Abstract
The protozoan Phytomonas serpens (class Kinetoplastea) is an important phytoparasite that has gained medical importance due to its similarities to Trypanosoma cruzi, the etiological agent of Chagas disease. The present work describes the first proteome analysis of P. serpens. The parasite was separated into cytosolic and high density organelle fractions, which, together with total cell extract, were subjected to LC-MS/MS analyses. Protein identification was conducted using a comprehensive database composed of genome sequences of other related kinetoplastids. A total of 1,540 protein groups were identified among the three sample fractions. Sequences from Phytomonas sp. in the database allowed the highest number of identifications, with T. cruzi and T. brucei the human pathogens providing the greatest contribution to the identifications. Based on the proteomics data obtained, we proposed a central metabolic map of P. serpens, which includes all enzymes of the citric acid cycle. Data also revealed a new range of proteins possibly responsible for immunological cross-reactivity between P. serpens and T. cruzi.
Introduction
Phytomonas serpens is a flagellated protozoan (class Kinetoplastea, order Trypanosomatida [1]) of considerable agricultural importance as a phytoparasite of tomato crops. This protozoan, which is transmitted by the vector Phthia picta, retains a promastigote form during its entire life cycle. To date, little is known about P. serpens in terms of its’ biology, life cycle, or how the species has adapted to life inside plants [2–4].
The members of the class Kinetoplastea are peculiar organisms that differ from most other eukaryotes in a number of biological features. Notable differences include the presence of organelles such as glycosomes and the existence of a single mitochondrion bearing a complex array of DNA called kinetoplast [5]. Trypanosomatids also possess peculiar characteristics during cell division, including closed mitosis, an absence of chromosome condensation and replication of DNA at the periphery of the nucleus [6–8]. Furthermore, Phytomonas species lack most of the known hemeproteins and do not require heme groups for the transport of electrons along the respiratory chain and for other important metabolic processes [9].
A number of previous studies have applied proteomic strategies to better understand the peculiar biology of members of the kinetoplastids, with reports for Trypanosoma cruzi, Trypanosoma brucei and Leishmania [10–18]. Currently, liquid chromatography-tandem mass spectrometry (LC-MS/MS) has become the gold standard method to analyze proteins from complex biological samples, since it allows the identification and the quantification of thousands of proteins in a single experiment [19]. Moreover, data from LC-MS/MS experiments can have a positive impact on poorly annotated genomes by contributing to the curation process [20]. Such a strategy has been applied to Leishmania donovani [21].
P. serpens has gained importance in medical research due to its similarities to Trypanosoma cruzi, the etiological agent of Chagas disease. Previous studies have shown that chagasic patients display antibodies which are able to recognize P. serpens antigens [22–24]. Furthermore, intraperitoneal and oral route inoculation of mice with P. serpens was reported to promote some protection against T. cruzi infection. Consequently, authors claimed that oral immunization with P. serpens might constitute an alternative vaccination approach to T. cruzi infection [24].
In the present work, we carried out a proteomic analysis of P. serpens via LC-MS/MS and employed databank sequences from all kinetoplastids available in UNIPROT for protein identification. We were able to identify 4,387 proteins with at least one unique peptide. Moreover, P. serpens proteome characterization has enabled the generation of useful data which may provide support to research on the biology and the mechanisms of pathogenicity in kinetoplastids.
Material and methods
Cell culture and fractionation
P. serpens (strain 9T) was grown in brain heart infusion broth (BHI) medium (Acumedia, Lansing, Michigan) at 27 °C. Parasite cells (5 x 108) collected in logarithmic growth phase were fractionated using a protocol based on a previously described report [25]. Parasites were centrifuged for 10 min at 5,000 g then washed three times in phosphate buffered saline (PBS). The pellet was resuspended in eight volumes of hypotonic buffer TENM2 (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 1 mM MgCl2, 1 mM MnCl2, 5 mM β-mercaptoethanol) and cell turgidity was confirmed by optical microscopy. Nonidet P40 (0.5% (v/v) final concentration) and protease inhibitors (cOmplete Mini, Roche, Meylan, France) were added to the cells, which were then lysed using a Dounce homogenizer. Cell lysis was followed by optical microscopy. Osmolarity of the lysate was reestablished by adding 12.5% (v/v) of 2 M sucrose (0.25 M final sucrose concentration). The sample was then transferred to a conical centrifuge tube containing 5 ml of 0.58 M sucrose in TENM2 and centrifuged at 2,000 g for 10 min. The top layer contained the cytosol (CYT), with the pellet containing the high-density organelles (HDO).
Protein digestion
CYT and HDO fractions, as well as total P. serpens cells (TOTAL), were trypsin digested using the filter-aided sample preparation (FASP) protocol adapted from [26]. Briefly, each sample was solubilized in LB1 lysis buffer (4% (w/v) SDS, 0.02 M TEAB, 0.1 M DTT, pH 7.9) and heated at 90 °C for 10 min. Samples were then submitted to sonication using a GE50T ultrasonic processor (Cole–Parmer, Chicago, USA). For that, 3 x 10 s cycles at 40% maximal power were employed. Samples were then centrifuged at 16,000 g for 15 min and proteins present in the supernatant were quantified using Qubit Protein Assay (Thermo Fisher Scientific, Waltham, USA).
Aliquots from each sample containing 30 μg of protein were diluted in 200 μl of UA (8 M Urea, 0.02 M TEAB, pH 8.5) in a 30 kDa filter unit (Sartorius, Goettingen, Germany) and centrifuged at 14,000 g for 15 min. The filter unit was then washed with 200 μl of UA, centrifuged at 14,000 g for 15 min and the flow-through discarded. Subsequently, 100 μl of IAA (0.05 M iodoacetamide in UA) solution was added and the system was incubated in a Thermomixer mod. 22331 (Ependorff, Hamburg, Germany) at 600 rpm for 1 min followed by 20 min at 21°C without mixing. Filter units were then centrifuged at 14,000 g for 10 min. IAA excess was removed with 100 μl of UA and 14,000 g centrifugation for 15 min. A volume of 100 μl of 0.02 M TEAB pH 7.9 was added to the filter unit and centrifuged at 14,000 g for 10 min. This step was repeated one more time, followed by the addition of 90 μl of 0.02 M TEAB, pH 7.9, containing trypsin (1:100 enzyme: protein ratio). The filter units were mixed at 600 rpm in the Thermomixer for 1 min and incubated in a wet chamber at 37°C for 18 h. The resulting tryptic peptides were collected by addition of 210 μl of water, followed by centrifugation as described above. The sample was acidified with 7.5 μl of 20% TFA and desalted using C18 Ultra-Micro Spin columns (Harvard Apparatus, Holliston, MA, USA). The resulting samples were dried in a vacuum centrifuge.
