Abstract
Argonaute 2 bound mature microRNA (Ago2‐miRNA) complexes are key regulators of the wound inflammatory response and function in the translational processing of target mRNAs. In this study, we identified four wound inflammation‐related Ago2‐miRNAs (miR‐139‐5p, miR‐142‐3p, miR‐142‐5p, and miR‐223) and show that miR‐223 is critical for infection control. miR‐223 Y/− mice exhibited delayed sterile healing with prolonged neutrophil activation and interleukin‐6 expression, and markedly improved repair of Staphylococcus aureus‐infected wounds. We also showed that the expression of miR‐223 was regulated by CCAAT/enhancer binding protein alpha in human neutrophils after exposure to S. aureus peptides. Treatment with miR‐223 Y/−‐derived neutrophils, or miR‐223 antisense oligodeoxynucleotides in S. aureus‐infected wild‐type wounds markedly improved the healing of these otherwise chronic, slow healing wounds. This study reveals how miR‐223 regulates the bactericidal capacity of neutrophils at wound sites and indicates that targeting miR‐223 might be of therapeutic benefit for infected wounds in the clinic.
Keywords: inflammation, miR‐223, neutrophil, skin wound healing, Staphylococcus aureus
Subject Categories: Immunology, Skin
Introduction
After tissue damage to the adult skin, there always follows a robust recruitment of inflammatory cells, including innate immune cells, neutrophils, and macrophages, into the wounded tissues to kill and phagocytose invading microbes as well as secreting bioactive substances that aid tissue repair (Eming et al, 2017). Healing of the damaged tissue is accomplished by the concerted actions of re‐epithelialization and wound angiogenesis and by migration and subsequent contraction of fibroblasts that lay down granulation tissue by deposition of collagen and other matrix components (Eming et al, 2014). These contributing cell behaviors are thought to be partly governed by signals from the inflammatory cell influx (Eming et al, 2007). Following collagen deposition, a scar develops at the healed wound site (Eming et al, 2014). Interestingly, skin wounds of embryos and late gestation fetuses (before embryonic day 15 in mice and end of second trimester in humans) can result in almost perfect repair without scarring and these early wounds are associated with a markedly reduced wound inflammatory response (Hopkinson‐Woolley et al, 1994), suggesting that inflammation might cause skin fibrosis (Martin & Leibovich, 2005). Indeed, neonatal PU.1‐deficient (PU.1 −/−) mice, which possess no neutrophils, macrophages, mast cells, or T cells, exhibit rapid repair and scarless healing in contrast to wild‐type (WT) siblings (Martin et al, 2003; Cooper et al, 2005). We previously showed that knockdown of osteopontin, a wound upregulated inflammation‐dependent gene, by antisense oligodeoxynucleotides (AS ODNs) at wound sites reduced scarring and improved healing in vivo (Mori et al, 2008). A similar effect was observed for wounds by knockdown of the transcription factor Foxo1 (Mori et al, 2014). In general, these findings suggest that gene therapies based on dampening wound inflammatory responses (reducing inflammatory cell influx, blocking pro‐fibrotic signals from inflammatory cells once they arrive at the wound site, or enhancing the resolution of inflammation; Cash et al, 2014) might provide novel molecular therapeutic targets.
MicroRNAs (miRNAs) of the small, non‐coding RNA family are approximately 20–25 nucleotides of single‐stranded RNA that regulate translational processing of target mRNAs (Winter et al, 2009). miRNAs are degraded by selective loading into an RNA‐induced silencing complex (RISC) with Argonaute (Ago) as its core. Ago family members (Ago1‐4) are ubiquitously expressed, but Ago2 is the most highly expressed and exhibits miRNA silencer activity in mice (Liu et al, 2004). Many studies have now indicated that a number of diverse miRNAs are involved with, and regulate, inflammatory responses in humans and mice (O'Connell et al, 2012). Furthermore, the specific miRNAs miR‐21 (Wang et al, 2012), miR‐130a (Pastar et al, 2012), and miR‐132 (Li et al, 2015) contribute to the healing of skin wounds. miRNA profiling using next‐generation sequencing (NGS) has already proved successful in identifying novel miRNAs from several tissues and organisms (Tam et al, 2014; Ma et al, 2015). However, methods to purify Ago2‐miRNA complexes from wound sites and miRNA library construction to perform NGS from these tissues have not previously been reported.
miR‐223 Y/− mice have an expanded granulocytic compartment resulting from a cell‐autonomous increase in the number of granulocyte progenitors (Johnnidis et al, 2008). Moreover, miR‐223 is overexpressed in rheumatoid arthritis patients (Fulci et al, 2010). Together, these studies suggest that inflammation‐related miRNAs, particularly miR‐223, might be key regulators of the inflammatory response and/or its subsequent resolution during skin tissue repair; however, the pathways involved have not been comprehensively characterized.
In this study, we developed a unique purification system to isolate functional miRNA‐Ago2 complexes from wounded skin tissues by immunoprecipitation (IP) using an anti‐Ago2 antibody (Ab), followed by the construction of libraries to perform NGS and identify candidate wound inflammation‐related miRNAs. Using this approach, we identified several inflammation‐dependent miRNAs including miR‐139‐5p, miR‐142‐3p, miR‐142‐5p, and miR‐223. Of these, miR‐223 was the most highly expressed in wound sites during the inflammatory phase and so the present study focused on the molecular mechanisms of miR‐223 in skin wound healing. Our wound healing studies showed the delayed repair of aseptic wounds in miR‐223 Y/− mice was associated with enhanced neutrophil activation and interleukin‐6 (Il6) expression. However, if wounds were infected with Staphylococcus aureus, then miR‐223 Y/− mice showed considerably enhanced repair compared with WT infected wounds, and either transplanting miR‐223 Y/− neutrophils or the delivery of miR‐223 AS ODNs to infected WT wounds rescued the impaired wound healing phenotype. The expression level of miR‐223 in human neutrophils was regulated by CCAAT/enhancer binding protein alpha (C/EBPα) after exposure to S. aureus peptides. Thus, miR‐223 is a potential therapeutic target for the treatment of skin wounds infected with S. aureus.
Results
Over 300 Ago2‐miRNAs are expressed during skin wound healing
To comprehensively identify wound‐induced mature miRNAs involved in skin wound healing, a 4‐mm‐diameter wound was made in WT mouse dorsal skin, and 6‐mm‐diameter unwounded and wound sites were harvested on day 1, 3, 7, 10, and 14 after injury as described previously (Mori et al, 2008, 2014). Because mature Ago2‐miRNA complexes bind to target mRNAs and inhibit their translation, we isolated Ago2‐miRNA complexes rather than total miRNAs. We purified Ago2‐miRNA complexes from skin wound tissues by IP with Ago2 Ab, followed by library generation and NGS with the Illumina platform (Appendix Fig S1). Known murine miRNA sequencing reads accounted for 76.6–91.0% of all sequence reads in our small RNA libraries that were highly enriched for mature miRNA sequences (Appendix Table S1). We identified over 300 known murine miRNA categories expressed during skin wound healing (Dataset EV1), demonstrating the high efficiency of our NGS procedure for identifying functional miRNAs from skin wound tissues.
miR‐139‐5p, miR‐142 family members, and miR‐223 are wound inflammation‐related miRNAs
As a partial filter to identify inflammation‐related miRNAs in skin wound healing, miRNAs in the inflammatory phase (day 1 after injury) were arranged by rank of upregulation and compared with intact skin (Appendix Table S2). miRNA expression levels were confirmed by qPCR (Fig 1). miR‐147, miR‐223 (miR‐223‐3p), miR‐129‐3p, miR‐129‐5p, miR‐139‐5p, miR‐21* (miR‐21‐3p), miR‐340‐5p, miR‐142‐3p, and miR‐142‐5p expressions at wound sites on day 1 after injury were significantly increased compared with intact skin.
Figure 1. Identification and expression of nine candidate inflammation‐related miRNAs in skin wound healing.
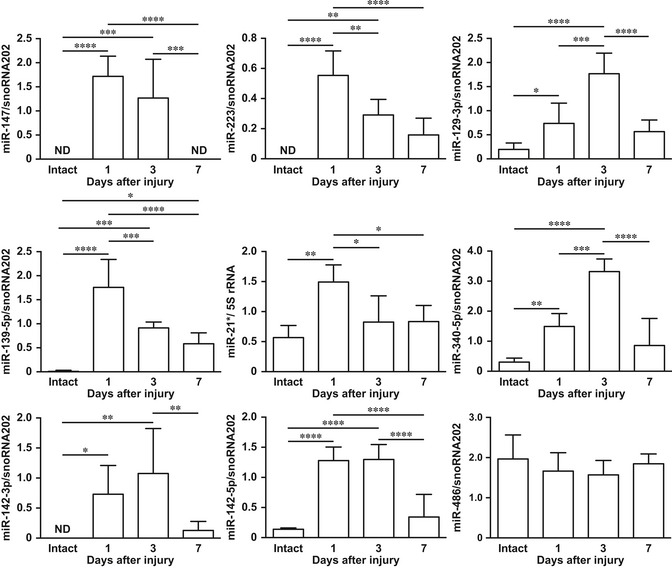
Temporal expression of murine miR‐147, miR‐223, miR‐129‐3p, miR‐139‐5p, miR‐21*, miR‐340‐5p, miR‐142‐3p, miR‐142‐5p, and miR‐486 in skin wound healing measured by qPCR relative to snoRNA202 or 5S rRNA (n = 4–6). All values represent the mean ± SD. Tukey's multiple comparison tests were used to generate the P‐values indicated in the Figure. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
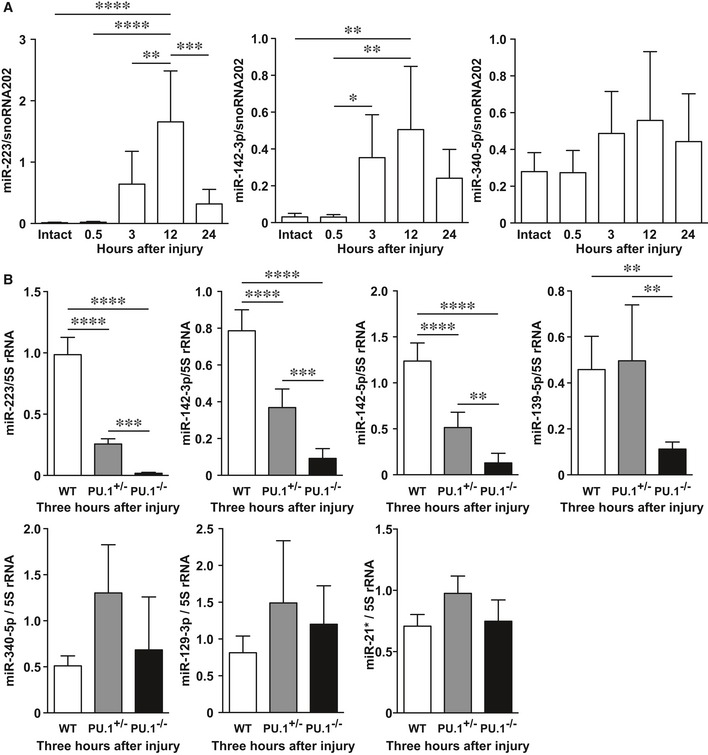
In WT neonatal mice, miR‐223 and miR‐142‐3p expression levels were markedly increased, peaking at 12 h after injury compared with intact skin, and were subsequently significantly decreased by 24 h (Fig 2A) indicating a temporal association with the inflammatory phase of healing. To confirm that these were inflammation‐related miRNAs expressed during skin wound healing, we made a 1‐cm incisional wound in the dorsal skin of WT neonatal mice and compared miR expression with heterozygous PU.1‐deficient (PU.1 +/−) and PU.1 −/− mice. qPCR analysis showed miR‐223, miR‐142‐3p, miR‐142‐5p, and miR‐139‐5p expression levels at wound sites in PU.1 −/− mice were significantly decreased compared with WT mice (Fig 2B), indicating that these four miRs are inflammation‐related miRNAs expressed during skin wound healing.
Figure 2. miR‐139‐5p, miR‐142‐3p, miR‐142‐5p, and miR‐223 are independently expressed by wound inflammation.
-
ATemporal expression of miR‐223, miR‐142‐3p, and miR‐340‐5p in skin wound healing of neonate mice using qPCR (n = 6).
-
BqPCR analysis indicates the expression levels of inflammation‐related miRNAs at 3 h in wound sites of WT (n = 5), PU.1 +/− (n = 6), and PU.1 −/− (n = 6) neonatal mice.
