Abstract
The lizards are evolutionarily the closest vertebrates to humans that demonstrate the ability to regenerate entire appendages containing cartilage, muscle, skin, and nervous tissue. We previously isolated PAX7-positive cells from muscle of the green anole lizard, Anolis carolinensis, that can differentiate into multinucleated myotubes and express the muscle structural protein, myosin heavy chain. Studying gene expression in these satellite/progenitor cell populations from A. carolinensis can provide insight into the mechanisms regulating tissue regeneration. We generated a transcriptome from proliferating lizard myoprogenitor cells and compared them to transcriptomes from the mouse and human tissues from the ENCODE project using XGSA, a statistical method for cross-species gene set analysis. These analyses determined that the lizard progenitor cell transcriptome was most similar to mammalian satellite cells. Further examination of specific GO categories of genes demonstrated that among genes with the highest level of expression in lizard satellite cells were an increased number of genetic regulators of chondrogenesis, as compared to mouse satellite cells. In micromass culture, lizard PAX7-positive cells formed Alcian blue and collagen 2a1 positive nodules, without the addition of exogenous morphogens, unlike their mouse counterparts. Subsequent quantitative RT-PCR confirmed up-regulation of expression of chondrogenic regulatory genes in lizard cells, including bmp2, sox9, runx2, and cartilage specific structural genes, aggrecan and collagen 2a1. Taken together, these data suggest that tail regeneration in lizards involves significant alterations in gene regulation with expanded musculoskeletal potency.
Introduction
Lizards are evolutionarily the closest vertebrate group to humans with the ability to regenerate a complex appendage i.e., an entire tail (Koshiba-Takeuchi et al., 2009; Eckalbar et al., 2012; Gilbert et al., 2013). The regenerated lizard tail is structurally complex with de novo generated musculoskeletal tissues such as, skeletal muscle groups, tendons, a hyaline cartilage endoskeleton, as well as vasculature, peripheral and sensory nerves, and skin (Fisher et al., 2012; Hutchins et al., 2014). Mammals have some regenerative capacity of appendages, limited to digit tip formation in neonatal mice and humans under age two (Yu et al., 2010). Neonatal mice can also regenerate limited damage to heart ventricular muscle during the first week of life (Porrello et al., 2011; Darehzereshki et al., 2015).
Tail regeneration in A. carolinensis likely occurs through a stem cell mediated process, rather than dedifferentiation, as occurs during epimorphic regeneration in salamanders (Fisher et al., 2012; Hutchins et al., 2014). After an initial phase of wound healing in the lizard tail, the appendage regrows with an unique architecture quite distinct from the original tail (Fisher et al., 2012; Ritzman et al., 2012). Key differences include; the development of a cartilage tube endoskeleton, instead of segmented vertebrae, and axial muscle groups that run the length of the tail instead of segmental vertebral muscles (Fisher et al., 2012; Hutchins et al., 2014). Regeneration of a multi-tissue structure such as the tail requires pools of proliferative stem cells capable of differentiating into different lineages. Regeneration capable species employ distinct strategies to generate these stem cell populations. In urodele amphibians, dedifferentiation of injured tissue results in proliferative, lineage restricted progenitors (Kragl et al., 2009). Another source is activation of resident tissue-specific stem cells that migrate to the site of injury. For example, in the axolotl limb, it has been shown that amputation activates PAX7 positive satellite cells from adjacent muscle (Sandoval-Guzmán et al., 2014). Finally, dedifferentiated cells and stem cells can also transdifferentiate and change their fate to contribute to more than one tissue (Jopling et al., 2011).
Studies of skeletal muscle repair in response to injury in mammals have provided considerable insight into the signaling pathways associated with satellite cell activation, proliferation, and differentiation during repair. In response to acute damage, the myofibers are repaired by resident PAX7 positive satellite cells (Lepper et al., 2011; Sambasivan et al., 2011). Mammalian satellite cells are limited in their function to the repair of existing myofibers (Chen and Goldhamer, 2003; Dhawan and Rando, 2005; Wang and Rudnicki, 2011; Relaix and Zammit, 2012). There are cells present in a similar niche on the muscle fibers of anoles (Kahn and Simpson, 1974). In our previous study, we isolated these cells from the skeletal muscle of A. carolinensis lizards and demonstrated that they expressed pax7 and could be induced to fuse into multinucleated myosin heavy chain (MHC) positive myotubes (Hutchins et al., 2014).
Several previous studies have profiled the transcriptomes of satellite cells in mammalian species such as the mouse (Ryall et al., 2015), human (Charville et al., 2015), pig (Jeong et al., 2013), and cow (Lee et al., 2014). However, comparison of gene expression across vertebrate species remains a bioinformatic challenge due to difficulties in identifying orthologous genes and differences in baseline gene expression. A useful framework for comparing transcriptome-wide expression profiles across species is based on testing whether a gene set that is specifically expressed in a species is shared with similar tissues or cell types in other species (Djordjevic et al., 2016).
Our earlier transcriptomic analysis of the regenerating A. carolinensis tail demonstrated that there were 326 differentially expressed genes along the proximal-distal axis, many of which are involved in the development of the skeletal system and muscle (Hutchins et al., 2014). In this study, we characterized the lizard muscle progenitor cells using XGSA cross species comparison of their transcriptome with that of mammalian cell types and tissues, including satellite cells (Djordjevic et al., 2016). Our data demonstrated that the transcriptomic profile of the A. carolinensis muscle progenitor cells was most similar to mammalian satellite cells. It is likely that changes in the regulation of gene expression underlies the ability to regenerate appendages, thus we also compared expression of musculoskeletal and TGFβ/BMP pathway genes between mouse and lizard satellite cells using this same bioinformatics approach. We found genes that regulate myogenesis and chondrogenesis showed highly ranked expression in lizard satellite cells. Further, we demonstrated that the lizard cells display greater phenotypic plasticity and formed Alcian blue positive, collagen 2a1 expressing, chondrogenic nodules unlike mouse satellite cells.
Materials and Methods
Animals:
CD-1 mice (Mus musculus) were bred and housed in a vivarium at Arizona State University (ASU) on a 10 hr light:14 hr dark schedule with ad libitum access to food and water. Adult A. carolinensis lizards were purchased from Charles D. Sullivan, Inc. (Nashville, TN) or Marcus Cantos Reptiles (Fort Myers, FL) and housed as described previously (Fisher et al., 2012). ASU is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AALAC). All procedures were carried out in compliance with the ASU Institutional Animal Care and Use Committee and AALAC under approved research protocols.
Cell culture and Isolation
Muscle progenitor cells were isolated from 12 week old CD-1 mice and adult A. carolinensis lizards, as previously described (George et al., 2013); Hutchins et al., 2014). Briefly, hind limb quadriceps femoris muscles were excised, trimmed of fat and connective tissue, finely minced to a pulp. For lizard cells, skeletal muscle was stripped out of all limbs and proximal tail and trimmed of connective tissue and treated as above. The muscle tissue was digested with 1.25 mg of protease XIV (Sigma-Aldrich, St. Louis, MO) for one hour at 37°C, triturated to extract single cells and passed through a 100 μm nylon mesh. The cell suspension was spun down at 1500 g for 10 mins and preplated in DMEM (Corning, Corning NY), 2% HS (Atlanta Biologicals, Flowery Branch, GA) and 100 μg/ml Primocin (Invivogen, San Diego, CA) to remove fibroblasts and other debris for 2 hours. Satellite cells were grown on Matrigel (BD Biosciences, Bedford, MA) in Hams F-10 (Corning, Corning, NY), 20% FBS (Atlanta Biologicals), 10 ng/ml bFGF (BD Biosciences) and 100 μg/ml Primocin (Invivogen). Cells were cultured at 37°C for mouse or 30°C for lizard muscle progenitor cells in 5% CO2 in a humidified chamber. Cells from each isolation were plated onto Matrigel coated coverslips and PAX7 expression was determined by immunofluorescence (IF). The average percentage of PAX7 positive cells isolated was 98.8+/−1.2% for mouse and 98.6+/−2.2% s.d. for lizard cells, consistent with our previous data (George et al., 2013; Hutchins et al., 2014; data not shown).
Micromass culture
Satellite cells were seeded at 5×107 cells/mL in 20 μL medium (1×106 cells/micromass) in Matrigel coated 24 well plates. After a 1 hour incubation at 37°C for mouse or 30°C for lizard muscle progenitor cells, the wells were flooded with growth medium (Ham’s F-10 supplemented with 20% FBS, 10 ng/ml bFGF and Primocin) or chondrogenic differentiation medium (Ham’s F-10 supplemented with 10% FBS, 1% ITS, 50 nM ascorbate-2-phosphate, 1 nM TGFβ1 (Preprotech, Rocky Hill, NJ), and Primocin, for 7 days. Medium was changed every 3 days.
RNA Isolation and quantitative RT-PCR (QRT-PCR)
Cells were lysed in Trizol (Invitrogen, Carlsbad, CA) for RNA isolation, per the manufacturer’s protocols. For these studies, three biological replicate experiments were performed. RNA was treated with DNase I and quantified by Nanodrop prior to cDNA synthesis using SuperScriptIII reverse transcriptase (Invitrogen). For each sample, 2 μg of RNA were used for cDNA synthesis. The cDNA was quantified using transcript specific, intron spanning primers and real time PCR with Sybergreen (Eurogentec, Fremont, USA) on an ABI 7900 HT thermocycler using a 384 well format in 10μl reactions. Products from each primer set were sequenced and analyzed by BLAST (NCBI) to verify their identity. Primer efficiency was determined using a standard curve. For each transcript, three biological replicates were assayed in triplicate. All samples were normalized to the Gapdh transcript and relative gene expression was calculated using ΔΔCq analysis (Haimes and Kelley, 2010). Primer sequences for mouse and lizard genes are in Table S1.
