Abstract
Transient receptor potential mucolipin 1 (TRPML1), a lysosomal channel, maintains the low pH and calcium levels for lysosomal function. Several small molecules modulate TRPML1 activity. ML-SA1, a synthetic agonist, binds to the pore region and phosphatidylinositol-3,5-bisphosphate (PtdIns(3,5)P2), a natural lipid, stimulates channel activity to a lesser extent than ML-SA1; moreover, PtdIns(4,5)P2, another natural lipid, prevents TRPML1-mediated calcium release. Notably, PtdIns(3,5)P2 and ML-SA1 cooperate further increasing calcium efflux. Here we report the structures of human TRPML1 at pH 5.0 with PtdIns(3,5)P2, PtdIns(4,5)P2, or ML-SA1 and PtdIns(3,5)P2, revealing a unique lipid-binding site. PtdIns(3,5)P2 and PtdIns(4,5)P2 bind to the extended helices of S1, S2, and S3. The phosphate group of PtdIns(3,5)P2 induces Y355 to form a π-cation interaction with R403, moving the S4–S5 linker, thus allosterically activating the channel. Our structures and electrophysiological characterizations reveal an allosteric site and provide molecular insight into how lipids regulate TRP channels.
Transient receptor potential mucolipin 1 (TRPML1) is a lysosomal channel which maintains the low pH and calcium levels for lysosomal function. Here authors use structural biology and electrophysiology to show how lipids bind and allosterically activate TRPML1.
Introduction
TRP channels, which include six sub-families, are widely regulated by various sensations and ligands1. Of these ligands, the TRP family has demonstrated regulation by more than 50 endogenous lipids2. TRPML1 regulates lysosomal calcium signaling, lipid trafficking, and autophagy-related processes3–5. Loss-of-function TRPML1 mutants cause a neurodegenerative disorder, Mucolipidosis type IV (MLIV)6–8. While recent structural determinations provide insight into the regulatory mechanisms of TRPs by small molecules9–12, there is still limited structural detail showing how specific lipids regulate activity. Determination of the structural regulation of TRP channels by endogenous lipid ligands can provide a wealth of insight into the physiological function of these channels13.
In previous studies, PtdIns(3,5)P2, a low-abundance phosphoinositol in late endosomes and lysosomes14, stimulates the opening of TRPML1, specifically at low pH12,15,16. In contrast, PtdIns(4,5)P2, a similarly structured phosphoinositol lipid abundant in the plasma membrane, decreases the TRPML1-mediated calcium release on the cell surface16. Notably, human fibroblasts derived from patients with Niemann-Pick C disease (a lysosomal storage disease caused by dysfunctional lipid trafficking), exhibit decreased TRPML1 activity15, implying a correlation between lysosomal abnormality and TRPML1 behaviors. Therefore, structural insights into TRPML1 regulation by lipid ligands represent a strong opportunity to study the lipid-mediated regulatory mechanism among TRP channels.
Recently, several groups reported structures of mammalian TRPML1 and TRPML311,12,17–20. TRPML1 forms a classic tetramer: each subunit contains six transmembrane helices (S1–S6), two pore helices (PH1 and PH2) and a ~30 kD lumenal/extracellular domain (Fig. 1a). The extensions of S1–S3 (labeled as IS1–IS3), including several basic amino acids, present an unique feature that presumably binds to PtdIns(3,5)P2 in the membrane16–18. However, there is no atomic detail for the interaction between the PtdInsP2 and TRPMLs.
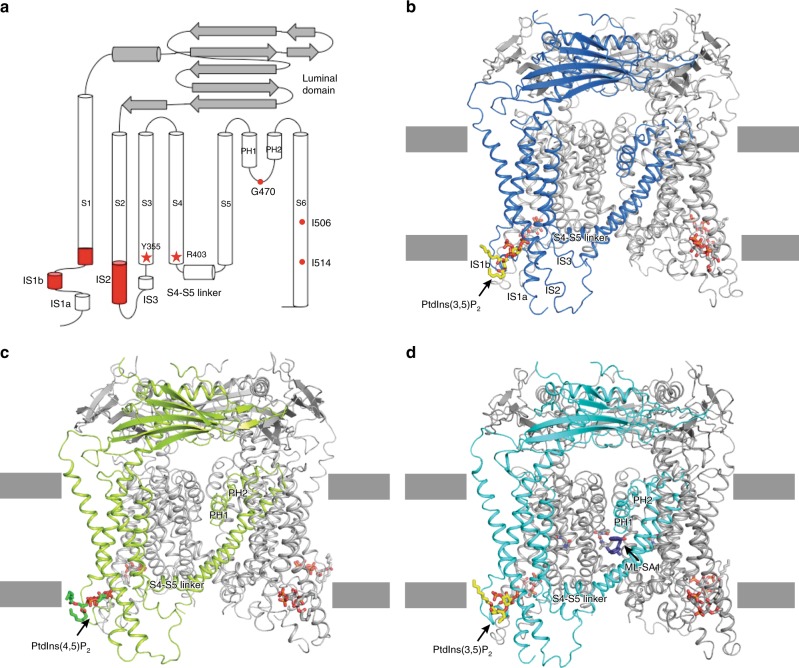
Fig. 1.
