Abstract
Background:
New sources of insulin-secreting cells are strongly in demand for treatment of diabetes. Induced pluripotent stem cells (iPSCs) have the potential to generate insulin-producing cells (iβ). However, the gene expression profile and secretory function of iβ still need to be validated in comparison with native β cells.
Methods:
Two clones of human iPSCs, reprogrammed from adult fibroblasts through integration-free Sendai virus, were differentiated into iβ and compared with donor pancreatic islets and EndoC-βH1, an immortalized human β cell line.
Results:
Both clones of iPSCs differentiated into insulin+ cells with high efficiency (up to 20%). iβ were negative for pluripotency markers (Oct4, Sox2, Ssea4) and positive for Pdx1, Nkx6.1, Chromogranin A, PC1/3, insulin, glucagon and somatostatin. iβ basally secreted C-peptide, glucagon and ghrelin and released insulin in response either to increasing concentration of glucose or a depolarizing stimulus. The comparison revealed that iβ are remarkably similar to donor derived islets in terms of gene and protein expression profile and similar level of heterogeneity. The ability of iβ to respond to glucose instead was more related to that of EndoC-βH1.
Discussion:
We demonstrated that insulin-producing cells generated from iPSCs recapitulate fundamental gene expression profiles and secretory function of native human β cells.
Keywords: induced pluripotent stem cells, diabetes, β cells
Introduction
Type 1 diabetes (T1D) is an autoimmune disorder in which the immune system attacks and destroys insulin producing islet cells in the pancreas. Therefore, T1D is characterized by deficient insulin production and persistently high blood glucose levels that are not restored to a fully physiological extent by treatment with exogenous insulin. As a consequence, prolonged glucose dysmetabolism can lead to a number of secondary complications including eye, kidney, nerve, and heart disease1,2. β cell replacement is potentially able to restore normoglycemia and ward off complications, as demonstrated by pancreas/islet transplantation, but scarcity of donors and the need for lifelong immunosuppression limit its broad application to T1D patients3,4. Pluripotent stem cells have shown the ability to differentiate in vitro into insulin producing cells, following the stages of fetal pancreatic organogenesis5–8, and could then represent an infinite source of new β cells for transplantation. Currently, pancreatic progenitors obtained from the differentiation of embryonic stem cell lines are already being transplanted into patients with T1D in a phase 1/2 clinical trial in the USA and Canada (NCT02239354 and NCT03163511).
Induced pluripotent stem cells (iPSCs) show the same plasticity of ESC, but can be derived from patient’s somatic cells, without ethical issues9–12. iPSCs are able to differentiate into insulin producing cells, although previous reports adopted different protocols and showed variable efficiency6,7,12–14. In addition, previous studies did not perform an accurate quality assessment of β cell derivatives in comparison with human β cell, an issue of particular relevance in light of the current push towards clinical application.
We recently published that human iPSCs, reprogrammed from fetal fibroblasts with retroviral vectors, can generate insulin-producing cells, engraft and secrete insulin in vivo15. In the present study, we show that two clones of iPSCs, reprogrammed from adult fibroblasts through integration-free Sendai virus delivery of Yamanaka’s factors, are able to generate β cells with a phenotype and function that recapitulate fundamental features of organ donor derived pancreatic islets and EndoC-βH1, an immortalized human β cell line.
Materials and Methods
iPSC Reprogramming, Characterization, Validation of Pluripotency and Multilineage Differentiation Potential
Two human iPSC clones (#5 and #9) were generated by reprogramming healthy subject fibroblasts with the Sendai virus technology (CytoTune-iPS Sendai Reprogramming Kit, ThermoFisher, Waltham, MA, USA)16. Clone #9 is characterized and banked at ISENET, Milan, Italy (www.isenet.it), clone #5 characterization is described below. Written informed consent was obtained from the donor for anonymized information to be published in this article.
HiPSC clones were individually picked and expanded on a feeder layer in mTeSR1 medium (STEMCELL Technologies, Vancouver, Canada). Cells were maintained in mTeSR1 on hESC-qualified Matrigel (BD Biosciences, Franklin Lakes, NJ, USA), dissociated with 0.5 mM EDTA (Ambion, Waltham, MA, USA) for passages and routinely tested for mycoplasma.
iPSCs cultured on Matrigel ES (BD) were treated with 0.2 μg/ml colchicine for 16 hours and sent to Integrated System Engineering (ISENET, Milan, Italy) for karyotype analysis using a Q-banding on 400 bands and by array cytogenetic hybridization (aCGH) of 600,00 probes with a median probe spacing of 41 kb (Agilent Technologies, Santa Clara, CA, USA).
Cells were tested for pluripotency markers by immunofluorescence (OCT4, NANOG, SSEA3, SSEA4 and SOX2) and by flow cytometry (SSEA4-FITC, TRA1-60-APC, TRA1-81-APC, Millipore, Burlington, MA, USA). For immunofluorescence, iPSC colonies were fixed in 4% paraformaldehyde, blocked with 5% goat serum in PBS and permeabilized with 0.3% TritonX-100 for nuclear staining in presence of primary antibody (Table 1) at 4°C overnight. Samples were exposed to secondary antibodies (1:1000, Thermo Scientific, Table 2) 1 hour and half in the dark. Nuclei were counterstained with DAPI diluted in PBS (1:10000, Sigma-Aldrich, Saint Louis, MO, USA) and mounted on coverslips with Faramount aqueous mounting solution (DAKO, Agilent). Images were taken with a confocal microscope (Leica SP5, Wetzlar, Germany).
Table 1.
List of Antibodies used in immunofluorescence (IF), Flow cytometry (FC) and immunohistochemistry (IHC).
| Marker | Host | Brand | Clone | Diluition | Application |
|---|---|---|---|---|---|
| SSEA3, surface | Rat | Millipore | MC-631 | 1:150 | IF |
| SSEA4, surface | Mouse | Millipore | MC-813-70 | 1:150 | IF, FC |
| TRA1-60, surface | Mouse | Millipore | TRA1-60 | 1:150 | FC |
| TRA1-81, surface | Mouse | Millipore | TRA1-81 | 1:150 | FC |
| OCT3/4, nuclear | Mouse | Santa Cruz | C-10 | 1:100 | IF |
| NANOG, nuclear | Rabbit | Abcam | Polyclonal | 1:100 | IF |
| PAX6, nuclear | Rabbit | BIOLEGEND | Polyclonal | 1:200 | IF |
| αSMA-1, cytoplasm | Mouse | SIGMA | 1A4 | 1:200 | IF |
| aFP, cytoplasm | Mouse | R&D | 189502 | 1:50 | IF |
| SOX17, nuclear | Goat | R&D | Polyclonal | 1:25 | IF |
| FOXA2, nuclear | Mouse | Santa Cruz | A-12 | 1:50 | IF |
| T (Brachyury), nuclear | Goat | R&D | Polyclonal | 1:25 | IF |
| Glucagon, cytoplasm | Mouse | Novocastra | Polyclonal | 1:200 | IHC |
| Insulin, cytoplasm | Mouse | Novocastra | 2D11-H5 | 1:100 | IHC |
| SOX2, nuclear | Mouse | R&D | 245610 | 1:500 | IHC |
| PDX1, nuclear | Rabbit | Abcam | EPR3358 | 1:5000 | IHC |
| NKX6.1, nuclear | Mouse | R&D | 631438 | 1:100 | IF |
| Glucagon, cytoplasm | Mouse | Sigma | K79bB10 | 1:100 | IF |
| Insulin, cytoplasm | Guinea pig | Dako | Polyclonal | 1:200 | IF |
| SSEA4, surface | Mouse | Abcam | MC813 | 1:100 | IF |
| CHG-a, cytoplasm | Rabbit | Abcam | Polyclonal | 1:100 | IF |
| PC1/3, cytoplasm | Mouse | Abcam | Monoclonal | 1:150 | IF |
| Somatostatin, cytoplasm | Rat | Abcam | 8.F.258 | 1:100 | IF |
Table 2.
