Abstract
Background
One important metabolic engineering strategy is to localize the enzymes close to their substrates for improved catalytic efficiency. However, localization configurations become more complex the greater the number of enzymes and substrates is involved. Indeed, optimizing synthetic pathways by localizing multiple enzymes remains a challenge. Terpenes are one of the most valuable and abundant natural product groups. Phytoene, lycopene and β-carotene serve as common intermediates for the synthesis of many carotenoids and derivative compounds, which are hydrophobic long-chain terpenoids, insoluble in water and usually accumulate in membrane compartments.
Results
While β-ionone synthesis by β-carotene cleavage dioxygenase PhCCD1 and astaxanthin synthesis by β-carotene ketolase (CrtW) and β-carotene hydroxylase (CrtZ) differ in complexity (single and multiple step pathways), the productivity of both pathways benefited from controlling enzyme localization to the E. coli cell membrane via a GlpF protein fusion. Especially, the astaxanthin synthesis pathway comprises both CrtW and CrtZ, which perform four interchangeable reactions initiated from β-carotene. Up to four localization strategies of CrtW and CrtZ were exhaustively discussed in this work, and the optimal positioning strategy was achieved. CrtW and CrtZ were linked using a flexible linker and localized to the membrane via a GlpF protein fusion. Enzymes in the optimal localization configuration allowed a 215.4% astaxanthin production increase.
Conclusions
This work exploits a localization situation involving membrane-bound substrates, intermediates and multiple enzymes for the first time, and provides a workable positioning strategy to solve problems in similar circumstances.
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1270-1) contains supplementary material, which is available to authorized users.
Keywords: Localization, Astaxanthin, β-Ionone, Carotenoids, Escherichia coli
Background
Terpenes, and especially the carotenoids, are one of the most valuable and abundant natural product groups [1, 2]. Metabolic engineering of microbial factories for the production of carotenoids has been a hotspot for decades, and numerous achievements have led to the advancement of this field [2, 3]. While many carotenoids and derivative compounds were produced heterologously, this remains challenging since the common intermediates phytoene, lycopene and β-carotene (Fig. 1) are hydrophobic long-chain terpenoids, insoluble in water, and usually accumulate in the membrane compartments [4–6]. Due to the specific localization of hydrophobic substrates, spatial optimization of their metabolism has attracted much attention in the metabolic engineering community, with some notable results. For example, an eight and 20-fold improvement in the production of valencene and amorphadiene was achieved by mitochondrial targeting of valencene and amorphadiene synthase in yeast, respectively [7]. Localizing the Ehrlich pathway into yeast mitochondria increased isobutanol production by 260%, whereas overexpression of the same pathway in the cytoplasm only improved yields by 10% [8]. Lv et al. proposed a dual metabolic engineering of cytoplasmic and mitochondrial acetyl-CoA utilization to boost isoprene synthesis in S. cerevisiae, which increased isoprene production 2.1- and 1.6-fold relative to the recombinant strains with solely mitochondrial or cytoplasmic engineering, respectively [9]. On the other hand, there were also excellent works concerning optimization of bacteria enzyme localization. For instance, using the protein scaffolds to recruit enzymes in a metabolic pathway, Dueber et al. acquired 77-fold improvement of product titer of mevalonate with low enzyme expression and reduced metabolic load [10]. Lee et al. exhibited the great potential of cytoscaffold for enzyme organization [11]. Furthermore, in addition to protein scaffolds, DNA scaffolds [12, 13] and lipid-containing scaffolds [14] are also applied to improve the pathway efficiency. In addition to scaffolds, enzymes were also located to bacterial microcompartments (MCPs), such as ethanolamine utilization (Eut) [15] and 1,2-propanediol utilization (Pdu) MCPs for similar purposes [16]. The key point of these strategies was to localize the enzymes as close as possible to their substrates. However, localization configurations become more complex when more enzymes and reaction steps are involved. Thus, optimizing synthetic pathways by positioning multiple enzymes remains a challenge.
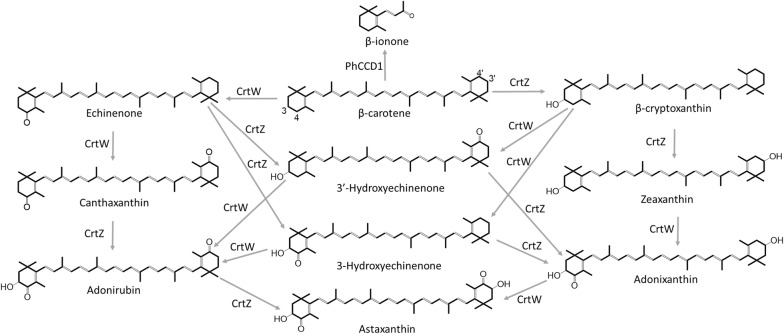
Fig. 1.
