Abstract
In mammals, haptoglobin (Hp) is an acute-phase plasma protein that binds with high affinity to hemoglobin (Hb) released by intravascular hemolysis. The resultant Hp-Hb complexes are bound and cleared by the scavenger receptor CD163, limiting Hb-induced oxidative damage. In this study, we show that Hp is a divergent member of the complement-initiating MASP family of proteins, which emerged in the ancestor of jawed vertebrates. We demonstrate that Hp has been independently lost from multiple vertebrate lineages, that characterized Hb-interacting residues of mammals are poorly conserved in non-mammalian species maintaining Hp, and that the extended loop 3 region of Hp, which mediates CD163 binding, is present only in mammals. We show that the Hb-binding ability of cartilaginous fish (nurse shark, Ginglymostoma cirratum; small-spotted catshark, Scyliorhinus canicula; and thornback ray, Raja clavata) and teleost fish (rainbow trout, Oncorhynchus mykiss) Hp is species-specific, and where binding does occur it is likely mediated through a different structural mechanism to mammalian Hp. The continued, high-level expression of Hp in cartilaginous fishes where Hb-binding is not evident signals that Hp has (an)other, yet unstudied, role(s) in these species. Previous work indicates that mammalian Hp also has secondary, immunomodulatory functions that are independent of Hb-binding; our work suggests these may be remnants of evolutionary more ancient functions, retained after Hb-removal became the primary role of Hp in mammals.
Keywords: Cartilaginous fish, complement, haptoglobin, hemoglobin, MASP
Introduction
Haptoglobin (Hp) is an acute-phase plasma protein first discovered in 1938 by Polonovski & Jayle (1). While there is evidence that Hp has immunomodulatory effects (2–4), its principal recognized function in mammals is the sequestration of free hemoglobin (Hb) released into the bloodstream by damaged erythrocytes (5). Hp binds almost irreversibly to Hb with the resultant complex being cleared by the scavenger receptor CD163 present on monocytes and macrophages (6). In this way Hp, aided by the structurally-unrelated, heme-binding protein, hemopexin (Hx) (7), limits Hb-induced oxidative damage (supplemental fig 1a).
In most species examined, Hp is produced as a pro-protein containing one or two complement control protein (CCP) domains and a C-terminal serine protease (SP) domain. Mammalian pro-Hp is cleaved between the CCP and SP domain in the endoplasmic reticulum by C1r-like protein (C1r-LP) to generate the mature, disulphide-linked, protein (8). The enzymatically-inactive SP domain (or β-chain) mediates both Hb and CD163 binding (9, 10), while the CCP domains (comprising the α-chain) dictate the oligomerization state of Hp in the blood. For example, the human Hp gene exists in two major allelic forms, designated Hp1 and Hp2; the Hp1 allele has a lone CCP and forms disulphide-bonded dimers, while the Hp2 allele, which seemingly arose through the non-homologous recombination of two Hp1 genes after the divergence of Homo from other primates (11), has two CCP domains and forms higher-order multimers (10, 12). In contrast, teleost fish Hp (HpL) has a short (20 aa) peptide containing a cleavage site for a subtilisin-like pro-protein convertase in place of CCP domains and is monomeric in blood (9) (supplemental fig 1b).
While many evolutionary relationships have been proposed for Hp, the weight of evidence suggests it arose via partial duplication of a MASP family member (9, 13). The members of this family, MASPs 1–3, C1r, and C1s, are initiators of the complement system and play key roles in immune protection. MASP family members are also produced as pro-proteins, composed of two CUB, one EGF, two CCP and an SP domain; cleavage between the second CCP and SP domain followed by covalent association of the two chains via a disulphide bond leads to the mature form of each protein. MASP family members are classified into two types based upon their SP domains: MASP-1 has an active site serine encoded by a TCN codon, while in all other MASP family members this serine is encoded by AGY (13–15).
Beyond mammals and teleost fishes, information on Hp is sparse. While chicken and goose have lost Hp, with the structurally-unrelated PIT54 apparently substituting its Hb-binding function, other birds retain both molecules (9). Further, despite reports of an unknown Hb-binding protein in amphibian and reptile serum, Hp is absent from the genomes of Xenopus and anole lizard (9, 14, 16). While the cartilaginous fishes (sharks, skates, rays and chimaera) are the earliest branching vertebrate taxon to share true orthologs of the tetrameric Hb of mammals (17) it is proposed that Hp arose in the common ancestor of teleost fishes and tetrapods (9, 14). However, during analysis of blood plasma from nurse sharks (Ginglymostoma cirratum) we identified one of the major plasma proteins as the shark ortholog of Hp. Subsequently, we re-examined the evolutionary origins and distribution of Hp across vertebrate phylogeny, alongside its sequence-level evolution and associated capacity for Hb binding. Our analyses pinpoint how, and when, Hp became a major player in Hb clearance via CD163 and suggest that in some species Hp has roles that are independent of Hb-binding.
Materials & methods
Animal maintenance and sampling
Wild-caught nurse sharks (Ginglymostoma cirratum) were maintained in artificial seawater at approx. 28°C in indoor tanks at the Institute of Marine & Environmental Technology, Baltimore, USA. Captive-bred small-spotted catsharks (Scyliorhinus canicula) were maintained in artificial seawater at approx. 12°C in indoor tanks at the University of Aberdeen, UK. Little skate (Leucoraja erinacea) and thornback ray (Raja clavata) samples were obtained from wild-caught animals, and rainbow trout (Oncorhynchus mykiss) samples from purchased, farm-raised fish. Animals were anesthetized with MS-222 (0.16 g/L seawater) before bleeds were harvested from the caudal vein into 1/10 blood volume of 1000 U/ml porcine heparin reconstituted in shark-modified PBS (mammalian PBS supplemented with 15 ml 5M NaCl and 100 ml 3.5M urea per L) or, for trout, mammalian PBS, then spun at 300 g for 10 min to isolate blood plasma and packed RBCs. All procedures were conducted in accordance with University of Maryland School of Medicine IACUC protocols and the UK Home Office ‘Animals and Scientific Procedures Act 1986; Amendment Regulations 2012’.
Nurse shark plasma fractionation
Nurse shark plasma was fractionated by passage over a high-prep 16/60 Sephacryl S-300 high resolution size exclusion column (GE healthcare life sciences Ltd.) equilibrated in 0.5M NaCl, 0.05M NaPO4 (pH 7.2) running buffer, as previously described (18); samples from each fraction were run non-reducing on 5% SDS-PAGE gels in SGT buffer. For 2D analysis lane 7 from the non-reducing gel was cut out and placed in the well of a 12% SDS-PAGE gel. Excess 2X reducing Laemmli sample buffer (LSB) was added to the well, incubated for 5 min, then the gel run as above. Bands were visualized following Coomassie blue staining.
