Abstract
Surface-enhanced Raman spectroscopy (SERS) was developed here for the non-invasive detection of the hepatitis B virus (HBV). Chronic hepatitis B virus (HBV) infection is a primary health problem in the world and may further develop into cirrhosis and hepatocellular carcinoma (HCC). SERS measurement was applied to two groups of serum samples. One group included 93 HBV patients and the other group included 94 healthy volunteers as control subjects. Tentative assignments of the Raman bands in the measured SERS spectra have shown the difference of the serum SERS spectra between HBV patients and healthy volunteers. The differences indicated an increase in the relative amounts of L-arginine, Saccharide band (overlaps with acyl band), phenylalanine and tyrosine, together with a decrease in the percentage of nucleic acid, valine and hypoxanthine in the serum of HBV patients compared with those of healthy volunteers. For better analysis of the spectral data, the first-order derivation was applied to the SERS data. Furthermore, principal components analysis (PCA), combined with linear discriminant analysis (LDA), were employed to distinguish HBV patients from healthy volunteers and to realize the diagnostic sensitivity of 78.5% and 91.4%, and specificity of 75% and 83% for SERS and the first order derivative SERS spectrum, respectively. These results suggest that derivative analysis could be an effective method to improve the classification of SERS spectra belonging to different groups. This exploratory work demonstrated that first-order derivative serum SERS spectrum combined with PCA-LDA has great potential for improving the detection of HBV.
1. Introduction
Chronic hepatitis B virus (HBV) infection is a serious global health problem and may further develop into cirrhosis and liver carcinoma. According to the report from World Health Organization (WHO) in 2015, around 240 million people were chronically infected with HBV worldwide [1]. HBV diagnosis is crucial for HBV control and prevention. HBV surface antigen (HBsAg) is a widely recognized serological marker and is usually employed for the detection of acute or chronic HBV infection. However the screening of blood or human organ donors and the supervision of individuals are at a risk of acquiring or transmitting HBV [2]. Moreover, there are still false-negative results using commercial HBV tests [3]. The primary infection in incubation period, low levels of the HBV carriers, S gene mutants and variants and HCV/HDV co-infection may affect HBV replication and/or HBsAg expression, leading to a failure of diagnosis [4] and subsequently an increased risk of HBV infection due to blood transfusion or liver transplantation. Therefore, the development of an alternative HBV detection technology has significant clinical value for accurate diagnosis of HBV and the safety of liver transplantation [5] or blood transfusion [6].
Raman spectroscopy (RS) based on molecular vibration of inelastic light scattering can detect and identify trace amount of chemical and biochemical species based on their unique vibrational signatures [7]. RS can provide fingerprinting type information on the structure and conformation of macromolecules. Furthermore, the distinct chemical and molecular features of biological molecules can be easily identified and quantified without additional marks [8]. The changes in these “molecular fingerprints” can provide biological information on the pathogenic aspects [9]. RS has been widely used in biomedical researches such as proteins, nucleic acids and other biological molecules related to important diseased infection and transformation [10–12]. Nevertheless, due to the inherently small cross section of Raman scattering and the strong auto fluorescence background, the weak Raman scattering signals are very hard to detect, which hinders its further clinical applications [13].
Fortunately, the surface-enhanced Raman spectroscopy (SERS) is capable of enhancing Raman signal by placing the interest molecules close proximity to the roughened surface of noble metal nanostructures [14]. The weak Raman signals can be enhanced by about 13 or even up to 15 orders of magnitude, which makes it an ultrasensitive technology even at single molecular level. The common view is that the electromagnetic field enhancement and the chemical enhancement, through surface plasmons and charge transfer, both contribute to SERS sensitivity [15]. SERS technique has been widely applied in biomedical studies including detection of DNA/RNA, protein, cell, tissue and body fluid such as serum/blood plasma and saliva samples. Recently, our group has published multiple studies on body fluid analysis based on SERS technology. For example, Lin et al. made use of serum SERS for the diagnosis of colorectal cancer [16]. Feng et al. studied blood plasma from gastric cancer patients based on SERS [17]. Wang et al. investigated the application of serum albumin and the globulin analysis for hepatocellular carcinoma detection [18]. However, there is few report on applying surface-enhanced Raman spectroscopy of human serum to assess the potential for HBV detection, which is very important for blood transfusion and liver transplantation. Moreover, if we analyze the serum SERS data directly as we did before, the statistical result will be unsatisfactory for discrimination HBV patients from normal group. Thus, the development of a more powerful data preprocessing methods that could help to identify Raman spectra belong to HBV patients would be of significant clinical value for blood SERS analysis. For HBV diagnosis based on SERS spectra, there have been some reports about HBV-DNA and HBV antibodies via label SERS detection [19–21], but these label methods are time consuming and more often require complex process.
For the first time, we explored the feasibility of a label-free method based on serum SERS spectroscopy with derivative preprocessing methods for HBV detection. Both SERS and first-order derivative SERS data were analyzed by PCA-LDA and the performance of these two data processing methods were compared. This preliminary work may offer a non-invasive, convenient and accurate diagnostic method for HBV detection.
2. Experiments and methods
2.1 Preparation of Ag NPs
Silver (Ag) nanoparticle solution was prepared using the process developed by Leopold and Lendl etc [22]. Briefly, 4.5 ml of sodium hydroxide (0.1 M) and 5 ml of hydroxylamine hydrochloride (0.06 M) are uniformly mixed. Then 90 mL of silver nitrate aqueous solution (0.0011 M) were immediately added to the mixture accompanied with intense stirring, until the solution turn gray color. The resulting colloid showed a milky gray color. The silver colloid can stabilize for one week. 1 ml silver nanoparticles solution was concentrated by centrifugation for 10000 rpm at 10 minutes, 0.95 ml supernatant was removed and the final concentration was used late on with the plasma samples. The concentration of silver nanoparticles was about 1.12 × 109 /ml. And the SERS enhancement factor of 4.42 × 106 can be calculated for Ag colloids. The same batch of silver was used in this research.
