Escherichia coli ST95 is a globally disseminated clone frequently associated with bloodstream infections and neonatal meningitis. However, the ST95 lineage is defined by low levels of drug resistance amongst clinical isolates, which normally provides for uncomplicated treatment options. Here, we provide the first detailed genomic analysis of an E. coli ST95 isolate that has both high virulence potential and resistance to multiple antibiotics. Using the genome, we predicted its virulence and antibiotic resistance mechanisms, which include resistance to last-line antibiotics mediated by the plasmid-borne mcr-1 gene. Finding an ST95 isolate resistant to nearly all antibiotics that also has a high virulence potential is of major clinical importance and underscores the need to monitor new and emerging trends in antibiotic resistance development in this important global lineage.
KEYWORDS: Escherichia coli, antibiotic resistance, genome analysis
ABSTRACT
Resistance to last-line polymyxins mediated by the plasmid-borne mobile colistin resistance gene (mcr-1) represents a new threat to global human health. Here we present the complete genome sequence of an mcr-1-positive multidrug-resistant Escherichia coli strain (MS8345). We show that MS8345 belongs to serotype O2:K1:H4, has a large 241,164-bp IncHI2 plasmid that carries 15 other antibiotic resistance genes (including the extended-spectrum β-lactamase blaCTX-M-1) and 3 putative multidrug efflux systems, and contains 14 chromosomally encoded antibiotic resistance genes. MS8345 also carries a large ColV-like virulence plasmid that has been associated with E. coli bacteremia. Whole-genome phylogeny revealed that MS8345 clusters within a discrete clade in the sequence type 95 (ST95) lineage, and MS8345 is very closely related to the highly virulent O45:K1:H4 clone associated with neonatal meningitis. Overall, the acquisition of a plasmid carrying resistance to colistin and multiple other antibiotics in this virulent E. coli lineage is concerning and might herald an era where the empirical treatment of ST95 infections becomes increasingly more difficult.
IMPORTANCE Escherichia coli ST95 is a globally disseminated clone frequently associated with bloodstream infections and neonatal meningitis. However, the ST95 lineage is defined by low levels of drug resistance amongst clinical isolates, which normally provides for uncomplicated treatment options. Here, we provide the first detailed genomic analysis of an E. coli ST95 isolate that has both high virulence potential and resistance to multiple antibiotics. Using the genome, we predicted its virulence and antibiotic resistance mechanisms, which include resistance to last-line antibiotics mediated by the plasmid-borne mcr-1 gene. Finding an ST95 isolate resistant to nearly all antibiotics that also has a high virulence potential is of major clinical importance and underscores the need to monitor new and emerging trends in antibiotic resistance development in this important global lineage.
INTRODUCTION
Polymyxins B and E (colistin) have been used in veterinary and human medicine for over 50 years. They have broad-spectrum activities against Gram-negative bacteria and are effective against most Enterobacteriaceae. Unfortunately, colistin is associated with both nephrotoxicity and neurotoxicity (1), and due to these adverse effects, it has seen limited use in human medicine. However, colistin has now emerged as an effective therapeutic against carbapenem-resistant Enterobacteriaceae (CRE) (2), carbapenem-resistant Acinetobacter baumannii (CRAB) (3), and Pseudomonas species (4), for which treatment options are limited (5). This overreliance on colistin for treatment of these extensively resistant infections has seen the emergence of CRE, CRAB, and Pseudomonas isolates resistant to colistin (6–10). Colistin resistance is typically mediated by chromosomal mutations resulting in modifications to lipopolysaccharide (LPS), the target site of the polymyxins, and a reduction in polymyxin affinity. These chromosomal mutations are only vertically transmissible, and until recently, the polymyxins remained one of the last classes of antibiotic where resistance was not spread horizontally from cell to cell (9–12).
In 2015, Liu et al. described for the first time a plasmid-borne transmissible colistin resistance gene, mcr-1 (13). The mcr-1 gene belongs to the phosphoethanolamine transferase family of enzymes, which function by catalyzing the 4′-phosphoethanolamine (PEA) modification of lipid A, a component of LPS (13, 14). To date, mcr-1 has been identified on both broad-host-range and narrow-host-range plasmids of different replicon types, including IncI2, IncX4, IncP, IncHI1, and IncHI2 (13, 15–17). Worryingly, carriage of mcr-1 is often associated with cocarriage of other drug resistance genes, including those for carbapenemases (18–20) and extended-spectrum β-lactamases (18, 20, 21). This coassociation of mcr-1 with other drug resistance genes is a significant step toward the emergence of pandrug resistance in the Enterobacteriaceae.
The mcr-1 gene has been identified in a number of different bacterial species, but to date, carriage of mcr-1 is most frequently associated with Escherichia coli (22). In a study of historical E. coli isolates in China, the emergence of the mcr-1 gene was traced back to the 1980s, which coincides with the introduction of colistin as a growth enhancer in food production (23). The study, which screened 1,611 E. coli strains of chicken origin collected from farms in China between 1970 and 2014, found that the proportion of mcr-1-positive E. coli isolates increased exponentially from 5.2% in 2009 to 30% in 2014 (23). Similar proportions of mcr-1-positive E. coli strains have been observed in surveillance studies of food and production animals globally (13, 22, 24). The carriage rates of mcr-1 are much lower in human E. coli isolates than in those from mammals and birds (13, 25). However, as extraintestinal pathogenic E. coli (ExPEC) strains that colonize humans and animals are highly similar (26–28), there is enormous zoonotic potential for mammals and birds to act as reservoirs of infection and transmit the mcr-1 gene to humans (29, 30).
E. coli sequence type 95 (ST95) is a global pandemic clone of ExPEC. In contrast to other pandemic clones, such as E. coli ST131, ST95 isolates are characterized by a low incidence of multidrug resistance (MDR) (31–33). For example, ST95 clinical isolates had the lowest level of antibiotic resistance in comparisons among the 10 most prevalent uropathogenic E. coli (UPEC) sequence types (32, 33). Similar low levels of drug resistance have been identified in ST95 clinical isolates from the United States, Canada, and France (34–37). A highly virulent O45:K1:H7 subclone of ST95 accounts for one-third of all neonatal meningitis cases in France (38). A key feature of this subclone is a ColV-like virulence plasmid (39).