N-terminal and internal protein sequencing of the unknown plasma proteins
Protein samples were run on gels pre-run for 2 h at 3 mA in SGT with 430 μl of 10 mM reduced glutathione stock added per 100 ml buffer in the upper reservoir. The buffer was removed and replaced with fresh SGT containing 100 μl of 100 mM sodium thioglycolate stock per 100 ml prior to sample loading. The resultant gels were pre-equilibrated then blotted onto methanol wetted and equilibrated Immulon-PSQ PVDF membrane (Millipore) in CAPS transfer buffer for 2 h at 250 mA in a wet blotting system. Membranes were stained with amido black and air-dried before sending to the Microchemical and Proteomics core facility at Emory University for N-terminal sequencing by Edman degradation or internal sequencing following tryptic digestion. Returned peptide sequences are presented in table 1.
Table 1: N-terminal and internal protein sequencing results for the unknown nurse shark plasma proteins.
Lower case letters denote the probable amino acid at that position; x denotes the amino acid could not be determined. v/i denotes that a mixture of these amino acids was found at that position. Bold denotes a probable N-liked glycosylation site.
| Peptide | Sequence |
|---|---|
| P1 N-terminal | VVGGHLVHNGATPxTVLMLGPsgtv |
| P2 N-terminal | DHVETDHSKVHCGVPVxIThghY |
| internal 14 | VVCGRPIVPLEQhrq |
| internal 16 | DAYVYR |
| internal 17 | VVCGRPv/iWLEQH |
| internal 23 | LWEDVHFSNHIMPAclpah |
| internal 28 | VYVGIEDAR |
| internal 34 | NTDLGYEFPTxexv |
| internal 36 | wIDGIIHpk |
Cloning of cartilaginous fish Hp
N-terminal amino acid sequences for nurse shark P1 and P2 were reverse translated and used to design two degenerate 3’ RACE primers, GcP1-degenF and GcP2-degenF. These primers were used on RACE primed cDNA synthesized from 2.5 μg of nurse shark total RNA using the SMARTER RACE cDNA amplification kit (Clonetech), as per the manufacturer’s instructions. A band of ~300 bp was obtained with the primer GcP2-degenF with liver RNA allowing design of the gene specific primer GcP2-F2 to perform nested 3’ RACE PCR. Using this approach, we amplified a transcript of ~1100 bp which encoded peptide sequences from both P1 and P2. The 5’ end of the transcript was amplified using a nested 5’RACE approach with the primers GcP2-R1 and GcP2-R2. A single band was obtained using the primers GcP1/2-F1 & GcP1/2-R1 and the full-length sequence confirmed. Oligo-dT primed cDNA was prepared from liver RNA of little skate and small-spotted catshark using Illustra ready-to-go RT-PCR beads (GE healthcare life sciences Ltd.) according to the manufacturer’s instructions. To confirm the full-length little skate sequence the primers LeHp-F1 and LeHp-R1 were utilized, while for catshark the primers ScHp-F1 and ScHp-R1 were used. All primer sequences are detailed in table 2. Full-length Hp sequences can be accessed at GenBank using the following accession numbers: nurse shark, HM566086; little skate, JN564036; and small-spotted catshark, MG747494.
Table 2:
Sequences of primers used in this work. Standard nucleotide ambiguity codes are used, i indicates the presence of inosine.
| Primer | Sequence |
|---|---|
| GcP1-degenF | GTNCAYAAYGGiGCiACiCC |
| GcP2-degenF | AARGTNCAYTGYGGiGTiCCiG |
| GcP2-F2 | GCACAGACGATAATCAGTGG |
| GcP2-R1 | GGCCATGGTCCACGGCAG |
| GcP2-R2 | CCACTGATTATCGTCTGTGC |
| GcP1/2-F1 | CCCTCTCCCTCCAGCTGGTAC |
| GcP1/2-R1 | CACTGCGGGTGAATGATGCCGTCG |
| ScHp-F1 | ATGCTTCTCACAAAGATGTTCACTGTGG |
| ScHp-R1 | ACTCAATGTGCGGCCATGGTTTCC |
| LeHp-F1 | ATGTGGTTCCTCGTGTTAAACG |
| LeHp-R1 | TCAGTTATGTTCTATGACGTTGTTGATCC |
Hb-precipitation of plasma proteins
Hb-sepharose was prepared by lysing packed RBCs in 10 volumes of PBS at a 1/10 dilution then spun at 13K rpm for 10 min to remove cell debris. The RBC lysates were run on reducing SDS-PAGE against purchased ‘purified’ human Hb, imaged on an Azure biosystems c500 imaging system then images subject to densitometric analysis using ImageJ to assess their Hb content; Hb constituted >95% of the total protein in the human and catshark samples, >80% for nurse shark, and >70% for rainbow trout. Supernatants were diluted in 10 volumes of binding buffer and conjugated to activated cyanogen bromide sepharose (Sigma-Aldrich) according to manufacturer’s instructions. The Hb-sepharose was washed extensively to remove loosely bound Hb then equilibrated overnight in mammalian PBS (human and trout) or shark-modified PBS (all cartilaginous fish samples). Plasma samples were diluted 1/10 in PBS and 1 ml added to 200 μl species-matched Hb-sepharose then incubated for 1 h at room temperature with rotation. The Hb-sepharose was washed x1, x5 or x10 with PBS containing 0.05% tween20 (PBST). Following the final wash all supernatant was carefully aspirated, 50 μl of 2x reducing LSB was added to the sepharose and the resultant slurry boiled for 5 min to remove all bound proteins. For IMAC-precipitation 1 ml of plasma diluted 1/10 in PBS was added to 100 μl Ni2+-sepharose (Qiagen). The sepharose was washed x4 with 1 ml PBST, x2 with 1 ml 20 mM imidazole and x4 with PBST before the addition of reducing LSB and boiling as above. Boiled slurries were zip-spun and supernatants run on 4–12% Novex Bis-Tris gels (Invitrogen) in MOPS buffer. Bands were visualized by Coomassie blue staining. Bands of interest were excised from the gel with a clean scalpel then sent to the University of Aberdeen Proteomics Facility for protein identification via LC-MS/MS following in-gel tryptic digestion.
Comparative genomic searches
Tblastn/blastp searches of genomic/transcriptomic/protein sequence data available for species of interest (supplemental table 1) were performed using the well characterized human, pig, and pufferfish Hp protein sequences at a relaxed E-value cut-off of 10. Resulting hits were then blast searched against the NCBI nr protein database, and those which had top hits to Hp or other MASP family members were retained for further analysis. These were examined for typical characteristics of Hp (no active site serine, no CUB or EGF domains), and added to phylogenetic analysis of the MASP family and Hp. Loss of Hp was inferred where we could identify non-Hp MASP family members in a species but not Hp itself, despite Hp being the query for the search. Additional searches of taxa where Hp was found to be missing were also performed using the most closely related species with a newly identified Hp.
Phylogenetic analyses
All multiple sequence alignments (MSAs) were generated using PRANK with the “+F” parameter specified (19). All available libraries in TCS (20) were used to assess alignments for positional reliability, with columns scoring below 2 discarded to alleviate phylogenetic noise as a result of potential misalignment. IQ-TREE was used to identify the best-fitting model of amino acid substitution for each alignment (21).