RNA-Sequencing and Transcriptomic Analysis
RNA-Seq analysis of A. carolinensis satellite cells (3 biological replicates) has been described previously by our group (Hutchins et al., 2014) and the data are deposited in the NIH Sequence Read Archive (SRR1502189, SRR1502190, and SRR1502191; BioProject PRJNA253971). RNA-Seq data of mouse C57Bl/6J satellite cells (Ryall et al., 2015; SRA accessions SRR1726676, SRR1726677) were supplemented by our RNA-Seq analysis of mouse CD1 satellite cells isolated as described above, using protocols outlined in Hutchins et al., 2014. Transcript reads were mapped to A. carolinensis (AnoCar2.0) or mouse (GRCm38) Ensembl annotated genomes with HISAT2 v2.0.1 using default parameters (Kim et al., 2015). Gene level read counts were generated using HTSeq v0.6.0 in intersection-nonempty mode (Anders et al., 2015). For lizard and mouse satellite cell transcriptomes, Reads Per Kilobase of transcript per Million mapped reads (RPKM) were generated using edgeR (Robinson et al., 2010). For human satellite cells, Fragments Per Kilobase of transcript per Million mapped reads (FPKMs) were generated from two biological replicates analyzed by RNA-Seq (Charville et al., 2015) using TopHat and Cuffnorm (Trapnell et al., 2009; 2010). For comparison with reported gene expression profiles for a library of different tissues, ENCODE transcriptome profiles summarized as FPKMs were obtained for human (hg19; 139 tissues) and mouse (mm9; 94 tissues) (ENCODE Project Consortium, 2012 doi: 10.1038/nature11247; Mouse ENCODE Consortium et l., 2012 DOI: 10.1186/gb-2012-13-8-418). Gene symbols follow the recommendations of the International Committee on Standardized Genetic Nomenclature for Mice and the Anolis Gene Nomenclature Committee. In instances where the homologous genes in both mouse and lizard species are being referenced, the mouse nomenclature was used.
Cross-species gene set analysis
For each RNA-Seq experiment containing RPKM or FPKM values, rank products were calculated for each gene using all available replicates (Breitling et al., 2004). We identified the 1,500 most highly expressed genes in each cell type or tissue type. We then remove genes that are deemed highly expressed in more than 10% of the cell types, as these genes are likely ubiquitously expressed genes. The remaining genes are considered specifically expressed in each cell type or tissue type. We call this collection of gene sets the cell-type specific gene sets. Using the human and mouse ENCODE data collections, we generated a compendium of cell-type specific gene sets in humans and mice. Using a similar procedure, we used our lizard RNA-Seq data to generate a highly expressed gene set. We compared the lizard gene set against the human and mouse cell-type compendia using XGSA. A p-value is generated to represent whether the lizard gene set has significant overlap with a mouse or human cell-type-specific gene set.
To confirm that our gene set analysis is robust against our choice of parameters, we repeated the same analysis with a range of parameters (number of top highly expressed genes: 500, 1,000, 1,500, 2,000; % of cells containing marker gene: 1%, 2%, 3%, 5%, 10%, 15%, and 20%).
Alcian Blue Staining
Micromasses were fixed with 0.1% glutaraldehyde after 7 days in culture, and stained with 1% wt/vol Alcian Blue (VWR, Radnor, PA) in 0.1 M HCl overnight, at 4°C. Excess stain was washed off with 0.1 M HCl, followed by PBS.
Immunocytochemistry and Immunofluorescence
Monoclonal anti-Col2A1 antibody, II-II6B3 (RRID: AB_528165), which was deposited to the DHSB by T.F. Linsenmayer (DSHB, Iowa City, IA) and biotinylated goat anti-mouse IgG/anti-rabbit IgG (RRID: AB_2336187) (BA-1400, Vector Laboratories, Burlingame, CA) were used for immunocytochemistry (ICC). Micromasses were fixed in 4% formaldehyde and permeabilized with 0.5% Triton X-100 PBS solution. After blocking non-specific binding with 10% goat serum (Invitrogen), micromasses were incubated overnight at 4°C with primary antibody diluted in 1% serum, washed with PBS, and subsequently incubated for 1 hour at 37°C with secondary antibody in PBS + 1% serum. HRP-Streptavidin solution was added for 30 minutes, followed by PBS washes and DAB substrate incubation (Broad Spectrum HRP Histostain, Invitrogen).
To visualize nuclei within myotubes, fixed micromasses were stained with DAPI (Biotium, Fremont, CA), per manufacturer’s instructions.
Transverse vibratome sections of 120 days post-autotomy (DPA) regenerated lizard tails were obtained and incubated with the anti-Col2A1 antibody. Briefly, sections were washed in 0.2% Triton X-100 PBS, blocked in 5% goat serum, and incubated overnight at 4°C with anti-Col2A1 antibody diluted in 5% serum. Following Triton-PBS washes, sections were incubated with FITC conjugated anti-mouse IgG secondary antibody (RRID: AB_259378) (F-0257, Sigma-Aldrich) diluted in 5% serum overnight at 4°C. Fluoro-Gel (Electron Microscopy Sciences, Hatfield, PA) was used to mount the sections. All images were obtained on a Nikon Eclipse TE2000-U microscope, using the 488 nm filter for immunofluorescence (IF). Images were adjusted for contrast and color balance. The surface area of nodules was quantified using the “freehand tool” and “set scale” features of ImageJ that allows a known length to be assigned a specific number of pixels.
Results
XGSA analysis of transcriptomes
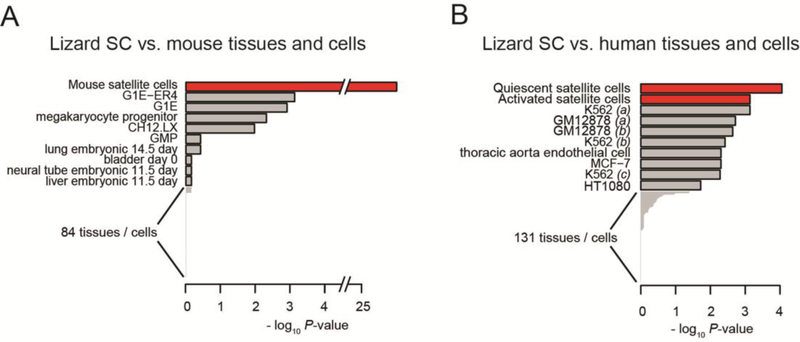
Satellite cells were isolated from mouse and lizard limb skeletal muscle and RNA was isolated from PAX7 positive cells, an established marker of satellite cells in mammals (Zammit et al., 2006; Lepper et al., 2011; Sambasivan et al., 2011). Analysis of genes expressed in lizard satellite cells was carried out using RNA-Seq transcriptomic analysis (Hutchins et al., 2014). Based on our previous data, we wanted to determine the similarity of the lizard PAX7 positive cells to the satellite cell population in the mouse and human. We carried out this comparison using XGSA, a statistical method for cross-species gene set analysis (Djordjevic et al., 2016). We compared highly expressed genes in the lizard satellite cell transcriptome to a compendium of cell-type-specific gene sets including 94 tissues from the mouse ENCODE project (Yue et al., 2014). This comparison demonstrated that out of the tissues examined, the lizard PAX7 positive cell has a significant similarity with the mouse satellite cell based on expression of cell-type-specific marker genes (p-value < 1.9 × 10–26). It has more overlap with mouse satellite cell markers than markers of any other mouse cell types studied here (Fig. 1A), and this result is robust against analysis parameters (Supplementary Fig.1A). Similarly, we compared lizard satellite cells with 139 tissues from the human ENCODE project and identified the greatest similarity with activated and quiescent human satellite cells (Fig. 1B; Supplementary Fig. 1B).
Figure 1. XGSA analyses comparing the transcriptome from lizard satellite cells to multiple tissues from the mouse and human ENCODE projects.
XGSA comparison of marker genes with 94 mouse (A), and 139 human (B) tissues reveals that the lizard satellite cell transcriptome (Hutchins et al., 2014) displayed highest similarity with mouse and human satellite cells. Depicted are the top 10 most similar for each species comparison. For complete comparisons see Supplemental Figure 1. Parentheses after sample names are used to differentiate replicate transcriptomes.
Gene rank comparison of mouse and lizard satellite cell transcriptomes.
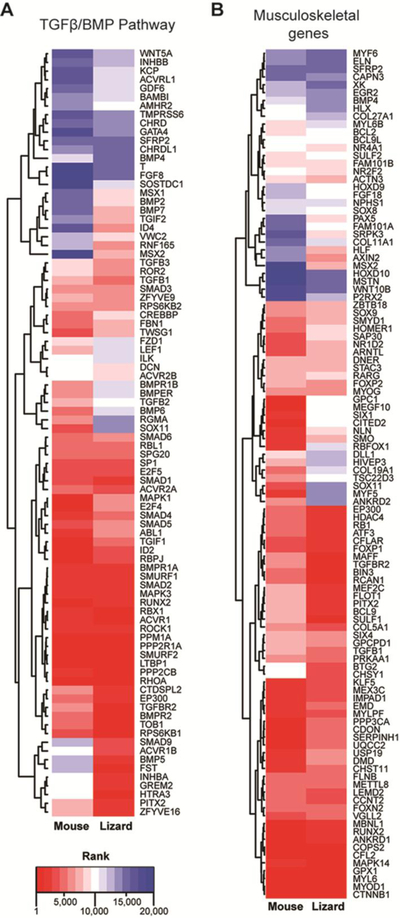
Bone morphogenic protein (BMP) and transforming growth factor β (TGFβ) signaling pathways have important regulatory roles both in embryonic myogenesis and postnatal muscle regeneration (Wang et al., 2010; Ono et al., 2010; Lee et al., 2010; 2012; Friedrichs et al., 2011; McFarlane et al., 2011; Winbanks et al., 2013; Sartori et al., 2014; George et al., 2015). We examined the differences in the expression of the TGFβ/BMP pathway genes by comparing the relative rankings of 105 TGFβ/BMP pathway genes (KEGG category mmu04350 and GO term GO:0030509) from lizard and mouse satellite cells (Fig. 2A). Genes with high differential expression, higher rankings in lizard (closer to 1.0), as compared to a lower ranking in mouse (closer to 20,000), included Bmp2, Bmp5, Bmp7, Dcn, Fst, Id4, Inhba, Pitx2, Smad9, Tgif2, Zfyve16, Msx1, Msx2, Rnf165, Grem2, and Sostdc1. Follistatin (Fst) and Pitx2 induce proliferation of satellite cells in regenerating muscle (Amthor et al., 2004; L’honore et al., 2007; Gilson et al., 2009; Lozano-Velasco et al., 2011). Msx1 regulates the cellularization of myofibers, i.e., the conversion of multinucleated skeletal muscle fibers into mononuclear cells, during amphibian limb regeneration. Msx1 also regulates cell cycle re-entry of the resulting myoblasts. Ectopic expression of either Msx1 or Msx2 in mouse myofibers induced cellularization and inhibited myoblast differentiation (Odelberg et al., 2000; Echeverri and Tanaka, 2002; Kumar et al., 2004; Yilmaz et al., 2015).
