Abstract
Background
Histone deacetylases (HDACs) function as key epigenetic factors in repressing the expression of genes in multiple aspects of plant growth, development and plant response to abiotic or biotic stresses. To date, the molecular function of HDACs is well described in Arabidopsis thaliana, but no systematic analysis of this gene family in soybean (Glycine max) has been reported.
Results
In this study, 28 HDAC genes from soybean genome were identified, which were asymmetrically distributed on 12 chromosomes. Phylogenetic analysis demonstrated that GmHDACs fall into three major groups previously named RPD3/HDA1, SIR2, and HD2. Subcellular localization analysis revealed that YFP-tagged GmSRT4, GmHDT2 and GmHDT4 were predominantly localized in the nucleus, whereas GmHDA6, GmHDA13, GmHDA14 and GmHDA16 were found in both the cytoplasm and nucleus. Real-time quantitative PCR showed that GmHDA6, GmHDA13, GmHDA14, GmHDA16 and GmHDT4 were broadly expressed across plant tissues, while GmHDA8, GmSRT2, GmSRT4 and GmHDT2 showed differential expression across various tissues. Interestingly, we measured differential changes in GmHDACs transcripts accumulation in response to several abiotic cues, indicating that these epigenetic modifiers could potentially be part of a dynamic transcriptional response to stress in soybean. Finally, we show that the levels of histone marks previously reported to be associated with plant HDACs are modulated by cold and heat in this legume.
Conclusion
We have identified and classified 28 HDAC genes in soybean. Our data provides insights into the evolution of the HDAC gene family and further support the hypothesis that these genes are important for the plant responses to environmental stress.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1454-7) contains supplementary material, which is available to authorized users.
Keywords: Histone deacetylases, Subcellular localization, Gene expression, Abiotic stresses, Soybean
Background
Throughout their life course, plants are frequently exposed to suboptimal environmental conditions that cause adverse effects on their growth and development. Abiotic stress is one of the major causes of agricultural losses in the world [1, 2]. Soybean (Glycine max) is an important global crop desirable for its high protein content and oil. More than 50% of globally consumed edible oil is contributed by soybeans while the proteins of this legume are highly desirable for food and feed applications [3]. However, the production of soybeans is greatly affected by abiotic stresses such as salt, drought, cold, heat, water submergence and heavy metals [3]. Recently, studies have demonstrated that epigenetic processes play vital regulatory roles in plant abiotic stress responses [4–7]. Chromatin structure and gene expression changes in response to abiotic stress are controlled through epigenetic mechanisms such as histone modification and chromatin remodeling. Among them, histone acetylation is regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs).
In the past decades, a large number of HDACs have been identified and characterized in plants, which can be grouped into three different families: the RPD3/HDA1, the SIR2, and the HD2. Members of the RPD3/HDA1 and the SIR2 families are proteins homologous to the yeast Reduced Potassium Dependency 3 (RPD3)/HDA1 and Silent Information Regulator 2 (SIR2), respectively, whereas the HD2 family was originally characterized in maize and appears to be present only in plants [8–10]. Members of the SIR2 family have a catalytic domain that requires nicotine adenine dinucleotide (NAD) as a cofactor [11], while members of the RPD3/HDA1 family share HDAC domain sequence homology and require a Zn2+ cofactor for deacetylase activity [9]. The HD2 family proteins contain a conserved pentapeptide motif (MEFWG) at the N-terminus and are considered to be zinc-dependent HDACs [12].
The Arabidopsis HDACs have been well characterized. The genome of this model plant encodes 18 HDACs distributed into the three aforementioned HDAC families [10, 13]. Increasing evidence suggests that AtHDACs play essential roles in regulating multiple aspects of plant growth and development [4, 14, 15]. AtHDA6 is a member of the RPD3/HDA1 family that was originally reported to play an important role in coordinating downstream gene silencing and maintenance of DNA methylation [16, 17]. It was also demonstrated that HDA6 interacted with a number of protein partners, such as FLOWERING LOCUS D (FLD), ASYMMETRIC LEAVES 1 (AS1), TOPLESS (TPL), and JASMONATE ZIM-DOMAIN (JAZ) to regulate flowering, leaf development, circadian transcription and hormonal responses by modulating the transcription of their respective target genes through histone deacetylation [18–21]. Other members of the RDP3/HDA1 family, such as AtHDA5, AtHDA7, AtHDA9, AtHDA14, AtHDA15, AtHDA18 and AtHDA19, were also reported to play important roles in plant growth and development [22–37]. For example, hda19 mutants displayed various developmental abnormalities, such as early senescence, suppression of apical dominance, flower defects, and male and female sterility, indicative of the importance of AtHDA19 for proper vegetative development [22, 24, 31]. HDA5 forms a protein complex with HDA6, FLD and MSI1-like WD40 repeat 4 (FVE/MSI4) in regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression through histone deacetylation [34]. The BRI1-EMS-SUPPRESSOR1 (BES1)-TPL-HDA19 repressor complex is required for deacetylation of ABA-insensitive 3 (ABI3) gene in controlling early seedling development [32]. Additionally, members of the HD2 family play crucial roles in plant development [38–42]. For instance, silencing of HD2A in Arabidopsis resulted in aborted seed development [38], while overexpression of HD2A caused morphological defects of leaves and flowers, delayed flowering and aborted seed development [39]. It was demonstrated that the SIR2 family protein AtSRT2 plays a negative role in plant basal defense against the pathogen PstDC3000, and the expression of AtSRT2 was repressed in response to the pathogen infection [43]. In rice (Oryza sativa), OsSRT1 is required for repressing the expression of starch metabolism genes during seed development [44].
In Arabidopsis, AtHDACs play vital roles in plant responses to abiotic and biotic stresses [4, 45–53]. It was reported that AtHDACs were responsive to various environmental cues at the transcription level [13], for example, AtHDA6 and AtHDA19 are induced by JA treatment [45], whereas the expression of HD2A, HD2B, HD2C, and HD2D is repressed by ABA and NaCl [49, 54]. AtHDA9 was shown to negatively regulate high salt and drought responses [53]. On the other hand, AtHDA6, AtHD2C, and AtHD2D confer plant resistance under high salt, drought, cold or freezing conditions [49–51, 55]. Recently, emerging evidence indicates that hda19 plants exhibit tolerance to high salinity stress, while hda5/14/15/18 plants exhibit hypersensitivity to salt stress. This suggests that AtHDA19 and AtHDA5/14/15/18 control plant responses to salt stress in different pathways [53]. While, different members of the HDAC family have been found to mediate distinct aspects of the plant growth and development as well as its abiotic and biotic stress responses, the characterization of HDAC genes in soybean has not been reported yet.
In this study, we have identified 28 HDAC coding genes in the soybean genome. YFP-tagged transient expression assays confirmed the subcellular localization of GmHDACs. While tissue-specific and stress-responsive expression patterns for nine representative genes were determined using quantitative RT-PCR. In addition, the levels of histone acetylation and methylation were analyzed under cold and heat treatments. Together, our results shed light on the involvement of GmHDAC genes in various aspects of plant growth and development including the response to abiotic stress.
Results
Identification and phylogenetic analysis of soybean histone deacetylases
The protein sequences of AtHDACs were used as queries to conduct sequence homology searches against the Soybase database (https://soybase.org/). In total, 28 independent soybean GmHDACs were identified. (Table 1). The complete open reading frames (ORFs) of the retrieved GmHDAC genes ranged from 582 to 1971 bp, while predicted proteins ranged from 193 to 656 amino acids with calculated molecular weights from 21.37 to 72.99 kDa and isoelectric points from 4.29 to 9.40 (Table 1). Further bioinformatics analyses indicated that GmHDACs were potentially localized in several organelles such as the nucleus, cytoplasm, chloroplast and mitochondria (Table 1).
Table 1.