Listo of secondary antibodies.
| Species | Fluorophore | Host | Brand | Code | Diluition |
|---|---|---|---|---|---|
| Guinea pig | Alexa Fluor-488® | Goat | Thermo Scientific | A-11073 | 1:500 |
| Mouse | Alexa Fluor-546® | Goat | Thermo Scientific | A-11003 | 1:500 |
| Rabbit | Alexa Fluor-546® | Goat | Thermo Scientific | A-11035 | 1:500 |
| Rat | Alexa Fluor-546® | Goat | Thermo Scientific | A-11081 | 1:500 |
For flow cytometry, colonies were dissociated into single cells with Accutase (STEMCELL Technologies), then cells were directly stained with SSEA4, TRA1-60, TRA1-81 (Table 1) and results acquired using a FACS Canto flow cytometer and the FACS Diva software. Results were re-analyzed with the FlowJo software version 9 (FlowJo LLC, Ashland, Oregon, USA).
To assess iPSC ability to differentiate in the three germinative lineages, iPSC colonies were incubated with Dispase solution (Gibco, Waltham, MA, USA) for 10–15 min at 37°C to promote colony lifting. Cell aggregates (embryoid bodies (EB)) were maintained in differentiation medium consisting of DMEMF-12, 20% Knock-out serum replacement, 20 μM β-mercaptoethanol, 1% sodium pyruvate, 2 mM L-glutamine, 2 mM nonessential amino acids (NEAA), 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Gibco) for 5 days in presence of ROCK inhibitor (STEMCELL Technologies). EB were plated on Matrigel coated 13 mm coverslips and incubated for 2 days. The EB immunofluorescence analysis was conducted as described above with specific primary antibodies (Table 1).
Differentiation of iPSC into Pancreatic Cells
Human iPSCs were differentiated into insulin-producing cells following a protocol established for pluripotent stem cells6, with slight modifications. Differentiation was initiated in adhesion when the iPSC cultures reached 70−80% confluence. Cells were imaged during the differentiation steps using an EVOS microscope (Life Technologies).
The following culture media were used for differentiation:
M1 medium: MCDB131 (Gibco) + 8 mM D-(+)-Glucose (Sigma) + 1.23 g/l NaHCO3 (Sigma) + 2% BSA (Sigma) + 0.25 mM Vitamin C (Sigma) + 1% Pen/Strep (Lonza, Basel, Switzerland) + 1% L-glutamine (Lonza);
M2 medium: MCDB131 + 20 mM D-Glucose + 1.754 g/l NaHCO3 + 2% BSA + 0.25 mM Vitamin C + Heparin 10mg/ml (Sigma) + 1% Pen/Strep + 1% L-glutamine.
All media were filter-sterilized through a 0.22 mm bottle top filter (Corning, New York State, USA). For sequential culture medium changes, small molecules and growth factors were added to the base media immediately before daily exchange. Media switches were as follows:
days 0–3: STEMdiff™ Definitive Endoderm Kit (STEMCELL) used following manufacturer instructions;
days 4–6: M1 medium + 50 ng/ml KGF (Peprotech, London, UK) + 1:50.000 ITS-X (Invitrogen, Carlsbad, CA, USA);
days 7, 8: M1 medium + 50 ng/ml KGF + 0.25 mM Sant1 (Sigma) + 2 μM Retinoic acid (RA) (Sigma) + 500 nM PdBU (Millipore) + 1:200 ITS-X + 200 nM LDN193189 (only Day 7) (Sigma);
days 9–13: M1 medium + 50 ng/ml KGF + 0.25 mM Sant1 + 100 nM RA + 1:200 ITS-X;
days 14–18: M2 medium + 0.25 mM Sant1 + 100 nM RA + 1 mM XXI (Millipore) + 10 mM Alk5i II (Selleckchem, Munich, Germany) + 1 mM L-3,30,5-Triiodothyronine (T3) (Sigma) + 20 ng/ml Betacellulin (R&D, Minneapolis, MN, USA) + 1:200 ITS-X.
Human Islets and EndoC-βH1
Human pancreatic islet (HI) preparations were isolated from heart-beating cadaveric organ donors as previously described17 in the Pancreatic Islet Processing Unit of the Diabetes Research Institute (DRI) at the San Raffaele Scientific Institute in Milan, Italy. The use of human specimens (islet preparations discarded from clinical use) was approved by the Institutional Review Board under the ‘European Consortium for Islet Transplantation (ECIT) human islet distribution program’ supported by the Juvenile Diabetes Research Foundation (JDRF) (3-RSC-2016-160-I-X)18. Islet purity was assessed as the percentages of endocrine clusters positive to dithizone staining (range: 80–90%).
The genetically engineered human pancreatic β cell line EndoC-βH119 was grown in DMEM low glucose (1 g/l) (Gibco), 2% BSA (Sigma), 50 μM 2-mercaptoethanol (Sigma), 10 mM nicotinamide (Sigma), 5.5 μg/ml transferrin (Gibco), 6.7 ng/ml sodium selenite (Sigma), 1% Pen/Strep.
Molecular Analysis
Total RNA was extracted with the mirVana Isolation Kit (Ambion) and quantified by spectrophotometry (Epoch, Gen5 software; BioTek, Winooski, VT). After DNAse (Invitrogen) treatment, 2 µg of RNA were reverse transcribed with SuperScript III RT (Invitrogen), according to the manufacturer’s instructions (Invitrogen). Predesigned gene-specific primer and probe sets from TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, USA; listed in Supplementary Table 1) were used for gene expression study. A predesigned TaqMan low-density array panel (Applied Biosystems) of 40 markers of β cells was also used for gene expression analysis (assays are listed in Supplementary Table 2) of 6/7 preparations of clone #5 and 4/5 of clone #9. PCRs were performed in a 7900 Real-Time PCR System (Applied Biosystems). Gene expression levels were normalized using GAPDH (2−ΔCt method) and are reported as fold change over undifferentiated iPSC and compared with HI and EndoC-βH1 (ΔΔCt method, mean ± SEM). Low-density array clustering analysis was performed with the Morpheus software (https://software.broadinstitute.org/morpheus). Values of mean difference (MD) between groups were calculated as difference between the log of arithmetic means of the two groups (βH1-Hi-iβ v. iPSC or βH1-Hi v. iβ):
assuming that random variables X and Y into two groups were independently and identically distributed. Positive or negative signs were then attributed according to up- or down-expression of genes between groups.