2.2 Preparation of the human serum sample
Human serum samples were collected from 187 individuals, including 93 patients which have been clinical confirmed with HBV and 94 healthy volunteers in the Quanzhou Blood Center. The patients and volunteers have signed an application in scientific research before blood sample collection and they have similar ethnic and socioeconomic backgrounds. After 12 hours of overnight fasting, a single peripheral blood samples were obtained from the study subjects between 7:00-10:00 A.M. with the use of coagulant. Blood cells were removed by centrifugation at 1000 rpm for 10 min to obtain the blood serum. Each of the serum samples was divided into two aliquots: 3mL for clinical test and another 3mL for SERS measurement. The 2.5 μL blood serum was mixed with the silver nanoparticle solution at 1:1 ratio. The pipette tip was used to create a mixture as homogeneous as possible for serum SERS on pure aluminum plate. The samples were measured immediately after natural drying at room temperature.
2.3 SERS measurement
A confocal Raman spectrometer (Renishaw, Great Britain) equipped with a 785 nm diode laser (laser power 100 mW) was used for measuring the SERS spectroscopy in the range of 400~1800 cm−1. The 520 cm−1 band of a silicon wafer was used for frequency calibration. The SERS spectrum of each serum samples was acquired within 10 s for integration using a microscope with a Leica 20 × objective. The incident power on the serum sample was approximately 2 mW. The Raman spectrometer resolution was 2 cm−1. The spectra were obtained from a sample in three different positions and the mean spectrum was used for further analysis. Moreover, the software package WIRE 2.0 was employed for SERS spectral acquisition and analysis. To get pure serum SERS spectra in the wave number of 400~1800 cm−1, the Vancouver Raman Algorithm was used to eliminate the signals of background noise and smooth from the raw SERS spectra [23]. Then all background-subtracted SERS spectra were normalized by the integrated area under the curve in the range of 400~1800 cm−1.
2.4 Statistical analysis
Principal component analysis (PCA) is a statistical technique for simplifying Raman spectral data sets and determining principal components (PCs) that best explain the differences in the observations. Linear discriminate analysis (LDA) determines the discriminant function line that maximizes the variance in the data between groups while minimizing the variance between members of the same group. The information of PCA of the serum SERS spectra is included in a finite set of PCs, which reduces the number of variables to be utilized in the LDA model. Then, an independent-sample t-test is utilized to identify the three most diagnostically significant PCs (P < = 0.05).
In bayesian statistics, the posterior probability of a random event or an uncertain proposition is the conditional probability that is assigned after the relevant evidence or background is taken into account. Similarly, the posterior probability distribution is the probability distribution of an unknown quantity, treated as a random variable, conditional on the evidence obtained from an experiment or survey. “Posterior”, in this context, means after taking into account the relevant evidence related to the particular case being examined. The posterior probability of classification results is applied to evaluate the discrimination of PCA-LDA algorithm.
The classification results of SERS data with PCA-LDA were estimated with the leave-one-out cross-validation (LOOCV) procedures. In LOOCV procedures, only one random sample is retained as the validation data for testing the model, and the remaining n-1 samples are used as training data. The cross-validation process is then repeated n times, with each of the n samples used exactly once as the validation data. The n results from the cross-validation (the folds) can then be averaged to produce a single estimation to measure the performance of the model. The advantage of this method is that almost all of the samples in each turn are used to train the model, so it is closest to the distribution of the original samples, so that the results are more reliable. Besides, there are no random factors that affect the experimental data and ensure that the experimental process can be replicated. LOOCV was automated in the PCA-LDA process. PCA-LDA statistic method was performed by SPSS 13.0 software package (SPSS Inc Chicago). The PCA-LDA statistic method for SERS spectra have already become a promising tool in biomedical research [24,25].
Derivatives have the capability to extract subtle spectral features and have been used in spectroscopy analysis for decades. Derivative spectrum is an analytical tool which differentiates normal spectrum by mathematical transformation of spectral curve into a derivative (first or higher derivatives) and enhances the detectability of subtle spectral features. The method for data processing of derivative spectrum has already been reported in some aspects of UV spectra and near infrared spectra [26,27]. Moreover, the first-order derivative spectrum has been used to analyze Raman spectral of Chinese medicine, organic material and so on [28–30]. But the application of derivative spectrum in SERS spectra statistical analysis has not been reported. To improve the serum SERS diagnosis, the first-order derivation was applied to the data normalization of our SERS by the Matlab software. The detailed sequence of derivative processing is as follows: 1) spectra were eliminated the signals of background noise; 2) spectra were normalized by the integrated area under the curve; 3) the first derivative calculation of spectral data was performed; 4) spectra was smoothed by mean filtering algorithm. Mean filtering algorithm is a method which can be used to smooth the data and further eliminate the noise in Raman signal. The mean filtering algorithm firstly needs to define a range (r), then the mean filter filters data by replacing every value by the mean value in its range (r) neighborhood. These data processing methods can be conveniently implemented in the tool box corresponding to MATLAB software. The usage of a smoothed derivative spectrum in the statistical analysis can decrease the detrimental effect on the signal-to-noise ratio. Derivative spectrum is a useful technique in extracting information from overlapping bands of serum SERS spectra and exploring subtle spectral features.