The overall structures of hTRPML1 with different ligands. a The secondary structure of hTRPML1. The major structural elements are labeled and the key residues for the PtdInsP2 activation or in the pore regions are indicated. b The structure of hTRPML1 with PtdIns(3,5)P2 (yellow sticks). c The structure of hTRPML1 with PtdIns(4,5)P2 (green sticks). d The structure of hTRPML1 with PtdIns(3,5)P2 (yellow sticks) and ML-SA1 (blue sticks)
In this manuscript, we report three structures of human TRPML1 with (1) PtdIns(3,5)P2, (2) PtdIns(4,5)P2, and (3) ML-SA1 with PtdIns(3,5)P2 at 3.5–3.7-Å resolution, revealing a lipid-binding site to allosterically activate the channel, distinct from the previously reported site in TRPV19,21. These structures along with electrophysiological characterizations reveal an allosteric site and illuminate how lipids regulate TRP channels.
Results
Assembly of TRPML1 with distinct ligands for cryo-EM study
To investigate the role of inositides in TRPML1 activation, we first assembled the human TRPML1 protein in nanodiscs containing PtdIns(3,5)P2 and used cryo-EM to detect the conformation of TRPML1 with PtdIns(3,5)P2. Unfortunately, the particles failed to reconstitute a structure at atomic resolution. To address this, we purified the protein in a low pH digitonin buffer and independently incubated with either PtdIns(3,5)P2, PtdIns(4,5)P2, or PtdIns(3,5)P2 plus ML-SA1 to capture TRPML1 in its various ligand-bound states. The resulting three structures (PtdIns(3,5)P2, PtdIns(4,5)P2, and PtdIns(3,5)P2 with ML-SA1; Fig. 1b–d and Supplementary Table 1) revealed a distinct inositide binding pocket for TRPML1 and yielded structural insight into one possible mechanism of channel regulation.
Structure of PtdIns(3,5)P2-bound TRPML1
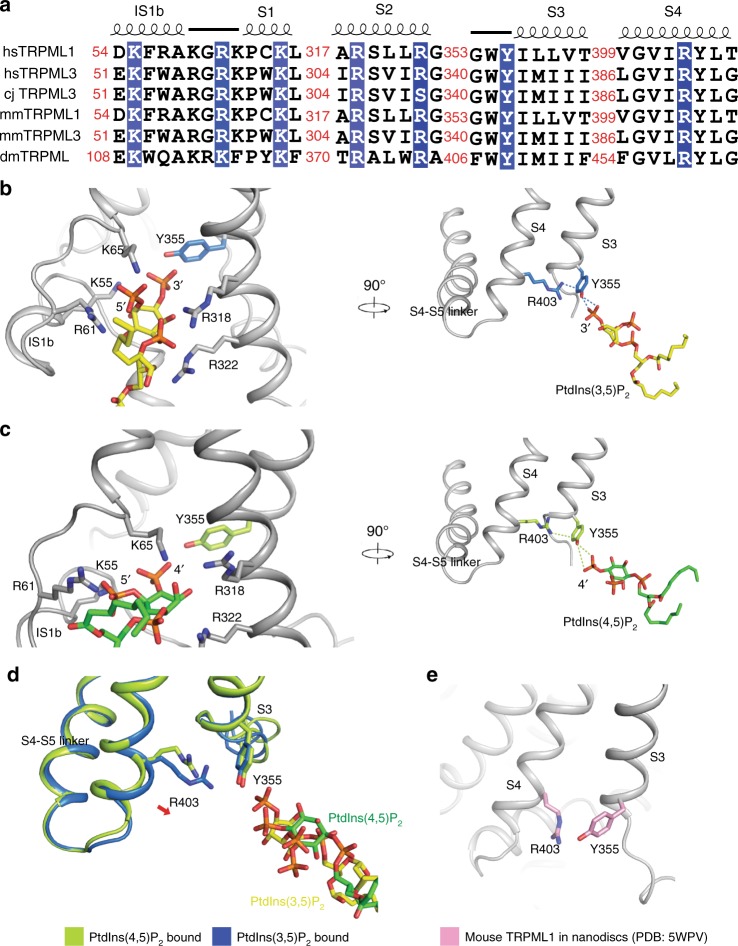
The cryo-EM structure of PtdIns(3,5)P2 bound TRPML1 is detected in a closed conformation at 3.5 Å resolution (Supplementary Fig. 1). An electron density is observed in the cavity created by extensions of S1, S2, and S3 (Supplementary Fig. 2). Our previously reported ML-SA1 bound structure in the absence of PtdIns(3,5)P2 lacked an extra density in this area at a similar resolution, leading us to putatively identify this density as PtdIns(3,5)P212. Single channel recordings of mouse TRPML1 activity indicated that the open probability of TRPML1 remained quite low when the PtdIns(3,5)P2 was bound, with less than half of the channel population presented in an open conformation18. This finding suggests that it may be difficult to capture significant populations of PtdIns(3,5)P2-mediated opening of TRPML1 for cryo-EM structural determinations. K55, R61, and K65 in S1, and R318 and R322 in S2 are involved in PtdIns(3,5)P2 binding (Fig. 2a). Mutations on these corresponding residues in TRPML317 and the deletion of IS1 and IS2 in mouse TRPML118 reduces the response of TRPMLs to PtdIns(3,5)P2. In addition to the residues in S1 and S2, the side chain density of Y355 in the PtdIns(3,5)P2-bound structure is adjacent to the density of the phosphate group of PtdIns(3,5)P2, implying a strong binding between these two groups (Fig. 2b and Supplementary Fig. 2). This interaction enriches the negative change of the aromatic ring plane of Y355, which recruits the head of R403 in S4 forming a π-cation bond (Fig. 2b). As R403 locates in the C-terminus of S4, it might induce a shift to move the S4–S5 linker away from the pore center.
