Abstract
Acute myeloid leukemia (AML) is associated with the sequential accumulation of acquired genetic alterations. Although at diagnosis cytogenetic alterations are frequent in AML, roughly 50% of patients present an apparently normal karyotype (NK), leading to a highly heterogeneous prognosis. Due to this significant heterogeneity, it has been suggested that different molecular mechanisms may trigger the disease with diverse prognostic implications. We performed whole-exome sequencing (WES) of tumor-normal matched samples of de novo AML-NK patients lacking mutations in NPM1, CEBPA or FLT3-ITD to identify new gene mutations with potential prognostic and therapeutic relevance to patients with AML. Novel candidate-genes, together with others previously described, were targeted resequenced in an independent cohort of 100 de novo AML patients classified in the cytogenetic intermediate-risk (IR) category. A mean of 4.89 mutations per sample were detected in 73 genes, 35 of which were mutated in more than one patient. After a network enrichment analysis, we defined a single in silico model and established a set of seed-genes that may trigger leukemogenesis in patients with normal karyotype. The high heterogeneity of gene mutations observed in AML patients suggested that a specific alteration could not be as essential as the interaction of deregulated pathways.
Introduction
Acute myeloid leukemia (AML) is associated with the sequential accumulation of acquired genetic alterations and epigenetic changes in hematopoietic stem cells that alter processes involved in proliferation, differentiation and self-renewal. At diagnosis, cytogenetic alterations are frequent in AML patients [1,2]. Nevertheless, at least 50% of patients present an apparently normal karyotype (NK) being its prognosis highly heterogeneous. These patients are consequently classified in the cytogenetic intermediate-risk (IR) category. However, their significant heterogeneity suggests that there are different molecular mechanisms involved in the development of the disease as well as related to different prognostic implications [3–5]. The recent progress of next-generation sequencing (NGS) technologies has allowed for the identification of a growing number of novel mutations in leukaemias [6–9]. Their application has led to broaden the list of genes that seems to be involved in the pathogenesis of such disorders. Consequently, it has reflected their complexity and described patterns of cooperation and exclusion between different genes and cellular pathways involved in the mechanisms of leucemogenesis. In recent years, The Cancer Genome Atlas (TCGA) has collected data from exomic or genomic sequencing, expression, methylation, and genotyping of 200 adult patients with AML, identifying 23 different frequently mutated genes [10]. These genes were grouped into 8 different categories by their cellular function, detecting at least one mutation per group in 99% of the studied AML patients. Recently, Papaemmanuil et al. defined 3 additional molecular subgroups in AML patients with prognostic implications [11]. However, clinicians only rely on the presence of alterations of 3 well established molecular markers, NPM1, CEBPA or FLT3-ITD, for the diagnosis classification of patients with AML-IR [12].
To extend our knowledge on subtle genetic alterations involved in AML-NK, in this study, we have performed whole-exome sequencing (WES) of tumor-normal matched samples on a selected discovery cohort of de novo AML patients. To this aim, we studied samples of leukemia cells from adults under the age of 60 years who lacked cytogenetic abnormalities and well known molecular features that is, wild-type NPM1, CEBPA, and FLT3–ITD. Genes identified from this analysis were validated on an independent cohort by targeted resequencing. Finally, network analysis [13] was performed in silico to assess their putative role as driver genes.
Material and methods summary
We performed whole-exome sequencing (WES) of tumor-normal matched samples from 7 de novo AML-NK patients without mutations in NPM1, CEBPA and FLT3–ITD (“discovery cohort”). WES data were analyzed using an in-house bioinformatics pipeline [14] to compare the coding sequence of matched samples, filter out germline variants and identify somatically acquired deleterious changes. The variants detected were confirmed in both samples of each patient by targeted sequencing using an Ion AmpliSeq™ analysed in an Ion Proton™ System. By using the SureDesign Tool (Agilent) for NGS we developed a custom design targeting hotspot regions of 55 genes found mutated in the discovery cohort. Furthermore, complete coding sequence of extra 32 genes (reported to be mutated in at least 2% of patients from previous AML studies [10]) were also included. This design was tested in 100 additional de novo AML-IR patients (“validation cohort”). Variants were selected according to VAF≥1%, its absence in the healthy population (UCSC Common SNPs; MAF < 0.01) and its putative effect on the protein (excluding synonymous mutations). Finally, network enrichment analysis [13–16] was used to assess the candidate genes on the basis of their connectivity, mutational recurrence and co-occurrence and cancer related functionalities.
This study was approved by the Research Ethics Board of IISLAFE (No.2012/0175) and informed consent in accordance with the Declaration of Helsinki was obtained before taking sample for genetic and genomic research.
Further Material and Methods details are provided in the Extended Experimental Procedures at S1 File.
Results
Whole exome sequencing
We characterized a discovery cohort of 7 AML-NK de novo patients from whom paired diagnosis and complete remission DNA samples were available. Clinical characteristics of the patients at diagnosis are summarized in S1 Table. The target coverage average in both diagnostic and remission samples and the mean of reads by genomes was 228.97(±125.78)x and 32944(±4192), respectively (S2 and S3 Tables). A total of 402,412 variant positions were detected in all sequenced exons from all the samples. After an accurate filtering of the sequencing data [14], a total of 102 candidate somatic variants (94 missense SNVs and 8 small indels) were found (S3 Table). The distribution of the number of variants per exome showed an average of 30 mutations per sample (range 22–37). Recurrent mutations or common mutated genes were not observed within the seven cases. Targeted resequencing of both samples of the discovery cohort confirmed 64/102 deleterious somatic variants in 55 genes. Among them, 57 were missense mutations and 8 indels, which stood for an average of 9.2 mutations by sample (range 3–22). Of these 55 candidate genes, 26 were previously reported as mutated in AML patients in previous studies [10, 11]. Detailed information is provided at S3 Table.