Immunocytochemistry
For immunocytochemistry iPSC clusters were fixed in PFA 4% (Sigma) and cytospinned for thin-layer cell preparation. Islet clusters were embedded in agarose and paraffin and 3 μm sections were cut with a microtome. Samples were processed routinely for histology. The peroxidase-antiperoxidase immunohistochemistry method (Labvision, Thermo Scientific) was used for detection with the antibodies listed in Table 1. Immunostained slides were acquired using an AperioScanscope (Leica), which allows the scanning and digitalization of the slide by multiple vertical scans at 40× magnification, and analyzed with the Aperio Image Scope software (Leica).
Cytofluorimetric Analysis
Human iPSC and EndoC-βH1 were stained with the Live/Dead stain (Molecular Probes) to exclude dead cells from the analysis. Intracellular staining required cell permeabilization (Cytofix/Phosflow™ perm buffer III, Becton Dickinson, BD, Franklin Lakes, NJ, USA). Cells were then stained using the following monoclonal antibodies (mAbs): 40/Oct3 Alexa Fluor647® anti-OCT3/4; 658A5 Alexa Fluor488® anti-PDX-1; R11-560 PE anti-NKX6.1; T56-706 Alexa Fluor647® anti-Insulin (BD). Analysis was carried out on a FACS Canto flow cytometer using the FACS Diva software. Results were analyzed with the FlowJo software version 9 (FlowJo LLC).
Immunofluorescence
For immunofluorescence human iPSC were differentiated in 4-Well Culture Slide (Falcon, Corning). HI were embedded in agarose, fixed with 4% paraformaldehyde, included in paraffin and 3 μm sections were cut with a microtome. Sections were de-paraffinized using xylene, and rehydrated in a serial dilution of absolute alcohol. Cells were permeabilized when necessary for intracellular staining (PermWash 0.2% Triton X-100 in PBS) and stained using the antibodies listed in Tables 1 and 2. Images were acquired using Confocals PerkinElmer UltraVIEW ERS microscope (PerkinElmer Life Sciences, Waltham, MA, USA) and deconvolved with Huygens Professional version 17.04 (Scientific Volume Imaging), using the Classic Maximum Likelihood Estimation (CMLE) algorithm, with signal-to-noise ratio (SNR): 100 iterations.
Hormone Secretion
Hormone levels in iPSC culture supernatants were measured at each differentiation stage using the Bio-Plex Pro™ human diabetes kit (BioRad, Hercules, CA, USA). Samples were assayed according to the manufacturer’s instructions, and the plates were read on a Luminex xMAP instrument (BioRad). The acquisition and analysis of the samples were performed with the Bio-Plex Manager 6.0 software (BioRad).
Dynamic Islet Perifusion
A high-capacity, automated perifusion system (BioRep® Perifusion V2.0.0) was used to dynamically stimulate cell secretion. A low pulsatility peristaltic pump was used to push HEPES-buffered solution (125 mM NaCl, 5.9 mM KCl, 2.56 mM CaCl2, 1 mM MgCl2, 25 mM HEPES, 0.1% BSA, pH 7.4) through a sample container harboring 50 iPSC clusters or 20 HI immobilized in Bio-Gel P-4 Gel (BioRad), or 700,000 EndoC-βH1 cells. Cells were stabilized with a slow-flow perifusion rate (30 µl/min) with low glucose (2 mM) for 60 minutes. A combined stimulus (11 mM glucose with or without 3-isobutyl-1-methylxanthine - IBMX) was then added for 20 minutes at a flow rate of 100 µl/min. Following a third step with 2 mM glucose for 20 minutes, cells were exposed to 30 mM KCl for 20 minutes and then to 2 mM glucose for another 20 minutes. The perifusates were collected every minute by an automated fraction collector designed for a multiwell plate format. Cells and perfusion solutions were kept at 37°C in a built-in temperature controlled chamber, and collected perifusates were stored at -20°C.
Quantification of Insulin Release
Insulin released in the perifusates was measured with an ELISA Kit (Mercodia, Uppsala, SW) following the manufacturer’s instructions, using an ELISA Reader (MicroPlate Reader, Model 680, BioRad).
Statistical Analysis
Nonparametric test (Kruskal Wallis with Dunn’s multiple comparison test) was used to compare groups and a 2-tailed P value less than 0.05 or 0.01 was considered significant. Analysis of data was performed using the Prism software (GraphPad Prism 5).
Results
IPSC Gene Expression Profile During In-vitro Differentiation Compared to Mature β Cells
IPSC lines (clone #5 and clone #9) were obtained from fibroblasts of a healthy adult donor, reprogrammed to iPSC by delivery of Yamanaka’s factors with Sendai virus vectors.
Clone #9 was previously characterized (HIPSCTR4#9, www.isenet.it). Regarding clone #5, its genetic stability was confirmed by karyotype analysis via chromosome Q-banding in the metaphase (Fig. 1S A) and by comparative genomic hybridization (CGH) array, showing no significant genomic alterations with the exception of three CNVs, not corresponding to known polymorphic variants and with no associated clinical phenotype (Fig. 1S B). Pluripotency requirements were assessed by flow cytometry staining (SSEA4, TRA1-60, TRA1-80, Fig. 1S C) and by immunofluorescence (OCT3/4, NANOG, TRA1-60, TRA1-80, SOX2, Fig. 1S D), confirming the staminal phenotype of the cells. Sendai virus vectors were rapidly cleared after reprogramming Figure 2S. The differentiation capability of iPSC towards cells of the three germinal layers was demonstrated by EB formation in vitro. Plated aggregates stained positively for endodermal (SOX17, alpha-FP, FOXA2), mesodermal (SMA1, T) and ectodermal (PAX6) markers (Fig. 1S).
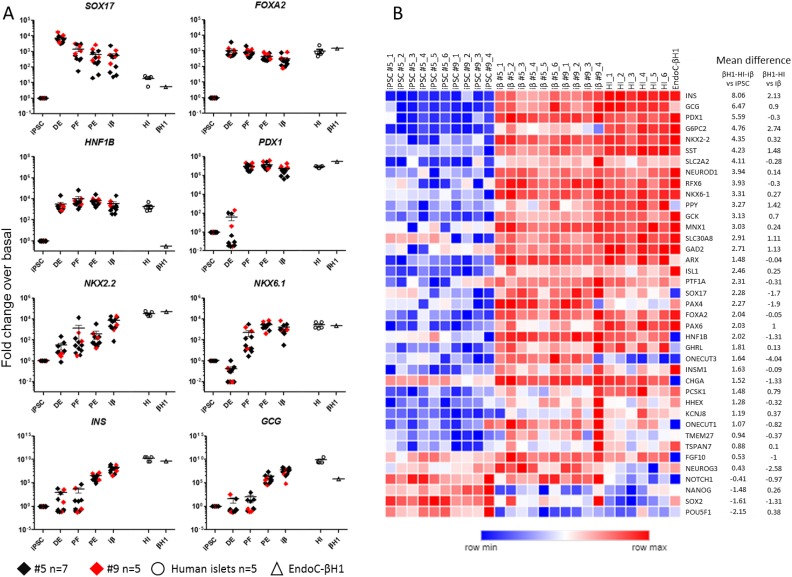
Fig 1.
