Summary
Aging is accompanied by a pervasive collapse of proteostasis, while reducing general protein synthesis promotes longevity across taxa. Here, we show that the eIF4E isoform IFE-2 is increasingly sequestered in mRNA processing (P) bodies during aging and upon stress in Caenorhabditis elegans. Loss of the enhancer of mRNA decapping EDC-3 causes further entrapment of IFE-2 in P bodies and lowers protein synthesis rates in somatic tissues. Animals lacking EDC-3 are long lived and stress resistant, congruent with IFE-2-deficient mutants. Notably, neuron-specific expression of EDC-3 is sufficient to reverse lifespan extension, while sequestration of IFE-2 in neuronal P bodies counteracts age-related neuronal decline. The effects of mRNA decapping deficiency on stress resistance and longevity are orchestrated by a multimodal stress response involving the transcription factor SKN-1, which mediates lifespan extension upon reduced protein synthesis. Our findings elucidate a mechanism of proteostasis control during aging through P body-mediated regulation of protein synthesis in the soma.
Keywords: aging, eukaryotic initiation factor 4E, mRNA decapping, mRNA translation, processing bodies, protein synthesis, proteostasis, stress response
Graphical Abstract

Highlights
-
•
Processing bodies (PBs) accumulate in somatic tissues during aging in C. elegans
-
•
Downregulation of mRNA decapping in PBs promotes stress resistance and longevity
-
•
Somatic eIF4E localizes to PBs upon stress and during aging
-
•
mRNA decapping influences eIF4E association with PBs and protein synthesis in the soma
Rieckher et al. uncover a mechanism modulating protein synthesis during aging in the Caenorhabditis elegans soma, through association of the mRNA translation initiation factor eIF4E with mRNA processing (P) bodies (PBs). Effects on longevity are mediated by attenuation of global protein synthesis in somatic tissues via the sequestration of eIF4E in PBs.
Introduction
Mature eukaryotic mRNAs can cycle between polysomes and discrete cytoplasmic foci known as processing (P) bodies (PBs) and stress granules (SGs). Non-translating mRNAs that enter PBs assemble into various messenger ribonucleoprotein complexes (mRNPs) associated with components of the mRNA decay machinery or translational repressors (Brengues and Parker, 2007, Decker and Parker, 2012). Notably, only select mRNAs are capable of reentering translation (Arribere et al., 2011). Over the past decade, studies in yeast, worms, flies, and mammalian cells have culminated in the identification of key PB components, such as proteins involved in mRNA decapping, 5′-3 mRNA degradation, non-sense-mediated mRNA decay (NMD), microRNA (miRNA), small interfering RNA (siRNA)-mediated gene silencing, and viral cap snatching, among others (Decker and Parker, 2012, Mir et al., 2008, Müller-McNicoll and Neugebauer, 2013). Intriguingly, identification of the molecular composition of PBs isolated from human epithelial cells suggests a role for PBs in mRNA storage rather than degradation (Hubstenberger et al., 2017).
Several proteins, including the Enhancer of decapping (Edc3) and the decapping activator Dcp1, function to increase the rate of mRNA decapping in vivo (Kshirsagar and Parker, 2004). Edc3, which belongs to the Lsm16 family of proteins, consists of three functional domains, namely, an N-terminal divergent LSm (Sm-like) domain followed by a low-complexity linker, an FDF domain containing a highly conserved FDF amino acid motif, and a C-terminal YjeF-N domain, which is conserved among yeast, Drosophila, and human (Tritschler et al., 2007). The N-terminal LSm domain mediates binding to Dcp1, localization of Edc3 to PBs, and its interaction with Dcp2. FDF motifs recruit particular RNA helicases that function in translational repression and mRNA decapping, such as the Dead box helicase Dhh1/DDX6 in yeast and humans, while the YjeF-N domain is required for self-association, contributing to PB assembly (Decker et al., 2007, Ling et al., 2008, Tritschler et al., 2008, Tritschler et al., 2007, Tritschler et al., 2009). Interestingly, Edc3 depletion does not prevent PB formation in Drosophila S2 cells, indicating that perhaps other mechanisms contribute to PB assembly in metazoans (Eulalio et al., 2007). Accumulating evidence suggests that the Dcp1/Dcp2/Edc3 protein complex competes with the cap-binding protein eIF4E, which is the only translation initiation factor accumulating in PBs both in mammalian cells and Drosophila neurons. Moreover, the interaction between eIF4E and the eIF4E transporter (eIF4E-T), also found in PBs, inhibits translation by blocking eIF4G recruitment (Andrei et al., 2005, Ferraiuolo et al., 2005). Thus, PB functions and mRNA translation may, at least in some cases, compete with each other (Franks and Lykke-Andersen, 2008).
Downregulation of mRNA translation delays aging in diverse organisms, ranging from yeast to mammals (Gems et al., 2002, McElwee et al., 2007, Partridge and Gems, 2002, Tavernarakis, 2007, Tavernarakis, 2008). In the nematode Caenorhabditis elegans, loss of a specific eIF4E isoform (IFE-2) that functions in somatic tissues reduces global protein synthesis and extends lifespan (Hansen et al., 2007, Pan et al., 2007, Syntichaki et al., 2007). eIF4E localizes to SGs and PBs in yeast and in mammalian cells. These two types of cytoplasmic foci physically interact and often exchange material in a process defined as docking. Accumulating evidence supports a role for PBs in the control of translation and mRNA degradation (Brengues et al., 2005, Decker and Parker, 2012). Furthermore, emerging findings suggest that mRNA metabolism factors can modulate stress resistance and aging in C. elegans (Rousakis et al., 2014) and indicate that aging causes aggregation of RNA-binding proteins (RBPs) inside SGs, correlating with decreased organismal fitness. The heat shock transcription factor HSF-1 reportedly regulates RBP aggregation, while reduced insulin/IGF-1 signaling restores their dynamics (Lechler et al., 2017). Despite this, the mechanisms underlying the effects of mRNA metabolism factors on aging remain largely unknown. Here, we report a pivotal role of PBs in modulating protein synthesis across somatic tissues in C. elegans. Specifically, we elucidate a molecular mechanism controlling mRNA translation in the soma, via EDC-3-mediated sequestration of eIF4E in PBs during aging. Given the importance of protein synthesis dynamics to cellular protein homeostasis (Labbadia and Morimoto, 2014), our results establish PBs as crucial regulators of proteostasis in C. elegans. We further show that the downregulation of mRNA decapping activates cellular homeostatic mechanisms that enhance stress resistance and promote longevity. Altogether, these findings provide significant insight into how components of the mRNA degradation pathway may regulate aging by directly interfering with key mRNA translation factors.
Results
The Somatic eIF4E Isoform IFE-2 Associates with PBs during Aging and under Stress
To investigate the role of PBs in regulating eIF4E availability, we tested the association of IFE-2 with PB-specific components. We used a double-transgenic strain carrying a full-length IFE-2::GFP fusion, driven by the endogenous ife-2 promoter (pife-2IFE-2::GFP; Syntichaki et al., 2007), and a PB-specific DCAP-1::DsRed reporter fusion under the control of the dcap-1 promoter. IFE-2::GFP and DCAP-1::DsRed foci, at least partially, colocalized across various tissues, especially in the head region of animals (Figure 1A). In addition, various monitoring methods and time-lapse recordings revealed increasing colocalization of DCAP-1 and IFE-2 during heat shock in most somatic tissues in a qualitative (Figures 1A and S1; Video S1) or quantitative (Figures 1B, S2A, S2B; Video S2) manner. Notably, we also detected SGs and PBs in close proximity or partly overlapping, indicative of docking between the two types of granules (Figures 1A, S2A, and S2B). Similarly, the PB-specific enhancer of decapping, EDC-3, colocalized with IFE-2 under normal conditions (Figure S2C). Colocalization of IFE-2 and EDC-3 increased significantly upon heat stress across tissues, including neurons (Figure S2D). Taken together, these results indicate that the soma-specific eIF4E isoform IFE-2 associates with PBs, and the extent of colocalization increases upon heat stress.
Figure 1.
IFE-2 Colocalizes with PBs under Stress Conditions and during Aging
(A) Representative images of transgenic animals co-expressing pife-2IFE-2::GFP and the PB-specific marker pdcap-1DCAP-1::DsRed before and after heat shock at 35°C for 1.5 hr. The anterior part of the animal is shown; inlays indicate higher magnification of the pharyngeal region and the hypodermal tissue. Scale bars, 25 μm. White arrows indicate possible docking between PBs and SGs.
(B) Two-dimensional (2D) intensity histograms with regression lines and Pearson’s coefficient (ρx,y) as quantitative representation of IFE-2::GFP and DCAP-1::DsRed colocalization within regions of interest in the pharynx of the animals (compare A) before and after heat shock.
(C–E) Quantification of DsRed-tagged PB-specific signal in the pharyngeal region of transgenic animals during aging for plsm-3LSM-3::DsRed (C), pedc-3EDC-3::DsRed (D), and PB number (E) for animals expressing pdcap-1DCAP-1::DsRed. Error bars denote SEM; n = 10 animals per time point and trial. ∗p < 0.01, ∗∗p < 0.001, and ∗∗∗p < 0.0001 (unpaired t test).
(F) Expression of the full-length pife-2IFE-2::GFP reporter decreases during aging. Error bars show SEM; n = 40 animals per experiment. ∗p < 0.01, ∗∗p < 0.001, and ∗∗∗p < 0.0001 (unpaired t test).
(G) Percentage of animals forming SGs in various tissues during aging at day 3, day 8, and day 15 of adulthood. Error bars represent SEM; n > 50 animals per experiment. ∗p < 0.01, ∗∗p < 0.001, and ∗∗∗p < 0.0001 (unpaired t test to measure statistical significance between day 8 or 15 in reference to day 3, respectively).
See also Figures S1 and S2 and Videos S1 and S2.
Frame rate is 30 s/scan, each scan contains maximum intensity information of 20 z stacks of 1 μm depth each.
The movie was produced with the IMARIS software (see Materials and Methods). Frame rate is 30 s/scan, each scan contains information of at least 20 z stacks of 0.3 μm depth each.
Previous work showed that PBs serve as sites for mRNA degradation or storage under physiological conditions (Aizer et al., 2014). Their formation is induced in response to various environmental stressors and conditions that alter mRNA degradation or translation initiation processes, influencing RNA homeostasis (Decker and Parker, 2012). In line with this, we found that the intensity of the DCAP-1::DsRed signal was significantly increased in nematodes subjected to prolonged heat shock (9 hr at 35°C), starvation (24 hr food deprivation), and hypoxia (6 hr in a hypoxia chamber) but not in animals treated with the mitochondrial toxin sodium azide (NaN3; 20 mM, 30 min incubation; Figure S3A). Furthermore, knockdown of the eat-3 gene encoding a mitochondrial dynamin family member homologous to human OPA1, which is essential for resistance of C. elegans to free radicals (Kanazawa et al., 2008), caused a substantial increase in PB formation during adulthood (Figure S3B).
