Abstract
Type 2 diabetes mellitus (T2DM) is a major contributor to peripheral artery disease (PAD), especially in cases that advance to critical limb ischemia (CLI). Accumulating evidence indicates that miRNAs play an important role in the development of PAD and T2DM. Due to the limited value of current diagnostic methods for CLI in T2DM patients, we compared the miRNA expression profiles of Chinese T2DM patients with or without CLI to find out whether distinctive miRNAs could serve as potential diagnostic biomarkers. We statistically identified 7 miRNAs (hsa-miR-200b-3p, hsa-miR-2115-3p, hsa-miR-431-5p, hsa-miR-486-5p, hsa-miR-210-3p, hsa-miR-1264, hsa-miR-323b-5p) which were up-regulated in the CLI group, whereas other 4 miRNAs (hsa-miR-5579-3p, hsa-miR-665, hsa-miR-4285, hsa-miR-500a-3p) were down-regulated. Our validation test suggested a relatively high diagnostic accuracy of serum hsa-miR-323b-5p levels for the detection of CLI in T2DM patients, with a sensitivity of 62.67% and a specificity of 80.65%. The area under the curve (AUC) for miR-323b-5p + confounding risk factors was 0.94 (95% CI: 0.884–0.994, P < 0.001), which was higher than that for miR-323b-5p. Taken together, our results indicate that circulating hsa-miR-323b-5p could be a promising serum biomarker for the diagnosis of critical limb ischemia in type 2 diabetic patients.
Introduction
Peripheral artery disease (PAD) is a progressive atherosclerotic disorder that affected almost 155 million people worldwide in 20151. With progressive aging of the global population, the prevalence of PAD in the ≥75 year-old age group has increased from 12.5% in 2003 to 18.5% in 20122, resulting in increased risks of hospitalization and mortality3,4. Critical limb ischemia (CLI) is the most severe stage of PAD, and can lead to ischemic rest pain, tissue ulcers, gangrene and death5,6. In fact, more than 60% of CLI patients will be dead 5 years after diagnosis7.
Type 2 diabetes mellitus (T2DM) is a major contributor to CLI8. It has been reported that the risk of major cardiovascular events increases by 26% for each 1% increase in HbA1c levels9. Epidemiological studies have revealed that diabetic patients are fivefold more likely to develop CLI than non-diabetic patients10,11, while the mortality rate is more than 3 times higher due to the high incidence of cardiovascular disease events12–14. It has been suggested that the main mechanisms underlying the formation of lower limb emboli in diabetic patients involve oxidative stress, persistent inflammation and damaged endothelial tissues15–17. However, the exact mechanisms remain unclear.
Moreover, studies conducted in Germany revealed that approximately 2.7–4% of symptomatic PAD patients go on to develop CLI after 1-year18 and more than 40% of these CLI patients will suffer lower limb amputations19,20; 70% of these patients are diabetic18. Therefore, it is necessary to diagnose, predict and treat CLI at an early stage in PAD patients in order to decrease the limb loss and mortality rates. Currently, several methods are used to diagnose CLI, such as the ankle-brachial index (ABI), color Doppler ultrasound examination, and transcutaneous oxygen pressure19,21,22. However, these tests are only partially useful in diabetics with PAD. For example, researchers have shown that the risk factors associated with a reduced ABI can vary and are different in patients with or without diabetes23; thus, the diagnostic value of ABI is limited24. Therefore, investigation of novel biomarkers for early diagnosis and prognosis, as well as more effective therapies, would benefit diabetic patients with PAD.
MicroRNAs (miRNAs) are small, non-coding RNA molecules (containing about 22 nucleotides) that regulate gene expression and mediate complex biological processes25. It has been reported that multiple pathologic conditions, including CVD26, DM27, cancers28 and PAD29, are associated with altered miRNAs. These molecules are being studied to better understand the pathophysiology of many diseases, and as potential biomarkers for diagnosis30,31. Previous studies have shown that the expression of several miRNAs, such as let 7e, miR-27b, miR-130a and miR-210, is significantly different between patients with lower limb atherosclerosis and age-matched controls29,32,33. However, the miRNA profile of diabetic patients with PAD is still unclear, especially when CLI is present. Therefore, in the present study we investigated whether the expression of miRNAs in T2DM patients with or without CLI was significantly different and evaluated the diagnostic value of miRNAs for the diagnosis of CLI in patients with T2DM.
Materials and Methods
Subjects
A total of 66 T2DM patients were recruited from the Central Hospital of Wuhan from May 1st 2015 to May 1th 2017, including 31 subjects with CLI (CLI group) and 35 subjects without CLI (non-CLI group). Exclusion criteria included type 1 diabetes, cancer, severe liver or kidney failure, and receiving therapy for any chronic inflammatory disease. All participants were Han Chinese and thus of the same genetic background. This study was approved by the Ethics Committee of the Central Hospital of Wuhan and was conducted in accordance with the principles of the Declaration of Helsinki. All samples were collected after obtaining a written informed consent from the participants.
T2DM was diagnosed according to current American Diabetes Association criteria34. Briefly, T2DM was defined as fasting glucose levels ≥17.0 mmol/L, or a 2-h plasma glucose value ≥ 11.1 mmol/L during an oral glucose tolerance test or glycated hemoglobin levels >6.5%. CLI was diagnosed based on ABI < 0.9 and lower extremity arterial steno is >50% by color Doppler ultrasound examination35,36. Patients with ABI values equal to or greater than 0.9 and lower than 1.3, and with a normal lower extremity arterial pressure were defined as non-CLI.