Gene expression analysis of iPSC differentiation into insulin-producing cells. (A) Gene expression analysis by Taqman of markers of definitive endoderm (FOXA2 and SOX17), posterior foregut (HNF1b and PDX1), pancreatic endoderm (NKX2.2 and NKX6.1) and endocrine cells (INS and GCG). Normalized gene expression levels of clone #5 (black squares, n=7) and clone #9 (red squares, n=5) during the steps of differentiation are reported as fold change referred to undifferentiated iPSC and compared with human islets (HI, empty circles, n=5) and EndoC-βH1 cell line (empty triangle) (mean ± SEM). (B) Heatmap showing the expression level of 39 genes characteristic of β cell identity, function and development in undifferentiated iPSC (clone #5 n=6 and clone #9 n=4), iβ (clone #5 n=6 and clone #9 n=4), HI (n=6) and EndoC-βH1. Samples are displayed in columns and genes in rows. Data are clustered using hierarchical analysis based on Spearman correlation for genes and k-means for samples. The bar below the heatmap is color coded according to the level of expression (Log10 2⁁-ΔCt). Mean difference of iPSC versus cluster composed by iβ-HI-EndoC-βH1 and iβ versus cluster of HI-EndoC-βH1 of each gene is reported.
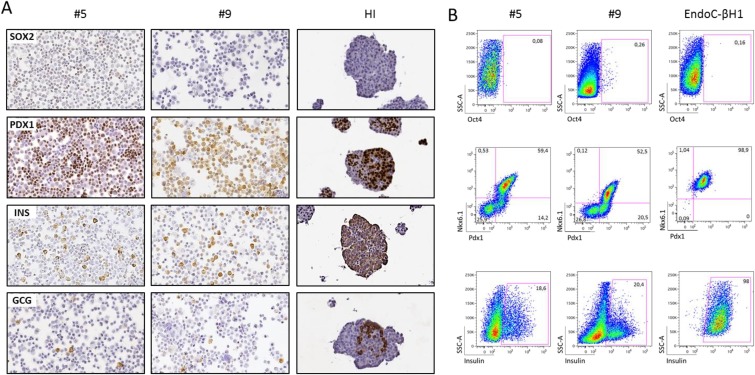
Fig 2.
Proteic expression analysis of iPSC-derived insulin-producing cells (iβ). (A) Protein expression analysis by immunocytochemistry of SOX2, PDX1, Insulin and Glucagon in cytospin preparations of iβ derived from iPSC clone #5 and #9 and of paraffin embedded HI. Magnification 20x. (B) Protein expression analysis by flow cytometry of markers of pluripotency (OCT4), pancreatic endoderm (PDX1, NKX6.1) and endocrine cells (insulin) in iβ and EndoC-βH1. SSC-A: side scatter. Gate delimitates positive events. Percentages of positive cells of a representative experiment are reported.
iPSC were differentiated into pancreatic β cells using a modified version of the Pagliuca et al. protocol12. The main modifications consist of: (a) culture of iPSC in adhesion throughout the five steps of the differentiation process, (b) the use of a commercial kit for definitive endoderm (DE) specification20 and (c) a shorter duration of the differentiation protocol (up to 20 days). In the second step, CXCR4+ DE cells (>90% in flow cytometry, data not shown) were induced to become posterior foregut (PF) by adding KGF, while cells transitioned to pancreatic endoderm (PE) following addition of retinoic acid and inhibition of the Sonic Hedgehog (Shh) signaling pathway. Finally, using a combination of factors that affect signaling of pathways like TGFβ, thyroid hormone, and γ-secretase, PE cells turned into pancreatic endocrine cells (induced pluripotent stem cell-derived β cells or iβ). Gene expression of the two iPSC clones was analyzed at each stage of differentiation and compared with the median of 5 preparations of human islets (purity >80%) and with the human β cell line EndoC-βH119 (Fig 1A). Genes and transcription factors related to pancreatic development were selectively upregulated at specific stages: SOX17 and FOXA2, DE markers, were upregulated during the first stage (iPSC versus DE p<0.001 for both genes), while HNF1B and PDX1 reached their higher expression level at the stage of posterior foregut (iPSC versus PF p<0.001 for both genes). The expression of the transcription factors NKX2.2 and NKX6.1 increased, as expected, during the last steps of differentiation (iPSC versus iβ p<0.001 for both genes) and reached levels comparable to HI and EndoC-βH1. iβ derived from iPSC expressed high levels of the INS and GCG pancreatic hormones (iPSC versus iβ p<0.001 for both genes). In particular, iβ expressed insulin mRNA levels very close to donor pancreatic islets (iβ v. HI p=ns), with iβ clone #5 and #9 expressing only 0.011 and 0.024 fold less insulin compared to HI (Fold change: 2⁁-(ΔCtiβ-ΔCtiPSC)/2⁁-(ΔCtHI-ΔCtiPSC)), respectively. Similarly, compared to the pure β cell line EndoC-βH1, clone #5 and #9 derived iβ cells expressed respectively only 0.045 and 0.095 fold less insulin (iβ versus EndoC-βH1 p=ns). The gene expression signature corresponding to pancreas development and to mature pancreatic β cells was also analyzed by low density array qRT-PCR in undifferentiated iPSC and iβ, in comparison with 6 preparations of HI (purity >80%) and EndoC-βH1. As shown in Fig. 1B, the cluster analysis identified two main groups, one consisting of undifferentiated iPSC and the other of iβ + HI + EndoC-βH1, confirming the similarity of the iβ gene expression profile to that of pancreatic β cells. A comparison between iβ and HI+EndoC-βH1 revealed that: (a) genes of pluripotency like NANOG, SOX2, POU5F1 and NOTCH1 were markedly reduced in iβ, (b) transcription factors and developmental genes like FGF10, PTF1a, ONECUT1, PDX1, NKX2.2, NKX6.1, PAX6, FOXA2, ARX, NEUROD1, MNX1, ISL1, HHEX and RFX6 were expressed by iβ at levels similar to those in HI-EndoC-βH1 (median fold change difference between -1 and 1), (c) pancreatic hormones were highly expressed in iβ with similar or slightly lower levels compared to HI-EndoC-βH1 (INS 2.13, SST 1.48, GCG 0.9, PPY 1.42, GHRL 0.13 mean difference of HI-EndoC-βH1 v. iβ); β cell specific autoantigens GAD2, TSPAN7 and SLC30A8 were also expressed in iβ. Finally, genes involved in insulin secretion and glucose sensing pathways like SLC2A2, GCK, TMEM27, PCSK1, KCNJ8 and INSM1 were expressed in iβ at levels comparable to HI-EndoC-βH1 (median fold change difference between -1 and 1). NEUROG3, PAX4, SOX17, ONECUT3 and HNF1b resulted instead more expressed in iβ than in mature β cells, suggesting the presence of still developing endocrine cells. EndoC-βH1 express very low levels of non-β cell specific genes as GCG, PPY, GHRL and genes of pancreas development as ARX, SOX17, HNF1b, FGF10 and ONECUT1.