Bayesian phylogenetic analyses were performed in BEAST v1.8 (22), specifying a Yule speciation prior, an uncorrelated lognormal relaxed molecular clock model, the best-fitting amino acid substitution (LG+I+G, JTT+I+G, and WAG+I+G for SP, C1r-LP, and CCP datasets respectively) and a random start tree. Two Markov chain Monte Carlo runs were executed for each analysis with chain lengths of 5, 10, and 20 million states each for the Hp CCP and C1r-LP, MASP CCP, and SP datasets respectively. In all cases chains were sampled every 1000 states before being assessed for sufficient sampling (effective sample sizes for all parameters >200) as well as convergence and mixing in Tracer v1.6 (http://beast.bio.ed.ac.uk/Tracer). Tree files were combined specifying a 20% burn-in using LogCombiner v1.8 and maximum clade credibility trees were then generated in TreeAnnotator v1.8. For the SP domain dataset Bayesian analyses were repeated as specified above but using less well-fitting models of amino acid substitution (LG+G, WAG+I+G, JTT+I+G) to determine if model perturbation would affect the placement of Hp within the MASP family.
Results
Identification of Hp in shark plasma
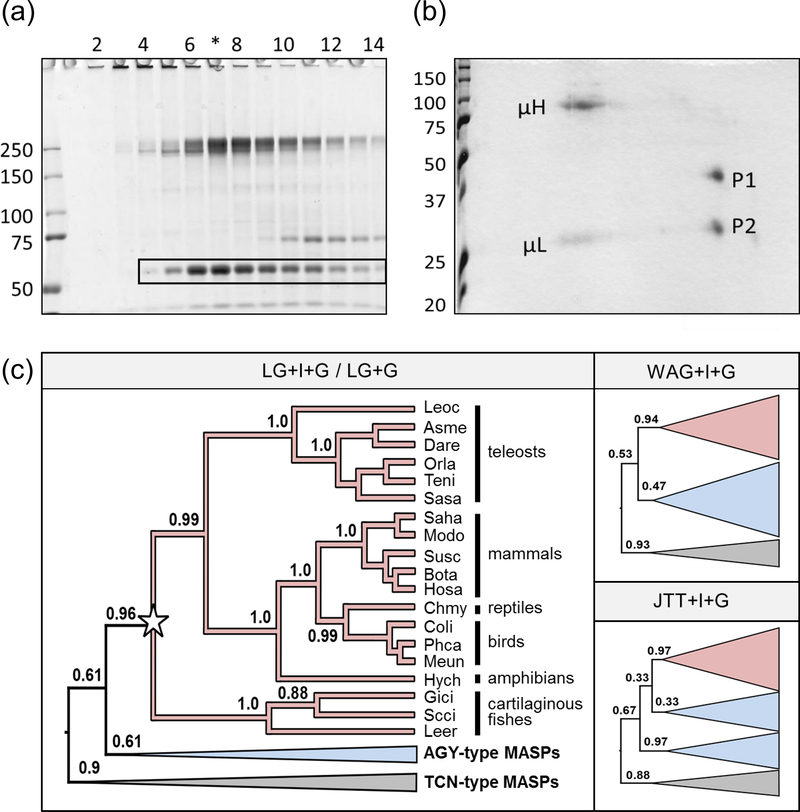
During size exclusion chromatography (SEC) of nurse shark plasma (18) we observed that one of the major plasma proteins eluted from the column at a molecular mass of ~250 kD, but on non-reducing SDS-PAGE had a molecular mass of ~60 kDa (boxed in figure 1a). Under reducing conditions this protein further resolved into two bands of ~35 and ~25 kDa (figure 1b; P1 and P2) indicating that the unknown protein was a disulphide-bonded heterodimer that forms non-covalently-associated oligomers in plasma. Immunoprecipitation of iodinated plasma proteins with immunoglobulin (Ig)-specific monoclonal antibodies showed the proteins were not associating with Igs present in the same fractions. P1 and P2 were submitted for N-terminal and internal protein sequencing, then the peptide sequences returned (table 1) subjected to blast analysis. A single hit, a 20 aa peptide sequence from a similarly sized, heterodimeric, sandbar shark (Carcharinus plumbeus) plasma protein (23), was returned using the P2 N-terminal sequence as the query.
Figure 1: Identification of unknown shark plasma proteins as Hp.
(a) Non-reducing SDS-PAGE of SEC-fractionated shark plasma showing the unknown protein at ~60 kDa (boxed). (b) When lane 7 (*) was cut from the gel and run in the second dimension under reducing conditions this band resolved into two spots of ~35 kDa (P1) and 25 kDa (P2). The heavy (μH) and light (μL) chains of IgM were also resolved. (c) Bayesian relaxed-clock rooting analyses of Hp (red clades) and the MASP family (blue clades AGY-type, grey clades TCN-type) strongly indicate that we have cloned cartilaginous fish Hp and, further, that Hp is a MASP family member irrespective of the model of amino acid substitution used (LG+I+G was the best-fitting model, and this was also tested without the invariant sites parameter i.e. LG+G. The less-well fitting, but commonly used, WAG and JTT models were also tested, along with the +I+G parameters). Four letter abbreviations for genus and species are used as detailed in supplemental table 1. Full tree topologies, posterior probabilities, species names, and accession numbers for (b) and (c) are presented in supplemental figure 2a.
Next, degenerate 5’ RACE primers were designed from the N-terminal sequences and a partial sequence obtained from liver RNA with the P2 primer; gene-specific RACE primers were then designed to complete the sequence. A product of ~1100 bp was identified that encoded both P1 and P2 within a single transcript of 429 amino acids. BLAST searches using this sequence identified Hp and MASP family members as the top hits, however analysis of conserved domains showed the unknown protein, with no CUB or EGF domains, was structurally most like Hp. tblastn searches using this nurse shark MASP/Hp homolog recovered similar transcripts for little skate (Leucoraja erinacea) in skatebase (24) and small-spotted catshark (Scyliorhinus canicula) in our in-house transcriptome (Redmond et al, manuscript submitted); both transcripts were amplified by RT-PCR from liver RNA from their respective species and confirmed by Sanger sequencing.
Phylogenetic analysis of Hp and the MASP family revealed the presence of a monophyletic Hp clade in jawed vertebrates that included our cartilaginous fish sequences, with branching patterns matching closely to expected species phylogeny (figure 1c). Critically, unlike previous studies (9, 14), we probabilistically determined the best-fitting amino acid substitution model and incorporated a relaxed molecular clock model to infer the root of the tree (25) as an appropriate outgroup is not known. The root fell between a MASP-1 clade (TCN) and a Hp + AGY-type MASP clade (figure 1c; supplemental figure 2a). This strongly indicates Hp is a MASP-family member that arose in the common ancestor of jawed vertebrates. Analyses using less well-fitting amino acid substitution models corroborated this finding, but the relationship of the AGY-type MASPs with Hp varied by model (figure 1c).
