Abstract
Posttranscriptional repression of UDP-glucuronosyltransferase (UGT) 1A expression by microRNAs (miRNAs) may be an important mechanism underlying interindividual variability in drug glucuronidation. Furthermore, the UGT1A 3’-UTR shared by all UGT1A enzymes is polymorphic, containing three linked SNPs (rs10929303, rs1042640, and rs8330) that could influence miRNA binding. The aim of this study was to identify the complete complement of miRNAs that could regulate UGT1A expression through binding to the reference and/or common variant UGT1A 3’-UTR. Luciferase reporter plasmids containing either the reference or variant UGT1A 3’-UTR were screened against a 2,048 human miRNA library to identify those miRNAs that decrease luciferase activity by at least 30% when co-transfected into HEK293 cells. Four novel miRNAs (miR-103b, miR-141–3p, miR-200a-3p, and miR-376b-3p) were identified that repressed both reference and variant UGT1A 3’-UTR, while two other miRNAs selectively repressed the reference (miR-1286) or variant (miR-21–3p) 3’-UTR. Deletion and mutagenesis studies confirmed the binding site location for each miRNA. rs8330 disrupted miR-1286 binding to the reference UGT1A 3’-UTR, while rs10929303 enhanced miR-21–3p binding to the variant 3’-UTR. Transfection of miR-21–3p, miR-103b, miR-141–3p, miR-200a-3p, and miR-376b-3p mimics into LS180 human intestinal cells showed repression of UGT1A1 and UGT1A6 mediated glucuronidation and mRNA without affecting UGT2B7 activity or mRNA. Furthermore, transfection of miR-21–3p, miR-141–3p, and miR-200a-3p into primary human hepatocytes, repressed UGT1A1 activity and mRNA without affecting CYP3A activity. Finally, miR-21–3p and miR-200a-3p expression were negatively correlated with UGT1A6 activity and mRNA in human liver samples. Thus, UGT1A is regulated by multiple miRNAs with some showing allele-dependent effects.
Keywords: UDP-glucuronosyltransferase, UGT1A, glucuronidation, microRNA, functional genomics
1. Introduction
The UDP-glucuronosyltransferases (UGTs) are a family of conjugation enzymes consisting of 19 functional enzymes in humans that are subdivided by genetic similarity into three subfamilies, UGT1A, UGT2A, and UGT2B. The UGT1A subfamily includes 9 functional enzymes in humans that are all encoded by a single gene through differential mRNA splicing of a unique first exon to common exons 2 to 5 that share the same 3’-untranslated region (UTR) [1]. The UGTs are critical for the efficient elimination of the majority of the top 200 prescribed drugs in the United States, ranked second only to the cytochrome P450 enzymes [2]. Furthermore, the UGT1A subfamily enzymes are involved in the metabolism of more than half of the most frequently prescribed drugs that are eliminated by glucuronidation [2]. Examples of clinically important drugs that are eliminated by UGT1A-mediated glucuronidation include those used to treat cancer (irinotecan) [3], HIV (raltegravir) [4], and organ rejection (mycophenolic acid) [5]. All these drugs have a narrow therapeutic index and demonstrate high interindividual variability in elimination [6–8] that can greatly complicate dosing and predispose some individuals to drug toxicity or inefficacy.
Previous studies in our laboratory and others have identified multiple genetic polymorphisms that are associated with the observed variability in UGT1A-mediated glucuronidation [1, 9–11]. Recently we identified a contributor to variability in acetaminophen glucuronidation, specifically a single nucleotide polymorphism (rs8330) in the 3’-UTR of the UGT1A gene, which is associated with altered susceptibility to acetaminophen hepatotoxicity in overdose patients [12]. However, only a relatively modest fraction of this variability is explained by the genetic variants discovered so far. For example, the well-known UGT1A1*28 polymorphism explains less than 27% of the observed interindividual variability in irinotecan glucuronidation in vivo [13]. Consequently, other trans-acting factors could regulate UGT1A expression, and explain this variability.
Recently, microRNAs (miRNAs) have emerged as crucial suppressive regulators of gene expression. miRNAs are a class of small non-coding RNAs that control gene expression via imperfect base pairing to the 3’-UTR of target genes [14]. Accumulating evidence demonstrates that miRNAs can modulate the expression of drug disposition genes, including cytochrome P450 enzymes, UGTs, and transporters [15]. Recent work has shown that miR-491–3p can bind directly to the UGT1A 3’-UTR and levels of miR-491–3p are negatively correlated with UGT1A3 and UGT1A9 transcript levels in human liver [16]. Studies in our laboratory have also shown that miR-375 levels in human liver are negatively correlated with UGT1A-mediated acetaminophen glucuronidation [17]. However, the mechanism of UGT1A regulation by miR-375 is indirect, through suppression of arylhydrocarbon receptor (AhR) transcription factor expression.
Despite these advances, there are still large gaps in our knowledge regarding the role of miRNAs in the regulation of the UGT1A enzyme subfamily. Importantly, it is unclear whether miRNAs other than miR-491–3p are involved in the direct regulation UGT1A expression. While most miRNAs have a quantitatively small (usually less than 2-fold) effect on gene expression, many genes are regulated by multiple miRNAs resulting in an additive or, in some instances, a synergistic effect [18]. Furthermore, the UGT1A 3’-UTR contains three common linked SNPs (rs10929303, rs1042640, and rs8330), which have the potential to either disrupt or enhance miRNA binding [12]. Consequently, the aim of the current study was to identify the complete complement of miRNAs that could regulate UGT1A expression through binding to the reference and/or common variant 3’-UTR. Our comprehensive discovery and validation approach involved initially a functional genomics screen using a UGT1A 3’-UTR luciferase reporter and a miRNA mimic library representing all of the available human miRNAs sequenced so far. This was followed by identification and functional validation of the miRNA response element in the 3’-UTR by a combination of in silico analysis, luciferase reporter construct assays, and evaluating miRNA mimic effects on UGT expression and activity in transfected human derived cells. Lastly, we confirmed the association of miRNA expression with UGT1A activity in human liver bank samples. The ultimate goal of this work is to elucidate the contribution of miRNA-regulated pathways to interindividual variability in UGT1A-mediated drug disposition.
2. Materials and methods
2.1. Chemicals and reagents.
Uridine diphosphate glucuronic acid (UDPGA), diazepam, and alamethicin were obtained from Sigma-Aldrich (St. Louis, MO). Ezetimibe and ezetimibe-D4 were purchased from Toronto Research Chemical (Toronto, ON, Canada). Morphine sulfate, serotonin, and midazolam were obtained from Cerilliant Corporation (Round Rock, TX). Formic acid, methanol, and acetonitrile (ACS grade) were purchased from Avantor Performance Materials, Inc. (Center Valley, PA). Acetic acid and ammonium formate (ACS grade) were obtained from Fisher Scientific (Grand Island, NY). The miRNA mimic library (miRIDIAN microRNA Mimic Library, v.19), the miRNA mimics for miR-21–3p, miR-103b, miR-141–3p, miR-200a-3p, and miR-376b-3p, the miRNA mimic negative controls (Negative controls 1 and 2), and the short interfering RNA directed against the UGT1A 3’-UTR [positive control (siUGT1A), 5’-CCU AGU CAU UUC CAA ACU UUU-3’] were purchased from GE Dharmacon (Lafayette, CO). The pMIR-REPORT plasmid was from Life Technologies (Grand Island, NY) and the Renilla luciferase vector pRL-CMV was purchased from Promega (Madison, WI). All synthesized DNA oligos used in this study were obtained from Integrated DNA Technologies, Inc. (Coralville, IA). Lipofectamine RNAiMAX was from Life Technologies and the transfection reagent DharmaFECT Duo was purchased from GE Dharmacon.
2.2. Cell lines and culture conditions.
HEK293 cells were cultured in DMEM (Life Technologies) supplemented with 10% FBS (Life Technologies). LS180 cells were maintained in DMEM (Life Technologies) supplemented with 10% FBS, 1 mM pyruvic acid (Life Technologies), and 0.1 mM MEM non-essential amino acids (Life Technologies). All cells were grown in a humidified chamber at 37°C and 5% CO2.
2.3. Construction of reporter plasmids.
The pMIR-UGT1A and pMIR-UGT1A-variant firefly luciferase plasmids containing the entire UGT1A 3′-UTR reference and major variant haplotypes (reference: CCC and variant: TGG for rs10929303, rs1042640, and rs8330, respectively) had been constructed as reported previously [12]. For the generation of the firefly luciferase plasmids pMIR-UGT1A rs10929303 and pMIR-UGT1A rs8330 we used the KAPA HiFi Polymerase (Kapa Biosystems, Inc., Wilmington, MA) together with the pMIR-UGT1A plasmid and primers rs10929303 F/R and rs8330 F/R (Table 1). For the generation of all deletion constructs we used the KAPA HiFi Polymerase together with the pMIR-UGT1A firefly luciferase plasmid and the respective set of primers (Table 1). pMIR firefly luciferase plasmids containing the putative miR-200a microRNA response element (MRE) triplicated in forward (miR-200a-3x F) and reverse (miR-200a-3x R) orientation were de novo synthesized by Blue Heron (Bothell, WA). All of the other triplicated constructs were generated using synthesized 5’-end phosphorylated oligonucleotides that were annealed with their complementary sequence (Table 1) and cloned into the pMIR-REPORT vector at the HindIII sites directly downstream of the firefly luciferase gene. All plasmids used in this study were verified by Sanger sequencing performed at the Washington State University Genomics Core.
Table 1.