Frequency and distribution of 87 genes in a validation cohort of AML patients
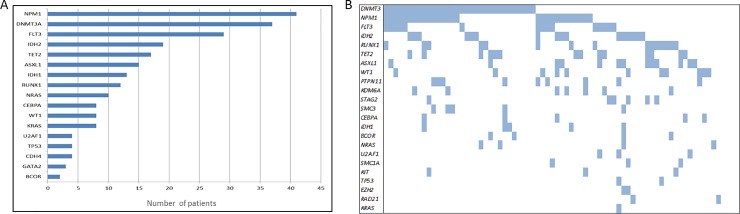
We further examined the mutation frequency of our candidate genes in an independent validation cohort of 100 de novo AML patients with intermediate cytogenetic risk. Details of the validation cohort are provided in S4 Table. Overall, mean target coverage was 260±106x, with an absolute median coverage of 220x at positions where mutations were identified. After processing the raw sequencing data with the procedures described in [14] we identified a total of 158 high-confidence different variants. Mutations were predominantly missense substitutions (53%), frameshift indels (34%), and nonframeshift indels (6%), in splice site regions (4%), nonsenses (2%), or ncRNA (1%) (S5 Table). A mean of 4.89 mutations per sample (range 0–10) were detected. Variants were detected affecting 73 different genes, being 28 of these variants concurrently harboured in more than one patient. Among the mutated genes, 35 showed mutations in at least 7% of the patients (range 7–34%), being the most frequent mutated genes NPM1, DNMT3A, FLT3, TET2 and ASXL1 (S5 Table). In silico analysis showed that most of the observed mutations were reported in the COSMIC database as mutations implicated in cancer. In addition, as we expected, recurrent hotspots previously reported were detected in genes such as DNMT3A, NPM1, NRAS, KRAS, FLT3, IDH2 and IDH1 (S5 Table). Finally, we analysed the co-occurrence of the identified mutations in our validation cohort, and identified that patients with DNMT3A, NPM1 and FLT3 did not usually harboured other mutations. In contrast, patients with DNMT3A and NPM1 appeared simultaneously with genes such as IDH2, PTPN11 or RUNX1. Finally, patients lacking mutations in DNMT3A showed a pattern that included a miscellany of altered genes (Fig 1).
Fig 1. Distribution of selected mutations along the different affected genes of the validation cohort.
A) Number of mutated samples by gene according to the described mutation filtering protocol. Only recurrent genes were included. B) Co-occurrence of all somatic mutations.
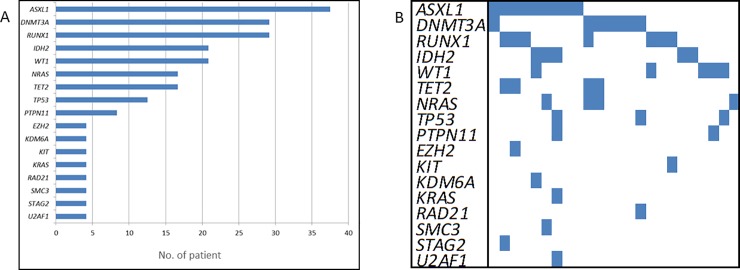
We found somatic mutations with deleterious effect in 100% of patients of the “validation cohort” (n = 24) carrying the same molecular features (i.e. without mutations in NPM1, CEBPA and FLT3–ITD) than the “discovery cohort”. Among them, 75% carried mutations in ASXL1, DNMT3A and/or RUNX1. Other genes such as DNAH9, IDH2, WT1, DNAH8, NRAS or TET2 were also recurrently mutated within this set of patients (Fig 2A). In addition, although DNMT3A mutations tend to appear as isolated alterations, ASXL1 and RUNX1 mutations mainly co-occur among them and with mutations in other genes (Fig 2B). In both cases, given the nature of these genes, the epigenetic regulation might be compromised. Moreover, we extended our analysis to the mutations described in AML patients from the TCGA cohort with the same molecular features (n = 20). Within TCGA patients, the most frequently mutated genes were DNMT3A, RUNX1, IDH2, TET2, and NRAS. Taking into account both sets of patients, we obtained an input of 44 samples with normal karyotype lacking mutations in NPM1, CEBPA, and FLT3-ITD, who harbor mutations in: DNMT3A (32%), RUNX1 (28%), ASXL1 (23%), IDH2 (21%), NRAS (17%), and TET2 (17%) genes.
Fig 2. Distribution of selected mutations along the different affected genes among validation cohort with same molecular features than the discovery cohort.
A) Number of mutated samples by gene according to the described mutation filtering protocol. Only recurrent genes were included. B) Co-occurrence of all somatic mutations.
