Summary
The farnesoid X receptor (FXR) functions as a bile acid (BA) sensor coordinating cholesterol metabolism, lipid homeostasis, and absorption of dietary fats and vitamins. However, BAs are poor reagents for characterizing FXR functions due to multiple receptor independent properties. Accordingly, using combinatorial chemistry we evolved a small molecule agonist termed fexaramine with 100-fold increased affinity relative to natural compounds. Gene-profiling experiments conducted in hepatocytes with FXR-specific fexaramine versus the primary BA chenodeoxycholic acid (CDCA) produced remarkably distinct genomic targets. Highly diffracting cocrystals (1.78Å) of fexaramine bound to the ligand binding domain of FXR revealed the agonist sequestered in a 726Å3 hydrophobic cavity and suggest mechanistic basis for the initial step in the BA signaling pathway. The discovery of fexaramine will allow us to unravel the FXR genetic network from the BA network and selectively manipulate components of the cholesterol pathway that may be useful in treating cholesterol-related human diseases.
Introduction
In vertebrates, the liver and the intestine maintain lipid homeostasis by regulating acquisition, synthesis, and metabolism of cholesterol (Chawla et al., 2000). Excess dietary cholesterol is either converted into bile acids in the liver or undergoes biliary excretion into the intestine (Chiang, 2002). The nuclear hormone receptor (NHR) farnesoid X receptor (FXR, NRIH4) has been implicated in the regulation of both of these metabolic processes.FXR is expressed in the liver and intestine as well as other cholesterol-rich tissues such as the adrenal gland.Knockout mice deficient in FXR expression display de fects in bile acid (BA) homeostasis when exposed to dietary stresses, including elevated serum BA, reduced bile acid pools, and reduced fecal BA secretion (Sinal et al., 2000). In the liver, the rate-limiting step for the conversion of excess cholesterol into BAs is catalyzed by the cytochrome p450 enzyme cholesterol 7-α hydroxylase (CYP7A1). A second enzyme, sterol 12-α hydroxylase (CYP8B), is key for regulating the ratio of cholic acid (CA) to chenodeoxycholic acid (CDCA) dur ing BA biosynthesis (Kerr et al., 2002; Wang et al., 2002;Edwards et al., 2002). In mammals, expression of these genes is indirectly regulated by FXR via the NHR homo log SHP (Lu et al., 2000; Goodwin et al., 2000). Physiolog ical concentrations of specific BAs bind and activate FXR, the most potent being CDCA, a major primary BA found in human bile (Makishima et al., 1999; Parks et al., 1999; Wang et al., 1999). Activation enables FXR to act as a transcriptional sensor for BAs, in directly repressing the transcription of both CYP7A and CYP8B genes by increasing the levels of the inhibitory nuclear receptor SHP. The ability of SHP to bind and inhibit the liver receptor homolog (LRH-1), a NHR required for CYP7A gene expression, allows FXR activation to exert a negative influence on cholesterol metabolism (Lu et al., 2000; Goodwin et al., 2000).
FXR belongs to a family of ligand-inducible transcrip tion factors whose members share two structurally con served domains: a central DNA binding domain that targets the receptor to specific DNA sequences, and a ligand binding domain (LBD) that binds small lipophilic hormones (Evans, 1988). The LBD functions as the mo lecular switch. Binding of the appropriate hormone to the LBD brings about conformational changes that re sult in the release of bound corepressor proteins and the recruitment of coactivator proteins to the site of DNA binding culminating in the transcription of target genes.The regulation of NHR transcription factors by small lipophilic hormones makes this family of transcription factors ideal targets for the design and synthesis of small molecule probes (Blumberg and Evans, 1998).
The current hypothesis that FXR senses BA levels and mediates the transcriptional repression of genes responsible for the conversion of excess cholesterol into BAs as well as the induction of genes necessary for BA transport makes FXR an attractive pharmacological target. The availability of high-affinity synthetic agonists for FXR is a critical step required for the validation of FXR as a drug target and the further elaboration of the physiological functions of FXR. Here we describe the discovery and structure/function characterization of a FXR agonist termed fexaramine, which is structurally distinct from natural BA ligands, and a synthetic ligand GW4064 (Maloney et al., 2000). Multiple mRNA expression experiments using high-density oligonucleotide arrays with the three currently available and chemically distinct classes of FXR agonists led to the elucidation of several gene targets in the liver. Surprisingly, cluster analysis revealed that CDCA, fexaramine, and GW4064 have distinct regulating profiles. In addition, the high-resolution crystal structure of active FXR bound to fexar amine was determined, thus providing an important chemo-architectural foundation of this receptor. This three-dimensional template also allowed us to model the interaction of BAs with FXR, providing a molecular explanation of how they interact.
Results
Identification and Development of Small Molecule Ligands for FXR
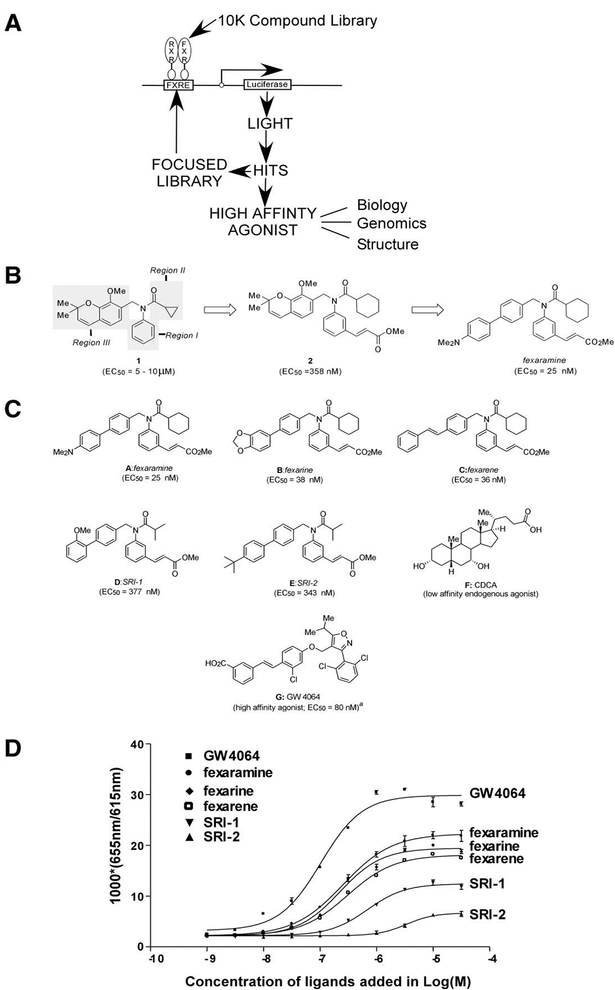
Chemical probes are especially powerful tools for dissecting protein structure and function. As a first step to facilitate a chemical genetic approach to FXR function, a high throughput screen was initiated for a synthetic agonist (Figure 1A). In a 384-well format, cells cotrans fected with FXR and RXR expression vectors and cognate reporter were screened against a recently constructed combinatorial library of ≈10,000 benzopyran-based compounds (Nicolaou et al., 2000a, 2000b, 2000c). The reporter vector contains a hormone response element controlling activation of a minimal eukaryotic promoter driving expression of a luciferase reporter gene. The initial screen identified several candidate compounds, possessing EC50 values ranging from 5–10 μM and whose prototypical structure (1) is shown in Figure 1B. Candidate compounds were retested and checked for crossreactivity for the retinoid X receptor (RXR). None of the identified compounds had the ability to bind to or activate RXR. Systematic optimization of regions I and II of the prototypical structure through multiple rounds of screening using smaller focused chemical li braries defined the requisite features of these domains for high-affinity binding to FXR. Specifically, we found that incorporation of the 3-methylcinnamate moiety in region I and the cyclohexyl amide unit in region II re sulted in a more than 10-fold enhancement in cellular potency, as demonstrated by compound 2 (EC50= 358 nM) (Figure 1B). Further exploration of region III through replacement of the parent benzopyran unit with styrenyl and biaryl moieties (2) yielded compounds with even higher potency (Nicolaou et al., 2003). This intelligence gathering facilitated the rational design of a focused library centered on region III of 94 new compounds synthesized on a solid support (Figure 2). Screening of this targeted library led to the discovery of several highly potent ligands including: A (coined fexaramine, EC50 = 25 nM), B (coined fexarine, EC50 = 38 nM), and C (coined fexarene, EC50 [H11005] 36 nM), and many specific compounds with lower potency including D (coined SRI-1, EC50 = 377 nM) and E (coined SRI-2, EC50 = 343 nM) (Figure 1C).Most notably, these compounds possess structurally distinct chemical scaffolds compared to any known nat ural and synthetic ligands for FXR, the BA chenodeoxycholic acid (CDCA) ,and GW4064 (Figure 1C, F and G).We found GW4064 to have EC50 values of [H11015]90 nM, comparable to the reported value. The compounds shown in Figure 1C (A, B, C, D, and E) were utilized for the biological studies described below. Information per taining to the chemical synthesis and screening of the library will be discussed elsewhere.
