Abstract
Oxyvinylglycines are a family of nonproteinogenic amino acids featuring an essential vinyl ether conferring mechanism-based inhibition of pyridoxal phosphate enzymes. The gene clusters for a few oxyvinylglycines are known, yet the biosynthetic origin of the vinyl ether is elusive. The in vitro biosynthesis of methoxyvinylglycine or L-2-amino-4-methoxy-trans-3-butenoic acid (AMB) is reported. It is shown that AMB is made from glutamate as an alanyl-AMB dipeptide and the rationale is provided for the N-term Ala. Using a chemical capture method, the order and timing of the modifications on non-ribosomal peptide synthetase (NRPS)-bound substrates was determined, including a cryptic hydroxylation of the Glu β-carbon. Eliminating this hydroxy group likely generates a key α,-β dehydroamino acid intermediate that facilitates decarboxylation. This work sheds light on vinyl ether biosyn-thesis and uncovers new NRPS chemistry.
Keywords: biosynthesis, enzymes, natural products, nonribosomal peptide synthetase, oxyvinylglycines
Nonproteinogenic amino acids (NAAs) are widely produced by bacteria, animals, and plants for purposes beyond building proteins. Over 500 NAAs have been identified, vastly outnumbering the 20 proteinogenic amino acids.[1] Free NAAs are known for their activities as antimetabolites, which mimic the structures of essential metabolites, inhibiting the enzymes that modify these metabolites.[2] Bioactivities of NAAs are imparted by their diverse functional groups, including olefins, aldehydes, ketones, and more rarely alkynes, aziridines, and diazo groups.[1] Understanding how these groups are installed can reveal useful enzymatic transformations and facilitate the discovery of new bioactive compounds.[1,3]
Oxyvinylglycines are a unique family of NAAs featuring a core vinyl ether and a variety of alkoxy substituents (Figure 1A). This family acts as mechanism-based inhibitors of the eliminase subgroup of pyridoxal phosphate (PLP)-dependent enzymes by forming a stable adduct with enzyme-bound PLP.[4] The electronics of the vinyl ether are key to their inhibition: after oxyvinylglycines form the enzyme-bound PLP adduct via transamination, the vinyl group is conjugated with PLP. The vinyl oxygen can donate its lone pair to the conjugated system, stabilizing the inhibitory adduct (Supporting Information, Figure S1).[4]
Figure 1.
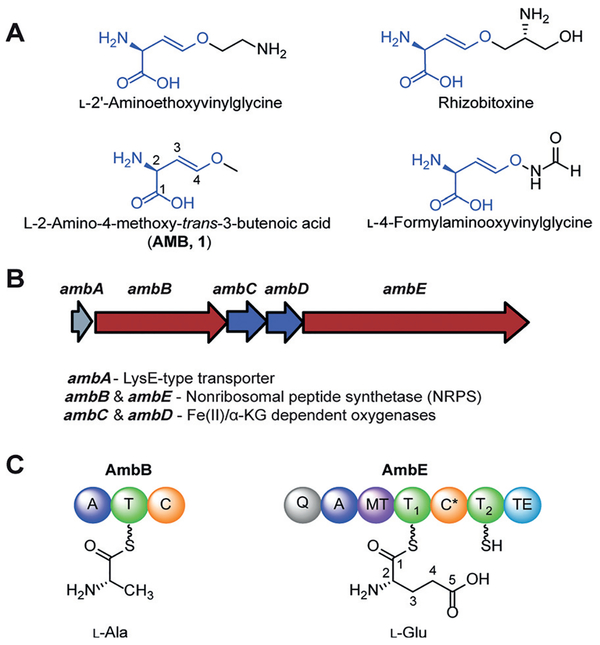
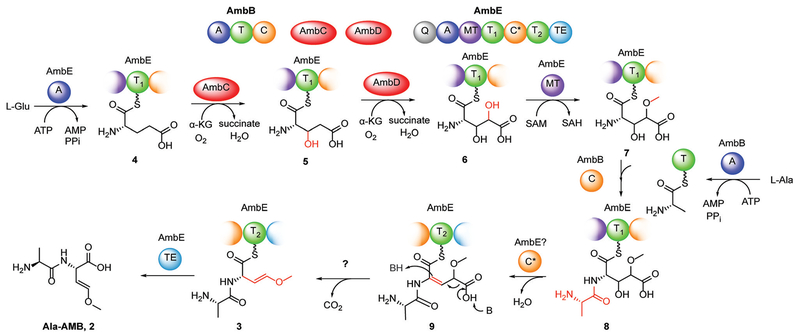
Oxyvinylglycines and amb gene cluster. A) Structures of representative oxyvinylglycines including AMB (1). B) amb biosynthetic gene cluster. C) NRPS domains of AmbB and AmbE, and amino acid preference. A =adenylation, T=thiolation, C =condensation, MT =O-methyl transferase, C* =a noncanonical condensation domain, TE =thioesterase, Q= domain of unknown function.
