Abstract
Context
Skeletal muscle endocannabinoids and sphingolipids (particularly sphingomyelins) are inversely associated with sleeping energy expenditure (SLEEP) in humans. The endocannabinoid system may increase sphingolipid synthesis via cannabinoid receptor-1.
Objective
To investigate in human skeletal muscle whether endocannabinoids are responsible for the effect of sphingomyelins on SLEEP.
Design
Muscle endocannabinoid [anandamide (AEA), 2-arachidonoylglycerol (2-AG)], endocannabinoid congeners [oleoylethanolamide (OEA), palmitoylethanolamide (PEA)], and sphingomyelin content were measured with liquid chromatography/mass spectrometry. SLEEP was assessed in a whole-room indirect calorimeter. Mediation analyses tested whether the inverse associations between sphingomyelins and SLEEP depended on endocannabinoids and endocannabinoid-related OEA and PEA.
Setting
Inpatient study.
Participants
Fifty-three Native Americans who are overweight.
Main Outcome Measure
SLEEP.
Results
AEA (r = 0.45, P = 0.001), 2-AG (r = 0.47, P = 0.0004), OEA (r = 0.27, P = 0.05), and PEA (r = 0.53, P < 0.0001) concentrations were associated with the total sphingomyelin content. AEA, OEA, and PEA correlated with specific sphingomyelins (SM18:1/23:0, SM18:1/23:1, and SM18:1/26:1) previously reported to be determinants of SLEEP in Native Americans (all r > 0.31, all P < 0.03). Up to half of the negative effect of these specific sphingomyelins on SLEEP was accounted for by AEA (all P < 0.04), rendering the direct effect by sphingomyelins per se on SLEEP negligible (P > 0.05).
Conclusions
In skeletal muscle, AEA is responsible for the sphingomyelin effect on SLEEP, indicating that endocannabinoids and sphingomyelins may jointly reduce human whole-body energy metabolism.
Mediation analyses investigated whether endocannabinoids and sphingomyelins jointly affect human energy metabolism. Anandamide is responsible for the sphingomyelin effect on human energy expenditure.
Participating in cellular signaling pathways, the endocannabinoid system (ECS) is a well-recognized effector of human energy homeostasis, and its dysregulation has been implicated in metabolic diseases (1–3). Because of their intolerability, clinical use of ECS antagonists for obesity therapy has not been established (1). Nevertheless, the ECS remains an intriguing peripheral target for pharmacological treatment of people with obesity (1, 2, 4). The ECS tone within a biological system refers to ECS activity primarily as a result of endocannabinoid levels within that system (2). Systemic reduction of ECS tone in humans and rodents increased energy expenditure (EE) (5–7), and lower skeletal muscle ECS activity increased insulin sensitivity and glucose uptake (8–10). These studies point to a negative effect of the ECS on skeletal muscle energy metabolism and the thermogenic response to glucose (11, 12). Consistent with this, we reported that skeletal muscle anandamide (AEA) and 2-arachidonoylglycerol (2-AG) are important determinants of daily EE in humans, explaining a large amount of the interindividual variance in EE, particularly sleeping energy expenditure (SLEEP) (4). Furthermore, AEA and 2-AG are commonly produced together with their congeners, oleoylethanolamide (OEA) and palmitoylethanolamide (PEA), and, as opposed to the negative effect of endocannabinoids on EE, higher OEA and PEA concentrations in skeletal muscle may instead lead to higher EE because, in vitro, OEA and PEA increase mitochondrial activity (13, 14) and lipolysis in skeletal muscle (15). Human adipose tissue OEA concentration was reported to be positively associated with fat oxidation rate (4). Thus, comparable to endocannabinoids, OEA and PEA may be feasible effectors of human EE via an effect on mitochondrial activity.
Sphingolipids are a different class of lipids shown to be major determinants of SLEEP (16). Specifically, human skeletal muscle ceramide and sphingomyelin concentrations were negatively associated with SLEEP and positively associated with weight gain in Native Americans who are overweight (16). Possible cellular mechanisms underlying this link include sphingolipid-induced reduction in mitochondrial respiratory activity due to increased uncoupling (17–19).
It has been proposed that the ECS may affect intracellular sphingolipid content (20–23), suggesting a possible crosstalk between these two entities that might explain their mutual effects on EE. For instance, a lower ECS tone in diet-induced insulin-resistant mice was associated with decreased hepatic ceramide synthesis (21). This finding corroborates the hypothesis that a higher ECS tone may lead to greater intracellular ceramide content (20, 22, 23), thus reducing mitochondrial respiratory activity and ultimately whole-body energy metabolism (20, 24, 25). The ECS may increase sphingolipid synthesis via gene expression regulation of the enzyme serine palmitoyltransferase (SPT), which catalyzes the rate-limiting step of ceramide and sphingomyelin synthesis (21, 23, 26). Accordingly, in vitro and mouse models have shown that a higher ECS tone upregulates SPT activity (21–23) and the ceramide content in hepatocytes (21), possibly by increased serine palmitoyltransferase long chain subunit 3 (SPTLC3) expression (one of the SPT regulatory subunits) (21, 27). This effect was mediated by cannabinoid receptor-1 (CB1, CNR1) (21).
Sphingomyelins are functionally closely related to ceramides (16, 28–30). In comparison with the latter, however, the former play a more dominant role in human EE regulation (16). Given the in vitro and animal model evidence that the ECS increases sphingomyelin synthesis, and to understand the relative importance of these lipid entities in human EE regulation, we hypothesized that the content of endocannabinoids AEA and 2-AG, as well as their congeners OEA and PEA, in human skeletal muscle is associated with sphingomyelin content, supporting their biological crosstalk; and endocannabinoids, OEA, and PEA mediate the effect of sphingomyelins on SLEEP, corroborating the active role of ECS in sphingomyelin synthesis. To further confirm the interplay between the ECS and sphingomyelins in human skeletal muscle, we also assessed the relationship between CNR1 and SPTLC3 mRNA expression in this tissue.
Subjects and Methods
Subjects and clinical assessment
Data from 53 Native Americans of at least half Southwestern heritage who, between February 1995 and March 2009, participated in an ongoing study to identify risk factors for diabetes and weight gain (clinicaltrials.gov, NCT00340132) were included in the current analyses. Having provided written informed consent, healthy individuals were admitted to our clinical research unit for 8 days and placed on a weight-maintaining balanced diet to ensure weight stability within 1% of admission weight for the entire length of the inpatient stay (31). Subjects were nondiabetic, as determined by 75-g oral glucose tolerance test (Beckman Instruments, Fullerton, CA, or Autoanalyzer, Technicon, Tarrytown, NY) (32). Body fat-free mass (FFM), fat mass (FM), and percentage body fat were assessed with dual-energy X-ray absorptiometry (DPX-1; Lunar Radiation Corp., Madison, WI) (33). Skeletal muscle tissue was acquired from the vastus lateralis muscle by percutaneous needle biopsy under local anesthesia (1% lidocaine). Samples were snap-frozen and stored at −80°C until analysis.
The study was approved by the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Diseases.
