Short abstract
MiR-206 is a remarkable miRNA because it functions as a suppressor miRNA in rhabdomyosarcoma while at the same time, as previously showed, it can act as an oncomiRNA in SMARCB1 immunonegative soft tissue sarcomas. The aim of this study was to investigate the effect of miR-206 on its several target genes in various human tumorous and normal cell lines. In the current work, we created miR-206-overexpressing cell lines (HT-1080, Caco2, iASC, and SS-iASC) using permanent transfection. mRNA expression of the target genes of miR-206 (SMARCB1, ACTL6A, CCND1, POLA1, NOTCH3, MET, and G6PD) and SMARCB1 protein expression were examined with quantitative real-time polymerase chain reaction, immunoblotting, immunocytochemistry, and flow cytometry. MiRNA inhibition was used to validate our results. We found a diverse silencing effect of miR-206 on its target genes. While an overall tendency of downregulation was noted, expression profiles of individual cell lines showed large variability. Only CCND1 and MET were consistently downregulated. MiR-206 had an antiproliferative effect on a normal human fibroblast cell line. A strong silencing effect of SMARCB1 in miR-206 transfected SS-iASC was most likely caused by the synergic influence of the SS18-SSX1 fusion protein and miR-206. In the same cell line, a moderate decrease of SMARCB1 protein expression could be observed with immunocytochemistry and flow cytometry. In the most comprehensive analysis of miR-206 effects so far, a modest but significant downregulation of miR-206 targets on the mRNA level was confirmed across all cell lines. However, the variability of the effect shows that the action of this miRNA is largely cell context-dependent. Our results also support the conception that the oncomiR effect of miR-206 on SMARCB1 plays an important but not exclusive role in SMARCB1 immunonegative soft tissue sarcomas so it can be considered important in planning the targeted therapy of these tumors in the future.
Impact statement
Mir-206 is a very unique microRNA because it can act as a suppressor miRNA or as an oncomiRNA depending on the tumor tissue. In SMARCB1 negative soft tissue sarcomas miR-206 is overexpressed, so thus in epithelioid and synovial sarcomas it functions as an oncomiRNA. MiR-206 has diverse silencing effects on its target genes. We found that the action of miR-206 is largely cell context dependent. The oncomiR role of miR-206 is crucial but not exclusive in SMARCB1 negative soft tissue sarcomas and miR-206 has an antiproliferative effect on a normal human fibroblast cell line. Expressions of miR-206 targets observed in tumors can only be reproduced in the corresponding tumorous cell lines. This is the first study which examined the permanent effect of miR-206 on its target genes in normal, tumor, and genetically engineered cell lines.
Keywords: miR-206, SMARCB1, synovial sarcoma, permanent microRNA transfection, epigenetic regulation, relative gene expression
Introduction
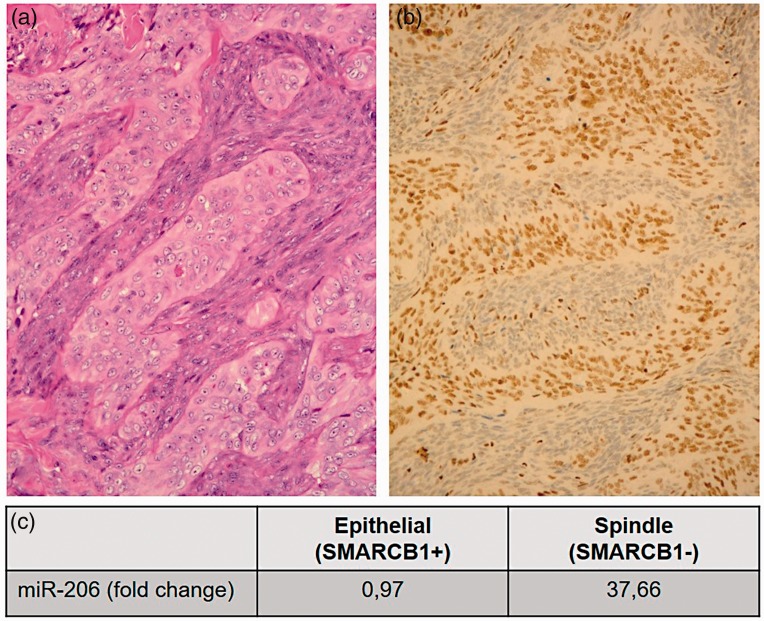
The SWI/SNF chromatin-remodeling complex plays an important role in cell proliferation, differentiation, and DNA repair. The complex contains an ATPase domain called BRG1 or BRM, and three core subunits, BAF47 or SMARCB1, BAF155 or SMARCC1, and BAF170 or SMARCC2. In addition to these four obligate subunits, the complex is composed of other variable subunits depending on the tissue microenvironment.1,2 SMARCB1 is considered as a classical tumor suppressor gene and its lack can cause tumor formation.2 SMARCB1 deficiency occurs in several soft tissue tumors including synovial sarcoma (SS).3 Kadoch and Crabtree demonstrated that the SS18-SSX fusion protein competes with the wild-type (WT) SS18 and SMARCB1. This competition results in an altered BAF complex, incorporating SS18-SSX while excluding WT SS18 and SMARCB1, and reduced protein expression of SMARCB1 due to proteasomal degradation.4 Complete loss of SMARCB1 is known in malignant rhabdoid tumor (MRT) and epithelioid sarcoma (ES). In MRT, the lack of SMARCB1 expression can be caused by homozygous inactivation (biallelic mutation and/or deletion).3 In ES, the absence of SMARCB1 expression can be induced by genetic and/or epigenetic alterations.3 In our previous work, we demonstrated that in monoallelically deleted ES cases, the loss of SMARCB1 nuclear immunopositivity can be induced by overexpressing three microRNAs (miRNA) (miR-206, miR-381, and miR-671–5p). MiR-765 was also overexpressed, but could not silence SMARCB1 expression in a functional test on cell lines.5 In a successive study, we demonstrated that in biphasic SS (wherein both spindle and epithelial cell components comprise the SS18-SSX fusion gene), the spindle cell component is characterized by weak or negative immunostaining for SMARCB1 of nuclei, while the epithelial cells retain their SMARCB1 protein expression (Figure 1(a) and (b)).6 We also showed that the three functionally active miRNAs (miR-206, miR-381, and miR-671–5p) were overexpressed and displayed higher expression in the spindle cells than in the epithelial cell component, whereas the ES-specific miR-765 had the same low expression value in both cell types (Figure 1(c)).6
Figure 1.