Terminally Differentiated Cells Express Pancreatic Protein Markers
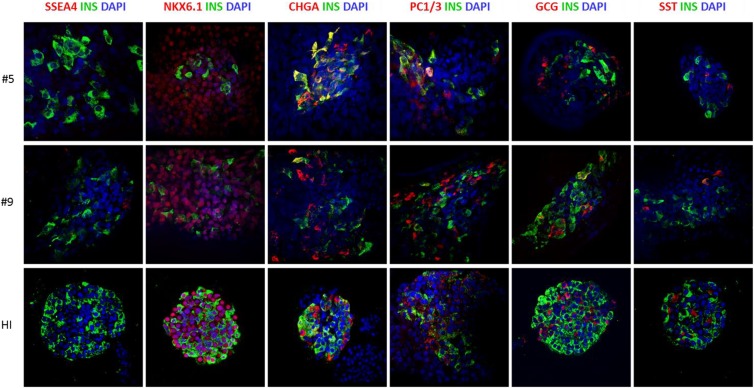
Expression of pancreatic protein markers was analyzed at the final stage of differentiation of the two iPSC clones and compared with HI and EndoC-βH119. By immunocytochemistry, no residual SOX2 pluripotent cells were observed, while iβ cells from both clones were positive for PDX1 and insulin and, in some cells, also glucagon. As expected, HI were composed by insulin and glucagon positive cells that were mostly PDX1 positive; no SOX2 positive cells were observed (Fig. 2A). We then determined quantitatively the composition of terminally differentiated cells by flow cytometry: iβ resulted negative for OCT4, positive for PDX1 (clone #5 n=8 differentiation experiments, mean ± SEM: 77.2 ± 2.9%; clone #9 n=6, 68.1 ± 8.8%), NKX6.1 (clone #5 n=8, 42.9 ± 5.1%; clone #9 n=6, 39.2 ± 8.7%) and insulin (clone #5 n=8, 11.0 ± 2.3%; clone #9 n=6 11.5 ± 3.8%). Co-expression analysis for PDX1 and NKX6.1 revealed the coexistence of cells at different developmental stages: indeed, virtually all NKX6.1 positive cells were also PDX1 positive (clone #5 n=3 differentiation experiments, mean ± SEM: 46.3 ± 4.5%; clone #9 n=4: 35.6 ± 8.6% PDX1+/NKX6.1+), while a proportion of cells was PDX1+/NKX6.1- (clone #5 n=3, 27.4 ± 6.3%; clone #9 n=4: 23.4 ± 7.1%). EndoC-βH1 cells were 98.9% PDX1/NKX6.1 double positive and 98% insulin positive (Fig. 2B). Finally, the expression of key β cell markers in iPSC-derived insulin positive cells was analyzed by immunofluorescence: insulin positive iβ cells were negative for pluripotency marker such as SSEA4, co-expressed the β cell specific transcription factor NKX6.1 and the neuroendocrine markers ChgA and PC1-3. Some cells of the terminal differentiated cell population resulted poly-hormonal and in particular few cells co-expressed either insulin and glucagon or insulin and somatostatin, suggesting a partially immature phenotype. Anyway, the vast majority of iβ were mono-hormonal cells. Control staining of HI showed that β cells were positive for NKX6.1, ChgA, PC1-3, negative for SSEA4, and did not co-stain with glucagon and somatostatin (Fig. 3 and Fig. 2S A-C).
Fig 3.
Proteic expression analysis of iPSC-derived insulin-producing cells (iβ). Immunofluorescence of insulin (green) in iβ and HI in co-staining with SSEA-4, NKX6.1, Chromogranin-A, PC1/3, Glucagon and Somatostatin (red). Nuclei stained in blue (DAPI). Magnification x40.
Terminally Differentiated Cells Secrete Insulin at Basal Level and in Response to Stimuli
The release of insulin, glucagon and ghrelin was measured in the supernatants of unstimulated cells during differentiation, then compared to HI and EndoC-βH1. All three hormones were absent during the stages from pluripotency to pancreatic progenitor cells. At the stage of iβ, cells started to release hormones in the culture medium (c-peptide 2361.8±1156.9 pg/ml, glucagon 524.6±159.9 pg/ml and ghrelin 506.1±169.5 pg/ml). HI secreted 1.4, 22.5 and 1.5 fold more c-peptide, glucagon and ghrelin than iβ, respectively. EndoC-βH1 cells secreted 3.1 fold more c-peptide than iβ but less ghrelin (0.4 fold) and no glucagon. This indicates that iβ show a certain degree of cell heterogeneity which make them closer to donor islets (low release of glucagon and ghrelin) but they also contain cells with a clear β cell phenotype (significant release of c-peptide) (Fig 4A).
Fig 4.
Secretory capacity of iPSC-derived insulin-producing cells (iβ). (A) Unstimulated levels of C-peptide, glucagon and ghrelin in the supernatant of iPSC at different stages (DE, PF, PE and iβ) of differentiation into insulin producing cells (n=15), in comparison with HI (n=4) and EndoC-βH1 (n=4), measured by Luminex. Mean±SEM. **P<0.01 and ***P<0.001. ns=not statistically significant. (B) Dynamic insulin secretion by terminally differentiated iPSC (left, n=4), HI (middle, n=6) and EndoC-βH1 (right, n=4), upon sequential stimulation with 0.5 mM glucose (basal), 11 mM glucose (stimulus) with (dashed line) or without (not dashed line) IBMX, 0.5 mM glucose and 30 mM KCl, measured by Elisa. Insulin levels are expressed as pm/ml.
iβ were then challenged with glucose and KCl in a dynamic perifusion assay to compare their insulin secretion profile with that of HI and EndoC-βH1 (Fig. 4B). iβ and EndoC-βH1 poorly responded to a glucose stimulus, while HI promptly released insulin. The addition to glucose of the phosphodiesterase inhibitor IBMX, promoted insulin secretion by HI and induced a response also in iβ and EndoC-βH1. A depolarizing stimulus with KCl resulted in an insulin secretory response in all tested β cells. These data suggest that iβ have the capacity to secrete insulin, even if they show the behavior of an immature β cell, partially recapitulating the EndoC-βH1 phenotype. Basal level of insulin secretion was different among the different sources of β cells (iβ 5,4±2 pm/ml, HI 221±32 pm/ml, EndoC-βH1 1076±175 pm/ml) and consistent with the much lower percentage of insulin positive cells in the iβ group.