List of primers used in this study. Underlined are shown the HindIII restriction sites used for cloning in the pMIR-REPORT vector. Bold are shown the respective MREs for each miRNA. All oligonucleotides used for the generation of the triplicate and deletion constructs were 5’-end phosphorylated.
| Primer | Sequence (5’ – 3’) | Comment |
|---|---|---|
| ΔMRE miR-1286 F | TCATAGGTGCCACCTTGTGTGTTTAAAGAAGGG | Used to generate the pMIR-UGT1A Δ1286 construct. |
| ΔMRE miR-1286 R | ACCATTATTGGTTAAGGATCAATTGCAACCATC | |
| miR-1286 MRE 3x wt F |
AGCTTCAGTCCTCATCTCTGTCCTGCTTCATAGGTG GAACCCCAGTCCTCATCTCTGTCCTGCTTCATAGGTG GAACCCCAGTCCTCATCTCTGTCCTGCTTCATAGGTGA |
Used to generate the 1286 3x F/R and 1286 3x wt F/R constructs. |
| miR-1286 MRE 3x wt R |
AGCTTCACCTATGAAGCAGGACAGAGATGAG GACTGGGGTTCCACCTATGAAGCAG GACAGAGATGAGGACTGGGGTTCCA CCTATGAAGCAGGACAGAGATGAGGACTGA |
|
| miR-1286 MRE 3x var F |
AGCTTCAGTCCTCATCTCTGTCGTGCTTCATAGGTG GAACCCCAGTCCTCATCTCTGTCGTGCTTCATAGGTG GAACCCCAGTCCTCATCTCTGTCGTGCTTCATAGGTGA |
Used to generate the 1286 3x var F/R constructs. |
| miR-1286 MRE 3x var R |
AGCTT
CACCTATGAAGCACGACAGAGATG AGGACTGGGGTTCCACCTATGAAG CACGACAGAGATGAGGACTGGGGTT CCACCTATGAAGCACGACAGAGATGAGGACTGA |
|
| ΔMRE miR-103b F | AGTTTACTTTTTTTCTGATGTTTCCTACAAC | Used to generate the pMIR-UGT1A Δ103b construct. |
| ΔMRE miR-103b R | ATTTTGGGTTTTTATCAAATTCAGCTCCATTTG | |
| miR-103b MRE 3x F | AGCTTACAGCTATGAAGTGCTGGGCA CGCCTTACAGCTATGAAGTGCTGGGCA CGCCTTACAGCTATGAAGTGCTGGGCAA | Used to generate the 103b 3x F/R constructs. |
| miR-103b MRE 3x R |
AGCTTTGCCCAGCACTTCATAGCTGTAAGGCGTGC CCAGCACTTCATAGCTGTAAGGCGTGCCCA GCACTTCATAGCTGTA |
|
| ΔMRE miR-141–3p F | AATTCATTTTATTCTTATTAAGGAAATACTTTGC | Used to generate the pMIR-UGT1A Δ141 construct. |
| ΔMRE miR-141–3p R | TGGAAATGACTAGGGAATGGTTCAAAATTTTACC | |
| miR-141–3p MRE 3x F |
AGCTTAACTTGAAAACAGAATCAGTGTTATATGTC AACTTGAAAACAGAATCAGTGTTATATGTCAACTTGA AAACAGAATCAGTGTTATATGTCA |
Used to generate the 141 3x F/R constructs. |
| miR-141–3p MRE 3x R |
AGCTTGACATATAACACTGATTCTGTTTTCAAGTTGACA TATAACACTGATTCTGTTTTCAAGTTGACATATAACACTG ATTCTGTTTTCAAGTTA |
|
| ΔMRE miR-200a-3p F | AAATTCATTTTATTCTTATTAAGGAAATAC | Used to generate the pMIR-UGT1A Δ200a construct. |
| ΔMRE miR-200a-3p R | GTTTGGAAATGACTAGGGAATGGTTC | |
| ΔMRE miR-376b-3p F | TTGGTGGGTGGTGTATTTGAGAAGATAATCATTG | Used to generate the pMIR-UGT1A Δ376b construct. |
| ΔMRE miR-376b-3p R | CCAAAGCAAGAAATCATATGCTGTTCTCAGTGC | |
| miR-376b-3p MRE 3x F |
AGCTTGGAAAAAGAATGATGCTATGAAA TATGTCGGAAAAAGAATGATGCTATGAAA TATGTCGGAAAAAGAATGATGCTATGAAAA |
Used to generate the 376b 3x F/R constructs. |
| miR-376b-3p MRE 3x R |
AGCTTTTTCATAGCATCATTCTTTTTCCG ACATATTTCATAGCATCATTCTTTTTCCGACAT ATTTCATAGCATCATTCTTTTTCCA |
|
| rs10929303 F | GAATATGTATCGTGCCCCCTCTGGTGTCTTTGATCAGGATG | Used to generate the pMIR-UGT1A rs10929303 construct. |
| rs10929303 R | CATCCTGATCAAAGACACCAGAGGGGGCACGATACATATTC | |
| miR-21–3p MRE 3x wt F |
AGCTTGCCCCCTCCGGTGTTAGACA GCCCCCTCCGGTGTTAGACAGCCCCCTCCGGTGTA |
Used to generate the 21 3x wt F/R constructs. |
| miR-21–3p MRE 3x wt R |
AGCTTACACCGGAGGGGGCTGTCTAACA CCGGAGGGGGCTGTCTAACACCGGAGGGGGCA |
|
| miR-21–3p MRE 3x var F |
AGCTTGCCCCCTCTGGTGTTAGACA GCCCCCTCTGGTGTTAGACA GCCCCCTCTGGTGTA |
Used to generate the 21 3x var F/R constructs. |
| miR-21–3p MRE 3x var R |
AGCTTACACCAGAGGGGGCTGTCTA ACACCAGAGGGGGCTGTCTAA CACCAGAGGGGGCA |
|
| rs8330 F | GGTCAGTCCTCATCTCTGTCGTGCTTCATAGGTGCCACC | Used to generate the pMIR-UGT1A rs8330 construct. |
| rs8330 R | GGTGGCACCTATGAAGCACGACAGAGATGAGGACTGACC |
2.4. Luciferase reporter assays.
For all luciferase assays HEK293 cells were co-transfected with a firefly luciferase construct (1.5 ng/well), the pRL-CMV Renilla luciferase control vector (0.5 ng/well) and the miRNA mimic or one of the mimic controls (negative or positive) at 30 nM final concentration using DharmaFECT Duo (0.05 μL/well). Transfections were performed in 384-well flat clear bottom white optical plates (Corning, NY) in quadruplicate. Briefly for each well to be transfected the transfection reagent was diluted in Opti-MEM Reduced Serum Medium without antibiotics, incubated 20 min at room temperature, followed by the addition of miRNA mimics and a subsequent incubation of 30 min at room temperature. HEK293 cells were harvested at ~80–90% of confluency, diluted in Opti-MEM, mixed with the transfection reagent-plasmid/miRNA complexes and plated at ~5 × 103 cells/well. Cells were harvested 24 hr after transfection and firefly activities relative to Renilla luciferase activities were measured with the SpectraMax i3 microplate reader (Molecular Devices, Sunnyvale, CA) using the Dual-Glo Luciferase Assay System (Promega).
For the miRNA mimic library screen we calculated for each 384-well plate the average of the relative luciferase values of cells transfected with the miRNA mimics, the negative mimic controls, and cells transfected only with the firefly luciferase constructs, and the pRL-CMV vector. Subsequently the relative luciferase values for each miRNA mimic were normalized to the average luciferase value of each plate and miRNA mimics were classified according to the changes in the normalized relative luciferase activity. For all subsequent luciferase experiments, the relative luciferase activity (firefly/Renilla ratio) of cells co-transfected with a miRNA mimic and a firefly luciferase construct were first normalized to the average luciferase activity values of cells transfected only with the firefly luciferase construct and cells transfected with the respective construct and the mimic negative controls (set at a value of 100%, control).
2.5. Transfection of LS180 cells and primary human hepatocytes and RNA extraction.
LS180 cells were transfected in 12-well and 6-well plates with miRNA mimics or the controls (positive or negative) at 30 nM in duplicate. Briefly, for each well 3 μL (12-well format) or 5 μL (6-well format) of the RNAiMAX transfection reagent was diluted in 100 μL or 500 μL respectively of Opti-MEM, incubated for 10 min at room temperature, miRNA mimics added and incubated for another 20 min at room temperature. LS180 cells were harvested at ~80–90% of confluency and diluted in Opti-MEM to ~8 × 105 cells/well (12-well format) or ~1.5 × 106 cells/well (6-well format). Cells were mixed with the miRNA mimics-transfection reagent complexes, plated, and incubated at 37°C in 5% CO2. All experiments were performed in triplicate.
Cryopreserved human hepatocytes from three donors (demographic details provided in Table 2) were obtained commercially from Life Technologies. Hepatocytes were plated in collagen-coated 96-well plates according to the manufacturer’s instructions. The cells were maintained (37°C at 5% CO2) in incubation medium. After 72 hr the sandwich-cultured human hepatocytes were transfected with miRNA mimics or the controls (positive or negative) at 30 nM in triplicate. Briefly for each well to be transfected 0.3 μL of RNAiMAX was diluted in 125 μL of William’s E Medium supplemented with the Maintenance Supplement Pack (Life Technologies), incubated 10 min at room temperature, followed by the addition of miRNA mimics and a subsequent incubation of 20 min at room temperature. The miRNA mimics-transfection reagent complexes were then added to the human hepatocytes. 48 hr after transfection, total RNA was extracted from LS180 cells and human hepatocytes with Trizol (Life Technologies).
Table 2.
Demographic characteristics of the hepatocyte donors.
| Lot number | Age (yrs) | Sex | Medications |
|---|---|---|---|
| Hu1540 | F | 59 | Not available |
| Hu1587 | F | 43 | Vitamin D, multivitamin, Caltrate plus |
| Hu1574 | M | 70 | Lipitor, Lisinopril, aspirin, Tamsulosin |
2.6. Reverse-transcriptase quantitative real-time PCR (qPCR).
Total RNA was isolated from primary human hepatocytes, and LS180 cells using Trizol (Life Technologies). Mature miRNAs were isolated from individual liver samples of our human liver bank (n=25) using the mirVana™ miRNA Isolation Kit, with phenol (Life Technologies) according to the manufacturers’ instructions. The source of the liver bank tissues have been published recently [1, 12]. These de-identified liver samples (Table 3) were obtained from publicly available sources, including the National Disease Research Interchange (Philadelphia, PA) and the Liver Tissue Procurement and Distribution Service (Minneapolis, MN). The use of these tissues for this study was approved by the Washington State University Institutional Review Board. mRNA and mature miRNA concentrations were determined by qPCR using reverse transcribed total RNA. For mRNA, cDNAs were first synthesized from total RNA using TaqMan Reverse Transcription Reagents (Life Technologies). qPCR was performed using TaqMan Gene Expression Master Mix and Taqman primer-probe mixes from Life Technologies, including UGT1A1 (Hs02511055_s1), UGT1A4 (Hs01655285_s1), UGT1A6 (Hs01592477_m1), UGT1A9 (Hs02516855_sH), UGT2B7 (Hs00426592_m1), CYP3A4 (Hs00604506_m1), and GAPDH (Hs02758991_g1). Mature miRNAs were also quantified from total RNA after reverse transcription with miRNA-specific primers and MultiScribe Reverse Transcriptase (Life Technologies). qPCR was performed using TaqMan Universal Master Mix II, no UNG and Taqman miRNA assays from Life Technologies, including miR-21–3p (ID# 002438), miR-103b (ID# 121115_mat), miR-141–3p (ID# 000463), miR-200a-3p (ID# 000502), miR-376b-3p (ID# 001102), miR-1286 (ID# 002773), miR-152–3p (ID# 000475), and miR-23b (ID# 002126). All assays were performed in triplicate. Gene expression was compared to an endogenous internal control (GAPDH for mRNA or the geometric mean of miR-152–3p/miR-23b for miRNA as described previously [19]). The ΔCt values for each miRNA were calculated this way for all of the samples in our human liver bank (n=25). Subsequently these values were normalized to the liver sample with the highest amount for each miRNA. These normalized values were then used to determine the correlation between the miRNA amounts in human liver samples and the activity of the UGT1A isoforms. For the determination of mRNA concentrations in transfected LS180 cells or human hepatocytes, the ΔCt values of the negative mimic controls and non-transfected cells were averaged (control). These values were subsequently used to calculate fold change in the expression levels of target genes (ΔΔCt method) in cells transfected with miRNA mimics.