3. Results
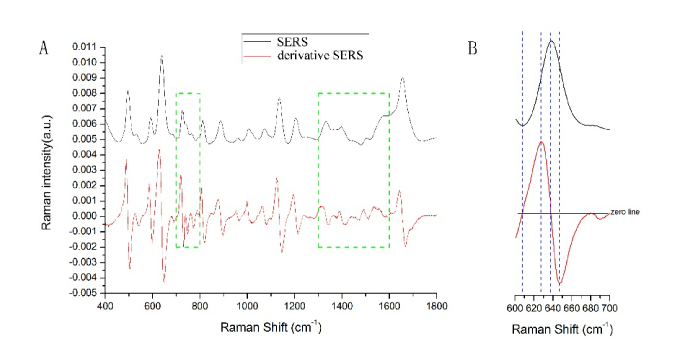
Figure 1(A) and 1(B) showed the comparison of SERS and first-order derivative SERS spectrum from a healthy volunteer in the range of 400~1800 cm−1 and amplification in the wave number of 600~700 cm−1. Among them, the black and red lines are SERS and derivative SERS spectrum zoomed in fifteen times, respectively. Derivative SERS spectrum consists of two zeros and positive, negative peaks corresponding to SERS spectrum on the peaks, troughs and two sides of the inflection point, which is the maximum slope, as shown in Fig. 1(B). Hence, we can observe that the number of spectral peaks in derivative SERS spectrum have obvious increased compared to SERS spectrum. Those weak spectral peaks hidden by stronger peaks became dateable by derivative SERS spectrum. So first-order derivative SERS spectra can be a beneficial complement for SERS spectra analysis. Especially, the increased spectral peaks of derivative SERS spectrum are more obvious in the two boxes marked with green dotted lines, as shown in Fig. 1(A). And some small SERS peaks hidden by strong bands were displayed after first-order derivatives. These results demonstrated that derivative SERS spectrum has some advantages over SERS spectrum for the classifications of HBV serum.
Fig. 1.
Representative serum SERS (black line) and derivative SERS (red line) of healthy volunteers in the range of 400~1800 cm−1 (A) and in the range of 600~700cm−1 (B). The increased spectral peaks of derivative SERS spectrum are obvious in the two boxes marked with green dotted lines.
3.1 SERS spectrum
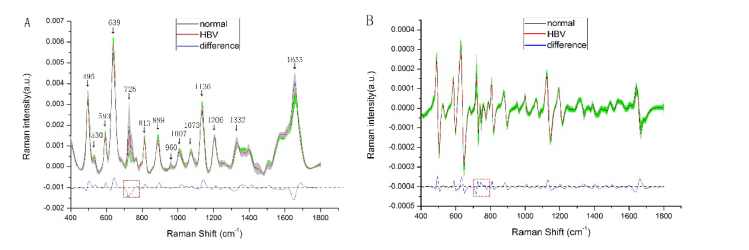
Figure 2(A) compares the mean SERS spectrum obtained from 94 serum samples of healthy volunteers (black line) and 93 HBV patients’ serum samples (red line). Shown at the top are the average normalized SERS spectra of healthy volunteers and HBV serum samples and the standard deviations was overlaid in the average spectra as shaded color fill. Shown at the bottom are their corresponding difference spectrum (HBV minus healthy) which are difference in normalized spectral intensity between HBV and healthy volunteers’ serum. It is clearly observed that SERS spectrum of the serum samples in healthy volunteers and HBV patients have obvious differences. The normalized intensities of HBV serum SERS peaks located at 495, 639, 813, 889, 1007, 1073, 1136 and 1206 cm−1 are greater than normal serum, while SERS bands at 725, 960, 1332 and 1655 cm−1 are lower in HBV serum samples. The most obvious differences can be found in the peaks at 495, 639, 725, 1136 and 1655 cm−1.
Fig. 2.
Comparison of the mean SERS spectrum (A) and first-order derivative SERS spectrum (B) of normal serum sample (black line, n = 94) and HBV serum sample (red line, n = 93). The shaded areas represent the standard deviations. Also shown at the bottom is the difference spectrum.
3.2 Derivative SERS spectrum
The average derivative SERS spectrum of serum sample of HBV group (red line, n = 93) and healthy volunteers group (black line, n = 94) are shown in Fig. 2(B), together with their difference spectra (blue line, HBV minus normal). Intensity standard deviations are reported as gray and green shaded areas. We can clearly observed that the difference of derivative SERS spectrum between HBV patients and healthy volunteers was significantly increased compared to that of SERS spectrum, due to the increase of spectral peaks and the appearance of negative peaks in derivative SERS spectrum. For example, the difference spectrum of derivative spectrum is changed from one negative peak (at 725 cm−1) in SERS spectrum to three negative peaks (at 721, 774, 799 cm−1) and two positive peaks (at 725, 747 cm−1) as shown in the red dashed box (in Fig. 2) of 700~800 cm−1.
