Abstract
Quorum sensing is a cell-cell communication mechanism mediated by chemical signals that leads to differential gene expression in response to high population density. Salmonella is unable to synthesize the autoinducer-1 (AI-1), N-acyl homoserine lactone (AHL), but is able to recognize AHLs produced by other microorganisms through SdiA protein. This study aimed to evaluate the fatty acid and protein profiles of Salmonella enterica serovar Enteritidis PT4 578 throughout time of cultivation in the presence of AHL. The presence of N-dodecanoyl-homoserine lactone (C12-HSL) altered the fatty acid and protein profiles of Salmonella cultivated during 4, 6, 7, 12 and 36 h in anaerobic condition. The profiles of Salmonella Enteritidis at logarithmic phase of growth (4 h of cultivation), in the presence of C12-HSL, were similar to those of cells at late stationary phase (36 h). In addition, there was less variation in both protein and fatty acid profiles along growth, suggesting that this quorum sensing signal anticipated a stationary phase response. The presence of C12-HSL increased the abundance of thiol related proteins such as Tpx, Q7CR42, Q8ZP25, YfgD, AhpC, NfsB, YdhD and TrxA, as well as the levels of free cellular thiol after 6 h of cultivation, suggesting that these cells have greater potential to resist oxidative stress. Additionally, the LuxS protein which synthesizes the AI-2 signaling molecule was differentially abundant in the presence of C12-HSL. The NfsB protein had its abundance increased in the presence of C12-HSL at all evaluated times, which is a suggestion that the cells may be susceptible to the action of nitrofurans or that AHLs present some toxicity. Overall, the presence of C12-HSL altered important pathways related to oxidative stress and stationary phase response in Salmonella.
1. Introduction
Quorum sensing is a mechanism of cell-cell communication involved in transcription regulation in response to signaling molecules that accumulate according to high cell density [1, 2]. Quorum sensing regulates a range of phenotypes in bacteria such as production of virulence factors, biofilm formation, protease production, swarming motility, pigment production, bioluminescence, among others [3]. In Salmonella, this mechanism can be mediated by three types of autoinducers (AI), denominated AI-1, AI-2 and AI-3 [4–8]. The signaling molecules known as AI-1 are N-acyl homoserine lactones (AHLs) [4, 6, 9, 10], the AI-2 molecules are (2R, 4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R-THMF) derived from (S)-4,5-dihydroxy-2,3-pentanedione (DPD) [11, 12] and the AI-3 are molecules produced by the gastrointestinal microbiota or hormones produced by the host such as norepinephrine and epinephrine [6, 13–18]. In addition, indole is a cell-cell communication molecule used by several Gram-positive and Gram-negative bacteria playing important biological roles such as spore formation, drug resistance, virulence, plasmid stability, and biofilm formation [8, 19, 20]. However, high concentrations of indole seem to inhibit AHL mediated signaling in Salmonella enterica serovar Typhimurium [21]. In addition to the described molecules, other types of signaling cues have been described for different microbes in the literature such as quinolones, diketopiperazines, hydroxyketones, gamma-butyro-lactones, among others [3].
The cell-cell communication mechanism mediated by AI-1 in the Proteobacteria phylum is composed of a pair of proteins named LuxI (acyl homoserine lactone synthase) and LuxR (transcriptional factor) and its homologous proteins. In contrast, some Proteobacteria belonging to the Enterobacteriaceae family, such as Salmonella spp. and Escherichia coli, do not synthesize AHL since they lack a LuxI homologue [9, 10, 22]. However, a homologue of LuxR, known as SdiA, is present in these bacteria and allows the detection of AHLs synthesized by other microorganisms such as Aeromonas hydrophila and Yersinia enterocolitica, leading to gene regulation [23, 24]. These AIs are able to enter and leave the cell by diffusion or through efflux pumps depending on the type of AHL [1, 25–27]. The AHL molecules bind to the N-terminal domain of the SdiA protein, altering the binding affinity of its C-terminal domain to the DNA and, consequently, regulating the expression of target genes [2, 28–31].
It has been shown that AHLs regulate the expression of the rck operon (resistant to complement killing), which codes for pefI, srgD, srgA, srgB, rck and srgC genes, and it is found in plasmids influencing virulence of Salmonella Typhimurium [9, 10, 32, 33]. Campos-Galvão et al. [34] also showed that the hilA, invA and invF genes present in the Pathogenicity Islands 1 (SPI-1) of S. enterica serovar Enteritidis PT4, and glgC, fliF, lpfA and fimF genes, involved in biofilm formation, were up-regulated in the presence of N-dodecanoyl homoserine lactone (C12-AHL).
A global analysis was carried out on the influence of AHL on the abundance of proteins and the levels of extracellular organic acids of Salmonella Enteritidis [35]. It was shown that the abundance of proteins involved in translation (PheT), transport (PtsI), metabolic processes (TalB, PmgI, Eno and PykF) and response to stress (HtpG and Adi) increased while the abundance of other proteins related to translation (RplB, RplE, RpsB and Tsf), transport (OmpA, OmpC and OmpD) and metabolic processes (GapA) decreased at 7 h of cultivation in the presence of AI-1. Additionally, there were changes in the formate consumption. It was hypothesized that these observed changes are correlated with entry into the stationary phase of growth. In other organisms, the effect of AHLs in anticipating the stationary phase responses was confirmed by global analysis, such as the transcriptome of Pseudomonas aeruginosa [36] and Burkholderia thailandensis [37], as well as the metabolomes of Burkholderia glumae, Burkholderia pseudomallei and B. thailandensis [38].
Besides the global changes in intracellular metabolites described above, other phenotypes are altered when Salmonella is grown in the presence of AIs. For instance, Nesse et al. [39] showed that invasion of HEp-2 cells by Salmonella Typhimurium was increased in the presence of N-hexanoyl homoserine lactone (C6-HSL) and N-octanoyl homoserine lactone (C8-HSL) at 37 °C. The addition of C8-HSL in cells of S. enterica serovar Typhi containing plasmid pRST98, which harbors the virulence gene rck, increased adhesion to HeLa cells after 1 h at 37 °C in the presence of 5% CO2 gas [40]. Biofilm formation by Salmonella Enteritidis PT4 578 in polystyrene surface was positively regulated by C12-HSL after 36 h in anaerobiosis, even though no growth changes were observed in planktonic cells [34, 41]. Similarly, Bai and Rai [42] showed that biofilm formation on polystyrene was increased by Salmonella Typhimurium when cultivated with N-butyryl homoserine lactone (C4-HSL) e C6-HSL.
On the other hand, a cell free supernatant (CFS) rich in AHLs, AI-2 and other unknown compounds of Y. enterocolitica and Serratia proteamaculans altered growth of different phagetypes of Salmonella Enteritidis and Salmonella Typhimurium under aerobiosis [43]. Similarly, the CFS of P. aeruginosa containing AHLs and different metabolites decreased growth of nine serovars of S. enterica [44]. Conversely, the CFS of Hafnia alvei containing AHLs, as well as the addition of synthetic N-3-oxo-hexanoyl homoserine lactone (3-oxo-C6-HSL) to the growth medium in aerobiosis did not influence biofilm formation by Salmonella Typhimurium [45]. It is noteworthy that in these studies, the CFS of different bacteria contained metabolites other than AHLs which might have interfered in the detection of the AIs subtle effects in the cells.
According to Atkinson and Williams [29], the absence of an AHL synthase in Salmonella and E. coli can be related to ecological aspects, since the bacterium would avoid the information transfer to other microorganisms present in the medium and there would be no energy expenditure with the synthesis of these signaling molecules. However, these bacteria would be favored by the environmental information provided by others through the AHLs detection. Thus, what remains uncertain is the real advantage for Salmonella to use cell-cell communication mediated by AI-1. According to Di Cagno et al. [46], the global analyses of the proteome and transcriptome should help elucidate the influences of chemical signals on the cellular physiology. However, most studies on AHL signaling in Salmonella were targeted at specific genes, as previously mentioned [9, 10, 32, 34], while overall changes in Salmonella physiology in the presence of AHLs is still largely unknown.
The aim of this work was to evaluate the altered physiological aspects of Salmonella, such as the cells fatty acid and protein profiles during growth in the presence of AHL. This is the first work that carefully examines the AHL effect throughout growth in anaerobic conditions and the first to show that levels of thiol and proteins related to the oxidation-reduction stress response are altered by a quorum sensing signal in Salmonella, suggesting that these cells may have a greater potential to resist oxidative stress.
2. Materials and methods
2.1. Bacterial strain
S. enterica serovar Enteritidis phage type 4 (PT4) 578 (GenBank: MF066708.1), isolated from chicken meat, was provided by Fundação Oswaldo Cruz (FIOCRUZ, Rio de Janeiro, Brazil). Culture was stored at -20 °C in Luria-Bertani (LB) broth [47] supplemented with 20% (v/v) of sterilized glycerol.
2.2. Inoculum preparation
Inoculum was prepared according to Almeida et al. [35, 41]. Tryptone soy broth (TSB; Sigma-Aldrich, India) was prepared with CO2 under O2-free conditions, dispensed in anaerobic bottles that were sealed with butyl rubber stoppers and then autoclaved (anaerobic TSB). Before each experiment, cells were cultivated in 20 mL of anaerobic TSB for 24 h at 37 °C. Then, 1.0 mL was transferred into 10 mL of anaerobic TSB and incubated at 37 °C. After 4 h of incubation, exponentially growing cells were harvested by centrifugation at 5000 g at 4 °C for 10 min (Sorvall, USA), washed with 0.85% saline, and the pellet resuspended in 0.85% saline. The inoculum was standardized to 0.1 of optical density at 600 nm (OD600nm) (approximately 107 CFU/mL) using a spectrophotometer (Thermo Fisher Scientific, Finland).
2.3. Preparation of HSL solution
The autoinducer-1, N-dodecanoyl-DL-homoserine lactone (C12-HSL; PubChem CID: 11565426; Fluka, Switzerland) was suspended in acetonitrile (PubChem CID: 6342; Merck, Germany) at a concentration of 10 mM and further diluted to a working solution of 10 μM in acetonitrile. Control experiment was performed using acetonitrile with final concentration in the media less than 1% (v/v) to avoid interference in the growth and response of Salmonella to C12-HSL [9].
2.4. Fatty acid profile analysis of Salmonella
2.4.1. Saponification, methylation and extraction of fatty acids
An aliquot of 10 mL of the standardized inoculum was added into anaerobic bottles containing 90 mL of anaerobic TSB supplemented with 50 nM of C12-HSL or the equivalent volume of acetonitrile as control and then, incubated at 37 °C. After 4, 6, 7, 12 and 36 h of incubation, OD600nm was determined. Concomitantly, 10 mL of the culture was centrifuged at 5000 g at 4 °C for 15 min (Sorvall, USA). The cells in the pellet were resuspended in 1 mL of sterilized distilled water, and once again, centrifuged at 5000 g at 4 °C for 10 min. The pellet was transferred to glass tubes free from fatty acids and afterwards, the fatty acids were saponified, methylated, and extracted by the procedure of the Sherlock® Analysis Manual (version 6.2; MIDI, USA) [48].
2.4.2. Analysis and identification of fatty acids
The cellular fatty acid composition was determined by 7890A gas chromatograph equipped with flame ionization detector (Agilent Technologies, USA). Afterwards, the fatty acids were identified and quantified in the Sherlock® Microbial Identification System software by the procedure of the Sherlock® Analysis Manual (version 6.2; MIDI, USA) using the Calibration Standard 1 (Microbial ID Part # 1200-A) containing the straight chain C9:0 to C20:0 fatty acid methyl esters [48].
2.4.3. Statistical analysis
Experiments were carried out in three biological replicates. The values of the triplicates were used for Principal Component Analysis (PCA) using RStudio software (version 1.0.143; USA). All data were subjected to analysis of variance (ANOVA) followed by Tukey’s test using the Statistical Analysis System and Genetics Software® [49]. A p-value < 0.05 was considered to be statistically significant.
2.5. Protein profile analysis of Salmonella
2.5.1. Extraction and quantification of proteins
An aliquot of 10 mL of the standardized inoculum was added into anaerobic bottles containing 90 mL of anaerobic TSB supplemented with 50 nM of C12-HSL or the equivalent volume of acetonitrile as control and then, incubated at 37 °C. After 4, 6, 7, 12 and 36 h of incubation, OD600nm was determined and 10 mL of the culture was centrifuged at 5000 g at 4 °C for 15 min (Sorvall, USA). The cells in the pellet were resuspended in 1 mL of sterilized distilled water, transferred to 1.5 mL microtubes and once again centrifuged at 9500 g at 4 °C for 30 min (Brikmann Instruments, Germany). The pellet was resuspended in 1 mL of Tris-HCl 50 mM, pH 8.0 added of 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mM dithiothreitol (DTT). Next, the mixture was kept in ice for 1 min, then 1 min in ultrasound bath (154 W, 37 KHz; Unique, Brazil) and this cycle was repeated five times. Posteriorly, five cycles of 1 min in ice, 1 min in vortex and 8 min in ultrasound bath were performed. The mixture was centrifuged at 9500 g at 4 °C for 30 min, the supernatant containing the intracellular proteins was transferred to 1.5 mL microtube and stored at −20 °C. The pellet was resuspended in 100 μL of ammonium bicarbonate 50 mM and 1 mL of 2:1 trifluoroethanol:chloroform (TFE:CHCl3) was added. Then, the mixture was subjected again to five cycles on ice and ultrasound bath and five cycles on ice, vortex and ultrasound bath as previously described. The mixture was centrifuged at 9500 g at 4 °C for 4 min to obtain three phases. The upper phase, composed by proteins soluble in TFE, was transferred to 1.5 mL microtubes and was dried in SpeedVac (Genevac, England). The supernatant with intracellular proteins was transferred to microtube containing the proteins soluble in TFE. The protein extract was precipitated with 10% (w/v) trichloroacetic acid (TCA) and kept on ice for 30 min and after that, the material was centrifuged at 9500 g at 4 °C for 10 min. The supernatant was discarded and the precipitate washed three times with cold acetone. After evaporation of the residual acetone at room temperature, the precipitate was resuspended in 400 μL of ammonium bicarbonate 50 mM. Proteins were quantified using Coomassie blue dye [50] and the protein extracts were stored at -20 °C.
2.5.2. In-solution protein digestion
The trypsin digestion of proteins in solution was performed according to Villen and Gygi [51], with modifications. An aliquot of the extract containing 10 μg of protein was transferred to 1.5 mL microtube and the final volume was adjusted to 150 μL with ammonium bicarbonate 50 mM and 150 μL of urea 8 M were added. The disulfide bonds of the proteins were reduced by 5 mM DTT for 25 min at 56 °C and the protein mixture was cooled to room temperature. Next, the alkylation was carried out by adding 14 mM iodoacetamide and followed by incubation for 30 min at room temperature in the dark. The free iodoacetamide was quenched by adding 5 mM DTT with further incubated for 15 min at room temperature in the dark. The urea concentration in the protein mixture was reduced to 1.6 M by diluting it in 1:5 in ammonium bicarbonate 50 mM with the addition of 1 mM calcium chloride. An aliquot containing 20 ng of trypsin (Sequencing grade modified trypsin; Promega, USA) in ammonium bicarbonate 50 mM was added to a final ratio of 1:50 of trypsin:protein and it was incubated for 16 h at 37 °C. After the incubation, the solution was cooled to room temperature and the enzymatic reaction was quenched with 1% (v/v) trifluoroacetic acid (TFA) until pH 2.0. After centrifugation at 2500 g for 10 min at room temperature, the supernatant containing the peptides was collected.