Classical hematoxylin and eosin (H&E) staining (a) and SMARCB1 immunhistochemistry (b) of a biphasic synovial sarcoma. Epithelial components show positive reaction with SMARCB1 immunostaining, while the spindle cell components are negatively stained (b). miR-206 is highly overexpressed (37,66-fold) in the spindle cell components compared to the epithelial components, measured with q-RT-PCR. Relative miR-206 level was normalized to endogenous RNU6B. As calibrator, normal liver tissue was used (c). (A color version of this figure is available in the online journal.)
MiR-206 is a very noteworthy miRNA because it functions as a suppressor miRNA in rhabdomyosarcoma (RMS),7 osteosarcoma,8 chondrosarcoma,9 breast cancer, melanoma, and lung cancers,10–12 while at the same time it can act as an oncomiRNA in ES, SS, and in other SMARCB1 immunonegative soft tissue tumors.5,6 This fact indicates the important role of tissue microenvironment. In skeletal muscle, miR-206 promotes cell differentiation.13 Functional studies have identified several target genes of miR-206.14,15 After creating cell lines with permanent miR-206 expression, our purpose was to investigate the expression levels of miR-206 target genes. We examined 6 miR-206 targets beside SMARCB1, namely ACTL6A, CCND1, POLA1, NOTCH3, MET, G6PD, and SNAI1. Albeit the latter is not a direct target of miR-206, it was selected due to its crucial role in epithelial-to-mesenchymal transition (EMT). ACTL6A (also known as BAF53A) is a variable domain of the SWI/SNF chromatin-remodeling complex and it is an oncogene.16 CCND1 as an oncogene promotes progression from G1 to S phase through cyclin-dependent kinase 4/6 (CDK4/6).17 POLA1 gene encodes the catalytic subunit p180 of DNA Polymerase α I (DNA Pol α I).18 NOTCH3 as an oncogene plays an important role in the NOTCH signaling pathway and is overexpressed in many tumors.19 MET as a proto-oncogene encodes the Met tyrosine kinase receptor and plays an important role in skeletal muscle differentiation.20 G6PD as an oncogene encodes a glucose-6-phosphate dehydrogenase enzyme active in the pentose-phosphate pathway.21 SNAI1 is an EMT marker gene which is overexpressed when epithelial cells transform into mesenchymal cells, while E-cadherin expression decreases – and vice versa – when mesenchymal cells transform into epithelial cells, SNAI1 is downregulated, and E-cadherin expression increases.22
In the present work, we created cell lines constitutively expressing miR-206 because in our previous transient miRNA transfection experiments, miR-206 was the most effective at silencing SMARCB1.5 In addition to the formerly used three human cell lines (HT-1080, Caco2 and HDFα),5 two new human cell lines, iASC (immortalized adipose tissue-derived mesenchymal stem cell) and SS-iASC (SS18-SSX1-carrying immortalized adipose tissue-derived mesenchymal stem cell) were involved in this study.23,24 SS-iASC is a novel SS18-SSX1-carrying immortalized adipose tissue-derived mesenchymal stem cell line previously established by our group. Immunocytochemistry showed that the SS18-SSX1 transcript successfully induced mesenchymal-to-epithelial transformation (MET) in SS-iASC.24 The overall aim of this study was to investigate the effect of miR-206 on several target genes in various human tumorous and normal cell lines, and to examine the combined effect of SS18-SSX1 and miR-206. According to our best knowledge, this is the first work where these miR-206 target genes are chosen to be examined in these cell lines.
Materials and methods
Cell culture
Two human tumor cell lines (HT-1080 and Caco2), a normal cell line (HDFα with retained SMARCB1 expression), an immortalized adipose tissue-derived mesenchymal stem-cell line (iASC)23 and the above-mentioned SS-iASC cell line24 were cultured for our transfection experiments. HT-1080 human fibrosarcoma cells were acquired from the American Type Culture Collection (ATCC number: CCL-121) and grown in RPMI-1640 medium (Biosera, Kansas, MO, USA) supplemented with 10% fetal bovine serum (FBS, Biosera) and gentamicin (Sandoz Hungária Kft., Budapest, Hungary, 160 µg/mL). Caco2 human colon adenocarcinoma cells (ATCC number: HTB-37) were cultured in minimum essential medium (MEM, HyClone, GE Healthcare Life Sciences, Logan, UT, USA) with L-Glutamine (Sigma-Aldrich, St Louis, MO, USA) completed with 20% FBS, 1% non-essential amino acids (Biosera), and antibiotics mentioned above. Human dermal fibroblast adult (HDFα) cell line (Thermo Fisher Scientific, Waltham, MA, USA) was grown in medium 106 supplemented with low serum growth supplement (LSGS, Invitrogen by Thermo Fisher Scientific) in the absence of antibiotics and antimycotics. iASC and SS-iASC were cultured in Dulbecco’s Modified Eagles Medium/F12 (DMEM/F12, Sigma-Aldrich) supplemented with 10% FBS (Biosera), L-Glutamine (Sigma-Aldrich), fibroblast growth factor-basic (bFGF, Gibco by Thermo Fisher Scientific 0.001 µg/mL), and antibiotics mentioned above. Cells were cultured in 25/75 cm2 tissue culture flasks (Sigma-Aldrich) at 37°C under a humidified atmosphere containing 5% v/v CO2. Media was refreshed every other day, and cells were subcultured at a confluency of 80%.
miRNA transfection
Expression plasmid for human miRNA MIR206 (accession number: MI0000490) and as a negative control empty vector pCMV-MIR (accession number: pCMVMIR) (OriGene Technologies, Rockville, MD, USA) was permanently transfected into the above-mentioned five cell lines with the Neon™ Transfection System (Invitrogen by Thermo Fisher Scientific) according to the manufacturer’s protocol. The OriGene miRNA precursor involves a 60–70 nucleotides long pre-miRNA sequence with a 250–300 nucleotides long up- and downstream flanking sequence. The expression of miRNA precursor is driven by CMV promoter and with human growth factor I poly(A) tailing signal. The miRNA plasmids also contain green fluorescent protein (GFP) as a reporter for transfection monitoring and also include Neomycin selection marker for stable cell establishment. The empty vector pCMV-MIR was used as a negative control with no detectable effects on known miRNA function. Cells prepared for transfection were washed with phosphate-buffered saline (PBS) and dissociated with trypsin. For one reaction, generally 4 × 105 cells were centrifuged at 800g for 10 min and washed with PBS. The pellet was suspended in a suitable volume of transfection resuspension buffer R and mixed with 1 µg of miRNA plasmid. The miRNA amount could not exceed 10% of total volume used. A 10-µL aliquot was placed in the electroporation Neon Pipette Tips and pulsed. The parameters for electroporation were selected from the cell line database of Invitrogen. After electroporation, cells were placed into a single well of a six-well tissue plate containing 2 mL of antibiotic-free media. Twenty-four hours later, effectivity of the transfection was examined with (epi)fluorescence microscope using GFP monitoring. MiRNA MIR206-transfected, empty vector pCMV-MIR-transfected, and non-transfected cells were treated with Geneticin® selective antibiotic (Gibco by Thermo Fisher Scientific, 0.05 mg/µL) for two weeks. Transfection of Anti-miR™ miRNA Inhibitor (against hsa-miR-206, AM17000), FAM-labeled Anti-miR™ Negative Control (catalog number: AM17012), and as positive control Anti-miR™ hsa-let-7c miRNA Inhibitor (catalog number: 4392431) (Ambion by Life Technologies) into the aforementioned miR-206-transfected and non-transfected cell lines was done with the Neon™ Transfection System (Invitrogen by Thermo Fisher Scientific) according to the manufacturer’s protocol as described above. The FAM-labeled Anti-miR negative control was used as a negative control. Anti-miR™ hsa-let-7c miRNA inhibitor was chosen as positive control because it effectively inhibits endogenous let-7 miRNA. The let-7 miRNA downregulates HMGA2 mRNA. Anti-miR™ hsa-let-7c miRNA inhibitor blocks endogenous let-7c miRNA, resulting in increased levels of HMGA2 mRNA, which can be detected by qRT-PCR. After 24 h of transfection, cells were harvested for RNA extraction.