Figure 2. Gene rank comparison of mouse and lizard satellite cell transcriptomes.
Heatmaps of genes involved in the TGFβ/BMP signaling pathway (A) and musculoskeletal development (B) show differential rankings between mouse and lizard satellite cells. Genes with the highest rank (highest level of expression) are closer to 1, and those with the lowest rank (lowest expression in that species) are closer to 20,000.
Our data demonstrated that the expression of Bmp2, Bmp5, and Bmp7, ranked considerably higher in satellite cells of the lizard as compared to mouse (Fig. 2A), and these factors are important regulators of chondrogenesis and osteogenesis (King et al., 1994; Tsuji et al., 2006; Knippenberg et al., 2006; Snelling et al., 2010; Shen et al., 2010; Shu et al., 2011). While mouse and human satellite cells and myoblasts typically only differentiate into skeletal muscle, high levels of BMP ligands can induce these cells to undergo osteogenesis and chondrogenesis in vitro (Katagiri et al., 1994; Asakura et al., 2001; Wada et al., 2002; Shea et al., 2003). Other genes with lower expression levels in lizard satellite cells, as compared to mouse, included Bmp6, Bmpr1b, Bmper, Fzd1, Lef1, and Sox11. Bmpr1b, also known as Alk6, encodes a receptor specific for BMP7 and GDF5 (Yi et al., 2000; Zhao et al., 2002; Yoon et al., 2005). Sox11 is expressed in condensing chondrocytes embryonically in mice (Cameron et al., 2009). Bmper, BMP-binding endothelial regulator or crossveinless-2, can enhance or inhibit BMP signaling (Zhang et al., 2010). BMP6 regulates iron metabolism and is co-expressed in developing cartilage with BMP2 (Corradini et al., 2011; Luo et al., 2016).
We observed that the transcriptomic profile of lizard satellite cells was marked by the co-expression of genes involved in not only myogenesis, but also chondrogenesis and osteogenesis (Fig. 2A). Focusing on genes in the gene ontology (GO) terms ‘skeletal muscle’ and ‘cartilage development and differentiation’, we found that expression of key genes involved in satellite cell activation and proliferation, and muscle differentiation ranked higher in lizard as compared to mouse, including Mef2c, Pitx2, Srpk3, Hdac4, Axin2, Bcl9, Nr2f2, and Sulf1 (Fig. 2B; (Molkentin et al., 1995; Brack et al., 2008; 2009; Gill et al., 2010; Raines et al., 2015). Nr2f2, an orphan nuclear receptor is up-regulated in activated satellite cells, and inhibits differentiation of stem cells, including those in the skeletal muscle, cartilage, and bone marrow lineages (Gao et al., 2016; Zhu et al., 2016). The myogenic regulatory factors Myod1, myogenin (Myog), Yap1, and Six4 were expressed at comparable levels in both species, but Myf5 was expressed at a much lower level in lizard cells (Fig. 2B). Other genes with known regulatory roles in muscle development and satellite cell function that demonstrated a lower level of expression in lizard satellite cells when compared to mouse included Dll1, Six1, Ankrd2, Cited2, Gpc1, Fzd1, Megf10, and Smo (Fig. 2B). Among these, Megf10, Ankrd2, Fzd1, and Smo induce differentiation of satellite cells (Zhao and Hoffman, 2004; Holterman et al., 2007; Brack et al., 2008). Together these data suggest that these myoprogenitors express the myogenic transcriptional program but are not differentiating, consistent with their status as proliferating single cells.
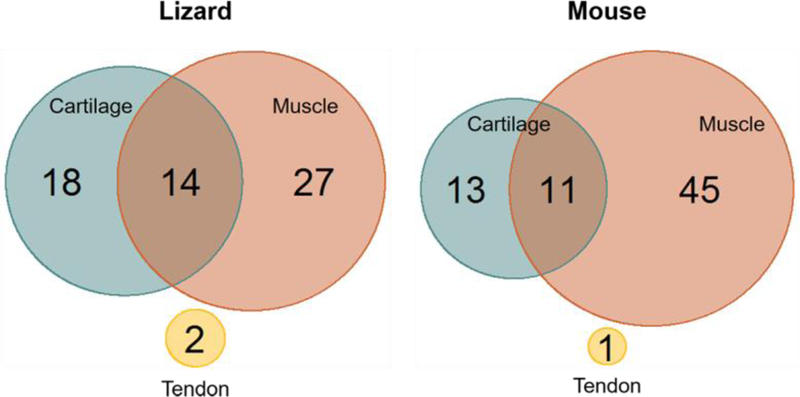
This analysis demonstrated that many genes involved in chondrogenesis had high levels of expression in lizard, but not mouse, satellite cells including Chsy1, Axin2, Tgfbr2, Fam101a, Fgf18, Col5a1, and Msx2. Fgf18 and Chsy1 induce proliferation of early chondrocytes (Liu et al., 2007; Wilson et al., 2012). Msx2 is important for regeneration in amphibians and osteogenesis in mice. Consistently, Col27a1, Bmp4, and Bmpr1b which are involved in cartilage differentiation (Plumb et al., 2011; Shu et al., 2011; Lim et al., 2015), have lower expression levels in lizard, as compared to mouse, satellite cells (Fig. 2B). Based on the observed higher ranking of cartilage specific genes in the lizard transcriptome, we identified genes from the ranked lists (supplemental tables 2 and 3) whose expression level ranked between 1 and 5000 (highest levels of expression) with roles in muscle development, differentiation, and regeneration, cartilage development and differentiation or tendon development (Fig. 3). We found 61 lizard genes and 70 mouse genes ranked in the respective transcriptomes with these functions (Fig. 3). Of these genes, 49.2% had a role in muscle development, differentiation, and regeneration in lizard cells whereas in proliferating mouse satellite cells 65.6% of the genes fell into this category. Further, 30.5% of lizard genes and 20.3% of mouse genes were involved in cartilage differentiation and development. Lastly, 20.3% of lizard genes and 14.1% of mouse genes had roles in both processes (Fig. 3). Our XGSA analyses showed that these cells were most similar to mammalian satellite cells, but there are significant differences and taken together these data suggest that lizard satellite cells may have increased potential.
Figure 3. Comparison of highly differentially ranked musculoskeletal genes.
A comparison of the functions of genes whose rank fell between 1 and 5,000 (top 20%) for lizard and mouse satellite cells. In lizard cells 44.3% of the genes were involved in skeletal muscle development and differentiation, whereas in mouse cells this was 63.4% of the top ranked genes. Cartilage specific genes were expressed in both populations, 29.5% and 18.3% of top ranked lizard and mouse genes, respectively. There are genes that have roles in both skeletal muscle and cartilage development and they represented 23% of lizard and 15.5% of mouse genes. Both cell types expressed a limited number of tendon specific genes at high levels. These data indicated that genes with the highest level of expression in lizard satellite cells represent fewer myogenic genes and an increase in genes involved in chondrogenesis.
Analysis of chondrogenic potential by culture in micromass
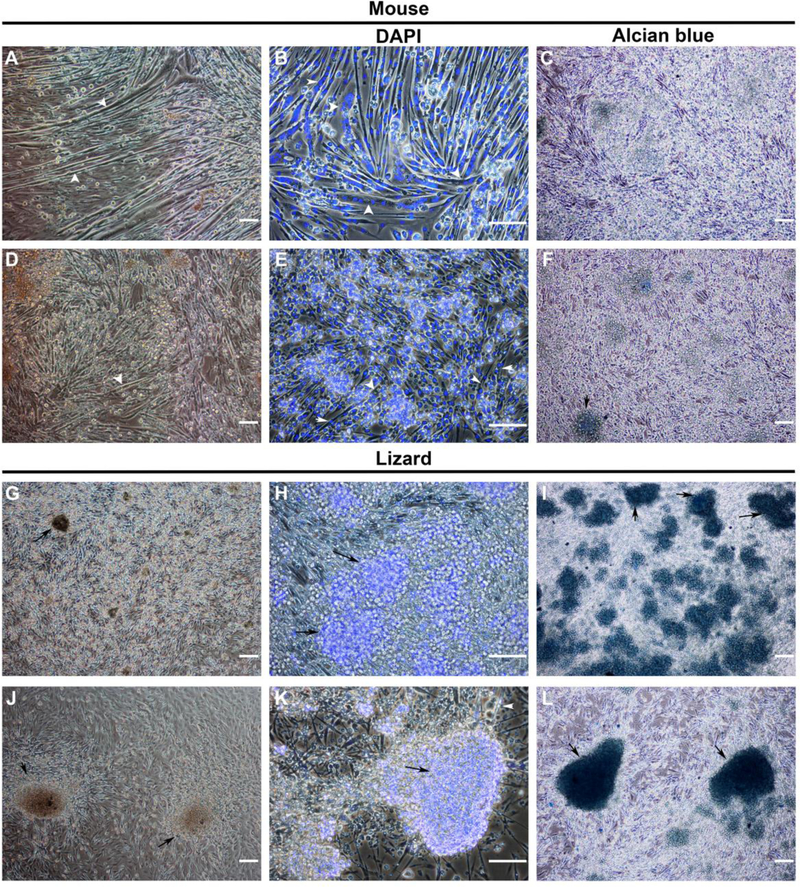
One of the most striking observations from our analyses were the very high ranks of expression of bmp2, bmp5, and bmp7, along with many other cartilage-promoting genes in the lizard satellite cells (Figs. 2 and 3). These observations indicated that these cells might have expanded potential and could adopt the chondrogenic lineage. To examine this, lizard and mouse satellite cells were cultured at high density in micromasses in either growth medium (Ham’s F10, 10% FBS, 10 ng/mL bFGF) or chondrogenic differentiation medium (Ham’s F10, 1% FBS, 6.25 mg/mL insulin, 10 ng/mL TGFβ1, 50 nM ascorbate 2 phosphate) without the addition of any morphogens, such as BMPs, and morphology was assessed at 7 days. Mouse satellite cells formed myotubes under either culture condition (Fig. 4A, D), although they were less extensive in chondrogenic differentiation medium with fewer nuclei per myotube (Fig. 4, compare B to E). Lizard satellite cells formed many large nodular structures comprised of small cells when cultured in chondrogenic medium (Fig. 4J, K) and many smaller nodules when cultured in growth medium (Fig. 4G, H). The lizard cells formed few small myotubes in both conditions, interestingly, they mostly remained as single cells (Fig. 4H, K). To demonstrate that the nodules were indeed chondrogenic, micromasses were stained with Alcian blue to detect cartilage specific glycosaminoglycans. Lizard satellite cells cultured in micromass formed Alcian blue positive nodules in both growth and chondrogenic media (Fig. 4I, L). In growth medium, the micromasses averaged 30.8 nodules (Table 1). In chondrogenic medium, there were fewer nodules, an average of 18 per micromass, but the average surface area was twice that of nodules formed in growth conditions (Table 1). Mouse micromasses demonstrated no Alcian blue positive nodules in growth medium and we detected less than one nodule per micromass in chondrogenic medium (Fig. 4C, F; Table 1).