2.5.3. Peptide desalting
The supernatant containing the peptides was desalted according to Rappsilber et al. [52], with modifications. Two membranes of octadecyl (C18; 3M EMPORE, USA) were packed in each stage tip to load 10 μg of digested proteins. The stage tip was conditioned with 100 μL of 100% (v/v) methanol and was equilibrated with 100 μL 0.1% (v/v) of formic acid. After that, the supernatant containing the peptides was loaded two times onto the stage tip and this was washed 10 times with 100 μL of 0.1% (v/v) of formic acid. The peptides were eluted with 200 μL of 80% (v/v) acetonitrile containing 0.1% (v/v) of formic acid. In each previous step, the stage tip was centrifuged at 400 g for 2 min (Eppendorf, Germany). The eluate was dried in SpeedVac (Thermo Fisher Scientific, Finland), resuspended in 22.5 μL of 0.1% (v/v) formic acid and stored at -20 °C.
2.5.4. Mass spectrometric analysis
The solution containing the peptides was centrifuged at 9000 g for 5 min and 15 μL of the supernatant was transferred to vials. An aliquot of 4.5 μL of peptides, equivalent to 2 μg of protein, was separated by C18 (100 mm x 100 mm) RP-nanoUPLC (nanoAcquity; Waters, USA) coupled to a Q-Tof Premier mass spectrometer (Waters, USA) with nanoelectrospray source at a flow rate of 0.6 μL.min-1. The gradient was 2–90% (v/v) acetonitrile in 0.1% (v/v) formic acid over 45 min. The nanoelectrospray voltage was set to 3.0 kV, a cone voltage of 50 V and the source temperature was 80 °C. The instrument was operated in the ‘top three’ mode, in which one MS spectrum is acquired followed by MS/MS of the top three most-intense peaks detected. After MS/MS fragmentation, the ion was placed on exclusion list for 60 s.
2.5.5. Identification and quantification of proteins
The spectra were acquired using software MassLynx (version 4.1; Waters) and the .raw data files were converted to a peak list format .mgf without summing the scans by the Mascot Distiller software (version 2.3.2.0; Matrix Science, United Kingdom) and searched against the knowledgebase UniProtKB using the Mascot software (version 2.4.0; Matrix Science, United Kingdom). For the search, the following parameters were considered: taxonomy Salmonella and all entries (separately), monoisotopic mass, trypsin, allowing up to one missed cleavage site, peptide charge +2, +3 and +4, fixed modification for carbamidomethylation of cysteine residues and variable modification for oxidation of methionine residues, peptide and MS/MS tolerance equal to 0.1 Da [53–55], ESI-QUA-TOF for instrument. The proteins identifications by Mascot software were validated and the proteins were quantified by Scaffold software (version 4.7.2; Proteome Software, USA). For the protein validation, peptide and protein identifications were accepted if they could be established at higher than 90% probability as specified by the Peptide Prophet algorithm [56] and by the Protein Prophet algorithm [57], respectively. In addition, the proteins should contain at least one identified peptide. The false discovery rates (FDR) for identification of proteins and peptides using decoy method by Scaffold software should to be less than or equal to 1% (FDR ≤ 1%) [54, 58, 59]. For the protein quantitation, the quantitative value of total ion current (TIC) of each protein was normalized by sum total of TIC. For the unidentified proteins, a TIC value of 0.05 was adopted (cut off of method sensibility).
2.5.6. Statistical analysis
Experiments were carried out in three biological replicates. The logarithms of normalized TIC values of the triplicates were used for PCA as well as the logarithm of normalized mean of TIC values of the triplicates of each protein which were used to construct the heatmap and dendrogram using RStudio software. The statistical difference of normalized TIC values among the samples were calculated by T-test with correction for multiple hypotheses by FDR using RStudio software. The fold changed was calculated as the ratio of the normalized mean of TIC values of the treatment with C12-HSL by the control and the result was shown as logarithm in base two (Log2 FC). The proteins with p-value less than 0.05 (p-value < 0.05) or negative logarithm of p-value more than 1.301 (-Log10 p > 1.301) and fold changed less than 0.667-fold or more than 1.500-fold or Log2 FC less than -0.585 or more than 0.585 (Log2 FC < -0.585 or > 0.585) were considered differentially abundant proteins [60]. Moreover, when the protein was not detected in one of the treatments, the p-value was not considered and the fold-changed was calculated by adopting a TIC value of 0.05 (cut off of method sensibility) for the sample in which the protein was not identified, being this protein also considered differentially abundant.
2.5.7. Bioinformatics analysis
The Gene Ontology (GO) annotations of the process and function of proteins were acquired with the tool QuickGO, implemented by the European Bioinformatics Institute (http://www.ebi.ac.uk/QuickGO/). Then, the Protein-Protein Interaction (PPI) network was generated for some proteins of Salmonella Typhimurium LT2 using the STRING database version 10.5 (http://string-db.org/, [61, 62]). The confidence levels of the PPI were considered in relation to the average local clustering coefficient: interactions at low confidence or better (coefficient ≥ 0.150), interactions at medium confidence or better (coefficient ≥ 0.400), interactions at high confidence or better (coefficient ≥ 0.700) and interactions at highest confidence (coefficient ≥ 0.900), available at http://string-db.org/ [61, 62].
2.6. Quantification of free cellular thiol
2.6.1. Extraction of free cellular thiol
The pellet obtained as described in item 2.5.1. was resuspended in 250 μL of sterilized distilled water. Next, the mixture was kept on ice for 1 min, 1 min mixed by vortex for 1 min and treated with ultrasound (400 W, 20 KHz; Sonics & Materials Inc., USA) for 30 s; and this cycle was repeated five times. The mixture was then centrifuged at 9500 g at 4 °C for 15 min and the supernatant containing the free cellular thiol was used immediately.
2.6.2. Quantification of free cellular thiol
The quantification of free cellular thiol was performed according to Ellman [63] and Riddles et al. [64], with modifications. For each 25 μL of the supernatant or standard, 5 μL of 0.4% (w/v) 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB or Ellman’s reagent; Sigma, USA) in buffer sodium phosphate 0.1 M pH 8.0 containing 1mM EDTA and 250 μL of the buffer sodium phosphate were added in microplate. The microplate was incubated at room temperature for 15 min and the absorbance at 412 nm measured by using a spectrophotometer (Thermo Fisher Scientific, Finland). The free cellular thiol was quantified by using cysteine hydrochloride monohydrate (Sigma, USA) as standard at concentrations from 0.0 to 1.5 mM. The obtained equation was as follows: absorbance = (0.9421 x concentration) + 0.0432, with R2 = 0.9936. Next, the quantification of free cellular thiol was normalized by OD600nm.
2.6.3. Statistical analysis
Experiments were carried out in four biological replicates. The statistical analysis was performed according to item 2.4.3, except that the PCA was not performed.
3. Results and discussion
3.1. HSL alters the fatty acid profile of Salmonella throughout time
Analysis of the fatty acid profile of Salmonella cells, both in the absence and presence of C12-HSL, showed alterations in the composition at different time points. The largest change appears to be at the 4 h time point with four fatty acids presenting altered abundance at this time of cultivation. Of these fatty acids, 17:0 cyclo ɷ7c and 19:0 cyclo ɷ8c had their abundance decreased in the presence of C12-HSL, while two unresolved mixtures of 16:1 ɷ6c/16:1 ɷ7c and 18:1 ɷ6c/18:1 ɷ7c had their abundance increased (Table 1 and S1 Table).
Table 1. Fatty acid profile of Salmonella Enteritidis PT4 578 anaerobically cultivated in TSB at 37 °C in the presence or absence of C12-HSL.
| Classification | Fatty acids | Time (h) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 6 | 7 | 12 | 36 | |||||||
| Control | C12-HSL | Control | C12-HSL | Control | C12-HSL | Control | C12-HSL | Control | C12-HSL | ||
| Saturated | 12:00 | 4.80A | 4.72A | 4.69AB | 4.61AB | 4.46C | 4.65AB | 4.31C | 4.38B | 4.50BC | 4.50AB |
| 14:00 | 6.68E | 6.85E | 9.21D | 9.21D | 9.82C | 10.21C | 11.04B | 11.46B | 11.84A | 12.10A | |
| 16:00 | 35.17D | 34.76C | 36.28aC | 35.66bBC | 36.71B | 36.68AB | 37.72A | 37.63A | 38.09A | 37.87A | |
| 18:00 | 0.58 | 0.74 | 0.50 | 0.48 | 0.53a | 0.47b | 0.47 | 0.46 | 0.47 | 0.47 | |
| 19:00 | 0.49A | 0.50 | 0.52A | 0.49 | 0.44AB | 0.48 | 0.34B | 0.41 | 0.46AB | 0.48 | |
| Cyclopropane | 17:0 cyclo ɷ7c | 15.52aB | 14.43bB | 16.64AB | 17.18A | 16.87A | 16.86A | 17.40A | 16.89A | 16.38AB | 16.22A |
| 19:0 cyclo ɷ8c | 11.21aC | 9.42bB | 15.18B | 15.45A | 16.74A | 15.88A | 16.87A | 16.34A | 14.47B | 14.54A | |
| Monounsaturated | 16:1 ɷ6c/16:1 ɷ7c | 2.87bA | 3.56aA | 0.80B | 0.82B | 0.75B | 0.70B | 0.53B | 0.57B | 0.77B | 0.70B |
| 17:1 ɷ7c | 0.00C | 0.00B | 0.31aA | 0.26bA | 0.24B | 0.30A | 0.23B | 0.27A | 0.28AB | 0.30A | |
| 18:1 ɷ6c/18:1 ɷ7c | 12.28bA | 14.93aA | 3.10B | 3.31B | 2.21C | 2.30C | 1.39D | 1.47D | 1.27D | 1.23D | |
| 18:1 ɷ7c 11-methyl | 0.76AB | 0.71 | 0.93A | 0.83 | 0.76AB | 0.84 | 0.57B | 0.69 | 0.83A | 0.86 | |
| Polyunsaturated | 20:2 ɷ6,9c | 0.58 | 0.50 | 0.84 | 0.71 | 0.58 | 0.69 | 0.75 | 0.57 | 0.69 | 0.67 |
| Hydroxy | 18:1 2OH | 0.00C | 0.00B | 0.35A | 0.31A | 0.24AB | 0.27A | 0.20B | 0.24A | 0.29AB | 0.30A |
| Unresolved | 14:0 3OH/16:1 iso I | 8.05BC | 7.97BC | 9.51A | 9.67A | 8.74AB | 8.68B | 7.41bC | 7.70aC | 8.58B | 8.66BC |
| 18:0 anteiso/18:2 ɷ6,9c | 1.01A | 0.91 | 0.89AB | 0.78 | 0.70B | 0.77 | 0.60B | 0.71 | 0.86AB | 0.85 | |
| 19:1 ɷ6c/19:1 ɷ7c/19:0 cyclo | 0.00C | 0.00B | 0.26A | 0.22A | 0.21AB | 0.23A | 0.18B | 0.21A | 0.21AB | 0.24A | |
The comparisons can be drawn among treatments or throughout time. Mean followed by different superscript lowercase letters in the same line (among treatments at the same time) and followed by different superscript uppercase letters in the columns (throughout time for each treatment, separately) differs at 5% probability (p-value < 0.05) by Tukey’s test. Where a letter is not shown, no statistical difference among samples was observed;
Main results discussed in the text are shown in bold.
The concentration of the fatty acids 19:00 and 18:1 ɷ7c 11-methyl, as well as, an unresolved mixture of the 18:0 anteiso/18:2 ɷ6,9c did not change throughout time of cultivation in the presence of C12-HSL, but changed in the control (Table 1). Additionally, after 6 h of cultivation, there was no difference in the concentration of the fatty acids 17:0 cyclo ɷ7c, 19:0 cyclo ɷ8c, 17:1 ɷ7c and 18:1 2OH, as well as, an unresolved mixture of the 19:1 ɷ6c/19:1 ɷ7c/19:0 cyclo in the treatment with C12-HSL (Table 1). Interestingly, the maintenance of the cyclopropane fatty acids 17:0 cyclo ɷ7c and 19:0 cyclo ɷ8c suggests that the cells are prepared for a possible stress condition. This hypothesis is strengthened by the fact that the cyclopropane fatty acids 17:0 cyclo and 19:0 cyclo are formed by transmethylation of cis monounsaturated fatty acids 16:1 ɷ7c and 18:1 ɷ7c, respectively, when the cell enters into stationary phase [65]. The level of cyclopropane fatty acids increases during the stationary phase due to the increased expression of the synthase that produces this fatty acid and is mediated by sigma S factor (σS) [66, 67]. According to several studies, this modification may help reduce the impact of environmental stresses such as starvation, oxidation, acid and heavy metal addition, organic compound toxicity, increased temperature and pressure because it increases the stability and fluidity of the membrane, and at the same time reduces its permeability against toxic compounds [66–73]. An example of the effect of fatty acid modifications on bacterial cell membrane related to a stress condition was presented by Guckert et al. [68] who observed an increased proportion of cyclopropane fatty acids during nutrient deprivation in Vibrio cholerae.
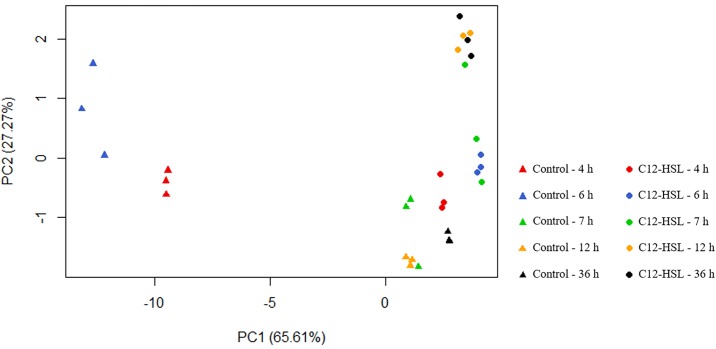
The PCA was used in order to evaluate the variations among triplicates and to the understanding of the global difference of levels and types of fatty acids among samples stimulated by C12-HSL (Fig 1). The distance of the dots in the PCA figure (Fig 1) is proportional to the difference between the fatty acid composition of the experimental groups. Based on this information, there is a high dispersion between the global profile of fatty acids identified in the absence and presence of C12-HSL, which is represented by the distance between control and treatment at the same time. However, the graphic shows that the distance between control and treatment is higher at the 4 h and 6 h time points, with the dispersion tending to reduce over time as both groups tend to cluster more closely. On the other hand, within the treatment with C12-HSL, the identified fatty acids tended to be less dispersed throughout growth. Thus, the fatty acid profile of the cells in the logarithmic (4 h) and stationary (36 h) phases of growth were more similar to each other in the presence of AI-1 than in its absence.