Measurement of skeletal muscle lipids
Combined liquid chromatography/mass spectrometry was used to measure concentrations of sphingomyelins, endocannabinoids, and endocannabinoid congeners OEA and PEA in a single set of analysis, respectively (4, 34). For sphingomyelins, extraction of lipids from muscle tissue was performed with a 3-mL 2:1 chloroform/methanol mixture with internal sphingomyelin standard. Then, organic phases were collected and, after a nitrogen drying process, dissolved in methanol/chloroform (3:1, vol/vol). For endocannabinoids and their congeners, frozen tissues were homogenized in methanol (1 mL/100 mg tissue) containing internal endocannabinoid standards. Extraction of lipids was performed with chloroform/water (2:1, vol/vol), and organic extract was collected by centrifugation (3000g for 15 minutes at 4°C) with consequent fractionation (Silica Gel G, 60-Å 230–400 Mesh ASTM; Whatman, Clifton, NJ).
Sphingomyelins, endocannabinoids, OEA, and PEA were identified by liquid chromatography/mass spectrometry (1110-LC system; Agilent Technologies, Inc., Palo Alto, CA) and separated on a Poroshell/XDB Eclipse C18 column (Agilent Technologies) at 30°C and 40°C, respectively. A linear gradient of methanol in water (containing 5 mM ammonium acetate and 0.25% acetic acid for sphingomyelins) was applied at a flow rate of 1 mL/min. Mass spectrometry detection was kept in the positive ionization mode with capillary voltage at 4.5/3 kV and capillary exit at 151/120 to 140 V for sphingomyelins, endocannabinoids, OEA, and PEA, respectively. Nitrogen was used as a drying gas (350°C, flow rate 10/13 L/min for sphingomyelins and endocannabinoids, respectively). For identification of sphingomyelin species, fragmentation patterns during mass spectrometry and liquid chromatography retention rates were compared with standards (Avanti Polar Lipids, Plymouth Meeting, PA). For endocannabinoids and their congeners, Na+ adducts were monitored in the selective ion-monitoring mode.
Because of incomplete lipid measurement, in subjects with valid SLEEP measurement, 30 subjects had complete assessment of endocannabinoids, OEA, and PEA, whereas 29 subjects had complete measurements of sphingolipid content in muscle.
Metabolic assessment
SLEEP, as a proxy for basal metabolism, was derived from 24-hour EE assessment in a whole-room indirect calorimeter during energy balance: 24-hour EE was estimated as the average EE during all 15-minute sampling intervals from 8:00 am until 7:45 am of the next morning and extrapolated to 24 hours. Measurement of spontaneous physical activity by radar systems (35) allowed the calculation of SLEEP, estimated as the energy expended overnight between 11:30 pm and 5:00 am of the next day when spontaneous physical activity was <1.5% (36). Of 53 subjects with lipid assessment in skeletal muscle, 30 were informative for SLEEP. Because we previously showed that sphingomyelin content was a strong determinant of SLEEP in this population (16), mediation analyses focused on SLEEP.
Microarray analysis
Microarray analysis was performed in skeletal muscle samples of a subset of 106 healthy, nondiabetic subjects (of at least half Southwestern heritage; Table 1) participating in our phenotyping study from March 1992 to February 2005.
Table 1.
Subject Characteristics
| All Subjects (n = 53) | Men (n = 35) | Women (n = 18) | |
|---|---|---|---|
| Age, y | 29.7 ± 8.13 | 30.5 ± 8.65 | 28.2 ± 6.99 |
| Body weight, kg | 94.3 ± 17.9 | 96.3 ± 17.6 | 90.5 ± 18.5 |
| Body mass index, kg/m2 | 33.1 ± 6.74 | 32.2 ± 6.70 | 34.9 ± 6.62 |
| Body fat, % | 33.3 ± 7.15 | 29.5 ± 5.22 | 40.7 ± 3.84 |
| FM, kg | 31.7 ± 9.83 | 29.0 ± 9.05 | 37.0 ± 9.33 |
| FFM, kg | 62.6 ± 11.9 | 67.3 ± 9.87 | 53.5 ± 10.1 |
| Fasting plasma glucose, mg/dL | 87.8 ± 8.99 | 86.3 ± 9.84 | 90.7 ± 6.31 |
| 2-h plasma glucose, mg/dL | 125.2 ± 27.9 | 119.9 ± 26.5 | 135.3 ± 28.5 |
| SLEEP, kcal/d | 1750 ± 289 | 1834 ± 276 | 1605 ± 263 |
| (n = 30) | (n = 19) | (n = 11) |
Data are reported as mean ± SD.
Microarray analysis was used to quantify mRNA expression levels and, specifically, allowed CNR1 gene expression assessment in skeletal muscle. CNR1 codes for the endocannabinoid receptor CB1, which is involved in skeletal muscle metabolic regulatory processes and biosynthesis of sphingolipids (21–23, 37). Additionally, gene expression of the SPT subunit shown to be affected by an altered ECS tone in hepatocytes (SPTLC3) was assessed (21). These analyses allowed correlation assessment between muscle CNR1 and SPTLC3 gene expression. For detailed description of SPTLC3 and CNR1 expression assessment, see the Supplemental Materials.
Statistical analysis
SAS Enterprise Guide 7.1 (SAS Institute, Inc., Cary, NC) was used for all statistical analyses. Nonnormally distributed data were log10-transformed to ensure a Gaussian distribution. The Shapiro-Wilk test was used to test for normality of data with a P threshold of 0.05. The Pearson correlation index (r) was used to quantify the association between continuous variables. The residuals of SLEEP (residual SLEEP) were calculated via linear regression analysis after adjustment for FFM, the strongest predictor of SLEEP as previously reported (38), and used in the mediation analyses. The regression equation for the current study group was
and residuals were calculated as SLEEPmeasured − SLEEPpredicted. Data are presented as mean ± SD. A two-sided α of 0.05 was set.
To test our hypothesis (i.e., whether the endocannabinoids AEA and 2-AG, as well as OEA and PEA, are associated with global sphingomyelin content in skeletal muscle), z scores for each detected sphingomyelin were calculated and averaged to derive an overall sphingomyelin score (SMcont) for each subject:
where n is the total number of individual sphingomyelins (Table 2). To obtain a global sphingomyelin score, z scores were calculated for each sphingomyelin to account for different ranges of sphingomyelin concentrations within skeletal muscle. SMcont was considered in correlation analyses with endocannabinoids and their congeners, and in case of a significant association with SMcont, post hoc correlation analyses were conducted for single sphingomyelin compounds, particularly those previously reported to be the strongest determinants of residual SLEEP (SM18:1/23:0, SM18:1/23:1, and SM18:1/26:1) (16).
Table 2.
