane microdomains. J Cell Sci. 2000;113(Pt 10):1771–1781. [DOI] [PubMed] [Google Scholar]
- 33.Shigetomi K, Ono Y, Inai T, Ikenouchi J. Adherens junctions influence tight junction formation via changes in membrane lipid composition. J Cell Biol. 2018;217:2373–2381. doi: 10.1083/jcb.201711042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Francis SA, Kelly JM, McCormack J, Rogers RA, Lai J, Schneeberger EE, et al. Rapid reduction of MDCK cell cholesterol by methyl-beta-cyclodextrin alters steady state transepithelial electrical resistance. Eur J Cell Biol. 1999;78:473–484. [DOI] [PubMed] [Google Scholar]
- 35.Van Itallie CM, Gambling TM, Carson JL, Anderson JM. Palmitoylation of claudins is required for efficient tight-junction localization. J Cell Sci. 2005;118:1427–1436. doi: 10.1242/jcs.01735. [DOI] [PubMed] [Google Scholar]
- 36.Shigetomi K, Ikenouchi J. Regulation of the epithelial barrier by post-translational modifications of tight junction membrane proteins. J Biochem.2017. [DOI] [PubMed] [Google Scholar]
- 37.Ling K, Bairstow SF, Carbonara C, Turbin DA, Huntsman DG, Anderson RA. Type I gamma phosphatidylinositol phosphate kinase modulates adherens junction and E-cadherin trafficking via a direct interaction with mu 1B adaptin. J Cell Biol. 2007;176:343–353. doi: 10.1083/jcb.200606023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Sayegh TY, Arora PD, Ling K, Laschinger C, Janmey PA, Anderson RA, et al. Phosphatidylinositol-4,5 bisphosphate produced by PIP5KIgamma regulates gelsolin, actin assembly, and adhesion strength of N-cadherin junctions. Mol Biol Cell. 2007;18:3026–3038. doi: 10.1091/mbc.e06-12-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinto Da Silva P, Kachar B. On tight-junction structure. Cell. 1982;28:441–450. [DOI] [PubMed] [Google Scholar]
- 40.Van Venetie R, Verkleij AJ. Analysis of the hexagonal II phase and its relations to lipidic particles and the lamellar phase. A freeze-fracture study. Biochim Biophys Acta. 1981;645:262–269. [DOI] [PubMed] [Google Scholar]
- 41.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 42.Lee DB, Jamgotchian N, Allen SG, Abeles MB, Ward HJ. A lipid-protein hybrid model for tight junction. Am J Physiol Renal Physiol. 2008;295:F1601–12. doi: 10.1152/ajprenal.00097.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lingaraju A, Long TM, Wang Y, Austin JR 2nd, Turner JR. Conceptual barriers to understanding physical barriers. Semin Cell Dev Biol. 2015;42:13–21. doi: 10.1016/j.semcdb.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]