Fig 1. PCA analysis of fatty acids from Salmonella Enteritidis PT4 578 anaerobically cultivated in TSB at 37 °C in the presence or absence of C12-HSL.
PCA of the percentages of each triplicate of fatty acid profile of Salmonella in the absence (control) and presence of C12-HSL.
The data also indicate that the fatty acid composition of Salmonella cells growing with C12-HSL for 4 h is similar to cells cultured in the absence of this autoinducer for 7 h (Fig 1). At this time of cultivation (7 h), the culture is already in the stationary phase of growth and the maintenance of the cyclopropane fatty acids suggest that the cells are prepared to support stressful conditions, as previously discussed. These results suggest that AHL signaling in Salmonella induces a profile of fatty acids which is typical to the stationary phase of growth, even in logarithmic phase of growth when the cell density was relatively low. According to Schuster et al. [74], this early preparation of the cells has a high fitness cost, but leads to more resistance should a stress condition arrive.
3.2. HSL alters the protein profile of Salmonella throughout time
The validation of proteins identifications showed percentages of FDR less than or equal to 0.8% for proteins (FDR ≤ 0.8%) and less than or equal to 0.14% for peptides (FDR ≤ 0.14%) (Table 2). These FDR values were lower than those accepted by Liu et al. [58] and Tran et al. [59], which admitted an FDR lower than 1% (FDR < 1.0%) for proteins and peptides of Salmonella Typhimurium.
Table 2. False discovery rates (FDR) for identification of proteins and peptides using decoy method by Scaffold software.
| Time (h) | FDR (%) | |
|---|---|---|
| Proteins | Peptides | |
| 4 | 0.7 | 0.14 |
| 6 | 0.7 | 0.06 |
| 7 | 0.0 | 0.00 |
| 12 | 0.7 | 0.14 |
| 36 | 0.8 | 0.05 |
FDR = False discovery rate.
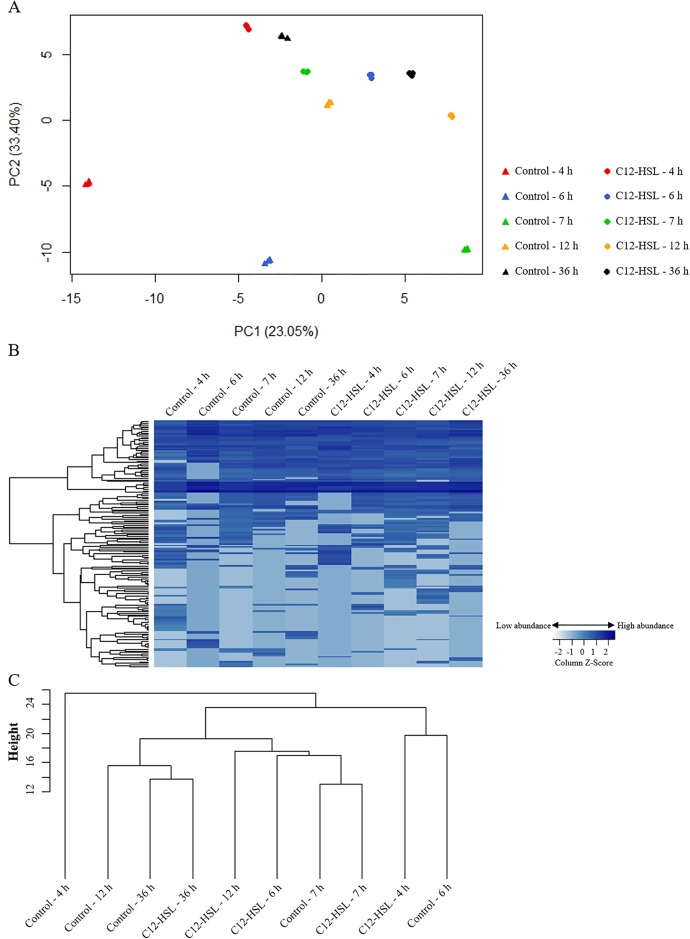
The protein composition at different time points is shown on Fig 2 and S2 Table. The PCA of the proteomic data showed that the variation among replicates was much lower than the variation among times (Fig 2A). There was a similar dispersion of the protein profile such as that observed with the fatty acid analysis, with a higher dispersion of protein and fatty acid profiles in the absence of C12-HSL (Fig 2A). The identified proteins in cells treated with C12-HSL tended to be less dispersed throughout growth than in the absence of the quorum sensing molecule (Fig 2A). Cluster analysis by agglomerative hierarchical methods of the proteins at the different time points was prepared generating a heatmap and a dendrogram that separated the proteins and the samples into two major clades, respectively (Fig 2B and 2C, respectively). Based on the color intensity variability in each column of the heatmap, the protein profile of Salmonella in the absence of C12-HSL at the 4 h time point presented more variations and, this was confirmed by the dendrogram (Fig 2C) that discriminates this sample from the others as a single branch in the tree.
Fig 2. PCA, heatmap and dendrogram analyses of proteins from Salmonella Enteritidis PT4 578 anaerobically cultivated in TSB at 37 °C in the presence or absence of C12-HSL.
(A) PCA of the logarithm of normalized TIC values of each triplicate of protein profile in the absence (control) and presence of C12-HSL. (B) Heatmap of the logarithm of normalized mean of TIC values of the triplicates for each identified protein. Each row corresponds to a unique protein, and each column the mean of the triplicate values. (C) Dendrogram of the logarithm normalized mean of TIC values of the triplicates for each identified protein. The height of the arms is proportional to the difference in the abundance profile of the proteins.
Interestingly, the PCA, the heatmap and the dendrogram analyses together showed that the addition of C12-HSL at the beginning of Salmonella cultivation resulted in smaller protein profile variations throughout the time of cultivation when compared with the control. Thus, the protein profile of the cells in the logarithmic (4 h) and stationary (36 h) phases of growth were more similar to each other in the presence of AI-1 than in its absence as well as the fatty acid profile previously shown. Based on these similarities, these results suggest that AI-1 can anticipate a stationary phase response. Moreover, these results corroborate our previous observation, in which the differentially abundant proteins and organic acids of Salmonella cultivated for 7 h in the presence of C12-HSL correlated with entry into the stationary phase of growth, mainly in relation to nitrogen and amino acid starvation as well as acid stress [35]. Schuster et al. [36] showed that genes with expression influenced by the growth phase in P. aeruginosa were repressed by quorum sensing during the late logarithmic and stationary phases. Goo et al. [38] showed that quorum sensing anticipates and influences the survival to the stress of the stationary phase of B. glumae, B. pseudomallei and B. thailandensis. The survival of these bacteria requires the activation of cellular enzymes through the quorum sensing mechanism for production of excreted oxalate, which serves to counteract ammonia-mediated alkaline toxicity during stationary phase [38]. In addition, the genes involved in transcription and translation of B. pseudomallei in stationary phase were co-regulated by RpoS protein (RNA polymerase sigma factor) and quorum sensing [75].
The statistical analyses of the identified proteins are shown in Table 3 and S3 Table. The results showed that at 4 and 6 h of incubation in presence of C12-HSL, a higher percentage of differentially abundant proteins was observed in comparison with other times of cultivation (Table 4). In addition, more proteins had their abundance decreased at 4 h in the presence of AHL (50.0%), while an opposite trend was observed at 6 h (54.8%). The greatest number of differentially abundant proteins at the initial times can be due to the addition of AI-1 at the beginning of the cultivation. Growth and cell size are altered by intrinsic and extrinsic factors during the adaptation phase to an environmental condition and, consequently, alter the cellular components resulting in a greater fluctuation in protein abundance at initial times of cultivation [76, 77]. In AHL-producing bacteria, such as B. thailandensis and P. aeruginosa, many genes were regulated at the end of the logarithmic and beginning of the stationary phase of growth due to the accumulation of signaling molecules in the medium [36, 37, 78]. Schuster et al. [36] showed that genes involved in carbohydrate utilization or nutrient transport were the most repressed by quorum sensing during the late logarithmic and stationary phases of P. aeruginosa.
Table 3. Statistical analysis of the abundance of proteins identified from Salmonella Enteritidis PT4 578 anaerobically cultivated in TSB at 37 °C in the presence or absence of C12-HSL.
| Protein | Protein name | Gene | Process | Time (h) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 6 | 7 | 12 | 36 | |||||||||
| Log2 FC | -Log10 p | Log2 FC | -Log10 p | Log2 FC | -Log10 p | Log2 FC | -Log10 p | Log2 FC | -Log10 p | ||||
| ATP synthase subunit delta | Q7CPE5 | atpH | Biosynthetic | -11.566 | 1.060 | 8.687 | 3.470 | 1.157 | 2.760 | ND | ND | ND | ND |
| Acetyl-CoA carboxylase, BCCP subunit | Q7CPM1 | accB | Biosynthetic | 2.049 | 0.841 | ND | ND | -10.547 | 1.542 | ND | ND | -1.505 | 1.241 |
| Acyl carrier protein | P0A6B1 | acpP | Biosynthetic | 1.930 | 1.139 | -0.409 | 0.737 | -0.439 | 0.714 | -0.277 | 1.882 | -0.452 | 1.297 |
| ATP synthase subunit beta | Q7CPE2 | atpD | Biosynthetic | ND | ND | ND | ND | -7.974 | 1.357 | ND | ND | ND | ND |
| Cell division protein FtsZ | Q8ZRU0 | ftsZ | Cell Division | ND | ND | ND | ND | -7.378 | 1.542 | 8.561 | 1.296 | ND | ND |
| Cell division protein ZapB | Q8ZKP1 | zapB | Cell Division | -9.554 | 2.451 | 10.650 | 2.037 | -0.305 | 0.317 | -0.300 | 0.554 | 0.912 | 1.133 |
| Protein phosphatase CheZ | P07800 | cheZ | Chemotaxis | -8.243 | 4.181 | 2.733 | 0.940 | 3.359 | 0.506 | 0.029 | 0.023 | 9.365 | 1.026 |
| Aspartate ammonia-lyase | Q7CPA1 | aspA | Metabolic process | ND | ND | ND | ND | 6.377 | 1.508 | ND | ND | ND | ND |
| Arginine decarboxylase | Q8ZKE3 | adi | Metabolic process | ND | ND | ND | ND | ND | ND | ND | ND | -8.168 | 1.714 |
| Shikimate kinase 1 | P63601 | aroK | Metabolic process | -10.245 | 2.324 | 7.418 | 1.665 | 0.677 | 0.288 | 1.517 | 1.882 | ND | ND |
| 2-iminobutanoate/2-iminopropanoate deaminase | Q7CP78 | ridA | Metabolic process | 2.789 | 1.273 | -3.083 | 3.151 | -0.235 | 0.865 | -2.437 | 0.866 | -0.781 | 1.249 |
| Autonomous glycyl radical cofactor | Q7CQ05 | grcA | Metabolic process | 0.816 | 1.306 | 0.940 | 2.063 | 0.511 | 1.542 | 0.072 | 0.162 | -0.513 | 1.714 |
| Pyruvate formate lyase I, induced anaerobically | Q7CQU1 | pflB | Metabolic process | ND | ND | 11.573 | 0.825 | -0.159 | 1.542 | ND | ND | ND | ND |
| Lactoylglutathione lyase | P0A1Q2 | gloA | Metabolic process | -6.833 | 0.914 | ND | ND | ND | ND | ND | ND | ND | ND |
| Enolase | P64076 | eno | Metabolic process | -1.074 | 1.640 | -0.551 | 0.328 | -0.417 | 0.506 | 1.104 | 0.668 | 0.180 | 0.140 |
| Glyceraldehyde-3-phosphate dehydrogenase | P0A1P0 | gapA | Metabolic process | ND | ND | ND | ND | ND | ND | 9.405 | 1.107 | ND | ND |
| Phosphoglycerate kinase | P65702 | pgk | Metabolic process | -0.310 | 0.812 | 0.108 | 0.113 | 0.746 | 1.988 | -0.487 | 1.719 | 0.562 | 1.987 |
| Triosephosphate isomerase | Q8ZKP7 | tpiA | Metabolic process | -11.938 | 2.806 | -11.340 | 2.011 | -0.216 | 0.169 | 8.235 | 0.996 | ND | ND |
| Adenylate kinase | P0A1V4 | adk | Metabolic process | 0.221 | 0.329 | 1.820 | 1.810 | 0.091 | 0.215 | -0.451 | 1.037 | -1.271 | 1.260 |
| Deoxyribose-phosphate aldolase | Q8ZJV8 | deoC | Metabolic process | -7.031 | 0.839 | ND | ND | -0.630 | 0.925 | ND | ND | 7.796 | 0.767 |
| Pyrimidine/purine nucleoside phosphorylase | Q8ZRE7 | ppnP | Metabolic process | ND | ND | ND | ND | ND | ND | 8.356 | 1.530 | ND | ND |
| Acetate kinase | P63411 | ackA | Metabolic process | -8.530 | 2.435 | 9.037 | 2.391 | 0.800 | 0.511 | 8.469 | 1.882 | ND | ND |
| Ribose-5-phosphate isomerase A | P66692 | rpiA | Metabolic process | -8.886 | 3.352 | ND | ND | 0.426 | 0.481 | ND | ND | -0.871 | 1.020 |
| 6,7-dimethyl-8-ribityllumazine synthase | P66038 | ribH | Metabolic process | -10.296 | 2.100 | 8.466 | 2.208 | -0.989 | 0.842 | -0.515 | 1.415 | 9.901 | 1.243 |
| Flagellar hook protein FlgE | P0A1J1 | flgE | Motility | ND | ND | ND | ND | -8.339 | 1.048 | ND | ND | -6.615 | 1.054 |