Figure 4. Micromass culture of lizard satellite cells results in condensation of cells into Alcian blue positive chondrogenic nodules.
Mouse and lizard satellite cells were cultured in micromasses of 1×106 cells on Matrigel coated 24 well plates for 7 days in either growth (A-C, G-I) or chondrogenic differentiation medium (D-F, J-L) without added BMPs. Mouse satellite cells formed myotubes with many nuclei, as detected by DAPI nuclear stain, in growth medium (A, B, white arrows) and smaller myotubes in chondrogenic differentiation medium (D, E white arrows), but no defined nodules were noted. Lizard satellite cells in growth medium differentiated into a few small myotubes, and smaller nodules with undefined edges that stained with Alcian blue (G-I, black arrows). Large Alcian blue positive nodules were found in micromasses cultured in differentiation medium, consistent with the onset of chondrogenesis in vitro (J-L, black arrows). Photomicrographs are representative, magnification is 100X except for photomicrographs of DAPI stained micromasses, which are 200X, bars = 100μm, n=6 micromasses for each condition.
Table 1. Average number, and surface area, of nodules in micromass.
The average number, and surface area, of nodules in day 7 micromasses was quantified using ImageJ (n=6 micromasses for each experimental condition).
| Growth Medium (GM) | Chondrogenic Medium(DM) | |
|---|---|---|
| Average nodule number (average nodule area) | ||
| Mouse | 0 | 0.67 (0.029 mm2) |
| Lizard | 30.8 (0.027 mm2) | 18 (0.048 mm2) |
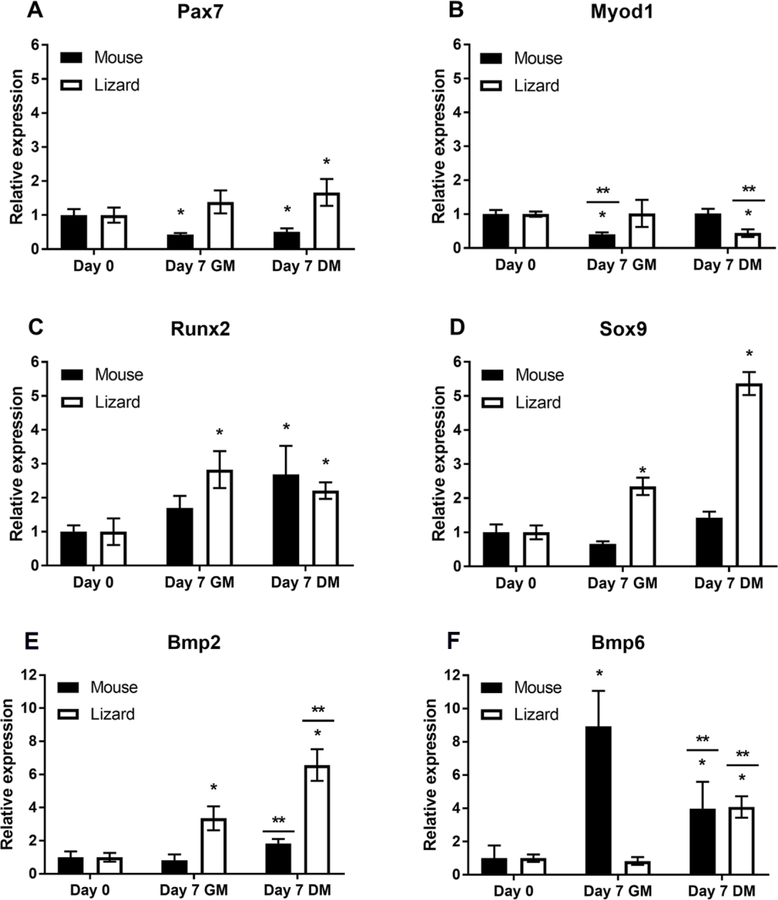
The expression of chondrogenic and myogenic marker genes in lizard and mouse satellite cells cultured in growth or chondrogenic medium was examined by qRT-PCR. CDNA was generated from total RNA isolated from high density micromass cultures after 7 days. As can be seen in Fig. 5, mouse cells significantly down-regulated Pax7 expression, consistent with myotube differentiation, while this gene was up-regulated in lizard satellite cells, significantly so in differentiation medium. This might reflect an increase in the number of satellite cells, as there were many single cells present at day 7 that did not participate in formation of myotubes or nodules (Fig. 4). Analysis of MyoD (Myod1), a myogenic basic helix-loop-helix transcription factor (bHLH), that is up-regulated in actively proliferating satellite cells and myoblasts (Yin et al., 2013), revealed a small but significant decrease in expression in mouse satellite cells in growth medium (Fig. 5). Conversely, Myod1 expression demonstrated a small but significant down-regulation in lizard satellite cells cultured in chondrogenic medium, consistent with the onset of chondrogenesis. Additionally, we found that Runx2, a transcription factor important for proliferation and differentiation of chondrocytes and skeletal morphogenesis (Fujita et al., 2004; Takarada et al., 2013), was up-regulated in cells from both species. This occurred in both growth and chondrogenic differentiation conditions, suggesting that high cell density may be sufficient to trigger its expression. Interestingly, Sox9, a transcription factor necessary for chondrogenesis (Wright et al., 1995; Akiyama et al., 2002; Asou et al., 2002; Leung et al., 2011) was only significantly up-regulated in lizard cells cultured in micromass, and its expression level increased under conditions that favored chondrogenesis (Fig. 5).
Figure 5. Expression of lineage-specific regulatory genes in lizard and mouse satellite cells cultured in micromass.
Micromasses of mouse and lizard satellite cells were cultured for 7 days in growth (GM) or chondrogenic differentiation media (DM) and gene expression was determined by gene specific qRT-PCR. Mouse satellite cells demonstrated decreased Pax7 expression in micromass culture, while anole cells up-regulated this gene (A). Both species of satellite cells expressed Myod1 and Runx2 (B,C), but only lizard cells significantly up-regulated the chondrogenesis regulator sox9 (D). Lizard satellite cells up-regulated bmp2 expression significantly in GM, and further increased expression significantly in DM (E). Murine satellite cells demonstrated no significant change in the expression of Bmp2 when compared to Day 0. However, DM samples were significantly up-regulated compared to GM (E). Lizard satellite cells significantly up-regulated bmp6 only when cultured in chondrogenic differentiation medium, while mouse cells demonstrated significant up-regulation of Bmp6 expression in growth medium as well (F). Graphs are as labeled. Data are expressed as relative gene expression and are the result of 3 biological replicates, each done in triplicate. Statistical analysis was done by one way ANOVA; * indicates p<0.05 when compared to Day 0 and ** denotes that DM and GM for that species are significantly different from each other (p<0.05).
Given the differences in baseline expression of BMP pathway genes between lizard and mouse satellite cells, including elevated Bmp2 and decreased Bmp6, we investigated whether further differences arose during chondrogenic differentiation in micromass culture. For lizard satellite cells, bmp2 transcription was significantly up-regulated under growth conditions, and culture in chondrogenic differentiation medium further significantly increased the level of its expression (Fig. 5). Bmp6 is expressed in differentiating hypertrophic chondrocytes, but unlike Bmp2, it does not play a role in regulating chondrogenesis, it is involved in regulating iron metabolism (Camaschella, 2009). Bmp6 expression was not up-regulated in lizard satellite cells in growth medium, but was increased in differentiation medium. Bmp6 was up-regulated in mouse satellite cells when cultured at high density and in chondrogenic medium (Fig. 5).
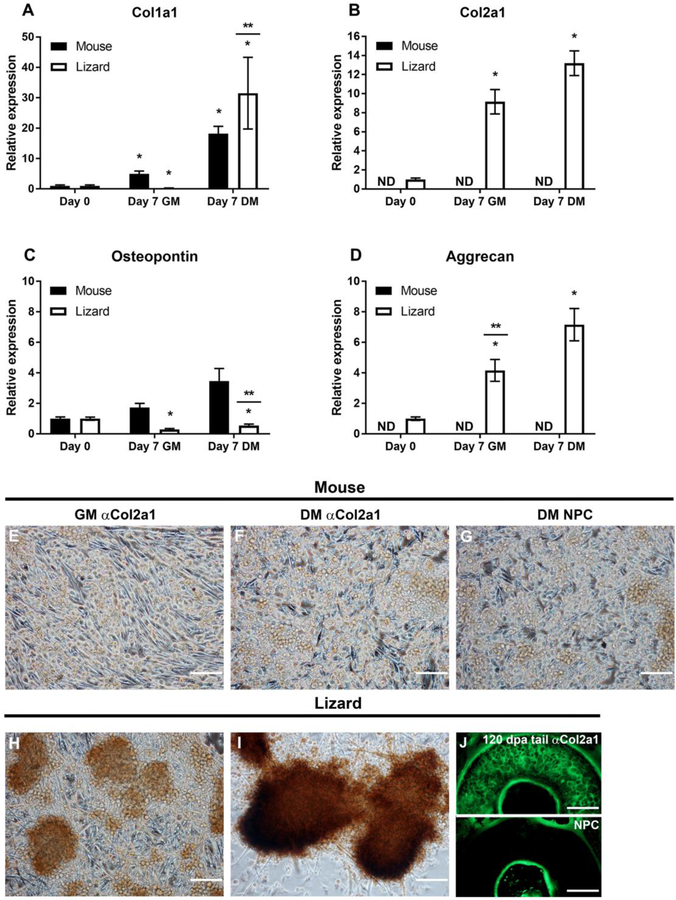
Compared to other tissues, cartilage is hypocellular with few nuclei relative to the extensive extracellular matrix. We further examined the expression of extracellular matrix proteins by satellite cells in micromass culture. Collagen I (Col1a1) is expressed in muscle, tendon, and bone (Reddi et al., 1977), whereas collagen 2A1 (Col2a1) is specific to cartilage. Both mouse and lizard cells significantly up-regulated Col1a1 in micromasses, but only the lizard cells expressed and significantly up-regulated cartilage specific Col2a1 (Fig. 6). Aggrecan (Acan) is a cartilage specific extracellular matrix protein that is expressed during chondrogenic differentiation (Lauing et al., 2014), and expression of this gene was significantly up-regulated in the lizard micromasses. Acan was not detected in mouse micromasses (Fig. 6). Osteopontin, or secreted phosphoprotein 1 (Spp1), is expressed by a variety of cell types and has many roles, among which are muscle regeneration, inflammatory responses, and bone calcification (Pagel et al., 2013; Singh et al., 2014). Interestingly, while murine satellite cells up-regulated expression of this gene, the lizard satellite cells significantly down-regulated its expression in chondrogenic medium (Fig. 6). We examined collagen 2A1 expression using ICC in micromasses at day 7. Cells were fixed in 4% paraformaldehyde and an anti-collagen2a1 antibody and protein detected with biotinylated anti-mouse secondary antibody and HRP-streptavidin and the DAB substrate. Consistent with the qRT-PCR observations, mouse micromasses did not express collagen 2A1 (Fig. 6, compare G to E, F), whereas lizard nodules demonstrated strong expression of this protein (Fig. 6H, I). Taken together our data demonstrates that the lizard cells express genes involved in chondrogenesis and can differentiate in culture by up-regulating the expression of BMP genes, cartilage specific transcription factors, and extracellular matrix genes.
