Abstract
Introduction
The therapeutic response to statins has a high interindividual variability with respect to reductions in plasma LDL-cholesterol (c-LDL) and increases in HDL cholesterol (c-HDL). Many studies suggest that there is a relationship between the rs20455 KIF6 gene variant (c.2155T> C, Trp719Arg) and a lower risk of cardiovascular disease in patients being treated with statins.
Aim
The aim of this study was to investigate whether or not the c.2155T> C KIF6 gene variant modulates the hypercholesteremic effects of treatment with simvastatin, atorvastatin, or rosuvastatin.
Materials and methods
This was a prospective, observational and multicenter study. Three hundred and forty-four patients who had not undergone prior lipid-lowering treatment were recruited. Simvastatin, atorvastatin or rosuvastatin were administered. Lipid profiles and multiple clinical and biochemical variables were assessed before and after treatment.
Results
The c.2155T> C variant of the KIF6 gene was shown to influence physiological responses to treatment with simvastatin and atorvastatin. Patients who were homozygous for the c.2155T> C variant (CC genotype, ArgArg) had a 7.0% smaller reduction of LDL cholesterol levels (p = 0.015) in response to hypolipidemic treatment compared to patients with the TT (TrpTrp) or CT (TrpArg) genotype. After pharmacological treatment with rosuvastatin, patients carrying the genetic variant had an increase in c-HDL that was 21.9% lower compared to patients who did not carry the variant (p = 0.008).
Conclusion
Being a carrier of the c.2155T> C variant of the KIF6 gene negatively impacts patient responses to simvastatin, atorvastatin or rosuvastatin in terms of lipid lowering effect. Increasing the intensity of hypolipidemic therapy may be advisable for patients who are positive for the c.2155T> C variant.
Introduction
Statins are drugs that specifically inhibit 3-hydroxy-3-methyl-glutaril-CoA reductase, a rate limiting enzyme of the pathway responsible for the synthesis of endogenous cholesterol. Among the lipid-lowering drugs, statins are the ones most often employed to prevent atherosclerosis [1]. In 2015, simvastatin and atorvastin were the third and fifth most prescribed generic drugs in Spain, respectively [2]. However, the therapeutic response to these drugs has a high interindividual variability: the reduction in plasma LDL-cholesterol (c-LDL) resulting from drug intake ranges from 20% to 60%. A similar variability is observed with respect to increases in plasma HDL cholesterol (c-HDL) [3]. The impact genetic variants may have on the effectiveness of statins is currently being investigated [4, 5, 6], with the aim of developing personalized and cost-effective medications.
Many studies have described a relationship between the rs20455 variant of the KIF6 gene (c.2155T> C) and differences in the response to treatment with statins. The c.2155T> C variant is a missense mutation that leads to substitution of an arginine for a tryptophan at position 719 of the KIF6 protein (NP_659464.3: p. Trp719Arg).
KIF6 is a member of the superfamily of kinesins, which are proteins that mediate the intracellular transport of organelles, complex proteins, and mRNAs. Those who carry variant C are at a higher risk of major cardiovascular events (MACE) if they do not receive hypolipidemic treatment [7]. Paradoxically, this allele is also a predictor of better outcomes with statin-based treatments [8, 9]. Specifically, variant C-carriers have been found to be less likely to suffer MACE when treated with pravastatin [10, 11, 12] or atorvastatin [9, 13] than their counterparts carrying the TT (TrpTrp) genotype. However, some reports also suggest that the association of this variant both with MACE and with the response to statins might not exist [14, 15, 16, 17]. Importantly, this SNP is highly prevalent in Europe: 37% of Europeans are carriers [18].
In light of these facts, it is important to clarify what influence this polymorphism might have on the efficacy of statin therapies most commonly employed in our clinical practice, such as atorvastatin, simvastatin or rosuvastatin. Until now, most published studies have focused only on the association between this variant and a lower risk of cardiovascular episodes. However, the effects of statins should be studied more directly. Changes in patient lipid profiles should be measured after treatment begins. This type of study better reflects the realities of clinical practice, in which medical decisions about changes in statin therapy are based primarily on the results of lipid profiling [19].
Objective
The objective of this study was to examine how the c.2155T> C variant of the KIF6 gene influences the hypolipidemic and hypocholesterolemic responses to simvastatin, atorvastatin, and rosuvastatin. Responses were quantified by measuring differences in the plasma concentration of c-LDL, Non-HDL cholesterol (c-Non-HDL), and c-HDL after treatment.
Materials and methods
Population selection
The c.2155T> C variant of the KIF6 gene was chosen as the object of study as part of an ongoing line of research into the genetic factors that influence patient responses to statins. Of particular interest were genetic variants of genes that play a role in the pharmacodynamics and metabolism of these drugs. This was a prospective, observational, and multicenter study. Non-medicated patients attending several Spanish Primary Health Care Units and Hospital Cardiovascular Risk Units with high-LDL concentration were managed as usual by their physicians. As the design of this study was observational, statin drugs, if necessary, were prescribed in conditions of normal medical practice and according solely to the patients’ physicians criteria. The patients who were excluded from the study were those who: 1) had chronic liver disease; 2) had familial hypercholesterolemia due to mutations in their LDLR, APOB, LDLRAP1 or PSCK9 genes; 3) suffered dysbetalipoproteinemia; 4) were receiving antidepressant, antiepileptic, immunosuppressants, antiretroviral or other lipid-lowering treatments 5) were suffering autoimmune diseases; 6) had discontinued treatment or were suspected of not adhering to prescriptions, which was verified during the follow-up visit through the interview and the medical criteria 7) had statin intolerance, which is defined as the inability to tolerate a dose of statin required to reduce MACE sufficiently from their baseline risk and could result from different statin related side effects including: muscle symptoms, headache, sleep disorders, dyspepsia, nausea, rash, alopecia, erectile dysfunction, gynecomastia, and/or arthritis [20] 8) had hypothyroidism; 9) were consuming 6 or more drugs (polymedication); 10) were already participating in a clinical trial.