Fig. 2.
The molecular details of hTRPML1 bound to PtdInsP2. a The sequence alignment of the key residues for PtdInsP2 recognition among multiple TRPMLs. b The interaction details of hTRPML1 bound to PtdIns(3,5)P2. c The interaction details of hTRPML1 bound to PtdIns(4,5)P2. d The structural comparison of hTRPML1 with PtdIns(3,5)P2 or PtdIns(4,5)P2. e Interaction details of R403 and Y355 (pink) in mouse TRPML1 closed conformation in nanodiscs (PDB: 5WPV)
Structure of PtdIns(4,5)P2-bound TRPML1
In order to compare the binding sites of PtdIns(3,5)P2 and PtdIns(4,5)P2, we determined the structure of PtdIns(4,5)P2 bound TRPML1 at 3.6 Å resolution (Supplementary Figs. 3, 4). Supporting our proposed inositide binding site, PtdIns(4,5)P2 binds to the same area of TRPML1 as PtdIns(3,5)P2 (Fig. 2c). However, the 3′ phosphate group of PtdIns(3,5)P2 is closer to Y355 than the 4′ phosphate group in the PtdIns(4,5)P2-bound structure (Fig. 2d). This difference may cause the charge of the tyrosine ring in the PtdIns(4,5)P2-bound structure to be less negative than in the PtdIns(3,5)P2-bound structure, preventing the strong π-cation interaction with R403 (Fig. 2d). Y355 and R403 in the structure of TRPML1 in nanodiscs do not form a π-cation interaction in the absence of PtdIns(3,5)P218 (Fig. 2e). From these observations we hypothesize that one potential structural mechanism behind the differential regulation of TRPML1 by PtdIns(4,5)P2 and PtdIns(3,5)P2 may lie in the 3′ phosphate supporting the formation of a π-cation interaction between Y355 and R403. Previous studies showed that PtdIns(4,5)P2 can decrease the ion efflux of TRPML1 by competing with PtdIns(3,5)P2 binding to the channel16,18. This is consistent with our structural observation that PtdIns(3,5)P2 and PtdIns(4,5)P2 bind the same area. PtdIns(4,5)P2 may serve as an agonist blocker on the cell surface, preventing the other lipid agonist from entering the lipid-binding site, therefore keeping the channel inactive. In normal cells, PtdIns(3,5)P2 and PtdIns(4,5)P2 are not abundant on the same membrane, and the activity of TRPML1 cation efflux depends on its cellular location.
Validation of PtdIns(3,5)P2-mediated TRPML1 activation
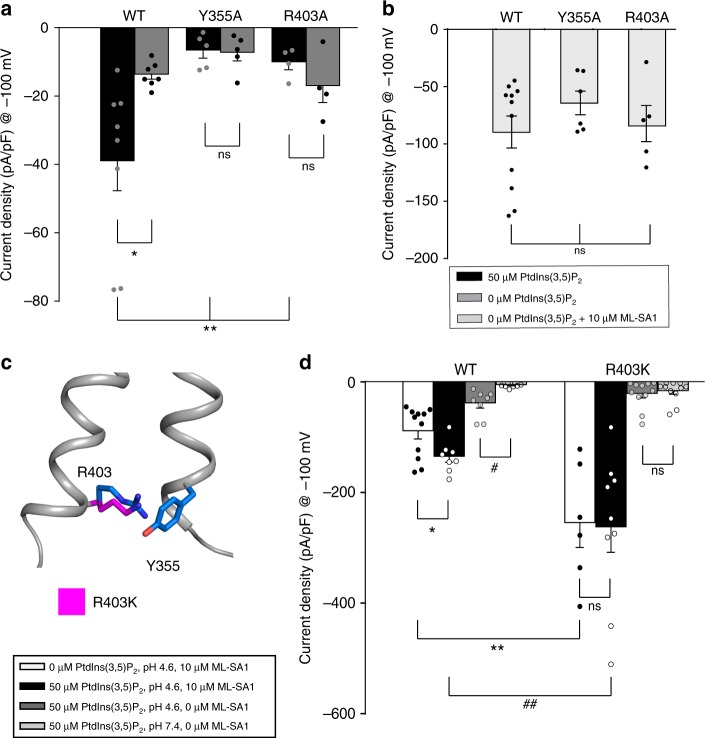
To test our observation and hypothesis, we generated two eGPF-fused TRPML1-L/A22 mutations, TRPML1-Y355A-LA and TRPML1-R403A-L/A. The four leucine-to-alanine mutations in TRPML1-L/A have been reported to increase cell surface localization for subsequent electrophysiological characterization via whole-cell patch clamp22. With PtdIns(3,5)P2 perfused in the cytoplasmic solution, there is a significant increase in current density for HEK cells expressing WT-L/A at low pH (Fig. 3a). However, both the Y355A-L/A and R403A-L/A mutants show no significant increase in channel activity at low pH when 50 µM PtdIns(3,5)P2 is present in the pipette (Fig. 3a).
Fig. 3.