Oxyvinylglycines are produced by a variety of bacteria and exhibit potent bioactivity against plants and bacteria.[5] For example, 2′-aminoethoxyvinylglycine (AVG) and rhizobitoxine inhibit 1-aminocyclopropane-1-carboxylate synthase, a key PLP-dependent enzyme in the biosynthesis of the plant growth hormone, ethylene.[4a,6] Consequently, AVG was developed as the active ingredient of the commercial plant growth regulator, ReTain (Valent). Methoxyvinylglycine, also known as L-2-amino-4-methoxy-trans-3-butenoic acid (AMB, 1), inhibits the growth of bacteria, including Bacillus subtilis, Escherichia coli, phytopathogen Erwinia amylovora, and the human pathogen Staphylococcus aureus.[4b,5a] Despite the potent biological activities of oxyvinylglycines, their biosyntheses are only beginning to be understood,[7] and the chemical transformations responsible for the vinyl ether formation remain unknown.
AMB was first isolated from Pseudomonas aeruginosa PAO1,[8] and the biosynthetic gene cluster of AMB (amb) was identified by transposon mutagenesis.[7a] The amb cluster comprises a transporter gene ambA and four enzyme-encoding genes ambB, ambC, ambD, and ambE (Figure 1B). This cluster has also been connected to a quorum-sensing molecule IQS,[9] potentially via regulatory mechanisms. AmbB and AmbE are mono- and di-modular nonribosomal peptide synthetases (NRPSs), respectively. AmbC and AmbD share sequence homology with non-heme iron, α-ketoglutarate (α-KG)-dependent oxygenases (Fe/α-KG oxygenases). AmbB activates and loads L-Ala onto its thiolation (T) domain and the second T domain of AmbE (AmbE-T2 or T2) upon inactivation of AmbB T domain; AmbE activates and loads L-Glu onto its first T domain (AmbE-T1 or T1) (Figure 1C).[7b] Inactivation of the AmbE thioesterase domain led to the accumulation of an AmbE-T2-tethered Ala-AMB-Ala tri-peptide.[7b] This initial study left many open questions concerning AMB biosynthesis, including the functions of the tailoring enzymes AmbC and AmbD, and the chemical transformations responsible for converting the γ-carboxylate of Glu into the methoxy vinyl ether.
Herein, we reconstitute the full biosynthesis of AMB in vitro and report the characterization of the biosynthetic enzymes in the amb pathway. We demonstrate that the amb gene cluster is responsible for synthesizing an alanyl-AMB dipeptide and identify the modifications that craft AMB from Glu. We also provide evidence for an important 2,3-dehydro-amino acid intermediate and propose the involvement of an unusual NRPS condensation domain in AMB biosynthesis.