Endocannabinoid, OEA, PEA, and Sphingolipid Concentrations in Human Skeletal Muscle
| Endocannabinoids | All Subjects (n = 53) | Men (n = 35) | Women (n = 18) |
|---|---|---|---|
| AEA, pmol/g | 1.5 ± 1.0 | 1.5 ± 1.1 | 1.5 ± 0.7 |
| 2-AG, nmol/g | 691.4 ± 358.4 | 737.9 ± 314.1 | 600.9 ± 427.2 |
| OEA, pmol/g | 23.3 ± 15.3 | 19.8 ± 7.6 | 30.2 ± 22.9 |
| PEA, pmol/ga | 33.8 ± 8.5 | 32.9 ± 8.6 | 35.7 ± 8.1 |
| Sphingomyelins, pmol/g | |||
| SM18:1/16:0 | 26.7 ± 12.6 | 28.4 ± 13.8 | 23.4 ± 9.4 |
| SM18:1/18:0 | 22.5 ± 5.5 | 23.4 ± 5.5 | 20.9 ± 5.0 |
| SM18:1/20:0 | 262.0 ± 52.7 | 261.3 ± 48.1 | 263.4 ± 62.1 |
| SM18:1/22:0 | 120.5 ± 32.5 | 129.0 ± 32.0 | 104.1 ± 27.3 |
| SM18:1/22:1 | 35.1 ± 10.3 | 35.1 ± 9.6 | 35.1 ± 11.9 |
| SM18:1/23:0 | 7.3 ± 2.5 | 7.4 ± 2.8 | 7.3 ± 1.9 |
| SM18:1/23:1 | 5.0 ± 2.4 | 4.9 ± 2.3 | 5.1 ± 2.6 |
| SM18:1/24:0 | 15.7 ± 7.1 | 15.5 ± 7.3 | 16.1 ± 7.0 |
| SM18:1/24:1 | 30.7 ± 11.4 | 32.3 ± 11.7 | 27.4 ± 10.2 |
| SM18:1/24:2 | 17.8 ± 5.8 | 18.7 ± 6.1 | 16.0 ± 4.7 |
| SM18:1/25:1 | 9.4 ± 4.3 | 9.0 ± 3.3 | 10.0 ± 5.9 |
| SM18:1/26:1 | 5.1 ± 3.0 | 4.8 ± 2.5 | 5.6 ± 3.7 |
| SM18:0/16:0 | 6.5 ± 2.6 | 6.6 ± 2.8 | 6.3 ± 2.4 |
| SM18:0/18:0 | 9.0 ± 3.2 | 8.7 ± 2.8 | 9.5 ± 3.9 |
| SM18:0/24:0 | 11.9 ± 5.8 | 12.3 ± 6.5 | 11.1 ± 4.2 |
| SM18:0/24:1 | 15.1 ± 9.8 | 17.4 ± 10.1 | 10.8 ± 7.9 |
n = 52 (35 men, 17 women) because of the exclusion of one subject with highly leveraged PEA concentrations.
After correlation analyses, mediation analyses based on hierarchical multivariable regression models (39, 40) were carried out to quantify the effects of endocannabinoids on the relationships between sphingomyelins and residuals of SLEEP in n = 29 subjects. In our mediation framework, the total effect of sphingomyelin (independent variable) on residual SLEEP (dependent variable) was partitioned into an endocannabinoid-specific effect exerted by AEA, 2-AG, OEA, or PEA; and a direct effect of sphingomyelins per se independent of endocannabinoids, OEA, or PEA. The Sobel test was used to determine the significance of the endocannabinoid-specific effect (41), to test whether endocannabinoids or their congeners accounted for the association between sphingomyelins and residual SLEEP. Results of the Sobel test were confirmed by bootstrapping (data not shown). First, a mediation analysis was conducted considering overall SMcont as independent variable; subsequently, in case of significant endocannabinoid-specific effect by AEA, 2-AG, OEA, or PEA on residual SLEEP, the three sphingomyelin compounds (SM18:1/23:0, SM18:1/23:1, and SM18:1/26:1) previously reported to be strongest determinants of residual SLEEP (16), were also included in the mediation analysis. Path coefficients are reported as mean ± SE with their significance.
To assess whether studied groups were different considering subjects with complete skeletal muscle lipid assessment, as well as complete SLEEP and endocannabinoid (n = 30) or sphingolipid (n = 29) assessment, respectively, unpaired t tests were performed. Subgroup comparison included age, sex, FFM, FM, percentage body fat, SLEEP, and skeletal muscle lipid content. Sensitivity analyses comparing the overall group of 53 subjects with lipid assessment in skeletal muscle and 106 subjects with microarray analysis performed included age, sex, FFM, FM, and percentage body fat.
Results
The characteristics of the study group are reported in Table 1, and concentrations of detected endocannabinoids and their congeners, as well as sphingolipid compounds in skeletal muscle, are reported in Table 2. FFM was a significant predictor of SLEEP (β = 19.0 ± 3.0 kcal/d/kg, r2 = 0.59, P < 0.0001) in 30 subjects with complete EE assessment. On average, the lipid (n = 53) and microarray (n = 106; Supplemental Table 1) study cohorts did not differ by age, sex, FM, FFM, and percentage body fat (all P > 0.05). In sensitivity analyses, concentrations of endocannabinoids and their congeners, sphingomyelin content, age, sex, FFM, FM, and percentage body fat did not differ between subjects with skeletal muscle lipid measurements (n = 53) and those who had valid SLEEP assessment (n = 30 for subjects with endocannabinoid, OEA, and PEA measurements and n = 29 for subjects with sphingolipid measurements; all P > 0.05). In the latter two groups, SLEEP was not different (P > 0.05).
Relationships between sphingomyelins, endocannabinoids, and gene expression levels
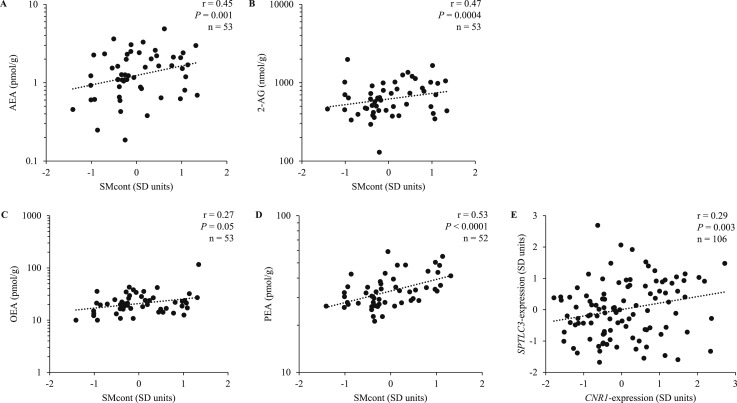
AEA (r = 0.45, P = 0.001), 2-AG (r = 0.47, P = 0.0004), OEA (r = 0.27, P = 0.05), and PEA (r = 0.53, P < 0.0001) concentrations in skeletal muscle were associated with SMcont (Fig. 1A–1D, respectively). Further exploration of these associations with individual sphingomyelin compounds (Table 2) showed that investigated endocannabinoids and their congeners OEA and PEA positively correlated with several specific sphingomyelins (0.28 ≤ r ≤ 0.59; Supplemental Table 2). Importantly, AEA was associated with SM18:1/23:0 (r = 0.39, P = 0.004), SM18:1/23:1 (r = 0.39, P = 0.005), and SM18:1/26:1 (r = 0.39, P = 0.004), the three sphingomyelin compounds reported to be the strongest determinants of residual SLEEP in Native Americans (16). Conversely, 2-AG did not correlate with any of these three sphingomyelin species (all P > 0.05). Both OEA and PEA were associated with SM18:1/23:1 (r = 0.31, P = 0.03 and r = 0.35, P = 0.01, respectively) and SM18:1/26:1 (r = 0.39, P = 0.01 and r = 0.39, P = 0.005, respectively) also (Supplemental Table 2).