Figure 1. Chemical Structures of Identified and Known FXR Ligands and Their In Vitro Binding Affinities.
(A) A schematic diagram of the ligand discovery phase and characterization scheme of identified high-affinity agonists. (B) Selected regions of interest for SAR evaluation of prototypical structure lead compounds 1. Region I, right-hand aromatic system; Region II, Acyl group region; Region III, left-hand benzopyran ring system. Compound 2 was produced by systematic optimization of Regions I and II. Fexaramine was selected from a final 94 membered combinatorial library of Region III. (C) Structures of lead compounds (and their EC50 values in a cell-based assay) selected for further biological evaluation as FXR agonists. A, fexaramine (EC50 = 25 nM); B, fexarine (EC50 = 38 nM); C, fexarene (EC50 = 36 nM); D, SRI-1 (EC50 = 377 nM); E, SRI-2 (EC50 = 343 nM). The identified compounds are structurally distinct from known FXR agonists F, CDCA a physiological low-affinity ligand, and the high-affinity ligand G, GW4064 (EC50 = 80 nM). (D) Identified compounds fexaramine, fexarine, fexarene, SRI-1, and SRI-2 are agonists for FXR in vitro. A FRET-based ligand binding assay was carried out in agonist mode using GW4064 as the control ligand. Increasing amounts of the compounds were added as indicated. Binding reactions contained 8 nM europium-labeled GST-FXR-LBD fusion protein and 16 nM allophycocyanin-labeled SRC-1 receptor binding peptide. Results are expressed at 1000*(665 nm/615 nm).
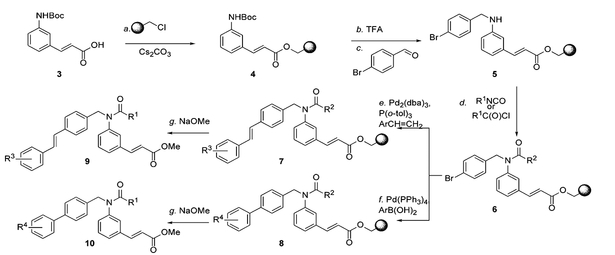
Figure 2. Solid Phase Synthesis of a 94 Membered Focused Library of Biaryl and Stilbene Cinnamates.
Reagents and conditions: (a) 2.0 equiv of 3, 1.0 equiv of Merrifield Resin (0.91 mmol/g), 2.0 equiv of Cs2CO3, 0.5 equiv of TBAI, DMF, 55°C, 24 hr; (b) 20% TFA in CH2Cl2, 25°C, 1 hr; (c) 10.0 equiv of 4-bromobenzaldehyde, 0.05 equiv of AcOH, THF:MeOH (2:1), 25°C, 1 hr; then, 8.0 equiv of NaCNBH3, THF:MeOH (2:1), 25°C, 2 hr; (d) for R1COCl: 30.0 equiv of R1COCl, 40.0 equiv of Et3N, 1.0 equiv of 4-DMAP, CH2Cl2, 25°C, 12 hr; for R1NCO: 30.0 equiv of R1NCO, 40.0 equiv of Et3N, 1.0 equiv of 4-DMAP, DMF, 65°C, 60 hr; (e) 8.0 equiv of styrene, 10.0 equiv of Et3N, 0.5 equiv of Pd2(dba)3, 1.5 equiv of P(o-tol)3, DMF, 90°C, 48 hr; (f) 5.0 equiv of boronic acid, 3.0 equiv Cs2CO3, 0.5 equiv of Pd(PPh3)4, DMF, 90°C, 24 hr; (g)10.0 equiv of NaOMe, Et2O:MeOH (10:1), 25°C, 20 min. AcOH, acetic acid; 4-DMAP, 4-dimethylaminopyridine; DMF, N,N-dimethylformamide; Et, ethyl; Me, methyl; Pd(PPh3)4, tetrakis(triphenylphosphine)palladium(0); Pd2(dba)3, tris(dibezylideneacetone)dipalladium(0); P(o-tol)3, tri-o-tolylphosphine; TBAI, tetrabutylammonium iodide; TEA, triethylamine; TFA, trifluoroacetic acid; THF, tetrahydrofuran.
Compounds Identified Activate FXR in Both In Vitro and In Vivo Assays
Nuclear receptors respond to agonist by recruiting transcriptional activators to an allosteric sensitive site in the LBD. To determine whether the “fexa-class” of compounds could promote the association of FXR with co- activators in vitro, we employed the fluorescence resonance energy transfer (FRET)-based coactivator binding assay (Makishima et al., 1999; Urizar et al., 2002). This assay relies on an agonist-induced interaction between the receptor and its coactivator. The fluorescent tagging of both the receptor and a peptide containing the recep tor binding domain of the steroid receptor coactivator SRC-1 (LXXLL) allows the measurement of agonist- induced receptor-peptide interaction by FRET. Recruitment of the SRC-1 peptide to the FXR LBD was only observed in the presence of the agonists fexaramine, fexarine, fexarene, SRI-1, SRI-2, and GW4064 (Figure 1D). GW4064 demonstrated the strongest FRET signal producing an EC50 value of 100 nM followed by fexara- mine (EC50 = 255 nM), fexarine (EC50 = 222 nM), and fexarene (EC50 = 255 nM). Weaker signals were seen with lead compounds SRI-1 and SRI-2.
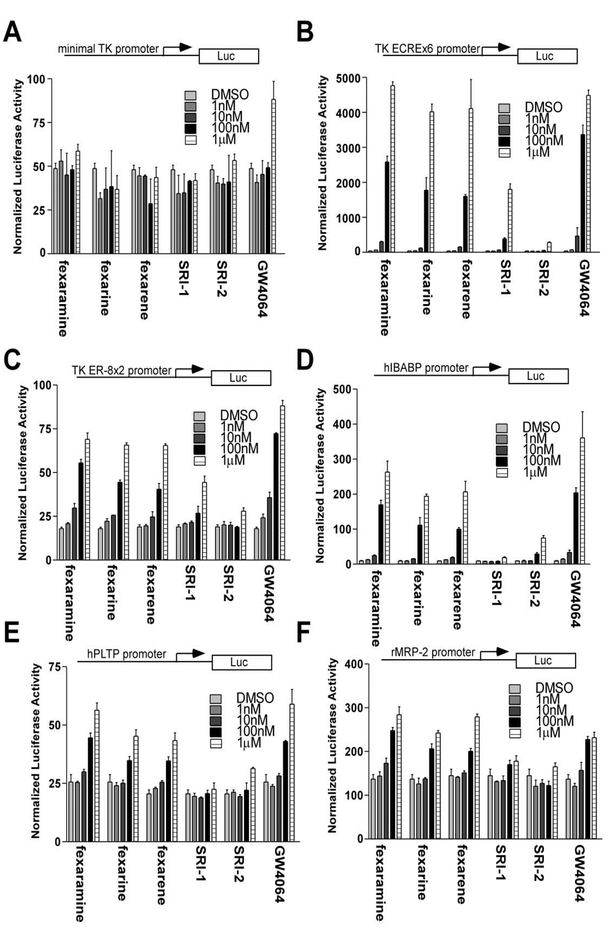
The functional properties of each of these compounds to activate FXR in six different cell-based reporter assays were explored. The recently described synthetic compound GW4064 was used as a control in these ex periments. CV-1 cells were transiently transfected with an expression plasmid for murine FXR and human RXR with a thymidine kinase (TK) minimal promoter reporter vector containing either no copies (Figure 3A) or six copies (Figure 3B) of the ecdysone response element (ECRE). ECRE is a well-characterized FXR response ele ment (FXRE) (Blumberg et al., 1998). In addition, two copies of the recently identified FXRE everted repeat separated by 8 nucleotides (ER-8) (Figure 3C) were also analyzed (Kast et al., 2002). The transfected cells were then treated with increasing concentrations of fexara-mine, fexarine, fexarene, SRI-1, SRI-2, or GW4064. The results depicted in Figures 3B and3C show that fexara- mine, fexarine, fexarene, and GW4064 elicit robust activation of both of the FXREs (ECRE, 100-fold; ER-8, 4-fold) with a maximal activity in each case achieved at 1 μM. Compounds SRI-1 and SRI-2, although structurally similar to fexaramine, showed substantially reduced activity. While the fexa-compounds showed no activity when using the minimal TK promoter lacking FXREs, GW4064 displayed a weak but reproducible activation (2-fold) (Figure 3A). These studies were repeated in liver (HEPG2) and kidney (HEK 293) cell lines with similar results (data not shown).
