Abstract
Chromosomal inversions often contribute to local adaptation across latitudinal clines, but the underlying selective mechanisms remain poorly understood. We and others have previously shown that a clinal inversion polymorphism in Drosophila melanogaster, In(3R)Payne, underpins body size clines along the North American and Australian east coasts. Here we ask whether this polymorphism also contributes to clinal variation in other fitness-related traits, namely survival traits (lifespan, survival upon starvation, and survival upon cold shock). We generated homokaryon lines, either carrying the inverted or standard chromosomal arrangement, isolated from populations approximating the endpoints of the North American cline (Florida, Maine), and phenotyped the flies at two growth temperatures (18°C, 25°C). Across both temperatures, high-latitude flies from Maine lived longer and were more stress resistant than low-latitude flies from Florida, as previously observed. Interestingly, we find that this latitudinal pattern is partly explained by the clinal distribution of the In(3R)P polymorphism, which is at ~50% frequency in Florida but absent in Maine: inverted karyotypes tended to be shorter-lived and less stress resistant than uninverted karyotypes. We also detected an interaction between karyotype and temperature on survival traits. Since In(3R)P influences body size and multiple survival traits, it can be viewed as a ‘supergene’, a cluster of tightly linked loci affecting multiple complex phenotypes. We conjecture that the inversion cline is maintained by fitness trade-offs and balancing selection across geography; elucidating the mechanisms whereby this inversion affects alternative, locally adapted phenotypes across the cline is an important task for future work.
Keywords: Inversion, supergene, clines, adaptation, temperature, life history, survival, D. melanogaster
Introduction
Since the pioneering work of Dobzhansky, many lines of evidence suggest that chromosomal inversion polymorphisms play a major role in climatic adaptation to altitudinal and latitudinal gradients, so-called clines (e.g., Dobzhansky, 1937; 1943; 1947a, b; Wright & Dobzhansky, 1946; Leumeunier & Aulard, 1992; Hoffmann et al., 2004; Kirkpatrick & Barton, 2006; Hoffmann & Rieseberg, 2008; Schaeffer, 2008; Kirkpatrick & Kern, 2012; Kapun et al., 2016a; and references therein). However, while inversions have been statistically associated with many traits (e.g., Sperlich & Pfriem, 1986; Etges, 1989; De Jong & Bochdanovits, 2003; Hoffmann et al., 2004; Lowry & Willis, 2010), little is known about associations between inversions and fitness-related traits, thus limiting our understanding of how these adaptive polymorphisms are maintained by selection (e.g., Hoffmann & Rieseberg, 2008).
The latitudinal frequency clines of inversion polymorphisms in Drosophila melanogaster, often observed in a parallel fashion on multiple continents, provide an excellent opportunity to address this problem (e.g., Mettler et al., 1977; Knibb et al., 1981; Knibb, 1982; Leumeunier & Aulard, 1992; De Jong & Bochdanovits, 2003; Hoffmann & Weeks, 2007). For example, a large (~8 Mb), cosmopolitan inversion polymorphism on the right arm of the third chromosome, In(3R)Payne (also called In(3R)P), varies clinally along latitudinal gradients on several continents, most prominently along the Australian and North American east coasts (e.g., Mettler et al., 1977; Inoue & Watanabe, 1979; Stalker, 1980; Knibb et al., 1981; Knibb, 1982; Das & Singh, 1991; Anderson et al. 2005; Matzkin et al., 2005; Fabian et al., 2012; Kapun et al., 2014; Rane et al., 2015; Kapun et al., 2016a). This inversion, which is of tropical African origin and approximately 130,000-150,000 years old (Corbett-Detig, 2012), has long been thought to be an important driver of climatic adaptation in D. melanogaster (e.g., De Jong & Bochdanovits, 2003; Weeks et al., 2002; Anderson et al., 2005; Hoffmann & Weeks, 2007; Rane et al., 2015; Kapun et al., 2016a, 2016b).
Potentially consistent with a role of this inversion in climate adaptation, the inverted karyotype of In(3R)P exhibits intermediate-to-high frequencies at low latitudes (i.e., in subtropical to tropical climates) but is rare or absent at high latitudes (i.e., in temperate, seasonal climates) on all continents or subcontinents examined so far (see references above). Along the North American east coast, for example, the inverted arrangement has a frequency of ~50% in Florida but its frequency decreases along the cline to ~0% in Maine (e.g., Mettler et al., 1977; Knibb, 1982; Fabian et al., 2012; Kapun et al., 2014, 2016a). Recent evidence suggests that the Australian and North American clines in In(3R)P are adaptively maintained by spatially varying selection (Rane et al., 2015; Kapun et al., 2016a). Although genetic patterns of clinal variation can be severely confounded by admixture and secondary contact with ancestral populations (Bergland et al. 2016; also see discussion in Flatt, 2016), the North American cline in In(3R)P seems to be maintained non-neutrally and independent of population structure and admixture (Kapun et al., 2016a).
Interestingly, several major fitness-related traits also exhibit strong clinality across latitude (e.g., De Jong & Bochdanovits, 2003; Hoffmann & Weeks, 2007; Adrion et al., 2015; and references therein), and it is thus tempting to speculate that the clinal behavior of In(3R)P (or potentially that of other clinal inversions) might underlie – or contribute to – these life-history clines. For instance, flies from North American high-latitude populations are characterized by increased body size, reduced wing loading, reduced fecundity, prolonged lifespan, increased resistance to starvation, cold and heat stress, and the plastic ability to undergo reproductive dormancy in response to cool temperature and short photoperiod, as compared to flies from low latitude (e.g., Coyne & Beecham, 1987; Azevedo et al., 1998; de Jong & Bochdanovits, 2003; Hoffmann et al., 2005; Schmidt et al., 2005a, b; Schmidt & Paaby, 2008; Fabian et al., 2015; Mathur & Schmidt, 2017). This suggests a pattern of climatic adaptation whereby harsh winters and seasonal stress at high latitudes favor stress resistance and overwintering ability (dormancy), along with correlated (e.g., pleiotropic) responses in terms of larger body size, increased lifespan and reduced fecundity (e.g., Schmidt & Paaby, 2008; Paaby & Schmidt, 2009; Flatt et al., 2014; Paaby et al., 2014). Yet, whether clinal inversion polymorphisms such as In(3R)P contribute to this pattern of phenotypic climatic adaptation is largely unclear (e.g., De Jong & Bochdanovits, 2003; Rako et al., 2006; Hoffmann et al., 2004; Hoffmann & Weeks, 2007; Hoffmann & Rieseberg, 2008; Kapun et al., 2016a, b).
Consistent with a contribution of In(3R)P to clinal trait differentiation, we and others have previously found that the latitudinal cline in this inversion contributes to the body size cline along the Australian (Weeks et al., 2002; Rako et al., 2006; Kennington et al., 2007) and North American (Kapun et al., 2016b) east coasts. However, little is known about whether In(3R)P also affects other traits that covary with latitude. In Australian populations, for example, Anderson et al. (2003) found an association between susceptibility to cold and In(3R)P, but a subsequent study by Rako et al. (2006) failed to find a clear effect. Similarly, for the North American cline, we also failed to detect an association between In(3R)P and chill coma recovery (Kapun et al., 2016b). Moreover, neither study found an effect of In(3R)P on developmental time (Rako et al., 2006; Kapun et al., 2016b). Thus, whether In(3R)P contributes to clinal variation in fitness-related traits other than body size is not clear. While it is possible that In(3R)P predominantly – or exclusively – affects size and not other traits, this seems rather unlikely, for two reasons: (1) the majority of significantly clinally varying SNPs in the genome reside in the region spanned by this inversion, and (2) many of these clinal SNPs within In(3R)P are located in genes that are known from studies of mutants and transgenes to affect life-history traits (Kapun et al., 2016a, b; also see Fabian et al., 2012).