Overview of histone deacetylases genes identified in soybean
| Gene ID | DNA attributes | Protein attributes | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene namea | Gene locusb | Accession numberc | Chromosome | ORF (bp) | No. of exons | Length (aa) | MW (kDa) | PI | Localizationd |
| GmHDA1 | Glyma.01 g245100 | XP_003517607.1 | 1 | 1494 | 7 | 497 | 56.28 | 5.30 | nucl, cyto |
| GmHDA2 | Glyma.04 g000200 | XP_003543935.1 | 4 | 1494 | 7 | 497 | 55.96 | 5.04 | cyto, nucl |
| GmHDA3 | Glyma.04 g187000 | XP_006578653.1 | 4 | 1398 | 6 | 465 | 52.18 | 5.28 | cyto, chlo |
| GmHDA4 | Glyma.04 g187100 | KRH63610.1 | 4 | 594 | 2 | 197 | 22.26 | 6.31 | cyto |
| GmHDA5 | Glyma.05 g012900 | KRH56687.1 | 5 | 1254 | 14 | 417 | 45.28 | 7.39 | nucl, cyto |
| GmHDA6 | Glyma.05 g021400 | XP_003524633.1 | 5 | 1971 | 14 | 656 | 73.04 | 5.25 | cyto, nucl |
| GmHDA7 | Glyma.05 g040600 | XP_003525556.1 | 5 | 1431 | 6 | 476 | 53.30 | 5.24 | nucl |
| GmHDA8 | Glyma.05 g192600 | XP_014631275.1 | 5 | 1263 | 9 | 420 | 45.54 | 6.09 | chlo, nucl |
| GmHDA9 | Glyma.06 g000100 | XP_003526730.1 | 6 | 1494 | 7 | 497 | 55.95 | 5.06 | nucl, cyto |
| GmHDA10 | Glyma.06 g178700 | KRH54335.1 | 6 | 705 | 1 | 234 | 26.74 | 8.88 | chlo, cyto |
| GmHDA11 | Glyma.11 g000300 | XP_006590384.1 | 11 | 1494 | 7 | 497 | 56.12 | 5.22 | nucl, cyto |
| GmHDA12 | Glyma.11 g187800 | XP_003538135.1 | 11 | 1290 | 14 | 429 | 48.94 | 4.98 | cyto, nucl |
| GmHDA13 | Glyma.12 g086700 | XP_003539814.1 | 12 | 1290 | 14 | 429 | 48.94 | 5.06 | cyto, mito |
| GmHDA14 | Glyma.12 g188200 | XP_003540263.1 | 12 | 1146 | 3 | 381 | 41.18 | 5.44 | cyto, chlo |
| GmHDA15 | Glyma.17 g078000 | KRH03123.1 | 17 | 1971 | 14 | 656 | 72.99 | 5.35 | cyto, nucl |
| GmHDA16 | Glyma.17 g085700 | XP_003549603.1 | 17 | 1419 | 6 | 472 | 52.92 | 5.26 | nucl, chlo |
| GmHDA17 | Glyma.17 g120900 | XP_006600776.1 | 17 | 1632 | 17 | 543 | 59.6 | 5.91 | nucl, cyto |
| GmHDA18 | Glyma.17 g229600 | XP_003550277.1 | 17 | 1047 | 13 | 348 | 38.50 | 6.29 | cyto, nucl |
| GmSRT1 | Glyma.04 g210000 | XP_003522478.1 | 4 | 1182 | 11 | 393 | 43.46 | 9.40 | chlo, mito |
| GmSRT2 | Glyma.06 g156000 | XP_003528059.2 | 6 | 1179 | 11 | 392 | 43.21 | 9.32 | chlo, mito |
| GmSRT3 | Glyma.08 g330200 | KHN11152.1 | 8 | 1302 | 13 | 433 | 48.09 | 9.11 | chlo, nucl |
| GmSRT4 | Glyma.18g076300 | XP_003551434.1 | 18 | 1440 | 14 | 479 | 53.19 | 9.15 | nucl, cyto |
| GmHDT1 | Glyma.03 g190700 | NP_001240859.1 | 3 | 867 | 10 | 288 | 31.46 | 4.64 | nucl |
| GmHDT2 | Glyma.11 g189500 | XP_006591094.1 | 11 | 870 | 9 | 289 | 30.77 | 4.80 | nucl |
| GmHDT3 | Glyma.12 g084700 | KRH25160.1 | 12 | 900 | 9 | 299 | 31.77 | 4.75 | nucl |
| GmHDT4 | Glyma.12 g181400 | KRH26575.1 | 12 | 924 | 10 | 307 | 33.31 | 4.9 | nucl |
| GmHDT5 | Glyma.13 g319500 | KRH22734.1 | 13 | 582 | 6 | 193 | 21.37 | 4.29 | chlo, cyto |
| GmHDT6 | Glyma.19 g191000 | XP_003554417.1 | 19 | 882 | 9 | 293 | 31.91 | 4.61 | nucl |
aSystematic designation given to soybean histone deacetylase genes
bAccession number of Soybase (http://soybase.org/) locus ID
cAccession numbers of protein sequence available at NCBI (http:// www.ncbi.nlm.nih.gov/)
dSubcellular Localization of soybean histone deacetylases supported by WoLF PSORT (http://www.genscript.com/psort/wolf_psort.html)
CDS, coding sequence; No., number; MW, molecular weight; PI, isoelectric point
Nuc, nuclear; Cyto, cytoplasm; Chl, chloroplast; Mito, mitochondrion
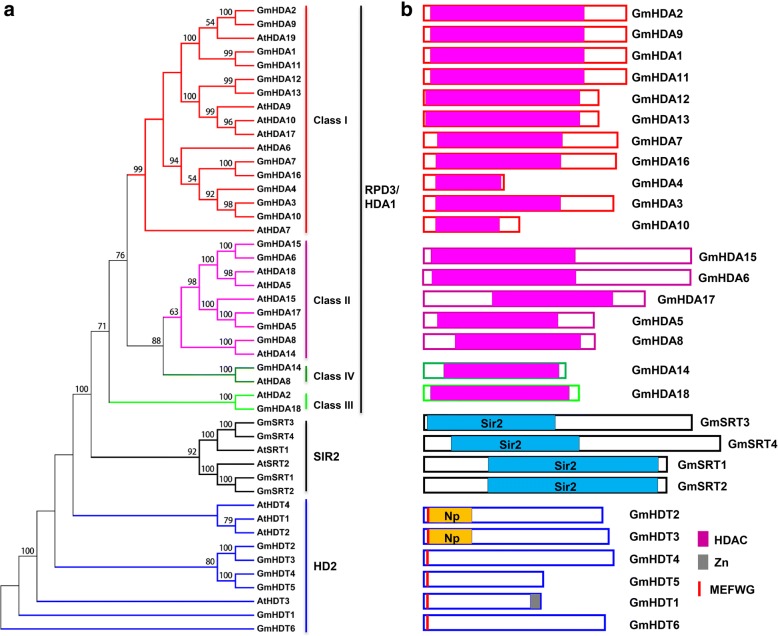
To evaluate the evolutionary relationship of plant histone deacetylases, a phylogenetic analysis was performed using the protein sequences of HDACs from soybean and Arabidopsis. The phylogenetic tree indicated that the 28 newly uncovered soybean HDACs are grouped into three types characterized by distinctive protein structures (Table 1 and Fig. 1). In soybean, type I (RPD3/HDA1 family) HDACs consist of 18 members, named GmHDA1 to GmHDA18 based on their coordinates on soybean chromosome (Table 1). All 18 members of this type have a characteristic histone deacetylase domain (Interpro: IPR003084) (Fig. 1b) and can be further divided into four classes based on sequence similarity (Fig. 1a). Class I encompassed 11 GmHDACs, Class II included 5 members while classes III and IV each contained one GmHDAC (Fig. 1a). The phylogenetic analysis also demonstrated that soybean has four type II (SIR2 family) HDACs with highly conserved Sir2 domains (Table 1 and Fig. 1). Finally, six plant-specific HDACs (type III: HD2 family) with conserved N-terminal MEFWG amino acid regions (Fig. 1b), which is required for transcriptional repression followed by a central acidic region rich in glutamic and/or aspartic acid [9] were identified in soybean (Table 1 and Fig. 1). A C2H2 zinc finger domain in the C-terminus portion of GmHDT1 was also detected, which might indicate that this protein has high DNA-binding affinity or could mediate protein-protein interactions. Additionally, both GmHDT2 and GmHDT3 may bind to nucleoplasmins through their N-terminal nucleophosmin domains (Fig. 1b).
Fig. 1.