Here, we describe the first complete genome of an mcr-1-positive E. coli isolate (MS8345) of human origin. MS8345 was isolated from a patient in Qatar with a subarachnoid hemorrhage and was resistant to multiple antibiotics. We show that MS8345 contains two large plasmids associated with resistance and virulence, respectively, and is phylogenetically related to strains within a discrete clade in the ST95 lineage (E. coli phylogroup B2) that cause meningitis and severe avian infection. The acquisition of plasmid-borne colistin resistance in this highly virulent E. coli lineage is of major concern to global health.
RESULTS
Identification and characterization of an mcr-1-positive E. coli strain.
A single E. coli strain (0.37% of a total of 267 E. coli strains) from a diverse collection of Gram-negative bacterial pathogens in the Gulf Cooperation Council states of the Middle East was identified to possess the mcr-1 gene. The strain, designated MS8345, was nonsusceptible to colistin, polymyxin B, multiple β-lactams (ampicillin, cefazolin, ceftriaxone, ceftazidime, cefepime), and most non-β-lactams (gentamicin, tobramycin, fluoroquinolones, trimethoprim, and trimethoprim-sulfamethoxazole). MS8345 was susceptible to meropenem, nitrofurantoin, the β-lactamase cefoxitin, and to the β-lactam/β-lactamase inhibitors amoxicillin-clavulanic acid and piperacillin-tazobactam (Table 1).
TABLE 1.
MICs of antibiotics for MS8345a
| Antimicrobial | MIC (mg/liter) | Interpretation | Gene |
|---|---|---|---|
| Ampicillin | ≥32 | R | TEM-1B |
| Amoxicillin-clavulanic acid | 8 | S | NA |
| Ticarcillin-clavulanic acid | 32 | R | |
| Piperacillin-tazobactam | ≤4 | S | NA |
| Cefazolin | ≥64 | R | CTX-M-1 |
| Cefoxitin | ≤4 | S | NA |
| Ceftazidime | 4 | R | CTX-M-1 |
| Ceftriaxone | ≥64 | R | CTX-M-1 |
| Cefepime | 2 | R | CTX-M-1 |
| Colistin | 8b | R | mcr-1 |
| Meropenem | ≤0.25 | S | NA |
| Amikacin | ≤2 | S | NA |
| Gentamicin | ≥16 | R | aac(3)-IIa |
| Tobramycin | 4 | R | aac(3)-IIa |
| Ciprofloxacin | ≥4 | R | gyrA |
| Norfloxacin | 8 | R | gyrA |
| Nitrofurantoin | ≤16 | S | NA |
| Trimethoprim | ≥16 | R | drfA |
| Trimethoprim-sulfamethoxazole | ≥320 | R | drfA |
| Polymyxin B | 4b | R | mcr-1 |
R, resistant; S, susceptible; NA, not applicable.
Tested by broth microdilution; all other MICs were obtained with Vitek2.
MS8345 is closely related to a highly virulent clonal lineage associated with neonatal meningitis.
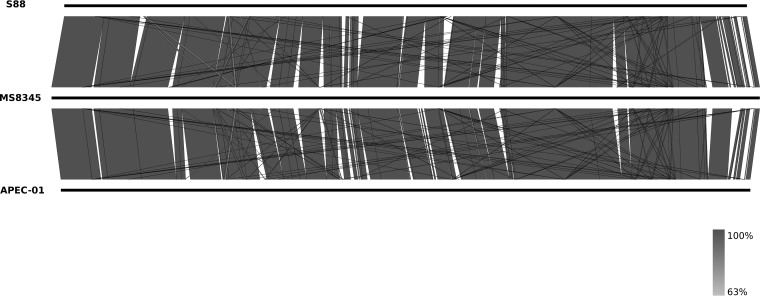
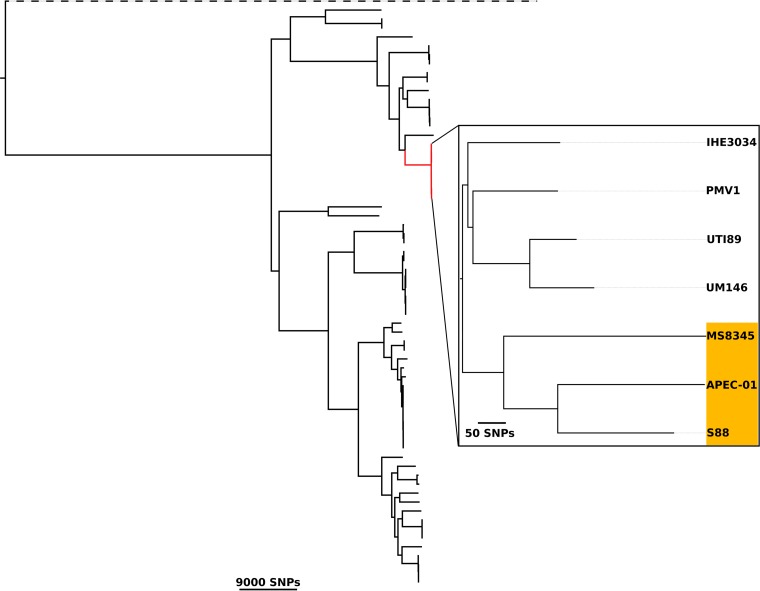
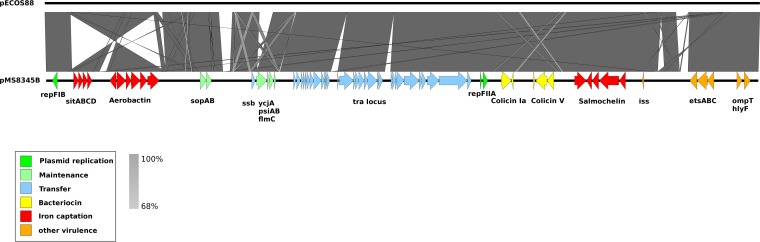
The complete genome of E. coli MS8345 comprised a single circular chromosome 5,220,996 bp in length with an average G+C content of 50.5% and two circular plasmids: a 241,164-bp multidrug resistance (MDR) plasmid (pMS8345A) containing mcr-1 and a 133,283-bp virulence plasmid (pMS8345B). In silico multilocus sequence typing (MLST) identified MS8345 as ST95. MS8345 was serotyped as O2:K1:H4 and possesses the fimH27 allele, placing it in the recently defined ST95 subgroup E (40). Pairwise genome comparisons revealed MS8345 to be highly similar to the human neonatal meningitis E. coli (NMEC) isolate S88 (GenBank accession number CU928161) and the avian pathogen APEC-O1 (GenBank accession number CP000468) (Fig. 1). Phylogenetic analysis demonstrated clustering of MS8345 with S88 and APEC-O1 in a clade discrete from the other completely sequenced ST95 strains (Fig. 2). The majority of ST95 complete genomes do not contain MDR plasmids, but several have been found to harbor a virulence plasmid (Fig. 2). Plasmid pMS8345B is highly similar to the ColV-like virulence plasmid pS88 (GenBank accession number CU928146) from E. coli S88 and carries an identical complement of virulence factors and iron uptake systems, specifically, etsABC, ompT, hlyF, the sitABCD operon, salmochelin (iroBCDEN), and aerobactin (iucABCD and iutA) (Fig. 3) (39).
