Abstract
Opisthorchis viverrini, a parasitic trematode, was recategorized as a group 1 biological carcinogen because it causes opisthorchiasis, which may result in cholangiocarcinoma. A new strategy for controlling opisthorchiasis is needed because of issues such as drug resistance and reinfection. Triosephosphate isomerase (TIM), a key enzyme in energy metabolism, is regarded as a potential drug target and vaccine candidate against various pathogens. Here, we determined the crystal structures of wild-type and 3 variants of TIMs from O. viverrini (OvTIM) at high resolution. The unique tripeptide of parasite trematodes, the SAD motif, was located on the surface of OvTIM and contributed to forming a 310-helix of the following loop in a sequence-independent manner. Through thermal stability and structural analyses of OvTIM variants, we found that the SAD motif induced local structural alterations of the surface and was involved in the overall stability of OvTIM in a complementary manner with another parasite-specific residue, N115. Comparison of the surface characteristics between OvTIM and Homo sapiens TIM (HsTIM) and structure-based epitope prediction suggested that the SAD motif functions as an epitope.
Introduction
Opisthorchis viverrini, a prevalent human liver fluke, is a parasitic trematode that is widely distributed in Southeast Asia1. After infection, O. viverrini resides in the bile ducts and causes minor symptoms such as inflammation and loss of appetite. Thus, the risk of opisthorchiasis has been underestimated and the disease has been regarded as minor. Many in vivo studies have shown that opisthorchiasis is a cause of cholangiocarcinoma (CCA)2–4. Additionally, recent studies showed that the excretory-secretory products (ESP) of Clonorchis sinensis, which is a related fluke that is also involved in cholangiocarcianoma, are related to oncogenesis5. In fact, the International Agency of Cancer Research reclassified O. viverrini as a group 1 biological carcinogen in 20096.
Since praziquantel (PZQ) was developed in the 1970s, chemotherapy involving PZQ has been a major strategy for controlling zoonotic helminthiasis such as opisthorchiasis, schistosomiasis, clonorchiasis, and echinococcosis. Although the overall prevalence and infection levels of zoonotic helminthiasis was slowly reduced, new issues have arisen. Because of its powerful efficacy and safety, PZQ was used exclusively for approximately three decades, which resulted in the appearance of resistance and reinfection. For example, reinfection by O. viverrini after PZQ treatment was reported in northeast Thailand in 20167. Furthermore, it was reported that repeated administration of PZQ elevated the risk of CCA8. For Schistosoma mansoni, a parasitic flatworm that causes schistosomiasis, the development of drug resistance has been suggested several times at the laboratory and worldwide levels9–12. Therefore, a new strategy for controlling and preventing helminthic diseases should be established, such as developing additional anthelminthic drugs and vaccines.
Triosephosphate isomerase (TIM, EC 5.3.1.1) is a key enzyme in glycolysis and catalyzes the reversible isomerization between dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (GAP). It was reported that chronic hemolytic anemia and severe neuromuscular disease was found in the TIM deficiency patients13 and DHAP was markedly accumulated in the erythrocytes of the patients14. Given that glycolytic enzymes are essential for maintaining life, some reports have suggested that the indispensability of glycolytic enzymes can be used in developing a drug or vaccine to overcome not only clonorchiasis15–17 but also other zoonotic helminthiasis18–20. It was reported that a unique tripeptide existed in TIMs from parasitic flatworms but not in those from other species, including non-pathogenic free-living planarians and vertebrates20,21. Although the reason why SXD/E tripeptide (X represents Ala, Ile, or Lys) exists in only parasitic flatworms is unclear, this motif clearly differs between the pathogen and its host. Therefore, molecular structural identification of the pathogen-specific SXD/E motif in OvTIM may be useful for vaccine development. Here, we determined the crystal structure of wild-type and 3 variants of OvTIMs at high resolution and confirmed the different formation of the following rigid secondary structure compared to that of human TIM. Additionally, we identified the stabilizing function of residue N115 and performed structural analysis of the surface characteristics of OvTIM to assess whether the SAD motif functions as an epitope.
Results
Overall structure of OvTIM
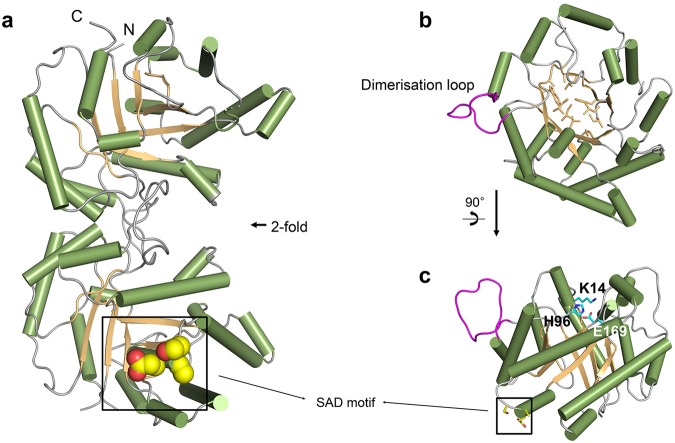
We determined the crystal structure of OvTIM and other variants at high resolution by molecular replacement. The amino acid residues except for residues 175–179 in OvTIM showed clear electron density. Two homodimers in the asymmetric unit were identical with an r.m.s. deviation Cα of 0.22 Å. The overall structure of OvTIM is similar to that of other known TIMs in that it contains 12 α-helices, three 310-helices, and eight parallel β-sheets, forming a classical α/β barrel fold (Fig. 1 and Supplementary Fig. S1). The hydrophobic core of the inner β-barrel is stabilized by hydrophobic residues such as F8, L41, A43, A63, W91, I93, I125, V165, I211, and L234. The inner β-barrel is protected from solvents by a bundle of α-helices (Fig. 1b,c). The solvent-accessible area of each subunit and dimer interface area are 19,728 Å2 and 1,609.8 Å2, respectively. The loop between α2 and β3 interacts with 2 loops of another protomer (β1 and α1, α3 and β4) to participate in dimerization of OvTIM. The residues involved in the dimerization and catalytic activity are highly conserved among species (Fig. 2a,b). The loops in which catalytic residues are located form dimerization interface with the dimerization loop of another protomer, indicating that OvTIM shares a common mechanism with triose phosphate isomerization based on the dimer formation. The catalytic triad (K14, H96, and E169) is in the upper entrance of the β-barrel, while the SAD motif is on the surface opposite the catalytic site and exposed to the outside of the protein, as expected.