Plasma collection and RNA extraction
Peripheral whole blood (5 mL) was collected in ethylenediaminetetraacetic acid (EDTA) tubes and stored at −80 °C until analysis. RNA was extracted from plasma using a miRNeasy serum/plasma kit (TIANGEN: catalog number DP503, China), according to the manufacturer’s instructions. RNA quality was determined by means of an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA). RNA was quantified using a Nanodrop 2000 Spectrophotometer (Thermo Scientific, Wilmington, USA).
miRNAs microarray hybridization
RNA samples were obtained from T2DM patients with CLI (n = 4) or without CLI (n = 4), matched for age, gender, and diabetes duration. The isolated total RNA from each subject was labeled using the miRCURYTM Power labeling kit (Exiqon, Inc., Vedbaek, Denmark) and hybridized to the miRCURYTM Array v16.0, according to the standard protocol. Hybridized arrays were analyzed by means of the GenePix 4000B microarray scanner (Molecular Devices, LLC., Sunnyvale, CA, USA), and the images were digitized with the Array-Pro image analysis software (Media Cybernetics, Silver Spring, MD, USA). Raw fluorescence data were processed using GenePix Pro 6.0 software (Molecular Devices, LLC., Sunnyvale, CA, USA). Individual spot fluorescence data were normalized based on the background and positive control fluorescence intensities.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Quantification of miRNAs was performed through a two-step reaction process: reverse transcription (RT) and PCR. cDNA synthesis was carried out by using a reverse transcriptase kit (TIANGEN; catalog number: KR211, China). miRNAs were quantified using SYBR Green (TIANGEN; catalog number FP411, China), as previously described37. The qRT-PCR reactions were carried out by heating at 95 °Cfor 1 min, followed by 40 cycles of 95 °C for 10 sec, 60 °C for 30 sec, and 70 °C for 10 sec. Duplicate assays were performed for each sample.
Target Prediction and Gene Ontology (GO) Analysis
Potential miRNA targets were predicted using the TargetScan human 7.038 (http://targetscan.org) and mirDB39 (http://www.mirdb.org/) web servers, and overlapping results from the two servers were considered the final predicted targets. Gene Ontology (GO) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analyses were conducted using the DAVID (Database for Annotation, Visualization and Integrated Discovery) bioinformatics resource.
Statistics
Statistical analysis was performed using SPSS (version 18.0) and GraphPad Prism (version 5.0) software. Continuous variables were expressed as the mean ± SEM or as the median (interquartile range), and categorical variables were compared by means of Fisher’s exact test or the χ2 test. The independent t-test or Fisher-Pitman Permutation test were used to compare normally distributed data, while the non-parametric Mann-Whitney U-test or Exact Mann-Whitney rank sum test was used to compare data with a non-normal distribution. P values were calculated using the Benjamini and Hochberg method to correct for multiple testing. Volcano plots (log fold-change versus negative log (base 10) P-values) were used to display the different biomarker distributions between CLI patients and non-CLI controls. Binary logistic regression analysis was performed to determine independent predictors of CLI in T2DM patients. All correlations were analyzed using the Pearson method or nonparametric Spearman method depending on the distribution of the data. Receiver operating characteristic (ROC) curves were used to evaluate the potential diagnostic value of each miRNA based on its ability to discriminate between T2DM patients with or without CLI. P < 0.05 was considered statistically significant.
Results
Clinical Characteristics
The study consisted of two groups: T2DM patients with CLI (n = 35), and T2DM patients without CLI (n = 31). The clinical data of all the study subjects and of the miRNA array cohort group are compared in Table 1. T2DM duration, hypertension, smoking, ABI, FPG, HbA1c, TC and Cr parameters were significantly different between T2DM patients with or without CLI (P < 0.05, exact P value shown in Table 1). Specifically in the miRNA array cohort group, there were significant differences in terms of ABI and TC (P = 0.007 and 0.048, respectively), but no differences were detected in other parameters (P > 0.05, exact P value shown in Table 1).
Table 1.
General characteristic of the study subjects.
| miRNA array cohort | P | validation cohor | P | |||
|---|---|---|---|---|---|---|
| CLI | Non - CLI | CLI | Non - CLI | |||
| No. of subjects | 4 | 4 | — | 27 | 31 | — |
| Age, y | 65.00 ± 1.73 | 65.50 ± 1.55 | 0.837 | 64.30 ± 2.73 | 61.0, (56.0–69.0) | 0.790‡ |
| T2D Duration, y | 10.00 ± 1.41 | 12.25 ± 1.80 | 0.363 | 15.48 ± 1.47 | 9.0, (8.0–14.0) | 0.004 ‡ |
| Women, % | 2, (50.00) | 2, (50.00) | 1.000 | 14, (51.85) | 16, (51.61) | 0.986† |
| Hyperlipidemia, % | 0, (0) | 2, (50.00) | 0.429 | 10, (37.04) | 10, (32.26) | 0.702† |
| Hypertension, % | 2, (50.00) | 2, (50.00) | 1.000 | 16, (59.26) | 9, (29.03) | 0.020 † |
| Smoking, % | 2, (50.00) | 2, (50.00) | 1.000 | 15, (55.56) | 8, (25.81) | 0.021 † |
