Abstract
Bax is a Bcl-2 protein critical for apoptosis induction. In healthy cells, Bax is mostly a monomeric, cytosolic protein, while upon apoptosis initiation it inserts into the outer mitochondrial membrane, oligomerizes, and forms pores that release proapoptotic factors like Cytochrome c into the cytosol. The structures of active Bax and its homolog Bak are only partially understood and the topology of the proteins with respect to the membrane bilayer is controversially described in the literature. Here, we systematically review and examine the protein–membrane, protein–water, and protein–protein contacts of the nine helices of active Bax and Bak, and add a new set of topology data obtained by fluorescence and EPR methods. We conclude based on the consistent part of the datasets that the core/dimerization domain of Bax (Bak) is water exposed with only helices 4 and 5 in membrane contact, whereas the piercing/latch domain is in peripheral membrane contact, with helix 9 being transmembrane. Among the available structural models, those considering the dimerization/core domain at the rim of a toroidal pore are the most plausible to describe the active state of the proteins, although the structural flexibility of the piercing/latch domain does not allow unambiguous discrimination between the existing models.
Subject terms: Mitochondrial proteins, Biophysical chemistry
Facts
Bax and Bak are executioners in the intrinsic apoptotic pathway inducing mitochondrial outer membrane permeabilization (MOMP).
Bax activation needs a multistep conformational transition from a soluble, monomeric protein to a membrane-embedded homo-oligomer. Bax homo-oligomers assemble into multimers of dimers. Active Bak forms membrane-embedded homo-oligomers starting from membrane-embedded monomers.
Inactive Bax and Bak have a globular fold. The active forms are composed of a dimerization/core domain that enables the formation of globular dimers and flexible piercing/latch domains involved in dimer–dimer contacts.
Bax and Bak can be activated by activator BH3-only proteins like cBid or Bim and inhibited by pro-survival Bcl-2 proteins.
Open questions
How do Bax or Bak oligomers align with respect to the membrane in order to form the functional pore?
How many dimers are necessary to form one pore?
Does the piercing/latch domain adopt a unique structure like the dimerization/core domain or can it only be described as an ensemble of flexible structures or as a disordered protein region?
Which of the suggested models of membrane-embedded Bax or Bak is plausible considering all available data?
Introduction
Apoptosis is a form of programmed cell death pivotal for mammals and regulated by Bcl-2 proteins [1–4]. Aberrant apoptosis causes severe diseases like cancer or autoimmune, as well as neurodegenerative diseases [5]. Therefore, Bcl-2 proteins are important targets for drug development [1, 6], and the first drug directly targeting the Bcl-2 proteins (Venetoclax) was recently approved.
Bax and its homolog Bak are crucial for mitochondrial outer membrane (MOM) permeabilization [3, 7]. The knockout of both their genes drastically reduces the sensitivity of cells to apoptosis and is either lethal or leads to severe deformities in mice [8, 9]. Development of drugs targeting Bax/Bak is an important scientific goal [10], but it is challenging as the structures of the active, membrane-embedded proteins are unknown and the multistep conformational transitions are difficult to understand [11–14] (Fig. 1a).
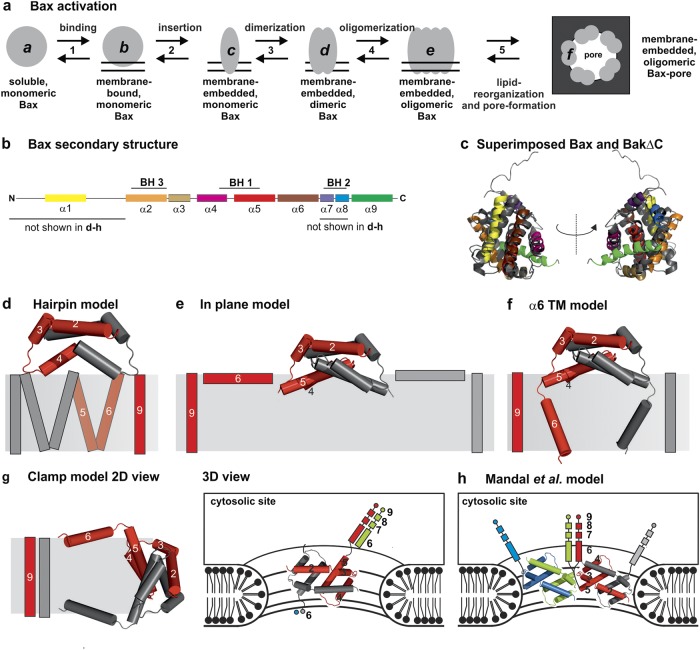
Fig. 1.
Topology models of Bax and Bak. a Sketch of the conformational changes of Bax during the transformation from the soluble, monomeric, inactive state to the membrane-embedded, oligomeric, pore-forming state. The protein can exist at least in six conformational states (a–f). b Schematic view of the secondary structure of Bax with the BH domains highlighted. c Superposition of the structures of soluble, monomeric Bax [15] (color code as in b) and BakΔNΔC (dark gray) [16]. d–g 2D representation of four published topology models of membrane-embedded Bax/Bak dimers. One monomer is shown in gray and the other in red. The structure of helices 2–5 is based on the PDB: 4BDU [25]. d Hairpin model based on Annis et al. [49]. e “In-plane” model as proposed by Westphal et al. [47]. f, g Models of Bax based on a structural model suggested by Bleicken et al. [30]: model considering a planar bilayer (panel f, α6 TM model) and the “clamp” model in the context of a toroidal pore (panel g, left: 2D model, right: 3D model with possible contacts to other helices 6–9 of adjacent dimers in green and blue). h Model suggested by Mandal et al. [50]. Two dimers are shown, one in gray and red, and the second in blue and green
The structures of the soluble, monomeric forms of Bax and BakΔC are solved and show a similar fold [15, 16] (Fig. 1b, c). In healthy cells, the monomeric conformations of Bax and Bak are reversibly shuttling to the mitochondria with different rates [17] (Fig. 1a, states a and b). Thereby, inactive Bax is mainly cytosolic and loosely associated at the MOM, whereas Bak is mainly MOM bound and part of a large protein complex containing the voltage-dependent anion channel 2 (VDAC2) [17–20]. Proapoptotic stimuli shift the conformational equilibria toward the membrane-bound form (Fig. 1a, state b) [17, 21].
To permeabilize the MOM, a transient interaction between Bax (or Bak) and an activator-type BH3-only protein takes place [14, 22–26], followed by rearrangements at the N and C terminus and a partial opening of the hairpin between helixes 5 and 6 [12, 23, 25, 27–30], leading to deep membrane insertion of the protein (Fig. 1a, state c). Afterward, homodimers are formed (Fig. 1a, state d) that assemble into higher-order oligomers [13, 14, 30, 31] (Fig. 1a, state e). Thereby, helices 2–5 (called the “dimerization” or “core” domain) form one very stable, symmetric dimer interface [25, 30, 32, 33], whereas a dimer–dimer interface is formed by helices 6–9 (called “piercing” or “latch” domain) [30, 34–39]. Membrane insertion of Bax induces changes in the membrane that enable the formation and stabilization of toroidal pores [30, 32, 40–45] (Fig. 1a, state f). No clear differences between the membrane-embedded conformations of Bax and Bak oligomers are reported, and the existence of hetero-oligomers [39, 46] suggest structural overlap.
Several structural and topology models of Bax and Bak have been suggested [30, 34, 39, 47–50]. Most models present monomers or dimers in the membrane, but fail to explain how the pore is formed. The latter information is addressed only by two available models [30, 50]. Why is it difficult to reconcile the available data into a coherent structural model of active Bax (Bak) in the context of a MOM pore? We envisage three main reasons: (i) Bax dimers assemble into an ensemble of oligomeric structures, which are not unique in size; (ii) the “piercing/latch” domain is flexible, which makes structure determination very challenging; and (iii) the membrane is deformed by the protein, which complicates the assignment of the helix topology.
The oldest topology model of Bax suggested that helices 5 and 6 form a tight hairpin [49] (Fig. 1d). It was challenged by cross-link experiments demonstrating hairpin opening upon Bax membrane insertion [25] and it was ruled out by X-ray structures and electron paramagnetic resonance (EPR) studies [25, 30, 33, 50]. Models suggesting an asymmetric or “daisy chain” oligomerization process [48] were also contradicted by these data. Kluck and coworkers performed water accessibility measurements on different regions in the Bax/Bak [37, 39, 47, 51, 52], and proposed the “in-plane model”, with helices 5 and 6 lying on an intact membrane (Fig. 1e). Based on the EPR experiments [30], we published a structural model of active Bax, in which helices 2–5 form a stable dimeric core in agreement with the X-ray data, the 5/6 helix hairpin partially open and helices 1 and 7–9 are disordered with respect to the core. Based on the structural model, we suggested two plausible topology models: one with helix 6 being the transmembrane (Fig. 1f) and another, with two helices 6 clamping the membrane to stabilize the rim of a toroidal pore (“clamp” model in Fig. 1g) [30]. Mandal et al. suggested a model of Bak, with the dimerization/core domain at the rim of the pore and the two helices 6 lying on one leaflet of the bilayer [50] (Fig. 1h). Although all existing models share certain aspects, they differ by aspects like the type of contact (peripheral or transmembrane) of helix 5 or 6 with the membrane, the angle between helices 5 and 6, or whether the C2 symmetry of the dimerization/core domain is kept until the C terminus.