Discussion
This study describes the characterization of the expression profile and insulin secretory capacity of iPSC derived insulin positive β cells in comparison to that of native human islets and a human pure β cell line.
We previously reported our studies on the differentiation into insulin producing cells of iPSC reprogrammed with retroviral vectors15. In the present work, we used two clones of iPSC obtained from skin fibroblasts of a healthy donor using the Sendai virus to deliver Yamanaka’s factors. Compared to iPSC generated using retroviral vectors, the generation of iPSC using Sendai represents a safer and more efficient strategy, since this RNA vector carries no risk of integrating into the host genome and is characterized by a high-infection efficiency, resulting in high rates of reprogramming16. The pluripotency and genomic stability of the two clones used in this study were extensively characterized in terms of viability, mycoplasma infection, microsatellite-based identity matching to the parent cell line, and karyotype analysis. Currently, one of the two clones is already stored in a biobank (http://www.isenet.it/) and is available for future studies.
Different protocol exist for the differentiation of pluripotent cells into pancreatic cells. While all share a common core set of steps, it is increasingly evident5–7 that there is a need to introduce adjustments and modifications depending on the starting pluripotent cell line21,22. In this study, we applied the protocol described by Pagliuca et al. with ad hoc modifications. In particular, for the induction of the differentiation to definitive endoderm, a critical step for efficient pancreatic differentiation usually achieved by supplying a high concentration of Activin A23, we instead adopted a commercially available differentiation system (STEMdiff™ Definitive Endoderm Kit)20. This resulted in a homogeneous and confluent monolayer of individualized DE cells at the end of the induction, which were strongly positive for stage-specific markers. These cells were then either grown in adhesion or induced to form clusters in suspension for functional tests.
To perform a robust ascertainment of successful pancreatic terminal differentiation, we analyzed also the intermediate steps of the pancreas specification, ruling out an aspecific up-regulation of the insulin gene in response to stress24, and investigated at both the mRNA and protein level multiple pancreatic β cell markers, demonstrating not only the presence of β cell phenotype but also characterizing its function and maturity.
In our iPSC-derived cells, throughout differentiation we observed the sequential up-regulation of HNF1b and PDX1, followed by NKX2.2 and NKX6.1, and finally insulin and glucagon. Conversely, at the end of the differentiation, iPSC-derived cells iβ were negative for pluripotency markers (OCT4, NANOG, SSEA4 and SOX2) and positive for PDX1, NKX6.1, insulin and glucagon by qRT-PCR, immunocytochemistry and flow cytometry. Moreover, an immunofluorescence analysis allowed us to determine that iβ cells were mostly mono-hormonal with only a few cells double positive for glucagon/insulin or somatostatin/insulin. The data for hormones’ basal secretion confirmed that among our terminally differentiated cells a large majority is constituted by insulin secreting β cells with only a minor fraction of cells secreting glucagon and ghrelin, suggesting that the minor heterogeneity is possibly due to cell immaturity. We have no evidence of the presence of exocrine cells after differentiation; we speculate that it is likely that few exocrine cells are present in final cell product, considering the widely shared differentiation path between endocrine and exocrine cell.
To verify the functional capacity of iβ, we decided to use a dynamic perifusion assay. In fact, the classical static incubation test with glucose, reported by most of the publications describing β cell generation25–28, does not assess the physiological release of insulin by β cells upon stimulation and does not provide information on the timing of the secretory response and its recovery. Overall, our iβ secreted insulin in response to a dynamic glucose challenge and we did not observe significant differences in the differentiation potential of the two clones analyzed, as expected since their derivation from a single donor29,30.
The terminal cell product of our differentiation has all the features of a β cell, including insulin secretion, both basal and under glycemic stimulus. Viacyte group utilizes a different strategy, which include the production of pancreatic progenitors in vitro from embryonic stem cells and then the spontaneous maturation into insulin-secreting cells in vivo weeks after transplantation8,31. Other experiences however, like that of Pagliuca et al6,12, which will soon become a clinical trial with Semma Therapeutics, are more oriented to the use of cells terminally differentiated to insulin-secreting β cells. The advantages of this last approach are the possibility to qualify the β cell before the transplant and the immediate function of the graft after transplantion, without delay of maturation.
Furthermore, the field of cell regeneration from stem cells requires the identification of a reference target cell to perform a quality assessment of differentiation products. Therefore, to corroborate our results on β cell differentiation and to bring iβ a step closer to a clinical application, we performed a comparison between iβ and two sources of control β cells: pancreatic islets from organ donors and the immortalized β cell line EndoC-βH1.
In the field of β cell replacement, pancreatic islets from organ donors constitute the most intuitive reference tissue. Donor derived pancreatic islets are infused in T1D patients when obtained with a purity of at least 80% of endocrine tissue, corresponding to around 35% of β cells at best32. While it has been proposed that islet heterogeneous composition might have beneficial effects, with non β cell components favoring post-transplant islets survival and function, we decided to compare our final cell product also with EndoC-βH1 cells, a human β cell line, resembling primary pancreatic β cells19,33. To generate the cell line, human fetal pancreatic buds were transduced with a lentiviral vector that expressed SV40LT under the control of the insulin promoter and grafted into SCID mice to allow the formation of mature insulinomas; the resulting β cells were then transduced with human telomerase reverse transcriptase, grafted into other SCID mice, and finally expanded in vitro.
Our analysis of β cell identity markers highlighted a remarkable similarity between iβ and both control β cells, with iβ, HI and EndoC-βH1 expressing similar levels of key β cell transcription factors, genes involved in secretion machinery, and of autoantigens: an observation with relevant implications for β cell replacement in T1D patients. However, albeit not unexpectedly, both immunofluorescence and flow cytometric analyses set iβ closer to HI than to EndoC-βH1, because of the heterogeneous expression of some endocrine markers.
At the functional level, iβ cells responded to increasing concentration of glucose and to depolarizing stimuli in a similar fashion to HI and EndoC-βH1, although with expectedly lower basal levels of released insulin (approximately 50 fold less than HI and 200 fold than EndoC-βH1, respectively) considering the comparatively smaller number of fully differentiated cells. The kinetic of response of iβ cells to a secretory stimulus, while showing a double-peak response without an initial delay like in HI, was more related to that of EndoC-βH1, because of the similar need for IBMX potentiation to achieve a response to glucose19.
In summary, our data report that the iβ cellular product obtained at the end of iPSC differentiation is very similar to donor human islets in terms of gene and protein expression as well as cell composition, with both islets and iβ being cellular aggregates composed of cells with different functions. At the level of insulin secretion instead, iβ contained a variable proportion of cells sharing secretory features typical of EndoC-βH1 and showing an only partially mature secretory function, suggesting that the efficiency of differentiation and maturation of stem cell-derived β cells is in need of further improvement.
Our study, while confirming that iPSC-derived β cells have the potential to constitute an unlimited, adjustable, and efficient source of new β cells for the cell therapy of T1D, indicates that the comparison with more than one reference tissue is necessary for proper assessment of the quality of the differentiation product. This assessment is crucial both for devising optimal cell therapy strategies in T1D and for in vitro β cell studies aimed at understanding experimental islet biology, disease pathogenesis, and drug discovery.