3.3 PCA-LDA statistical analysis
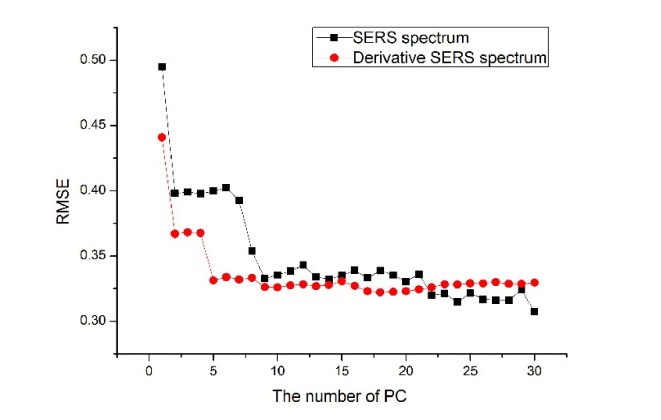
To test the classification ability of serum SERS between HBV patients and healthy volunteers, PCA combined with the LDA (PCA-LDA) multivariate statistical method was utilized to develop diagnostic algorithms. An independent-sample the test on all the PC scores showed that there are three most significant PCs (PC1, PC2 and PC5) that are diagnostically significant (P < 0.05) for discriminating normal and HBV groups in SERS spectrum and derivative SERS spectrum. The percent of variances of the three selected PCs were 27.2% for PC1, 16.2% for PC2, and 5.9% for PC5 calculated from the SERS data set. And the percent of variances of the three selected PCs were 15.9% for PC1, 9.1% for PC2, and 2.7% for PC5 calculated from the derivative SERS data set. This data processing method was applied to build diagnostic models and algorithms for serum SERS spectra. We can further draw the curve for the relationship between the number of PCs and the root mean square error of cross validation (RMSE) for correct classification of normal and HBV patients in Fig. 3. As shown in Fig. 3, with increasing the number of PCs, the RMSE decreased gradually, and the area of corresponding ROC curve was increased. When the PC number reached PC8 in SERS spectrum and PC5 in derivative SERS spectra, the RMS error tends to a stable minimum. However, to avoid overtraining problem, we only used the three most significant PCs (PC1, PC2 and PC5) for LDA analysis and ROC curve.
Fig. 3.
The relationship between the number of PC and the root mean square error of cross validation (RMSE) for correct classification of normal and HBV patients.
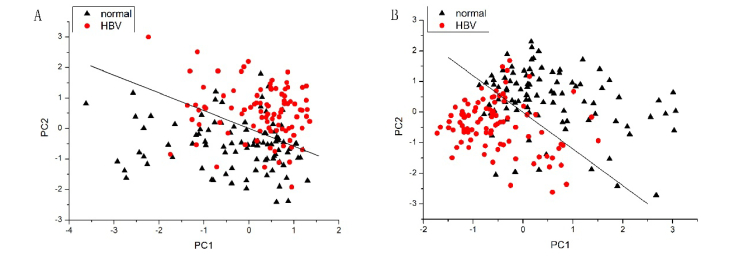
Direct comparisons of normal and HBV serum group based on SERS and derivative SERS data are shown in Figs. 4(A) and 4(B). The lines (PC2 = −0.58 PC1 + 0.0365) in Fig. 4(A) and another line (PC2 = −1.196 PC1-0.00549) in Fig. 4(B) as diagnostic algorithms classify HBV from normal serum samples. Normal and HBV serum are largely clustered into two separate groups based on the combinations of significant PCs (PC1 and PC2), and the corresponding separation lines differentiate HBV from a normal serum sample with the sensitivity of 75.3% (70/93), 84.9% (79/93) and the specificity of 77.7% (73/94), 80.8% (76/94) for SERS and derivative SERS data, respectively.
Fig. 4.
Scatter plots of the PCA result for HBV group (red circles) and the healthy volunteer group (black triangles) with (A) PC1, PC2 in SERS spectrum and (B) PC1, PC2 in first order derivative SERS spectrum as the two axes. The diagnostic lines (PC2 = −0.580 PC1 + 0.0365 in Fig. 4(A); PC2 = −1.196 PC1-0.00549 in Fig. 4(B)) classify HBV from normal serum sample with the sensitivity of 75.3% (70/93), 84.9% (79/93) and the specificity of 77.7% (73/94), 80.8% (76/94) for SERS and derivative SERS data, respectively.
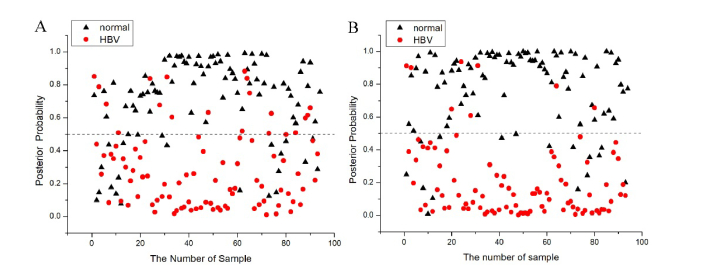
To further improve the diagnosis of SERS spectrum and derivative SERS spectrum, all the three most significant PCs (PC1, PC2 and PC5) of SERS spectrum and derivative SERS spectrum were loaded into the LDA model for serum classification. Figure 5 shows the cross-validated prediction results (posterior probabilities) belonging to normal (black triangles) and HBV (red circles) serum sample as calculated for (A) SERS spectrum and (B) derivative SERS spectrum, respectively. The classification results of serum SERS with PCA-LDA, estimated with the leave-one-out cross-validation procedures, showed that the diagnostic sensitivity and specificity for classifying HBV from the serum SERS analysis are 78.5%, 91.4% and 75.5%, 83.0%, respectively.
Fig. 5.
Scatter plots of the posterior probability of normal (black triangles) and HBV (red circles) serum sample as calculated for (A) SERS spectrum, (B) first-order derivative spectrum, respectively, using the PCA-LDA together with leave-one-out cross-validation method. The posterior probability corresponding to the dashed separation line is 0.5.