FIG 1.
Pairwise whole-genome nucleotide comparison of E. coli MS8345, S88, and APEC-O1. Black bars represent the chromosome of each strain, and the gray shading represents regions of nucleotide sequence identity (63% to 100%) determined by BLASTn analysis. The figure was prepared using Easyfig (66).
FIG 2.
Phylogenetic tree of ST95 E. coli isolates. Maximum-likelihood phylogenetic tree of MS8345 and 65 additional complete E. coli chromosomes built using 111,631 core single-nucleotide polymorphisms (SNPs). The ST95 clonal complex is highlighted in red. The enlarged area displays a high-resolution maximum-likelihood phylogenetic tree of 7 ST95 isolates grouped into two discrete subclades in which MS8345 clusters with S88 and APEC-O1. Strains carrying a Col-like (ColV or ColBM) virulence plasmid are highlighted in orange. The tree was rooted using Escherichia fergusonii (dashed branch). Scale bars indicate branch lengths in numbers of SNPs. Phylogeny was visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
FIG 3.
Pairwise nucleotide comparison of the ColV-like virulence plasmids pS88 and pMS8345B. Protein-coding genes involved in replication, plasmid transfer, maintenance, and virulence are represented by colored arrows (as indicated in the key). The results of a BLASTn comparison between the two plasmids are shown with gray or yellow for (reverse-complement) shading between 68% and 100% nucleotide identities. Yellow indicates a match. Several large insertion or deletion events are evident between both plasmids as is an inversion in MS8345 that reversed the order of the sitABCD and aerobactin genes.
The mcr-1 colistin resistance gene is borne on a large MDR IncHI2 plasmid.
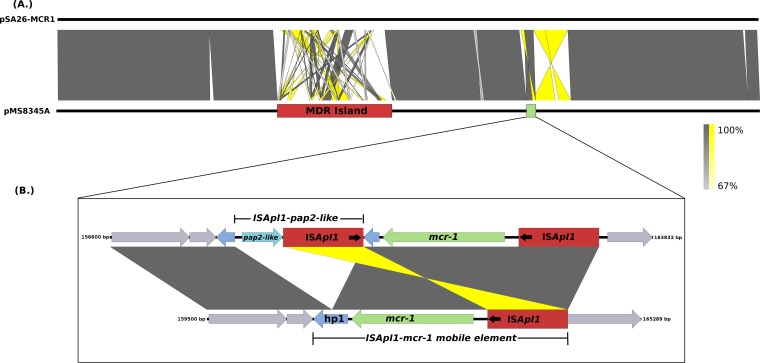
The mcr-1 gene was located on pMS89345A, a 241-kb IncHI2 plasmid sharing high sequence identity (99% nucleotide identity, 89% sequence length) with the mcr-1-positive plasmid pSA26-MCR1 (GenBank accession number KU743384). pSA26-MCR1 was identified in the carbapenem-resistant (blaNDM-1-positive) E. coli ST68 strain SA26 isolated in Saudi Arabia and is highly similar (99% nucleotide identity, 84% sequence length) to the IncHI2 mcr-1-positive plasmid pHNSHP45-2 (GenBank accession number KU341381) from the Chinese pig isolate SHP45 (41) and to mcr-1-negative plasmids carried in human, poultry, and pig Salmonella isolates from China (99% nucleotide identity, 86% sequence length) (15, 41). The major difference between pMS8345A and pSA26-MCR1 is the structure and content of a large MDR region carried on both plasmids (coordinates 74,782 to 112,611 and 73,858 to 112,218, respectively). The MDR region of pMS8345A contains eight resistance genes that are not present in the MDR region of pSA26-MCR1 and includes a single copy of blaCTX-M-1 carried on an ISEcp1 mobile element. In pMS8345A and pSA26-MCR1, mcr-1 is not a component of the MDR region, but instead, the mcr-1 mobile element ISApal1-mcr-1 (42) is inserted 46,815 bp and 47,761 bp downstream (Fig. 4A). The ISApal1-mcr-1 elements of pMS8345A and pSA26-MCR1 are identical (100% nucleotide sequence identity); however, in pSA26-MCR1, the gene encoding the hypothetical protein hp1, downstream of mcr-1, has been disrupted by insertion of a second ISApl1 element carrying a putative pap2-like phosphatase (15) (Fig. 4B). No additional mcr-type genes were identified.
FIG 4.
Pairwise nucleotide comparison of mcr-1-positive multidrug resistance plasmids pSA26-MCR1 and pMS8345. (A) A comparison of mcr-1-positive plasmids from Qatar and Saudi Arabia reveal them to have highly similar backbones. However, both plasmids differ considerably in their resistance gene complement, clearly visible as a large, highly variable MDR region characterized by numerous insertions, deletions, inversions, and rearrangements. The MDR island is represented by a red rectangle and the mcr-1 region by a green rectangle. (B) Comparison of the ISApl1-mcr-1 mobile elements from pMS8345A and pSA26-MCR1. ISApl1 mobile elements are represent by red rectangles. Black arrows indicate the orientation of the insertion sequence (IS) elements, and protein-coding regions are represented by the colored arrows.
Additional resistance genes.
The MDR island of pMS8345A carries 15 additional resistance genes and encodes three putative multidrug efflux pumps, which together provide resistance to multiple classes of antibiotics, including aminoglycosides, β-lactams, macrolides, sulfonamides, tetracycline, and trimethoprim. In addition, a further 14 resistance genes were identified on the MS8345 chromosome. Chromosomally borne resistance genes were distributed across four discrete locations and included two nearly identical copies of an ∼50-kb genomic island (GI) (53,078 bp and 52,977 bp, respectively) bearing sul1, aadA1, and erm(B). Finally, mutations in gyrA (resulting in S83L and D87N) and parC (S80I) that lead to fluoroquinolone resistance were also identified. In silico resistance profiling of MS8345 correlates with the observed phenotypic resistances reported via Vitek (Table 1). A complete list of resistance genes carried by MS8345 is reported in Table 2.