Skin wound healing is delayed in miR‐223 Y/− mice
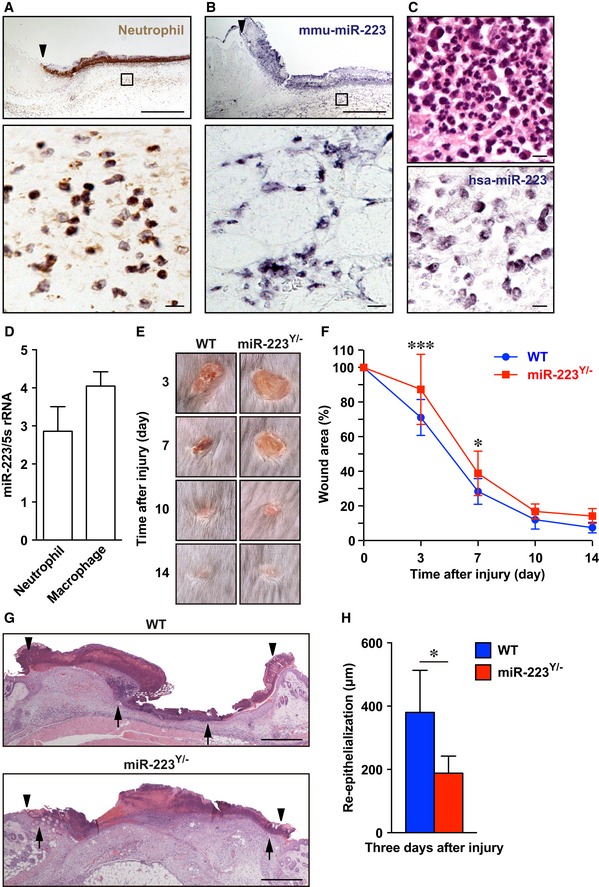
Of all inflammation‐related miRNAs, miR‐223 was the most highly expressed during the inflammatory phase at wound sites and so the present study focused on the molecular mechanisms of miR‐223 in skin wound healing (Appendix Fig S2). To determine which cells expressed miR‐223 during skin wound healing, we performed immunohistochemistry (IHC) and in situ hybridization (ISH). Wound‐infiltrated neutrophils at day 1 in the wound site of WT mice predominantly expressed miR‐223 (Fig 3A and B), and this was similar in human skin‐inflamed sites (Fig 3C). We used qPCR to confirm that murine wound‐infiltrating neutrophils (1 day after wounding) and macrophages (3 days after wounding) isolated by immunoaffinity selection with anti‐Ly‐6G (neutrophil marker) and anti‐CD11b (macrophage marker) Abs (Tanaka et al, 2017) expressed miR‐223 at the wound sites (Fig 3D). Thus, miR‐223 is predominantly expressed in neutrophils and macrophages at skin wound sites.
Figure 3. miR‐223 Y/− mice show delayed aseptic skin wound healing.
-
AIHC of neutrophils (Ly‐6G and Ly‐6C) at day 1 wound sites. Arrowhead indicates the wound margin. Area indicated with a rectangle is shown at a higher magnification below. Scale bars: 50 μm (bottom) and 500 μm (top).
-
BISH of Mus musculus (mmu) miR‐223 showing that wound‐infiltrated neutrophils were predominantly present in the wound sites of adult WT mice at day 1 after injury. Arrowhead indicates the wound margin. Area indicated with a rectangle is shown at a higher magnification below. Scale bars: 50 μm (bottom) and 500 μm (top).
-
CNeutrophils express Homo sapiens (hsa) miR‐223 in human skin‐inflamed sites. Representative H&E staining (top) and ISH of miR‐223 (bottom) showing that miR‐223‐expressing neutrophils were predominantly present in patient skin‐inflamed sites (n = 15, Appendix Table S3). Scale bars: 50 μm.
-
DqPCR analysis shows miR‐223 was expressed by wound‐infiltrating neutrophils (day 1) and macrophages (day 3) (n = 3).
-
ERepresentative photo images of the gross appearance of excisional wounds in WT (left) and miR‐223 Y/− (right) mice.
-
FProportion of the wound area remaining open relative to initial wound area at each time point in WT (n = 14) and miR‐223 Y/− (n = 13) mice.
-
GH&E staining of re‐epithelialization at day 3 after injury (wound margin (arrowheads) and the leading edge of epithelia (arrows)). Scale bar: 500 μm.
-
HMeasurement of epithelial tongue at day 3 after injury in WT (n = 8) and miR‐223 Y/− (n = 5) mice.
To clarify the pathophysiological role of miR‐223 in skin wound healing, we made dorsal skin wounds in miR‐223 Y/− mice. Gross examination showed that wound closure was significantly delayed in miR‐223 Y/− mice compared with WT mice (Fig 3E and F). We investigated re‐epithelialization using histological analysis and observed that epithelial wound tongues in miR‐223 Y/− mice at day 3 after injury were significantly shorter (188.3 ± 24.11 μm, P = 0.0115) than in WT mice (380.1 ± 47.08 μm; Fig 3G and H) confirming that miR‐223 Y/− mice showed delayed wound re‐epithelialization.
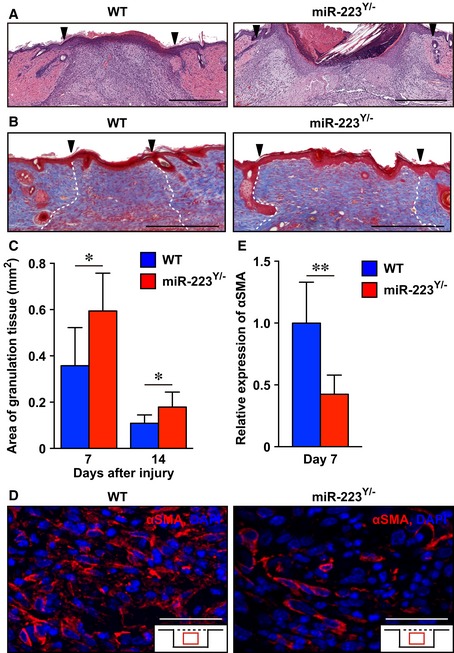
We investigated the area of granulation tissue at day 7 and day 14 at wound sites in miR‐223 Y/− mice and found them to be significantly increased (day 7; 0.60 ± 0.16 mm2, day 14; 0.18 ± 0.064 mm2) compared with WT mice (day 7; 0.36 ± 0.16 mm2, day 14; 0.11 ± 0.036 mm2; Fig EV1A–C). We examined the localization and expression level of α‐smooth muscle actin (αSMA), a marker of contractile myofibroblasts (Gabbiani et al, 1971), using IHC. The expression of αSMA at day 7 in granulation tissues of miR‐223 Y/− mice was markedly decreased (58%) compared with WT mice, even though the localization of αSMA‐expressing cells at day 7 in granulation tissues of miR‐223 Y/− mice was not altered when compared with WT mice (Fig EV1D and E). Collectively, miR‐223 Y/− mice showed delayed skin wound healing and an increased scar area.
Figure EV1. Attenuation of wound contraction and increased scar area in miR‐223 Y/− mice.
-
ARepresentative images using H&E staining of wound sites at day 7 in WT (n = 8) and miR‐223 Y/− (n = 6) mice. Scale bars: 500 μm.
-
BRepresentative images of Masson's Trichrome staining of collagen fibers (blue) in skin wound tissues of WT (n = 7) and miR‐223 Y/− (n = 8) mice at day 14 after injury. Scar sites were visualized at the mid‐point of the wound (indicated by dotted line). Nuclei are stained black, and cytosol is stained red. Scar formation is clearly recognizable by day 14 after injury. Scale bars: 500 μm.
-
CMeasurement of granulation tissue area at wound sites from WT (day 7; n = 8, day 14; n = 7) and miR‐223 Y/− (day 7; n = 6, day 14; n = 8) mice.
-
DRepresentative images of αSMA‐positive wound‐infiltrated myofibroblasts at day 7 in wound sites from WT and miR‐223 Y/− mice (n = 7). Scale bars: 50 μm.
-
EQuantification of expression of αSMA at day 7 in wound sites from WT and miR‐223 Y/− mice (n = 7).
MiR‐223 regulates neutrophils in the acute inflammatory responses at wound sites
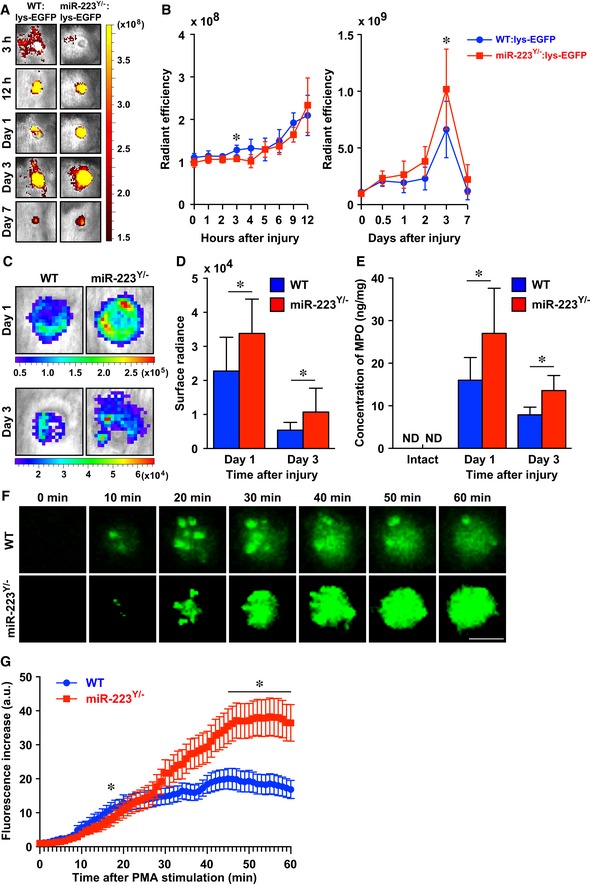
Because we found miR‐223 was expressed in neutrophils at wound sites and miR‐223 Y/− mice exhibited significantly delayed skin repair, we investigated whether miR‐223 regulates neutrophil functions in acute inflammatory responses, in ways which might impact on the recruitment of macrophages leading to a further chronic inflammatory response. To investigate how miR‐223 might regulate the duration of the inflammatory response during skin wound healing in vivo, we examined neutrophil migration into wound sites during repair using lysozyme M (lys)‐enhanced green fluorescent protein (lys‐EGFP) mice, in which most myelomonocytic cells, especially mature neutrophil granulocytes, specifically express EGFP (Faust et al, 2000). In vivo live imaging analysis using these mice allowed semi‐quantifiable evaluation of the spatiotemporal recruitment of neutrophils to skin wound sites, because EGFP fluorescence intensity is directly correlated with the number of neutrophils at wound sites (Kim et al, 2008; Tanaka et al, 2017). At 3 h, neutrophils began to accumulate at the margins of wounds in WT:lys‐EGFP mice (by measuring EGFP fluorescence intensity) with the intensity peaking at day 3 after injury (Fig 4A and B). In contrast, no migration or accumulation of neutrophils was observed in miR‐223 Y/−:lys‐EGFP mice at 3 h after injury; however, after 12 h the number of infiltrated neutrophils was greater than that of the WT neutrophil influx. At day 3, the number of neutrophils measured by the intensity of EGFP in miR‐223 Y/−:lys‐EGFP mice was significantly increased compared with WT:lys‐EGFP mice.
Figure 4. miR‐223 regulates neutrophil activation at wound sites.
-
ARepresentative in vivo fluorescent images of EGFP‐expressing neutrophils at skin wound sites. Active fluorescent imaging on color scale overlaid on a gray scale image of wounds sites.
-
BRecruitment of neutrophils at skin wound sites measured by in vivo fluorescence. All values indicate the mean radiant efficiency (photon/sec/cm2/sr)/(uW/cm2) (n = 3).
-
CRepresentative in vivo MPO activity bioluminescence imaging on color scale overlaid on a gray scale image of wound sites.
-
DMeasurements of MPO activity in vivo at wound sites of WT and miR‐223 Y/− mice. All values are the mean surface radians (photon/sec/cm2/sr) (day 1; n = 9, day 3; n = 12).
-
EMPO concentrations measured by ELISA revealed MPO levels of miR‐223 Y/− mice were significantly increased compared with WT mice (intact skin; n = 4, day 1; n = 5 (miR‐223 Y/−) or n = 8 (WT), day 3; n = 6 (miR‐223 Y/−) or n = 5 (WT)).
-
FRepresentative live in vitro fluorescence imaging of ROS production in WT and miR‐223 Y/−‐derived neutrophils (Movies EV1 and EV2). Scale bar: 10 μm.
-
GTemporal ROS production in neutrophils from WT (32 neutrophils from 3 mice, blue) and miR‐223 Y/− (36 neutrophils from 3 mice, red) mice were measured at 60 min after PMA stimulus.
We next examined the function of neutrophils in vivo as an index of myeloperoxidase (MPO) activation, a marker for neutrophil activation at wound sites (Mori et al, 2014). In vivo live imaging analysis using luminescence was used to monitor dynamic changes in neutrophil activation at skin wound sites. We found that MPO activation was markedly increased at wound sites of miR‐223 Y/− mice compared with WT mice at day 1 and day 3 after injury, which correlated with the amount of MPO at skin wound sites (Fig 4C–E).
2‐[6‐(4′‐amino)phenoxy‐3H‐xanthen‐3‐on‐9‐yl]benzoic acid (APF) is a fluorescent probe that selectively binds to reactive oxygen species (ROS) such as hydroxyl radicals, peroxynitrite, and hypochlorite produced by MPO in neutrophils (Setsukinai et al, 2003). To dissect the time‐dependent activation of neutrophils, we performed live cell imaging analysis using confocal microscopy. We observed that ROS production in peripheral blood neutrophils (PBNs) derived from miR‐223 Y/− mice was markedly delayed after incubation with phorbol myristate acetate (PMA) for 30 min (Fig 4F and G; Movies EV1 and EV2). Interestingly, miR‐223 Y/− PBNs subsequently exhibited marked hyperactivation at 60 min compared with WT PBNs (14.6 ± 2.4 in WT PBNs versus 26.1 ± 4.3 in miR‐223 Y/− PBNs, P = 0.0113), suggesting miR‐223 might be involved in neutrophil activation.