Table 3.
Demographics of liver donors (n=25).
| Donor identity | Age | Gender | Ethnicitya | Cause of death | Smoking | Alcoholb | Other drug exposurec | rs8330 / rs10929303d |
|---|---|---|---|---|---|---|---|---|
| LV2 | 8 | F | A | Head trauma | No | No | +/- | |
| LV3 | 5 | M | C | Meningitis | No | No | Dopamine, penicillin | +/+ |
| LV4 | 24 | F | C | Auto accident | Dopamine | +/+ | ||
| LV5 | 20 | M | C | Head trauma | Yes | No | Nafcillin, mannitol, furosemide, erythromycin, nystatin, neomycin | +/+ |
| LV6 | 35 | F | C | Head trauma | No | No | None | +/+ |
| LV7 | 21 | M | A | Head trauma | Yes | Yes | DDAVP | +/+ |
| LV16 | 45 | F | H | Stroke | No | No | Dopamine | +/+ |
| LV17 | 6 | M | C | Auto accident | No | No | Dopamine | +/+ |
| LV23 | 74 | M | C | Colon cancer | +/+ | |||
| LV25 | 49 | F | C | Stroke | Yes | No | Diazepam, furosemide | -/- |
| LV30 | 40 | F | C | Head trauma | Yes | Yes | Mannitol, furosemide. phentolamine | +/+ |
| LV32 | 45 | F | C | Stroke | Yes | Yes | Prednisone, synthroid, phenytoin, naproxen, diazepam | +/- |
| LV36 | 43 | M | C | Stroke | No | No | Metoprolol, dexamethasone, hydrocortisol | +/+ |
| LV39 | 36 | M | C | Head trauma | Yes | Yes | Cefazolin, norcuronium, phytonadione, enalapril, metoclopramide | +/+ |
| LV40 | 34 | M | C | Gun shot wound | Yes | No | Crack cocaine, dopamine, norcuronium, fentanyl, midazolam | +/+ |
| LV41 | 43 | M | C | Head trauma | No | Yes | Dopamine, furosemide, neosynephrine, acetaminophen | +/- |
| LV43 | 24 | M | C | Blunt trauma | No | No | +/+ | |
| LV46 | 49 | M | C | Stroke | Yes | Yes | Occasional crack cocaine and marijuana | +/+ |
| LV48 | 68 | F | C | Stroke | No | No | Synthroid, celecoxib, fluoxetine, atenolol, omeprazole | +/+ |
| LV49 | 21 | M | C | Head trauma | Yes | No | Crack cocaine, marijuana | -/- |
| LV50 | 37 | M | C | Head trauma | Yes | Yes | Marijuana | +/- |
| LV51 | 62 | M | C | Stroke | No | No | Dopamine, norepinephrine, insulin, DDAVP, cefasolin | -/- |
| LV53 | 54 | M | C | Brain abscess | No | No | Simvastatin, acetaminophen, alprazolam, flurazepam, venlafaxine | +/+ |
| LV54 | 63 | M | C | Stroke | No | Yes | Dopamine, norepinephrine, nitroprusside | +/- |
| LV55 | 46 | M | C | Stroke | No | No | +/- |
: C, Caucasian; A, African-American; H, Hispanic.
: History of consumption of more than 14 drinks per week.
: DDAVP, desmopressin acetate.
: +/+, wild type homozygote; +/-, heterozygote; -/-, variant homozygote.
2.7. SNP genotyping analysis of UGT1A 3’-UTR in LS180 cells and primary human hepatocytes.
DNA from LS180 cells and sandwich-cultured human hepatocytes was extracted using a spin column kit (QIAamp DNA Blood Mini Kit, Qiagen, Germantown, MD). DNA samples were then genotyped using a real-time PCR instrument (CFX96 Touch, Bio-Rad, Hercules, CA) by allele discrimination assay (C_7607429_10, Applied Biosystems TaqMan SNP Genotyping Assay, Thermo Fisher Scientific, Waltham, MA). We demonstrated previously that the three SNPs present in the UGT1A 3’-UTR (rs10929303, rs1042640, and rs8330) are linked [12], thus the only variant assayed was UGT1A−3’UTR rs8330.
2.8. Glucuronidation assays.
UGT1A1, UGT1A6, and UGT2B7 selective glucuronidation activities were measured using ezetimibe, serotonin, and morphine as probe substrates, respectively [20, 21]. The procedure followed for LS180 cells (all activities) and sandwich-cultured human hepatocytes in 96-well format (ezetimibe glucuronidation only) was the same as we have described previously [17]. The normalized activity values of non-transfected cells and cells transfected with the mimic negative controls were averaged (control). Activity values of cells transfected with miRNA mimics were presented relative to the control. The concentration of metabolites was normalized to the total number of cells per well (ezetimibe and morphine glucuronides) or to protein concentration per assay (serotonin glucuronide).
2.9. CYP3A activity assay.
CYP3A activity in sandwich-cultured primary human hepatocytes (96-well format) was measured 48 hr after transfection using midazolam as the probe substrate. In brief, the culture medium was aspirated, the cells were rinsed twice with pre-warmed phosphate-buffered saline, and then incubated (37°C, 5% CO2) with serum-free William’s E Medium containing midazolam (2 μM) for 1 hr. The supernatant was collected and mixed with an equal volume of ice-cold acetonitrile containing the internal standard diazepam (0.5 μM final concentration) for measurement of 1’-hydroxymidazolam (1-OH MDZ) formation. After centrifugation at 15,000 g for 10 min, samples were dried down in a centrifugal vacuum, reconstituted in 40 μL of 0.1% formic acid in water/acetonitrile (90:10) and analyzed by HPLC-MS/MS. Gradient elution for 1’-hydroxymidazolam was performed using a mobile phase system consisting of (A) 0.1% formic acid in water, (B) acetonitrile and a flow rate of 0.4 mL/min. The gradient conditions were 10% B for 1.5 min, increasing to 70% B over 2.5 min, holding at 70% B for 3 min, increasing to 90% B over 0.05 min, holding at 90% B for 0.95 min, returning to 10% B over 0.05 min and holding at 10% B for 1.95 min. Positive ion (M/Z+) mass transitions monitored included 342→324 for 1’-hydroxymidazolam and 285→257 for diazepam.
All incubations were performed in triplicate. The normalized activity values of non-transfected cells and cells transfected with the mimic negative controls were averaged (control). Activity values of cells transfected with miRNA mimics were presented relative to the control. The concentration of 1’-hydroxymidazolam was normalized to the total number of cells per well.
2.10. Statistical analysis.
Statistical analysis was performed with a two-tailed Student’s t test using SigmaPlot Version 12 (Systat Software, Inc., San Jose, CA). Correlation analysis was performed by a Spearman’s rank method. A p value of less than 0.05 was considered statistically significant.
3. Results
3.1. A genome-wide miRNA mimic library screen identifies 5 novel miRNAs that decrease UGT1A-dependent firefly luciferase activity.
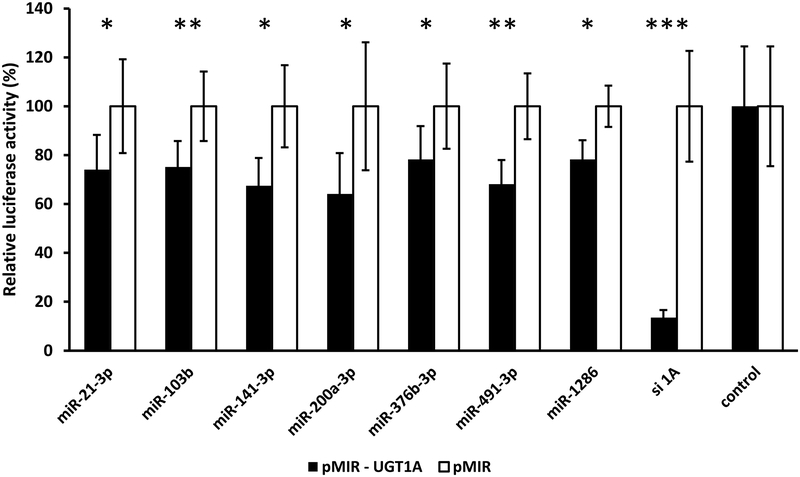
To identify candidate miRNAs involved in the regulation of UGT1A expression via direct binding to the common UGT1A 3’-UTR, we used a commercially available miRNA mimic library (miRIDIAN microRNA Mimic Library, v.19) that contained all human miRNAs sequenced at that time (2,048 miRNAs). We transiently co-transfected pMIR-UGT1A luciferase reporter plasmid and the pRL-CMV vector (Renilla transfection control) together with the miRNA mimics or the respective positive (siUGT1A) and negative controls into HEK293 cells. This cell line is ideal for the heterologous expression of UGT1A since the mRNA levels of all UGT1A subfamily members in HEK293 cells are very low [22]. Subsequently we determined the effect of each miRNA mimic on UGT1A−dependent luciferase activity, and classified them according to the degree of decrease of the relative UGT1A−mediated luciferase activity. We chose to study further the miRNAs from the initial library screen that showed at least 30% decrease in the relative UGT1A−dependent luciferase activity (30 miRNAs listed in Table 4). Although miR-491–3p caused 33% decrease in reporter activity, we did not study this miRNA further since a recent report has already demonstrated that it can regulate UGT1A 3’-UTR expression [16]. After counter-screening with the empty pMIR-REPORT vector to exclude effects unrelated to the presence of the UGT1A-3’UTR sequence, only five miRNAs (miR-103b, miR-141–3, miR-200a-3p, miR-376b-3p, and miR-1286) were found to significantly and specifically reduce the luciferase activity of cells transfected with pMIR-UGT1A luciferase reporter (Figure 1).