Here, we examine whether the clinal In(3R)P polymorphism affects three major survival traits in North American populations of D. melanogaster: adult lifespan, survival upon starvation, and cold resistance (measured as survival upon cold shock). All three traits are known to vary clinally in North America as a function of latitude (or of high-latitude vs. low-latitude genotypes) (e.g., Schmidt et al., 2000; Schmidt et al., 2005a, b; Schmidt & Paaby, 2008; Paaby et al., 2014; Mathur & Schmidt, 2017). In agreement with previous phenotypic results, we find that high-latitude flies from Maine live longer and are more stress resistant than low-latitude flies from Florida, consistent with the idea that selection at high latitude favors genotypes and phenotypes that confer improved survival and somatic maintenance (Paaby & Schmidt, 2009; Flatt et al., 2013; Paaby et al., 2014). Interestingly, we observe that the clines in these traits are partly driven by the clinal frequency gradient in In(3R)P: on average flies carrying the In(3R)P inversion from Florida live shorter and are less stress resistant than flies from Florida or Maine which possess the uninverted chromosomal segment. Our findings support the hypothesis that life-history clines are maintained by fitness trade-offs across geography and that clinal inversions, such as In(3R)P, make an important contribution to this phenomenon.
Materials and methods
Fly stocks and maintenance
We isolated third-chromosome isochromosomal (homokaryon) lines, either carrying two copies of the inverted In(3R)P arrangement or two copies of the uninverted (standard) arrangement, from two areas approximating the endpoints of the North American cline (low latitude: Florida [Homestead and Jacksonville]; and high latitude: Maine [Bowdoin]), as previously described (see Kapun et al., 2016b for details of sampling locations and isolation methods). Across the cline In(3R)P has a frequency of ~50% in Florida but is absent in Maine, so that flies from high-latitude populations are fixed for the uninverted arrangement (e.g., Mettler et al., 1977; Knibb, 1982; Fabian et al., 2012; Kapun et al., 2014, 2016a, b). Wild-type chromosomes were isogenized using a compound balancer for the second and third chromosomes (SM6b; TM6B; Bloomington Drosophila Stock Center [BDSC] stock #5687) in an ebony (e1) mutant background (cf. Kapun et al., 2016b). From Florida, where both the inverted and uninverted segments segregate, we isolated 9 isochromosomal lines carrying In(3R)P (‘Florida inverted’, FI) and 9 lines possessing the standard arrangement (‘Florida standard’, FS); from Maine, where the inverted segment is absent, we isolated 9 lines with the standard arrangement only (‘Maine standard’, MS). Prior to phenotyping assays, which were performed at two growth temperatures (see below), lines were kept under common garden conditions for three generations (~21°C, 10h:14h light:dark [LD], ~50% relative air humidity [RH]).
Phenotype assays
We measured three clinally varying survival traits on the homokaryon lines: lifespan, survival upon starvation, and survival upon cold shock (see below). Assays were performed at two growth temperatures (18°C,25°C), at 12:12h LD and 60% RH, on a cornmeal/sugar/yeast/agar diet. To obtain (F1) flies for phenotypic assays, we let ~20-25 females and males mate and lay eggs into vials containing 8 mL of medium at room temperature (n = 46 vials for each of the 27 lines [= 3 karyotypes × 9 lines]; total = 1242 vials). Depending on their fecundity, females in each vial were allowed to lay eggs for up to ~24 hours; egg density was inspected regularly by eye and adjusted to ~40-50 eggs per vial. Vials were then transferred to their respective developmental temperature treatment (i.e., 18°C vs. 25°C; 23 replicate vials per temperature and isochromosomal line).
For lifespan assays, we collected cohorts of newly eclosed adult females and males within a 24-h period. Flies were sexed and counted under light CO2 anesthesia and transferred to 1L demography cages (see Tatar et al., 2001 for details of cage design) 24 hours after eclosion. Each cage was initiated with 75 females and 75 males. We set up 2 replicate cages per line and temperature (n = 2 cages × 27 lines × 2 temperatures = 108 cages; 108 cages × 150 flies = 16,200 flies in total). Every second day at 25°C and every third day at 18°C, we changed food vials and removed and recorded dead flies until all flies in the experiment had died. Flies that got stuck to the food medium were censored from analysis.
For assays of survival upon starvation, we used 5-7 day-old mated individuals. Eclosing adults were collected in 48-hour cohorts and maintained in mixed-sex groups for 4 days in their respective thermal treatments. 24 hours prior to initiating the assay, flies were sexed under light CO2 anesthesia and transferred to fresh vials containing 10 individuals per vial and sex. On the day of the experiment, flies were transferred to food-free vials, containing 0.5% agar/water solution. For each line, temperature and sex, we used 5 replicate vials (n = 5 vials × 27 lines × 2 temperatures × 2 sexes = 540 vials, each with 10 flies; total = 5400 flies). Age at death was scored in 8-hour intervals until all flies had died.
We used an identical experimental design for measuring survival upon cold shock; again, we used 5 replicate vials per group (n = 5 vials × 27 lines × 2 temperatures × 2 sexes = 540 vials, each with 10 flies; total = 5400 flies). On the day of the experiment, 5-7 day-old mated flies were transferred to media-free vials and the vials dipped immediately into −4°C cold, salted ice water for 90 minutes. (Depending on acclimation, several hours of exposure to temperatures between −2°C and −5°C typically result in >50% mortality; cf. Hoffmann, 2010.) After cold shock, flies were transferred to petri dishes (60 × 15 mm) with 2 mL fly food in one corner and left at room temperature for recovery. Survival was scored after 24 hours; flies that were alive after 24 hours were censored from analysis.
Note that all assays of fitness components were performed on mated flies, for two reasons. First, unmated flies live dramatically longer than mated flies – such a massive physiological effect might have masked the more subtle karyotypic differences we aimed to observe. Second, the majority of work on phenotypic clines in D. melanogaster, including our work on In(3R)P (Kapun et al., 2016b), has been done on mated flies so that – for the sake of comparison – we decided to not assay unmated flies.
All phenotypic raw data have been deposited at Dryad: doi:10.5061/dryad.3vb89dj.
Statistical analysis
The primary interest of our analysis was to determine the effects of In(3R)P karyotype (inverted vs. uninverted arrangement) upon survival traits; effects of clinality/geography (i.e., Florida vs. Maine) were of secondary interest. However, the biology of this system is such that the effects of karyotype vs. geography cannot be completely disentangled: since In(3R)P is polymorphic (with a ~50:50% frequency of inverted vs. standard arrangement) in Florida but not in Maine, where only the uninverted arrangement is present (i.e., the frequency of the inversion is ~0%), one cannot use a fully factorial, orthogonal design to analyze data for this inversion polymorphism when considering the cline ends. Analyzing the cline ends is however important since this allows investigating the extent of life-history clinality, and the potential contribution of the inversion to it, across the entire latitudinal gradient. Importantly, even though the inversion is absent in Maine, this design nonetheless permits clear inferences regarding the effects of karyotype (also see Kapun et al., 2016b): significant differences between FI and FS and between FI and MS, with no difference between FS and MS, imply a clear main effect of In(3R)P karyotype. On the other hand, a situation in which all three pairwise comparisons (FI vs. FS; FI vs. MS; FS vs. MS) are different implies that inverted vs. standard arrangements differ in their effects, yet that the two uninverted arrangement types from Florida and Maine differ too, perhaps due to an effect of geography (Florida vs. Maine). In this scenario, it is not possible to clearly separate the effects of karyotype vs. geography; nevertheless, a significant difference between FI and FS would indicate an effect of In(3R)P karyotype. In both cases, the inference would be that In(3R)P karyotype affects the trait of interest. Lastly, a scenario in which FI = FS and where both FI and FS are significantly different from MS might imply – assuming parsimony – a main effect of geography/clinal differentiation independent of In(3R)P karyotype.
