Abstract
The composition of the human microbiome is considered a major source of inter-individual variation in immunity, and by extension, susceptibility to diseases. Although intestinal bacteria have been the major focus of research, diverse communities of viruses that infect microbes and the animal host cohabitate the gastrointestinal tract, which collectively constitute the gut virome. Although viruses are typically investigated as pathogens, recent studies highlight a relationship between the host and animal viruses in the gut that is more akin to host-microbiome interactions and includes both beneficial and detrimental outcomes for the host. These viruses are likely sources of immune variation, both locally and extra-intestinally. In this review we describe the components of the gut virome, in particular mammalian viruses, and their ability to modulate host responses during homeostasis and disease.
Introduction
The human body harbors diverse populations of infectious entities, collectively known as the microbiome, that interact with each other and with the host to influence health and disease. While most commonly studied are the bacterial members of the microbiome, there are vast numbers of viruses present in the human body. Together, these viruses form the virome. Comprehensive annotation of the human virome is confounded by the staggering diversity of viruses detected at multiple anatomical sites that can have ssRNA, dsRNA, ssDNA or dsDNA genomes. Despite this challenge, recent advances in sequencing and analysis of metagenomic data have facilitated the discovery of new viruses and improved our ability to catalog viral communities in an unbiased manner (1, 2). These pioneering efforts reveal substantial intestinal virome diversity between individuals likely due to differences in bacterial composition and diet (3, 4). Studies comparing the virome between individuals have contributed to the growing evidence that differential exposure to viruses influences host physiology, either to the detriment or benefit of the host, much like the bacterial microbiome.
Although the study of virology has generally focused on disease-causing animal viruses, a large fraction of the virome is comprised of bacteriophages and endogenous retroviral elements. Approximately 1015 bacteriophages exist in the human intestine (5). 108-109 virus-like particles (VLPs) are found in a gram of human stool (6). The majority of these contain a DNA genome. Among the DNA viruses that can be matched to an annotated genome, 99% are bacteriophages and the remaining 1% are animal viruses such as anellovirus, parvovirus, adenovirus, and papillomavirus (6). Intrapersonal bacteriophage abundance is mostly stable over time but does show rapid sequence diversification (7). The predominant classifiable bacteriophage species in the gut are the dsDNA Caudovirales and ssDNA Microviridae (8). However, one uncharacterized dsDNA bacteriophage known as crAssphage is present in 73% of fecal metagenomes and predicted to infect Bacteroides species that are prevalent in the human gut (9, 10). In addition to directly influencing microbiome population dynamics by killing their bacterial hosts during lytic release of viral particles, bacteriophages that integrate into bacterial genomes contribute to the coding potential of the microbiome to indirectly influence the physiology of the animal host (8).
Endogenous retroviruses (ERVs) resemble present day exogenous retroviruses but are integrated in the host genome and transferred vertically between generations. They are estimated to comprise 8% of the human genome (11). The syncytin proteins that mediate placental development are derived from ERV env genes, and ERVs have dispersed interferon-inducible enhancer elements throughout mammalian genomes, suggesting that retroviral integration played a substantial role in mammalian evolution (12). Although most ERVs have accumulated many changes to their sequence over time that have rendered them defective, there are a limited number of ERVs with the potential to produce viral products that activate immune response or promote tumorigenesis (12–15). ERVs can also facilitate insertional mutagenesis and chromosomal rearrangements that affect cellular gene expression (16). The virome can also include plant viruses, likely introduced through food, and viruses that infect archaea and eukaryotic members of the microbiome such as fungi (mycobiome). One study showed that 97% of VLPs from the healthy human gut that harbor an RNA genome represent pathogenic plant viruses, with the remaining 3% belonging to animal viruses (17). How these viruses affect animal hosts is unknown.
The remainder of the virome consists of RNA and DNA animal viruses that are not integrated into the germ-line. At any given time, an individual human harbors multiple animal viruses, many of which establish chronic infections (18–20). The prevalence of animal viruses that cause transient infections, also considered part of the virome, can be more difficult to investigate, especially if the infection is asymptomatic. In contrast to serological methods that capture the infectious history of an individual (21), metagenomic studies may miss the contribution of a virus that is no longer present in a patient or diseased tissue. Additionally, chronic infections are often difficult to detect because certain viruses can exist in a quiescent state (latency), and the immune system may restrict replication to levels that are undetectable by conventional methods. In one of the few longitudinal virome studies performed to date, fecal samples from healthy human infants were shown to harbor RNA and DNA animal viruses belonging to 16 distinct families during the first 24 months of life (22). By adulthood, a typical individual will have been infected by at least 10 different viruses, with some individuals showing evidence of infection by 50-100 viral species (21).
The traditional paradigm of host-pathogen interactions, where infection by an individual agent directly produces immediate disease, fails to fully capture our relationship with many of these animal viruses. The necessity of the host machinery for their life cycle suggests that these viruses are unlikely to be silent passengers. Recent studies highlight how the gastrointestinal tract is an important site for virus-microbiome and virus-host interactions that likely contribute to inter-individual variation in immunity and disease susceptibility (Figure 1). Therefore, in this article we will focus on the impact of intestinal animal viruses as modifiers of the immune system. We will first review the pathways involved in recognition and responding to intestinal infection, and provide evidence of functional interactions between animal viruses and the bacterial microbiome. We will next discuss the beneficial and detrimental impact of intestinal infections by viruses beyond their role as pathogens. At the end, we use examples of how knowledge gained from the study of viral infections at non-intestinal sites can guide future research into the gut virome.
Figure 1. Virus-microbiome and virus-host genome interactions in immune variation.
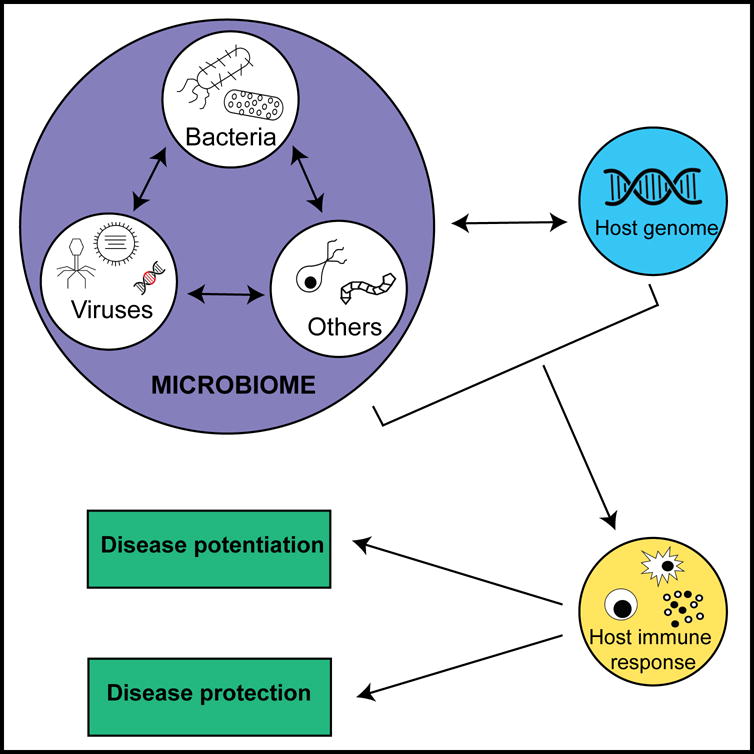
The host microbiome is a complex network of viruses, bacteria and other organisms (fungi, archaea, protozoans and helminths) that reside in the human body. The virome is comprised of animal viruses, bacteriophages and endogenous retroviruses. The gastrointestinal tract is inhabited by vast numbers of viruses and is an important site for virus-microbiome interactions and virus-host genome interactions. Intestinal bacteria interact with the virome by harboring bacteriophages and facilitating infection of barrier cells by animal viruses. Although typically investigated as pathogens, this review highlights how animal viruses in the gut serve as immune modulators that potentially explains inter-individual differences in disease susceptibility. The responses induced by various virus-microbiome and virus-host genome interactions likely alter the magnitude and function of the immune response to either the detriment or benefit of the host leading to either potentiation or protection from disease.