Emerging findings suggest that regulation of mRNA metabolism deteriorates during aging (Cookson, 2012). However, a direct cause-and-effect relationship between mRNA metabolism and aging has not been established. To investigate the potential role of PBs in the context of mRNA metabolism and aging, we generated transgenic animals expressing the PB components EDC-3, DCAP-1, and LSM-3 tagged with DsRed. Temporal analysis of LSM-3, EDC-3, and DCAP-1 abundance revealed a substantial increase in PB size and number during aging (Figures 1C–1E). Moreover, we found that the levels of IFE-2::GFP protein fusion decreased with age (Figure 1F).
eIF4E localizes predominantly in SGs that are formed mainly in response to stress in mammalian cells (Kedersha et al., 2002, Kimball et al., 2003) and C. elegans (Rousakis et al., 2014). To examine SG formation and dynamics in somatic C. elegans tissues, animals expressing the pife-2IFE-2::GFP reporter fusion were subjected to various stress insults, including hypoxia, heat, NaN3, and starvation. We found that the number of animals (day 1 of adulthood) containing IFE-2::GFP SGs increased significantly under heat stress and hypoxia (Figure S3C) compared with unstressed animals, which is in line with a previous report indicating that hypoxia induces SG formation in embryonic Drosophila muscles (van der Laan et al., 2012). By contrast, NaN3 and starvation did not substantially enhance the formation of IFE-2-specific SGs. Taken together, our findings suggest that in response to specific stresses, eIF4E localizes to SGs in somatic C. elegans cells.
Similar to PB formation, SG assembly is markedly induced with age (Lechler et al., 2017). In line with this, we found that the number of animals exhibiting IFE-2::GFP granules in various somatic cells, especially in muscles, the canal cell, and intestinal cells, significantly increased during aging (Figures 1G and S3D–S3K). We also tested whether depletion of specific PB components alters cellular eIF4E/IFE-2 protein levels. Loss of DCAP-1, DCAP-2, or EDC-3 resulted in increased IFE-2 levels (Figure S3L). This increase was not due to transcriptional induction of ife-2, as expression of an ife-2 transcriptional reporter remained unaffected (Figure S3M). Together, these observations suggest that enzymes of the decapping complex act post-transcriptionally to modulate eIF4E abundance in somatic tissues.
mRNA Decapping in Somatic PBs Modulates Aging
Although the levels of PB components have been shown to increase with age (Figures 1C–1E), these alterations do not establish a causative relationship between PB function and longevity. To address this issue, we focused on the scaffolding enzyme EDC-3, which is crucial for the assembly of the mRNA degradation complex and PB formation, as previously shown in yeast and mammalian cells (Figure 2A; Tritschler et al., 2009). We performed a temporal analysis of EDC-3::DsRed abundance and localization in animals co-expressing GFP under the control of the pan-neuronal unc-119 promoter during aging. We observed that edc-3 is broadly expressed during embryogenesis (data not shown). Its expression is maintained in several cell types, including pharyngeal muscles, intestinal cells, neurons, and hypodermic cells, at early post-embryonic stages. Interestingly, EDC-3 foci, indicative of PBs, were specifically detected in neuronal and intestinal cells during adulthood (Figure 2B).
Figure 2.
Downregulation of mRNA Degradation Components Extends the Lifespan of Otherwise Wild-Type Animals but Does Not Affect the Lifespan of IFE-2-Depleted Mutants
(A) Scheme displaying highly conserved key factors of bulk mRNA degradation as studied in various species including yeast, mammals, and C. elegans. Edc3 is a central scaffolding protein, responsible for the recruitment of the Dcp2 decapping enzyme and its coactivator Dcp1, the formation of the mRNA decay complex and contributes to PB assembly. Schematic adapted from Decker and Parker (2012).
(B) Adult animal expressing pedc-3EDC-3::DsRed and the pan-neuronal reporter punc-119GFP. Inlays are also shown at higher magnification. Arrows indicate localization of pedc-3EDC-3::DsRed in neurons. Asterisks indicate areas of increased zoom. Scale bars, 25 μm.
(C) Animals overexpressing EDC-3 (pedc-3EDC-3, pedc-3EDC-3::DsRed) under its endogenous promoter are short lived compared with wild-type nematodes. RNAi-mediated knockdown of edc-3 extends lifespan, though to a lesser extent than the edc-3 (ok1427) deletion allele.
(D) Loss of the PB-specific factor LSM-3 leads to a significant lifespan extension, while the overexpression of plsm-3LSM-3::DsRed significantly shortens lifespan compared with the wild-type.
(E) Downregulation of edc-3 further prolongs the lifespan of daf-2(e1370), dietary-restricted eat-2(ad465), and clk-1(e2519) mutants, compared with genetically identical control animals fed with bacteria harboring the RNAi empty vector.
(F) Knockdown of edc-3 does not further increase the lifespan of long-lived mutants harboring the ife-2(ok306) allele. ife-2(ok306);edc-3 (ok1427) double mutants do not show significant lifespan extension compared with ife-2(ok306) mutants.
(G) Downregulation of ife-2 by RNAi partially suppresses the short-lived phenotype of EDC-3-overexpressing animals.
Next, we examined the effects of loss or overexpression of EDC-3 on C. elegans lifespan. Knockdown of edc-3 via RNAi extends lifespan at 20°C. Consistently, we observed a substantial increase in the lifespan of edc-3(ok1427) mutants harboring a deletion in the edc-3 locus (Figure 2C). The corresponding mutant protein lacks 202 amino acids, including the conserved FDF domain of EDC-3 (Figure S4A). EDC-3 deficiency had a similar effect on lifespan at 25°C (Figure S4B). Notably, edc-3 mutation caused a more pronounced lifespan extension compared with RNAi-mediated edc-3 knockdown (Figure 2C), most probably because of inefficient silencing of the gene in C. elegans neurons, which are known to be largely refractory to RNAi (Tavernarakis et al., 2000). By contrast, overexpression of either the native EDC-3 (pedc-3EDC-3) or a full-length DsRed fusion (pedc-3EDC-3::DsRed), driven by the endogenous edc-3 promoter, markedly shortened the lifespan of wild-type animals (Figure 2C). Similarly, we observed that animals carrying a mutation in the PB component LSM-3 that is part of the deadenylation complex, which functions prior to decapping (Figure 2A), are long lived. Conversely, LSM-3-overexpressing animals exhibited a short lifespan phenotype (Figure 2D), corroborating the notion that downregulation of specific mRNA degradation components promotes longevity in C. elegans.
Because Dcp2 interacts with Edc3 in yeast and in mammalian cells (Decker and Parker, 2012, Decker et al., 2007), we tested whether the Dcp2 C. elegans homolog, DCAP-2, might also influence aging. We found that RNAi-mediated knockdown of dcap-2 had no significant effect on the lifespan of wild-type worms. Consistent with this result, the lifespan of dcap-2(ok2023) deletion mutants was not altered (Figure S4C). Interestingly, RNAi-mediated knockdown of edc-3 did not cause significant modification of the life expectancy of dcap-2 mutants, while dcap-2 knockdown significantly reduced the lifespan of edc-3 mutants (Figure S4C; Table S1). Together, these findings raise the possibility that the decapping complex components EDC-3 and DCAP-2 might interdependently modulate aging in C. elegans.
We further investigated whether PB functions interface with pathways known to influence lifespan in C. elegans (Kenyon, 2010). We found that downregulation of edc-3 further enhanced the longevity of animals carrying mutations in the insulin/IGF-1-like receptor homolog DAF-2 (Figure 2E). In addition, knockdown of edc-3 further increased the lifespan of dietary-restricted eat-2 mutants (Lakowski and Hekimi, 1998) (Figure 2E). Similarly, EDC-3 depletion extended the lifespan of long-lived mitochondrial clk-1 mutants that are defective in the biosynthesis of ubiquinone, an essential component of the electron transport chain (Felkai et al., 1999) (Figure 2E). Together, these observations suggest that EDC-3 functions independently of insulin/IGF-1 signaling, dietary restriction, or mitochondrial energy metabolism to regulate lifespan. In sharp contrast, EDC-3 deficiency failed to further enhance the longevity of animals depleted for the soma-specific eIF4E isoform IFE-2 (Hansen et al., 2007, Pan et al., 2007, Syntichaki et al., 2007) (Figure 2F), supporting the idea that loss of EDC-3 modulates lifespan via eIF4E. Indeed, knockdown of ife-2, at least partially, counterbalanced the detrimental outcome of EDC-3 overexpression on lifespan (Figure 2G). Moreover, we observed that dcap-2 knockdown shortened the lifespan of IFE-2-deficient animals, whereas ife-2 knockdown significantly increased the lifespan of dcap-2(ok2023) mutants (Figure S4D), suggesting that DCAP-2 and EDC-3 share redundant functions in aging modulation.
To gain insight into the mechanism underlying the effects of PBs on aging, we considered whether EDC-3 deficiency indirectly increases longevity by interfering with animal morphology or behavior. Neither edc-3-mutant nor edc-3 RNAi-treated animals showed significant differences in ultradian rhythmic or other behaviors previously implicated in aging, including pharyngeal pumping of bacterial food into the intestine, sinusoidal locomotion, dauer formation, developmental timing, and internal organ morphology (gonad, intestine, musculature, and pharynx), while edc-3 mutants displayed a slightly reduced egg-laying rate and shortening of the body, compared with wild-type animals (Figure S4E).
mRNA Decapping Influences Longevity by Regulating Protein Synthesis
Having established that the abundance of PBs in somatic C. elegans tissues increases during aging, we sought to explore the contribution of mRNA decapping factors to age-related accumulation of PBs. We found that the number of DCAP-1-specific PBs as well as the levels of DCAP-1::DsRed signal markedly increased in both wild-type and EDC-3-deficient animals with age (Figures 3A and S4F). Moreover, edc-3 mutants fail to form large PBs late in adulthood (Figure S4G). Congruently, IFE-2 levels significantly decreased during aging; this decrease was attenuated in edc-3 mutant animals (Figure 3B). Our findings suggest a functional interaction between EDC-3 and IFE-2. To test the hypothesis that EDC-3 depletion influences aging by directly modulating translation initiation in somatic cells via IFE-2 availability, we monitored protein synthesis rates in whole animals or specific somatic tissues in vivo, using fluorescence recovery after photobleaching (FRAP; Kourtis and Tavernarakis, 2009a, Kourtis and Tavernarakis, 2017, Syntichaki et al., 2007). Protein synthesis rates were assessed in wild-type animals and edc-3(ok1427) mutants expressing GFP either throughout somatic tissues under the control of the ife-2 gene promoter (pife-2GFP; at L4 larval stage and day 5 of adulthood) or specifically in neurons under the control of the pan-neuronal unc-119 gene promoter (punc-119GFP; at days 2 and 7 of adulthood). We found that fluorescence recovery was significantly faster in wild-type animals compared with edc-3(ok1427) mutants (Figures 3C–3E), suggesting that alterations in PB function and EDC-3-mediated mRNA turnover modulate aging by impinging on mRNA translation initiation.
Figure 3.
EDC-3 Deficiency Promotes IFE-2 Entrapment in P Bodies and Reduces Protein Synthesis in Somatic C. elegans Tissues during Aging
(A) The number of DCAP-1::DsRed-labeled PBs increases in edc-3 mutants compared with wild-type at older age. Error bars denote SEM; n = 25 animals per trial. ∗p < 0.01, ∗∗p < 0.001, and ∗∗∗p < 0.0001 (unpaired t test between the edc-3 mutant and the age-matched wild-type or wild-type day 10 and day 17 compared with wild-type at day 3).