Figure 1.
Relationships between (A) AEA, (B) 2-AG, (C) OEA, and (D) PEA and the overall sphingomyelin content in skeletal muscle (SMcont), calculated as the average of individual sphingomyelin z scores reported in Table 2. For PEA, n = 52 because one subject had highly leveraged values for these measures (association between SMcont and PEA r = 0.54, P < 0.0001 upon inclusion). (E) The positive relationship between SPTLC3 and CNR1 gene expression levels in skeletal muscle. Gene expression levels were batch- and sex-standardized and reported in SD units. Pearson partial correlations are reported.
In our larger study subgroup with gene expression data (n = 106), CNR1 and SPTLC3 gene expression levels in skeletal muscle were positively correlated (r = 0.29, P = 0.003; Fig. 1E).
Endocannabinoids account for the sphingomyelin effect on SLEEP
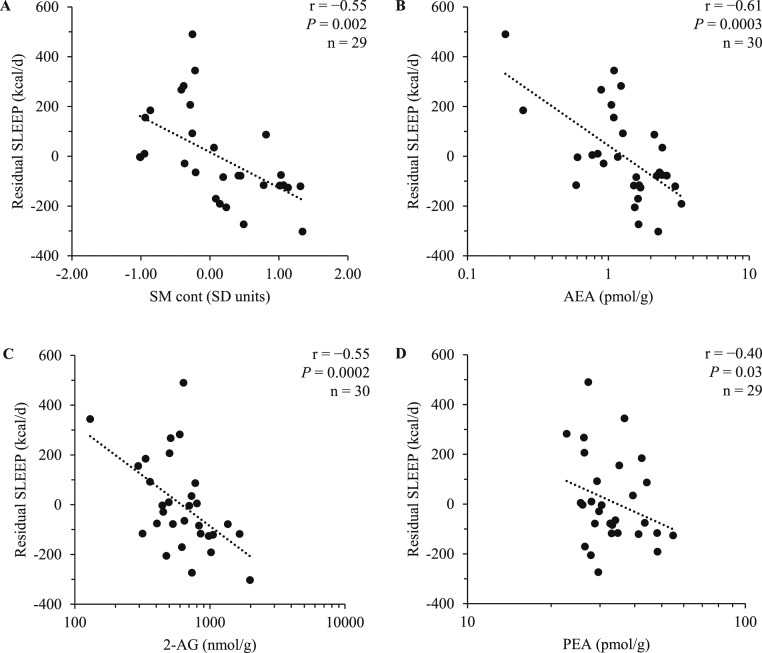
Mediation analyses were performed to test whether, for AEA, the effects of SMcont, SM18:1/23:0, SM18:1/23:1, and SM18:1/26:1 on residual SLEEP were accounted by AEA as an antecedent mediator. Conditions to perform mediation analyses for AEA were met, given the positive association of these lipids with SMcont and specific sphingomyelin compounds, as well as reported associations of sphingomyelins (16) and AEA (4) with residual SLEEP, respectively. Notably, both SMcont (r = −0.55, P = 0.002; Fig. 2A), AEA (r = −0.61, P = 0.0003; Fig. 2B), and 2-AG (r = −0.55, P = 0.0002; Fig. 2C) were inversely associated with residual SLEEP in the current study population, confirming our previous reports (4, 16).
Figure 2.
Correlation of the average (A) sphingomyelin content (SMcont) in skeletal muscle, (B) AEA, (C) 2-AG, and (D) PEA, with sleeping EE adjusted for FFM (residual SLEEP). For correlation of SMcont with residual SLEEP, 29 subjects were included because of incomplete assessment of sphingomyelins. For correlation of PEA with residual SLEEP, 29 subjects were included because of one subject with highly leveraged PEA measurement. As previously shown (4), the association of AEA and residual SLEEP was still significant upon exclusion of two subjects with lower AEA content (all P < 0.05). Pearson correlations and P values are reported.
The effect of SMcont on residual SLEEP accounted by AEA made up ~43% (−61.9 ± 31.9 kcal/d per SD unit increase in SMcont, P = 0.04) of the total effect of SMcont on residual SLEEP (−142.8 ± 41.6 kcal/d per SD unit increase in SMcont, P = 0.002). This effect due to AEA accounted for 77% of the effect of SMcontper se on residual SLEEP (−80.9 ± 44.1 kcal/d per SD unit increase in SMcont), which was negligible after we accounted for AEA (P = 0.09).
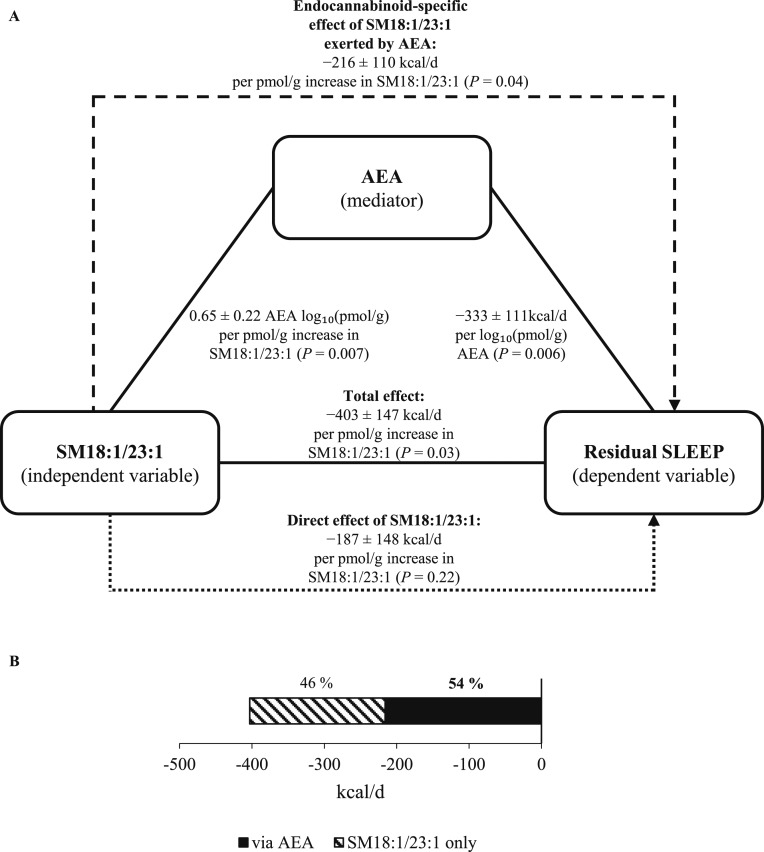
Because the effect of SMcont on residual SLEEP was dependent on AEA, post hoc mediation analyses were performed for the three specific sphingomyelin species (SM18:1/23:0, SM18:1/23:1, and SM18:1/26:1) that strongly associated with residual SLEEP (16). Figure 3A shows the mediation scheme for SM18:1/23:1. The endocannabinoid-specific effect of SM18:1/23:1 on residual SLEEP exerted by AEA accounted for 54% (−215.9 ± 109.7 kcal/d per pmol/g increase in SM18:1/23:1, P = 0.04, Fig. 3B) of the total effect of this sphingomyelin on residual SLEEP (−402.6 ± 146.7 kcal/d per pmol/g, P = 0.03). The effect of SM18:1/23:1 on residual SLEEP accounted by AEA was 1.2 times greater than the effect of this sphingomyelin per se, which was insignificant (P = 0.22). Similar results were obtained for SM18:1/23:0 and SM18:1/26:1 (see Supplemental Materials).