RNA isolation
Transfected cells were cultured in six-well tissue plate or 25 cm2 tissue culture flasks for 24 or 48–72 h before isolation of total RNA using the PureLink RNA Mini Kit (Invitrogen by Thermo Fisher Scientific) according to the manufacturer’s protocol. We removed residual DNA by DNAse treatment as an optional step. The isolated RNA’s yield and purity were estimated by NanoDrop 1000 (NanoDrop Technologies, Houston, TX, USA). The isolated RNA samples were stored at −80°C until further experiments.
Reverse transcription
For real-time PCR quantification of miR-206 expression, the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) was used to synthetize first-strand complementary DNA (cDNA). The reactions were performed at 16°C for 30 min then at 42°C for another 30 min and finally for enzyme inactivation, it was hold at 85°C for 5 min. For real-time PCR analysis of miR-206 target gene expressions, the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) was used to reverse-transcribe the RNA samples. After initial incubation at 25°C for 10 min, the reaction was performed at 37°C for 120 min which was followed by final inactivation at 85°C for 5 min. Reverse transcription reactions were run in a Verity® 96-Well Thermal Cycler PCR instrument. Until next experiments, cDNA samples were stored at −20°C.
Real-time PCR quantification of miR-206
Expression level of hsa-miR-206 (assay ID 000510) was measured in the miR-206-transfected or empty vector-transfected and non-transfected cell lines using TaqMan miRNA assays (Applied Biosystems) following the supplier’s protocol. Reactions were run in triplicate in a 96-well plate on a Lightcycler® 480 Instrument (Roche Applied Science, Indianapolis, IN, USA). After initial denaturation at 95°C for 10 min, 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min were performed. RNU6B (assay ID 0010093, PN 4427975) was chosen as endogenous control based on supplier application note (endogenous controls for real-time quantification of miRNA using TaqMan MiRNA assays (Applied Biosystems)). As a calibrator, non-transfected and empty vector-transfected cells were used. Expression levels were calculated by the ΔΔCT method.25
Real-time PCR analysis of miR-206 targets
SMARCB1 (assay ID: Hs00992522_m1), ACTL6A (assay ID: Hs00895061_m1), CCND1 (assay ID: Hs00765553_m1), POLA1 (assay ID: Hs00415835_m1), NOTCH3 (assay ID: Hs01128537_m1), SNAI1 (assay ID: Hs00195591_m1), MET (assay ID: Hs01565576_m1), G6PD (assay ID: Hs00166169_m1), HMGA2 (assay ID: Hs00171569_m1) mRNA expression was analyzed by qRT-PCR; 50 ng of cDNA was used as template in 20-µL PCR reactions with 2X TaqMan Universal PCR master mix (Applied Biosystems). The thermal parameters were the same as described above in part of real-time PCR quantification of miR-206. Relative quantification of gene expression was measured four times in triplicate using 20X TaqMan Assays on a Lightcycler® 480 Instrument (Roche Applied Science) by ΔΔCT method, using GAPDH (part no.: 4352338E) as an endogenous reference control. As a calibrator, non-transfected and empty vector-transfected cells were used.
Western blot analysis of SMARCB1 after stable miR-206 transfection
Cells were homogenized with a lysis buffer containing 20 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.05% Triton X-100, 0.5% protease inhibitor cocktail (P8340, Sigma-Aldrich), 2 mM Na3VO4, and 10 mM NaF. Protein concentrations were measured by Bradford method; 30 µg of total proteins were mixed with loading buffer containing beta-mercaptoethanol and denatured at 99°C for 5 min. Denatured samples were loaded onto a 10% SDS-polyacrylamide gel and separated for 40 min at 200 V. Proteins were transferred to a PVDF membrane with overnight blotting at 4°C at constant 75 mA. Blotting efficiency was checked by Ponceau staining. Blocking was carried out with 5% non-fat dry milk dissolved in tris-buffered saline (TBS) for 1 h. Next, the membrane was incubated overnight with primary anti-SMARCB1/BAF47 Antibody (clone: D9C2, 1:1000, Cell Signaling Technology, Danvers, MA, USA). After washing with TBST (TBS + 0.05% Tween 20), the appropriate secondary antibody dissolved in 1% non-fat dry milk (TBS) was applied for 1 h at room temperature. After washing, immunoreactions were visualized using SuperSignal West Pico Chemiluminescent Substrate kit (Thermo Fisher Scientific). Bands were detected with Kodak Image Station 4000MM (Kodak, Rochester, NY, USA).
SMARCB1 immunostaining
Cells were grown on coverslips. After methanol fixation miR-206-transfected, empty vector-transfected and non-transfected cells were immunostained with Novolink™ Max Polymer Detection System (Novocastra, Newcastle Upon Tyne, UK), according to the manufacturer’s instruction. Following peroxidase and protein block solutions (5–5 min), the samples were incubated at room temperature for 1 h with the purified mouse anti-BAF47 (anti-SMARCB1) antibody (clone 25; 1:50; BD Transduction Laboratories, San Diego, CA, USA). After incubation with the primary antibody, the samples were incubated with post primary solution for 30 min and then incubated with the polymer for 30 min (both provided with the Novolink™ Max Polymer Detection System). The reaction was stained with diaminobenzidine (DAB) followed by hematoxylin counterstaining. The slides were then dehydrated in ethanol series, cleared in xylene, and mounted with micromount (Surgipath, Richmond, IL, USA). For SMARCB1, the stained cells were scored as having nuclear expression intact or absent.