Figure 6. Extracellular matrix gene expression in micromass cultures demonstrate that lizard cells express cartilage-specific genes.
Micromasses were cultured for 7 days in growth (GM) or chondrogenic differentiation media (DM). Gene expression was determined by gene specific qRT-PCR. Collagen 1a1 (Col1a1), was up-regulated in both species in DM (A), while collagen 2a1 (col2a1), a cartilage specific collagen, was detected in lizard satellite cells only (B). Osteopontin (Opn), was increased in mouse cells but not significantly, it was significantly down-regulated in lizard cells (C). Aggrecan (acan) expression was detected only in lizard micromasses, where it was significantly up-regulated in GM and further increased in DM (D). qRT-PCR data are expressed as relative gene expression and are the result of 3 biological replicates, each done in triplicate. Statistical analysis was done by one way ANOVA; * indicates p<0.05 when compared to Day 0 and ** denotes that DM and GM for that species are significantly different from each other (p<0.05); ND= not detected. Protein expression was determined by ICC, using a monoclonal anti-COL2A1 antibody and anti-mouse-HRP and the DAB substrate to detect the protein. Mouse satellite cells exhibit background staining that is indistinguishable from the no primary antibody control (NPC) sample (E-G). Chondrogenic nodules express COL2A1 protein in lizard micromasses in both culture conditions, although the staining is more robust in DM (H, I). To show the specificity of the anti-COL2A1 antibody in lizard, IF was done with an anti-mouse-FITC secondary antibody on transverse sections of regenerating lizard tails 120 DPA. The matrix of the cartilage is evident compared to the NPC control (J).
Discussion
Transcriptomic and histological analyses of regeneration in the A. carolinensis tail point to a stem cell-mediated process (Fisher et al., 2012; Hutchins et al. 2014). Our previous work demonstrated that lizard skeletal muscle contains PAX7 positive progenitor cells and RNA-Seq analysis showed that the regenerating tail expressed marker genes of activated satellite cells and myoblasts (Hutchins et al., 2014). Using the cross-species analytical tool, XGSA, we carried out comparative transcriptomic analysis of proliferating satellite cells from the lizard with mouse and human tissues from the ENCODE project. As might be expected, we found the closest match with satellite cells (Fig. 1). However, the expression level of genes associated with chondrogenesis and osteogenesis were higher in lizard satellite cells compared with their mouse counterparts (Figs. 2, 3).
BMP signaling induces different responses depending on ligand concentrations and exposure times. BMPs and their inhibitors define where and when muscle formation occurs ( Re’em-Kalma et al., 1995; Hirsinger et al., 1997; Reshef et al., 1998). During skeletal muscle regeneration, low concentrations of BMP2, 4, and 7 maintain proliferation of satellite cells and myoblasts (Amthor et al., 1998; Ozeki et al., 2007; Ono et al., 2010; Wang et al., 2010; Friedrichs et al., 2011; Sartori et al., 2013;), whereas, high levels of BMP2, 4, 5, or 7 inhibit myogenesis, induce chondrogenesis, and ultimately, osteogenesis of satellite cells, C2C12 myoblasts, C3H10T1/2 MSCs, and other MSCs, with continued exposure in culture (Katagiri et al., 1994; Schmitt et al., 2003; Shea et al., 2003; Bandyopadhyay et al., 2006; Knippenberg et al., 2006; Ozeki et al., 2007; Friedrichs et al., 2011; Takács et al., 2013; Liao et al., 2014; Zhou et al., 2016). Proliferating lizard satellite cells expressed bmp2, 5, and 7 at high levels, unlike their murine counterparts (Fig. 2). A significant observation was the endogenous up-regulation of bmp2 by these cells in micromass culture and a further increase in its expression under conditions that favored cartilage differentiation, indicating the lizard satellite cells can adopt the chondrogenic fate.
In mammals, BMP2 induces the expression of the transcriptional activators, Runx2 and Sox9, that mediate chondrogenesis (Katagiri et al., 1994; Denker et al., 1999; Haas and Tuan, 1999; Schmitt et al., 2003; Shea et al., 2003; Hashimoto et al., 2008; Kwon et al., 2013; Liao et al., 2014; Zhou et al., 2016). RUNX2 is a transcription factor regulating the differentiation of mesenchymal cells to chondrocytes, and chondrogenesis is disrupted in Runx2−/− mice (Komori et al., 1997; Inada et al., 1999; Takarada et al., 2013). SOX9 is a transcription factor that regulates differentiation of embryonic cartilage and induces chondrogenic differentiation of myogenic cells (Wright et al., 1995; Akiyama et al., 2002; Cairns et al., 2012; Liao et al., 2014; Zhou et al., 2016). In micromass culture, the lizard satellite cells concomitantly up-regulated expression of bmp2, runx2, and sox9 significantly, and formed dense nodules (Figs. 4–6). Expression of the cartilage specific matrix proteins, col2a1 and acan, that are also SOX9 target genes, by lizard satellite cells was consistent with the onset of chondrogenesis (Fig. 6). Cartilage specific matrix protein expression in lizard satellite cells was also confirmed by Alcian blue staining (Fig. 4) and detection of collagen 2A1 by ICC (Fig. 6). We found that mouse satellite cells in micromass culture did not initiate chondrogenesis (Figs. 4–6). These data indicate that lizard satellite cells can enter a different musculoskeletal lineage by endogenously up-regulating the expression of the BMP morphogens and cartilage specific proteins and point to important differences in the regulation of expression of these genes in the lizard. The up-regulation of cartilage specific regulators by lizard satellite cells, indicates that the PAX7 positive satellite cells in fact have greater potency as stem cells and are able to differentiate along multiple musculoskeletal lineages to form cartilage, and skeletal muscle and potentially tendons. Further research is required to confirm and elucidate the mechanisms underlying this process.
The expression of Msx1 and Msx2 was ranked higher in lizard compared to mouse satellite cells. These transcriptional repressors play important roles in embryonic chondrogenesis in mammals (Odelberg et al., 2000; Satokata et al., 2000; Ishii et al., 2005). In amphibians, these proteins are important for dedifferentiation of muscle during limb regeneration and are expressed in the blastema (Echeverri and Tanaka, 2002). When C2C12 myotubes in culture expressed Msx1 or Msx2 from inducible viral promoters, these proteins caused delamination of single cells from the myotubes. The expression of myogenic marker genes was decreased in these cells, but less than 10 percent re-entered the cell cycle (Odelberg et al., 2000; Yilmaz et al., 2015). The expression of these two genes in proliferating lizard satellite cells may indicate that they are necessary for maintenance of proliferation, or Msx1 and Msx2 may have another role in the increased plasticity of the lizard satellite cells.
Not surprisingly, our data also showed that lizard satellite cells demonstrated changes in gene expression that are consistent with decreased fibrosis, important for successful regeneration. For example, Decorin (Dcn) encodes an extracellular matrix protein that decreases the fibrotic response during muscle regeneration (McCroskery et al., 2005; Li et al., 2007; Li et al., 2008) and is expressed at a higher level in lizard as compared to mouse satellite cells. Similarly, Tgfb2 and Fzd1, are both implicated in fibrosis in dystrophic and aged skeletal muscle (Brack et al., 2007; Biressi et al., 2014), and were expressed at much lower levels in lizard than in mouse myoprogenitors.
Taken together, these data suggest that some lizard satellite cells have increased musculoskeletal potential governed by changes in the regulation of gene expression, as only the tail is regenerated. Further inquiry into these changes will not only shed light on the mechanisms of lizard tail regeneration, but also provide a means for approaching improved tissue engineering of mammalian muscle and cartilage from satellite cells. Understanding the regeneration strategy employed by reptiles could provide important genetic information regarding specific pathways or genes whose regulation could be altered to improve the potential therapeutic uses of satellite cells in mammals.
Supplementary Material
Highlights.
Bioinformatic comparisons demonstrate that the transcriptome of PAX7+ lizard cells is most similar to those of satellite cells of mice and humans.
Lizard satellite cells express genes involved in chondrogenesis, myogenesis, and BMP/TGFβ signaling genes.
Lizard satellite cells can condense into Alcian blue and collagen 2A1 positive nodules and express chondrogenic genes in micromass culture without added morphogens.
Acknowledgements
We thank the Department of Animal Care and Technologies at Arizona State University for assistance in maintaining the lizard colony and Stephen Pratt for statistical consultation. This work was supported by funding from the National Center for Research Resources and the Office of Research Infrastructure Programs (ORIP) grant R21 RR031305, National Institute of Arthritis, Musculoskeletal, and Skin Diseases grant R21 AR064935 of the National Institutes of Health, Arizona Biomedical Research Commission grant 1113 and Burroughs Wellcome Fund Collaborative Research Travel Grant to KK. XGSA bioinformatic analysis was supported by a Career Development Fellowship (1105271) from the National Health and Medical Research Council Australia and a Future Leader Fellowship from the National Heart Foundation of Australia to JWKH. Computational analysis was supported by allocations from Arizona State University Research Computing. The funding sources listed had no role in the study design, collection, analysis, nor interpretation of the data; in the writing of the paper; nor the decision to submit the article for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors have no interests to declare. This work has not been published previously, nor is it under consideration for publication elsewhere. This publication has been approved by all authors.