A schematic diagram of the astaxanthin and β-ionone synthesis pathways. The complex astaxanthin synthesis pathway comprises two enzymes, β-carotene ketolase (CrtW) and β-carotene hydroxylase (CrtZ), which perform four interchangeable reactions initiated from β-carotene, the substrate of the reaction
The apocarotenoid β-ionone, which is derived from β-carotene, is notably valuable in the flavoring industry due to its characteristic rose-like aroma [17]. The carotenoid cleavage dioxygenase (CCD) PhCCD1 from Petunia hybrida, encoded by the CCD1 gene (PhCCD1), was used for heterologous β-ionone production [18]. The CCD enzymes are usually cytoplasmic proteins [19, 20] and there was no predicted transmembrane region on PhCCD1 based on the information in ExPASy and UniProt [21]. Moreover, our previous work has shown that most β-carotene accumulated with cell membrane compartment in E. coli [22]. This means that it may be located far away from its substrate, the membrane-bound β-carotene, which would obviously decrease its catalytic efficiency. Thus, PhCCD1 can be localized to various compartments of the E. coli cell to investigate the relationship between the location of PhCCD1 and its catalytic efficiency (Figs. 2a, b, 3).
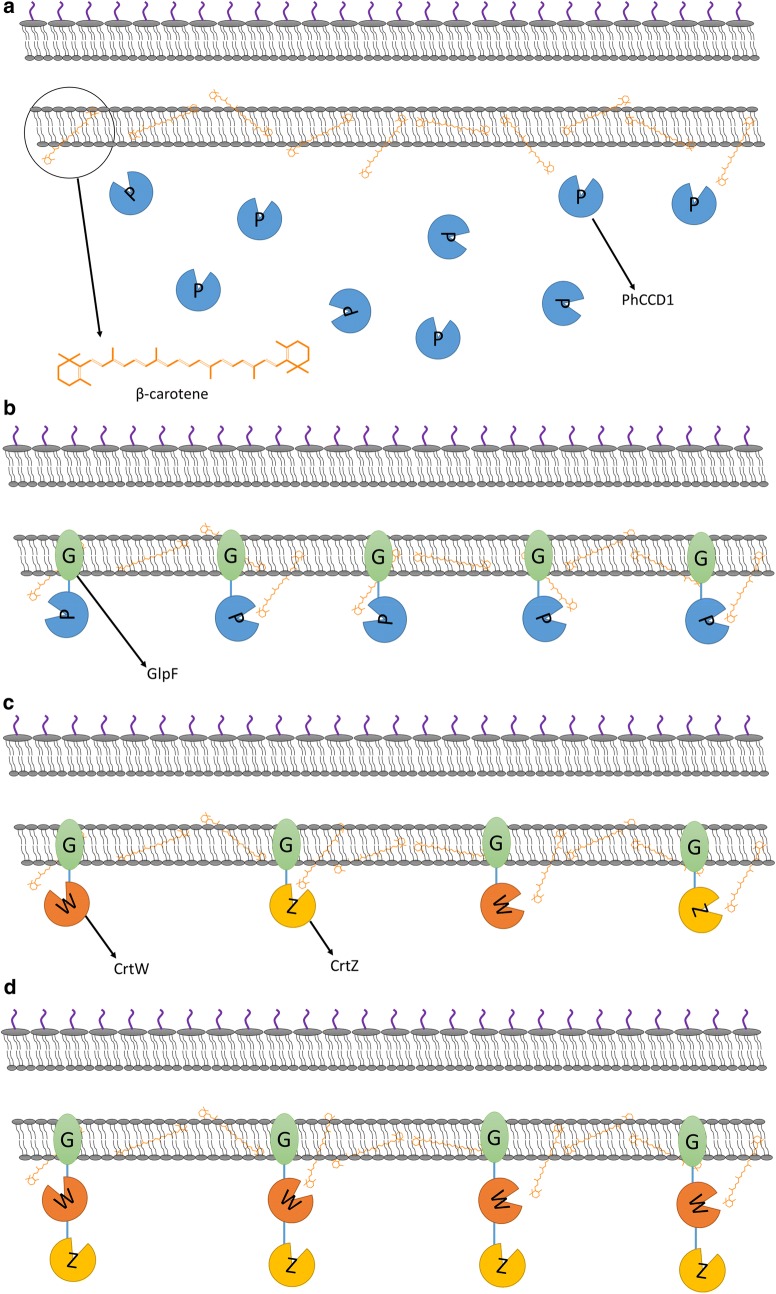
Fig. 2.
Enzyme locations in the host strain producing the β-carotene substrate. a Soluble cytoplasmic enzymes (PhCCD1) in the cells. b PhCCD1 localized to the membrane compartment by fusion with GlpF. c CrtW and CrtZ both localized to the membrane separately. d GlpF fused to the fusion protein CrtW-CrtZ to target it to the membrane
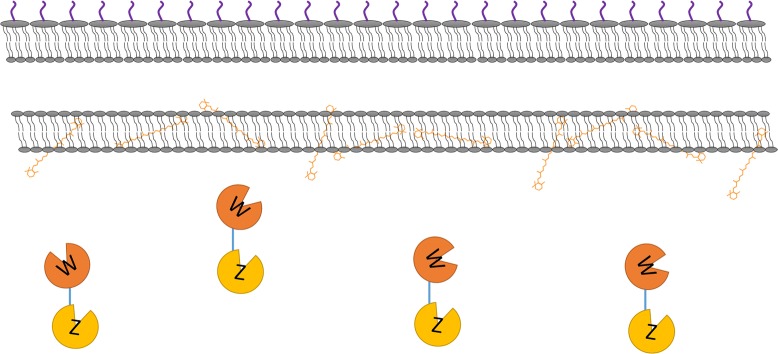
Fig. 3.