TABLE 2.
Antibiotic resistance genes identified in E. coli MS8345
| Gene | Locus tag | Requirement(s) for resistance phenotype |
Location | Coordinates | Resistance to antibiotic(s) |
|---|---|---|---|---|---|
| gyrA | MS8345_02349 | S83L D87N | Chromosome | 2417780–2420407 | Fluoroquinolones |
| sul1 | MS8345_03203 | Chromosome | 3283535–3284374 | Sulfonamide | |
| aadA1 | MS8345_03205 | Chromosome | 3284879–3285658 | Spectinomycin, streptomycin | |
| erm(B) | MS8345_03209 | Chromosome | 3288275–3289012 | Erythromycin | |
| parC | MS8345_03375 | S80I | Chromosome | 3452849–3455107 | Fluoroquinolones |
| blaTEM–1B | MS8345_04039 | Chromosome | 4136995–4137855 | Penicillin | |
| sul2 | MS8345_04043 | Chromosome | 4141072–4141887 | Sulfonamide | |
| strA | MS8345_04044 | Chromosome | 4141948–4142751 | Aminoglycosides | |
| strB | MS8345_04045 | Chromosome | 4142751–4143587 | Aminoglycosides | |
| tetR | MS8345_04057 | Chromosome | 4151677–4152354 | Tetracycline | |
| tetA | MS8345_04061 | Chromosome | 4152433–4153632 | Tetracycline | |
| dfrA1 | MS8345_04149 | Chromosome | 4244602–4245075 | Trimethoprim | |
| aadA1 | MS8345_04151 | Chromosome | 4245752–4246540 | Spectinomycin, streptomycin | |
| erm(B) | MS8345_04728 | Chromosome | 4844585–4845322 | Erythromycin | |
| aadA1 | MS8345_04732 | Chromosome | 4847939–4848718 | Spectinomycin, streptomycin | |
| sul1 | MS8345_04733 | Chromosome | 4849219–4849887 | Sulfonamide | |
| ampC | MS8345_04780 | Chromosome | 4888466–4889599 | Cephalosporins | |
| arr-2 | MS8345_A00270 | pMS8345A | 76547–76999 | Rifampin | |
| ere(A) | MS8345_A00271 | pMS8345A | 77275–78501 | Erythromycin | |
| aadA1 | MS8345_A00272 | pMS8345A | 78587–79378 | Spectinomycin, streptomycin | |
| blaCTX-M-1 | MS8345_A00277 | pMS8345A | 83441–84274 | β-Lactams | |
| aac(3)-Iia | MS8345_A00004 | pMS8345A | 87010–87870 | Aminoglycosides | |
| tmrB | MS8345_A00005 | pMS8345A | 87883–88425 | Tunicamycin | |
| strA | MS8345_A00011 | pMS8345A | 91706–92509 | Aminoglycosides | |
| strB | MS8345_A00012 | pMS8345A | 92509–93345 | Aminoglycosides | |
| blaTEM-1B | MS8345_A00015 | pMS8345A | 94194–95054 | Penicillin | |
| mph(A) | MS8345_A00018 | pMS8345A | 96389–97294 | Macrolides | |
| sul1 | MS8345_A00025 | pMS8345A | 102605–103444 | Sulfonamide | |
| dfra7 | MS8345_A00027 | pMS8345A | 104015–104716 | Trimethoprim | |
| tetX | MS8345_A00031 | pMS8345A | 107415–108551 | Tetracycline | |
| sul1 | MS8345_A00032 | pMS8345A | 108707–109546 | Sulfonamide | |
| aadA1 | MS8345_A00034 | pMS8345A | 110050–110904 | Spectinomycin, streptomycin | |
| mcr-1 | MS8345_A00099 | pMS8345A | 160818–162443 | Colistin, polymyxin B |
DISCUSSION
The recent discovery of the transmissible, plasmid-borne colistin resistance gene mcr-1 poses a significant threat to global human health. Since the mcr-1 gene was first identified in China in 2015 (13), multidrug-resistant human and animal bacterial isolates carrying the mcr-1 gene have been reported in over 25 countries throughout Asia, Europe, the Middle East, North Africa, and North America (22). Here, we describe the first report of mcr-1-mediated colistin resistance in an ExPEC strain of the pandemic ST95 complex from the Persian Gulf region.
Genomic analysis of MS8345 revealed that it is highly similar to the neonatal meningitis strain S88 and the avian pathogenic strain APEC-O1. Notably, these three strains are phylogenetically more related to one another than to other ST95 strains for which a complete genome is available. S88 is a representative of the highly virulent O45:K1:H7 clone, which accounts for one-third of all neonatal meningitis cases in France (38) and carries a ColV-like virulence plasmid (pS88) (39) almost identical to plasmid pMS8345B in MS8345. Although MS8345 was isolated from respiratory secretions and attempts to culture it from the patient’s bloodstream and cerebrospinal fluid were unsuccessful, the extraordinary similarity between these two strains, despite their different O and H types, suggests that they may be equally virulent.
A key feature of the ST95 lineage is the low frequency of MDR among clinical isolates. In England, in a survey of the nine most common uropathogenic E. coli ST clonal lineages, ST95 clinical isolates were identified as having the lowest levels of resistance (32, 33). Similar low levels of drug resistance have been identified in ST95 clinical isolates from the United States, Canada, and France (34–37). However, despite low levels of resistance, ST95 remains a significant cause of extraintestinal E. coli infections worldwide. Low levels of antibiotic resistance provide for uncomplicated treatment options, but increasing levels of resistance among the ST95 strains pose a significant risk for treatment failure. Consequently, the emergence of ST95 isolates resistant to multiple antibiotics, including β-lactams and carbapenems, is worrying (37). In this study, in silico antimicrobial resistance profiling of MS8345 revealed it to carry 31 resistance genes, an unusually high number for this clone compared to the number carried by other ST95 strains, which typically have fewer than five resistance genes and remain susceptible to most antibiotics (31–33). Multidrug resistance in MS8345 is attributed to a large complement of acquired resistance genes carried on the chromosome and on the MDR plasmid pMS8345A. High levels of resistance, combined with an extensive virulence profile, characterized MS8345 as a significant outbreak threat and a likely reservoir of plasmid-mediated mcr-1 trafficking in clinical environments and in the community, emphasizing the need for continuing surveillance.