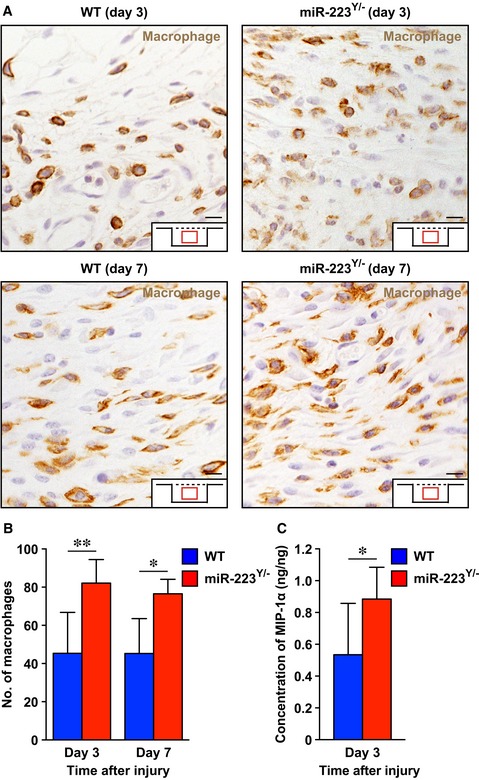
Macrophages infiltrate wound sites in the later inflammatory phase after neutrophil migration (Eming et al, 2014). IHC for F4/80, a macrophage marker (Austyn & Gordon, 1981), revealed that macrophage numbers were significantly increased in the wound sites of miR‐223 Y/− mice at day 3 and 7 after injury (Fig EV2A and B). In agreement with the altered macrophage infiltration time course, macrophage inflammatory protein (MIP)‐1α was significantly increased at day 3 after injury in the wound sites of miR‐223 Y/− mice (Fig EV2C). Collectively, our in vivo and in vitro analyses indicate that miR‐223 might regulate the acute inflammatory response at wound sites and subsequently affect macrophage infiltration into wound sites.
Figure EV2. Macrophage infiltration of wound sites in miR‐223 Y/− and WT mice.
-
ARepresentative images of F4/80‐positive macrophages at wound sites from WT (day 3; n = 6, day 7; n = 5) and miR‐223 Y/− (day 3; n = 6, day 7; n = 4) mice. Scale bars: 50 μm.
-
BQuantification of F4/80‐positive macrophages at wound sites from WT (day 3; n = 6, day 7; n = 5) and miR‐223 Y/− (day 3; n = 6, day 7; n = 4) mice.
-
CMeasurements of MIP‐1α at wound sites using ELISA (n = 6).
miR‐223 regulates Il6 expression at wound sites
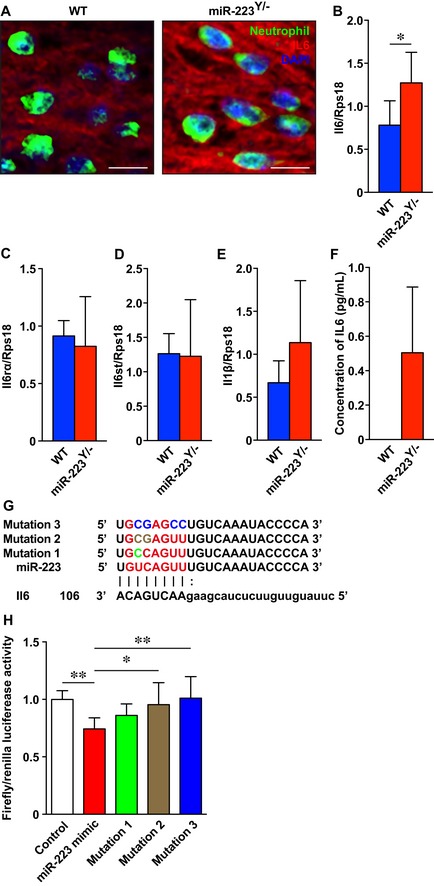
We investigated the molecular mechanisms of transcriptome regulation by miR‐223 at wound sites using NGS mRNA‐Sequencing (mRNA‐Seq) of day 1 skin wound samples from miR‐223 Y/− and WT mice (Dataset EV2). We screened potential miR‐223 target mRNAs using bioinformatics and our NGS data using a fold‐change cutoff of 1.5 (Appendix Fig S3). We identified 2 miR‐223 target mRNAs, Il6 (WT: 13.3, miR‐223 Y/−: 31.3; 2.36‐fold increase in expression level) and Piccolo (Pclo), a presynaptic cytomatrix protein (WT: 0.025, miR‐223 Y/−: 0.043, 1.71‐fold increase). Il6 is a classic proinflammatory cytokine gene, and IL‐6 protein is produced locally at human skin wound sites 30 min after injury (Grellner et al, 2000). IHC revealed that wound‐infiltrating neutrophils predominantly expressed and secreted IL‐6 in WT and miR‐223 Y/− wound sites at day 1 (Fig 5A) and was significantly overexpressed in miR‐223 Y/− wound sites at day 1 compared with WT mice (Fig 5B). However, expression levels of IL‐6 receptor‐α (Il6ra), an IL‐6 signal transducer (Il6st) that is also a candidate miR‐223 target mRNA, and the proinflammatory cytokine gene Il1b were not altered (Fig 5C–E). Interestingly, non‐stimulated bone marrow‐derived miR‐223 Y/− neutrophils, but not WT neutrophils, constitutively secreted IL‐6 in vitro (Fig 5F). To determine whether miR‐223 seed sequences dependently bind to Il6 target mRNAs in cells, we designed three miR‐223 point mutation mimics (Fig 5G). The miR‐223 mimic bound to the Il6 3′‐untranslated regions (UTRs) and was seed sequence dependent compared with the control and two miR‐223 point mutation mimics with 2 and 4 point mutations in the seed sequence, respectively (Fig 5H). These data suggest that miR‐223 might directly regulate IL‐6 translation and secretion from neutrophils at wound sites.
Figure 5. Neutrophil IL‐6 secretion is regulated by miR‐223 at wound sites.
-
AIHC for IL‐6 (red) and neutrophils (green) shows neutrophils express IL‐6 at day 1 after injury in WT and miR‐223 Y/− mice. Nuclei were counterstained with DAPI. Secreted IL‐6 was deposited around neutrophils and in wound sites. Scale bar: 10 μm.
-
B–EqPCR analysis indicates the expression levels of Il6 (B), Il6ra (C), Il6st (D), and Il1b (E) at day 1 in wound sites of WT and miR‐223 Y/− mice (n = 6).
-
FELISA reveals non‐stimulated bone marrow‐derived miR‐223 Y/− neutrophils constitutively secreted IL‐6 (n = 3). Non‐stimulated bone marrow‐derived WT neutrophils were not detected.
-
GAlignment of seven miR‐223 seed sequences and the corresponding seed sequences in Il6 mRNAs (red color and capital letters, respectively). Point mutations in each mutation mimic are shown as green, brown, and blue, respectively. The digit indicates the 3′‐UTR position.
-
HmiR‐223 binds to Il6 3′‐UTRs with high affinity. A luciferase reporter vector encoding 3′‐UTRs was co‐transfected with miR‐223 mimics into 3T3 cells. A decrease in luciferase activity indicates binding of the miR‐223 mimic to the 3′‐UTR of the target sequence (n = 6‐8).
miR‐223 Y/− neutrophils contribute to improved healing of Staphylococcus aureus‐infected wounds
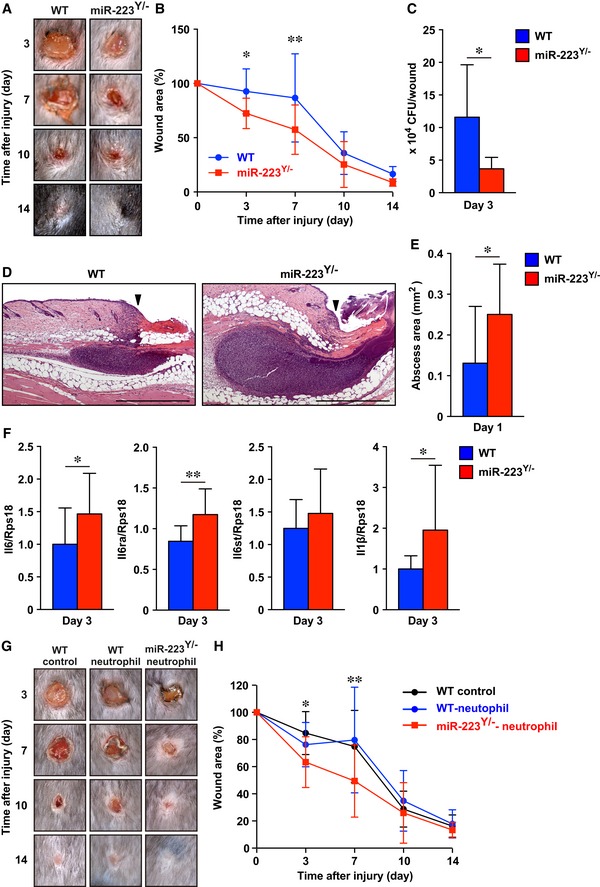
Staphylococcus aureus is the most common cause of skin infection in humans and is frequently detected in severe chronic skin wound sites in the clinic (Salgado‐Pabon & Schlievert, 2014). Long‐term S. aureus infection can lead to methicillin‐resistant S. aureus. Our studies showed that miR‐223 Y/− neutrophils were hyperactivated and because they expressed IL‐6, which was important for S. aureus clearance in a subcutaneous infection model using Il6 −/− mice (Hruz et al, 2009), we investigated whether miR‐223 Y/− neutrophils preferentially kill bacteria at wound sites. Wounds in WT and miR‐223 Y/− mice were inoculated with S. aureus (1 × 108 colony‐forming units [CFU]/10 μl), which led to impaired wound healing in WT mice; these wounds were still not healed by day 7 after injury, with overt healing only initiated at day 10 to day 14. By contrast, miR‐223 Y/− mice showed significantly enhanced healing of S. aureus‐infected wounds at day 3 and 7 compared with controls (Fig 6A and B), accompanied by enhanced re‐epithelialization, reduced total wound area and pathological postinfectious necrotic lesions without significant alteration in area of granulation tissue, and the expression of αSMA in granulation tissues at day 7, but not at day 14 (Fig EV3). However, the percentage of granulation tissue area within the total wound area of miR‐223 Y/− mice at day 7 was significantly increased (72.2% ± 23.1%, P = 0.0494) compared with WT mice (35.4% ± 33.1). The colonization of S. aureus at day 3 in the wound sites of miR‐223 Y/− mice was significantly reduced (3.67×104 ± 7149 CFU/wound) compared with WT mice (1.16 × 105 ± 32,849 CFU/wound; Fig 6C).
Figure 6. miR‐223 Y/− neutrophils improve Staphylococcus aureus‐infected skin wound healing.
-
ARepresentative photo images of the gross appearance of S. aureus‐infected wounds in WT (left) and miR‐223 Y/− (right) mice.
-
BProportion of the wound area remaining open relative to initial wound area at each time point in S. aureus‐infected WT (n = 11, blue) and miR‐223 Y/− (n = 13, red) mice.
-
CAmount of S. aureus in wound sites of miR‐223 Y/− at day 3 were significantly decreased (n = 6).
-
DRepresentative images of H&E staining of abscess formation at day 1 wound sites. Arrowheads indicate wound margin. Scale bars: 500 μm.
-
EMeasurement of abscess area of 1 day infected wounds (WT; n = 11, miR‐223 Y/−; n = 12).
-
FqPCR analysis showed Il6, Il6ra, and Il1b expressions in S. aureus‐infected wound sites of miR‐223 Y/− mice were significantly increased (WT; n = 16, miR‐223 Y/−; n = 15).
-
GRepresentative photo images of the gross appearance of non‐transplant WT control (left), WT neutrophil‐transplanted (middle), and miR‐223 Y/− neutrophil‐transplanted (right) wounds.
-
HProportion of the wound area remaining open relative to initial wound area at each time point. Note that wound closure of miR‐223 Y/− neutrophil‐transplanted sites was significantly faster than non‐transplant and WT neutrophil‐transplanted wounds (WT control n = 11, WT neutrophil n = 12, miR‐223 Y/− neutrophil n = 13).
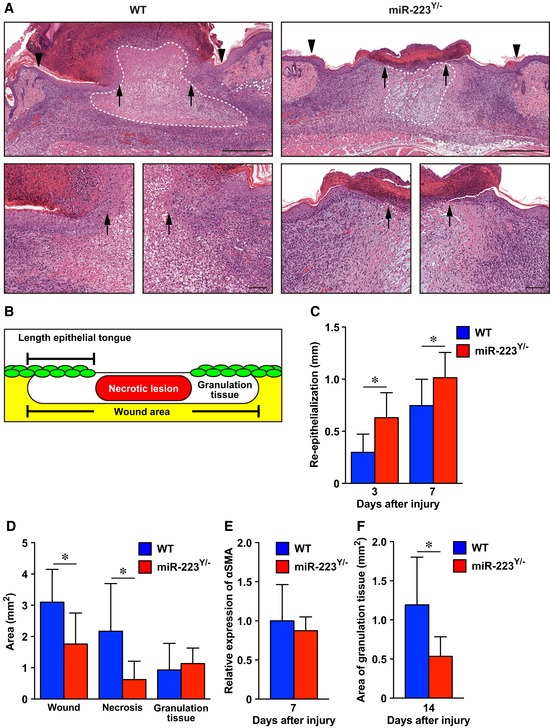
Figure EV3. miR‐223 Y/− mice show accelerated Staphylococcus aureus‐infected skin wound healing.