Figure 3.
(A) Example of the mediation analysis of the sphingomyelin effect on residual SLEEP accounted for by AEA. Residual SLEEP was calculated after adjustment for FFM via linear regression analysis. Path coefficients (±SE) are shown for the associations between SM18:1/23:1 (independent variable) and AEA (mediator), SM18:1/23:1 and SLEEP (dependent variable), and AEA and residual SLEEP, and were derived by linear regression analysis. The endocannabinoid-specific effect of SM18:1/23:1 on residual SLEEP exerted by AEA was calculated by multiplication of the path coefficients for the associations between SM18:1/23:1 and AEA and between AEA and residual SLEEP, respectively. The Sobel test was used to test the significance of the endocannabinoid-specific effect. Mediation analysis was performed in 29 subjects with complete SLEEP, endocannabinoid, and sphingolipid measurements. (B) Percentage of the total effect of SM18:1/23:1 on residual SLEEP as accounted by the endocannabinoid-specific (i.e., via AEA) vs direct (i.e., SM18:1/23:1 only, independent of AEA) effect.
2-AG did not substantially account for the associations between SMcont, SM18:1/23:0, SM18:1/23:1, and SM18:1/26:1 with residual SLEEP (all P > 0.05). With regard to endocannabinoid congeners, only PEA (r = −0.40, P = 0.03; Fig. 2D), but not OEA (r = −0.05, P = 0.80), was associated with residual SLEEP; however, the relationship between PEA and residual SLEEP became insignificant (P = 0.08) after we controlled for AEA. Because of the association between PEA and SMcont, SM18:1/23:1, and SM18:1/26:1, as well as the association between PEA (but not OEA) with residual SLEEP, mediation analyses were also performed for PEA as an antecedent mediator. No mediation effect was observed when PEA was introduced as an antecedent mediator for the relationship between SMcont, SM18:1/23:1, and SM18:1/26:1 with residual SLEEP (all P > 0.05).
Discussion
The current study investigated whether in skeletal muscle the endocannabinoids AEA and 2-AG and their congeners OEA and PEA influence the previously reported inverse association between sphingomyelin content and SLEEP in Native Americans of Southwestern heritage. In agreement with in vitro and rodent studies describing a biological link between the ECS and cellular sphingolipid content (20–23), we showed that AEA, 2-AG, OEA, and PEA concentrations positively correlated with the sphingomyelin content in skeletal muscle. Additionally, the concentrations of specific sphingomyelins previously shown to account for a large amount of the interindividual variance in SLEEP in this same ethnic group (16) were all associated with AEA but not with 2-AG. Importantly, the results of our mediation analyses indicate that AEA substantially contributes (54%) to the negative association between these sphingomyelins and SLEEP. Supporting the assumption that the ECS may increase sphingolipid synthesis via the CB1 receptor (21–23), CNR1 expression in skeletal muscle positively correlated with the expression of SPTLC3, the same SPT subunit reported to respond to ECS tone in rodent hepatocytes (21). Taken together, our results indicate that a decrease in whole-body EE as part of the peripheral effect of endocannabinoids acts through sphingomyelins and that, in skeletal muscle, AEA substantially contributes to and is responsible for the sphingomyelin effect on sleeping EE.
Endocannabinoids and sphingolipids are lipid signaling molecules that regulate human energy metabolism (4, 10, 16, 30, 42). In in vitro studies of rodent and human skeletal muscle, sphingolipids were shown to interfere with mitochondrial oxidative activity and integrity (17, 18). Endocannabinoids may exert their influence on EE via a negative regulatory effect on skeletal muscle glucose uptake. In rodent studies, administration of inverse agonists to the CB1 receptor led to an increase in skeletal muscle glucose uptake and to improvement in insulin sensitivity (8, 9), findings consistent with impaired insulin signaling observed in isolated human skeletal muscle tissue upon incubation with AEA (10). Skeletal muscle is the major organ responsible for serum glucose disposal (11) and, as part of FFM, a contributor to human EE (11, 35). Obese subjects with hyperglycemia experience a lower rate of carbohydrate oxidation and higher rate of fat oxidation (43), and with impairment of glucose tolerance the thermogenic response to glucose was shown to decrease in humans (12). The previously reported inverse association of skeletal muscle endocannabinoid content with EE (4) might therefore be explained by impairment of glucose metabolism caused by a higher ECS tone.
AEA correlated with sphingomyelins in skeletal muscle and mediated the sphingolipid effect on SLEEP, an effect not seen for 2-AG. Although we recently reported inverse associations between both skeletal muscle AEA and 2-AG with EE (4), AEA and 2-AG were also independent predictors of EE, indicating that these lipid species may affect EE via different pathways (4): AEA has distinct cellular functions in skeletal muscle, including a negative effect on insulin-dependent glucose uptake (10), thus explaining a link between EE and AEA. For 2-AG, however, less is known about its role in skeletal muscle metabolism (2, 44) and it may be that 2-AG is involved in pathways with a more indirect effect on metabolism such as skeletal muscle cell differentiation (44).
The mediation effect of AEA on the inverse association of sphingomyelins with SLEEP provides insight into potential mechanisms underlying the association between endocannabinoids and SLEEP. Although further studies (particularly in skeletal muscle) are warranted to elucidate the underlying biological mechanisms, these results indicate that endocannabinoids and sphingomyelins act in concert to regulate EE and are in line with previous reports of a cellular link between these lipid moieties (20–23). As seen for sphingolipids, previous studies reported a negative effect of the ECS on mitochondrial respiration (20, 45). Reduction of mitochondrial respiratory activity and disruption of mitochondrial integrity attributed to sphingolipid action in skeletal muscle was proposed to involve inhibition of the key positive mitochondrial regulator of mitochondrial respiratory activity protein kinase B (20, 46, 47). Importantly, AEA-mediated inhibition of protein kinase B activation is dependent on sphingolipids (20, 21). This common pathway may explain the mediation effect of AEA on the negative association between sphingomyelins and SLEEP. Supporting an impact of these subcellular mechanisms on whole-body EE, higher ECS tone and sphingolipid accumulation in metabolically active tissues are positively associated with obesity in humans (1, 42, 48), and in our previous studies we showed that the skeletal muscle contents of both endocannabinoids and sphingolipid compounds are strong determinants of human EE (4, 16). Interestingly, a higher ECS tone increases cellular sphingolipid levels in several tissues (20–23), possibly via increased activity of SPT, the central enzyme in sphingolipid synthesis (26), by the CB1 receptor (20, 21).