Immune responses to enteric viruses
Unlike bacteria, viruses need to infect host cells within the gastrointestinal tract to support their propagation. Target cells include the one-layer thick epithelial cells that serve the dual function of facilitating nutrient exchange and a physical barrier against invasion (23). Dendritic cells (DCs) and macrophages within the lamina propria (tissue underlying the epithelium) and gut-associated lymphoid tissue (GALT, such as Peyer’s patches) also commonly encounter viruses (24). Nucleic acid derived from enteric viruses are sensed by these cells through many of the same pattern recognition receptors (PRRs) that are important at other sites. These include endosomal toll-like receptors (TLRs) that signal through MYD88 and TRIF, and the cytosolic sensors retinoic acid inducible gene–I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) that signal through MAVS, to stimulate the expression of type I (IFN-I) and type III (IFN-III) interferons (24). Both RIG-I and MDA5 are necessary for optimal antiviral responses to rotavirus, a dsRNA virus that infects the small intestinal epithelium to cause diarrheal disease in children (25). Sensing of ssRNA noroviruses, which also cause gastroenteritis in humans, can occur through MDA5, TLR7, and TLR3 in myeloid cells (26, 27). Although best known for responding to bacteria, recent studies suggest that cytosolic Nod-like receptors (NLRs) have an intestine-specific role in restricting viruses. Mice deficient in NLRP6, which can serve as a co-factor for RNA helicase DHX15 to signal through MAVS, display a blunted IFN-I/III response and are susceptible to oral infection by encephalomyocarditis virus (ECMV) and murine norovirus (MNV) (28). The multiple pathways involved in norovirus recognition may reflect inter-strain differences or their ability to infect broad cell types including myeloid cells, lymphocytes, and the specialized sensory epithelial cell known as tuft cells (29–32). Inhibiting the ability of MNV to engage these rare tuft cells prevents infection, highlighting the importance of an exquisitely specific cell tropism and remarkable adaptation of enteric viruses (30). Rotavirus RNA is sensed by NLRP9b and another RNA helicase DHX9 in the intestinal epithelium and is essential for inflammasome-mediated cell death (pyropotosis). Given that rotaviruses antagonize IFN signaling, this pathway may explain why other PRRs are inadequate for controlling this virus (33).
Despite the ability of certain viruses to evade IFN responses, numerous studies highlight the essential role of these cytokines during intestinal infection. IFN-I (IFNα and IFNβ) binds the interferon-α/β receptor (IFNAR1 and IFNAR2 complex) while IFN-III (IFNλ) binds the interferon lambda receptor (IFNLR1 and IL10R2 complex) to induce an antiviral gene expression program. Although the exact effector mechanisms in the gut remain obscure, they are presumably similar to other sites of infection and involve interferon stimulated genes (ISGs) that reduce viral replication (e.g., modification of viral RNA) and induce a refractory state in neighboring uninfected cells. IFNAR1 is broadly expressed on a variety of cell types, and is necessary for preventing systemic dissemination of MNV, rotavirus and reovirus (34). In contrast, the relatively restricted expression of IFNLR1 to epithelial cells suggests it has a more defined role in controlling mucosal viral replication (35). IFNλ, in combination with IL-22 production by group 3 innate lymphoid cells (ILC3s), effectively controls intestinal rotavirus infection (36). IFNλ signaling in epithelial cells is also required to regulate fecal shedding and viral replication in mice infected with MNV and reovirus (37). IFNs also promote adaptive immune responses (38), which are critical for control of enteric viruses (39–42). Suboptimal CD8+ T cell responses and avoidance of CD8+ T cell detection are associated with MNV persistence (43, 44). Furthermore, successful vaccination against rotavirus directly correlates with IgA production (45). Although we emphasize the role of cytokines downstream of PRRs in subsequent sections as a common means by which viruses affect host physiology, understanding how adaptive immunity functions or fails to control enteric viruses remains an important topic with direct relevance to vaccine efforts.
Boosting innate immunity may be an effective strategy for overcoming insufficient antiviral immunity in the gut. Treatment with bacterially-derived flagellin prevents and cures chronic rotavirus infection of mice by triggering IL-22 and IL-18 production through TLR5 and NLRC4, respectively (46). For noroviruses, effective antivirals and vaccines currently do not exist, and there is increased concern that persistent norovirus infection may be contributing to morbidity in immunocompromised individuals or facilitating transmission (47, 48) Remarkably, administration of recombinant IFNλ is sufficient to clear infection of a persistent strain of MNV independently of the adaptive immune response (29, 49). As we discuss throughout this article, the induction of IFNs is a hallmark of viral infection, and likely mediates many of the consequences of enteric viruses on host physiology.
Impact of bacteria on enteric virus infection
Depletion of intestinal bacteria often reduces the replication of enteric viruses, as the bacterial microbiome is known to facilitate viral infection and modify anti-viral immune responses (50, 51). Optimal poliovirus infectivity is dependent on the stabilization of virions upon binding to bacterial surface polysaccharides (52, 53). Binding to bacteria also promotes poliovirus attachment to target cells, which can enhance viral fitness by facilitating co-infection with multiple virions and genetic recombination events between viral strains (54). For similar reasons, intestinal titers and pathology are reduced following infection of antibiotics-treated Ifnar1−/− mice with reovirus (52). During vertical transmission of mouse mammary tumor virus (MMTV) through maternal milk, LPS bound to virions from the mother stimulates the production of the immunosuppressive cytokine IL-10 in the pups to allow the establishment of infection (55). Treatment of mice with antibiotics also decreases intestinal MNV replication (32, 56, 57). Here, bacteria reduce the efficacy of IFNλ-mediated viral clearance and enhance infection of B cells (32, 56). During rotavirus infection, bacteria reduce systemic and intestinal anti-rotavirus IgA titers to support increased rotavirus replication (58). These studies indicate that animal viruses that infect the gastrointestinal tract have adapted to the presence of bacteria.
Beneficial impact of viruses in the gut
The importance of the microbiome to the intestinal environment is apparent in germ-free mice, which display numerous intestinal and immune abnormalities due to the lack of microbial communities (59). Germ-free mice and mice treated with antibiotics also show increased susceptibility to models of intestinal damage, peanut allergy, allergic asthma and bacterial infections (60–64). Although in some cases intestinal bacteria are sufficient to modulate these responses, in many models a role for the virome cannot be ruled out, especially given that antiviral signaling has a prominent role in diseases involving the gut. Non-hematopoietic expression of MAVS is required to protect mice against colitis following intestinal injury by dextran sodium sulfate (DSS) (65). Similarly, IFN-I signaling following stimulation of the MAVS and RIG-I pathways improves intestinal barrier function and protects mice from graft-vs-host disease (GVHD), a complication that occurs following allogeneic stem cell transplantation (66). In another example, administration of a TLR7 agonist enhances colonization resistance to vancomycin-resistant Enterococcus (VRE), a common hospital-acquired opportunistic pathogen, by stimulating dendritic cells that induce IL-22 production by ILC3s (27). Another example where intestinal viruses potentially promote colonization resistance was observed during fecal microbiome transplantation in patients harboring Clostridium difficile. Surprisingly, filtrated feces (to remove the bacterial component and retain viruses) have the same efficacy as un-filtrated feces in treating the patients (67). It is possible that the active component of the fecal microbiome transplantation (FMT) consists of bacteriophages because patients with C. difficile infection had altered bacteriophage abundance and richness compared to healthy controls, and successful transplantation was associated with transfer of Caudovirales species (68). These and other observations indirectly suggest that enteric viral infections fortify the intestinal barrier in certain situations via triggering beneficial immune responses or influences the bacterial microbiome.
Mechanistic experiments in mice, especially with MNV infections, provide formal evidence that viruses can function as a subset of the microbiome in a manner analogous to intestinal bacteria (Table I). Many MNV strains establish persistent infection in the intestine, frequently in the absence of obvious symptoms. Infection by a persistent strain of MNV compensates for the absence of bacteria in germ-free mice by restoring intestinal morphology and promoting lymphocyte differentiation (57). In addition, MNV protects antibiotics-treated mice from DSS-induced intestinal injury in a manner dependent on IFNAR1 (57). MNV can also protect antibiotics-treated mice from pathology during superinfection with the intestinal bacterial pathogen Citrobacter rodentium and reduces colonization by VRE (27, 57). The recent discovery that MNV infects tuft cells, which coordinates type 2 immune responses and mucus production, suggests that infection by this virus could directly influence the function of the intestinal epithelium and warrants further investigation (30). It is possible that these effects extend beyond the gastrointestinal tract because MNV infection protects mice from lung injury following infection with Pseudomonas aeruginosa (69), and MNV restores serum immunoglobin in germ-free mice to levels observed in conventional mice (57). The observation that norovirus RNA is detected in up to 16% of healthy humans reinforces the need to understand how enteric viral infections impact host biology when they are not causing diarrheal disease (70).
Table I.