Phylogenetic tree and domain architecture of HDACs in soybean. a Phylogenetic tree of HDAC proteins in soybean and Arabidopsis. The phylogenetic tree was generated using the MEGA 5.0 software and the Maximum Parsimony method with the following parameters: bootstrap analysis of 1000 replicates and partial deletion. The numbers at the nodes indicate the bootstrap values. b The schematic diagrams show the domain organization of these proteins according to analysis by NCBI Batch-CD, SMART and PFAM searches. Different domains are indicated by the use of different colors. The proteins belonging to each family are grouped together
Chromosomal localization and duplications of HDAC genes in soybean
The chromosomal localization of the 28 GmHDAC genes was determined by their genomic distributions on soybean chromosomes. The 28 GmHDAC genes were asymmetrically distributed on 12 chromosomes (Fig. 2). Chromosome 4, 5, 12 and 17 each contain the largest number of HDAC genes with four each, followed by chromosome 6 and 11 with three genes each, whereas only one HDAC gene was present on each of chromosomes 1, 3, 8, 13, 18, and 19. No HDAC genes were found on chromosome 2, 7, 9, 10, 14, 15, and 20 (Fig. 2). Moreover, the HDAC gene density per chromosome was uneven with the highest densities of HDACs at proximal regions of chromosomes 5 and 17 and the distal region of chromosome 4 (Fig. 2).
Fig. 2.
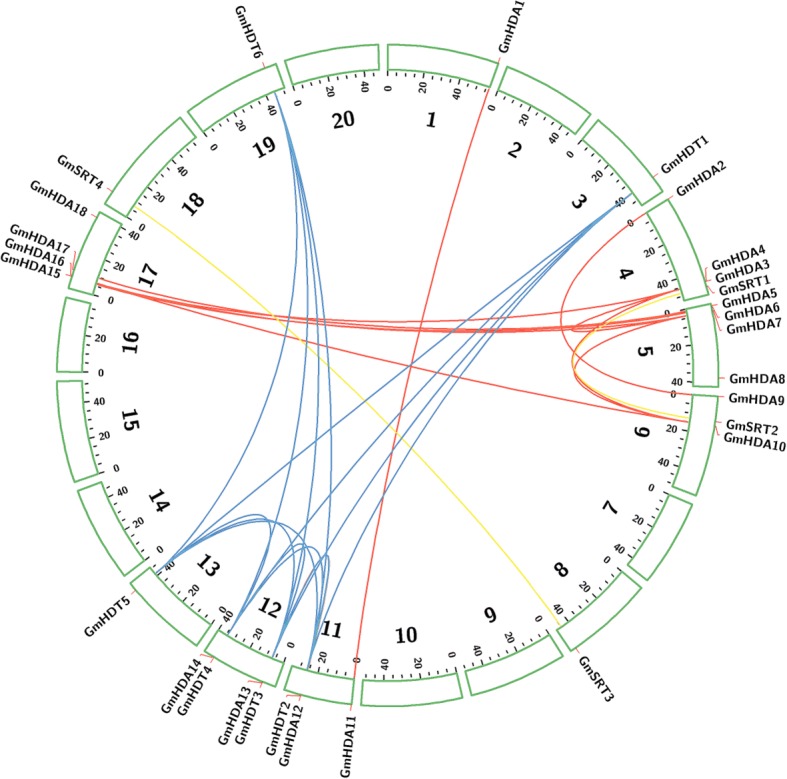
Chromosomal localization and duplication of HDAC genes in soybean. Each box refers to a chromosome, and chromosome numbers are shown beside each chromosome. The scale is in megabase. The approximate location of each soybean HDAC gene is indicated by a short orange line. Colored lines in the circle indicate the linkage group with segmentally duplicated HDAC gene pairs, and segmental duplication regions were determined using the Plant Genome Duplication Database
Tandem duplication and segmental genome duplication are major contributors to the generation and maintenance of gene families. If two paralogous genes are physically close together, we conclude that they probably arose through tandem duplication [56]. It was found that only one gene set (GmHDA3 and GmHDA4) was tandemly distributed on chromosome 4 and was separated by 4407 bp. However, only 23.8% and 29.7% of sequence identity at the nucleotide and protein level were respectively observed between them, indicating that GmHDA3 and GmHDA4 have not undergone tandem amplification during evolution. We further investigated whether segmental duplications contributed to the expansion of HDAC gene family in soybean. It is noteworthy that 27 duplication sets of HDAC genes on the same block were identified. These duplicated genes pairs clustered into a discrete clade in the phylogenetic tree and shared a high degree of identity at the protein level within each pair (Figs. 1 and 2, Additional file 1: Table S1). To evaluate the selection mode of the 27 duplicated gene pairs of HDAC in soybean, we calculated the nonsynonymous/synonymous substitution ratios (Ka/Ks). According to the literature a Ka/Ks ratio above 1 indicates positive selection while ratios below and equal to 1 respectively indicate purifying and neural selection [57]. As shown in the Additional file 1: Table S1, the Ka/Ks values of all duplicated gene pairs were less than 1, suggesting that these duplicates likely have been subjected to purifying selection.
Subcellular localization of GmHDACs
Prediction analysis indicated that GmHDACs exhibit various patterns of subcellular localization (Table 1). To further determine the subcellular localization of GmHDACs, full-length cDNAs were fused to the Yellow Fluorescent Protein (YFP) driven by the CaMV 35S promoter and transiently expressed in protoplasts of Arabidopsis suspension culture cells. As shown in Fig. 3, four RPD3/HDA1 family members, GmHDA6, 13, 14 and 16 localized in both the cytosol and nucleus. Consistent with the predicted localization using bioinformatics programs, GmSRT4 and two members of HD2 family (GmHDT2 and GmHDT4) localized in the nucleus, mainly in the nucleolus (Fig. 3).
Fig. 3.
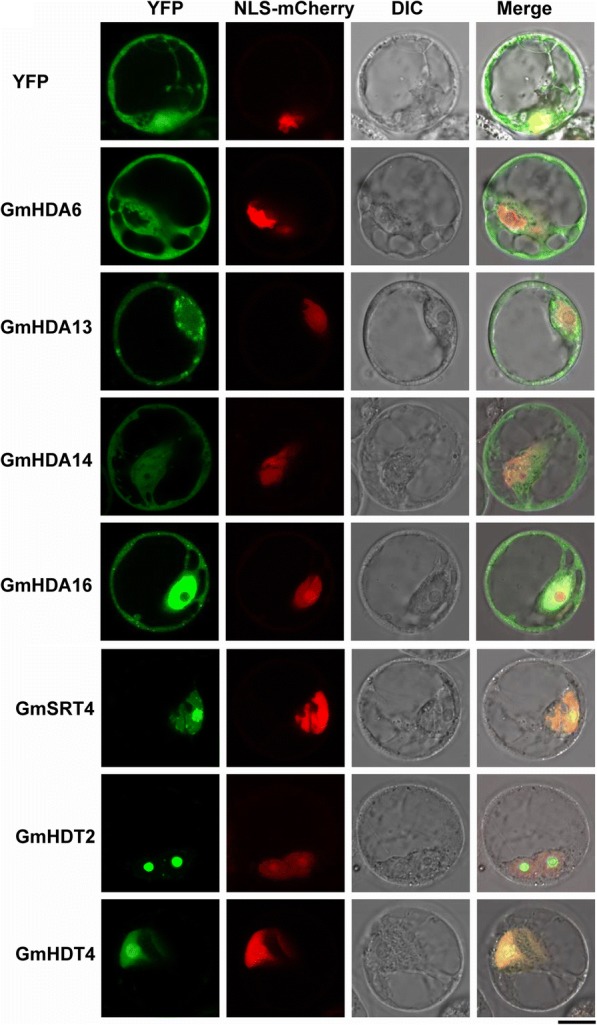
Subcellular localization of GmHDACs. GmHDACs-EYFP fusion constructs were used to determine the subcellular localization of GmHDACs. NLS-mCherry was used as a nuclear marker. Fluorescence images of YFP and mCherry were captured with confocal laser scanning microscopy and are shown in green and red, respectively (scale bars, 10 μm)
Tissue and organ specific expression of HDAC genes in soybean
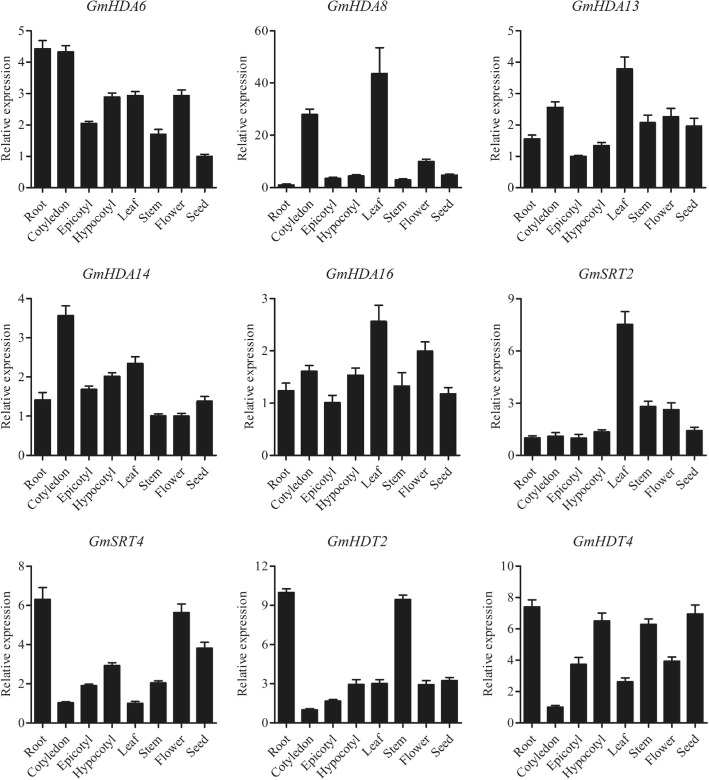
To investigate the tissue and organ specific expression profiles of HDAC genes in soybean, quantitative RT-PCR assays for nine representative HDAC genes from different families were conducted in different tissues and developmental stages. As shown in Fig. 4, GmHDA6, GmHDA13, GmHDA14 and GmHDA16 were ubiquitously expressed at high levels in most of the organs examined. GmHDA8 showed high expression in cotyledon and leaf, as well as relatively low expression in other tissues. Specific high transcript accumulation of GmSRT2 was observed in the leaves, while the expression in the other organs remained low. Unlike GmSRT2, GmSRT4 mRNA levels were high in roots and flower tissues, moderate in hypocotyls and seeds, and lowest in the leaves. In addition, the GmHDT2 transcripts were abundant in roots and stems, whilst relatively low in other tissues. Finally, GmHDT4 was ubiquitously expressed at high levels in most of the organs examined, except for the cotyledons.