CB1 receptor antagonism in vitro inhibits de novo ceramide synthesis in hepatocytes, probably via modulation of SPTLC3 expression (21). We observed an association between SPTLC3 and CNR1 expression, supporting a comparable relationship between the ECS and sphingolipids in skeletal muscle. Although assumptions about protein content and activity based on mRNA expression levels must be considered carefully, higher SPTLC3 mRNA levels correlate with sphingolipid accumulation (21, 27), and for CNR1, increased gene expression increases CB1 receptor availability (49, 50). Therefore, the association between skeletal muscle SPTLC3 and CNR1 expression might reflect a role for the ECS in muscle sphingolipid synthesis. Although studies indicate that endocannabinoids increase cellular sphingolipid content via the CB1 receptor (21, 24), which in turn would reduce whole-body energy metabolism (51), the current association between SPTLC3 and CNR1 gene expression must be confirmed by future functional studies in skeletal muscle.
Ceramide studies provide the bulk of evidence that sphingolipids reduce mitochondrial activity via lipid signaling (18, 20, 29, 30, 42). It has previously been proposed that both ceramides and sphingomyelins might be involved in similar signaling pathways (16, 28–30), consistent with the tight link between these two sphingolipid subgroups (22, 29, 30). Ceramides participating in cellular signaling are created mainly by hydrolysis of sphingomyelins (22, 29), and, conversely, most newly synthesized ceramides are transformed into sphingomyelins (30). Accordingly, alongside ceramides, the sphingomyelin content in skeletal muscle was previously shown to be a determinant of human EE, particularly SLEEP (4). In that study, however, sphingomyelins appeared to have a stronger negative effect on SLEEP compared with ceramides, rendering them candidates for further exploration regarding their role in energy balance regulation (16, 28–30). Additionally, sphingomyelins play a central role in pathways leading to ceramide accumulation, as mediated by the ECS via sphingomyelin hydrolysis (22) or by SPT-dependent de novo synthesis (21, 22). Thus, an ECS-induced increase in ceramide synthesis and a subsequent ceramide-mediated decrease in mitochondrial respiratory activity may also involve sphingomyelins (20, 24–26). Our present results are consistent with a key role for sphingomyelins in EE control and with an influence of the ECS on skeletal muscle sphingomyelin content.
Studies using murine cell models proposed that OEA may activate lipolysis in skeletal muscle via stimulation of peroxisome proliferator-activated receptor α (15). Possibly via stimulation of the transient receptor potential cation channel subfamily V member 1 in skeletal muscle (13, 52), OEA and PEA might improve mitochondrial function in a calcium-dependent way (13). These mechanisms might explain how, via an increase in mitochondrial activity, greater content of OEA and PEA in skeletal muscle may ultimately lead to higher EE (16, 17, 37). Although in our study both OEA and PEA correlated with skeletal muscle sphingomyelins and an inverse association between PEA and SLEEP was observed, both lipids did not mediate the sphingomyelin effect on SLEEP. For OEA, a possible explanation may be that, in humans, this lipid plays a more dominant role in regulating lipolytic activity in adipose tissue but not in skeletal muscle (4). The association of PEA with SLEEP suggests that it could act as a lipid regulator of human EE, although this association was no longer significant after we controlled for AEA. Importantly, the lack of a mediation effect by PEA on the relationship between SM and SLEEP indicates that sphingomyelins and PEA may not interact to regulate whole-body EE in our study group.
One assumption of the current study is that the skeletal muscle endocannabinoid and sphingomyelin content in the vastus lateralis muscle are representative of other type 1 muscle fibers. To our knowledge, comparisons of the muscle endocannabinoid and sphingomyelin content across the human body are not available. However, human skeletal muscle (vastus lateralis muscle), subcutaneous adipose tissue, and plasma sphingomyelin content are correlated (16), supporting the idea of a common distribution of sphingomyelins across tissues. Furthermore, our cohort is composed by Native Americans of Southwestern heritage, which may limit the generalizability of our results to other populations, although previous metabolic studies from this cohort have been confirmed in other populations (53). The homogeneity of our study group is a strength because our measures are less prone to confounders. We acknowledge that functional studies of the endocannabinoid influence on sphingomyelin synthesis in relation to energy metabolism and mitochondrial activity are still needed to elucidate the underlying cellular mechanisms.
To conclude, in Native Americans of Southwestern heritage, the endocannabinoid and sphingomyelin contents in skeletal muscle are correlated. Mediation analyses demonstrated that AEA is responsible for the inverse association between sphingomyelins and SLEEP, indicating that these lipid entities probably interact in regulating SLEEP in this population. Therefore, because the ECS remains a promising target for obesity treatment, the link between the ECS and sphingomyelins in human energy balance control might provide a salient pathway for future pharmacological interventions.
Supplementary Material
Acknowledgments
The authors thank all volunteers for their participation in our clinical study.
Financial Support: Intramural Research Program of the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases.
Clinical Trial Information: ClinicalTrials.gov no. NCT00340132 (registered 18 August 1982).
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- 2-AG
2-arachidonoylglycerol
- AEA
anandamide
- CB1
cannabinoid receptor-1
- CNR1
cannabinoid receptor-1
- ECS
endocannabinoid system
- EE
energy expenditure
- FFM
fat-free mass
- FM
fat mass
- OEA
oleoylethanolamide
- PEA
palmitoylethanolamide
- SLEEP
sleeping energy expenditure
- SPT
serine palmitoyltransferase
- SPTLC3
serine palmitoyltransferase long chain subunit 3
References
- 1. Mazier W, Saucisse N, Gatta-Cherifi B, Cota D. The endocannabinoid system: pivotal orchestrator of obesity and metabolic disease. Trends Endocrinol Metab. 2015;26(10):524–537. [DOI] [PubMed] [Google Scholar]
- 2. Silvestri C, Di Marzo V. The endocannabinoid system in energy homeostasis and the etiopathology of metabolic disorders. Cell Metab. 2013;17(4):475–490. [DOI] [PubMed] [Google Scholar]