Examples in which intestinal viruses contribute to protection against or potentiation of diseases.
| Virus | Model | Outcome | Mechanism | Reference |
|---|---|---|---|---|
| Murine norovirus (MNV) | Germ-free and broad-spectrum antibiotics-treated wild type mice | Restoration of intestinal architecture, immune cell populations, and resistance to chemically-induced colitis | Dependent on IFN-I | Kernbauer et al., 2014 (49) |
| Ampicillin-treated wild type mice | Colonization resistance to vancomycin-resistant enterococcus | Increased IL-22+ ILC3 | Abt et al., 2016 (19) | |
| Atg16L1 mutant mice | Crohn’s like pathology in the small intestine | Paneth cell necroptosis due to virally-induced TNFα | Cadwell et al., 2010 (72) Matsuzawa-Ishimoto et al., 2017 (73) |
|
| IL-10−/− mice | Intestinal inflammation | Dependent on bacteria | Basic et al., 2014 (75) | |
| H. bilis-infected MDR1a−/− mice | Intestinal inflammation | Unknown | Lencioni et al., 2008 (74) | |
| Wild type mice | Protection from lung damage following P. aeruginosa infection | Unknown | Thepaut et al., 2015 (61) | |
| Reovirus | DQ8 transgenic mice and humans | Celiac disease manifestations | Suppression of peripheral Tregs and promotion of IRF1 and TH1 immunity to dietary antigen | Bouziat et al., 2017 (68) |
| Caudovirales bacteriophage | Humans | Crohn’s disease | Virus-microbiome interaction | Norman et al., 2015 (78) |
| Successful treatment of C. difficile by FMT | Transfer of species from healthy donors | Zuo et al., 2017 (59) | ||
| Picobirnaviridae | Humans | Sever enteric GVHD | Unknown | Legoff et al., 2017 (79) |
| Coxsackievirus B | NOD mice | Accelerated autoimmune diabetes onset | Virus spread to the pancreas and local IFN-I response | Reviewed in Jean-Baptiste et al., 2017 (82) |
| Rotavirus | NOD mice | Accelerated autoimmune diabetes onset | IFN-I dependent bystander activation of lymphocytes in the pancreatic lymph nodes | Pane et al., 2014 (87) Pane et al., 2016 (88) |
| Circovirus | Humans | Protection from T1D | Unknown | Zhao et al., 2017 (92) |
| Adenovirus and anellovirus | Human | HIV disease progression | Unknown | Monaco et al., 2016 (95) |
Animal models and observations in patients provide evidence for a role of intestinal viruses in modulating susceptibility to a range of disease conditions including intestinal inflammation (inflammatory bowel diseases, celiac disease and opportunistic colonization by antibiotic-resistance bacteria) and extra-intestinal disorders (T1D, lung infections, and HIV). Mechanisms frequently involve cytokines produced in response to viral infection that act on surrounding tissue or induce the mobilization of lymphoid cells. The outcome can be beneficial or detrimental to the host depending on whether a heightened state of immunity is desirable (e.g., protection against an infection versus fueling a chronic inflammatory disease). Abbreviations: IFN-1, type I interferon; ILC3, type 3 innate lymphoid cell; IRF1, interferon regulatory factor 1; FMT, fecal microbiome transplantation; NOD, non-obese diabetic; T1D, type 1 diabetes.
Other animal viruses may also promote intestinal homeostasis. Treatment of mice with a cocktail of antivirals increases the severity of DSS-induced colitis, while treatment with inactivated rotavirus or TLR3/7 agonists reduces disease (71). In this case, the protective effect of TLR ligation was attributed to IFN-I expression by plasmacytoid DCs. This response to viruses may be conserved in humans because TLR3 and TLR7 gene variants are associated with increased severity of inflammatory bowel disease (IBD) in patients (71). Murine cytomegalovirus (CMV), a herpesvirus that chronically infects a variety of tissues and cell types, promotes turnover of the epithelium in multiple organs including the intestine. This effect was attributed to epithelial proliferation induced by the ISG Apol9a/b expressed by macrophages downstream of elevated IFN-I and was shown to enhance intestinal wound healing (72). When taken together, these studies show that IFN-I and other antiviral responses induce factors that promote intestinal epithelial health in addition to those that inhibit viral replication. Whether IFNs in the gut are beneficial to the host may be context-specific and not without controversy (24). A major future direction is to elucidate the specific mechanisms of action downstream of IFN signaling in the models described above.
Negative impact of viruses in the gut
Excess IFN-I production and other antiviral responses in the gut can potentiate disease (Table I). Until recently, IFN-I in combination with antiviral drugs was standard treatment for chronic hepatitis C virus (HCV) infection and was associated with significant toxicity, including gastrointestinal illness. There is also evidence from case studies to suggest that IFN-I therapy may potentiate the development of celiac disease, an autoimmune disorder that mainly occurs in individuals harboring HLA-DQ2 or DQ8 alleles where inappropriate responses to gluten leads to intestinal damage (73). This side effect of antiviral therapy is consistent with the observation that patients with celiac disease display increased levels of IFN-I production by intestinal DCs that promote Th1 responses in the gut (74). It is therefore unsurprising that virus infections have long been suspected to be involved in the development of celiac disease (75). A compelling recent study demonstrated that celiac disease may be caused by reoviruses, dsRNA viruses that commonly infect humans and typically associated with mild or undetectable disease (76). In an animal model, reovirus infection blocked the differentiation of peripheral regulatory T cells through IFN-I and enhanced dietary antigen-specific Th1 responses through the transcription factor interferon regulatory factor 1 (IRF1) (76). Patients with celiac disease were also more likely to have higher anti-reovirus antibody titers, which was associated with higher expression of IRF1 in the small intestinal mucosa (76). Therefore, enteric viruses that are otherwise tolerated may induce serious intestinal disease in susceptible individuals.
The paradigm of virus-plus-susceptibility gene interaction was initially demonstrated in experiments with MNV, and reinforces the concept that viruses function as members of the gut microbiome. A common variant of ATG16L1, a gene that mediates the cellular degradative pathway of autophagy, is associated with increased susceptibility to a form of IBD known as Crohn’s disease. IBD is widely considered a disorder originating from a perturbed microbiome (77). Atg16L1 mutant mice and Crohn’s disease patients harboring the ATG16L1 risk allele display morphological defects in Paneth cells (78), antimicrobial epithelial cells in the small intestine that are essential for preventing inflammation (79). In the Atg16L1 mutant mice, the Paneth cell defects and other inflammatory pathologies were dependent on infection by MNV (78, 80). In this model, loss of Atg16L1 in the intestinal epithelium sensitizes Paneth cells to necroptosis mediated by TNFα produced in response to viral infection (81). MNV also accelerates the onset of intestinal inflammation in mice deficient in the toxin transporter MDR1a that are colonized by Helicobacter bilis (82) and IL-10-deficient mice (83). Thus, immune responses to an otherwise beneficial or innocuous virus can contribute to intestinal disease when combined with genetic susceptibility.
Although host responses to MNV and Paneth cell properties are likely conserved between mice and humans, further evidence is required to support the role of IFN-I or the virome in Crohn’s disease. A number of other viruses, including enterovirus, have been linked to Crohn’s disease (84, 85). In a virome study, expansion of the Caudovirales bacteriophages in the gut was observed in Crohn’s disease patients (86). Here, bacteriophage expansion was associated with decreased bacterial diversity suggesting that virus-microbiome interactions contribute to disease pathogenesis (86). A similar expansion of bacteriophage diversity is observed in patients with colorectal cancer and specific bacteriophage signatures can delineate patients in early or late stage and those with reduced survival (87). Also, a gut virome analysis of patients displaying GVHD with intestinal involvement revealed a marked increase in animal viruses with a DNA genome and bacteriophage richness (88). In particular, the presence of a dsRNA Picobirnaviridae species is predictive of a severe enteric disease (88). The combined approach of metagenomics through deep sequencing and targeted investigation of specific viral agents (like reovirus and celiac disease) may be necessary to explore the contribution of viruses to Crohn’s disease, GVHD, and other complex inflammatory disorders that are likely influenced by multiple infectious and genetic factors.
Impact of intestinal viruses beyond the gastrointestinal tract
The impact of the intestinal virome may extend beyond the gastrointestinal tract to influence autoimmune diseases (Table I). One explanation for such extra-intestinal effects of viruses is the spread of infectious particles or viral RNA/DNA from the intestine to other body sites. Indeed, the transit of antigens between the gastrointestinal tract and pancreatic lymph nodes has been suggested as a mechanism for the effects of environmental agents on type 1 diabetes (T1D), an autoimmune disease where insulin-producing β cells are destroyed by pancreas-infiltrating autoreactive lymphocytes (89). Polymorphisms in MDA5 (IFIH1) and an IFN-I gene expression signature are associated with disease onset, supporting a role for viruses in disease progression (90, 91). Also, the appearance of autoantibodies in patients correlates with infection by coxsackievirus B1 (CVB1), a +ssRNA virus that belongs to a diverse and prevalent group of picornaviruses that transmit fecal-orally (92). Infection with CVB3 and CVB6, which may provide cross-protection against CVB1, reduces the risk of T1D development in pre-diabetic children (92). Similar protection against virus-induced T1D is induced by CVB vaccination of mice (93). Although direct causation has not been established, CVB is believed to induce T1D by chronically infecting the pancreas and altering the local immune response (91). Rotavirus infection is also associated with progression to T1D (94). In the non-obese diabetic (NOD) mouse model of T1D, oral infection of adult mice with rotavirus leads to IFN-I-dependent bystander activation of lymphocytes in the pancreatic lymph nodes and acceleration of T1D onset, likely due to spread of infectious virus to the mesenteric and pancreatic lymph nodes (95–98). In contrast, neonatal infection of NOD mice with rotavirus or reovirus delays the onset of disease suggesting that timing of virus infections impacts the course of autoimmunity (99, 100).