Table 4.
miRNAs identified to decrease the UGT1A−3’-UTR-mediated luciferase reporter activity by at least 30% through library screening. Decrease in luciferase activity is represented relative to the average luciferase activities of non-transfected HEK-293 cells and cells transfected with the miRNA mimic negative controls. In bold are the 5 novel candidate miRNAs identified in this study. Underlined is miR-491–3p which had been identified in a previous study by an in silico approach. Other listed miRNAs were not evaluated further since they showed a similar amount of inhibition of luciferase activity in a counter-screen of cells transfected with luciferase reporter lacking the UGT1A−3’UTR and so were considered false positives.
| miRNA | Decrease in luciferase activity relative to the miRNA negative control (%) |
|---|---|
| miR-1262 | 62.1 |
| miR-30c-1-3p | 56.7 |
| miR-30c-2-3p | 56.3 |
| miR-200a-3p | 53.5 |
| miR-208a | 51.5 |
| miR-4664-3p | 50.4 |
| miR-1271-5p | 47.5 |
| miR-4701-3p | 46.2 |
| miR-4781-5p | 43.2 |
| miR-103b | 42.6 |
| miR-1257 | 40.6 |
| miR-103a-2-5p | 40.6 |
| miR-3591-3p | 39.3 |
| miR-4648 | 37.7 |
| miR-129-5p | 36.8 |
| miR-204-5p | 36.1 |
| miR-630 | 35.1 |
| miR-1272 | 34.6 |
| miR-4738-5p | 34.5 |
| miR-4715-3p | 34.1 |
| miR-4740-5p | 33.8 |
| miR-1286 | 33.7 |
| miR-374b-5p | 33.6 |
| miR-5089-5p | 33.5 |
| miR-491-3p | 32.8 |
| miR-5001-5p | 31.0 |
| miR-190a | 30.9 |
| miR-376b-3p | 30.8 |
| miR-141-3p | 30.5 |
| miR-4731-3p | 30.3 |
Figure 1.
miRNAs identified through a functional genomics library screen that reduce UGT1A 3’-UTR luciferase reporter (pMIR-UGT1A) activity by at least 30% without affecting activity of the empty vector (pMIR). Shown are relative luciferase activities (firefly/Renilla ratio normalized to the pMIR control ratio) of HEK293 cells co-transfected with the pMIR-UGT1A or pMIR and with each miRNA mimic, a positive control (siUGT1A: short interfering RNA directed against the UGT1A 3’-UTR) or the mimic negative control. Bars represent mean ± one S.D. from three independent experiments performed in quadruplicate. * p < 0.05, ** p < 0.01, *** p < 0.001.
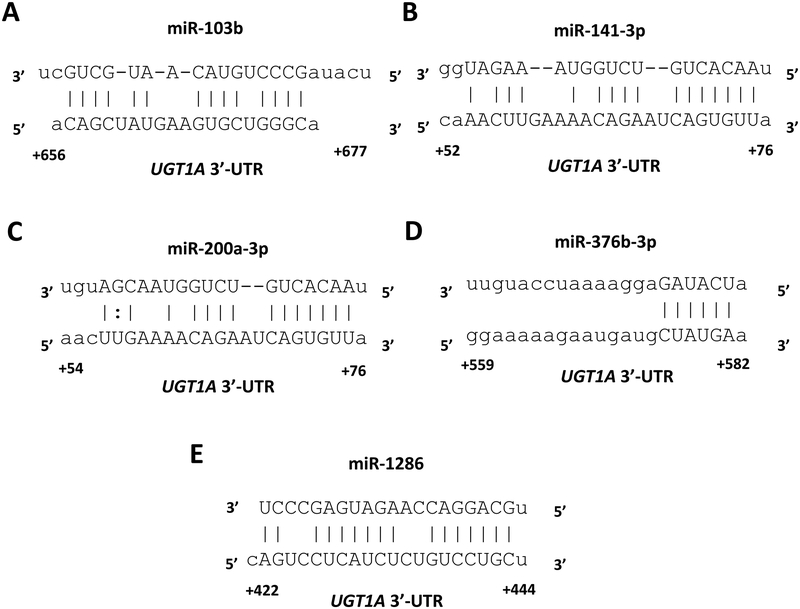
3.2. The UGT1A 3’-UTR contains functional microRNA response elements (MREs) for 5 novel candidate miRNAs.
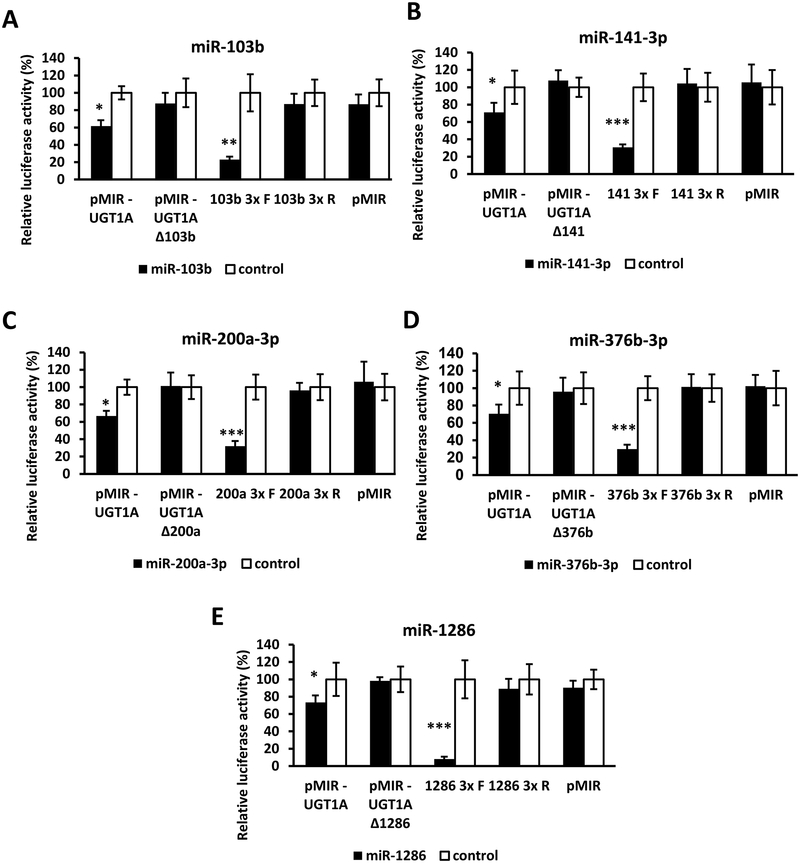
miRanda and RNAhybrid are well-verified online bioinformatics programs that predict the location of MREs in the 3’-UTRs of target mRNAs [23–25]. Using these programs, we identified putative MREs in the UGT1A 3’-UTR for each of the 5 candidate miRNAs (Figure 2). To confirm the functionality of the 5 candidate MREs in the common UGT1A 3’-UTR we generated firefly reporter constructs that carried the full length UGT1A 3’-UTR with a deletion (Δ) of the predicted MREs (pMIR-UGT1A-ΔmiR-103b, -ΔmiR-141–3p, -ΔmiR-200a-3p, -ΔmiR-376b-3p and -ΔmiR-1286, plasmids). We also cloned each predicted MRE sequence as a triplicate in the forward or reverse (negative control) orientation in the pMIR-REPORT firefly luciferase vector. We then transiently co-transfected the full length, the deletion, the triplicate constructs, or the empty pMIR-REPORT vector with miRNA mimics (miR-103b, miR-141–3p, miR-200a-3p, miR-376b-3p, or miR-1286) or the miRNA mimic negative controls into HEK293 cells and quantified the luciferase activity of each construct. Since miR-141–3p and miR-200a-3p were found to share overlapping UGT1A 3’-UTR MREs (see Figure 2B and 2C), we used the same deletion and triplicate MRE constructs to test the effects of both the miR-200a-3p and miR-141–3p mimics.
Figure 2.
MicroRNA response elements identified in the UGT1A 3’-UTR (reference sequence) by in silico analysis for 5 miRNAs (A-F) shown to reduce UGT1A 3’-UTR luciferase reporter activity by at least 30% without affecting an empty luciferase reporter. Nucleotide positions in the mRNA are indicated relative to the last nucleotide of the stop codon.
As shown in Figure 3 (left set of columns in each panel A-E), all miRNA mimics tested reduced the luciferase activity of constructs carrying the full length UGT1A 3’-UTR (pMIR-UGT1A). However, these reductions were abrogated when the respective MRE was deleted (Figure 3; second set of columns from left in each panel A-E). Moreover, cells transfected with the miRNA mimics and pMIR constructs carrying the MREs in the forward orientation showed a significant decrease in the luciferase activity compared with the control, while those with MREs in the reverse orientation showed no miRNA mimic effect (Figure 3; third and fourth set of columns from left in each panel A-E). As expected, none of the miRNA mimics affected luciferase activity of the empty pMIR-REPORT vector (Figure 3; right set of columns in each panel A-E).
Figure 3.
Functional validation of novel miRNA response elements (MREs; shown in Figure 2) in the UGT1A 3’-UTR (reference sequence). For each miRNA evaluated (panels A – E) luciferase reporter constructs were generated containing either the full length UGT1A 3’-UTR, a deletion (Δ) of the predicted MRE, or the triplicated MRE in the forward or reverse (negative control) orientation. Shown are relative luciferase activities (firefly/Renilla ratio normalized to the miRNA control) of HEK293 cells co-transfected with the luciferase reporter and either the miRNA mimic or the mimic negative control. Bars represent mean ± one S.D. from three independent experiments performed in quadruplicate. * p < 0.05, ** p < 0.01, *** p < 0.001.