Enzyme locations in the host strain producing the β-carotene substrate. CrtW and CrtZ linked by a flexible eight-amino acid linker
Astaxanthin is one of the strongest antioxidants in nature [1], and has a tremendous potential for applications in healthcare and pharmaceuticals [2, 23]. The heterologous synthesis pathway of astaxanthin comprises two enzymes, β-carotene ketolase (CrtW) from Brevundimonas sp. SD212 [24, 25] and β-carotene hydroxylase (CrtZ) from Pantoea agglomerans [26], which perform four interchangeable reactions initiated from β-carotene. Although CrtW and CrtZ might contain transmembrane regions according to the predicted structures from ExPASy and UniProt [21], there was no information regarding their original location, let alone their location in the heterologous hosts. Thus, the optimal localization of the astaxanthin synthesis pathway enzymes is a more complex problem than that of the single-step β-ionone synthesis pathway.
In this study, we applied E. coli localizing tags to position PhCCD1 into different cell compartments to investigate how the localization of the enzyme could affect its catalytic efficiency. Moreover, the localization situation of CrtW and CrtZ was studied to determine an optimal configuration for maximizing the production of astaxanthin.
Methods
Strains, media and growth conditions
The strains used in this study are listed in Table 1. During strain construction, cultures were grown aerobically at 37 °C in Lysogeny broth (per liter: 10 g Difco tryptone, 5 g Difco yeast extract and 5 g NaCl, referred to as LB in the following), or in LB with 2% glycerin for fermentation. For seed cultures, single colonies were picked from the plates and used to inoculate 15 × 100 mm tubes containing 4 mL of LB with 34 mg/L chloramphenicol, and grown at 37 °C and 250 rpm overnight. The resulting seed culture was subsequently used to inoculate a 100-mL flask containing 10 mL of fermentation medium with 34 mg/L chloramphenicol to an initial OD600 of 0.05, and grown at 30 °C and 250 rpm. After 48 h of growth, the cells were collected for the measurement of the products. IPTG was added after 6 h of inoculation to 0.1 mM final concentration when CAR025 was cultured. For β-ionone production, 1 mL dodecane was added to the cultures to capture this volatile product.
Table 1.
Strains and plasmids in this study
| Strains/plasmids | Relevant characteristics | Source/notes |
|---|---|---|
| Strains | ||
| CAR010 | CAR005 [27], mRSL-4::ispG, mRSL-14::ispH | Unpublished |
| CAR003 | CAR010, ∆crtX | Unpublished |
| CAR025 | CAR005 [27], ispG-mRSL-4, ispH-mRSL-14, replacing the promoter of crtEYIB with trc promoter | [22] |
| Plasmids | ||
| pSC101 | Low copy plasmid | Unpublished |
| pACYC184-M | cat; replace tet with lacI and Ptrc of pTrc99A-M | [27] |
| pACYC184M2-Pm46 | cat; kan; expression vector; replace lacI and Ptrc of pAYCA184-M with FRT-Km-FRT::M1-46 | Unpublished |
| pSC102 | Low copy plasmid, ori and repA from pSC101, M1-46 promoter, cat from pACYC184-M2-Pm46 | Unpublished |
| pYL002 | crtW in pSC102 | Unpublished |
| pYL501 | crtW and crtZ in pSC102 | Unpublished |
| pGlpF-CrtW | glpF fused with crtW in pSC102 | This study |
| pCrtZ | crtZ in pSC102 | This study |
| pGlpF-CrtZ | glpF fused with crtZ in pSC102 | This study |
| pGlpF-CrtW/GlpF-CrtZ | glpF fused with crtW and glpF fused with crtZ in pSC102 | This study |
| pCrtW-CrtZ | crtW fused with crtZ in pSC102 | This study |
| pGlpF-CrtW-CrtZ | glpF fused with crtW and crtZ in pSC102 | This study |
| pPhCCD1 | PhCCD1 in pACYC184M2-Pm46 | This study |
| pGlpF-PhCCD1 | glpF fused with PhCCD1 in pACYC184M2-Pm46 | This study |
| pMBP-PhCCD1 | MBP fused with PhCCD1 in pACYC184M2-Pm46 | This study |
| pSPompA-PhCCD1 | The signal peptide sequence of ompA fused with PhCCD1 in pACYC184M2-Pm46 | This study |
Plasmid construction
The backbone fragments were amplified from plasmid pACYC184-M [27], pSC101, or their derivatives. The codon-optimized PhCCD1 gene of Petunia hybrida was synthesized by Genewiz (Suzhou, China). The gene fragments GG-PhCCD1 and GG-2PhCCD1 were amplified from the plasmid carrying PhCCD1 using the primer pairs phccd1-F/phccd1-R and 2phccd1-F/phccd1-R, respectively. Fragments containing crtW and crtZ were amplified from pYL501. PCRs were performed using PrimeSTAR® HS DNA Polymerase (Takara, Japan) and carried out in a thermocycler using the following program: 98 °C for 10 s, X °C for 5 s (step 2), 72 °C for Y min (step 3), repeat step 1–3 for 30 cycles, which X means the Tm of primers minus 5, Y equates the length (Kb) of PCR products. The PCR products were subjected to DpnI digestion (10 U, 16 h, 37 °C) and gel purification. The primers used in this study are summarized in Additional file 1: Table S1. Part of primers is designed by j5 DNA Assembly Design Software for scarless ligation [28]. All the primers were designed with linker sequences for Type II restriction enzymes for DNA assembly. The oligonucleotide primers were purchased from GENEWIZ (Suzhou, China).