(B) IFE-2::GFP intensity decreases substantially with age in both wild-type and edc-3-mutant background but remains higher in animals carrying the edc-3 (ok1427) allele. Error bars denote SEM; n = 25 animals per trial. ∗p < 0.01, ∗∗p < 0.001, and ∗∗∗p < 0.0001 (unpaired t test between the edc-3 mutant and the age-matched wild-type or wild-type at day 10 and day 17 compared with wild-type at day 3 of adulthood).
(C–E) Fluorescence recovery after photobleaching (FRAP) studies to measure de novo protein synthesis at various time points. The percentage of fluorescence recovery is calculated on the basis of average pixel intensity measured before bleaching (100%) and directly after bleaching the signal to about 20%–40% of the original intensity. Error bars in all studies denote SD. ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.0001 (multiple t tests with the Holm-Sidak method, in reference to the control with alpha = 5.000%).
(C) Fluorescence recovery is monitored in wild-type animals and edc-3 mutants expressing the pan-neuronal reporter punc-119GFP. Percentage of fluorescence recovery in the area of the nerve ring is plotted against time for 2-day-old and 7-day-old adult animals (n = 24 animals).
(D) Recovery of the fluorescent signal in all somatic tissues of wild-type and edc-3-mutant animals expressing pife-2GFP, with or without cycloheximide treatment, at L4 stage and day 5 of adulthood (n = 10 animals).
(E) FRAP study in animals fed with control (empty vector), edc-3, or ife-2 RNAi, at L4 stage and day 5 of adulthood (n = 10 animals).
(F) IFE-2::GFP sequestration in PBs is increased in EDC-3-deficient animals compared with wild-type controls as quantified by mean voxel intensity over time in 2-day-old and 6-day-old nematodes. Animals are constantly exposed to 34°C. Error bars denote SEM; n = 5 animals.
(G) IFE-2::GFP sequestration in PBs at 5.5 min after constant exposure to heat stress at 34°C is markedly decreased in animals subjected to hsf-1 RNAi. Error bars denote SEM; n = 5 animals. ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.0001 (unpaired t test to compare edc-3(ok1427) mutant background with the control).
See also Figure S4.
To investigate the mechanism by which EDC-3 influences eIF4E availability to modulate longevity, we monitored IFE-2::GFP granulation and colocalization with PBs, following heat stress at 34°C, over the course of 5.5 min in a three-dimensional (3D) time-lapse experiment (Figure S1B; Video S2; STAR Methods). IFE-2::GFP localization to PBs was significantly higher in edc-3 mutants, compared with wild-type animals, at day 2 of adulthood and, to a lesser extent, at day 6 of adulthood (Figure 3F). In addition, we measured eIF4E sequestration at 0 and 5.5 min in edc-3(ok1427) mutant and wild-type animals, upon downregulation of key lifespan and stress response regulators such as SKN-1, HSF-1, and DAF-16. At day 2 of adulthood, no significant alterations were detected, and edc-3 mutants generally displayed higher eIF4E localization to PBs, compared with wild-type (Figure 3G). In 6-day-old adults, eIF4E sequestration in PBs of edc-3 mutants was reduced to wild-type levels, specifically upon hsf-1 knockdown (Figure 3G). Notably, downregulation of the C. elegans mTOR homolog let-363, which lowers protein synthesis rates and prolongs lifespan (Hansen et al., 2007), increased eIF4E localization to PBs in edc-3 mutants during adulthood (Figure 3G). Altogether, these findings indicate that EDC-3 regulates the sequestration of eIF4E in PBs, thereby controlling its availability. This sequestration upon heat stress depends on the activity of HSF-1.
A Transcriptional Network Mediates the Effects of mRNA Decapping on Lifespan and Stress Resistance
Key signal transduction pathways regulating lifespan converge on specific transcription factors that influence the expression of diverse genes, ultimately producing longevity effects. Central among these transcription regulators are the forkhead transcription factor DAF-16 (Hsu et al., 2003), the Nrf 1 and 2 (Nuclear factor-erythroid-related factors 1 and 2) homolog SKN-1 (An and Blackwell, 2003), and the heat shock transcription factor HSF-1 (Morley and Morimoto, 2004, Seo et al., 2013). To determine whether impairment of PB function modulates aging by altering gene transcription, we examined the effects of DAF-16, SKN-1, and HSF-1 depletion on edc-3-mutant animals. Knockdown of daf-16 abrogated longevity instigated by EDC-3 deficiency (Figure 4A). Similarly to daf-16, knockdown of hsf-1 abolished longevity conferred by depletion of PB components (Figure 4B). In agreement with previous studies postulating an involvement of HSF-1 in the lifespan extension upon reducing general protein synthesis (Howard et al., 2016), downregulation of hsf-1 abrogated the lifespan extension mediated by loss of IFE-2 (Figure 4C). In addition, SKN-1 depletion shortened the lifespan of both wild-type and edc-3-mutant animals (Figure 4D). SKN-1 has previously been shown to mediate oxidative stress resistance and longevity under conditions of reduced protein synthesis (Wang et al., 2010). Thus, we considered whether hormetic signaling via reactive oxygen species (ROS) engages SKN-1 upon edc-3 knockdown to extend lifespan (Kourtis and Tavernarakis, 2011). To test this hypothesis, we treated wild-type and edc-3(ok1427) mutant animals with N-acetylcysteine (NAC), an antioxidant and ROS scavenger (Benrahmoune et al., 2000). We found that lifespan extension caused by edc-3 deletion is largely mitigated by NAC treatment (Figure 4E). The lifespan of wild-type controls remained unaffected in these conditions. Importantly, although NAC ameliorated the detrimental effect of skn-1 knockdown on longevity of edc-3(ok1427) mutants, it did not ameliorate the short lifespan of animals subjected to skn-1 RNAi (Figure 4F). We also found that skn-1 knockdown markedly increased the number of PBs across all tissues in aged edc-3 mutants (day 8 of adulthood), while NAC treatment reversed this effect (Figure 4G). Similarly, we observed a slight increase of DCAP-1 foci in both wild-type and edc-3 mutant worms subjected to daf-16 RNAi (Figure S5A). Moreover, HSF-1 was also required for temperature-dependent PB accumulation in both wild-type animals and edc-3 mutants (20°C versus 25°C; Figure S5B). Notably, the levels of DCAP-1::DsRed were substantially elevated upon heat shock in edc-3(ok1427) mutants compared with wild-type animals at day 1 of adulthood (Figure S5C). Supporting a role of key transcription factors in mediating the effects of PB components on longevity, both daf-16 and skn-1 knockdown significantly shortened the lifespan of dcap-2(ok2023) mutants (Figure S5D).
Figure 4.
Longevity and Oxidative Stress Resistance in EDC-3-Depleted Animals Depend on SKN-1, DAF-16, and HSF-1 Transcription Factors
(A) Knockdown of daf-16 shortens the lifespan of both wild-type and edc-3-mutant animals.
(B) Knockdown of hsf-1 shortens the lifespan of edc-3 mutants below that of wild-type animals.
(C) Downregulation of hsf-1 reduces the lifespan of ife-2 mutants to levels of respective controls.
(D) Knockdown of skn-1 reduces the lifespan of edc-3 mutants below wild-type levels.
(E) N-acetylcysteine (NAC) diminishes longevity conferred by EDC-3 deficiency.
(F) Survival curves of EDC-3-deficient animals subjected to skn-1 knockdown, compared with skn-1-deficient worms, with or without NAC treatment. The log rank (Mantel-Cox) test shows a significant difference between edc-3(ok1427);skn-1(RNAi)-NAC and edc-3(ok1427);skn-1(RNAi)+NAC (∗∗∗p < 0.0001), while the lifespan of wild-type treated with skn-1(RNAi) with or without NAC is not significantly different (p = 0.6534).
(G) Quantification of DCAP-1::DsRed foci in wild-type and edc-3-mutant animals at day 8 of adulthood. Error bars show SEM; n = 25 animals per experiment. ∗p < 0.05 and ∗∗∗p < 0.001 (one-way ANOVA). N-acetylcysteine (NAC) treatment suppresses the increased abundance of PBs caused by skn-1 knockdown.
(H) Knockdown of edc-3 increases the lifespan of short-lived mev-1 mutants that are sensitized to oxidative stress.
(I) Survival of 7-day-old, EDC-3-deficient nematodes exposed to the oxidative phosphorylation inhibitor sodium azide. Error bars denote SEM; n > 80 per trial; ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.0001 (unpaired t test).
(J) Survival of 7-day-old, EDC-3-depleted nematodes under oxidative stress induced by the herbicide paraquat. Error bars represent the SEM; n > 80 per trial; ∗p < 0.05, ∗∗p < 0.005, and ∗∗∗p < 0.0001 (unpaired t test).
Having established that eIF4E protein levels steadily decline during aging (Figure 1F) and that downregulation of key mRNA decapping components influences eIF4E abundance in somatic C. elegans tissues (Figure S3L), we set out to examine whether these changes are mediated by DAF-16, SKN-1, or HSF-1, which are known for their roles in proteostasis maintenance. Although depletion of SKN-1 or DAF-16 did not alter IFE-2 abundance, HSF-1 deficiency triggered accumulation of the IFE-2::GFP protein and upregulation of ife-2 transcription in aged animals (day 8 of adulthood; Figures S5E and S5F). In addition to increasing IFE-2 protein levels, HSF-1 depletion caused extensive IFE-2 granulation in somatic tissues (Figure S5G). Taken together, these findings indicate that HSF-1 modulates both PB accumulation and eIF4E abundance during aging in somatic tissues.
Stress response mechanisms are known to interface with conserved signaling pathways and interventions that influence aging (Kirkwood and Austad, 2000). Intrigued by the effects of PB perturbations on longevity, we investigated whether interfering with PB function alters stress resistance in C. elegans. Nematodes deficient for the mitochondrial integral membrane protein cytochrome b MEV-1, which is part of the respiratory chain complex II (ubiquinol-cytochrome c reductase), experience oxidative stress and are short lived (Ishii et al., 2011). Knockdown of edc-3 increased the lifespan of mev-1 mutant animals (Figure 4H). Similarly, edc-3 knockdown ameliorated the short-lived phenotype of gas-1 mutants (Kayser et al., 2004), deficient for the 49 kDa iron-sulfur subunit of the mitochondrial electron transport chain complex I (Table S1). In addition, EDC-3 deficiency increased resistance to the chemical compounds NaN3 and paraquat (PQ) that are potent oxidative stress inducers (Figures 4I and 4J). Taken together, these results indicate that EDC-3 depletion enhances oxidative stress resistance during adulthood.