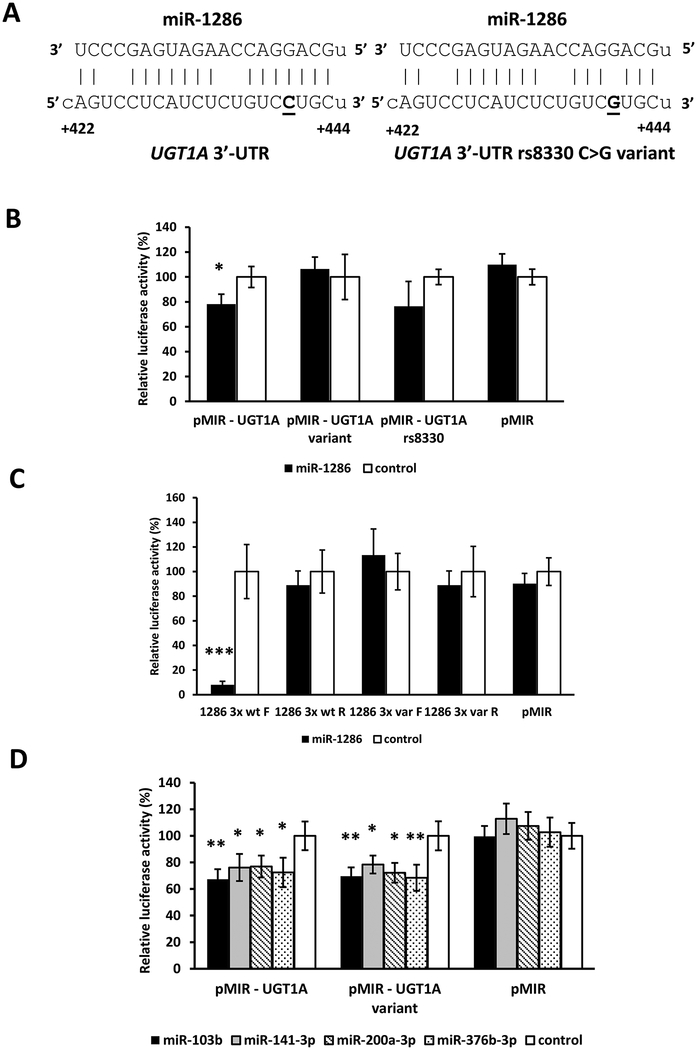
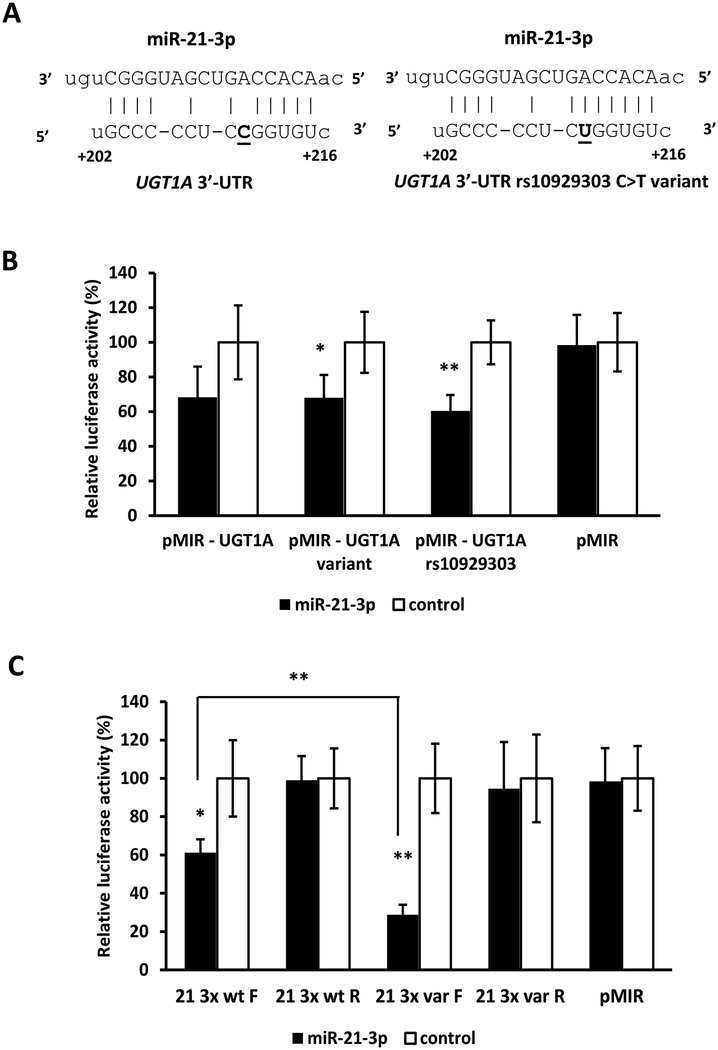
3.3. The miR-1286 MRE in the UGT1A 3’-UTR is destroyed by the rs8330 G>C SNP.
Of the 5 novel MREs we identified in the UGT1A 3’-UTR, one of these (miR-1286 MRE) contained one of the three SNPs (rs10929303, rs1042640, rs8330) that comprise the most common UGT1A 3’-UTR allelic variant. As shown in Figure 4A, the variant C allele of the rs8330 SNP was predicted to disrupt perfect base-pairing of the reference G allele of the rs8330 SNP in the miR-1286 “seed sequence” region. Consequently, to determine whether rs8330 could affect the function of the miR-1286 MRE, HEK293 cells were transfected with luciferase constructs carrying the full length reference UGT1A 3’-UTR sequence (pMIR-UGT1A), the rs10929303 T>C, rs1042640 C>G, and rs8330 C>G triple variant UGT1A 3’-UTR sequence (pMIR-UGT1A-variant), the rs8330 G>C only variant UGT1A 3’-UTR sequence (pMIR-UGT1A-rs8330), or the empty pMIR-REPORT vector, as well as the miR-1286 mimic or negative controls. As shown in Figure 4B, the miR-1286 mimic significantly reduced (by 30%, p = 0.03) luciferase activity of cells transfected with the reference UGT1A 3’-UTR, but had no effect on cells transfected with the UGT1A 3’-UTR with all 3 SNPs, or with only the rs8330 G>C variant, suggesting specific disruption of the miR-1286 MRE.
Figure 4.
Effect of the most common UGT1A 3’-UTR variant allele (comprised of three linked SNPs: rs10929303, rs8330, and rs1042640) on the binding of miRNAs to the reference UGT1A 3’-UTR. A) The rs8330 C>G SNP (bold and underlined) is located in the seed sequence of the miR-1286 miRNA response element (MRE) and predicted to disrupt binding of miR-1286 to the variant UGT1A 3’-UTR. B) miR-1286 decreases luciferase activity of the reference UGT1A 3’-UTR not the variant UGT1A 3’-UTR (all 3 SNPs) or the rs8330 UGT1A 3’-UTR (single SNP). C) miR-1286 decreases luciferase activity of the forward (F) triplicate reference (wt) miR-1286 MRE reporter but not of the triplicate variant (var) miR-1286 MRE reporter, or of the reverse orientation (R) reference or variant miR-1286 MRE reporters. D) All of the other miRNAs evaluated (miR-103b, miR-141–3p, miR-200a-3p and miR-376b-3p) decrease luciferase activities of both the reference and variant UGT1A 3’-UTR constructs. Shown are relative luciferase activities (firefly/Renilla ratio normalized to the miRNA control) of HEK293 cells co-transfected with the luciferase reporter and either the miRNA mimic or the mimic negative control. Bars represent mean ± one S.D. from three independent experiments performed in quadruplicate. * p < 0.05, ** p < 0.01, *** p < 0.001.
The effect of the rs8330 G>C nucleotide change on the miR-1286 MRE was further confirmed by evaluating luciferase constructs containing the reference sequence (G allele) of the miR-1286 MRE triplicated in forward and reverse orientation (miR-1286 3x wt F and miR-1286 3x wt R), and constructs with the variant (C allele) miR-1286 MRE triplicated in forward and reverse orientation (miR-1286 3x var F and miR-1286 3x var R). As seen in Figure 4C, we observed significantly reduced luciferase activity (by more 70%) for the forward construct with the reference miR-1286 MRE (miR-1286 3x wt F). This decrease was abrogated in the forward construct with the variant miR-1286 MRE (miR-1286 3x F variant). The miR-1286 mimic had no effect on any of the reverse constructs or the empty vector.
Finally, we evaluated whether the repressive effect of the other 4 microRNAs (miR-103b, miR-141–3p, miR-200a-3p, and miR-376b-3p) on the reference UGT1A 3’UTR (pMIR-UGT1A) was influenced by any of the 3 SNPs present in the variant UGT1A 3’UTR (pMIR-UGT1A-variant). As shown in Figure 4D, luciferase activities were reduced by miR-103b, miR-141–3p, miR-200a-3p, and miR-376b-3p in both pMIR-UGT1A (reference) and pMIR-UGT1A-variant by the same extent indicating these SNPs do not affect regulation by these other miRNAs.
3.4. A miR-21–3p MRE is created by the rs10929303 C>T SNP in the UGT1A variant allele 3’-UTR.
We next sought to determine whether the three common SNPs present in the variant UGT1A 3’-UTR could create one or more novel MREs. We used the same miRNA mimic library (miRIDIAN microRNA Mimic Library, v.19) to perform a second genome-wide miRNA screen to identify miRNAs that selectively repress the variant UGT1A 3’-UTR reporter (pMIR-UGT1A variant) compared with the reference UGT1A 3’-UTR reporter (pMIR-UGT1A). Twenty eight miRNAs were initially identified that decreased pMIR-UGT1A-variant luciferase activity by at least 20% more than the effect of the same miRNA on luciferase activity of the reference pMIR-UGT1A construct in the first genome-wide screen. validation of the effects of these miRNAs in repeated independent experiments, only miR-21–3p was found to reduce significantly the luciferase activity of cells transfected with pMIR-UGT1A-variant without affecting the reference pMIR-UGT1A activities. We used a more stringent effect cutoff value in this screen (20%) than in the initial screen of the wild-type UGT1A 3’-UTR reporter (30%) since the use of a 30% cutoff failed to identify any miRNAs that selectively inhibited the variant UGT1A 3’-UTR reporter.
Using RNAhybrid we identified a putative miR-21–3p MRE in the UGT1A 3’-UTR variant sequence that was not present in the UGT1A 3’-UTR reference sequence since it contained the reference C allele (rather than the variant T allele) of the rs10929303 SNP which disrupted the “seed sequence” of the miR-21–3p MRE (Figure 5A). The functionality of this novel miR-21–3p MRE and specificity of the disruptive effect of this SNP was initially evaluated by generating a full-length UGT1A 3’-UTR reporter construct that contained only the rs10929303 SNP variant T allele (pMIR-UGT1A rs10929303) and comparing the effect of the miR-21–3p mimic on this construct as well as the reference pMIR-UGT1A and pMIR-UGT1A-variant constructs. As shown in Figure 5B, the miR-21–3p mimic decreased luciferase activities in both the triple variant (pMIR-UGT1A-variant) and single variant (pMIR-UGT1A rs10929303) constructs. Although the miR-21–3p mimic also appeared to decrease luciferase activity in the reference pMIR-UGT1A construct, this difference did not achieve statistical significance (p > 0.05). Finally, the effect of the rs10929303 C>T nucleotide change on the miR-21–3p MRE was further confirmed by evaluating luciferase constructs containing the reference sequence (C allele) of the miR-21–3p MRE triplicated in forward and reverse orientation (miR-21–3p 3x wt F and miR-21–3p 3x wt R), and constructs with the variant (T allele) miR-21–3p MRE triplicated in forward and reverse orientation (miR-21–3p 3x var F and miR-21–3p 3x var R). Interestingly, as seen in Figure 5C, we observed significantly reduced luciferase activity (by about 40%) for the forward construct with the reference miR-21–3p MRE (miR-21–3p 3x wt F). This decrease was even greater (to almost 75%; p<0.01 versus miR-21–3p 3x wt F) in the forward construct with the variant miR-21–3p MRE (miR-21–3p 3x F variant). The miR-21–3p mimic had no effect on any of the reverse constructs or the empty vector.
Figure 5.
Effect of the rs10929303 SNP on the binding of miR-21–3p to the UGT1A 3’-UTR. A) The rs10929303 C>T SNP (bold and underlined) is located in the seed sequence of the miR-21–3p miRNA response element (MRE) and predicted to enhance binding of miR-21–3p to the variant UGT1A 3’-UTR (right) compared to the reference UGT1A 3’-UTR (left). B) miR-21–3p decreases luciferase activity of the variant UGT1A 3’-UTR (all 3 SNPs) and the rs10929303 UGT1A 3’-UTR (single SNP) but not the reference UGT1A 3’-UTR. C) miR-21–3p decreases luciferase activity of the triplicate variant (var) miR-21–3p MRE reporter more than the reference (wt) miR-21–3p MRE reporter in the forward (F) orientation, but has no effect on the reverse orientation (R) reporters. Shown are relative luciferase activities (firefly/Renilla ratio normalized to the miRNA control) of HEK293 cells co-transfected with the luciferase reporter and either the miRNA mimic or the mimic negative control. Bars represent mean ± one S.D. from three independent experiments performed in quadruplicate. * p < 0.05, ** p < 0.01.