-
AH&E staining of re‐epithelialization at day 7 after injury (wound margin (arrowheads) and the leading edge of epithelia (arrows)). Dotted line indicates pathological necrotic lesion. Scale bars: 500 μm (top) and 100 μm (bottom).
-
BSchematic indicating the measurements derived from histological sections.
-
CMeasurement of the epithelial tongue at day 3 and day 7 after injury in WT (day 3; n = 4, day 7; n = 8) and miR‐223 Y/− (day 3; n = 5, day 7; n = 6) mice.
-
DMeasurement of the total wound area, necrotic lesion area, and granulation tissue area at day 7 after injury in WT neutrophil‐transplanted and miR‐223 Y/− neutrophil‐transplanted wounds (n = 6).
-
EQuantification of the expression of αSMA at day 7 in S. aureus‐infected wound sites from WT (n = 6) and miR‐223 Y/− mice (n = 7).
-
FMeasurements of the granulation tissue area at day 14 in WT (n = 7) or miR‐223 Y/− wound sites (n = 5).
Abscess formation is a defensive reaction of tissues to prevent the spread of infecting bacteria to other sites and previous studies showed it was indispensable for promoting healing in S. aureus‐infected wounds (Kobayashi et al, 2015). Our histological observations demonstrated a significantly increased abscess size in day 1 wounds in miR‐223 Y/− mice (0.25 ± 0.04 mm2) compared with WT mice (0.13 ± 0.04 mm2; Fig 6D and E).
We also investigated the expression levels of neutrophil‐derived Il6 and Il1b that are positively linked to S. aureus clearance (Hruz et al, 2009; Cho et al, 2012) by qPCR, and found that they were both markedly increased in S. aureus‐infected wound sites of miR‐223 Y/− mice at day 3 compared with equivalent wound sites of WT mice (Fig 6F).
Our studies described above suggest the potential beneficial effects of cell transplantation therapy using miR‐223 Y/− neutrophils engrafted in S. aureus‐infected wound sites. To test the feasibility of such a therapy, we applied bone marrow‐derived neutrophils (1 × 106 neutrophils in 50 μl of saline/wound, number of neutrophils was 1:100) to S. aureus‐infected WT skin wound sites. We confirmed that transplanted WT neutrophils remained in S. aureus‐infected skin wound sites up to 3 days after transplantation (Appendix Fig S4A). miR‐223 Y/− neutrophil‐treated skin wound sites showed significantly improved healing accompanied by a reduced total wound area and increased the expression of αSMA in granulation tissues at day 7 and a reduced area of granulation tissue at 7 and 14 days when compared with non‐treated or WT neutrophil‐treated S. aureus‐infected wound sites (Fig 6G and H; Appendix Fig S4B–E). This indicated that miR‐223 Y/− neutrophils might have therapeutic benefit for S. aureus‐infected skin wound healing.
Acute knockdown of miR‐223 using AS ODNs at Staphylococcus aureus‐infected wounds sites improves healing
Our data provide experimental evidence that blocking miR‐223 activity may improve S. aureus‐infected skin wound healing. Because molecular targeting by the delivery of AS ODN to wounds knocked down specific mRNAs at skin wound sites (Mori et al, 2006, 2008, 2014), we wondered whether a similar approach might knockdown miR‐223 expression. To test this hypothesis and investigate whether the acute knockdown of miR‐223 activity might be a feasible therapeutic strategy for improving the healing of infected wounds, we designed and optimized locked nucleic acid (LNA)‐modified miR‐223‐specific AS ODN in vitro (Fig 7A and B).
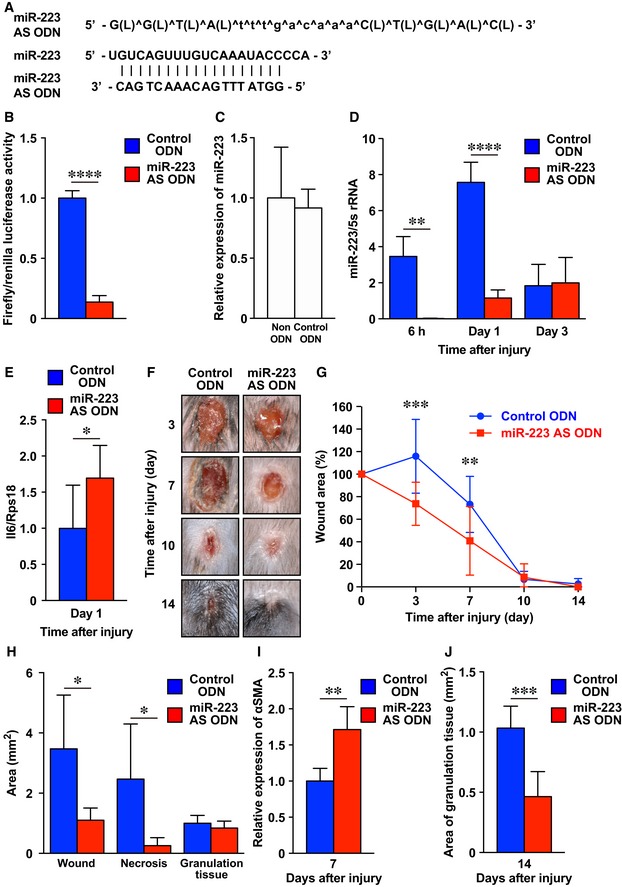
Figure 7. miR‐223 AS ODN improves Staphylococcus aureus‐infected skin wound healing.
-
AL, and ^ indicate LNA, and phosphorothioated ODN, respectively. Alignment of miR‐223 AS ODN sequences and the corresponding sequences in mature miR‐223.
-
BConfirmation of miR‐223 AS ODN binding activity in vitro. miR‐223 AS ODN binds to the mature miR‐223 region with a high affinity. A luciferase reporter vector encoding the mature miR‐223 was co‐transfected with miR‐223 AS ODN into 3T3 cells. A decrease in luciferase activity indicates binding of the control ODN to the target sequence (n = 4).
-
CExpression of miR‐223 in PB gel or control ODN‐treated S. aureus‐infected wound sites using qPCR (n = 4).
-
DTemporary expression of miR‐223 in control ODN or miR‐223 AS ODN‐treated S. aureus‐infected wound sites using qPCR (n = 4).
-
EExpression of Il6 at day 1 in control ODN or miR‐223 AS ODN‐treated S. aureus‐infected wound sites using qPCR (n = 4).
-
FRepresentative photo images of the gross appearances of control and miR‐223 AS ODN‐treated wounds at various time points after wounding (n = 7).
-
GThe proportion of the wound area remaining open relative to the initial S. aureus‐infected wound area at each time point after the injury in control ODN versus miR‐223 AS ODN‐treated wounds (n = 7).
-
HMeasurement of total wound area, necrotic lesion area, and granulation tissue area at day 7 after injury in control ODN or miR‐223 AS ODN‐treated S. aureus‐infected wound sites (n = 6).
-
IQuantification of the expression of αSMA at day 7 in control ODN (n = 5) or miR‐223 AS ODN‐treated S. aureus‐infected wound sites (n = 6).
-
JMeasurement of the area of granulation tissues at day 14 in control ODN or miR‐223 AS ODN‐treated S. aureus‐infected wound sites (n = 6)
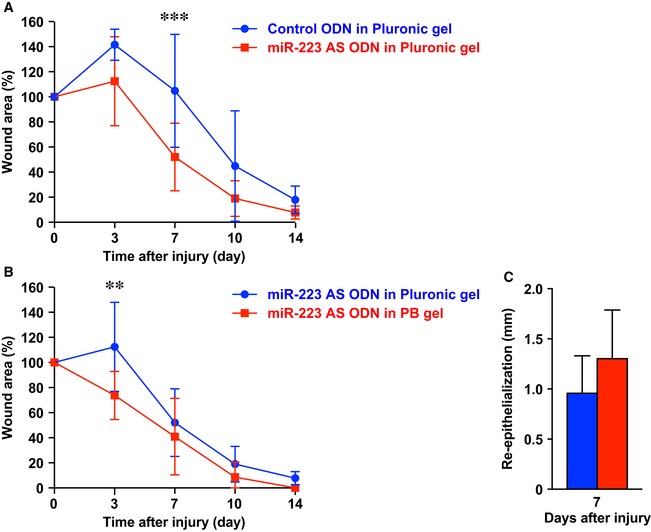
We prepared a novel poloxamer P407/P188 binary thermosensitive hydrogel (PB gel) that is highly effective for ODN delivery to tissues. The characteristics of PB gel include its liquid form at room temperature but rapid solidification at body temperature, efficient release of 18‐mer ODNs by 1 h (R. Takahashi et al, to be submitted elsewhere), and 30% increased efficiency compared with poloxamer Pluronic gel, which is currently a standard tool for ODN delivery to wound sites (Mori et al, 2006, 2008, 2014; Fig EV4A and B). We confirmed that the expression of miR‐223 in control ODN (with a sequence predicted to be non‐binding to other mRNAs and miRNAs)‐treated S. aureus‐infected skin wound sites was not altered compared with control PB gel‐treated S. aureus‐infected skin wound sites (Fig 7C). We applied miR‐223‐specific AS ODN in PB gel versus control ODN in PB gel to S. aureus‐infected skin wound sites and confirmed that the expression of miR‐223 in miR‐223 AS ODN‐treated S. aureus‐infected skin wound sites was significantly decreased (6 h, 0.022 ± 0.013; day 1, 1.16 ± 0.45) compared with control ODN‐treated S. aureus‐infected skin wound sites (6 h, 3.46 ± 1.10; day 1, 7.57 ± 1.12; Fig 7C and D). The expression of IL‐6 at day 1 in miR‐223 AS ODN‐treated S. aureus‐infected skin wound sites was significantly increased compared with control ODN‐treated S. aureus‐infected skin wound sites (Fig 7E). Macroscopic analysis of wounds at day 3 and day 7 after injury indicated that wound closure in miR‐223 AS ODN‐treated S. aureus‐wound sites was markedly accelerated (day 3; 73 ± 19%, day 7; 41 ± 30%) compared with control ODN‐treated S. aureus‐infected wounds (day 3; 115 ± 33%, day 7; 73 ± 24%; Fig 7F and G). Histological analysis indicated that the total wound area and necrotic lesions in miR‐223 AS ODN‐treated S. aureus‐infected wound sites at day 7 were markedly decreased compared with controls and were accompanied by the enhanced expression of αSMA in granulation tissues (Fig 7H and I). The percentage of granulation tissue contribution to total wound area of miR‐223 AS ODN‐treated S. aureus‐infected wound sites was significantly increased (80.9 ± 17.7%, P = 0.0017) compared with control ODN‐treated S. aureus‐infected wound sites (36.4% ± 18.6). By 14 days postwounding, the area of granulation tissue in miR‐223 AS ODN‐treated S. aureus‐infected wound sites was significantly reduced compared with controls (Fig 7J). Our data suggest that the acute downregulation of miR‐223 at wound sites using miR‐223 AS ODN accelerated skin healing in S. aureus‐infected wounds as effectively as miR‐223‐deficient neutrophil transplantation, as described earlier.
Figure EV4. miR‐223 AS ODN in PB gel‐treated Staphylococcus aureus‐infected skin wound sites are improved compared with Pluronic gel.
-
AThe proportion of the wound remaining open relative to the initial S. aureus‐infected wound area at each time point after the injury in control ODN in Pluronic gel versus miR‐223 AS ODN in Pluronic gel‐treated wounds (n = 8).
-
BThe proportion of the wound remaining open relative to the initial S. aureus‐infected wound area at each time point after injury in PB gel (n = 7) versus Pluronic gel (n = 8).
-
CMeasurement of the epithelial tongue at day 7 after injury in control ODN in PB gel and miR‐223 AS ODN in PB gel‐treated wounds (n = 6).
IL‐6 secretion from Staphylococcus aureus‐recognizing human neutrophils is regulated by C/EBPα‐mediated miR‐223 expression
To clarify the molecular mechanisms of the miR‐223/IL‐6 secretion pathway in human neutrophils after infection with S. aureus, we established a miR‐223 knockdown human promyelocytic leukemia HL‐60 cell line that could be differentiated to neutrophils upon treatment with DMSO using miR‐223 AS ODN. As expected, the expression of miR‐223 in DMSO‐treated HL‐60 cells increased as they progressed in their differentiation to mature neutrophils (Fig 8A) because miR‐223 contributed to neutrophil development (Johnnidis et al, 2008).
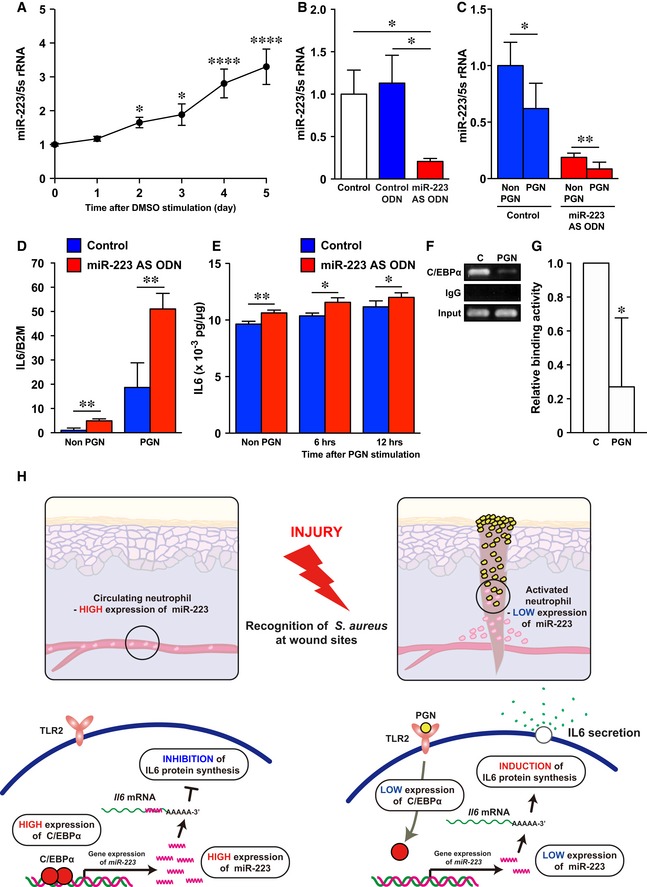
Figure 8. Human neutrophil IL‐6 secretion is regulated by the C/EBPα‐miR‐223 signaling pathway stimulated with Staphylococcus aureus PGN .