1.1 LC/MS-MS analyses
Protein digests of each sample were loaded (3 μg of total peptide) onto a Reprosil-Pur 120 C18-AQ in-house packed trap column (5 μm particle size, 5.0 cm length, 100 μm inner diameter, 360 μm outer diameter) using an UltiMate 3000 Nano LC (Dionex, Amsterdam, The Netherlands). The trap column was washed for 5 min with solvent A (0.1% (v/v) formic acid, 2% (v/v) acetonitrile). Peptides were eluted onto a C18 Reprosil-Pur 120 C18-AQ (3 μm particle size, 23 cm length, 75 μm inner diameter, 360 μm outer diameter) in-house packed analytical column at a flow rate of 230 nL.min−1. The gradient comprised 10–35% of solvent B (0.1% (v/v) formic acid, 95% (v/v) acetonitrile) for 155 min. Peptides were electrosprayed into an LTQ-Orbitrap Elite mass spectrometer (Thermo Fischer Scientific, Bremen, Germany) via a nanospray probe (Thermo Scientific, Germany) with a spray voltage of 3.02 kV and transfer capillary temperature set to 275 °C. The mass spectrometer was operated in Data Dependent Acquisition (DDA) mode using Xcalibur 2.2 software (Thermo Scientific). The acquisition cycle consisted of a survey scan from 300–1650 m/z at 120,000 resolution (full width at half maximum) at m/z 400 using one microscan in the Orbitrap, followed by fragmentation of the 15 most intense multiply charged precursors using higher energy collision induced dissociation (HCD) under normalized collision energy of 35. Fragmentation was also performed by collision induced dissociation (CID) fragmentation of the 20 most intense multiply charged precursors in new LC-MS/MS runs. The ion selection threshold for MS/MS was set to 3,000 counts using a precursor isolation window of 2 amu, while dynamic exclusion was set at 30 s.
Bioinformatics
Data files were analyzed using the software PEAKS (Version 7; Bioinformatics Solutions Inc., Waterloo, Ontario, Canada). A local database was employed comprising available Kinetoplastea parasite protein sequences for more than 200 different species (http://www.uniprot.org; release oct_2015) including two Phytomonas spp. (HART1 from group H and EM1 from group D), Trypanosoma cruzi (CL Brener), Trypanosoma brucei and Leishmania infantum (S1 Database). Criteria for protein identification included the detection of at least one unique tryptic peptide and false-positive discovery rate (FDR) of less than 1%. Tolerance filters of 0.5 Da for precursor ions and 10 ppm for fragment ions were employed. Carbamidomethylation of cysteine was considered a fixed modification, while acetylation of the N-terminal and methionine oxidation were set as variable modifications.
Gene ontology analyses were performed in Blast2GO (version 4.1.9) and DAVID bioinformatics resources 6.7 (http://david.abcc.ncifcrf.gov/) using the software default parameters.
Results and discussion
Most proteomic studies concerning kinetoplastids have been supported by the availability of complete and annotated genome sequences of human pathogens such as T. cruzi, T. brucei, Leishmania and several other organisms [13,27–31]. In contrast, the present study was performed to analyse the proteome of P. serpens, a phytopathogenic kinetoplastid. Although the P. serpens genome was previously sequenced in order to enable identification of heme-containing proteins in this parasite [9], the genome sequence unfortunately did not provide annotation of protein coding genes, which is crucial for proteomic analysis. Given this, we compiled a comprehensive reference database composed of genome sequences for closely related species, from T. cruzi, T. brucei, L. infantum and a compilation of other species belonging to the Kinetoplastea class (S1 Database).