References
- 1.Akiyama H, Chaboissier M-C, Martin JF, Schedl A, and de Crombrugghe B (2002). The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev 16, 2813–2828. 10.1073/pnas.1522054113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amthor H, Christ B, Weil M, and Patel K (1998). The importance of timing differentiation during limb muscle development. Curr Biol 8, 642–652. [DOI] [PubMed] [Google Scholar]
- 3.Amthor H, Nicholas G, McKinnell I, Kemp CF, Sharma M, Kambadur R, and Patel K (2004). Follistatin complexes Myostatin and antagonises Myostatin-mediated inhibition of myogenesis. Dev Biol 270, 19–30. 10.1016/j.ydbio.2004.01.046 [DOI] [PubMed] [Google Scholar]
- 4.Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asakura A, Rudnicki MA, and Komaki M (2001). Muscle satellite cells are multipotential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation 68, 245–253. 10.1046/j.1432-0436.2001.680412. [DOI] [PubMed] [Google Scholar]
- 6.Asou Y, Nifuji A, Tsuji K, Shinomiya K, Olson EN, Koopman P, and Noda M (2002). Coordinated expression of scleraxis and Sox9 genes during embryonic development of tendons and cartilage. J Orthop Res 20, 827–833. 10.1016/S0736-0266(01)00169-3 [DOI] [PubMed] [Google Scholar]
- 7.Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, and Tabin CJ (2006). Genetic Analysis of the Roles of BMP2, BMP4, and BMP7 in Limb Patterning and Skeletogenesis. PLoS Genet 2, e216–15. 10.1002/btpr.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biressi S, Miyabara EH, Gopinath SD, Carlig PMM, and Rando TA. (2014). A Wnt-TGFβ2 axis induces a fibrogenic program in muscle stem cells from dystrophic mice. Sci Transl Med 6(267),267ar176. 10.1126/scitranslmed.3008411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brack AS, Conboy IM, Conboy MJ, Shen J, and Rando TA (2008). A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell 2, 50–59. 10.1016/j.stem.2007.10.006 [DOI] [PubMed] [Google Scholar]
- 10.Brack AS, Murphy-Seiler F, Hanifi J, Deka J, Eyckerman S, Keller C, Aguet M, and Rando TA (2009). BCL9 is an essential component of canonical Wnt signaling that mediates the differentiation of myogenic progenitors during muscle regeneration. Dev Biol 335, 93–105. 10.1016/j.ydbio.2009.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitling R, Armengaud P, Amtmann A, and Herzyk P (2004). Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Letters 573, 83–92. 10.1016/j.febslet.2004.07.055 [DOI] [PubMed] [Google Scholar]
- 12.Cairns DM, Liu R, Sen M, Canner JP, Schindeler A, Little DG, and Zeng L (2012). Interplay of Nkx3.2, Sox9 and Pax3 Regulates Chondrogenic Differentiation of Muscle Progenitor Cells. PLoS ONE 7, e39642–17. 10.1371/journal.pone.0039642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camaschella C (2009). BMP6 orchestrates iron metabolism. Nat Genet 41, 386–388. 10.1038/ng0409-386 [DOI] [PubMed] [Google Scholar]
- 14.Cameron TL, Belluoccio D, Farlie PG, Brachvogel B, and Bateman JF (2009). Global comparative transcriptome analysis of cartilage formation in vivo. BMC Dev Biol 9, 20 10.1186/1471-213X-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charville GW, Cheung TH, Yoo B, Santos PJ, Lee GK, Shrager JB, and Rando TA (2015). Ex Vivo Expansion and In Vivo Self-Renewal of Human Muscle Stem Cells. Stem Cell Rep 5, 621–632. 10.1016/j.stemcr.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JCJ, and Goldhamer DJ (2003). Skeletal muscle stem cells. Reprod Biol Endocrinol 1, 101 10.1186/1477-7827-1-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corradini E, Meynard D, Wu Q, Chen S, Ventura P, Pietrangelo A, and Babitt JL (2011). Serum and liver iron differently regulate the bone morphogenetic protein 6 (BMP6)‐SMAD signaling pathway in mice. Hepatology 54, 273–284. 10.1002/hep.24359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darehzereshki A, Rubin N, Gamba L, Kim J, Fraser J, Huang Y, Billings J, Mohammadzadeh R, Wood J, Warburton D, et al. (2015). Differential regenerative capacity of neonatal mouse hearts after cryoinjury. Dev Biol 399, 91–99. 10.1016/j.ydbio.2014.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denker AE, Haas AR, Nicoll SB, and Tuan RS (1999). Chondrogenic differentiation of murine C3H10T1/2 multipotential mesenchymal cells: I. Stimulation by bone morphogenetic protein-2 in high-density micromass cultures. Differentiation 64, 67–76. 10.1046/j.1432-0436.1999.6420067.x [DOI] [PubMed] [Google Scholar]
- 20.Dhawan J, and Rando TA (2005). Stem cells in postnatal myogenesis: molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol 15, 666–673. 10.1016/j.tcb.2005.10.007 [DOI] [PubMed] [Google Scholar]
- 21.Djordjevic D, Kusumi K, and Ho JWK (2016). XGSA: A statistical method for cross-species gene set analysis. Bioinformatics 32, i620–i628. 10.1093/bioinformatics/btw428 [DOI] [PubMed] [Google Scholar]
- 22.Echeverri K, and Tanaka EM (2002). Mechanisms of muscle dedifferentiation during regeneration. Semin Cell Dev Biol 13, 353–360. 10.1016/S1084-9521(02)00091-5 [DOI] [PubMed] [Google Scholar]
- 23.Eckalbar WL, Lasku E, Infante CR, Elsey RM, Markov GJ, Allen AN, Corneveaux JJ, Losos JB, DeNardo DF, Huentelman MJ, et al. (2012). Somitogenesis in the anole lizard and alligator reveals evolutionary convergence and divergence in the amniote segmentation clock. Dev Biol 363, 308–319. 10.1016/j.ydbio.2011.11.021 [DOI] [PubMed] [Google Scholar]
- 24.Fisher RE, Geiger LA, Stroik LK, Hutchins ED, George RM, DeNardo DF, Kusumi K, Rawls JA, and Wilson-Rawls J (2012). A Histological Comparison of the Original and Regenerated Tail in the Green Anole, Anolis carolinensis. Anat Rec (Hoboken) 295, 1609–1619. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrichs M, Wirsdöerfer F, Flohé SB, Schneider S, Wuelling M, and Vortkamp A (2011). BMP signaling balances proliferation and differentiation of muscle satellite cell descendants. BMC Cell Biol 12, 26 10.1186/1471-2121-12-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujita T, Azuma Y, Fukuyama R, Hattori Y, Yoshida C, Koida M, Ogita K, and Komori T (2004). Runx2 induces osteoblast and chondrocyte differentiation and enhances their migration by coupling with PI3K-Akt signaling. J Cell Biol 166, 85–95. 10.1083/jcb.200401138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukushima K, Badlani N, Usas A, Riano F, Fu F, and Huard J (2001). The use of an antifibrosis agent to improve muscle recovery after laceration. Am J Sports Med 29, 394–402. [DOI] [PubMed] [Google Scholar]
- 28.Gao G, Zhang X-F, Hubbell K, and Cui X (2016). NR2F2 regulates chondrogenesis of human mesenchymal stem cells in bioprinted cartilage. Biotechnol Bioeng 1–9. 10.1002/bit.26042 [DOI] [PubMed]
- 29.George RM, Biressi S, Beres BJ, Rogers E, Mulia AK, Allen RE, Rawls A, Rando TA, and Wilson-Rawls J (2013). Numb-deficient satellite cells have regeneration and proliferation defects. Proc Natl Acad Sci USA 110, 18549–18554. 10.1073/pnas.1311628110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert EAB, Payne SL, and Vickaryous MK (2013). The anatomy and histology of caudal autotomy and regeneration in lizards. Physiol Biochem Zool 86,631–644. 10.1086/673889 [DOI] [PubMed] [Google Scholar]
- 31.Gill R, Hitchins L, Fletcher F, and Dhoot GK (2010). Sulf1A and HGF regulate satellite-cell growth. J Cell Sci 123, 1873–1883. 10.1242/jcs.061242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilson H, Schakman O, Kalista S, Lause P, Tsuchida K, and Thissen JP (2009). Follistatin induces muscle hypertrophy through satellite cell proliferation and inhibition of both myostatin and activin. AJP: Endocrinol Metab 297, E157–E164. 10.1152/ajpendo.00193.2009 [DOI] [PubMed] [Google Scholar]
- 33.Haas AR, and Tuan RS (1999). Chondrogenic differentiation of murine C3H10T1/2 multipotential mesenchymal cells: II. Stimulation by bone morphogenetic protein-2 requires modulation of N-cadherin expression and function. Differentiation 64, 77–89. 10.1046/j.1432-0436.1999.6420077.x [DOI] [PubMed] [Google Scholar]
- 34.Haimes J, and Kelley M (2010). Demonstration of a deltadeltaCq calculation method to compute relative gene expression from qPCR data. Thermo Scientific Tech Note 1–4.