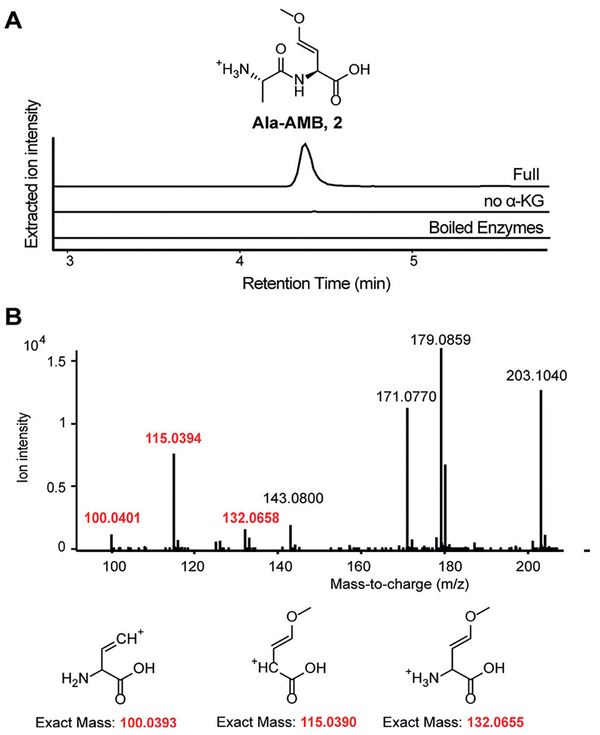
We first sought to reconstitute AMB biosynthesis in vitro. We cloned and purified all four amb biosynthetic enzymes and conducted a “one-pot assay” by incubating these enzymes with their predicted substrates and cofactors. AmbB, AmbC, AmbD, and AmbE were incubated with substrates Glu and Ala and cofactors ATP, S-adenosyl-methionine (SAM), MgCl2, Fe2+, and α-KG. This assay yielded a product that corresponds to an alanyl-AMB dipeptide (2) rather than an Ala-AMB-Ala tripeptide, based on analysis by liquid chromatography coupled high-resolution mass spectrometry (LCHRMS; Figure 2A; Supporting Information, Figure S2). Ala-AMB was enzymatically synthesized on a 2 mg-scale and derivatized using 9-fluorenylmethyloxycarbonyl chloride (Fmoc-Cl; Supporting Information, Figure S3). Partially purified Fmoc-Ala-AMB was analyzed by tandem MS, 1H, and (1H, 1H) COSY NMR. The data support the proposed structure (Figure 2B; Supporting Information, Figures S4–6, Table S2). The coupling constants of the vinyl hydrogens are consistent with those of the naturally occurring E isomer of AMB (Supporting Information, Figure S5, Table S2), suggesting in vitro synthesized Ala-AMB has E stereochemistry.[10] Although we could not assign the stereochemistry of the AMB portion to D or L, we expect it to be L as found in the natural product. Omission of α-KG, each cofactor, or each individual enzyme also abolished formation of Ala-AMB (Figure 2A; Supporting Information, Figure S7), confirming that each enzyme and cofactor is necessary for the biosyn-thesis of Ala-AMB.
Figure 2.
Alanyl-AMB (2) is the product of in vitro reconstitution of amb enzymes. A) Extracted ion chromatograms (EICs) of 2 (m/z 203.1026 [M+H]+) from LC-HRMS in the one-pot assay and negative controls. B) Fragmentation pattern of Fmoc-Ala-AMB from tandem mass spectrometry. Fragments in red indicate that the alanine portion of Ala-AMB is attached to Fmoc. Complete analysis of all fragments is described in the Supporting Information, Figure S2.