Fig. 4.
Tissue-specific expression patterns of GmHDAC genes. The x-axis represents different tissues or organs. The y-axis shows the gene expression levels after normalization to reference gene GmCYP2. Roots, cotyledons, epicotyls, and hypocotyls of 5-day-old plants and leaves, stems of 14-day-old plants, young flowers and seeds were collected for total RNA isolation. Quantitative RT-PCR was performed using gene-specific primers. Data are the mean ± SEM of three independent experiments
Soybean seeds are one of the most important agricultural commodities, being a prime source of oil, protein, and carbohydrate. To investigate the expression profiles of GmHDAC genes during soybean seed development and maturation, we obtained publicly available transcriptome data from the Soybase database (https://soybase.org/). As shown in Additional file 2: Fig. S1, nine GmHDAC genes displayed differential expression patterns in developing seeds. GmHDA8, GmHDA14, GmHDT2 and GmHDT4 were highly expressed throughout seed development. However, the expression of GmSRT4 was only detected at the early stages of seed development, whereas GmHDA13 was highly expressed during the middle stage. GmHDA6 transcripts accumulated progressively early in developing seeds but their abundance decreased markedly during the seed maturation process. Additionally, the transcripts of GmHDA16 were most abundant in 21 DAF (days after flowering) seeds but scarce in 28 DAF seeds (Additional file 2: Fig. S1).
GmHDACs are involved in various abiotic stress responses
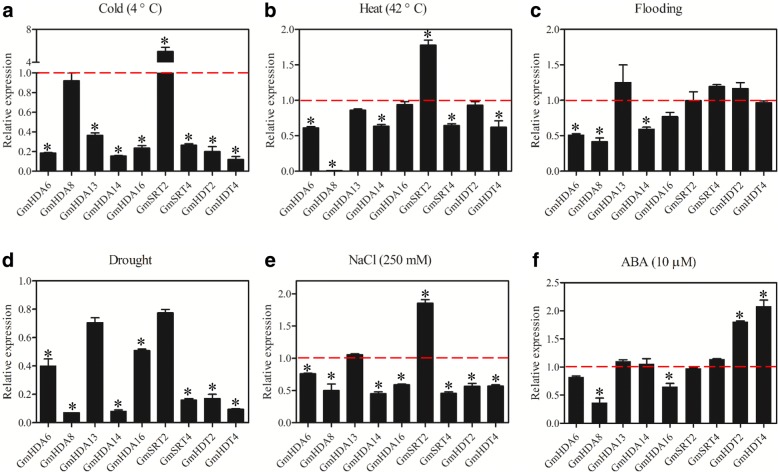
Evidence suggests that HDACs play important functions in plant response to various abiotic stresses. To study the potential roles of GmHDACs in abiotic stress responses, we performed expression analysis of GmHDACs under various abiotic stress conditions using quantitative RT-PCR. As shown in Fig. 5, our results demonstrated that GmHDAC genes significantly responded to various abiotic stress treatments. When exposed to cold, the expression of seven GmHDAC genes (GmHDA6, 13, 14, 16, GmSRT4, GmHDT2 and GmHDT4) was strongly repressed, while GmSRT2 expression was significantly induced. On the other hand, GmHDA8 was not significantly changed in response to low temperature (Fig. 5a). Following a heat shock treatment, five GmHDAC genes (GmHDA6, 8, 14, GmSRT4 and GmHDT4) became down-regulated, GmSRT2 was up-regulated, while the expression of GmHDA13, 16 and GmHDT2 were unchanged (Fig. 5b). Notably, the accumulation level of the GmHDA8 transcript was nearly completely suppressed under heat treatment (Fig. 5b). Furthermore, the differential expression patterns of the GmHDAC genes under flooding stress were observed. Four RPD3/HDA1 family genes, GmHDA6, 8, 14 and 16 were down-regulated, while GmHDA13, GmSRT4 and GmHDT2 were slightly up-regulated and GmSRT2 and GmHDT4 did not respond to the flooding treatment (Fig. 5c). The expression levels of all nine GmHDACs examined decreased following a drought stress treatment with GmHDA8, GmHDA14, GmSRT4, GmHDT2 and GmHDT4 being the most impacted (Fig. 5d). Finally, high salt treatment significantly induced GmSRT2 expression, but repressed the expression of GmHDA6, 8, 14, 16, GmSIR4, GmHDT2 and GmHDT4, and did not modulate the expression of GmHDA13 (Fig. 5e).
Fig. 5.
Expression profiles of GmHDAC genes under abiotic stresses. 10-day old plants were treated with cold (4 °C), heat (42 °C), flooding, air-drought, 250 mM NaCl and 10 μM ABA for 12 h and the leaves were harvested for qRT-PCR analysis of gene expression. The x-axis presents different genes. The y-axis shows expression levels relative to the control, which was set to 1.0. Data are the mean ± SEM of three independent experiments. *, P-value < 0.05, Student’s t-test
Finally, we evaluated the responses of GmHDACs to abscisic acid (ABA) treatment, which is the most important stress-protective phytohormone. Two HD2 family genes (GmHDT2 and GmHDT4) were induced and two RPD3/HDA1 family genes (GmHDA8 and GmHDA16) were markedly suppressed by our ABA treatment. On the other hand, the expression of five other HDAC genes (GmHDA6, 13, 14, GmSRT2 and GmSRT4) was only slightly modulated (Fig. 5f). In addition, status of H3 acetylation following our cold and heat treatments was analyzed. As shown in Fig. 6, the level of H3ac decreased under cold treatment, but increased after heat treatment. Interestingly, status of H3K4me2 and H3K4me3 were also modulated by the both cold and heat treatments. High levels of H3K4me2 and low levels of H3K4me3 were observed following our cold treatment. Meanwhile, the level of H3K4me2 was up-regulated in response to heat stress, while this treatment only had a mild effect on H3K4me3 levels (Fig. 6).
Fig. 6.