3.5. Expression of candidate miRNAs in human liver bank samples.
qPCR was then used to determine which of the 6 candidate miRNAs (the 5 miRNAs identified in Table 4 and miR-21–3p) are expressed in human liver. Mean (± SD) threshold cycle (Ct) values for each miRNA were determined using RNA extracted from 25 of our human liver samples. Relatively high transcript levels (corresponding to low Ct values) were observed for miR-200a-3p (28.9 ± 1.7) and miR-21–3p (30.3 ± 0.9). Much lower transcript levels were observed for miR-141–3p (34.0 ± 0.9), miR-103b (35.4 ± 5.6) and miR-376b-3p (37.6 ± 1.4), while miR-1286 showed no measurable expression in any of the liver samples tested (Ct values over 40).
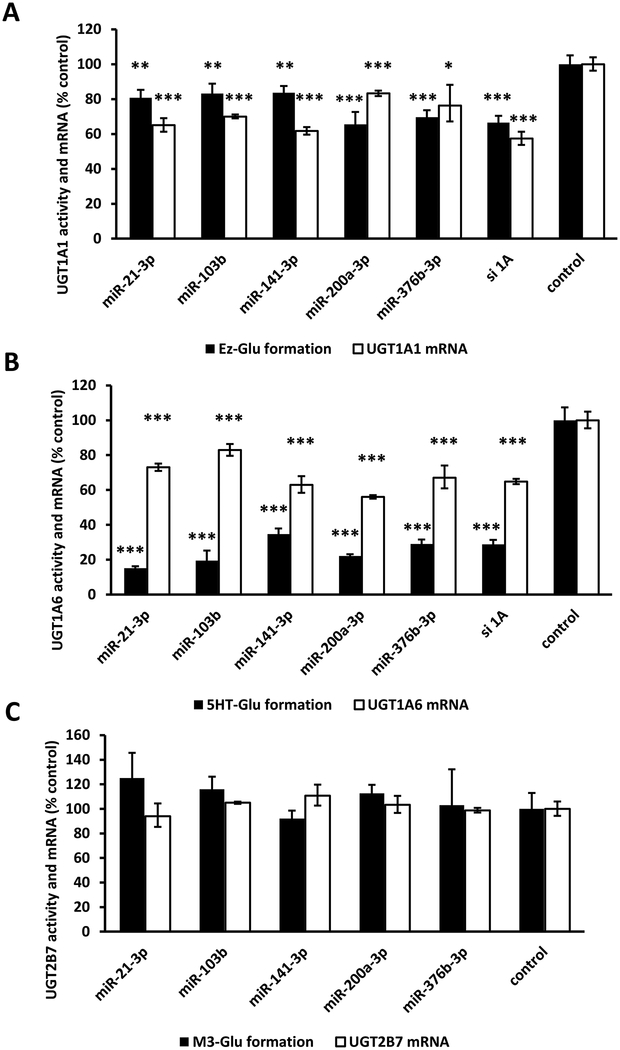
3.6. Effect of candidate miRNAs on UGT1A1, UGT1A6, and UGT2B7 activity and mRNA in LS180 cells.
Next, we examined the effects of 5 remaining candidate miRNAs (miR-21–3p, miR-103b, miR-141–3p, miR-200a-3p, and miR-376b-3p) on endogenous UGT expression in human LS180 adenocarcinoma cells, a model cell line that we have previously used to study UGT1A regulation [17]. We did not examine the effects of miR-1286 further in these studies since this miRNA did not appear to be expressed, at least in amounts that could be readily quantified by qPCR, in human liver (above). Genotyping analysis showed that these LS180 cells were homozygous for the UGT1A-3’-UTR reference (wild-type) allele. Cells were transfected with miRNA mimics or negative controls, and changes in UGT1A1, UGT1A6, and UGT2B7 isoform selective glucuronidation activities and UGT mRNA concentrations were determined. Glucuronidation of ezetimibe and serotonin were used as selective markers of UGT1A1 and UGT1A6 activity (respectively), while morphine-3-glucuronide formation was used to evaluate UGT2B7 function. UGT2B7 activity and mRNA served as negative controls since none of the miRNAs evaluated were predicted to interact with the UGT2B7 3’-UTR. As shown in Figure 6A, UGT1A1 activity and mRNA concentration were moderately reduced (by 20–40%) in cells transfected with each of the miRNA mimics when compared with the control cells. Furthermore, we observed moderate reductions in UGT1A6 mRNA (by 20–40%) and large reductions (by 65–85%) in UGT1A6 activity in cells transfected with each of the miRNA mimics compared to control cells (Figure 6B). Finally, UGT2B7 activity or mRNA concentrations were not affected by any of these miRNAs (Figure 6C).
Figure 6.
Effect of miRNA mimic overexpression on UGT1A1 (A), UGT1A6 (B), and UGT2B7 (C) selective activities and mRNA levels in LS180 human intestinal cells. miRNA mimics or the mimic negative controls were transfected into LS180 cells at 30 nM concentration. 48 hr after transfection, glucuronidation activities (UGT1A1: ezetimibe glucuronidation, Ez-Glu; UGT1A6: 5-hydroxytryptamine glucuronidation, 5HT-Glu; UGT2B7: morphine 3-glucuronidation, M3-Glu) were measured and total RNA isolated for the measurement of expression levels of the respective genes. Glucuronidation activity values and mRNA levels were compared with the average values of cells transfected with the mimic negative controls and non-transfected cells (control). Bars represent mean ± one S.D. from three independent experiments performed in quadruplicate. * p < 0.05, ** p < 0.01, *** p < 0.001.
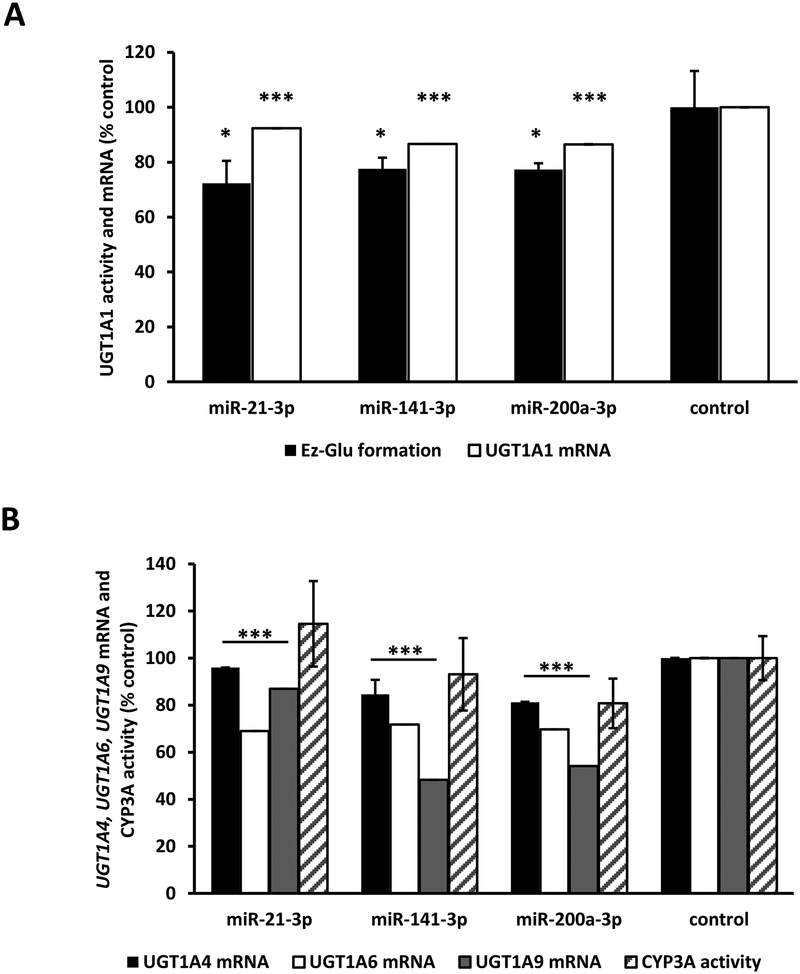
3.7. Effect of candidate miRNAs on UGT1A activity and mRNA in sandwich-cultured primary human hepatocytes.
We then determined whether the candidate miRNAs could regulate endogenous UGT1A gene expression in primary human hepatocytes from three different donors. Genotyping analysis indicated that all three donors were heterozygous for the UGT1A−3’-UTR variant allele. We chose miR-21–3p, miR-141–3p, and miR-200a-3p for this study since these miRNAs showed moderate to high expression (qPCR Ct values ≤ 35) in our human liver bank samples (see data above). Hepatocytes were transiently transfected with miRNA mimics or the miRNA mimic negative controls and the effects on UGT1A1 and CYP3A activities and UGT1A1, UGT1A4, UGT1A6, and UGT1A9 mRNA concentrations were determined. Ezetimibe glucuronidation and midazolam 1’-hydroxylation were used to indicate UGT1A1 and CYP3A activities, respectively. CYP3A activity was evaluated as negative control, since none of the miRNAs evaluated were predicted to interact with the CYP3A4 3’-UTR. As shown in Figure 7A, UGT1A1 activities were significantly reduced (by 20–30%) in hepatocytes transfected with each of the miRNA mimics compared with control cells. UGT1A1 mRNA concentrations also showed small but statistically significant decreases (by 5–10%) in human hepatocytes transfected with each of the miRNA mimics. Furthermore, as shown in Figure 7B, UGT1A4 showed small decreases (by 5–20%), while UGT1A6 showed moderate decreases (by 30–35%), and UGT1A9 showed the largest decreases (up to 55% for miR-141–3p and miR-200a-3p) following transfection of hepatocytes with the evaluated miRNAs. As expected, CYP3A activities were not affected by any of the miRNA mimics tested (Figure 7B).
Figure 7.
Effect of miRNA mimic overexpression on UGT1A1 and CYP3A enzyme activities, and UGT1A1, UGT1A4, UGT1A6, UGT1A9, and CYP3A4 mRNA levels in sandwich-cultured primary human hepatocytes. miRNA mimics or the mimic negative controls were transfected into primary human hepatocytes at 30nM concentration. 48hr after transfection enzyme activities (UGT1A1: ezetimibe glucuronidation, Ez-Glu; CYP3A: midazolam 1-hydroxylation) were measured and total RNA isolated for the measurement of expression levels of the respective genes. Enzyme activity values and mRNA levels are compared with the average values of cells transfected with the mimic negative controls and non-transfected cells (control). Bars represent mean ± one S.D. from three independent experiments performed in quadruplicate. * p < 0.05, *** p < 0.001.