| ABI | 0.46 ± 0.10 | 1.06 ± 0.03 | 0.007 | 24.10 (22.70–25.80) | 1.11 ± 0.01 | 0.000 ‡ |
| BMI, kg/m2 | 23.80 ± 1.21 | 25.98 ± 1.99 | 0.386 | 24.95 ± 0.57 | 25.46 ± 0.52 | 0.512 |
| CAD, % | 2, (50.00) | 2, (50.00) | 1.000 | 8, (29.63) | 8, (25.81) | 0.745† |
| Statin use, % | 0, (0) | 1, (25.00) | 1.000 | 6, (22.22) | 4, (12.90) | 0.349† |
| Antihypertensive treatment use, % | 2, (50.00) | 1, (25.00) | 1.000 | 8, (29.63) | 5, (16.13) | 0.219† |
| FPG, mmol/L | 8.27 ± 2.69 | 7.87 ± 1.52 | 0.899 | 9.60 (8.65–14.36) | 7.27 (6.14–9.05) | 0.001 ‡ |
| HbA1c, % | 10.35 ± 1.15 | 7.48 ± 0.31 | 0.509 | 8.90 (7.90–9.60) | 6.90 (6.50–9.00) | 0.001 ‡ |
| HDL-C, mmol/L | 0.97 ± 0.08 | 1.06 ± 0.06 | 0.372 | 1.08 ± 0.05 | 1.16 (0.90–1.32) | 0.281‡ |
| LDL-C, mmol/L | 2.14 ± 0.16 | 3.10 ± 0.41 | 0.100 | 2.63 ± 0.17 | 2.63 ± 0.15 | 0.987 |
| TG, mmol/L | 1.29 ± 0.33 | 1.37 ± 0.45 | 0.901 | 1.48 (1.07–1.86) | 1.29 (0.92–1.84) | 0.847‡ |
| TC, mmol/L | 3.51 ± 0.23 | 4.69 ± 0.42 | 0.048 | 2.80 (2.55–3.68) | 4.30 ± 0.18 | 0.006 ‡ |
| Apo A, g/L | 1.09 ± 0.07 | 1.19 ± 0.04 | 0.248 | 1.16 (1.03–1.29) | 1.15 (1.06–1.35) | 0.172‡ |
| Apo B, g/L | 0.80 ± 0.06 | 0.96 ± 0.10 | 0.194 | 0.92 ± 0.04 | 0.86 ± 0.04 | 0.265 |
| Cr, μmol/L | 55.50 ± 6.36 | 72.75 ± 9.58 | 0.184 | 75.00 (58.00–107.00) | 65.00 (55.00–72.00) | 0.044 ‡ |
| BUN, mmol/L | 4.95 ± 0.93 | 6.24 ± 1.06 | 0.393 | 5.60 (4.60–7.24) | 6.11 ± 0.40 | 0.374‡ |
| UA, mmol/L | 268.00 ± 10.96 | 262.25 ± 39.80 | 0.897 | 327.97 ± 13.45 | 323.76 ± 18.77 | 0.860 |
| Total bilirubin, mol/L | 11.63 ± 1.48 | 10.03 ± 1.71 | 0.586 | 12.51 ± 0.97 | 12.50 ± 0.87 | 0.995 |
| hsCRP, mg/dL | 1.24 ± 0.69 | 0.64 ± 0.25 | 0.446 | 0.35 (0.19–1.36) | 0.64 (0.26–2.30) | 0.402‡ |
| ALT, U/L | 18.00 ± 7.53 | 22.00 ± 5.02 | 0.674 | 14.60 (8.00–26.00) | 14.00 (10.90–23.00) | 0.343‡ |
| AST, U/L | 21.75 ± 3.78 | 22.50 ± 2.72 | 0.877 | 19.00 (13.00–31.00) | 19.28 ± 1.25 | 0.136‡ |
ABI, Ankle brachial index; BMI, body mass index; CAD, coronary artery disease; FPG, fasting plasma glucose; HbA1c, glycosylated hemoglobin; HDL-C, high density lipoprotein cholesterol; LDL-C, low density lipoprotein cholesterol; TG, triglyceride; TC, total cholesterol; Apo A, Apolipoprotein A; Apo B, Apolipoprotein B; Cr, serum creatinine; BUN, blood urea nitrogen; UA, uric acid; hsCRP, high-sensitivity C-reactive protein; ALT, alanine aminotransferase; AST, aspartate aminotransferase. Data is expressed as mean ± SEM, median (interquartile range) or percentage. Significant values are marked in italic. Continuous variales were compared by the Fisher-Pitman Permutation test, and categorical variables were compared by Fisher’s exact test in the miRNA array cohort, however, the validation cohort was analyzed using †χ2 test or ‡Mann-Whitney U-test, and the others were analyzed using t-tests.
Identification of different miRNA profiles in T2DM patients with and without CLI
To identify miRNAs that were differentially expressed between the two groups, we employed miRNA microarrays. The serum levels of eleven miRNAs were significantly different between diabetes mellitus patients with CLI (n = 4) and without CLI (n = 4) (Fig. 1). Briefly, 7 miRNAs, including hsa-miR-200b-3p, hsa-miR-2115-3p, hsa-miR-431-5p, hsa-miR-486-5p, hsa-miR-210-3p, hsa-miR-1264, hsa-miR-323b-5p were up-regulated in the CLI group (mean ratio = 1.57–4.18, P < 0.05; exact P value shown in Table 2), whereas 4 other miRNAs, including hsa-miR-5579-3p, hsa-miR-665, hsa-miR-4285, hsa-miR-500a-3p were down-regulated (mean ratio = 0.04–0.53, P < 0.05; exact P value shown in Table 2).
Figure 1.
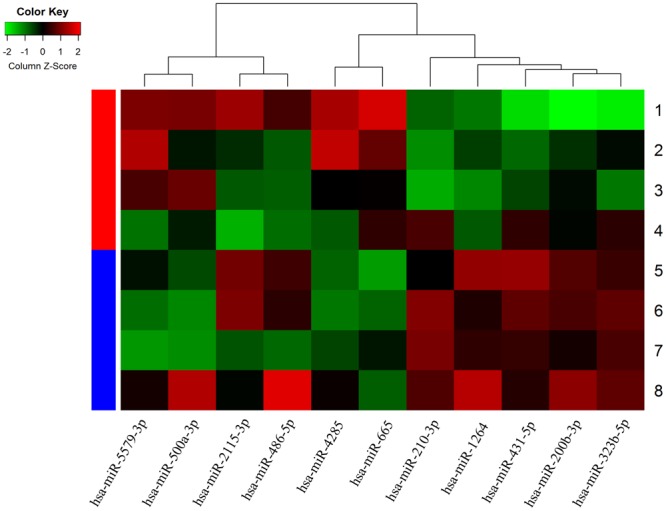
Hierarchical cluster analysis of 4 T2DM patients with CLI and 4 T2DM patients without CLI, based on differentially expressed miRNAs. Red indicates relative up-regulation, and green indicates relative down-regulation.
Table 2.
Differentially expression miRNAs identified by microarray analysis in plasma between T2DM with CLI and without CLI controls.
| Differentially expression type | microRNA name | Mean ratio | P value |
|---|---|---|---|
| Up-regulated | hsa-miR-200b-3p | 2.6211 | 0.016 |
| hsa-miR-2115-3p | 1.8945 | 0.040 | |
| hsa-miR-431-5p | 4.1855 | 0.042 | |
| hsa-miR-486-5p | 1.5782 | 0.029 | |
| hsa-miR-210-3p | 3.7626 | 0.036 | |
| hsa-miR-1264 | 2.3110 | 0.011 | |
| hsa-miR-323b-5p | 2.0134 | 0.034 | |
| Down-regulated | hsa-miR-5579-3p | 0.0474 | 0.038 |
| hsa-miR-665 | 0.3571 | 0.026 | |
| hsa-miR-4285 | 0.5322 | 0.044 | |
| hsa-miR-500a-3p | 0.2741 | 0.044 |
Validation of the miRNA microarray results by quantitative RT-PCR
Based on the microarray results and on their recently reported function, two miRNAs were selected for further validation: (1) miR-323b-5p, which is a newly described miRNA. Scarce information exists about this miRNA in terms of disease prediction, although it has been reported to be associated with an increased risk of gestational diabetes mellitus in pregnant Chinese Han women40; (2) miR-500a-3p, which is known to be significantly down-regulated in the plasma of patients with advanced atherosclerotic lesions41 and is also down-regulated in diabetic cardiomyopathy in the Akita mouse model42.