P. serpens cells were firstly fractionated into a cytosolic (CYT) and a high-density organelle fraction (HDO). This fractionation into two different subproteomes was conducted as a strategy to decrease sample complexity for LC-MS/MS analysis, a step usually performed to increase the likelihood of identifying proteins sometimes underrepresented in global proteomic assays, as previously demonstrated [32]. In addition, total cell extracts (TOTAL) were also subjected to proteomic analyses. The overall strategy outlined in Fig 1 resulted in the identification of 2,949 proteins in CYT; 2,976 in HDO; and 3,807 in TOTAL (S1 Table, MS/MS raw data files are available at PeptideAtlas, dataset identifier PASS01214). These proteins were distributed in 1,540 different protein groups. Detailed information about the peptides identified in each experimental condition is available in S2 Table.
Fig 1. Cell fractionation and proteomic strategy analyses.
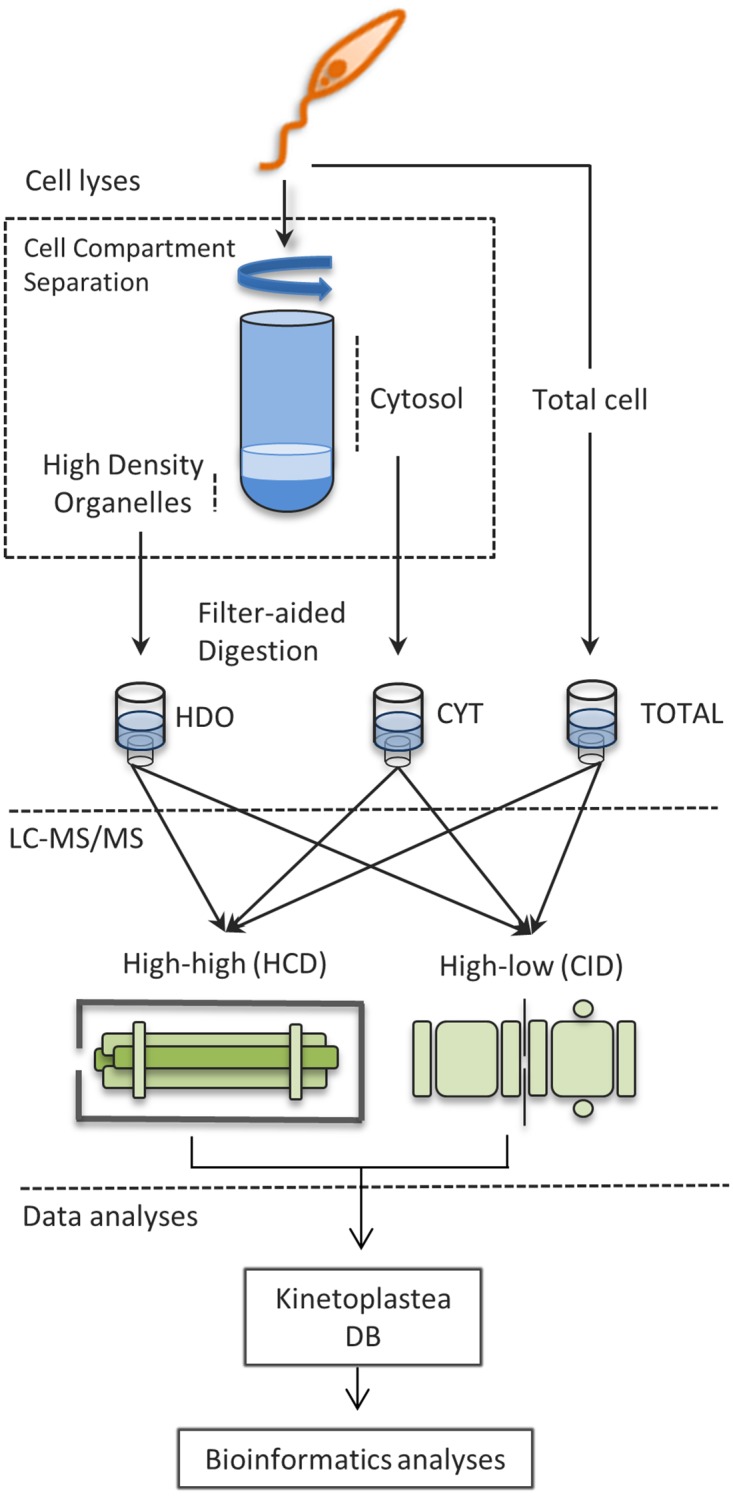
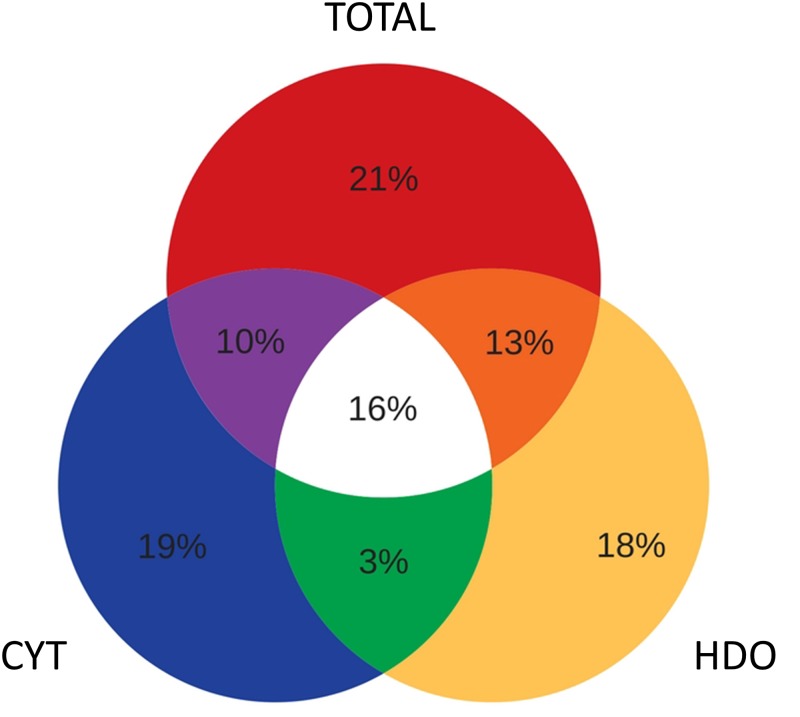
Fig 2 illustrates the distribution of the identified proteins among CYT, HDO and TOTAL. The Venn diagram shows that around 18% of the proteins were exclusively identified within the CYT fraction, 18% in HDO and 20% in TOTAL. Interestingly, 58% of the proteins were identified in just one of the fractions (CYT, HDO or TOTAL), showing that the approach used here succeeded in providing a larger proteome data set. In addition, we observed that the average sequence coverage of proteins identified in CYT and HDO (19.2% and 18.5% respectively) was higher than in the TOTAL fraction (16.8%). This highlights that the fractionation improved the coverage of the identifications.
Fig 2. Venn diagram showing the distribution of identified proteins in TOTAL (total cell proteome), CYT (cytosol) and HDO (high density organelles) fractions.
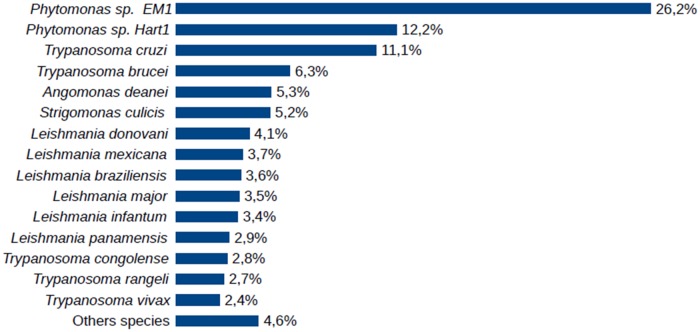
Among the organisms used in construction of the local database, Phytomonas sp EM1 and HART1 provided more protein hits in the identification process than the other species (Fig 3). This result was expected since they agree with previous phylogenetic studies based on HSP90 sequence homology [30]. However, although this phylogenetic study showed Phytomonas serpens closer to Leishmanias [30], T. cruzi and T. brucei were the human pathogens that provided more protein matches.
Fig 3. Distribution of proteins identified in the P. serpens proteome in terms of the sequence databank species.
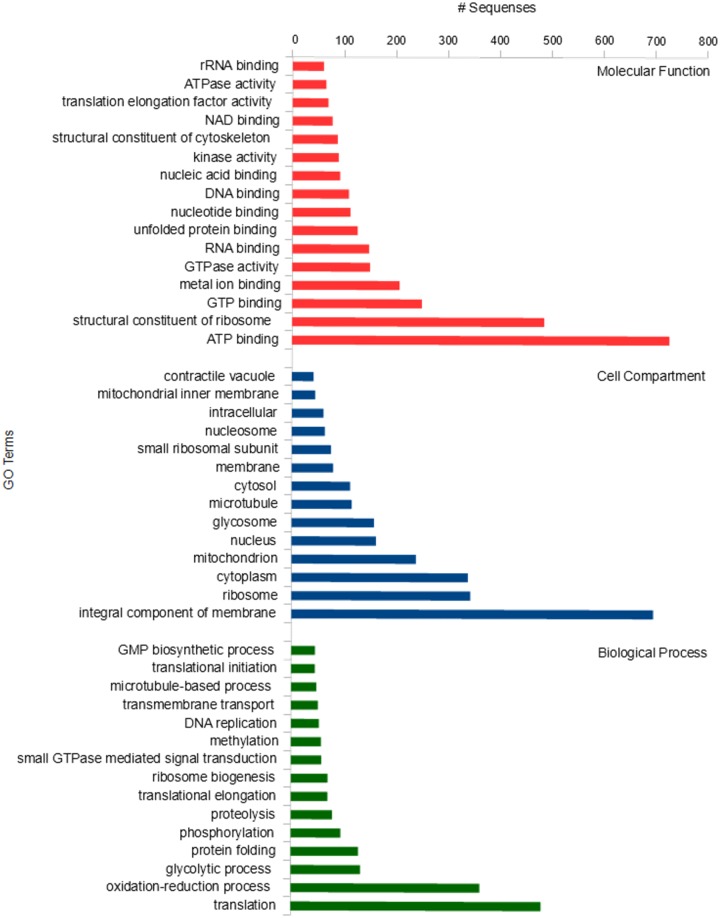
We performed functional annotation of all the identified proteins using Blast2GO, with 73% of proteins grouped in GO terms related to molecular function, biological process or cellular component (Fig 4). The most abundant molecular function terms were those related to binding activities, including ATP, GTP, metal ion, RNA, unfolded protein, nucleotide, DNA, nuclei acid and NAD binding proteins. Overall, Blast2GO classified 148 proteins under RNA binding, which can be considered very relevant for kinetoplastids [33], as gene expression is post-transcriptionally regulated in these organisms [34].