To provide a coherent description of Bax/Bak topology in the context of the pore, we compare the existing accessibility data of Bax and Bak and add two new experimental datasets using fluorescence spectroscopy and EPR techniques. Despite some incongruencies, most topology data are consistent with the following picture: the N terminus, helix 1, and the loop1/2 are highly water exposed and dislodged from the dimerization/core domain. The dimerization/core domains of two Bax or Bak monomers form a symmetric, stable dimer, in which helices 2 and 3 are water accessible, whereas helices 4 and 5 are in peripheral membrane contact. Helices 6–8 are also in peripheral membrane contact, and helix 9 is transmembrane. Finally, we integrate the topology data with the available structural data on Bax and Bak, and propose the most plausible organization of membrane-embedded, active Bax/Bak in the context of a toroidal pore.
Sequence conservation between Bax and Bak hinders the use of point mutations
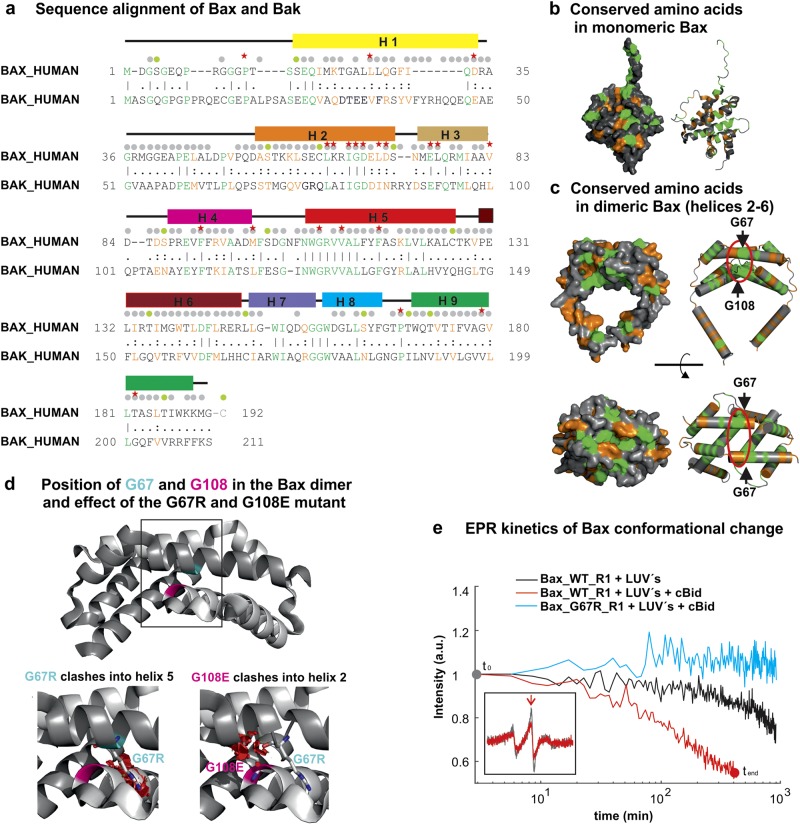
We found it difficult to integrate all existing water and lipid accessibility data into a unique topology model due to some inconsistencies (see Supplementary Information). Besides the different techniques used, the discrepancies are likely due to structure and/or function alterations introduced by the point mutations [53–55] mostly used for these studies. To lay the basis for our discussion, we need to address the high degree of conservation of the primary sequences of human Bax and Bak (Fig. 2a). Conserved and identical amino acids are highlighted in the three-dimensional (3D) structures of the monomeric and dimeric Bax (Fig. 2b, c). In the monomeric form, conserved amino acids are spread over the whole structure, whereas in the membrane-embedded Bax dimers, a highly conserved region forming the intra-dimer interface is detectable (Fig. 2c). Crucial for this interface are two highly conserved glycines (G67 and G108 in Bax; G82 and G126 in Bak). Replacement of either one to a larger amino acid can be tolerated in the monomeric conformation, but provokes a loss-of-function phenotype in the cells [12, 56, 57], which is likely caused by steric clashes preventing dimer formation (Fig. 2d).
Fig. 2.
Sequence and structural homology of Bax and Bak. a Sequence alignment of the amino acids of human Bax (isoform alpha) and human Bak (isoform 1) using the tool EMBOSS Strecher [93]. Identical amino acids are green and conserved ones are orange. Bax helices H1–9 are indicated. Gray dots: mutants used previously to study Bax/Bak topology; green dots: mutants used here. Red stars: mutations with a negative effect on protein activity (loss-of-function mutations), protein oligomerization (loss-of-dimerization mutations), or on the ability to adopt the soluble, the membrane-embedded, or both Bax/Bak conformations. More information is given in Supplementary Tables 1 and 2. b, c Surface and cartoon representation of the structure of monomeric (b, PDB: 1F16) and membrane-embedded dimeric Bax (c, helices 2–6, based on Bleicken et al. [30]). Identical amino acids in Bax and Bak are green and conserved ones are orange. The positions of G67 and G108 are indicated by black arrows in the dimeric structure. d Zoom-in structure of the core domain of Bax dimers with G67 and G108 are highlighted. In the membrane-embedded dimeric structure, G67 and G108 are so buried that mutations to R or E amino acids (for 67 and 108, respectively) provoke steric clashes (highlighted in red, using Pymol), which likely inhibits dimer formation. e Kinetics of Bax conformational change at 37 °C derived by continuous-wave EPR using the two natural cysteines (C62 and C126) spin-labeled with MTSL (R1 side chain) on the Bax wild type and the G67R mutant (in the presence or absence of cBid and large unilamellar vesicles (LUV’s) mimicking the MOM). Inset: EPR spectra of spin-labeled Bax wild type at the beginning (gray) and at the end (red) of the kinetics
As an example of the possible detrimental effects of point mutations, we show how the G67R mutation affects Bax activation. BaxWT and BaxG67R were labeled with a spin label at the two native cysteines (labeled side chains are indicated by R1), and continuous-wave EPR spectra were detected over time (technique introduced in Bleicken et al. [32]). In case of BaxWT_R1, membrane insertion and oligomerization induced clear spectral changes (Fig. 2e), whereas under the same experimental conditions, BaxG67R_R1 showed no changes. Thus, BaxG67R lost the ability to be activated. Interestingly, Bax is an auto-active molecule [32, 58, 59], and even in absence of cBid we could detect a spectral change in BaxWT_R1, which was absent in the G67R variant (Fig. 2e). Additionally, G67R is located in the proposed interaction area with Bid [25], which might also affect protein activation. However, as BaxG67R_R1 is not auto-active, we can conclude that the mutation inhibits Bax conformational change, explaining the loss-of-function phenotype in cells [12, 56, 57, 60].
Several point mutants provoking a loss-of-function or a change in fold or localization are described for Bax and Bak [37, 49, 51, 61–65] (see Supplementary Information). 21 of 26 loss-of-function mutations are placed at highly conserved positions (red asterisks in Fig. 2a), demonstrating that mutations at conserved sites can largely affect Bax/Bak fold and function, and therefore must be chosen and analyzed with great care. However, roughly 80% of the Bax/Bak primary sequence and therefore many conserved residues were mutated for topology studies (Fig. 2a gray dots [30, 32, 34–39, 47, 49, 50–52, 61, 65–72]), which can at least partly explain the observed inconsistencies.
To systematically analyze the available topology data, we first assert whether the point mutant of interest is on a conserved amino acid, whether loss/gain-of-function mutations are described, and whether the published topology data are consistent (Supplementary Tables 1 and 2). Additionally, we included new topology data on Bax using the singly labeled mutants proven to be functional in the liposome content release assays [30, 32, 73] and well folded both into the water-soluble and the membrane-embedded forms [30, 74].
Methods used in topology studies
The Bax/Bak topology models are mainly based on two accessibility methods: IASD (4-acetamido-4'-((iodoacetyl)amino)silbene-2-2'-disulfonic acid) labeling [34, 37, 39, 47, 49] and EPR-based methods [32, 50, 67] (Supplementary Figure 1a,b and Supplementary Table 1 and 2). We found that the data are not fully consistent even when the same technique is used, for example, IASD data on Bax L122C showed 10% accessibility in Annis et al. [49], 60% in Zhang et al. [34], and 80% in Westphal et al. [47].