Procedures and interventions
Data collection
Patient demographic and clinical information were collected during initial consultations. No treatments were administered during this consultation; however, lipid metabolism was evaluated. This evaluation included measurements of triglycerides, total cholesterol, c-Non-HDL, c-HDL, and c-LDL. The Friedewald formula was used when triglyceride concentrations did not exceed 2.3 mmoL/L. An identical evaluation of lipid metabolism was made during the final consultations, which occurred approximately 3 months after treatment was initiated.
Genetic analysis
DNA extraction from blood samples was performed with the Maxwell 16 Blood DNA Purification Kit (catalog# AS1010) and the Maxwell 16 System Kit (catalog#AS1010) (Promega, Madison, USA). Real-time polymerase chain reaction (RT-PCR) was used to detect the c.2155T> C variant of the KIF6 gene and also the 521T> C variant of the SLCO1B1 gene. The amplification was performed using all-specific Applied Biosystems Taqman probes labelled with fluorochrome in a 7500 Real Time PCR System thermocycler (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
The following independent control variables were analyzed: age, sex, diabetes, hypertension, diastolic blood pressure, systolic blood pressure, exercise, intensity of exercise, history of smoking, current tobacco use, history of alcohol consumption, current alcohol consumption, body mass index, initial concentrations of c-LDL, c-Non-HDL and c-HDL, detection of lipoprotein a, prior ischemia, family history of ischemia, and carrying the SLCO1B1 gene variant rs4149056 [21]. Models were also fitted according to dose. Treatment intensity was evaluated qualitatively (low, moderate, and high) according to the clinical practice guide of the American College of Cardiology [22]. The statistically significant control variables (p<0.05) were selected for each model according to a simple linear regression with the lipid concentration variations. An explanatory quantitative multiple linear regression analysis was carried out in order to discern the relationship between the differences in patient c-LDL, c-Non-HDL and c-HDL measurements that were made during the two consultations and detection of the c.2155T > C variant. Each statin treatment group was analyzed in this way.
The relationship between lipid concentration variations and being a carrier of the c.2155T> C variant was assessed using additive models. When significant differences were observed, these were grouped according to the most appropriate model for each case (recessive or dominant).
Statistical analyses were performed with SPSS v.17 (SPSS Inc., Chicago, IL). The level of significance was set at p<0.05.
Ethical and confidentiality issues
Informed consent was received from study participants during their initial consultation. In compliance with regulation #SAS/3470/2009, this study was classified by the Pharmacoepidemiology Division of the Medicines and Health Products Agency of Spain (ref. PR169/14) and approved by the institutional review boards of each participating center: Comité Ético de Investigación Clínica (CEIC) del Hospital Universitario de Bellvitge, CEIC Hospital universitario Miguel Servet and CEIC Corporació Sanitària Parc Taulí.
Results
Population selection
From June 2014 to February 2017, 344 patients were recruited prospectively. However, 92 of these were later excluded. Of the excluded, 33 had had prior hypolipidemic treatment, 3 had statin intolerance related with muscle complications (myopathy and rhabdomyolysis), 9did not adhere to the treatment regimen, 20 did not participate in follow-ups, 11 had been diagnosed, through genetic testing, with familial hypercholesterolemia, 2 had hypothyroidism, 1 had an autoimmune disease. Another thirteen were excluded for miscellaneous reasons. Consequently, only the data from 252 of total number patients recruited for the study was analyzed.
Descriptive statistics
The clinical and demographic data of the patients is found in Table 1.
Table 1. Patient clinical, demographic, biochemical and treatment characteristics depending on the c.2155T>C (Trp719Arg) KIF6 genotypes.
| Clinical and Demographic Variables | TT (TrpTrp) | TC (TrpArg) | CC (ArgArg) | p value |
| Sex (% male) | 48.1 | 51.9 | 57.9 | 0.553 |
| Age (years) | 53 (50 to 55) | 54 (51 to 56) | 57 (53 to 61) | 0.201 |
| Body Mass Index | 26.7 (25.8 to27.7) | 27.3 (26.5 to 28.0) | 27.5 (26.0 to 29.0) | 0.574 |
| Tobacco Use (% yes) | 37.7 | 21.7 | 19.4 | 0.025* |
| Personal history of tobacco use (% yes) | 71.0 | 58.5 | 55.2 | 0.092 |
| Diabetes mellitus (% yes) | 18.8 | 13.2 | 24.2 | 0.267 |
| Current alcohol consumption (% yes) | 33.3 | 24.3 | 30 | 0.429 |
| Personal history of alcohol consumption (% yes) | 27.1 | 21.3 | 27.6 | 0.621 |
| MACEs (% yes) | 13.6 | 19.8 | 28.9 | 0.049* |
| Family history of MACEs (% yes) | 38.6 | 38.5 | 35.7 | 0.960 |
| Exercise (% yes) | 55.4 | 59 | 65.6 | 0.616 |
| Intensity of Exercise (none/low/moderate/high) % | 33.3/21.6/21.6/23.5 | 34.6/17.3/17.3/30.9 | 34.8/8.7/8.7/47.8 | 0.417 |
| Arterial hypertension (% yes) | 27.2 | 30.5 | 36.8 | 0.563 |
| Systolic Blood Pressure (SBP) (mmHg) | 129.3 (125.8 to 132.8) | 131.5 (128.0 to 135.0) | 132.8 (127.7 to 138.0) | 0.535 |
| Diastolic Blood pressure (DBP) (mmHg) | 79.8 (77.4 to 82.1) | 79.0 (76.6 to 81.2) | 79.2 (75.2 to 83.1) | 0.891 |
| Biochemical and Treatment Variables | TT (TrpTrp) | TC (TrpArg) | CC (ArgArg) | p value |
| Lipoprotein A (reference value: 0–0.3 g/L) | 0.4 (0.3 to 0.5) | 0.5 (0.4 to 0.6) | 0.5 (0.2 to 0.7) | 0.364 |
| Serum cholesterol; initial (mmol/L) | 7.3 (7.1 to 7.6) | 7.1 (6.9 to 7.3) | 7.2 (6.7 to 7.6) | 0.562 |
| Serum cholesterol LDL; initial (mmol/L) | 5.1 (4.8 to 5.3) | 4.9 (4.7 to 5.1) | 4.9 (4.5 to 5.3) | 0.627 |
| Serum cholesterol no HDL; initial (mmol/L) | 5.8 (5.6 to 6.0) | 5.7 (5.5 to 5.9) | 5.6 (5.2 to 6.0) | 0.650 |
| Serum triglycerides; initial (mmol/L) | 1.9 (1.6 to 2.17) | 2.1 (1.8 to 2.4) | 1.7 (1.4 to 2.1) | 0.343 |
| Treatment with atorvastatin (%) | 38.3 | 51.1 | 42.1 | 0.338 |
| Treatment with simvastatin (%) | 50.6 | 37.4 | 42.1 | |
| Treatment with rosuvastatin (%) | 11.1 | 11.5 | 15.8 | |
| Intensity of Treatment (low/medium/high) % | (12.3/63/24.7) | (11.5/59.5/29) | (10.5/44.7/44.7) | 0.271 |
Continuous variables are expressed as averages and as 95% confidence intervals (CI95%). Categorical variables are expressed in percentages.