Fig 4. GO terms of proteins found in the P. serpens proteome relateing to molecular function, cellular component and biological activity.
Gene ontology analysis classified 697 proteins as integral components of the membrane, this being the cell compartment with highest number of hits. In part, the identification of a large number of those proteins might be explained by the method of choice used for protein digestion. Previous studies have shown that filter-aided sample preparation (FASP) enables an increase in the identification of membrane proteins when compared with other digestion protocols [26] Moreover, gene ontology analysis also revealed proteins associated with other cellular structures such as ribosome, mitochondrion, nucleus and glycosome. For instance, 159 proteins were assigned to glycosome–a spherical cellular structure found in trypanosomatids–, which is a special type of peroxisome and contains the main enzymes of the glycolytic pathway [35].
The investigation of P. serpens metabolism is vital to understand its pathogenicity to plants and for comparative analysis to other trypanosomatid flagellates that share evolutionary relationships with this phytopathogen. In general, trypanosomatids produce and secrete still-reduced carbon compounds from glucose catabolism (e.g. pyruvate, ethanol, acetate, alanine) even under aerobic conditions, instead of oxidizing glucose completely to CO2 and water [36].
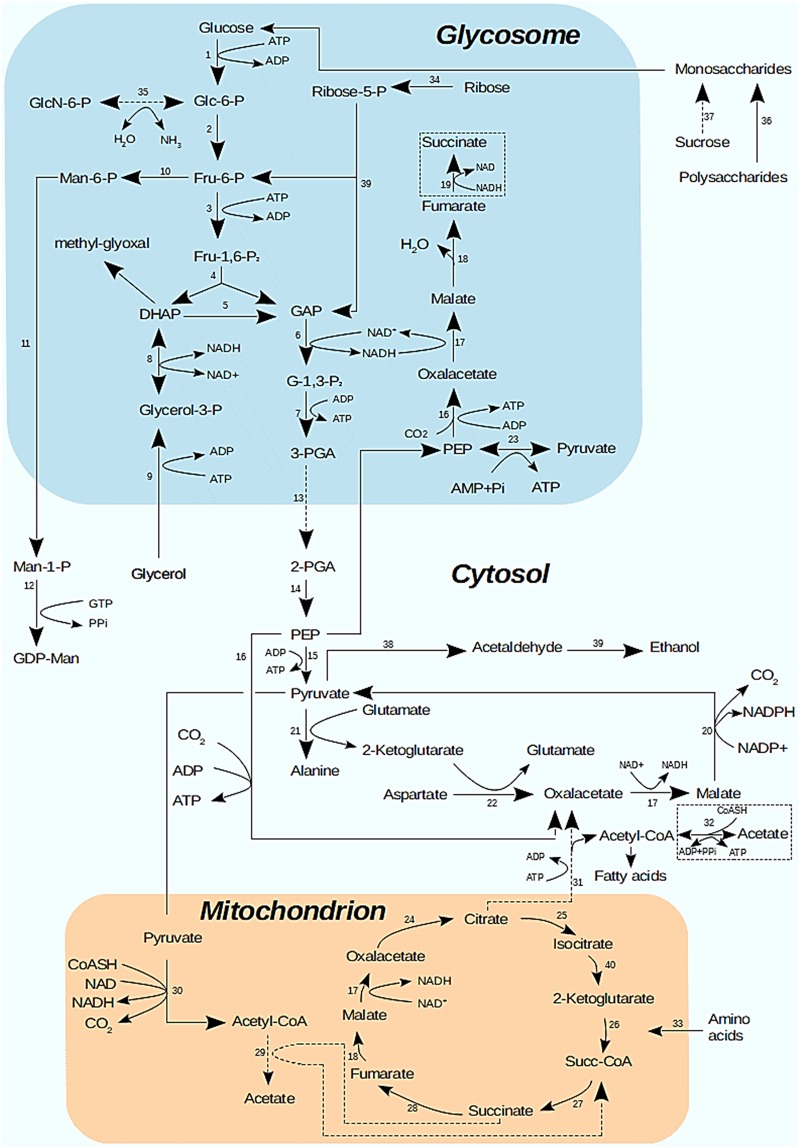
Many enzymes of metabolic pathways located in the glycosome, in the mitochondrion and in the cytosol, including glycolysis and citric acid cycle, were observed in the P. serpens proteome. These proteins allowed us to propose a central metabolic map (Fig 5) for P. serpens based on the core metabolism pathways previously published for two Phytomonas sp. (EM1 and HART1) and Leishmania major [30].
Fig 5. Proposed P. serpens metabolic map based on the present proteomic data as compared to those of Phytomonas sp. EM1 and HART1 [30].
Dashed boxes show enzymes found in the present P. serpens proteome analysis but not in Phytomonas sp EM1 and HART1 genome [30]. Dashed arrows correspond to enzymes found in Phytomonas sp EM1 and HART1 genome but not in the present work. Solid arrows represent enzymes found in P. serpens proteome and in Phytomonas sp EM1 and HART1 genome. Enzymes: 1, hexokinase; 2, glucose-6-phosphate isomerase; 3, 6-phosphofructokinase; 4, fructose-bisphosphate aldolase; 5, triosephosphate isomerase; 6, glyceraldehyde-3-phosphate dehydrogenase; 7, phosphoglycerate kinase; 8, glycerol-3-phosphate dehydrogenase; 9, glycerol kinase; 10, mannose-6-phosphate isomerase; 11, phosphomannomutase; 12, mannose-1-phosphate guanyltransferase, 13, phosphoglycerate mutase; 14, enolase; 15, pyruvate kinase; 16, phosphoenolpyruvate carboxykinase; 17, malate dehydrogenase; 18, fumarate hydratase; 19, NADH-dependent fumarate reductase; 20, malic enzyme; 21, alanine aminotransferase; 22, aspartate aminotransferase; 23, pyruvate phosphate dikinase; 24, citrate synthase; 25, isocitrate dehydrogenase; 26, 2-oxoglutarate dehydrogenase; 27, succinate-CoA ligase; 28, succinate dehydrogenase; 29, acetate: succinate CoA transferase, 30, pyruvate dehydrogenase; 31, citrate lyase; 32, acetyl-CoA synthetase; 33, amino acid oxidation pathway; 34, ribokinase; 35, glucosamine-6-phosphate deaminase; 36, glucoamylase; 37, invertase; 38, pyruvate descarboxylase, 39, alcohol dehydrogenase, 40. isocitrate dehydrogenase.