Characterization of cartilaginous fish Hp
All identified cartilaginous fish Hp orthologs have two CCP domains and an SP domain with an incomplete catalytic triad (H-D-R/H instead of H-D-S; figure 2a). Although CCP1 and CCP2 of classical MASPs grouped into separate clades (figure 2b, supplemental figure 2b), both CCPs from human and cow Hp fell within the CCP2 clade (figure 2c, supplemental figure 2c), fitting with their known evolutionary histories (26). Further, both cartilaginous fish CCPs fell within the CCP2 clade, indicating an independent CCP domain duplication event after their split with bony vertebrates, but before the emergence of elasmobranchs. Thus, Hp of the jawed vertebrate ancestor likely had a single CCP domain (CCP2), that has independently duplicated in several vertebrate lineages.
Figure 2: Sequence features of cartilaginous fish Hp.
(a) MSA of nurse shark (Gici), little skate (Leer) and small-spotted catshark (Scca) Hp with the two human (Hosa) Hp alleles (Genbank accession numbers; Hp1 NP_005134.1; Hp2 NP_001119574.1) and trout (Onmy) Hp-like (sequence from 9); CCP and serine protease (SP) domains are shaded and the pro-Hp cleavage motif underlined. The cysteines responsible for interchain bonding are marked by dots above the sequence and the unpaired cysteines that facilitate Hp oligomerization in mammals are highlighted in yellow. Cysteine pairs that form disulphide bonds in mammalian Hp are indicated by solid lines above the alignment while additional disulphide bonds predicted in cartilaginous fish Hp are indicated by dashed lines below the alignment. The charged residues important for the electrostatic docking of human Hp oligomers are indicated by +/− above the alignment. The residues which form the active site in other SP domains are indicated by white lettering on black, and the conserved Asp located at the base of the active-site cavity is marked with an asterisk. Residues identified as important in Hp-Hb complex formation [30] in mammals are shaded red, while those important for CD163 binding [32] are shaded blue. (b) Phylogenetic analysis of MASP CCP domains; regardless of the MASP family member or species from which they are derived, the CCP1 and CCP2 domains (shown in blue and red respectively on the structural schematic for a typical MASP family member) segregate into distinct clades. (c) However, both CCP domains in human, cow, shark, and skate Hp fall within the CCP2 group indicating independent domain duplication events in these lineages (marked by stars). Full tree topologies, posterior probabilities, species names, and accession numbers for (a) and (b) are presented in supplemental figures 2b and 2c respectively.
With two CCP domains, cartilaginous fish Hp has the same structure as human Hp2 (12, 27) and, according to its SEC profile, likewise forms multimers in plasma. However, as this interaction is disrupted under non-reducing conditions, oligomerization must occur via non-covalent forces rather than disulphide bonding, as in human Hp. Sequence comparison indicates that the unpaired Cys found at the N-terminus of each CCP domain in human Hp (highlighted in yellow in figure 2a) are paired in cartilaginous fish Hps (and classical MASPs; dotted lines on figure 2a), making it unavailable for intermolecular bonding. In human Hp, electrostatic docking occurs between the inter-CCP linker and tip of the N-terminal CCP domain of the two molecules before disulfide bond formation (key residues marked with +/− in figure 2a) (27); although these residues differs between the species, from our SEC data it appears that salt bonds maintain the quaternary structure of shark Hp, despite the harsh (high-salt, high-urea) conditions in vivo.
Both nurse shark and catshark Hp have a pro-protein convertase site akin to mammalian Hp (and not a subtilisin-like site as in teleost fish Hp), followed by an Ile-(Val/Leu/Ile)-Gly-Gly motif which is thought to drive a conformational change in the SP domains of trypsin-like serine proteases through interaction with a conserved Asp (marked with a star below the alignment in figure 2a) at the base of the active-site cavity (27) post-cleavage. Neither the cleavage site, nor I-(V/L/I)-G-G motif were found in little skate Hp suggesting the pro-protein is not processed in the same manner (or at all) in this species. However, the Cys pair required to link the cleaved chains in the mature protein (marked with dots in figure 2a) is present in all species.
Recurrent losses of Hp during vertebrate evolution
To better understand the evolution of Hp, we screened an array of sequence datasets for orthologs (see supplemental table 1 for full species list) and identified none within jawless fishes or non-vertebrate chordates. While we could not identify Hp in the elephant shark (a chimera) genome, its presence in sharks and skates indicates this to be a secondary loss. Further, the presence of Hp across vertebrate phylogeny was sporadic; for example, while conserved in ray-finned fish, including holosteans and teleosts, we did not identify Hp in coelacanth. Additionally, although Hp was missing from multiple anuran (frog) transcriptomes, it was identified in a salamander transcriptome. In reptiles, we identified Hp in turtle and crocodile, but not lizard or snake (squamate) datasets. Our data also indicates that the previously reported loss of Hp in birds is likely limited to the Galloanserae (land and water fowl). Although Hp is the primary route for Hb-sequestration and recycling in humans, the high frequency of loss events in other vertebrate lineages led us to question the role of Hp outside mammals.
Hp-Hb binding is species-dependent
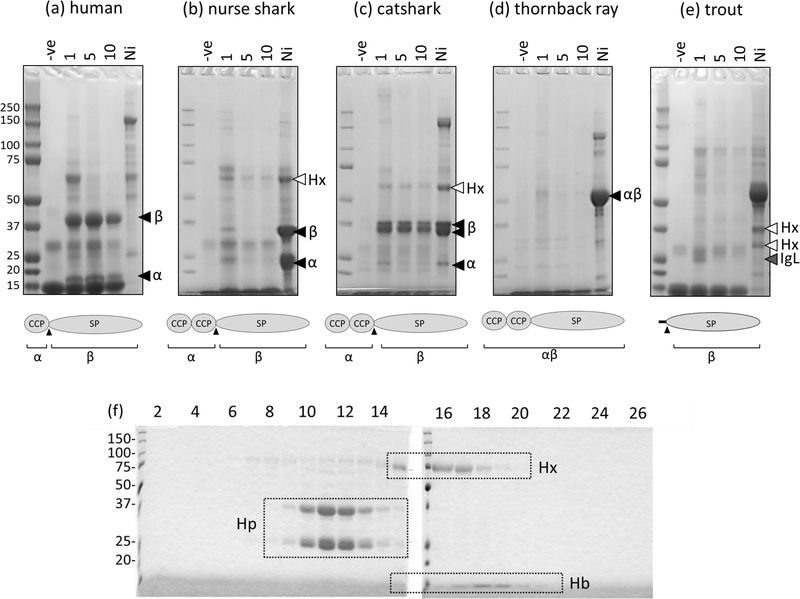
To test the Hb-binding ability of cartilaginous fish Hp we obtained blood from three species; nurse shark, small-spotted catshark, and thornback ray (Raja clavata). Red cell pellets were lysed in hypotonic buffer and the released Hb covalently coupled to activated cyanogen bromide sepharose, which was then used as a matrix for precipitation of proteins from species-matched blood plasma. Following 1, 5, or 10 washes, bound proteins were eluted by boiling the sepharose in reducing Laemmli sample buffer prior to SDS-PAGE; low intensity bands, representing leached Hb, were observed in all no-plasma controls (figure 3, -ve lane). As it was shown previously that shark Hp could be enriched from plasma by immobilized metal-ion affinity chromatography (IMAC) (23, 28) we also incubated plasma from each species with Ni2+-sepharose and washed 10 times before elution as above. Human Hp did not bind to Ni2+-sepharose but two strong bands representing the dissociated Hp α- and β-chains were observed following incubation with species-matched Hb-sepharose, regardless of pre-elution wash stringency (figure 3a). Given that the ability of an interaction to withstand dissociation by elution correlates well with affinity measured by other means (e.g. equilibrium dialysis or BIAcore (29, 30)), this result was consistent with the previously reported high affinity interaction of human Hp with dimeric Hb (10, 31).