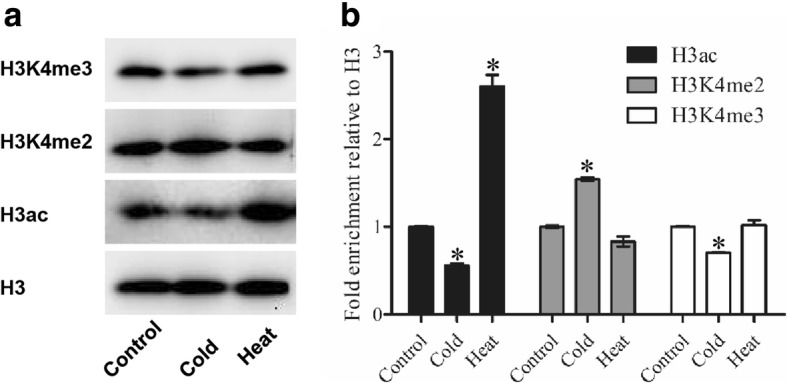
Levels of histone H3ac, H3K4me2, and H3K4me3 under cold and heat treatments. a Western blot showing the H3ac, H3K4me2, and H3K4me3 status in soybean leaves treated with cold and heat. 10-day-old seedlings were treated under cold (4 °C) and heat (42 °C) conditions for 12 h and the leaves were sampled for total protein extraction. b Quantification of western blot results. Signal intensities were measured using the ImageJ software and normalized to the loaded amount of H3. Values are expressed as fold change over control treatment. Shown is the mean ± SEM of three independent experiments. *, P-value < 0.05, Student’s t-test
Discussion
Studies have shown that HDACs play critical roles in multiple aspects of plant development and response to various environmental cues by regulating gene expression through histone deacetylation [4, 14, 15]. Genome-wide identification and characterization of HDACs have previously been reported in several plant species, including Arabidopsis, rice, grape (Vitis vinifera) and tomato (Solanum lycopersicum) etc. [10, 58–61]. However, the involvement of HDACs in the response to environmental cues has not been documented in soybean. In this study, 28 HDACs were identified and characterized in soybean, with respect to their tissue-specific expression profiles, subcellular localizations and abiotic stress responsive expression patterns. The 28 GmHDACs are divided into three families: RPD3/HDA1, SIR2 and HD2 and are unevenly distributed on 12 soybean chromosomes (Figs. 1 and 2). The number of HDAC gene in soybean is larger than those in Arabidopsis, rice and tomato and compared to other plant species, gene expansion of HDACs in soybean is evident [10, 58, 60, 61]. For example, Arabidopsis, rice and tomato only have two SIR2 genes, while soybean contains four (Fig. 1a). Tandem amplification and segmental duplication of chromosomal regions are likely the main contributors for gene extension during evolution [56]. In total, 27 duplication pairs of HDAC genes were identified but no tandem gene sets were found (Fig. 2, Additional file 1: Table S1), indicating that segmental duplication events during evolution are most likely the major drivers of HDAC gene expansion in soybean.
In humans, RPD3 type Class I HDACs are localized exclusively in the nucleus, whereas class II HDACs are shuttled between the cytoplasm and the nucleus [62]. In Arabidopsis, previous studies demonstrated that RPD3 type Class II HDACs, HDA5, HDA8, and HDA14 are localized in the cytoplasm, whereas HDA15 is localized exclusively in the nucleus. In addition, AtHDA15 was shown to shuttle from the cytoplasm to the nucleus is response to light [63]. Moreover, RPD3 type Class I HDA6 and HDA19 also localized in the nucleus [45, 64, 65]. In this study, we found that GmHDA6, GmHDA13, GmHDA14 and GmHDA16 were localized in both the nucleus and cytoplasm (Fig. 3), suggesting a possible shuttling process between these compartments. Similar to the behavior of the Arabidopsis HD2 proteins [39], the two members of soybean HD2 family were localized in the nucleus (Fig. 3). SIR2 proteins were reported to occupy discrete subcellular compartments in plants. In addition, OsSRT1 appears to be mainly localized in the nucleus [66], while SlSRT2 localized in both the nucleus and cytoplasm [60] and OsSRT2 and AtSRT2 are found in the mitochondria [67]. In this study, we demonstrated that GmSRT4 localized in the nucleus (Fig. 3). Together, the different subcellular localization patterns of GmHDACs suggest that they might be differentially regulated and may have distinct roles in soybean. However, it is worth noting that the subcellular localization assays described in this study were conducted with Arabidopsis cells so it is entirely possible that soybean-specific interactions could affect the localization pattern in situ.
Increasing evidence suggests that the tissue-specificity and stress-responsiveness of HDAC genes play critical roles in plant development and environmental responses [14, 15, 68]. In our study, quantitative-RT PCR was employed to investigate the tissue-specific and stress-responsive expression patterns of nine typical GmHDACs (Figs. 4 and 5). GmHDA13 was highly expressed in all tissues and developmental stages tested (Fig. 4), which is similar to its close homologous gene AtHDA6. Previous studies have shown that AtHDA6 plays essential roles in various aspects of plant growth and development, such as leaf development and flowering, jasmonate and ethylene signaling, and abiotic stress responses [17–20, 48, 49, 65, 69, 70]. GmHDA13 may thus have functions similar to those of AtHDA6 considering their close evolutionary relationship and their similar expression patterns. GmHDA8, the closest homolog of AtHDA14, is preferentially expressed in cotyledons and leaves (Fig. 4), suggesting a potential role in regulating cotyledon and leaf development. Notably, the expression level of GmSRT2 in leaves was much higher than in other organs, indicating that GmSRT2 might play an important role in regulating leaf development (Fig. 4). In rice, OsSRT1 was reported to regulate leaf senescence and cell death [66, 71], however, the functions of SIR2 proteins in plant growth and development remain largely unknown.
Microarray analysis demonstrated that several RPD3 type HDACs were repressed under high salt and drought treatments in rice [72]. Furthermore, the expression of SlHDACs was induced under salt, dehydration, and low/high temperature treatments, suggesting that SlHDACs might function in different stress responses in tomato [61]. In this study, we found these nine GmHDACs responded to various abiotic stress treatments, including low/high temperature, flooding, drought, NaCl and ABA (Fig. 5). Interestingly, the expression of most genes were repressed under the stress conditions examined, whilst GmSRT2 was significantly induced both by cold and heat treatments (Fig. 5), indicating potentially distinct HDAC gene functions in response to different environmental cues. In Arabidopsis, ABA and NaCl treatments are known to repress the gene expression of AtHD2A, AtHD2B, AtHD2C, and AtHD2D [49]. AtHD2C interacts with AtHDA6 and regulates ABA-responsive gene expression by histone deacetylation [49]. Recently, Han et al. (2016) demonstrated that AtHD2D could confer tolerance to abiotic stresses, including drought, salt, and cold stresses in Arabidopsis. In addition, the transcription of HD2 type HDACs is modulated under salt, ABA, and PEG stress treatment in rice [73]. In the present study, GmHDT2 and GmHDT4 were down-regulated following a NaCl treatment and induced by the application of ABA, which is in support of previous findings (Fig. 5e, f). Furthermore, it has been shown that the expression of HDACs and the levels of histone H3K9ac, H4ac and H4K5ac were reduced by low temperature in maize [74]. Similarly, we demonstrated that cold treatment can repress the level of histone H3 acetylation (H3ac) in soybean while a heat treatment could increased H3ac (Fig. 6), indicating that histone acetylation may play pivotal roles in the plant responses to both cold and heat stress. Interestingly, the status of H3K4me2 and H3K4me3 were also modulated by the cold and heat treatments, indicating histone methyltransferases (HMTs) or histone demethylases (HDMs) may also be involved in these processes (Fig. 6).
Conclusions
Our work identified 28 HDAC genes in soybean, which can be divided into RPD3/HDA1, SIR2, and HD2 families. Segmental duplication events during evolution were the major driver of HDAC gene expansion in soybean. Subcellular localization indicated that GmHDA6, GmHDA13, GmHDA14 and GmHDA16 were localized in both the nucleus and cytoplasm, while GmSRT4, GmHDT2 and GmHDT4 were found solely in the nucleus. Nine typical GmHDAC genes were differentially expressed in all tissues examined and all of them were stress-responsive. Interestingly, our results indicate that global histone acetylation and methylation levels were affected in response to cold and heat stress treatments in soybean, indicating that histone modifiers, such as HDACs might be involved in the response of this plant to abiotic stress. Further research is required to determine the function and molecular mechanisms of GmHDACs in plant responses to abiotic stress, which will provide tools for the improvement of soybean productivity.