Whilst most enzymes described in the core metabolic pathway of Phytomonas sp. were also found in the P. serpens proteome, we also identified a NADH-dependent fumarate reductase and an acetyl-CoA syntetase, which have not been observed in the Phytomonas sp. (EM1 and HART1) genomes. The enzymes phosphoglycerate mutase, citrate lyase, glucosamine-6-phosphate deaminase and acetate: succinate CoA transferase deaminase could not be identified in the present work. However, we managed to find sequences which probably code for homologs of phosphoglycerate mutase (85% identity), citrate lyase (56% identity), glucosamine-6-phosphate deaminase (73% identity) but not for acetate: succinate CoA transferase in the non-annotated P. serpens genome sequence [9] (S1 Fig). Interestingly, enzymes found in the EM1 genome but absent from HART1 (i.e. malic enzyme and phosphomannomutase) were identified in our study. This is in agreement with positioning in a phylogenetic tree of trypanosomatids based on HSP90 sequence homology that places P. serpens closer to EM1 than to HART1 [30].
Chaumont and collaborators [37] demonstrated, using enzymatic and NMR measurements, that the major end-products of glucose catabolism of Phytomonas sp. isolated from Euphorbia characias under aerobic conditions were acetate, ethanol and carbon dioxide. The pathways related to the production of ethanol and carbon dioxide were identified in this work. Although the enzyme acetate: succinate CoA transferase, that converts acetyl-CoA to acetate, was not found here, acetate could be produced from acetyl CoA by acetyl-CoA synthetase. In this sense, all enzymes of glycolysis, apart from phosphoglycerate mutase, were identified (Fig 5). Also, our data revealed pyruvate/indolepyruvate decarboxylase, a key enzyme in alcoholic fermentation previously characterized in P. serpens [38], and alcohol dehydrogenase. These results ratify that ethanol is produced in aerobic conditions, and this metabolic route is an alternative and necessary route to reoxidize part of the NADH produced in the highly demanding glycolytic pathway [38]. The absence of invertases in the protein lists suggests that P. serpens, as with Phytomonas sp. EM1 and HART1 [30], does not have the capacity to convert sucrose to glucose and fructose.
Phytomonas isolated from Euphorbia characias were shown to contain high activities of enzymes involved in the hydrolysis of polysaccharides into monosaccharides [39]. However, no amylases, amylomaltases, invertases or carboxymethyl cellulases were identified in the present work. Porcel and collaborators [30] investigated Phytomomas sp. putative secreted proteins (containing a secretion signal peptide, no transmembrane domains and no GPI anchors) involved in carbohydrate degradation. They found a sequence containing a glycoside hydrolase family 31 domain in both isolates (HART1 and EM1) and a secreted beta-fructofuranosidase in HART1 only. However, expression data did not show translation of any proteins likely to be involved in plant cell degradation. The authors pointed out that Phytomonas does not need to degrade cell walls to penetrate the host, since the parasite is directly injected in the plant phloem by the insect vector.
The citric acid cycle in Phytomonas has been described as nonfunctional, as the mitochondria are not capable of oxidizing 2-ketoglutarate, succinate and proline [39]. However, a recent study showed that 2-ketoglutarate dehydrogenase, succinate dehydrogenase and proline oxidation pathway enzymes are present in the Phytomonas sp (EM1 and HART1 isolates) genome [30]. Surprisingly, all the enzymes belonging to the citric acid cycle have been found in the P. serpens proteome, raising once again the question as to whether the mitochondrion is metabolically inactive, as it was once proposed for Phytomonas sp. [37], or if it is active, but only under certain conditions.
Phytomonas serpens is an eukaryote able to survive in the absence of heme-proteins, as it does not require heme for electron transport in the respiratory chain, protection against oxidative stress or desaturation of fatty acids [9]. As expected, cytochromes of the electron transport chain were not found in this proteome data set. However, the lanosterol 14-alpha-demethylase (ortholog of T. brucei), a heme-protein involved in the synthesis of sterols, such as cholesterol and ergosterol [40] is available in the proteome. The presence of lanosterol 14-alpha-demethylase in P. serpens had been previously described [9].
The absence of cytochrome-mediated respiration in P. serpens results in a limited mitochondrial role in energy metabolism of this phytoparasite [41]. Still, P. serpens has a short mitochondrial electron transport chain based on an alternative oxidase (salicylhydroxamic acid sensitive), which transfers reducing equivalents from glycolytic NADH to oxygen via glycerol-3-phosphate dehydrogenase and the ubiquinone pool [39,42–44]. P. serpens NADH-ubiquinone oxidoreductases were previously characterized [45], and also identified in this work (S1 Table).
As previously mentioned, T. cruzi is the human pathogen with the largest number of orthologous proteins with P serpens. Given this, P. serpens–a phytoparasite harmless to humans–has been attracting medical interest due its immunological cross-reactivity with T. cruzi, the etiological agent of Chagas disease [24,46,47]. The immunological cross-reactivity between P. serpens and T. cruzi has been well described, as the two species share common antigens. BALB/c mice immunization by intraperitoneal or oral route with P. serpens induces protective immunity against T. cruzi infection [24]. Moreover, the protective immunity provided by oral immunization is associated with enhanced NO production during the acute phase of T. cruzi infection [23] Also, immunization is able to attenuate thrombocytopenia and leukopenia during acute infection in mice [47]. In contrast, antibodies present in the sera of humans affected by Chagas disease react with 22 different P. serpens antigens [22], supporting the immunological cross reacting between both species.
The present P. serpens proteome analysis identified a new range of proteins, which may contribute to the process of immunological cross-reactivity. Among these proteins, some have already been described as candidates for potential vaccines against infections caused by kinetoplastids, such as the KMP-11 (kinetoplastid membrane protein-11) protein family [48,49].
The development of an efficient human vaccine against T. cruzi infection has been prevented by difficulties such as controversy about its genetic complexity and a limited set of engineering techniques for genome manipulation. Therefore, the development of a prophylactic vaccine able to reduce the parasite burden in humans and its reservoirs has become a challenge [50]. As mentioned before, P. serpens has potential for the development of vaccines against T. cruzi infection [22–24,46]. As an intracellular parasite, T. cruzi promotes the presentation of MHC class I epitopes by mammalian infected cells. [51]. A proteomic and immunoinformatics analysis using trypomastigote forms of T. cruzi predicted a total of 296 proteins as being able to produce major histocompatibility complex (MHC) class I epitopes [52]. Here, we showed that fourteen of those predicted proteins are found in the P. serpens proteome and, consequently, could perhaps originate the same MHC I epitopes (Table 1). Therefore, our results revealed proteins that could support the immunological cross-reactivity between P. serpens and T. cruzi. The possible use of these proteins in the development of a P. serpens-based safe vaccine should result in considerable advancement in the treatment of Chagas disease.