Due to the constraint that the effects of karyotype and geography must be analyzed jointly, we created a compound grouping factor K (‘karyotype’, partly confounded by geography) with three levels (‘Florida inverted’, FI; ‘Florida standard’, FS; and ‘Maine standard’, MS) and used pairwise comparisons between the levels of K in order to infer the effects of karyotype, geography or both (see below). The second factor that entered our analyses was temperature T (18°C vs. 25°C). We analyzed our survival (mortality) data in two ways. First, we used Cox (proportional hazards) regression to fit the fixed effects of K, T and the interaction K × T. This ‘global’ approach gave us main effects for K (averaged across temperatures) and T (averaged across levels of K) and indicated whether – importantly – there are significant K × T (i.e., genotype by environment) interactions. Second, to dissect the source of significance of the effects of K and/or K × T, we employed Kaplan-Meier survival analysis with generalized Wilcoxon (χ2) tests (GWT) to perform pairwise comparisons (i.e., FI vs. FS; FI vs. MS; FS vs. MS) for each temperature and sex separately, followed by Bonferroni correction for multiple testing. These pairwise comparisons thus serve as post-hoc tests for the Cox models. Analyses were performed in JMP v.11.2.0 (SAS, Raleigh, NC, USA).
We also provide a preliminary analysis of the potential relationship between size (wing area) and lifespan and of the effects of karyotype on the multivariate combination of life-history traits (see Supporting Information for details).
Results
The In(3R)P polymorphism contributes to clinality of lifespan
We first analyzed the effects of karyotype and temperature and their interaction on lifespan (Fig. 1, Fig. S1). For both females and males, Cox regression revealed effects of karyotype (likelihood ratio test [LRT]; females: χ2(2) = 387.8, males: χ2(2) = 359.0, both P < 0.0001), temperature T (females: χ2(1) = 4301.9, males: χ2(1) = 2893.4, both P < 0.0001) and the K × T interaction (females: χ2(2) = 19.3, males: χ2(2) = 31.2, both P < 0.0001). This analysis, together with pairwise generalized Wilcoxon χ2 tests [GWT], showed that Florida inverted (FI) flies lived shorter than both Florida standard (FS) and Maine standard (MS) flies (Fig. 1, Table 1, Fig. S1), implying a clear effect of In(3R)P karyotype on adult survival. Moreover, at 18°C – but not at 25°C – FS flies lived shorter than MS flies (Fig. 1, Table 1, Fig. S1). These results indicate that both inversion karyotype (inverted flies live shorter than uninverted flies from both Florida and Maine) and geography (at least at 18°C, uninverted flies from Maine live longer than both inverted and uninverted flies from Florida) affect lifespan (Fig. 1, Table 1, Fig. S1). (A preliminary analysis of size data from our experiment suggests that karyotypic differences in lifespan might partly be explained by effects of In(3R)P on size; see Supporting Information.) With regard to temperature, flies lived longer at 18°C than at 25°C (see significant effect of T in Cox regression above; and GWT, females: χ2(1) = 3490.4, males: χ2(1) = 2262.5, both P < 0.0001) (Fig. 1, Fig. S1). At 18°C females lived longer than males, but we failed to find such a sex difference at 25°C (GWT, 18°C: χ2(1) = 115.7, P < 0.0001, 25°C: χ2(1) = 0.122, P = 0.73) (Fig. 1, Fig. S1). Thus, In(3R)P has a negative impact upon adult survival, and lifespan exhibits clinal differentiation, with high-latitude flies overall living longer than flies from low-latitude.
Fig. 1. In(3R)P shortens lifespan and lifespan varies clinally.
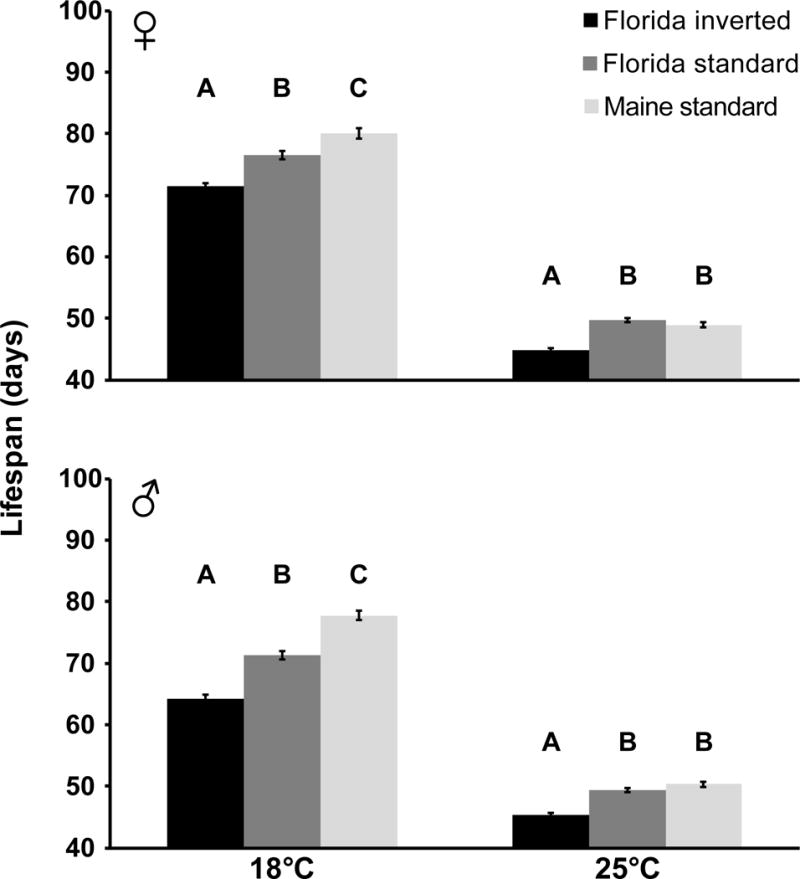
Effects of the In(3R)P inversion polymorphism on adult lifespan (days) in females and males. The bar plots show means and standard errors. Black bars: Florida inverted (FI); dark grey bars: Florida standard (FS); light grey bars: Maine standard (MS). Results for pairwise comparisons among karyotypes with generalized Wilcoxon (χ2) tests are shown in letters: groups that do not contain the same letter are significantly different from each other (P<0.05). The In(3R)P inversion shortens lifespan as compared to uninverted karyotypes; lifespan is clinally differentiated, with high-latitude flies from Maine living longer than flies from low-latitude. See Results and Table 1 for details; for survival curves see Fig. S1.
Table 1.
Analysis of lifespan
| Sex | Temperature | Direction | χ2 | P | n |
|---|---|---|---|---|---|
| Female | 18°C | FI < FS | 35.86 | <0.0001† | 2609 (2639) |
| FI < MS | 137.31 | <0.0001† | 2591 (2644) | ||
| FS < MS | 28.52 | <0.0001† | 2520 (2571) | ||
|
| |||||
| 25°C | FI < FS | 98.75 | <0.0001† | 2500 (2549) | |
| FI < MS | 71.53 | <0.0001† | 2478 (2539) | ||
| FS = MS | 0.65 | 0.42 | 2508 (2566) | ||
|
| |||||
| Male | 18°C | FI < FS | 85.72 | <0.0001† | 2437 (2455) |
| FI < MS | 247.67 | <0.0001† | 2514 (2547) | ||
| FS < MS | 37.26 | <0.0001† | 2453 (2484) | ||
|
| |||||
| 25°C | FI < FS | 100.39 | <0.0001† | 2565 (2600) | |
| FI < MS | 109.59 | <0.0001† | 2537 (2580) | ||
| FS = MS | 0.32 | 0.57 | 2512 (2558) | ||
The columns show the directionality of lifespan effects for each pairwise comparison between the three karyotypes (FI = Florida inverted, FS = Florida standard, MS = Maine standard), grouped by sex and temperature. χ2 test statistics and P-values are from generalized Wilcoxon tests. Significant effects are in bold; significance after Bonferroni correction is indicated by † (α′= 0.05/3 = 0.016). n represents the number of dead individuals; the total cohort size is shown in parenthesis. See Results and Figs. 1 and S1 for further details.