Candidate gene prioritization
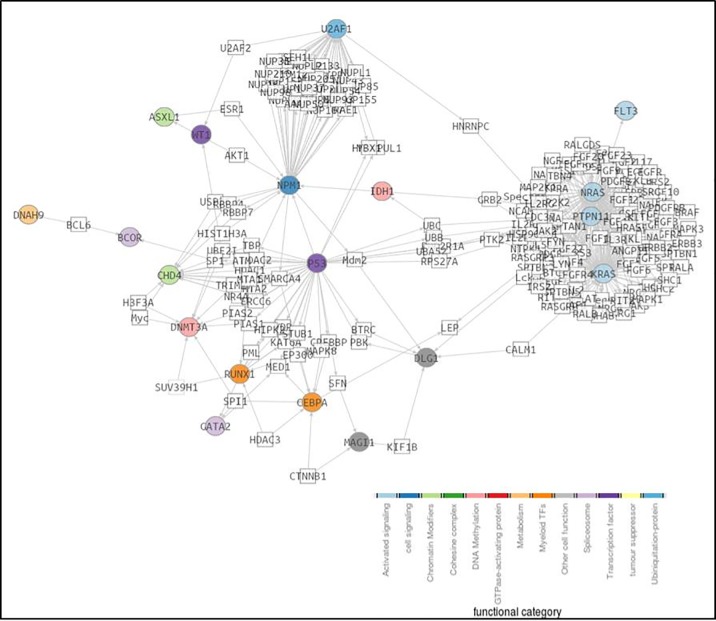
Focusing on the deleterious variants found in our AML patients, a gene prioritization analysis was performed in order to define an in silico model based on the physical interaction, regulation, functionality and cell pathway alterations caused by each mutation. The analysis included the interactome of the 28 candidate genes selected by harboring recurrent mutations and displaying SIFT values in the range of the deleteriousness, with significant intrinsic mutation rates (NPM1, DNMT3A, NRAS, PTPN11, IDH2, KRAS, WT1, IDH1, RUNX1, U2AF1, CEBPA, TP53, CHD4, PCDHA6, GATA2, ASXL1, DLG1, BCOR, PKD1L2, SIPA1L2, MAGI1, FAM70B, FCGBP, TET2, DNAH9, TEKT4, FLT3). We identified a network in which 19 out of the 28 genes were found to be significantly more connected that the random expectation (P = 0.02) (Fig 3). Moreover, 13 genes (NPM1, DNMT3A, NRAS, PTPN11, IDH2, KRAS, WT1, IDH1, RUNX1, U2AF1, CEBPA, TP53, CHD4) also displayed an accumulation of mutations significantly higher that the corresponding healthy controls taken from the 1000 genomes repository (P≤0.01) [17]. Focusing on these genes, we found an average of 2 altered genes by patient. Although not always the same pathway was altered, the DNA methylation pathway prevailed above the others. Indeed, more than half of the patients had alterations in the genes involved in this pathway. Moreover, the co-occurrence of mutations in genes involved in less frequent pathways (i.e. signalling activation, chromatin modification, transcription factors and cohesine complex) may trigger the leukemogenesis (Fig 4).
Fig 3. Network-based analysis (SNOW; Babelomics) applied to 28 selected genes.
The network was complemented with the co-occurrence relationships, in order to summarize the two kind of significant results. Significant network-based analysis genes are coloured depending on their biological role and circle shaped. Intermediate genes were painted in white and square shaped. While grey edges represent protein-protein interaction, relationships, broad orange dashed lines describe significant co-occurrences.
Fig 4. Distribution of mutations according to their functional category among the validation cohort.
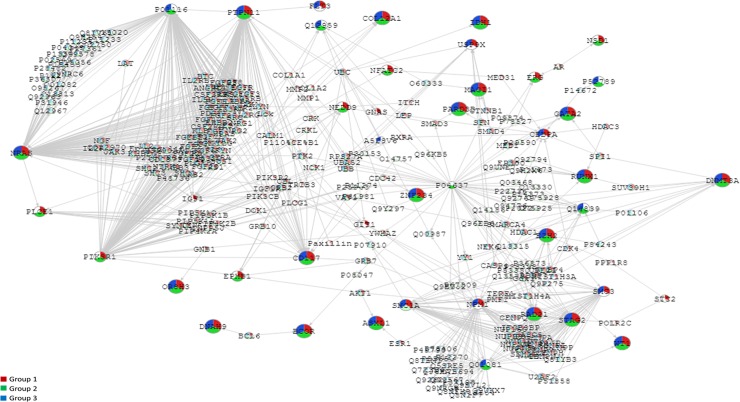
In addition, according to the in silico model, we were able to classify patients in 3 different sets: patients with NK and/or well- known gene mutations that is, NPM1, CEBPA, and/or FLT3–ITD (Group 1, n = 56); patients with NK and lacking mutations in NPM1, CEBPA, and FLT3–ITD (Group 2, n = 22); and IR patients with cytogenetic abnormalities and/or known molecular features, NPM1, CEBPA, and/or FLT3–ITD (Group 3, n = 22). According to these, we found 3 different pathways rather similar between them. Comparing all the pathways, we defined a single in silico model including the alterations detected in our sets of patients (Fig 5). Focusing on group 1 and 2, we established a set of seed-genes that might be potentially involved in leukemogenesis due to not being present in patients harboring cytogenetic abnormalities (Group 3), such as NSD1 (12%), PLCE1 (9%), NFATC2 (5%), EPHB1 (5%), ERG (4%), NEDD9 (3%) and PIK3R1 (3%). Pathways affected by the presence of these mutations might have a similar effect in the cell than IR-cytogenetic alterations.
Fig 5. Network-based analysis (SNOW; Babelomics) applied to 28 selected genes.
The network was complemented with the co-occurrence relationships, in order to summarize the two kind of significant results. Significant network-based analysis genes are coloured depending their categorical group: patients with NK and/or well- known gene mutations (Group 1, red), patients with NK and without mutations in NPM1, CEBPA and FLT3–ITD (Group 2, green) and IR patients with cytogenetic abnormalities and/or known molecular features (Group 3, blue). Grey edges represent protein-protein interaction, relationships, broad orange dashed lines describe significant co-occurrences.
Clinical correlations
Variables included in the regression analyses were age, gender, WBC count, platelet count, percentage of bone marrow and peripheral blood blasts, cytogenetic risk group, MRC code, ECOG, mutational status of NPM1 (exon 12), FLT3, CEBPA, and all those genes analyzed by NGS. The follow-up of the patients was updated on December 2017, and all follow-up data were censored at that time point. The median follow-up of surviving patients was 52 months (range, 26 to 164).
No significant correlation was found between the mutations detected and the clinical or biological features studied. In addition, our results did not show statistically significant pattern of cooperating mutations in the studied group of genes. However, results from the elastic net cox regression regarding overall survival, displayed a complex pattern of associations between the different mutations and risk of death, with 26 different variables and age interacting. Hazard ratios are presented in Table 1 and partial dependence plots for the different variables are depicted in S1 Fig. With reference to event-free survival values, a higher risk of adverse event was associated to mutations in DNAH8, TET2, RUNX1, and WT1; whilst a lower risk of adverse event was correlated to the presence of mutations in KDM6A. Hazard ratios are presented in Table 2. To facilitate the interpretation of these results, partial dependence plots for the different variables are provided in S2 Fig.