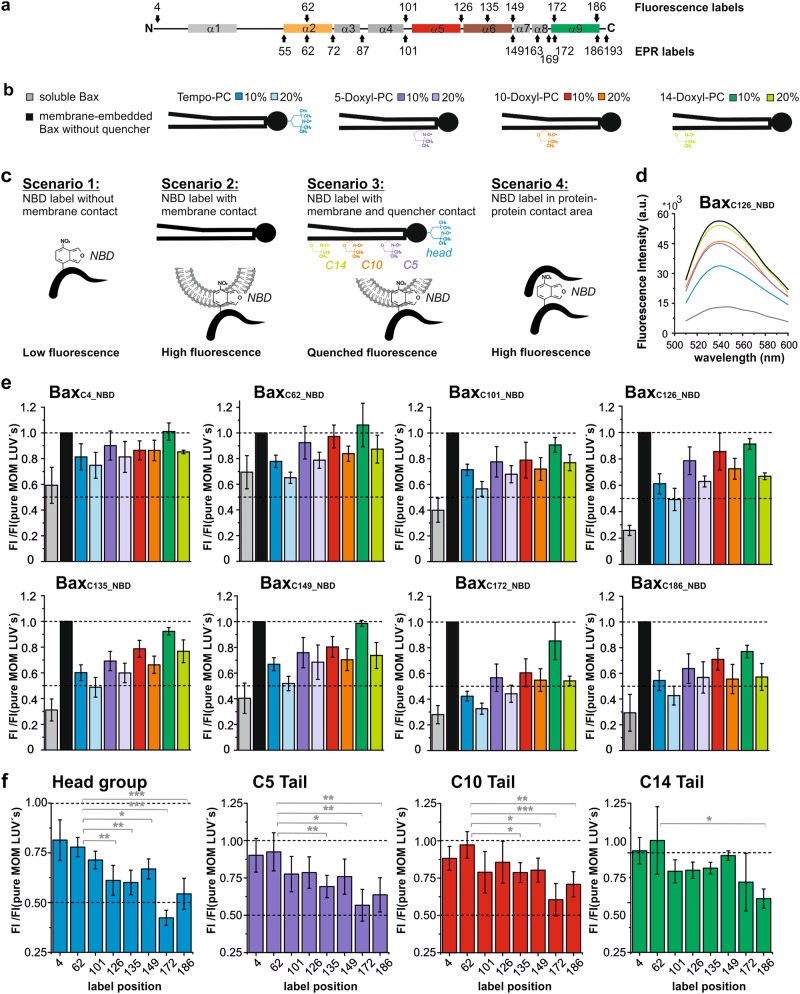
Thus, in the interest of reproducibility, we add here two independent methods to address protein topology based on fluorophores and spin labels bound to cysteines introduced into Bax (Figs. 3 and 4). The fluorescence-based method probes the contact between the fluorophore NBD (nitrobenzoxadiazole) with water or lipid molecules (reviewed in Ladokhin [75]). NBD fluorescence is quenched by water, whereas it is high in protein–protein or protein–membrane contact areas. To discriminate between the latter two possibilities, we introduced quenchers (in form of doxyl groups) at the lipid headgroup or at different positions in the lipid tail. This allowed to measure the immersion depth of the NBD into the membrane bilayer (Fig. 3 and Supplementary Figure 2). Recently, this technique was used on Bax by another group [76].
Fig. 3.
Fluorescence quenching experiments to probe Bax topology. a Bax secondary structure with positions used to attach fluorescence or EPR labels highlighted. b Color coding scheme of soluble and membrane-embedded Bax and positions of the TEMPO or doxyl group in the PC derivatives used in the following panels. c Sketch of four possible quenching conditions producing low or high fluorescence signals (details in Supplementary Figure 2a. d). Example of fluorescence emission spectra of the NBD at position 126 in Bax (excitation at 478 nm). e, f Bar diagrams summarizing the fluorescence emission spectra results of the NBD-labeled Bax variants. In gray is the control signal of the soluble Bax. Prior to the measurement, samples were incubated for 60 min at 37 °C with or without cBid and liposomes of different compositions (color code shown in panel b; excitation at 478 nm; emission at 541 nm). Mean values and error bars represent the standard deviation of six independent repetitions (except for the lipid mixture containing the doxyl group at position C14 with only four independent repetitions, data shown in Supplementary Figure 2b). The asterisk in (f) refers to the level of significance based on a two-tailed t-test *p ≤ 0.02; **p ≤ 0.001; ***p ≤ 0.0001 calculated by GraphPad Prism
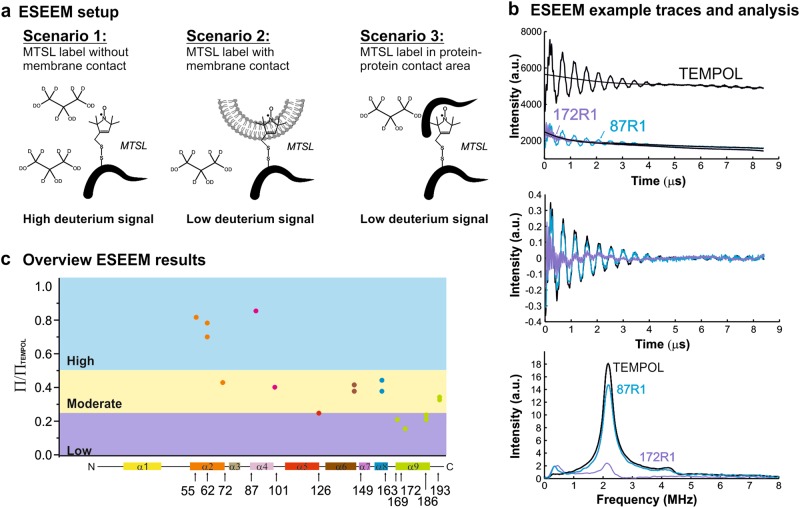
Fig. 4.
ESEEM experiments to probe Bax topology. a Sketch of three possible scenarios in which the spin label has different contact to the deuterium atoms introduced with 10% v/v deuterated glycerol in the bulk water, and respective strength of the ESEEM signals. b Examples of 3-pulse ESEEM traces on water-soluble TEMPOL (black) as as positive control for a completely water-exposed label; Bax87R1 (blue) and Bax172R1 (red). The latter two show, respectively, the highest and lowest deuterium signals among the mutants tested. Upper plot: Raw 3-pulse ESEEM traces (the 100 μM TEMPOL trace was divided by three for better comparison). Middle plot: background-corrected data with zero filling and Hamming. Bottom plot: magnitude of the Fourier transform of the ESEEM trace. The peak at around 2 MHz is the deuterium signal (all mutants are shown in Supplementary Figure 3). c Comparison of the normalized deuterium peak intensities for all tested Bax variants. Values falling in the blue area indicate a high, in the yellow a moderate and in red a low water accessibility of the spin label
The second method is based on electron spin echo envelope modulation (ESEEM) experiments (Fig. 4 and Supplementary Figure 3). ESEEM is a pulse-EPR technique, which detects interactions between the unpaired electron of a protein-attached spin label and nearby deuterons, which can be added via deuterated glycerol in the buffer [77–80]. When deuterium nuclei are close to the unpaired electron ( < 0.8 nm), a deuterium frequency modulation (around 2.3 MHz) is present in the ESEEM traces with an amplitude proportional to the concentration of nearby deuterons. Thus, a high-modulation amplitude indicates high water exposure of the spin-labeled site, whereas a low value correlates with shielding of the water molecules by lipids or protein contacts.
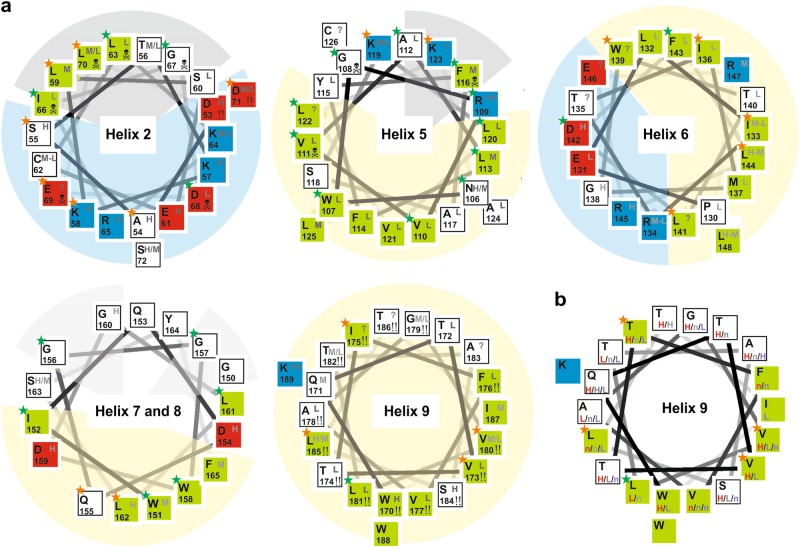
An overview on all accessibility data available in literature and those obtained in this work is presented in the following paragraphs and summarized in Fig. 5a (as well as Supplementary Figure 4 and Supplementary Tables 1 and 2). We classified the data into high, moderate, or low water accessibility (H, M, L). However, this classification depends on the positive and negative controls performed in each study, and therefore, the accessibility of one amino acid may vary between high and medium (or medium and low) and still be considered reproducible. In cases where independent studies vary between H or M or M and L, we classify the accessibility in Fig. 5a as high to medium (H–M) or medium to low (M–L), respectively. However, if the data vary between H and L, we consider the data as not consistent and the the amino acid will be marked by a question mark symbol (?) in Fig. 5a.
Fig. 5.