The DNA fragments were assembled using the Golden Gate DNA assembly method [29, 30]. 100 ng of the vector fragment and equimolar amounts of other DNA parts were mixed in a 20 μL Golden Gate reaction with 1 μL BsaI-HF, 1 μL T4 ligase (New England Biolabs, USA) and 1× T4 ligase buffer. The reaction was carried out in a thermocycler using the following program: 37 °C for 5 min, 37 °C for 5 min (step 2), 16 °C for 10 min (step 3), repeat step 2 − 3 for 20 cycles, 16 °C for 20 min, 37 °C for 30 min, 75 °C for 6 min, and 4 °C hold. 1.5 μL of the resulting reaction solution was used to transform 80 μL of competent cells (Table 1) to obtain the assembled plasmids.
Measurement of β-ionone and astaxanthin concentrations
To obtain the produced β-ionone, culture samples were centrifuged for 5 min at 3186×g to separate the dodecane phase as the upper layer, which was filtered through a 0.22 μm organic nylon filter. Quantitation of β-ionone was performed on a 7890B gas chromatography system (Agilent Technologies, USA) coupled with a flame ionization detector, using a 19091J-413 HP-5 capillary column (30 m × 0.32 mm id, 0.25 μm film thickness; J&W Scientific, Agilent Technologies, USA). Injection of the samples was performed in splitless mode at 250 °C. The oven program comprised 80 °C for 1 min, ramp at 10 °C/min to 120 °C, and 3 °C/min to 240 °C. The concentrations of β-ionone were calculated using a calibration curve comprising 66.25–1060 mg/L of an authentic β-ionone standard purchased from Sigma (Sigma-Aldrich, St. Luis, MO, USA).
To obtain the produced astaxanthin (including precursors of zeaxanthin, canthaxanthin, and β-carotene), a 2-mL culture was centrifuged at 16,200×g for 3 min to obtain the cell pellet, which was washed with sterile water and centrifuged at 16,200×g for 3 min to obtain the cleaned pellet. Subsequently, 750 μL of the extraction solution (acetonitrile/methanol/dichloromethane, 21:21:8, v/v/v) was added to the pellet and ultrasonicated in the ice bath for 30 min, after which the resulting lysate was centrifuged at 16,200×g for 3 min. The resulting supernatant was transferred to a fresh centrifuge tube and another 750 μL of extraction solution was added to perform the extraction again. The supernatants of both extractions were combined, centrifuged at 16,200×g for 3 min, and the resulting combined supernatant filtered through a 0.22 μm organic nylon filter before astaxanthin content was measured by high-performance liquid chromatography (HPLC). The HPLC was performed on a Series 1200 system (Agilent Technologies, Agilent, USA) with a variable wavelength detector and a Symmetry C18 column (250 mm × 4.6 mm, 5 μm, Waters Ireland). The column was eluted at a rate of 0.8 mL/min with a linear gradient from 80% solvent D (acetonitrile/methanol/dichloromethane, 21:21:8, v/v/v) and 20% solvent B (methanol/water, 1:9, v/v) to 100% solvent D for 18 min, followed by a gradient from 100% solvent D to 80% solvent D and 20% solvent B for 7 min. The final solvent ratio was kept for 10 min to re-equilibrate the column. The content of the carotenoids including astaxanthin, zeaxanthin, canthaxanthin, and β-carotene was determined from the HPLC peak area at 476 nm. Concentrations of carotenoids were calculated using a calibration curve comprising an authentic reference standard purchased from Sigma (Sigma-Aldrich, St. Luis, MO, USA).
Results and discussion
Targeting PhCCD1 to different cell compartments for optimal catalytic efficiency
Since PhCCD1 is a cytoplasmic protein and the substrate β-carotene is membrane bound, its position in the heterologous host might not be optimal. In this study, E. coli localizing tags were applied to target PhCCD1 to different cell compartments to find the optimal location for its catalytic activity. We fused GlpF [31, 32] to PhCCD1 to position it in the inner membrane, to the E. coli maltose-binding protein (MBP, without the signal peptide) to position it in the cytoplasm and increase its solubility [33], and to the signal peptide of OmpA to position it in the periplasmic space [34]. All the proteins or polypeptide were fused to the N-terminal of PhCCD1. Plasmids carrying these fusion proteins were introduced into the β-carotene producing strain CAR003, and the resulting transformants were subjected to shake-flask fermentation. As can be seen in Fig. 4, the position of PhCCD1 had tremendous impact on the production of β-ionone. While the strains with PhCCD1 in the cytoplasm and periplasmic space had similar β-ionone yields as the control with no positioning tags, the strain with the membrane-localized PhCCD1 had tremendously improved production, reaching 386.0% of the control. Since PhCCD1 catalyzes the production of β-ionone from β-carotene in a simple one-step reaction, the results suggest that β-carotene is mainly associated with the membrane compartment, which means that localizing other β-carotene-converting enzymes to the membrane is a feasible strategy to improve their catalytic efficiency. As a corollary, the results also suggest that β-carotene content in either the cytoplasm or the periplasmic space is low, and the efficiency of β-carotene-converting enzymes is, therefore, diminished in these cell compartments. This might be true for most reactions with relatively large hydrophobic substrates.