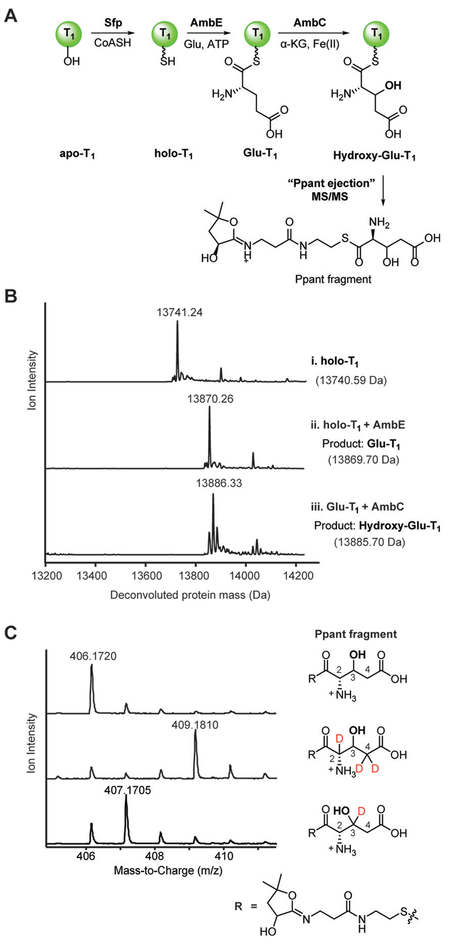
With active enzymes in hand, we set out to characterize the functions of each biosynthetic enzyme. We hypothesized that key modifications may occur when Glu is tethered to AmbE-T1. The truncated T1 domain was expressed, purified, and converted into its holo form using the phosphopantetheine (Ppant) transferase Sfp (Figure 3A,B-i). T1 can be loaded with L-Glu in trans by full length AmbE, generating Glu covalently bound to T1 (Glu-T1; Figure 3A,B-ii). Incubation with AmbC resulted in a modified protein peak with a 16 Da mass increase, suggesting incorporation of an oxygen atom onto the structure, likely via hydroxylation, a common reaction for Fe/α-KG oxygenases (Figure 3A,B-iii).[11] A tandem MS-based Ppant-ejection assay[12] on the AmbC-modified T1 confirmed incorporation of an oxygen to form a hydroxy-Glu intermediate (Figure 3A,C).
Figure 3.
AmbC catalyzes a cryptic hydroxylation at C3 of AmbE-T1-bound Glu. A) Multistep reaction to modify the standalone T1 domain of AmbE, AmbE-T1. Ppant fragment formation is shown. B) Deconvoluted protein mass spectra for i) holo-AmbE-T1, ii) holo-AmbE-T1 loaded with Glu (Glu-T1), iii) AmbC hydroxylation product of Glu-T1. Expected and observed masses are shown for each species. C) Mass spectra of Ppant ejection fragments from assays containing l-Glu (top), 2,4,4-D3-l-Glu (middle) and 3,3-D2-l-Glu (bottom) as substrates.
We investigated the regiochemistry of the AmbC-mediated oxidation by loading deuterium labeled Glu onto T1. First, triply deuterated Glu, 2,4,4-D3-L-Glu, was loaded onto T1 and modified by AmbC. Ppant ejection of the product indicated that all three deuteriums were retained in the product (Figure 3C), suggesting AmbC abstracts a hydrogen and hydroxylates at C3 instead of at C2 or C4. To rule out the possibility that deuterium labeling altered the kinetics of hydroxylation, thus redirecting hydroxylation to a different position,[13] we synthesized 3,3-D2-L-Glu and loaded it onto T1 (Supporting Information, Figures S19,S20). The resulting product retained only one deuterium (Figure 3C), consistent with AmbC hydroxylation at C3. These results demonstrate that AmbC preferentially hydroxylates C3 of Glu-T1, installing a cryptic hydroxy group that is later removed during biosynthesis.