Figure 1.
Overall structure of OvTIM. (a) Overall dimeric structure of OvTIM. The bundle of α-helices and β-barrel of OvTIM are represented as green and orange cartoon models, respectively. The SAD motif is denoted as a yellow sphere model. (b) The top view and (c) rotation view of the monomer of OvTIM are displayed. Hydrophobic residues forming the β-barrel fold are represented as a stick model. The catalytic triad, SAD motif, and dimerization loop are highlighted as cyan, yellow, and magenta, respectively.
Figure 2.
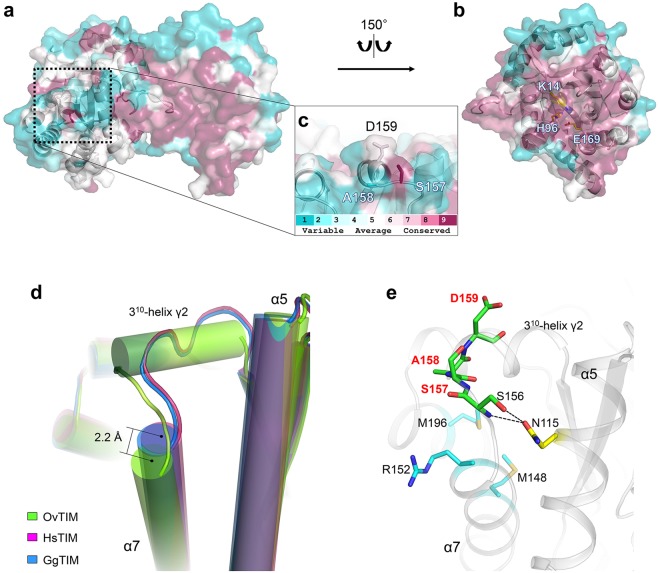
Structural analysis of SAD motif. (a) Degree of residue conservation of TIM among 7 parasitic trematodes (O. viverrini (accession code: A0A074Z863), Clonorchis sinensis (accession code: G7YDG0), Schistosoma haematobium (A5A6F9), Schistosoma japonicum (Q27775), Schistosoma mansoni (P48501), Schistosoma turkestanicum (Q45XG1), and Taenia solium (Q9GTX8)) is represented as color variations in a surface model from cyan (lowest conserved residue) to wine (highest conserved residue). (b) Rotated view of the OvTIM monomer is shown, and residues forming the catalytic triad are represented as yellow sticks. (c) An enlarged view of the SAD motif. (d) Superimposition of OvTIM (green) structure with Homo sapiens TIM (PDB code: 4POC) and Gallus gallus TIM (GgTIM) (PDB code: 8TIM) is represented in magenta and blue, respectively. The corresponding region of the SAD motif in OvTIM is designated as dark green. (e) The intermolecular interaction of the region around the SAD motif. The bulky side chains are highlighted as a cyan stick model.
Structural analysis of the SAD motif
According the multiple sequence alignment results, Ser, the first residue in the SXD/E motif, is almost conserved among 7 parasitic trematodes except for Clonorchis sinensis TIM (CsTIM) (Fig. 2c and Supplementary Fig. S1). The second position of the SXD/E motif is mainly occupied by a hydrophobic residue except for Taenia solium TIM (TsTIM) and CsTIM, while an acidic amino acid (Asp, Glu) is in the third position21. In OvTIM, a specific SAD motif (S157–D159) is located between α7 and β6, inducing the formation of a subsequent 310-helix γ2 (D159–I164), although the corresponding region is not conserved in either vertebrate TIMs (Fig. 2d and Supplementary Fig. S2). Two in silico structures of TIMs from S. mansoni20 and Fasciola hepatica22 was reported. The overall fold was well predicted to OvTIM, r.m.s. deviation of 0.44 Å (SmTIM) and 0.47 Å (FhTIM) for Cα, respectively. However, the back-bone assignment and parasite-specific 310-helix induced by SXD/E motifs of in silico structures were not matched to the SAD motif in OvTIM (Supplementary Fig. S3). The side chains of the SAD motif that protrude into the solvent region are not well-defined, while surrounding hydrophobic residues such as L155, M160, and W161 are directed towards the core of the protein to take part in the hydrophobic interaction (Fig. 2e). Additionally, the N115 in α5 is another characteristic feature of OvTIM. In parasitic trematode TIMs, polar residues such as Asn, Glu, Gln, and Lys are found rather than Ala as in vertebrate TIMs (Supplementary Fig. S1). The side chain of N115 forms hydrogen bonds with the main chain and side chain of S156, stabilizing the loop located between α7 and γ2. The residues forming the hydrophobic core surrounded by α7, γ2, and α9 are also substituted with bulky amino acids such as M148, R152, and M196 in OvTIM. This substitution in OvTIM resulted in tilting of the end part of α7 toward the solvent region by ~2.2 Å because of steric hindrance of the substituted bulky side chain and N115 compared to that of the corresponding region of Homo sapiens TIM (HsTIM) (Fig. 2d,e). Therefore, formation of short 310-helix γ2 caused by the SAD motif, the stabilizing function of N115, and tilting of α7 cause unique local alterations in the surface structure in OvTIM, distinguishing it from vertebrate TIMs.