Given that SKN-1 is required for lifespan extension and stress resistance caused by edc-3 downregulation, we tested whether impairment of the decapping complex promotes SKN-1-dependent transcriptional changes. To this end, we assessed the expression of gst-4, which encodes a putative glutathione-requiring prostaglandin D synthase and is a known target of SKN-1 (Wang et al., 2010), upon knockdown of dcap-1, dcap-2, or edc-3. We found that interfering with the decapping complex causes a significant upregulation of pgst-4GFP transcriptional reporter (Figure S5H). In addition, we also tested DAF-16 activation using its target gene sod-3 (superoxide dismutase; Libina et al., 2003). Knockdown of dcap-1, dcap-2, or edc-3 increased the levels of psod-3GFP transcriptional fusion (Figure S5I). To establish a more direct link between mRNA decapping impairment and stress resistance, we used the fluorescent hydrogen peroxide (H2O2) sensor protein HyPer (Belousov et al., 2006) for monitoring ROS levels. We found that ROS production increased upon downregulation of key mRNA decapping genes (Figure S5J). Finally, we observed that EDC-3::DsRed fluorescence increased upon heat shock (at 35°C for 1.5 hr; Figure S5K), and EDC-3-depleted animals were more thermotolerant than wild-type worms (Figures S5L and S5M). Altogether, these findings indicate that mRNA decapping modulates aging and stress responses through a multilayered mechanism relying on the activity of SKN-1, HSF-1, and, albeit to a lesser extent, DAF-16.
mRNA Decapping Activity in Neurons Regulates Aging
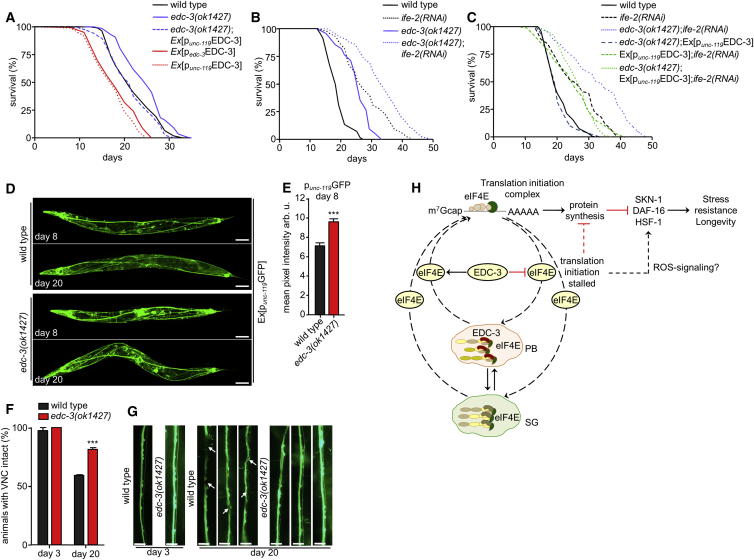
Our findings demonstrated that edc-3 is predominantly expressed in neurons during adulthood (Figure 2B), and RNAi-mediated knockdown had milder effects on lifespan compared with edc-3 deletion allele (Figure 2C), raising the possibility that EDC-3 function in the nervous system modulates aging in C. elegans. To test this hypothesis, we created transgenic wild-type and edc-3(ok1427)-mutant animals, expressing edc-3 under the control of the pan-neuronal unc-119 promoter. In both cases, neuron-specific expression of EDC-3 markedly reduced lifespan. Moreover, neuron-specific expression of edc-3 was sufficient to recapitulate the lifespan shortening effect of ubiquitous edc-3 overexpression (Figure 5A). In addition, ife-2(RNAi) markedly increased the lifespan of edc-3 mutants (Figure 5B). In contrast, edc-3(RNAi) failed to increase further the lifespan of IFE-2-depleted animals, as previously mentioned (Figure 2F), which is consistent with the notion that EDC-3 functions primarily in neurons, in which RNAi might not act efficiently to knock down edc-3. These observations suggest that IFE-2 acts downstream or more likely in parallel with EDC-3 to regulate aging. To delineate further the interplay between IFE-2 and EDC-3, we tested whether lack of IFE-2 affects the lifespan of EDC-3-overexpressing animals. Interestingly, we found that ife-2 knockdown increased the lifespan of animals overexpressing EDC-3 in neurons (Figure 5C) and suppressed the short-lived phenotype caused by neuronal expression of EDC-3 in edc-3-mutant animals (Figure 5C). Taken together, these findings suggest that IFE-2 might function in neuronal and non-neuronal tissues, while EDC-3 predominantly acts in the nervous system to modulate longevity.
Figure 5.
EDC-3 Functions in the Nervous System to Modulate Aging
(A) Pan-neuronal expression of punc-119EDC-3 abrogates longevity of the edc-3(ok1427)-mutant strain.
(B) RNAi-mediated knockdown of ife-2 increases the lifespan of animals bearing the edc-3(ok1427) mutation.
(C) Pan-neuronal expression of punc-119EDC-3 in edc-3 mutants abrogates lifespan extension caused by downregulation of ife-2.
(D) Representative confocal images of 8- and 20-day-old wild-type and edc-3(ok1427)-mutant animals expressing the pan-neuronal punc-119GFP reporter. Scale bars, 100 μm.
(E) Corresponding quantification of fluorescence intensity at day 8 of adulthood. Error bars represent the SEM; n = 40 animals per experiment. ∗∗∗p < 0.0001 (unpaired t test).
(F) Quantification of defects in the ventral nerve cord (VNC) of wild-type and edc-3 mutants bearing the punc-119GFP transgene during aging. Wild-type worms display a higher frequency of ventral nerve cord defects compared with edc-3 mutants at day 20 of adulthood. Error bars denote SEM; n = 25 animals per experiment; ∗∗∗p < 0.0001 (unpaired t test between edc-3[ok1427] and the age-matched control).
(G) Representative images of the VNC in wild-type and edc-3 mutant worms at day 3 and day 20 of adulthood. Scale bars, 10 μm.
(H) Proposed model for PB-mediated control of protein synthesis in the soma and its causative role in the modulation of aging. The PB-specific component EDC-3 influences mRNA translation by modulating mRNA decapping. Properly functioning decapping limits eIF4E localization to PBs. By contrast, stalled translation initiation complexes accumulating in EDC-3-deficient PBs and SGs cause eIF4E sequestration, lowering its availability for translation initiation. In turn, protein synthesis reduction leads to an orchestrated activation of the transcription factors SKN-1, HSF-1, and DAF-16, possibly via ROS signaling, mediating gene expression changes that increase longevity and stress resistance.
We next examined the morphology and integrity of the nervous system in wild-type and edc-3 mutant background using a GFP reporter driven by the pan-neuronal unc-119 promoter and found that its expression was higher in edc-3 mutants at older age compared with wild-type nematodes (Figures 5D and 5E). Moreover, neural morphology and overall integrity are better maintained during aging in edc-3 mutants compared with age-matched wild-type animals (Figures 5F and 5G). We focused on the ventral nerve cord (VNC), which stretches along the whole body length of the adult animal (White et al., 1976), and performed a systematic temporal analysis of VNC neuronal phenotypes, including apparent breaks, waving, and acute bends of axons. We observed that EDC-3 deficiency attenuates age-dependent deterioration of the VNC (Figures 5F and 5G). We also found that EDC-3::DsRed foci accumulate mainly in neurons and the intestine (Figures S6A–S6C) and increasingly colocalize with IFE-2::GFP, primarily in neuronal tissues, including the VNC, head and tail neurons, during aging (Figures S6D–S6G). Together these findings suggest that sequestration of eIF4E in neuronal PBs contributes to the maintenance of nervous system integrity with increasing age.
Discussion
Our study elucidates a molecular mechanism modulating protein synthesis in the C. elegans soma via association of the mRNA translation initiation factor eIF4E with PBs (Figure 5H). We show that the somatic eIF4E isoform IFE-2 localizes to PBs during aging and under stress conditions. Accumulating evidence suggests that blocking mRNA degradation, particularly at the stage of decapping, results in increased PB formation (Sheth and Parker, 2003). Consistent with this notion, we find that EDC-3 deficiency increases PB formation preferentially in old animals or animals subjected to stress. In addition, deletion or depletion of the enhancer of decapping EDC-3, among other PB components, increases the levels of IFE-2 in the soma, triggers the formation of IFE-2-containing granules, and enhances IFE-2 sequestration in PBs. Moreover, we find that the rate of protein synthesis in somatic tissues is reduced under these conditions. The effects of EDC-3 on stress resistance and longevity mirror the cytoplasmic availability of IFE-2. Combined, these observations indicate that PB formation directly impinges on mRNA translation regulation in the soma, which is central to the maintenance of proteostasis during aging.
Previous studies have shown that reduced mRNA translation, as a consequence of IFE-2 loss, protects from oxidative stress and increases lifespan in C. elegans (Hansen et al., 2007, Pan et al., 2007, Syntichaki et al., 2007). In line with this notion, knockdown of specific PB components, such as EDC-3 and LSM-3, enhances heat and oxidative stress resistance and extends lifespan. Conversely, EDC-3 or LSM-3 overexpression shortens lifespan. Together, these data suggest that PB components have a dose-dependent effect on lifespan, which is consistent with previous findings demonstrating that overexpression of the dendritic PB component TDP-1/TDP-43 (Wang et al., 2008) results in age-dependent proteotoxicity and short lifespan, while TDP-1 depletion increases lifespan in worms (Vaccaro et al., 2012), which is further supported by studies in flies (Estes et al., 2011) and mice (Xu et al., 2010). It is worth noting that despite the evolutionary conservation of key PB components, the effects of their overexpression or depletion on their formation and function vary depending on the specific component and the species.
Moreover, we find that the ROS scavenger NAC suppresses lifespan extension caused by the edc-3 mutation, indicating that mitohormesis likely mediates the effects of EDC-3 depletion on stress resistance and aging in C. elegans. Besides, PB formation is significantly enhanced upon SKN-1 depletion in EDC-3-deficient animals. This increase is suppressed by NAC treatment, suggesting a role for PBs in cellular ROS signaling that impinges on the regulation of organismal aging.
Interestingly, we found that lifespan extension caused by EDC-3 deficiency depends on the multimodal activity of the stress-induced transcription factors SKN-1, DAF-16, and HSF-1. The finding that edc-3 knockdown extends the lifespan of various longevity mutants (daf-2, eat-2), but requires their downstream effectors (DAF-16, HSF-1, SKN-1) for its beneficial effects, suggests that EDC-3 deficiency triggers a stress response through multiple longevity effectors that are all required for extended longevity.
Previous work has implicated HSF-1 in the regulation of various RNA granule components, including SG proteins. Moreover, DAF-16 has been shown to prevent age-dependent SG aggregation (Lechler et al., 2017). In line with these findings, loss of HSF-1 suppresses PB formation and IFE-2 sequestration in PBs under heat stress conditions specifically during late adulthood (day 6) as opposed to early adulthood (day 2). The aging process is associated with a marked deterioration of vital stress response factors, resulting in increased susceptibility to stress insults. For example, the regulatory role of DAF-16 in the DNA damage response declines during aging (Mueller et al., 2014). Young animals are more resistant to various stresses, including heat shock. The effectiveness of their response to heat in early adulthood (at day 2) might rely on the redundant activity of regulatory factors, such as DAF-16, that most likely mediate adaptation to heat stress (Hsu et al., 2003) and modulate IFE-2 sequestration in PBs, thus compensating for the absence of HSF-1 (Lechler et al., 2017). Given that the heat shock response attenuates with age, HSF-1-mediated adaptive responses to heat and other stress insults may be more crucial for enhancing proteostasis late in life, when other proteostasis surveillance systems fail. In this sense, the role of HSF-1 in mediating IFE-2 sequestration in PBs, thereby controlling protein synthesis and enhancing proteostasis, is more important in late adulthood (day 6) compared with early adulthood (day 2).