Flow cytometry
Expression of nuclear SMARCB1 protein in iASC, iASC-206, SS-iASC, SS-iASC-206, Caco2, and Caco2–206 cell suspensions was assessed by flow cytometry. After fixation and permeabilization (IntraStain, Dako-Agilent, CA, USA) of 5 × 105 cells, cells were incubated with the purified mouse anti-BAF47 (anti-SMARCB1) antibody (clone 25; 1:25; BD Transduction Laboratories) at room temperature for 20 min, followed by washing; the bound primary antibody was visualized by goat anti-mouse IgG (H + L) – Alexa Flour® 647 secondary antibody (cat. no.: A-21235; 1:500; Invitrogen by Thermo Fisher Scientific) applied in dark at room temperature for 20 min; 20,000 stained cells were analyzed by FACSCalibur bench-top flow cytometer (BD Biosciences, San Jose, CA, USA). Data analysis was carried out using FlowJo (v10) (Ashland, OR, USA). Debris and dead cells were excluded using forward and side-scattered light scattergrams and mean fluorescence intensity (MFI) of the living cells was determined.
Statistical analysis
To calculate the significance of gene expression changes (up- or downregulation), we used non-parametric one-sample Wilcoxon signed rank test. Due to the small sample sizes, normal distribution could not be assumed; hence, parametric one-sample t-test was not applicable. Significance level was set to 0.05. We performed analysis using real statistics excel add-in Resource Pack (http://www.real-statistics.com/).
Results
Transfection efficiency and expression of miR-206
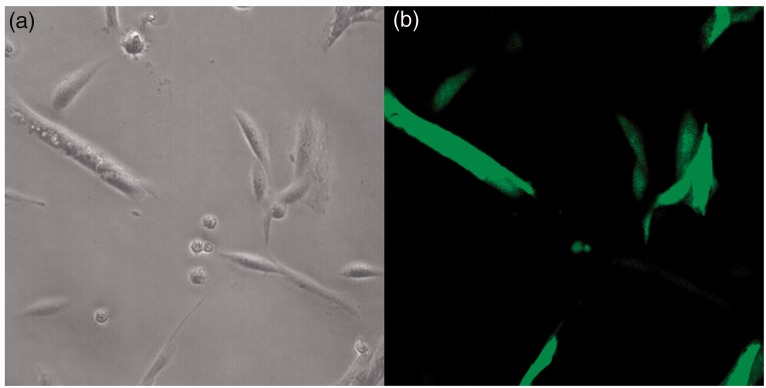
Twenty-four hours after the electroporation, cells were examined for GFP expression and as Figure 2 shows, the efficiency of the transfection was estimated to be 75–80% in the transfected cell lines. Interestingly, HDFα-206 cells were not growing as fast as the other transfected cell lines during the two weeks of antibiotic selection and after one month they died. MiR-206 was confirmed to be overexpressed in the stably transfected cell lines. Relative expression levels of miR-206 were 138-fold in SS-iASC-206, 1493-fold in iASC-206, 222-fold in Caco2–206, and 360-fold in HT-1080–206, respectively, compared to their non-transfected and empty vector-transfected controls.
Figure 2.
Images of SS-iASC-206 as observed under inverted phase contrast microscope (a) and fluorescence microscope (b). Efficiency of permanent miR-206 transfection was determined after 24 h. GFP (green fluorescent protein) signals were detectable in 75–80% of the permanently transfected cells. (A color version of this figure is available in the online journal.)
Relative gene expression of miR-206 targets before and after inhibition
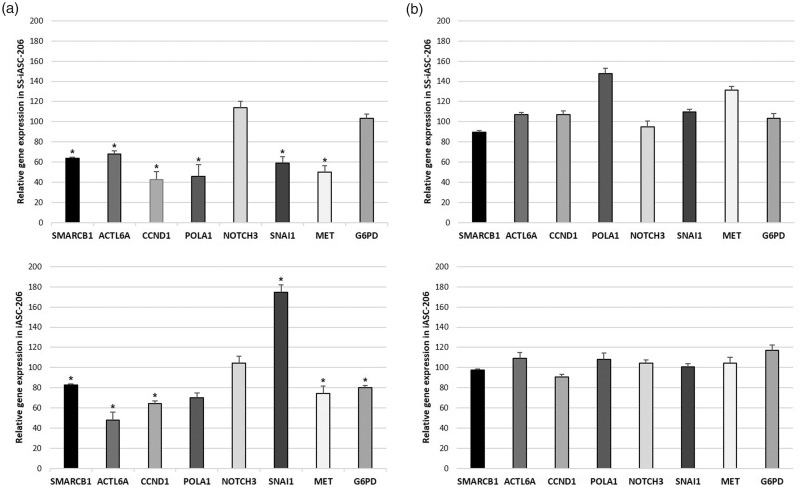
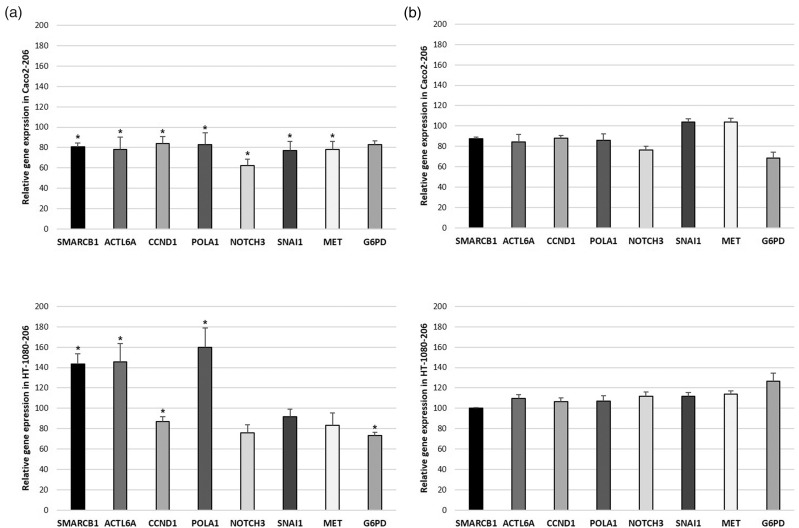
Figures 3(a) and (b) and 4(a) and (b) show the relative expression levels of the seven selected miR-206 target genes and the EMT marker SNAI1. Results significant at P < 0.05 are listed below. SMARCB1 expression was found to be downregulated to 64% in SS-iASC-206, to 83% in iASC-206, and to 81% in Caco2–206, but upregulated to 143% in HT-1080–206. ACTL6A was found to be downregulated to 68% in SS-iASC-206, to 48% in iASC-206, and to 78% in Caco2–206, while in HT-1080–206, it was overexpressed to 143%. CCND1 expression was found to be decreased to 42% in SS-iASC-206, to 64% in iASC-206, to 84% in Caco2–206, and to 87% in HT-1080–206. POLA1 was downregulated to 46% in SS-iASC-206 and to 83% in Caco2–206. However, in HT-1080–206, POLA1 was found to be overexpressed to 160%. NOTCH3 expression decreased to 62% in Caco2–206 only. MET expression was found to be downregulated to 50% in SS-iASC-206, to 74% in iASC-206, and to 78% in Caco2–206. G6PD expression was decreased to 80% in iASC-206 and to 73% in HT-1080–206. SNAI1 was overexpressed to 175% in iASC-206; however, it was downregulated to 59% in SS-iASC-206 and to 77% in Caco2–206. The means of the relative expression levels of the target genes with corresponding standard deviation values, P values, and levels of significance are represented in Table 1. MiR inhibition experiments with the permanently transfected cell lines verified our gene silencing results, since specific inhibition of miR-206 restored the expression levels of most target genes to around 100% of baseline (Figures 3(b) and 4(b)).