Local structural alteration and overall stability affected by the SAD motif and N115
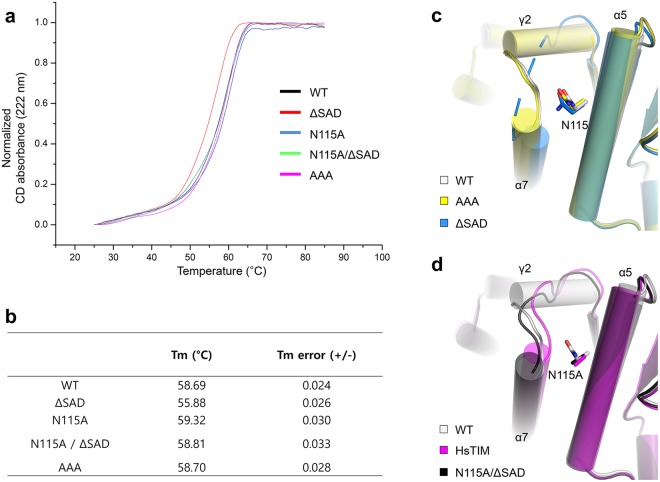
To estimate the effect of the parasite unique tripeptide on the overall protein stability of OvTIM, we performed circular dichroism to measure the thermal stability of various OvTIM variants (Fig. 3a,b). We constructed four variants, including a SAD deletion variant (ΔSAD), variant in which the SAD region was substituted with AAA (AAA), variant in which N115 was replaced with Ala (N115A), and double variant (N115A_ΔSAD). First, the thermal shift of the melting curve of the AAA variant was negligible, indicating that the side chains of the SAD motif that protrude into the solvent region do not contribute to protein stability. Interestingly, the ΔSAD variant exhibited a lower protein thermal stability, with a Tm of ~3 °C, compared to that of the wild-type. In contrast, the Tm of the N115A variant was slightly elevated, with a Tm of approximately 0.63 °C above the wild-type Tm, while the Tm of N115A_ΔSAD was restored to that of the wild-type. Structural analysis of variants was also carried out to evaluate the effects of the unique structural alteration in OvTIM (Fig. 3c,d). In the ΔSAD variant, the corresponding region (K153–W158) was not defined except for the molecule of the region which is stabilized via an interaction with other symmetry molecules, while the AAA variant showed a similar structure as the wild-type. The electron density of N115 of ΔSAD was not defined and normalized B factor of N115 in ΔSAD structure and wild-type structure were 42.71 Å2 and 16.01 Å2, respectively. These results show that the hydrogen bond between N115 and the main chain of S156 no longer forms because of deletion of the SAD motif. Consequently, the remaining steric hindrance of N115 pushes the α7 so that α7 exhibits a more shifted orientation than that of the wild-type. The α7 of N115A_ΔSAD variant moves to the original position of the wild-type and exhibits a stable conformation because all other steric hindrance caused by N115 does not exist. The overall shape of the designated region of the N115A_ΔSAD variant is similar to that of HsTIM but showed a unique difference with an α7 shift that resulted from the bulky side chains (M148, R152, and M196). Therefore, the SAD motif and intramolecular interaction of N115 as well as the bulky side chains are major stabilizing factors in OvTIM.
Figure 3.
Thermostability of OvTIM and variants and comparative analysis. (a) Thermal stability of wild-type and variants (AAA, ∆SAD, N115A, and N115A_∆SAD). (b) Melting temperature of variants based on the CD absorbance at 222 nm. (c) Superimposition of wild-type, AAA, and ∆SAD variant structures. N115 residues from wild-type, AAA, and ∆SAD are represented as a stick model. The disordered region of ∆SAD variant is represented as a dash. (b) Superimposition of the OvTIM, HsTIM, and ∆SAD_N115A structures.
Comparison of surface characteristics between OvTIM and HsTIM
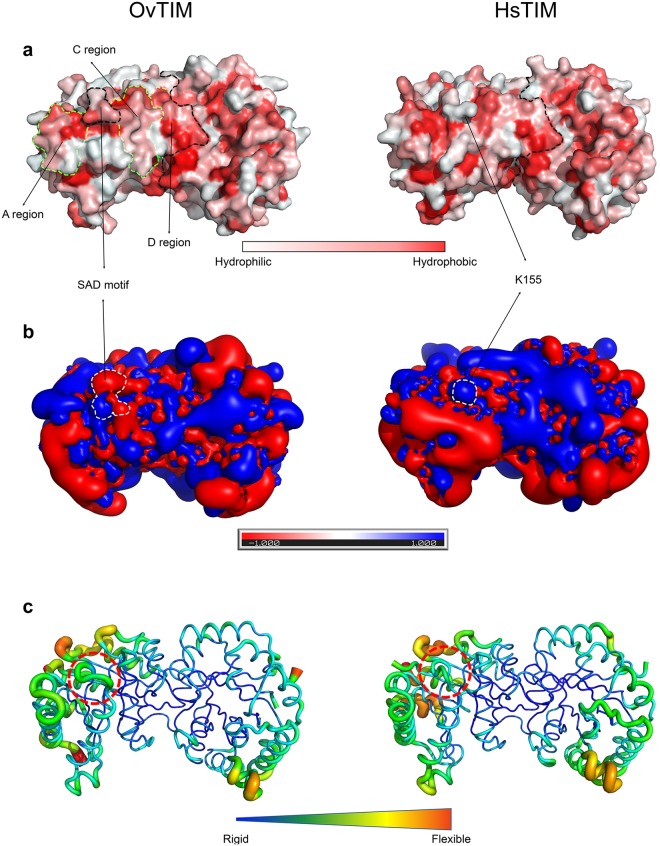
The positions of Cα in OvTIM and HsTIM are identical, with an r.m.s.d. = 1.08 Å, showing great similarity. However, the surface characteristics are quite different between the two enzymes (Fig. 4). The total solvent-accessible surface area of OvTIM (19,728 Å2) is larger than that of HsTIM (18,931 Å2), while the hydrophobic surface area of OvTIM (5,355 Å2) is smaller than that of HsTIM (5,630 Å2). Interestingly, the cluster of hydrophobic residues in OvTIM results in formation of a long hydrophobic patch. The patch can be divided to 4 parts (Fig. 4a): a region which in the helix-turn-helix motif between α9 and α10, SAD motif region, B region located on the end part of α5, and C region on the exterior surface of the dimerization loop. In contrast, the K155 residue of HsTIM that is present rather than the SAD motif interrupts the consecutive hydrophobic patch. The distribution of the electrostatic fields of OvTIM is also distinct from that of HsTIM (Fig. 4b). Both electrostatic fields of HsTIM tend to be clustered exclusively, while OvTIM shows a relatively even electrostatic field distribution. Additionally, the SAD motif region shows another characteristic feature related to its charge distribution compared to the K155 region of HsTIM. The K155 region, which shows a moderate positive electrostatic field, is located between two strong electrostatic fields. The SAD motif, which has a more protruding D159 than the K155 of HsTIM, exhibits strong negative electrostatic fields. The distribution degree of the temperature factor in both TIMs is universal in that the external surface residues show relatively high flexibility (Fig. 4c). The side chain atoms of the SAD motif (45.4 Å2) showed a slightly high temperature factor compared to the corresponding region in HsTIM (30.9 Å2).