Importantly, neuron-specific function of EDC-3 is sufficient to suppress longevity in edc-3-mutant animals. Furthermore, we find that the depletion of EDC-3 robustly suppresses protein synthesis in neurons contributing to the maintenance of neuronal integrity during aging. Indeed, PBs are known to regulate RNA transport and local protein synthesis in neurons to modulate neuronal plasticity (Fritzsche et al., 2013, Hirokawa, 2006, Mitsumori et al., 2017). In this context, we observe a significant decrease in body size in edc-3 mutants. Previous studies have shown that the food-dependent body size change in worms is closely related to protein synthesis that eventually determines the protein content of the cells (So et al., 2011). In line with this, edc-3 mutants have reduced protein synthesis rates that are likely responsible for their smaller body size compared with wild-type animals. Additionally, the nervous system plays a critical role in the regulation of organismal body size, and this effect is likely mediated by the amine neurotransmitter dopamine in C. elegans in response to sensory cues. Specifically, it has been shown that dopamine negatively regulates body size by suppressing octopamine signaling (Funayama and Ishikawa, 2007, Nagashima et al., 2016).
Together, our findings establish a causative link between PBs and aging through a molecular mechanism modulating protein synthesis. In summary, our study elucidates a pivotal role for PBs in controlling the availability of eIF4E, eventually shaping stress responses and organismal survival. The tight evolutionary conservation and ubiquitous expression of PB components and regulatory factors involved suggest that similar mechanisms may regulate protein synthesis in somatic tissues during aging or under stress, across different taxa.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Sodium azide | Merck | S2002; CAS: 26628-22-8 |
| Paraquat | Merck | 36541; CAS: 75365-73-0 |
| N-Acetyl-L-cysteine | Merck | A7250; CAS: 616-91-1 |
| Cycloheximide | Merck | 01810; CAS: 66-81-9 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain N2: wild type | Caenorhabditis Genetics Center | WB strain: N2 |
| C. elegans: Strain RB1311: edc-3(ok1427)I | Caenorhabditis Genetics Center | WB strain: RB1311; Wormbase: WBVar00092641 |
| C. elegans: Strain RB1641: dcap-2(ok2023)IV | Caenorhabditis Genetics Center | WB strain: RB1641; WormBase: WBVar00093213 |
| C. elegans: Strain KX15: ife-2(ok306)X | Caenorhabditis Genetics Center | WB strain: KX15; WormBase: WBVar00091604 |
| C. elegans: Strain RB2609: lsm-3(ok3635)IV | Caenorhabditis Genetics Center | WB strain: RB2609; WormBase: WBVar00094590 |
| C. elegans: Strain CB1370: daf-2(e1370)III | Caenorhabditis Genetics Center | WB strain: CB1370; WormBase: WBVar00143949 |
| C. elegans: Strain CB4876: clk-1(e2519)III | Caenorhabditis Genetics Center | WB strain: CB4876; Wormbase: WBVar00144863 |
| C. elegans: Strain DA465: eat-2(ad465)II | Caenorhabditis Genetics Center | WB strain: DA465; Wormbase: WBVar00000014 |
| C. elegans: Strain CW152: gas-1(fc21)X | Caenorhabditis Genetics Center | WB strain: CW152; Wormbase: WBVar00145317 |
| C. elegans: Strain TK22: mev-1(kn1)III | Caenorhabditis Genetics Center | WB strain: TK22; WormBase: WBVar00088257 |
| C. elegans: Strain CF1553: muIs84 [pAD7 (psod-3GFP)] | Caenorhabditis Genetics Center | WB strain: CF1553; WormBase: CF1553 |
| C. elegans: Strain CF1580: daf-2 (e1370)III;muIs84 | Caenorhabditis Genetics Center | WB strain: CF1580; WormBase: CF1580 |
| C. elegans: Strain CL2166: dvIs19 [pAF15 (gst-4::GFP::NLS)] | Caenorhabditis Genetics Center | WB strain: CL2166; WormBase: CL2166 |
| C. elegans: Strain JV1: [prpl-17HyPer; UNC-119 (+)] | Caenorhabditis Genetics Center | WB strain: JV1; WormBase: JV1 |
| C. elegans: N2;Ex[pife-2IFE-2::GFP;pRF4] | Syntichaki et al., 2007 | N/A |
| C. elegans: N2;Ex[pife-2GFP;pRF4] | Syntichaki et al., 2007 | N/A |
| C. elegans: edc-3(ok1427)III;ife-2(ok306)X | This paper | N/A |
| C. elegans: N2;Ex[pedc-3EDC-3::DsRed;pRF4] | This paper | N/A |
| C. elegans: N2;Ex[pdcap-1DCAP-1::DsRed;pRF4] | This paper | N/A |
| C. elegans: N2;Ex[plsm-3LSM-3::DsRed;pRF4] | This paper | N/A |
| C. elegans: N2;Ex[pedc-3EDC-3::DsRed;pife-2IFE-2::GFP;pRF4] | This paper | N/A |
| C. elegans: edc-3(ok1427);Ex[pdcap-1DCAP-1::DsRed;pife-2IFE-2::GFP;pRF4] | This paper | N/A |
| C. elegans: N2;Ex[pdcap-1DCAP-1::DsRed;pife-2IFE-2::GFP;pRF4] | This paper | N/A |
| C. elegans: N2;Ex[pedc-3EDC-3::DsRed;punc-119GFP;pRF4] | This paper | N/A |
| C. elegans: edc-3 (ok1427);Ex[punc-119GFP;pRF4] | This paper | N/A |
| C. elegans: edc-3 (ok1427);Ex[pedc-3EDC-3+3′UTR;pRF4] | This paper | N/A |
| C. elegans: N2;Ex[pedc-3EDC-3+3′UTR;pRF4] | This paper | N/A |
| C. elegans: edc-3 (ok1427);Ex[punc-119EDC-3+3′UTR;pRF4] | This paper | N/A |
| C. elegans: N2;Ex[punc-119EDC-3+3′UTR;pRF4] | This paper | N/A |
| Oligonucleotides | ||
| Primer: lsm-3 promoter forward GATTCTAGAGTGCACCAAGC TCTCAAC |
This paper | N/A |
| Primer: lsm-3 promoter reverse GATACCGGTCTTTCTTCTCAA TCTGCTTG |
This paper | N/A |
| Primer: lsm-3 gene forward GATACCGGTATGGCCACCGAAA AGAAAG |
This paper | N/A |
| Primer: lsm-3 gene reverse GATACCGGTAGGGATGCACGAA TCGGTG |
This paper | N/A |
| Primer: dcap-1 gene forward GTAATACGACTCACTATAGGGC | This paper | N/A |
| Primer: dcap-1 gene reverse GGACCGGTCCATCGATATTTAG ACGCCGATTG |
This paper | N/A |
| Primer: edc-3 promoter forward TTCTTCTAAGGCCGATTTCACAAC | This paper | N/A |
| Primer: edc-3 promoter reverse CGACAAATTGGGAAAAATAATTG | This paper | N/A |
| Primer: edc-3 gene forward ATGGATGACAAACTCATTGGAAGC | This paper | N/A |
| Primer: edc-3 gene reverse ACCGGTCCATTGGATTGTGAAAG TCTGACAAG |
This paper | N/A |
| Primer: edc-3 gene +3′UTR forward AGAGCGGCCGCCGGTGC TAAAAGTAGACGTGGTTG |
This paper | N/A |
| Primer: edc-3 gene +3′UTR reverse AGAGCGGCCGCATGGAT GACAAACTCATTGGAAGC |
This paper | N/A |
| Primer: unc-119 promoter forward CTCTCAAAATTCAGTTTTTT AAACCAC |
This paper | N/A |
| Primer: unc-119 promoter reverse ATATGCTGTTGTAGCTGAA AATTTTG |
This paper | N/A |
| Primer: edc-3 RNAi forward CAGGAGAATGATGACCAGTACTATG | This paper | N/A |
| Primer: edc-3 RNAi reverse GGAAAGTTGTAGAGAATGCGGTAG | This paper | N/A |
| Primer: dcap-2 RNAi forward CACGAATTCCGAATACCCC | This paper | N/A |
| Primer: dcap-2 RNAi reverse CCGCTCGAGTAACGAGACCAA GTACCG |
This paper | N/A |
| Primer: hsf-1 RNAi forward ACACTGCAGATGCAGCCAACAGG | This paper | N/A |
| Primer: hsf-1 RNAi reverse TGTCTCGAGGATCGTGGTCCTTC | This paper | N/A |
| Primer: skn-1 RNAi forward TCAGTTCACCATCGTCCAACACCTC | This paper | N/A |
| Primer: skn-1 RNAi reverse TGTCGTGACGATCCGTGCGTC | This paper | N/A |
| Primer: daf-16 RNAi forward CACTGATCTTTCAAGCCG | This paper | N/A |
| Primer: daf-16 RNAi reverse CTTGTGCAAGAGTTAACCG | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pRF4: rol-6(su1006 dm), injection marker | Mello et al., 1991 | WormBase: pRF4 |
| Plasmid: pPD95.77 | Fire lab C. elegans Vector Kit 1995, Addgene | Addgene: #1495 |
| Plasmid: pcol-12DsRed | Wong et al., 2007 | N/A |
| Plasmid: pife-2IFE-2::GFP | This paper | N/A |
| Plasmid: pBluescript II KS+ dcap-1 gene | Min Han lab | N/A |
| Plasmid: plsm-3LSM-3::DsRed | This paper | N/A |
| Plasmid: pdcap-1DCAP-1::DsRed | This paper | N/A |
| Plasmid: pedc-3EDC-3::DsRed | This paper | N/A |
| Plasmid: punc-119GFP | This paper | N/A |
| Plasmid: pedc-3EDC-3+3′UTR | This paper | N/A |
| Plasmid: punc-119EDC-3+3′UTR | This paper | N/A |
| Plasmid: L4440 | Fire lab C. elegans Vector Kit 1995, Addgene | Addgene: #1654 |
| Plasmid: dcap-1(RNAi) | This paper | N/A |
| Plasmid: dcap-2(RNAi) | This paper | N/A |
| Plasmid: edc-3(RNAi) | This paper | N/A |
| Plasmid: daf-16(RNAi) | Syntichaki et al., 2007 | N/A |
| Plasmid: hsf-1(RNAi) | This paper | N/A |
| Plasmid: skn-1(RNAi) | Palikaras et al., 2015 | N/A |
| Plasmid: ife-2(RNAi) | Syntichaki et al., 2007 | N/A |
| Plasmid: eat-3(RNAi) | Artal-Sanz and Tavernarakis, 2009 | N/A |
| Software and Algorithms | ||
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Fiji processing package (with Colocalization Analysis tool Coloc2) | Schindelin et al., 2012 | https://fiji.sc |
| Adobe Photoshop | Adobe | https://www.adobe.com/de/ |
Contact for Reagent and Resource Sharing
Please contact Nektarios Tavernarakis at tavernarakis@imbb.forth.gr for reagent and resource sharing.