3.8. miR-21–3p and miR-200a-3p are negatively correlated with UGT1A6 activity in human liver.
Finally, we sought to determine whether any of the candidate miRNAs were correlated with decreased hepatic glucuronidation activities for the major UGT1A enzymes expressed in human liver. We had previously determined hepatic glucuronidation activities for probe substrates that are selective for the UGT1A1 (estradiol), UGT1A4 (trifluoperazine), UGT1A6 (serotonin), and UGT1A9 (propofol) enzymes using samples from our human liver bank (results reported in [1]). We then determined miRNA concentrations of miR-21–3p, miR-103b, miR-141–3p, miR-200a-3p, and miR-376b-3p (normalized to the geometric mean of miR-152–3p/miR-23b concentrations) and correlated these values with the UGT1A activities by Spearman rank correlation. As shown in Table 5, a significant negative correlation was observed between hepatic UGT1A6 activity and transcript concentrations of both miR-21–3p and miR-200a-3p (Rho= −0.408, p < 0.05, for both). However, none of the other associations evaluated between UGT1A activities and miRNA concentrations reached statistical significance.
Table 5.
Correlation between hepatic miRNA transcript levels and glucuronidation activities of various human UGT1A isoforms measured in matched microsomal and RNA samples isolated from a human liver bank (n=25). Shown are the Spearman correlation coefficients (Rho) and the associated P values.
| miRNA | UGT1A1 activity | UGT1A4 activity | UGT1A6 activity | UGT1A9 activity | ||||
|---|---|---|---|---|---|---|---|---|
| Rho | p | Rho | p | Rho | p | Rho | p | |
| miR-21–3p | −0.376 | 0.07 | −0.150 | 0.475 | −0.408 | 0.048 | −0.295 | 0.16 |
| miR-103b | −0.132 | 0.538 | −0.169 | 0.43 | 0.077 | 0.719 | −0.096 | 0.653 |
| miR-141–3p | −0.073 | 0.734 | −0.1 | 0.639 | −0.36 | 0.0839 | −0.148 | 0.49 |
| miR-200a-3p | −0.376 | 0.069 | −0.153 | 0.475 | −0.408 | 0.048 | −0.296 | 0.16 |
| miR-376b-3p | −0.151 | 0.48 | −0.33 | 0.114 | −0.138 | 0.52 | 0.053 | 0.802 |
4. Discussion
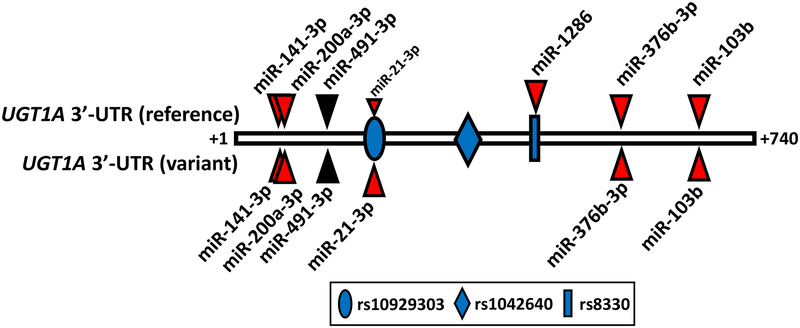
The most important outcome of this study was the discovery and validation of six novel MREs for miR-21–3p, miR-103b, miR-141–3p, miR-200a-3p, miR-376b-3p, and miR-1286 in the reference and/or common variant UGT1A 3’-UTR through an integrated functional genomics approach. The location of each MRE in relation to the SNPs defining the common variant UGT1A 3’-UTR are shown in Figure 8, as well as the MRE for miR-491–3p, which had been previously discovered by an in silico screen [16].
Figure 8.
Locations of known miRNA response elements (MREs) in the reference (top) and variant (bottom) UGT1A 3’-UTR. Indicated are the positions (from nucleotides +1 to +740 downstream of the UGT1A stop codon) of the six MREs validated in this study (red triangles) and the MRE for miR-491–3p, which had been reported previously [16]. Also indicated are the positions of the three linked SNPs (rs10929303, rs1042640, and rs8330) that comprise the variant UGT1A 3’-UTR allele. The rs10929303 SNP was found to substantially increased binding of miR-21–3p to the variant allele, while the rs8330 SNP abolished binding of miR-1286 to the variant 3’-UTR.
Of the six novel miRNAs we identified, two had MREs that were either allele-specific or allele-selective since they contained common SNPs located within the predicted miRNA seed sequence that affected miRNA binding. miR-1286 was allele-specific in that it bound only to the more common reference 3’-UTR sequence, while miR-21–3p was allele-selective since it bound to both alleles, but preferentially to the rarer variant sequence. In prior work we had found some evidence for UGT1A−3’UTR allelic imbalance in human liver RNA with lower amounts of reference versus variant mRNA associated with the rs8330 SNP that is located within the miR-1286 MRE [12]. Consequently, selective binding of miR-1286 to the reference allele could explain the observed allelic imbalance. We were unable to demonstrate significant expression of miR-1286 in our human liver samples using a commercial qPCR assay. Although expression of miR-1286 has been reported in cancer tissues, expression in normal human tissues is unclear. Publicly available RNA-seq data (http://www.gtexportal.org) indicates that miR-1286 is expressed in liver, lung, colon, breast, and cerebellum, although the absolute amount is unclear. Consequently, additional studies are needed to verify our tentative negative expression finding of miR-1286 in liver, including alternate methods to qPCR to quantitate miRNA levels.
Relatively high levels of miR-21–3p and miR-200a-3p were found in liver, and both of these were significantly correlated with hepatic UGT1A6 activity. Although a weaker but borderline non-significant correlation was observed with UGT1A1 activity, no correlation was observed with UGT1A4 or UGT1A9 activities. This lack of correlation with UGT1A1, UGT1A4, and UGT1A9 activities may be a consequence of additional regulatory mechanisms acting through the unique 5’-enhancer region of these UGTs (but not UGT1A6) that masks the effects of these miRNAs on the 3’-UTR shared by all these UGTs. The number of human livers available to us for study was somewhat limited (n = 25) and so it is also possible that evaluation of a larger number may reveal stronger correlations. Interestingly, our studies with LS180 cells and primary human hepatocytes also suggested differences in sensitivity of the different UGT1A isoforms to inhibition of mRNA expression and/or activity by the studied miRNAs, with UGT1A6 generally showing the greatest effect. Since our luciferase expression data suggested that the rs10929303 variant should enhance the effect of miR-21–3p on UGT1A expression, we would expect a stronger correlation between miR-21–3p levels and UGT1A activities in livers with this variant. Although preliminary analysis suggested a stronger negative correlation coefficient for heterozygous livers (data not shown), there were too few variant livers (6 heterozygous, 3 variant) out of the 25 total livers with complete data to reach a definitive conclusion.
In a previous study, we identified an alternate mechanism by which the UGT1A−3’-UTR variants could influence gene expression [12]. Specifically we found that the rs8330 SNP created an exon splice enhancer element that promotes splicing of exon 4 to the canonical exon 5a over an alternative exon 5b. Transcripts containing exon 5b encode for C-terminal truncated UGT1A isoforms that lack a transmembrane domain and act as suppressors of UGT enzyme activity. Consequently, rs8330 may enhance gene expression and enzyme activity through multiple mechanisms including enhanced splicing and reduced miRNA repression.
Results from different groups have shown that miR-21–3p is also involved in the regulation of transcription factors peroxisome proliferator activated receptor alpha (PPARα) [26] and hepatocyte nuclear factor 4 alpha (HNF4α) [27]. Moreover, expression of PPARα was shown to be regulated by miR-141–3p [28], while transcription factor Nrf2 was regulated by both miR-141–3p and miR-200a [29, 30]. Each of these transcription factors (PPARα, HNF4α, and Nrf2) are also known to regulate the hepatic expression of various UGT1A subfamily members [31–33]. Furthermore, hepatic mRNA levels of these transcriptional regulators are correlated with mRNA levels of the UGT1A isoforms [34]. Taken together these results suggest that these miRNAs (miR-21–3p, miR-141–3p, and miR-200a) probably regulate UGT1A expression through both direct (via the UGT1A 3’-UTR) and indirect (via transcription factors) mechanisms. Conversely, we have recently reported that miR-375 regulates UGT1A expression via an indirect effect by regulating Ah receptor transcription factor levels [17]. Consequently, further studies are required to elucidate the complex network of miRNAs and transcription factors responsible for regulating the UGT1A subfamily members.
A limitation of the current study was that we used a 30% cutoff for luciferase reporter inhibition in the initial library screen to focus our efforts on those miRNAs most likely to impact UGT1A expression. This threshold was sensitive enough to identify miR-491–3p (33% inhibition), which to our knowledge was the only other miRNA with a functionally validated MRE [16]. This cutoff also allowed us to minimize the significant amount of counter-screening that was needed since approximately 80% of candidate miRNAs were subsequently found to inhibit reporters lacking the UGT1A 3’-UTR (i.e. false positives). However, it is possible that some of the miRNAs evaluated in our initial screen that showed less than 30% inhibition of reporter activity could also be important regulators of UGT1A regulation. A recent report (published after our screening studies were completed) showed a small but statistically significant decrease (by 20%) in UGT1A−3’UTR luciferase reporter activity in HEK293 cells transfected with a miR-548d-5p precursor, but no effect on a reporter containing the cytochrome P450 3A4 3’UTR [35]. A review of our initial library screening data showed that miR-548d-5p, decreased UGT1A-3’UTR luciferase reporter activity by 28% (i.e. just below our threshold). They went on to show decreases in UGT1A1 mRNA levels (by nearly 50%) and glucuronidation activity (by over 30%) in HepG2 cells transfected with miR-548d-5p. However, they did not identify or functionally confirm the location of the miR-548d-5p MRE(s) within the UGT1A−3’UTR. Consequently, further studies are ongoing in our laboratory to evaluate whether additional miRNAs that we identified below the 30% threshold can regulate UGT1A−3’UTR expression, as well as to develop library screening approaches that minimize the number of false positives resulting from nonspecific effects on the empty luciferase reporter.
5. Acknowledgements.
This work was supported by the National Institutes of Health through National Institute of General Medical Sciences Grant NIH-R01-GM102130 (to M.H.C.) and the National Institute of Diabetes and Digestive and Kidney Diseases Grant N01-DK-7–0004 (to the Liver Tissue Cell Distribution System, University of Minnesota and University of Pittsburgh). Dr. Court was also supported by the William R. Jones endowment to Washington State University College of Veterinary Medicine.