Figure 3: SDS-PAGE of Hb-precipitated proteins obtained from (a) human, (b) nurse shark, (c) thornback ray, (d) small-spotted catshark, and (e) rainbow trout plasma.
Proteins were eluted from species matched Hb-sepharose incubated without plasma (-ve) or with plasma following 1, 5, or 10 washes with species matched PBS containing 0.05% tween20 (PBST) and run under reducing conditions on 4–12% gradient gels. The structure of Hp in each species is illustrated under each SDS-PAGE image, and their expected products upon reduction indicated underneath. Plasma incubated with Ni2+-sepharose (Ni) proves that Hp (black arrowheads indicate the SP domain-containing β-chain and CCP-containing α-chain) is present in all cartilaginous fish plasma samples, however only human and catshark Hp are significantly enriched on species-matched Hb. White arrowheads indicate full-length or partially degraded forms of the heme-binding protein Hx and the grey arrowhead identifies dissociated trout immunoglobulin light chain (IgL). All band identities were confirmed by LC-MS/MS. (f) Size-exclusion chromatography (SEC) confirms that nurse shark Hp does not bind species-matched Hb. Hp and Hx were IMAC-purified from nurse shark plasma and mixed with a limited amount of species-matched Hb. The mixture was passed over an S300 SEC column and the resulting fractions subject to reducing SDS-PAGE to assess their protein content. Fraction numbers are indicated above the lanes, boxes indicate the presence of Hb (which also gave the fraction a red color), Hp, or hemopexin (Hx) in each fraction.
Confirming previous observations (28) three bands were precipitated from nurse shark plasma with Ni2+-sepharose with those at 25 and 35 kDa being confirmed by LC-MS/MS as Hp α- and β-chains respectively (black arrowheads figure 3b). Neither of these bands was enriched on Hb-sepharose except under the lowest stringency wash conditions where faint bands were observed. The band at ~70 kDa (white arrowhead figure 3b) is Hx (28), indicating that our pull-down was successful, but that nurse shark Hp binds Hb with low affinity. Only a single band of ~50 kDa was observed for Hp following Ni2+-sepharose pull-down of thornback ray plasma suggesting that, like little skate Hp, this species also lacks a convertase cleavage motif and so is present in plasma in an unprocessed form. Regardless, there was no significant enrichment of this band on species-matched Hb-sepharose, even under the lowest stringency wash conditions (figure 3d). To ensure that the lack of Hp binding was not a consequence of the Hb-sepharose coupling procedure we mixed IMAC-enriched nurse shark plasma proteins with a limited amount of species-matched Hb, and the resultant mix was passed over an S300 SEC column. Fractions were then subject to SDS-PAGE to assess their protein content. Confirming our pull-down results, nurse shark Hb was found in the same fractions as Hx, not Hp (figure 3f). Contrasting with nurse shark and thornback ray, Hp from small-spotted catshark did bind Hb-sepharose (figure 3c). Several bands were enriched on both Ni2+-sepharose and Hb-sepharose; LC-MS/MS revealed the band at ~60 kDa (white arrowhead) is a degradation product of catshark Hx while those below (black arrowheads) are the α- and β-chains of catshark Hp.
Information on Hb-binding by Hp in other non-mammalian lineages is sparse, however Wicher and Fries previously reported successful precipitation of pufferfish (Takifugu rubripes) Hp on immobilized Hb from common carp (Cyprinus carpio) (9). Conversely, we found that even under our least stringent wash conditions, rainbow trout (Oncorhynchus mykiss) Hp was not enriched on species matched Hb (figure 3e; lacking CCP domains the expected molecular mass of trout pro-Hp is ~35 kDa, mature Hp ~30 kDa). However, the only bands approximating this size were determined by LC-MS/MS to be degradation products of trout Hx (also known as WAP65; white arrowheads) and Ig light chains (grey arrowhead).
Known Hb-interacting residues are poorly conserved outside mammals
Structural analysis of the mammalian Hp-Hb complex identified a number of residues important for Hb binding (10, 32). Specifically, residues within loop D, at the end of loop 3 and within loop 1 (using the nomenclature of Perona & Craik (33)) form extensive interactions with Hb, contributing significantly to the high affinity binding of the two molecules. MSA of Hp SP domains from across phylogeny (supplemental figure 3) indicates these Hb-interacting residues are poorly preserved between mammals and other vertebrates. This implies that distinct molecular interactions are used for complex formation in non-mammalian species where Hp does bind Hb. Interestingly, we observed that cartilaginous fish Hp is enriched for histidine (mean±SD: 9.1±1.7% His across the SP domain, n3 sequences) when compared to mammalian (2.9±0.5%, n6), reptile (4.4±1.0%, n3), bird (1.7±0.3%, n6), and teleost (2.1±0.9%, n7) orthologs, as well as other cartilaginous fish MASP family members (2.8±0.8%, n14; see supplemental table 1 for further details). His-enrichment likely accounts for the ability of cartilaginous fish Hp to bind Ni2+-sepharose (a property not possessed by mammalian or teleost Hp; figure 3a and 3e) and suggests Hb-binding by catshark Hp may occur via His-coordination of Hb’s Fe2+ ion.
The CD163 binding region of Hp arose in mammals
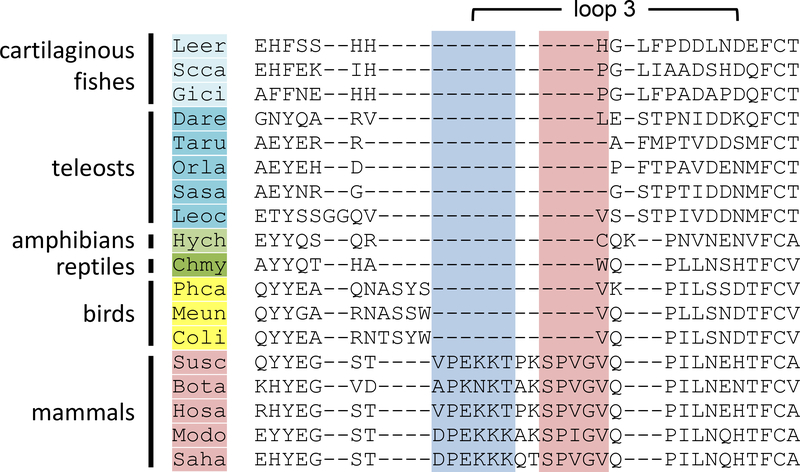
Loop 3 of human Hp is extended when compared to all other SPs (34, 35), forming a protrusion that binds the scavenger receptor CD163, allowing the removal of circulating Hp-Hb complexes (34). The MSA of vertebrate Hp SP domains, generated using PRANK to accurately predict indels (19), revealed that loop 3 is very short or missing in all vertebrate lineages besides mammals (figure 4), being more like that of MASPs and other SPs. This suggests that the entire loop 3 extension was acquired as a single insertion during early mammalian evolution and was present in the common therian ancestor. Importantly, as mutation of individual residues within loop 3 of human Hp abolishes CD163 binding (34), it is unlikely that Hb-complexed Hp proteins lacking loop 3 are removed from the blood by the same mechanism.