Experimental Model and Subject Details
C. elegans strains and genetics
All experiments were performed in C. elegans (for strain details see Key Resources Table). We followed standard procedures for maintenance of C. elegans strains and transgenic lines (Brenner, 1974). Animals were grown at 20°C unless noted otherwise. The following strains were used for this study (summarized in Key Resources Table): N2: Wild-type Bristol isolate, RB1311: edc-3(ok1427)I, RB1641: dcap-2(ok2023)IV, KX15: ife-2(ok306)X, RB2609: lsm-3(ok3635)IV, CB1370: daf-2(e1370)III, CB4876: clk-1(e2519)III, DA465: eat-2(ad465)II, CW152: gas-1(fc21)X, TK22: mev-1(kn1)III. To image DAF-16 transcription factor target activation we used: CF1553: muIs84 [pAD7 (psod-3GFP)] and CF1580: daf-2 (e1370)III;muIs84. To image SKN-1 target activation we used: CL2166:dvIs19 [pAF15 (gst-4::GFP::NLS)] and ife-2 (ok306);dvIs19. To detect H2O2 enrichment in vivo, we used JRIS1: [prpl-17HyPer; UNC-119 (+)]. To study the possible interactions of aging pathways we created edc-3(ok1427)III;ife-2(ok306)X mutants. Overexpression lines for this study were created as extrachromosomal arrays through microinjection (Rieckher et al., 2009). To study PB or SG localization and colocalization we established: N2;Ex[pedc-3EDC-3::DsRed;pRF4], N2;Ex[pife-2IFE-2::GFP;pRF4] (Syntichaki et al., 2007), N2;Ex[pdcap-1DCAP-1::DsRed;pRF4], N2;Ex[plsm-3LSM-3::DsRed;pRF4], N2;Ex[pedc-3EDC-3::DsRed;pife-2IFE-2::GFP;pRF4],edc-3(ok1427);Ex[pdcap-1DCAP-1::DsRed;pife-2IFE-2::GFP;pRF4], which was outcrossed with N2 to create N2;Ex[pdcap-1DCAP-1::DsRed;pife-2IFE-2::GFP;pRF4]. To monitor neuron-specific fluorescence and colocalization with PBs we created: N2;Ex[pedc-3EDC-3::DsRed;punc-119GFP;pRF4], edc-3(ok1427);Ex[punc-119GFP;pRF4], which was outcrossed with N2 to create N2;Ex[punc-119GFP;pRF4]. To study the effects of EDC-3 overexpression on aging, we created edc-3(ok1427);Ex[pedc-3EDC-3+3′UTR;pRF4] and N2;Ex[pedc-3EDC-3+3′UTR;pRF4]. To examine the neuron-specific role of EDC-3 in aging we established edc-3(ok1427);Ex[punc-119EDC-3+3′UTR;pRF4] and N2;Ex[punc-119EDC-3+3′UTR;pRF4].
Method Details
Molecular cloning
All primers used for molecular cloning are listed in the Key Resources Table. To investigate the expression patterns of PB-specific genes we first constructed a DsRed reporter. GFP was removed from plasmid vector pPD95.77 using the AgeI/EcoRI restriction enzymes. DsRed was isolated as an AgeI/EcoRI fragment from vector pcol-12DsRed (Wong et al., 2007), and inserted into the corresponding sites of pPD95.77, thereby creating pPD95.77 (DsRed). The PB-specific gene lsm-3 is located in C. elegans operon CEOP4548. To construct plsm-3LSM-3::DsRed we designed two primer pairs to individually amplify the gene and endogenous promoter from genomic DNA: “lsm-3 promoter forward” and “lsm-3 promoter reverse” for the promoter region covering 880bp upstream of the first gene mdt-9 in the operon and “lsm-3 gene forward” and “lsm-3 gene reverse “ for the 566bp full sequence of lsm-3. Both PCR fragments were subcloned into pCRII-TOPO vector (Invitrogen). The lsm-3 promoter was excised as an AgeI/XbaI fragment and inserted into pPD95.77 (DsRed). Finally, the lsm-3 genomic sequence was cloned into AgeI site between the promoter and the DsRed. The resulting construct was injected into gonads of wild-type animals together with pRF4, a plasmid carrying the rol-6(su1006) dominant transformation marker. The PB reporter pdcap-1DCAP-1::DsRed was created from a pBluescript II KS+ containing the whole dcap-1 gene and promoter by amplification with “dcap-1 gene forward” and “dcap-1 gene reverse.” The resulting 2300bp fragment was digested with AgeI/XbaI and inserted into pPD95.77 (DsRed). The final construct was co-injected with pRF4 into gonads of wild-type animals. To generate pedc-3EDC-3::DsRed full length DsRed reporter fusion, the primers “edc-3 promoter forward” and “edc-3 promoter reverse” were used to amplify a 1429 bp fragment encompassing the putative promoter sequence of edc-3 gene. The PCR fragment was initially cloned into pCR-II TOPO vector (Invitrogen) and after digestion with XbaI/BamHI it was inserted into plasmid vector pPD95.77 (DsRed). The primers “edc-3 gene forward” and “edc-3 gene reverse” were used to amplify an AgeI-flanked fragment containing the edc-3 genomic sequence. The 2550bp PCR generated fragment was initially cloned into pCR-II TOPO vector (Invitrogen). The resulting plasmid was digested with ApaI, blunted with Klenow (NEB) and recut with AgeI. The ApaI blunted/AgeI fragment was gel purified and inserted downstream of the edc-3 promoter sequence into BamHI blunted/AgeI sites of plasmid pPD95.77-DsRed. The final construct was injected into gonads of wild-type animals together with pRF4. To study the neuron-specific role of EDC-3 in aging we created the pan-neuronal reporter punc-119GFP. We used the primer pair “unc-119 promoter forward” and “unc-119 promoter reverse” to amplify a 1990bp sequence upstream of unc-119 gene. The resulting fragment was initially subcloned into pCRII-TOPO and subsequently isolated as an AgeI/XbaI fragment and inserted into pPD95.75 at the corresponding sites. The resulting construct was injected together with pRF4 into gonads of edc-3 (ok1427) mutant worms and outcrossed with wild-type to study general expression levels when decapping is compromised. We further co-injected the construct with pedc-3EDC-3::DsRed and pRF4 into gonads of wild-type animals to study colocalization of EDC-3 with the neuronal system. For SG and PB colocalization studies, we used the construct pife-2IFE-2::GFP (Syntichaki et al., 2007) and co-injected it with either pdcap-1DCAP-1::DsRed or pedc-3EDC-3::DsRed and the transformation marker pRF4 into edc-3 (ok1427) mutants or wild-type animals, respectively. To study the effects of EDC-3 global and neuronal overexpression on aging we constructed pedc-3EDC-3+3′UTR and punc-119EDC-3+3′UTR. For expression under its endogenous promoter, we used pCRII-TOPO containing edc-3 promoter and EDC-3::DsRed and excised EDC-3::DsRed through flanking NotI restriction sites. The edc-3 gene including its 3′UTR were amplified from genomic DNA with the primer pair “edc-3 gene +3′UTR forward” and “edc-3 gene +3′UTR reverse” containing NotI restriction sites. The resulting fragment was NotI digested and inserted downstream of either edc-3 promoter or unc-119 promoter in pCRII-TOPO. The final plasmids were injected into gonads of wild-type and edc-3(ok1427) mutant animals. We generated several RNAi constructs for PB-specific genes including edc-3, dcap-1, dcap-2 and ife-2 using C. elegans genomic DNA as template and the appropriate primer sets. For edc-3 RNAi, a 1440bp fragment was amplified with the primer pair “edc-3 RNAi forward” and “edc-3 RNAi reverse,” subcloned into pCRII-TOPO, excised with EcoRI and finally ligated into pL4440. For the dcap-1 RNAi construct, a 1000bp fragment was excised from pBluescript II KS+ containing dcap-1 ORF with EcoRI/XhoI and cloned into the corresponding sites of pL4440. For dcap-2 RNAi, the primer pair “dcap-2 RNAi forward” and “dcap-2 RNAi reverse” was used to generate an 800bp fragment, which was cloned into L4440 after digestion with XbaI/BamHI. Preparation of the ife-2 RNAi construct was described previously (Syntichaki et al., 2007). To study whether hsf-1, skn-1 and daf-16 are epistatic to mutations in genes acting in the decapping pathway, we created RNAi constructs for each of the transcription factors. For skn-1 RNAi, a 4079bp fragment was PCR amplified using the following primers: “skn-1 RNAi forward” and “skn-1 RNAi reverse.” The resulting fragment was subcloned into pCRII-TOPO, and after digestion with EcoRI, it was inserted into L4440 (Palikaras et al., 2015). For daf-16 RNAi, a 1.2kb fragment resulted from amplification with “daf-16 RNAi forward” and “daf-16 RNAi reverse” primers, was initially subcloned into pCRII-TOPO and after digestion with HindIII/XhoI it was ligated into L4440 vector (Syntichaki et al., 2007). The hsf-1 RNAi construct was generated by amplifying a 1000bp fragment through the primers “hsf-1 RNAi forward” and “hsf-1 RNAi reverse.” The PCR generated fragment was digested with PstI/XhoI and cloned into the corresponding sites of pL4440. To investigate whether mitochondrial integrity affects PB formation, we used a previously described eat-3 RNAi construct (Artal-Sanz and Tavernarakis, 2009). All the RNAi constructs were transformed into HT115 (DE3) E. coli bacteria, deficient for RNase III. To characterize the edc-3(ok1427) deletion allele, total RNA was extracted from the RB1311: edc-3(ok1427)I strain using Trizol (Life Technologies). cDNA was synthesized using the Accuscript high fidelity 1st strand synthesis kit (Stratagene) and the edc-3 gene-specific primers according to the manufacturer’s instructions. edc-3 cDNA was amplified using 2μl of the experimental first strand cDNA reaction, the Pfu Ultra DNA polymerase of Stratagene and the following pair of primers: “edc-3 gene forward” and “edc-3 gene reverse.” The ∼1095bp PCR generated fragment was cloned into pCRII-TOPO vector after adding 3′ A overhangs and sequenced using SP6 and T7 primers.
Lifespan analysis
Lifespan analyses were performed at 20°C unless noted otherwise. Synchronous animal populations were generated by egg-laying experiments with 10 to 15 gravid adults per plate, which were removed after 5 hours. For RNAi lifespan experiments worms were placed on RNAi plates containing 1mM IPTG and seeded with HT115(DE3) bacteria transformed with either the L4440 vector or the test RNAi construct. For assays evaluating the effect of hormesis on aging, N-acetyl-cysteine (NAC, Invitrogen) was added into NGM media at a final concentration of 10mM. Progeny were grown through the L4 larval stage and then transferred to fresh plates in groups of 15-17 worms per plate for a total of 150–170 individuals per experiment. The day after egg laying was used as t = 0 (days from hatching), unless indicated otherwise. Animals were transferred to fresh plates every 2–3 days and examined every second day for touch-provoked movement and pharyngeal pumping, until death. Worms that died due to internally hatched eggs, an extruded gonad or desiccation due to crawling on the edge of the plates, were incorporated into the dataset as censored. Each survival assay was repeated at least twice and Figures represent typical assays. The percentage of animals remaining alive is plotted against animal age. Lifespan assays were carried out at 20°C unless otherwise stated. Full lifespan statistics are summarized in the Table S1. Statistical analysis of lifespan assays was performed using the Prism software package (GraphPad Software), which applies the method of Kaplan and Meier and the log-rank (Mantel–Cox) test to evaluate differences between survivals and the corresponding p values.