Methods
Plant materials and treatments
Seedlings of soybean (Glycine max) were grown at 25 °C in a growth cabinet under a light intensity of 10,000 lx and a 16 h photoperiod. To investigate tissue or organ specific expression, roots, cotyledons, epicotyls, hypocotyls were harvested from 5-day-old seedlings, while leaves and stems were collected from 2-week-old seedlings. Flowers were collected when they were in full bloom. Seeds were collected 2 weeks after flowering. For the stress treatments, 10-day-old seedlings were exposed to different stress conditions and collected under the light period. Cold and heat stresses were performed by transferring soybean plants grown under control conditions into chambers set at 4 °C or 42 °C for the indicated period of time. The flooding treatment was performed by completely submerging seedlings under water at 25 °C. Dehydration was induced by removing plants from the pots and by placing them on filter paper at 25 °C. Soybean seedlings were exposed to salt stress by removing them from soil and by soaking them in a solution containing 250 mM NaCl. For ABA treatment, leaf tissues of the soybean plants were sprayed with 10 μM ABA solution. Each treatment was performed on five similar plants (same leaf number, leaf size and plant height) and seedlings without treatment were used as control. After exposure to these stresses for a period of 12 h, the plant tissues were harvested and immediately frozen in liquid nitrogen for further gene expression and protein immunoblotting experiments.
Identification of HDAC genes in soybean
To identify potential HDACs in soybean, we used the protein sequences of AtHDACs as queries to run the BLASTP program against the soybean database (https://soybase.org/). After removing the duplicates from all the captured sequences, we initially retrieved the recognizable domains using BLAST-based NCBI conserved domain searches (https://www.ncbi.nlm.nih.gov/Structure/cdd/ wrpsb.cgi). We then further analyzed these domains using the HMMER-based Simple Modular Architecture Research Tool database (http://smart.embl-heidelberg.de/) and the Pfam software program (http://pfam.xfam.org/search).
Phylogenetic tree construction
All the HDAC protein sequences from soybean and Arabidopsis were aligned using ClustalW, and the alignment was imported in MEGA5.0 for phylogenetic analysis. The phylogenetic trees were generated using the Maximum Parsimony method with partial deletion. The bootstrap value was assessed with 1000 replicates [75].
Chromosomal localization and gene duplication analyses
The gene location and chromosome number for each soybean HDAC gene was retrieved from the Phytozome 12 Glycine max Wm82.a2.v1 database (www.phytozome.net). The Circos software was used to determine the chromosomal localization and generate the duplication image of the GmHDAC genes [76]. GmHDAC genes in duplicated genomic regions and the Ka/Ks values for the 27 duplicated gene pairs were obtained from Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/) to evaluate the contribution of tandem and segmental genome duplication to the expansion of the HDAC gene family over evolutionary time. Thus, homologous genes on the same duplicated chromosomal blocks were set as segmental duplication, while two paralogs physically close together were defined as tandem duplications. Ka/Ks values superior to 1 indicate positive selection while values below 1 and equal to 1 respectively indicate purifying selection and neural selection [57].
RNA isolation and quantitative real-time RT-PCR
Total RNAs were isolated from the plant samples using the Hi-Pure Plant RNA Mini Kit (Magen, Guangzhou, China). The first-strand cDNA was generated using the TransScript One-Step gDNA Removal and cDNA synthesis SuperMix (TransGen, Beijing, China). The manufacturers’ instructions were followed in each case. Quantitative RT-PCR assays were performed on three biological replicates on a LightCycler 480 system (Roche, Basel, Switzerland) using TransStart Green qPCR SuperMix Kit (TransGen, Beijing, China). The reaction conditions were as follows: 95 °C for 1 min, followed by 50 cycles of 95 °C for 10 s, and 60 °C for 30 s. The soybean CYP2 gene was used as the internal control, and the relative expression levels of genes were calculated using the 2−ΔΔCT method [77]. Primer sequences are listed in Additional file 3: Table S2.
Subcellular localization assays
The subcellular localizations of GmHDACs were first predicted using the pSORT web server (http://www.genscript.com/psort/wolf_psort.html) and confirmed by YFP-tagged transient expression assays in Arabidopsis suspension cultured cells. The full-length coding sequence of GmHDACs was introduced into pSAT6-EYFP-N1 to generate pSAT6-GmHDACs-EYFP, containing a GmHDACs-EYFP fusion construct under the control of the CaMV 35S promoter. The fusion constructs and nuclear localization marker NLS-mCherry were co-transfected into protoplast cells for in vivo protein targeting. The protoplast isolation and transient expression were conducted as described previously [78]. After transfection, the protoplasts were incubated at 22 °C for 12 h in the dark, and the distribution of the fusion protein was determined using a confocal fluorescence microscope.
Protein immunoblotting
Soybean leaves sampled from control, cold (4 °C) and heat (42 °C) treated seedlings were ground to a powder in liquid nitrogen and mixed with 1 mL of ice-cold extraction buffer (0.1 M Tris-HCl, pH 8.0, 10% glycol, 3% SDS, 0.05% beta-mercaptoethanol). The samples were then boiled for 5 min, centrifuged at 13,000 rpm for 10 min and the supernatants collected as the total protein fractions. The protein were mixed with a loading dye and loaded on 12% polyacrylamide gels. After electrophoresis and transfer to a PVDF membrane, the samples were immunoblotted with the following commercial antibodies: anti-H3 (Millipore, 05–499), anti-H3ac (Millipore, 06–599), anti-H3K4me2 (Millipore, 07–030) and anti-H3K4me3 (Millipore, 04–745). The experiments were carried out three times and the Image J software was used to quantify the relative protein levels.
Statistical analyses
A Student’s t-test (two tail, unpaired, equal variance) was used to determine the statistical significance of the differential transcripts abundance patterns and protein accumulation levels between treatments and their corresponding controls. Differential expression data were regarded as statistically significant and were marked by * only when passing the t-test with a P-value of < 0.05.
Additional files
Table S1. Segmental duplication events of soybean genes during evolution. (DOC 67 kb)
Figure S1. Expression profiles of GmHDAC genes in developmental seeds. The transcript profiling data of soybean seeds was extracted from the publicly-available Soybase database (https://www.soybase.org/) for heatmap generation. The colors indicate expression intensity (red, high expression; black, low expression; grey, no expression). (JPG 538 kb)
Table S2. Primers used in this study. (DOC 50 kb)
Acknowledgements
We thank editors and reviewers for the careful reading and valuable comments. We apologize to researchers whose studies are not cited due to space limitations.
Funding
This work was financially supported by Youth Innovation Promotion Association, Chinese Academy of Sciences (2017399), Guangdong Natural Science Foundation (2018A030313350),the Strategic Priority Research Program of Chinese Academy of Sciences (XDA13020603), Innovation Training Programs for Undergraduates (CAS), the National Natural Science Foundation of China (31671467 and 31770048) and the China 1000-Talents Plan for young researchers (C83025).
Availability of data and materials
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ABA
Abscisic acid
- bp
base pair
- CaMV
Cauliflower mosaic virus
- cDNA
complementary deoxyribonucleicacid
- DAF
days after flowering
- gDNA
genomic deoxyribonucleicacid
- kDa
kilo Dalton
- NLS
nuclear localization signal
- RNA
ribonucleicacid
- RT-PCR
Real time-polymerase chain reaction
- YFP
Yellow fluorescent protein
Authors’ contributions
ML, CG and JL conceived and designed the experiments; CY, WS, HC, LC, YX, XZ, CL, CC, JZ, QL and ML performed the experiments; ML, CY and CG analysed the data; ML and CY wrote the manuscript; CY, LC, YX, XZ, JC and ML revised the manuscript; all authors read and approved the final manuscript.
Ethics approval and consent to participate
The plant materials were collected from South China Botanical Garden, CAS. The experimental research on plants, including collection of plant material, was complied with the institutional, national, or international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chao Yang, Email: 798916640@qq.com.
Wenjin Shen, Email: gzswj@163.com.
Hongfeng Chen, Email: h.f.chen@scbg.ac.cn.
Liutian Chu, Email: chuliushegan@outlook.com.