Figure 3. Characterization of Identified FXR Ligands in Cell-Based Assays.
CV-1 cells were cotransfected with pCMX-mFXR and pCMX-hRXR and a reporter gene containing the (A) minimal TK promoter, (B) TK-ECREx6 promoter, (C) TK-ER8×2 promoter, (D) hI-BABP promoter, (E) hPLTP promoter, or (F) rMRP-2 promoter. Increasing amounts (1 nM to 1 μM) of the compounds fexaramine, fexarine, fexarene, SRI-1, SRI-2, and GW4064 were added to the cells 24 hr posttransfection. Activation of the luciferase reporter gene was measured in relative light units with β-galactosidase activity as a control for transfection efficiency and presented as normalized luciferase units. Ligand response data were derived from triplicate points and two independent experiments and presented as the mean ±SE; n = 6.
In addition to synthetic reporters we examined the ability of fexa-compounds to activate physiological promoters of known FXR target genes in a transient trans fection cell-based assay. Three promoters, including I-BABP (Grober et al., 1999), phospholipid transfer pro tein (PLTP) (Laffitte et al., 2000; Urizar et al., 2000), and multidrug resistance-related protein 2 (MRP-2) genes (Kast et al., 2002), were tested. The I-BABP and PLTP promoters contain one copy of an inverted repeat se quence AGGTCA with a one base spacing (IR-1) while MRP-2 contains an ER-8 element. The results obtained and shown in Figures 3D (hI-BABP promoter), 2E (hPLTP promoter), and 2F (rMRP-2 promoter) were similar to the previously described experiments carried out with multiple FXRE copies. Again, a dose-dependent maxi mum efficacy of the fexaramine, fexarine, fexarene, and GW4064 compounds was observed at 1 μM concentra tion while SRI-1 and SRI-2 showed minimal activity. The most robust activation (28-fold) was seen on the I-BABP promoter with only modest (2- to 3-fold) induction of the PLTP and MRP-2 promoters.
Crossreactivity with Other Nuclear Receptors
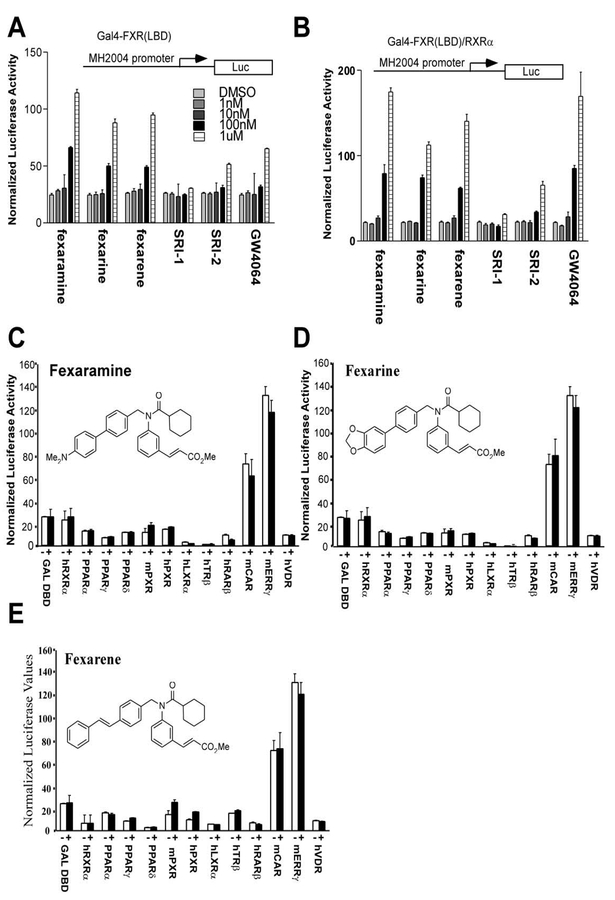
Cell-based transcriptional activation assays using chi meric NHR constructs were employed to measure the selectivity of compounds for FXR’s LBD relative to other NHRs (Forman et al., 1995). In these assays the yeast GAL4 DBD is connected to the LBD of the respectiveNHR. These activator constructs were cotransfected into cells with a thymidine kinase (TK) minimal promoter reporter vector containing four copies of the GAL4 binding site. The transiently transfected cells were then ti trated with the specified small molecule regulators of FXR. In Figures 4A and4B we show that fexaramine, fexarine, fexarene, and GW4064 all activate the chimeric FXR construct in the presence and absence of RXR. Interestingly, fexaramine, fexarine, and fexarene are more efficacious ligands for FXR than GW4064 in the absence of RXR, suggesting interesting mechanistic differences between the modes of activation of the two chemically distinct classes of compounds. Addition of RXR had no effect on the activation potential of fexara- mine, fexarine, and fexarene in this assay. Compounds SRI-1 and SRI-2 again showed little or no activity consis tent with all of the previous results. In terms of potential crossreactivity, fexaramine, fexarine, and fexarene were highly selective for FXR. Notably, no transcriptional ac tivity was observed when using other chimeric NHR con structs including hRXRα, hPPARαγδ, mPXR, hPXR, hLXRα, hTRβ, hRARβ, mCAR, mERRγ, and hVDR (Figures 4C, 4D, and 4E).
Figure 4. Crossreactivity Studies of the Identified FXR Ligands with Other Nuclear Hormone Receptors.
CV-1 cells were cotransfected with a reporter gene containing the (A) MH2004 promoter that contains four GAL4 binding sites with pCMXGAL4-FXR LBD chimeric expression construct or (B) MH2004 promoter with pCMXGAL4-FXR LBD/RXRα constructs and treated with increasing amounts of the compounds fexaramine, fexarine, fexarene, SRI-1, SRI-2, and GW4064. (C, D, and E) CV-1 cells were transiently transfected with the indicated plasmids and treated with either DMSO or 10 μM of the compounds fexaramine, fexarine, and fexarene. Reporter activity was normalized to the internal control, and the data were plotted as fold activation relative to untreated cells. All transfections contained CMX-gal as an internal control.