Fig. 4.
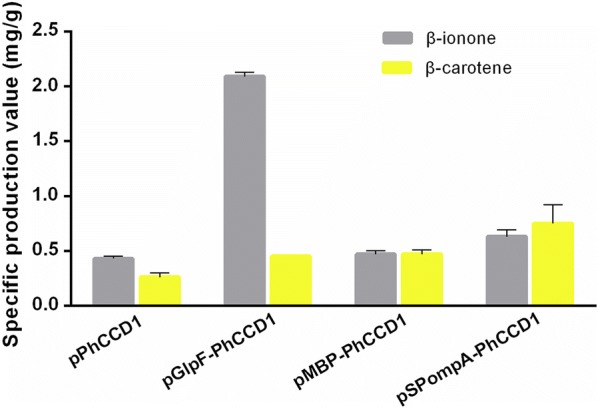
The specific β-ionone and β-carotene production with differently localized PhCCD1
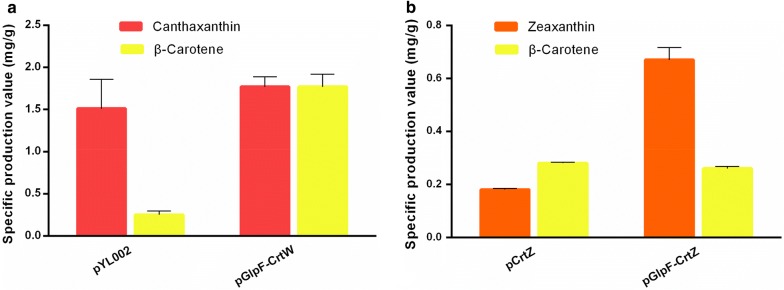
Targeting CrtW and CrtZ to the membrane for improved canthaxanthin and zeaxanthin production
As illustrated in Fig. 1, canthaxanthin and zeaxanthin are not only intermediates of astaxanthin, but also high-value natural products themselves [35–38]. Both are derivatives of β-carotene and each is produced by a single enzyme via a two-step reaction. With such reactions, we have proved that targeting the enzymes to the membrane was able to improve their catalytic efficiency. Thus, GlpF was fused to the N-terminal of CrtW and CrtZ, and expressed in the β-carotene-producing E. coli CAR003 to produce canthaxanthin and zeaxanthin, respectively (Fig. 5). While the yield of canthaxanthin was only 17.2% higher than that of the control strain with no GlpF fusion (Fig. 5a) which is not a significant increase due to the larger standard deviation of control, the yield of zeaxanthin was significantly increased, reaching 272.2% of the control (Fig. 5b). Nevertheless, localizing CrtW and CrtZ generally increased their catalytic efficiency, similar to PhCCD1. This observation further indicated that the membrane-localization strategy may be universally applicable to β-carotene-converting enzymes, and may be even to other enzymes with large hydrophobic substrates.
Fig. 5.
The specific canthaxanthin and zeaxanthin production by membrane-localized CrtW (a), and CrtZ (b)
Optimizing the localization of astaxanthin-synthesis enzymes for improved production
The heterologous synthesis pathway of astaxanthin comprises two enzymes, CrtW and CrtZ, which perform four interchangeable reactions initiated from β-carotene (Fig. 1). Although we have demonstrated that localizing β-carotene-converting enzymes to the membrane could improve their catalytic efficiency, optimal localization of the astaxanthin synthesis pathway enzymes remained a more complex problem. As illustrated in Fig. 2c, d, there are at least three localization configurations for the two enzymes CrtW and CrtZ and their membrane-bound substrate. They can be both localized to the membrane separately (Fig. 2c), linked and localized to the membrane (Fig. 2d) or just linked without the membrane-targeting tag (Fig. 3). The localization situation of CrtW and CrtZ, therefore, needed to be discussed in detail to find an optimal configuration for maximizing the production of astaxanthin.