Figure 3.
Expression levels of miR-206 target genes. (a) The left column charts represent the SMARCB1, ACTL6A, CCND1, POLA1, NOTCH3, SNAI1, MET, and G6PD mRNA expression in the miR-206 transfected SS-iASC-206 and iASC-206 cell lines. (b) The right column charts illustrate these gene expressions after inhibiting miR-206. Bars represent means ± SD; *P < 0.05.
Table 1.
Summarized data of the target gene expressions and statistical analysis.
| Cell lines | Target genes | Mean valuea | SDb | P | Significance |
|---|---|---|---|---|---|
| SS-iASC-206 | SMARCB1 | 64 | 1.51 | 0.034 | S |
| ACTL6A | 68 | 2.86 | 0.034 | S | |
| CCND1 | 42 | 8.33 | 0.022 | S | |
| POLA1 | 46 | 11.28 | 0.022 | S | |
| NOTCH3 | 114 | 6.34 | 0.054 | NS | |
| MET | 50 | 6.48 | 0.022 | S | |
| G6PD | 103 | 4.21 | 0.143 | NS | |
| SNAI1 | 59 | 6.38 | 0.022 | S | |
| iASC-206 | SMARCB1 | 83 | 0.87 | 0.034 | S |
| ACTL6A | 48 | 7.65 | 0.022 | S | |
| CCND1 | 64 | 2.94 | 0.034 | S | |
| POLA1 | 70 | 4.95 | 0.054 | NS | |
| NOTCH3 | 104 | 6.91 | 0.137 | NS | |
| MET | 74 | 7.53 | 0.034 | S | |
| G6PD | 80 | 1.77 | 0.034 | S | |
| SNAI1 | 175 | 7.43 | 0.022 | S | |
| Caco2-206 | SMARCB1 | 81 | 3.32 | 0.034 | S |
| ACTL6A | 78 | 12.31 | 0.034 | S | |
| CCND1 | 84 | 6.82 | 0.034 | S | |
| POLA1 | 83 | 11.45 | 0.034 | S | |
| NOTCH3 | 62 | 6.32 | 0.022 | S | |
| MET | 78 | 8.12 | 0.034 | S | |
| G6PD | 83 | 3.35 | 0.054 | NS | |
| SNAI1 | 77 | 9.29 | 0.034 | S | |
| HT-1080-206 | SMARCB1 | 143 | 10.02 | 0.022 | S |
| ACTL6A | 146 | 17.71 | 0.022 | S | |
| CCND1 | 87 | 4.93 | 0.034 | S | |
| POLA1 | 160 | 19.25 | 0.022 | S | |
| NOTCH3 | 76 | 7.57 | 0.054 | NS | |
| MET | 83 | 12.06 | 0.054 | NS | |
| G6PD | 73 | 3.04 | 0.034 | S | |
| SNAI1 | 91 | 7.63 | 0.054 | NS |
S: significant; NS: not significant.
aMean value of the relative gene expression levels of miR-206 target genes measured by qRT-PCR.
bStandard deviation value.
Figure 4.
Expression levels of miR-206 target genes. (a) The left column charts represent the SMARCB1, ACTL6A, CCND1, POLA1, NOTCH3, SNAI1, MET, and G6PD mRNA expression in the miR-206-transfected Caco2–206 and HT-1080–206 cell lines. (b) The right column charts illustrate these gene expressions after inhibiting miR-206. Bars represent means ± SD; *P < 0.05.
SMARCB1 protein expression
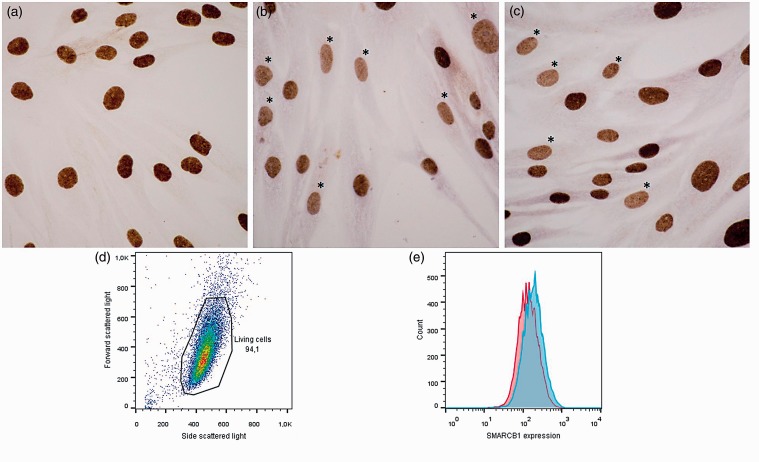
Western blot analysis did not detect any significant change of SMARCB1 protein expression in the miR-206-transfected cell lines compared to the non-transfected and the empty vector-transfected cells (results not shown). However, in the miR-206-transfected SS-iASC-206 cell line, SMARCB1 immunocytochemical reactions showed a focal decreased nuclear stain compared with the evenly strong nuclear stain of SS-iASC cell line (Figure 5(a) to (c)). Flow cytometry evaluation also demonstrated a modest decrease of SMARCB1 protein expression in SS-iASC-206 compared to SS-iASC, confirming the immunocytochemical results (Figure 5(d) and (e)).
Figure 5.