Induction of Endogenous FXR Target Genes by the Identified Compounds in Colon and Liver Cell Lines
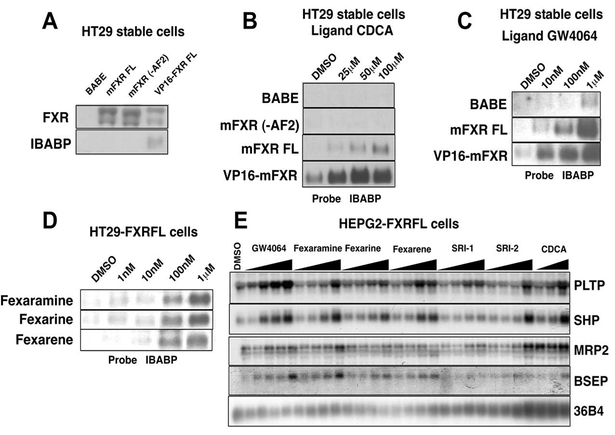
The liver and intestinal organ systems are major sites of FXR regulation in response to physiological BA pro duction. To determine whether the receptor and the fexa-compounds can regulate an endogenous genetic network, it was first necessary to establish relevant cell lines. Accordingly, we infected human colon cells HT29 (FXR null until differentiated) with a retroviral vector that expresses either FXR and the puromycin-resistant gene or the puromycin-resistant gene alone (pBABE). Puro mycin-resistant cells were isolated, and pooled cell populations were propagated that harbored either the vector alone (HT29-BABE), overexpressed FXR full-length (HT29-FXRFL), a nonfunctional FXR truncated at the AF2 region (HT29-FXR–AF2), or a constitutively active FXR that has the VP16 activation domain fused to the N terminus of the protein (HT29-VP16-FXR). The successful establishment of the various stable cell lines was verified via Northern blot analysis of FXR message levels in the rescued cell lines (Figure 5A). As expected, HT29-BABE control lines show no FXR mRNA expression.
Figure 5. Gene Induction of Identified FXR Targets by Distinct Fexa-Compounds.
(A) Total RNA (20 μg) isolated from HT29 stable cells was used for Northern blot analysis. cDNA probes for mFXR and hI-BABP were hybridized to the blot. Blots were normalized to β-actin expression levels. (B and C) HT29 stable cells were then treated overnight with increasing amounts of CDCA (B) and GW4064 (C) as indicated. Total RNA (20 μg) from treated cells was used for Northern blot analysis. cDNA probes for hI-BABP were prepared and hybridized to the blot. Blots were normalized to β -actin. (D) HT29-FXRFL stable cells were cultured until confluence. Cells were then treated overnight with increasing amounts of fexaramine, fexarine, or fexarene as indicated. Total RNA (20 μg) was then isolated using Trizol and used for Northern blot analysis. cDNA probe for human I-BABP was prepared and hybridized to the blot. Blots were normalized to β -actin. (E) HEPG2-FXRFL stable cells were cultured until confluence. Cells were then treated with increasing amounts of fexaramine, fexarine, fexarene, SRI-1, SRI-2, GW4064 (10 nM, 100 nM, 1 μM, 10 μM), and CDCA (1 μM, 10 μM, 100 μM). Twenty micrograms total RNA was then isolated using Trizol and was used for Northern blot analysis. cDNA probes for human PLTP, SHP, MRP-2, and BSEP were prepared and hybridized to the blot. Blots were normalized to 36B4 expression.
The ability of FXR stable cell lines to induce target genes was assessed by isolating total RNA from cells treated overnight with increasing amounts of CDCA or GW4064. Northern blot analysis of the HT29-FXRFL cell line showed robust, concentration-dependent induction of I-BABP mRNA by both CDCA and GW4064 (Figures 5B and 5C). Maximal activation of the I-BABP gene by CDCA was observed at 100 μM while only 1 μM of GW4064 was needed to achieve the same level of induction. No induction of I-BABP mRNA levels was observed in control HT29-BABE or HT29-FXR–AF2 cell lines. As expected, constitutive expression was seen in the HT29-VP16-FXR cell line and was superinduced by the addition of either CDCA or GW4064. Next, total RNA fromHT29 stable cells treated overnight with fexaramine, fexarine, or fexarene was probed for I-BABP gene expres sion (Figure 5D). All induced expression of the I-BABP mRNA in the HT29-FXRFL with similar profiles to GW4064. These observations verify the utility of generating colon cell model system for studying FXR target genes.
In addition to the intestinal cells we have in parallel developed a model hepatocyte cell system that stablyexpresses the FXR gene (Laffitte et al., 2000; Kast et al., 2002). As above, confluent HEPG2-FXR cells were treated overnight with increasing concentrations of fex aramine, fexarine, fexarene, SRI-1, SRI-2, or the control ligands GW4064 and CDCA. Total RNA was isolated, and the expression of the FXR target genes SHP, MRP-2,BSEP, and PLTP was measured by Northern blot analysis (Figure 5E). The control ligands CDCA and GW4064 showed similar patterns of induction to what has been previously reported. Of our fexa-compounds, fexara- mine was the most effective inducer of target genes, although strong and comparative inductions were also observed with fexarine and fexarene. Interestingly, al though GW4064 showed slightly better induction of the FXR target genes PLTP and SHP, fexaramine-matched GW4064 induced activation of the BSEP and MRP-2 genes. Interestingly, even the weak compound SRI-1 displayed remarkably effective induction of PLTP al though it weakened on other genes. These results dem onstrate that the fexa-compounds can be used to iden tify FXR-dependent target genes in liver and intestinal cell lines. The somewhat ([H11015]10[H11003]) reduced sensitivity of the hepatic cells may reflect the ability of the liver hepa tocytes to mount a xenobiotic response or may be af fected by cell-specific permeability characteristics of the compounds. For these reasons a multiplicity of in ducers can be valuable in determining the FXR regula tory network.
Gene Profiling of FXR Agonist in Primary Hepatocyte Cells
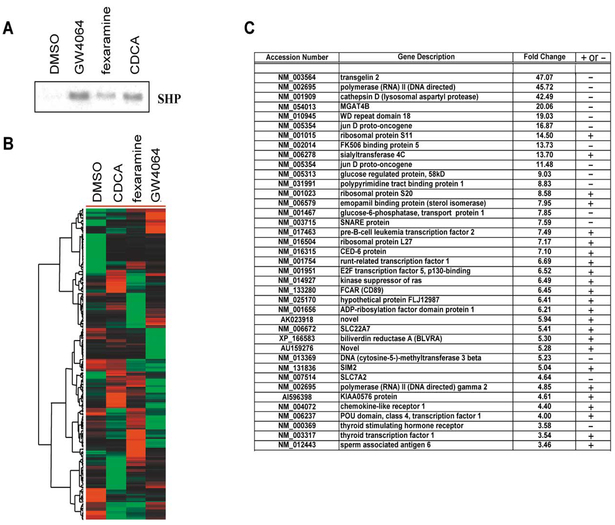
Having established fexaramine as a potent FXR-specific agonist in both cell culture systems, we next compared fexaramine’s gene activation profile with CDCA and GW4064 in human primary hepatocytes. Hepatocytes were treated with either DMSO (control group), fexara-mine (10 μM), CDCA (100 μM), or GW4064 (10 μM), and total RNA was isolated at 6 and 12 hr time points. Prior to gene profiling experiments, the samples were verified by Northern blot analysis for induction of SHP, a known FXR target gene (Figure 6A). Subsequently, biotinylated cRNAs prepared from mRNA samples were independently labeled and hybridized to duplicate sets of high- density microarrays (U-133A set, Affymetrix, Palo Alto, CA). A total of 222 transcripts were identified whose expression changed relative to the DMSO control using a paired Student’s t test and each of the three agonists.These genes were then subjected to hierarchal clustering and visualized (Cluster and Treeview, Mike Eisen).The most surprising observation was the distinct expression profiles of the ≈30,000 genes seen using the chemically distinct FXR agonists (Figure 6B). Indeed, relatively few genes were observed whose expression profiles changed in a similar fashion using all three agonists. As mentioned above, this may be due in part to CDCA as a physiological BA, exerting a multiplicity of effects via non-FXR pathways. For example, the recent knockout of SHP reveals that BAs act through at least two pathways to mediate repression of the CYP7A en zyme. In addition, a small subset of genes (Figure 6C) exhibited 3-fold changes in expression when using any of the three FXR ligands (genes below the 3-fold cutoff like SHP [2-fold] are not documented but have being actively investigated). This list suggests additional roles for FXR in the bilirubin biosynthetic pathway (BLVRA, 5-fold), thyroid metabolism (TSHR, 3-fold; thyroid tran scription factor 1, 3-fold), and amino acid transport (SCL7A2, 4-fold). Confirmations of gene induction by FXR agonists of many of the target genes reported in this list were done by Northern blot analysis and will be the subject of a more detailed paper.