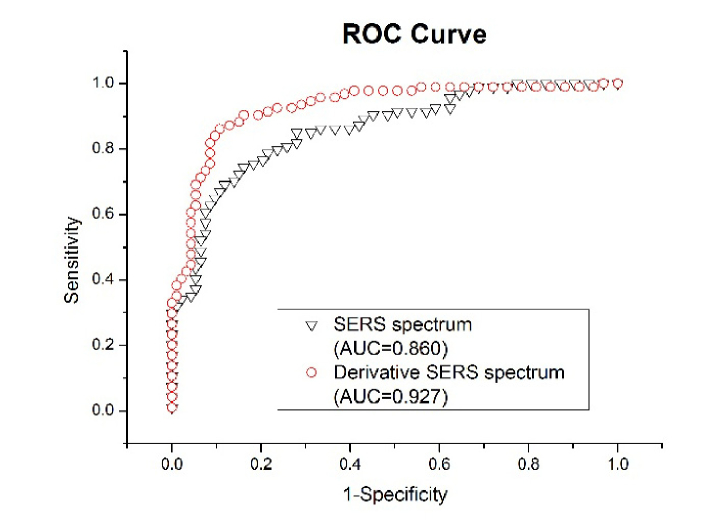
To estimate the performance of the PCA-LDA for HBV detection, receiver operating characteristic (ROC) curves were generated. Figure 6 shows that the receiver operating characteristic (ROC) curves of the discrimination result for the PCA-LDA based on the spectral classification of serum SERS spectrum and derivative SERS spectrum. The integration area under the ROC curves was 0.860 and 0.927 based on the PCA-LDA multivariate analysis for SERS and derivative SERS data, respectively. These results demonstrated that derivative SERS spectra can provide a better diagnostics accuracy than the routine SERS data.
Fig. 6.
The receiver operating characteristic (ROC) curve of the discrimination result for the PCA-LDA based on the spectral classification of SERS and first order derivative SERS spectrum.
4. Discussions
Figure 2(A) showed that SERS spectrum of serum are dominated by many vibrational modes of various biomolecules such as proteins, saccharide and nucleic acids, which may change in quantity and composition associated with HBV transformation and infection. To better understand the relationship between the molecular basis and SERS spectrum of human serum, the SERS bands are compared with known serum Raman bands. Table 1 presents tentative assignments for the observed SERS bands, according to the literature [16,17,24,31–36]. For example, the SERS bands of hypoxanthine (725 cm−1) showed lower signals for HBV patients compared to the normal.
Table 1. Peak positions and tentative assignments of major vibrational bands observed in serum samples.
| Peak position (cm−1) | Major assignments |
|---|---|
|
| |
| 495 | L-arginine ( + ) |
| 530 | Cholesterol ester |
| 639 | Tyrosine ring breathing ( + ) |
| 725 | Hypoxanthine (-) |
| 813 | C-C stretching (collagen assignment) |
| 889 | Saccharide band (overlaps with acyl band) ( + ) |
| 960 | ν (C-C) valine (-) |
| 1007 | Phenylalanine ( + ) |
| 1073 | Tyrosine ( + ) |
| 1136 | Phenylalanine ( + ) |
| 1206 | Tyrosine (collagen assignment) ( + ) |
| 1332 | Nucleic acid (-) |
| 1655 | Amide I, α-helix (-) |
( + ) indicates an increase and (-) indicates an decrease in spectra intensity.
The prominent SERS peak at 1655 cm−1 can be attributed to the amide I band of proteins in the α-helix conformation and human serum albumin (HSA) which is a principal extracellular transport protein. The SERS bands of L-arginine (493 cm−1) in the serum of HBV patients show higher percentage signals than those in normal serum, suggesting a increase in the percentage of certain amino acids contents relative to the total SERS active components in the serum of HBV patients. The SERS peak at 1332 cm−1 due to the C-H vibration of nucleic acid bases exhibited a lower signal in HBV serum, indicating a decrease in the percentage of the nucleic acid bases content relative to the total SERS active components in the serum of HBV patients. The SERS bands of saccharide (889 cm−1) are higher and the SERS band of valine (960 cm−1) is lower in HBV patient serum, due to the disorder of saccharide metabolism, and the process of glycolysis is inhibited. These findings agreed with previous studies [37,38]. The SERS peak at 1007 (phenylalanine), 1136 (phenylalanine), 639 (tyrosine), 1073 (tyrosine) and 1206 cm−1 (tyrosine) are higher in HBV serum sample compared to healthy serum sample, and this phenomenon has also been reported in some hepatocellular carcinoma cancer (HCC) studies based on SERS technology [35,39].
4.1 Statistical analysis for classification of HBV
To compare the performance of the two data processing methods, PCA-LDA was applied to the analysis of SERS and derivative SERS spectrum. PCA is a statistical method that can extract features of SERS spectra and single out the characteristic variables that represent the difference between HBV patients and healthy volunteer groups. Figure 4(A) and 4(B) showed the PC scores calculated from SERS and derivative SERS spectrum, respectively. We can see that they were distributed in separate areas except for a small overlap. Additionally, the effect of the separation was better in Fig. 4(B). Further, all the three significant PCs (PC1, PC2, PC5 in SERS spectrum and PC1, PC2, PC5 in derivative SERS spectrum) were used for performing LDA model. The diagnostic accuracy appears different between SERS and derivative spectrum, which are more clearly showed in Figs. 5(A) and 5(B). Utilizing a discrimination threshold of 0.5 in the PCA-LDA model, the diagnostic accuracy for detecting HBV is obviously higher in derivative SERS spectrum than in SERS spectrum. The diagnostic accuracy was increased by 10.2% after first-order derivative transformation in this work. Receiver operating characteristic analysis (Fig. 6) further confirms that derivative SERS spectrum was more robust and powerful than that of SERS spectrum in distinguishing HBV from normal serum sample integrated with the diagnostic algorithms (PCA-LDA).