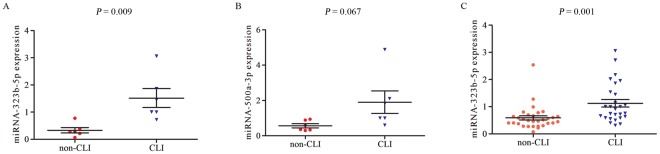
We initially tested the expression levels of these two miRNAs in a small number of diabetic subjects with CLI (n = 6) and without CLI (n = 6). The results showed that the expression of miR-323b-5p was significantly upregulated in the CLI group when compared with the non-CLI group (P = 0.009, Fig. 2A). In contrast, there was no significant difference in miR-500a-3p expression between the two groups (P = 0.067, Fig. 2B). Therefore, we tested the expression levels of miR-323b-5p in a larger number of subjects (n = 27 in the CLI group and n = 31 in the non-CLI group). The results showed that miR-323b-5p expression was significantly higher (1.9 fold) in T2DM patients with CLI than in T2DM patients without CLI (P = 0.001, Fig. 2C).
Figure 2.
Validation of miRNAs expression by qRT-PCR. Initial assessment of (A) miRNA-323b-5p and (B) miRNA-500a-3p expression in six CLI and six non-CLI patients. (C) Validation of miRNA-323b-5p expression in a larger number of CLI (n = 27) and non-CLI (n = 31) patients. The p-values were calculated by means of the Mann-Whitney U-test. CLI: T2DM patient with CLI, non-CLI: T2DM patient without CLI.
Association between miR-323b-5p levels and baseline clinical parameters with T2DM and CLI
In order to investigate the association between miR-323b-5p levels and CLI in relation to baseline factors, binary logistic regression analysis was conducted.
After adjusting for T2D duration, hypertension, smoking, FPG, HbA1c, TC, Cr, statins treatment and antihypertensive treatment, increased miR-323b-5p levels were found to be associated with an increased odds ratio (OR) (OR = 8.170) for T2DM with CLI (P = 0.032) (Table 3).
Table 3.
Binary logistic regression analysis for the risk factors of CLI in T2DM patients.
| OR | 95% CI for OR | P | OR* | 95% CI for OR* | P * | |
|---|---|---|---|---|---|---|
| T2D Duration | 1.173 | 1.044–1.318 | 0.007 | ∕ | ∕ | ∕ |
| Hypertension | 3.556 | 1.194–10.588 | 0.032 | ∕ | ∕ | ∕ |
| Smoking | 3.594 | 1.189–10.862 | 0.023 | ∕ | ∕ | ∕ |
| FPG | 1.360 | 1.108–1.668 | 0.003 | ∕ | ∕ | ∕ |
| HbA1c | 1.765 | 1.185–2.628 | 0.005 | ∕ | ∕ | ∕ |
| TC | 0.528 | 0.324–0.861 | 0.010 | ∕ | ∕ | ∕ |
| Cr | 1.019 | 1.000–1.038 | 0.048 | ∕ | ∕ | ∕ |
| Statin treatment | 0.519 | 0.129–2.077 | 0.519 | ∕ | ∕ | ∕ |
| Antihypertensive treatment | 0.457 | 0.129–1.617 | 0.224 | ∕ | ∕ | ∕ |
| miRNA-323b-5p | 6.756 | 1.644–27.770 | 0.008 | 8.170 | 1.029–64.864 | 0.032 |
FPG, fasting plasma glucose; HbA1c, glycosylated hemoglobin; TC, total cholesterol; Cr, serum creatinine. All study subjects were included in the analysis. Significant values are marked in italic.
Correlation between miR-323b-5p levels and clinical parameters
Next, we examined the correlation between miR-323b-5p levels and clinical parameters, such as T2D duration, hypertension, smoking, FPG, HbA1c, TC, Cr, statin treatment and antihypertensive treatment in T2DM patients with CLI (Table 4). Our results indicate that miR-323b-5p expression levels were positively correlated with smoking (r = 0.275, P = 0.037), FPG (r = 0.285, P = 0.030) and HbA1c (r = 0.335, P = 0.010). However, no significant correlation was found between the levels of miR-323b-5p and T2D duration, TC, Cr, statins treatment or antihypertensive treatment (P > 0.05, exact P value shown in Table 4).
Table 4.
Spearman Rho correlations between plasma miRNA-323b-5p level and CLI risk factors in T2DM patients.
| Variable | miRNA-323b-5p | |
|---|---|---|
| T2D Duration | rho | 0.251 |
| p | 0.059 | |
| Hypertension | rho | 0.201 |
| p | 0.131 | |
| Smoking | rho | 0.275 |
| p | 0.037 | |
| FPG | rho | 0.285 |
| p | 0.030 | |
| HbA1c | rho | 0.335 |
| P | 0.010 | |
| TC | rho | −0.255 |
| p | 0.053 | |
| Cr | rho | 0.055 |
| p | 0.682 | |
| Statin use | rho | −0.205 |
| p | 0.124 | |
| Antihypertensive treatment | rho | −0.079 |
| p | 0.555 |
CI, confidence interval; FPG, fasting plasma glucose; HbA1c, glycosylated hemoglobin; TC, total cholesterol; Cr, serum creatinine. *Adjusted for T2D Duration, hypertension, smoking, FPG, HbA1c, TC, Cr. All study subjects were included in the analysis. Significant values are marked in italic.