Figure 6. Gene Array Studies with FXR Ligands Fexaramine, GW4064, and CDCA.
(A) Primary mouse hepatocytes were treated for 6 or 12 hr with either vehicle alone or 100 μM CDCA, 10 μM fexaramine, or 10 μM GW4064, as indicated. Total RNA was isolated using Trizol, and 10 μg total RNA was used for Northern blot analysis. The human SHP probe was prepared and hybridized to the blot. To ensure constant loading of total RNA to the blot, GAPDH was also hybridized as a control (data not shown). (B) Clustergram of genes whose expression pattern is altered by FXR agonist treatment. Genes were identified using a paired Student’s t test and DMSO treatment as the control group. Transcripts (222) were identified meeting a criteria of a change of at least 0.005 and a ≥2-fold change with respect to DMSO. Data was imported into the cluster, and the genes were subjected to hierarchal clustering. The output was visualized using Treeview. Red coding indicates induction relative to other conditions, green indicates repression, and black indicates no change. (C) Table of genes that were commonly repressed or activated in primary human hepatocytes by CDCA, fexaramine, or GW4064 compared with DMSO treatment. Fold changes indicated represent the average change over the three treatments. – indicates repressed gene message while + indicates an increase in gene expression.
Structural Basis of FXR-Mediated Fexaramine Recognition
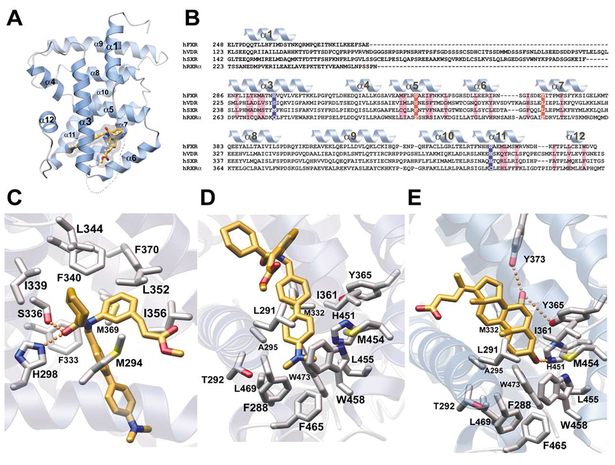
To understand the molecular determinants of ligand binding as well as to gain insight into the physical prop erties of the active FXR receptor, we solved and refined the crystal structure of the ligand binding domain (LBD) of human FXR (amino acids 248–472) in a complex withfexaramine to 1.78Å resolution. The hFXR-LBD adopts a 12 α-helix bundle as seen in all NHR LBD structures described to date (RXRα [Egea et al., 2000], PXR/SXR[Watkins et al., 2001], PPAR[H9253] [Xu et al., 2001], and RORβ[Stehlin et al., 2001]) (Figures 7A and7B). The most significant difference between other NHRs (RXR, VDR, and PPARs) and FXR is in the replacement of the βturn found following helix 5 with a more pronounced helix 6 in FXR (Figure 7A).
Figure 7. Crystal Structure of FXR Bound to Fexaramine.
(A) Structure of hFXR-LBD. Residues 248 to 270 and 286 to 476 of hFXR-LBD in complex with the high-affinity agonist fexaramine. The α1 helices are shown in blue, and the ligand is shown in gold embedded in a transparent van der Waals surface. The structural elements are numbered according to the canonical structure for the LBD of nuclear receptors (NSB ref for canonical label). (B) Sequence alignment of FXR, VDR, SXR, and RXRα1 LBDs. The secondary structural elements of the hFXR-LBD are shown above the FXR sequence in blue and are labeled accordingly (see Figure 1A). Hydrophobic residues involved in binding fexaramine are highlighted in violet. Polar interactions are shown in blue and red. (C) Close-up of the first set of interactions with fexaramine. The hexyl group protrudes out into solution while making weak van der Waals contact with I339 and L344. The fexaramine carbonyl oxygen participates in two hydrogen bonding interactions (H298 and S336). The methyl ester aliphatic chain makes van der Waals contacts with Met294, Leu352, and I356. No charged interactions are seen in contact with the methyl ester moiety itself. (D) Close-up of the second set of interactions with fexaramine. The double benzyl rings make van der Waals contact with 15 residues. The majority of the ligand binding pocket is hydrophobic and partially aromatic in nature. (E) Close-up of a proposed model for complexation of CDCA by FXR-LBD. CDCA was modeled on the experimentally derived orientation of fexaramine. CDCA’s two hydroxyl groups are pointed toward the side chains of Y365 and H451 to putatively participate in favorable hydrogen bonding. This positions the CDCA carboxyl group in the same orientation as the fexaramine hexyl group, suggesting that it protrudes from the protein or makes contacts with the insertion domain region. Notably, glycine and taurine bile acid conjugates could be accommodated in this orientation, which affords steric accommodation of the cognate tails.
In addition, the 15 residue insertion region between helices 1 and 3 is completely disordered in the FXR crystal structure (Figures 7A and7B). RXR α, which most closely resembles FXR in both primary sequence and length of the insertion region, has an additional helix (helix 2) in this position in the absence of ligand that unfolds and becomes disordered upon binding of 9-cisretinoic acid (Egea et al., 2000). This region of RXR α has been proposed to act as a molecular spring which accommodates the large conformational movements of helix 3 upon ligand binding. The insertion region may serve a similar role in hFXR, facilitating helix 3 rearrangements upon ligand binding. In the PPARs, this region contains a helix 2, and this region is the proposed ligand access site for the small molecule binding pocket. In SXR (Watkins et al., 2001) and VDR (Rochel et al., 2000) the insertion domain region is significantly longer (Figure 7B). Analysis of root-mean-square deviations (rmsd) between the apo and ligand bound structures of SXR and VDR revealed no significant differences, suggesting that a shorter insertion domain region may be responsible for regulating large rearrangements of helix 3. Significantly, the activation function-2 domain (AF2 or helix 12), essential for transcriptional activation of the receptor, is packed against the body of FXR, positioned between helices 3 and 4 (Figure 7A). This compact conformation is a signature feature that en ables stable interactions between NHRs and their co-activator partners (Xu et al., 2001). By analogy, coactiva tors would bind in the hydrophobic pocket formed by helices 3, 4, 5, and 12. This extended and complemen tary pocket would interact with the hydrophobic face of the LXXLL helix located within coactivator proteins.
The ligand binding cavity of the hFXR-LBD is predominantly hydrophobic in nature and is formed by 25 amino acid side chains (Figures 7C and7D). The binding pocket has a volume of 726 Å3 that is smaller than that seen in SXR (1150 Å3 ) (Watkins et al., 2001) but larger than that of RXR α (439 Å3 ) (Egea et al., 2000) (Figure 7E). Fexara- mine is sequestered between helices 3 and 7 and makes significant contacts with helices 5, 6, 11, and 12 (Figure 7B). Interactions between FXR and fexaramine can be divided into two sets. The first set of interactions stabilizes the position of fexaramine’s hexyl ring, the outer most first benzene ring, as well as the methyl ester moiety. The hexyl group makes minimal van der Waals contacts with Ile339 and Leu344 (helix 5), while Phe333 (helix 5), and Met369 and Phe370 (helix 7) create a hy drophobic surface behind fexaramine’s central nitrogen and single benzyl group. Met294 (helix 3) as well as Leu352 and Ile356 (helix 6) stabilize the aliphatic linker between the first benzene ring and the methyl ester moiety (Figure 7C). The methyl ester group occupies a neutral groove between helices 3 and 6 and is stabilized by two hydrogen bonds from the N∈2 proton of His298 (helix 3) and the hydroxyl moiety of Ser336 (helix 5) to the amide carbonyl oxygen of fexaramine.
The second group of protein-small molecule interactions stabilizes the biaryl rings and the dimethyl amine moiety of fexaramine. Phe288, Leu291, Thr292, and Ala295 (helix 3) form a hydrophobic surface on one side, while Ile361 (helix 6 and loop 7) and His451, Met454, Leu455, and Trp458 (helix 11) form a hydrophobic sur face on the other side of fexaramine’s double ring structure. Phe465 (helix 11 and loop 12) and Leu469 and Trp473 (helix 12) bridge the hydrophobic surface from the helix 11 region to helix 3 creating a deep hydrophobic pocket that is filled by the biaryl moiety (Figure 7D).