- 3. Matias I, Di Marzo V. Endocannabinoids and the control of energy balance. Trends Endocrinol Metab. 2007;18(1):27–37. [DOI] [PubMed] [Google Scholar]
- 4. Heinitz S, Basolo A, Piaggi P, Piomelli D, Jumpertz von Schwartzenberg R, Krakoff J. Peripheral endocannabinoids associated with energy expenditure in Native Americans of southwestern heritage. J Clin Endocrinol Metab. 2018;103(3):1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Addy C, Wright H, Van Laere K, Gantz I, Erondu N, Musser BJ, Lu K, Yuan J, Sanabria-Bohórquez SM, Stoch A, Stevens C, Fong TM, De Lepeleire I, Cilissen C, Cote J, Rosko K, Gendrano IN III, Nguyen AM, Gumbiner B, Rothenberg P, de Hoon J, Bormans G, Depré M, Eng WS, Ravussin E, Klein S, Blundell J, Herman GA, Burns HD, Hargreaves RJ, Wagner J, Gottesdiener K, Amatruda JM, Heymsfield SB. The acyclic CB1R inverse agonist taranabant mediates weight loss by increasing energy expenditure and decreasing caloric intake. Cell Metab. 2008;7(1):68–78. [DOI] [PubMed] [Google Scholar]
- 6. Liu YL, Connoley IP, Wilson CA, Stock MJ. Effects of the cannabinoid CB1 receptor antagonist SR141716 on oxygen consumption and soleus muscle glucose uptake in Lep(ob)/Lep(ob) mice. Int J Obes. 2005;29(2):183–187. [DOI] [PubMed] [Google Scholar]
- 7. Kunz I, Meier MK, Bourson A, Fisseha M, Schilling W. Effects of rimonabant, a cannabinoid CB1 receptor ligand, on energy expenditure in lean rats. Int J Obes. 2008;32(5):863–870. [DOI] [PubMed] [Google Scholar]
- 8. Lindborg KA, Teachey MK, Jacob S, Henriksen EJ. Effects of in vitro antagonism of endocannabinoid-1 receptors on the glucose transport system in normal and insulin-resistant rat skeletal muscle. Diabetes Obes Metab. 2010;12(8):722–730. [DOI] [PubMed] [Google Scholar]
- 9. Lindborg KA, Jacob S, Henriksen EJ. Effects of chronic antagonism of endocannabinoid-1 receptors on glucose tolerance and insulin action in skeletal muscles of lean and obese Zucker rats. Cardiorenal Med. 2011;1(1):31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eckardt K, Sell H, Taube A, Koenen M, Platzbecker B, Cramer A, Horrighs A, Lehtonen M, Tennagels N, Eckel J. Cannabinoid type 1 receptors in human skeletal muscle cells participate in the negative crosstalk between fat and muscle. Diabetologia. 2009;52(4):664–674. [DOI] [PubMed] [Google Scholar]
- 11. Abdul-Ghani MA, DeFronzo RA. Pathogenesis of insulin resistance in skeletal muscle. J Biomed Biotechnol. 2010;2010:476279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weyer C, Bogardus C, Pratley RE. Metabolic factors contributing to increased resting metabolic rate and decreased insulin-induced thermogenesis during the development of type 2 diabetes. Diabetes. 1999;48(8):1607–1614. [DOI] [PubMed] [Google Scholar]
- 13. Luo Z, Ma L, Zhao Z, He H, Yang D, Feng X, Ma S, Chen X, Zhu T, Cao T, Liu D, Nilius B, Huang Y, Yan Z, Zhu Z. TRPV1 activation improves exercise endurance and energy metabolism through PGC-1α upregulation in mice. Cell Res. 2012;22(3):551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaneko Y, Szallasi A. Transient receptor potential (TRP) channels: a clinical perspective. Br J Pharmacol. 2014;171(10):2474–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guzmán M, Lo Verme J, Fu J, Oveisi F, Blázquez C, Piomelli D. Oleoylethanolamide stimulates lipolysis by activating the nuclear receptor peroxisome proliferator-activated receptor α (PPAR-α). J Biol Chem. 2004;279(27):27849–27854. [DOI] [PubMed] [Google Scholar]
- 16. Heinitz S, Piaggi P, Vinales KL, Basolo A, Spranger J, Piomelli D, Krakoff J, Jumpertz von Schwartzenberg R. Specific skeletal muscle sphingolipid compounds in energy expenditure regulation and weight gain in Native Americans of southwestern heritage. Int J Obes. 2017;41(10):1585–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perreault L, Newsom SA, Strauss A, Kerege A, Kahn DE, Harrison KA, Snell-Bergeon JK, Nemkov T, D’Alessandro A, Jackman MR, MacLean PS, Bergman BC. Intracellular localization of diacylglycerols and sphingolipids influences insulin sensitivity and mitochondrial function in human skeletal muscle. JCI Insight. 2018;3(3):96805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Smith ME, Tippetts TS, Brassfield ES, Tucker BJ, Ockey A, Swensen AC, Anthonymuthu TS, Washburn TD, Kane DA, Prince JT, Bikman BT. Mitochondrial fission mediates ceramide-induced metabolic disruption in skeletal muscle. Biochem J. 2013;456(3):427–439. [DOI] [PubMed] [Google Scholar]
- 19. Yang G, Badeanlou L, Bielawski J, Roberts AJ, Hannun YA, Samad F. Central role of ceramide biosynthesis in body weight regulation, energy metabolism, and the metabolic syndrome. Am J Physiol Endocrinol Metab. 2009;297(1):E211–E224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lipina C, Irving AJ, Hundal HS. Mitochondria: a possible nexus for the regulation of energy homeostasis by the endocannabinoid system? Am J Physiol Endocrinol Metab. 2014;307(1):E1–E13. [DOI] [PubMed] [Google Scholar]
- 21. Cinar R, Godlewski G, Liu J, Tam J, Jourdan T, Mukhopadhyay B, Harvey-White J, Kunos G. Hepatic cannabinoid-1 receptors mediate diet-induced insulin resistance by increasing de novo synthesis of long-chain ceramides. Hepatology. 2014;59(1):143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guzmán M, Galve-Roperh I, Sánchez C. Ceramide: a new second messenger of cannabinoid action. Trends Pharmacol Sci. 2001;22(1):19–22. [DOI] [PubMed] [Google Scholar]
- 23. Gómez del Pulgar T, Velasco G, Sánchez C, Haro A, Guzmán M. De novo-synthesized ceramide is involved in cannabinoid-induced apoptosis. Biochem J. 2002;363(Pt 1):183–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gustafsson K, Christensson B, Sander B, Flygare J. Cannabinoid receptor-mediated apoptosis induced by R(+)-methanandamide and Win55,212-2 is associated with ceramide accumulation and p38 activation in mantle cell lymphoma. Mol Pharmacol. 2006;70(5):1612–1620. [DOI] [PubMed] [Google Scholar]
- 25. Velasco G, Galve-Roperh I, Sánchez C, Blázquez C, Haro A, Guzmán M. Cannabinoids and ceramide: two lipids acting hand-by-hand. Life Sci. 2005;77(14):1723–1731. [DOI] [PubMed] [Google Scholar]
- 26. Huwiler A, Kolter T, Pfeilschifter J, Sandhoff K. Physiology and pathophysiology of sphingolipid metabolism and signaling. Biochim Biophys Acta. 2000;1485(2-3):63–99. [DOI] [PubMed] [Google Scholar]
- 27. Hornemann T, Penno A, Rütti MF, Ernst D, Kivrak-Pfiffner F, Rohrer L, von Eckardstein A. The SPTLC3 subunit of serine palmitoyltransferase generates short chain sphingoid bases. J Biol Chem. 2009;284(39):26322–26330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chakraborty M, Jiang X-C. Sphingomyelin and its role in cellular signaling. Adv Exp Med Biol. 2013;991:1–14. [DOI] [PubMed] [Google Scholar]