* indicates statistical significance.
Simvastatin, atorvastatin, and rosuvastatin were prescribed to 106, 116 and 30 patients, respectively. The genotype frequencies of this sample fulfilled the Hardy-Weinberg equilibrium and their distribution matched those found in the HapMap of Europe [14], (Table 2).
Table 2. Genotype distribution of the genetic variants investigated, expressed in percentages.
| c.2155T>C (Trp719Arg) gene KIF6 | Genotype distribution (%) | p | ||
|---|---|---|---|---|
| HapMAP | TT (37.2) | CT (51.3) | CC (11.5) | |
| Simvastatin+Atorvastatin+Rosuvastatin | TT (32.4) | CT (52.4) | CC (15.2) | 0.37 |
| n = 81 | n = 131 | n = 38 | ||
| Simvastatin | TT (38.7) | CT (46.2) | CC (15.1) | 0.56 |
| n = 41 | n = 49 | n = 16 | ||
| Atorvastatin | TT (28.4) | CT (57.7) | CC (13.7) | 0.18 |
| n = 31 | n = 67 | n = 16 | ||
| Rosuvastatin | TT (30.0) | CT (50.0) | CC (20.0) | 0.38 |
| n = 9 | n = 15 | n = 6 | ||
p: probability of statistical significance as calculated using the Chi squared test.
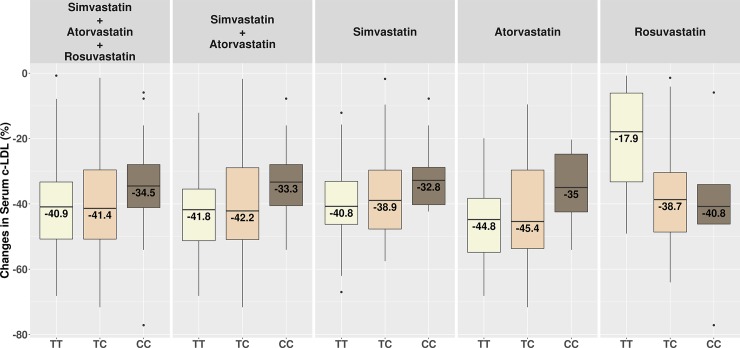
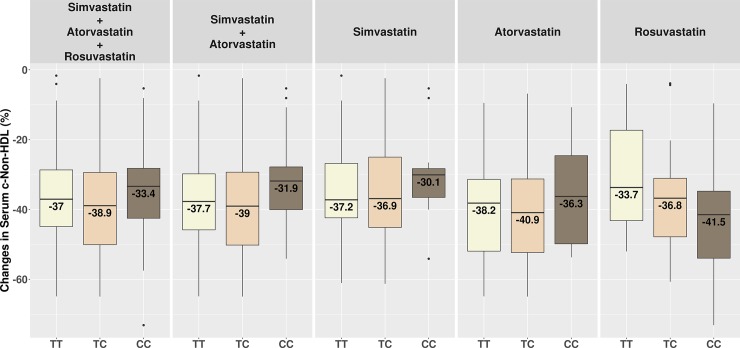
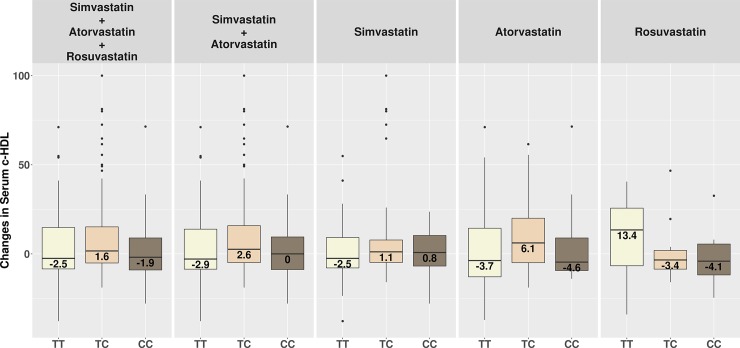
Figs 1, 2 and 3 show changes in serum c-LDL, c-Non-HDL and c-HDL after treatment stratified by genotypes.