Modeling of Bile Acids into the Ligand Binding Cavity of hFXR
Under conditions tested neither GW4064 nor CDCA formed stable cocrystals for X-ray analysis. However, the structure of the activated form of the FXR LBD allows us to explore how BAs might bind and activate the receptor. We initially modeled CDCA into the FXR binding pocket by overlaying its steroidal backbone onto the biaryl group in fexaramine (Figure 7E). The model sug gested that potential hydrogen bonds could occur between CDCA’s hydroxyl groups and Tyr365, Tyr373, and His451 on helices 7 and 11. These interactions were subsequently used to refine the modeled orientation of the ligand. In the resultant model, hydrophobic interactions with CDCA are predicted to secure helix 3 in an orientation similar to that seen in the complex with fexar amine. This model also provides an explanation for the partial activation of FXR by lithocholic acid (LCA) and deoxycholic acid (DCA) (Makishima et al., 1999). These BAs lack one of the two hydroxyl groups (the αOH at position 7) found in CDCA, and therefore, both partial agonists are predicted to interact significantly only with the helix 7. These variant BAs would therefore not bridge helix 3 to helix 7 as securely as CDCA, which in turn would affect the rigidity of helix 12. In addition, although the inhibitory BA ursodeoxycholic acid (UDCA) has two hydroxyl groups, these moieties are orientated in a trans rather than cis relationship that would likely orientate UDCA to create a more open ligand binding pocket. This arrangement, in turn, may force a suboptimal orientation of helix 12 and result in partial inhibition of the coactiva tor interaction.
Modeling of the recently identified synthetic BA ago nist 6- α-ethyl-chenodeoxycholic acid (6-ECDCA) onto the positional coordinates for the CDCA model further supports the validity of the model and suggests a mech anism for its efficacy as well (Pellicciari et al., 2002). 6-ECDCA differs from CDCA by an additional aliphatic moiety at the 6 α position. The ethyl substituent at this position would be predicted to fit snugly into a hy drophobic pocket formed by Met332 and Phe333 from helix 5. Furthermore, it was demonstrated that either a methyl substituent or a bulkier group at this position reduced efficacy (Pellicciari et al., 2002). This model would predict a less than optimal interaction of a methyl substituent with FXR since the smaller methyl group does not fill the hydrophobic pocket as well as the larger ethyl group. The resultant loss of binding energy through a decrease in contact surface area would result in a loss of efficacy. Bulkier substituents would also be unfavorable, as they would surpass the 0.3 Å limit allowed for in van der Waals overlap resulting in a significant repulsive force in the FXR ligand binding site.
Fexaramine is a much stronger activator of FXR-medi ated transcriptional activity than even FXR’s most po tent natural ligand. Our model suggests that fexara- mine’s potency appears to be mediated using two mechanistic paths. First, the fexaramine methyl ester group provides a significant number of contacts with helix 3 that are absent in our model of CDCA binding. The methyl ester aliphatic chain effectively bridges helix 3 with helix 6 through van der Waals contacts. FXR further stabilizes helix 3 against the remainder of the structure via interactions between Asn297 from helix 3 and Arg335 from helix 5, in addition to interactions from Asn286 (helix 3) and Arg354 (helix 6). The second mecha nism seems to be a function of fexaramine’s length which by comparison to fexarene and fexarine and the BAs suggests that the sequential hydrophobic ring structures of these compounds penetrate deeper into the ligand binding pocket to increase the number of stable contacts with the LBD’s binding pocket. The larger volume of fexaramine (461Å3 ) compared to CDCA (339 Å 3 ) more effectively fills the ligand binding cavity. Analysis of buried surface area in the absence and presence of fexaramine reveals an additional 9Å2 of buried hydrophobic surface when fexaramine is bound. This corresponds to an increase of approximately 1 kJ/M in stabilizing energy. Fexaramine also appears to make direct contact with helix 12 to enhance rigidity and pre sumably stabilize coactivator binding.
Discussion
We describe the development of a synthetic natural product-like molecule, fexaramine, which binds FXR and facilitates analysis of complex physiologic events such as cholesterol metabolism. To that end, fexaramine has revealed an FXR-specific genomic profile distinct from CDCA and played a critical role in obtaining a high- resolution structure of the activated receptor.
Biological and functional studies were undertaken to further characterize fexaramine activity. In vitro assays established that fexaramine and related ligands robustly recruited the coactivator SRC-1 peptide to FXR in a manner comparable to that of GW4064. Cell-based in vivo assays with FXR response elements and natural promoters of known target genes demonstrated that these ligands potently activate FXR in a concentration- dependent manner. Crossreactivity experiments re vealed the specificity of this fexa-class of ligands. Unlike the fexa-compounds, GW4064 required cotransfection of RXR to achieve maximal efficacy in the chimeric GAL4DBD-FXR-LBD protein. This suggests that the in vivo binding of GW4064 to FXR may preferentially recog nize the FXR/RXR heterodimer. Induction of known tar get genes in both intestinal and liver cell systems dem onstrated the usefulness of the identified compounds in studying FXR target genes. In intestinal cells, treatment with fexaramine robustly induced the I-BABP gene in a concentration-dependent manner with efficacy similar to GW4064. Likewise, in the HEPG2 liver cell system, strong induction of target genes SHP, PLTP BSEP, and MRP-2 was achieved at comparable concentrations of fexaramine and GW4064.
Having confirmed the biological specificity and effi cacy of fexaramine, we then investigated FXR target genes. The structural dissimilarity of the FXR agonists discussed here was reflected in the effects these com pounds had on global gene regulation. Each molecule had a distinct signature of gene expression, in addition to a smaller subset of genes that were affected similarly by all three ligands (Figures 6C and5E). The differences between the synthetic compounds could be due to ef fects mediated by heterogeneous FXR-containing com plexes due to structural differences in the ligand, differ ential clearance rates in primary liver hepatocytes, or through non-FXR-mediated signaling mechanisms. In contrast, the distinct profile seen by CDCA is likely a consequence of non-FXR dependent activation of the xenobiotic receptor PXR and the c-Jun N-terminal ki nase JNK (Wang et al., 2002). In addition, it has been shown that partial agonists such as tamoxifen or raloxi fene for human estrogen receptor can display strikingly different gene expression profiles. We suggest that the differences between the fexa-compounds and GW4064 might be because of their individual association with the LBD achieving specificity by aggregating with distinct cofactor complexes. This has yet to be shown for any synthetic compound but is consistent with the known mechanism of action. This exemplifies the difficulties of investigating NHR function using a natural ligand and highlights the need for specific synthetic ligands to iso late NHR’s dependent pathways. However, even high- affinity synthetic compounds may have nonspecific effects. This potential limitation indicates the utility of employing multiple synthetic ligands to accurately discern the common core signaling pathways.
To complete this study, we next analyzed the FXR protein using a series of detailed structural studies. CDCA and other BAs, which bind to FXR with relatively low affinity, failed to promote receptor crystallization while, in contrast, fexaramine cocrystallizes with FXR. This provided not only a high-resolution structure but also enabled the subsequent modeling of CDCA with a high degree of confidence into the ligand binding pocket of FXR. This model provides a molecular explanation for the selectivity of BAs on FXR and highlights the importance of position and orientation of the hydroxyl groups at positions 3 and 7 in binding affinity. This model provides a possible rationale for the beneficial effects of UDCA in the treatment of primary biliary cirrhosis. Although UDCA has two hydroxyl groups that could potentially form hydrogen bonds with FXR in the ligand binding cavity, their trans configuration creates a more open ligand binding pocket that would destabilize helix 12 and thereby inhibit activation of the receptor.
The body of work presented in this paper was the product of combining chemical, genetic, and structural approaches to the analysis of FXR. In doing so we have not only gained a valuable chemical probe, fexaramine, to study the mechanism of receptor signaling, but we have also begun to unravel the FXR genetic network from the BA network and gained the ability to manipulate selective components of the pathway.