miR-323b-5p: Target prediction, function and pathway analysis
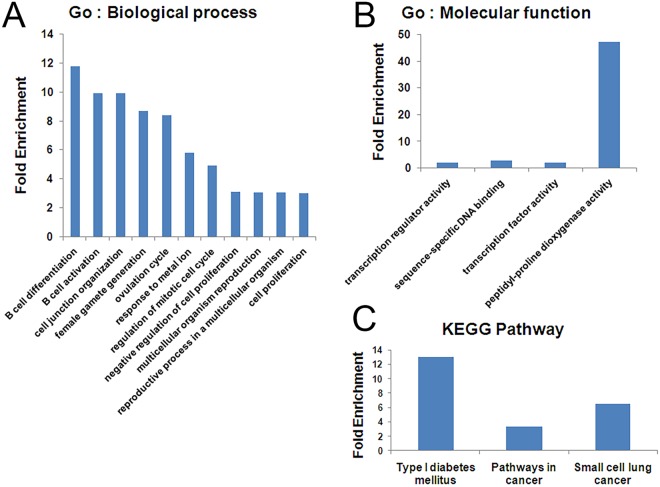
To investigate the possible biological function of miR-323b-5p, we conducted GO and KEGG pathway analyses using the DAVID online analysis tool. Eleven GO: biological process terms (GO: bp), 4 GO: molecular function terms (GO: mf) and 3 KEGG pathways were identified (Fig. 3). The most significantly enriched GO: bp terms were “GO: 0030183~B cell differentiation”, “GO: 0042113~B cell activation” and “GO: 0034330~cell junction organization” (Fig. 3A). The most enriched GO: mf term was “GO: 0031543~peptidyl-proline dioxygenase activity” (Fig. 3B). On the other hand, two significantly enriched KEGG pathways were identified, including hsa04940: type I diabetes mellitus and hsa05200: pathways in cancer (P < 0.05) (Fig. 3C). The exact GO and KEGG pathway analysis information, including GO IDs, pathway IDs, P values, fold enrichments and target genes, is shown in Tables S1 and S2.
Figure 3.
Functional analysis of miRNA-323b-5p targets in T2DM patients with CLI. (A) Significantly enriched GO: biological process terms. (B) GO: molecular function terms. (C) KEGG pathways (p value < 0.05).
In addition, we predicted miR-323b-5p target genes by means of TargetScan and mirDB software. There were 105 and 2244 predicted target genes by Targetscan and miRDB, respectively. Only 92 genes overlapped between the two types of software (Table S3).
Evaluation of miR-323b-5p for the diagnosis of T2DM patients with CLI
We explored the potential value of miR-323b-5p as a biomarker for the diagnosis of CLI in T2DM patients by means of (ROC) curve analysis. As shown in Fig. 4, the area under the curve (AUC) for miR-323b-5p was 0.78 (95% confidence intervals (CI): 0.667–0.903, P < 0.001) and the optimal cut-off point for miR-323b-5p was 0.71. The ROC curve analysis indicated that miR-323b-5p showed a relatively high diagnostic accuracy for the diagnosis of CLI in T2DM patients, with a Youden index of 0.47, a sensitivity of 62.67% (95% Cl: 0.460–0.835) and a specificity of 80.65% (95% Cl: 0.625–0.926). The AUC for miR-323b-5p + confounding risk factors (including T2DM duration, hypertension, smoking, FPG, HbA1c, TC, Cr, statin treatment and antihypertensive treatment) was 0.94 (95% CI: 0.884–0.994, P < 0.001), which was higher than that for miR-323b-5p (95% CI: 0.024–0.0285, P = 0.021) and for confounding risk factors (95% CI: −0.066–0.114, P = 0.602) (Fig. 4).
Figure 4.
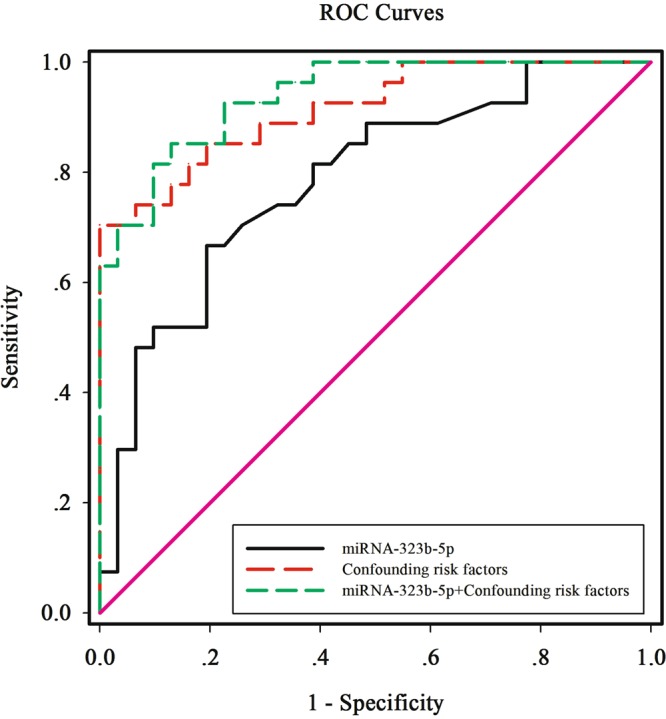
ROC curve analysis of the miRNA-323b-5p cut off point for the presence of CLI in T2DM patients. The AUC for miRNA-323b-5p was 0.78 (p < 0.001), the identified miR-323b-5p cut off point was 0.71, the Youden index was 0.47, the sensitivity was 62.67% and the specificity was 80.65%. The AUC for miRNA-323b-5p + confounding risk factors was 0.94 (p < 0.001). The confounding risk factors included T2DM duration, hypertension, smoking, FPG, HbA1c, TC, Cr, stain treatment and antihypertensive treatment. FPG, fasting plasma glucose; HbA1c, glycosylated hemoglobin; TC, total cholesterol; Cr, serum creatinine.
Discussion
It is widely believed that diabetes mellitus is a critical risk factor for PAD and death43, due to increased arterial calcification and endothelial dysfunction44. Studies have shown that calcification or reduced arterial wall elasticity can affect the accuracy of ABI measurements and lead to false results, especially in CLI patients45,46. For example, there were no significant differences in the ABI values of 269 Japanese CLI patients with or without diabetes47. Therefore, due to the limitations of current diagnostic methods for CLI in diabetes patients and the high risk of amputation, identification of accurate and sensitive biomarkers is of the utmost importance.
Increasing evidence suggests that miRNAs play a significant role in the diagnosis of T2DM or as therapeutic targets48–50 and may also regulate endothelial cell function and angiogenesis51,52. In this study, we found significant differences in the serum miRNA expression profiles of Han Chinese T2DM patients with or without CLI. Seven miRNAs were up-regulated and four miRNAs were down-regulated in T2DM patients with CLI (Fig. 1). ROC curve analysis indicated that one up-regulated miRNA, hsa-miR-323b-5p, could be a specific biomarker for CLI in T2DM (Fig. 4, P < 0.001).