Figure 4: Comparison of the Hp loop 3 region across phylogeny.
MSA of the loop 3 region across vertebrate phylogeny. Four letter abbreviations for genus and species are as detailed in table S3. Residues identified as important in Hp-Hb complex formation [30] in mammals are shaded red, while those important for CD163 binding [32] are shaded blue.
Discussion
Mammalian Hp binds to Hb with extremely high affinity, allowing the resultant complex to be removed via CD163, thus limiting oxidative damage following intravascular hemolysis. Here we provide multiple lines of evidence that Hp acquired this role late in its evolutionary history.
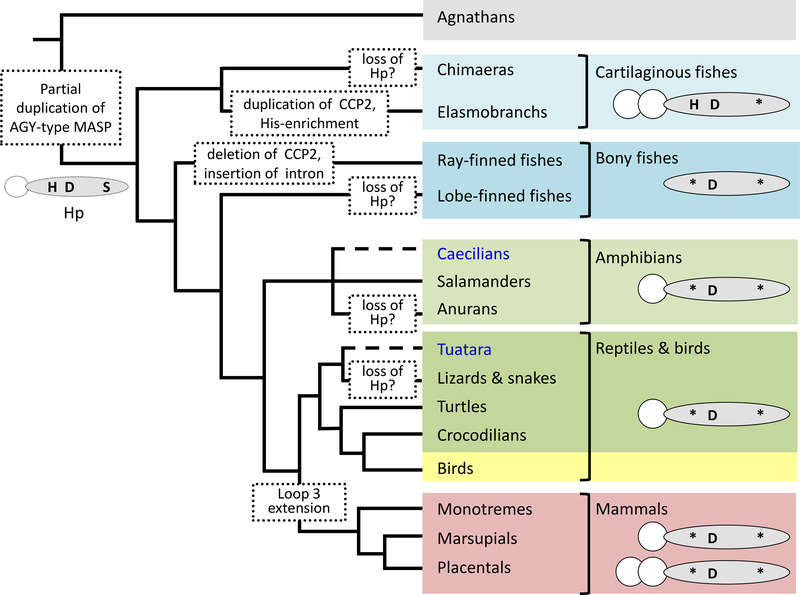
Our analyses indicate that the proto-Hp (CCP-SP structure) arose sometime before the emergence of extant jawed vertebrate lineages, via partial duplication of an AGY-type MASP family member (figure 5). Mutation of the active site serine would have rendered the proto-Hp unable to cleave other MASP-family members, while the absence of CUB and EGF domains would likely have excluded its incorporation into C1 or MASP complexes (36). However, the maintenance of the pro-protein convertase site (and lack of C1r-LP demonstrated herein) suggests the new molecule was still cleaved by complement system initiators, most likely C1r. Extant cartilaginous fishes have extremely high levels (>20 mg/ml) of IgM in their plasma (37); perhaps the equally-high levels of Hp expressed by members of this lineage (regardless of whether Hb-binding is evident or not) help prevent complement over-activation through the antibody-mediated, C1r-initiated, classical pathway.
Figure 5: Phylogeny and proposed evolution of Hp.
From the phylogeny of Hp we propose that a partial gene duplication of an AGY-type MASP gave rise to the proto-Hp, composed of one SP domain (shaded oval), and a single CCP domain (CCP2; white circle), prior to the emergence of the jawed vertebrates. This ‘proto-Hp’ likely carried a complete catalytic triad (H-D-S) in its SP domain, however mutation (*) of the serine (S) residue soon after would have rendered the molecule proteolytically inactive. In cartilaginous fishes the CCP domain was duplicated to give a CCP-CCP-SP structure, enabling formation of higher-order Hp oligomers, and became enriched for histidine. Bony fishes, in contrast, lost their CCP domain meaning their Hp cannot oligomerize and is found as a monomer in their blood. While some groups (Caecilians and Tuatara; highlighted in blue text) could not be assessed due to a lack of genomic data, from datasets currently available it appears Hp has been lost in several lineages (notably the anuran amphibians, scaled reptiles, and possibly also chimera and lobe-finned fishes). Additional, independent, CCP duplication events occurred in several mammalian lineages (e.g. human, cow, and deer). The extended loop 3 region required for high affinity binding to both Hb and the scavenger receptor CD163 is only found in mammals; this implies that even if Hp in other vertebrate groups can bind free Hb it is unlikely that the resultant complex is removed by the same mechanism.
Following their split from a common ancestor with bony vertebrates, the Hp of cartilaginous fish underwent CCP domain duplication (to regain a CCP-CCP-SP structure) and became markedly enriched for the amino acid histidine. The selective force behind this His-enrichment is unknown but, intriguingly, in the case of catshark Hp may confer the ability to bind-Hb. We predict that in this instance binding occurs through coordination of Hb’s Fe2+ ion, rather than the non-covalent interactions that dictate mammalian Hp-Hb binding. A similar, cross-reactivity has been observed for other His-rich proteins; for example, mammalian HRG (His-rich glycoprotein) binds a wide-range of targets that include divalent metal ions and heme (38), with binding to both being negated by chemical modification of the His residues (39). It is noteworthy that, in our hands, trout Hp did not show Hb- (or Ni2+−) binding. This contradicts the observation of Wicher and Fries with pufferfish Hp (9), and suggests that Hb-binding is also species-specific in bony fishes. How Hb-binding is mediated in teleost species, lacking many of the Hb-interacting residues of mammalian Hp or the his-enrichment of cartilaginous fish Hp, requires further investigation. Further, the mechanism used to safely detoxify any Hp-Hb complex formed in vivo in non-mammals remains unclear, given that it likely cannot be removed from the circulation by CD163. Some of these questions will hopefully be answered as crystal structures of Hp from different non-mammalian vertebrates become available.