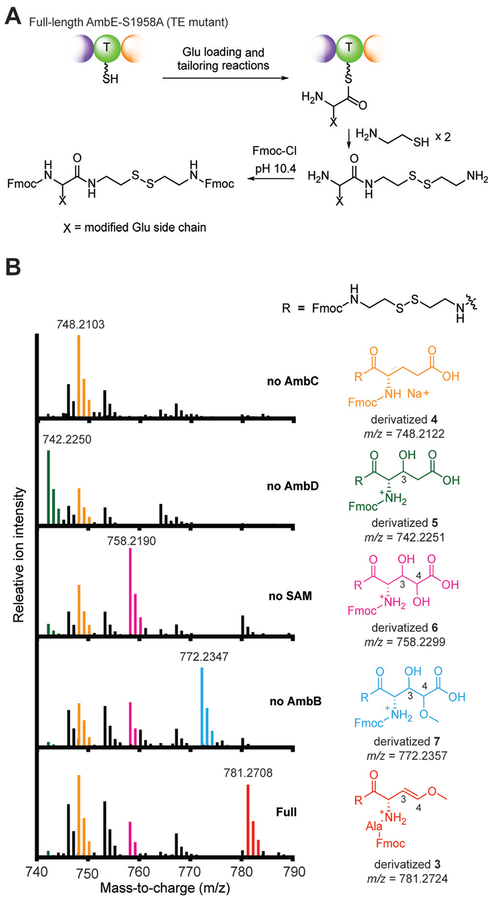
We next mapped the order and timing of the transformations necessary to generate Ala-AMB from 3-hydroxy- Glu. We found that AmbD does not modify Glu-T1 or the hydroxylated Glu-T1 product of AmbC (Supporting Information, Figure S8), suggesting that full length AmbE may be necessary for the activity of AmbD. Thus, we adopted a chemical strategy to capture the biosynthetic intermediates attached to the full length NRPSs, which involves the use of cysteamine as a thioester reactive probe.[14] The thioesterase (TE) domain of AmbE was inactivated by mutating the conserved Ser to Ala (AmbE S1958A) to prevent hydrolytic release of intermediates or products. Indeed, use of AmbE S1958A in the one-pot assay abolished production of Ala- AMB (Supporting Information, Figure S9), but allowed various intermediates to accumulate on the NRPSs. We captured these NRPS-bound intermediates using cysteamine and further derivatized them using Fmoc-Cl for improved retention and detection by LC-HRMS (Figure 4A). Using this strategy, we detected NRPS-bound Ala-AMB (3) as the major product accumulated on AmbE S1958A (Figure 4B), confirming that Ala-AMB is the final product of the NRPS assembly line. A small amount of Ala-AMB-Ala was captured (Supporting Information, Figure S10), indicating Ala-AMB-Ala is a minor product under these conditions. No Ala-AMB-Ala was detected in the one pot assay with wild-type AmbE (Supporting Information, Figure S3). Thus, the tripeptide may be a shunt product generated by AmbE S1958A. When AmbC was excluded, unmodified Glu (4) was exclusively detected on the assembly line, further demonstrating that AmbC is responsible for the first modification. Exclusion of AmbD led to the detection of monohydroxy-Glu (5), suggesting that AmbD catalyzes the second modification after AmbC hydroxylation. Omission of SAM led to detection of dihydroxy-Glu (6), suggesting that O-methylation occurs after tandem hydroxylations by AmbC and AmbD. Lastly, omission of Ala or AmbB resulted in nearly identical spectra showing the accumulation of a dihydroxylated and methylated Glu (7), but not AMB, suggesting that condensation with Ala occurs after the hydroxylations and O-methylation, but prior to AMB formation.
Figure 4.
Chemical capture of biosynthetic intermediates reveals order and timing of biosynthesis. A) Cysteamine cleavage of thioester-linked intermediates and subsequent derivatization by Fmoc-Cl. B) Mass spectra of intermediates captured in the one-pot assays using AmbE S1958A. MS peaks are colored to match the corresponding compounds captured under different assay conditions. Both the sodium and proton adducts are observed for derivatized 4, but only the sodium adduct is shown for clarity. Extended mass range is shown in the Supporting Information, Figure S10.
The above data suggest that AmbD may hydroxylate at C4 to install the enol ether oxygen of AMB. To confirm the regiochemistry of AmbD hydroxylation, we performed the chemical capture experiment with 2,4,4-D3-Glu and 4,4-D2- Glu. When AmbD was excluded from a one-pot assay with AmbE S1958A, the isotopic labeling patterns of captured products are consistent with C3-hydroxylation by AmbC (Supporting Information, Figures S11,S12). When SAM was excluded, the accumulated product of AmbD-dihydroxy-Glu (D2-6)-lost a single deuterium from both 2,4,4-D3-Glu and 4,4-D2-Glu (Supporting Information, Figures S11,S12). These results indicate that AmbD hydroxylates Glu at C4 and supports the hypothesis that AmbD installs the oxygen ultimately incorporated in the enol ether.