Supplemental Material
Supplemental Material, CT-2091_Supplementary_Figure_1S for Differentiation of Sendai Virus-Reprogrammed iPSC into β Cells, Compared with Human Pancreatic Islets and Immortalized β Cell Line by Silvia Pellegrini, Fabio Manenti, Raniero Chimienti, Rita Nano, Linda Ottoboni, Francesca Ruffini, Gianvito Martino, Philippe Ravassard, Lorenzo Piemonti, and Valeria Sordi in Cell Transplantation
Supplemental Material
Supplemental Material, CT-2091_Supplementary_Figure_2S for Differentiation of Sendai Virus-Reprogrammed iPSC into β Cells, Compared with Human Pancreatic Islets and Immortalized β Cell Line by Silvia Pellegrini, Fabio Manenti, Raniero Chimienti, Rita Nano, Linda Ottoboni, Francesca Ruffini, Gianvito Martino, Philippe Ravassard, Lorenzo Piemonti, and Valeria Sordi in Cell Transplantation
Supplemental Material
Supplemental Material, CT-2091_Supplementary_Figure_3Sa for Differentiation of Sendai Virus-Reprogrammed iPSC into β Cells, Compared with Human Pancreatic Islets and Immortalized β Cell Line by Silvia Pellegrini, Fabio Manenti, Raniero Chimienti, Rita Nano, Linda Ottoboni, Francesca Ruffini, Gianvito Martino, Philippe Ravassard, Lorenzo Piemonti, and Valeria Sordi in Cell Transplantation
Supplemental Material
Supplemental Material, CT-2091_Supplementary_Figure_3Sb for Differentiation of Sendai Virus-Reprogrammed iPSC into β Cells, Compared with Human Pancreatic Islets and Immortalized β Cell Line by Silvia Pellegrini, Fabio Manenti, Raniero Chimienti, Rita Nano, Linda Ottoboni, Francesca Ruffini, Gianvito Martino, Philippe Ravassard, Lorenzo Piemonti, and Valeria Sordi in Cell Transplantation
Supplemental Material
Supplemental Material, CT-2091_Supplementary_Figure_3Sc for Differentiation of Sendai Virus-Reprogrammed iPSC into β Cells, Compared with Human Pancreatic Islets and Immortalized β Cell Line by Silvia Pellegrini, Fabio Manenti, Raniero Chimienti, Rita Nano, Linda Ottoboni, Francesca Ruffini, Gianvito Martino, Philippe Ravassard, Lorenzo Piemonti, and Valeria Sordi in Cell Transplantation
Supplementary Material
Acknowledgments
We thank Advanced Light and Electron Microscopy BioImaging Center (ALEMBIC) of San Raffaele Institute for confocal immunofluorescence images.
Footnotes
Author Contributions: Lorenzo Piemonti, Valeria Sordi equally contributed to the work.
Silvia Pellegrini: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing.
Fabio Manenti: Collection and assembly of data, data analysis and interpretation.
Raniero Chimienti: Data analysis and interpretation.
Rita Nano: Provision of study material.
Linda Ottoboni: Collection of data, provision of study material.
Francesca Ruffini: Collection of data, provision of study material.
Gianvito Martino: provision of study material, final approval of manuscript.
Philippe Ravassard: Provision of study material.
Lorenzo Piemonti: Conception and design, data analysis and interpretation, final approval of manuscript.
Valeria Sordi: Conception and design, data analysis and interpretation, manuscript writing.
Ethical Approval: The use of human islets was approved by the Institutional Review Board under the ‘European Consortium for Islet Transplantation (ECIT) human islet distribution program’.
Statement of Human and Animal Rights: The use of human islets was approved by the Institutional Review Board under the ‘European Consortium for Islet Transplantation (ECIT) human islet distribution program’ supported by the Juvenile Diabetes Research Foundation (JDRF) (3-RSC-2016-160-I-X).
Statement of Informed Consent: For iPSC lines, written informed consent was obtained from the donor.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research and/or authorship of this article: supported by the European Commission (H2020 grant 681070), Fondazione Diabete Ricerca ONLUS (Società Italiana di Diabetologia-SID) and Eli Lilly Italia (grant ‘Sostegno alla ricerca sul diabete 2017’).
ORCID iD: Valeria Sordi  http://orcid.org/0000-0003-0179-1679
http://orcid.org/0000-0003-0179-1679
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Atkinson MA, Eisenbarth GS, Michels AW. Type 1 diabetes. Lancet. 2014;383(9911):69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Belle TL, Coppieters KT, von Herrath MG. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol Rev. 2011;91(1):79–118. [DOI] [PubMed] [Google Scholar]
- 3. Shapiro AMJ, Pokrywczynska M, Ricordi C. Clinical pancreatic islet transplantation. Nat Rev Endocrinol. 2017;13(5):268–277. [DOI] [PubMed] [Google Scholar]
- 4. Hering BJ, Clarke WR, Bridges ND, et al. Clinical islet transplantation consortium. phase 3 trial of transplantation of human islets in type 1 diabetes complicated by severe hypoglycemia. Diabetes Care. 2016;39(7):1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. D’Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol. 2006;24(11):1392–1401. [DOI] [PubMed] [Google Scholar]
- 6. Pagliuca FW, Millman JR, Gü M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA. Generation of functional human pancreatic beta cells in vitro. Cell. 2014;159:428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O’Dwyer S, Quiskamp N, Mojibian M, Albrecht T, Yang YHC, Johnson JD, Kieffer TJ. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32(11):1121–1133. [DOI] [PubMed] [Google Scholar]
- 8. Agulnick AD, Ambruzs DM, Moorman MA, Bhoumik A, Cesario RM, Payne JK, Kelly JR, Haakmeester C, Srijemac R, Wilson AZ, Kerr J, Frazier MA, Kroon EJ, D’Amour KA. Insulin-producing endocrine cells differentiated in vitro from human embryonic stem cells function in macroencapsulation devices in vivo. Stem Cells Transl Med. 2015;4(10):1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. [DOI] [PubMed] [Google Scholar]
- 10. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. [DOI] [PubMed] [Google Scholar]
- 11. Maehr R, Chen S, Snitow M, Ludwig T, Yagasaki L, Goland R, Leibel RL, Melton DA. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci USA. 2009;106(37):15768–15773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Millman JR, Xie C, Dervort AV, Gürtler M, Pagliuca FW, Melton DA. Generation of stem cell-derived b-cells from patients with type 1 diabetes. Nat Commun. 2016;7:11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tateishi K, He J, Taranova O, Liang G, D’Alessio AC, Zhang Y. Generation of insulin-secreting islet-like clusters from human skin fibroblasts. J Biol Chem. 2008;283(46):31601–31607. [DOI] [PubMed] [Google Scholar]