Fig 1. Changes in serum c-LDL after treatment stratified by genotypes.
TT = homozygous TrpTrp; TC = heterozygous TrpArg; CC = homozygous ArgArg.
Fig 2. Changes in serum c-Non-HDL after treatment stratified by genotypes.
TT = homozygous TrpTrp; TC = heterozygous TrpArg; CC = homozygous ArgArg.
Fig 3. Changes in serum c-HDL after treatment stratified by genotypes.
TT = homozygous TrpTrp; TC = heterozygous TrpArg; CC = homozygous ArgArg.
Tables 3, 4 and 5 contain the results for each drug as well as the dependent variables.
Table 3. Multiple regression analysis that correlates the effect of the variant KIF6 on the LDL cholesterol concentration adjusted by statistically significant covariates.
| Changes in Serum LDL cholesterol concentration after treatment; (%) | ||||
|---|---|---|---|---|
| Treatment | Model | Variables | B + CI (95%) | P |
| Simvastatin+Atorvastatin+Rosuvastatin | Additive | Genotypes: TT, TC, and CC | 2.8 (-0.2 to 5.9) | 0.070 |
| Intensity of qualitative treatment | -4.7 (-8.1 to -1.3) | 0.008* | ||
| Initial concentration of c-LDL | -2.4 (-4.3 to -0.6) | 0.006* | ||
| Simvastatin+ Atorvastatin | Additive | Genotypes: TT, TC, and CC | 4.0 (1.0 to 6.8) | 0.010* |
| Intensity of qualitative treatment | -6.0 (-9.2 to -2.6) | 4.1x10-4* | ||
| Initial concentration of c-LDL | -2.4 (-4.2 to -0.6) | 0.008* | ||
| Recessive | KIF6 (CC compared to T) | 7.0 (1.3 to 12.6) | 0.015* | |
| Intensity of qualitative treatment | -6.0 (-9.2 to -2.6) | 4.1x10-4* | ||
| Initial concentration of c-LDL | -2.4 (-4.2 to -0.6) | 0.008* | ||
| Simvastatin | Additive | Genotypes: TT, TC, and CC | 3.8 (-0.04 to 7.7) | 0.053 |
| Intensity of qualitative treatment | -6.0 (-11.8 to -0.03) | 0.039* | ||
| Atorvastatin | Additive | Genotypes: TT, TC, and CC | 4.4 (-0.2 to 9.1) | 0.064 |
| Intensity of qualitative treatment | -6.6 (-11.7 to -1.5) | 0.011* | ||
| Initial concentration of c-LDL | -2.9 (-5.3 a -0.5) | 0.016* | ||
| Rosuvastatin | Additive | Genotypes: TT, TC, and CC | -7.0 (-20.2 a 6.0) | 0.275 |
| Prior MACE | -24.1 (-4.,7 to -2.5) | 0.031* | ||
B+CI (95%) = coefficient B + a 95% confidence interval
* indicates statistical significance.
Table 4. Multiple regression analysis that correlates the effect of the variant KIF6 on the Non-HDL cholesterol concentration adjusted by statistically significant covariates.
| Changes in Serum Non-HDL Cholesterol concentration after treatment; (%) | ||||
|---|---|---|---|---|
| Treatment | Model | Variables | B + CI (95%) | P |
| Simvastatin+Atorvastatin+Rosuvastatin | Additive | Genotypes: TT, TC, and CC | 0.8 (-1.7 to 3.4) | 0.535 |
| Intensity of qualitative treatment | -4.4 (-7.2 to -1.5) | 0.003* | ||
| Initial concentration of c-Non-HDL | -2.6 (-4.0 to -1.2) | 2.6x10-4* | ||
| Simvastatin+ Atorvastatin | Additive | Genotypes: TT, TC, and CC | 1.7 (-0.9 to 4.4) | 0.200 |
| Intensity of qualitative treatment | -5.3 (-8.2 to 2.3) | 4.0x10-4* | ||
| Initial concentration of c-LDL | -2.6 (-4.0 to -1.2) | 2.2x10-4* | ||
| Recessive | KIF6 (CC compared to T) | 5.1 (0.1 to 10.1) | 0.045* | |
| Intensity of qualitative treatment | -5.4 (-8.2 to -2.5) | 2.8x10-4* | ||
| Initial concentration of c-LDL | -2.6 (-4.0 to -1.2) | 2.6x10-4* | ||
| Simvastatin | Additive | Genotypes: TT, TC, and CC | 1.2 (-2.2 to 4.7) | 0.504 |
| Initial concentration of c-Non-HDL | -3.4 (-5.4 to -1.3) | 0.001* | ||
| Atorvastatin | Additive | Genotypes: TT, TC, and CC | 3.6 (-0.4 to 7.6) | 0.070 |
| Intensity of qualitative treatment | -6.2 (-10.6 to -1.8) | 0.006* | ||
| Initial concentration of c-Non-HDL | -2.8 (-4.7 to -1) | 0.004* | ||
| Age | -0.3 (-0.5 to 0.1) | 0.001* | ||
| Recessive | KIF6 (CC compared to T) | 9.03 (2.1 to 16.5) | 0.012* | |
| Intensity of qualitative treatment | -6.8 (-11.1 to -2.4) | 0.003* | ||
| Initial concentration of c-Non-HDL | -2.8 (-4.7 to -1.0) | 0.003* | ||
| Age | 0.35 (0.15 to 0.54) | 0.001* | ||
| Rosuvastatin | Additive | Genotypes: TT, TC, and CC | -3.4 (-12.9 to 6.0) | 0.463 |
| Prior MACE | -15.4 (-33-3 to 2.5) | 0.089 | ||
B+CI (95%) = coefficient B + a 95% confidence interval
* indicates statistical significance.