- 35.Hashimoto N, Kiyono T, Wada MR, Umeda R, Goto Y-I, Nonaka I, Shimizu S, Yasumoto S, and Inagawa-Ogashiwa M (2008). Osteogenic properties of human myogenic progenitor cells. Mech Dev 125, 257–269. 10.1016/j.mod.2007.11.004 [DOI] [PubMed] [Google Scholar]
- 36.Hirsinger E, Duprez D, Jouve C, Malapert P, Cooke J, and Pourquié O (1997). Noggin acts downstream of Wnt and Sonic Hedgehog to antagonize BMP4 in avian somite patterning. Development 124, 4605–4614. [DOI] [PubMed] [Google Scholar]
- 37.Holterman CE, Le Grand F, Kuang S, Seale P, and Rudnicki MA (2007). Megf10 regulates the progression of the satellite cell myogenic program. J Cell Biol 179, 911–922. 10.1083/jcb.200709083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hutchins ED, Markov GJ, Eckalbar WL, George RM, King JM, Tokuyama MA, Geiger LA, Emmert N, Ammar MJ, Allen AN, et al. (2014). Transcriptomic Analysis of Tail Regeneration in the Lizard Anolis carolinensis Reveals Activation of Conserved Vertebrate Developmental and Repair Mechanisms. PLoS ONE 9, e105004 10.1371/journal.pone.0105004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, Himeno M, Sato M, Yamagiwa H, Kimura T, Yasui N, et al. (1999). Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev Dyn 214, 279–290. [DOI] [PubMed] [Google Scholar]
- 40.Ishii M, Han J, Yen H-Y, Sucov HM, Chai Y, and Maxson RE (2005). Combined deficiencies of Msx1 and Msx2 cause impaired patterning and survival of the cranial neural crest. Development 132, 4937–4950. 10.1242/dev.02072 [DOI] [PubMed] [Google Scholar]
- 41.Jeong JY, Kim JM, Rajesh RV, and Suresh S (2013). Transcriptional Profiling of Differentially Expressed Genes in Porcine Satellite Cell. Reprod Dev Biol 37(4) 233–245. 10.12749/RDB.2013.37.4.233 [DOI] [Google Scholar]
- 42.Jopling C, Boue S, and Belmonte JCI (2011). Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol 12, 79–89. 10.1038/nrm3043 [DOI] [PubMed] [Google Scholar]
- 43.Kahn EB, and Simpson SB (1974). Satellite cells in mature, uninjured skeletal muscle of the lizard tail. Dev Biol 37, 219–223. 10.1016/0012-1606(74)90181-X [DOI] [PubMed] [Google Scholar]
- 44.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, and Suda T (1994). Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol 127, 1755–1766. 10.1083/jcb.127.6.1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Meth 12, 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King JA, Marker PC, Seung KJ, and Kingsley DM (1994). BMP5 and the molecular, skeletal, and soft-tissue alterations in short ear mice. Dev Biol 166, 112–122. 10.1006/dbio.1994.1300 [DOI] [PubMed] [Google Scholar]
- 47.Knippenberg M, Helder MN, Zandieh Doulabi B, Wuisman PIJM, and Klein-Nulend J (2006). Osteogenesis versus chondrogenesis by BMP-2 and BMP-7 in adipose stem cells. Biochem Biophys Res Commun 342, 902–908. 10.1016/j.bbrc.2006.02.052 [DOI] [PubMed] [Google Scholar]
- 48.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. (1997). Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764. [DOI] [PubMed] [Google Scholar]
- 49.Koshiba-Takeuchi K, Mori AD, Kaynak BL, Cebra-Thomas J, Sukonnik T, Georges RO, Latham S, Beck L, Beck L, Henkelman RM, et al. (2009). Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature 461, 95–98. 10.1038/nature08324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, and Tanaka EM (2009). Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature 460, 60–65. 10.1038/nature08152 [DOI] [PubMed] [Google Scholar]
- 51.Kumar A, Velloso CP, Imokawa Y, and Brockes JP (2004). The Regenerative Plasticity of Isolated Urodele Myofibers and Its Dependence on Msx1. 2, PLoS Biol 2(8), e218–e219. 10.1371/journal.pbio.0020218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwon S-H, Lee T-J, Park J, Hwang J-E, Jin M, Jang H-K, Hwang NS, and Kim B-S (2013). Modulation of BMP-2-Induced Chondrogenic Versus Osteogenic Differentiation of Human Mesenchymal Stem Cells by Cell-Specific Extracellular Matrices. Tiss Eng Part A 19, 49–58. 10.1089/ten.tea.2012.0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.L’honore A, Coulon V, Marcil A, Lebel M, Lafrance-Vanasse J, Gage P, Camper S, and Drouin J (2007). Sequential expression and redundancy of Pitx2 and Pitx3 genes during muscle development. Dev Biol 307, 421–433. 10.1016/j.ydbio.2007.04.034 [DOI] [PubMed] [Google Scholar]
- 54.Lauing KL, Cortes M, Domowicz MS, Henry JG, Baria AT, and Schwartz NB (2014). Aggrecan is required for growth plate cytoarchitecture and differentiation. Dev Biol 396, 224–236. 10.1016/j.ydbio.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee EJ, Malik A, Pokharel S, Ahmad S, Mir BA, Cho KH, Kim J, Kong JC, Lee D-M, Chung KY, et al. (2014). Identification of genes differentially expressed in myogenin knock-down bovine muscle satellite cells during differentiation through RNA sequencing analysis. PLoS ONE 9, e92447 10.1371/journal.pone.0092447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee S-J, Huynh TV, Lee Y-S, Sebald SM, Wilcox-Adelman SA, Iwamori N, Lepper C, Matzuk MM, and Fan C-M (2012). Role of satellite cells versus myofibers in muscle hypertrophy induced by inhibition of the myostatin/activin signaling pathway. Proc Natl Acad Sci USA 109, E2353–E2360. 10.1073/pnas.1206410109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S-J, Lee Y-S, Zimmers TA, Soleimani A, Matzuk MM, Tsuchida K, Cohn RD, and Barton ER (2010). Regulation of Muscle Mass by Follistatin and Activins. Mol Endocrinol 24, 1998–2008. 10.1210/me.2010-0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lepper C, Partridge TA, and Fan CM (2011). An absolute requirement for Pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639–3646. 10.1242/dev.067595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leung VYL, Gao B, Leung KKH, Melhado IG, Wynn SL, Au TYK, Dung NWF, Lau JYB, Mak ACY, Chan D, et al. (2011). SOX9 Governs Differentiation Stage-Specific Gene Expression in Growth Plate Chondrocytes via Direct Concomitant Transactivation and Repression. PLoS Genet 7, e1002356–16. 10.1371/journal.pgen.1002356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X, McFarland DC, and Velleman SG (2008). Extracellular matrix proteoglycan decorin-mediated myogenic satellite cell responsiveness to transforming growth factor-β1 during cell proliferation and differentiation. Domest Anim Endocrinol 35, 263–273. 10.1016/j.dom [DOI] [PubMed] [Google Scholar]
- 61.Li Y, Li J, Zhu J, Sun B, Branca M, Tang Y, Foster W, Xiao X, and Huard J (2007). Decorin Gene Transfer Promotes Muscle Cell Differentiation and Muscle Regeneration. Mol Ther 15, 1616–1622. 10.1016/S0002-9440(10)63188-4 [DOI] [PubMed] [Google Scholar]
- 63.Liao J, Hu N, Zhou N, Lin L, Zhao C, Yi S, Fan T, Bao W, Liang X, Chen H, et al. (2014). Sox9 Potentiates BMP2-Induced Chondrogenic Differentiation and Inhibits BMP2-Induced Osteogenic Differentiation. PLoS ONE 9, e89025–13. 10.1371/journal.pone.0089025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lim J, Tu X, Choi K, Akiyama H, Mishina Y, and Long F (2015). BMP-Smad4 signaling is required for precartilaginous mesenchymal condensation independent of Sox9 in the mouse. Dev Biol 400, 132–138. 10.1016/j.ydbio.2015.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Z, Lavine KJ, Hung IH, and Ornitz DM (2007). FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Dev Biol 302, 80–91. 10.1016/j.ydbio.2006.08.071 [DOI] [PubMed] [Google Scholar]
- 66.Lozano-Velasco E, Contreras A, Crist C, Hernández-Torres F, Franco D, and Aránega AE (2011). Pitx2c modulates Pax3+/Pax7+ cell populations and regulates Pax3 expression by repressing miR27 expression during myogenesis. Dev Biol 357, 165–78. 10.1016/j.ydbio.2011.06.039 [DOI] [PubMed] [Google Scholar]
- 67.Luo X, Luo Z, Zhang Z, Yang H, Lai B, Yao Q, Xiao L, and Wang N (2016). Homocysteine upregulates hepcidin expression through BMP6/SMAD signaling pathway in hepatocytes. Biochem. Biophys. Res. Commun 471, 303–308. 10.1016/j.bbrc.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 68.McCroskery S, Thomas M, Platt L, Hennebry A, Nishimura T, McLeay L, Sharma M, and Kambadur R (2005). Improved muscle healing through enhanced regeneration and reduced fibrosis in myostatin-null mice. J Cell Sci 118, 3531–3541. 10.1242/jcs.02482 [DOI] [PubMed] [Google Scholar]
- 69.McFarlane C, Hui GZ, Amanda WZW, Lau HY, Lokireddy S, Xiaojia G, Mouly V, Butler-Browne G, Gluckman PD, Sharma M, et al. (2011). Human myostatin negatively regulates human myoblast growth and differentiation. Am J Physiol, Cell Physiol 301, C195–C203. 10.1152/ajpcell.00012.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Molkentin JD, Black BL, Martin JF, and Olson EN (1995). Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83, 1125–1136. [DOI] [PubMed] [Google Scholar]
- 71.Odelberg SJ, Kollhoff A, and Keating MT (2000). Dedifferentiation of mammalian myotubes induced by msx1. Cell 103, 1099–1109. [DOI] [PubMed] [Google Scholar]