| Flagellar hook-associated protein 3 | P16326 | flgL | Motility | -6.794 | 1.152 | ND | ND | ND | ND | ND | ND | ND | ND |
| Flagella synthesis protein FlgN | P0A1J7 | flgN | Motility | -9.157 | 1.970 | ND | ND | -0.528 | 0.735 | -0.138 | 0.125 | 8.395 | 4.679 |
| Flagellar hook-associated protein 2 | B5R7H2 | fliD | Motility | ND | ND | ND | ND | 7.292 | 2.329 | ND | ND | ND | ND |
| Flagellin | B5R7H3 | fljB | Motility | 0.012 | 0.034 | -0.297 | 1.298 | 0.179 | 1.542 | -0.076 | 0.940 | 0.316 | 1.016 |
| [2FE-2S] ferredoxin | Q7CQ13 | fdx | Oxidation-reduction | -0.511 | 0.404 | ND | ND | 10.435 | 2.995 | 1.162 | 1.696 | -8.019 | 1.577 |
| Flavodoxin 1 | Q8ZQX1 | fldA | Oxidation-reduction | ND | ND | ND | ND | ND | ND | 9.103 | 1.504 | 9.307 | 1.054 |
| Glutaredoxin 1 | P0A1P8 | grxA | Oxidation-reduction | ND | ND | 10.883 | 0.737 | ND | ND | ND | ND | -0.839 | 0.547 |
| Glutaredoxin 3 | Q7CPH7 | grxC | Oxidation-reduction | ND | ND | ND | ND | ND | ND | ND | ND | 8.107 | 1.078 |
| Hydrogenase-3, iron-sulfur subunit | Q7CPY1 | hycB | Oxidation-reduction | -7.993 | 1.393 | ND | ND | ND | ND | ND | ND | ND | ND |
| Protease involved in processing C-terminal end of HycE | Q8ZMJ3 | hycI | Oxidation-reduction | ND | ND | ND | ND | 0.469 | 0.605 | -1.598 | 1.076 | 0.655 | 0.668 |
| Glutaredoxin | Q7CQK9 | ydhD | Oxidation-reduction | -1.224 | 0.976 | 9.653 | 1.513 | -0.180 | 1.357 | 1.196 | 1.179 | -0.311 | 1.054 |
| Alkyl hydroperoxide reductase subunit C | P0A251 | ahpC | Oxidation-reduction | -0.080 | 0.233 | 0.592 | 1.393 | -0.625 | 1.154 | 0.305 | 0.488 | -0.400 | 0.430 |
| Thioredoxin dependent thiol peroxidase | Q7CQ23 | bcp | Oxidation-reduction | ND | ND | ND | ND | ND | ND | ND | ND | 8.506 | 1.054 |
| Thiol:disulfide interchange protein DsbC | P55890 | dsbC | Oxidation-reduction | ND | ND | -8.491 | 0.940 | ND | ND | ND | ND | ND | ND |
| Oxygen-insensitive NAD(P)H nitroreductase | P15888 | nfsB | Oxidation-reduction | 8.325 | 4.181 | 9.274 | 1.111 | 0.590 | 1.542 | 9.311 | 1.719 | 1.708 | 2.151 |
| Superoxide dismutase [Fe] | P0A2F4 | sodB | Oxidation-reduction | ND | ND | ND | ND | 1.417 | 1.979 | -1.254 | 0.738 | ND | ND |
| Superoxide dismutase [Cu-Zn] 1 | P0CW86 | sodC1 | Oxidation-reduction | 11.567 | 2.324 | 0.060 | 0.082 | -0.093 | 0.098 | 0.866 | 1.107 | 0.151 | 0.747 |
| Putative thiol-alkyl hydroperoxide reductase | Q7CR42 | STM0402 | Oxidation-reduction | -0.931 | 3.817 | 1.287 | 1.810 | -0.538 | 1.542 | -0.294 | 0.689 | -0.746 | 0.767 |
| Putative thiol-disulfide isomerase and thioredoxin | Q8ZP25 | STM1790 | Oxidation-reduction | -9.429 | 1.436 | 10.287 | 2.011 | 0.629 | 2.192 | 1.026 | 1.719 | -0.532 | 1.000 |
| Probable thiol peroxidase | Q8ZP65 | tpx | Oxidation-reduction | -9.378 | 1.761 | 1.394 | 1.606 | 0.131 | 0.434 | 0.191 | 0.911 | 0.295 | 0.968 |
| Thioredoxin 1 | P0AA28 | trxA | Oxidation-reduction | 0.284 | 0.274 | -0.518 | 0.812 | 0.792 | 1.542 | -1.247 | 1.525 | -0.567 | 1.836 |
| Arsenate reductase | Q8ZN68 | yfgD | Oxidation-reduction | -9.881 | 1.761 | 9.480 | 1.565 | -0.553 | 1.154 | -0.854 | 1.152 | -0.491 | 0.668 |
| Fimbrial protein | P12061 | sefA | Pathogenesis | 1.178 | 3.123 | -0.109 | 0.677 | -0.236 | 3.658 | -0.321 | 1.359 | -0.228 | 0.747 |
| Non-specific acid phosphatase | P26976 | phoN | Pathogenesis | ND | ND | -9.715 | 2.323 | ND | ND | ND | ND | ND | ND |
| Major outer membrane lipoprotein 1 | Q7CQN4 | lpp1 | Pathogenesis | ND | ND | -8.366 | 1.046 | ND | ND | ND | ND | ND | ND |
| Ecotin | Q8ZNH4 | eco | Pathogenesis | 1.515 | 1.561 | 10.403 | 1.629 | 0.246 | 3.740 | -0.399 | 0.571 | 1.421 | 3.339 |
| Peptidyl-prolyl cis-trans isomerase | Q8ZLL6 | fkpA | Protein folding | 1.695 | 2.381 | 0.213 | 1.810 | -0.265 | 1.501 | 0.945 | 1.415 | -0.773 | 1.304 |
| Peptidyl-prolyl cis-trans isomerase | Q8XFG8 | ppiB | Protein folding | -8.726 | 2.358 | 7.501 | 1.665 | ND | ND | ND | ND | ND | ND |
| Peptidyl-prolyl cis-trans isomerase | Q8ZLL4 | slyD | Protein folding | 0.401 | 1.152 | 1.340 | 1.072 | 0.691 | 1.542 | 0.560 | 0.883 | -0.362 | 0.404 |
| Trigger fator | P66932 | tig | Protein folding | -1.402 | 1.238 | 0.665 | 0.451 | 0.708 | 1.105 | -0.426 | 1.485 | -0.730 | 1.971 |
| Chaperone protein DnaK | Q56073 | dnaK | Protein folding | -2.246 | 1.390 | 0.407 | 0.928 | -0.040 | 0.142 | 0.166 | 0.289 | -0.627 | 0.809 |
| 60 kDa chaperonin | P0A1D3 | groL | Protein folding | 0.758 | 0.661 | -0.350 | 0.139 | 2.690 | 4.349 | -1.241 | 0.765 | -0.108 | 0.067 |
| 10 kDa chaperonin | P0A1D5 | groS | Protein folding | 0.969 | 1.188 | 0.433 | 0.865 | 0.037 | 0.178 | 0.076 | 0.585 | 0.182 | 1.840 |
| Protein GrpE | Q7CPZ4 | grpE | Protein folding | -9.471 | 1.122 | 7.826 | 0.799 | 8.528 | 0.842 | ND | ND | -1.209 | 1.972 |
| Small heat shock protein IbpA | Q7CPF1 | ibpA | Protein folding | ND | ND | ND | ND | ND | ND | ND | ND | -7.675 | 1.054 |
| Chaperone protein Skp | P0A1Z2 | skp | Protein folding | ND | ND | ND | ND | -10.241 | 1.299 | 9.656 | 0.990 | ND | ND |
| Chaperone SurA | Q7CR87 | surA | Protein folding | -7.111 | 2.324 | ND | ND | ND | ND | ND | ND | ND | ND |
| Iron-sulfur cluster insertion protein ErpA | Q7CR66 | erpA | Protein maturation | ND | ND | -7.504 | 1.598 | ND | ND | ND | ND | ND | ND |
| Fe/S biogenesis protein NfuA | Q8ZLI7 | nfuA | Protein maturation | 2.317 | 1.761 | 0.908 | 0.593 | -0.996 | 1.048 | -1.033 | 1.106 | -0.700 | 0.767 |
| Iron-sulfur cluster assembly scaffold protein IscU | Q7CQ11 | nifU | Protein maturation | 9.913 | 1.176 | -1.624 | 1.512 | -8.446 | 1.151 | 9.074 | 1.271 | ND | ND |
| S-ribosylhomocysteine lyase | Q9L4T0 | luxS | Quorum sensing | -0.240 | 0.614 | 11.588 | 1.665 | 0.421 | 0.914 | 0.761 | 1.076 | 0.039 | 0.097 |
| Single-stranded DNA-binding protein 1 | P0A2F6 | ssb | Response to stress | -7.772 | 1.761 | ND | ND | 8.250 | 1.368 | ND | ND | ND | ND |
| UPF0234 protein YajQ | Q8ZRC9 | yajQ | Response to stress | -7.420 | 2.808 | 8.948 | 0.625 | ND | ND | ND | ND | ND | ND |
| Universal stress protein G | P67093 | uspG | Response to stress | ND | ND | 9.407 | 2.079 | 0.013 | 0.039 | 1.175 | 1.107 | 0.198 | 1.026 |
| Putative outer membrane protein | Q7CPS4 | ygiW | Response to stress | ND | ND | 6.498 | 1.320 | -7.298 | 2.011 | -6.702 | 1.296 | ND | ND |
| Putative molecular chaperone (Small heat shock protein) | Q8ZPY6 | STM1251 | Response to stress | ND | ND | ND | ND | ND | ND | 9.601 | 1.882 | 0.157 | 0.078 |
| RNA polymerase-binding transcription factor DksA | P0A1G5 | dksA | Transcription | ND | ND | ND | ND | 7.810 | 2.307 | 8.954 | 0.727 | -7.468 | 2.038 |
| Cold shock-like protein CspC | P0A9Y9 | cspC | Transcription | 11.820 | 1.185 | 0.813 | 0.987 | 2.846 | 2.329 | -1.023 | 0.571 | -0.194 | 0.767 |
| RNA chaperone, negative regulator of cspA transcription | Q7CQZ5 | cspE | Transcription | ND | ND | -8.997 | 1.858 | ND | ND | ND | ND | -8.747 | 1.840 |
| Transcriptional repressor of emrAB operon | Q7CPY9 | emrR | Transcription | -7.967 | 1.060 | 7.639 | 2.079 | 7.947 | 1.745 | 7.095 | 1.485 | ND | ND |
| Transcriptional repressor of iron-responsive genes (Fur family) (Ferric uptake regulator) | Q7CQY3 | fur | Transcription | 0.175 | 0.574 | 8.426 | 1.998 | -0.134 | 0.248 | 0.544 | 1.076 | 0.459 | 0.253 |
| Transcription elongation factor GreA | P64281 | greA | Transcription | -1.164 | 1.352 | 10.895 | 1.810 | -0.544 | 0.776 | -0.501 | 1.754 | 0.006 | 0.011 |
| DNA-binding protein H-NS | P0A1S2 | hns | Transcription | -3.346 | 2.381 | -1.089 | 0.898 | 0.174 | 0.248 | 0.042 | 0.022 | -0.278 | 0.809 |
| DNA-binding protein HU-alpha | P0A1R6 | hupA | Transcription | 1.714 | 2.363 | 0.140 | 0.593 | 0.646 | 1.729 | 0.940 | 0.990 | -0.633 | 1.826 |
| Virulence transcriptional regulatory protein PhoP | P0DM78 | phoP | Transcription | -10.751 | 1.390 | 8.923 | 1.046 | 0.758 | 2.854 | 0.030 | 0.019 | 0.955 | 5.924 |
| Regulator of nucleoside diphosphate kinase | Q7CQZ7 | rnk | Transcription | -7.467 | 2.747 | ND | ND | -0.012 | 0.019 | -6.467 | 1.037 | ND | ND |
| DNA-directed RNA polymerase subunit alpha | P0A7Z7 | rpoA | Transcription | -0.255 | 2.435 | 2.402 | 1.998 | -0.135 | 0.851 | -1.706 | 0.561 | -1.425 | 1.297 |
| DNA-directed RNA polymerase subunit omega | P0A803 | rpoZ | Transcription | -1.482 | 1.796 | 8.487 | 1.998 | -1.524 | 2.995 | 0.738 | 0.554 | -0.963 | 2.151 |
| DNA-binding protein | B5RBI8 | SG2019 | Transcription | -7.989 | 2.381 | ND | ND | 6.415 | 2.307 | 0.535 | 0.328 | ND | ND |
| Transcriptional regulator SlyA | P40676 | slyA | Transcription | ND | ND | ND | ND | -0.341 | 0.142 | -8.648 | 1.327 | -2.417 | 1.026 |
| DNA-binding protein StpA | P0A1S4 | stpA | Transcription | 0.152 | 0.125 | ND | ND | ND | ND | 8.362 | 3.372 | ND | ND |
| Nucleoid-associated protein YbaB | P0A8B8 | ybaB | Transcription | 0.454 | 0.416 | 0.312 | 0.255 | -0.286 | 0.349 | -9.610 | 1.037 | -0.817 | 1.249 |
| Transcription modulator YdgT | Q7CQK5 | ydgT | Transcription | -9.106 | 2.324 | ND | ND | ND | ND | ND | ND | ND | ND |
| Inorganic pyrophosphatase | P65748 | ppa | Transcription | -10.551 | 1.376 | -2.442 | 0.737 | 0.540 | 0.598 | 0.730 | 0.480 | -0.911 | 2.335 |
| Modulator of Rho-dependent transcription termination | Q8ZRN4 | rof | Transcription | ND | ND | ND | ND | 0.720 | 1.072 | -0.815 | 0.616 | -0.751 | 0.981 |
| Regulator of ribonuclease activity A | P67651 | rraA | Transcription | ND | ND | ND | ND | 7.518 | 2.329 | ND | ND | ND | ND |
| Stringent starvation protein B | Q7CPN4 | sspB | Transcription | ND | ND | ND | ND | 7.192 | 2.063 | ND | ND | ND | ND |
| Peptide deformylase | Q8ZLM7 | def | Translation | -8.092 | 2.806 | ND | ND | -8.530 | 1.503 | -8.086 | 1.719 | ND | ND |
| Ribosome recycling fator | P66738 | rrf or frr | Translation | -2.277 | 3.208 | 0.989 | 1.558 | 0.444 | 0.506 | 1.071 | 1.330 | 0.602 | 1.026 |
| Stationary-phase-induced ribosome-associated protein | Q7CQJ0 | sra | Translation | ND | ND | -9.400 | 2.414 | -9.252 | 1.151 | 0.305 | 0.238 | -2.258 | 1.840 |
| Ribosome associated fator | Q7CQ00 | yfiA | Translation | ND | ND | -7.943 | 2.079 | ND | ND | -8.664 | 2.295 | -9.657 | 1.387 |
| Translation initiation factor IF-1 | P69226 | infA | Translation | ND | ND | ND | ND | 8.689 | 0.617 | 7.127 | 1.719 | ND | ND |
| Translation initiation factor IF-3 | P33321 | infC | Translation | -10.818 | 1.790 | 8.352 | 1.512 | ND | ND | -7.574 | 1.754 | -7.938 | 2.417 |
| 50S ribosomal protein L1 | P0A2A3 | rplA | Translation | -0.903 | 1.148 | -8.794 | 2.134 | 9.181 | 1.039 | ND | ND | ND | ND |
| 50S ribosomal protein L3 | P60446 | rplC | Translation | 9.334 | 2.324 | ND | ND | ND | ND | ND | ND | ND | ND |
| 50S ribosomal protein L6 | P66313 | rplF | Translation | 0.593 | 1.957 | -0.186 | 0.328 | ND | ND | 7.572 | 0.841 | -9.383 | 1.241 |