Table 5. Multiple regression analysis that correlates the effect of the variant KIF6 on the HDL cholesterol concentration adjusted by statistically significant covariates.
| Change in Serum HDL Cholesterol concentration after treatment; (%) | ||||
|---|---|---|---|---|
| Treatment | Model | Variables | B + CI (95%) | P |
| Simvastatin+Atorvastatin+Rosuvastatin | Additive | Genotypes: TT, TC, and CC | 0.3 (-3.8 to 4.6) | 0.860 |
| Initial concentration of c-HDL | -26.6 (-33.4 to -19.8) | 3.4x10-13* | ||
| Simvastatin+ Atorvastatin | Additive | Genotypes: TT, TC, and CC | 1.0 (-3.5 to 5.6) | 0.661 |
| Initial concentration of c-HDL | -28.7 (-36.1 to -21.3) | 7.1x10-13* | ||
| Simvastatin | Additive | Genotypes: TT, TC, and CC | 1.4 (-4.8 to7.7) | 0.648 |
| Initial concentration of c-HDL | -31.9 (-42.2 to -21.6) | 1.39x10-8* | ||
| Atorvastatin | Additive | Genotypes: TT, TC, and CC | 0.6 (-6.3 to 7.5) | 0.857 |
| Initial concentration of c-HDL | -25.4 (-36.4 to -14.4) | 1.18x10-5* | ||
| Rosuvastatin | Additive | Genotypes: TT, TC, and CC | -11.3 (-21.5 to -1.2) | 0.030* |
| Current tobacco use | -36.3 (-60.1 a -12.5) | 0.004* | ||
| Dominant | KIF6 (C compared to TT) | -21.9 (-37.6 to -6.2) | 0.008* | |
| Current tobacco use | -41.9 (-65.7 to 18.0) | 0.001* | ||
B+CI (95%) = coefficient B + a 95% confidence interval
* indicates statistical significance.
Intensity of qualitative treatment and the initial concentration of each lipid were found as significant control variables in the case of atorvastatin, simvastatin and all patients studied together. Concerning atorvastatin and c-Non-HDL variation, the age was also included. Regarding rosuvastatin, prior MACE was found as significant variable for the c-LDL and c-Non-HDL variation, and current tobacco use in the case of c-HDL variation.
Concerning patients that had been treated with simvastatin, an almost statistically significant relationship was observed between being a carrier of the c.2155T> C variant of KIF6 and changes in c-LDL after treatment (p = 0.053). Treatment responses were less pronounced among those patients who were homozygous for the mutation (CC, ArgArg) than among those who were homozygous for the non-mutated gene (TT, TrpTrp). A similar trend was observed with patients who had been treated with atorvastatin. In this case, there was a statistically significant relationship between the c.2155T> C variant of KIF6 and the c-Non-HDL variation after treatment (p = 0.012). There was also a clear tendency for the presence or absence of the variant to be associated with c-LDL variation after treatment (p = 0.06). However, an association between the c.2155T> C variant and c-LDL and c-Non-HDL variation after treatment was not observed in the group of patients treated with rosuvastatin (p = 0.353 and p = 0.454, respectively)
When all 252 recruited patients were included in the statistical analyses and the control variables were adjusted for, a clear tendency for the c.2155T> C variant and the reduction in plasma c-LDL after treatment was found. (Additive model: p = 0.07; B + IC95% = 2.8 (-0.2 to 5.9)). When only taking into account patients who underwent treatment with simvastatin and atorvastatin, this relationship was even more pronounced, and a statistically significant relationship was found: (additive model: p = 0.010; B + IC95% = 4 (1 to 6.8); recessive model: p = 0.015; B + IC95% = 7 (1.3 to 12.6)).
There was also a statistically significant relationship between being a carrier of the variant c.2155T> C and a weaker response to rosuvastatin in comparison to non-carriers, with higher levels of c-HDL being observed after treatment (p = 0.03). However, no statistically significant relation was found between the variant and c-HDL level changes in the simvastatin and atorvastatin treatment groups (p = 0.648 and p = 0.857, respectively).
Based on these results, we can conclude that being a carrier of the c.2155T> C variant of the KIF6 gene negatively impacts patient responses to statin treatments. A less pronounced decrease in c-LDL in the case of simvastatin and atorvastatin and less pronounced increase in c-HDL in the case of rosuvastatin are observed with respect to non-carriers.
Discussion
Statins are highly effective prophylactics against arteriosclerosis. Nevertheless, a high proportion of patients consuming statins develop some type of cardiovascular disease. These outcomes suggest that genetic factors may influence patient responses to treatment with statins [23]. Numerous genes, such as SLCO1B1, CETP, ABCA, HMGCR, and CYP3A4 are currently being investigated [24, 25] as possible factors affecting statin therapy outcomes. Of these, there has been a particular focus on KIF6 gene and its variant c.2155T> C (Trp719Arg).
A member of the kinesin superfamily, the KIF6 gene encodes the protein KIF6. Kinesins are proteins which mediate the intracellular transport of organelles, complex proteins and mRNAs. This gene is ubiquitously expressed in coronary arteries and other vascular tissue [26].
The effects of the c.2155T> C variant of KIF6 on the ability of statins to reduce the risk of cardiovascular events has been documented, but the results are contradictory. Several studies carried out by Iakouvova et al. [10, 11, 12, 13] concluded that patients that are carriers for the variant (TC (TrpArg) + CC (ArgArg)) are less likely, compared to non-carriers, to suffer from coronary heart disease when treated with pravastatin or atorvastatin. Meanwhile, a meta-analysis by Shiffman et al. [27] concluded that the variant affected responses to pravastatin treatment. Nevertheless, these conclusions are based on data from a single research group and therefore should be confirmed in other studies carried out independently by other groups.
Importantly, this association was not confirmed in other studies of simvastatin [28], rosuvastatin [29] and atorvastatin therapies [30].