Yingchao Xu, Email: 602265475@qq.com.
Xiaochen Zhou, Email: l.icky@qq.com.
Chuanliang Liu, Email: lclbeyond@126.com.
Chunmiao Chen, Email: 756207537@qq.com.
Jiahui Zeng, Email: 1037476680@qq.com.
Jin Liu, Email: gjinliu@pku.edu.cn.
Qianfeng Li, Email: qfli@yzu.edu.cn.
Caiji Gao, Email: gaocaiji@163.com.
Jean-Benoit Charron, Email: jean-benoit.charron@mcgill.ca.
Ming Luo, Email: luoming@scbg.ac.cn.
References
- 1.Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218(1):1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- 2.Qin F, Shinozaki K, Yamaguchi-Shinozaki K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011;52(9):1569–1582. doi: 10.1093/pcp/pcr106. [DOI] [PubMed] [Google Scholar]
- 3.Deshmukh R, Sonah H, Patil G, Chen W, Prince S, Mutava R, Vuong T, Valliyodan B, Nguyen HT. Integrating omic approaches for abiotic stress tolerance in soybean. Front Plant Sci. 2014;5:244. doi: 10.3389/fpls.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo M, Liu X, Singh P, Cui Y, Zimmerli L, Wu K. Chromatin modifications and remodeling in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2012;1819(2):129–136. doi: 10.1016/j.bbagrm.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Zong W, Zhong X, You J, Xiong L. Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol Biol. 2013;81(1):175–188. doi: 10.1007/s11103-012-9990-2. [DOI] [PubMed] [Google Scholar]
- 6.Kim JM, Sasaki T, Ueda M, Sako K, Seki M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front Plant Sci. 2015;6:114. doi: 10.3389/fpls.2015.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehdi S, Derkacheva M, Ramstr M, Kralemann L, Bergquist J, Hennig L. The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. Plant Cell. 2016;28(1):42–54. doi: 10.1105/tpc.15.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lusser A, Brosch G, Loidl A, Haas H, Loidl P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science. 1997;277(5322):88–91. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]
- 9.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 10.Pandey R, Mler A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30(23):5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20(21):2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 12.Lee WK, Cho MH. Telomere-binding protein regulates the chromosome ends through the interaction with histone deacetylases in Arabidopsis thaliana. Nucleic Acids Res. 2016;44(10):4610–4624. doi: 10.1093/nar/gkw067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alinsug MV, Yu CW, Wu K. Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biol. 2009;9(1):37. doi: 10.1186/1471-2229-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma X, LV S, Zhang C, Yang C. Histone deacetylases and their functions in plants. Plant Cell Rep. 2013;32(4):465–478. doi: 10.1007/s00299-013-1393-6. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Yang S, Zhao M, Luo M, Yu CW, Chen CY, Tai R, Wu K. Transcriptional repression by histone deacetylases in plants. Mol Plant. 2014;7(5):764–772. doi: 10.1093/mp/ssu033. [DOI] [PubMed] [Google Scholar]
- 16.Aufsatz W. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. The EMBO Journal. 2002;21(24):6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Probst AV, Fagard M, Proux F, Mourrain P, Boutet S, Earley K, Lawrence RJ, Pikaard CS, Murfett J, Furner I, et al. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell. 2004;16(4):1021–1034. doi: 10.1105/tpc.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu CW, Liu X, Luo M, Chen C, Lin X, Tian G, Lu Q, Cui Y, Wu K. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 2011;156(1):173–184. doi: 10.1104/pp.111.174417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Z, An F, Feng Y, Li P, Xue L, A M JZ, Kim JM, To TK. Li W, et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci. 2011;108(30):12539–12544. doi: 10.1073/pnas.1103959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo M, Yu CW, Chen FF, Zhao L, Tian G, Liu X, Cui Y, Yang JY, Wu K. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet. 2012;8(12):e1003114. doi: 10.1371/journal.pgen.1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci. 2013;110(2):761–766. doi: 10.1073/pnas.1215010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian L, Chen ZJ. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci. 2001;98(1):200–205. doi: 10.1073/pnas.98.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu CR, Liu C, Wang YL, Li LC, Chen WQ, Xu ZH, Bai SN. Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proc Natl Acad Sci. 2005;102(40):14469–14474. doi: 10.1073/pnas.0503143102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312(5779):1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- 25.Krogan NT, Hogan K, Long JA. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development. 2012;139(22):4180–4190. doi: 10.1242/dev.085407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran HT, Nimick M, Uhrig RG, Templeton G, Morrice N, Gourlay R, DeLong A, Moorhead GBG. Arabidopsis thaliana histone deacetylase 14 (HDA14) is an α-tubulin deacetylase that associates with PP2A and enriches in the microtubule fraction with the putative histone acetyltransferase ELP3. Plant J. 2012;71(2):263–272. doi: 10.1111/j.1365-313X.2012.04984.x. [DOI] [PubMed] [Google Scholar]
- 27.Cigliano RA, Cremona G, Paparo R, Termolino P, Perrella G, Gutzat R, Consiglio MF, Conicella C. Histone deacetylase AtHDA7 is required for female gametophyte and embryo development in Arabidopsis. Plant Physiol. 2013;163(1):431–440. doi: 10.1104/pp.113.221713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim W, Latrasse D, Servet C, Zhou DX. Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem Biophys Res Commun. 2013;432(2):394–398. doi: 10.1016/j.bbrc.2012.11.102. [DOI] [PubMed] [Google Scholar]
- 29.Liu C, Li LC, Chen WQ, Chen X, Xu ZH, Bai SN. HDA18 affects cell fate in Arabidopsis root epidermis via histone acetylation at four kinase genes. Plant Cell. 2013;25(1):257–269. doi: 10.1105/tpc.112.107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Chen CY, Wang K-C, Luo M, Tai R, Yuan L, Zhao M, Yang S, Tian G, Cui Y, et al. PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell. 2013;25(4):1258–1273. doi: 10.1105/tpc.113.109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Tan B, Luo M, Li Y, Liu C, Chen C, Yu CW, Yang S, Dong S, Ruan J, et al. HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell. 2013;25(1):134–148. doi: 10.1105/tpc.112.096313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryu H, Cho H, Bae W, Hwang I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat Commun. 2014;5:4138. doi: 10.1038/ncomms5138. [DOI] [PubMed] [Google Scholar]
- 33.Chen CY, Wu K, Schmidt W. The histone deacetylase HDA19 controls root cell elongation and modulates a subset of phosphate starvation responses in Arabidopsis. Sci Rep. 2015;5:15708. doi: 10.1038/srep15708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo M, Tai R, Yu CW, Yang S, Chen CY, Lin WD, Schmidt W, Wu K. Regulation of flowering time by the histone deacetylase HDA5 in Arabidopsis. Plant J. 2015;82(6):925–936. doi: 10.1111/tpj.12868. [DOI] [PubMed] [Google Scholar]
- 35.Gu D, Chen CY, Zhao M, Zhao L, Duan X, Duan J, Wu K, Liu X. Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res. 2017;45(12):7137–7150. doi: 10.1093/nar/gkx283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang Y, Liu X, Liu X, Li Y, Wu K, Hou X. Arabidopsis NF-YCs mediate the light-controlled hypocotyl elongation via modulating histone acetylation. Mol Plant. 2017;10(2):260–273. doi: 10.1016/j.molp.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Gao MJ, Li X, Huang J, Gropp GM, Gjetvaj B, Lindsay DL, Wei S, Coutu C, Chen Z, Wan XC, et al. SCARECROW-LIKE15 interacts with HISTONE DEACETYLASE19 and is essential for repressing the seed maturation programme. Nat Commun. 2015;6:7243. doi: 10.1038/ncomms8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu K, Tian L, Malik K, Brown D, Miki B. Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 2000;22(1):19–27. doi: 10.1046/j.1365-313x.2000.00711.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhou C, Labbe H, Sridha S, Wang L, Tian L, Latoszek-Green M, Yang Z, Brown D, Miki B, Wu K. Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 2004;38(5):715–724. doi: 10.1111/j.1365-313X.2004.02083.x. [DOI] [PubMed] [Google Scholar]