Table 1. P. serpens proteins that could potentially generate MHC I epitopes against T. cruzi trypomastigotes.
| Cell fraction | Accession number | -10logP | Coverage (%) | #Peptides | #Unique | Avg. Mass | Description |
|---|---|---|---|---|---|---|---|
| TOTAL | Q8STF3|Q8STF3_TRYCR | 306.15 | 69 | 51 | 1 | 49700 | Beta tubulin 1.9 |
| TOTAL | Q01530|Q01530_TRYCR | 213.92 | 19 | 16 | 1 | 69548 | Major paraflagellar rod protein |
| CYT | Q4DZ98|Q4DZ98_TRYCC | 219.27 | 19 | 15 | 2 | 46444 | Enolase putative |
| CYT | Q4CLA1|Q4CLA1_TRYCC | 334.24 | 64 | 49 | 1 | 49799 | Alpha tubulin putative |
| TOTAL | Q4CLA1|Q4CLA1_TRYCC | 330.48 | 66 | 47 | 2 | 49799 | Alpha tubulin putative |
| HDO | Q4DZ41|RS3A2_TRYCC | 76.03 | 13 | 2 | 1 | 29855 | 40S ribosomal protein S3a-2 |
| CYT | Q4DYK2|Q4DYK2_TRYCC | 167.07 | 13 | 7 | 2 | 34955 | ADP ATP carrier protein 1 mitochondrial putative |
| TOTAL | Q4DYK2|Q4DYK2_TRYCC | 151.00 | 16 | 6 | 1 | 34955 | ADP ATP carrier protein 1 mitochondrial putative |
| HDO | Q4D3P5|Q4D3P5_TRYCC | 71.81 | 7 | 4 | 1 | 52144 | Hexokinase |
| CYT | I6LE92|I6LE92_TRYCR | 83.31 | 8 | 2 | 1 | 41969 | Actin |
| TOTAL | I6LE92|I6LE92_TRYCR | 122.33 | 18 | 4 | 1 | 41969 | Actin |
| TOTAL | Q4CVR9|Q4CVR9_TRYCC | 206.32 | 21 | 13 | 2 | 70990 | Heat shock 70 kDa protein mitochondrial putative |
| HDO | Q4D7Y8|Q4D7Y8_TRYCC | 86.01 | 14 | 2 | 2 | 20677 | ADP-ribosylation factor 1 putative |
| HDO | Q4D5K2|Q4D5K2_TRYCC | 190.46 | 40 | 10 | 3 | 28375 | 60S ribosomal protein L2 putative |
| CYT | Q4D1S0|Q4D1S0_TRYCC | 146.21 | 15 | 4 | 1 | 55554 | Vacuolar ATP synthase subunit B putative |
| HDO | Q4CUL0|Q4CUL0_TRYCC | 68.55 | 6 | 1 | 1 | 24144 | 40S ribosomal protein S3 putative |
| TOTAL | Q4CUL0|Q4CUL0_TRYCC | 128.75 | 21 | 5 | 1 | 24144 | 40S ribosomal protein S3 putative |
| CYT | Q4DWG6|Q4DWG6_TRYCC | 79.75 | 4 | 2 | 1 | 59186 | Chaperonin containing T-complex protein putative |
Furthermore, the present homology-based P. serpens proteome analyses generated information relating to biological features, including its’ metabolism, and should contribute to the annotation and assembly of the genome [53]. In addition, this work may lead to a better understanding of the biology, biochemistry and evolutionary history of P. serpens.
Supporting information
(XLSX)
(XLSX)
Phytomonas sp HART1 with the non-annotated P. serpens genome sequence [9] using tblastn NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&PROG_DEF=blastn&BLAST_SPEC=Assembly&ASSEMBLY_NAME=GCA_000331125.1.).
(TIF)
Local database comprising available Kinetoplastea parasite protein sequences for more than 200 different species (http://www.uniprot.org; release oct_2015) including two Phytomonas spp. (HART1 from group H and EM1 from group D), Trypanosoma cruzi (CL Brener), Trypanosoma brucei and Leishmania infantum.
(RAR)
Acknowledgments
The authors acknowledge the receipt of financial support from the Brazilian National Council for Scientific and Technological Development (CNPq -grant number 423006/2016-9), Coordination for the Improvement of Higher Education Personnel (CAPES), Financing Agency for Studies and Projects (FINEP). The authors also would like to thank Prof. Robert Miller (University of Brasilia) and Prof. Consuelo M. R. Lima (University of Brasilia) for diligent proofreading of this manuscript.
Data Availability
All relevant data are within the paper, Supporting Information files or PeptideAtlas (dataset identifier PASS01214).
Funding Statement
The work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grants 423006/2016-9 and 407855/2013), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo a Pesquisa do Estado de Goiás (FAPEG) and Financiadora de Estudos e Projetos (FINEP, grants 0439/11 and 0694/13 to M.V.S.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cavalier-Smith T. Higher classification and phylogeny of Euglenozoa. Eur J Protistol. Elsevier GmbH; 2016;56: 250–276. 10.1016/j.ejop.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 2.Hoare C a., Wallace FG. Developmental Stages of Trypanosomatid Flagellates: a New Terminology. Nature. 1966;212: 1385–1386. 10.1038/2121385a0 [DOI] [Google Scholar]
- 3.JANKEVICIUS JV, JANKEVfCIUS SI, CAMPANER M, CONCHON I, MAEDA LA, TEIXEIRA MMG, et al. Life Cycle and Culturing of Phytomonas serpens (Gibbs), a Trypanosomatid Parasite of Tomatoes. J Protozool. 1989;36: 265–271. 10.1111/j.1550-7408.1989.tb05361.x [DOI] [Google Scholar]
- 4.Jaskowska E, Butler C, Preston G, Kelly S. Phytomonas: trypanosomatids adapted to plant environments. PLoS Pathog. 2015;11: e1004484 10.1371/journal.ppat.1004484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Souza W. The basic biology T cruzi. Curr Pharm Des. 2002;8: 1–17.11812246 [Google Scholar]
- 6.Solari AJ. Mitosis and genome partition in trypanosomes. Biocell. 1995. pp. 65–84. [PubMed] [Google Scholar]
- 7.Solari AJ. The 3-dimensional fine structure of the mitotic spindle in Trypanosoma cruzi. Chromosoma. 1980;78: 239–255. 10.1007/BF00328395 [DOI] [PubMed] [Google Scholar]
- 8.Elias MCQB, Faria M, Mortara RA, Motta MCM, De Souza W, Thiry M, et al. Chromosome localization changes in the Trypanosoma cruzi nucleus. Eukaryot Cell. 2002;1: 944–953. 10.1128/EC.1.6.944-953.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koreny L, Sobotka R, Kovarova J, Gnipova A, Flegontov P, Horvath A, et al. Aerobic kinetoplastid flagellate Phytomonas does not require heme for viability. Proc Natl Acad Sci U S A. 2012;109: 3808–3813. www.pnas.org/cgi/doi/10.1073/pnas.1201089109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paba J, Santana J, Teixeira A, Fontes W, Sousa M, Ricart C. Proteomic analysis of the human pathogen Trypanosoma cruzi. Proteomics. 2004;4: 1052–1059. 10.1002/pmic.200300637 [DOI] [PubMed] [Google Scholar]