- 14. Kunisada Y, Tsubooka-Yamazoe N, Shoji M, Hosoya M. Small molecules induce efficient differentiation into insulin-producing cells from human induced pluripotent stem cells. Stem Cell Res. 2012;8(2):274–284. [DOI] [PubMed] [Google Scholar]
- 15. Pellegrini S, Ungaro F, Mercalli A, Melzi R, Sebastiani G, Dotta F, Broccoli V, Piemonti L, Sordi V. Human induced pluripotent stem cells differentiate into insulin-producing cells able to engraft in vivo. Acta Diabetol. 2015;52(6):1025–1035. [DOI] [PubMed] [Google Scholar]
- 16. Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, Takada N, Inoue M, Hasegawa M, Kawamata S, Nishikawa SI. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci USA. 2011;108(34):14234–14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ricordi C, Lacy PE, Finke EH, Olack BJ, Scharp DW. Automated method for isolation of human pancreatic islets. Diabetes. 1988;37(4):413–420. [DOI] [PubMed] [Google Scholar]
- 18. Nano R, Bosco D, Kerr-Conte JA, Karlsson M, Charvier S, Melzi R, Ezzouaoui R, Mercalli A, Hwa A, Pattou F, Korsgren O, Berney T, Piemonti L. Human islet distribution programme for basic research: activity over the last 5 years. Diabetologia. 2015;58(5):1138–1140. [DOI] [PubMed] [Google Scholar]
- 19. Ravassard P, Hazhouz Y, Pechberty S, Bricout-Neveu E, Armanet M, Czernichow P, Scharfmann R. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J Clin Invest. 2011;121(9):3589–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carpentier A, Nimgaonkar I, Chu V, Xia Y, Hu Z, Liang TJ. Hepatic differentiation of human pluripotent stem cells in miniaturized format suitable for high-throughput screen. Stem Cell Res. 2016;16(3):640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26(3):313–315. [DOI] [PubMed] [Google Scholar]
- 22. Kilpinen H, Goncalves A, Leha A, et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546(7658):370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23(12):1534–1541. [DOI] [PubMed] [Google Scholar]
- 24. Haigis MC, Yankner BA. The aging stress response. Mol Cell. 2010;40(2):333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Russ HA, Parent AV, Ringler JJ, Hennings TG, Nair GG, Shveygert M, Guo T, Puri S, Haataja L, Cirulli V, Blelloch R, Szot GL, Arvan P, Hebrok M. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 2015;34(13):1759–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu S, Russ HA, Wang X, Zhang M, Ma T, Xu T, Tang S, Hebrok M, Ding S. Human pancreatic beta-like cells converted from fibroblasts. Nat Commun. 2016;7:10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cogger KF, Sinha A, Sarangi F, McGaugh EC, Saunders D, Dorrell C, Mejia-Guerrero S, Aghazadeh Y, Rourke JL, Screaton RA, Grompe M, Streeter PR, Powers AC, Brissova M, Kislinger T, Nostro MC. Glycoprotein 2 is a specific cell surface marker of human pancreatic progenitors. Nat Commun. 2017;8(1):331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yabe SG, Fukuda S, Takeda F, Nashiro K, Shimoda M, Okochi H. Efficient generation of functional pancreatic β-cells from human induced pluripotent stem cells. J Diabetes. 2017;9(2):168–179. [DOI] [PubMed] [Google Scholar]
- 29. Kajiwara M, Aoi T, Okita K, Takahashi R, Inoue H, Takayama N, Endo H, Eto K, Toguchida J, Uemoto S, Yamanaka S. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc Natl Acad Sci USA. 2012;109(31):12538–12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kyttälä A, Moraghebi R, Valensisi C, Kettunen J, Andrus C, Pasumarthy KK, Nakanishi M, Nishimura K, Ohtaka M, Weltner J, Van Handel B, Parkkonen O, Sinisalo J, Jalanko A, Hawkins RD, Woods NB, Otonkoski T, Trokovic R. Genetic variability overrides the impact of parental cell type and determines iPSC differentiation potential. Stem Cell Reports. 2016;6(2):200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, Agulnick AD, D’Amour KA, Carpenter MK, Baetge EE. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol. 2008;26(4):443–452. [DOI] [PubMed] [Google Scholar]
- 32. Pisania A, Weir GC, O’Neil JJ, Omer A, Tchipashvili V, Lei J, Colton CK, Bonner-Weir S. Quantitative analysis of cell composition and purity of human pancreatic islet preparations. Lab Investig. 2010;90(11):1661–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scharfmann R, Pechberty S, Hazhouz Y, von Bülow M, Bricout-Neveu E, Grenier-Godard M, Guez F, Rachdi L, Lohmann M, Czernichow P, Ravassard P. Development of a conditionally immortalized human pancreatic β cell line. J Clin Invest. 2014;124(5):2087–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, CT-2091_Supplementary_Figure_1S for Differentiation of Sendai Virus-Reprogrammed iPSC into β Cells, Compared with Human Pancreatic Islets and Immortalized β Cell Line by Silvia Pellegrini, Fabio Manenti, Raniero Chimienti, Rita Nano, Linda Ottoboni, Francesca Ruffini, Gianvito Martino, Philippe Ravassard, Lorenzo Piemonti, and Valeria Sordi in Cell Transplantation
Supplemental Material, CT-2091_Supplementary_Figure_2S for Differentiation of Sendai Virus-Reprogrammed iPSC into β Cells, Compared with Human Pancreatic Islets and Immortalized β Cell Line by Silvia Pellegrini, Fabio Manenti, Raniero Chimienti, Rita Nano, Linda Ottoboni, Francesca Ruffini, Gianvito Martino, Philippe Ravassard, Lorenzo Piemonti, and Valeria Sordi in Cell Transplantation
Supplemental Material, CT-2091_Supplementary_Figure_3Sa for Differentiation of Sendai Virus-Reprogrammed iPSC into β Cells, Compared with Human Pancreatic Islets and Immortalized β Cell Line by Silvia Pellegrini, Fabio Manenti, Raniero Chimienti, Rita Nano, Linda Ottoboni, Francesca Ruffini, Gianvito Martino, Philippe Ravassard, Lorenzo Piemonti, and Valeria Sordi in Cell Transplantation
Supplemental Material, CT-2091_Supplementary_Figure_3Sb for Differentiation of Sendai Virus-Reprogrammed iPSC into β Cells, Compared with Human Pancreatic Islets and Immortalized β Cell Line by Silvia Pellegrini, Fabio Manenti, Raniero Chimienti, Rita Nano, Linda Ottoboni, Francesca Ruffini, Gianvito Martino, Philippe Ravassard, Lorenzo Piemonti, and Valeria Sordi in Cell Transplantation
Supplemental Material, CT-2091_Supplementary_Figure_3Sc for Differentiation of Sendai Virus-Reprogrammed iPSC into β Cells, Compared with Human Pancreatic Islets and Immortalized β Cell Line by Silvia Pellegrini, Fabio Manenti, Raniero Chimienti, Rita Nano, Linda Ottoboni, Francesca Ruffini, Gianvito Martino, Philippe Ravassard, Lorenzo Piemonti, and Valeria Sordi in Cell Transplantation