Table 1. Coefficients and their corresponding Hazard Ratios for overall survival based on predictive factors in 100 de novo AML samples.
| Variable | Coefficient | HR |
|---|---|---|
| Age | 0.017 | 1.018 |
| FLT3-ITD | 0.150 | 1.162 |
| KRAS | 0.009 | 1.010 |
| PTPN11 | 0.001 | 1.001 |
| DNAH8 | 0.093 | 1.097 |
| IGF1 | -0.026 | 0.975 |
| EZH2 | 0.263 | 1.301 |
| SMC3 | -0.009 | 0.991 |
| RAD21 | 0.386 | 1.471 |
| DNMT3A | 0.015 | 1.015 |
| IDH2 | 0.047 | 1.048 |
| TET2 | 0.179 | 1.196 |
| SIPA1L2 | -0.027 | 0.974 |
| GPR6 | 0.114 | 1.121 |
| PKD1L2 | 0.187 | 1.205 |
| RUNX1 | 0.060 | 1.062 |
| NPM1 | -0.134 | 0.874 |
| KCNU1 | -0.114 | 0.892 |
| FAM69B | 0.038 | 1.038 |
| NLRP5 | -0.164 | 0.848 |
| PCDHA7 | 0.511 | 1.667 |
| ZCCHC1 | -0.025 | 0.975 |
| CCNL2 | -0.014 | 0.986 |
| TP53 | 0.376 | 1.457 |
| WT1 | 0.165 | 1.179 |
| PIK3R | -0.123 | 0.884 |
| OTUD7A | 0.301 | 1.352 |
Table 2. Hazard ratios for event free survival based on predictive factors in 100 de novo AML samples.
| Variable | Coefficient | HR |
|---|---|---|
| DNAH8 | 0,045 | 1,046 |
| KDM6A | -0,005 | 0,995 |
| TET2 | 0,057 | 1,059 |
| RUNX1 | 0,008 | 1,008 |
| WT1 | 0,147 | 1,158 |
Discussion
This study shows a comprehensive analysis of AML, combining WES with the assessment of somatic mutations by a custom next-generation sequencing panel of targeted genes with network enrichment analysis. In the “discovery cohort”, we identified and validated 64 deleterious somatic variants in 55 genes that were further targeted sequencing in the “validation cohort” of patients along with 32 extra genes previously reported to play a significant role in AML pathogenesis [10]. After bioinformatic filtering of variants, we identified likely deleterious mutations in 73 genes, where 35 were previously reported as recurrent mutated in AML [10, 11]. Network enrichment analysis identified 13 recurrently mutated genes involved in AML-IR pathogenesis. Among them, we defined a single in silico model and established a set of seed-genes that may trigger leukemogenesis.
To identify leukemia-specific somatic alterations that could cooperate in the development and progression of leukemia, WES analysis was performed in 7 tumor-normal matched samples. As a normal paired sample, we examined DNA from saliva or from bone marrow at complete remission of each single case (defined according to the recommendations of Cheson et al.). To prevent false-positive calls in WES, all stringent filtered variants were resequenced by means of target NGS in paired samples of each patient using a different NGS platform. Among them, 64 deleterious variants were confirmed to be true. Although our protocol took into account several sequencing artifacts as strand bias and other measurable effects derived from poorly mapped reads, some of the not validated mutations could be false positives inherent in the technology used. Furthermore, the issue to confidently identifying indels or the presence of low frequency alleles could explain some false negative calls at the validation resequencing. As a whole, these technical limitations might imply an underestimation of candidate mutated genes in the “discovery cohort”.
In a first step, we identified 55 candidate genes, where 26 were previously reported as mutated in AML patients in previous studies [10]. The identification of previously reported driver mutations validated the robustness of our approach. Then, we extended the analyses through the examination of our candidate genes in an independent validation cohort of 100 de novo AML-IR patients. For this propose, a custom panel of 87 genes was designed. Although custom panels require a previous design stage and are optimized by the laboratory, their main advantage is the flexibility they offer, since you can include the regions of your interest and change this design over time (add or remove regions) depending on the needs of each pathology. As no germline matched sample was available from each patient, we implemented a germinality test previously described by our group [14]. In order to filter out the majority of germline variants, we compared the mutations found in our patients against the healthy 1000G cohort and other public variant population databases. Finally, we found a mean of 4.89 mutations per sample (range 0–10) affecting 73 different genes of which 35 genes were recurrently mutated in more than one patient. Recurrent hotspots in genes such as DNMT3A, NRAS, KRAS and IDH1/2 were also present in several samples. In our whole validation cohort, the most frequent mutated genes were NPM1, DNMT3A, FLT3, TET2, and ASXL1. These results agreed with those previously reported [10, 11]. When we focussed our analysis on those patients from the validation cohort with same molecular features than the discovery cohort (n = 24), we found somatic mutations with deleterious effect in 100% of patients, mainly in ASXL1, DNMT3A and/or RUNX1. There were other mutated genes within this set of patients such as DNAH9, IDH2, WT1, DNAH8, NRAS or TET2. However, we found that almost 25% of the patients carried DNMT3A mutations as an only alteration whilst 13% presented ASXL1 and RUNX1 in a simultaneous manner. Although this association was established evaluating a subset of 24 patients, such observations might suggest that the alterations harboured by this set of patients could derive in an aberrant epigenetics regulation. In line with recent reports, highlighting the complex nature of genomic aberrations in leukemia [4, 6, 7], probably the mechanisms that trigger the leukemogenic process could have a common consequence. Functional validation studies will be required to assess the importance of these mutations for AML pathogenesis when well established mutations are not present.