TRPML1 channel activation for WT-L/A and Inositide binding mutants. a Current densities at −100 mV for various TRPML1 constructs with or without cytoplasmic PtdIns(3,5)P2 reveal significant stimulation for WT but not the inositide pore binding mutants Y355A and R403A when 50 µM PtdIns(3,5)P2 is present in the pipette solution. (*p = 0.038; **p < 0.005); (n = 8, 7, 5, 5, 4, 4, resp). b Stimulation with the synthetic agonist ML-SA1 (10 µM); (n = 11, 6, 5 resp). c Structural view of the R403 and Y355 residues (blue) and the R403K mutant (magenta). d Current densities at −100 mV for various WT-L/A and R403K TRPML1 mutant with or without 50 µM PtdIns(3,5)P2 or 10 µM ML-SA1 at pH 4.6 and 7.4. The R403K mutation leads to a significant increase in current densities when ML-SA1 is applied regardless of the presence of inositide yet does not significantly increase channel current densities when ML-SA1 is not present. (*p = 0.023; #p = 0.001; **p < 0.001; ##p = 0.008); (n = 11, 8, 8, 6, 9, 6, 11, 11, resp). Values are mean ± s.e.m
Earlier reports describe a significant reduction of PtdIns(3,5)P2 mediated TRPML1 current at higher pH in the presence of 1 mM Ca2+20. Among all the channels tested, only WT-L/A channels demonstrated this significant reduction in current density at pH 7.4 (Supplementary Figure 5a) further supporting our observation of PtdIns(3,5)P2 mediated TRPML1 function. To verify channel function for all WT-L/A and inositide binding mutant channels, application of the synthetic agonist ML-SA1 was used to open the channel in the absence of perfused PtdIns(3,5)P2 (Fig. 3b and Supplementary Fig. 5b). All expressed constructs were stimulated by 10 μM ML-SA1 with current densities 2–3 times higher than for WT-LA channels with PtdIns(3,5)P2 application alone, similar to our previously reported findings12 and providing validation of proper channel surface expression and function. This data coupled with our structural observations and previous work on TRPML3 and PtdIns(3,5)P217, suggest an alternate site for inositide binding separate from ML-SA1 that can allosterically affect channel opening. Notably, since an R403 mutation causes MLIV, it is likely that this mutant cannot be activated by PtdIns(3,5)P2.
Previous studies showed that PtdIns(3,5)P2 and ML-SA1 cooperate to stimulate TRPML1 considerably more than either agonist alone12,15. To investigate whether or not the π-cation interaction is involved in this potentiation, we utilized the ability of lysine to form a stronger π-cation interaction than the endogenous arginine23. Compared with arginine, lysine has a shorter side chain that presents less flexibility and an amine group with a more compressed positive charge, theoretically making it easier to capture the aromatic ring of Y355 and form a stronger π-cation interaction (Fig. 3c).
We expressed this mutant and compared the current with WT channels with and without PtdIns(3,5)P2 or ML-SA1. In the presence of 10 μM ML-SA1, current densities of both WT and R403K were greatly enhanced (Fig. 3d and Supplementary Fig. 5a). Cytoplasmic supplementation of PtdIns(3,5)P2 significantly potentiates WT current, but not R403K current. Regardless of the presence or absence of cytoplasmic PtdIns(3,5)P2, the R403K stimulation with ML-SA1 is significantly higher than WT. Interestingly, in the absence of ML-SA1, the R403K mutant was not significantly stimulated by PtdIns(3,5)P2 at pH 4.6, unlike WT channels (Fig. 3d and Supplementary Fig. 5a). This leads us to speculate whether the PtdIns(3,5)P2 interaction may induce additional confirmations not directly supported by R403 and Y355 interactions. This could help explain why the open probability of the PtdIns(3,5)P2 stimulated channel remains low as well as the difficulty in detecting an open conformation with only inositide binding18. We hypothesize that the gain of function mutation R403K does not necessarily support inositide signaling in the absence of ML-SA1. Instead the mutation may enhance the π-cation interaction supporting robust stimulation when ML-SA1 is applied regardless of PtdIns(3,5)P2.
Structure of PtdIns(3,5)P2/ML-SA-1-bound TRPML1
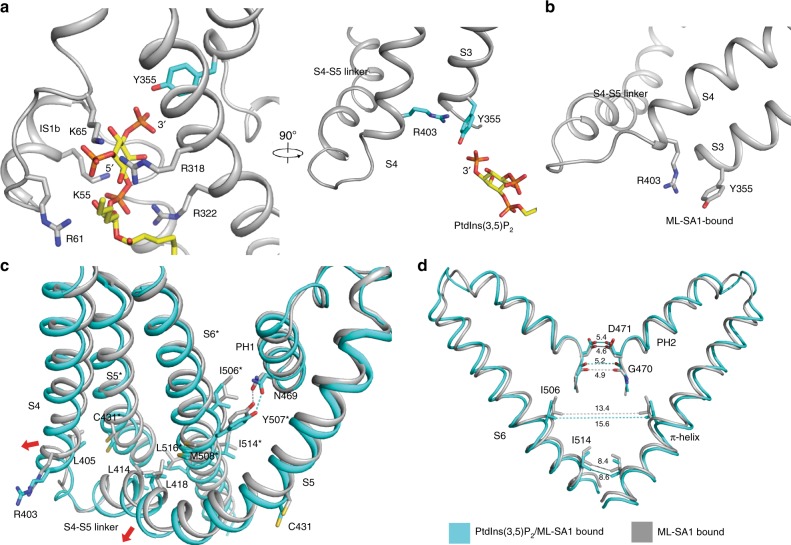
To capture the mechanism of ML-SA1 and PtdIns(3,5)P2 cooperation, the ML-SA1/PtdIns(3,5)P2-bound structure was determined at 3.7 Å resolution in an open conformation (Supplementary Figs. 6, 7). In this structure Y355 binds the 3′ phosphate group of PtdIns(3,5)P2 (Fig. 4a) and S1–S3 have a conformational shift due to PtdIns(3,5)P2 binding when compared to the ML-SA1 alone structure (Supplementary Fig. 8a). The comparison between the PtdIns(3,5)P2 /ML-SA1 and PtdIns(3,5)P2-bound structures shows that ML-SA1 binding causes the S3 and S4 of the PtdIns(3,5)P2 binding site to move away from the pore center (Supplementary Fig. 8b). In the ML-SA1-bound structure, the ring of Y355 does not align with the polar head of R403; consequently, there is no π-cation interaction between Y355 and R403 (Fig. 4b). Although, the