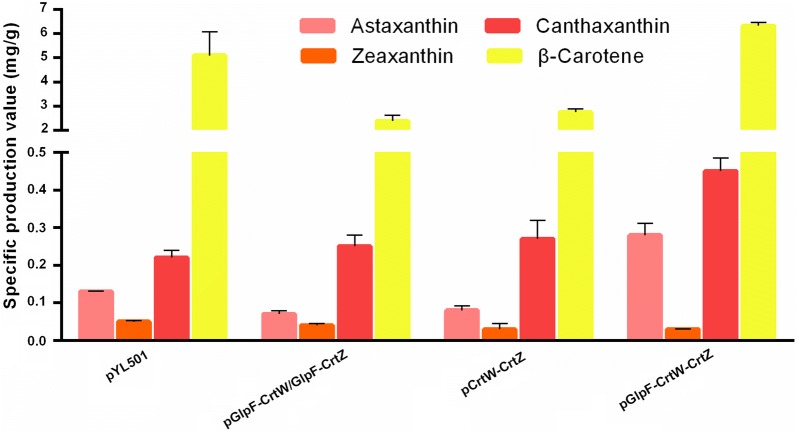
To localize both CrtW and CrtZ to the membrane (Fig. 2c), GlpF was fused to them individually. To link CrtW and CrtZ together, linker-2, a flexible eight-amino acid linker (Additional file 1: Table S1), was used to fuse them into one protein CrtW–CrtZ (Fig. 3). Furthermore, GlpF was fused to the fusion protein CrtW-CrtZ to position it to the membrane (Fig. 2d). CrtW and CrtZ constructs in these three localization configurations were expressed in the β-carotene-producing strain CAR025 to produce astaxanthin. As illustrated in Fig. 6, individually located CrtW and CrtZ (pGlpF–CrtW/GlpF–CrtZ) and their fusion protein CrtW–CrtZ (pCrtW–CrtZ) actually showed reduced astaxanthin specific production compared to the control strain CAR025 (pYL501), which having CrtW and CrtZ expressed in one operon with their own RBSs. By contrast, the membrane-targeted CrtW–CrtZ (pGlpF–CrtW–CrtZ) had increased catalytic efficiency and achieved a yield 215.4% of that of the control strain. While the zeaxanthin yield of all groups was low, the canthaxanthin accumulation was higher than zeaxanthin, which may be due to the lower enzyme activity of CrtW than CrtZ. In addition, β-carotene accumulation was much higher than the both, which was probably also caused by difference in enzyme activities.
Fig. 6.
The specific production of astaxanthin, canthaxanthin, zeaxanthin and β-carotene with differently localized CrtW and CrtZ
These interesting results demonstrated that different localization strategies actually had tremendous impact on the pathway efficiency. While two of the three configurations we tested decreased the catalytic efficiency, it was serendipitous to obtain one with significantly improved efficiency. In the configuration with separately membrane-bound CrtW and CrtZ, their efficiency might be increased individually. However, since the products of CrtW and CrtZ each serve as the substrates for the other enzyme (Fig. 1), the separately localized CrtW and CrtZ on the membrane might still be distant from their respective substrates for astaxanthin production (Fig. 2c). Furthermore, CrtW and CrtZ were not able to move as freely to bind their substrates as in the control strain, where they are in the cytoplasm and hence may have had decreased efficiency due at least in part to lower mobility. While the fusion protein CrtW–CrtZ should have the advantage of close proximity to each other (Fig. 3), the scarce β-carotene content in the cytoplasm could have diminished such effects. In the optimal configuration with CrtW and CrtZ linked and localized to the membrane together (Fig. 2d), theoretically, the substrates for all reaction steps were brought to the vicinity of the corresponding enzymes (Fig. 1), which resulted in the observed drastically improved catalytic efficiency.
Conclusion
There were few reports regarding the optimization of synthetic pathways by localization of multiple enzymes, especially when the substrates were membrane bound. In this work, E. coli localizing tags were applied to position PhCCD1 into different cell compartments, and the optimal location for its catalytic efficiency was found to be in the membrane, as expected. The results suggested that β-carotene was mainly associated with the membrane compartment, and, hence, targeting β-carotene-converting enzymes to the membrane is a feasible strategy to improve their catalytic efficiency. As a corollary, the results suggest that the β-carotene content in either the cytoplasmic or periplasmic compartments is too low for efficient enzymatic conversion, which might also be true for other large hydrophobic substrates.
How the localization situation of CrtW and CrtZ affects astaxanthin production was exhaustively investigated in this work, and an optimal positioning strategy was found, whereby CrtW and CrtZ were linked with a flexible linker and localized to the membrane via a GlpF protein fusion. Enzymes in the optimal localization configuration allowed a 215.4% increase of astaxanthin production. This work exploits a localization situation involving membrane-bound substrates, intermediates and multiple enzymes for the first time and provides a workable positioning strategy to solve problems in similar circumstances.
Additional files
Additional file 1: Table S1. Oligonucleotides used in this study.
Additional file 2. Plasmid maps and DNA sequences.
Authors’ contributions
YL planned and performed the experiments, analyzed and interpreted the data. BC and ZX supervised the study, designed the experiments and analyzed and interpreted the results. YL wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
We provide supporting and necessary data for publication of the article. All supporting data is present in the article and the additional documents. Specifically, plasmid maps and DNA sequences in GenBank format are provided in Additional file 2. The strains and plasmids associated with this work will be physically available upon request from the authors.
Consent for publication
I hereby give the journal of Biotechnology for Biofuels the right and permission to publish this article.
Ethics approval and consent to participate
Not applicable.
Funding
This research was financially supported by the Key Research Program of the Chinese Academy of Science (KFZD-SW-215, ZDRW-ZS-2016-3), National Natural Science Foundation of China (31522002, 31770105), and Tianjin Science and Technology Program (15PTCYSY00020).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- RBS
ribosome-binding site
- DCW
dry cell weight
- MEP
2C-methyl-d-erythritol-4-phosphate
Contributor Information
Changhao Bi, Email: bi_ch@tib.cas.cn.
Xueli Zhang, Email: zhang_xl@tib.cas.cn.