- 40.Farhi J, Tian G, Fang H, Maxwell D, Xing T, Tian L. Histone deacetylase HD2D is involved in regulating plant development and flowering time in Arabidopsis. Plant Signal Behav. 2017;12(7):e1300742. doi: 10.1080/15592324.2017.1300742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Huchen, Torres-Garcia Jesus, Latrasse David, Benhamed Moussa, Schilderink Stefan, Zhou Wenkun, Kulikova Olga, Hirt Heribert, Bisseling Ton. Plant-Specific Histone Deacetylases HDT1/2 Regulate GIBBERELLIN 2-OXIDASE2 Expression to Control Arabidopsis Root Meristem Cell Number . The Plant Cell. 2017;29(9):2183–2196. doi: 10.1105/tpc.17.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ueno Y, Ishikawa T, Watanabe K, Terakura S, Iwakawa H, Okada K, Machida C, Machida Y. Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in LEAVES of Arabidopsis. Plant Cell. 2007;19(2):445–457. doi: 10.1105/tpc.106.042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C, Gao F, Wu J, Dai J, Wei C, Li Y. Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol. 2010;51(8):1291–1299. doi: 10.1093/pcp/pcq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Lu Y, Zhao Y, Zhou DX. OsSRT1 is involved in rice seed development through regulation of starch metabolism gene expression. Plant Sci. 2016;248:28–36. doi: 10.1016/j.plantsci.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Zhou C, Zhang L, Duan J, Miki B, Wu K. HISTONE DEACETYLASE19 is involved in Jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell. 2005;17(4):1196–1204. doi: 10.1105/tpc.104.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim KC, Lai Z, Fan B, Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008;20(9):2357–2371. doi: 10.1105/tpc.107.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen LT, Wu K. Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal Behav. 2010;5(10):1318–1320. doi: 10.4161/psb.5.10.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.To TK. Nakaminami K, Kim JM, Morosawa T, Ishida J, Tanaka M, Yokoyama S, Shinozaki K, Seki M. Arabidopsis HDA6 is required for freezing tolerance. Biochem Biophys Res Commun. 2011;406(3):414–419. doi: 10.1016/j.bbrc.2011.02.058. [DOI] [PubMed] [Google Scholar]
- 49.Luo M, Wang YY, Liu X, Yang S, Lu Q, Cui Y, Wu K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot. 2012;63(8):3297–3306. doi: 10.1093/jxb/ers059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung JH, Park JH, Lee S, To TK. Kim JM, Seki M, Park CM. The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell. 2013;25(11):4378–4390. doi: 10.1105/tpc.113.118364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buszewicz D, Archacki R, Palusiński A, Kotliński M, Fogtman A, Iwanicka-Nowicka R, Sosnowska K, Kuciński J, Pupel P, Olędzki J, et al. HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 2016;39(10):2108–2122. doi: 10.1111/pce.12756. [DOI] [PubMed] [Google Scholar]
- 52.Zheng Y, Ding Y, Sun X, Xie S, Wang D, Liu X, Su L, Wei W, Pan L, Zhou DX. Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J Exp Bot. 2016;67(6):1703–1713. doi: 10.1093/jxb/erv562. [DOI] [PubMed] [Google Scholar]
- 53.Ueda M, Matsui A, Tanaka M, Nakamura T, Abe T, Sako K, Sasaki T, Kim JM, Ito A, Nishino N, et al. The distinct roles of class I and II RPD3-like histone deacetylases in salinity stress response. Plant Physiol. 2017;175:1760–1773. doi: 10.1104/pp.17.01332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sridha S, Wu K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 2006;46(1):124–133. doi: 10.1111/j.1365-313X.2006.02678.x. [DOI] [PubMed] [Google Scholar]
- 55.Han Z, Yu H, Zhao Z, Hunter D, Luo X, Duan J, Tian L. AtHD2D gene plays a role in plant growth, development, and response to abiotic stresses in Arabidopsis thaliana. Front Plant Sci. 2016;7:310. doi: 10.3389/fpls.2016.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 58.Fu W, Wu K, Duan J. Sequence and expression analysis of histone deacetylases in rice. Biochem Biophys Res Commun. 2007;356(4):843–850. doi: 10.1016/j.bbrc.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 59.Aquea F, Timmermann T, Arce-Johnson P. Analysis of histone acetyltransferase and deacetylase families of Vitis vinifera. Plant Physiol Biochem. 2010;48(23):194–199. doi: 10.1016/j.plaphy.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 60.Zhao L, Lu J, Zhang J, Wu PY, Yang S, Wu K. Identification and characterization of histone deacetylases in tomato (Solanum lycopersicum) Front Plant Sci. 2014;5:760. doi: 10.3389/fpls.2014.00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo JE, Hu Z, Guo X, Zhang L, Yu X, Zhou S, Chen G. Molecular characterization of nine tissue-specific or stress-responsive genes of histone deacetylase in tomato (Solanum lycopersicum) J Plant Growth Regul. 2017;36(3):566–577. doi: 10.1007/s00344-016-9660-8. [DOI] [Google Scholar]
- 62.Gray SG, Ekstr TJ. The human histone deacetylase family. Exp Cell Res. 2001;262(2):75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 63.Alinsug MV, Chen FF, Luo M, Tai R, Jiang L, Wu K. Subcellular localization of class II HDAs in Arabidopsis thaliana: nucleocytoplasmic shuttling of HDA15 is driven by light. PLoS One. 2012;7(2):e30846. doi: 10.1371/journal.pone.0030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006;20(10):1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu K, Zhang L, Zhou C, Yu CW, Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot. 2008;59(2):225–234. doi: 10.1093/jxb/erm300. [DOI] [PubMed] [Google Scholar]
- 66.Huang L, Sun Q, Qin F, Li C, Zhao Y, Zhou DX. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in Rice. Plant Physiol. 2007;144(3):1508–1519. doi: 10.1104/pp.107.099473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.König AC, Hartl M, Pham PA, Laxa M, Boersema PJ, Orwat A, Kalitventseva I, Pl hinger M, Braun HP, Leister D, et al. The Arabidopsis class II Sirtuin is a lysine deacetylase and interacts with mitochondrial energy metabolism. Plant Physiol. 2014;164(3):1401–1414. doi: 10.1104/pp.113.232496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan L, Liu X, Luo M, Yang S, Wu K. Involvement of histone modifications in plant abiotic stress responses. J Integr Plant Biol. 2013;55(10):892–901. doi: 10.1111/jipb.12060. [DOI] [PubMed] [Google Scholar]
- 69.Tanaka M, Kikuchi A, Kamada H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008;146(1):149–161. doi: 10.1104/pp.107.111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen LT, Luo M, Wang YY, Wu K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot. 2010;61(12):3345–3353. doi: 10.1093/jxb/erq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang C, Zhang H, Wan J, Wu Y, Li K, Jin C, Chen W, Wang S, Wang W, Zhang H, et al. Control of leaf senescence by an MeOH-Jasmonates Cascade that is epigenetically regulated by OsSRT1 in Rice. Mol Plant. 2016;9(10):1366–1378. doi: 10.1016/j.molp.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 72.Hu Y, Qin F, Huang L, Sun Q, Li C, Zhao Y, Zhou DX. Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem Biophys Res Commun. 2009;388(2):266–271. doi: 10.1016/j.bbrc.2009.07.162. [DOI] [PubMed] [Google Scholar]
- 73.Zhao J, Zhang J, Zhang W, Wu K, Zheng F, Tian L, Liu X, Duan J. Expression and functional analysis of the plant-specific histone deacetylase HDT701 in rice. Front Plant Sci. 2014;5:764. doi: 10.3389/fpls.2014.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu Y, Zhang L, Zhao L, Li J, He S, Zhou K, Yang F, Huang M, Jiang L, Li L. Trichostatin a selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLoS One. 2011;6(7):e22132. doi: 10.1371/journal.pone.0022132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 78.Miao Y, Jiang L. Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat Protoc. 2007;2:2348. doi: 10.1038/nprot.2007.360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Segmental duplication events of soybean genes during evolution. (DOC 67 kb)
Figure S1. Expression profiles of GmHDAC genes in developmental seeds. The transcript profiling data of soybean seeds was extracted from the publicly-available Soybase database (https://www.soybase.org/) for heatmap generation. The colors indicate expression intensity (red, high expression; black, low expression; grey, no expression). (JPG 538 kb)
Table S2. Primers used in this study. (DOC 50 kb)
Data Availability Statement
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.