density of the R403 side chain is not clear in the density map (Supplementary Fig. 7), structural comparison still suggests that the shift of R403 may trigger a movement of the S4–S5 linker and channel pore, since the S4–S5 linker moves 2–3 Å away from the pore (Fig. 4c). When PtdIns(3,5)P2 binds TRPML1, R403 induces a shift of L405, L414, and L418, which have hydrophobic interactions with C431 in the S5, and L516 and M508 in the S6 of neighboring subunit, allosterically forcing the S5 and S6 away from the pore center (Fig. 4c). The movement of L516 can facilitate the opening of I514 in the lower gate; the shift of M508 causes the π-helix opening by I506; and Y507 can form a hydrophilic interaction with N469, opening the selectivity filter (Fig. 4d). Although PtdIns(3,5)P2 cannot directly bind to the S4–S5 linker, where the agonists of TRPV1 bind and allosterically affect the opening of TRPV121, the allosteric activation mechanism of TRPV1 and TRPML1 still present similar behavior: the movement of the S4–S5 affects the opening of the channel pore. Interestingly, in mouse TRPML1-nanodisc structures, this linker presents two distinct conformations, presumably reflecting a different allosteric regulation of the S4–S5 linker in the lipid environment18.
Fig. 4.
The putative mechanism of PtdIns(3,5)P2 and ML-SA1 cooperation. a PtdIns(3,5)P2 induces the π-cation interaction of Y355 and R403 in PtdIns(3,5)P2/ML-SA1 bound structure. b The molecular detail of Y355 and R403 in the ML-SA1 bound structure (PDB: 5WJ9). c Structural comparison of both agonists bound (cyan) and the ML-SA1 bound (gray) hTRMPL1 structures. d The comparison of the pore region of both agonists bound and the ML-SA1 bound (gray) hTRMPL1 structures
Discussion
In this manuscript, we report three human TRPML1 structures with different ligands. TRPML1 employs its extensions of IS1–IS3 to recognize the PtdInsP2 binders using basic amino acids. PtdIns(3,5)P2 induces Y355 in S3 to form a π-cation interaction with R403 in S4 through the binding of its 3′ phosphate group to the hydroxyl group of Y355. This π-cation interaction affects the movement of the S4–S5 linker, a common allosteric site among TRP channels. Compared with ML-SA1, which directly forces the pore open, this stimulation is indirect and mild, explaining the low proportion of TRPML1 channels that are active with PtdIns(3,5)P2. The fact that the R403K mutant appears to facilitate ML-SA1 pore opening but does not significantly affect PtdInsP2 mediated channel opening poses an interesting hypothesis. It is possible that the inositide binding site allosterically regulates channel opening in several different ways. As noted by earlier reports, the idea that both PtdInsP2 bind to the same pocket but have significantly different effects on channel opening could represent a way to control channel activity through localization within different lipid enriched compartments. In this manner endogenous TRPML1 is kept relatively inactive at the surface of the cell due to an abundance of PtdIns(4,5)P2. As TRPML1 is trafficked to the lysosome, the membrane is enriched with PtdIns(3,5)P2. Concurrently, PtdIns(4,5)P2 is displaced by PtdIns(3,5)P2 allowing the formation of the observed π-cation interaction. In this conformation, the channel is more susceptible to pore opening by agonists like ML-SA1. However, this does not rule out the possibility of additional open conformations supported by PtdIns(3,5)P2 binding. As the R403K mutation does not increase channel activity, it is also possible these conformations may not rely on the π-cation formed between R403 and Y355.
The structures of other TRP channels have revealed several lipid-binding sites among the different channels. The structure of TRPML3 in the closed conformation with amphipols shows several sterol-like molecules attached near the pore region including our observed ML-SA1-binding pocket (Supplementary Fig. 9a). However, there is no lipid density in the extensions of IS1–IS317. The structure of TRPV6 in the open conformation reveals a lipid bound to the S4–S5 linker, the allosteric site of the channel, presumably stimulating channel opening10 (Supplementary Fig. 9b). Similar to TRPV6, the structure of TRPV1 in nanodiscs reveals an associated lipid bound to the pore region consistent with the ML-SA1-binding pocket of TRPML1 and a phosphatidylinositol lipid was observed in the S4–S5 linker of TRPV1 although it is also in a closed conformation9 (Supplementary Fig. 9c). Recently, the structure of TRPV5 with PtdIns(4,5)P2 in an open conformation has been reported revealing a unique binding site between the N-linker, S4–S5 linker and S6 helix of TRPV524 (PDB: 6DMU). In our reported structures, there is no lipid density in the S4–S5 linker implying this site may not be used for the channel activation among TRPML family. Instead of the S4–S5 linker, the allosteric site for TRPML activation resides in the extensions of IS1–IS3, which capture the PtdInsP2 lipid.