A recent prospective study of infants at risk for T1D performed a longitudinal virome analysis and showed that increased bacteriophage diversity predicts a lack of progression to disease (101). Increased bacteriophage diversity correlated with changes in abundance of specific bacterial taxa, which may be related to the extensive literature using NOD mice demonstrating that the composition of the bacterial microbiome is an important factor in disease development (102). The same study also found an enrichment of sequences belonging to Circoviridae in the controls compared with individuals who develop T1D, raising the possibility that these group of poorly characterized small ssDNA animal viruses are protective (101). Although these studies implicate multiple viruses in disease pathogenesis, an exact mechanistic role for viruses in humans has yet to be established and requires additional research.
Metagenomic studies of HIV+ individuals have been particularly informative in that they reveal the presence of a dynamic gut virome in a disease state. Low peripheral CD4 cells counts leads to the development of AIDS which is marked by increased susceptibility to secondary infection and other immunopathologies. The gut is a major site of HIV replication, HIV-specific immune responses and pathology (103). Enteric adenoviruses and anelloviruses are increased in HIV+ patients with low peripheral CD4+ T cell counts (104). Similar expansion of the virome is observed in primates infected with simian immunodeficiency virus (SIV), with intestinal adenovirus associated with increased enteritis and parvovirus viremia associated with increased progression to AIDS (105). It is possible that this increased presence of viruses contributes to AIDS in a manner similar to the proposed role of the bacterial microbiome, where depletion of T cells in the gut disrupt the barrier, leading to the systemic dissemination of bacterial products that fuel chronic and pathological immune activation (106). Given that the majority of humans by the time they reach adulthood become transiently or chronically infected by the animal viruses discussed in this section (Anelloviridae, Adenoviridae, and Picornaviridae), careful analyses of the gut virome in other immune-related disorders is warranted.
Lessons from extra-intestinal virus infection
When considering how the enteric virome might influence inter-individual variation, it is worth examining the known mechanisms by which viruses at other sites of the body alter the immune system. Detailed immunological studies in mice have supported a role for the IFN response, but also highlight other pathways (Table II). Viruses that are traditionally considered to display localized infection and disease can provoke systemic responses, such as influenza A virus that induces hepatitis and intestinal damage without local infection (107, 108). Liver damage induced by influenza A virus is caused by the accumulation of pathogen-specific CD8+ T cells and intestinal damage is the result of an altered bacterial microbiome and increased IL-17a (107, 108). Therefore, an important possibility to examine is whether enteric viruses select for T cells that exert pathological outcomes once they migrate to other sites.
Table II.
Mechanisms by which extra-intestinal viruses contribute to protection against or potentiation of diseases.
| Virus | Route of Infection | Model | Outcome | Mechanism | Reference |
|---|---|---|---|---|---|
| Cytomegalovirus (CMV) | Intraperitoneal | Wild type mice | Resistant to infection with the bacterial pathogens Listeria monocytogenes and Yersinia pestis | Latent infection, IFNγ expression and activation of systemic macrophages | Barton et al., 2007 (100) |
| Intestinal proliferation | IFN-I mediated Apol9a/b expression | Sun et al., 2015 (64) | |||
| Resistance to Influenza A infection | Mediated by IFNγ expression | Furman et al., 2015 (101) | |||
| Gammaherpesvirus 68 (γHV-68) | Intranasal | HOIL-1−/− mice | Rescues lethality to Listeria monocytogenes | Mediated by increased proinflammatory cytokines | MacDuff et al., 2015 (103) |
| Intraperitoneal | Wild type mice | Resistant to infection with the bacterial pathogens Listeria monocytogenes and Yersinia pestis | Latent infection, IFNγ expression and activation of systemic macrophages | Barton et al., 2007 (100) | |
| Intranasal | Wild type mice | Protection from allergic asthma | Alteration of alveolar macrophage subsets | Machiels et al., 2017 (104) | |
| Influenza A virus | Respiratory | Wild type mice | Collateral liver damage | Accumulation of virus-specific CD8+ T cells in the liver | Polakos et al., 2006 (98) |
| Intestinal damage | Microbiota-dependent expression of IL-17a in the intestine | Wang et al., 2014 (99) | |||
| Rhinovirus C | Respiratory | Humans | Exacerbated asthma | Use of a specific receptor, CDHR3 | Bizzintino et al., 2011 (114) Bochkov et al., 2015 (115) |
| Respiratory | Humans | Cystic Fibrosis | Unknown | Goffard et al., 2014 (110) | |
| Pegivirus | Blood | Humans | HIV disease protection | Unknown | Williams et al., 2004 (108) |
Examples discussed in this article by which extra-intestinal viruses have unexpected immunomodulatory effects on the host are listed. As with intestinal viruses, these viruses can protect against infectious and non-infectious challenges, while simultaneously increasing susceptibility to autoimmune or inflammatory diseases. These examples highlight the ability of viruses to affect tissue outside their replicative niche, sometimes long after the initial infection, and suggest that enteric viral infections may have similar consequences for the host. Abbreviations: CDHR3, cadherin-related family member 3
Latent viral infections may be a particularly potent modulator of host responses. Gammaherpesvirus 68 (γHV-68) or CMV infection in mice enhances macrophage and NK cell activation, and improves the outcome of secondary infection with Listeria monocytogenes, Yersinia pestis and influenza A (109–111). In an animal model of primary immune-deficiency, latent γHV-68 infection rescues survival following L. monocytogenes infection by inducing an inflammatory reaction that compensates for inadequate cytokine levels, suggesting that deleterious mutations can be masked by an individual’s virome (112). γHV-68 also protects against allergic asthma by altering the composition of macrophage subsets in the lung (113). In this case, the effect of viral infection was not dependent on latent infection and occurred during a developmental window. These findings in animal models are supported by elegant human cohort studies taking advantage of monozygotic twins with discrepancies in their history of exposure to infectious agents. CMV infection was identified as a particularly significant environmental variable that influences a broad range of immune parameters (114). Further, young adults previously exposed to CMV show a superior antibody and CD8+ T cell response to influenza A vaccination (110). Thus, exposure to viruses can explain inter-individual heterogeneity when other factors fail to provide an adequate explanation.
The blood virome of healthy individuals include herpesviruses, anelloviruses, papillomaviruses, polyomaviruses, adenoviruses, parvoviruses and pegivirus (115, 116). Although it is unclear whether the presence of these viruses in the blood is consequential, the inverse relationship between pegivirus and HIV disease progression suggests that deeper investigation of the blood virome will be fruitful (117). In contrast, there is a wealth of examples demonstrating that respiratory viruses cause or exacerbate chronic lung diseases. In cystic fibrosis patients who display altered mucus production due to mutations in a chloride channel, disease is associated with the presence of a core group of bacteriophages that infect bacterial species persistent in lungs (118). Cystic fibrosis patients also show increased susceptibility to infection with rhinoviruses, which is linked to poor recovery of lung function following flares (119–121). Respiratory viruses are also linked to asthma (122). Patients have impaired IFN-I responses following rhinovirus infection, and rhinovirus C (RV-C) in particular is detectable in a significant proportion of children with moderate to severe asthma (122, 123). Consistent with this observation, the Y529 variant of cadherin-related family member 3 (CDHR3) that leads to increased binding of the virus to the lung epithelium confers susceptibility to RV-C-associated asthma (124, 125). As gene variants such as the loss of function allele of fucosyltransferase 2 (FUT2) can determine whether intestinal viruses bind cells (126, 127), an important area of research will be to examine how heritable factors affect the gut virome.
To summarize, virus-host interactions at extra-intestinal sites inform areas of future enteric virome research in the following ways. First, a number of these studies suggest that viruses that cause local infections such as in the lung have long range immunomodulatory effects. Therefore, examining the presence of viruses in affected tissues may not be sufficient, and a role for the gut virome should be considered. Second, the effect of a virus is not always apparent directly subsequent to virus exposure. This is clearly an important consideration for blood transfusions as viruses not routinely screened prior to blood donation could be transmitted to patients and have consequences for future disease pathogenesis. Perhaps similar concerns apply to FMTs that are routinely performed for C. difficile treatment and being considered for many other conditions. Finally, studies with CMV highlight the effect of viruses on the lymphocyte compartment and how adaptive immunity to subsequent antigens (i.e., not the original virus) may be altered. Molecular mimicry and bystander effects have been discussed extensively in other contexts (18), but are rarely considered downstream possibilities of intestinal virus infections in healthy individuals.