Summary on Bax/Bak topology data using helix wheel projections. a Helix wheel representation of helices 2 and 5–9 of Bax showing amino acid type (one letter code, upper left corner of the box) and its position (lower left corner of the box). The putative environment of the helix is indicated by the background color behind the wheel (blue—water contact; yellow—contact with the membrane; gray—protein–protein contact based on the X-ray structure of Czabotar et al. [25]; light gray—putative protein–protein contact). The hydrophobicity of each amino acid is indicated by the color of the box: blue—positively charged; red—negatively charged; green—hydrophobic; white—other types). Amino-acid conservation highlighted by green (identical amino acid) or orange (conserved amino acid) stars on the upper left corner of the box. The water accessibility of each amino acid based on the available data ([49, 37, 39, 47, 34, 50, 68, 52, 76] and our data) is indicated by the gray letters in the upper right corner of the box (classified as high (H), high to moderate (H/M), moderate (M), moderate to low (M/L) and low (L) water accessibility). The symbol “?” indicates accessibility data inconsistencies. Amino acids with known mutation-induced loss-of-function are highlighted by the “skull” symbol (mutations to be avoided), whereas the symbol “!!” indicate that mutants are described that affect protein function or subcellular localization. b Helix wheel representation of helix 9 highlighting positions for which zero-length cross-link data exist (red: Iyer et al. [37]; gray: Uren et al. [39]; violet: Zhang et al. [34]). The data are classified into three groups: n: no cross-link detected; L: cross-link possible with low efficiency; H: cross-link with high efficiency. Inconsistencies are found for most positions
The N-terminal region before helix 2 is water exposed and dynamic in membrane-embedded Bax
Surprisingly, 7 of the 15 flexible N-terminal amino acids of Bax [15] are identical to Bak, with 6 being glycine and prolines. In helix 1, 11 out of 19 amino acids are conserved and in the 1–2 loop 6 out of 18 (Fig. 2a). Based on this degree of conservation, it seems reasonable to suggest that the N-terminal region before helix 2 has a functional relevance. Indeed, dislodging the N terminus and helix 1 from the protein core is described to be an early step in Bax/Bak activation [12, 25, 29, 30, 68] and N-terminal truncations; as well as isoforms with altered N terminus, or the P13A mutant all affect the Bax subcellular localization and functionality [81-83].
All accessibility data agree that the N-terminal region is highly water exposed in the active conformation [28, 39, 49, 50, 67] and BaxC4NBD showed no indication of membrane contact in our experiments (Fig. 3e, f and Supplementary Figure 2).
It is controversially discussed, whether the region before helix 2 is involved in inter-monomer contacts. Cross-link experiments on activated Bak suggested inter-monomer contacts [39], whereas Förster resonance energy transfer (FRET) and double electron electron resonance (DEER) experiments on activated Bax showed long, broad distance distribution between helices 1 [30, 72]. Additionally, the short distance between helices 1 and 2 in monomeric Bax becomes longer and more broadly distributed upon Bax activation [30] and the 1–2 loop becomes accessible [26, 28]. Thus, the N-terminal region of membrane-embedded Bax/Bak is water exposed upon Bax/Bak activation and likely flexible with respect to the dimerization/core domain, explaining the discrepancies between FRET, DEER, and cross-link data.
The dimerization/core domain of Bax (helices 2–5) is peripherally interacting with the membrane
The dimerization/core domain of two Bax (or Bak) monomers forms a very stable symmetric dimer in the membrane-embedded conformation [25, 30, 33, 50]. Based on the available crystal structures, two distinct surfaces are formed by helices 2 and 3 and helices 4 and 5. The latter surface harbors several aromatic and hydrophobic amino acids likely facilitating membrane contact [25, 30, 33, 47].
We probed different positions within the dimerization/core domain by ESEEM or fluorescence experiments: three positions within helix 2 (BaxC55R1, BaxC62R1 BaxC72R1, BaxC62NBD), one after helix 3 (BaxC87R1), one in the loop between 4 and 5 (BaxC101R1, BaxC101NBD), and one at the end of helix 5 (BaxC126R1, BaxC126NBD). ESEEM data showed that BaxC55R1, BaxC62R1, and BaxC87R1 were highly water accessible, whereas Bax C72R1, BaxC101R1, and BaxC126R1 were moderately accessible (Fig. 4c and Supplementary Figure 3), suggesting that none of the investigated positions are membrane embedded. In line with this, the fluorescence data showed BaxC62NBD water exposed with some contact to the lipid headgroups, but no lipid tail contact, whereas BaxC101NBD and BaxC126NBD showed a strong quenching by the doxyl group attached to the lipid headgroup, and some quenching by doxyl groups at the tail positions C5 and even C10 (Fig. 3e, f and Supplementary Figure 2). These results are in line with helix 5 being in peripheral membrane contact [25, 30, 47], whereas helices 2 and 3 being water exposed.
All published data on helix 3 and the following loop are consistent with high water accessibility. In contrast, the data on helix 2 (harboring the BH3 domain) are not fully consistent and several loss-of-function mutations are described in the region (Fig. 5a). The IASD studies of Kluck lab showed eight highly water exposed, three > 50% water exposed, and one (A79C) < 50% water exposed positions in helix 2 [39, 47]. In contrast, Zhang et al. [34] studied five positions and found them all to be < 50% water exposed, whereas EPR studies suggested four positions involved in protein–protein contacts [32, 50, 67]. Finally, Flores-Romero et al. [76] detected a low water and a low lipid accessibility for two positions in helix 2, also in line with a protein contact area. An example of data inconsistencies is the following: E69C (in Bax, D84C in Bak) was found to be 100% water accessible in Uren et al. [39], 10% accessible in Zhang et al. [34], and a third study described the D84G mutation to have a loss-of-function phenotype [65] (more examples in the Supplementary Tables). The observed inconsistencies together with the existent loss-of-function mutants show that point mutations in helix 2 can strongly affect the protein fold or function.
The situation for helices 4 and 5 (harboring the BH1 domain) is similar. 20 out of 39 amino acids are conserved and five point mutations provoke a loss-of-function phenotype (Fig. 5a). Kluck lab studied 10 mutants in helix 4 and the loop 4/5 of Bak. Most positions were highly water exposed, with one (A107C) being inaccessible. The accessibility results of the other groups mainly agree with the high water accessibility, with the exception of Flores-Romero et al. [76]. The data on helix 5 are less consistent, and especially mutations on the conserved amino acids N106, V111, F116, and L122 are problematic (Fig. 5a and Supplemental Information). We tried to place a cysteine within helix 5, but none of the tested mutants was correctly labeled, folded, and functional (data not shown). Thus, similar to helix 2, mutations in helix 5 should be done with great care.
In summary, the dimerization/core domain is involved in tertiary or quaternary protein contacts. Helices 2 and 3 have mostly water-exposed surfaces, whereas helices 4 and 5 are in contact with the membrane headgroups, but they are not deeply embedded in the bilayer.
Helix 6 is in peripheral contact with the lipid headgroups
Helix 6 and the short loops next to it have 8 of 22 amino acids conserved. This helix is described to be involved in homodimer contacts [35, 36, 47], and in the published topology models it is either transmembrane or in peripheral membrane contact (Fig. 1d–h).
Here we studied the accessibility of BaxC135NBD (in the middle of helix 6) and BaxC149NBD, as well as BaxC149R1 (immediately after helix 6). BaxC149R1 showed a moderate water accessibility (Fig. 4c and Supplementary Figure 3), but due to the position at the end of helix 6, this result can be in line with both peripheral or transmembrane contact. However, BaxC135NBD and BaxC149NBD are found to be both significantly quenched by doxyl groups attached to the lipid headgroups and at position C5 and C10, but not C14 (Fig. 3e, f). As position C135 is in the middle of helix 6, this strongly suggests that helix 6 is in peripheral membrane contact and not transmembrane.
The published accessibility data on helix 6 are not fully consistent. Helix 6 was intensively studied by EPR and all tested amino acids were reported to be in membrane contact [50, 67], whereas IASD results showed several highly water accessible amino acids in the area [39, 47, 69]. Controversial results exist especially on positions 135, 139, and 146 in Bax (153, 157, and 164 in Bak), which is interesting as all three amino acids are on the same side of helix 6 (Fig. 5a). Therefore, it is tempting to speculate that these amino acids are involved in the inter-dimer interface and may be therefore difficult to exchange. Published cross-link data (using zero-distance linkers) agree with this interpretation, but alternative orientations of the interacting helices are also possible [35, 36, 47]. In summary, helix 6 is in peripheral membrane contact and is involved in inter-dimer contacts in active Bax and Bak.
The BH2 domain (helices 7 and 8) of Bax and Bak
The least studied part of Bax/Bak is the region between helix 6 and 9 harboring the BH2 domain. Only few accessibility data are available and show highly or moderately water-exposed residues (Fig. 5a). As the flanking helices 6 and 9 are in peripheral membrane contact and transmembrane, respectively, helices 7 and 8 should have at least some membrane contact. We successfully place one spin label (C163R1) in this region and we detected by ESEEM a moderate water accessibility (Fig. 4c and Supplementary Figure 3).