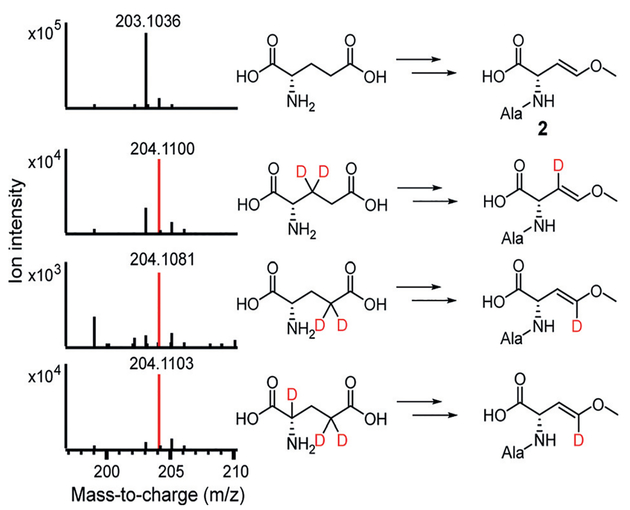
Use of the deuterium-labeled substrates in the one-pot assay containing wild-type AmbE reveals additional late-stage modifications in Ala-AMB biosynthesis. Retention of a single deuterium in both 3,3-D2-Glu and 4,4-D2-Glu was observed in Ala-AMB, consistent with AmbC and AmbD hydroxylations at C3 and C4, respectively (Figure 5). Surprisingly, only one deuterium from 2,4,4-D3-Glu is retained in Ala-AMB (Figure 5), indicating that the C2 deuterium is also removed during biosynthesis. The loss of the C2 deuterium is also observed in the chemical capture experiment of one-pot assay using 2,4,4-D3-Glu and AmbE S1958A (Supporting Information, Figure S11). To investigate this surprising result, we inactivated the T2 domain in the second module of AmbE (S1819A). Use of AmbE S1819A abolished Ala-AMB production in the one-pot assay (Supporting Information, Figure S9). Using the S1819A mutant to capture NRPS-bound molecules, we identified a new intermediate, alanyl-3-hydroxy-4-methoxy-Glu (8; Figure 6; Supporting Information, Figure S13). The loss of the Glu C2 deteurium in Ala-AMB suggests that 8 undergoes dehydration to eliminate the C2 deuterium and C3 hydroxy, yielding a potential 2,3-dehydro intermediate (9) (Figure 6). The double bond in 9 is conjugated with the thioester and can facilitate decarboxylation by stabilizing the carbanion intermediate. Subsequent protonation at C2 would generate Ala-AMB.
Figure 5.
AMB biosynthesis likely involves a 2,3-dehydro-amino acid intermediate. Mass spectra of deuterium-labeled Ala-AMB produced in the one-pot assays using wild-type AmbE and multiple deuterium-labeled Glu as substrates.
Figure 6.
Proposed biosynthetic pathway of Ala-AMB (2).
Based on these results, we propose the biosynthetic pathway for Ala-AMB depicted in Figure 6. Ala and Glu are activated and loaded onto T domains by AmbB and AmbE, respectively. AmbC catalyzes a cryptic hydroxylation on C3 of Glu, followed by AmbD hydroxylation on C4. The C4 hydroxy is methylated by the O-methyltransferase (MT) domain of AmbE, and then condensed with Ala by the AmbB C domain to form the dipeptide 8. The modifications up to this point occur at the T1 site of AmbE. The cryptic C3 hydroxy is eliminated to form 9, which can undergo subsequent decarboxylation and isomerization to generate the methoxy enol ether in 3. The decarboxylation and vinyl shift likely occur on the T2 domain of AmbE. Lastly, hydrolysis by the thioesterase domain of AmbE releases Ala-AMB.