- 11.Paba J, Ricart CAO, Fontes W, Santana JM, Teixeira ARL, Marchese J, et al. Proteomic analysis of Trypanosoma cruzi developmental stages using isotope-coded affinity tag reagents. J Proteome Res. 2004;3: 517–524. 10.1021/pr034075o [DOI] [PubMed] [Google Scholar]
- 12.Parodi-Talice A, Durán R, Arrambide N, Prieto V, Piñeyro MD, Pritsch O, et al. Proteome analysis of the causative agent of Chagas disease: Trypanosoma cruzi. Int J Parasitol. 2004;34: 881–886. 10.1016/j.ijpara.2004.05.002 [DOI] [PubMed] [Google Scholar]
- 13.Atwood JA, Weatherly DB, Minning TA, Bundy B. The Trypanosoma cruzi Proteome. Science (80-). 2005;309: 473–476. 10.1126/science.1110289 [DOI] [PubMed] [Google Scholar]
- 14.Van Deursen FJ, Thornton DJ, Matthews KR. A reproducible protocol for analysis of the proteome of Trypanosoma brucei by 2-dimensional gel electrophoresis. Mol Biochem Parasitol. 2003;128: 107–110. 10.1016/S0166-6851(03)00042-2 [DOI] [PubMed] [Google Scholar]
- 15.Cirovic O, Ochsenreiter T. Whole proteome analysis of the protozoan parasite Trypanosoma brucei using stable isotope labeling by amino acids in cell culture and mass spectrometry. Methods Mol Biol. 2014;1188: 47–55. 10.1007/978-1-4939-1142-4_4 [DOI] [PubMed] [Google Scholar]
- 16.Nugent PG, Karsani SA, Wait R, Tempero J, Smith DF. Proteomic analysis of Leishmania mexicana differentiation. Mol Biochem Parasitol. 2004;136: 51–62. 10.1016/j.molbiopara.2004.02.009 [DOI] [PubMed] [Google Scholar]
- 17.Oliveira DM, Gouveia JJS, Diniz NB, Pacheco ACL, Vasconcelos EJR, Diniz MC, et al. Pathogenomics analysis of Leishmania spp.: flagellar gene families of putative virulence factors. OMICS. 2005;9: 173–193. 10.1089/omi.2005.9.173 [DOI] [PubMed] [Google Scholar]
- 18.Cuervo P, de Jesus JB, Junqueira M, Mendonça-Lima L, González LJ, Betancourt L, et al. Proteome analysis of Leishmania (Viannia) braziliensis by two-dimensional gel electrophoresis and mass spectrometry. Mol Biochem Parasitol. 2007;154: 6–21. 10.1016/j.molbiopara.2007.03.013 [DOI] [PubMed] [Google Scholar]
- 19.Pontes AH, de Sousa M V. Mass Spectrometry-Based Approaches to Understand the Molecular Basis of Memory. Front Chem. 2016;4 10.3389/fchem.2016.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nesvizhskii AI. Proteogenomics: concepts, applications and computational strategies. Nat Methods. Nature Publishing Group; 2014;11: 1114–1125. 10.1038/nmeth.3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pawar H, Sahasrabuddhe NA, Renuse S, Keerthikumar S, Sharma J, Kumar GSS, et al. A proteogenomic approach to map the proteome of an unsequenced pathogen—Leishmania donovani. Proteomics. 2012;12: 832–844. 10.1002/pmic.201100505 [DOI] [PubMed] [Google Scholar]
- 22.Graça-de Souza VK, Monteiro-Góes V, Manque P, Souza TACB, Corrêa PRC, Buck GA, et al. Sera of chagasic patients react with antigens from the tomato parasite Phytomonas serpens. Biol Res. 2010;43: 233–241. doi: /S0716-97602010000200011 [PubMed] [Google Scholar]
- 23.Pinge-Filho P, Peron JPS, De Moura TR, Menolli RA, Graça VK, Estevão D, et al. Protective immunity against Trypanosoma cruzi provided by oral immunization with Phytomonas serpens: Role of nitric oxide. Immunol Lett. 2005;96: 283–290. 10.1016/j.imlet.2004.09.010 [DOI] [PubMed] [Google Scholar]
- 24.Breganó JW, Picão RC, Graça VK, Menolli RA, Jankevicius SI, Filho PP, et al. Phytomonas serpens, a tomato parasite, shares antigens with Trypanosoma cruzi that are recognized by human sera and induce protective immunity in mice. FEMS Immunol Med Microbiol. 2003;39: 257–264. 10.1016/S0928-8244(03)00256-6 [DOI] [PubMed] [Google Scholar]
- 25.Dos Santos Junior A de CM, Kalume DE, Camargo R, Gómez-Mendoza DP, Correa JR, Charneau S, et al. Unveiling the trypanosoma Cruzi nuclear proteome. PLoS One. 2015;10 10.1371/journal.pone.0138667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wisniewski JR, Zougman A, Nagaraj N, Mann M, Wi JR. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6: 377–362. [DOI] [PubMed] [Google Scholar]
- 27.Berriman M. The Genome of the African Trypanosome Trypanosoma brucei. Science (80-). 2005;309: 416–422. 10.1126/science.1112642 [DOI] [PubMed] [Google Scholar]
- 28.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, et al. The genome of the kinetoplastid parasite, Leishmania major. Science (80-). 2005;309: 436–442. 10.1126/science.1112680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran A-N, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science (80-). 2005;309: 409–415. 10.1126/science.1112631 [DOI] [PubMed] [Google Scholar]
- 30.Porcel BM, Denoeud F, Opperdoes F, Noel B, Madoui MA, Hammarton TC, et al. The Streamlined Genome of Phytomonas spp. Relative to Human Pathogenic Kinetoplastids Reveals a Parasite Tailored for Plants. PLoS Genet. 2014;10 10.1371/journal.pgen.1004007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alcolea PJ, Alonso A, Larraga V. Proteome profiling of leishmania infantum promastigotes. J Eukaryot Microbiol. 2011;58: 352–358. 10.1111/j.1550-7408.2011.00549.x [DOI] [PubMed] [Google Scholar]
- 32.W S, Fahy E, Ghosh S. Global organellar proteomics. TRENDS Biotechnol. 2003;21: 1–7. [DOI] [PubMed] [Google Scholar]
- 33.D’Orso I, De Gaudenzi JG, Frasch ACC. RNA-binding proteins and mRNA turnover in trypanosomes. Trends Parasitol. 2003;19: 151–155. 10.1016/S1471-4922(03)00035-7 [DOI] [PubMed] [Google Scholar]
- 34.Araújo P, Teixeira S. Regulatory elements involved in the post-transcriptional control of stage-specific gene expression in Trypanosoma cruzi: a review. Mem Inst Oswaldo Cruz. 2011;106: 257–266. [DOI] [PubMed] [Google Scholar]