-
AqPCR analysis was used to determine the expression levels of miR‐223 in dHL‐60 (n = 3).
-
BmiR‐223 AS ODN decreased miR‐223 expression in dHL60 (control; n = 5, control ODN and miR‐223 AS ODN; n = 6).
-
CDownregulation of miR‐223 expression in dHL60 cells at 6 h after stimulation with PGN (n = 6).
-
DqPCR analysis indicates the expression levels of Il6 relative to beta‐2‐microglobulin (B2m) in non‐stimulated and PGN‐stimulated (6 h) dHL60 cells were markedly increased compared with controls (n = 3) after miR‐223 knockdown.
-
EELISA revealed non‐stimulated and PGN‐stimulated dHL60 cells constitutively secreted IL‐6 (n = 3–6).
-
FDecreased C/EBPα binding activity to the miR‐223 promotor after PGN stimulation (PGN) in dHL‐60 cells compared with non‐stimulated dHL‐60 cells (C, control) revealed by ChIP assay. Full scans for electropherogram in Fig EV5C.
-
GQuantification of anti‐C/EBPα Ab‐ChIP relative to non‐stimulated dHL‐60 cells using ChIP‐qPCR (n = 3).
-
HA model summarizing the interplay between S. aureus, C/EBPα, miR‐223, and IL‐6 secretion in neutrophils at wound sites.
Next, we stimulated the differentiation HL‐60 (dHL‐60) cells with S. aureus‐derived peptidoglycan (PGN), a ligand for Toll‐like receptor (TLR)‐2 (Takeuchi et al, 1999), and investigated the expression pattern of miR‐223 in neutrophils after the recognition of S. aureus. The expression of miR‐223 in dHL‐60 cells stimulated with PGN at 6 h was significantly deceased (38%) compared with non‐stimulated dHL‐60, to a similar degree when treated with miR‐223 AS ODN, indicating miR‐223 expression is affected by PGN even though miR‐223 was knocked down by miR‐223 AS ODN (Fig 8B and C). Furthermore, even when miR‐223 expression is strongly suppressed by miR‐223 AS ODN, if PGN stimulation (S. aureus recognition) is received, positive feedback occurs; thus, through increased IL‐6 production, infection control becomes more effective.
Il6 mRNA expression was markedly increased in non‐PGN (4.9‐fold increase) and PGN‐stimulated (2.7‐fold increase) miR‐223 AS ODN‐treated dHL‐60 cells compared with controls (Fig 8D). The concentration of IL‐6 protein was significantly increased in conditioned media from miR‐223 AS ODN‐treated dHL‐60 cells compared with controls at each time point and this corresponded to Il6 mRNA levels (Fig 8E).
Finally, to investigate how miR‐223 expression is regulated in human neutrophils infected with S. aureus, we investigated expression of transcription factors C/EBPα (Fazi et al, 2005; Fukao et al, 2007), RUNX1 (Fazi et al, 2007), and PU.1 (Fukao et al, 2007) that regulate miR‐223 expression using qPCR. The expression of CEBPA, but not RUNX1 and PU.1, in dHL‐60 was significantly decreased after 6‐h stimulation with PGN compared with non‐stimulated dHL‐60 cells (Fig EV5A). Finally, we confirmed the physical interaction between C/EBPα and the miR‐223 promotor locus in dHL‐60 cells after PGN stimulation because of the decreased levels of C/EBPα‐ miR‐223 promotor locus interaction compared with non‐stimulated dHL‐60 cells using a chromatin immunoprecipitation (ChIP)‐qPCR assay (Fig 8F and G; Fig EV5B and C).
Figure EV5. Expression of CEBPA,RINX1, and PU.1 .
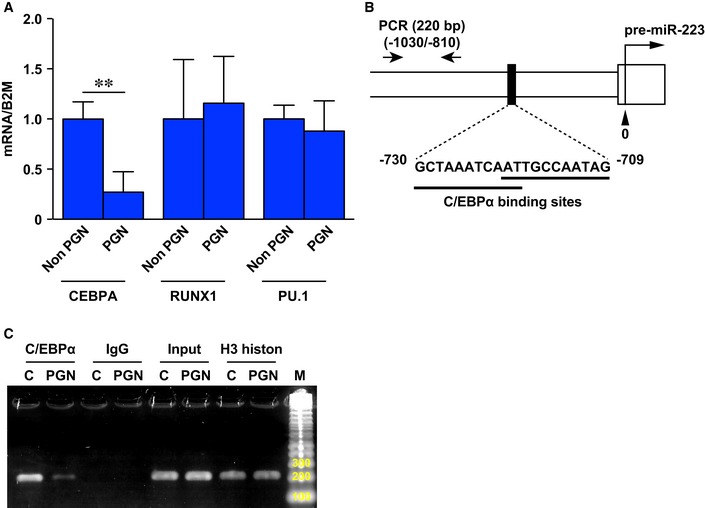
-
AGene expression of CEBPA, RINX1, and PU.1 in dHL‐60 after stimulation with PGN for 6 h measured by qPCR relative to beta‐2‐microglobulin (B2m) (n = 3).
-
BSchematic representation of the genomic composition of the pre‐miR‐223 locus and C/EBPα binding sites. ChIP‐PCR was performed with PCR primers #1 and #2. The location of primers (−1,030/−810 with respect to the 5′ end of the pre‐miR‐223) is shown in the lower panel together with sequence for the C/EBPα binding sites.
-
CRepresentative image of ChIP‐PCR production amplified by primers (220 bp) on 2% agarose gel. Anti‐C/EBPα Ab‐ChIP amplification in non‐stimulated dHL‐60 cells was markedly increased compared with 6‐h PGN‐stimulated dHL‐60 cells. Control amplification was carried out using anti‐IgG isotype control Ab‐ChIP (negative control), anti‐H3 histone Ab‐ChIP (procedure positive control), and input chromatin.
Non‐activated, mature neutrophils, such as those in the circulation, highly express miR‐223 (Fig 8H), which is induced by C/EBPα and leads to the inhibition of Il6 translation. By contrast, activated neutrophils exposed to S. aureus downregulate miR‐223 expression concomitant with decreased C/EBPα and the subsequent induction of Il6 translation.
Discussion
In the present study, we established novel Ago2‐miRNAs purification methods using NGS from skin wound tissues and identified skin wound‐induced miRNAs that might bind to target key wound mRNAs. We identified four miRNA candidates that appear to be wound inflammation‐related by comparison of WT with PU.1 −/− mouse wounds. Of these, we demonstrated that miR‐223 Y/− mice exhibited delayed sterile skin wound healing but that in S. aureus‐infected wounds, the addition of miR‐223 Y/− neutrophils, or knockdown of miR‐223 with AS ODNs significantly improved healing. The miR‐223/Il6 pathway is regulated by C/EBPα and might have a critical role in S. aureus clearance. The present data provide a novel molecular insight into the function of miR‐223 in skin wound healing and suggest the potential therapeutic targeting of this miR in conditions such as infected skin ulcers.
Previously, over 30 inflammation and immune response‐related miRNAs have been identified including miR‐142‐3p, miR‐142‐5p, and miR‐223 and our wound studies support a role for these miRNAs in aspects of the wound inflammatory response (O'Connell et al, 2012; Marques‐Rocha et al, 2015). Recently, miR‐139‐5p −/− mice were shown to develop colitis‐associated tumorigenesis (Mao et al, 2015) and thus represent a potential clinical biomarker for cancer (Zhang et al, 2015). Our findings suggest that miR‐139‐5p is also associated with inflammatory responses and tissue repair.
Previous reports have shown that miR‐142 family members and miR‐223 may play critical roles in in vivo immune homeostasis. miR‐223 regulates granulocyte development and function (Johnnidis et al, 2008), and using miR‐142 −/− mice, several groups revealed that miR‐142 regulates the hematopoietic development and function of mast cells, megakaryocytes, and lymphocytes (Mildner et al, 2013; Chapnik et al, 2014; Yamada et al, 2014; Sun et al, 2015). We further investigated the function of miR‐142 in skin wound healing by analyzing a newly‐developed miR‐142 knockout mouse and showed that this miR regulates neutrophil migration at skin wound sites via the small GTPase regulation of its actin cytoskeleton (Tanaka et al, 2017).
In the current study, we identified a role for miR‐223 in the inflammatory phase of murine skin wound healing. Furthermore, we report the miR‐223 seed sequence is critical for the regulation of IL‐6 expression and secretion. Recent studies have indicated roles for miR‐223 in numerous inflammation‐related diseases, such as tuberculosis, inflammatory bowel disease, diabetes, and cancers (Chuang et al, 2015; Liu et al, 2015). Human and mouse studies by Fukao et al (2007) suggest miR‐223 might fine‐tune the expression of numerous genes during inflammatory episodes. A recent study reported that miR‐223 does not bind to Il6 mutation sites, suggesting miR‐223 directly targets Il6 in myeloid cells (Dorhoi et al, 2013). Over twenty miR‐223 targets associated with immunity have been validated in humans and mice (Haneklaus et al, 2013). In addition, the downregulation of miR‐223 promoted Il6 and Il1b transcription in a RAW264.7 macrophage cell line (Chen et al, 2012). Thus, miR‐223 might participate in trafficking pathways including the synthesis, storage, and secretion of bioactive substances and de novo synthesis of proinflammatory cytokines through transcriptional regulation (Sheshachalam et al, 2014).
It was reported that IL6 −/− mice exhibited delayed aseptic skin wound healing (Gallucci et al, 2000; Lin et al, 2003) and recombinant murine IL‐6 improved skin wound healing in glucocorticoid‐treated mice (Gallucci et al, 2001). In contrast, excess IL‐6 causes inflammatory diseases; therefore, IL‐6 receptor antibody (Tocilizumab) has been used as therapeutic agent against Castleman disease and rheumatic diseases in the clinic (Rubbert‐Roth et al, 2018; Yoshizaki et al, 2018). Taken together, it appears that precise regulation of IL‐6 levels is necessary at inflamed sites to regulate inflammation and tissue repair.
It is well established that neutrophils kill microbes through various strategies including the release of ROS generated by NADPH‐dependent oxidase (Ellson et al, 2006). Previous studies reported that miR‐223 Y/− neutrophils exhibited increased H2O2 production and were efficient at Candida albicans killing in vitro (Johnnidis et al, 2008). miR‐223 expression was markedly increased in the inflamed pelvic sites of individuals suffering from chlamydial genital infection (Yeruva et al, 2014), indicating miR‐223‐expressing cells might be involved in many infectious diseases. We wondered whether miR‐223‐expressing cells might play a role in S. aureus‐infected wounds. Indeed, we found that S. aureus‐infected miR‐223 Y/− murine wounds healed faster than in WT mice during the early inflammation phase. Moreover, Il6 and Il1b expressions, which contribute to S. aureus clearance in skin wound sites (Hruz et al, 2009; Cho et al, 2012) and in keratitis (Hume et al, 2006), were significantly higher in miR‐223 Y/− wounds. We mirrored the efficacy of cell transplantation therapy against S. aureus‐infected skin wounds using purified miR‐223 Y/− neutrophils, by delivery of miR‐223 AS ODN knockdown of the same miRNA which similarly resulted in a better and faster healing of infected wounds in mice and has more immediate therapeutic applicability. Re‐epithelialization and granulation tissue formation at the aseptic wound sites of miR‐223 Y/− mice were markedly delayed in contrast to the S. aureus‐infected skin wound sites of miR‐223 Y/−, miR‐223 Y/− neutrophil‐transplanted, and miR‐223 AS ODN‐treated groups, which were significantly improved compared with each control. This is likely a consequence of increased acute inflammatory responses (secondary effect), because miR‐223 was not expressed in wound‐infiltrated fibroblasts or keratinocytes.
In this study, we found that activating human neutrophils by exposure to S. aureus peptide downregulated miR‐223 associated with the attenuation of C/EBPα that is required for S. aureus clearance in vivo. It has previously been reported that murine neutrophils produced IL‐6 after PGN stimulation (Strassheim et al, 2005). C/EBPα expression was downregulated by PGN and lipopolysaccharide in microglial cells, whereas Il6 and Il1b did not affect C/EBPα expression (Ejarque‐Ortiz et al, 2007). Indeed, the TLRs‐C/EBPα‐miR‐223 signaling pathway is likely to be the predominant early phase signaling pathway driving bacteria clearance, rather than cytokine signaling.
Because we showed that miR‐223 is also upregulated in human inflamed tissues, it is tempting to speculate that these therapeutic approaches may also translate to human chronic wounds. Chronic wounds affect 6.5 million patients annually, and 25 billion USD is spent in the United States to combat this (Sen et al, 2009). Infected chronic wounds frequently show a poor response to treatment and are difficult to cure; therefore, there is an enormous economic and social impact worldwide. We hope that a deeper understanding of miR‐223 molecular mechanisms may aid the development of novel drugs and cell‐based therapies to enhance the healing of chronically infected wounds.