Experimental Procedures
Constructs
The pCMX expression plasmids and luciferase reporter plasmids have been described elsewhere (Blumberg et al., 1998; Kast et al., 2002). The hPLTP-luc promoter was kindly provided by Dr. Dennis Dowhan, and the hIBABP-luc promoter was created from a plasmid provided by Dr. Philippe Besnard.
Residues 248 to 476 of human FXR LBD were PCR amplified and subcloned into the BamHI or NcoI/BamHI sites of pGEX and pHIS, respectively, to generate protein expression vectors (Jez et al., 2000).
The retroviral plasmids were constructed by cloning FXRFL, FXRAF2, and VP16-FXR cDNAs into the BamHI site of the established pBABE retroviral backbone vector. Viral extracts were established using published procedures and used to infect HT29 colon cells. After exposure for 24 hr, cells were selected by the addition of 4 μg/ml of puromycin. Cells that survived this selection procedure were then pooled and analyzed for the expression of the FXR gene.
All constructs were verified by sequencing to confirm identity and reading frame. Detailed information regarding each construct is available upon request.
Transfections
CV-1 and HEPG2 cells were grown in DMEM supplemented with 10% FBS, 50 U/ml penicillin G, and 50 μg/ml streptomycin sulfate at 37 °C in 7% CO2. CV-1 cells (60%–70% confluence, 48-well plate) were cotransfected with 16.6 ng of the appropriate expression vec tor, 100 ng of reporter plasmid, and 100 ng of pCMX-LacZ in 200 μl of DMEM containing 10% FBS by the Lipofectamine 2000 procedure (Invitrogen, Carlsbad, CA). After 24 hr, the medium was replaced, and cells were harvested and assayed for luciferase activity 36–48 hr after transfection. The luciferase activity was normalized by β-galactosidase activity. Each transfection was performed in tripli cate and repeated at least three times.
Solid Phase Synthesis of Small Molecule Combinatorial Libraries and Ligands
The synthesis of the chemical compounds was carried out on solid phase supports in parallel as summarized in Figure 2. In brief, Boc- protected cinnamic acid (1) was immobilized on Merrifield resin using Cs2CO3 to afford conjugate (2). The Boc group was removed by treatment with 20% (v/v) TFA (for abbreviations see legend to Figure 2) in CH2Cl2, and the resultant resin-bound amine was reduc tively alklylated with 4-bromobenzaldehyde in the presence of NaCNBH3 to yield amino resin (3). Resin (3) was acylated with one of three acyl groups to give amide or urea resins (4). The acylated resins (4) were then subjected to either a Heck coupling (Pd2[dba]3, P[o-tol]3, Et3N]) with thirteen substituted styrenes or a Suzuki cou pling (Pd[PPh3]4, Cs2CO3) with eighteen boronic acids to yield stil bene resins (5) and biaryl resins (6), respectively. Cleavage of the resulting compounds from resins (5) and (6) with NaOMe yielded methyl cinnamates (7) and (8). Analysis of the library by LCMS after purification showed the average purity of these compounds to be > 95%. Further details of the chemistry involved in this project can be found elsewhere (Nicolaou et al., 2003).
RNA Isolation and Northern Blot Hybridization Unless otherwise indicated, HepG2- or HT29-derived cell lines were cultured in medium containing superstripped FBS for 24 hr before the addition of a ligand solution or DMSO (vehicle) for an additional 24–48 hr. Total RNA was isolated using Trizol reagent and was resolved (20 μg/lane) on a 1% agarose, 2.2 M formaldehyde gel, transferred to a nylon membrane, and UV crosslinked. cDNA probes were radiolabeled with [α−32P]dCTP using the high-prime labeling kit. Membranes were hybridized using the QuikHyb hybridization solution according to the manufacturer’s protocol (Stratagene).Blots were normalized for loading with control ribosomal 18 S cDNA or 36B4 protein probes. The RNA levels were quantified using a PhosphorImager in addition to being exposed to X-ray film.
Protein Expression and Purification
The plasmid pHIS8–3-hFXR LBD (residues 248 to 476) was transformed into E. coli strain BL21 (DE3), and cells were grown at 37 °C until an OD600nm of 1.0. Expression was induced by adding isopropyl-1-thio-β-D-galactopyrauoside to 0.1 mM, and cells were grown for an additional 6 hr at 20°C. Bacteria were harvested by centrifugation at 8000 × g, and pellets were stored at ‐70°C. Cell pellets were thawed and resuspended in 50 mM Tris-Cl (pH 8.0), 500 mM NaCl, 10 mM imidazole (pH 8.0), 10% (v/v) glycerol, 1% (v/v) Tween 20, and 10 mM β -mercaptoethanol (β -ME) at 4°C. Resuspended cells were sonicated, and lysates were centrifuged at 100,000 × g at 4°C. Supernatants were purified by Ni2+-chelation chromatography (QIAGEN, Valencia, CA). After washing bound protein sample was eluted using 50 mM Tris-Cl (pH 8.0), 500 mM NaCl, 250 mM imidazole (pH 8.0), 10% (v/v) glycerol, and 10 mM β -ME. The N-terminal octa histidine tag was removed by thrombin (Sigma, St. Louis, MO) digestion during dialysis against 50 mM Tris (pH 8.0), 500 mM NaCl, and 10 mM dithiothretiol (DTT) at 4°C for 24 hr. The dialyzed and cleaved sample was purified using a Superdex 200 26/60 gel filtration column (Amersham Biosciences, Piscataway, NJ) equilibrated in dialysis/ thrombin cleavage buffer. Peak fractions were collected and dia lyzed against 5 mM Tris-HCL (pH 8.0) 62.5 mM NaCl, and 1 mM DTT, concentrated to 15 mg/ml using a Centricon 10 (Amicon, Bed ford, MA), and stored at –70°C. Selenomethionine-substituted protein (SeMet) was obtained from E. coli grown in minimal media using the methionine pathway inhibition methods (Doublié, 1997) and was purified similarly to the native FXR-LBD.
Crystallization and Structure Determination
Fexaramine was solubilized in dimethylsulfoxide (DMSO) to a final concentration of 10 mM; hFXR-LBD (15 mg/ml) was incubated with fexaramine at a 1:2 molar ratio. Crystals of the hFXR-LBD/fexara- mine mixture were grown by the hanging drop vapor diffusion method at 4°C by mixing 1.0μl of hFXR-LBD/fexaramine complex with 1.0 μl of a reservoir solution containing 15%–20% (w/v) PEG 8000, 100 mM HEPES-Na+ (pH 7.5), 0.2 M MgCl2, 1 mM DTT. Crystals of selenomethionine-substituted hFXR-LBD were grown similarly using 10 mM DTT. Crystals were stabilized in 10%–15% (v/v) glycerol, 20% (w/v) PEG 8000, 0.2 M MgCl2, 100 mM HEPES-Na+ (pH7.5), and 10 mM DTT and rapidly frozen in a 100 K stream of nitrogen gas. MAD data to 2.1Å were collected around the Se edge at the European Synchrotron Radiation Facility (ESRF, Grenoble, France) on beamline FIP (BM30A). Native data to 1.78Å were collected at the Stanford Synchrotron Radiation Laboratory, beamline 9–1. All data were processed with DENZO and SCALEPACK (Otwinowski and Minor, 1997). The crystals contain one molecule per asymmetric unit (52.9% solvent) and belong to the space group P212121 (a = 36.656, b = 56.776, c = 117.646, α = 9, β = 9, γ = 90.0˚). Three wavelength MAD data were scaled to the λ3. Seven of nine Se sites were located and MAD phasing was accomplished using SOLVE (Terwilliger and Berendzen, 1992), and density modification was carried out with RESOLVE (Terwilliger, 2000). The initial model was built into the experimental electron density maps displayed in O (Jones et al., 1991). The resulting model was positionally refined against all of the high-resolution native data set using the default bulk solvent model in CNS with maximum likelihood targets (Brünger et al., 1998). The structure of the FXR-LBD/fexaramine complex was refined to a Rcryst and a Rfree value of 23.0% and 27.5%, respectively, using all data extending to 1.78Å resolution. The R factor = Σ |Fobs ‐ Fcalc| / Σ Fobs, where summation is over the data used for refinement and the Rfree was calculated using 5% of the reflection data chosen and excluded from refinement. The model consists of residues 248 to 270 and 286 to 475 of human FXR, 1 fexaramine molecule, and 340 water molecules. PROCHECK (Laskowski et al., 1993) revealed a total of 92% of the residues in the most favored region of the Ramachandran plot and 8% in the additionally allowed region. Main chain and side chain structural parameters were consistently better than average (overall G value of 0.16).