Conclusions
Significant progress has been made in the last decade towards understanding enteric viruses beyond their role as pathogens. While we continue to perform essential research into the pathogenic role of viruses and develop antivirals and vaccines, we can no longer ignore the possibility that they function as components of the microbiome. Like bacteria, the effects that viruses have are critically dependent on their tissue location, microenvironment and host. These factors will directly influence whether the virus acts beneficially, detrimentally or remains neutral for the host. With recent advances in metagenomics coupled with techniques that enrich in sensitivity (1, 2, 128), we can now preform large human studies with the aim of linking changes in specific viral populations with disease pathogenesis. These studies can then support the development of more defined animal and cell culture studies that address the mechanisms that individual viruses use to contribute to these phenotypes. This research will certainly lead to the discovery of novel ways in which viruses interact with the host that we can potentially harness for disease prevention and therapies. It may even be possible to engineer enteric viruses with desirable traits, much like current attempts at administering oncolytic viruses as adjuvants for cancer therapy (129–131).
Acknowledgments
K.C. is supported by US National Institute of Health (NIH) grants HL123340, R01 DK093668, R01 DK103788, and AI121244, Faculty Scholar grant from the Howard Hughes Medical Institute, Stony Wold-Herbert Fund, Merieux Institute, and Rainin Foundation. K.C. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases. J.A.N. received support through the Sir Keith Murdoch Fellowship and Vilcek Fellowship.
References
- 1.Krishnamurthy SR, Wang D. Origins and challenges of viral dark matter. Virus Res. 2017;239:136–142. doi: 10.1016/j.virusres.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Zhang YZ, Shi M, Holmes EC. Using Metagenomics to Characterize an Expanding Virosphere. Cell. 2018;172:1168–1172. doi: 10.1016/j.cell.2018.02.043. [DOI] [PubMed] [Google Scholar]
- 3.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carding SR, Davis N, Hoyles L. Review article: the human intestinal virome in health and disease. Aliment Pharmacol Ther. 2017;46:800–815. doi: 10.1111/apt.14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim MS, Park EJ, Roh SW, Bae JW. Diversity and abundance of single-stranded DNA viruses in human feces. Appl Environ Microbiol. 2011;77:8062–8070. doi: 10.1128/AEM.06331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc Natl Acad Sci U S A. 2013;110:12450–12455. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manrique P, Dills M, Young MJ. The Human Gut Phage Community and Its Implications for Health and Disease. Viruses. 2017;9:141. doi: 10.3390/v9060141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, Boling L, Barr JJ, Speth DR, Seguritan V, Aziz RK, Felts B, Dinsdale EA, Mokili JL, Edwards RA. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yutin N, Makarova KS, Gussow AB, Krupovic M, Segall A, Edwards RA, Koonin EV. Discovery of an expansive bacteriophage family that includes the most abundant viruses from the human gut. Nat Microbiol. 2018;3:38–46. doi: 10.1038/s41564-017-0053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan FP. Human endogenous retroviruses in health and disease: a symbiotic perspective. J R Soc Med. 2004;97:560–565. doi: 10.1258/jrsm.97.12.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurst TP, Magiorkinis G. Activation of the innate immune response by endogenous retroviruses. J Gen Virol. 2015;96:1207–1218. doi: 10.1099/jgv.0.000017. [DOI] [PubMed] [Google Scholar]
- 14.Stauffer Y, Marguerat S, Meylan F, Ucla C, Sutkowski N, Huber B, Pelet T, Conrad B. Interferon-alpha-induced endogenous superantigen. a model linking environment and autoimmunity. Immunity. 2001;15:591–601. doi: 10.1016/s1074-7613(01)00212-6. [DOI] [PubMed] [Google Scholar]
- 15.Goff SP. Operating under a Gag order: a block against incoming virus by the Fv1 gene. Cell. 1996;86:691–693. doi: 10.1016/s0092-8674(00)80141-5. [DOI] [PubMed] [Google Scholar]
- 16.Kassiotis G, Stoye JP. Making a virtue of necessity: the pleiotropic role of human endogenous retroviruses in cancer. Philos Trans R Soc Lond B Biol Sci. 2017:372. doi: 10.1098/rstb.2016.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SW, Hibberd ML, Liu ET, Rohwer F, Ruan Y. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadwell K. The virome in host health and disease. Immunity. 2015;42:805–813. doi: 10.1016/j.immuni.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 20.Popgeorgiev N, Temmam S, Raoult D, Desnues C. Describing the silent human virome with an emphasis on giant viruses. Intervirology. 2013;56:395–412. doi: 10.1159/000354561. [DOI] [PubMed] [Google Scholar]
- 21.Xu GJ, Kula T, Xu Q, Li MZ, Vernon SD, Ndung’u T, Ruxrungtham K, Sanchez J, Brander C, Chung RT, O’Connor KC, Walker B, Larman HB, Elledge SJ. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348:aaa0698. doi: 10.1126/science.aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D, Holtz LR. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramanan D, Cadwell K. Intrinsic Defense Mechanisms of the Intestinal Epithelium. Cell Host Microbe. 2016;19:434–441. doi: 10.1016/j.chom.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metzger RN, Krug AB, Eisenacher K. Enteric Virome Sensing-Its Role in Intestinal Homeostasis and Immunity. Viruses. 2018;10:146. doi: 10.3390/v10040146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broquet AH, Hirata Y, McAllister CS, Kagnoff MF. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J Immunol. 2011;186:1618–1626. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 26.McCartney SA, Thackray LB, Gitlin L, Gilfillan S, Virgin HW, Colonna M. MDA-5 recognition of a murine norovirus. PLoS Pathog. 2008;4:e1000108. doi: 10.1371/journal.ppat.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abt MC, Buffie CG, Susac B, Becattini S, Carter RA, Leiner I, Keith JW, Artis D, Osborne LC, Pamer EG. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci Transl Med. 2016;8:327ra325. doi: 10.1126/scitranslmed.aad6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang P, Zhu S, Yang L, Cui S, Pan W, Jackson R, Zheng Y, Rongvaux A, Sun Q, Yang G, Gao S, Lin R, You F, Flavell R, Fikrig E. Nlrp6 regulates intestinal antiviral innate immunity. Science. 2015;350:826–830. doi: 10.1126/science.aab3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, Wilen CB, Orvedahl A, McCune BT, Kim KW, Orchard RC, Peterson ST, Nice TJ, Baldridge MT, Virgin HW. Norovirus Cell Tropism Is Determined by Combinatorial Action of a Viral Non-structural Protein and Host Cytokine. Cell Host Microbe. 2017;22:449–459 e444. doi: 10.1016/j.chom.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilen CB, Lee S, Hsieh LL, Orchard RC, Desai C, Hykes BL, Jr, McAllaster MR, Balce DR, Feehley T, Brestoff JR, Hickey CA, Yokoyama CC, Wang YT, MacDuff DA, Kreamalmayer D, Howitt MR, Neil JA, Cadwell K, Allen PM, Handley SA, van Lookeren Campagne M, Baldridge MT, Virgin HW. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science. 2018;360:204–208. doi: 10.1126/science.aar3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grau KR, Roth AN, Zhu S, Hernandez A, Colliou N, DiVita BB, Philip DT, Riffe C, Giasson B, Wallet SM, Mohamadzadeh M, Karst SM. The major targets of acute norovirus infection are immune cells in the gut-associated lymphoid tissue. Nat Microbiol. 2017;2:1586–1591. doi: 10.1038/s41564-017-0057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, Tibbetts SA, Wallet SM, Karst SM. Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 2014;346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu S, Ding S, Wang P, Wei Z, Pan W, Palm NW, Yang Y, Yu H, Li HB, Wang G, Lei X, de Zoete MR, Zhao J, Zheng Y, Chen H, Zhao Y, Jurado KA, Feng N, Shan L, Kluger Y, Lu J, Abraham C, Fikrig E, Greenberg HB, Flavell RA. Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature. 2017;546:667–670. doi: 10.1038/nature22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingle H, Peterson ST, Baldridge MT. Distinct Effects of Type I and III Interferons on Enteric Viruses. Viruses. 