Figure 5a shows a helix wheel projection of helices 7 and 8 forming one single helix. This helix has one highly conserved hydrophobic surface (Q155, L162, W151, and W158 in Bax) possibly in contact with the membrane, whereas on the opposite side of the helix has a motif with two conserved glycines and two small amino acids (glycines in Bax and alanines in Bak) that may hint toward the presence of a protein–protein interface. Based on all data, we surmise that helices 7 and 8 are in peripheral membrane contact and might be involved in inter-dimer contacts.
Helix 9 of Bax is a flexible transmembrane helix
Fusions of helix 9 of Bax or Bak to the green fluorescent protein (or a FLAG tag) are reported to localize at the MOM [64, 70, 84], which suggested that helix 9 is transmembrane and anchors Bax/Bak to the MOM. Our accessibility data corroborate this notion: BaxC172_NBD and BaxC186_NBD (positioned at the N and C terminus of helix 9) showed a large (3- to 4-fold) increase in fluorescence intensity upon membrane insertion and the presence of doxyl groups at the lipid's headgroup region or along the tail quenched by their fluorescence (Fig. 3e, f and Supplementary Figure 1e and 2b). BaxC186_NBD was even significantly quenched by the doxyl group at position C14 in the hydrophobic center of the membrane (Fig. 3f). Accordingly, BaxC169R1 (before helix 9), BaxC172R1, and BaxC186R1 showed a low water accessibility by ESEEM, whereas BaxC193R1 (last C-terminal amino acid of the protein) shows a moderate water accessibility (Fig. 4c and Supplementary Figure 3).
The published accessibility data are not fully consistent, which is likely due to the fact that many mutations in helix 9 affect the intracellular localization of Bax and Bak [12, 37, 70] (Fig. 5a and Supplementary Table 1) and therefore may interfere with the interpretation of the accessibility data. Additionally, helix 9 is involved in dimer–dimer contacts within the Bax and Bak oligomers [30, 34, 37]. Two groups studied the contacts using zero-distance cross-linkers [34, 37, 39], which should allow to identify the interaction interface and indeed Zhang et al. [34] suggested one. However, drawing the cross-link results of different groups into a helix wheel representation revealed data inconsistencies (Fig. 5b), making it impossible to define a unique interaction interface. Thus, helix 9 is transmembrane and involved in inter-dimer contacts.
Structural organization of membrane-embedded Bax in context of the toroidal pore
The accessibility data on membrane-bound Bax suggest the following topology model: the N terminus, helix 1, and the loop1/2 are highly water exposed, flexible, and dislodged from the dimerization/core domain. Helices 2–5 of two monomers form the symmetric dimer interface with one water accessible surface formed by helices 2 and 3 and one hydrophobic surface formed by helices 4 and 5, likely in peripheral membrane contact. Helix 6, 7, and 8 are in peripheral membrane contact and helix 9 is transmembrane. Moreover, helices 6–9 are involved in inter-dimer contacts.
Intriguingly, these data perfectly agree with the “in-plane model” [47] when sketched on a intact membrane (Fig. 6a). However, they are also fully in line with the “clamp model” [30] and the model suggested by Mandal et al. [50], which consider the toroidal pore formed by membrane-bound Bax oligomers. The available data completely exclude the “hairpin model” [49] and the “helix 6 TM model” [30].
Fig. 6.
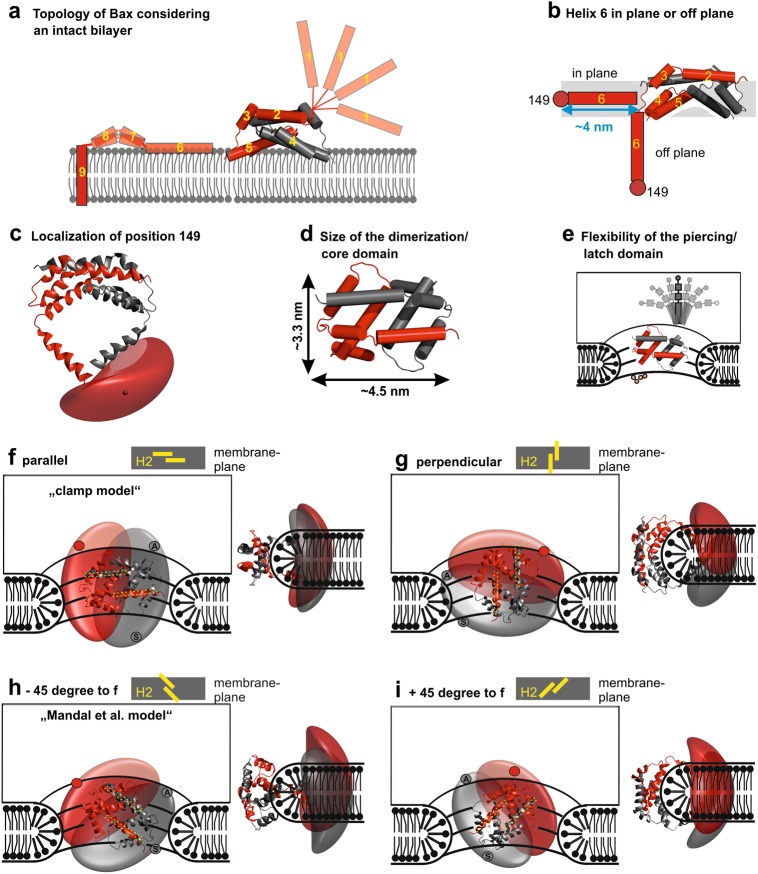
Structural organization of membrane-embedded Bax in context of a toroidal pore. a Model of the water/membrane accessibility of the nine Bax helices (one monomer shown in red; helix 2–4 of a second dimerization/core domain shown in gray) on a lipid bilayer. b Scheme of an “in-plane” or “off plane” helix 6 with respect to the dimerization/core domain of Bax. Color code as in panel a. Position 149 is indicated with a dark red sphere and the plane of the dimerization/core domain as gray rectangle. c Dimerization/core domain and recalculated off-plane localization of residue 149 (red cloud) for the red monomer. The dark red localization surface represents the top 50% of the probability density. The point of highest probability density is represented as a small dark dot. d Sketch of the dimensions of a Bax dimer. e Schematic model of a Bax dimer at the rim of a toroidal pore showing the flexibility of the piercing/latch domain with respect to the dimerization/core domain. f–i 3D topology models of the dimerization/core domain of two Bax monomers (red and gray) within one Bax dimer together with the localization cloud of position 149 in the context of a toroidal pore. Two views are presented (front and side view). For all models, the end of helix 6 is in a different plane than the membrane. The four models vary in how the two interacting helices 2 (yellow bars on the top of each panel) in the dimerization domain are oriented with respect to the membrane plane: f “clamp” model, with helix 2 parallel to the membrane plane; g perpendicular orientation; h “Mandal” model with helix 2 rotated by −45° with respect to the membrane plane; i +45° orientation. The red dot indicates one possible localization of helix 6 belonging to the red monomer. The gray dot labeled with “S” (symmetric) indicates where helix 6 of the gray monomer would be keeping the C2 symmetry. The gray dot labeled with “A” (asymmetric) indicates where helix 6 of the gray monomer would be when the C2 symmetry axis is broken after helix 5
One experimentally testable difference between the “in-plane” [47], the “clamp” [30], and the “Mandal” model [50] is the localization of helix 6 with respect to the dimerization/core domain. In the “in-plane model”, the end of helix 6 (position 149 in Bax) is in the same plane as the dimerization/core domain, whereas the other two models place it on a different plane (Fig. 6b). Information about the localization of helix 6 with respect to the protein core was obtained by DEER via distance constraints between position 149 and six positions within the dimerization/core domain [30]. Using a multilateration approach, which is in a nutshell a GPS localization on a nanometer length scale, the end of helix 6 was located in a plane about 4 nm below the center of mass of the dimerization/core domain [30], and thus “off plane” (Fig. 6b). The probability to find position 149 in the same plane as the dimerization/core domain was found to be negligible [30].
We would like to highlight that analyzing the DEER traces on membrane-embedded Bax oligomers is challenging because it is impossible to completely suppress inter-monomer distances, and the distance constraints toward the piercing/latch domain were broadly distributed due to the flexibility of this domain [30]. However, as the localization of position 149 with respect to the membrane plane is a key difference between models, we recalculated its localization probability using a larger standard deviation of the main distances of the already published constraints [30], supported by new DEER traces measured for pairs 101–149, 87–149, and 62–149 (Supplementary Figure 5). Based on the new calculations, the probability cloud for position 149 has a larger size, but the top 50% probability density still strongly supports a helix 6 kinked out of the membrane plane (Fig. 6c). Thus, we can conclude that the “in-plane model” may account for a minor population of the Bax oligomer or a transiently populated pre-pore state, whereas in the majority of active Bax molecules helix 6 is “off” the membrane plane.
The dimerization/core domain has has a crescent shape (Fig. 6d [25]) with one hydrophobic surface (helices 4 and 5) that based on its size and shape fits well to the curved rim of a toroidal pore, as suggested by the “clamp” and the “Mandal” models. The main difference between these two models is that the C2 symmetry of the dimerization/core domain is assumed to exist throughout the protein in the first model [30], whereas it is broken after helix 5 in the second [50]. Both models are plausible, considering inherent protein flexibility between the dimerization/core domain and the piercing/latch domain suggested for active Bax [30] and Bak [39], which increases toward the C terminus (Fig. 6e).