SMARCB1 immunocytochemistry and flow cytometry analysis of SS-iASC and SS-iASC-206. (a) Positive SMARCB1 staining of nuclei of not transfected SS-iASC cell line. (b and c) Between the strongly SMARCB1-immunopositive nuclei, decreased SMARCB1 protein expression was observed in individual nuclei of SS-iASC-206; (*) marks the decreased nuclear SMARCB1 expression. (d) Measurement of SMARCB1 expression by flow cytometry: gating strategy on the living cells using forward and side scattered light scattergram. (e) Fluorescence intensity of SMARCB1 in SS-iASC-206 and SS-iASC cell populations labeled with Alexa Flour® 647 conjugated goat anti-mouse IgG. The red histogram demonstrates miR-206 transfected SS-iASC-206, while the blue histogram shows the not transfected SS-iASC population. (A color version of this figure is available in the online journal.)
Discussion
MiR-206, as part of the myomiR family, plays a substantial role in muscle differentiation26 and can act as a tumor suppressor miRNA in several human tumors.27–29 Based on our previous findings using transient transfection,5 in the present study, we set out to examine the effect of permanent miR-206 transfection on various cell lines and assess how this small RNA molecule could affect the expression of SMARCB1 and other miR-206 target genes. To verify our data, we used an miRNA inhibition strategy. Based on the literature data, we chose six target genes of miR-206, the expression levels of which were examined following permanent miR-206 transfection. The gene silencing effects of miR-206 on these targets (namely ACTL6A, CCND1, POLA1, NOTCH3, MET, and G6PD) had been proven in some tumor tissues and cell lines,7,14,17,26,30,31 but until now we had no information about the effects of miR-206 on non-tumorous cells or other tumor cell lines in which miR-206 acts as an oncomiRNA. From the examined miR-206 target genes, one is a tumor suppressor gene (SMARCB1), one is the coding gene of the subunit p180 of DNA Polymerase α I (POLA1), and the others are oncogenes (ACTL6A, CCND1, NOTCH3, MET, G6PD). We also examined an EMT marker (SNAI1). The lack of SMARCB1 protein expression was observed in RT, ES, and in the spindle cell component of SS.4,32,33 Genetic and epigenetic regulation of SMARCB1 expression was also demonstrated in a variety of SMARCB1-immunonegative soft tissue sarcomas.5,6 The examined oncogenes are overexpressed in several tumor tissues or cell lines.7,26,31,34,35 In RMS cell lines, ACTL6A,7 MET,35 and G6PD31 are overexpressed. Upon increasing the level of miR-206 in the tumor cells, target genes were downregulated,7,31,35 tumor growth was impaired,7 cells finished their myogenic program,35 and the activity of pentose-phosphate pathway was decreased.31 CCND1 is overexpressed in clear cell renal cell carcinoma, hence increasing miR-206 level in these cells led to cell cycle arrest at the G0/G1 phase, indicating a major contribution of miR-206 to cell cycle regulation.34 Introducing miR-206 into myoblasts decreases the expression of POLA1 and therefore inhibits DNA synthesis26; hence, miR-206 has an antiproliferative effect here. NOTCH3 is a direct target of miR-206 and is overexpressed in HeLa cells. Increased miR-206 induces apoptosis in HeLa cells via cross-talk with NOTCH3.30
Following the permanent transfections, we were only able to analyze the results of four cell lines (iASC-206, SS-iASC-206, Caco2–206, and HT-1080–206) because HDFα cells were arrested in growth and eventually died. In C2C12 myoblasts, CCND1 and p180 subunits of DNA polymerase α were shown to be downregulated by miR-206 during differentiation, triggering G1 arrest, and drove myoblasts toward differentiation instead of proliferation.17,26 In our hands, miR-206 had a similar antiproliferative effect on HDFα cells which probably is the single most relevant action of miR-206 on normal cell lines.
After permanent transfection and antibiotic selection, stable overexpression of miR-206 was confirmed by qRT-PCR in the remaining four cell lines, and expression levels of miR-206 target genes were estimated using the same method.
In SS-iASC-206, the gene expression level of SMARCB1 decreased to 64% upon miR-206 transfection in comparison to iASC-206 where the same value was 83%; the difference most likely reflects an effect of the chimeric SS18-SSX1 fusion protein. The more pronounced silencing effect of miR-206 on SMARCB1 in SS-iASC-206 indicates a synergic influence of SS18-SSX1 fusion protein and miR-206. This synergic effect was proven in our previous work using microdissection in biphasic SS, thus separating the spindle cells with miR-206 overexpression from the epithelial cell with an unchanged miR-206 level. The difference is obvious in SMARCB1 protein levels, emphasizing the role of miR-206 (Figure 1) However, it is important to recognize that our SS-iASC cell line, even harboring the SS18-SSX1 fusion gene, is not a SS cell line which explains why we could not attain the same protein decrease in our cell line and also underlines the importance of different cell association.
SMARCB1, ACTL6A, and POLA1 were significantly downregulated in SS-iASC-206, iASC-206, and Caco2–206, while in HT-1080–206 these targets of miR-206 were overexpressed. These findings clearly showing the importance of the tumor tissue type and raising the possibility that miR-206 has no silencing effect on SMARCB1, ACTL6A, and POLA1 in HT-1080 fibrosarcoma cell line.
Interestingly, CCND1 and MET were the only oncogenes on which miR-206 invariably acted as a suppressor, so thus these findings collectively suggest a general suppressor function of miR-206 on CCND1 and MET in the four transfected cell lines.
NOTCH3 was significantly downregulated in Caco2–206, while changes in expression were not significant in SS-iASC-206, iASC-206, or HT-1080–206; hence, miR-206 had a significant effect on NOTCH3 only in Caco2–206.
G6PD was significantly downregulated after miR-206 transfection in iASC-206, Caco2–206, and HT-1080–206, but did not change in SS-iASC-206.
Biphasic SSs contain nests of glandular epithelium. These biphasic SSs harbor the SS18-SSX1 fusion gene instead of SS18-SSX2.36 As Saito et al. described, the fusion protein SS18-SSX1 interferes with SNAI1, the strongest repressor of E-cadherin promoter. They found in SSs that SS18-SSX1 can overcome the transcriptional repression of E-cadherin by SNAI1 which triggers mesenchymal-to-epithelial transition in this tumor.36 Saito et al. established that the derepression effect of SS18-SSX depends on the endogenous SNAI1 level in the cell line examined, i.e. with higher SNAI1 levels, SS18-SSX is able to release E-cadherin from repression more efficiently. Our miR-206 experiments showed that SNAI1 was downregulated in SS-iASC-206, while overexpressed in iASC-206. The SNAI1 downregulation observed in SS-iASC-206 can probably be accounted for by the combined effect of the fusion gene and miR-206. We have already seen SNAI1 downregulation in SS-iASC compared to iASC, but it was not as large as in the miR-206-transfected cells (data not shown).