In addition, others, such as Chen S. et al [31] have recently observed that patients undergoing statin therapy were at significantly reduced risk for MACE in the case of all three Trp719Arg polymorphisms. Furthermore, subjects carrying (TC) TrpArg or CC+CT (ArgArg+TrpArg) suffered from a greater incidence of MACE when being treated with statins than TT (TrpTrp).
As far as we know, our study is the first attempt to study the relationship between the KIF6 gene variant and the effects of statins on changes on lipid profiles using a prospective and observational design. In addition, the analyses of these effects have been adjusted against control variables. Importantly, normal clinical procedures were reproduced: medical decisions about therapeutic treatments were based primarily on changes in patient lipid profiles.
According to our results, patients homozygous for variant C did not respond as well to treatment with simvastatin and atorvastatin as did the patients who were homozygous or heterozygous for variant T. The same tendency was observed with rosuvastatin when measuring increases in c-HDL levels. Considering these results, it should be noted that while Rosticci et al [32], did not observe a relationship between the KIF6 variant and statin therapy efficacy, this group did observe a relationship between the variant and c-HDL levels.
However, this relationship was not observed for rosuvastatin when measuring changes in c-LDL and c-Non-HDL. This discrepancy could be explained by the fact that there were a low number of rosuvastatin cases included in the study. Rosuvastatin is usually prescribed to high risk populations when other statins have already been found to be insufficient. Therefore, there were fewer of rosuvastatin cases because this prospective and observational study only included individuals who had not undergone prior hypocholesterolemic treatment.
In a study carried out by Angelini et al. [33] it was shown that there was no relationship between the KIF6 variant and the effects of statins on patient lipid profiles. Although an observed trend says that subjects carrying CC (ArgArg) do not respond as well to treatment as TT (TrpTrp) and TC (TrpArg) subjects. However, in contrast to ours, the study of Angelini et al. was retrospective in nature, rather than prospective. In addition, the Angelini et al. study did not adjust for dose intensity or type of statin.
Moreover, the dependent variable was qualitative: the sole criteria it depended on was whether or not a clinical target of c-LDL ≤ 3.4 (130 mg / dL) mmol / L was reached or not [34].
Importantly, our results could be consistent with the latest information from the literature. We found that there was a statistically significant correlation between being a sufferer of MACE and being a carrier of c.2155T> C variant of KIF6 when taking into consideration all patients prior to treatment with statins (p = 0.049). This outcome is consistent with the current literature [35, 36, 37].
In addition, we have demonstrated that, while, c-LDL levels are reduced in all the three genotypes in response to treatment with simvastatin and atorvastatin, c-LDL levels are reduced to a lesser degree in the case of the variant.
Moreover, there is a general consensus that carriers benefit more from statin therapy in terms of a lower risk of suffering from MACE. However, the reason for this relationship may not lie in the fact that c-LDL levels decrease to a greater extent in non-carriers than in carriers.
The mechanism through which KIF6 influences statin therapy outcomes is not well understood. However, our study is an important step to clarifying the relationship between the c.2155T> C variant of KIF6 and responses to statin treatment. If our results can be confirmed, we could hypothesize that the carriers undergoing statin treatment suffer less from MACEs due to the pleiotropic effects of these drugs rather than a reduction in c-LDL. This hypothesis is consistent with the theory posited by Iakoubova et al. [13]. In the PROVE IT-TIMI 22 study, no statistically significant relationship was found between the variant and a reduction of c-LDL levels in response to atorvastatin or pravastatin, however, this group found that carriers benefited significantly more from intensive statin therapy than non-carriers and hypothesized that the mechanism could be the early plaque-stabilizing effect of the intensive treatment regimen, a pleiotropic effect that has been proposed to explain the early benefit from statin therapy that appears not to be due to LDL [38, 39]. However, functional studies of the KIF6 gene will be required to a better understand of the mechanism between c.2155T> C variant and statin treatment.
A key outcome of our study is that we have demonstrated that carriers of variant C have a 7% smaller decrease in c-LDL or c-Non-HDL levels in response to simvastatin or atorvastatin. The fact that the less common variant C is associated with a weaker response to treatment is especially relevant. If this relationship were to be confirmed, patients carrying variant C could be considered candidates for intensive treatment with stronger statins or complementary therapies. Complementary therapies include the drug Ezetimibe, or dietary supplements, such as omega-3.
Conclusions
The c.2155T> C variant of the KIF6 gene influences patient responses to treatment with simvastatin and atorvastatin. Patients homozygous for the variant (CC, ArgArg) had a smaller decrease in the c-LDL and c-Non-HDL. Therefore, these patients did not respond as well to hypolipidemic therapies as patients who were homozygous TT (TrpTrp) or heterozygous TC (TrpArg). The c.2155T> C variant is associated with a less pronounced increase in c-HDL upon rosuvastatin treatment.
These findings, if confirmed, may have an impact on the type of therapies selected for patients carrying the genetic variant. For example, the intensity of statin therapy could be increased, or complementary therapies, such as Ezetimibe or dietary supplements could be employed to treat carriers.
Acknowledgments
We thank Emili Corbella-Inglés, Ferran Trias-Vilagut, Marta Fanlo- Maresma, Hannia Elena Lafuente-González and Enric Juncadella-García, who provided insight and expertise that greatly assisted the research.
The present work was performed as part of the Biochemistry, Molecular Biology and Biomedicine doctoral program of Cristina Ruiz-Iruela at Universitat Autònoma de Barcelona (Barcelona, Spain).
Data Availability
All relevant data are within the manuscript.