Figure 4.
Comparison of surface characteristics between OvTIM and HsTIM. (a) Hydrophobicity of the 4 hydrophobic regions comprising the long hydrophobic patch in OvTIM is represented as dotted lines. (b) Electrostatic field, the SAD motif in OvTIM and K155 in HsTIM are designated as white dotted lines. (c) Temperature factor, the SAD motif in OvTIM and K155 in HsTIM are designated as red dotted circles.
Structure-based epitope prediction of OvTIM
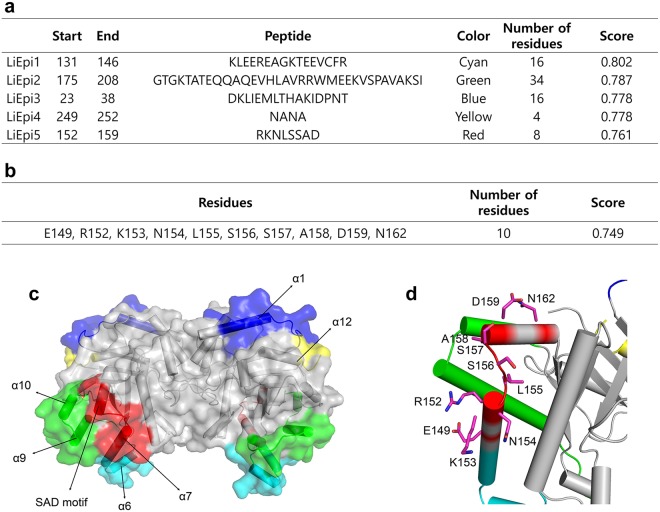
We predicted the epitope on the OvTIM using ElliPro, which is a modified prediction program developed by Thornton to identify epitope regions that protrude from the globular surface of a protein23 (Supplementary Fig. S4). Among all possible continuous epitopes, a total of 5 potential epitopes with a score above 0.70 were predicted to be located at the surface region of OvTIM (Fig. 5a,c). The predicted linear epitopes are broadly classified into 2 regions; LiEpi3 which includes α1 and LiEpi4 which includes the end of α12 are located at the C-terminal part of OvTIM. The remaining linear epitopes clustered near the SAD motif, including α6 (LiEpi1), α9 and α10 (LiEpi2), and the SAD motif and α7 (LiEpi5). In contrast, only the SAD motif is included in the discontinuous epitope, was predicted by a modified method based on the protrusion index (S) and distance (R) calculation of the residue’s center of mass (Fig. 5b). In addition to the SAD motif, peripheral hydrophilic residues such as E149, R152, K153, N154, and N162 collaboratively participate in the protrusion (Fig. 5d).
Figure 5.
Structure-based epitope prediction of OvTIM. (a,b) Predicted linear epitope (a) and discontinuous epitope (b) from the ElliPro results. For precise results, the dimer structure of OvTIM was used, and possible epitopes with a score over 0.7 are listed. (c) The results of the predicted linear epitope in (a) are designated on the surface model of OvTIM. (d) The residues predicted to form discontinuous epitope are represented as a stick model.
Discussion
In this study, we determined and analyzed the structure of OvTIM, which showed high similarity with other known TIMs, suggesting a common intrinsic mechanism. As previously reported15–20, inhibiting the parasitic glycolytic enzyme is advantageous. Since the similarity of the active site and reaction mechanism between the parasite and its host makes it challenging to develop drugs that selectively inhibit the parasitic enzyme, it is important to identify a new pathogen-specific inhibitory mechanism that is not shared with its host. It was reported that thiol-reactive reagents induced decreased affinity for the substrates by chemical modification of C222 of TIM from Giardia lamblia (GlTIM) that was located in the non-catalytic region24,25. Interestingly, the corresponding region of the C221 of OvTIM shows the similar environment with that of GlTIM, surrounding hydrophobic pocket is well conserved (Supplementary Fig. S5). Therefore, we may suggest that the inhibitory strategy based on thiol-reactive compound could be also possible in the case of OvTIM. The SAD motif, which is a parasite-specific region, is not included in the catalytic and conserved region. Furthermore, the efficiency of OvTIM ΔSAD to isomerase DHAP to GAP showed slightly lower than that of wild-type in the quick activity check assay (Supplementary Fig. S6). Considering them, the SAD motif may be another potential region of the parasite-specific inhibitory strategy.