- 29. Milhas D, Clarke CJ, Hannun YA. Sphingomyelin metabolism at the plasma membrane: implications for bioactive sphingolipids. FEBS Lett. 2010;584(9):1887–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tafesse FG, Huitema K, Hermansson M, van der Poel S, van den Dikkenberg J, Uphoff A, Somerharju P, Holthuis JC. Both sphingomyelin synthases SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. J Biol Chem. 2007;282(24):17537–17547. [DOI] [PubMed] [Google Scholar]
- 31. Ferraro R, Boyce VL, Swinburn B, De Gregorio M, Ravussin E. Energy cost of physical activity on a metabolic ward in relationship to obesity. Am J Clin Nutr. 1991;53(6):1368–1371. [DOI] [PubMed] [Google Scholar]
- 32. Seino Y, Nanjo K, Tajima N, Kadowaki T, Kashiwagi A, Araki E, Ito C, Inagaki N, Iwamoto Y, Kasuga M, Hanafusa T, Haneda M, Ueki K; Committee of the Japan Diabetes Society on the Diagnostic Criteria of Diabetes Mellitus . Report of the committee on the classification and diagnostic criteria of diabetes mellitus. J Diabetes Investig. 2010;1(5):212–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mazess RB, Barden HS, Bisek JP, Hanson J. Dual-energy x-ray absorptiometry for total-body and regional bone-mineral and soft-tissue composition. Am J Clin Nutr. 1990;51(6):1106–1112. [DOI] [PubMed] [Google Scholar]
- 34. Realini N, Solorzano C, Pagliuca C, Pizzirani D, Armirotti A, Luciani R, Costi MP, Bandiera T, Piomelli D. Discovery of highly potent acid ceramidase inhibitors with in vitro tumor chemosensitizing activity. Sci Rep. 2013;3(1):1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ravussin E, Lillioja S, Anderson TE, Christin L, Bogardus C. Determinants of 24-hour energy expenditure in man. Methods and results using a respiratory chamber. J Clin Invest. 1986;78(6):1568–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schutz Y, Ravussin E, Diethelm R, Jequier E. Spontaneous physical activity measured by radar in obese and control subject studied in a respiration chamber. Int J Obes. 1982;6(1):23–28. [PubMed] [Google Scholar]
- 37. Cavuoto P, McAinch AJ, Hatzinikolas G, Cameron-Smith D, Wittert GA. Effects of cannabinoid receptors on skeletal muscle oxidative pathways. Mol Cell Endocrinol. 2007;267(1-2):63–69. [DOI] [PubMed] [Google Scholar]
- 38. Piaggi P, Thearle MS, Bogardus C, Krakoff J. Lower energy expenditure predicts long-term increases in weight and fat mass. J Clin Endocrinol Metab. 2013;98(4):E703–E707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. MacKinnon DP. Introduction to Statistical Mediation Analysis. London, UK: Routledge; 2008. [Google Scholar]
- 40. Piaggi P, Thearle MS, Krakoff J, Votruba SB. Higher daily energy expenditure and respiratory quotient, rather than fat-free mass, independently determine greater ad libitum overeating. J Clin Endocrinol Metab. 2015;100(8):3011–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sobel ME. Asymptotic confidence intervals for indirect effects in structural equation models. Sociol Methodol. 1982;13:290–312. [Google Scholar]
- 42. Choi S, Snider AJ. Sphingolipids in high fat diet and obesity-related diseases. Mediators Inflamm. 2015;2015:520618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Felber J-P, Ferrannini E, Golay A, Meyer HU, Theibaud D, Curchod B, Maeder E, Jequier E, DeFronzo RA. Role of lipid oxidation in pathogenesis of insulin resistance of obesity and type II diabetes. Diabetes. 1987;36(11):1341–1350. [DOI] [PubMed] [Google Scholar]
- 44. Iannotti FA, Silvestri C, Mazzarella E, Martella A, Calvigioni D, Piscitelli F, Ambrosino P, Petrosino S, Czifra G, Bíró T, Harkany T, Taglialatela M, Di Marzo V. The endocannabinoid 2-AG controls skeletal muscle cell differentiation via CB1 receptor-dependent inhibition of Kv7 channels. Proc Natl Acad Sci USA. 2014;111(24):E2472–E2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Athanasiou A, Clarke AB, Turner AE, Kumaran NM, Vakilpour S, Smith PA, Bagiokou D, Bradshaw TD, Westwell AD, Fang L, Lobo DN, Constantinescu CS, Calabrese V, Loesch A, Alexander SP, Clothier RH, Kendall DA, Bates TE. Cannabinoid receptor agonists are mitochondrial inhibitors: a unified hypothesis of how cannabinoids modulate mitochondrial function and induce cell death. Biochem Biophys Res Commun. 2007;364(1):131–137. [DOI] [PubMed] [Google Scholar]
- 46. Hajduch E, Balendran A, Batty IH, Litherland GJ, Blair AS, Downes CP, Hundal HS. Ceramide impairs the insulin-dependent membrane recruitment of protein kinase B leading to a loss in downstream signalling in L6 skeletal muscle cells. Diabetologia. 2001;44(2):173–183. [DOI] [PubMed] [Google Scholar]
- 47. Hajduch E, Turban S, Le Liepvre X, Le Lay S, Lipina C, Dimopoulos N, Dugail I, Hundal HS. Targeting of PKCzeta and PKB to caveolin-enriched microdomains represents a crucial step underpinning the disruption in PKB-directed signalling by ceramide. Biochem J. 2008;410(2):369–379. [DOI] [PubMed] [Google Scholar]
- 48. Blachnio-Zabielska A, Baranowski M, Zabielski P, Gorski J. Effect of high fat diet enriched with unsaturated and diet rich in saturated fatty acids on sphingolipid metabolism in rat skeletal muscle. J Cell Physiol. 2010;225(3):786–791. [DOI] [PubMed] [Google Scholar]
- 49. Mukhopadhyay B, Liu J, Osei-Hyiaman D, Godlewski G, Mukhopadhyay P, Wang L, Jeong W-I, Gao B, Duester G, Mackie K, Kojima S, Kunos G. Transcriptional regulation of cannabinoid receptor-1 expression in the liver by retinoic acid acting via retinoic acid receptor-γ. J Biol Chem. 2010;285(25):19002–19011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Proto MC, Gazzerro P, Di Croce L, Santoro A, Malfitano AM, Pisanti S, Laezza C, Bifulco M. Interaction of endocannabinoid system and steroid hormones in the control of colon cancer cell growth. J Cell Physiol. 2012;227(1):250–258. [DOI] [PubMed] [Google Scholar]
- 51. Herling AW, Kilp S, Elvert R, Haschke G, Kramer W. Increased energy expenditure contributes more to the body weight-reducing effect of rimonabant than reduced food intake in candy-fed wistar rats. Endocrinology. 2008;149(5):2557–2566. [DOI] [PubMed] [Google Scholar]
- 52. Di Marzo V, De Petrocellis L. Endocannabinoids as regulators of transient receptor potential (TRP) channels: a further opportunity to develop new endocannabinoid-based therapeutic drugs. Curr Med Chem. 2010;17(14):1430–1449. [DOI] [PubMed] [Google Scholar]
- 53. Stefan N, Stumvoll M, Weyer C, Bogardus C, Tataranni PA, Pratley RE. Exaggerated insulin secretion in Pima Indians and African-Americans but higher insulin resistance in Pima Indians compared to African-Americans and Caucasians. Diabet Med. 2004;21(10):1090–1095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.