It is apparent, however, that Hp underwent significant changes during early mammalian evolution, with further gene duplications (giving rise to the primate Hpr proteins (40)), the emergence of C1r-LP (supplemental figure 4) allowing cleavage of pro-Hp prior to secretion (8), as well as extension of the loop 3 region of the SP domain (supplemental figure 3). The loop 3 extension enabled Hp to bind Hb with high affinity by increasing the electrostatic pairing and surface area of interaction between the two molecules, as well as enabling the removal of the resultant Hp-Hb complex by the scavenger receptor CD163 (41) that had also newly emerged (42). Intriguingly, our data suggests the Hb-Hp-CD163 axis arose around the time RBCs became enucleated. While loss of their nuclei allowed RBCs to increase intracellular Hb levels, and hence aerobic capacity (43), it also prevented them from producing proteins for the maintenance and repair of their cell membranes. Thus they rupture in large numbers even under normal physiological conditions (44). The need to compensate for increased levels of free Hb would provide a strong selective force for the emergence of a more-efficient binding and clearance mechanism; the role appropriated by Hp in mammals. In contrast, ectothermic vertebrates, with lower counts of less fragile, nucleated RBCs (45, 46), within lower pressure circulatory systems, likely have much lower rates of RBC lysis. Further, as the subunits of other vertebrate Hbs are bound together more strongly than those of mammals (47, 48) we would expect less dissociation following hemolysis, and consequently only small amounts of dimeric Hb avaliable for capture by Hp (49). In such species, the heme-scavenger Hx, alone or in combination with other, more passive, systems (e.g. membrane-anchored scavenger receptors (50)), should be sufficient to cope with the expected lower levels of free Hb. Indeed, contrasting the high frequency of Hp loss events, Hx has been almost universally retained across vertebrate phylogeny (supplemental table 1), supporting the idea that it is the main route of Hb-sequestration and detoxification in many non-mammalian species.
Although the role(s) performed by Hp in non-mammal vertebrates awaits further investigation, given Hp’s origin as a member of the MASP family we anticipate they will be immune related. Indeed, this hypothesis is supported by the fact that even mammalian Hp has been ascribed additional, immunomodulatory functions, beyond the removal of Hb (e.g. regulation of endotoxin-induced inflammation (3), and supression of T-cell proliferation and Th2 cytokine release (4)). Our work suggests these could be evolutionary more ancient functions, retained after Hb-removal became the primary role of Hp in mammals. Future work should aim to better understand the extent of Hp’s functional repertoire, especially if off-target effects are to be minimised upon the clinical application of human Hp (e.g. to limit organ damage following blood transfusion or sickle-cell crisis (as reviewed in 51)).
Supplementary Material
Acknowledgements
Our thanks to E. Bryan Buckingham at the University of Maryland School of Medicine and the staff at the University of Aberdeen Proteomics Facility for their technical assistance.
2 Funding: This work was supported by NIH grant RR06603 awarded to MFF, Royal Society research grant RG130789 awarded to HD, and a PhD studentship awarded by the Centre for Genome-Enabled Biology & Medicine, University of Aberdeen, to AR. The authors declare that no competing interests exist.
3 Nonstandard abbreviations:
- Hb
hemoglobin
- Hp
haptoglobin
- IMAC
immobilized-metal affinity chromatography
- MASP
mannan-binding lectin-associated serine protease
- MSA
multiple sequence alignment
- SEC
size-exclusion chromatography
- SP
serine protease
References
- 1.Polonovski M, and Jayle M. 1938. Existence in the blood plasma of a substance hastening the peroxydasic activity of the hemoglobin. Cr Soc Biol 129: 457–460. [Google Scholar]
- 2.Huntoon KM, Wang Y, Eppolito CA, Barbour KW, Berger FG, Shrikant PA, and Baumann H. 2008. The acute phase protein haptoglobin regulates host immunity. J. Leukoc. Biol 84: 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arredouani MS, Kasran A, Vanoirbeek JA, Berger FG, Baumann H, and Ceuppens JL. 2005. Haptoglobin dampens endotoxin-induced inflammatory effects both in vitro and in vivo. Immunology 114: 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arredouani M, Matthijs P, Van Hoeyveld E, Kasran A, Baumann H, Ceuppens JL, and Stevens E. 2003. Haptoglobin directly affects T cells and suppresses T helper cell type 2 cytokine release. Immunology 108: 144–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy AP, Asleh R, Blum S, Levy NS, Miller-Lotan R, Kalet-Litman S, Anbinder Y, Lache O, Nakhoul FM, Asaf R, Farbstein D, Pollak M, Soloveichik YZ, Strauss M, Alshiek J, Livshits A, Schwartz A, Awad H, Jad K, and Goldenstein H. 2010. Haptoglobin: basic and clinical aspects. Antioxid. Redox. Signal 12: 293–304. [DOI] [PubMed] [Google Scholar]
- 6.Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SK, and Moestrup SK. 2001. Identification of the haemoglobin scavenger receptor. Nature 409: 198–201. [DOI] [PubMed] [Google Scholar]
- 7.Smith A, and McCulloh RJ. 2015. Hemopexin and haptoglobin: allies against heme toxicity from hemoglobin not contenders. Front Physiol 6: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wicher KB, and Fries E. 2004. Prohaptoglobin is proteolytically cleaved in the endoplasmic reticulum by the complement C1r-like protein. Proc. Natl. Acad. Sci. U. S. A 101: 14390–14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wicher KB, and Fries E. 2006. Haptoglobin, a hemoglobin-binding plasma protein, is present in bony fish and mammals but not in frog and chicken. Proc. Natl. Acad. Sci. U. S. A 103: 4168–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen CB, Torvund-Jensen M, Nielsen MJ, de Oliveira CL, Hersleth HP, Andersen NH, Pedersen JS, Andersen GR, and Moestrup SK. 2012. Structure of the haptoglobin-haemoglobin complex. Nature 489: 456–459. [DOI] [PubMed] [Google Scholar]
- 11.Maeda N, Yang F, Barnett DR, Bowman BH, and Smithies O. 1984. Duplication within the haptoglobin Hp2 gene. Nature 309: 131–135. [DOI] [PubMed] [Google Scholar]
- 12.Wejman JC, Hovsepian D, Wall JS, Hainfeld JF, and Greer J. 1984. Structure and assembly of haptoglobin polymers by electron microscopy. J. Mol. Biol 174: 343–368. [DOI] [PubMed] [Google Scholar]
- 13.Tosi M, Duponchel C, Meo T, and Couture-Tosi E. 1989. Complement genes C1r and C1s feature an intronless serine protease domain closely related to haptoglobin. J Mol Biol 208: 709–714. [DOI] [PubMed] [Google Scholar]
- 14.Wicher KB, and Fries E. 2010. Evolutionary aspects of hemoglobin scavengers. Antioxid. Redox. Signal 12: 249–259. [DOI] [PubMed] [Google Scholar]
- 15.Endo Y, Takahashi M, Nakao M, Saiga H, Sekine H, Matsushita M, Nonaka M, and Fujita T. 1998. Two lineages of mannose-binding lectin-associated serine protease (MASP) in vertebrates. J. Immunol 161: 4924–4930. [PubMed] [Google Scholar]