The effects of In(3R)P on starvation survival depend on temperature
Next, we examined whether In(3R)P affects survival upon starvation. We found significant effects of karyotype (Cox LRT; females: χ2(2) = 174.9, males: χ2(2) = 93.2, both P < 0.0001), temperature (females: χ2(1) = 253.3, males: χ2(1) = 660.2, both P < 0.0001), and – for females – of the K × T interaction (females: χ2(2) = 18.2, P < 0.0001; males: χ2(2) = 4.1, P = 0.13) (Fig. 2, Fig. S2). Interestingly, at 18°C FI flies were more starvation resistant than FS flies for both sexes, whereas at 25°C this pattern was reversed for females, without a significant difference in males (Fig. 2, Table 2, Fig. S2). Overall, across both temperatures, high-latitude MS flies were more starvation resistant than low-latitude FI and FS flies, incidating an effect of clinality (Fig. 2, Table 2, Fig. S2). Survival upon starvation was greater for flies reared at 18°C than at 25°C (see significant effect of T in Cox model above; GWT, females: χ2(1) = 252.4, males: χ2(1) = 848.7, both P < 0.0001), and females were more resistant than males at both temperatures (GWT, 18°C: χ2(1) = 938.6, 25°C: χ2(1) = 1339.1, both P<0.0001) (Fig. 2, Fig. S2). Together, these results show that in Florida the effects of In(3R)P karyotype on starvation survival depend on temperature, and that high-latitude flies from Maine are more starvation resistant than low-latitude flies from Florida.
Fig. 2. In(3R)P affects starvation in a temperature-dependent manner.
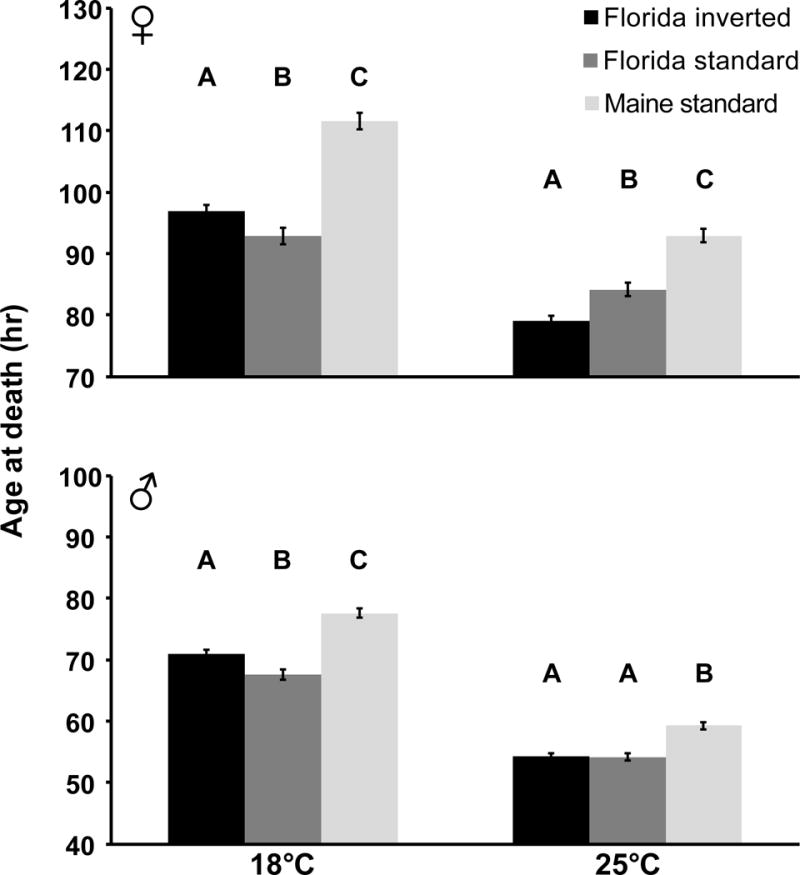
Effects of In(3R)P on age at death (hours) upon starvation in females and males. Shown are means and standard errors. Black bars: Florida inverted (FI), dark grey bars: Florida standard (FS), light grey bars: Maine standard (MS). Results for pairwise comparisons among karyotypes with generalized Wilcoxon (χ2) tests are shown in letters: groups that do not contain the same letter are significantly different from each other (P<0.05). At 18°C Florida inverted flies survive starvation better than uninverted flies, whereas this pattern is reversed for females at 25°C. Morevoer, high-latitude flies from Maine are overall more resistant than low-latitude flies from Florida. See Results and Table 2 for details; for survival curves see Fig. S2.
Table 2.
Analysis of survival upon starvation
| Sex | Temperature | Direction | χ2 | P | n |
|---|---|---|---|---|---|
| Female | 18°C | FI > FS | 5.35 | 0.02 | 881 |
| FI < MS | 72.94 | <0.0001† | 878 | ||
| FS < MS | 90.88 | <0.0001† | 899 | ||
|
| |||||
| 25°C | FI < FS | 10.14 | 0.0014† | 899 | |
| FI < MS | 88.68 | <0.0001† | 900 | ||
| FS < MS | 25.99 | <0.0001† | 899 | ||
|
| |||||
| Male | 18°C | FI > FS | 10.44 | 0.0012† | 900 |
| FI < MS | 34.72 | <0.0001† | 897 | ||
| FS < MS | 78.31 | <0.0001† | 897 | ||
|
| |||||
| 25°C | FI = FS | 0.15 | 0.70 | 898 | |
| FI < MS | 30.44 | <0.0001† | 896 | ||
| FS < MS | 31.60 | <0.0001† | 894 | ||
The columns show the directionality of survival upon starvation for each pairwise comparison between the three karyotypes (FI = Florida inverted, FS = Florida standard, MS = Maine standard), grouped by sex and temperature. χ2 test statistics and P-values are from generalized Wilcoxon tests. Significant effects are in bold; significance after Bonferroni correction is indicated by † (α′ = 0.05/3 = 0.016). n represents the total cohort size, i.e. the number of dead individuals (no flies were censored in this assay). See Results and Figs. 2 and S2 for further details.
In(3R)P confers increased mortality to cold shock
To examine whether the In(3R)P inversion also contributes to cold tolerance we investigated the mortality of flies upon 24 hours of exposure to cold shock at −4°C (Fig. 3). In both females and males, karyotypes did overall not differ in cold-shock mortality (Cox LRT; females: χ2(2) = 2.1, P = 0.39; males: χ2(2) ≈ 0, P = 1.0), but there was a significant K × T interaction for females (females: χ2(2) = 9.2, P = 0.01; in males, the interaction term could not be fit since at 18°C all males survived and were censored from analysis). Temperature affected cold-shock survival in both sexes (Cox LRT; females: χ2(1) = 350.7, males: χ2(1) = 1529.8, both P < 0.0001). Pairwise comparisons between the three karyotypes with GWT showed that at 25°C FI inverted females survived cold shock less well than both FS and MS uninverted females; at the same time, uninverted high-latitude females from Maine survived cold shock better than both low-latitude karyotypes from Florida (Fig. 3, Table 3). In contrast, we found no differences among karyotypes at 18°C and for males at both temperatures (Fig. 3, Table 3). For both females and males, flies survived cold shock better at 18°C than at 25°C (see significant effect of T in Cox regression above; GWT, females: χ2(1) = 652.2, males: χ2(1) = 1875.1, both P < 0.0001) (Fig. 3). At 25°C females tended to survive cold shock better than males (GWT, 25°C: χ2 (1) = 41.0, P<0.0001; since at 18°C all males were censored for analysis we did not compare the two sexes at this temperature) (Fig. 3). In(3R)P thus confers increased mortality to cold shock, and uninverted female flies from Maine tend to survive acute cold exposure better than low-latitude flies from Florida, at least at 25°.