Acknowledgments
We thank Drs. Peter Ordentlich and David Egan for their helpful advice and Drs. Ann Atkins, Ruth Yu, and Joyce Havstad for proof reading. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology. This work was supported by the How ard Hughes Medical Institute. M.D. and R.M.E. acknowledge funding support through the NIH NURSA orphan receptor program, grant number U19DK62434–01. K.C.N. thanks the National Institutes of Health (NIH) and the Skaggs Institute for Chemical Biology for fund ing support. J.P.N. received funding from the NIH, grant number CA54418. P.A.E. received funding from the NIH, grant numbers HL30568 and HL68445.
Footnotes
Accession Numbers
The Protein Data Bank code for the FXR-feraramine X-ray structure is 1OSH.pdb, and the code for the FXR-CDCA model is 1OSK.pdb.
References
- Blumberg B, and Evans RM (1998). Orphan nuclear receptors—new ligands and new possibilities. Genes Dev. 12, 3149–3155. [DOI] [PubMed] [Google Scholar]
- Blumberg B, Sabbagh W Jr., Juguilon H, Bolado J Jr., van Meter CM, Ong ES, and Evans RM (1998). SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 12, 3195–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chawla A, Saez E, and Evans RM. (2000). “Don’t know much bile- ology”. Cell. 103, 1–4. [DOI] [PubMed] [Google Scholar]
- Chiang JY (2002). Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr. Rev. 23, 443–463. [DOI] [PubMed] [Google Scholar]
- Doublié S (1997). Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530. [PubMed] [Google Scholar]
- Edwards PA, Kast HR, and Anisfeld AM (2002). BAREing it all: the adoption of LXR and FXR and their roles in lipid homeostasis. J. Lipid Res. 43, 2–12. [PubMed] [Google Scholar]
- Egea PF, Mitschler A, Rochel N, Ruff M, Chambon P, and Moras D (2000). Crystal structure of the human RXR αligand-bind ing domain to its natural ligand: 9-cis retinoic acid. EMBO J. 19, 2592–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM. (1988). The steroid and thyroid hormone receptor super- family. Science 240, 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Per lmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, et al. (1995). Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81, 687–693. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, et al. (2000). A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6, 517–526. [DOI] [PubMed] [Google Scholar]
- Grober J, Zaghini I, Fujii H, Jones SA, Kliewer SA, Willson TM, Ono T, and Besnard P (1999). Identification of a bile acid- responsive element in the human ileal bile acid-binding protein gene.Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J. Biol. Chem. 274, 29749–29754. [DOI] [PubMed] [Google Scholar]
- Jez JM, Ferrer J-L, Bowman ME, Dixon RA, and Noel JP (2000). Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39, 890–902. [DOI] [PubMed] [Google Scholar]
- Jones AT, Zou J-Y, Cowan S, and Kjeldgaard M (1991). Im proved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr.A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfeld AM, Stoltz CM, Tontonoz P, Kliewer S, Willson TM, and Edwards PA (2002). Regulation of multidrug resistance-associated protein 2 (ABCC2) by the nuclear receptors pregnane X receptor, farnesoid X-activated receptor, and constitutive androstane receptor. J. Biol.Chem. 277, 2908–2915. [DOI] [PubMed] [Google Scholar]
- Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, Shan B, Russell DW, and Schwarz M (2002). Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell 2, 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD, and Edwards PA (2000). Identification of the DNA binding specific ity and potential target genes for the farnesoid X-activated receptor.J. Biol. Chem. 275, 10638–10647. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, and Thornton JM (1993). PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Au werx J, and Mangelsdorf DJ (2000). Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6, 507–515. [DOI] [PubMed] [Google Scholar]
- Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, and Shan B (1999). Identification of a nuclear receptor for bile acids. Science.284, 1362–1365. [DOI] [PubMed] [Google Scholar]
- Maloney PR, Parks DJ, Haffner CD, Fivush AM, Chandra G, Plunket KD, Creech KL, Moore LB, Wilson JG, Lewis MC, et al. (2000). Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med. Chem. 43, 2971–2974. [DOI] [PubMed] [Google Scholar]
- Nicolaou KC, Pfefferkorn JA, Roecker AJ, Cao G-Q, Bar luenga S, and Mitchell HJ (2000a). Natural product-like combina torial libraries based on privileged structutres. 1. General principles and solid-phase synthesis of benzopyrans. J. Am. Chem. Soc. 122, 9939–9953. [Google Scholar]
- Nicolaou KC, Pfefferkorn JA, Mitchell HJ, Roecker AJ, Bar luenga S, Cao G-Q, Affleck RL, and Lillig JE (2000b). Natural product-like combinatorial libraries based on privileged structures.2. Construction of a 10000-membered benzopyran library by di rected split-and-pool chemistry using nanokans and optical encoding. J. Am. Chem. Soc. 122, 9954–9967. [Google Scholar]
- Nicolaou KC, Pfefferkorn JA, Barluenga S, Mitchell HJ, Roecker AJ, and Cao G-Q (2000c). Natural product-like combinatorial libraries based on privileged structutres. 3. The “libraries from libraries” principle for diversity enhancement of benzopyran libraries. J. Am. Chem. Soc. 122, 9968–9976. [Google Scholar]
- Nicolaou KC Evans RM, Roecker AJ, Hughes R, Downes M, and Pfefferkorn JA (2003). Discovery of four classes of potenet, non-steroidal FXR agonists originating from natural product-like li braries. Org. Biomol. Chem. 1, 908–920. [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, and Lehmann JM (1999). Bile acids: natural ligands for an orphan nuclear receptor. Science 284, 1365–1368. [DOI] [PubMed] [Google Scholar]
- Pellicciari R, Fiorucci S, Camaioni E, Clerici C, Costantino G, Maloney PR, Morelli A, Parks DJ, and Willson TM (2002). 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J. Med. Chem. 45, 3569–3572. [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, and Gonzalez FJ (2000). Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744. [DOI] [PubMed] [Google Scholar]
- Stehlin C, Wurtz JM, Steinmetz A, Greiner E, Schule R, Moras D, and Renaud JP (2001). X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. EMBO J. 20, 5822–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochel N, Wurtz JM, Mitschler A, Klaholz B, and Moras D (2000). The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol. Cell 5, 173–179. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC (2000). Maximum likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, and Berendzen J (1992). Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Cystallogr. 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urizar NL, Dowhan DH, and Moore DD (2000). The farnesoid X-activated receptor mediates bile acid activation of phospholipid transfer protein gene expression. J. Biol. Chem. 275, 39313–39317. [DOI] [PubMed] [Google Scholar]
- Urizar NL, Liverman AB, Dodds DT, Silva FV, Ordentlich P, Yan Y, Gonzalez FJ, Heyman RA, Mangelsdorf DJ, and Moore DD (2002). A natural product that lowers cholesterol as an antagonist ligand for FXR. Science 296, 1703–1706. [DOI] [PubMed] [Google Scholar]
- Wang H, Chen J, Hollister K, Sowers LC, and Forman BM (1999). Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 3, 543–553. [DOI] [PubMed] [Google Scholar]
- Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, and Moore DD (2002). Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell 2, 721–731. [DOI] [PubMed] [Google Scholar]
- Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, Kliewer SA, and Redinbo MR (2001). The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science 292, 2329–2333. [DOI] [PubMed] [Google Scholar]
- Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, Oplinger JA, Kliewer SA, Gampe RT Jr., McKee DD, et al. (2001). Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc. Natl. Acad. Sci. USA 98, 13919–13924. [DOI] [PMC free article] [PubMed] [Google Scholar]