Protein synthesis assays (Fluorescence recovery after photobleaching, FRAP)
To determine the role of mRNA degradation components in protein synthesis control, we slightly modified a previously established FRAP assay that combines immobilization of animals on agar pads for photobleaching without anesthetics and monitoring of fluorescent recovery on OP50 seeded plates (Kourtis and Tavernarakis, 2009a). Transgenic animals carrying the pan-neuronal fluorescence reporter punc-119GFP were grown to the desired age. For each bleaching procedure 4-5 worms were transferred into a drop of M9 on freshly prepared 2% agarose pads. Reduction of movement of animals occurred within 1-2min due to absorbance of M9 into the agar. Once immobilization was complete, final positioning of worms was corrected by the use of a hairpin. The pad was placed under a 40x lens of a Zeiss epifluorescence microscope Axio Imager.Z2 without coverslip. After focusing and recording a reference image (t = before bleaching), photobleaching of the area of interest was performed for 90 s at 30% luminescent power of the fluorescent illumination source (Fl illumination System X-Cite 120 XL FL PC, 120W metal halogenide lamp) and fully opened iris, until the fluorescence signal was quenched to approx. 20%–40% intensity relative to t = before bleaching. A reference image (t = after bleaching) after photobleaching was recorded and worms were allowed to recover for 5min in a drop of M9 buffer. Worms were then transferred to individual NGM plates seeded with a drop of OP50 in the center and an image was taken under a 10x lens of the epifluorescence microscope, which counts as t = 0 for the recovery after photobleaching. Transgenic animals carrying the somatic fluorescence reporter pife-2GFP were bleached directly on OP50 seeded NGM plates using a Zeiss Axio Imager.M2 with 100% luminescent power with the 25x objective for 7.5min. Images on plate were recorded every 1-2 hours for at least 6 hours at a fluorescence dissection microscope (Axio Zoom, Zeiss). To assure that only healthy animals were scored for the assay, individuals were carefully observed for indications of damage such as locomotion or egg-laying defects, and survival over the next 2-3 days. The recorded images were then processed by the imaging software package Fiji (ImageJ) and mean pixel intensity of the area of interest was determined for each time point. The fluorescence recovery was calculated based on the reference images. Animals treated with the protein synthesis inhibitor cycloheximide were used as a positive control.
Time-lapse microscopy and quantification of eIF4E sequestration in PBs
To record live formation and co-aggregation of SGs and PBs under constant heat stress, we used N2;Ex[pdcap-1DCAP-1::DsRed;pife-2IFE-2::GFP;pRF4] animals, which were immobilized with nanobeads as published previously (Kim et al., 2013). Imaging was performed with a confocal laser scanning microscope Leica TCS SP8 with climate chamber for environmental control, which was adjusted to 34°C. Movies of 20 Z stacks in 1 μm steps, recorded every 30 s over a time span of at least 5 minutes were then generated. We used the IMARIS (Bitplane) software package to generate 3D/4D reconstructions of Z stack recordings of pharyngeal region of at least 5 animals per genetic background, which were imaged sequentially (Figure S1A and S1B). All PBs within the field of view were detected, rendered and subsequently IFE-2::GFP localization within PBs was calculated via IMARIS and summarized as mean voxel intensity.
PB and SG monitoring and quantification of colocalization
A full protocol for in vivo monitoring of PBs and SGs is available at (Rieckher and Tavernarakis, 2017). Briefly, transgenic lines generated for quantification of PB or SG formation in C. elegans were grown on OP50 seeded NGM plates (or RNAi plates) to the desired age. Individual animals were transferred into a drop of M9 buffer containing 10mM levamisole on a microscope slide and sealed with a cover slide. Worms were imaged at a Zeiss epifluorescence microscope Axio Imager Z2 under a 10x lens (EC Plan-Neofluar, numerical aperture 0.3) with focus on the anteroposterior center of each animal. At least 25 whole worms were recorded per transgenic line, per experiment. Alternatively, animals were imaged under a 25x lens (LCI Plan-Neofluar, numerical aperture 0.8, Immersion Corrected DIC) of a LSM710 Zeiss confocal microscope, Axio-observer Z1 controlled by the ZEN software package (ZEN2011). Here, we focused on the head region and 10-15 single plane images in 5 μm steps of at least 10 worms were recorded and quantified individually or as the resulting maximum intensity projection image. To quantify PB formation, we used the Fiji (ImageJ) software and measured either PB intensity (mean pixel intensity) or PB number (particle analysis with appropriate threshold assignment). To quantify SG formation during aging (Figure 1G) or upon stress insults (Figure S3C), we counted the total amount of animals forming SGs across the indicated tissues (compare representative images in Figure S3D to S3K). To qualitatively monitor PB and SG formation and colocalization, we imaged 4 to 5 sections under the 25x lens at the Zeiss confocal microscope covering the full body length, recording 10-15 single planes in 3 μm steps. Single planes or maximum intensity projections were thereafter assembled into a whole worm image using Adobe Photoshop. Colocalization (Figures 1B and S2) analysis of confocal z stacks was achieved via the Coloc2 function of ImageJ that produces 2D histograms, the regression line for colocalization events and calculates the Pearson’s coefficient (ρx,y) that describes the correlation of the two components (−1 perfect anti-correlation, 0 for no correlation, +1 for perfect correlation). The 2D histograms contain the two intensity values for each pixel plotted against each other. The brightness of the color reflects the amount of correlating pixels of the respective color channels. The cloud in the histogram indicates colocalization events, which are expressed by a linear regression and correlation coeffient (https://imagej.net/Colocalization_Analysis).
Stress assays
We performed multiple stress assays to assess PB and SG formation in transgenic lines. To evaluate the influence of heat stress on PB and SG formation, we placed the animals on pre-warmed OP50 seeded plates and incubated them for 3, 6 or 9 hours at 35°C followed by immediate imaging. The influence of heat stress on survival was quantified by exposing age-synchronized animals (2-day-old adults) to 37°C on pre-warmed plates for 4h or 12h, respectively, after which we counted survival (Figure S5L), or continued quantifying survival in a lifespan (Figure S5M). To examine the effect of starvation, we grew animals under physiological conditions to day 2 of adulthood, transferred them to empty NGM plates, followed by monitoring 24 hours later. To examine effects of hypoxia, we used a hypoxic chamber and applied a low oxygen concentration following previously described methodology (Troulinaki and Tavernarakis, 2012). Briefly, a population of 2-day-old adults was exposed to low oxygen conditions (< 0.5%) at 25°C for 6h on OP50 seeded NGM plates. Animals were imaged immediately thereafter. Immediate (acute) response to oxidative stress insults was tested by incubating worms for 30min in 20mM sodium azide (NaN3, Sigma) or 100mM paraquat (PQ, Sigma), followed by monitoring. To monitor chronic effects of oxidative stress, we grew worms to day 2 or day 7 of adulthood, respectively. We then transferred worms to plates with freshly applied drugs NaN3 (150 μM) or PQ (5mM). These concentrations assured that a several-day lasting response would occur prior to death, which was scored every day thereafter. Following this protocol, we compared wild-type worms grown on control or edc-3 RNAi and edc-3 mutants. Transgenic animals were imaged 2 days after exposure to the drugs.
Quantification of neuronal integrity
We used transgenic animals carrying the pan-neuronal promoter punc-119GFP to quantify neuronal integrity. Images of at least 10 individuals were collected by confocal scanning (10-12 z stacks per animal) at an LSM710 (Zeiss) microscope. Images were summarized in mean intensity projections and subsequently scored for neuronal defects, with focus on the VNC.
Behavioral assays
For the phenotypic analysis presented in Figure S4E, we quantified pumping rates by monitoring the grinder movement in the pharyngeal region of 15 individuals per genotype under a stereoscope, for 30sec each. Dauer formation was induced by placing 10 adults to OP50 seeded NGM plates. 10 days later, after the food source was exhausted, the amount of dauer formation was determined by counting an area of defined size for starvation-induced dauers, which are easily distinguishable at a stereoscope. Locomotion was quantified by observing the movement of the animals on plate while counting the sinus waves of 10 animals, for 2 min each. The body size was determined by measuring the length of animals on pictures taken at the Zeiss AxioImager.Z2 with a 10x objective, applying the Fiji (ImageJ) length-measurement of free hand selection tool. Egg-laying quantification was applied by placing three individuals (synchronized day-1-adults) on an OP50 seeded NGM plate for 3 hours. The animals were then removed and eggs were counted. All behavioral assays were performed in three biological replicates and data were summarized in relation to measurements performed in the wild-type.
Quantification and Statistical Analysis
Statistical analyses were carried out using the Prism software package (GraphPad Software Inc., San Diego, USA) and the Microsoft Office 2003 Excel software package (Microsoft Corporation, Redmond, WA, USA). The statistical tests applied for each experiment are specified in the Figure legends.
Acknowledgments
We thank Min Han (University of Colorado, Boulder) for the pBluescript II Ks+ dcap-1 plasmid, George Mountoufaris for pdcap-1DCAP-1::DsRed, and Jonathan Ewbank (Centre d’Immunologie de Marseille-Luminy) for pcol-12DsRed. We thank Aggela Pasparaki for valuable support with confocal microscopy and Georgia Tsikala for help with time-lapse microscopy. We kindly thank the CECAD imaging facility for feedback on microscopy procedures. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was funded by grants from the European Research Council (ERC-GA695190, MANNA; ERC-GA737599, NeuronAgeScreen) and the European Commission 7th Framework Programme to N.T. B.S. acknowledges funding from the Deutsche Forschungsgemeinschaft (CECAD, SFB 829, SFB 670, and KFO 286), the European Research Council (ERC Starting grant 260383), Marie Curie (FP7 ITN CodeAge 316354, aDDRess 316390, MARRIAGE 316964), Deutsche Krebshilfe (70112899), and the COST action (BM1408, GENiE).
Author Contributions
M.R., M.M., A.P., and N.T. designed and performed experiments. M.R., M.M., B.S., and N.T. analyzed data and wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: October 2, 2018
Footnotes
Supplemental Information includes six figures, one table, and two videos and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.09.009.