2018;10:46. doi: 10.3390/v10010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pott J, Stockinger S. Type I and III Interferon in the Gut: Tight Balance between Host Protection and Immunopathology. Front Immunol. 2017;8:258. doi: 10.3389/fimmu.2017.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez PP, Mahlakoiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F, Gronke K, Ryffel B, Hoelscher C, Dumoutier L, Renauld JC, Suerbaum S, Staeheli P, Diefenbach A. Interferon-lambda and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol. 2015;16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baldridge MT, Lee S, Brown JJ, McAllister N, Urbanek K, Dermody TS, Nice TJ, Virgin HW. Expression of Ifnlr1 on Intestinal Epithelial Cells Is Critical to the Antiviral Effects of Interferon Lambda against Norovirus and Reovirus. J Virol. 2017;91:e02079–16. doi: 10.1128/JVI.02079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwasaki A, Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chachu KA, LoBue AD, Strong DW, Baric RS, Virgin HW. Immune mechanisms responsible for vaccination against and clearance of mucosal and lymphatic norovirus infection. PLoS Pathog. 2008;4:e1000236. doi: 10.1371/journal.ppat.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chachu KA, Strong DW, LoBue AD, Wobus CE, Baric RS, Virgin HWT. Antibody is critical for the clearance of murine norovirus infection. J Virol. 2008;82:6610–6617. doi: 10.1128/JVI.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokoyama CC, Loh J, Zhao G, Stappenbeck TS, Wang D, Huang HV, Virgin HW, Thackray LB. Adaptive immunity restricts replication of novel murine astroviruses. J Virol. 2012;86:12262–12270. doi: 10.1128/JVI.02018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franco MA, Greenberg HB. Immunity to rotavirus infection in mice. J Infect Dis. 1999;179(Suppl 3):S466–469. doi: 10.1086/314805. [DOI] [PubMed] [Google Scholar]
- 43.Tomov VT, Osborne LC, Dolfi DV, Sonnenberg GF, Monticelli LA, Mansfield K, Virgin HW, Artis D, Wherry EJ. Persistent enteric murine norovirus infection is associated with functionally suboptimal virus-specific CD8 T cell responses. J Virol. 2013;87:7015–7031. doi: 10.1128/JVI.03389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomov VT, Palko O, Lau CW, Pattekar A, Sun Y, Tacheva R, Bengsch B, Manne S, Cosma GL, Eisenlohr LC, Nice TJ, Virgin HW, Wherry EJ. Differentiation and Protective Capacity of Virus-Specific CD8(+) T Cells Suggest Murine Norovirus Persistence in an Immune-Privileged Enteric Niche. Immunity. 2017;47:723–738 e725. doi: 10.1016/j.immuni.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westerman LE, McClure HM, Jiang B, Almond JW, Glass RI. Serum IgG mediates mucosal immunity against rotavirus infection. Proc Natl Acad Sci U S A. 2005;102:7268–7273. doi: 10.1073/pnas.0502437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang B, Chassaing B, Shi Z, Uchiyama R, Zhang Z, Denning TL, Crawford SE, Pruijssers AJ, Iskarpatyoti JA, Estes MK, Dermody TS, Ouyang W, Williams IR, Vijay-Kumar M, Gewirtz AT. Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science. 2014;346:861–865. doi: 10.1126/science.1256999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bok K, Green KY. Norovirus gastroenteritis in immunocompromised patients. N Engl J Med. 2012;367:2126–2132. doi: 10.1056/NEJMra1207742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz S, Vergoulidou M, Schreier E, Loddenkemper C, Reinwald M, Schmidt-Hieber M, Flegel WA, Thiel E, Schneider T. Norovirus gastroenteritis causes severe and lethal complications after chemotherapy and hematopoietic stem cell transplantation. Blood. 2011;117:5850–5856. doi: 10.1182/blood-2010-12-325886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nice TJ, Baldridge MT, McCune BT, Norman JM, Lazear HM, Artyomov M, Diamond MS, Virgin HW. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Z, Gewirtz AT. Together Forever: Bacterial-Viral Interactions in Infection and Immunity. Viruses. 2018;10 doi: 10.3390/v10030122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karst SM. The influence of commensal bacteria on infection with enteric viruses. Nat Rev Microbiol. 2016;14:197–204. doi: 10.1038/nrmicro.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson CM, Jesudhasan PR, Pfeiffer JK. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe. 2014;15:36–46. doi: 10.1016/j.chom.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, Pfeiffer JK. Bacteria Facilitate Enteric Virus Co-infection of Mammalian Cells and Promote Genetic Recombination. Cell Host Microbe. 2018;23:77–88 e75. doi: 10.1016/j.chom.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, Diamond MS, Ivanova Y, Artyomov M, Virgin HW. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science. 2015;347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uchiyama R, Chassaing B, Zhang B, Gewirtz AT. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J Infect Dis. 2014;210:171–182. doi: 10.1093/infdis/jiu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hernandez-Chirlaque C, Aranda CJ, Ocon B, Capitan-Canadas F, Ortega-Gonzalez M, Carrero JJ, Suarez MD, Zarzuelo A, Sanchez de Medina F, Martinez-Augustin O. Germ-free and Antibiotic-treated Mice are Highly Susceptible to Epithelial Injury in DSS Colitis. J Crohns Colitis. 2016;10:1324–1335. doi: 10.1093/ecco-jcc/jjw096. [DOI] [PubMed] [Google Scholar]
- 61.Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK, Tjota MY, Seo GY, Cao S, Theriault BR, Antonopoulos DA, Zhou L, Chang EB, Fu YX, Nagler CR. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herbst T, Sichelstiel A, Schar C, Yadava K, Burki K, Cahenzli J, McCoy K, Marsland BJ, Harris NL. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 63.Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 65.Li XD, Chiu YH, Ismail AS, Behrendt CL, Wight-Carter M, Hooper LV, Chen ZJ. Mitochondrial antiviral signaling protein (MAVS) monitors commensal bacteria and induces an immune response that prevents experimental colitis. Proc Natl Acad Sci U S A. 2011;108:17390–17395. doi: 10.1073/pnas.1107114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fischer JC, Bscheider M, Eisenkolb G, Lin CC, Wintges A, Otten V, Lindemans CA, Heidegger S, Rudelius M, Monette S, Porosnicu Rodriguez KA, Calafiore M, Liebermann S, Liu C, Lienenklaus S, Weiss S, Kalinke U, Ruland J, Peschel C, Shono Y, Docampo M, Velardi E, Jenq RR, Hanash AM, Dudakov JA, Haas T, van den Brink MRM, Poeck H. RIG-I/MAVS and STING signaling promote gut integrity during irradiation- and immune-mediated tissue injury. Sci Transl Med. 2017;9:eaag2513. doi: 10.1126/scitranslmed.aag2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ott SJ, Waetzig GH, Rehman A, Moltzau-Anderson J, Bharti R, Grasis JA, Cassidy L, Tholey A, Fickenscher H, Seegert D, Rosenstiel P, Schreiber S. Efficacy of Sterile Fecal Filtrate Transfer for Treating Patients With Clostridium difficile Infection. Gastroenterology. 2017;152:799–811 e797. doi: 10.1053/j.gastro.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 68.Zuo T, Wong SH, Lam K, Lui R, Cheung K, Tang W, Ching JYL, Chan PKS, Chan MCW, Wu JCY, Chan FKL, Yu J, Sung JJY, Ng SC. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2017;67:634–643. doi: 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thepaut M, Grandjean T, Hober D, Lobert PE, Bortolotti P, Faure K, Dessein R, Kipnis E, Guery B. Protective role of murine norovirus against Pseudomonas aeruginosa acute pneumonia. Vet Res. 2015;46:91. doi: 10.1186/s13567-015-0239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Phillips G, Tam CC, Rodrigues LC, Lopman B. Prevalence and characteristics of asymptomatic norovirus infection in the community in England. Epidemiol Infect. 2010;138:1454–1458. doi: 10.1017/S0950268810000439. [DOI] [PubMed] [Google Scholar]