Carriage of mcr-1 in human isolates has so far been rare, with less than 2% of clinical Enterobacteriaceae isolates in China and ≤0.2% of clinical E. coli isolates in Europe testing positive (13, 25). Low rates of mcr-1 carriage in human isolates might reflect the traditionally low levels of colistin usage in hospitals. In contrast, colistin is widely used to control diarrheal diseases in poultry and pigs (43). In 2018, the use of colistin in veterinary products is estimated to increase by ∼500% from 1992 usage levels, with China being the largest user, consuming an estimated 12,000 tonnes in 2015 (13, 22). High rates of colistin use in animal production is almost certainly a strong driver of selective pressure for colistin resistance in animal isolates. Indeed, the rate of mcr-1 carriage in animals and in animal meat products is significantly greater than carriage rates in human isolates (22). For example, in a survey of three chicken farms in Tunisia, up to 83% of birds were estimated to be mcr-1 positive (24). Notably, poultry and retail chicken meat are recognized as reservoirs of ExPEC in humans (30, 36, 44). A study of serogroup O45 ST95 ExPEC from Spain found human and avian isolates to be highly homogeneous (30), and a study of serotype O1, O2, and O18 APEC strains in China showed that APEC O1:K1 and O2:K1 strains are a major cause of colibacillosis in domestic and wild birds (35, 39, 45, 46) and can cause septicemia and meningitis in mammalian infection models (47). Here, we have shown that one of the closest relatives of MS8345 is the E. coli avian pathogen APEC-O1, highlighting the zoonotic potential of these bacteria and the potential impact of continued antibiotic misuse in animal production.
Recent reports describing mcr-1-positive plasmids from Enterobacteriaceae have shown that the ISApal1-mcr-1 element contains a putative pap2 gene located immediately downstream of mcr-1 (hypothetical protein MS8345_A200 in pMS8345A) (48, 49). The PAP2-like family of phosphatases is capable of modifying lipid A by replacing the negatively charged phosphate groups with a positively charged amine group (50). Changing the charge on LPS is a recognized strategy employed by some bacteria to increase resistance to cationic antimicrobial peptides, such as the polymyxins (51). Although MS8345_A200 does display some homology to putative PAP2 family proteins from other species (BLASTp, 86% amino acid identity, 55% query coverage), it does not possess any functional domains associated with this superfamily and at best represents a nonfunctional fragment of pap-2. However, over the course of our study, we identified an intact pap2-like gene associated with an ISApal1 element (ISApal1–pap2-like), carried on the mcr-1-positive plasmid pA26-MCR1, which has inserted into a hypothetical protein highly similar to MS8345_A200 (15). Whether this pap2-like gene (unannotated, coordinates 158363 to 158905) is functional is currently unknown; however, its potential impact on colistin sensitivity provides an intriguing avenue for further research.
In summary, we provide the first report of an mcr-1-positive isolate of the E. coli ST95 lineage. Using long-read sequencing data enabled us to resolve the complete genome sequence, including the precise context of mcr-1 on an IncHI2 plasmid and the resistance gene profile across three MDR genomic regions. The emergence of colistin resistance in this highly virulent ExPEC lineage is of serious concern to global human health.
MATERIALS AND METHODS
MCR-1 real-time PCR screening.
A total of 694 isolates, comprising Acinetobacter baumannii (n = 130), Klebsiella pneumoniae (n = 162), E. coli (n = 267), Pseudomonas aeruginosa (n = 128), Citrobacter freundii (n = 3), and Enterobacter cloacae (n = 4) isolates mainly from the Gulf Cooperation Council states of the Middle East, were tested by PCR. Isolates were prepared using a simple heat denaturation step to release nucleic acids, as previously described (52). Heat-denatured suspensions were pooled (10 isolates per pool) for PCR testing. Isolate pools were then simultaneously tested by two different real-time PCR assays: one using the QuantiTect SYBR Green PCR kit (Qiagen, Australia) as the basis for the reaction mix with previously described primers (CLR5-F and CLR5-R) (13) and the other using the QuantiTect Probe PCR kit with primers (ACAATCTCGGCTTTGTGCTGA and CGATACGATGATAACAGCGTGGT) and a TaqMan probe (FAM-TGCTCTTTGGCGCGATGCTACT-DQ, where FAM is 6-carboxyfluorescein and DQ is dark quencher) designed as part of this study. PCR assays were run simultaneously and returned identical results. All isolates from any pool providing a positive result were then tested individually. From this screening exercise, we identified a single E. coli isolate (MS8345) that contained the mcr-1 gene.
Case record.
MS8345 (also designated HZ-QTR-HMC-19) is an extended-spectrum β-lactamase (ESBL)-producing E. coli strain isolated from respiratory secretions of a 58-year-old male of Nepali origin, admitted to an intensive-care unit in Qatar with subarachnoid hemorrhage. His admission was complicated by hydrocephalus, requiring insertion of an extraventricular drain, and severe sepsis with multiorgan dysfunction following gastrointestinal perforation. However, he had no clear signs of ventilator-associated pneumonia at the time that MS8345 was isolated. During his admission, he had exposure to vancomycin, piperacillin-tazobactam, meropenem, and caspofungin but did not receive any treatment with polymyxins. He survived and was repatriated after 2 months in the hospital.
Antibiotic resistance phenotypic testing.
Susceptibility testing to 18 different antibiotics was performed using automated broth microdilution (Vitek 2 card AST-N246; bioMérieux), and the presence of ESBL production was confirmed by clavulanate synergy with ceftriaxone and ceftazidime according to EUCAST standards. MICs of colistin (Beta Pharma Co. Ltd., Shanghai, China) and polymyxin B (Beta Pharma Co. Ltd.) were determined using broth microdilution according to the CLSI guideline. Twofold dilutions of colistin and polymyxin B ranging from 0 to 128 mg/liter were made in cation-adjusted Mueller-Hinton broth (CAMHB) (Oxoid, Hampshire, UK). Bacterial suspensions were prepared by suspending colonies from nutrient agar (School of Biomedical Sciences Media Unit, Monash University, Australia) in normal saline to match a 0.5 McFarland standard (DensiCHEK; bioMérieux). The suspensions were further diluted in CAMHB to yield ∼106 CFU/ml. Antibiotic solution (100 µl) was added to 100 µl of diluted inoculum in 96-well microtiter plates (Techno Plas, St. Marys, South Australia), and MICs were determined at 20 h after incubation at 37°C. Quality control using P. aeruginosa ATCC 27853 was included as recommended by the CLSI.
Genome sequencing and assembly.