We confirmed that the SAD motif induced formation of the 310-helix γ2, which is a loop structure in vertebrate TIMs. The side chain of the SAD motif protruded into the solvent region and no significant Tm change was observed in the AAA variant, indicating that local structural alterations and the stability of OvTIM are related to the number of amino acids of the SAD motif and not the exact sequence of the SAD motif. We also found that the N115 residue, which is conserved as a bulky and hydrophilic residue in parasitic flatworms, is involved in forming the unique parasitic surface structure via hydrogen bonding with S156. The overall stability of OvTIM was also affected by the SAD motif and N115 in a complementary manner in that the Tm decrease in the ΔSAD variant was rescued by N115A substitution.
In contrast to the sequence-independent contribution of the SAD motif to the surface structure change and stability, the side chains of the SAD motif, which are exposed to the solvent region, are still open to participate in the host-parasite interaction. It has been reported that CsTIM and fructose-1,6-bisphosphate aldolase (EC 4.1.2.13) of O. viverrini, which were identified by immunoscreening with human CCA serum26,27, exist not only in the cytosol but also in the ESP28. Additionally, cell surface display of TIMs in various pathogens was observed15,19,29–31. In Schistosoma mansoni, a monoclonal antibody recognizing SmTIM was identified and specifically inhibited the reactivity of SmTIM18. These reports indicate the importance of glycolytic enzymes in zoonotic helminths and other pathogens in the host-parasite interaction as well as energy metabolism. From this perspective, we performed a surface characteristic comparison of OvTIM and HsTIM because the difference between a parasite and its host is a major factor affecting the host-parasite interaction. The distribution degrees of hydrophobicity, electrostatic field, and flexibility are major factors in the protein-protein interaction and antigen-antibody complex32–34. As a result, we identified a long hydrophobic patch in OvTIM, electrostatic field difference between OvTIM and HsTIM, and relatively high flexibility of the SAD motif. In fact, the hydrophobic patch, the center of which is the SAD motif, is surrounded by polar or charged residues, which is a major characteristic of the epitope. Moreover, we predicted using ElliPro23 that the SAD motif of OvTIM is a discontinuous epitope that is directly recognized by B-cell receptors. Together with the high flexibility and hydrophobicity of α9, the flexibility and high protruding index of the SAD motif suggest that the SAD motif forms an interface with the complementary-determining region of the antibody paratope. The linear epitope range from K148 to V161 in HsTIM, which includes the corresponding region of the SAD motif in OvTIM, was reported35 (epitope ID 438866) in Immune Epitope Database and analysis resource (IEDB). The difference in the charge distribution between the SAD motif in OvTIM and the reported epitope in HsTIM is included in another selection mechanism between the parasite and its host. Analysis of the surface characteristics and the fact that OvTIM is included in the ESP support that the SAD motif is an epitope. In conclusion, the results of this study provide a new pathway for vaccine development against O. viverrini and another strategy for overcoming drug resistance and reinfection.
Methods
Cloning and expression of OvTIM
cDNA encoding triose phosphate isomerase from O. viverrini was synthesized and amplified by PCR5. The amplified DNA was digested with the restriction nucleases NdeI and XhoI and cloned into the vector pET17b, which contained a 6 His-tag and thrombin site. The recombinant plasmid was transformed into Escherichia coli BL21 (DE3) pLysS cells for expression tests under various conditions. The soluble portion of the target protein was observed, and the target was scaled-up to increase expression. The clone was grown at 37 °C in LB medium until the OD600 reached 0.6–0.8. The target protein was induced by adding isopropyl β-D-1-thiogalactopyranoside at a final concentration of 1 mM at 18 °C for 18 h. The cells were harvested by centrifugation at 4 °C at 4,392 × g for 20 min, and the pellet was stored at −70 °C.
Purification of OvTIM
The frozen pellet was resuspended in buffer A (20 mM Tris, pH 8.9, 50 mM NaCl, 2 mM β-mercaptoethanol) and disrupted by sonication on ice for 25 min. Cell debris was pelleted by centrifugation (24,878 × g for 60 min). The filtered supernatant was purified using a HisTrap HP column (GE Healthcare, Little Chalfont, UK). The recombinant protein bound on the column was eluted with a linear imidazole concentration gradient of buffer B (20 mM Tris, pH 8.5, 50 mM NaCl, 500 mM imidazole, 2 mM β-mercaptoethanol). The purity of the target protein was determined by SDS-PAGE, and fractions were pooled for further purification and 6x His-tag of recombinant protein was not cleaved. The protein solution was concentrated to 1 mL using Amicon Ultra centrifugal filters and then diluted with buffer C (20 mM Tris, pH 8.5, 2 mM DTT) up to 10 times to lower the salt concentration. The diluted protein solution was loaded onto a HiTrap Q HP column (GE Healthcare), and a linear NaCl gradient in buffer D (20 mM Tris, pH 8.5, 1 M NaCl, 2 mM dithiothreitol) was applied. Because two different surface ionic characteristics of OvTIM were observed in the SDS-PAGE results, each protein was concentrated separately. Size-exclusion chromatography was performed for each protein using a HiLoad 26/60 Superdex-200 prep-grade column (GE Healthcare) in final buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 2 mM DTT). The purified proteins were finally concentrated to about 80 and ~40 mg/mL, respectively, and then stored at −70 °C.