The detection of 8 on T1 as the most advanced intermediate suggests that a subsequent 2,3-dehydration occurs on the second, partial module of AmbE. This module contains a non-canonical C domain (C* domain, Figure 1C). Phylogenetic analysis revealed that AmbE-C* belongs to a unique group of C domains (Supporting Information, Figure S14), which are also found in the biosynthetic clusters of the natural products such as microcystin and nodularin.[15] Interestingly, both microcystin and nodularin contain 2,3-dehydro-amino acids resulting from serine or threonine, suggesting that the C* domain may catalyze 2,3-dehydration in NRPS pathways, a distinct strategy from the biosynthesis of ribosomal peptides such as lanthipeptides and thiopeptides.[16] In AMB biosynthesis, this C* domain may be responsible for the concurrent elimination of the cryptic C3-hydroxy and C2-hydrogen to generate the 2,3-dehydro-amino acid intermediate (Figure 6). The precise role of the C* domain, as well as decarboxylation of the 2,3-dehydro intermediate will be the focus of future studies.
We revealed that AMB is synthesized as part of an Ala-AMB dipeptide. An NRPS-bound Ala-AMB-Ala tripeptide was previously proposed as a late-stage biosynthetic intermediate,[7b] but we found that the Ala-AMB dipeptide is the final, released product of the amb pathway. We propose that the N-term Ala serves dual functions in biosynthesis and self-protection. In the biosynthesis of Ala-AMB, because N-term 2,3-dehydroamino acids are known to undergo isomerization and hydrolysis to form α-keto acids,[16] condensation with an N-term Ala stabilizes the 2,3-dehydroamino acid intermediate and prevents this off-pathway reaction. This N-term Ala may also play a self-protective role for P. aeruginosa by masking the α-amino group of AMB that is required for mechanism-based inhibition of PLP enzymes by oxyvinylglycines.[4a,17] The N-term Ala may be removed by a P. aeruginosa protease before the export of AMB by the LysE-type amino acid transporter AmbA.
In summary, we have reconstituted the in vitro biosynthesis of the oxyvinylglycine AMB. Our work reveals the orchestrated biosynthesis by two NRPSs and two Fe/α-KG oxygenases to morph Glu into a potent vinyl ether-containing antibiotic. Using a combination of deuterium labeling, chemical capture, and mass spectrometry, we determine the regiochemistry and timing of the modifications that occur on NRPS-bound substrates. We identify a cryptic hydroxylation in AMB biosynthesis and provide evidence for the removal of this hydroxy by a 2,3-dehydration mechanism. We propose that the 2,3-dehydration in nonribosomal peptide synthesis is catalyzed by a unique group of condensation domains. This work elucidates an important biosynthetic pathway for installing the vinyl ether in NAAs and sheds light on the self-protection mechanisms of oxyvinylglycines. Further structural and activity studies of the amb enzymes will reveal important insights on the function of C* domains and the interplay between NRPSs and their tailoring enzymes.
Supplementary Material
Acknowledgements
The authors thank Dr. Albert Bowers and Dr. Gary Pielak for carefully reading the manuscript, Kevin Santa Maria for assistance with bioinformatics, Sam Pellock for cloning of the amb genes, Steffen Good for helpful discussions, Andrew Chan for assistance with LC-HRMS analysis, and Dr. Rita Tamayo for access to the anaerobic chamber. This work is supported by National Institutes of Health (R00 GM099904), National Science Foundation (CHE1654678), the Rita Allen Foundation, and the University of North Carolina at Chapel Hill.
Footnotes
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: https://doi.org/10.1002/anie.201713419.
Conflict of interest
The authors declare no conflict of interest.