Likewise, exploratory in silico approaches were used to provide a reliable approach to prioritize genes that would undergo additional analysis. Candidate genes were selected according to the recurrence of mutations and their SIFT values in the range of the deleteriousness as it has been previously reported [14]. Network analysis resulted in a significantly connected subnetwork in which 19 of 28 genes were significantly more connected that the random expectation. Amidst, 13 genes (NPM1, DNMT3A, NRAS, PTPN11, IDH2, KRAS, WT1, IDH1, RUNX1, U2AF1, CEBPA, TP53, CHD4) also displayed an accumulation of mutations significantly higher that the corresponding healthy controls taken from the 1000 genomes repository. This result strongly suggests that all these genes are close in the interactome, as frequently occurs with genes of the same disease [18,19]. The epigenetic regulation is affected in these patients directly or through the interactome neighborhood by specific combinations of mutations that contribute to the arising and maintenance of AML. When we classified patients in 3 different sets according to their harbored mutations [Group 1, (n = 56), Group 2, (n = 22), Group 3, (n = 22)] we established a set of seed-genes that could be potentially involved in leukemogenesis due to not being present in patients with cytogenetic abnormalities (ERG, NSD1, PLCE1, NFATC2, NEDD9, PIK3R1, and EPHB1). The consequence of these mutations might have a similar effect in the cell than those presents in IR-cytogenetic alterations. As a member of the ETS (erythroblast transformation-specific) family of transcription factors, ERG (ETS-related gene) regulates transcription probably by modifying the structure of the chromatin. In leukemia, ERG targets transcription factors such as GATA2, which is an important regulator of hematopoietic stem cell and megakaryocyte development and, RUNX1, which is involved in the development of normal hematopoiesis. Functional analyses conducted in mice and human CD34 normal and leukemic cells have demonstrated the role of ERG in the induction of early myeloid progenitors in leukemia stem cell through the activation of RAS pathway and Pim1 [20–24]. Several studies have established the prognosis impact of ERG expression on adult patients with AML, improving the molecular risk stratification of NK-AML [22, 25–29]. Although mutations in ERG have not been reported in previous AML studies, 222 different mutations affecting 211 patients in other cancers (n = 22) such as breast, brain or soft tissue have been described. NSD proteins are epigenetic regulators that methylate lysine side chains affecting chromatin organization. This protein is encoded by NSD1 gene and plays a pivotal role in childhood acute myeloid leukemia (AML). This gene has been described in a cryptic rearrangement with NUP98 in 15–20% of children with NK-AML. However, the oncogenic fusion transcript is rarely found in adults (~2%) [30–32]. In previous results, mutations in this histone methyltransferase has been detected at low frequency in AML (~3%) and its impact on the outcome of AML patients still remains unclear [10, 33, 34]. PLCE1 belongs to a phospholipase family that controls gene expression, cell growth and differentiation. A total of 665 different mutations have been reported in several types of cancer. In particular, mutations in PLCE1 have been described in ~3% of patients with AML [10]. NFATC2 is a member of a protein family that acts as a transcription factor in immune response participating in the maturation of peripheral lymphocytes [35–38]. Mutations (n = 326) in this gene have also been reported in 293 patients from 19 different types of cancer [10]. Particularly, several studies performed in ALL patients (n = 3308) have associated NFATC2 mutations with a poor outcome by increasing the risk of asparaginase hypersensitivity [39]. NEDD9 regulates tyrosine-kinase-based signaling complexes involved in multiple actions such as cell adhesion, migration and apoptosis, affecting cancer metastasis [40–43]. In addition, this gen has been related to BCR receptor in B- and T-cell and the inhibition of migration and dissemination of neoplastic myeloid cells [44–47]. Recently, Pallarès et al. have studied two independent cohorts of AML patients (n = 279) and established NEDD9 gene expression as a favorable prognosis factor in IR-AML patients subgroup [48]. Moreover, mutations in this gene have been associated with drug resistance in several solid tumors [10]. Gain-of-function mutations in PIK3R1 have been associated with an oncogenic activation of PIK signaling pathway which plays an important role in the regulation of FGFR family, PDGFRA, PDGFRB and KIT genes, among others [49, 50]. Previous reports have described mutations in PIK3R1 in leukemia (6.4%), establishing PIK3R1 as an actionable gen in AML [51, 52]. Finally, human and murine myeloid assays have shown a reduction of BCR-ABL transformation by inhibiting PI3K/AKT pathway [53–56]. Finally, EPHB1 is a receptor tyrosine kinase involved in normal hematopoietic development and in leukemogenesis [57]. Mutations causing a loss-of-function in this gene have been associated with an aggressive cancer phenotype [58, 59]. In AML, EPHB1 has been defined as a tumor suppressor that regulates DNA damage response system; therefore, mutations affecting this gene have been correlated with poor overall survival [60, 61]. Moreover, mutations disturbing EPHB1 methylation have been related with ALL pathogenesis [62]. Since mutations in these seed genes are very rare in previously published AML cohorts [10], they could trigger leukemogenesis only in this subset of AML patients which lack cytogenetic alterations and/or well-known molecular features that is, wild-type NPM1, CEBPA, and FLT3–ITD. For these reason, the impairment of a specific alteration could not be as essential as the interaction of several mutated genes belonging to functionally related categories in the development of AML. Further analysis will focus on the characterization larger and independent cohorts of patients with NK-AML to establish the frequency and their impact on the leukemogenesis. In addition, to improve our knowledge about their interactions, in vitro assays will be performed by gene editing technologies. With this purpose, observed mutations will be introduced in cell cultures to transform these results into data of clinical relevance that will be virtually used in the prognostic and therapeutic decision process.