References
- 1.Ajikumar PK, Tyo K, Carlsen S, Mucha O, Phon TH, Stephanopoulos G. Terpenoids: opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol Pharm. 2008;5:167. doi: 10.1021/mp700151b. [DOI] [PubMed] [Google Scholar]
- 2.Das A, Yoon SH, Lee SH, Kim JY, Oh DK, Kim SW. An update on microbial carotenoid production: application of recent metabolic engineering tools. Appl Microbiol Biotechnol. 2007;77:505–512. doi: 10.1007/s00253-007-1206-3. [DOI] [PubMed] [Google Scholar]
- 3.Schmidtdannert C. Engineering novel carotenoids in microorganisms. Curr Opin Biotechnol. 2000;11:255. doi: 10.1016/S0958-1669(00)00093-8. [DOI] [PubMed] [Google Scholar]
- 4.Doshi R, Nguyen T, Chang G. Transporter-mediated biofuel secretion. Proc Natl Acad Sci USA. 2013;110:7642–7647. doi: 10.1073/pnas.1301358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meckel RA, Kester AS. Extractability of carotenoid pigments from nonphotosynthetic bacteria with solvents and detergents: implications for the location and binding of the pigments. Microbiology. 1980;120:111–116. doi: 10.1099/00221287-120-1-111. [DOI] [Google Scholar]
- 6.Mathews MM, Sistrom WR. Intracellular location of carotenoid pigments and some respiratory enzymes in Sarcina lutea. J Bacteriol. 1959;78:778–787. doi: 10.1128/jb.78.6.778-787.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farhi M, Marhevka E, Masci T, Marcos E, Eyal Y, Ovadis M, Abeliovich H, Vainstein A. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab Eng. 2011;13:474–481. doi: 10.1016/j.ymben.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Avalos JL, Fink GR, Stephanopoulos G. Compartmentalization of metabolic pathways in yeast mitochondria improves the production of branched-chain alcohols. Nat Biotechnol. 2013;31:335–341. doi: 10.1038/nbt.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lv X, Wang F, Zhou P, Ye L, Xie W, Xu H, Yu H. Dual regulation of cytoplasmic and mitochondrial acetyl-CoA utilization for improved isoprene production in Saccharomyces cerevisiae. Nat Commun. 2016;7:12851. doi: 10.1038/ncomms12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD. Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 11.Lee MJ, Mantell J, Hodgson L, Alibhai D, Fletcher JM, Brown IR, Frank S, Xue WF, Verkade P, Woolfson DN, Warren MJ. Engineered synthetic scaffolds for organizing proteins within the bacterial cytoplasm. Nat Chem Biol. 2018;14:142–147. doi: 10.1038/nchembio.2535. [DOI] [PubMed] [Google Scholar]
- 12.Conrado RJ, Wu GC, Boock JT, Xu H, Chen SY, Lebar T, Turnsek J, Tomsic N, Avbelj M, Gaber R, et al. DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency. Nucleic Acids Res. 2012;40:1879–1889. doi: 10.1093/nar/gkr888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu LY, Qiu XY, Zhu LY, Wu XM, Zhang Y, Zhu QH, Fan DY, Zhu CS, Zhang DY. Spatial organization of heterologous metabolic system in vivo based on TALE. Sci Rep. 2016;6:26065. doi: 10.1038/srep26065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myhrvold C, Polka JK, Silver PA. Synthetic lipid-containing scaffolds enhance production by colocalizing enzymes. ACS Synth Biol. 2016;5:1396–1403. doi: 10.1021/acssynbio.6b00141. [DOI] [PubMed] [Google Scholar]
- 15.Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. Engineered protein nano-compartments for targeted enzyme localization. PLoS ONE. 2012;7:e33342. doi: 10.1371/journal.pone.0033342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jakobson CM, Slininger Lee MF, Tullman-Ercek D. De novo design of signal sequences to localize cargo to the 1,2-propanediol utilization microcompartment. Protein Sci. 2017;26:1086–1092. doi: 10.1002/pro.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Bustamante E, Sanchez S. Microbial production of C13-norisoprenoids and other aroma compounds via carotenoid cleavage. Crit Rev Microbiol. 2007;33:211–230. doi: 10.1080/10408410701473306. [DOI] [PubMed] [Google Scholar]
- 18.Lopez J, Essus K, Kim IK, Pereira R, Herzog J, Siewers V, Nielsen J, Agosin E. Production of beta-ionone by combined expression of carotenogenic and plant CCD1 genes in Saccharomyces cerevisiae. Microb Cell Fact. 2015;14:84. doi: 10.1186/s12934-015-0273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubio A, Rambla JL, Santaella M, Gomez MD, Orzaez D, Granell A, Gomez-Gomez L. Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativa are both involved in beta-ionone release. J Biol Chem. 2008;283:24816–24825. doi: 10.1074/jbc.M804000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frusciante S, Diretto G, Bruno M, Ferrante P, Pietrella M, Prado-Cabrero A, Rubio-Moraga A, Beyer P, Gomez-Gomez L, Al-Babili S. Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc Natl Acad Sci USA. 2014;111:12246–12251. doi: 10.1073/pnas.1404629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu T, Ye L, Zhao D, Li S, Li Q, Zhang B, Bi C, Zhang X. Membrane engineering—a novel strategy to enhance the production and accumulation of beta-carotene in Escherichia coli. Metab Eng. 2017;43:85–91. doi: 10.1016/j.ymben.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 23.Lee PC, Schmidt-Dannert C. Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Appl Microbiol Biotechnol. 2002;60:1–11. doi: 10.1007/s00253-002-1101-x. [DOI] [PubMed] [Google Scholar]