References
- [1].Walsh CT, O’Brien RV, Khosla C, Angew. Chem. Int. Ed 2013, 52, 7098–7124; Angew. Chem. 2013, 125, 7238 – 7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].González B, Pajares M. a. A., Hermoso JA, Alvarez L, Garrido F, Sufrin JR, Sanz-Aparicio J, J. Mol. Biol 2000, 300, 363–375. [DOI] [PubMed] [Google Scholar]
- [3].Du Y-L, He H-Y, Higgins MA, Ryan KS, Nat. Chem. Biol 2017, 13, 836–838. [DOI] [PubMed] [Google Scholar]
- [4].a) Capitani G, McCarthy DL, Gut H, Grütter MG, Kirsch JF, J. Biol. Chem 2002, 277, 49735–49742; [DOI] [PubMed] [Google Scholar]; b) Berkowitz DB, Charette BD, Karukurichi KR, McFadden JM, Tetrahedron: Asymmetry 2006, 17, 869–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Lee X, Azevedo MD, Armstrong DJ, Banowetz GM, Reimmann C, Environ. Microbiol. Rep 2013, 5, 83–89; [DOI] [PubMed] [Google Scholar]; b) Even-Chen Z, Mattoo AK, Goren R, Plant Physiol. 1982, 69, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yasuta T, Satoh S, Minamisawa K, Appl. Environ. Microbiol 1999, 65, 849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Lee X, Fox Á, Sufrin J, Henry H, Majcherczyk P, Haas D, Reimmann C, J. Bacteriol 2010, 192, 4251–4255; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rojas Murcia N, Lee X, Waridel P, Maspoli A, Imker HJ, Chai T, Walsh CT, Reimmann C, Front. Microbiol 2015, 6, 1–13; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Fernández M, Cuadrado Y, Aparicio JF, Martín JF, Microbiology 2004, 150, 1467–1474; [DOI] [PubMed] [Google Scholar]; d) Yasuta T, Okazaki S, Mitsui H, Yuhashi K-I, Ezura H, Minamisawa K, Appl. Environ. Microbiol 2001, 67, 4999–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Scannell JP, Pruess DL, Demny TC, Sello LH, Williams T, Stempel A, J. Antibiot 1972, 25, 122–127. [DOI] [PubMed] [Google Scholar]
- [9].Dandekar A. a., Greenberg EP, Nat. Chem. Biol 2013, 9, 292–293. [DOI] [PubMed] [Google Scholar]
- [10].a) Keith DD, Tortora J. a., Yang R, Hy E, J. Org. Chem 1978, 43, 3711–3713; [Google Scholar]; b) Sufrin JR, Lombardini JB, Keith DD, Biochem. Biophys. Res. Commun 1982, 106, 251–255. [DOI] [PubMed] [Google Scholar]
- [11].Hausinger RP, Crit. Rev. Biochem. Mol. Biol 2004, 39, 21–68. [DOI] [PubMed] [Google Scholar]
- [12].Dorrestein PC, Bumpus SB, Calderone CT, Garneau-Tsodikova S, Aron ZD, Straight PD, Kolter R, Walsh CT, Kelleher NL, Biochemistry 2006, 45, 12756–12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Price JC, Barr EW, Glass TE, Krebs C, Bollinger JM, J. Am. Chem. Soc 2003, 125, 13008–13009; [DOI] [PubMed] [Google Scholar]; b) Matthews ML, Neumann CS, Miles L. a., Grove TL, Booker SJ, Krebs C, Walsh CT, Bollinger JM, Proc. Natl. Acad. Sci. USA 2009, 106, 17723–17728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Belecki K, Townsend CA, J. Am. Chem. Soc 2013, 135, 14339–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].a) Tillett D, Dittmann E, Erhard M, von Döhren H, Bçrner T,Neilan B. a., Chem. Biol 2000, 7, 753–764; [DOI] [PubMed] [Google Scholar]; b) Moffitt MC, Neilan B. a., Appl. Environ. Microbiol 2004, 70, 6353–6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Velásquez JE, Zhang X, van der Donk WA, Chem. Biol 2011, 18, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rando RR, Relyea N, Cheng L, J. Biol. Chem 1976, 251, 3306–3312. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.