Three major limitations to our study are the relative small sample size, their selection bias and normal-matched samples. We have analyzed an initial cohort of 7 patients by WES and 100 patients by a targeted gene-panel using NGS. As a consequence, to perform a study of a specific group of patients, there were not randomly included in our study. For this reason, it has been difficult to set up a significant independent association for any gene mutation using univariate or multivariate analysis. However, by conducting an elastic net cox regression, we found a complex pattern of associations between the different mutations and risk of death, consistent with previous reports [10–12]. In this regard, we used regularized regression methods in our statistical analyses to reduce over-fitting in small samples and deal appropriately with analyses were the number of variables is high relative to the number of observations [63, 64]. Therefore, more solid evidence based on larger series with long-term follow-up is needed to correlate our results with other clinical data and to clarify the prognostic impact of each mutation. In addition, the use of bone marrow sample from complete remission as “normal” matched control could have led us to discard somatic variants persistent at complete remission. The same for the saliva sample, which could be contaminated with leucocytes. Therefore, we could underestimate candidate mutated genes in our discovery cohort. However, we were interested in those mutations that trigger leukemogenesis, which should not be present when patients achieve complete remission.
In summary, this report describes a complete analysis of AML patients with NK combining WES, targeted NGS and network enrichment approaches. As a result, in 100 AML patients we have described mutations in 73 genes, 35 of them recurrently mutated. Additionally, we have defined a functional module in the interactome of AML pointed 13 recurrent mutated genes significantly involved in the pathogenesis of AML. After classify patients according to their mutations, we defined a single in silico model and established a set of seed-genes that may trigger leukemogenesis. These results led us to hypothesize that the perturbation caused in biological key functions as a consequence of gene mutations might be at least as important as the combination of mutations harbored in each patient. Finally, it is clear that the understanding of the events occurred at the clonal hematopoiesis in this disease is still limited and the future potential of discovery of new pathways interactions is high.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
This study was supported in part by research funding from FEDER funds (CIBERONC (CB16/12/00284)), “Red Temática de Investigación Cooperativa en Cancer” grant (RD12/0036/0014,); “Instituto de Salud Carlos III” grants PI12/01047, PI13/01640, PI13/02837, PT13/0010/0026, PT13/0001/0007, PIE13/00046, PI16/00665 and PI16/011113; from the “Consellería de Educación, Cultura y Deporte” AC15/00068 and PROMETEOII/2015/008 and supported by grant SAF2017-88908-R from the Spanish Ministry of Economy and Competitiveness (MINECO). We would like to thank the Biobank La Fe for providing all the samples.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported in part by research funding from FEDER funds (CIBERONC (CB16/12/00284)), “Red Temática de Investigación Cooperativa en Cancer” grant (RD12/0036/0014,); “Instituto de Salud Carlos III” grants PI12/01047, PI13/01640, PI13/02837, PT13/0010/0026, PT13/0001/0007, PIE13/00046, PI16/00665 and PI16/011113; from the “Consellería de Educación, Cultura y Deporte” AC15/00068 and PROMETEOII/2015/008 and supported by grant SAF2017-88908-R from the Spanish Ministry of Economy and Competitiveness (MINECO).
References
- 1.Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev 2004;18:115–36. 10.1016/S0268-960X(03)00040-7 [DOI] [PubMed] [Google Scholar]
- 2.Mrózek K, Marcucci G, Paschka P, Whitman SP, Bloomfield CD. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood 2007;109:431–48. 10.1182/blood-2006-06-001149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valk PJM, Verhaak RGW, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 2004;350:1617–28. 10.1056/NEJMoa040465 [DOI] [PubMed] [Google Scholar]
- 4.Bullinger L, Döhner K, Bair E, Fröhling S, Schlenk RF, Tibshirani R, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med 2004; 350:1605–16. 10.1056/NEJMoa031046 [DOI] [PubMed] [Google Scholar]
- 5.Radmacher MD, Marcucci G, Ruppert AS, Mrózek K, Whitman SP, Vardiman JW, et al. Independent confirmation of a prognostic gene-expression signature in adult acute myeloid leukemia with a normal karyotype: a Cancer and Leukemia Group B study. Blood 2006;108:1677–83. 10.1182/blood-2006-02-005538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chuang HY, Lee E, Liu YT, Lee D, Ideker T: Network-based classification of breast cancer metastasis. Mol Syst Biol 2007, 3:140 10.1038/msb4100180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol 2009, 27:199–204. 10.1038/nbt.1522 [DOI] [PubMed] [Google Scholar]
- 8.Stoll G, Surdez D, Tirode F, Laud K, Barillot E, Zinovyev A,et al. Systems biology of Ewing sarcoma: a network model of EWS-FLI1 effect on proliferation and apoptosis. Nucleic Acids Res 2013, 41:8853–8871. 10.1093/nar/gkt678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel VN, Gokulrangan G, Chowdhury SA, Chen Y, Sloan AE, Koyuturk M, et al. Network signatures of survival in glioblastoma multiforme. PLoS Comput Biol 2013, 9:e1003237 10.1371/journal.pcbi.1003237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013. May 30;368(22):2059–74. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med. 2015. September 17;373(12):1136–52. 10.1056/NEJMra1406184 [DOI] [PubMed] [Google Scholar]