| 50S ribosomal protein L9 | Q8ZK80 | rplI | Translation | 0.306 | 2.454 | -0.381 | 1.665 | 0.147 | 0.183 | -0.995 | 1.359 | -0.361 | 1.026 |
| 50S ribosomal protein L10 | P0A297 | rplJ | Translation | -0.425 | 0.870 | 9.051 | 2.011 | 0.557 | 0.637 | 1.216 | 1.396 | 9.329 | 1.220 |
| 50S ribosomal protein L11 | P0A7K0 | rplK | Translation | 0.564 | 1.152 | -9.100 | 1.288 | 9.783 | 0.846 | ND | ND | -8.354 | 1.971 |
| 50S ribosomal protein L7/L12 | P0A299 | rplL | Translation | 1.043 | 1.148 | 0.832 | 1.665 | 0.408 | 0.615 | -1.167 | 0.990 | 0.692 | 0.668 |
| 50S ribosomal protein L15 | P66073 | rplO | Translation | -10.938 | 4.151 | ND | ND | ND | ND | ND | ND | ND | ND |
| 50S ribosomal protein L17 | Q7CPL7 | rplQ | Translation | -0.782 | 1.990 | 3.526 | 4.757 | 0.467 | 0.466 | 9.427 | 0.677 | -0.123 | 0.055 |
| 50S ribosomal protein L18 | Q7CPL6 | rplR | Translation | 1.375 | 2.324 | ND | ND | -0.275 | 0.235 | 9.705 | 1.182 | ND | ND |
| 50S ribosomal protein L19 | P0A2A1 | rplS | Translation | -0.533 | 0.420 | ND | ND | ND | ND | 10.169 | 0.909 | ND | ND |
| 50S ribosomal protein L24 | P60626 | rplX | Translation | -0.441 | 0.873 | 11.243 | 3.470 | 1.216 | 0.773 | -0.730 | 1.107 | 0.442 | 0.305 |
| 50S ribosomal protein L25 | Q7CQ71 | rplY | Translation | -0.776 | 1.230 | 7.944 | 1.598 | 0.984 | 1.542 | 0.793 | 0.382 | -0.605 | 0.707 |
| 50S ribosomal protein L28 | P0A2A5 | rpmB | Translation | -1.746 | 2.358 | 9.704 | 1.095 | 0.537 | 0.297 | 8.210 | 1.025 | ND | ND |
| 50S ribosomal protein L29 | P66170 | rpmC | Translation | -1.525 | 1.640 | 8.702 | 1.677 | 1.191 | 0.481 | -0.900 | 2.043 | 0.539 | 0.280 |
| 50S ribosomal protein L30 | P0A2A7 | rpmD | Translation | ND | ND | ND | ND | ND | ND | ND | ND | -9.619 | 2.532 |
| 50S ribosomal protein L31 | P66191 | rpmE | Translation | ND | ND | ND | ND | ND | ND | 10.159 | 1.074 | ND | ND |
| 50S ribosomal protein L33 | P0A7P2 | rpmG | Translation | ND | ND | 8.313 | 2.037 | ND | ND | 7.849 | 1.076 | ND | ND |
| 50S ribosomal protein L34 | P0A7P8 | rpmH | Translation | -7.903 | 2.324 | ND | ND | ND | ND | 9.446 | 1.053 | ND | ND |
| 30S ribosomal protein S1 | Q7CQT9 | rpsA | Translation | -3.033 | 4.181 | ND | ND | 1.901 | 1.525 | -1.215 | 0.505 | ND | ND |
| 30S ribosomal protein S2 | P66541 | rpsB | Translation | -7.096 | 1.761 | ND | ND | ND | ND | ND | ND | ND | ND |
| 30S ribosomal protein S4 | O54297 | rpsD | Translation | -0.643 | 0.937 | ND | ND | ND | ND | 8.864 | 0.990 | ND | ND |
| 30S ribosomal protein S5 | P0A7W4 | rpsE | Translation | -1.050 | 0.941 | -8.356 | 1.776 | ND | ND | ND | ND | ND | ND |
| 30S ribosomal protein S6 | P66593 | rpsF | Translation | -9.552 | 1.680 | 6.762 | 1.521 | 0.510 | 0.344 | ND | ND | ND | ND |
| 30S ribosomal protein S7 | P0A2B3 | rpsG | Translation | -0.345 | 1.761 | ND | ND | -0.539 | 0.666 | ND | ND | ND | ND |
| 30S ribosomal protein S8 | P0A7X0 | rpsH | Translation | -1.382 | 2.297 | 2.600 | 2.902 | 0.735 | 1.542 | 0.112 | 0.285 | -2.163 | 2.532 |
| 30S ribosomal protein S10 | P67904 | rpsJ | Translation | 0.303 | 0.245 | -0.520 | 0.495 | 0.125 | 0.142 | -1.227 | 0.585 | ND | ND |
| 30S ribosomal protein S11 | O54296 | rpsK | Translation | 1.423 | 1.139 | ND | ND | ND | ND | ND | ND | ND | ND |
| 30S ribosomal protein S13 | Q8ZLM1 | rpsM | Translation | 0.283 | 0.242 | ND | ND | ND | ND | ND | ND | ND | ND |
| 30S ribosomal protein S14 | P66409 | rpsN | Translation | -0.353 | 0.568 | 7.881 | 1.810 | -0.451 | 2.036 | 2.427 | 0.990 | ND | ND |
| 30S ribosomal protein S18 | Q8ZK81 | rpsR | Translation | -12.161 | 2.208 | ND | ND | ND | ND | ND | ND | ND | ND |
| 30S ribosomal protein S19 | P66491 | rpsS | Translation | 0.149 | 0.404 | ND | ND | ND | ND | ND | ND | ND | ND |
| 30S ribosomal protein S20 | P0A2B1 | rpsT | Translation | 1.295 | 4.181 | 1.915 | 1.348 | 0.834 | 1.072 | 1.105 | 0.776 | -1.148 | 1.054 |
| Elongation factor P | P64036 | efp | Translation | -9.275 | 1.432 | ND | ND | ND | ND | ND | ND | ND | ND |
| Elongation factor Ts | P64052 | tsf | Translation | -0.212 | 0.451 | 1.346 | 1.596 | 0.609 | 1.154 | -0.872 | 1.754 | -2.134 | 2.151 |
| Elongation factor Tu | P0A1H5 | tufA | Translation | -3.968 | 1.957 | 2.390 | 2.077 | -0.543 | 0.598 | -0.425 | 0.147 | -0.140 | 0.096 |
| Histidine-binding periplasmic protein | P02910 | hisJ | Transport | -9.017 | 1.957 | -1.073 | 0.737 | -0.934 | 0.522 | 9.351 | 1.331 | 0.646 | 0.920 |
| PTS system glucose-specific EIIA component | P0A283 | crr | Transport | -0.304 | 0.841 | -2.267 | 3.151 | -1.073 | 1.822 | -0.399 | 1.485 | 0.063 | 0.257 |
| Multiphosphoryl transfer protein | P17127 | fruB | Transport | ND | ND | -7.368 | 1.542 | ND | ND | ND | ND | ND | ND |
| Phosphocarrier protein HPr | P0AA07 | ptsH | Transport | 8.437 | 1.230 | 7.848 | 3.151 | 0.504 | 0.481 | 1.948 | 0.711 | -9.520 | 0.737 |
| Glutamine high-affinity transporter | Q7CQW0 | glnH | Transport | ND | ND | ND | ND | ND | ND | -7.105 | 1.102 | ND | ND |
| Ferritin | Q8ZNU4 | ftn | Transport | -10.668 | 2.044 | 9.884 | 2.902 | 1.148 | 0.945 | ND | ND | 10.071 | 1.054 |
| Protein-export protein SecB | Q7CPH8 | secB | Transport | -7.460 | 1.409 | 10.905 | 1.252 | ND | ND | ND | ND | ND | ND |
| Outer membrane protein A | P02936 | ompA | Transport | 1.039 | 2.044 | 0.063 | 0.113 | -0.312 | 0.482 | 0.140 | 0.207 | 0.693 | 1.987 |
| Outer membrane channel | Q8ZLZ4 | tolC | Transport | ND | ND | ND | ND | 10.139 | 0.594 | ND | ND | -0.744 | 0.767 |
| BssS protein Family | A0A1V9AFN8 | ABA47_0691 | Unclassified | -8.706 | 2.381 | ND | ND | 0.807 | 0.663 | 0.364 | 0.253 | ND | ND |
| Uncharacterized protein | A0A1F2JWR0 | HMPREF 3126_08675 |
Unclassified | ND | ND | ND | ND | ND | ND | -9.642 | 1.719 | ND | ND |
| Uncharacterized protein | A0A1R2IBX3 | R567_04560 | Unclassified | ND | ND | ND | ND | 8.671 | 0.842 | ND | ND | -7.007 | 0.967 |
| Uncharacterized protein | B5RBG9 | SG1997 | Unclassified | -7.908 | 1.098 | -9.302 | 1.665 | ND | ND | 7.916 | 0.779 | ND | ND |
| Putative periplasmic protein | Q8ZPY8 | STM1249 | Unclassified | ND | ND | ND | ND | 1.332 | 1.094 | 7.565 | 1.882 | 9.917 | 1.431 |
| UPF0253 protein YaeP | P67551 | yaeP | Unclassified | ND | ND | ND | ND | 8.765 | 1.985 | ND | ND | ND | ND |
| Putative cytoplasmic protein | Q8ZQ41 | yccJ | Unclassified | ND | ND | ND | ND | 9.189 | 0.663 | 9.850 | 1.719 | -9.249 | 2.067 |
| Uncharacterized protein | Q7CQR0 | ycfF | Unclassified | ND | ND | ND | ND | ND | ND | 7.127 | 1.107 | ND | ND |
| Putative cytoplasmic protein | Q7CQJ6 | ydfZ | Unclassified | -1.164 | 1.244 | 0.884 | 2.134 | 0.262 | 2.329 | -0.047 | 0.221 | -0.048 | 0.063 |
| Putative cytoplasmic protein | Q7CQB7 | yecF | Unclassified | -9.069 | 0.706 | ND | ND | ND | ND | ND | ND | 6.772 | 1.342 |
| UPF0265 protein YeeX | P67605 | yeeX | Unclassified | -12.867 | 2.813 | 10.455 | 1.629 | 1.293 | 0.890 | -0.540 | 0.696 | 8.884 | 1.714 |
| Putative cytoplasmic protein | Q7CQ33 | yfcZ | Unclassified | 1.467 | 1.409 | -0.175 | 0.344 | 0.415 | 1.314 | -0.656 | 1.562 | 0.010 | 0.029 |
| Putative cytoplasmic protein | Q8ZKH3 | yjbR | Unclassified | ND | ND | ND | ND | ND | ND | 7.963 | 1.687 | ND | ND |
| Uncharacterized protein | I3W485 | - | Unclassified | ND | ND | 8.155 | 1.028 | ND | ND | 8.028 | 0.859 | ND | ND |
ND = not detected;
Log2 FC = logarithm in base two of fold changed (ratio of the normalized mean of TIC values of the treatment with C12-HSL by the control);
-Log10 p = negative logarithm of p-value;
Gray background and bold number = increased abundance of protein in C12-HSL and detected in both treatments (Log2 FC > 0.585 and -Log10 > 1.301);
Gray background = increased abundance of protein in C12-HSL and detected only in the treatment with C12-HSL (Log2 FC > 0.585);
Yellow background and bold number = decreased abundance of protein in C12-HSL and detected in both treatments (Log2 FC < -0.585 and -Log10 > 1.301);
Yellow background = decreased abundance of protein in C12-HSL and detected only in the control treatment (Log2 FC < -0.585).
Table 4. Number and percentage of differentially abundant proteins in comparison to the total proteins identified from Salmonella Enteritidis PT4 578 anaerobically cultivated in TSB at 37 °C in the presence C12-HSL.
| Time (h) | Proteins identified | ||||||
|---|---|---|---|---|---|---|---|
| Abundance increased | Abundance decreased | Differentially abundant | Total | ||||
| Number | % | Number | % | Number | % | Number | |
| 4 | 17 | 15.5 | 55 | 50.0 | 72 | 65.5 | 110 |
| 6 | 51 | 54.8 | 16 | 17.2 | 67 | 72.0 | 93 |
| 7 | 31 | 28.7 | 11 | 10.2 | 42 | 38.9 | 108 |
| 12 | 36 | 34.0 | 14 | 13.2 | 50 | 47.2 | 106 |
| 36 | 16 | 17.8 | 23 | 25.6 | 39 | 43.4 | 90 |
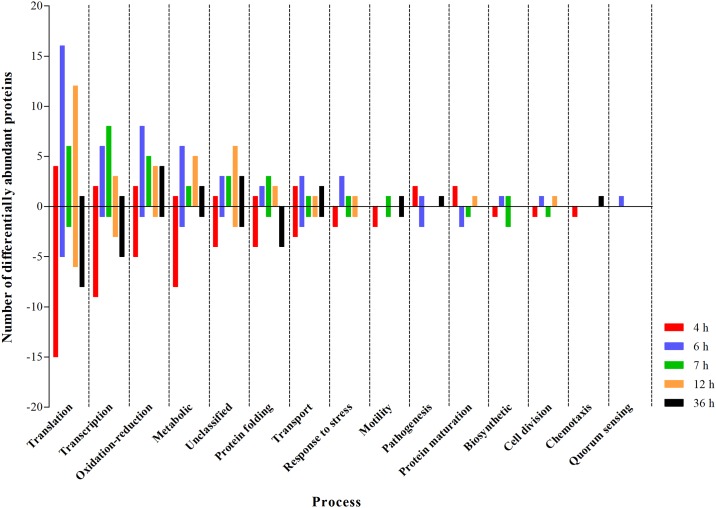
The differentially abundant proteins were grouped in order to perform an analysis of enrichment of biological processes based on GO annotations, as shown in Fig 3. The proteins related to translation, transcription, oxidation-reduction, metabolic, protein folding, transport processes as well as unclassified proteins, had their abundance affected by C12-HSL in all the studied time points.
Fig 3. Number of differentially abundant proteins grouped according to the process on Gene Ontology (GO) annotations (European Bioinformatics Institute).
The Y-axis represents the number of differentially abundant proteins: above zero the number of proteins in which the abundance increased in the presence of C12-HSL compared to the control and below zero represents the proteins in which the abundance decreased in the presence of C12-HSL.
An important identified protein is LuxS (S-ribosylhomocysteine lyase), which produces the autoinducer-2 signaling molecule (Table 3 and Fig 3). In our experimental conditions, this protein was identified only in the presence of 50 nM of C12-HSL at 6 h of cultivation, that is, at the end of the logarithmic phase of growth of Salmonella. This result suggests that there could be a cross response between quorum sensing mechanisms mediated by AI-1 and AI-2 in Salmonella depending on the growth phase. Interactions among the different mechanisms of quorum sensing present in P. aeruginosa have been described leading to a hierarchical activation of these mechanisms [79–81] and also to the synthesis of inductive and inhibitory molecules [82]. The existence of multiple arrangements between the different mechanisms of quorum sensing might play an important role in processing of environmental cues and thus, dictating necessary and robust collective responses [83]. Additional studies should be performed in Salmonella to confirm this possible connection.