Materials and Methods
Mice and wound model
All experiments were conducted according to the provisions of the Ethics Review Committee for Animal Experimentation at Nagasaki University. Mice were maintained in a barrier facility (temperature; 22–25°C, 12‐h light/dark cycle) under specific pathogen‐free conditions. Mice were fed ad libitum with Charles River‐LPF diet (360 kcal/100 g; 13% fat calories, 26% protein calories, and 61% carbohydrate calories [Oriental Yeast, Tokyo, Japan]). miR‐223 Y/− mice were generated as described previously (Johnnidis et al, 2008). The miR‐223 locus is located on the X chromosome and is transcribed independently of any known genes, so that miR‐223 Y/− hemizygous male mice are completely deficient in mature miR‐223 expression. WT male mice (6–12 weeks) and miR‐223 Y/− male mice (B6.Cg‐Ptprc a Mir223 tm1Fcam/J, 6–12 weeks [The Jackson Laboratory, Bar Harbor, ME, USA]) were anaesthetized and 2 or 4 full‐thickness excisional wounds (4‐mm biopsy punch; Kai Industries, Gifu, Japan) were aseptically made to the shaved dorsal skin. Generation of PU.1 −/− mice was described previously (McKercher et al, 1996). One‐day‐old pups received anesthetic and full‐thickness 1‐cm incisional wounds were performed on the dorsal skin; then, the wounds were harvested.
Faust et al (2000) generated lys‐EGFP mice as described previously. To generate male lys‐EGFP‐expressing miR‐223 Y/− mice, male miR‐223 Y/− mice were crossed with female lys‐EGFP mice to produce lys‐EGFP heterozygous‐expressing male miR‐223 Y/− and female miR‐223 +/− mice. A second cross generated male lys‐EGFP homozygous‐expressing miR‐223 Y/− mice. Mice genotypes were defined by PCR as previously described (Faust et al, 2000; Johnnidis et al, 2008).
Staphylococcus aureus type strain (NBRC 100910) was obtained from the National Institute of Technology and Evaluation (Tokyo, Japan). In the S. aureus‐infected group, mice were locally inoculated with S. aureus (1 × 108 CFU per 10 μl in saline) at pre‐skin wound sites followed by making the wound.
For wound area analysis, digital images of wound areas were measured by Photoshop CC (Adobe Systems, San Jose, CA, USA) and the mean wound area was calculated from two or four wounds from a single mouse (Mori et al, 2008, 2014; Tanaka et al, 2017).
Extraction and purification for miRNA from skin wounds
Wounds in mice were harvested with a 6‐mm biopsy punch, and miRNAs were extracted/purified using the microRNA Isolation Kit, Mouse Ago2 (Wako Pure Chemical Industries Ltd, Osaka, Japan) according to the manufacturer's instructions. Briefly, 4 wound tissues were placed in cell lysis buffer and homogenized for 5 min, incubated on ice for 15 min, and centrifuged at 14,000 g for 15 min at 4°C. Debris in supernatants was eliminated using an Ultrafree‐MC 0.45‐μm filter (Merck Millipore, Darmstadt, Germany). Filtrated supernatants were immunoprecipitated with anti‐mouse Ago2 Ab for 3 h at 4°C. Samples were washed with cell lysis buffer 3 times, and miRNAs were eluted by elusion solution. miRNAs were purified and precipitated by standard phenol/chloroform extraction and ethanol precipitation methods.
MiRNA library construction and high‐throughput whole‐miRNAs sequencing
Cloning of miRNAs was performed using a small RNA Cloning Kit (Takara Bio, Shiga, Japan) according to the manufacturer's instructions (Appendix Fig S1). Briefly, purified‐miRNAs were treated with Bacterial Alkaline Phosphatase to dephosphorylate the 3′ end of miRNAs at 37°C for 1 h, then purified and precipitated by standard phenol/chloroform extraction and ethanol precipitation methods. Dephosphorylated‐3′ ends of miRNAs were bound to biotinylated‐3′ adaptors using T4 RNA ligase at 15°C for 1 h, reacted with MAGNOTEX‐SA (Takara Bio), a streptavidin conjugated‐magnetic bead and then washed. Magnet bead‐conjugated miRNAs were phosphorylated at the miRNA 5′ end using T4 polynucleotide kinase at 37°C for 30 min, bound with 5′ adaptors using T4 RNA ligase at 15°C for 1 h, and then washed. Finally, miRNAs were transcribed to cDNA and amplified using ExTaq Hot Start Version (Takara Bio). Specific primers were as follows: (Forward) 5′‐AAAGATCCTGCAGGTGCGTCA‐3′ and (Reverse) 5′‐GTCTCTAGCCTGCAGGATCGATG‐3′. Amplification was performed for 15 cycles with annealing at 60°C for 30 s, extension at 72°C for 30 s, and a final extension for 3 min. First PCR products were amplified with the same conditions as for the 1st PCR methods. Second PCR products (approximately 65 bp) were separated by electrophoresis on a 10% TBE gel stained with SYBR Gold nucleic acid gel stain (Invitrogen, Carlsbad, CA, USA). Second PCR products were extracted from the gel and purified using a small RNA gel Extraction kit (Takara Bio) according to the manufacturer's instructions. Purified 2nd PCR products were amplified using ExTaq Hot Start Version (Takara Bio) with the same conditions as for the 1st PCR methods. Specific primers were as follows: (Forward) 5′‐AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTAAAGATCCTGCAGGTGCGTCA‐3′ and (Reverse) 5′‐CAAGCAGAAGACGGCATACGAGCTCTTCCGATCTGTCTCTAGCCTGCAGGATCGATG‐3′.
Third PCR products (approximately 156 bp) were separated by electrophoresis on a 10% TBE gel stained with SYBR Gold nucleic acid gel stain (Invitrogen). Target PCR products were extracted from gels and purified using a small RNA gel Extraction kit (Takara Bio) according to the manufacturer's instructions. Purified miRNA libraries were subjected to Solexa sequencing system (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. miRNA‐Seq data reported are available in the DNA Data Bank of Japan (DDBJ, http://trace.ddbj.nig.ac.jp/DRASearch/) under accession no. DRA004094.
Analysis of miRNA sequencing data
All individual sequence reads with base quality scores were trimmed and eliminated from ineffective sequencing in the initial data. Resulting sets of unique sequence reads were mapped onto the mouse genome with reference to Langmead et al (2009) using the UCSC Genome Bioinformatics Site. To identify sequence tags originating from known ncRNA (miRNA, other species miRNA, piRNA, rRNA, tRNA, snRNA, snoRNA, scRNA, miscRNA) and transcripts, we used miRBase (http://www.mirbase.org/index.shtml), NCBI Entrez Nucleotide database (http://www.ncbi.nlm.nih.gov/sites/entrez/query.fcgi?db=Nucleotide), and Reference Sequences (ftp://ftp.ncbi.nih.gov/), as well as Ensembl Genome Browser (ftp://ftp.ensembl.org/pub/release55/).
qPCR
Harvested tissues were homogenized by TissueLyzer II (Qiagen, Hilden, Germany), and total miRNAs and mRNAs were extracted/purified using an miRNeasy Mini Kit (Qiagen) and RNeasy Plus Universal Mini Kit (Qiagen), respectively, according to the manufacturers' instructions. Quantification of miRNAs was performed by TaqMan MicroRNA Assay (Applied Biosystems, Foster City, CA, USA) and the miRCURY LNA microRNA PCR system (Exiqon, Vedbaek, Denmark) using ABI PRISM 7900HT (Applied Biosystems). Quantification of mRNAs was performed with Thunderbird SYBR qPCR Mix (Toyobo Inc., Osaka, Japan) using ABI PRISM 7900HT (Applied Biosystems) (sequences of primers are shown in Appendix Table S6).
Histology
Harvested tissues were fixed in 4% paraformaldehyde (PFA) overnight and embedded in paraffin. All specimens were cut to 4‐μm‐thick sections and subjected to hematoxylin and eosin (H&E), ISH, and IHC (Ab information in Appendix Table S5). IHC and quantification of macrophages and αSMA expression were performed as previously described (Mori et al, 2006, 2008, 2014). Observations were made via digital whole slide scanning system (Aperio AT Turbo; Leica Microsystems, Tokyo, Japan) confocal microscopy (C2+ system; Nikon Corp., Tokyo, Japan)]. Aperio eSlide Manager (Leica Microsystems), NIS‐Elements C software version 4.13 (Nikon Corp.), AR software version 4.0 (Nikon Corp.), or IMARIS 7.7.2 (Bitplane, Zurich, Switzerland) were used for data analysis.
ISH
In situ hybridization was performed using microRNA ISH buffer set and miRCURY LNA Detection 5′‐ and 3′‐DIG‐labeled probes (Exiqon) according to the manufacturer's instructions. In brief, 4% PFA perfusion‐fixed tissues were embedded in paraffin. Six‐μm sections were deparaffinized and incubated with Proteinase K solution (Dako, Glostrup, Denmark) for 10 min at 37°C. After washing in PBS, sections were dehydrated. Hybridization was performed using 40 nM of miRNA probe in microRNA ISH buffer (Exiqon) at 55°C for 2 h. Sections were rinsed in 5× SSC at 55°C for 5 min, twice with 1× SSC at 55°C for 5 min, twice with 0.2× SSC at 55°C for 5 min, and with 0.2× SSC at room temperature for 5 min. Sections were treated with blocking solution (Nacalai Tesque Inc., Kyoto, Japan) for 1 h at room temperature and then were incubated with anti‐DIG Ab (1:800; Roche Diagnostics GmbH, Mannheim, Germany) in blocking solution (Nacalai Tesque Inc.) overnight at 4°C. Sections were developed using NTB/BCIP (Roche Diagnostics GmbH) at 30°C.
Isolation of neutrophils and macrophages from skin wounds and bone marrow
Neutrophils and macrophages were isolated with a MicroBead Kit (Miltenyi Biotech Inc., Bergisch Gladbach, Germany) according to the manufacturer's instructions. Cells were reacted with anti‐Ly‐6G Ab and anti‐CD11b Ab to isolate neutrophils and macrophages, respectively (Tanaka et al, 2017).
In vivo live imaging analysis of EGFP‐expressing neutrophil recruitment and MPO activity at wound sites
Neutrophil recruitment at skin wound sites was visualized and measured as previously described (Kim et al, 2008; Tanaka et al, 2017). In brief, WT:lys‐EGFP mice and miR‐223 Y/−:lys‐EGFP mice were wounded and fluorescence intensity was monitored with IVIS Lumina II System (PerkinElmer, Waltham, MA, USA). Fluorescence intensities were expressed as the mean radiant efficiency ((p/sec/cm2/sr)/(uW/cm2).
To visualize and measure MPO activity in vivo, mice were intraperitoneally injected with XenoLight RediJect Inflammation Probe (5 μl/g body weight; PerkinElmer) on day 1 and 3 after injury, and luminescence images were acquired at 10 min after probe injection with the IVIS Lumina II System (PerkinElmer). Luminescence intensities were expressed as the mean radiance (photons/second/cm2/steradian). Living Image software (PerkinElmer) was used to analyze the intensities of each wound.
Measurement of MPO, MIP‐1α, and IL‐6 protein concentrations
Extraction of total protein was performed as previously described (Mori et al, 2014). Briefly, harvested tissues were homogenized by TissueLyzer II (Qiagen) and T‐PER Reagent (Thermo Fisher Scientific, Waltham, MA, USA), consisting of proteinase and dephosphorylation inhibitor. Sample proteins were filtered using an Ultrafree‐MC 0.45‐μm filter (Millipore). Concentrations of MPO and MIP‐1α were measured with an MPO mouse ELISA kit (Abcam, Cambridge, UK) and a Bio‐Plex Pro mouse cytokine G1 23‐Plex panel (Bio‐Rad Laboratories, Inc. Hercules, CA, USA), respectively.
Murine neutrophils were isolated from bone marrow with a MicroBead Kit (Miltenyi Biotech Inc.) according to the manufacturer's instructions. Cells were cultured in RPMI 1640 at 37°C in 5% CO2 for 5 h. Concentrations of mouse IL‐6 in conditioned media were measured by Mouse IL‐6 Quantikine ELISA kit (R&D Systems, Minneapolis, MN, USA) according to the manufacturer's instructions.
Live imaging analysis of neutrophil ROS production in vitro
Measurement of hypochlorite production was detected using APF (Goryo Chemical Inc., Sapporo, Japan) according to the manufacturer's instructions. In brief, 1 drop of tail vein peripheral blood was added to 2 ml of live cell imaging solution (Gibco, Carlsbad, CA, USA), and stained with 10 μM of APF for 30 min at room temperature. APF‐loaded neutrophils were stimulated with 1 μl PMA, and fluorescence images were acquired every 1 min for 60 min using a confocal laser scanning unit microscope (C2+ system, Nikon Corp.) equipped with Plan Apo VC20x (0.75 NA), and the images were processed using IMARIS software (Bitplane).
RNA‐Seq
Wound tissues harvested at day 1 were homogenized by TissueLyzer II (Qiagen) followed by total RNA and polyA+ RNA purification using an RNeasy Plus Universal Mini Kit (Qiagen) and TruSeq Stranded Total RNA Library Prep Kit (Illumina), respectively. mRNA libraries were constructed with a TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) according to the manufacturer's instructions (TruSeq Stranded mRNA Sample Preparation Guide Rev.E). RNA‐Seq was performed with HiSeq 2500 (Illumina). RNA‐Seq data reported are available in the DNA Data Bank of Japan under accession no. DRA004092.