The favorable discrimination results achieved using derivative SERS spectra might be explained as follows. On one hand, it should be noted that derivative SERS spectrum has negative peaks, which does not exist in SERS spectrum. As shown in Fig. 1(B), one spectral peak in SERS spectrum was converted into one positive and one negative peak after first-order derivative transformation. Some weak spectral peaks hidden by stronger Raman bands in SERS spectrum became clearly observed in derivative SERS spectrum. These made the information contained in derivative SERS spectrum more abundant than that of SERS spectra. Thus, some biochemical changes, which are very small changes reflected in SERS spectrum but closely related to HBV, are amplified and highlighted by the derivative method. On the other hand, the increasing spectral peaks number and the enhanced weak SERS peaks were also reflected in the difference spectrum in derivative SERS spectrum in Fig. 2(B). The peaks' numbers for normal and HBV serum SERS spectra increased in the derivative spectrum. The positive and negative peaks of the derivative spectrum corresponded to the two sides of one SERS peak. The information of the weak peaks and two sides of SERS spectral peaks may play important roles in classifying HBV from normal serum samples using first-order derivative SERS spectral analysis. But these advantages of derivative SERS spectrum cannot be achieved by SERS spectral analysis. So the derivative transformation is a useful pre-processing strategy for the analysis of SERS spectrum in biomedical research. This exploratory work indicates that derivative SERS spectrum combined with PCA-LDA analysis has tremendous potential for the label free detection of HBV.
5. Conclusions
The routine method in our hospital employs the COBAS AmpliPre/TaqMan PCR system and the matching reagent COBAS AmpliPre/COBAS TaqMan HBV Test V2.0 to detect the HBV DNA. The linear range for this test is 2.0 −1.7x108 IU/ml. The average test time is around 4 hours. The serum SERS methods just need 10 mins for each sample, so our method is faster than the routine method in our hospital. The comparison of Raman bands in the SERS spectrum suggests the differences of serum SERS spectrum between HBV patients and healthy volunteers, such as an increase of the relative amounts of L-arginine, Saccharide band (overlaps with acyl band), phenylalanine, tyrosine and a decrease of the percentage of nucleic acid, valine, hypoxanthine were showed in the serum of HBV patients compared with those of healthy volunteers. High diagnostic sensitivity of 91.4% and specificity of 83.0% can be achieved for differentiating HBV from normal samples based on derivative SERS spectra using the PCA-LDA diagnostic algorithm, which has almost a 10.2% improvement in diagnostic accuracy compared with SERS spectra. This improvement may be attributed to the ability of derivative SERS spectra for exploring subtle features in the spectra and extracting more information from overlapping Raman bands of serum sample. This exploratory work demonstrates that the derivative SERS spectrum analysis in conjuction with PCA-LDA has great potential for the diagnosis of HBV. This work may offer an alternative method for the label free and noninvasive detection of HBV patients.
Funding
National Natural Science Foundation of China (Nos. 61575043, and U1605253); The Scientific Research Innovation Team Construction Program of Fujian Normal University (No. IRTL1702); the program for Changjiang Scholars and Innovative Research Team in University (No. IRT1115); New Century Excellent Talent In Fujian Province University (J1-1160); Natural Science Foundation of Fujian Province of China (No. 2016J01292 and 2015J01436).
Disclosures
The authors declare that there are no conflicts of interest related to this article.
References and links
- 1.Leong J., Lin D., Nguyen M. H., “Hepatitis B surface antigen escape mutations: Indications for initiation of antiviral therapy revisited,” World J. Clin. Cases 4(3), 71–75 (2016). 10.12998/wjcc.v4.i3.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ly T. D., Servant-Delmas A., Bagot S., Gonzalo S., Férey M. P., Ebel A., Dussaix E., Laperche S., Roque-Afonso A. M., “Sensitivities of four new commercial hepatitis B virus surface antigen (HBsAg) assays in detection of HBsAg mutant forms,” J. Clin. Microbiol. 44(7), 2321–2326 (2006). 10.1128/JCM.00121-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber B., “Recent developments in the diagnosis and monitoring of HBV infection and role of the genetic variability of the S gene,” Expert Rev. Mol. Diagn. 5(1), 75–91 (2005). 10.1586/14737159.5.1.75 [DOI] [PubMed] [Google Scholar]