Figures 6f–i show how Bax/Bak may arrange in the context of the toroidal pore, with the dimerization/core domain placed at the rim of it. The localization of position 149 is shown as cloud in the same color as the the corresponding dimerization/core domain. Each subfigure shows two different pore views: one highlighting the orientation of the dimerization/core domain and one the localization of 149. The four subfigures vary by the angle between helix 2 and the membrane plane as illustrated at the top of each subfigure. Figure 6f represents the “clamp” model, in which the two interacting helices 2 of each dimer are parallel with respect to the membrane plane, whereas Fig. 6g shows the perpendicular orientation. The two orientations at +45 or −45 degrees are shown in Fig. 6h, i (with the −45 degrees view presenting the “Mandal” model). In each proposed orientation, the 149 clouds allow helix 6 to be in peripheral membrane contact, in agreement with the topology data. Moreover, we checked whether symmetric (C2 symmetry conserved) and asymmetric (C2 symmetry broken) helix 6 orientations are plausible. Thereby, helix 6 of the red monomer was always placed on top of the upper membrane leaflet (red dot), whereas helix 6 of the second gray monomer was either on the lower leaflet or upper leaflet depending if the C2 symmetry was kept or broken (the gray dot with”S” represents helix 6 in a scenario keeping the C2 symmetry, whereas the gray dot with”A” represents the asymmetric counterpart). Surprisingly and owing to the large localization probability, both arrangements are plausible for all four subfigures. However, to allow the asymmetric conformation, the 5–6 loop in one of the two monomers needs to be stretched (see Fig. 1h).
In summary, the most plausible models are the “clamp” and the “Mandal” models, as well as other models with the core/piercing domain at the rim of the pore. Due to the flexibility of the piercing/latch domain, further model discrimination is currently not possible.
Biological implications and outlook
Besides agreeing with the available data, the favored topology models can explain how Bax and Bak enable the release of Cytochrome c and much larger complexes like mitochondrial DNA [85, 86] into the cytosol, which is a long-standing question in the apoptotic field. Bax and Bak are suggested to form stable toroidal pores [40–42, 44, 87] that are meta-stable lipid arrangement requiring proteins to be stabilized. Based on the suggested models in Fig. 6f–i, Bax and Bak could stabilize a toroidal pore by at least four mechanisms known to stabilize highly curved membranes [88–90]. First, each dimer placed on the pore rim has >20 hydrophobic and aromatic amino acids in helices 4 and 5 to stabilize the lipid structure, and the crescent-like shape of the rigid core/dimerization domain could act as scaffolding chaperone, further stabilizing the pore. Second, each amphipathic helix 6 in the dimer would stabilize the adjoining lipids. Third, the transmembrane helix 9 would anchor the whole structure into the membrane, further stabilizing the lipid arrangement. Fourth, the formation of higher-order oligomers [13, 30, 32, 35, 36, 91] would potentiate the stabilizing effect.
Inter-dimer contacts are most likely built by contacts between piercing/latch domains [30, 34, 35, 37, 92]. The observation that this domain is flexible (disordered) with respect to the dimerization/core domain may provide clues on how Bax/Bak oligomers adapt their size to allow pore growth upon increasing protein concentration [40, 42] and over time [85] or how Bcl-xL can reduce the size of Bax oligomers [13, 58] and act as an inhibitor.
The next key step to fully understand the structure of the Bax/Bak pores is to address whether the C2 symmetry of the dimerization/core domain is kept into the piercing/latch domain or if heterogeneous symmetric and non-symmetric configurations exist in the membrane. If the symmetry holds, two helices 9 within one dimeric unit must be antiparallel, whereas a break in the symmetry allows parallel arrangements. Notably, parallel arrangements of helix 9 exist in the oligomers [30, 34, 37, 92], but discriminating whether they arise from intra- or inter-dimer contacts has not yet been possible and will remain challenging, considering the high flexibility of the piercing/latch domain.
Even when the structure(s) of the active oligomers will be completely unveiled, more challenging questions will emerge. How many Bax/Bak dimers are minimally needed to form a pore? How are oligomer size and pore properties quantitatively related? How interactions with anti-apoptotic Bcl-2 proteins or other regulators affect the structure of the membrane-embedded Bax/Bak oligomer and the pore structure?
Electronic supplementary material
Acknowledgements
This work was supported by the DFG Priority Program SPP1601 “New Frontiers in Sensitivity in EPR Spectroscopy” (EB and TEA), the DFG grant BO 3000/5-1 and INST 130/972-1 FUGG (EB), the European Research Council (ERC-2012-StG 309966; SB, CS, and AJG-S), the Forschergruppe 2036 (SB, CS, and AJG-S), and the Cluster of Excellence RESOLV (EXC 1069; SB and EB) funded by the Deutsche Forschungsgemeinschaft. The ESEEM data were recorded in the laboratory of Robert Bittl, FU Berlin. EB would like to thank Gunnar Jeschke and ETH Zurich for the Matlab script used for ESEEM data analysis and the home-made Q-band resonator.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M. Piacentini
Contributor Information
Stephanie Bleicken, Email: stephanie.bleicken@rub.de.
Enrica Bordignon, Email: enrica.bordignon@rub.de.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0184-6) contains supplementary material, which is available to authorized users.
References
- 1.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 2.Martinou JC, Youle Richard J. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Saez AJ. The secrets of the Bcl-2 family. Cell Death Differ. 2012;19:1733–40. doi: 10.1038/cdd.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 5.Strasser A, Cory S, Adams JM. Deciphering the rules of programmed cell death to improve therapy of cancer and other diseases. EMBO J. 2011;30:3667–83. doi: 10.1038/emboj.2011.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delbridge AR, Grabow S, Strasser A, Vaux DL. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat Rev Cancer. 2016;16:99–109. doi: 10.1038/nrc.2015.17. [DOI] [PubMed] [Google Scholar]
- 7.Westphal D, Dewson G, Czabotar PE, Kluck RM. Molecular biology of Bax and Bak activation and action. Biochim Biophys Acta-Mol Cell Res. 2011;1813:521–31. doi: 10.1016/j.bbamcr.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Lindsten T, Ross AJ, King A, Zong WX, Rathmell JC, Shiels HA, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell. 2000;6:1389–99. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–30. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reyna DE, Garner TP, Lopez A, Kopp F, Choudhary GS, Sridharan A, et al. Direct activation of BAX by BTSA1 overcomes apoptosis resistance in acute myeloid leukemia. Cancer Cell. 2017;32:490–505. doi: 10.1016/j.ccell.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bleicken Stephanie, Garcia-Saez Ana J. Bcl-2 proteins: Unraveling the details of a complex and dynamic network. Molecular & Cellular Oncology. 2017;5(1):e1384880. doi: 10.1080/23723556.2017.1384880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, et al. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell. 2009;36:487–99. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subburaj Y, Cosentino K, Axmann M, Pedrueza-Villalmanzo E, Hermann E, Bleicken S, et al. Bax monomers form dimer units in the membrane that further self-assemble into multiple oligomeric species. Nat Commun. 2015;6:8042–52. [DOI] [PMC free article] [PubMed]
- 14.Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, et al. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–84. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki M, Youle RJ, Tjandra N. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 2000;103:645–54. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 16.Moldoveanu T, Liu Q, Tocilj A, Watson M, Shore G, Gehring K. The X-ray structure of a BAK homodimer reveals an inhibitory zinc binding site. Mol Cell. 2006;24:677–88. doi: 10.1016/j.molcel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Todt F, Cakir Z, Reichenbach F, Emschermann F, Lauterwasser J, Kaiser A, et al. Differential retrotranslocation of mitochondrial Bax and Bak. EMBO J. 2015;34:67–80. doi: 10.15252/embj.201488806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng EH, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science. 2003;301:513–7. doi: 10.1126/science.1083995. [DOI] [PubMed] [Google Scholar]