Fig. 3. At 25°C In(3R)P confers mortality upon cold shock.
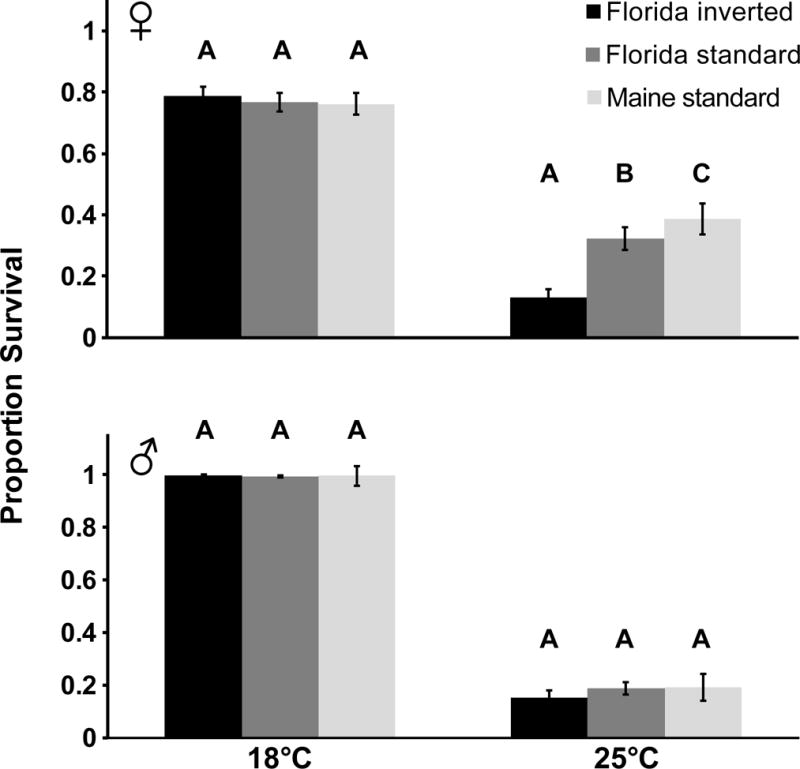
Effects of In(3R)P on the proportion of female and male flies surviving cold shock. Shown are means and standard errors. Black bars: Florida inverted (FI); dark grey bars: Florida standard (FS); light grey bars: Maine standard (MS). Results for pairwise comparisons among karyotypes with generalized Wilcoxon (χ2) tests are shown in letters: groups that do not contain the same letter are significantly different from each other (P<0.05). The In(3R)P inversion increases sensitivity to cold shock at 25°C. Generally, at 25°C, high-latitude females from Maine are more cold-shock resistant than low-latitude females from Florida. See Results and Table 3 for details.
Table 3.
Analysis of survival upon cold shock.
| Sex | Temperature | Direction | χ2 | P | n |
|---|---|---|---|---|---|
| Female | 18°C | FI = FS | 0.64 | 0.43 | 197 (887) |
| FI = MS | 0.88 | 0.35 | 199 (888) | ||
| FS = MS | 0.02 | 0.89 | 210 (893) | ||
|
| |||||
| 25°C | FI < FS | 47.39 | <0.0001† | 694 (897) | |
| FI < MS | 77.48 | <0.0001† | 664 (896) | ||
| FS < MS | 4.18 | 0.04 | 580 (899) | ||
|
| |||||
| Male | 18°C | - | - | - | 0 (896) |
| - | - | - | 0 (897) | ||
| - | - | - | 0 (895) | ||
|
| |||||
| 25°C | FI = FS | 2.01 | 0.16 | 737 (887) | |
| FI = MS | 2.35 | 0.14 | 734 (885) | ||
| FS = MS | 0.01 | 0.90 | 719 (886) | ||
The columns show the directionality of survival upon cold shock for each pairwise comparison between the three karyotypes (FI = Florida inverted, FS = Florida standard, MS = Maine standard), grouped by sex and temperature. χ2 test statistics and P-values are from generalized Wilcoxon tests. Significant effects are in bold; significance after Bonferroni correction is indicated by † (α′ = 0.05/3 = 0.016). n represents the number of dead individuals; the total cohort size is shown in parenthesis. Note that at 18°C all males survived 24 hours of cold shock and were thus all censored. See Results and Fig. 3 for further details.
The In(3R)P inversion might represent a life-history ‘supergene’
The fact that In(3R)P affects body size (e.g., Weeks et al. 2002; Rako et al., 2006; Kapun et al., 2016b) and, as shown above, several survival traits indicates that this inversion might represent a ‘supergene’, a set of tightly linked loci that affects multiple complex phenotypes (Schwander et al., 2014). Consistent with this idea we found that karyotype has a significant upon the multivariate life-history phenotype (i.e., a linear combination of size, lifespan, starvation resistance and cold-shock survival), using multivariate analysis of variance (MANOVA) (for details see Supporting Information).
Discussion
High-latitude flies are more long-lived and stress resistant
Populations of D. melanogaster in North America, and also on other continents, display gradients of phenotypic differentiation for fitness-related traits such as body size, fecundity, stress resistance, reproductive dormancy and longevity across latitude (e.g., Coyne & Beecham, 1987; de Jong & Bochdanovits, 2003; Hoffmann et al., 2005; Schmidt et al., 2005a, b; Schmidt & Paaby, 2008; Paaby et al., 2014; Fabian et al., 2015; Mathur & Schmidt, 2017). These patterns of clinal differentiation are hypothesized to be driven by differential selection pressures at high vs. low latitude (e.g., Paaby & Schmidt, 2009): genotypes that confer stress resistance and survival at the expense of reduced fecundity might be favored at high latitudes, where seasonal stressors such as cold and food shortage impose strong selection on somatic maintenance, whereas at low latitude selection might favor alternative genotypes that confer fast development and high fecundity at the expense of reduced stress resistance and survival. In support of this adaptive scenario, we observed that high-latitude flies from Maine lived longer and were more resistant to starvation and cold stress than low-latitude flies from Florida, consistent with previous observations along the North American cline (Schmidt et al., 2000; Schmidt et al., 2005a, b; Schmidt & Paaby, 2008; Paaby et al., 2014; Mathur & Schmidt, 2017).
While the genetic basis of latitudinal clines for survival traits is poorly understood, many strongly clinally varying single nucleotide polymorphisms (SNPs) are located in genes known to be important for the determination of adult lifespan and stress resistance, for example in the insulin/insulin-like growth factor signaling (IIS) pathway (see Fabian et al., 2012; Kapun et al., 2016a). This observation opens up an opportunity for identifying naturally segregating polymorphisms that affect lifespan and stress resistance (e.g., Flatt & Schmidt, 2009; Paaby & Schmidt, 2009).
Three examples serve to illustrate this point. A haplotype at the methuselah (mth) locus, a gene known from mutant studies to affect longevity and stress resistance, shows a 40% cline in frequency across the North American east coast that coincides with among-population differences in life-history traits including lifespan, even though phenotypic effects of this haplotype were not directly examined (Schmidt et al., 2000; Duvernell et al., 2003); subsequent work identified effects of wild-derived mth alleles on lifespan, fecundity and stress resistance (Paaby & Schmidt, 2008). Another example comes from a clinally varying indel polymorphism in the insulin-like receptor (InR) which affects lifespan and stress resistance in the predicted clinal direction, with the high-latitude genotype conferring improved stress resistance and survival (Paaby et al., 2010, 2014). Similarly, an amino acid polymorphism in the couch potato (cpo) gene explains clinal variation in the ability of flies to undergo reproductive dormancy (Schmidt et al., 2008), a plastic state associated with greatly improved stress resistance and lifespan (e.g., Schmidt & Paaby, 2008; Flatt et al., 2013).
