Abstract
Purpose
Multiple myeloma (MM) is a clonal bone marrow disease characterized by the neoplastic transformation of differentiated postgerminal B cells. It is a heterogeneous disease both at the genetic level and in terms of clinical outcome. Immunoglobulin M (IgM) MM is a rare subtype of myeloma. Similar to Waldenström macroglobulinemia (WM), patients with MM experience IgM monoclonal gammopathy; however, both diseases are distinct in terms of treatment and clinical behavior.
Materials and Methods
To shed light on the presentation of IgM MM, its prognosis, and its gene expression profiling, we identified and characterized 21 patients with IgM MM from our database.
Results
One of these patients presented with a rare IgM monoclonal gammopathy of undetermined significance that progressed to smoldering myeloma. The median survival of the 21 patients was 4.9 years, which was comparable to a matched group of patients with non-IgM MM with similar myeloma prognostic factors (age, gender, albumin, creatinine, anemia, lactate dehydrogenase, β2-microglobulin, cytogenetics abnormalities), but much less than the median survival reported for patients with WM (9 years). We identified a cluster of genes that differ in their expression profile between MM and WM and found that the patients with IgM MM displayed a gene expression profile most similar to patients with non-IgM MM, confirming that IgM MM is a subtype of MM that should be differentiated from WM.
Conclusion
Because the prognosis of IgM MM and WM differ significantly, an accurate diagnosis is essential. Our gene expression model can assist with the differential diagnosis in controversial cases.
INTRODUCTION
Multiple myeloma (MM) is considered a malignancy of postgerminal center long-lived plasma cells. Although rare, T-cell–independent antigen stimulation can occur in these patients, and when it does, it results in the production of immunoglobulin M (IgM)–secreting short-lived plasma cells and lymphoplasmacytes. IgM MM is an infrequent subtype of MM, with an estimated prevalence of 0.5%.1 Because of its rarity and similarity to Waldenström macroglobulinemia (WM), little is known about its clinical characteristics, genetics, and prognosis compared with WM and other MM subtypes.
The presence of serum monoclonal IgM and uncontrolled plasma cell proliferation is associated with both IgM MM and WM. In most cases, it is not difficult to clinically differentiate between these two diseases. WM is suspected in patients with lymphadenopathy, hepatosplenomegaly, hyperviscosity syndrome, and the presence of a monoclonal IgM in the serum.2 A panel of B-cell antigens (CD19, CD20, CD21, CD22, and CD24) is present on the WM cells, whereas the CD23 antigen is usually absent.3 The concentration of IgM varies widely in WM, and it is not possible to define a concentration that reliably distinguishes WM from other lymphoproliferative disorders.4 Alternatively, IgM MM is suspected in the presence of osteolytic lesions, bone pain, monoclonal protein in blood or urine, or immune paresis, and it is confirmed with a bone marrow biopsy along with the other established International Myeloma Working Group (IMWG) criteria.5 Patients with IgM MM tend to have plasmacytic differentiation with high expression of CD138 and cytoplasmic immunoglobulin.6 However, when patients present with IgM monoclonal gammopathy, osteolytic lesions, and lymphadenopathies or hepatosplenomegaly, untangling the MM-WM differential diagnosis can be challenging.5
One of the characteristics of MM is the rearrangement of its immunoglobulin heavy and light chain genes. The high load of somatic hypermutations in the immunoglobulin heavy chain (IgH) locus is consistent with its postgerminal antigen-driven B-cell origin.7 There are five main translocation chromosomes in MM, which seem to be mediated mostly by errors in the IgH switch recombination that occurs during B-cell maturation in germinal centers.8 Those translocations are t(4;14), t(6;14), t(11;14), t(14;16), and t(14;20), and they result in the overexpression of MMSET and FGFR3,9 CCND3,10 CCND1,9,11 MAF,12 and MAFB, respectively.13 Those aberrant chromosomal translocations are one of the central molecular hallmarks of MM. Thus, translocations involving the 14q32 region, for example, might represent clear-cut differences between MM and WM, and may be of diagnostic value in difficult patients.14
MATERIALS AND METHODS
Patients
Twenty-one patients diagnosed with IgM MM between 1993 and 2013 were identified in the University of Arkansas for Medical Sciences Myeloma Institute for Research and Therapy (MIRT) patient database. Patient data collected for this study included overall survival (OS), bone disease status (as defined by x-rays, positron emission tomography [PET] scans, and magnetic resonance imaging scans), gene expression profiles (GEPs), and laboratory values (hemoglobin, calcium, and creatinine).