Abbreviations:
- UGT
UDP-glucuronosyltransferase
- 3’-UTR
3’-untranslated region
- SNP
single nucleotide polymorphism
- miRNA
microRNA
- UDPGA
uridine diphosphate glucuronic acid
- qPCR
quantitative real-time PCR
- MRE
miRNA response element
- Ez-Glu
ezetimibe glucuronide
- 5HT-Glu
serotonin glucuronide
- M3-Glu
morphine-3 glucuronide
6. References
- [1].Court MH, Interindividual variability in hepatic drug glucuronidation: studies into the role of age, sex, enzyme inducers, and genetic polymorphism using the human liver bank as a model system, Drug Metab Rev. 42 (2010) 209–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, Ball SE, Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios, Drug Metab Dispos. 32 (2004) 1201–1208. [DOI] [PubMed] [Google Scholar]
- [3].Iyer L, King CD, Whitington PF, Green MD, Roy SK, Tephly TR, Coffman BL, Ratain MJ, Genetic predisposition to the metabolism of irinotecan (CPT-11). Role of uridine diphosphate glucuronosyltransferase isoform 1A1 in the glucuronidation of its active metabolite (SN-38) in human liver microsomes, J Clin Invest. 101 (1998) 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kassahun K, McIntosh I, Cui D, Hreniuk D, Merschman S, Lasseter K, Azrolan N, Iwamoto M, Wagner JA, Wenning LA, Metabolism and disposition in humans of raltegravir (MK-0518), an anti-AIDS drug targeting the human immunodeficiency virus 1 integrase enzyme, Drug Metab Dispos. 35 (2007) 1657–1663. [DOI] [PubMed] [Google Scholar]
- [5].Picard N, Ratanasavanh D, Premaud A, Le Meur Y, Marquet P, Identification of the UDP-glucuronosyltransferase isoforms involved in mycophenolic acid phase II metabolism, Drug Metab Dispos. 33 (2005) 139–146. [DOI] [PubMed] [Google Scholar]
- [6].Arab-Alameddine M, Fayet-Mello A, Lubomirov R, Neely M, di Iulio J, Owen A, Boffito M, Cavassini M, Gunthard HF, Rentsch K, Buclin T, Aouri M, Telenti A, Decosterd LA, Rotger M, Csajka C, Population pharmacokinetic analysis and pharmacogenetics of raltegravir in HIV-positive and healthy individuals, Antimicrob Agents Chemother. 56 (2012) 2959–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cecchin E, Innocenti F, D’Andrea M, Corona G, De Mattia E, Biason P, Buonadonna A, Toffoli G, Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan, J Clin Oncol. 27 (2009) 2457–2465. [DOI] [PubMed] [Google Scholar]
- [8].Levesque E, Delage R, Benoit-Biancamano MO, Caron P, Bernard O, Couture F, Guillemette C, The impact of UGT1A8, UGT1A9, and UGT2B7 genetic polymorphisms on the pharmacokinetic profile of mycophenolic acid after a single oral dose in healthy volunteers, Clin Pharmacol Ther. 81 (2007) 392–400. [DOI] [PubMed] [Google Scholar]
- [9].Court MH, Duan SX, von Moltke LL, Greenblatt DJ, Patten CJ, Miners JO, Mackenzie PI, Interindividual variability in acetaminophen glucuronidation by human liver microsomes: identification of relevant acetaminophen UDP-glucuronosyltransferase isoforms, J Pharmacol Exp Ther. 299 (2001) 998–1006. [PubMed] [Google Scholar]
- [10].Girard H, Court MH, Bernard O, Fortier LC, Villeneuve L, Hao Q, Greenblatt DJ, von Moltke LL, Perussed L, Guillemette C, Identification of common polymorphisms in the promoter of the UGT1A9 gene: evidence that UGT1A9 protein and activity levels are strongly genetically controlled in the liver, Pharmacogenetics. 14 (2004) 501–515. [DOI] [PubMed] [Google Scholar]
- [11].Girard H, Thibaudeau J, Court MH, Fortier LC, Villeneuve L, Caron P, Hao Q, von Moltke LL, Greenblatt DJ, Guillemette C, UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver, Hepatology. 42 (2005) 448–457. [DOI] [PubMed] [Google Scholar]
- [12].Court MH, Freytsis M, Wang X, Peter I, Guillemette C, Hazarika S, Duan SX, Greenblatt DJ, Lee WM, The UDP-glucuronosyltransferase (UGT) 1A polymorphism c.2042C>G (rs8330) is associated with increased human liver acetaminophen glucuronidation, increased UGT1A exon 5a/5b splice variant mRNA ratio, and decreased risk of unintentional acetaminophen-induced acute liver failure, J Pharmacol Exp Ther. 345 (2013) 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Innocenti F, Kroetz DL, Schuetz E, Dolan ME, Ramirez J, Relling M, Chen P, Das S, Rosner GL, Ratain MJ, Comprehensive pharmacogenetic analysis of irinotecan neutropenia and pharmacokinetics, J Clin Oncol. 27 (2009) 2604–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chekulaeva M, Filipowicz W, Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells, Curr Opin Cell Biol. 21 (2009) 452–460. [DOI] [PubMed] [Google Scholar]
- [15].Yu AM, Tian Y, Tu MJ, Ho PY, Jilek JL, MicroRNA Pharmacoepigenetics: Posttranscriptional Regulation Mechanisms behind Variable Drug Disposition and Strategy to Develop More Effective Therapy, Drug Metab Dispos. 44 (2016) 308–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dluzen DF, Sun D, Salzberg AC, Jones N, Bushey RT, Robertson GP, Lazarus P, Regulation of UDP-glucuronosyltransferase 1A1 expression and activity by microRNA 491–3p, J Pharmacol Exp Ther. 348 (2014) 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Papageorgiou I, Freytsis M, Court MH, Transcriptome association analysis identifies miR-375 as a major determinant of variable acetaminophen glucuronidation by human liver, Biochem Pharmacol. 117 (2016) 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Schmitz U, Lai X, Winter F, Wolkenhauer O, Vera J, Gupta SK, Cooperative gene regulation by microRNA pairs and their identification using a computational workflow, Nucleic Acids Res. 42 (2014) 7539–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lamba V, Ghodke-Puranik Y, Guan W, Lamba JK, Identification of suitable reference genes for hepatic microRNA quantitation, BMC Res Notes. 7 (2014) 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ghosal A, Hapangama N, Yuan Y, Achanfuo-Yeboah J, Iannucci R, Chowdhury S, Alton K, Patrick JE, Zbaida S, Identification of human UDP-glucuronosyltransferase enzyme(s) responsible for the glucuronidation of ezetimibe (Zetia), Drug Metab Dispos. 32 (2004) 314–320. [DOI] [PubMed] [Google Scholar]
- [21].Krishnaswamy S, Duan SX, Von Moltke LL, Greenblatt DJ, Court MH, Validation of serotonin (5-hydroxtryptamine) as an in vitro substrate probe for human UDP-glucuronosyltransferase (UGT) 1A6, Drug Metab Dispos. 31 (2003) 133–139. [DOI] [PubMed] [Google Scholar]
- [22].Nakamura A, Nakajima M, Yamanaka H, Fujiwara R, Yokoi T, Expression of UGT1A and UGT2B mRNA in human normal tissues and various cell lines, Drug Metab Dispos. 36 (2008) 1461–1464. [DOI] [PubMed] [Google Scholar]
- [23].Betel D, Wilson M, Gabow A, Marks DS, Sander C, The microRNA.org resource: targets and expression, Nucleic Acids Res. 36 (2008) D149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS, MicroRNA targets in Drosophila, Genome Biol. 5 (2003) R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R, Fast and effective prediction of microRNA/target duplexes, RNA. 10 (2004) 1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kida K, Nakajima M, Mohri T, Oda Y, Takagi S, Fukami T, Yokoi T, PPARalpha is regulated by miR-21 and miR-27b in human liver, Pharm Res. 28 (2011) 2467–2476. [DOI] [PubMed] [Google Scholar]
- [27].Wirsing A, Senkel S, Klein-Hitpass L, Ryffel GU, A systematic analysis of the 3’UTR of HNF4A mRNA reveals an interplay of regulatory elements including miRNA target sites, PLoS One. 6 (2011) e27438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hu W, Wang X, Ding X, Li Y, Zhang X, Xie P, Yang J, Wang S, MicroRNA-141 represses HBV replication by targeting PPARA, PLoS One. 7 (2012) e34165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shi L, Wu L, Chen Z, Yang J, Chen X, Yu F, Zheng F, Lin X, MiR-141 Activates Nrf2-Dependent Antioxidant Pathway via Down-Regulating the Expression of Keap1 Conferring the Resistance of Hepatocellular Carcinoma Cells to 5-Fluorouracil, Cell Physiol Biochem. 35 (2015) 2333–2348. [DOI] [PubMed] [Google Scholar]
- [30].Yang JJ, Tao H, Hu W, Liu LP, Shi KH, Deng ZY, Li J, MicroRNA-200a controls Nrf2 activation by target Keap1 in hepatic stellate cell proliferation and fibrosis, Cell Signal. 26 (2014) 2381–2389. [DOI] [PubMed] [Google Scholar]
- [31].Aueviriyavit S, Furihata T, Morimoto K, Kobayashi K, Chiba K, Hepatocyte nuclear factor 1 alpha and 4 alpha are factors involved in interindividual variability in the expression of UGT1A6 and UGT1A9 but not UGT1A1, UGT1A3 and UGT1A4 mRNA in human livers, Drug Metab Pharmacokinet. 22 (2007) 391–398. [DOI] [PubMed] [Google Scholar]
- [32].Barbier O, Girard H, Inoue Y, Duez H, Villeneuve L, Kamiya A, Fruchart JC, Guillemette C, Gonzalez FJ, Staels B, Hepatic expression of the UGT1A9 gene is governed by hepatocyte nuclear factor 4alpha, Mol Pharmacol. 67 (2005) 241–249. [DOI] [PubMed] [Google Scholar]
- [33].Yueh MF, Huang YH, Hiller A, Chen S, Nguyen N, Tukey RH, Involvement of the xenobiotic response element (XRE) in Ah receptor-mediated induction of human UDP-glucuronosyltransferase 1A1, J Biol Chem. 278 (2003) 15001–15006. [DOI] [PubMed] [Google Scholar]
- [34].Liu W, Ramirez J, Gamazon ER, Mirkov S, Chen P, Wu K, Sun C, Cox NJ, Cook E Jr., Das S, Ratain MJ, Genetic factors affecting gene transcription and catalytic activity of UDP-glucuronosyltransferases in human liver, Hum Mol Genet. 23 (2014) 5558–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Douglas SR, Shenoda BB, Qureshi RA, Sacan A, Alexander GM, Perreault M, Barrett JE, Aradillas-Lopez E, Schwartzman RJ, Ajit SK, Analgesic Response to Intravenous Ketamine Is Linked to a Circulating microRNA Signature in Female Patients With Complex Regional Pain Syndrome, J Pain. 16 (2015) 814–824. [DOI] [PubMed] [Google Scholar]