Genomic DNA (gDNA) from MS8345 was sequenced on a PacBio RSII instrument (The Doherty Institute for Infection & Immunity, The University of Melbourne) using a single SMRT cell, a 15-kb insert library, and the P6 polymerase and C4 sequencing chemistry. De novo assembly of the raw PacBio sequencing data were done using the hierarchical genome assembly process (HGAP version 2) and quiver (53) from the SMRT Analysis software suite (version 2.3.0 [http://www.pacb.com/devnet/]) with default parameters. Following de novo assembly, the completeness of the chromosome and plasmids was visually verified using Contiguity (https://github.com/mjsull/Contiguity) (54). The complete chromosome and plasmids were then subjected to a polishing phase, during which the raw PacBio sequencing reads were mapped back onto the assembled circular contigs (BLASR [55] and quiver) to validate the assembly and resolve any remaining errors. gDNA from MS8345 was also prepared as Nextera XT libraries and sequenced on an Illumina NextSeq sequencer at the Australian Centre for Ecogenomics. The raw Illumina sequencing reads were used to resolve 1,046 single-nucleotide insertion and deletion errors associated with homopolymer tracts. Illumina reads were aligned to the complete genome of MS8345 using bwa version 0.7.12 (56), and a corrected consensus was called using Pilon version 1.18 (57).
Multilocus sequencing typing.
In silico sequencing typing was performed using MLST version 2.8 (https://github.com/tseemann/mlst) and the E. coli typing scheme available from PubMLST (https://pubmlst.org/) (58).
In silico serotyping.
Determination of O and H antigens was performed using SerotypeFinder version 1.1 (59). K antigen was determined using Kaptive (60) and an in-house database of E. coli capsule genes.
Genome annotation and comparative genomics.
In silico functional annotation of MS8345 was performed using prokka (Prokka, Prokaryotic Genome Annotation System, http://vicbioinformatics.com/). Identification of antimicrobial resistance genes was performed using ResFinder version 2.0 (61). Additional screening for antimicrobial resistance genes was performed by screening the raw Illumina reads against the ARG-ANNOT database (62) using srst2 (63), and chromosomal genes associated with antibiotic resistance were manually inspect for point mutations known to contribute to a resistance phenotype (e.g., gyrA). Chromosome and plasmid comparisons were performed using BLASTn (64), the Artemis comparison tool (65), and Easyfig (66).
Phylogeny.
MS8345 belonged to ST95. To determine the phylogenetic relationship of MS8345 to other ST95 isolates, we carried out phylogenomic analysis. Briefly, the complete genomes of MS8345, UTI89 (GenBank accession number CP000243), S88 (GenBank accession number CU928161), APEC-O1 (GenBank accession number CP000468), IHE3034 (GenBank accession number CP001969), PMV1 (GenBank accession number HG428755), UM146 (GenBank accession number CP002167), and other E. coli isolates were aligned using Parsnp (67). Recombinant regions were filtered from the alignment using Gubbins v2.1.0 (68), and core single-nucleotide polymorphisms (SNPs) were determined. A maximum-likelihood tree was estimated using RAxML (69) under the GTRGAMMA nucleotide substitution rate model.
Ethics approval.
The permission for publication was granted by the Medical Research Centre at HMC (reference number MRC/0765/2017).
Accession number(s).
The complete genome of MS8345 (chromosome and plasmids) has been deposited in GenBank under the accession numbers CP025401, CP025402, and CP025403. PacBio and Illumina sequence read data have been deposited in the Sequence Read Archive (SRA) under the accession numbers SRR6364639 and SRR6364638, respectively.
ACKNOWLEDGMENTS
Part of this work was generously supported by funds raised during the philanthropic Carder Polo Cup under the patronage and support of the Royal Brisbane and Women’s Hospital Foundation, Superbug Slayers Polo Initiative, SuperbugsFree movement, Queensland Polo Association, and Alsace Polo. This work was also supported by grants from the National Health and Medical Research Council (NHMRC) of Australia. D.L.P., D.M.W., J.L., M.A.S., and S.A.B. are supported by NHMRC Fellowships. D.M.W. and E.T. report research funding from SpeeDx Pty Ltd. D.L.P. has received honoraria for advisory board participation and speaking at events sponsored by Achaogen, Merck, and GlaxoSmithKline.
B.M.F., H.M.Z., P.N.A.H., D.L.P., and S.A.B. conceived and planned the study. B.M.F. performed the analysis with input from D.M.W., L.R., E.I., N.S., A.D., M.A.M., K.C., E.T., L.S., H.H.Y., and J.L. B.M.F. produced an initial draft of the manuscript. B.M.F., H.M.Z., P.N.A.H., M.A.S., and S.A.B. wrote the final draft of the manuscript. All authors contributed to, reviewed, and approved the final draft of the manuscript.