In addition to diabetes, it is well established that in almost 70% of PAD patients there are additional risk factors, such as age, hypertension, dyslipidemia and smoking53, and this is consistent with our logistic regression analysis results. Our results showed significant differences between T2DM patients with and without CLI in terms of smoking, FPG, HbA1c, TC and Cr, miR-323b-5p (Table 3, P < 0.05). Additional correlation analysis results showed that the expression levels of miR-323b-5p were positively correlated with smoking (P = 0.037), FPG (P = 0.030) and HbA1c (P = 0.010) (Table 4). However, after adjusting for all risk factors including T2DM duration, hypertension, smoking, FPG, HbA1c, TC, Cr, statins treatments and antihypertensive treatment, miR-323b-5p remained a significant association with an increased odds ratio (OR) for diabetes with CLI. All these results suggest that miR-323b-5p is independently and strongly associated with T2DM patients with CLI.
MiR-323b-5p is a relatively new microRNA. So far, there are only a few studies investigating its functions in cell growth and proliferation54,55. However, various studies have shown that miRNAs which mediate cell differentiation and proliferation may also affect endothelial cell function and be associated with tissue ischemia56. For instance, miR let-7, which is aberrantly expressed in myocardial infarction, arrhythmia, angiogenesis, atherosclerosis, and hypertension57, has also been reported to inhibit Bcl-xl expression and to mediate ox-LDL-induced endothelial apoptosis through the regulation of cell proliferation, migration, autophagy and apoptosis58. Furthermore, Li and coworkers showed that a total of 84 miRNAs were differentially expressed between RAW 264.7 macrophages and foam cells induced by ox-LDL, and GO terms and KEGG pathways analyses revealed that the target genes of most of these miRNAs were enriched for cell differentiation59. Similarly, our GO and KEGG pathways analysis for miR-323b-5p showed significant enrichment in the biological processes of B cell differentiation, cell proliferation and apoptosis, suggesting the presence of common mechanisms with the miRNAs mentioned above. However, the biological function, bioinformatics analysis and target gene information reports are limited to miR-323b-5p in CLI patients with T2DM.
In the present study, we predicted 92 possible miR-323b-5p target genes in CLI patients with T2DM by means of TargetScan and mirDB software. However, only limited relevant information was found on these genes. A global gene expression dataset suggested that miR-323b-5p was mainly present in adipose tissue60. Stuart and coworkers showed that miR-323b-5p levels were increased in familial combined hyperlipidemia patients and that this miRNA could induce lipid accumulation in 3T3-L1 cells by reducing endogenous CDKN2B (cyclin-dependent kinase 4 2B) protein levels61, suggesting that CDKN2B may participate in adipose tissue metabolism. Furthermore, a recent study showed that CDKN2B RNA may indirectly regulate CAD-associated genes via targeting miR-92a62. Our results showed that CDKN2B was one of the 92 possible target genes of miR-323b-5p. Therefore, CDKN2B RNA could be a target gene of miR-323b-5p, although more studies would be needed to verify this assumption.
In summary, our study showed that eleven circulating miRNAs were significantly differentially expressed between T2DM patients with and without CLI. The validation study indicated that the diagnostic accuracy of serum hsa-miR-323b-5p was significantly higher than that of other miRNAs, suggesting that miR-323b-5p could be a potential biomarker for the diagnosis of CLI in diabetes patients. However, the exact role played by miR-323b-5p during the development of CLI is still unknown. Further investigations are required to clarify the underlying mechanisms as well as the biological functions of this miRNA.
Electronic supplementary material
Acknowledgements
This work was supported by the Natural Science Foundation of China (81603013 and 81800704), Health and Family Planning Commission of Hubei Province (WJ2017M183), Health and Family Planning Commission of Wuhan (WX18M02) and Basic Research of Wuhan Science and Technology Bureau (2017060201010204).
Author Contributions
Conceived and designed the experiments: A.P. Deng and J. Liu. Performed the experiments: B. Cheng, J.Y. Li, X.C. Li, Z.J. Wang. Analyzed the data: B. Cheng, J.Y. Li, A.P. Deng, X.F. Wang and J. Liu. Wrote the paper: B. Cheng and J.Y. Li.
Competing Interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Biao Cheng and Ju-yi Li contributed equally.
Contributor Information
Jue Liu, Email: 15562399@qq.com.
Ai-ping Deng, Email: dapyxb@163.com.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33310-4.
References
- 1.Disease GBD, Injury I, Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–1602. doi: 10.1016/S0140-6736(16)31678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalbaugh, C. A. et al. Peripheral Artery Disease Prevalence and Incidence Estimated From Both Outpatient and Inpatient Settings Among Medicare Fee-for-Service Beneficiaries in the Atherosclerosis Risk in Communities (ARIC) Study. J Am Heart Assoc6 (2017). [DOI] [PMC free article] [PubMed]
- 3.Criqui MH, et al. Mortality over a period of 10 years in patients with peripheral arterial disease. N Engl J Med. 1992;326:381–386. doi: 10.1056/NEJM199202063260605. [DOI] [PubMed] [Google Scholar]
- 4.McDermott MM. The magnitude of the problem of peripheral arterial disease: epidemiology and clinical significance. Cleve Clin J Med. 2006;73(Suppl 4):S2–7. doi: 10.3949/ccjm.73.Suppl_4.S2. [DOI] [PubMed] [Google Scholar]
- 5.European Stroke O, et al. ESC Guidelines on the diagnosis and treatment of peripheral artery diseases: Document covering atherosclerotic disease of extracranial carotid and vertebral, mesenteric, renal, upper and lower extremity arteries: the Task Force on the Diagnosis and Treatment of Peripheral Artery Diseases of the European Society of Cardiology (ESC) Eur Heart J. 2011;32:2851–2906. doi: 10.1093/eurheartj/ehr211. [DOI] [PubMed] [Google Scholar]
- 6.Vasa45 Suppl 95, 1–100 (2016).