- 4.Yang L., Song L. W., Fang L. L., Wu Y., Ge S. X., Li H., Yuan Q., Zhang J., Xia N. S., “Evaluation of a novel chemiluminescent microplate enzyme immunoassay for hepatitis B surface antigen detection,” J. Virol. Methods 228, 55–59 (2016). 10.1016/j.jviromet.2015.11.013 [DOI] [PubMed] [Google Scholar]
- 5.Samuel D., Muller R., Alexander G., Fassati L., Ducot B., Benhamou J. P., Bismuth H., “Liver transplantation in European patients with the hepatitis B surface antigen,” N. Engl. J. Med. 329(25), 1842–1847 (1993). 10.1056/NEJM199312163292503 [DOI] [PubMed] [Google Scholar]
- 6.Babes V. T., Cajal N., Filipescu Z., Milcov V., “Accidental transfusion of blood containing HBsAg or anti-HBs,” Virologie 28(3), 173–178 (1977). [PubMed] [Google Scholar]
- 7.Chan J., Fore S., Wachsmann-Hogiu S., Huser T., “Raman spectroscopy and microscopy of individual cells and cellular components,” Laser Photonics Rev. 2(5), 325–349 (2008). 10.1002/lpor.200810012 [DOI] [Google Scholar]
- 8.Fenn M. B., Xanthopoulos P., Pyrgiotakis G., Grobmyer S. R., Pardalos P. M., Hench L. L., “Raman Spectroscopy for Clinical Oncology,” Adv. Opt. Technol. 2011, 213783 (2011). 10.1155/2011/213783 [DOI] [Google Scholar]
- 9.Nagarajan S., Sopanen M., “Non-destructive method for strain imaging in an individual GaN nanorod by confocal Raman technique,” J. Phys. D Appl. Phys. 49(46), 465303 (2016). 10.1088/0022-3727/49/46/465303 [DOI] [Google Scholar]
- 10.Sdobnov A. Y., Tuchin V. V., Lademann J., Darvin M. E., “Confocal Raman microscopy supported by optical clearing treatment of the skin—influence on collagen hydration,” J. Phys. D Appl. Phys. 50(28), 285401 (2017). 10.1088/1361-6463/aa77c9 [DOI] [Google Scholar]
- 11.Seto K., Okuda Y., Tokunaga E., Kobayashi T., “Multiplex stimulated Raman imaging with white probe-light from a photonic-crystal fibre and with multi-wavelength balanced detection,” J. Phys. D Appl. Phys. 47(34), 345401 (2014). 10.1088/0022-3727/47/34/345401 [DOI] [Google Scholar]
- 12.Varade V., Honnavar G. V., Anjaneyulu P., Kramesh K. P., Menon R., “Probing disorder and transport properties in polypyrrole thin-film devices by impedance and Raman spectroscopy,” J. Phys. D Appl. Phys. 46(36), 365306 (2013). 10.1088/0022-3727/46/36/365306 [DOI] [Google Scholar]
- 13.Feng S., Li Z., Chen G., Lin D., Huang S., Huang Z., Li Y., Lin J., Chen R., Zeng H., “Ultrasound-mediated method for rapid delivery of nano-particles into cells for intracellular surface-enhanced Raman spectroscopy and cancer cell screening,” Nanotechnology 26(6), 065101 (2015). 10.1088/0957-4484/26/6/065101 [DOI] [PubMed] [Google Scholar]
- 14.Lin J., Huang Z., Feng S., Lin J., Liu N., Wang J., Li L., Zeng Y., Li B., Zeng H., Chen R., “Label-free optical detection of type II diabetes based on surface-enhanced Raman spectroscopy and multivariate analysis,” J. Raman Spectrosc. 45(10), 884–889 (2014). 10.1002/jrs.4574 [DOI] [Google Scholar]
- 15.Çulha M., “Raman spectroscopy for cancer diagnosis: how far have we come?” Bioanalysis 7(21), 2813–2824 (2015). 10.4155/bio.15.190 [DOI] [PubMed] [Google Scholar]
- 16.Lin D., Feng S., Pan J., Chen Y., Lin J., Sun L., Chen R., “Diagnostic potential for gold nanoparticle-based surface-enhanced Raman spectroscopy to provide colorectal cancer screening using blood serum sample,” In Photonics and Optoelectronics Meetings 2011: International Society for Optics and Photonics, 83290L (2012). [Google Scholar]
- 17.Feng S., Pan J., Wu Y., Lin D., Chen Y., Xi G., Lin J., Chen R., “Study on gastric cancer blood plasma based on surface-enhanced Raman spectroscopy combined with multivariate analysis,” Sci. China Life Sci. 54(9), 828–834 (2011). 10.1007/s11427-011-4212-8 [DOI] [PubMed] [Google Scholar]
- 18.Wang J., Feng S., Lin J., Zeng Y., Li L., Huang Z., Li B., Zeng H., Chen R., “Serum albumin and globulin analysis for hepatocellular carcinoma detection avoiding false-negative results from alpha-fetoprotein test negative subjects,” Appl. Phys. Lett. 103(20), 204106 (2013). 10.1063/1.4830047 [DOI] [Google Scholar]
- 19.Kamińska A., Witkowska E., Winkler K., Dzięcielewski I., Weyher J. L., Waluk J., “Detection of Hepatitis B virus antigen from human blood: SERS immunoassay in a microfluidic system,” Biosens. Bioelectron. 66, 461–467 (2015). 10.1016/j.bios.2014.10.082 [DOI] [PubMed] [Google Scholar]
- 20.Li M., Cushing S. K., Liang H., Suri S., Ma D., Wu N., “Plasmonic nanorice antenna on triangle nanoarray for surface-enhanced Raman scattering detection of hepatitis B virus DNA,” Anal. Chem. 85(4), 2072–2078 (2013). 10.1021/ac303387a [DOI] [PubMed] [Google Scholar]
- 21.Yao C. K., Liao J. D., Chang C. W., Lin J. R., “Spatially reinforced nano-cavity array as the SERS-active substrate for detecting hepatitis virus core antigen at low concentrations,” Sens. Actuators B Chem. 174, 478–484 (2012). 10.1016/j.snb.2012.08.037 [DOI] [Google Scholar]