Funding Statement
This research was supported by the Barcelona’s Professional Pharmacist Association (Col·legi de Farmacèutics de la Província de Barcelona, https://www.cofb.net) to CRI, Fundación José Luis Castaño (FENIN scholarship 2014) to BCE and Fundació Parc Taulí Institut Universitari UAB (Parc Taulí Recerca scholarship 2012) to BCE. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Chasman DI, Giulianini F, MacFadyen J, Barratt BJ, Nyberg F, Ridker PM. Genetic determinants of statin-induced low-density lipoprotein colesterol reduction: The Justification for the Use of Statins in Prevention: an Intervention Trial Evaluating Rosuvastatin (JUPITER) trial. Circ Cardiovasc Genet 2012; 5(2):257–64. 10.1161/CIRCGENETICS.111.961144 [DOI] [PubMed] [Google Scholar]
- 2.Ministerio de Sanidad, Servicios Sociales e Igualdad: National Healthcare system Anual Report, 2016. Available in www.msssi.gob.es.
- 3.Davidson MH, Toth PP. Comparative effects of lipid-lowering therapies. Progress in Cardiovascular Diseases 2004; 47(2),73–104. [DOI] [PubMed] [Google Scholar]
- 4.Kitzmiller JP, Mikulik EB, Dauki AM, Murkherjee C, Luzum JA. Pharmacogenomics of statins: understanding susceptibility to adverse effects. Pharmacogenomics and Personalized Medicine 2016; 9:97–106. 10.2147/PGPM.S86013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maxwell WD,Ramsey LB, Johnson SG, Moore KG,Shtutman M, Schoonover JH et al. Impact of Pharmacogenetics on Efficacy and Safety of Statin Therapy for Dyslipidemia. Pharmacotherapy 2017; 37: 1172–1190. 10.1002/phar.1981 [DOI] [PubMed] [Google Scholar]
- 6.Leusink M, Onland-Moret N, de Bakker P, de Boer A, Maitland-van der Zee AH. Seventeen years of statin pharmacogenetics: a systematic review. Pharmacogenomics. 2015; 17:15–158. [DOI] [PubMed] [Google Scholar]
- 7.Peng P, Lian J, Huang RS, Xu L, Huang Y, Ba Y, et al. Meta-Analyses of KIF6 Trp719Arg in Coronary Heart Disease and Statin Therapeutic Effect. PLoS ONE 2012: e50126 10.1371/journal.pone.0050126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Iakoubova OA, Shiffman D, Devlin JJ, Forrester JS, Superko HR. KIF6 Polymorphism as a Predictor of Risk of Coronary Events and of Clinical Event Reduction by Statin Therapy. American Journal of Cardiology 2010; 106:994–998. 10.1016/j.amjcard.2010.05.033 [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Sabatine M, Tong C, Ford I, Kirchgessner T, Packard C et al. Genetic variants in the KIF6 region and coronary event reduction from statin therapy. Human Genetics 2011; 129:17–23. 10.1007/s00439-010-0892-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iakoubova OA, Tong CH, Rowland CM, Kirchgessner TG, Young BA, Arellano AR et al. Association of the Trp719Arg polymorphism in kinesin-like protein 6 with myocardial infarction and coronary heart disease in 2 prospective trials: the CARE and WOSCOPS trials. Journal of the American College of Cardiology 2008; 51:435–443. 10.1016/j.jacc.2007.05.057 [DOI] [PubMed] [Google Scholar]
- 11.Shiffman D, Sabatine MS, Louie JZ, Kirchgessner TG, Iakoubova OA, Campos H et al. Effect of pravastatin therapy on coronary events in carriers of the KIF6 719Arg allele from the cholesterol and recurrent events trial. Journal of the American College of Cardiology 2010; 105:1300–1305. [DOI] [PubMed] [Google Scholar]
- 12.Iakoubova OA, Robertson M, Tong CH, Rowland CM, Catanese JJ, Blauw GJ et al. KIF6 Trp719Arg polymorphism and the effect of statin therapy in elderly patients: results from the PROSPER study by in European journal of cardiovascular prevention and rehabilitation. European Journal of Preventive Cardiology 2010; 17(4):455–461. [DOI] [PubMed] [Google Scholar]
- 13.Iakoubova OA, Sabatine MS, Rowland CM, Tong CH, Catanese JJ, Ranade K et al. Polymorphism in KIF6 gene and benefit from statins after acute coronary syndromes: results from the PROVE IT-TIMI 22 study. Journal of the American College of Cardiology 2008; 51(4):449–55. 10.1016/j.jacc.2007.10.017 [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Chen Z, Song H. Association between KIF6 rs20455 polymorphism and the risk of coronary heart disease (CHD): a pooled analysis of 50 individual studies including 40,059 cases and 64,032 controls. Lipids Health Disease. 2018. January 5; 17(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vatte C, Cyrus C, Al Shehri AM, Chathoth S, Almansori M, Al-Nafaie A et al. Investigation of KIF6 Trp719Arg gene polymorphism in a case-control study of coronary artery disease and non-fatal myocardial infarction in the Eastern Province of Saudi Arabia. Annals of Saudi Medicine. 2016. Mar-Apr; 36(2):105–11. 10.5144/0256-4947.2016.21.3.1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubacek JA, Vrablik M, Dlouha D, Stanek V, Gebauerova M, Adamkova V,et al. Gene variants at FTO, 9p21, and 2q36.3 are age-independently associated with myocardial infarction in Czech men. Clinica Chimica Acta. 2016. February 15; 454:119–23. [DOI] [PubMed] [Google Scholar]
- 17.Musunuru K. Lack of Association of KIF6 Genotype with Vascular Disease and Statin Response. Cardiovascular Genetics 2011; 4(4):467–8. 10.1161/CIRCGENETICS.111.960955 [DOI] [PubMed] [Google Scholar]
- 18.https://hapmap.ncbi.nlm.nih.gov/. (Accessed: 08-13-18).