REFERENCES
- 1.Akajagbor DS, Wilson SL, Shere-Wolfe KD, Dakum P, Charurat ME, Gilliam BL. 2013. Higher incidence of acute kidney injury with intravenous colistimethate sodium compared with polymyxin B in critically ill patients at a tertiary care medical center. Clin Infect Dis 57:1300–1303. doi: 10.1093/cid/cit453. [DOI] [PubMed] [Google Scholar]
- 2.Gales AC, Jones RN, Sader HS. 2011. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY Antimicrobial Surveillance Program (2006–09). J Antimicrob Chemother 66:2070–2074. doi: 10.1093/jac/dkr239. [DOI] [PubMed] [Google Scholar]
- 3.Gordon NC, Wareham DW. 2010. Multidrug-resistant Acinetobacter baumannii: mechanisms of virulence and resistance. Int J Antimicrob Agents 35:219–226. doi: 10.1016/j.ijantimicag.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 4.Hachem RY, Chemaly RF, Ahmar CA, Jiang Y, Boktour MR, Rjaili GA, Bodey GP, Raad II. 2007. Colistin is effective in treatment of infections caused by multidrug-resistant Pseudomonas aeruginosa in cancer patients. Antimicrob Agents Chemother 51:1905–1911. doi: 10.1128/AAC.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zowawi HM, Harris PN, Roberts MJ, Tambyah PA, Schembri MA, Pezzani MD, Williamson DA, Paterson DL. 2015. The emerging threat of multidrug-resistant Gram-negative bacteria in urology. Nat Rev Urol 12:570–584. doi: 10.1038/nrurol.2015.199. [DOI] [PubMed] [Google Scholar]
- 6.Bradford PA, Kazmierczak KM, Biedenbach DJ, Wise MG, Hackel M, Sahm DF. 2015. Correlation of beta-lactamase production and colistin resistance among Enterobacteriaceae isolates from a global surveillance program. Antimicrob Agents Chemother 60:1385–1392. doi: 10.1128/AAC.01870-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cannatelli A, Di Pilato V, Giani T, Arena F, Ambretti S, Gaibani P, D'Andrea MM, Rossolini GM. 2014. In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob Agents Chemother 58:4399–4403. doi: 10.1128/AAC.02555-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Napier BA, Band V, Burd EM, Weiss DS. 2014. Colistin heteroresistance in Enterobacter cloacae is associated with cross-resistance to the host antimicrobial lysozyme. Antimicrob Agents Chemother 58:5594–5597. doi: 10.1128/AAC.02432-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonnevend A, Ghazawi A, Hashmey R, Haidermota A, Girgis S, Alfaresi M, Omar M, Paterson DL, Zowawi HM, Pal T. 2017. Multihospital occurrence of pan-resistant Klebsiella pneumoniae sequence type 147 with an ISEcp1-directed blaOXA-181 insertion in the mgrB gene in the United Arab Emirates. Antimicrob Agents Chemother 61:e00418-17. doi: 10.1128/AAC.00418-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zowawi HM, Forde BM, Alfaresi M, Alzarouni A, Farahat Y, Chong TM, Yin WF, Chan KG, Li J, Schembri MA, Beatson SA, Paterson DL. 2015. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci Rep 5:15082. doi: 10.1038/srep15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayol A, Nordmann P, Brink A, Poirel L. 2015. Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob Agents Chemother 59:2780–2784. doi: 10.1128/AAC.05055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Turkoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 14.Wanty C, Anandan A, Piek S, Walshe J, Ganguly J, Carlson RW, Stubbs KA, Kahler CM, Vrielink A. 2013. The structure of the neisserial lipooligosaccharide phosphoethanolamine transferase A (LptA) required for resistance to polymyxin. J Mol Biol 425:3389–3402. doi: 10.1016/j.jmb.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Sonnevend A, Ghazawi A, Alqahtani M, Shibl A, Jamal W, Hashmey R, Pal T. 2016. Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int J Infect Dis 50:85–90. doi: 10.1016/j.ijid.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Webb HE, Granier SA, Marault M, Millemann Y, den Bakker HC, Nightingale KK, Bugarel M, Ison SA, Scott HM, Loneragan GH. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:144–145. doi: 10.1016/S1473-3099(15)00538-1. [DOI] [PubMed] [Google Scholar]
- 17.Zhang R, Huang Y, Chan EW, Zhou H, Chen S. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:291–292. doi: 10.1016/S1473-3099(16)00062-1. [DOI] [PubMed] [Google Scholar]
- 18.Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. 2016. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis 16:281. doi: 10.1016/S1473-3099(16)00006-2. [DOI] [PubMed] [Google Scholar]
- 19.Du H, Chen L, Tang YW, Kreiswirth BN. 2016. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis 16:287–288. doi: 10.1016/S1473-3099(16)00056-6. [DOI] [PubMed] [Google Scholar]
- 20.Yao X, Doi Y, Zeng L, Lv L, Liu J-H. 2016. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis 16:288–289. doi: 10.1016/S1473-3099(16)00057-8. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Seward CH, Wu Z, Ye H, Feng Y. 2016. Genomic insights into the ESBL and MCR-1-producing ST648 Escherichia coli with multi-drug resistance. Sci Bull (Beijing) 61:875–878. doi: 10.1007/s11434-016-1086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skov RL, Monnet DL. 2016. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill 21:30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155. [DOI] [PubMed] [Google Scholar]
- 23.Shen Z, Wang Y, Shen Y, Shen J, Wu C. 2016. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis 16:293. doi: 10.1016/S1473-3099(16)00061-X. [DOI] [PubMed] [Google Scholar]
- 24.Grami R, Mansour W, Mehri W, Bouallegue O, Boujaafar N, Madec JY, Haenni M. 2016. Impact of food animal trade on the spread of mcr-1-mediated colistin resistance, Tunisia, July 2015. Euro Surveill 21:30144. doi: 10.2807/1560-7917.ES.2016.21.8.30144. [DOI] [PubMed] [Google Scholar]
- 25.Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agerso Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM, Skov RL. 2015. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 20:30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085. [DOI] [PubMed] [Google Scholar]
- 26.Ewers C, Li G, Wilking H, Kiessling S, Alt K, Antao EM, Laturnus C, Diehl I, Glodde S, Homeier T, Bohnke U, Steinruck H, Philipp HC, Wieler LH. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int J Med Microbiol 297:163–176. doi: 10.1016/j.ijmm.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Mora A, Lopez C, Dabhi G, Blanco M, Blanco JE, Alonso MP, Herrera A, Mamani R, Bonacorsi S, Moulin-Schouleur M, Blanco J. 2009. Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiol 9:132. doi: 10.1186/1471-2180-9-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moulin-Schouleur M, Reperant M, Laurent S, Bree A, Mignon-Grasteau S, Germon P, Rasschaert D, Schouler C. 2007. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J Clin Microbiol 45:3366–3376. doi: 10.1128/JCM.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergeron CR, Prussing C, Boerlin P, Daignault D, Dutil L, Reid-Smith RJ, Zhanel GG, Manges AR. 2012. Chicken as reservoir for extraintestinal pathogenic Escherichia coli in humans, Canada. Emerg Infect Dis 18:415–421. doi: 10.3201/eid1803.111099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mora A, Viso S, Lopez C, Alonso MP, Garcia-Garrote F, Dabhi G, Mamani R, Herrera A, Marzoa J, Blanco M, Blanco JE, Moulin-Schouleur M, Schouler C, Blanco J. 2013. Poultry as reservoir for extraintestinal pathogenic Escherichia coli O45:K1:H7-B2-ST95 in humans. Vet Microbiol 167:506–512. doi: 10.1016/j.vetmic.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Gomi R, Matsuda T, Matsumura Y, Yamamoto M, Tanaka M, Ichiyama S, Yoneda M. 2016. Whole-genome analysis of antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in river water. Appl Environ Microbiol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibreel TM, Dodgson AR, Cheesbrough J, Fox AJ, Bolton FJ, Upton M. 2012. Population structure, virulence potential and antibiotic susceptibility of uropathogenic Escherichia coli from Northwest England. J Antimicrob Chemother 67:346–356. doi: 10.1093/jac/dkr451. [DOI] [PubMed] [Google Scholar]