Previous reports showed that other ion channels, such as GIRK225, Kir2.226, and TPC127, are also regulated by PtdInsP2. Compared with those structures, our structures reveal a binding site far away from either the pore or the S4–S5 linker that can open the channel remotely. It is possible that TRPML1 is evolutionarily suitable for binding PtdIns(3,5)P2 due to the high abundance of PtdIns(3,5)P2 in the lysosomal environment. Thus, our structural observations show an allosteric site among TRP channels that can host lipid ligands, and may reveal one molecular mechanism for lipid-mediated TRPMLs regulation.
Methods
Protein expression and purification
Human TRPML1 was cloned into pEG BacMam with an N-terminal Flag tag. The protein was expressed using baculovirus-mediated transduction of mammalian HEK293S GnTI− cells (ATCC). These cells tested negative for mycoplasma contamination. At 48 h post infection at 37 °C, cells were disrupted by sonication in buffer A (20 mM HEPES, pH 7.0, 150 mM NaCl) with 1 mM PMSF and 5 μg ml−1 each of leupeptin and aprotinin. After low-speed centrifugation at 3470 g, the resulting supernatant was incubated in buffer A with 1% (w/v) lauryl maltose neopentyl glycol (MNG, Anatrace) for 1 h at 4 °C. The lysate was centrifuged at 34,572 g and the supernatant was loaded onto an anti-Flag M2 affinity column (Sigma). After washing twice, the protein was eluted in 20 mM HEPES, pH 7.0, 150 mM NaCl, 0.1 mg ml−1 Flag peptide and 0.01% MNG, and then concentrated. The protein was purified by Superdex-200 size-exclusion chromatography (GE Healthcare) in a buffer containing 20 mM sodium acetate, pH 5.0, 150 mM NaCl and 0.06% (w/v) digitonin (Sigma). The peak fractions were collected and concentrated to 5–7 mg/ml for grid preparation. The mutated DNA constructs were generated using QuikChange Mutagenesis Kit (Agilent) (Supplementary Table 2).
Whole-cell patch clamp electrophysiology
The full-length human TRPML1 (L/A) was subcloned to pEGFP-C1 for electrophysiological assays. Whole-cell patch clamp recordings of cell electrical parameters were performed using Matlab based Capmeter v7.228,29 with a National Instruments digital acquisition board and an Axopatch 200B patch clamp amplifier. For current density comparisons, capacitance was monitored in real-time using square-wave voltage perturbations (20 mV; 0.5 kHz). Input resistances for the capillary pipettes were 2–10 MΩ, and the apparent cell resistances were 0.5–2 GΩ. External solutions were adjusted to RT (23–25 °C) in gravity-fed parallel solution lines with an outlet flow velocities of 2–5 mm/s allowing extracellular solution changes within 2–3 s. For TPRML1 current–voltage analysis, a voltage ramp was generated from −150 mV to 20 mV over 1.5 s. Voltage ramps were aligned and statistical analysis was performed at −100 mV. Boroscilicate glass pipettes were fire polished and back-filled with the cytoplasmic solutions containing in mM: 120 cesium methanesulfonate, 4 NaCl, 10 EGTA, 2 MgCl2, 20 HEPES, pH 7.2 with CsOH (~25 mM). A 5 mM stock solution of C8: PtdIns(3,5)P2 (Echelon) was prepared in ddH2O and aliquots were stored at −80 °C. For PtdIns(3,5)P2 stimulated recordings, a 1:100 dilution of lipid was added to the cytoplasmic solution and briefly sonicated. Extracellular solutions were set to pH 4.6 and contained in mM: 140 sodium gluconate, 5 KCl, 10 glucose, 10 HEPES, 10 MES, 1 MgCl2, 1 CaCl2, and 8 HCl. For control experiments at pH 7.4, bath solution contained in mM: 140 NaCl, 5 KCl, 10 glucose, 20 HEPES, 1 MgCl2, 1 CaCl2, and 8 NaOH. A total of 10 µM ML-SA1 was added to the appropriate bath solution immediately prior to recording. Low passage HEK293T (ATCC CRL-11268) cells used for electrophysiology were maintained in DMEM (GIBCO) with 10% FBS, penicillin-streptomycin, and l-glutamine (Sigma), and routinely monitored for mycoplasma infection using MycoSensor PCR kits (Agilent). HEK293T cells were transiently transfected with Lipofectamine 3000 (Invitrogen) and used within 48–72 h. The cells were deplated with trypsin (0.25%) and placed in a bath solution on an inverted Nikon TE2000U inverted microscope equipped with a 60X oil immersion, 1.45-NA objective. A Lambda DG-4 xenon epifluorescence power supply with Semrock FITC filters were used to detect cells expressing surface TRPML1-GFP mutants. Statistical analyses were performed in Matlab and SigmaPlot (SigmaStat) using either a Rank Sum Test or a Student's t-test as determined by the SigmaPlot Normality Test. Figures were created in GraphPad Prism 7.05. Current–voltage relations represent the mean of at least 4 voltage ramps, and agonist-induced current density changes represent the mean and s.e.m. of at least four independent cells per condition.