- 71.Yang JY, Kim MS, Kim E, Cheon JH, Lee YS, Kim Y, Lee SH, Seo SU, Shin SH, Choi SS, Kim B, Chang SY, Ko HJ, Bae JW, Kweon MN. Enteric Viruses Ameliorate Gut Inflammation via Toll-like Receptor 3 and Toll-like Receptor 7-Mediated Interferon-beta Production. Immunity. 2016;44:889–900. doi: 10.1016/j.immuni.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 72.Sun L, Miyoshi H, Origanti S, Nice TJ, Barger AC, Manieri NA, Fogel LA, French AR, Piwnica-Worms D, Piwnica-Worms H, Virgin HW, Lenschow DJ, Stappenbeck TS. Type I interferons link viral infection to enhanced epithelial turnover and repair. Cell Host Microbe. 2015;17:85–97. doi: 10.1016/j.chom.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cammarota G, Cuoco L, Cianci R, Pandolfi F, Gasbarrini G. Onset of coeliac disease during treatment with interferon for chronic hepatitis C. Lancet. 2000;356:1494–1495. doi: 10.1016/S0140-6736(00)02880-4. [DOI] [PubMed] [Google Scholar]
- 74.Di Sabatino A, Pickard KM, Gordon JN, Salvati V, Mazzarella G, Beattie RM, Vossenkaemper A, Rovedatti L, Leakey NA, Croft NM, Troncone R, Corazza GR, Stagg AJ, Monteleone G, MacDonald TT. Evidence for the role of interferon-alfa production by dendritic cells in the Th1 response in celiac disease. Gastroenterology. 2007;133:1175–1187. doi: 10.1053/j.gastro.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 75.Lerner A, Arleevskaya M, Schmiedl A, Matthias T. Microbes and Viruses Are Bugging the Gut in Celiac Disease. Are They Friends or Foes? Front Microbiol. 2017;8:1392. doi: 10.3389/fmicb.2017.01392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bouziat R, Hinterleitner R, Brown JJ, Stencel-Baerenwald JE, Ikizler M, Mayassi T, Meisel M, Kim SM, Discepolo V, Pruijssers AJ, Ernest JD, Iskarpatyoti JA, Costes LM, Lawrence I, Palanski BA, Varma M, Zurenski MA, Khomandiak S, McAllister N, Aravamudhan P, Boehme KW, Hu F, Samsom JN, Reinecker HC, Kupfer SS, Guandalini S, Semrad CE, Abadie V, Khosla C, Barreiro LB, Xavier RJ, Ng A, Dermody TS, Jabri B. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science. 2017;356:44–50. doi: 10.1126/science.aah5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843–854. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HWT. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, Glickman JN, Tschurtschenthaler M, Hartwig J, Hosomi S, Flak MB, Cusick JL, Kohno K, Iwawaki T, Billmann-Born S, Raine T, Bharti R, Lucius R, Kweon MN, Marciniak SJ, Choi A, Hagen SJ, Schreiber S, Rosenstiel P, Kaser A, Blumberg RS. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–276. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, Hubbard-Lucey VM, Cammer M, Neil J, Dewan MZ, Lieberman SR, Lazrak A, Marinis JM, Beal A, Harris PA, Bertin J, Liu C, Ding Y, van den Brink MRM, Cadwell K. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med. 2017;214:3687–3705. doi: 10.1084/jem.20170558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lencioni KC, Seamons A, Treuting PM, Maggio-Price L, Brabb T. Murine norovirus: an intercurrent variable in a mouse model of bacteria-induced inflammatory bowel disease. Comp Med. 2008;58:522–533. [PMC free article] [PubMed] [Google Scholar]
- 83.Basic M, Keubler LM, Buettner M, Achard M, Breves G, Schroder B, Smoczek A, Jorns A, Wedekind D, Zschemisch NH, Gunther C, Neumann D, Lienenklaus S, Weiss S, Hornef MW, Mahler M, Bleich A. Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm Bowel Dis. 2014;20:431–443. doi: 10.1097/01.MIB.0000441346.86827.ed. [DOI] [PubMed] [Google Scholar]
- 84.Hubbard VM, Cadwell K. Viruses, autophagy genes, and Crohn’s disease. Viruses. 2011;3:1281–1311. doi: 10.3390/v3071281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nystrom N, Berg T, Lundin E, Skog O, Hansson I, Frisk G, Juko-Pecirep I, Nilsson M, Gyllensten U, Finkel Y, Fuxe J, Wanders A. Human enterovirus species B in ileocecal Crohn’s disease. Clin Transl Gastroenterol. 2013;4:e38. doi: 10.1038/ctg.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, Stappenbeck TS, McGovern DP, Keshavarzian A, Mutlu EA, Sauk J, Gevers D, Xavier RJ, Wang D, Parkes M, Virgin HW. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, Li X, Szeto CH, Sugimura N, Yuen-Tung Lam T, Chi-Shing Yu A, Wang X, Chen Z, Chi-Sang Wong M, Ng SC, Chan MTV, Chan PKS, Leung Chan FK, Jao-Yiu Sung J, Yu J. Alterations in Enteric Virome Associate With Colorectal Cancer and Survival Outcomes. Gastroenterology. 2018 doi: 10.1053/j.gastro.2018.1004.1018. [DOI] [PubMed] [Google Scholar]
- 88.Legoff J, Resche-Rigon M, Bouquet J, Robin M, Naccache SN, Mercier-Delarue S, Federman S, Samayoa E, Rousseau C, Piron P, Kapel N, Simon F, Socie G, Chiu CY. The eukaryotic gut virome in hematopoietic stem cell transplantation: new clues in enteric graft-versus-host disease. Nat Med. 2017;23:1080–1085. doi: 10.1038/nm.4380. [DOI] [PubMed] [Google Scholar]
- 89.Turley SJ, Lee JW, Dutton-Swain N, Mathis D, Benoist C. Endocrine self and gut non-self intersect in the pancreatic lymph nodes. Proc Natl Acad Sci U S A. 2005;102:17729–17733. doi: 10.1073/pnas.0509006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smyth DJ, Cooper JD, Bailey R, Field S, Burren O, Smink LJ, Guja C, Ionescu-Tirgoviste C, Widmer B, Dunger DB, Savage DA, Walker NM, Clayton DG, Todd JA. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat Genet. 2006;38:617–619. doi: 10.1038/ng1800. [DOI] [PubMed] [Google Scholar]
- 91.Jean-Baptiste VSE, Xia CQ, Clare-Salzler MJ, Horwitz MS. Type 1 Diabetes and Type 1 Interferonopathies: Localization of a Type 1 Common Thread of Virus Infection in the Pancreas. EBioMedicine. 2017;22:10–17. doi: 10.1016/j.ebiom.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Laitinen OH, Honkanen H, Pakkanen O, Oikarinen S, Hankaniemi MM, Huhtala H, Ruokoranta T, Lecouturier V, Andre P, Harju R, Virtanen SM, Lehtonen J, Almond JW, Simell T, Simell O, Ilonen J, Veijola R, Knip M, Hyoty H. Coxsackievirus B1 is associated with induction of beta-cell autoimmunity that portends type 1 diabetes. Diabetes. 2014;63:446–455. doi: 10.2337/db13-0619. [DOI] [PubMed] [Google Scholar]
- 93.Stone VM, Hankaniemi MM, Svedin E, Sioofy-Khojine A, Oikarinen S, Hyoty H, Laitinen OH, Hytonen VP, Flodstrom-Tullberg M. A Coxsackievirus B vaccine protects against virus-induced diabetes in an experimental mouse model of type 1 diabetes. Diabetologia. 2018;61:476–481. doi: 10.1007/s00125-017-4492-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Honeyman MC, Coulson BS, Stone NL, Gellert SA, Goldwater PN, Steele CE, Couper JJ, Tait BD, Colman PG, Harrison LC. Association between rotavirus infection and pancreatic islet autoimmunity in children at risk of developing type 1 diabetes. Diabetes. 2000;49:1319–1324. doi: 10.2337/diabetes.49.8.1319. [DOI] [PubMed] [Google Scholar]
- 95.Graham KL, Sanders N, Tan Y, Allison J, Kay TW, Coulson BS. Rotavirus infection accelerates type 1 diabetes in mice with established insulitis. J Virol. 2008;82:6139–6149. doi: 10.1128/JVI.00597-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pane JA, Webster NL, Coulson BS. Rotavirus activates lymphocytes from non-obese diabetic mice by triggering toll-like receptor 7 signaling and interferon production in plasmacytoid dendritic cells. PLoS Pathog. 2014;10:e1003998. doi: 10.1371/journal.ppat.1003998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pane JA, Fleming FE, Graham KL, Thomas HE, Kay TW, Coulson BS. Rotavirus acceleration of type 1 diabetes in non-obese diabetic mice depends on type I interferon signalling. Sci Rep. 2016;6:29697. doi: 10.1038/srep29697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pane JA, Webster NL, Graham KL, Holloway G, Zufferey C, Coulson BS. Rotavirus acceleration of murine type 1 diabetes is associated with a T helper 1-dependent specific serum antibody response and virus effects in regional lymph nodes. Diabetologia. 2013;56:573–582. doi: 10.1007/s00125-012-2798-4. [DOI] [PubMed] [Google Scholar]