- 22.Leopold N., Lendl B., “A new method for fast preparation of highly surface-enhanced Raman scattering (SERS) active silver colloids at room temperature by reduction of silver nitrate with hydroxylamine hydrochloride,” J. Phys. Chem. B 107(24), 5723–5727 (2003). 10.1021/jp027460u [DOI] [Google Scholar]
- 23.Zhao J., Lui H., McLean D. I., Zeng H., “Automated autofluorescence background subtraction algorithm for biomedical Raman spectroscopy,” Appl. Spectrosc. 61(11), 1225–1232 (2007). 10.1366/000370207782597003 [DOI] [PubMed] [Google Scholar]
- 24.Lin D., Feng S., Pan J., Chen Y., Lin J., Chen G., Xie S., Zeng H., Chen R., “Colorectal cancer detection by gold nanoparticle based surface-enhanced Raman spectroscopy of blood serum and statistical analysis,” Opt. Express 19(14), 13565–13577 (2011). 10.1364/OE.19.013565 [DOI] [PubMed] [Google Scholar]
- 25.Ma J., Zhou H., Gong L., Liu S., Zhou Z., Mao W., Zheng R., “Distinction of gastric cancer tissue based on surface-enhanced Raman spectroscopy,” In Photonics Asia: International Society for Optics and Photonics, pp. 855328–855329 (2012). [Google Scholar]
- 26.Miclo L., Perrin E., Driou A., Mellet M., Linden G., “Determination of the ratios of the aromatic amino acid residues by first- or second-derivative UV spectrometry for a simple characterization of peptides,” Int. J. Pept. Protein Res. 46(2), 186–192 (1995). 10.1111/j.1399-3011.1995.tb01335.x [DOI] [PubMed] [Google Scholar]
- 27.Gallignani M., Garrigues S., de la Guardia M., “Direct determination of ethanol in all types of alcoholic beverages by near-infrared derivative spectrometry,” Analyst (Lond.) 118(9), 1167–1173 (1993). 10.1039/an9931801167 [DOI] [Google Scholar]
- 28.Lin W. S., Guo S. Z., Huang H., Chen R., Feng S. Y., Li Y. Z., Chen W. W., “[Infrared spectrum analysis of admixture decoction of herba Ephedrae with Ramulus cinnarnomi],” Guangpuxue Yu Guangpu Fenxi 29(7), 1847–1850 (2009). [PubMed] [Google Scholar]
- 29.Ma C., Xiao H., He L., “Surface‐enhanced Raman scattering characterization of monohydroxylated polymethoxyflavones,” J. Raman Spectrosc. 47(8), 901–907 (2016). 10.1002/jrs.4932 [DOI] [Google Scholar]
- 30.Meneghello M., Papadopoulou E., Ugo P., Bartlett P. N., “Using Electrochemical SERS to Measure the Redox Potential of Drug Molecules Bound to dsDNA—a Study of Mitoxantrone,” Electrochim. Acta 187, 684–692 (2016). 10.1016/j.electacta.2015.11.121 [DOI] [Google Scholar]
- 31.Movasaghi Z. M., Rehman S., Rehman I. U., “Raman spectroscopy of biological tissues,” Appl. Spectrosc. Rev. 50(1), 46–111 (2015). 10.1080/05704928.2014.923902 [DOI] [Google Scholar]
- 32.Austin L. A., Osseiran S., Evans C. L., “Raman technologies in cancer diagnostics,” Analyst (Lond.) 141(2), 476–503 (2016). 10.1039/C5AN01786F [DOI] [PubMed] [Google Scholar]
- 33.Bonifacio A., Dalla Marta S., Spizzo R., Cervo S., Steffan A., Colombatti A., Sergo V., “Surface-enhanced Raman spectroscopy of blood plasma and serum using Ag and Au nanoparticles: a systematic study,” Anal. Bioanal. Chem. 406(9-10), 2355–2365 (2014). 10.1007/s00216-014-7622-1 [DOI] [PubMed] [Google Scholar]
- 34.Premasiri W. R., Lee J. C., Ziegler L. D., “Surface-enhanced Raman scattering of whole human blood, blood plasma, and red blood cells: cellular processes and bioanalytical sensing,” J. Phys. Chem. B 116(31), 9376–9386 (2012). 10.1021/jp304932g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao R., Zhang X., Rong Z., Xiu B., Yang X., Wang C., Hao W., Zhang Q., Liu Z., Duan C., Zhao K., Guo X., Fan Y., Zhao Y., Johnson H., Huang Y., Feng X., Xu X., Zhang H., Wang S., “Non-invasive detection of hepatocellular carcinoma serum metabolic profile through surface-enhanced Raman spectroscopy,” Nanomedicine (Lond.) 12(8), 2475–2484 (2016). 10.1016/j.nano.2016.07.014 [DOI] [PubMed] [Google Scholar]
- 36.Feng S., Chen R., Lin J., Pan J., Chen G., Li Y., Cheng M., Huang Z., Chen J., Zeng H., “Nasopharyngeal cancer detection based on blood plasma surface-enhanced Raman spectroscopy and multivariate analysis,” Biosens. Bioelectron. 25(11), 2414–2419 (2010). 10.1016/j.bios.2010.03.033 [DOI] [PubMed] [Google Scholar]
- 37.Chung T. J., “Hepatitis B and glucose metabolism: a systematic review.” Hku Theses Online (2014).
- 38.Yang Y., Li C., Nie X., Feng X., Chen W., Yue Y., Tang H., Deng F., “Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis,” J. Proteome Res. 6(7), 2605–2614 (2007). 10.1021/pr070063h [DOI] [PubMed] [Google Scholar]
- 39.Lin J., Wang J., Xu C., Zeng Y., Chen Y., Li L., Huang Z., Li B., Chen R., “Differentiation of digestive system cancers by using serum protein-based surface-enhanced Raman spectroscopy,” J. Raman Spectrosc. 48(1), 16–21 (2017). 10.1002/jrs.4982 [DOI] [Google Scholar]