EM sample preparation and imaging
A protein sample was added to Quantifoil R1.2/1.3 400 mesh Au holey carbon grids (Quantifoil), blotted with Vitrobot Mark IV (FEI), and frozen in liquid ethane. For PtdIns(3,5)P2 or PtdIns(4,5)P2 bound protein, the protein in a buffer containing 20 mM sodium acetate, pH 5.0, 150 mM NaCl and 0.06% digitonin was incubated with 0.2 mM PtdIns(3,5)P2 or PtdIns(4,5)P2 (Echelon, dissolved in H2O as a 10 mM stock) on ice for 1 h before grid preparation and freezing. For PtdIns(3,5)P2/ML-SA1 bound protein, the protein in a buffer containing 20 mM sodium acetate, pH 5.0, 150 mM NaCl and 0.06% digitonin was incubated with 0.2 mM PtdIns(3,5)P2 and 0.3 mM ML-SA1 (Tocris Bioscience, dissolved in DMSO as a 20 mM stock) on ice for 1 h before grid preparation and freezing. The grids were imaged with a 300 keV Titan Krios (FEI) with a Gatan K2 Summit direct electron detector (Gatan). The data were collected at 1.07 Å per pixel with a dose rate of eight electrons per physical pixel per second. Images were recorded for 12-s exposure in 30 subframes to give a total dose of 84 electrons per Å2.
Imaging processing and 3D reconstruction
Dark subtracted images were first normalized by gain reference that resulted in a pixel size of 1.07 Å per pixel. Drift correction was performed using the program Unblur30. The contrast transfer function (CTF) was estimated using CTFFIND431. After automatic picking and manual micrograph inspection, the particles were extracted for subsequent 2D and 3D classification. Motion correction of individual particles was performed using the program alignparts_lmbfgs32. Using the structure of human TRPML1 (EMD-8840) low-pass filtered to 60 Å as the initial model, 3D classification was carried out in RELION33. The best classes of four structures, containing 50k~60k particles, provided a 5–6 Å map. Refinement was performed in FREALIGN34 using this best class as the initial model and all particles post motion correction to generate the final map for model construction.
Model construction
To obtain better side-chain densities for model building, we sharpened one half-map using BFACTOR.EXE (author: Nikolaus Grigorieff) with a resolution limit of 3.53 Å (PtdIns(3,5)P2 bound) and 3.57 Å (PtdIns(4,5)P2 bound) and 3.7 Å (ML-SA1/PtdIns(3,5)P2 bound), and a B-factor value of −100 Å2. To precisely determine the location of PtdInsP2, we took advantage of the stronger signal phosphate atoms have in the electron density compared to other atoms (oxygen, nitrogen, and carbon), and we increased signal to 8–10σ level to determine its head position. The entire model was built in COOT35.
Model refinement and validation
The model was refined in real space using PHENIX36 and also in reciprocal space using Refmac37,38. Structural factors were calculated from a half-map (working) using the program SFall38. Fourier shell correlations (FSCs) were calculated between the two half maps, the model against the half1 map, the half2 map, and full map40. Local resolutions were estimated using Blocres41. MolProbity42 was used to validate the geometries of the model. Structure figures were generated using PyMOL (http://www.pymol.org) and Chimera43.
Electronic supplementary material
Acknowledgements
This research was done in the memory of G. Blobel for his generosity and encouragement. The data were collected at the UT Southwestern Medical Center Cryo-EM Facility (funded in part by the CPRIT Core Facility Support Award RP170644); we thank D. Nicastro for the access of cryo-EM, and Z.G. Chen and D. Stoddard for assistance in data collection. We thank Z.X. Chen, D. Hilgemann, E. Coutavas, and E. Debler for help in manuscript preparation. M.F. wishes to thank S. Lin for years of support. This work was supported by the Endowed Scholars Program in Medical Science of UT Southwestern Medical Center and O’Donnell Junior Faculty Funds (to X.L.) and NIH grant P01 HL020948 (Tissue Culture Core); X.L. is Rita C. and William P. Clements, Jr. Scholar in Biomedical Research of UT Southwestern.
Authors contributions
P.S. and X.L. purified proteins and carried out cryo-EM work. M.F. carried out functional characterization of the TRPML1 channel by electrophysiology. All the authors analyzed the data and contributed to manuscript preparation. X.L. conceived the project and wrote the paper.
Data availability
The data supporting the findings of this manuscript are available from the corresponding author upon reasonable request. The 3D cryo-EM density maps of TRPML1 have been deposited in the Electron Microscopy Data Bank under the accession numbers EMDB-9000 (PI(3,5)P2-bound), EMDB-9001 (PI(4,5)P2-bound) and EMDB-9002 (PI(3,5)P2/ML-SA1-bound). Atomic coordinates for the atomic model of TRPML1 have been deposited in the Protein Data Bank under the accession numbers 6E7P (PI(3,5)P2-bound), 6E7Y (PI(4,5)P2-bound) and 6E7Z (PI(3,5)P2/ML-SA1-bound).
Competing interests
X.L. is a consultant of Casma Therapeutics. The remaining authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Michael Fine, Philip Schmiege.
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-06493-7.