Even though relative miR-206 expression was hundred- to thousand-fold in all permanently transfected cell lines, we saw only modest repression of the target genes. However, these results are supported by the findings of Mukherji et al.37 They found that the degree of repression by miRNAs is mild in a cell population and the repression varies greatly in single cells. Moreover, our results were validated with in vitro miR-206 inhibition experiments. In the four permanently transfected cell lines, the effect of miR-206 on its target genes was mostly reverted by miRNA inhibition.
Immunoblotting experiments showed no significant decrease in SMARCB1 protein expression in any of the cell lines (data not shown). However, more intriguing was our SMARCB1 immunocytochemistry and flow cytometry results. We could demonstrate a slight SMARCB1 protein expression decrease in individual nuclei of SS-iASC-206 intermingled with strong SMARCB1 positive nuclei at the same time (Figure 5(b) and (c)). These results suggest that in this subpopulation of individual cells, the SMARCB1 protein was expressed at a lower level compared to those that somehow overcame the downregulative effect of miR-206. It is also possible that on an individual cell level, both the chimeric gene and miR-206 transfection effect were differently represented. It is evident that permanent miR-206 overexpression in itself is not sufficient to abolish the SMARCB1 protein in SMARCB1 immunonegative sarcomas (with no biallelic damage of SMARCB1 gene), but definitely plays an important role in it – together with other gene-defects, like SS18-SSX fusion genes in SS. It would be interesting to test the silencing effect of miR-206 in a WT ES not carrying biallelic SMARCB1 deletion but to our knowledge, no such cell line is available on the market.
The mechanism of SMARCB1 silencing in different tumors is a subject of intensive research. As we showed in our previous transient miRNA experiments, a possible pathway to SMARCB1 silencing is miRNA overexpression.5,6 Here, we examined the influence of permanent miR-206 overexpression on its target genes in multiple cell lines. Prior to our experiments, no information was available on the effect of miR-206 on its target genes in tumor cell lines such as Caco2 or HT-1080, in normal cells such as HDFα, or in genetically engineered cell lines such as iASC or SS-iASC. We hypothesized that constitutive expression of miR-206 might successfully silence the target genes of miR-206, including SMARCB1 in tumorous, non-tumorous, and SS18-SSX1 transgenic cell lines. Our qRT-PCR results showed an overall tendency of downregulation of miR-206 target genes; however, in any individual cell line, the expression profile of the targets showed large variability. Permanent miR-206 transfection could silence SMARCB1 expression on the mRNA level (Figures 3(a) and 4(a)) but results in a slight protein level decrease and only on an individual cell level (Figure 5(b), (c) and (e)). These results suggest that the outcome of miR-206 overexpression depends on the cell line used. The expression changes of miR-206 targets observed in tumor tissues can only be reproduced in the corresponding tumorous cell lines, but not in other tumorous or normal cells. In a different context, miR-206 may fail to repress its target genes which is in line with the known fact that the effect of miRNAs is tissue- or tumor-dependent. We are not aware of a similarly comprehensive previous study to confirm this conclusion. Our results also clearly show that while miRNA alterations are excellent diagnostic tools, much more complex analysis is required to elucidate their mechanism of action in any given tumor type. Our results also support the conception that the oncomiR effect of miR-206 on SMARCB1 plays an important but not exclusive role in SMARCB1 immunonegative soft tissue sarcomas so it can be considered important in planning the targeted therapy of these tumors in the future.
Authors’ contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; DM carried out the miRNA transfection experiments, performed the qRT-PCR experiments, the immunocytochemical reactions and the flow cytometry investigation, collected the results and designed and drafted the manuscript. ZM performed the immunoblotting. AR helped with the preparation of figures. JS performed the statistical analysis. GSz helped with the analysis of flow cytometry. ZS conceived of the study and GP, PT and ZS participated in its design and helped to draft the manuscript. All authors have read and approved the final manuscript.
Declaration of Conflicting Of Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was financially supported by the Hungarian Scientific Research Found (OTKA) http://www.otka.hu/en, (No: K-112993); and the (NVKP_16–1-2016–0004) grant of the Hungarian National Research, Development and Innovation Office (NFKIH).
References
- 1.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene 2009; 28:1653–68 [DOI] [PubMed] [Google Scholar]
- 2.Kalimuthu SN, Chetty R. Gene of the month: SMARCB1. J Clin Pathol 2016; 69:484–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohashi K, Oda Y. Oncogenic roles of SMARCB1/INI1 and its deficient tumors. Cancer Sci 2017; 108:547–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 2013; 153:71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papp G, Krausz T, Stricker TP, Szendroi M, Sapi Z. SMARCB1 expression in epithelioid sarcoma is regulated by miR-206, miR-381, and miR-671-5p on Both mRNA and protein levels. Genes Chromosomes Cancer 2014; 53:168–76 [DOI] [PubMed] [Google Scholar]
- 6.Sapi Z, Papp G, Szendroi M, Papai Z, Plotar V, Krausz T, Fletcher CD. Epigenetic regulation of SMARCB1 By miR-206, -381 and -671-5p is evident in a variety of SMARCB1 immunonegative soft tissue sarcomas, while miR-765 appears specific for epithelioid sarcoma. A miRNA study of 223 soft tissue sarcomas. Genes Chromosomes Cancer 2016; 55:786–802 [DOI] [PubMed] [Google Scholar]
- 7.Taulli R, Foglizzo V, Morena D, Coda DM, Ala U, Bersani F, Maestro N, Ponzetto C. Failure to downregulate the BAF53a subunit of the SWI/SNF chromatin remodeling complex contributes to the differentiation block in rhabdomyosarcoma. Oncogene 2014; 33:2354–62 [DOI] [PubMed] [Google Scholar]
- 8.Bao YP, Yi Y, Peng LL, Fang J, Liu KB, Li WZ, Luo HS. Roles of microRNA-206 in osteosarcoma pathogenesis and progression. Asian Pac J Cancer Prev 2013; 14:3751–5 [DOI] [PubMed] [Google Scholar]