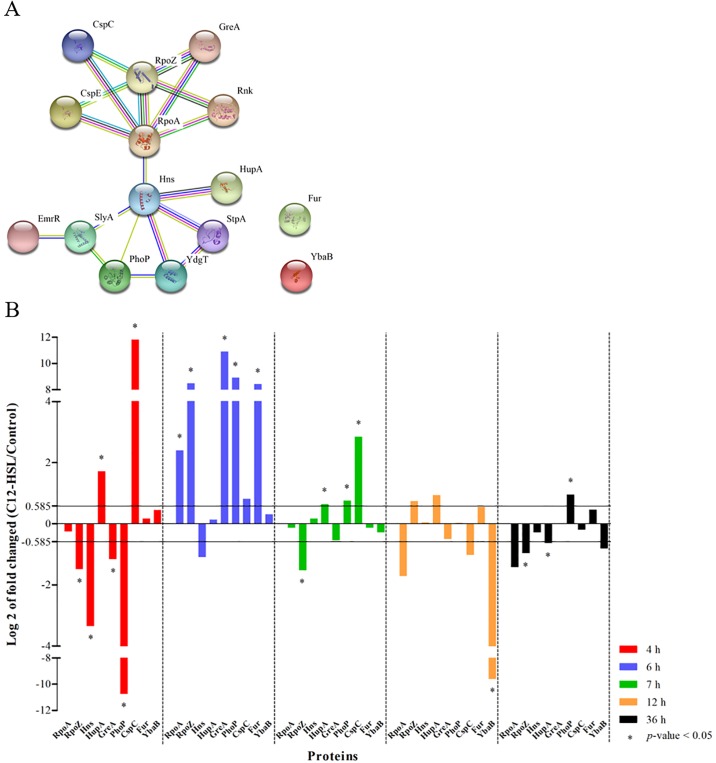
Among the identified proteins, a greater number was involved in transcription process showed a variation of their abundance in the presence of C12-HSL (Fig 3). Since transcription is an essential step in gene expression and the transcriptional regulation determines the molecular machinery for developmental plasticity, homeostasis and adaptation [84], a network of interaction between the proteins related to the transcription process and “regulation of transcription, DNA-templated” was generated (Fig 4A). The PPI network showed an average local clustering coefficient of 0.636 and a p-value of <1.06e-10 for enrichment, indicating that the interactions showed at medium confidence or better and, the proteins have more interactions among themselves than what would have been expected for a random set of proteins of similar size. This result also indicates that these proteins are, at least, partially biologically connected as a group [61, 62]. The RpoA (DNA-directed RNA polymerase subunit alpha) and Hns (DNA-binding protein H-NS) proteins were the central nodes of two networks that are connected (Fig 4A). RpoA had its abundance increased at 6 h of cultivation of Salmonella Enteritidis in the presence of C12-HSL (Fig 4B). Soni et al. [85] showed that the abundance of this protein decreased in late logarithmic phase of growth of Salmonella Typhimurium in the presence of AI-2. In addition, these proteins were identified at all the time points evaluated in this study (Fig 4B). Thus, the differential abundance of proteins related to transcription regulation may be responsible for the differences observed in the abundance of other proteins between control and treatment with C12-HSL and throughout the time of cultivation of Salmonella (Figs 3 and 4B).
Fig 4. Proteins related to transcription process and “regulation of transcription, DNA-templated” function of Salmonella Enteritidis PT4 578, anaerobically cultivated in TSB at 37 °C in the presence or absence of C12-HSL.
(A) The network of interactions among the proteins of the transcription process and “regulation of transcription, DNA-templated” function and (B) the logarithm in base two of fold changed of the proteins that were identified at all times and in at least one of the treatments.
3.3. Levels of thiol and proteins related to the oxidation-reduction process are altered by HSL in Salmonella
The proteins related to the oxidation-reduction process had their abundance affected by the presence of C12-HSL at all times of cultivation (Fig 3). At 4 h of cultivation with this signaling molecule, a greater number of these proteins had their abundance decreased in comparison to the control (Fig 3). Conversely, their abundance was increased at 6 h of cultivation. This might suggest that cells cultivated during this period in the presence of C12-HSL have greater potential to resist oxidative stress than cells cultivated in the absence of this molecule. The proteins related to the oxidative process can be considered crucial to maintenance of the cellular redox balance, as well as to anticipate resistance to a possible oxidative stress due to excessive production of reactive oxygen/nitrogen species (ROS/RNS) [86–88].
In other bacteria, quorum sensing has been associated to oxidative stress response. For instance, in P. aeruginosa, the expression of katA (catalase) and sodA (superoxide dismutase) genes and, concomitantly, the activities of the catalase and superoxide dismutase enzymes were up-regulated by quorum sensing [89]. In addition, Garcia-Contreras et al. [90] showed that resistance of P. aeruginosa to oxidative stress caused by the addition of hydrogen peroxide (H2O2) was enhanced due to quorum sensing, increasing the production of catalase and NADPH dehydrogenases. In the study herein, the SodC1 (Superoxide dismutase [Cu-Zn] 1) also had its abundance increased at 4 h of incubation (Log2 FC = 11.567), while SodB (Superoxide dismutase [Fe]) protein had its abundance increased at 7 h when in the presence of C12-HSL (Log2 FC = 1.417) (Table 3). The cell-cell communication system via BpsIR (homologous to LuxIR) and N-octanoyl-homoserine lactone (C8-HSL) also increased resistance to oxidative stress of B. pseudomallei, as well as the expression of the dpsA gene (DNA-binding protein from starved cells) [91]. The Dps is a non-specific DNA-binding protein involved in resistance to oxidative stress and it is an abundant protein in stationary phase of growth in E. coli [92, 93].
A network of interaction among the proteins of the oxidation-reduction process was generated (Fig 5A). The PPI network showed an average local clustering coefficient of 0.609 and a p-value of <1e-16 for enrichment. These results indicate that the interactions showed at medium confidence or better and, the proteins have more interactions among themselves than for a random set of proteins of similar size and also that these proteins are at least partially biologically connected as a group [61, 62].
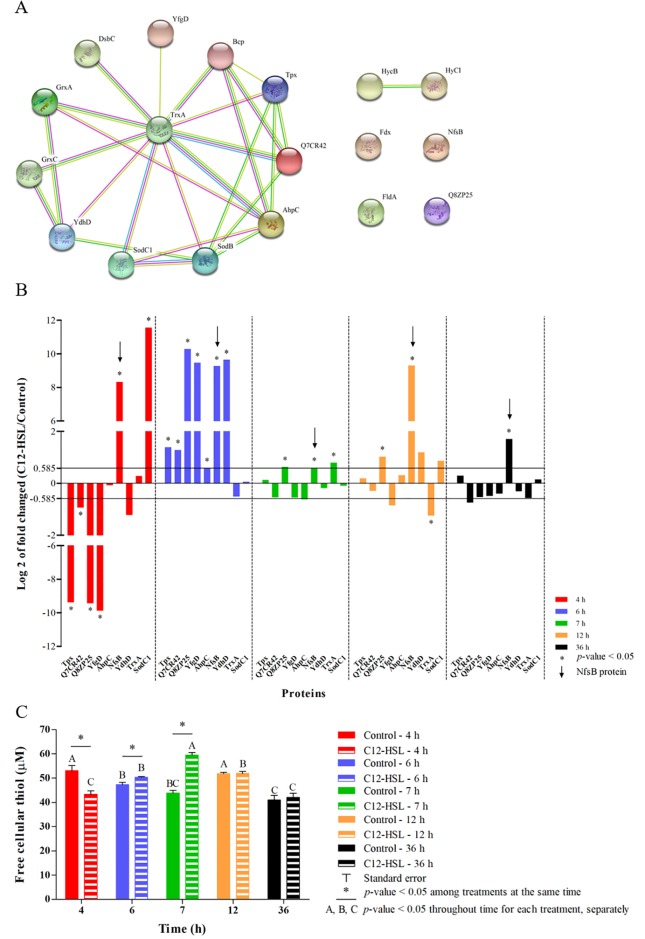
Fig 5. Identified proteins related to the oxidation-reduction process and quantification of free cellular thiol in Salmonella Enteritidis PT4 578 anaerobically cultivated in TSB at 37 °C in the presence or absence of C12-HSL.
(A) The network of interactions among the proteins of oxidation-reduction process and (B) the logarithm in base two of fold changed of the proteins that were identified at all time points and in at least one of the treatments, as well as (C) the quantification of free cellular thiol in the absence (control) and presence of C12-HSL.
The predicted network is an efficient tool to identify potential interactions between a large number of proteins identified by proteomics providing a biological meaning for the data. Furthermore, the centrality of proteins in this network is generally associated to the importance of this element to the biological process associated to the network [94, 95]. Interestingly, the TrxA protein (Thioredoxin 1) stands out as being a central node by interacting with most of the proteins used to generate the network (Fig 5A). In these experiments, this protein had its increased abundance at 7 h of cultivation in the presence of C12-HSL, but its abundance was lower at 12 h (Fig 5B). This protein is known to be essential to activate gene transcription of the pathogenicity island 2 (SPI-2) of Salmonella Typhimurium and consequently, for resistance during mice infection [96–98].
Thioredoxin is an oxidoreductase that participates in redox reactions by oxidation of its thiol active-sites which are then reduced by NADPH. It also exerts control over the activity of target proteins via reversible thiol-disulfide exchange reactions by the thioredoxin and glutaredoxin systems [88, 99–102]. This protein also has a regulatory mechanism independent of thiol redox activity, which thioredoxin interacts with other proteins and forms a functional complex [100]. Thiol, also known as mercaptan or sulfhydryl,—SH side chain of cysteine is susceptible to reactions with ROS or RNS species, giving rise to a range of post-translational oxidative modifications by thiol proteins, including reversible (intra-protein disulfides, inter-protein disulfides, S-sulfenation, S-nitrosation, S-thiolation, S-sulfhydration, S-sulfenamidation) and non-reversible hyper-oxidized (S-sulfination, S-sulfonation) redox states. In addition, in some cases it can alter the structure and activity of proteins that contain cysteine residues [87, 88, 103, 104].
From nine proteins that were identified at all cultivation times and used for the generation of the network (Fig 5A), eight are or have some relation to thiol proteins such as: Tpx (Probable thiol peroxidase), Q7CR42 (Putative thiol-alkyl hydroperoxide reductase), Q8ZP25 (Putative thiol-disulfide isomerase and thioredoxin), YfgD (Arsenate reductase), AhpC (Alkyl hydroperoxide reductase subunit C), NfsB (Oxygen-insensitive NAD(P)H nitroreductase), YdhD (Glutaredoxin) and TrxA (Thioredoxin 1) proteins. The thiol proteins: Tpx, Q7CR42, Q8ZP25 and YfgD decreased in abundance at 4 h of cultivation of Salmonella Enteritidis in the presence of the 50 nM C12-HSL (Fig 5B). On the other hand, at 6 h of cultivation, the Tpx, Q7CR42, Q8ZP25, YfgD, AhpC, NfsB and YdhD proteins increased in abundance, as well as Q8ZP25, NfsB and TrxA proteins at 7 h of cultivation (Fig 5B). In addition, more thiol proteins had their abundances altered at 4, 6 and 7 h of culture, which refer to the logarithmic phase up to the early stationary phase of growth compared to the times of 12 and 36 h where the cells were in stationary phase for a long time (Fig 5B).
The quantification of free cellular thiol showed a correlation with the abundance of thiol proteins at each time and treatment (Fig 5B and 5C). At 4 h of cultivation, a lower level of thiol was observed in the treatment with the quorum sensing molecule as well as a lower abundance of the thiol proteins in comparison to the control. Subsequently, at 6 and 7 h higher levels of thiol were observed in cells cultivated in presence of C12-HSL (Fig 5C and S4 Table). Then, at 12 and 36 h no differences in the levels of thiol were observed, correlating with the number of differentially abundant thiol proteins. In addition, for the same treatment throughout the time of cultivation of Salmonella, the levels of thiol increased up to 7 h of cultivation in the presence of C12-HSL and decreased after this time (Fig 5C). On the other hand, in the absence of this AHL, the level of thiol varied throughout time without a trend (Fig 5C). These results showed that quorum sensing alters not only the abundance of thiol proteins but also the levels of thiol, suggesting that resistance to possible oxidative stress can be mediated by the signaling molecule. This is the first time that the relationship between thiol proteins and levels of free cellular thiol with quorum sensing is reported.
Variations in the abundance of thiol proteins and levels of free cellular thiol due to the growth phase of Salmonella and the presence of acyl homoserine lactone can be related to changes in the structure of the SdiA protein (LuxR homologue) which could alter its ability to bind DNA and, consequently, activate transcription. On the other hand, the thiol proteins and thiol could prevent structural alterations of the SdiA protein caused by ROS/RNS. This rationale is possible because the cysteine residues (C) and their respective positions (C45, C122, C142, and C232) present in the SdiA protein of Salmonella Enteritidis PT4 578 could be susceptible to oxidative stress [105]. The C232 is the main conserved residue among LuxR family proteins [31] and it is involved in the interaction between the Ligand-binding domain (LBD) with DNA-binding domain (DBD) and DBD-DBD [30].
Kafle et al. [106] evaluated all cysteine residues of the LasR protein, a LuxR homologue, of P. aeruginosa to infer their redox sensitivity and to probe the connection between stress response and the activity of that protein. The C79 residue is important for ligand recognition and folding of this domain which further potentiates DNA binding, but it does not seem to be sensitive to oxidative stress when bound to its native ligand. The C201 and C203 residues in the DBD form a disulfide bond when treated with hydrogen peroxide, and this bond seems to disrupt the DNA binding activity of the transcription factor. Mutagenesis of either of these cysteines leads to expression of a protein that no longer binds DNA. Thus, these authors provided a possible mechanism for oxidative stress response by the cysteine residues of the LasR protein in P. aeruginosa and indicated that multiple cysteines within the protein can be useful targets for disabling its activity.
The presence of C12-HSL increased the abundance of thiol proteins, such as oxidoreductases, which can change the structure of this AHL and inactivate it. Some oxidoreductases synthesized by Rhodococcus erythropolis and Bacillus megaterium, as well as by eukaryotic cells, are able to inactivate AHLs by oxidation or reduction of their acyl side chain [107–110]. In fact, this is one of the known mechanisms of quorum quenching [107].
Finally, the NfsB protein (also named NfnB or NfsI) was the only protein that had its abundance increased at all cultivation times in the presence of C12-HSL compared to the control treatment in Salmonella (Fig 5B, indicated with a black arrow at each time point). This result suggests that Salmonella cultivated in the presence of this quorum sensing molecule can be susceptible to the action of nitrofurans or that C12-HSL has a certain toxicity or mutagenicity activity to the cell. The NfsB protein is a flavin mononucleotide-containing flavoprotein that can use either NADH or NADPH as a source of reducing power in order to reduce the nitro moiety of nitrofurans, yielding biologically inactive end products. This process occurs through a sequence of intermediates, including nitroso and hydroxylamine states, which are assumed to be responsible for toxicity [111, 112]. E. coli and Salmonella Typhimurium containing the nfsA and nfsB genes are more sensitive to nitrofurans, which are widely used as antimicrobial agents [113, 114]. Carroll et al. [115] also showed that the introduction of plasmids carrying the nfsA and nfsB genes into Salmonella Typhimurium increased sensibility to nitrofuran compounds with mutagenic potential.
Toxicity of AHLs has already been proven in many studies, especially those performed with eukaryotic cells. For instance, Gomi et al. [116] showed that 10 μg/mL of C12-HSL derived from Chromobacterium violaceum induced the production of tumor necrosis factor-α (TNF-α) and interleukin-1β (IL-1β) by mouse RAW264.7 cells and IL-8 by human THP-1 cells, while C4, C6, C7, C8, C10 and C14-HSL had no effect. The C12-HSL and C12-oxo-HSL also decreased the levels of putrescine in human epidermal cells (HaCat) and, consequently, decreased the rate of cell proliferation [117]. John et al. [118] showed that N-3-oxo-tetradecanoyl-homoserine lactone (3-oxo-C14-HSL) produced by Acinetobacter baumannii had a dose-dependent cytotoxic effect on human cervical cancer cells (HeLa), adenocarcinoma human alveolar basal epithelial cells (A549), Dukes’ type C colorectal adenocarcinoma cells (HCT15) and Dukes’ type B colorectal adenocarcinoma cells (SW480), with induction of apoptosis and reduced viability and proliferation of these cells in the presence of AHL. In addition, these authors showed that 3-oxo-C14-HSL was able to decrease the growth of Staphylococcus aureus.