- 13.Mitra K, Carvunis AR, Ramesh SK, Ideker T: Integrative approaches for finding modular structure in biological networks. Nat Rev Genet 2013, 14:719–732. 10.1038/nrg3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibáñez M, Carbonell-Caballero J, García-Alonso L, Such E, Jiménez-Almazán J, Vidal E, et al. The Mutational Landscape of Acute Promyelocytic Leukemia Reveals an Interacting Network of Co-Occurrences and Recurrent Mutations. PLoS One. 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minguez P, Gotz S, Montaner D, Al-Shahrour F, Dopazo J: SNOW, a web-based tool for the statistical analysis of protein-protein interaction networks. Nucleic Acids Res 2009, 37:W109–114. 10.1093/nar/gkp402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Shahrour F, Diaz-Uriarte R, Dopazo J: FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 2004, 20:578–580 10.1093/bioinformatics/btg455 [DOI] [PubMed] [Google Scholar]
- 17.1000 Genomes Project Consortium, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature 2010, 467:1061–1073. 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon M, Lee SJ, Reddy S, Rybak Y, Adem A, Libutti SK: Down-regulation of Filamin A interacting protein 1-like Is associated with promoter methylation and an invasive phenotype in breast, colon, lung and pancreatic cancers [corrected]. PLoS ONE 2013, 8:e82620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL: The human disease network. Proc Natl Acad Sci U S A 2007, 104:8685–8690. 10.1073/pnas.0701361104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg L, Tijssen MR, Birger Y, Hannah RL, Kinston SJ, Schütte J, et al. Genome-scale expression and transcription factor binding profiles reveal therapeutic targets in transgenic ERG myeloid leukemia. Blood. 2013. October 10;122(15):2694–703. 10.1182/blood-2013-01-477133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldus CD, Burmeister T, Martus P, Schwartz S, Gökbuget N, Bloomfield CD, et al. High expression of the ETS transcription factor ERG predicts adverse outcome in acute T-lymphoblastic leukemia in adults. J Clin Oncol. 2006;24(29):4714–4720. 10.1200/JCO.2006.06.1580 [DOI] [PubMed] [Google Scholar]
- 22.Marcucci G, Baldus CD, Ruppert AS, Radmacher MD, Mrózek K, Whitman SP, et al. Overexpression of the ETS-related gene, ERG, predicts a worse outcome in acute myeloid leukemia with normal karyotype: a Cancer and Leukemia Group B study. J Clin Oncol. 2005; 23(36):9234–9242. 10.1200/JCO.2005.03.6137 [DOI] [PubMed] [Google Scholar]
- 23.Metzeler KH, Dufour A, Benthaus T, Hummel M, Sauerland MC, Heinecke A, et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J Clin Oncol. 2009;27(30): 5031–5038. 10.1200/JCO.2008.20.5328 [DOI] [PubMed] [Google Scholar]
- 24.Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011; 17(9):1086–1093 10.1038/nm.2415 [DOI] [PubMed] [Google Scholar]
- 25.Xiao SJ, Shen JZ, Huang JL, Fu HY. Prognostic significance of the BAALC gene expression in adult patients with acutemyeloid leukemia: a meta-analysis. Molecular and clinical oncology 3(4): 880–888. 10.3892/mco.2015.562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwind S, Marcucci G, Maharry K, Radmacher MD, Mrózek K, Holland KB, et al. BAALC and ERG expression levels are associated with outcome and distinct gene and microRNA expression profiles in older patients with de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010. 116(25):5660–5669. 10.1182/blood-2010-06-290536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eid MA, Attia M, Abdou S, El-Shazly SF, Elahwal L, Farrag W, et al. BAALC and ERG expression in acute myeloid leukemia with normal karyotype: impact on prognosis. Int J Lab Hematol 32(2) [DOI] [PubMed] [Google Scholar]
- 28.Marcucci G, Maharry K, Whitman SP, Vukosavljevic T, Paschka P, Langer C, et al. High expression levels of the ETS-related gene, ERG, predict adverse outcome and improve molecular risk-based classification of cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 25(22):3337–3343. [DOI] [PubMed] [Google Scholar]
- 29.Pan J, Zhang Y, Zhao YL, Yang JF, Zhang JP, Liu HX, et al. Impact of clinical factors on outcome of leukemia patients with TLS-ERG fusion gene. Leukemia & lymphoma 58(7): 1655–1663. [DOI] [PubMed] [Google Scholar]
- 30.Hollink IH, van den Heuvel-Eibrink MM, Arentsen-Peters ST, Pratcorona M, Abbas S, Kuipers JE, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118:3645–3656. 10.1182/blood-2011-04-346643 [DOI] [PubMed] [Google Scholar]
- 31.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118(24):6247–6257. 10.1182/blood-2011-07-328880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soler G, Kaltenbach S, Dobbelstein S, et al. Identification of GSX2 and AF10 as NUP98 partner genes in myeloid malignancies. Blood Cancer J. 2013;3:e124 10.1038/bcj.2013.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan XJ, Xu J, Gu ZH, Broccardo C, Radford I, Mozziconacci MJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43(4):309–315. 10.1038/ng.788 [DOI] [PubMed] [Google Scholar]
- 34.Dolnik A, Engelmann JC, Scharfenberger-Schmeer M, Mauch J, Kelkenberg-Schade S, Haldemann B, et al. Commonly altered genomic regions in acute myeloid leukemia are enriched for somatic mutations involved in chromatin remodeling and splicing. Blood. 2012. November 1;120(18):e83–92 10.1182/blood-2011-12-401471 [DOI] [PubMed] [Google Scholar]
- 35.Serfling E, Berberich-Siebelt F, Chuvpilo S, Jankevics E, Klein-Hessling S, Twardzik T, et al. The role of NF-AT transcription factors in T cell activation and differentiation. Biochim Biophys Acta. 2000;1498: 1–18. [DOI] [PubMed] [Google Scholar]
- 36.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell.2002;109(suppl 2): S67–S79. [DOI] [PubMed] [Google Scholar]
- 37.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT.Genes Dev. 2003;17: 2205–2232. [DOI] [PubMed] [Google Scholar]
- 38.Amasaki Y, Adachi S, Ishida Y, Iwata M, Arai N, Arai K, et al. A constitutively nuclear form of NFATx shows efficient transactivation activity and induces differentiation of CD4(+)CD8(+) T cells. J Biol Chem. 2002;277:25640–25648. 10.1074/jbc.M201860200 [DOI] [PubMed] [Google Scholar]