- 99.Graham KL, O’Donnell JA, Tan Y, Sanders N, Carrington EM, Allison J, Coulson BS. Rotavirus infection of infant and young adult nonobese diabetic mice involves extraintestinal spread and delays diabetes onset. J Virol. 2007;81:6446–6458. doi: 10.1128/JVI.00205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wetzel JD, Barton ES, Chappell JD, Baer GS, Mochow-Grundy M, Rodgers SE, Shyr Y, Powers AC, Thomas JW, Dermody TS. Reovirus delays diabetes onset but does not prevent insulitis in nonobese diabetic mice. J Virol. 2006;80:3078–3082. doi: 10.1128/JVI.80.6.3078-3082.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhao G, Vatanen T, Droit L, Park A, Kostic AD, Poon TW, Vlamakis H, Siljander H, Harkonen T, Hamalainen AM, Peet A, Tillmann V, Ilonen J, Wang D, Knip M, Xavier RJ, Virgin HW. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc Natl Acad Sci U S A. 2017;114:E6166–E6175. doi: 10.1073/pnas.1706359114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pearson JA, Wong FS, Wen L. The importance of the Non Obese Diabetic (NOD) mouse model in autoimmune diabetes. J Autoimmun. 2016;66:76–88. doi: 10.1016/j.jaut.2015.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brenchley JM, Douek DC. HIV infection and the gastrointestinal immune system. Mucosal Immunol. 2008;1:23–30. doi: 10.1038/mi.2007.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Monaco CL, Gootenberg DB, Zhao G, Handley SA, Ghebremichael MS, Lim ES, Lankowski A, Baldridge MT, Wilen CB, Flagg M, Norman JM, Keller BC, Luevano JM, Wang D, Boum Y, Martin JN, Hunt PW, Bangsberg DR, Siedner MJ, Kwon DS, Virgin HW. Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome. Cell Host Microbe. 2016;19:311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, Stanley K, Kramer J, Macri SC, Permar SR, Schmitz JE, Mansfield K, Brenchley JM, Veazey RS, Stappenbeck TS, Wang D, Barouch DH, Virgin HW. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 107.Polakos NK, Cornejo JC, Murray DA, Wright KO, Treanor JJ, Crispe IN, Topham DJ, Pierce RH. Kupffer cell-dependent hepatitis occurs during influenza infection. Am J Pathol. 2006;168:1169–1178. doi: 10.2353/ajpath.2006.050875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang J, Li F, Wei H, Lian ZX, Sun R, Tian Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J Exp Med. 2014;211:2397–2410. doi: 10.1084/jem.20140625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin HWT. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 110.Furman D, Jojic V, Sharma S, Shen-Orr SS, Angel CJ, Onengut-Gumuscu S, Kidd BA, Maecker HT, Concannon P, Dekker CL, Thomas PG, Davis MM. Cytomegalovirus infection enhances the immune response to influenza. Sci Transl Med. 2015;7:281ra243. doi: 10.1126/scitranslmed.aaa2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.White DW, Keppel CR, Schneider SE, Reese TA, Coder J, Payton JE, Ley TJ, Virgin HW, Fehniger TA. Latent herpesvirus infection arms NK cells. Blood. 2010;115:4377–4383. doi: 10.1182/blood-2009-09-245464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.MacDuff DA, Reese TA, Kimmey JM, Weiss LA, Song C, Zhang X, Kambal A, Duan E, Carrero JA, Boisson B, Laplantine E, Israel A, Picard C, Colonna M, Edelson BT, Sibley LD, Stallings CL, Casanova JL, Iwai K, Virgin HW. Phenotypic complementation of genetic immunodeficiency by chronic herpesvirus infection. Elife. 2015;4 doi: 10.7554/eLife.04494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Machiels B, Dourcy M, Xiao X, Javaux J, Mesnil C, Sabatel C, Desmecht D, Lallemand F, Martinive P, Hammad H, Guilliams M, Dewals B, Vanderplasschen A, Lambrecht BN, Bureau F, Gillet L. A gammaherpesvirus provides protection against allergic asthma by inducing the replacement of resident alveolar macrophages with regulatory monocytes. Nat Immunol. 2017;18:1310–1320. doi: 10.1038/ni.3857. [DOI] [PubMed] [Google Scholar]
- 114.Brodin P, Jojic V, Gao T, Bhattacharya S, Angel CJ, Furman D, Shen-Orr S, Dekker CL, Swan GE, Butte AJ, Maecker HT, Davis MM. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moustafa A, Xie C, Kirkness E, Biggs W, Wong E, Turpaz Y, Bloom K, Delwart E, Nelson KE, Venter JC, Telenti A. The blood DNA virome in 8,000 humans. PLoS Pathog. 2017;13:e1006292. doi: 10.1371/journal.ppat.1006292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bernardin F, Operskalski E, Busch M, Delwart E. Transfusion transmission of highly prevalent commensal human viruses. Transfusion. 2010;50:2474–2483. doi: 10.1111/j.1537-2995.2010.02699.x. [DOI] [PubMed] [Google Scholar]
- 117.Williams CF, Klinzman D, Yamashita TE, Xiang J, Polgreen PM, Rinaldo C, Liu C, Phair J, Margolick JB, Zdunek D, Hess G, Stapleton JT. Persistent GB virus C infection and survival in HIV-infected men. N Engl J Med. 2004;350:981–990. doi: 10.1056/NEJMoa030107. [DOI] [PubMed] [Google Scholar]
- 118.Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goffard A, Lambert V, Salleron J, Herwegh S, Engelmann I, Pinel C, Pin I, Perrez T, Prevotat A, Dewilde A, Delhaes L. Virus and cystic fibrosis: rhinoviruses are associated with exacerbations in adult patients. J Clin Virol. 2014;60:147–153. doi: 10.1016/j.jcv.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dijkema JS, van Ewijk BE, Wilbrink B, Wolfs TF, Kimpen JL, van der Ent CK. Frequency and Duration of Rhinovirus Infections in Children With Cystic Fibrosis and Healthy Controls: A Longitudinal Cohort Study. Pediatr Infect Dis J. 2016;35:379–383. doi: 10.1097/INF.0000000000001014. [DOI] [PubMed] [Google Scholar]
- 121.Cousin M, Molinari N, Foulongne V, Caimmi D, Vachier I, Abely M, Chiron R. Rhinovirus-associated pulmonary exacerbations show a lack of FEV1 improvement in children with cystic fibrosis. Influenza Other Respir Viruses. 2016;10:109–112. doi: 10.1111/irv.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Edwards MR, Strong K, Cameron A, Walton RP, Jackson DJ, Johnston SL. Viral infections in allergy and immunology: How allergic inflammation influences viral infections and illness. J Allergy Clin Immunol. 2017;140:909–920. doi: 10.1016/j.jaci.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bizzintino J, Lee WM, Laing IA, Vang F, Pappas T, Zhang G, Martin AC, Khoo SK, Cox DW, Geelhoed GC, McMinn PC, Goldblatt J, Gern JE, Le Souef PN. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011;37:1037–1042. doi: 10.1183/09031936.00092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, Palmenberg AC, Gern JE. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci U S A. 2015;112:5485–5490. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Palmenberg AC. Rhinovirus C, Asthma, and Cell Surface Expression of Virus Receptor CDHR3. J Virol. 2017;91:e00072–17. doi: 10.1128/JVI.00072-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Currier RL, Payne DC, Staat MA, Selvarangan R, Shirley SH, Halasa N, Boom JA, Englund JA, Szilagyi PG, Harrison CJ, Klein EJ, Weinberg GA, Wikswo ME, Parashar U, Vinje J, Morrow AL. Innate Susceptibility to Norovirus Infections Influenced by FUT2 Genotype in a United States Pediatric Population. Clin Infect Dis. 2015;60:1631–1638. doi: 10.1093/cid/civ165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lopman BA, Trivedi T, Vicuna Y, Costantini V, Collins N, Gregoricus N, Parashar U, Sandoval C, Broncano N, Vaca M, Chico ME, Vinje J, Cooper PJ. Norovirus Infection and Disease in an Ecuadorian Birth Cohort: Association of Certain Norovirus Genotypes With Host FUT2 Secretor Status. J Infect Dis. 2015;211:1813–1821. doi: 10.1093/infdis/jiu672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Briese T, Kapoor A, Mishra N, Jain K, Kumar A, Jabado OJ, Lipkin WI. Virome Capture Sequencing Enables Sensitive Viral Diagnosis and Comprehensive Virome Analysis. MBio. 2015;6:e01491–01415. doi: 10.1128/mBio.01491-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov. 2015;14:642–662. doi: 10.1038/nrd4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Samson A, Scott KJ, Taggart D, West EJ, Wilson E, Nuovo GJ, Thomson S, Corns R, Mathew RK, Fuller MJ, Kottke TJ, Thompson JM, Ilett EJ, Cockle JV, van Hille P, Sivakumar G, Polson ES, Turnbull SJ, Appleton ES, Migneco G, Rose AS, Coffey MC, Beirne DA, Collinson FJ, Ralph C, Alan Anthoney D, Twelves CJ, Furness AJ, Quezada SA, Wurdak H, Errington-Mais F, Pandha H, Harrington KJ, Selby PJ, Vile RG, Griffin SD, Stead LF, Short SC, Melcher AA. Intravenous delivery of oncolytic reovirus to brain tumor patients immunologically primes for subsequent checkpoint blockade. Sci Transl Med. 2018;10:eaam7577. doi: 10.1126/scitranslmed.aam7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bourgeois-Daigneault MC, Roy DG, Aitken AS, El Sayes N, Martin NT, Varette O, Falls T, St-Germain LE, Pelin A, Lichty BD, Stojdl DF, Ungerechts G, Diallo JS, Bell JC. Neoadjuvant oncolytic virotherapy before surgery sensitizes triple-negative breast cancer to immune checkpoint therapy. Sci Transl Med. 2018;10:eaao1641. doi: 10.1126/scitranslmed.aao1641. [DOI] [PubMed] [Google Scholar]