It is possible that the NfsB protein can be used as a biomarker for the presence of the AI-1 in Salmonella, due to its greater abundance in the presence of this molecule in all the evaluated times. It is noteworthy that nitroreductases homologous to NfsA and NfsB are found in many members of the Enterobacteriaceae family [119].
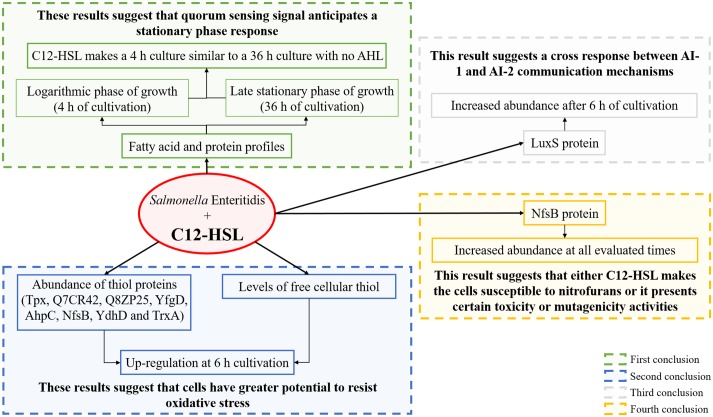
4. Conclusions
The fatty acid and protein profiles of Salmonella Enteritidis PT4 578 in logarithmic phase of growth at 4 h of cultivation in the presence of C12-HSL were similar to the profiles of cells in late stationary phase at 36 h, suggesting that quorum sensing signal anticipates a stationary phase response (Fig 6). Overall, the fatty acid and protein profiles varied less along the bacterium growth in the presence of AI-1. In addition, the presence of this signaling molecule increased the abundance of thiol proteins and the levels of free cellular thiol after 6 h of cultivation suggesting that the cells may be prepared for a possible oxidative stress (Fig 6). This increase may lead to modifications in the structure of the AHL or SdiA protein which, consequently, could alter its binding affinities to the DNA. More studies are needed in order to confirm this hypothesis.
Fig 6. Global response of Salmonella to the presence of C12-HSL.
The results related to a conclusion.
There was an increased abundance of LuxS protein in presence of C12-HSL which suggests a cross response between the quorum sensing mechanisms mediated by AI-1 and AI-2 in Salmonella (Fig 6), while the increased abundance of NfsB protein in this condition suggests that the cells can either be susceptible to the action of nitrofurans or this signaling molecule has a certain toxicity or mutagenicity to the cell (Fig 6). Further studies are needed in order to confirm the hypotheses generated in this work.
Supporting information
(XLSX)
- MM = molecular mass;
- pI = isoeletric point;
- TIC = total ion current;
- Mascot = Mascot software (version 2.4.0; Matrix Science, United Kingdom);
- Scaffold = Scaffold software (version 4.7.2; Proteome Software, USA).
(XLSX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Felipe Alves de Almeida was supported by a fellowship from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and this research has been supported by Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Financiadora de Estudos e Projetos (Finep), Sistema Nacional de Laboratórios em Nanotecnologias (SisNANO) and Ministério da Ciência, Tecnologia e Informação (MCTI). The authors acknowledge the Mass Spectrometry Facility at Brazilian Biosciences National Laboratory (LNBio), CNPEM, Campinas, Brazil and the Núcleo de Análise de Biomoléculas (Nubiomol) at Universidade Federal de Viçosa, Viçosa, Brazil for their support on mass spectrometry and data analysis. The authors thank Professor Marcos Rogério Tótola of Department of Microbiology at Universidade Federal de Viçosa for equipment and software for fatty acids analysis. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. 10.1128/jb.176.2.269-275.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reading NC, Sperandio V. Quorum sensing: the many languages of bacteria. FEMS Microbiol Lett. 2006;254:1–11. 10.1111/j.1574-6968.2005.00001.x [DOI] [PubMed] [Google Scholar]
- 3.Quecán BXV, Rivera MLC, Pinto UM. Bioactive phytochemicals targeting microbial activities mediated by quorum sensing In: Kalia V, editor. Biotechnological applications of quorum sensing inhibitors. Singapore: Springer; 2018. pp.397–416. [Google Scholar]
- 4.Ahmer BMM. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol Microbiol. 2004;52:933–945. 10.1111/j.1365-2958.2004.04054.x [DOI] [PubMed] [Google Scholar]
- 5.Smith JL, Fratamico PM, Novak JS. Quorum sensing: a primer for food microbiologists. J Food Prot. 2004;67:1053–1070. 10.4315/0362-028X-67.5.1053 [DOI] [PubMed] [Google Scholar]
- 6.Walters M, Sperandio V. Quorum sensing in Escherichia coli and Salmonella. Int J Med Microbiol. 2006;296:125–131. 10.1016/j.ijmm.2006.01.041 [DOI] [PubMed] [Google Scholar]
- 7.Ammor MS, Michaelidis C, Nychas GJE. Insights into the role of quorum sensing in food spoilage. J Food Prot. 2008;71:1510–1525. 10.4315/0362-028X-71.7.1510 [DOI] [PubMed] [Google Scholar]
- 8.Jayaraman A, Wood TK. Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu Rev Biomed Eng. 2008;10:145–167. 10.1146/annurev.bioeng.10.061807.160536 [DOI] [PubMed] [Google Scholar]
- 9.Michael B, Smith JN, Swift S, Heffron F, Ahmer BMM. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J Bacteriol. 2001;183:5733–5742. 10.1128/JB.183.19.5733-5742.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith JN, Ahmer BMM. Detection of other microbial species by Salmonella: expression of the SdiA regulon. J Bacteriol. 2003;185:1357–1366. 10.1128/JB.185.4.1357-1366.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taga ME, Semmelhack JL, Bassler BL. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella Typhimurium. Mol Microbiol. 2001;42:777–793. 10.1046/j.1365-2958.2001.02669.x [DOI] [PubMed] [Google Scholar]
- 12.Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, et al. Salmonella Typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004;15:677–687. 10.1016/j.molcel.2004.07.020 [DOI] [PubMed] [Google Scholar]
- 13.Walters M, Sircili MP, Sperandio V. AI-3 synthesis is not dependent on luxS in Escherichia coli. J Bacteriol. 2006;188:5668–5681. 10.1128/JB.00648-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asad S, Opal SM. Bench-to-bedside review: Quorum sensing and the role of cell-to-cell communication during invasive bacterial infection. Crit Care. 2008;12:236 10.1186/cc7101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bearson BL, Bearson SM. The role of the QseC quorum-sensing sensor kinase in colonization and norepinephrine-enhanced motility of Salmonella enterica serovar Typhimurium. Microb Pathog. 2008;44:271–278. 10.1016/j.micpath.2007.10.001 [DOI] [PubMed] [Google Scholar]
- 16.Rasko DA, Moreira CG, Li R, Reading NC, Ritchie JM, Waldor MK, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. 10.1126/science.1160354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. The QseC adrenergic signaling cascade in Enterohemorrhagic E. coli (EHEC). PLoS Pathog. 2009;5:e1000553 10.1371/journal.ppat.1000553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreira CG, Weinshenker D, Sperandio V. QseC mediates Salmonella enterica serovar Typhimurium virulence in vitro and in vivo. Infect Immun. 2010;78:914–926. 10.1128/IAI.01038-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev. 2010;34:426–444. 10.1111/j.1574-6976.2009.00204.x [DOI] [PubMed] [Google Scholar]
- 20.Kim J, Park W. Indole: a signaling molecule or a mere metabolic byproduct that alters bacterial physiology at a high concentration? J Microbiol. 2015;53:421–428. 10.1007/s12275-015-5273-3 [DOI] [PubMed] [Google Scholar]
- 21.Sabag-Daigle A, Soares JA, Smith JN, Elmasry ME, Ahmer BMM. The acyl homoserine lactone receptor, SdiA, of Escherichia coli and Salmonella enterica serovar Typhimurium does not respond to indole. Appl Environ Microbiol. 2012;78:5424–5431. 10.1128/AEM.00046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabag-Daigle A, Ahmer BMM. ExpI and PhzI are descendants of the long lost cognate signal synthase for SdiA. PLoS One. 2012;7:e47720 10.1371/journal.pone.0047720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyszel JL, Soares JA, Swearingen MC, Lindsay A, Smith JN, Ahmer BMM. E. coli K-12 and EHEC genes regulated by SdiA. PLoS One. 2010;5:e8946 10.1371/journal.pone.0008946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith JN, Dyszel JL, Soares JA, Ellermeier CD, Altier C, Lawhon SD, et al. SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS One. 2008;3:e2826 10.1371/journal.pone.0002826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaplan HB, Greenberg EP. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985;163:1210–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson JP, van Delden C, Iglewski BH. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol. 1999;181:1203–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Case RJ, Labbate M, Kjelleberg S. AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2008;2:345–349. 10.1038/ismej.2008.13 [DOI] [PubMed] [Google Scholar]
- 28.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Ann Rev Genet. 2009;43:197–222. 10.1146/annurev-genet-102108-134304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atkinson S, Williams P. Quorum sensing and social networking in the microbial world. J R Soc Interface. 2009;6:959–978. 10.1098/rsif.2009.0203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen Y, Nguyen NX, Rogers JL, Liao J, MacMillan JB, Jiang Y, et al. Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. mBio. 2015;6:e02429–e02438. 10.1128/mBio.02429-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almeida FA, Pinto UM, Vanetti MCD. Novel insights from molecular docking of SdiA from Salmonella Enteritidis and Escherichia coli with quorum sensing and quorum quenching molecules. Microb Pathog. 2016;99:178–190. 10.1016/j.micpath.2016.08.024 [DOI] [PubMed] [Google Scholar]
- 32.Ahmer BMM, van Reeuwijk J, Timmers CD, Valentine PJ, Heffron F. Salmonella Typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J Bacteriol. 1998;180:1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soares JA, Ahmer BMM. Detection of acyl-homoserine lactones by Escherichia and Salmonella. Curr Opin Microbiol. 2011;14:188–193. 10.1016/j.mib.2011.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campos-Galvão ME, Ribon AO, Araujo EF, Vanetti MCD. Changes in the Salmonella enterica Enteritidis phenotypes in presence of acyl homoserine lactone quorum sensing signals. J Basic Microbiol. 2015;56:493–501. 10.1002/jobm.201500471 [DOI] [PubMed] [Google Scholar]
- 35.Almeida FA, Pimentel-Filho NJ, Carrijo LC, Bento CBP, Baracat-Pereira MC, Pinto UM, et al. Acyl homoserine lactone changes the abundance of proteins and the levels of organic acids associated with stationary phase in Salmonella Enteritidis. Microb Pathog. 2017;102:148–159. 10.1016/j.micpath.2016.11.027 [DOI] [PubMed] [Google Scholar]
- 36.Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. 10.1128/JB.185.7.2066-2079.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majerczyk C, Brittnacher M, Jacobs M, Armour CD, Radey M, Schneider E, et al. Global analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J Bacteriol. 2014;196:1412–1424. 10.1128/JB.01405-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goo E, Majerczyk CD, An JH, Chandler JR, Seo YS, Ham H, et al. Bacterial quorum sensing, cooperativity, and anticipation of stationary-phase stress. Proc Natl Acad Sci USA. 2012;109:19775–19780. 10.1073/pnas.1218092109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nesse LL, Berg K, Vestby LK, Olsaker I, Djonne B. Salmonella Typhimurium invasion of HEp-2 epithelial cells in vitro is increased by N-acyl homoserine lactone quorum sensing signals. Acta Vet Scand. 2011;53:44–48. 10.1186/1751-0147-53-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z, Que F, Liao L, Zhou M, You L, Zhao Q, et al. Study on the promotion of bacterial biofilm formation by a Salmonella conjugative plasmid and the underlying mechanism. PLoS One. 2014;9:e109808 10.1371/journal.pone.0109808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Almeida FA, Pimentel-Filho NJ, Pinto UM, Mantovani HC, Oliveira LL, Vanetti MCD. Acyl homoserine lactone-based quorum sensing stimulates biofilm formation by Salmonella Enteritidis in anaerobic conditions. Arch Microbiol. 2017;199:475–486. 10.1007/s00203-016-1313-6 [DOI] [PubMed] [Google Scholar]
- 42.Bai AJ, Rai VH. Effect of small chain N-acyl homoserine lactone quorum sensing signals on biofilms of food-borne pathogens. J Food Sci Technol. 2016;53:3609–3614. 10.1007/s13197-016-2346-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dourou D, Ammor MS, Skandamis PN, Nychas GJE. Growth of Salmonella Enteritidis and Salmonella Typhimurium in the presence of quorum sensing signalling compounds produced by spoilage and pathogenic bacteria. Food Microbiol. 2011;28:1011–1018. 10.1016/j.fm.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 44.Wang HH, Ye KP, Zhang QQ, Dong Y, Xu XL, Zhou GH. Biofilm formation of meat-borne Salmonella enterica and inhibition by the cell-free supernatant from Pseudomonas aeruginosa. Food Control. 2013;32:650–658. 10.1016/j.foodcont.2013.01.047 [DOI] [Google Scholar]
- 45.Blana V, Georgomanou A, Giaouris E. Assessing biofilm formation by Salmonella enterica serovar Typhimurium on abiotic substrata in the presence of quorum sensing signals produced by Hafnia alvei. Food Control. 2017;80:83–91. 10.1016/j.foodcont.2017.04.037 [DOI] [Google Scholar]
- 46.Di Cagno R, De Angelis M, Calasso M, Gobbetti M. Proteomics of the bacterial cross-talk by quorum sensing. J Proteomics. 2011;74:19–34. 10.1016/j.jprot.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 47.Miller JH. Experiments in Molecular Genetics. New York: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 48.Sherlock® Analysis Manual. Microbial Identification System. 6.2 version. United States of America: MIDI, Inc.; 2012.