Since the In(3R)P polymorphism is the dominant driver of genotypic latitudinal clines in North America (Fabian et al., 2012; Kapun et al., 2016a), either due to direct or indirect selection (via genetic draft/‘hitchhiking’), it is interesting to ask whether the cline in In(3R)P might contribute to the phenotypic clines seen for survival traits. Addressing this question was the main purpose of our study.
In(3R)P contributes to latitudinal clinality of multiple survival traits
Recent evidence has shown that In(3R)P is adaptively maintained by spatially varying selection along the North American cline (Kapun et al., 2016a), but how this inversion polymorphism affects trait differentiation is poorly understood. In previous assays, we found that In(3R)P affects body size, consistent with observations from Australia (e.g., Rako et al., 2006), but developmental time, egg-to-adult survival, chill coma recovery, oxidative stress resistance, and lipid content were unaffected by this inversion (Kapun et al., 2016b). In agreement with our findings for North America, the Australian study by Rako et al. (2006) also failed to find effects of In(3R) on developmental time and chill coma recovery. Despite these negative results for traits beyond size, here we have found that the North American cline in In(3R)P underpins, at least to some extent, latitudinal clines in three survival traits, i.e. lifespan, starvation resistance and survival upon cold shock (cf. Schmidt et al., 2000; Schmidt et al., 2005a, b; Schmidt & Paaby, 2008; Paaby et al., 2014; Mathur & Schmidt, 2017). The effects of In(3R)P on these traits go in the predicted clinal direction, with flies from Maine and Florida possessing the uninverted arrangement being on average longer-lived and more stress resistant than flies from Florida which carry the inverted In(3R)P segment. Importantly, this establishes that the In(3R)P polymorphism affects – and harbors genetic variance for – multiple, clinally varying components of fitness. This is in line with the observation that the genomic region spanned by In(3R)P contains numerous clinally varying SNPs in genes known to affect body size, lifespan, stress resistance and other fitness-related traits (Fabian et al., 2012; Kapun et al., 2016a, b). For example, it is noteworthy in this context that both InR and cpo (see above) are located in the region spanned by In(3R)P.
Although work by Rako et al. (2006) and us (Kapun et al., 2016b) did not find an effect of In(3R)P on cold tolerance in terms of chill coma recovery (but see Anderson et al., 2003), our experiments here show that the inverted arrangement is associated with increased mortality in response to cold shock in females. This result is in good qualitative agreement with the data of Anderson et al. (2003), who found that cold-shock mortality is associated with a genetic marker (hsr-omega) that is in linkage disequilibrium (LD) with In(3R)P, and also with the earlier findings of Tucic (1979), who found a major effect of chromosome 3 on larval and adult cold tolerance. The fact that different measures of cold tolerance (chill coma recovery vs. cold-shock mortality) can give discordant results implies that the details of assay protocols used for measuring aspects of cold tolerance matter greatly (e.g., McDonald et al., 2004; Andersen et al., 2015). Importantly, Andersen et al. (2015) found that measures of the time to lethality at low temperature are not correlated with chill coma recovery time.
Thus, by experimentally isolating and phenotyping In(3R)P karyotypes from populations approximating the end points of the North American cline, our results suggest that this inversion makes a major contribution to the clinality of several survival traits. An open task for future work will be to regress means of survival traits for multiple populations spanning the cline against the population frequency of In(3R)P as a predictor, for this would allow to estimate the amount of phenotypic variance explained by In(3R)P.
In a preliminary analysis using MANOVA we also found evidence that In(3R)P affects the multivariate combination of life-history traits (i.e., a linear combination of a size proxy and the three survival traits; see Supporting Information), indicating that this inversion might represent a life-history ‘supergene’ (Schwander et al., 2014).
How is the In(3R)P polymorphism maintained?
Although In(3R)P is maintained by selection along the North American cline (Kapun et al., 2016a), the details of the underlying selective mechanisms remain unknown. Clearly, our results cannot fully explain how this polymorphism is maintained since the inverted segment seems to have predominantly negative effects on the measured fitness-components: inverted homokaryons were on average smaller, shorter-lived and less stress resistant than uninverted homokaryons (also see Kapun et al., 2016b). So, what are the fitness benefits that maintain the inverted karyotype at low latitude?
Four major considerations should be kept in mind. First, we have only phenotyped inverted vs. uninverted homokaryons but not any heterokaryons: because inversions might be maintained by overdominance or associative overdominance (e.g., Dobzhansky, 1970; Kirpatrick & Barton, 2006), it will be critical to phenotype In(3R)P heterokaryons in future work (cf. Rako et al., 2006). Second, inversion polymorphisms can be maintained by frequency-dependent selection. For example, Nassar et al. (1973) reported that In(3R)P might be subject to frequency-dependent selection under conditions of larval crowding. However, on theoretical grounds it is unclear how frequency-dependent selection would be able to maintain an inversion polymorphism for a long period of time, since even small amounts of recombination or gene conversion in heterokaryons will destroy LD between the genic target(s) of balancing selection and the inversion (Kirpatrick & Barton, 2006). Nonetheless, it would be interesting to reassess the findings of Nassar et al. (1973) and to directly investigate, for example, larval competitive ability as a function of In(3R)P karyotype. Third, there are several fitness-related traits that we have not measured on the homokaryons, including fecundity: since short-lived low-latitude flies are more fecund than long-lived high-latitude flies (Schmidt & Paaby, 2008), an important open question is whether In(3R)P affects fecundity and the trade-off between fecundity and lifespan. Putative effects of In(3R)P on fecundity and the fecundity-lifespan trade-off (or on other traits, such as larval competitive ability) might potentially help to explain how this inversion contributes to the maintenance of stable phenotypic clines across latitude. Fourth, the fitness components through which the In(3R)P polymorphism is maintained might be subject to genotype by environment interactions. In our assays, we phenotyped homokaryons at two growth temperatures and indeed found several karyotype by temperature interactions. Perhaps most interestingly, we observed that at 18°C Florida inversion homokaryons are significantly more resistant to starvation stress than Florida uninverted homokaryons, whereas this pattern was reversed for females at 25°C. However, this effect was very small; moreover, while the latitudinal temperature gradient is a major determinant of the cline in In(3R)P, other latitudinally varying environmental factors (e.g., precipitation, seasonality) seem to be important too (Kapun et al., 2016a). Determining how selection maintains In(3R)P will ultimately depend on a more detailed understanding of the environmental factors that affect this system.
Conclusions
Here we have asked whether a clinally varying chromosomal inversion polymorphism in D. melanogaster, In(3R)P, affects the clinal distribution of three fitness-related traits: adult lifespan, survival of starvation stress, and survival upon acute cold shock. Our central finding is that the cline in In(3R)P contributes to the latitudinal clines observed for these survival traits, in addition to effects of geography that are independent of this inversion (cf. Schmidt et al. 2000; Schmidt et al., 2005a, b; Schmidt & Paaby, 2008; Mathur & Schmidt, 2017). Together with the fact that In(3R)P underpins latitudinal clines in body size (e.g., Rako et al., 2006; Kapun et al., 2016b), our results thus suggest that this inversion might represent a clinally varying life-history ‘supergene’ (On the other hand, we cannot yet rule out that the effects on multiple complex traits are due to a single pleiotropic locus within the inversion.) In particular, our findings support the idea that life-history clines in D. melanogaster are maintained by fitness trade-offs across geography and that In(3R)P contributes to this maintenance. However, the precise nature of the evolutionary forces and fitness effects that keep this chromosomal inversion polymorphic in some places but not others remains to be elucidated.
Supplementary Material
Figure S1. Survival curves as a function of In(3R)P karyotype and temperature. Effects of In(3R)P and temperature (18°C vs. 25°C) on the proportion adult survival in females and males. The different curves represent Florida inverted (black), Florida standard (red), and Maine standard (blue). See Results, Fig. 1, and Table 1 for details.