- 16.Liang CC 1957. The formation of complexes between haemoglobins and plasma proteins in a variety of animals. Biochem J 66: 552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mumm DP, Atha DH, and Riggs A. 1978. The hemoglobin of the common sting-ray, Dasyatis sabina: structural and functional properties. Comp Biochem Physiol B 60: 189–193. [DOI] [PubMed] [Google Scholar]
- 18.Dooley H, and Flajnik MF. 2005. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur. J. Immunol 35: 936–945. [DOI] [PubMed] [Google Scholar]
- 19.Loytynoja A, and Goldman N. 2008. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320: 1632–1635. [DOI] [PubMed] [Google Scholar]
- 20.Chang JM, Di Tommaso P, Lefort V, Gascuel O, and Notredame C. 2015. TCS: a web server for multiple sequence alignment evaluation and phylogenetic reconstruction. Nucleic Acids Res 43: W3–6.25855806 [Google Scholar]
- 21.Nguyen LT, Schmidt HA, von Haeseler A, and Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drummond AJ, Suchard MA, Xie D, and Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vazquez-Moreno L, Porath J, Schluter SF, and Marchalonis JJ. 1992. Purification of a novel heterodimer from shark (Carcharhinus plumbeus) serum by gel-immobilized metal chromatography. Comp Biochem. Physiol B 103: 563–568. [DOI] [PubMed] [Google Scholar]
- 24.Wyffels J, King BL, Vincent J, Chen C, Wu CH, and Polson SW. 2014. SkateBase, an elasmobranch genome project and collection of molecular resources for chondrichthyan fishes. F1000Res 3: 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drummond AJ, Ho SY, Phillips MJ, and Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wicher KB, and Fries E. 2007. Convergent evolution of human and bovine haptoglobin: partial duplication of the genes. J. Mol. Evol 65: 373–379. [DOI] [PubMed] [Google Scholar]
- 27.Polticelli F, Bocedi A, Minervini G, and Ascenzi P. 2008. Human haptoglobin structure and function--a molecular modelling study. FEBS J 275: 5648–5656. [DOI] [PubMed] [Google Scholar]
- 28.Dooley H, Buckingham EB, Criscitiello MF, and Flajnik MF. 2010. Emergence of the acute-phase protein hemopexin in jawed vertebrates. Mol. Immunol 48: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macdonald RA, Hosking CS, and Jones CL. 1988. The measurement of relative antibody affinity by ELISA using thiocyanate elution. J Immunol Methods 106: 191–194. [DOI] [PubMed] [Google Scholar]
- 30.McCloskey N, Turner MW, and Goldblatt TD. 1997. Correlation between the avidity of mouse-human chimeric IgG subclass monoclonal antibodies measured by solid-phase elution ELISA and biospecific interaction analysis (BIA). J Immunol Methods 205: 67–72. [DOI] [PubMed] [Google Scholar]
- 31.Hwang PK, and Greer J. 1980. Interaction between hemoglobin subunits in the hemoglobin. haptoglobin complex. J Biol Chem 255: 3038–3041. [PubMed] [Google Scholar]
- 32.Nantasenamat C, Prachayasittikul V, and Bulow L. 2013. Molecular modeling of the human hemoglobin-haptoglobin complex sheds light on the protective mechanisms of haptoglobin. PLoS One 8: e62996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perona JJ, and Craik CS. 1997. Evolutionary divergence of substrate specificity within the chymotrypsin-like serine protease fold. J Biol Chem 272: 29987–29990. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen MJ, Petersen SV, Jacobsen C, Thirup S, Enghild JJ, Graversen JH, and Moestrup SK. 2007. A unique loop extension in the serine protease domain of haptoglobin is essential for CD163 recognition of the haptoglobin-hemoglobin complex. J Biol Chem 282: 1072–1079. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen MJ, and Moestrup SK. 2009. Receptor targeting of hemoglobin mediated by the haptoglobins: roles beyond heme scavenging. Blood 114: 764–771. [DOI] [PubMed] [Google Scholar]
- 36.Venkatraman Girija U, Gingras AR, Marshall JE, Panchal R, Sheikh MA, Gal P, Schwaeble WJ, Mitchell DA, Moody PC, and Wallis R. 2013. Structural basis of the C1q/C1s interaction and its central role in assembly of the C1 complex of complement activation. Proc Natl Acad Sci U S A 110: 13916–13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettinello R, and Dooley H. 2014. The immunoglobulins of cold-blooded vertebrates. Biomolecules 4: 1045–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poon IK, Patel KK, Davis DS, Parish CR, and Hulett MD. 2011. Histidine-rich glycoprotein: the Swiss Army knife of mammalian plasma. Blood 117: 2093–2101. [DOI] [PubMed] [Google Scholar]
- 39.Morgan WT 1981. Interactions of the histidine-rich glycoprotein of serum with metals. Biochemistry 20: 1054–1061. [DOI] [PubMed] [Google Scholar]
- 40.McEvoy SM, and Maeda N. 1988. Complex events in the evolution of the haptoglobin gene cluster in primates. J Biol Chem 263: 15740–15747. [PubMed] [Google Scholar]
- 41.Nielsen MJ, Andersen CB, and Moestrup SK. 2013. CD163 binding to haptoglobin-hemoglobin complexes involves a dual-point electrostatic receptor-ligand pairing. J Biol Chem 288: 18834–18841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herzig CT, Waters RW, Baldwin CL, and Telfer JC. 2010. Evolution of the CD163 family and its relationship to the bovine gamma delta T cell co-receptor WC1. BMC Evol Biol 10: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snyder GKS, B.A. 1999. Red Blood Cells: Centerpiece in the Evolution of the Vertebrate Circulatory System. Am Zool 39: 9. [Google Scholar]
- 44.Thomsen JH, Etzerodt A, Svendsen P, and Moestrup SK. 2013. The haptoglobin-CD163-heme oxygenase-1 pathway for hemoglobin scavenging. Oxid Med Cell Longev 2013: 523652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aldrich KJS, D.K.; Sievert LM; Sievert G 2006. Comparison of Erythrocyte Osmotic Fragility among Amphibians, Reptiles, Birds and Mammals. Transactions of the Kansas Academy of Science 109: 10. [Google Scholar]
- 46.Arnold JE 2005. Hematology of the sandbar shark, Carcharhinus plumbeus: standardization of complete blood count techniques for elasmobranchs. Vet. Clin. Pathol 34: 115–123. [DOI] [PubMed] [Google Scholar]
- 47.Edelstein SJ, McEwen B, and Gibson QH. 1976. Subunit dissociation in fish hemoglobins. J Biol Chem 251: 7632–7637. [PubMed] [Google Scholar]
- 48.Chiancone E, Vecchini P, Forlani L, Antonini E, and Wyman J. 1966. Dissociation of hemoglobin from different animal species into subunits. Biochim Biophys Acta 127: 549–552. [DOI] [PubMed] [Google Scholar]
- 49.Nagel RL, and Gibson QH. 1971. The binding of hemoglobin to haptoglobin and its relation to subunit dissociation of hemoglobin. J Biol Chem 246: 69–73. [PubMed] [Google Scholar]
- 50.Schaer DJ, Schaer CA, Buehler PW, Boykins RA, Schoedon G, Alayash AI, and Schaffner A. 2006. CD163 is the macrophage scavenger receptor for native and chemically modified hemoglobins in the absence of haptoglobin. Blood 107: 373–380. [DOI] [PubMed] [Google Scholar]
- 51.Andersen CBF, Stodkilde K, Saederup KL, Kuhlee A, Raunser S, Graversen JH, and Moestrup SK. 2017. Haptoglobin. Antioxid Redox Signal 26: 814–831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.