- 49.Ferreira DF. Sisvar: a computer statistical analysis system. Ciênc Agrotec. 2011;35:1039–1042. [Google Scholar]
- 50.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 51.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3:1630–1638. 10.1038/nprot.2008.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- 53.Aragão AZB, Belloni M, Simabuco FM, Zanetti MR, Yokoo S, Domingues RR, et al. Novel processed form of syndecan-1 shed from SCC-9 cells plays a role in cell migration. PLoS One. 2012;7:e43521 10.1371/journal.pone.0043521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyamoto KN, Monteiro KM, Caumo KS, Lorenzatto KR, Ferreira HB, Brandelli A. Comparative proteomic analysis of Listeria monocytogenes ATCC 7644 exposed to a sublethal concentration of nisin. J Proteomics. 2015;119:230–237. 10.1016/j.jprot.2015.02.006 [DOI] [PubMed] [Google Scholar]
- 55.Ghiraldi-Lopes LD, Campanerut-Sá PAZ, Meneguello JE, Seixas FAV, Lopes-Ortiz MA, Scodro RBL, et al. Proteomic profile of Mycobacterium tuberculosis after eupomatenoid-5 induction reveals potential drug targets. Future Microbiol. 2017;12:867–879. 10.2217/fmb-2017-0023 [DOI] [PubMed] [Google Scholar]
- 56.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- 57.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 58.Liu Y, Zhang Q, Hu M, Yu K, Fu J, Zhou F, et al. Proteomic analyses of intracellular Salmonella enterica serovar Typhimurium reveal extensive bacterial adaptations to infected host epithelial cells. Infect Immun. 2015;83:2897–2906. 10.1128/IAI.02882-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tran TK, Han QQ, Shi Y, Guo L. A comparative proteomic analysis of Salmonella Typhimurium under the regulation of the RstA/RstB and PhoP/PhoQ systems. Biochim Biophys Acta. 2016;1864:1686–1695. 10.1016/j.bbapap.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 60.Lago M, Monteil V, Douche T, Guglielmini J, Criscuolo A, Maufrais C, et al. Proteome remodelling by the stress sigma factor RpoS/σS in Salmonella: identification of small proteins and evidence for post-transcriptional regulation. Sci Rep. 2017;7:2127–2141. 10.1038/s41598-017-02362-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. [DOI] [PubMed] [Google Scholar]
- 64.Riddles PW, Blakeley RL, Zerner B. Ellman’s reagent: 5,5'-dithiobis(2-nitrobenzoic acid)-a reexamination. Anal Biochem. 1979;94:75–81. [DOI] [PubMed] [Google Scholar]
- 65.Law JH, Zalkin H, Kaneshiro T. Transmethylation of fatty acids. Biochim Biophys Acta. 1962;70:143–151. 10.1016/0006-3002(63)90734-0 [DOI] [Google Scholar]
- 66.Wang AY, Cronan JE. The growth phase-dependent synthesis of cyclopropane fatty acids in Escherichia coli is the result of an rpoS(katF)-dependent promoter plus enzyme instability. Mol Microbiol. 1994;11:1009–1017. 10.1111/j.1365-2958.1994.tb00379.x [DOI] [PubMed] [Google Scholar]
- 67.Pagán R, Mackey B. Relationship between membrane damage and cell death in pressure-treated Escherichia coli cells: differences between exponential- and stationary-phase cells and variation among strains. Appl Environ Microbiol. 2000;66:2829–2834. 10.1128/AEM.66.7.2829-2834.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guckert JB, Hood MA, White DC. Phospholipid ester-linked fatty acid profile changes during nutrient deprivation of Vibrio cholerae: increases in the trans/cis ratio and proportions of cyclopropyl fatty acids. Appl Environ Microbiol. 1986;52;794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grogan DW, Cronan JE. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol Mol Biol Rev. 1997;61:429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaur A, Chaudhary A, Kaur A, Choudhary R, Kaushik R. Phospholipid fatty acid—A bioindicator of environment monitoring and assessment in soil ecosystem. Curr Sci. 2005;89:1103–1112. [Google Scholar]
- 71.Zhang YM, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. 10.1038/nrmicro1839 [DOI] [PubMed] [Google Scholar]
- 72.Poger D, Mark AE. A ring to rule them all: the effect of cyclopropane fatty acids on the fluidity of lipid bilayers. J Phys Chem B. 2015;119:5487–5495. 10.1021/acs.jpcb.5b00958 [DOI] [PubMed] [Google Scholar]
- 73.Chen YY, Ganzle MG. Influence of cyclopropane fatty acids on heat, high pressure, acid and oxidative resistance in Escherichia coli. Int J Food Microbiol. 2016;222:16–22. 10.1016/j.ijfoodmicro.2016.01.017 [DOI] [PubMed] [Google Scholar]
- 74.Schuster M, Sexton DJ, Hense BA. Why quorum sensing controls private goods. Front Microbiol. 2017;8:885–900. 10.3389/fmicb.2017.00885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wongtrakoongate P, Tumapa S, Tungpradabkul S. Regulation of a quorum sensing system by stationary phase sigma factor RpoS and their co-regulation of target genes in Burkholderia pseudomallei. Microbiol Immunol. 2012;56:281–294. 10.1111/j.1348-0421.2012.00447.x [DOI] [PubMed] [Google Scholar]
- 76.Tsuru S, Ichinose J, Kashiwagi A, Ying BW, Kaneko K, Yomo T. Noisy cell growth rate leads to fluctuating protein concentration in bacteria. Phys Biol. 2009;6:036015 10.1088/1478-3975/6/3/036015 [DOI] [PubMed] [Google Scholar]
- 77.Shahrezaei V, Marguerat S. Connecting growth with gene expression: of noise and numbers. Curr Opin Microbiol. 2015;25:127–135. 10.1016/j.mib.2015.05.012 [DOI] [PubMed] [Google Scholar]
- 78.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185:2080–2095. 10.1128/JB.185.7.2080-2095.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.An D, Danhorn T, Fuqua WC, Parsek MR. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc Natl Acad Sci USA. 2006;103:3828–3833. 10.1073/pnas.0511323103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaufmann GF, Sartorio R, Lee SH, Rogers CJ, Meijler MM, Moss JA, et al. Revisiting quorum sensing: discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc Natl Acad Sci USA. 2005;102:309–314. 10.1073/pnas.0408639102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xavier JB, Kim W, Foster KR. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol Microbiol. 2011;79:166–179. 10.1111/j.1365-2958.2010.07436.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lazazzera BA. Quorum sensing and starvation: signals for entry into stationary phase. Curr Opin Microbiol. 2000;3:177–182. [DOI] [PubMed] [Google Scholar]
- 83.Prajapat MK, Saini S. Logic of two antagonizing intra-species quorum sensing systems in bacteria. Biosystems. 2018;165:88–98. 10.1016/j.biosystems.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 84.Balleza E, Lopez-Bojorquez LN, Martinez-Antonio A, Resendis-Antonio O, Lozada-Chavez I, Balderas-Martinez YI, et al. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol Rev. 2009;33:133–151. 10.1111/j.1574-6976.2008.00145.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Soni KA, Jesudhasan PR, Cepeda M, Williams B, Hume M, Russell WK, et al. Autoinducer AI-2 is involved in regulating a variety of cellular processes in Salmonella Typhimurium. Foodborne Pathog Dis. 2008;5:147–153. 10.1089/fpd.2007.0050 [DOI] [PubMed] [Google Scholar]
- 86.Jones DP. Redefining oxidative stress. Antioxid Redox Signal. 2006;8:1865–1879. 10.1089/ars.2006.8.1865 [DOI] [PubMed] [Google Scholar]
- 87.Rudyk O, Eaton P. Biochemical methods for monitoring protein thiol redox states in biological systems. Redox Biol. 2014;2:803–813. 10.1016/j.redox.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Landeta C, Boyd D, Beckwith J. Disulfide bond formation in prokaryotes. Nat Microbiol. 2018;3:270–280. 10.1038/s41564-017-0106-2 [DOI] [PubMed] [Google Scholar]
- 89.Hassett DJ, Ma JF, Elkins JG, McDermott TR, Ochsner UA, West SE, et al. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34:1082–1093. 10.1046/j.1365-2958.1999.01672.x [DOI] [PubMed] [Google Scholar]
- 90.Garcia-Contreras R, Nunez-Lopez L, Jasso-Chavez R, Kwan BW, Belmont JA, Rangel-Vega A, et al. Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheatin. ISME J. 2014;9:115–125. 10.1038/ismej.2014.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lumjiaktase P, Diggle SP, Loprasert S, Tungpradabkul S, Daykin M, Camara M, et al. Quorum sensing regulates dpsA and the oxidative stress response in Burkholderia pseudomallei. Microbiology. 2006;152:3651–3659. 10.1099/mic.0.29226-0 [DOI] [PubMed] [Google Scholar]
- 92.Almiron M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. [DOI] [PubMed] [Google Scholar]
- 93.Martinez A, Kolter R. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol. 1997;179:5188–5194. 10.1128/jb.179.16.5188-5194.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koschützki D, Schreiber F. Centrality analysis methods for biological networks and their application to gene regulatory networks. Gene Regul Syst Bio. 2008;2:193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cickovski T, Peake E, Aguiar-Pulido V, Narasimhan G. ATria: a novel centrality algorithm applied to biological networks. BMC Bioinformatics. 2017;18:239 10.1186/s12859-017-1659-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bjur E, Eriksson-Ygberg S, Aslund F, Rhen M. Thioredoxin 1 promotes intracellular replication and virulence of Salmonella enterica serovar Typhimurium. Infect Immun. 2006;74:5140–5151. 10.1128/IAI.00449-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peters SE, Paterson GK, Bandularatne ES, Northen HC, Pleasance S, Willers C, et al. Salmonella enterica serovar Typhimurium trxA mutants are protective against virulent challenge and induce less inflammation than the live-attenuated vaccine strain SL3261. Infect Immun. 2010;78:326–336. 10.1128/IAI.00768-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Song M, Kim JS, Liu L, Husain M, Vazquez-Torres A. Antioxidant defense by thioredoxin can occur independently of canonical thiol-disulfide oxidoreductase enzymatic activity. Cell Reports. 2016;14:2901–2911. 10.1016/j.celrep.2016.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jakob U, Muse W, Eser M, Bardwell JCA. Chaperone activity with a redox switch. Cell. 1999;96:341–352. 10.1016/S0092-8674(00)80547-4 [DOI] [PubMed] [Google Scholar]
- 100.Kumar JK, Tabor S, Richardson CC. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc Natl Acad Sci USA. 2004;101:3759–3764. 10.1073/pnas.0308701101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rietsch A, Bessette P, Georgiou G, Beckwith J. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol. 1997;179:6602–6608. 10.1128/jb.179.21.6602-6608.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. 10.1126/science.279.5357.1718 [DOI] [PubMed] [Google Scholar]
- 103.Danon A. Redox reactions of regulatory proteins: do kinetics promote specificity? Trends Biochem Sci. 2002;27:197–203. 10.1016/S0968-0004(02)02066-2 [DOI] [PubMed] [Google Scholar]
- 104.Reniere ML. Reduce, induce, thrive: bacterial redox sensing during pathogenesis. J. Bacteriol. 2018. 10.1128/JB.00128-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Campos-Galvão MEM, Leite TD, Ribon AO, Araujo EF, Vanetti MCD. A new repertoire of informations about the quorum sensing system in Salmonella enterica serovar Enteritidis PT4. Genet Mol Res. 2015;14:4068–4084. 10.4238/2015.April.27.22 [DOI] [PubMed] [Google Scholar]
- 106.Kafle P, Amoh AN, Reaves JM, Suneby EG, Tutunjian KA, Tyson RL, et al. Molecular insights into the impact of oxidative stress on the quorum-sensing regulator protein LasR. J Biol Chem. 2016;291:11776–11786. 10.1074/jbc.M116.719351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Uroz S, Chhabra SR, Cámara M, Williams P, Oger P, Dessaux Y. N-acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology. 2005;151:3313–3322. 10.1099/mic.0.27961-0 [DOI] [PubMed] [Google Scholar]
- 108.Chowdhary PK, Keshavan N, Nguyen HQ, Peterson JA, González JE, Haines DC. Bacillus megaterium CYP102A1 oxidation of acyl homoserine lactones and acyl homoserines. Biochemistry. 2007;46:14429–14437. 10.1021/bi701945j [DOI] [PubMed] [Google Scholar]
- 109.Bijtenhoorn P, Mayerhofer H, Müller-Dieckmann J, Utpatel C, Schipper C, Hornung C, et al. A novel metagenomic short-chain dehydrogenase/reductase attenuates Pseudomonas aeruginosa biofilm formation and virulence on Caenorhabditis elegans. PLoS One. 2011;6:e26278 10.1371/journal.pone.0026278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pietschke C, Treitz C, Forêt S, Schultze A, Künzel S, Tholey A, et al. Host modification of a bacterial quorum-sensing signal induces a phenotypic switch in bacterial symbionts. Proc Natl Acad Sci USA. 2017;114:E8488–E8497. 10.1073/pnas.1706879114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bryant DW, McCalla DR, Leeksma M, Laneuville P. Type I nitroreductases of Escherichia coli. Can J Microbiol. 1981;27:81–86. [DOI] [PubMed] [Google Scholar]
- 112.Michael NP, Brehm JK, Anlezark GM, Minton NP. Physical characterisation of the Escherichia coli B gene encoding nitroreductase and its over-expression in Escherichia coli K12. FEMS Microbiol Lett. 1994;124:195–202. [DOI] [PubMed] [Google Scholar]
- 113.García V, Montero I, Bances M, Rodicio R, Rodicio MR. Incidence and genetic bases of nitrofurantoin resistance in clinical isolates of two successful multidrug-resistant clones of Salmonella enterica serovar Typhimurium: Pandemic "DT 104" and pUO-StVR2. Microb Drug Resist. 2017;23:405–412. 10.1089/mdr.2016.0227 [DOI] [PubMed] [Google Scholar]
- 114.Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, et al. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol. 1998;180:5529–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carroll CC, Warnakulasuriyarachchi D, Nokhbeh MR, Lambert IB. Salmonella Typhimurium mutagenicity tester strains that overexpress oxygen-insensitive nitroreductases nfsA and nfsB. Mutat Res. 2002;501:79–98. 10.1016/S0027-5107(02)00018-0 [DOI] [PubMed] [Google Scholar]
- 116.Gomi K, Kikuchi T, Tokue Y, Fujimura S, Uehara A, Takada H, et al. Mouse and human cell activation by N-dodecanoyl-DL-homoserine lactone, a Chromobacterium violaceum autoinducer. Infection and Immunity. 2006;74:7029–7031. 10.1128/IAI.00038-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kristiansen S, Bjarnsholt T, Adeltoft D, Ifversen P, Givskov M. The Pseudomonas aeruginosa autoinducer dodecanoyl-homoserine lactone inhibits the putrescine synthesis in human cells. APMIS. 2008;116:361–371. 10.1111/j.1600-0463.2008.00966.x [DOI] [PubMed] [Google Scholar]
- 118.John J, Saranathan R, Adigopula LN, Thamodharan V, Singh SP, Lakshmi TP, et al. The quorum sensing molecule N-acyl homoserine lactone produced by Acinetobacter baumannii displays antibacterial and anticancer properties. Biofouling. 2016;32:1029–1047. 10.1080/08927014.2016.1221946 [DOI] [PubMed] [Google Scholar]
- 119.Rau J, Stolz A. Oxygen-insensitive nitroreductases NfsA and NfsB of Escherichia coli function under anaerobic conditions as lawsone-dependent Azo reductases. Appl Environ Microbiol. 2003;69:3448–3455. 10.1128/AEM.69.6.3448-3455.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
- MM = molecular mass;
- pI = isoeletric point;
- TIC = total ion current;
- Mascot = Mascot software (version 2.4.0; Matrix Science, United Kingdom);
- Scaffold = Scaffold software (version 4.7.2; Proteome Software, USA).
(XLSX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.