Crystallization
The initial crystallization conditions were identified by the sitting-drop vapor diffusion method using the commercial Index screening kit (Hampton Research, Aliso Viejo, CA, USA) in MRC 2 Well Crystallization Plates (Hampton Research). Next, 0.5 μL of solution that contained ~80 mg/ml of proteins was mixed with an equal volume of screening solution. Initial crystals were observed under these conditions (0.2 M magnesium chloride hexahydrate, 0.1 M Tris, pH 8.5, 25% polyethylene glycol 3350 at 293 K) 5 days later. A single crystal suitable for diffraction was obtained by mixing 1 μL of protein solution with an equal volume of reservoir solution (0.3 M magnesium chloride hexahydrate, 0.1 M Tris, pH 8.5, 22% polyethylene glycol 3350 at 20 °C) by the hanging-drop vapor diffusion method. The crystals were cryoprotected using 25% (v/v) glycerol before mounting, and then transferred to cryo-solution for a few seconds.
Data collection and structure determination
The diffraction data set for the crystal of OvTIM were collected at beamline 5 C of the Pohang Light Source (PLS, Pohang, South Korea) at a wavelength of 1.00001 Å. Images were indexed, integrated, and scaled using HKL200036. We carried out molecular replacement for phasing using the TIM of chicken (PDB entry 1TPH) as a model with the Phaser module in PHENIX37. Next, 95% of the residues were automatically built with the Autobuild module in PHENIX38, and building of the remaining residues was performed manually using Coot39. The model of OvTIM that was refined using the Refine module in PHENIX37 was validated using MolProbity40. The final values of Rwork and Rfree were 0.1618 and 0.1990, respectively. The other variants of OvTIM were determined by molecular replacement using the wild-type of OvTIM as a model. The statistics for data collection and refinement are provided in Table 1.
Table 1.
Statistics for X-ray data collection and refinement.
| OvTIM_wild-type | OvTIM_ΔSAD | OvTIM_AAA | OvTIM_N115A_ΔSAD | |
|---|---|---|---|---|
| Data collection | ||||
| X-ray source | PAL BL5C | Spring-8 BL44XU | PAL BL11C | PAL BL11C |
| Wavelength (Å) | 1.00001 | 0.90000 | 0.97942 | 0.97942 |
| Space group | C2221 | P21 | P212121 | P21 |
| Unit-cell parameters | ||||
| a, b, c (Å) | 96.53 206.62 97.47 | 73.74 91.92 75.78 | 68.84 90.37 102.27 | 74.04 92.56 76.08 |
| α, β, γ (°) | 90 90 90 | 90 109.128 90 | 90 90 90 | 90 109.39 90 |
| Resolution (Å) | 50.00–1.75 (1.81–1.75) | 50.00–1.75 (1.85–1.75) | 50.00–1.58 (1.61–1.58) | 50.00–1.80 (1.83–1.80) |
| Rmerge (%) | 10.5 (11.0) | 7.4 (42.5) | 16.9 (47.3) | 9.1 (27.3) |
| <I/σ(I)> | 64.05 (21.1) | 14.81 (3.26) | 15.15 (2.04) | 26.38 (5.03) |
| Completeness (%) | 98.8 (93.3) | 99.7 (99.2) | 98.7 (95.3) | 97.2 (95.1) |
| Average multiplicity | 5.6 (3.1) | 4.24 (4.19) | 7.9 (4.5) | 5.3 (4.0) |
| Observed reflections | 541,897 (28061) | 410,026 (64976) | 654,169 (17640) | 464,383 (17084) |
| Unique reflections | 96,825 (9052) | 96,733 (15516) | 82,985 (3920) | 87,968 (4271) |
| Refinement | ||||
| Rwork/Rfree (%) | 0.1640/0.2004 | 0.1599/0.1904 | 0.1706/0.1950 | 0.1466/0.1810 |
| Average B factor (Å2) | 21.192 | 19.162 | 18.469 | 21.271 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.007 | 0.006 | 0.007 | 0.009 |
| Bond angles (°) | 0.929 | 0.975 | 0.937 | 1.104 |
| Ramachandran plot (MolProbity±) | ||||
| Favored (%) | 97.41 | 97.18 | 98.17 | 97.60 |
| Allowed (%) | 2.59 | 2.82 | 1.83 | 2.40 |
| Outliers (%) | 0 | 0 | 0 | 0 |
| PDB code | 5ZFX | 5ZG4 | 5ZG5 | 5ZGA |
Site-directed mutagenesis
Four OvTIM variants, N115A, ∆SAD, AAA, and N115A-∆SAD, were produced using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). The variants were expressed and purified as described above for wild-type OvTIM.
Thermal shift assay
Each protein including the wild-type and variants were diluted to 0.5 mg/mL with buffer (20 mM HEPES, pH 7.5, 100 mM NaCl) prior to loading. The samples were loaded and then heated from 25 to 85 °C at 0.5 °C/min. The circular dichroism absorbance at 222 nm was recorded using a circular dichroism spectrophotometer (Jasco, Oklahoma City, OK, USA) and normalized to calculate the melting temperature of each protein.
Structural analysis
The amino acid sequences of TIMs were aligned via CLUSTALW41, and the results were processed using the ESPript42 server. The surface model of the residue conservation degree was constructed using the Consurf43 server. The solvent-accessible surface area and buried area were calculated using PISA44 and the surface area that was composed of hydrophobic residues was added to calculate hydrophobic surface area. Protein topology diagrams which show protein secondary structures and fold were drawn using the Pro-origami45 server. The hydrophobicity of surface residues was calculated using the Eisenberg hydrophobic scale46. To precisely compare surface temperature (B) factors between OvTIM and HsTIM, the temperature factors were normalized using a previously described equation47:
The electrostatic field and normalized temperature factor of proteins were calculated and visualized using PyMol48. All images describing the structures of proteins were made using PyMol48.