- 72.Ono Y, Calhabeu F, Morgan JE, Katagiri T, Amthor H, and Zammit PS (2010). BMP signalling permits population expansion by preventing premature myogenic differentiation in muscle satellite cells. Cell Death Diff 18, 222–234. 10.1016/j.ydbio.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ozeki N, Jethanandani P, Nakamura H, Ziober BL, and Kramer RH (2007). Modulation of satellite cell adhesion and motility following BMP2-induced differentiation to osteoblast lineage. Biochem. Biophys. Res. Commun 353, 54–59. 10.1016/j.bbrc.2006.11.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pagel CN, Wasgewatte Wijesinghe DK, Taghavi Esfandouni N, and Mackie EJ (2013). Osteopontin, inflammation and myogenesis: influencing regeneration, fibrosis and size of skeletal muscle. J. Cell Commun. Signal 8, 95–103. 10.1007/s12079-013-0217-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Plumb DA, Ferrara L, Torbica T, Knowles L, Mironov A, Kadler KE, Briggs MD, and Boot-Handford RP (2011). Collagen XXVII Organises the Pericellular Matrix in the Growth Plate. PLoS ONE 6, e29422–12. 10.1371/journal.pone.0029422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, and Sadek HA (2011). Transient Regenerative Potential of the Neonatal Mouse Heart. Science 331, 1078–1080. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raines AM, Magella B, Adam M, and Potter SS (2015). Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development. BMC Dev Biol 15, 1–15. 10.1186/s12861-015-0078-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Re’em-Kalma Y, Lamb T, and Frank D (1995). Competition between noggin and bone morphogenetic protein 4 activities may regulate dorsalization during Xenopus development. Proc Natl Acad Sci USA 92, 12141–12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reddi AH, Gay R, Gay S, and Miller EJ (1977). Transitions in collagen types during matrix-induced cartilage, bone, and bone marrow formation. Proc Natl Acad Sci USA 74, 5589–5592. 10.1016/j.bonr.2016.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Relaix F, and Zammit PS (2012). Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development 139, 2845–2856. 10.1242/dev.069088 [DOI] [PubMed] [Google Scholar]
- 81.Reshef R, Maroto M, and Lassar AB (1998). Regulation of dorsal somitic cell fates: BMPs and Noggin control the timing and pattern of myogenic regulator expression. Genes Dev 12, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ritzman TB, Stroik LK, Julik E, Hutchins ED, Lasku E, DeNardo DF, Wilson-Rawls J, Rawls JA, Kusumi K, and Fisher RE (2012). The Gross Anatomy of the Original and Regenerated Tail in the Green Anole (Anolis carolinensis). Anat Rec 295, 1596–1608. 10.1002/ar.22524 [DOI] [PubMed] [Google Scholar]
- 83.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ryall JG, Dell’Orso S, Derfoul A, Juan A, Zare H, Feng X, Clermont D, Koulnis M, Gutierrez-Cruz G, Fulco M, et al. (2015). The NAD+-Dependent SIRT1 Deacetylase Translates a Metabolic Switch into Regulatory Epigenetics in Skeletal Muscle Stem Cells. Cell Stem Cell 16, 171–183. 10.1016/j.stem.2014.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sambasivan R, Yao R, Kissenpfennig A, Van Wittenberghe L, Paldi A, Gayraud-Morel B, Guenou H, Malissen B, Tajbakhsh S, and Galy A (2011). Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656. 10.1242/dev.067587 [DOI] [PubMed] [Google Scholar]
- 86.Sandoval-Guzmán T, Wang H, Khattak S, Schuez M, Roensch K, Nacu E, Tazaki A, Joven A, Tanaka EM, and Simon A (2014). Fundamental Differences in Dedifferentiation and Stem Cell Recruitment during Skeletal Muscle Regeneration in Two Salamander Species. Stem Cell 14, 174–187. 10.1016/j.stem.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 87.Sartori R, Gregorevic P, and Sandri M (2014). TGFβ and BMP signaling in skeletal muscle: potential significance for muscle-related disease. Trends Endocrinol Metab 25, 464–471. 10.1016/j.tem.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 88.Sartori R, Schirwis E, Blaauw B, Bortolanza S, Zhao J, Enzo E, Stantzou A, Mouisel E, Toniolo L, Ferry A, et al. (2013). BMP signaling controls muscle mass. Nature 45, 1309–1318. 10.1038/ng.2772 [DOI] [PubMed] [Google Scholar]
- 89.Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, et al. (2000). Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet 24, 391–395. 10.1038/74231 [DOI] [PubMed] [Google Scholar]
- 90.Schmitt B, Ringe J, Häupl T, Notter M, Manz R, Burmester G-R, Sittinger M, and Kaps C (2003). BMP2 initiates chondrogenic lineage development of adult human mesenchymal stem cells in high-density culture. Differentiation 71, 567–577. 10.1111/j.1432-0436.2003.07109003.x [DOI] [PubMed] [Google Scholar]
- 91.Shea CM, Edgar CM, Einhorn TA, and Gerstenfeld LC (2003). BMP treatment of C3H10T1/2 mesenchymal stem cells induces both chondrogenesis and osteogenesis. J Cell Biochem 10.1002/jcb.10734 [DOI] [PubMed]
- 92.Shen B, Wei A, Whittaker S, Williams LA, Tao H, Ma DDF, and Diwan AD (2010). The role of BMP‐7 in chondrogenic and osteogenic differentiation of human bone marrow multipotent mesenchymal stromal cells in vitro. J Cell Biochem 109, 406–416. 10.1002/jcb.22412 [DOI] [PubMed] [Google Scholar]
- 93.Shu B, Zhang M, Xie R, Wang M, Jin H, Hou W, Tang D, Harris SE, Mishina Y, O’Keefe RJ, et al. (2011). BMP2, but not BMP4, is crucial for chondrocyte proliferation and maturation during endochondral bone development. J Cell Sci 124, 3428–3440. 10.1242/jcs.083659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Singh M, Dalal S, and Singh K (2014). Osteopontin: At the cross-roads of myocyte survival and myocardial function. Life Sci 118, 1–6. 10.1016/j.lfs.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takarada T, Hinoi E, Nakazato R, Ochi H, Xu C, Tsuchikane A, Takeda S, Karsenty G, Abe T, Kiyonari H, and Yonari Y (2013). An analysis of skeletal development in osteoblast-specific and chondrocyte-specific runt-related transcription factor-2 (Runx2) knockout mice. J Bone Miner Res 28(10),2064–2069. 10.1002/jbmr.1945 [DOI] [PubMed] [Google Scholar]
- 96.Takács R, Matta C, Somogyi C, Juhász T, and Zákány R (2013). Comparative Analysis of Osteogenic/Chondrogenic Differentiation Potential in Primary Limb Bud-Derived and C3H10T1/2 Cell Line-Based Mouse Micromass Cultures. Intl J Mol Sci 14, 16141–16167. 10.3390/ijms140816141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech 28, 516–520. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tsuji K, Bandyopadhyay A, Harfe BD, Cox K, Kakar S, Gerstenfeld L, Einhorn T, Tabin CJ, and Rosen V (2006). BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet 38, 1424–1429. 10.1038/ng1916 [DOI] [PubMed] [Google Scholar]
- 100.Wada MR, Inagawa-Ogashiwa M, Shimizu S, Yasumoto S, and Hashimoto N (2002). Generation of different fates from multipotent muscle stem cells. Development 129, 2987–2995. [DOI] [PubMed] [Google Scholar]
- 101.Wang H, Noulet F, Edom-Vovard F, Le Grand F, and Duprez D (2010). Bmp signaling at the tips of skeletal muscles regulates the number of fetal muscle progenitors and satellite cells during development. Dev Cell 18, 643–654. 10.1016/j.devcel.2010.02.008 [DOI] [PubMed] [Google Scholar]
- 102.Wang YX, and Rudnicki MA (2011). Satellite cells, the engines of muscle repair. Nat Rev Mol Cell Biol 13, 127–133. 10.1038/nrm3265 [DOI] [PubMed] [Google Scholar]
- 103.Wilson DG, Phamluong K, Lin WY, Barck K, Carano RAD, Diehl L, Peterson AS, Martin F, and Solloway MJ (2012). Chondroitin sulfate synthase 1 (Chsy1) is required for bone development and digit patterning. Dev Biol 363, 413–425. 10.1016/j.ydbio.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 104.Winbanks CE, Chen JL, Qian H, Liu Y, Bernardo BC, Beyer C, Watt KI, Thomson RE, Connor T, Turner BJ, et al. (2013). The bone morphogenetic protein axis is a positive regulator of skeletal muscle mass. J Cell Biol 203, 345–357. 10.1083/jcb.201211134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, and Koopman P (1995). The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet 9, 15–20. 10.1242/dev.01417 [DOI] [PubMed] [Google Scholar]
- 106.Yi SE, Daluiski A, Pederson R, Rosen V, and Lyons KM (2000). The type I BMP receptor BMPRIB is required for chondrogenesis in the mouse limb. Development 127, 621–630. [DOI] [PubMed] [Google Scholar]
- 107.Yilmaz A, Engeler R, Constantinescu S, Kokkaliaris KD, Dimitrakopoulos C, Schroeder T, Beerenwinkel N, and Paro R (2015). Ectopic expression of Msx2 in mammalian myotubes recapitulates aspects of amphibian muscle dedifferentiation. Stem Cell Res 15, 542–553. 10.1016/j.scr.2015.09.012 [DOI] [PubMed] [Google Scholar]
- 108.Yin H, Price F, and Rudnicki MA (2013). Satellite Cells and the Muscle Stem Cell Niche. Physiol Rev 93, 23–67. 10.1152/physrev.00043.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yoon BS, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, and Lyons KM (2005). Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci USA 102, 5062–5067. 10.1073/pnas.0500031102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu L, Han M, Yan M, Lee E-C, Lee J, and Muneoka K (2010). BMP signaling induces digit regeneration in neonatal mice. Development 137, 551–559. 10.1242/dev.042424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. (2014). A comparative encyclopedia of DNA elements in the mouse genome. Nature 515, 355–364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, and Beauchamp JR (2006). Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci 119, 1824–1832. 10.1242/jcs.02908 [DOI] [PubMed] [Google Scholar]
- 113.Zhang J-L, Patterson LJ, Qiu L-Y, Graziussi D, Sebald W, and Hammerschmidt M (2010). Binding between Crossveinless-2 and Chordin Von Willebrand Factor Type C Domains Promotes BMP Signaling by Blocking Chordin Activity. PLoS ONE 5, e12846–12. 10.1371/journal.pone.0012846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhao M, Harris SE, Horn D, Geng Z, Nishimura R, Mundy GR, and Chen D (2002). Bone morphogenetic protein receptor signaling is necessary for normal murine postnatal bone formation. J Cell Biol 157, 1049–1060. 10.1083/jcb.200109012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao P, and Hoffman EP (2004). Embryonic myogenesis pathways in muscle regeneration. Dev Dyn 229, 380–392. 10.1002/dvdy.10457 [DOI] [PubMed] [Google Scholar]
- 116.Zhou N, Li Q, Lin X, Hu N, Liao J-Y, Lin L-B, Zhao C, Hu Z-M, Liang X, Xu W, et al. (2016). BMP2 induces chondrogenic differentiation, osteogenic differentiation and endochondral ossification in stem cells. Cell Tissue Res 1–11. 10.1007/s00441-016-2403-0 [DOI] [PubMed]
- 117.Zhu N, Wang H, Wang B, Wei J, Shan W, Feng J, and Huang H (2016). A Member of the Nuclear Receptor Superfamily, Designated as NR2F2, Supports the Self-Renewal Capacity and Pluripotency of Human Bone Marrow-Derived Mesenchymal Stem Cells. Stem Cells Intl 2016, 1–11. 10.1155/2016/5687589 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.