- 7.Davies MG. Criticial limb ischemia: epidemiology. Methodist Debakey Cardiovasc J. 2012;8:10–14. doi: 10.14797/mdcj-8-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morley RL, Sharma A, Horsch AD, Hinchliffe RJ. Peripheral artery disease. BMJ. 2018;360:j5842. doi: 10.1136/bmj.j5842. [DOI] [PubMed] [Google Scholar]
- 9.Selvin E, et al. Meta-analysis: glycosylated hemoglobin and cardiovascular disease in diabetes mellitus. Ann Intern Med. 2004;141:421–431. doi: 10.7326/0003-4819-141-6-200409210-00007. [DOI] [PubMed] [Google Scholar]
- 10.Kannel WB. Risk factors for atherosclerotic cardiovascular outcomes in different arterial territories. J Cardiovasc Risk. 1994;1:333–339. doi: 10.1097/00043798-199412000-00009. [DOI] [PubMed] [Google Scholar]
- 11.Resnick HE, Valsania P, Phillips CL. Diabetes mellitus and nontraumatic lower extremity amputation in black and white Americans: the National Health and Nutrition Examination Survey Epidemiologic Follow-up Study, 1971–1992. Arch Intern Med. 1999;159:2470–2475. doi: 10.1001/archinte.159.20.2470. [DOI] [PubMed] [Google Scholar]
- 12.Dormandy J, Heeck L, Vig S. The fate of patients with critical leg ischemia. Semin Vasc Surg. 1999;12:142–147. [PubMed] [Google Scholar]
- 13.Jude EB, Eleftheriadou I, Tentolouris N. Peripheral arterial disease in diabetes–a review. Diabet Med. 2010;27:4–14. doi: 10.1111/j.1464-5491.2009.02866.x. [DOI] [PubMed] [Google Scholar]
- 14.Layden J, Michaels J, Bermingham S, Higgins B. & Guideline Development, G. Diagnosis and management of lower limb peripheral arterial disease: summary of NICE guidance. BMJ. 2012;345:e4947. doi: 10.1136/bmj.e4947. [DOI] [PubMed] [Google Scholar]
- 15.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 16.Dinh T, et al. Mechanisms involved in the development and healing of diabetic foot ulceration. Diabetes. 2012;61:2937–2947. doi: 10.2337/db12-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edsfeldt A, et al. Impaired fibrous repair: a possible contributor to atherosclerotic plaque vulnerability in patients with type II diabetes. Arterioscler Thromb Vasc Biol. 2014;34:2143–2150. doi: 10.1161/ATVBAHA.114.303414. [DOI] [PubMed] [Google Scholar]
- 18.Dohmen A, Eder S, Euringer W, Zeller T, Beyersdorf F. Chronic critical limb ischemia. Dtsch Arztebl Int. 2012;109:95–101. doi: 10.3238/arztebl.2012.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santilli JD, Santilli SM. Chronic critical limb ischemia: diagnosis, treatment and prognosis. Am Fam Physician. 1999;59:1899–1908. [PubMed] [Google Scholar]
- 20.Hirsch AT, et al. Peripheral arterial disease detection, awareness, and treatment in primary care. JAMA. 2001;286:1317–1324. doi: 10.1001/jama.286.11.1317. [DOI] [PubMed] [Google Scholar]
- 21.Ohtake T, et al. Impact of lower limbs’ arterial calcification on the prevalence and severity of PAD in patients on hemodialysis. J Vasc Surg. 2011;53:676–683. doi: 10.1016/j.jvs.2010.09.070. [DOI] [PubMed] [Google Scholar]
- 22.Rhee SY, Kim YS. Peripheral Arterial Disease in Patients with Type 2 Diabetes Mellitus. Diabetes Metab J. 2015;39:283–290. doi: 10.4093/dmj.2015.39.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bąk Ewelina, Marcisz Czesław, Kadłubowska Monika, Michalik Anna, Krawczyk Bożena, Dobrzyń-Matusiak Dorota, Krzemińska Sylwia, Fiałkowski Tomasz, Glądys Elżbieta, Drosdzol-Cop Agnieszka. Independent Factors of Changes of Ankle-Brachial Index in Peripheral Arterial Occlusive Disease in Elderly Patients with or without Diabetes. International Journal of Environmental Research and Public Health. 2016;13(11):1103. doi: 10.3390/ijerph13111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nam SC, et al. Factors affecting the validity of ankle-brachial index in the diagnosis of peripheral arterial obstructive disease. Angiology. 2010;61:392–396. doi: 10.1177/0003319709348295. [DOI] [PubMed] [Google Scholar]
- 25.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 26.Bayoumi, A. S., Aonuma, T., Teoh, J. P., Tang, Y. L. & Kim, I. M. Circular noncoding RNAs as potential therapies and circulating biomarkers for cardiovascular diseases. Acta Pharmacol Sin (2018). [DOI] [PMC free article] [PubMed]
- 27.Pordzik J, et al. The Potential Role of Platelet-Related microRNAs in the Development of Cardiovascular Events in High-Risk Populations, Including Diabetic Patients: A Review. Front Endocrinol (Lausanne) 2018;9:74. doi: 10.3389/fendo.2018.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bortoluzzi Stefania, Lovisa Federica, Gaffo Enrico, Mussolin Lara. Small RNAs in Circulating Exosomes of Cancer Patients: A Minireview. High-Throughput. 2017;6(4):13. doi: 10.3390/ht6040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereira-da-Silva T, et al. Circulating microRNA profiles in different arterial territories of stable atherosclerotic disease: a systematic review. Am J Cardiovasc Dis. 2018;8:1–13. [PMC free article] [PubMed] [Google Scholar]
- 30.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welten SM, Goossens EA, Quax PH, Nossent AY. The multifactorial nature of microRNAs in vascular remodelling. Cardiovasc Res. 2016;110:6–22. doi: 10.1093/cvr/cvw039. [DOI] [PubMed] [Google Scholar]
- 32.Stather PW, et al. Differential microRNA expression profiles in peripheral arterial disease. Circ Cardiovasc Genet. 2013;6:490–497. doi: 10.1161/CIRCGENETICS.111.000053. [DOI] [PubMed] [Google Scholar]
- 33.Signorelli SS, et al. Circulating miR-130a, miR-27b, and miR-210 in Patients With Peripheral Artery Disease and Their Potential Relationship With Oxidative Stress. Angiology. 2016;67:945–950. doi: 10.1177/0003319716638242. [DOI] [PubMed] [Google Scholar]
- 34.Marathe PH, Gao HX, Close KL. American Diabetes Association Standards of Medical Care in Diabetes 2017. J Diabetes. 2017;9:320–324. doi: 10.1111/1753-0407.12524. [DOI] [PubMed] [Google Scholar]