Figure S2. Starvation survival curves as a function of In(3R)P and temperature. Effects of In(3R)P and temperature (18°C vs. 25°C) on the proportion adult survival upon starvation in females and males. The different curves show Florida inverted (black), Florida standard (red), Maine standard (blue). See Results, Fig. 2 and Table 2 for details.
Acknowledgments
We thank the members of the Flatt and Schmidt labs for discussion and help in the laboratory. Our research was financially supported by the Swiss National Science Foundation (SNSF PP00P3_133641; PP00P3_165836; 310030E-164207 to TF), the National Institutes of Health (NIH R01GM100366 to PS), and the National Science Foundation (NSF DEB 0921307 to PS). We also thank two anonymous reviewers as well as Fred Mery and Wolf Blanckenhorn for helpful comments on our paper.
Footnotes
DR ESRA DURMAZ (Orcid ID : 0000-0002-4345-2264)
DR MARTIN KAPUN (Orcid ID : 0000-0002-3810-0504)
PROFESSOR THOMAS FLATT (Orcid ID : 0000-0002-5990-1503)
Supporting Information
Additional Supporting Information may be found in the online version of this article:
A preliminary analysis of trait relationships. Supporting results text describing a (i) preliminary analysis of the relationship between wing area, as a proxy of body size and lifespan and (ii) MANOVA on multivariate life-history phenotype (i.e., a linear combination of size, lifespan, starvation and cold survival).
References
- Adrion JR, Hahn MW, Cooper BS. Revisiting classic clines in Drosophila melanogaster in the age of genomics. Trends Genet. 2015;31:434–444. doi: 10.1016/j.tig.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JL, Manenti T, Sørensen JG, MacMillan HA, Loeschcke V, Overgaard J. How to assess Drosophila cold tolerance: chill coma temperature and lower lethal temperature are the best predictors of cold distribution limits. Func Ecol. 2015;29:55–65. [Google Scholar]
- Anderson AR, Collinge JE, Hoffmann AA, Kellett M, McKechnie SW. Thermal tolerance trade-offs associated with the right arm of chromosome 3 and marked by the hsr-omega gene in Drosophila melanogaster. Heredity. 2003;90:195–202. doi: 10.1038/sj.hdy.6800220. [DOI] [PubMed] [Google Scholar]
- Anderson AR, Hoffmann AA, McKechnie SW, Umina PA, Weeks AR. The latitudinal cline in the In(3R)Payne inversion polymorphism has shifted in the last 20 years in Australian Drosophila melanogaster populations. Mol Ecol. 2005;14:851–858. doi: 10.1111/j.1365-294X.2005.02445.x. [DOI] [PubMed] [Google Scholar]
- Azevedo R, James AC, McCabe J, Partridge L. Latitudinal variation of wing : thorax size ratio and wing-aspect ratio in Drosophila melanogaster. Evolution. 1998;52:1353–1362. doi: 10.1111/j.1558-5646.1998.tb02017.x. [DOI] [PubMed] [Google Scholar]
- Bergland AO, Tobler R, González J, Schmidt P, Petrov D. Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. Mol Ecol. 2016;25:1157–1174. doi: 10.1111/mec.13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Hartl DL. Population Genomics of Inversion Polymorphisms in Drosophila melanogaster. PLoS Genet. 2012;8:e1003056. doi: 10.1371/journal.pgen.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Beecham E. Heritability of Two Morphological Characters Within and Among Natural Populations of Drosophila melanogaster. Genetics. 1987;117:727–737. doi: 10.1093/genetics/117.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Singh BN. Genetic differentiation and inversion clines in Indian natural populations of Drosophila melanogaster. Genome. 1991;34:618–625. doi: 10.1139/g91-094. [DOI] [PubMed] [Google Scholar]
- De Jong G, Bochdanovits Z. Latitudinal clines in Drosophila melanogaster: body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. J Genet. 2003;82:207–223. doi: 10.1007/BF02715819. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. Columbia University Press; New York: 1937. [Google Scholar]
- Dobzhansky T. Genetics of natural populations. IX. Temporal Changes in the Composition of Populations of Drosophila pseudoobscura. Genetics. 1943;28:162–186. doi: 10.1093/genetics/28.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of natural populations. XIV A Response of Certain Gene Arrangements in the Third Chromosome of a Response of Certain Gene Arrangements in the Third Chromosome of Drosophila pseudoobscura to natural selection. Genetics. 1947a;32:142–160. doi: 10.1093/genetics/32.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Adaptive changes induced by natural selection in wild populations of Drosophila. Evolution. 1947b;1:1–16. [Google Scholar]
- Dobzhansky T. Genetics of the Evolutionary Process. Columbia University Press; New York: 1970. [Google Scholar]
- Duvernell DD, Schmidt PS, Eanes WF. Clines and adaptive evolution in the methuselah gene region in Drosophila melanogaster. Mol Ecol. 2003;12:1277–1285. doi: 10.1046/j.1365-294x.2003.01841.x. [DOI] [PubMed] [Google Scholar]
- Etges WJ. Chromosomal Influences on Life-History Variation Along an Altitudinal Transect in Drosophila robusta. Am Nat. 1989;133:83–110. [Google Scholar]
- Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlötterer C, et al. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol Ecol. 2012;21:4748–4769. doi: 10.1111/j.1365-294X.2012.05731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian DK, Lack JB, Mathur V, Schlötterer C, Schmidt PS, Pool JE, et al. Spatially varying selection shapes life history clines among populations of Drosophila melanogaster from sub-Saharan Africa. J Evol Biol. 2015;28:826–840. doi: 10.1111/jeb.12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T. Genomics of clinal variation in Drosophila: disentangling the interactions of selection and demography. Mol Ecol. 2016;25:1023–1026. doi: 10.1111/mec.13534. [DOI] [PubMed] [Google Scholar]
- Flatt T, Schmidt PS. Integrating evolutionary and molecular genetics of aging. Biochim Biophys Acta. 2009;1790:951–962. doi: 10.1016/j.bbagen.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Amdam GV, Kirkwood TBL, Omholt SW. Life-history evolution and the polyphenic regulation of somatic maintenance and survival. Quart Rev Biol. 2013;88:185–218. doi: 10.1086/671484. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA. Physiological climatic limits in Drosophila: patterns and implications. J Exp Biol. 2010;213:870–880. doi: 10.1242/jeb.037630. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. Revisiting the Impact of Inversions in Evolution: From Population Genetic Markers to Drivers of Adaptive Shifts and Speciation? Annu Rev Ecol Evol Syst. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Weeks AR. Climatic selection on genes and traits after a 100 year-old invasion: a critical look at the temperate-tropical clines in Drosophila melanogaster from eastern Australia. Genetica. 2007;129:133–147. doi: 10.1007/s10709-006-9010-z. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Sgrò CM, Weeks AR. Chromosomal inversion polymorphisms and adaptation. Trends Ecol Evol. 2004;19:482–488. doi: 10.1016/j.tree.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Shirriffs J, Scott M. Relative importance of plastic vs genetic factors in adaptive differentiation: geographical variation for stress resistance in Drosophila melanogaster from eastern Australia. Func Ecol. 2005;19:222–227. [Google Scholar]
- Inoue Y, Watanabe TK. Inversion polymorphisms in Japanese natural populations of D. melanogaster. Jpn J Genet. 1979;54:69–82. [Google Scholar]
- Kapun M, van Schalkwyk H, McAllister B, Flatt T, Schlötterer C. Inference of chromosomal inversion dynamics from Pool-Seq data in natural and laboratory populations of Drosophila melanogaster. Mol Ecol. 2014;23:1813–1827. doi: 10.1111/mec.12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapun M, Fabian DK, Goudet J, Flatt T. Genomic Evidence for Adaptive Inversion Clines in Drosophila melanogaster. Mol Biol Evol. 2016a;33:1317–1336. doi: 10.1093/molbev/msw016. [DOI] [PubMed] [Google Scholar]