- 19.Stone NJ et al. ACC/AHA Guideline on the Treatment of Blood Cholesterol to Reduce Atherosclerotic Cardiovascular Risk in Adults. ACC/AHA Blood Cholesterol Guideline 2013. [Google Scholar]
- 20.Banach M, Rizzo M, Toth PP, Farnier M, Davidson MH, Al-Rasadi K et al. Position paper. Statin intolerance-an attempt at a unified definition. Position paper from an International Lipid Expert Panel. Archives of Medical Science. 2015;11(1):1–23. 10.5114/aoms.2015.49807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey LB, Johnson SG, Caudle KE, Haidar CE, Voora D, Wilke RA,et al. The Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clinical pharmacology and therapeutics 2014; 96(4): 423–428. 10.1038/clpt.2014.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ACC/AHA Release Updated Guideline on the Treatment of Blood Cholesterol to Reduce ASCVD Risk. American Family Physician 2014. [Google Scholar]
- 23.Kolovou V, Kolovou G. Satins treatment under the umbrella of pharmacogenetics. Health Science Journal. 2015; 9:1–3. [Google Scholar]
- 24.Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA et al. Genome-Wide Association of Lipid-Lowering Response to Statins in Combined Study Populations. PLoS ONE. 2010; 5(3): e976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitzmiller JP, Binkley PF, Pandey SR, Suhy AM, Baldassarre D, Hartmann K. Statin pharmacogenomics: Pursuing biomarkers for predicting clinical outcomes. Discovery Medicine. 2013; 16(86): 45–5 [PMC free article] [PubMed] [Google Scholar]
- 26.https://www.ncbi.nlm.nih.gov/gene/221458. (Accessed: 08-13-18).
- 27.Shiffman D, Trompet S, Louie JZ, Rowland CM, Catanese JJ, Iakoubova OA, et al. Genome-Wide Study of Gene Variants Associated with Differential Cardiovascular Event Reduction by Pravastatin Therapy. PLoS ONE 2012; 7(5): e38240 10.1371/journal.pone.0038240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopewell J, Parish S, Clarke R, Armitage J, Bowman L, Hager J et al. No impact of KIF6 Genotype on vascular risk and statin response among 18,348 randomized patients in the heart protection study. Journal of the American College of Cardiology 2011; 57(20):2000–7. 10.1016/j.jacc.2011.02.015 [DOI] [PubMed] [Google Scholar]
- 29.Ridker PM, Macfadyen JG, Glynn RJ, Chasman DI. Kinesin-Like Protein 6 (KIF6) Polymorphism and the Efficacy of Rosuvastatin in Primary Prevention. Circulation Cardiovascular Genetics 2011; 4(3):312–317. 10.1161/CIRCGENETICS.110.959353 [DOI] [PubMed] [Google Scholar]
- 30.Arsenault BJ, Boekholdt SM, Hovingh GK, Hyde CL, DeMicco DA, Chatterjee A et al. The 719Arg variant of KIF6 and cardiovascular outcomes in statin-treated, stable coronary patients of the treating to new targets and incremental decrease in end points through aggressive lipid-lowering prospective studies. Circulation Cardiovascular Genetics 2012; 5(1):51–7. 10.1161/CIRCGENETICS.111.960252 [DOI] [PubMed] [Google Scholar]
- 31.Chen S, Xiang Q, Zhao X, Xie Q, Zhou S, Zhang Z et al. Impact of the 719Arg variant of KIF6 and major cardiovascular events on patients who received statins: a systematic review and meta-analysis. Current pharmaceutical design 2018; June 24. [DOI] [PubMed] [Google Scholar]
- 32.Rosticci M, Marullo L, Cicero AF, Magi R, Fischer K, Pervjakova N et al. 1a.11: Association of KIF6 and HMGCR loci with cardiometabolic phenotypes and response to statin therapy in the brisighella cohort. Journal of Hypertension. 2015. June;33 Suppl 1: e3–4. [Google Scholar]
- 33.Angelini S, Rosticci M, Massimo G, Musti M, Ravegnini G, Consolini N et al. Relationship between Lipid Phenotypes, Overweight, Lipid Lowering Drug Response and KIF6 and HMG-CoA Genotypes in a Subset of the Brisighella Heart Study Population. International Journal of Molecular Sciences. 2017; December 24;19(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Third Report of the National Cholesterol Education Program (NCEP). Expert Panel onDetection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III). Circulation 2002. [PubMed] [Google Scholar]
- 35.Ruiz-Ramos D, Hernández-Díaz Y, Tovilla-Zárate CA, Juárez-Rojop I, López-Narváez ML, González-Castro TB et al. The Trp719Arg polymorphism of the KIF6 gene and coronary heart disease risk: systematic review and meta-analysis. Hereditas 2015; 152:3 10.1186/s41065-015-0004-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamidizadeh L, Haji Hosseini Baghdad Abadi R, Babaee Baigi MA, Dastsooz H, Khazaei Nejhad A, Fardaei M. Impact of KIF6 Polymorphism rs20455 on Coronary Heart Disease Risk and Effectiveness of Statin Therapy in 100 Patients from Southern Iran. Archives of Iranian medicine. 2015; 18(10):683–687. [PubMed] [Google Scholar]
- 37.Iakoubova OA, Tong CH, Catanese J, Rowland CM, Luke MM, Tranquilli M et al. KIF6 719Arg Genetic Variant and Risk for Thoracic Aortic Dissection. AORTA (Stamford) 2016; 4(3):83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwartz GG, Olsson AG. The case for intensive statin therapy after acute coronary syndromes. American Journal of Cardiology 2005; 96:45F–53F. 10.1016/j.amjcard.2005.06.026 [DOI] [PubMed] [Google Scholar]
- 39.Halcox JP, Deanfield JE. Beyond the laboratory: clinical implications for statin pleiotropy. Circulation 2004; 109: II42–8. 10.1161/01.CIR.0000129500.29229.92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.