- 24.Nishida Y, Adachi K, Kasai H, Shizuri Y, Shindo K, Sawabe A, Komemushi S, Miki W, Misawa N. Elucidation of a carotenoid biosynthesis gene cluster encoding a novel enzyme, 2,2′-beta-hydroxylase, from Brevundimonas sp. strain SD212 and combinatorial biosynthesis of new or rare xanthophylls. Appl Environ Microbiol. 2005;71:4286–4296. doi: 10.1128/AEM.71.8.4286-4296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi SK, Nishida Y, Matsuda S, Adachi K, Kasai H, Peng X, Komemushi S, Miki W, Misawa N. Characterization of beta-carotene ketolases, CrtW, from marine bacteria by complementation analysis in Escherichia coli. Mar Biotechnol (NY) 2005;7:515–522. doi: 10.1007/s10126-004-5100-z. [DOI] [PubMed] [Google Scholar]
- 26.Scaife MA, Burja AM, Wright PC. Characterization of cyanobacterial beta-carotene ketolase and hydroxylase genes in Escherichia coli, and their application for astaxanthin biosynthesis. Biotechnol Bioeng. 2009;103:944–955. doi: 10.1002/bit.22330. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, Li Q, Sun T, Zhu X, Xu H, Tang J, Zhang X, Ma Y. Engineering central metabolic modules of Escherichia coli for improving β-carotene production. Metab Eng. 2013;17:42–50. doi: 10.1016/j.ymben.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Hillson NJ, Rosengarten RD, Keasling JD. j5 DNA assembly design automation software. ACS Synth Biol. 2012;1:14–21. doi: 10.1021/sb2000116. [DOI] [PubMed] [Google Scholar]
- 29.Engler C, Marillonnet S. Generation of families of construct variants using golden gate shuffling. Methods Mol Biol. 2011;729:167–181. doi: 10.1007/978-1-61779-065-2_11. [DOI] [PubMed] [Google Scholar]
- 30.Liang J, Chao R, Abil Z, Bao Z, Zhao H. FairyTALE: a high-throughput TAL effector synthesis platform. ACS Synth Biol. 2014;3:67–73. doi: 10.1021/sb400109p. [DOI] [PubMed] [Google Scholar]
- 31.Fu D, Libson A, Miercke LJW, Weitzman C, Nollert P, Krucinski J, Stroud RM. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 32.Neophytou I, Harvey R, Lawrence J, Marsh P, Panaretou B, Barlow D. Eukaryotic integral membrane protein expression utilizing the Escherichia coli glycerol-conducting channel protein (GlpF) Appl Microbiol Biotechnol. 2007;77:375–381. doi: 10.1007/s00253-007-1174-7. [DOI] [PubMed] [Google Scholar]
- 33.Raran-Kurussi S, Keefe K, Waugh DS. Positional effects of fusion partners on the yield and solubility of MBP fusion proteins. Protein Expr Purif. 2015;110:159–164. doi: 10.1016/j.pep.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamabhai M, Emrat S, Sukasem S, Pesatcha P, Jaruseranee N, Buranabanyat B. Secretion of recombinant Bacillus hydrolytic enzymes using Escherichia coli expression systems. J Biotechnol. 2008;133:50–57. doi: 10.1016/j.jbiotec.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Hulisz DT, Boles GL. Clinical review of canthaxanthin (6Tanning Pills’): before recommending this product for tanning long-term safety must be established. Am Pharm. 1993;33:44–46. doi: 10.1016/S0160-3450(15)30697-8. [DOI] [PubMed] [Google Scholar]
- 36.Ma L, Yan SF, Huang YM, Lu XR, Qian F, Pang HL, Xu XR, Zou ZY, Dong PC, Xiao X. Effect of lutein and zeaxanthin on macular pigment and visual function in patients with early age-related macular degeneration. Ophthalmology. 2012;119:2290–2297. doi: 10.1016/j.ophtha.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 37.Palozza P, Maggiano N, Calviello G, Lanza P, Piccioni E, Ranelletti FO, Bartoli GM. Canthaxanthin induces apoptosis in human cancer cell lines. Carcinogenesis. 1998;19:373. doi: 10.1093/carcin/19.2.373. [DOI] [PubMed] [Google Scholar]
- 38.Zhao L, Sweet BV. Lutein and zeaxanthin for macular degeneration. Am J Health-Syst Pharm. 2008;65:1232. doi: 10.2146/ahjp080052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Oligonucleotides used in this study.
Additional file 2. Plasmid maps and DNA sequences.
Data Availability Statement
We provide supporting and necessary data for publication of the article. All supporting data is present in the article and the additional documents. Specifically, plasmid maps and DNA sequences in GenBank format are provided in Additional file 2. The strains and plasmids associated with this work will be physically available upon request from the authors.