References
- 1.Venkatachalam K, Montell C. TRP channels. Annu. Rev. Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taberner FJ, Fernandez-Ballester G, Fernandez-Carvajal A, Ferrer-Montiel A. TRP channels interaction with lipids and its implications in disease. Biochim. Biophys. Acta. 2015;1848:1818–1827. doi: 10.1016/j.bbamem.2015.03.022. [DOI] [PubMed] [Google Scholar]
- 3.Zeevi DA, Frumkin A, Bach G. TRPML and lysosomal function. Biochim. Biophys. Acta. 2007;1772:851–858. doi: 10.1016/j.bbadis.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Wang W, Zhang X, Gao Q, Xu H. TRPML1: an ion channel in the lysosome. Handb. Exp. Pharmacol. 2014;222:631–645. doi: 10.1007/978-3-642-54215-2_24. [DOI] [PubMed] [Google Scholar]
- 5.Venkatachalam K, Wong CO, Zhu MX. The role of TRPMLs in endolysosomal trafficking and function. Cell Calcium. 2015;58:48–56. doi: 10.1016/j.ceca.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun M, et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum. Mol. Genet. 2000;9:2471–2478. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- 7.Altarescu G, et al. The neurogenetics of mucolipidosis type IV. Neurology. 2002;59:306–313. doi: 10.1212/WNL.59.3.306. [DOI] [PubMed] [Google Scholar]
- 8.Eichelsdoerfer JL, Evans JA, Slaugenhaupt SA, Cuajungco MP. Zinc dyshomeostasis is linked with the loss of mucolipidosis IV-associated TRPML1 ion channel. J. Biol. Chem. 2010;285:34304–34308. doi: 10.1074/jbc.C110.165480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGoldrick LL, et al. Opening of the human epithelial calcium channel TRPV6. Nature. 2018;553:233–237. doi: 10.1038/nature25182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, et al. Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states. Nat. Struct. Mol. Biol. 2017;24:1146–1154. doi: 10.1038/nsmb.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmiege, P., Fine, M., Blobel, G. & Li, X. Human TRPML1 channel structures in open and closed conformations. Nature 550, 366–370 (2017). [DOI] [PMC free article] [PubMed]
- 13.Schmiege, P., Fine, M. & Li, X. The regulatory mechanism of mammalian TRPML s revealed by cryo‐EM. FEBS J.285, 2579–2585 (2018). [DOI] [PMC free article] [PubMed]
- 14.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen D, et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat. Commun. 2012;3:731. doi: 10.1038/ncomms1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Li X, Xu H. Phosphoinositide isoforms determine compartment-specific ion channel activity. Proc. Natl Acad. Sci. USA. 2012;109:11384–11389. doi: 10.1073/pnas.1202194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirschi M, et al. Cryo-electron microscopy structure of the lysosomal calcium-permeable channel TRPML3. Nature. 2017;550:411–414. doi: 10.1038/nature24055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Q, et al. Structure of mammalian endolysosomal TRPML1 channel in nanodiscs. Nature. 2017;550:415–418. doi: 10.1038/nature24035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S, Li N, Zeng W, Gao N, Yang M. Cryo-EM structures of the mammalian endo-lysosomal TRPML1 channel elucidate the combined regulation mechanism. Protein Cell. 2017;8:834–847. doi: 10.1007/s13238-017-0476-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, et al. Structural basis of dual Ca(2+)/pH regulation of the endolysosomal TRPML1 channel. Nat. Struct. Mol. Biol. 2017;24:205–213. doi: 10.1038/nsmb.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–118. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vergarajauregui S, Puertollano R. Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic. 2006;7:337–353. doi: 10.1111/j.1600-0854.2006.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc. Natl Acad. Sci. USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes, T. E. T. et al. Structural insights on TRPV5 gating by endogenous modulators. Preprint at 10.1101/338798 (2018). [DOI] [PMC free article] [PubMed]
- 25.Wang W, Whorton MR, MacKinnon R. Quantitative analysis of mammalian GIRK2 channel regulation by G proteins, the signaling lipid PIP2 and Na+ in a reconstituted system. eLife. 2014;3:e03671. doi: 10.7554/eLife.03671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen SB, Tao X, MacKinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+channel Kir2.2. Nature. 2011;477:495–498. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.She J, et al. Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel. Nature. 2018;556:130–134. doi: 10.1038/nature26139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fine M, et al. Massive endocytosis driven by lipidic forces originating in the outer plasmalemmal monolayer: a new approach to membrane recycling and lipid domains. J. Gen. Physiol. 2011;137:137–154. doi: 10.1085/jgp.201010469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang TM, Hilgemann DW. Ca-dependent nonsecretory vesicle fusion in a secretory cell. J. Gen. Physiol. 2008;132:51–65. doi: 10.1085/jgp.200709950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohou A, Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubinstein JL, Brubaker MA. Alignment of cryo-EM movies of individual particles by optimization of image translations. J. Struct. Biol. 2015;192:188–195. doi: 10.1016/j.jsb.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grigorieff N. Frealign: an exploratory tool for single-particle Cryo-EM. Methods Enzymol. 2016;579:191–226. doi: 10.1016/bs.mie.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eyck LFT. Efficient structure-factor calculation for large molecules by the fast fourier transform. Acta Crystallogr.A. 1977;33:486–492. doi: 10.1107/S0567739477001211. [DOI] [Google Scholar]
- 40.Wang Z, et al. An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Nat. Commun. 2014;5:4808. doi: 10.1038/ncomms5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this manuscript are available from the corresponding author upon reasonable request. The 3D cryo-EM density maps of TRPML1 have been deposited in the Electron Microscopy Data Bank under the accession numbers EMDB-9000 (PI(3,5)P2-bound), EMDB-9001 (PI(4,5)P2-bound) and EMDB-9002 (PI(3,5)P2/ML-SA1-bound). Atomic coordinates for the atomic model of TRPML1 have been deposited in the Protein Data Bank under the accession numbers 6E7P (PI(3,5)P2-bound), 6E7Y (PI(4,5)P2-bound) and 6E7Z (PI(3,5)P2/ML-SA1-bound).