- 9.Wang CQ, Huang YW, Wang SW, Huang YL, Tsai CH, Zhao YM, Huang BF, Xu GH, Fong YC, Tang CH. Amphiregulin enhances VEGF-A production in human chondrosarcoma cells and promotes angiogenesis by inhibiting miR-206 via FAK/c-Src/PKCdelta pathway. Cancer Lett 2017; 385:261–70 [DOI] [PubMed] [Google Scholar]
- 10.Tian R, Liu T, Qiao L, Gao M, Li J. Decreased serum microRNA-206 level predicts unfavorable prognosis in patients with melanoma. Int J Clin Exp Pathol 2015; 8:3097–103 [PMC free article] [PubMed] [Google Scholar]
- 11.Ge X, Lyu P, Cao Z, Li J, Guo G, Xia W, Gu Y. Overexpression of miR-206 suppresses glycolysis, proliferation and migration in breast cancer cells via PFKFB3 targeting. Biochem Biophys Res Commun 2015; 463:1115–21 [DOI] [PubMed] [Google Scholar]
- 12.Chen QY, Jiao DM, Wu YQ, Chen J, Wang J, Tang XL, Mou H, Hu HZ, Song J, Yan J, Wu LJ, Chen J, Wang Z. MiR-206 inhibits HGF-induced epithelial-mesenchymal transition and angiogenesis in non-small cell lung cancer via c-Met/PI3k/Akt/mTOR pathway. Oncotarget 2016; 7:18247–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amirouche A, Jahnke VE, Lunde JA, Koulmann N, Freyssenet DG, Jasmin BJ. Muscle-specific microRNA-206 targets multiple components in dystrophic skeletal muscle representing beneficial adaptations. Am J Physiol Cell Physiol 2017; 312:C209–21 [DOI] [PubMed] [Google Scholar]
- 14.Cui Y, Xie S, Luan J, Zhou X, Han J. Identification of the receptor tyrosine kinases (RTKs)-oriented functional targets of miR-206 by an antibody-based protein array. FEBS Lett 2015; 589:2131–5 [DOI] [PubMed] [Google Scholar]
- 15.Ren XL, He GY, Li XM, Men H, Yi LZ, Lu GF, Xin SN, Wu PX, Li YL, Liao WT, Ding YQ, Liang L. MicroRNA-206 functions as a tumor suppressor in colorectal cancer by targeting FMNL2. J Cancer Res Clin Oncol 2016; 142:581–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krasteva V, Buscarlet M, Diaz-Tellez A, Bernard MA, Crabtree GR, Lessard JA. The BAF53a subunit of SWI/SNF-like BAF complexes is essential for hemopoietic stem cell function. Blood 2012; 120:4720–32 [DOI] [PubMed] [Google Scholar]
- 17.Alteri A, De Vito F, Messina G, Pompili M, Calconi A, Visca P, Mottolese M, Presutti C, Grossi M. Cyclin D1 is a major target of miR-206 in cell differentiation and transformation. Cell Cycle 2013; 12:3781–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chattopadhyay S, Bielinsky AK. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol Biol Cell 2007; 18:4085–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, Lu C, Fang T, Wang Y, Hu W, Qiao J, Liu B, Liu J, Chen N, Li M, Zhu R. Notch3 functions as a regulator of cell self-renewal by interacting with the beta-catenin pathway in hepatocellular carcinoma. Oncotarget 2015; 6:3669–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crepaldi T, Bersani F, Scuoppo C, Accornero P, Prunotto C, Taulli R, Forni PE, Leo C, Chiarle R, Griffiths J, Glass DJ, Ponzetto C. Conditional activation of MET in differentiated skeletal muscle induces atrophy. J Biol Chem 2007; 282:6812–22 [DOI] [PubMed] [Google Scholar]
- 21.Kuo W, Lin J, Tang TK. Human glucose-6-phosphate dehydrogenase (G6PD) gene transforms NIH 3T3 cells and induces tumors in nude mice. Int J Cancer 2000; 85:857–64 [DOI] [PubMed] [Google Scholar]
- 22.Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer 2016; 15:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatrai P, Szepesi A, Matula Z, Szigeti A, Buchan G, Madi A, Uher F, Nemet K. Combined introduction of Bmi-1 and hTERT immortalizes human adipose tissue-derived stromal cells with low risk of transformation. Biochem Biophys Res Commun 2012; 422:28–35 [DOI] [PubMed] [Google Scholar]
- 24.Mihaly D, Matula Z, Changchien YC, Papp G, Tatrai P, Sapi Z. First cloned human immortalized adipose derived mesenchymal stem-cell line with chimeric SS18-SSX1 gene (SS-iASC). Cancer Genet 2017; 216–217:52–60 [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25:402–8 [DOI] [PubMed] [Google Scholar]
- 26.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 2006; 174:677–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koshizuka K, Hanazawa T, Fukumoto I, Kikkawa N, Matsushita R, Mataki H, Mizuno K, Okamoto Y, Seki N. Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive miR-1 and miR-206 in head and neck squamous cell carcinoma. J Hum Genet 2017; 62:113–21 [DOI] [PubMed] [Google Scholar]
- 28.Amir S, Simion C, Umeh-Garcia M, Krig S, Moss T, Carraway KL, 3rd, Sweeney C. Regulation of the T-box transcription factor Tbx3 by the tumour suppressor microRNA-206 in breast cancer. Br J Cancer 2016; 114:1125–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W, Xu C, Wan H, Liu C, Wen C, Lu H, Wan F. MicroRNA-206 overexpression promotes apoptosis, induces cell cycle arrest and inhibits the migration of human hepatocellular carcinoma HepG2 cells. Int J Mol Med 2014; 34:420–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song G, Zhang Y, Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem 2009; 284:31921–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coda DM, Lingua MF, Morena D, Foglizzo V, Bersani F, Ala U, Ponzetto C, Taulli R. SMYD1 and G6PD modulation are critical events for miR-206-mediated differentiation of rhabdomyosarcoma. Cell Cycle 2015; 14:1389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohashi K, Tanaka Y, Kishimoto H, Yamamoto H, Yamada Y, Taguchi T, Iwamoto Y, Oda Y. Reclassification of rhabdoid tumor and pediatric undifferentiated/unclassified sarcoma with complete loss of SMARCB1/INI1 protein expression: three subtypes of rhabdoid tumor according to their histological features. Mod Pathol 2016; 29:1232–42 [DOI] [PubMed] [Google Scholar]
- 33.Modena P, Lualdi E, Facchinetti F, Galli L, Teixeira MR, Pilotti S, Sozzi G. SMARCB1/INI1 tumor suppressor gene is frequently inactivated in epithelioid sarcomas. Cancer Res 2005; 65:4012–9 [DOI] [PubMed] [Google Scholar]
- 34.Xiao H, Xiao W, Cao J, Li H, Guan W, Guo X, Chen K, Zheng T, Ye Z, Wang J, Xu H. miR-206 functions as a novel cell cycle regulator and tumor suppressor in clear-cell renal cell carcinoma. Cancer Lett 2016; 374:107–16 [DOI] [PubMed] [Google Scholar]
- 35.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest 2009; 119:2366–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito T, Nagai M, Ladanyi M. SYT-SSX1 and SYT-SSX2 interfere with repression of E-cadherin by snail and slug: a potential mechanism for aberrant mesenchymal to epithelial transition in human synovial sarcoma. Cancer Res 2006; 66:6919–27 [DOI] [PubMed] [Google Scholar]
- 37.Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet 2011; 43:854–9 [DOI] [PMC free article] [PubMed] [Google Scholar]