Catalytic activity measurement
The catalytic activity of wild-type OvTIM and ΔSAD variant to isomerase DHAP to GAP was measured using triose phosphate isomerase activity colorimetric assay kit (BioVision)49,50. Absorbance at 450 nm was plotted to NADH concentration for standard curve after seven NADH solutions which range from 0.05 mM to 0.4 mM were incubated with enzyme mix and developer for 10 mins. The absorbance of reaction mixtures containing 0.4 mM DHAP and 0.018 mM OvTIM dimer was recorded at 450 nm in every 10 secs. Total 2 sets of activity curves of wild-type OvTIM and ΔSAD variant were obtained and plotted after conversion of the absorbance to NADH concentration based on previously determined standard curve.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the National Research Foundation of Korea (2017R1A2B2005666 and 2018M3A9F3055925). S.K. was supported by Wiset program. We thank the supporting staff of beamline BL5C and BL11C of the Pohang Accelerator Light Source (Pohang, South Korea) and beamline BL44XU of Spring-8 (Hyogo, Japan) for providing help with data collection. We also acknowledge the Korean Basic Science Institute (Daejeon, Korea) for the use of a circular dichroism spectrophotometer.
Author Contributions
J.S. and K.Y.H. designed all experiments; M.R.L. cloned wild-type of OvTIM; J.S. purified and crystallized wild-type of OvTIM; S.K., S.E.K. and H.L. performed mutagenesis of OvTIM variants; J.S., S.K. and S.E.K. collected and processed the diffraction data of wild-type and OvTIM variants; J.S. determined the crystal structures of wild-type and OvTIM variants; J.S. and K.Y.H. analyzed the data and wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jonghyeon Son, Sulhee Kim and So Eun Kim contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33479-8.
References
- 1.Sadun EH. Studies on Opisthorchis viverrini in Thailand. Am. J. Hyg. 1955;62:81–115. doi: 10.1093/oxfordjournals.aje.a119772. [DOI] [PubMed] [Google Scholar]
- 2.Flavell DJ, Lucas SB. Promotion of N-nitrosodimethylamine-initiated bile duct carcinogenesis in the hamster by the human liver fluke, Opisthorchis viverrini. Carcinogenesis. 1983;4:927–930. doi: 10.1093/carcin/4.7.927. [DOI] [PubMed] [Google Scholar]
- 3.Kim YI. Liver carcinoma and liver fluke infection. Arzneimittelforschung. 1984;34:1121–1126. [PubMed] [Google Scholar]
- 4.Satarug S, et al. Relationships between the synthesis of N-nitrosodimethylamine and immune responses to chronic infection with the carcinogenic parasite, Opisthorchis viverrini, in men. Carcinogenesis. 1998;19:485–491. doi: 10.1093/carcin/19.3.485. [DOI] [PubMed] [Google Scholar]
- 5.Kim DW, et al. Transcriptional induction of minichromosome maintenance protein 7 (Mcm7) in human cholangiocarcinoma cells treated with Clonorchis sinensis excretory-secretory products. Mol. Biochem. Parasitol. 2010;173:10–16. doi: 10.1016/j.molbiopara.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Bouvard V, et al. A review of human carcinogens–Part B: biological agents. Lancet Oncol. 2009;10:321–322. doi: 10.1016/S1470-2045(09)70096-8. [DOI] [PubMed] [Google Scholar]
- 7.Saengsawang P, Promthet S, Bradshaw P. Reinfection by Opisthorchis Viverrini after Treatment with Praziquantel. Asian Pac. J. Cancer Prev. 2016;17:857–862. doi: 10.7314/APJCP.2016.17.2.857. [DOI] [PubMed] [Google Scholar]
- 8.Kamsa-Ard S, et al. Association between praziquantel treatment and cholangiocarcinoma: a hospital-based matched case-control study. BMC Cancer. 2015;15:776. doi: 10.1186/s12885-015-1788-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coles GC, Bruce JI. In vitro selection of drug resistant Schistosoma mansoni. Int. J. Parasitol. 1987;17:767–771. doi: 10.1016/0020-7519(87)90057-9. [DOI] [PubMed] [Google Scholar]
- 10.Coles GC, Kinoti GK. Defining resistance in Schistosoma. Parasitol. Today. 1997;13:157–158. doi: 10.1016/S0169-4758(97)89815-8. [DOI] [PubMed] [Google Scholar]
- 11.Danso-Appiah A, De Vlas SJ. Interpreting low praziquantel cure rates of Schistosoma mansoni infections in Senegal. Trends Parasitol. 2002;18:125–129. doi: 10.1016/S1471-4922(01)02209-7. [DOI] [PubMed] [Google Scholar]
- 12.Fallon PG, Doenhoff MJ. Drug-resistant schistosomiasis: resistance to praziquantel and oxamniquine induced in Schistosoma mansoni in mice is drug specific. Am. J. Trop. Med. Hyg. 1994;51:83–88. doi: 10.4269/ajtmh.1994.51.83. [DOI] [PubMed] [Google Scholar]
- 13.Vives-Corrons JL, et al. Triosephosphate isomerase deficiency with hemolytic anemia and severe neuromuscular disease: familial and biochemical studies of a case found in Spain. Hum. Genet. 1978;42:171–180. doi: 10.1007/BF00283637. [DOI] [PubMed] [Google Scholar]
- 14.Olah J, et al. Triosephosphate isomerase deficiency: consequences of an inherited mutation at mRNA, protein and metabolic levels. Biochem. J. 2005;392:675–683. doi: 10.1042/bj20050993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, et al. Surface display of Clonorchis sinensis enolase on Bacillus subtilis spores potentializes an oral vaccine candidate. Vaccine. 2014;32:1338–1345. doi: 10.1016/j.vaccine.2014.01.039. [DOI] [PubMed] [Google Scholar]
- 16.Chen T, et al. Sequence analysis and molecular characterization of Clonorchis sinensis hexokinase, an unusual trimeric 50-kDa glucose-6-phosphate-sensitive allosteric enzyme. PLoS One. 2014;9:e107940. doi: 10.1371/journal.pone.0107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T, et al. Advanced enzymology, expression profile and immune response of Clonorchis sinensis hexokinase show its application potential for prevention and control of clonorchiasis. PLoS Negl. Trop. Dis. 2015;9:e0003641. doi: 10.1371/journal.pntd.0003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harn DA, et al. A protective monoclonal antibody specifically recognizes and alters the catalytic activity of schistosome triose-phosphate isomerase. J. Immunol. 1992;148:562–567. [PubMed] [Google Scholar]