Assay for miRNA binding to the 3′‐UTR of mRNA
The 3′‐UTRs of miR‐223 targets were screened using Strand NGS software (Strand Genomics, San Francisco, CA, USA). Vectors were constructed with pmirGLO Dual‐Luciferase miRNA Target Expression Vector (Promega Corp., Madison, WI, USA) according to the manufacturer's procedures and as previously described (Tanaka et al, 2017). In brief, primers consisting of the 3′‐UTRs of predicted miR‐223 family target sequences and appropriate restriction sites were synthesized, annealed, and cloned downstream of the firefly luciferase reporter (luc2) gene in pmirGLO. Sequences were as follows: Il6 sense (78–106) 5′‐aaacCTTATGTTGTTCTCTACGAAGAACTGACAt‐3′; and Il6 antisense (106–78) 5′‐ctagaTGTCAGTTCTTCGTAGAGAACAACATAAGgttt‐3′. Capital and lowercase letters indicate the 3′‐UTR and restriction sites (PmeI and XbaI), respectively.
Double‐strand miR‐223 mimic and miR‐223 point mutation mimics were purchased from GeneDesign Inc. (Osaka, Japan). Sequences were as follows (underline indicates a mutation point): miR‐223‐3p 5′‐UGUCAGUUUGUCAAAUACCCCA‐3′; miR‐223‐3p point mutation (No. 1) 5′‐UGCCAGUUUGUCAAAUACCCCA‐3′; miR‐223‐3p point mutation (No. 2) 5′‐UGCGAGUUUGUCAAAUACCCCA‐3′; and miR‐223‐3p point mutation (No. 3) 5′‐UGCGAGCCUGUCAAAUACCCCA‐3′.
3T3 cells were co‐transfected with a miR‐223 mimic and reporter vector using Lipofectamine 3000 (Life Technologies). Luciferase activity was measured with a Dual‐Glo Luciferase Assay System (Promega Corp.) using the manufacturer's procedure.
Neutrophil transplantation
Neutrophils were isolated from bone marrow using a Neutrophil Isolation Kit (Miltenyi Biotech Inc.) according to the manufacturer's instructions. Isolated neutrophils (1 × 106 cells/50 μl in saline) were locally applied at day 1 after inoculating S. aureus to wound sites.
Generating miR‐223 AS ODN
To obtain mature miR‐223‐expressing vector, vectors were constructed with pmirGLO Dual‐Luciferase miRNA Target Expression Vector (Promega Corp) according to the manufacturer's procedures. In brief, oligos consisting of the miR‐223 sequences and appropriate restriction sites were synthesized, annealed, and cloned downstream of the luc2 gene in pmirGLO. Sequences were as follows: miR‐223 sense 5′‐aaacTGTCAGTTTGTCAAATACCCCAt‐3′; and miR‐223 antisense 5′‐ctagaTGGGGTATTTGACAAACTGACAgttt‐3′. Capital and lowercase letters indicate the mature miR‐223 and restriction sites (PmeI and XbaI), respectively.
Control ODN and miR‐223 AS ODN were originally designed and had sequences as follows: Control ODN 5′‐G(L)^5(L)^A(L)^G(L)^a^t^t^g^t^a^c^g^a^5(L)^T(L)^5(L)^A(L)^5(L)‐3′; and miR‐223 AS ODN 5′‐G(L)^G(L)^T(L)^A(L)^t^t^t^g^a^c^a^a^a^5(L)^T(L)^G(L)^A(L)^5(L)‐3′.
L, ^, and 5(L) indicate LNA, phosphorothioated ODN, and LNA‐modified 5‐methylcytosine, respectively (Fig 6A).
3T3 cells were co‐transfected with control ODN or miR‐223 AS ODN and the reporter vector using Lipofectamine 3000 (Life Technologies). Luciferase activity was measured with a Dual‐Glo Luciferase Assay System (Promega Corp.) using the manufacturer's procedure.
miR‐223 knockdown at skin wound sites using miR‐223 AS ODN
For in vivo experiments involving ODN delivery, ODNs [10 μM in 50 μl poloxamer P407/P188 (25%/4%; BASF, Ludwigshafen am Rhein, Germany) binary thermosensitive hydrogel and 30% Pluronic F‐127 gel (Sigma‐Aldrich, St. Louis, MO, USA), which acts as a slow release vehicle (Mori et al, 2006, 2008, 2014)] were topically applied at 1 day after inoculation with S. aureus.
Human skin sample
Human skin samples were harvested from Japanese patients at the time of surgery, and diagnosis was confirmed by routine pathological examination (Appendix Table S4). All experiments were conducted with the approval of the ethics committee of Nagasaki University Graduate School of Biomedical Sciences, in accordance with the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report.
Induction of neutrophilic differentiation and analysis for gene expression stimulation with S. aureus PGN
The HL‐60 cell line (RBRC‐RCB0041) was provided by RIKEN BRC through the National Bio‐Resource Project of MEXT, Japan. HL‐60 cell culture and neutrophilic differentiation were previously described (Shuto et al, 2007). Neutrophilic differentiation was induced by exposing HL‐60 cells to 1.3% DMSO for 5 day. At 3 day after exposure to 1.3% DMSO, HL‐60 cells were transfected with control or miR‐223 AS ODN using Lipofectamine 3000 (Life Technologies) in Opti‐MEM with 1.3% DMSO (Life Technologies). On 2 day after transfection, cells and conditioned media were harvested and applied to gene expression assays and ELISA (Human IL‐6 ELISA Kit, Sigma‐Aldrich, St. Louis, MO, USA).
ChIP‐qPCR assay
ChIP‐qPCR assay was performed with ChIP‐IT PBMC (Active Motif, Carlsbad, CA, USA) according to the manufacturer's instructions. Briefly, crosslinking of proteins to DNA was achieved by direct treatment of 1% formaldehyde to non‐stimulated or PGN‐stimulated dHL60 cells (1 × 107 cells) for 15 min at room temperature. Chromatin was sonicated with a Covaris M220 (Covaris Inc., Woburn, MA, USA). Sonication conditions were as follows: duty factor 5%, peak incident power 75, cycles per burst 200, water temperature 7°C, and duration 15 min. Sonicated chromatin was immunoprecipitated for 16 h at 4°C with anti‐C/EBPα Ab (GeneTex Inc., Irvine, CA, USA), anti‐Histone H3 Ab (Cell Signaling Technology Inc., Danvers, MA, USA), and anti‐IgG isotype control Ab (Appendix Table S5).
ChIP‐PCR was performed with GoTaq Master Mix and Polymerase (Promega Corp., Madison, WI, USA) according to the manufacturer's instructions. In brief, ChIP‐DNA fragments (0.25 μg/reaction) were amplified by PCR using the following primers that recognize the human C/EBPα binding miR‐223 promotor site as described (Fazi et al, 2005; Appendix Table S6): ChIP‐PCR primer (Forward) 5′‐GCCCTCTTTGTTGATGTGTC‐3′ and primer#1 (Reverse) 5′‐GGCAGCTATTAAAGTGCCCT‐3′ for the −1,030/−810 fragment with respect to the end of the pre‐miR‐223 sequence (210 bp). Annealing was performed at 55°C followed by 35 amplification cycles. PCR products were separated by electrophoresis on 2% agarose gels (Fig 8F; Fig EV5D). ChIP‐qPCR was performed with Thunderbird SYBR qPCR Mix (Toyobo Inc.) using an ABI PRISM 7900HT Sequence Detection System.
Statistical analysis
Data are shown as the means ± SD or SEM (Fig 4G only). The statistical significance of differences between means was assessed by ANOVA, followed by Tukey's test for multiple comparisons, Dunnett's multiple comparisons test, the Mann–Whitney U‐test or by an unpaired Student's t‐test followed by Welch's test when only two groups were analyzed. Multiple comparison tests were performed using two‐way ANOVA followed by Sidak multiple comparisons test or by Holm‐Sidak multiple comparisons test using GraphPad Prism software (GraphPad Software, San Diego, CA, USA) (Appendix Table S7).
Data availability
miRNA‐Seq data reported are available in the DNA Data Bank of Japan (DDBJ) under accession no. DRA004094 (http://ddbj.nig.ac.jp/DRASearch/submission?acc=DRA004094). RNA‐Seq data reported are available in the DDBJ under accession no. DRA004092 (http://ddbj.nig.ac.jp/DRASearch/submission?acc=DRA004092).
Author contributions
MK performed experiments using miR‐223 Y/− mice and wrote part of the manuscript. KT, TU, MO, SN, and ST performed qPCR and ISH analysis. SN performed ChIP assay. YM and YO performed experiments using PU.1 −/− mice. KT, SK, TN, and YS performed NGS. TK and SP managed the breeding of mice. KT and HY contributed to the generation of miR‐223 −/−:lys‐EGFP mice. KIk performed immunoblotting and ELISA analysis. RT, YK, and TH developed the PB gel. HT designed miR‐223 AS ODNs. HH and KIw prepared human samples. KT, PM, and IS advised on all experiments and edited the manuscript. RM designed the research, performed experiments, wrote, and edited the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
The paper explained.
Problem
A neutrophil influx is necessary for effective wound repair and essential for fighting off wound infection. However, an excessive inflammatory response is bad for healing. It is important to identify the regulators of wound inflammation that determine the extent and level of neutrophil activation to therapeutically target this response in sterile and infected wound scenarios.
Results
We identified a wound inflammation‐related miR, miR‐223, that appears to be a master regulator of neutrophil homeostasis and dampens activation in sterile wounds to prevent chronic inflammation. miR‐223 knockout (miR‐223 Y/−) mice exhibited impaired healing of chronic wounds, but markedly faster healing of Staphylococcus aureus‐infected wounds compared with infected wild‐type (WT) mice; moreover, cell transplantation therapy using miR‐223 Y/−‐derived neutrophils and miR‐223 antisense oligodeoxynucleotide knockdown in wounds markedly improved the healing of infected WT wounds.
Impact
Our findings suggest that targeting miR‐223 might be of therapeutic benefit to enhance the healing of infected wounds in the clinic.
For more information
miR‐139 in Mouse Genome Informatics (MGI): http://www.informatics.jax.org/marker/MGI:2676824; miR‐142 in MGI: http://www.informatics.jax.org/marker/MGI:2676827; and miR‐223 in MGI: http://www.informatics.jax.org/marker/MGI:2684360.
Supporting information
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Movie EV1
Movie EV2
Source Data for Expanded View
Review Process File
Source Data for Figure 8
Acknowledgements
We thank Drs. Thomas Graf (Gene Regulation, Stem Cells and Cancer Program, Centre for Genomic Regulation, Barcelona, Spain), Shintaro Hashimoto, and Masaki Honda (Kumamoto University, Japan) for providing lys‐EFGP mice. We appreciate comments from Dr. Eun Seong Hwang (University of Seoul, Korea) regarding the experiments. We thank Ms. Utako Kikutani for modifying the figure in Synopsis. This work was supported in part by the Japan Society for the Promotion of Science (Grant‐in‐Aid for Research Activity Start‐up, 20890258; Grants‐in‐Aid for Young Scientists (A), 21689049 and 24689069; Challenging Exploratory Research, 23650484 and 25560055 to R. M. and 26670773 to H. Y.; Grant‐in‐Aid for Scientific Research (B), 16H05493 to R. M.; Grants‐in‐Aid for Encouragement of Scientists, 15H00601 to M. d. K.; Grants‐in‐Aid for Young Scientists (B), 26861503 and 16K20361 to K. T., by The Takeda Science Foundation (R. M.), The Uehara Memorial Foundation (R. M.), The Nakatomi Foundation (R. M.), The Wellcome Trust (Senior Investigator Award 097791/Z/11/Z to P. M.), and The Royal Society (International Joint Project, R. M. and P. M.)).