- Kapun M, Schmidt C, Durmaz E, Schmidt PS, Flatt T. Parallel effects of the inversion In(3R)Payne on body size across the North American and Australian clines in Drosophila melanogaster. J Evol Biol. 2016b;29:1059–1072. doi: 10.1111/jeb.12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennington WJ, Hoffmann AA, Partridge L. Mapping Regions Within Cosmopolitan Inversion In(3R)Payne Associated With Natural Variation in Body Size in Drosophila melanogaster. Genetics. 2007;177:549–556. doi: 10.1534/genetics.107.074336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419–434. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Kern A. Where’s the Money? Inversions, Genes, and the Hunt for Genomic Targets of Selection. Genetics. 2012;190:1153–1155. doi: 10.1534/genetics.112.139899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb WR. Chromosome inversion polymorphisms in Drosophila melanogaster II. Geographic clines and climatic associations in Australasia, North America and Asia. Genetica. 1982;58:213–221. [Google Scholar]
- Knibb WR, Oakeshott JG, Gibson JB. Chromosome Inversion Polymorphisms in Drosophila melanogaster. I Latitudinal Clines and Associations between Inversions in Australasian Populations. Genetics. 1981;98:833–847. doi: 10.1093/genetics/98.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeunier F, Aulard S. Inversion polymorphism in Drosophila melanogaster. In: Krimbas CB, Powell JR, editors. Drosophila Inversion Polymorphism. CRC Press; Boca Raton: 1992. pp. 339–405. [Google Scholar]
- Lowry DB, Willis JH. A Widespread Chromosomal Inversion Polymorphism Contributes to a Major Life-History Transition, Local Adaptation, and Reproductive Isolation. PLoS Biol. 2010;8:e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur V, Schmidt PS. Adaptive patterns of phenotypic plasticity in laboratory and field environments in Drosophila melanogaster. Evolution. 2017;71:465–474. doi: 10.1111/evo.13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin LM, Merritt TJS, Zhu CT, Eanes WF. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics. 2005;170:1143–1152. doi: 10.1534/genetics.104.038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald SS, Rako L, Batterham P, Hoffmann AA. Dissecting chill coma recovery as a measure of cold resistance: evidence for a biphasic response in Drosophila melanogaster. J Insect Physiol. 2004;50:695–700. doi: 10.1016/j.jinsphys.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Mettler LE, Voelker RA, Mukai T. Inversion Clines in Populations of Drosophila melanogaster. Genetics. 1977;87:169–176. doi: 10.1093/genetics/87.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar R, Muhs HJ, Cook RD. Frequency-Dependent Selection at the Payne Inversion in Drosophila melanogaster. Evolution. 1973;27:558–564. doi: 10.1111/j.1558-5646.1973.tb00705.x. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Schmidt PS. Functional significance of allelic variation at methuselah, an aging gene in Drosophila. PLoS ONE. 2008;3:e1987. doi: 10.1371/journal.pone.0001987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, Schmidt PS. Dissecting the genetics of longevity in Drosophila melanogaster. Fly (Austin) 2009;3:29–38. doi: 10.4161/fly.3.1.7771. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Bergland AO, Behrman EL, Schmidt PS. A highly pleiotropic amino acid polymorphism in the Drosophila insulin receptor contributes to life-history adaptation. Evolution. 2014;68:3395–3409. doi: 10.1111/evo.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby AB, Blacket MJ, Hoffmann AA, Schmidt PS. Identification of a candidate adaptive polymorphism for Drosophila life history by parallel independent clines on two continents. Mol Ecol. 2010;19:760–774. doi: 10.1111/j.1365-294X.2009.04508.x. [DOI] [PubMed] [Google Scholar]
- Rako L, Anderson AR, Sgrò CM, Stocker AJ, Hoffmann AA. The association between inversion In(3R)Payne and clinally varying traits in Drosophila melanogaster. Genetica. 2006;128:373–384. doi: 10.1007/s10709-006-7375-7. [DOI] [PubMed] [Google Scholar]
- Rane RV, Rako L, Kapun M, Lee SF, Hoffmann AA. Genomic evidence for role of inversion 3RP of Drosophila melanogaster in facilitating climate change adaptation. Mol Ecol. 2015;24:2423–2432. doi: 10.1111/mec.13161. [DOI] [PubMed] [Google Scholar]
- Schaeffer SW. Selection in heterogeneous environments maintains the gene arrangement polymorphism of Drosophila pseudoobscura. Evolution. 2008;62:3082–3099. doi: 10.1111/j.1558-5646.2008.00504.x. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Paaby AB. Reproductive Diapause and Life-History Clines in North American Populations of Drosophila melanogaster. Evolution. 2008;62:1204–1215. doi: 10.1111/j.1558-5646.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Duvernell DD, Eanes WF. Adaptive evolution of a candidate gene for aging in Drosophila. Proc Natl Acad Sci USA. 2000;97:10861–10865. doi: 10.1073/pnas.190338897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PS, Matzkin L, Ippolito M, Eanes WF. Geographic Variation in Diapause Incidence, Life-History Traits, and Climatic Adaptation in Drosophila melanogaster. Evolution. 2005a;59:1721–1732. [PubMed] [Google Scholar]
- Schmidt PS, Paaby AB, Heschel MS. Genetic variance for diapause expression and associated life histories in Drosophila melanogaster. Evolution. 2005b;59:2616–2625. [PubMed] [Google Scholar]
- Schmidt PS, Zhu CT, Das J, Batavia M, Yang L, Eanes WF. An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2008;105:16207–16211. doi: 10.1073/pnas.0805485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander T, Libbrecht R, Keller L. Supergenes and complex phenotypes. Curr Biol. 2014;24:R288–R294. doi: 10.1016/j.cub.2014.01.056. [DOI] [PubMed] [Google Scholar]
- Sperlich D, Pfriem P. Chromosomal polymorphism in natural and experimental populations. In: Ashburner M, Carson HL, Thompson JR, editors. The Genetics and Biology of Drosophila. 3e. Academic Press; New York: 1986. pp. 257–309. [Google Scholar]
- Stalker HD. Chromosome Studies in Wild Populations of Drosophila melanogaster. II Relationship of Inversion Frequencies to Latitude, Season, Wing-Loading and Flight Activity. Genetics. 1980;95:211–223. doi: 10.1093/genetics/95.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Chien SA, Priest NK. Negligible senescence during reproductive dormancy in Drosophila melanogaster. Am Nat. 2001;158:248–258. doi: 10.1086/321320. [DOI] [PubMed] [Google Scholar]
- Tucic N. Genetic capacity for adaptation to cold resistance at different developmental stages in Drosophila melanogaster. Evolution. 1979;33:350–358. doi: 10.1111/j.1558-5646.1979.tb04688.x. [DOI] [PubMed] [Google Scholar]
- Weeks AR, McKechnie SW, Hoffmann AA. Dissecting adaptive clinal variation: markers, inversions and size/stress associations in Drosophila melanogaster from a central field population. Ecol Lett. 2002;5:756–763. [Google Scholar]
- Wright S, Dobzhansky Genetics of natural populations. XII Experimental Reproduction of Some of the Changes Caused by Natural Selection in Certain Populations of Drosophila pseudoobscura. Genetics. 1946;31:125–156. doi: 10.1093/genetics/31.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Survival curves as a function of In(3R)P karyotype and temperature. Effects of In(3R)P and temperature (18°C vs. 25°C) on the proportion adult survival in females and males. The different curves represent Florida inverted (black), Florida standard (red), and Maine standard (blue). See Results, Fig. 1, and Table 1 for details.
Figure S2. Starvation survival curves as a function of In(3R)P and temperature. Effects of In(3R)P and temperature (18°C vs. 25°C) on the proportion adult survival upon starvation in females and males. The different curves show Florida inverted (black), Florida standard (red), Maine standard (blue). See Results, Fig. 2 and Table 2 for details.