- 33.Lau SH, Reddy S, Cheesbrough J, Bolton FJ, Willshaw G, Cheasty T, Fox AJ, Upton M. 2008. Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J Clin Microbiol 46:1076–1080. doi: 10.1128/JCM.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manges AR, Tabor H, Tellis P, Vincent C, Tellier PP. 2008. Endemic and epidemic lineages of Escherichia coli that cause urinary tract infections. Emerg Infect Dis 14:1575–1583. doi: 10.3201/eid1410.080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bert F, Johnson JR, Ouattara B, Leflon-Guibout V, Johnston B, Marcon E, Valla D, Moreau R, Nicolas-Chanoine MH. 2010. Genetic diversity and virulence profiles of Escherichia coli isolates causing spontaneous bacterial peritonitis and bacteremia in patients with cirrhosis. J Clin Microbiol 48:2709–2714. doi: 10.1128/JCM.00516-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent C, Boerlin P, Daignault D, Dozois CM, Dutil L, Galanakis C, Reid-Smith RJ, Tellier PP, Tellis PA, Ziebell K, Manges AR. 2010. Food reservoir for Escherichia coli causing urinary tract infections. Emerg Infect Dis 16:88–95. doi: 10.3201/eid1601.091118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams-Sapper S, Diep BA, Perdreau-Remington F, Riley LW. 2013. Clonal composition and community clustering of drug-susceptible and -resistant Escherichia coli isolates from bloodstream infections. Antimicrob Agents Chemother 57:490–497. doi: 10.1128/AAC.01025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fluit AC, Terlingen AM, Andriessen L, Ikawaty R, van Mansfeld R, Top J, Cohen Stuart JW, Leverstein-van Hall MA, Boel CH. 2010. Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species. J Clin Microbiol 48:3979–3989. doi: 10.1128/JCM.01191-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peigne C, Bidet P, Mahjoub-Messai F, Plainvert C, Barbe V, Medigue C, Frapy E, Nassif X, Denamur E, Bingen E, Bonacorsi S. 2009. The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect Immun 77:2272–2284. doi: 10.1128/IAI.01333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon DM, Geyik S, Clermont O, O’Brien CL, Huang S, Abayasekara C, Rajesh A, Kennedy K, Collignon P, Pavli P, Rodriguez C, Johnston BD, Johnson JR, Decousser J-W, Denamur E. 2017. Fine-scale structure analysis shows epidemic patterns of clonal complex 95, a cosmopolitan Escherichia coli lineage responsible for extraintestinal infection. mSphere 2:e00168-17. doi: 10.1128/mSphere.00168-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhi C, Lv L, Yu L-F, Doi Y, Liu J-H. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:292–293. doi: 10.1016/S1473-3099(16)00063-3. [DOI] [PubMed] [Google Scholar]
- 42.Gao R, Hu Y, Li Z, Sun J, Wang Q, Lin J, Ye H, Liu F, Srinivas S, Li D, Zhu B, Liu YH, Tian GB, Feng Y. 2016. Dissemination and mechanism for the MCR-1 colistin resistance. PLoS Pathog 12:e1005957. doi: 10.1371/journal.ppat.1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kempf I, Fleury MA, Drider D, Bruneau M, Sanders P, Chauvin C, Madec JY, Jouy E. 2013. What do we know about resistance to colistin in Enterobacteriaceae in avian and pig production in Europe? Int J Antimicrob Agents 42:379–383. doi: 10.1016/j.ijantimicag.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Manges AR, Johnson JR. 2012. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin Infect Dis 55:712–719. doi: 10.1093/cid/cis502. [DOI] [PubMed] [Google Scholar]
- 45.Blanco JE, Blanco M, Mora A, Jansen WH, Garcia V, Vazquez ML, Blanco J. 1998. Serotypes of Escherichia coli isolated from septicaemic chickens in Galicia (northwest Spain). Vet Microbiol 61:229–235. doi: 10.1016/S0378-1135(98)00182-5. [DOI] [PubMed] [Google Scholar]
- 46.Mulvey MA, Schilling JD, Hultgren SJ. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun 69:4572–4579. doi: 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Ge X, Jiang J, Pan Z, Hu L, Wang S, Wang H, Leung FC, Dai J, Fan H. 2014. Comparative genomic analysis shows that avian pathogenic Escherichia coli isolate IMT5155 (O2:K1:H5; ST complex 95, ST140) shares close relationship with ST95 APEC O1:K1 and human ExPEC O18:K1 strains. PLoS One 9:e112048. doi: 10.1371/journal.pone.0112048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li A, Yang Y, Miao M, Chavda KD, Mediavilla JR, Xie X, Feng P, Tang YW, Kreiswirth BN, Chen L, Du H. 2016. Complete sequences of mcr-1-harboring plasmids from extended-spectrum-beta-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 60:4351–4354. doi: 10.1128/AAC.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang R, van Dorp L, Shaw LP, Bradley P, Wang Q, Wang X, Jin L, Zhang Q, Liu Y, Rieux A, Dorai-Schneiders T, Weinert LA, Iqbal Z, Didelot X, Wang H, Balloux F. 2018. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun 9:1179. doi: 10.1038/s41467-018-03205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Touze T, Tran AX, Hankins JV, Mengin-Lecreulx D, Trent MS. 2008. Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol Microbiol 67:264–277. doi: 10.1111/j.1365-2958.2007.06044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raetz CR, Reynolds CM, Trent MS, Bishop RE. 2007. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem 76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zowawi HM, Sartor AL, Balkhy HH, Walsh TR, Al Johani SM, AlJindan RY, Alfaresi M, Ibrahim E, Al-Jardani A, Al-Abri S, Al Salman J, Dashti AA, Kutbi AH, Schlebusch S, Sidjabat HE, Paterson DL. 2014. Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf cooperation council: dominance of OXA-48 and NDM producers. Antimicrob Agents Chemother 58:3085–3090. doi: 10.1128/AAC.02050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 54.Sullivan MJ, Ben Zakour NL, Forde BM, Stanton-Cook M, Beatson SA. 2015. Contiguity: contig adjacency graph construction and visualisation. PeerJ Preprints 3:e1037v1. [Google Scholar]
- 55.Chaisson MJ, Tesler G. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13:238. doi: 10.1186/1471-2105-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE. 2016. Identification of Klebsiella capsule synthesis loci from whole genome data. Microbial Genomics 2 doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 65.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 66.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]