- 19.Ma SB, Nguyen TN, Tan I, Ninnis R, Iyer S, Stroud DA, et al. Bax targets mitochondria by distinct mechanisms before or during apoptotic cell death: a requirement for VDAC2 or Bak for efficient Bax apoptotic function. Cell Death Differ. 2014;21:1925–35. doi: 10.1038/cdd.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazarou Michael, Stojanovski Diana, Frazier Ann E., Kotevski Aneta, Dewson Grant, Craigen William J., Kluck Ruth M., Vaux David L., Ryan Michael T. Inhibition of Bak Activation by VDAC2 Is Dependent on the Bak Transmembrane Anchor. Journal of Biological Chemistry. 2010;285(47):36876–36883. doi: 10.1074/jbc.M110.159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schellenberg B, Wang P, Keeble James A, Rodriguez-Enriquez R, Walker S, Owens TW, et al. Bax exists in a dynamic equilibrium between the cytosol and mitochondria to control apoptotic priming. Mol Cell. 2013;49:959–71. doi: 10.1016/j.molcel.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edwards AL, Gavathiotis E, LaBelle JL, Braun CR, Opoku-Nsiah KA, Bird GH, et al. Multimodal interaction with BCL-2 family proteins underlies the proapoptotic activity of PUMA BH3. Chem Biol. 2013;20:888–902. doi: 10.1016/j.chembiol.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–81. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shamas-Din A, Satsoura D, Khan O, Zhu W, Leber B, Fradin C, et al. Multiple partners can kiss-and-run: Bax transfers between multiple membranes and permeabilizes those primed by tBid. Cell Death Dis. 2014;5:e1277. doi: 10.1038/cddis.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czabotar Peter E, Westphal D, Dewson G, Ma S, Hockings C, Fairlie WD, et al. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–31. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 26.Moldoveanu T, Grace CR, Llambi F, Nourse A, Fitzgerald P, Gehring K, et al. BID-induced structural changes in BAK promote apoptosis. Nat Struct Mol Biol. 2013;20:589–97. doi: 10.1038/nsmb.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartron PF, Gallenne T, Bougras G, Gautier F, Manero F, Vusio P, et al. The first α helix of Bax plays a necessary role in its ligand-induced activation by the BH3-only proteins Bid and PUMA. Mol Cell. 2004;16:807–18. doi: 10.1016/j.molcel.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 28.Bleicken S, Zeth K. Conformational changes and protein stability of the pro-apoptotic protein Bax. J Bioenerg Biomembr. 2009;41:29–40. doi: 10.1007/s10863-009-9202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gavathiotis E, Reyna DE, Davis ML, Bird GH, Walensky LD. BH3-triggered structural reorganization drives the activation of proapoptotic BAX. Mol Cell. 2010;40:481–92. doi: 10.1016/j.molcel.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bleicken S, Jeschke G, Stegmueller C, Salvador-Gallego R, García-Sáez Ana J, Bordignon E. Structural model of active Bax at the membrane. Mol Cell. 2014;56:496–505. doi: 10.1016/j.molcel.2014.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu X, Brahmbhatt H, Mergenthaler P, Zhang Z, Sang J, Daude M, et al. A small-molecule inhibitor of Bax and Bak oligomerization prevents genotoxic cell death and promotes neuroprotection. Cell Chem Biol. 2017;24:493–506. doi: 10.1016/j.chembiol.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bleicken S, Classen M, Padmavathi PV, Ishikawa T, Zeth K, Steinhoff HJ, et al. Molecular details of Bax activation, oligomerization, and membrane insertion. J Biol Chem. 2010;285:6636–47. doi: 10.1074/jbc.M109.081539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brouwer JM, Westphal D, Dewson G, Robin AY, Uren RT, Bartolo R, et al. Bak core and latch domains separate during activation, and freed core domains form symmetric homodimers. Mol Cell. 2014;55:938–46. doi: 10.1016/j.molcel.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Subramaniam S, Kale J, Liao C, Huang B, Brahmbhatt H, et al. BH3-in-groove dimerization initiates and helix 9 dimerization expands Bax pore assembly in membranes. EMBO J. 2016;35:208–36. doi: 10.15252/embj.201591552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dewson G, Kratina T, Czabotar P, Day CL, Adams JM, Kluck RM. Bak activation for apoptosis involves oligomerization of dimers via their alpha6 helices. Mol Cell. 2009;36:696–703. doi: 10.1016/j.molcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 36.Dewson G, Ma S, Frederick P, Hockings C, Tan I, Kratina T, et al. Bax dimerizes via a symmetric BH3:groove interface during apoptosis. Cell Death Differ. 2012;19:661–70. doi: 10.1038/cdd.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer S, Bell F, Westphal D, Anwari K, Gulbis J, Smith BJ, et al. Bak apoptotic pores involve a flexible C-terminal region and juxtaposition of the C-terminal transmembrane domains. Cell Death Differ. 2015;22:1665–75. doi: 10.1038/cdd.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Zhu WJ, Lapolla SM, Miao YW, Shao YL, Falcone M, et al. Bax forms an oligomer via separate, yet interdependent, surfaces. J Biol Chem. 2010;285:17614–27. doi: 10.1074/jbc.M110.113456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uren RT, O’Hely M, Iyer S, Bartolo R, Shi MX, Brouwer JM, et al. Disordered clusters of Bak dimers rupture mitochondria during apoptosis. Elife 2017;6:e19944. [DOI] [PMC free article] [PubMed]
- 40.Bleicken S, Landeta O, Landajuela A, Basanez G, Garcia-Saez AJ. Proapoptotic Bax and Bak form stable protein-permeable pores of tunable size. J Biol Chem. 2013;288:33241–52. doi: 10.1074/jbc.M113.512087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian S, Wang W, Yang L, Huang HW. Structure of transmembrane pore induced by Bax-derived peptide: evidence for lipidic pores. Proc Natl Acad Sci USA. 2008;105:17379–83. doi: 10.1073/pnas.0807764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gillies LA, Du H, Peters B, Knudson CM, Newmeyer DD, Kuwana T. Visual and functional demonstration of growing Bax-induced pores in mitochondrial outer membranes. Mol Biol Cell. 2015;26:339–49. doi: 10.1091/mbc.E13-11-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schafer B, Quispe J, Choudhary V, Chipuk JE, Ajero TG, Du H, et al. Mitochondrial outer membrane proteins assist Bid in Bax-mediated lipidic pore formation. Mol Biol Cell. 2009;20:2276–85. doi: 10.1091/mbc.E08-10-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bleicken S, Hofhaus G, Ugarte-Uribe B, Schroder R, Garcia-Saez AJ. cBid, Bax and Bcl-xL exhibit opposite membrane remodeling activities. Cell Death Dis. 2016;7:e2121. doi: 10.1038/cddis.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Saez AJ, Coraiola M, Serra MD, Mingarro I, Muller P, Salgado J. Peptides corresponding to helices 5 and 6 of Bax can independently form large lipid pores. FEBS J. 2006;273:971–81. doi: 10.1111/j.1742-4658.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- 46.Mikhailov V, Mikhailova M, Degenhardt K, Venkatachalam MA, White E, Saikumar P. Association of Bax and Bak homo-oligomers in mitochondria. Bax requirement for Bak reorganization and cytochrome c release. J Biol Chem. 2003;278:5367–76. doi: 10.1074/jbc.M203392200. [DOI] [PubMed] [Google Scholar]
- 47.Westphal D, Dewson G, Menard M, Frederick P, Iyer S, Bartolo R, et al. Apoptotic pore formation is associated with in-plane insertion of Bak or Bax central helices into the mitochondrial outer membrane. Proc Natl Acad Sci USA. 2014;111:E4076–4085. doi: 10.1073/pnas.1415142111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pang Y-P, Dai H, Smith A, Meng XW, Schneider PA, Kaufmann SH. Bak Conformational changes induced by ligand binding: insight into BH3 domain binding and Bak homo-oligomerization. Sci Rep. 2012;2:257. [DOI] [PMC free article] [PubMed]
- 49.Annis MG, Soucie EL, Dlugosz PJ, Cruz-Aguado JA, Penn LZ, Leber B, et al. Bax forms multispanning monomers that oligomerize to permeabilize membranes during apoptosis. EMBO J. 2005;24:2096–103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mandal T, Shin S, Aluvila S, Chen HC, Grieve C, Choe JY, et al. Assembly of Bak homodimers into higher order homooligomers in the mitochondrial apoptotic pore. Sci Rep. 2016;6:30763. doi: 10.1038/srep30763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alsop AE, Fennell SC, Bartolo RC, Tan IK, Dewson G, Kluck RM. Dissociation of Bak alpha1 helix from the core and latch domains is required for apoptosis. Nat Commun. 2015;6:6841. doi: 10.1038/ncomms7841. [DOI] [PubMed] [Google Scholar]
- 52.Li MX, Tan IKL, Ma SB, Hockings C, Kratina T, Dengler MA, et al. BAK α6 permits activation by BH3-only proteins and homooligomerization via the canonical hydrophobic groove. Proc Natl Acad Sci USA. 2017;114:7629–34. doi: 10.1073/pnas.1702453114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logan J, Hiestand D, Daram P, Huang Z, Muccio DD, Hartman J, et al. Cystic fibrosis transmembrane conductance regulator mutations that disrupt nucleotide binding. J Clin Invest. 1994;94:228–36. doi: 10.1172/JCI117311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Studer RA, Dessailly BH, Orengo CA. Residue mutations and their impact on protein structure and function: detecting beneficial and pathogenic changes. Biochem J. 2013;449:581–94. doi: 10.1042/BJ20121221. [DOI] [PubMed] [Google Scholar]
- 55.Piel FB, Steinberg MH, Rees DC. Sickle cell disease. N Engl J Med. 2017;376:1561–73. doi: 10.1056/NEJMra1510865. [DOI] [PubMed] [Google Scholar]