- 39.Fernandez CA, Smith C, Yang W, Mullighan CG, Qu C, Larsen E, et al. Genome-wide analysis links NFATC2 with asparaginase hypersensitivity. Blood. 2015. July 2;126(1):69–75. 10.1182/blood-2015-02-628800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Neill GM, Seo S, Serebriiskii IG, Lessin SR, Golemis EA. A new central scaffold for metastasis: parsing HEF1/Cas-L/NEDD9. Cancer Res. 2007; 67:8975–9. 10.1158/0008-5472.CAN-07-1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh M, Cowell L, Seo S, O’Neill G, Golemis E. Molecular basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator of invasion, apoptosis and cell cycle. Cell Biochem Biophys. 2007; 48:54–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikonova AS, Gaponova AV, Kudinov AE, Golemis EA. CAS proteins in health and disease: an update. IUBMB Life. 2014; 66:387–95. 10.1002/iub.1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong J, Baquiran JB, Bonakdar N, Lees J, Ching YW, Pugacheva E, et al. NEDD9 stabilizes focal adhesions, increases binding to the extra-cellular matrix and differentially effects 2D versus 3D cell migration. PLoS One. 2012; 7:e35058 10.1371/journal.pone.0035058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamoto T, Seo S, Sakai R, Kato T, Kutsuna H, Kurokawa M, et al. Expression and tyrosine phosphorylation of Crk-associated substrate lymphocyte type (Cas-L) protein in human neutrophils. J Cell Biochem. 2008; 105:121–8. 10.1002/jcb.21799 [DOI] [PubMed] [Google Scholar]
- 45.Seo S, Nakamoto T, Takeshita M, Lu J, Sato T, Suzuki T, et al. Crk-associated substrate lymphocyte type regulates myeloid cell motility and suppresses the progression of leukemia induced by p210Bcr/Abl. Cancer Sci. 2011; 102:2109–17. 10.1111/j.1349-7006.2011.02066.x [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Bavarva JH, Wang Z, Guo J, Qian C, Thibodeau SN, et al. HEF1, a novel target of Wnt signaling, promotes colonic cell migration and cancer progression. Oncogene. 2011; 30:2633–43. 10.1038/onc.2010.632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seo S, Asai T, Saito T, Suzuki T, Morishita Y, Nakamoto T, et al. Crk-associated substrate lymphocyte type is required for lymphocyte trafficking and marginal zone B cell maintenance. J Immunol. 2005; 175:3492–501. [DOI] [PubMed] [Google Scholar]
- 48.Pallarès V, Hoyos M, Chillón MC, Barragán E, Conde MIP, Llop M, et al. NEDD9, an independent good prognostic factor in intermediate-risk acute myeloid leukemia patients. Oncotarget. 2017. June 16;8(44):76003–76014. doi: 10.18632/oncotarget.18537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quayle SN, Lee JY, Cheung LW, Ding L, Wiedemeyer R, Dewan RW, et al. Somatic mutations of PIK3R1 promote gliomagenesis. PLoS One 2012, 7:e49466 10.1371/journal.pone.0049466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheung LW, Hennessy BT, Li J, Yu S, Myers AP, Djordjevic B, et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov 2011, 1:170e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Easton J, Shao Y, Maciaszek J, Wang Z, Wilkinson MR, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. 2017. August;49(8):1211–1218. 10.1038/ng.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pritchard CC, Salipante SJ, Koehler K, Smith C, Scroggins S, Wood B, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014. January;16(1):56–67 10.1016/j.jmoldx.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skorski T., Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi JK, et al. 1997. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 16:6151–6161. 10.1093/emboj/16.20.6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skorski T., Kanakaraj P, Nieborowska-Skorska M, Ratajczak MZ, Wen SC, Zon G, et al. 1995. Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood. 86:726–736. [PubMed] [Google Scholar]
- 55.Kharas M.G., Deane JA, Wong S, O'Bosky KR, Rosenberg N, Witte ON, et al. 2004. Phosphoinositide 3-kinase signaling is essential for ABL oncogenemediated transformation of B-lineage cells. Blood. 103:4268–4275. 10.1182/blood-2003-07-2193 [DOI] [PubMed] [Google Scholar]
- 56.Sattler M., Mohi MG, Pride YB, Quinnan LR, Malouf NA, Podar K, et al. 2002. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell. 1:479–492. [DOI] [PubMed] [Google Scholar]
- 57.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600): 1912–1934. 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 58.Wang H, Wen J, Wang H, Guo Q, Shi S, Shi Q, et al. Loss of expression of EphB1 protein in serous carcinoma of ovary associated with metastasis and poor survival. Int J Clin Exp Pathol 2014;7:313–21. [PMC free article] [PubMed] [Google Scholar]
- 59.Wang JD, Dong YC, Sheng Z, Ma HH, Li GL, Wang XL, et al. Loss of expression of EphB1 protein in gastric carcinoma associated with invasion and metastasis. Oncology 2007;73:238–45. 10.1159/000127421 [DOI] [PubMed] [Google Scholar]
- 60.Wang H, Wen J, Wang H, Guo Q, Shi S, Shi Q, et al. Loss of expression of EphB1 protein in serous carcinoma of ovary associated with metastasis and poor survival. Int J Clin Exp Pathol 2014;7:313–21. [PMC free article] [PubMed] [Google Scholar]
- 61.Wang JD, Dong YC, Sheng Z, Ma HH, Li GL, Wang XL, et al. Loss of expression of EphB1 protein in gastric carcinoma associated with invasion and metastasis. Oncology 2007;73:238–45. 10.1159/000127421 [DOI] [PubMed] [Google Scholar]
- 62.Kuang SQ, Bai H, Fang ZH, Lopez G, Yang H, Tong W, et al. Aberrant DNA methylation and epigenetic inactivation of Eph receptor tyrosine kinases and ephrin ligands in acute lymphoblastic leukemia. Blood. 2010. March 25;115(12):2412–9 10.1182/blood-2009-05-222208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waldron L, Pintilie M, Tsao MS, Shepherd FA, Huttenhower C, Jurisica I. Optimized application of penalized regression methods to diverse genomic data. Bioinformatics. 2011. October 24;27(24):3399–406. 10.1093/bioinformatics/btr591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gui J, Li H. Penalized Cox regression analysis in the high-dimensional and low-sample size settings, with applications to microarray gene expression data. Bioinformatics. 2005. April 6;21(13):3001–8. 10.1093/bioinformatics/bti422 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.