- 35.Rooke TW, et al. 2011 ACCF/AHA Focused Update of the Guideline for the Management of Patients With Peripheral Artery Disease (updating the 2005 guideline): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2011;58:2020–2045. doi: 10.1016/j.jacc.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li JY, et al. Siglec-5 is a novel marker of critical limb ischemia in patients with diabetes. Sci Rep. 2017;7:11272. doi: 10.1038/s41598-017-11820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu J, et al. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int J Cancer. 2014;135:1187–1194. doi: 10.1002/ijc.28757. [DOI] [PubMed] [Google Scholar]
- 38.Agarwal, V., Bell, G. W., Nam, J. W. & Bartel, D. P. Predicting effective microRNA target sites in mammalian mRNAs. Elife4 (2015). [DOI] [PMC free article] [PubMed]
- 39.Wang X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics. 2016;32:1316–1322. doi: 10.1093/bioinformatics/btw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, et al. Association study of the miRNA-binding site polymorphisms of CDKN2A/B genes with gestational diabetes mellitus susceptibility. Acta Diabetol. 2015;52:951–958. doi: 10.1007/s00592-015-0768-2. [DOI] [PubMed] [Google Scholar]
- 41.Eken SM, et al. MicroRNA-210 Enhances Fibrous Cap Stability in Advanced Atherosclerotic Lesions. Circ Res. 2017;120:633–644. doi: 10.1161/CIRCRESAHA.116.309318. [DOI] [PubMed] [Google Scholar]
- 42.Chavali V, Tyagi SC, Mishra PK. Differential expression of dicer, miRNAs, and inflammatory markers in diabetic Ins2+/− Akita hearts. Cell Biochem Biophys. 2014;68:25–35. doi: 10.1007/s12013-013-9679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinecke H, et al. Peripheral arterial disease and critical limb ischaemia: still poor outcomes and lack of guideline adherence. Eur Heart J. 2015;36:932–938. doi: 10.1093/eurheartj/ehv006. [DOI] [PubMed] [Google Scholar]
- 44.Potier L, Abi Khalil C, Mohammedi K, Roussel R. Use and utility of ankle brachial index in patients with diabetes. Eur J Vasc Endovasc Surg. 2011;41:110–116. doi: 10.1016/j.ejvs.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 45.Foran TG, Sheahan NF, Cunningham C, Feely J. Pseudo-hypertension and arterial stiffness: a review. Physiol Meas. 2004;25:R21–33. doi: 10.1088/0967-3334/25/2/R02. [DOI] [PubMed] [Google Scholar]
- 46.Jackler JM. Asymptomatic cardiac lesions occurring in a viral disease–report of two cases. J Maine Med Assoc. 1960;51:58–60. [PubMed] [Google Scholar]
- 47.Takahara M, et al. Association of diabetes and hemodialysis with ankle pressure and ankle-brachial index in Japanese patients with critical limb ischemia. Diabetes Care. 2012;35:2000–2004. doi: 10.2337/dc11-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ortega FJ, et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37:1375–1383. doi: 10.2337/dc13-1847. [DOI] [PubMed] [Google Scholar]
- 49.Zhu H, Leung SW. Identification of microRNA biomarkers in type 2 diabetes: a meta-analysis of controlled profiling studies. Diabetologia. 2015;58:900–911. doi: 10.1007/s00125-015-3510-2. [DOI] [PubMed] [Google Scholar]
- 50.Liang YZ, et al. Identification of Neuroendocrine Stress Response-Related Circulating MicroRNAs as Biomarkers for Type 2 Diabetes Mellitus and Insulin Resistance. Front Endocrinol (Lausanne) 2018;9:132. doi: 10.3389/fendo.2018.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Araldi E, Suarez Y. MicroRNAs as regulators of endothelial cell functions in cardiometabolic diseases. Biochim Biophys Acta. 2016;1861:2094–2103. doi: 10.1016/j.bbalip.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caporali A, Emanueli C. MicroRNA regulation in angiogenesis. Vascul Pharmacol. 2011;55:79–86. doi: 10.1016/j.vph.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 53.Meijer WT, Grobbee DE, Hunink MG, Hofman A, Hoes AW. Determinants of peripheral arterial disease in the elderly: the Rotterdam study. Arch Intern Med. 2000;160:2934–2938. doi: 10.1001/archinte.160.19.2934. [DOI] [PubMed] [Google Scholar]
- 54.Qiu S, et al. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J Transl Med. 2013;11:10. doi: 10.1186/1479-5876-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Momen-Heravi F, Trachtenberg AJ, Kuo WP, Cheng YS. Genomewide Study of Salivary MicroRNAs for Detection of Oral Cancer. J Dent Res. 2014;93:86S–93S. doi: 10.1177/0022034514531018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou YF, et al. MicroRNA-30c-5p ameliorates hypoxia-reoxygenation-induced tubular epithelial cell injury via HIF1alpha stabilization by targeting SOCS3. Oncotarget. 2017;8:92801–92814. doi: 10.18632/oncotarget.21582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bao MH, et al. Let-7 in cardiovascular diseases, heart development and cardiovascular differentiation from stem cells. Int J Mol Sci. 2013;14:23086–23102. doi: 10.3390/ijms141123086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qin B, et al. MicroRNA let-7c inhibits Bcl-xl expression and regulates ox-LDL-induced endothelial apoptosis. BMB Rep. 2012;45:464–469. doi: 10.5483/BMBRep.2012.45.8.033. [DOI] [PubMed] [Google Scholar]
- 59.Li X, et al. Expression profiles of microRNAs in oxidized low-density lipoprotein-stimulated RAW 264.7 cells. In Vitro Cell Dev Biol Anim. 2018;54:99–110. doi: 10.1007/s11626-017-0225-3. [DOI] [PubMed] [Google Scholar]
- 60.Parts L, et al. Extent, causes, and consequences of small RNA expression variation in human adipose tissue. PLoS Genet. 2012;8:e1002704. doi: 10.1371/journal.pgen.1002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horswell SD, et al. CDKN2B expression in adipose tissue of familial combined hyperlipidemia patients. J Lipid Res. 2013;54:3491–3505. doi: 10.1194/jlr.M041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng M, An S, Li J. CDKN2B-AS may indirectly regulate coronary artery disease-associated genes via targeting miR-92a. Gene. 2017;629:101–107. doi: 10.1016/j.gene.2017.07.070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.