- 56.Meijerink JP, Mensink EJ, Wang K, Sedlak TW, Sloetjes AW, de Witte T, et al. Hematopoietic malignancies demonstrate loss-of-function mutations of BAX. Blood. 1998;91:2991–7. [PubMed] [Google Scholar]
- 57.Cheng EH, Levine B, Boise LH, Thompson CB, Hardwick JM. Bax-independent inhibition of apoptosis by Bcl-XL. Nature. 1996;379:554–6. doi: 10.1038/379554a0. [DOI] [PubMed] [Google Scholar]
- 58.Bleicken S, Hantusch A, Das KK, Frickey T, Garcia-Saez AJ. Quantitative interactome of a membrane Bcl-2 network identifies a hierarchy of complexes for apoptosis regulation. Nat Comm. 2017;8:73. doi: 10.1038/s41467-017-00086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Neill KL, Huang K, Zhang J, Chen Y, Luo X. Inactivation of prosurvival Bcl-2 proteins activates Bax/Bak through the outer mitochondrial membrane. Genes Dev. 2016;30:973–88. doi: 10.1101/gad.276725.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sedlak TW, Oltvai ZN, Yang E, Wang K, Boise LH, Thompson CB, et al. Multiple Bcl-2 family members demonstrate selective dimerizations with Bax. Proc Natl Acad Sci USA. 1995;92:7834–8. doi: 10.1073/pnas.92.17.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cartron PF, Arokium H, Oliver L, Meflah K, Manon S, Vallette FM. Distinct domains control the addressing and the insertion of Bax into mitochondria. J Biol Chem. 2005;280:10587–98. doi: 10.1074/jbc.M409714200. [DOI] [PubMed] [Google Scholar]
- 62.Zhou H, Hou Q, Hansen JL, Hsu YT. Complete activation of Bax by a single site mutation. Oncogene. 2007;26:7092–102. doi: 10.1038/sj.onc.1210517. [DOI] [PubMed] [Google Scholar]
- 63.Valentijn AJ, Upton JP, Bates N, Gilmore AP. Bax targeting to mitochondria occurs via both tail anchor-dependent and -independent mechanisms. Cell Death Differ. 2008;15:1243–54. doi: 10.1038/cdd.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schinzel A, Kaufmann T, Schuler M, Martinalbo J, Grubb D, Borner C. Conformational control of Bax localization and apoptotic activity by Pro168. J Cell Biol. 2004;164:1021–32. doi: 10.1083/jcb.200309013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dewson G, Kratina T, Sim HW, Puthalakath H, Adams JM, Colman PM, et al. To trigger apoptosis, Bak exposes its BH3 domain and homodimerizes via BH3:groove interactions. Mol Cell. 2008;30:369–80. doi: 10.1016/j.molcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 66.Ferrer PE, Frederick P, Gulbis JM, Dewson G, Kluck RM. Translocation of a Bak C-terminus mutant from cytosol to mitochondria to mediate cytochrome: implications for Bak and Bax apoptotic function. PLoS ONE. 2012;7:e31510. doi: 10.1371/journal.pone.0031510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oh KJ, Singh P, Lee K, Foss K, Lee S, Park M, et al. Conformational changes in BAK, a pore-forming proapoptotic Bcl-2 family member, upon membrane insertion and direct evidence for the existence of BH3-BH3 contact interface in BAK homo-oligomers. J Biol Chem. 2010;285:28924–37. doi: 10.1074/jbc.M110.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aluvila S, Mandal T, Hustedt E, Fajer P, Choe JY, Oh KJ. Organization of the mitochondrial apoptotic BAK pore: oligomerization of the BAK homodimers. J Biol Chem. 2013;289:2537–51. doi: 10.1074/jbc.M113.526806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weber K, Harper N, Schwabe J, Cohen GM. BIM-mediated membrane insertion of the BAK pore domain is an essential requirement for apoptosis. Cell Rep. 2013;5:409–20. doi: 10.1016/j.celrep.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nechushtan A, Smith CL, Hsu YT, Youle RJ. Conformation of the Bax C-terminus regulates subcellular location and cell death. EMBO J. 1999;18:2330–41. doi: 10.1093/emboj/18.9.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gahl RF, Tekle E, Tjandra N. Single color FRET based measurements of conformational changes of proteins resulting from translocation inside cells. Methods. 2014;66:180–7. doi: 10.1016/j.ymeth.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gahl RF, He Y, Yu S, Tjandra N. Conformational rearrangements in the pro-apoptotic protein, Bax, as it inserts into mitochondria: a cellular death switch. J Biol Chem. 2014;289:32871–82. doi: 10.1074/jbc.M114.593897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bleicken S, Wagner C, García-Sáez Ana J. Mechanistic differences in the membrane activity of Bax and Bcl-xL correlate with their opposing roles in apoptosis. Biophys J. 2013;104:421–31. doi: 10.1016/j.bpj.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fischer Axel W., Bordignon Enrica, Bleicken Stephanie, García-Sáez Ana J., Jeschke Gunnar, Meiler Jens. Pushing the size limit of de novo structure ensemble prediction guided by sparse SDSL-EPR restraints to 200 residues: The monomeric and homodimeric forms of BAX. Journal of Structural Biology. 2016;195(1):62–71. doi: 10.1016/j.jsb.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ladokhin AS. Measuring membrane penetration with depth-dependent fluorescence quenching: distribution analysis is coming of age. Biochim Biophys Acta. 2014;1838:2289–95. doi: 10.1016/j.bbamem.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flores-Romero H, Garcia-Porras M, Basanez G. Membrane insertion of the BAX core, but not latch domain, drives apoptotic pore formation. Sci Rep. 2017;7:16259. doi: 10.1038/s41598-017-16384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Volkov A, Dockter C, Bund T, Paulsen H, Jeschke G. Pulsed EPR determination of water accessibility to spin-labeled amino acid residues in LHCIIb. Biophys J. 2009;96:1124–41. doi: 10.1016/j.bpj.2008.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Segawa TF, Doppelbauer M, Garbuio L, Doll A, Polyhach YO, Jeschke G. Water accessibility in a membrane-inserting peptide comparing overhauser DNP and pulse EPR methods. J Chem Phys. 2016;144:194201. doi: 10.1063/1.4948988. [DOI] [PubMed] [Google Scholar]
- 79.Liu L, Mayo DJ, Sahu ID, Zhou A, Zhang R, McCarrick RM, et al. Determining the secondary structure of membrane proteins and peptides via electron spin echo envelope modulation (ESEEM) spectroscopy. Methods Enzymol. 2015;564:289–313. doi: 10.1016/bs.mie.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carmieli R, Papo N, Zimmermann H, Potapov A, Shai Y, Goldfarb D. Utilizing ESEEM spectroscopy to locate the position of specific regions of membrane-active peptides within model membranes. Biophys J. 2006;90:492–505. doi: 10.1529/biophysj.105.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cartron PF, Oliver L, Martin S, Moreau C, LeCabellec MT, Jezequel P, et al. The expression of a new variant of the pro-apoptotic molecule Bax, Baxpsi, is correlated with an increased survival of glioblastoma multiforme patients. Hum Mol Genet. 2002;11:675–87. doi: 10.1093/hmg/11.6.675. [DOI] [PubMed] [Google Scholar]
- 82.Cartron PF, Priault M, Oliver L, Meflah K, Manon S, Vallette FM. The N-terminal end of Bax contains a mitochondrial-targeting signal. J Biol Chem. 2003;278:11633–41. doi: 10.1074/jbc.M208955200. [DOI] [PubMed] [Google Scholar]
- 83.Upton JP, Valentijn AJ, Zhang L, Gilmore AP. The N-terminal conformation of Bax regulates cell commitment to apoptosis. Cell Death Differ. 2007;14:932–42. doi: 10.1038/sj.cdd.4402092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Setoguchi K, Otera H, Mihara K. Cytosolic factor- and TOM-independent import of C-tail-anchored mitochondrial outer membrane proteins. EMBO J. 2006;25:5635–47. doi: 10.1038/sj.emboj.7601438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McArthur K, Whitehead LW, Heddleston JM, Li L, Padman BS, Oorschot V, et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 2018;359:6378. [DOI] [PubMed]
- 86.Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–6. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 87.Fernandez-Marrero Y, Bleicken S, Das KK, Bachmann D, Kaufmann T, Garcia-Saez AJ. The membrane activity of BOK involves formation of large, stable toroidal pores and is promoted by cBID. FEBS J. 2017;284:711–24. doi: 10.1111/febs.14008. [DOI] [PubMed] [Google Scholar]
- 88.McMahon HT, Boucrot E. Membrane curvature at a glance. J Cell Sci. 2015;128:1065–70. doi: 10.1242/jcs.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 90.Stachowiak JC, Brodsky FM, Miller EA. A cost-benefit analysis of the physical mechanisms of membrane curvature. Nat Cell Biol. 2013;15:1019–27. doi: 10.1038/ncb2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Antonsson B, Montessuit S, Sanchez B, Martinou JC. Bax is present as a high molecular weight oligomer/complex in the mitochondrial membrane of apoptotic cells. J Biol Chem. 2001;276:11615–23. doi: 10.1074/jbc.M010810200. [DOI] [PubMed] [Google Scholar]
- 92.Andreu-Fernandez V, Sancho M, Genoves A, Lucendo E, Todt F, Lauterwasser J, et al. Bax transmembrane domain interacts with prosurvival Bcl-2 proteins in biological membranes. Proc Natl Acad Sci USA. 2017;114:310–5. doi: 10.1073/pnas.1612322114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, et al. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43(W1):W580–584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.