- 35.Opperdoes FR. The glycosome of trypanosomes and Leishmania. Biochem Soc Trans. 1990;18: 729–731. Available: http://www.ncbi.nlm.nih.gov/pubmed/2083659 [DOI] [PubMed] [Google Scholar]
- 36.Maugeri DA, Cannata JJB, Cazzulo J. Glucose metabolism in Trypanosoma cruzi. Essays Biochem. 2011;51: 15–30. 10.1042/bse0510015 [DOI] [PubMed] [Google Scholar]
- 37.Chaumont F, Schanck AN, Blum JJ, Opperdoes FR. Aerobic and anaerobic glucose metabolism of Phytomonas sp. isolated from Euphorbia characias. Mol Biochem Parasitol. 1994;67: 321–331. 10.1016/0166-6851(94)00141-3 [DOI] [PubMed] [Google Scholar]
- 38.Ienne S, Pappas G, Benabdellah K, González A, Zingales B. Horizontal gene transfer confers fermentative metabolism in the respiratory-deficient plant trypanosomatid Phytomonas serpens. Infect Genet Evol. Elsevier B.V.; 2012;12: 539–548. 10.1016/j.meegid.2012.01.016 [DOI] [PubMed] [Google Scholar]
- 39.Sanchez-Moreno M, Lasztity D, Coppens I, Opperdoes FR. Characterization of carbohydrate metabolism and demonstration of glycosomes in a Phytomonas sp. isolated from Euphorbia characias. Mol Biochem Parasitol. 1992;54: 185–199. 10.1016/0166-6851(92)90111-V [DOI] [PubMed] [Google Scholar]
- 40.Zhou W, Lepesheva GI, Waterman MR, Nes WD. Mechanistic analysis of a multiple product sterol methyltransferase implicated in ergosterol biosynthesis in Trypanosoma brucei. J Biol Chem. 2006;281: 6290–6296. 10.1074/jbc.M511749200 [DOI] [PubMed] [Google Scholar]
- 41.Maslov DA, Nawathean P, Scheel J. Partial kinetoplast-mitochondrial gene organization and expression in the respiratory deficient plant trypanosomatid Phytomonas serpens. Mol Biochem Parasitol. 1999;99: 207–221. 10.1016/S0166-6851(99)00028-6 [DOI] [PubMed] [Google Scholar]
- 42.Tielens AGM, Van Hellemond JJ. Differences in energy metabolism between Trypanosomatidae. Parasitol Today. 1998;14: 265–271. 10.1016/S0169-4758(98)01263-0 [DOI] [PubMed] [Google Scholar]
- 43.González-Halphen D, Maslov DA. NADH-ubiquinone oxidoreductase activity in the kinetoplasts of the plant trypanosomatid Phytomonas serpens. Parasitol Res. 2004;92: 341–346. 10.1007/s00436-003-1058-4 [DOI] [PubMed] [Google Scholar]
- 44.Nawathean P, Maslov DA. The absence of genes for cytochrome c oxidase and reductase subunits in maxicircle kinetoplast DNA of the respiration-deficient plant trypanosomatid Phytomonas serpens. Curr Genet. 2000;38: 95–103. 10.1007/s002940000135 [DOI] [PubMed] [Google Scholar]
- 45.Čermáková P, Verner Z, Man P, Lukeš J, Horváth A. Characterization of the NADH:ubiquinone oxidoreductase (complex I) in the trypanosomatid Phytomonas serpens (Kinetoplastida). FEBS J. 2007;274: 3150–3158. 10.1111/j.1742-4658.2007.05847.x [DOI] [PubMed] [Google Scholar]
- 46.Santos ALS, d’Avila-Levy CM, Elias CGR, Vermelho AB, Branquinha MH. Phytomonas serpens: immunological similarities with the human trypanosomatid pathogens. Microbes Infect. 2007;9: 915–921. 10.1016/j.micinf.2007.03.018 [DOI] [PubMed] [Google Scholar]
- 47.da Silva R V., Malvezi AD, da Augusto LS, Kian D, Tatakihara VLH, Yamauchi LM, et al. Oral Exposure to Phytomonas serpens Attenuates Thrombocytopenia and Leukopenia during Acute Infection with Trypanosoma cruzi. PLoS One. 2013;8: 1–9. 10.1371/journal.pone.0068299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu R, Bhaumik S, Basu JM, Naskar K, De T, Roy S. Kinetoplastid Membrane Protein-11 DNA Vaccination Induces Complete Protection against Both Pentavalent Antimonial-Sensitive and -Resistant Strains of Leishmania donovani That Correlates with Inducible Nitric Oxide Synthase Activity and IL-4 Generation: Ev. J Immunol. 2005;174: 7160–7171. 10.4049/jimmunol.174.11.7160 [DOI] [PubMed] [Google Scholar]
- 49.de Mendonça SCF, Cysne-Finkelstein L, de S Matos DC. Kinetoplastid membrane protein-11 as a vaccine candidate and a virulence factor in Leishmania. Front Immunol. 2015;6: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodríguez-Morales O, Monteón-Padilla V, Carrillo-Sánchez SC, Rios-Castro M, Martínez-Cruz M, Carabarin-Lima A, et al. Experimental Vaccines against Chagas Disease: A Journey through History. Journal of Immunology Research. 2015. 10.1155/2015/489758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tarleton RL, Grusby MJ, Postan M, Glimcher LH. Trypanosoma cruzi infection in MHC-deficient mice: further evidence for the role of both class I- and class II-restricted T cells in immune resistance and disease. Int Immunol. 1996;8: 13–22. 10.1093/intimm/8.1.13 [DOI] [PubMed] [Google Scholar]
- 52.Nakayasu E, Sobreira T, Torres R, Ganiko L, Oliveira P, Marques A, et al. Improved Proteomic Approach for the Discovery of Potential Vaccine Targets in. J Proteome Res. 2012;11: 237–246. 10.1021/pr200806s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prasad TSK, Mohanty AK, Kumar M, Sreenivasamurthy SK, Dey G, Nirujogi RS, et al. Integrating transcriptomic and proteomic data for accurate assembly and annotation of genomes. Genome Res. 2017;27: 133–144. 10.1101/gr.201368.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Phytomonas sp HART1 with the non-annotated P. serpens genome sequence [9] using tblastn NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&PROG_DEF=blastn&BLAST_SPEC=Assembly&ASSEMBLY_NAME=GCA_000331125.1.).
(TIF)
Local database comprising available Kinetoplastea parasite protein sequences for more than 200 different species (http://www.uniprot.org; release oct_2015) including two Phytomonas spp. (HART1 from group H and EM1 from group D), Trypanosoma cruzi (CL Brener), Trypanosoma brucei and Leishmania infantum.
(RAR)
Data Availability Statement
All relevant data are within the paper, Supporting Information files or PeptideAtlas (dataset identifier PASS01214).