EMBO Mol Med (2018) 10: e9024
See also: https://doi.org/10.15252/emmm.201809574 (September 2018)
References
- Austyn JM, Gordon S (1981) F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol 11: 805–815 [DOI] [PubMed] [Google Scholar]
- Cash JL, Bass MD, Campbell J, Barnes M, Kubes P, Martin P (2014) Resolution mediator chemerin15 reprograms the wound microenvironment to promote repair and reduce scarring. Curr Biol 24: 1406–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapnik E, Rivkin N, Mildner A, Beck G, Pasvolsky R, Metzl‐Raz E, Birger Y, Amir G, Tirosh I, Porat Z et al (2014) miR‐142 orchestrates a network of actin cytoskeleton regulators during megakaryopoiesis. eLife 3: e01964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Wang H, Liu Y, Song Y, Lai L, Han Q, Cao X, Wang Q (2012) Inducible microRNA‐223 down‐regulation promotes TLR‐triggered IL‐6 and IL‐1beta production in macrophages by targeting STAT3. PLoS One 7: e42971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JS, Guo Y, Ramos RI, Hebroni F, Plaisier SB, Xuan C, Granick JL, Matsushima H, Takashima A, Iwakura Y et al (2012) Neutrophil‐derived IL‐1beta is sufficient for abscess formation in immunity against Staphylococcus aureus in mice. PLoS Pathog 8: e1003047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TY, Wu HL, Chen CC, Gamboa GM, Layman LC, Diamond MP, Azziz R, Chen YH (2015) MicroRNA‐223 expression is upregulated in insulin resistant human adipose tissue. J Diabetes Res 2015: 943659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper L, Johnson C, Burslem F, Martin P (2005) Wound healing and inflammation genes revealed by array analysis of ‘macrophageless’ PU.1 null mice. Genome Biol 6: R5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorhoi A, Iannaccone M, Farinacci M, Fae KC, Schreiber J, Moura‐Alves P, Nouailles G, Mollenkopf HJ, Oberbeck‐Muller D, Jorg S et al (2013) MicroRNA‐223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J Clin Invest 123: 4836–4848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejarque‐Ortiz A, Tusell JM, Serratosa J, Saura J (2007) CCAAT/enhancer binding protein‐alpha is down‐regulated by toll‐like receptor agonists in microglial cells. J Neurosci Res 85: 985–993 [DOI] [PubMed] [Google Scholar]
- Ellson CD, Davidson K, Ferguson GJ, O'Connor R, Stephens LR, Hawkins PT (2006) Neutrophils from p40phox−/− mice exhibit severe defects in NADPH oxidase regulation and oxidant‐dependent bacterial killing. J Exp Med 203: 1927–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming SA, Krieg T, Davidson JM (2007) Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol 127: 514–525 [DOI] [PubMed] [Google Scholar]
- Eming SA, Martin P, Tomic‐Canic M (2014) Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med 6: 265sr266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming SA, Wynn TA, Martin P (2017) Inflammation and metabolism in tissue repair and regeneration. Science 356: 1026–1030 [DOI] [PubMed] [Google Scholar]
- Faust N, Varas F, Kelly LM, Heck S, Graf T (2000) Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood 96: 719–726 [PubMed] [Google Scholar]
- Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I (2005) A minicircuitry comprised of microRNA‐223 and transcription factors NFI‐A and C/EBPalpha regulates human granulopoiesis. Cell 123: 819–831 [DOI] [PubMed] [Google Scholar]
- Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo‐Coco F et al (2007) Epigenetic silencing of the myelopoiesis regulator microRNA‐223 by the AML1/ETO oncoprotein. Cancer Cell 12: 457–466 [DOI] [PubMed] [Google Scholar]
- Fukao T, Fukuda Y, Kiga K, Sharif J, Hino K, Enomoto Y, Kawamura A, Nakamura K, Takeuchi T, Tanabe M (2007) An evolutionarily conserved mechanism for microRNA‐223 expression revealed by microRNA gene profiling. Cell 129: 617–631 [DOI] [PubMed] [Google Scholar]
- Fulci V, Scappucci G, Sebastiani GD, Giannitti C, Franceschini D, Meloni F, Colombo T, Citarella F, Barnaba V, Minisola G et al (2010) miR‐223 is overexpressed in T‐lymphocytes of patients affected by rheumatoid arthritis. Hum Immunol 71: 206–211 [DOI] [PubMed] [Google Scholar]
- Gabbiani G, Ryan GB, Majne G (1971) Presence of modified fibroblasts in granulation tissue and their possible role in wound contraction. Experientia 27: 549–550 [DOI] [PubMed] [Google Scholar]
- Gallucci RM, Simeonova PP, Matheson JM, Kommineni C, Guriel JL, Sugawara T, Luster MI (2000) Impaired cutaneous wound healing in interleukin‐6‐deficient and immunosuppressed mice. FASEB J 14: 2525–2531 [DOI] [PubMed] [Google Scholar]
- Gallucci RM, Sugawara T, Yucesoy B, Berryann K, Simeonova PP, Matheson JM, Luster MI (2001) Interleukin‐6 treatment augments cutaneous wound healing in immunosuppressed mice. J Interferon Cytokine Res 21: 603–609 [DOI] [PubMed] [Google Scholar]
- Grellner W, Georg T, Wilske J (2000) Quantitative analysis of proinflammatory cytokines (IL‐1beta, IL‐6, TNF‐alpha) in human skin wounds. Forensic Sci Int 113: 251–264 [DOI] [PubMed] [Google Scholar]
- Haneklaus M, Gerlic M, O'Neill LA, Masters SL (2013) miR‐223: infection, inflammation and cancer. J Intern Med 274: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkinson‐Woolley J, Hughes D, Gordon S, Martin P (1994) Macrophage recruitment during limb development and wound healing in the embryonic and foetal mouse. J Cell Sci 107(Pt 5): 1159–1167 [DOI] [PubMed] [Google Scholar]
- Hruz P, Zinkernagel AS, Jenikova G, Botwin GJ, Hugot JP, Karin M, Nizet V, Eckmann L (2009) NOD2 contributes to cutaneous defense against Staphylococcus aureus through alpha‐toxin‐dependent innate immune activation. Proc Natl Acad Sci USA 106: 12873–12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume EB, Cole N, Garthwaite LL, Khan S, Willcox MD (2006) A protective role for IL‐6 in staphylococcal microbial keratitis. Invest Ophthalmol Vis Sci 47: 4926–4930 [DOI] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling‐Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD (2008) Regulation of progenitor cell proliferation and granulocyte function by microRNA‐223. Nature 451: 1125–1129 [DOI] [PubMed] [Google Scholar]
- Kim MH, Liu W, Borjesson DL, Curry FR, Miller LS, Cheung AL, Liu FT, Isseroff RR, Simon SI (2008) Dynamics of neutrophil infiltration during cutaneous wound healing and infection using fluorescence imaging. J Invest Dermatol 128: 1812–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi SD, Malachowa N, DeLeo FR (2015) Pathogenesis of Staphylococcus aureus abscesses. Am J Pathol 185: 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Wang A, Liu X, Meisgen F, Grunler J, Botusan IR, Narayanan S, Erikci E, Li X, Blomqvist L et al (2015) MicroRNA‐132 enhances transition from inflammation to proliferation during wound healing. J Clin Invest 125: 3008–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZQ, Kondo T, Ishida Y, Takayasu T, Mukaida N (2003) Essential involvement of IL‐6 in the skin wound‐healing process as evidenced by delayed wound healing in IL‐6‐deficient mice. J Leukoc Biol 73: 713–721 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua‐Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang R, Jiang J, Yang B, Cao Z, Cheng X (2015) miR‐223 is upregulated in monocytes from patients with tuberculosis and regulates function of monocyte‐derived macrophages. Mol Immunol 67: 475–481 [DOI] [PubMed] [Google Scholar]
- Ma X, Tang Z, Qin J, Meng Y (2015) The use of high‐throughput sequencing methods for plant microRNA research. RNA Biol 12: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Zou F, Yang L, Lin S, Li Y, Ma M, Yin P, Liang X, Liu J (2015) The loss of MiR‐139‐5p promotes colitis‐associated tumorigenesis by mediating PI3K/AKT/Wnt signaling. Int J Biochem Cell Biol 69: 153–161 [DOI] [PubMed] [Google Scholar]
- Marques‐Rocha JL, Samblas M, Milagro FI, Bressan J, Martinez JA, Marti A (2015) Noncoding RNAs, cytokines, and inflammation‐related diseases. FASEB J 29: 3595–3611 [DOI] [PubMed] [Google Scholar]
- Martin P, D'Souza D, Martin J, Grose R, Cooper L, Maki R, McKercher SR (2003) Wound healing in the PU.1 null mouse–tissue repair is not dependent on inflammatory cells. Curr Biol 13: 1122–1128 [DOI] [PubMed] [Google Scholar]
- Martin P, Leibovich SJ (2005) Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol 15: 599–607 [DOI] [PubMed] [Google Scholar]
- McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ et al (1996) Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 15: 5647–5658 [PMC free article] [PubMed] [Google Scholar]
- Mildner A, Chapnik E, Manor O, Yona S, Kim KW, Aychek T, Varol D, Beck G, Itzhaki ZB, Feldmesser E et al (2013) Mononuclear phagocyte miRNome analysis identifies miR‐142 as critical regulator of murine dendritic cell homeostasis. Blood 121: 1016–1027 [DOI] [PubMed] [Google Scholar]
- Mori R, Power KT, Wang CM, Martin P, Becker DL (2006) Acute downregulation of connexin43 at wound sites leads to a reduced inflammatory response, enhanced keratinocyte proliferation and wound fibroblast migration. J Cell Sci 119: 5193–5203 [DOI] [PubMed] [Google Scholar]
- Mori R, Shaw TJ, Martin P (2008) Molecular mechanisms linking wound inflammation and fibrosis: knockdown of osteopontin leads to rapid repair and reduced scarring. J Exp Med 205: 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori R, Tanaka K, de Kerckhove M, Okamoto M, Kashiyama K, Kim S, Kawata T, Komatsu T, Park S, Ikematsu K et al (2014) Reduced FOXO1 expression accelerates skin wound healing and attenuates scarring. Am J Pathol 184: 2465–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Baltimore D (2012) microRNA regulation of inflammatory responses. Annu Rev Immunol 30: 295–312 [DOI] [PubMed] [Google Scholar]
- Pastar I, Khan AA, Stojadinovic O, Lebrun EA, Medina MC, Brem H, Kirsner RS, Jimenez JJ, Leslie C, Tomic‐Canic M (2012) Induction of specific microRNAs inhibits cutaneous wound healing. J Biol Chem 287: 29324–29335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbert‐Roth A, Furst DE, Nebesky JM, Jin A, Berber E (2018) A review of recent advances using tocilizumab in the treatment of rheumatic diseases. Rheumatol Ther 5: 21–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado‐Pabon W, Schlievert PM (2014) Models matter: the search for an effective Staphylococcus aureus vaccine. Nat Rev Microbiol 12: 585–591 [DOI] [PubMed] [Google Scholar]
- Sen CK, Gordillo GM, Roy S, Kirsner R, Lambert L, Hunt TK, Gottrup F, Gurtner GC, Longaker MT (2009) Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen 17: 763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsukinai K, Urano Y, Kakinuma K, Majima HJ, Nagano T (2003) Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J Biol Chem 278: 3170–3175 [DOI] [PubMed] [Google Scholar]
- Sheshachalam A, Srivastava N, Mitchell T, Lacy P, Eitzen G (2014) Granule protein processing and regulated secretion in neutrophils. Front Immunol 5: 448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuto T, Furuta T, Cheung J, Gruenert DC, Ohira Y, Shimasaki S, Suico MA, Sato K, Kai H (2007) Increased responsiveness to TLR2 and TLR4 ligands during dimethylsulfoxide‐induced neutrophil‐like differentiation of HL‐60 myeloid leukemia cells. Leuk Res 31: 1721–1728 [DOI] [PubMed] [Google Scholar]
- Strassheim D, Kim JY, Park JS, Mitra S, Abraham E (2005) Involvement of SHIP in TLR2‐induced neutrophil activation and acute lung injury. J Immunol 174: 8064–8071 [DOI] [PubMed] [Google Scholar]
- Sun Y, Oravecz‐Wilson K, Mathewson N, Wang Y, McEachin R, Liu C, Toubai T, Wu J, Rossi C, Braun T et al (2015) Mature T cell responses are controlled by microRNA‐142. J Clin Invest 125: 2825–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S (1999) Differential roles of TLR2 and TLR4 in recognition of gram‐negative and gram‐positive bacterial cell wall components. Immunity 11: 443–451 [DOI] [PubMed] [Google Scholar]
- Tam S, de Borja R, Tsao MS, McPherson JD (2014) Robust global microRNA expression profiling using next‐generation sequencing technologies. Lab Invest 94: 350–358 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kim SE, Yano H, Matsumoto G, Ohuchida R, Ishikura Y, Araki M, Araki K, Park S, Komatsu T et al (2017) MiR‐142 is required for Staphylococcus aureus clearance at skin wound sites via small GTPase‐mediated regulation of the neutrophil actin cytoskeleton. J Invest Dermatol 137: 931–940 [DOI] [PubMed] [Google Scholar]
- Wang T, Feng Y, Sun H, Zhang L, Hao L, Shi C, Wang J, Li R, Ran X, Su Y et al (2012) miR‐21 regulates skin wound healing by targeting multiple aspects of the healing process. Am J Pathol 181: 1911–1920 [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11: 228–234 [DOI] [PubMed] [Google Scholar]
- Yamada Y, Kosaka K, Miyazawa T, Kurata‐Miura K, Yoshida T (2014) miR‐142‐3p enhances FcepsilonRI‐mediated degranulation in mast cells. Biochem Biophys Res Commun 443: 980–986 [DOI] [PubMed] [Google Scholar]
- Yeruva L, Myers GS, Spencer N, Creasy HH, Adams NE, Maurelli AT, McChesney GR, Cleves MA, Ravel J, Bowlin A et al (2014) Early microRNA expression profile as a prognostic biomarker for the development of pelvic inflammatory disease in a mouse model of chlamydial genital infection. MBio 5: e01241‐14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki K, Murayama S, Ito H, Koga T (2018) The role of interleukin‐6 in castleman disease. Hematol Oncol Clin North Am 32: 23–36 [DOI] [PubMed] [Google Scholar]
- Zhang HD, Jiang LH, Sun DW, Li J, Tang JH (2015) MiR‐139‐5p: promising biomarker for cancer. Tumour Biol 36: 1355–1365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Dataset EV1
Dataset EV2
Movie EV1
Movie EV2
Source Data for Expanded View
Review Process File
Source Data for Figure 8
Data Availability Statement
miRNA‐Seq data reported are available in the DNA Data Bank of Japan (DDBJ) under accession no. DRA004094 (http://ddbj.nig.ac.jp/DRASearch/submission?acc=DRA004094). RNA‐Seq data reported are available in the DDBJ under accession no. DRA004092 (http://ddbj.nig.ac.jp/DRASearch/submission?acc=DRA004092).