Supplemental Information
References
- Aizer A., Kalo A., Kafri P., Shraga A., Ben-Yishay R., Jacob A., Kinor N., Shav-Tal Y. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J. Cell Sci. 2014;127:4443–4456. doi: 10.1242/jcs.152975. [DOI] [PubMed] [Google Scholar]
- An J.H., Blackwell T.K. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei M.A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Lührmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J.A., Doudna J.A., Gilbert W.V. Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol. Cell. 2011;44:745–758. doi: 10.1016/j.molcel.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artal-Sanz M., Tavernarakis N. Prohibitin couples diapause signalling to mitochondrial metabolism during ageing in C. elegans. Nature. 2009;461:793–797. doi: 10.1038/nature08466. [DOI] [PubMed] [Google Scholar]
- Belousov V.V., Fradkov A.F., Lukyanov K.A., Staroverov D.B., Shakhbazov K.S., Terskikh A.V., Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- Benrahmoune M., Thérond P., Abedinzadeh Z. The reaction of superoxide radical with N-acetylcysteine. Free Radic. Biol. Med. 2000;29:775–782. doi: 10.1016/s0891-5849(00)00380-4. [DOI] [PubMed] [Google Scholar]
- Brengues M., Parker R. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2592–2602. doi: 10.1091/mbc.E06-12-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson M.R. Aging–RNA in development and disease. Wiley Interdiscip. Rev. RNA. 2012;3:133–143. doi: 10.1002/wrna.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Teixeira D., Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes P.S., Boehringer A., Zwick R., Tang J.E., Grigsby B., Zarnescu D.C. Wild-type and A315T mutant TDP-43 exert differential neurotoxicity in a Drosophila model of ALS. Hum. Mol. Genet. 2011;20:2308–2321. doi: 10.1093/hmg/ddr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felkai S., Ewbank J.J., Lemieux J., Labbé J.C., Brown G.G., Hekimi S. CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. EMBO J. 1999;18:1783–1792. doi: 10.1093/emboj/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo M.A., Basak S., Dostie J., Murray E.L., Schoenberg D.R., Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T.M., Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol. Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsche R., Karra D., Bennett K.L., Ang F.Y., Heraud-Farlow J.E., Tolino M., Doyle M., Bauer K.E., Thomas S., Planyavsky M. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 2013;5:1749–1762. doi: 10.1016/j.celrep.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Funayama R., Ishikawa F. Cellular senescence and chromatin structure. Chromosoma. 2007;116:431–440. doi: 10.1007/s00412-007-0115-7. [DOI] [PubMed] [Google Scholar]
- Gems D., Pletcher S., Partridge L. Interpreting interactions between treatments that slow aging. Aging Cell. 2002;1:1–9. doi: 10.1046/j.1474-9728.2002.00003.x. [DOI] [PubMed] [Google Scholar]
- Hansen M., Taubert S., Crawford D., Libina N., Lee S.-J., Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. mRNA transport in dendrites: RNA granules, motors, and tracks. J. Neurosci. 2006;26:7139–7142. doi: 10.1523/JNEUROSCI.1821-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard A.C., Rollins J., Snow S., Castor S., Rogers A.N. Reducing translation through eIF4G/IFG-1 improves survival under ER stress that depends on heat shock factor HSF-1 in Caenorhabditis elegans. Aging Cell. 2016 doi: 10.1111/acel.12516. Published online August 18, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A.-L., Murphy C.T., Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Hubstenberger A., Courel M., Benard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.B., Munier A., Fradet M. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell. 2017;68:144–157.e5. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Ishii T., Miyazawa M., Hartman P.S., Ishii N. Mitochondrial superoxide anion (O(2)(-)) inducible “mev-1” animal models for aging research. BMB Rep. 2011;44:298–305. doi: 10.5483/BMBRep.2011.44.5.298. [DOI] [PubMed] [Google Scholar]
- Kanazawa T., Zappaterra M.D., Hasegawa A., Wright A.P., Newman-Smith E.D., Buttle K.F., McDonald K., Mannella C.A., van der Bliek A.M. The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genet. 2008;4:e1000022. doi: 10.1371/journal.pgen.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser E.B., Sedensky M.M., Morgan P.G. The effects of complex I function and oxidative damage on lifespan and anesthetic sensitivity in Caenorhabditis elegans. Mech. Ageing Dev. 2004;125:455–464. doi: 10.1016/j.mad.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C.J. The genetics of ageing. Nat. Rev. 2010;464:505–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kim E., Sun L., Gabel C.V., Fang-Yen C. Long-term imaging of Caenorhabditis elegans using nanoparticle-mediated immobilization. PLoS ONE. 2013;8:e53419. doi: 10.1371/journal.pone.0053419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S.R., Horetsky R.L., Ron D., Jefferson L.S., Harding H.P. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 2003;284:C273–C284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- Kirkwood T.B., Austad S.N. Why do we age? Nature. 2000;408:233–238. doi: 10.1038/35041682. [DOI] [PubMed] [Google Scholar]
- Kourtis N., Tavernarakis N. Cell-specific monitoring of protein synthesis in vivo. PLoS ONE. 2009;4:e4547. doi: 10.1371/journal.pone.0004547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N., Tavernarakis N. Cellular stress response pathways and ageing: intricate molecular relationships. EMBO J. 2011;30:2520–2531. doi: 10.1038/emboj.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N., Tavernarakis N. Protein synthesis rate assessment by fluorescence recovery after photobleaching (FRAP) Bio Protoc. 2017;7:e2156. doi: 10.21769/BioProtoc.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar M., Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J., Morimoto R.I. Proteostasis and longevity: when does aging really begin? F1000Prime Rep. 2014;6:7. doi: 10.12703/P6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B., Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler M.C., Crawford E.D., Groh N., Widmaier K., Jung R., Kirstein J., Trinidad J.C., Burlingame A.L., David D.C. Reduced insulin/IGF-1 signaling restores the dynamic properties of key stress granule proteins during aging. Cell Rep. 2017;18:454–467. doi: 10.1016/j.celrep.2016.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N., Berman J.R., Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Ling S.H.M., Decker C.J., Walsh M.A., She M., Parker R., Song H. Crystal structure of human Edc3 and its functional implications. Mol. Cell. Biol. 2008;28:5965–5976. doi: 10.1128/MCB.00761-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J.J., Schuster E., Blanc E., Piper M.D., Thomas J.H., Patel D.S., Selman C., Withers D.J., Thornton J.M., Partridge L., Gems D. Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol. 2007;8:R132. doi: 10.1186/gb-2007-8-7-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO. J. 1991;12:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Duran W.A., Hjelle B.L., Ye C., Panganiban A.T. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc. Natl. Acad. Sci. U S A. 2008;105:19294–19299. doi: 10.1073/pnas.0807211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsumori K., Takei Y., Hirokawa N. Components of RNA granules affect their localization and dynamics in neuronal dendrites. Mol. Biol. Cell. 2017;28:1412–1417. doi: 10.1091/mbc.E16-07-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley J.F., Morimoto R.I. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller M.M., Castells-Roca L., Babu V., Ermolaeva M.A., Müller R.U., Frommolt P., Williams A.B., Greiss S., Schneider J.I., Benzing T. DAF-16/FOXO and EGL-27/GATA promote developmental growth in response to persistent somatic DNA damage. Nat. Cell Biol. 2014;16:1168–1179. doi: 10.1038/ncb3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-McNicoll M., Neugebauer K.M. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 2013;14:275–287. doi: 10.1038/nrg3434. [DOI] [PubMed] [Google Scholar]
- Nagashima T., Oami E., Kutsuna N., Ishiura S., Suo S. Dopamine regulates body size in Caenorhabditis elegans. Dev. Biol. 2016;412:128–138. doi: 10.1016/j.ydbio.2016.02.021. [DOI] [PubMed] [Google Scholar]
- Palikaras K., Lionaki E., Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521:525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- Pan K.Z., Palter J.E., Rogers A.N., Olsen A., Chen D., Lithgow G.J., Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Gems D. Mechanisms of ageing: public or private? Nat. Rev. Genet. 2002;3:165–175. doi: 10.1038/nrg753. [DOI] [PubMed] [Google Scholar]
- Rieckher M., Tavernarakis N. P-body and stress granule quantification in Caenorhabditis elegans. Bio Protoc. 2017;7:e2018. doi: 10.21769/BioProtoc.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckher M., Kourtis N., Pasparaki A., Tavernarakis N. Humana Press; Totowa, NJ: 2009. Transgenesis in Caenorhabditis elegans; pp. 21–39. [DOI] [PubMed] [Google Scholar]
- Rousakis A., Vlanti A., Borbolis F., Roumelioti F., Kapetanou M., Syntichaki P. Diverse functions of mRNA metabolism factors in stress defense and aging of Caenorhabditis elegans. PLoS ONE. 2014;9:e103365. doi: 10.1371/journal.pone.0103365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Meth. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo K., Choi E., Lee D., Jeong D.E., Jang S.K., Lee S.J. Heat shock factor 1 mediates the longevity conferred by inhibition of TOR and insulin/IGF-1 signaling pathways in C. elegans. Aging Cell. 2013;12:1073–1081. doi: 10.1111/acel.12140. [DOI] [PubMed] [Google Scholar]
- Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So S., Miyahara K., Ohshima Y. Control of body size in C. elegans dependent on food and insulin/IGF-1 signal. Genes Cells. 2011;16:639–651. doi: 10.1111/j.1365-2443.2011.01514.x. [DOI] [PubMed] [Google Scholar]
- Syntichaki P., Troulinaki K., Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N. Protein synthesis and aging: eIF4E and the soma vs. germline distinction. Cell Cycle. 2007;6:1168–1171. doi: 10.4161/cc.6.10.4230. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N. Ageing and the regulation of protein synthesis: a balancing act? Trends Cell Biol. 2008;18:228–235. doi: 10.1016/j.tcb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N., Wang S.L., Dorovkov M., Ryazanov A., Driscoll M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- Tritschler F., Eulalio A., Truffault V., Hartmann M.D., Helms S., Schmidt S., Coles M., Izaurralde E., Weichenrieder O. Divergent Sm fold in EDC3 proteins mediates DCP1 binding and P-body targeting. Mol. Cell. Biol. 2007;27:8600–8611. doi: 10.1128/MCB.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F., Eulalio A., Helms S., Schmidt S., Coles M., Weichenrieder O., Izaurralde E., Truffault V. Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol. Cell. Biol. 2008;28:6695–6708. doi: 10.1128/MCB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritschler F., Braun J.E., Eulalio A., Truffault V., Izaurralde E., Weichenrieder O. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol. Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Troulinaki K., Tavernarakis N. Endocytosis and intracellular trafficking contribute to necrotic neurodegeneration in C. elegans. EMBO J. 2012;31:654–666. doi: 10.1038/emboj.2011.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro A., Tauffenberger A., Ash P.E., Carlomagno Y., Petrucelli L., Parker J.A. TDP-1/TDP-43 regulates stress signaling and age-dependent proteotoxicity in Caenorhabditis elegans. PLoS Genet. 2012;8:e1002806. doi: 10.1371/journal.pgen.1002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan A.M., van Gemert A.M., Dirks R.W., Noordermeer J.N., Fradkin L.G., Tanke H.J., Jost C.R. mRNA cycles through hypoxia-induced stress granules in live Drosophila embryonic muscles. Int. J. Dev. Biol. 2012;56:701–709. doi: 10.1387/ijdb.103172al. [DOI] [PubMed] [Google Scholar]
- Wang I.F., Wu L.S., Chang H.Y., Shen C.K. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J. Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- Wang J., Robida-Stubbs S., Tullet J.M., Rual J.F., Vidal M., Blackwell T.K. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 2010;6:1–17. doi: 10.1371/journal.pgen.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.G., Southgate E., Thomson J.N., Brenner S. The structure of the ventral nerve cord of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976;275:327–348. doi: 10.1098/rstb.1976.0086. [DOI] [PubMed] [Google Scholar]
- Wong D., Bazopoulou D., Pujol N., Tavernarakis N., Ewbank J.J. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194. doi: 10.1186/gb-2007-8-9-r194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.F., Gendron T.F., Zhang Y.J., Lin W.L., D’Alton S., Sheng H., Casey M.C., Tong J., Knight J., Yu X. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J. Neurosci. 2010;30:10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frame rate is 30 s/scan, each scan contains maximum intensity information of 20 z stacks of 1 μm depth each.
The movie was produced with the IMARIS software (see Materials and Methods). Frame rate is 30 s/scan, each scan contains information of at least 20 z stacks of 0.3 μm depth each.