- 19.Wright MD, Davern KM, Mitchell GF. The functional and immunological significance of some schistosome surface molecules. Parasitol. Today. 1991;7:56–58. doi: 10.1016/0169-4758(91)90191-P. [DOI] [PubMed] [Google Scholar]
- 20.Zinsser VL, Farnell E, Dunne DW, Timson DJ. Triose phosphate isomerase from the blood fluke Schistosoma mansoni: biochemical characterisation of a potential drug and vaccine target. FEBS Lett. 2013;587:3422–3427. doi: 10.1016/j.febslet.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Chen B, Wen JF. The adaptive evolution divergence of triosephosphate isomerases between parasitic and free-living flatworms and the discovery of a potential universal target against flatworm parasites. Parasitol. Res. 2011;109:283–289. doi: 10.1007/s00436-010-2249-4. [DOI] [PubMed] [Google Scholar]
- 22.Zinsser VL, Hoey EM, Trudgett A, Timson DJ. Biochemical characterisation of triose phosphate isomerase from the liver fluke Fasciola hepatica. Biochimie. 2013;95:2182–2189. doi: 10.1016/j.biochi.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 23.Ponomarenko J, et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9:514. doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enriquez-Flores S, et al. Determining the molecular mechanism of inactivation by chemical modification of triosephosphate isomerase from the human parasite Giardia lamblia: a study for antiparasitic drug design. Proteins. 2011;79:2711–2724. doi: 10.1002/prot.23100. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez-Alcantara G, et al. Structural and functional perturbation of Giardia lamblia triosephosphate isomerase by modification of a non-catalytic, non-conserved region. PLoS One. 2013;8:e69031. doi: 10.1371/journal.pone.0069031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senawong G, Laha T, Loukas A, Brindley PJ, Sripa B. Cloning, expression, and characterization of a novel Opisthorchis viverrini calcium-binding EF-hand protein. Parasitol. Int. 2012;61:94–100. doi: 10.1016/j.parint.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, et al. Molecular identification, immunolocalization, and characterization of Clonorchis sinensis triosephosphate isomerase. Parasitol. Res. 2015;114:3117–3124. doi: 10.1007/s00436-015-4530-z. [DOI] [PubMed] [Google Scholar]
- 28.Zheng M, et al. Proteomic analysis of excretory secretory products from Clonorchis sinensis adult worms: molecular characterization and serological reactivity of a excretory-secretory antigen-fructose-1,6-bisphosphatase. Parasitol. Res. 2011;109:737–744. doi: 10.1007/s00436-011-2316-5. [DOI] [PubMed] [Google Scholar]
- 29.Ikeda R, Ichikawa T. Interaction of surface molecules on Cryptococcus neoformans with plasminogen. FEMS Yeast Res. 2014;14:445–450. doi: 10.1111/1567-1364.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furuya H, Ikeda R. Interaction of triosephosphate isomerase from Staphylococcus aureus with plasminogen. Microbiol. Immunol. 2011;55:855–862. doi: 10.1111/j.1348-0421.2011.00392.x. [DOI] [PubMed] [Google Scholar]
- 31.Pereira LA, et al. Analysis of the Paracoccidioides brasiliensis triosephosphate isomerase suggests the potential for adhesin function. FEMS Yeast Res. 2007;7:1381–1388. doi: 10.1111/j.1567-1364.2007.00292.x. [DOI] [PubMed] [Google Scholar]
- 32.Lijnzaad P, Argos P. Hydrophobic patches on protein subunit interfaces: characteristics and prediction. Proteins. 1997;28:333–343. doi: 10.1002/(SICI)1097-0134(199707)28:3<333::AID-PROT4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 33.Chothia C, Janin J. Principles of protein-protein recognition. Nature. 1975;256:705–708. doi: 10.1038/256705a0. [DOI] [PubMed] [Google Scholar]
- 34.Jones S, Thornton JM. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schellens IM, et al. Comprehensive Analysis of the Naturally Processed Peptide Repertoire: Differences between HLA-A and B in the Immunopeptidome. PLoS One. 2015;10:e0136417. doi: 10.1371/journal.pone.0136417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/s0076-6879(97)76066-x. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/s0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terwilliger TC, et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 2009;65:582–601. doi: 10.1107/s0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/s0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/s0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashkenazy H, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Stivala A, Wybrow M, Wirth A, Whisstock JC, Stuckey PJ. Automatic generation of protein structure cartoons with Pro-origami. Bioinformatics. 2011;27:3315–3316. doi: 10.1093/bioinformatics/btr575. [DOI] [PubMed] [Google Scholar]
- 46.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 47.Arafat Y, et al. Structural determinants of GAD antigenicity. Mol. Immunol. 2009;47:493–505. doi: 10.1016/j.molimm.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 48.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version1.8 (2015).
- 49.Yang M-J, Lee HW, Kim H. Enhancement of thermostability of Bacillus subtilis endoglucanase by error-prone PCR and DNA shuffling. Applied Biological Chemistry. 2017;60:73–78. doi: 10.1007/s13765-017-0254-3. [DOI] [Google Scholar]
- 50.Kim M-S, Woo M-H, Chang Y-H, Chung N, Kim J-S. Biochemical characterization of a noble xylanase from Paenibacillus sp. EC116. Applied Biological Chemistry. 2016;59:313–320. doi: 10.1007/s13765-016-0159-6. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.