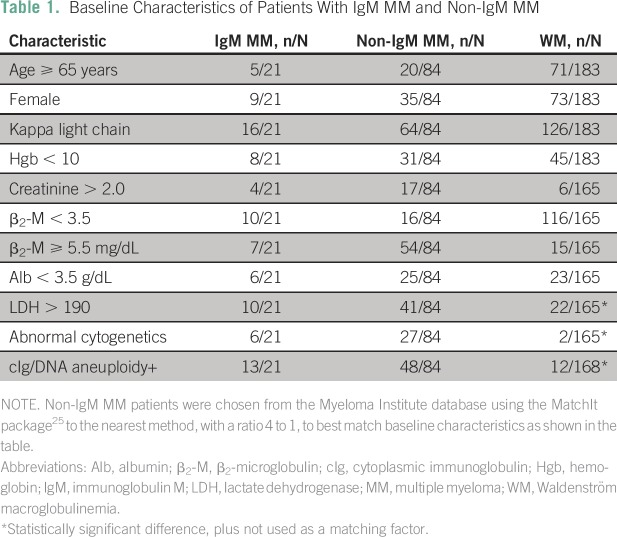
To determine the potential impact of possessing the IgM MM subtype on the prognosis of these patients, the survival of 21 patients with IgM MM was retrospectively compared with a historical control group of 158 patients with WM seen by MIRT15 and a group of 84 patients with non-IgM MM matched by important prognostic clinical factors: age,16 albumin and β2-microglobulin,17 creatinine,18 light chain type, serum lactate dehydrogenase,19 and abnormal cytogenetics.20 All patients were treated around the same time at MIRT (Table 1).
Table 1.
Baseline Characteristics of Patients With IgM MM and Non-IgM MM
A diagnosis of IgM MM was based on the morphologic and immunophenotypical findings of biopsy specimens, the presence of a monoclonal IgM, and the presence of typical clinical characteristics of MM (lytic bone lesions, hypercalcemia, renal failure), using IMWG criteria.5 In a similar fashion, the diagnosis of WM was based on the morphologic and immunophenotypical findings of biopsy specimens, the presence of a monoclonal IgM, and the presence of typical clinical characteristics of WM (hepatosplenomegaly, lymphadenopathy). However, the presence of bone disease per se was not exclusive to the MM diagnosis because verified patients with WM have been described with pathologically confirmed bone involvement.21-23
Statistical Analysis
Survival curves were estimated using the Kaplan-Meier method and compared using the log-rank test. Matching between IgM and non-IgM MM was performed using R software24 and the MatchIT package25 using “to the nearest” method, with a 1 to 4 ratio without replacement, taking into consideration the baseline characteristics listed in Table 1.
Patients with IgM MM, non-IgM MM, and WM with available GEPs from MIRT were compared. To find gene expressions that could best distinguish between WM and non-IgM MM diagnoses, we selected the top 1,000 probe sets that maximized their gene-specific ratio between these two groups to within-groups sums of squares on the basis of the method by Dudoit et al.26 Next, we performed unsupervised hierarchical clustering on all samples, including patients with IgM MM using the top 1,000 probe sets.
For better illustration of the results, we reduced the dimensionality of the top 1,000 probe sets in WM and non-IgM MM groups to three dimensions using principal component analysis27 and partial least squares.28 Then, a linear support vector machine model29 was applied on the WM and non-IgM MM groups, and the boundary plane was plotted. Next, the same transformation was applied to IgM MM samples and added to the corresponding three-dimensional plot. The Database for Annotation, Visualization, and Integrated Discovery (DAVID)30 was used for the functional enrichment analysis.
RESULTS
Patient Characteristics and Survival
Of the 21 confirmed patients with IgM MM, 13 presented at MIRT for initial diagnosis, whereas eight were diagnosed and treated elsewhere before seeking care at MIRT. Seven patients presented with each of stages I, II, and III of the international staging system for plasma cell myeloma.31 Osteolytic bone lesions and/or pathologic fractures were evident by x-ray and computed tomography scan in 15 patients. Either magnetic resonance imaging or PET scan detected active bone focal lesions in three patients. Bone lesions were not observed in the remaining three patients. There was no organomegaly evident in patients with an available PET/computed tomography scan at baseline, and only one patient had evidence of hilar and mediastinal lymphadenopathy along with calcified lung nodules. Elevated creatinine levels (> 2.0 mg/dL) were evident in four patients at the time of initial diagnosis. Disease characteristics of these patients and the WM and non-IgM MM control groups are listed in Table 1.
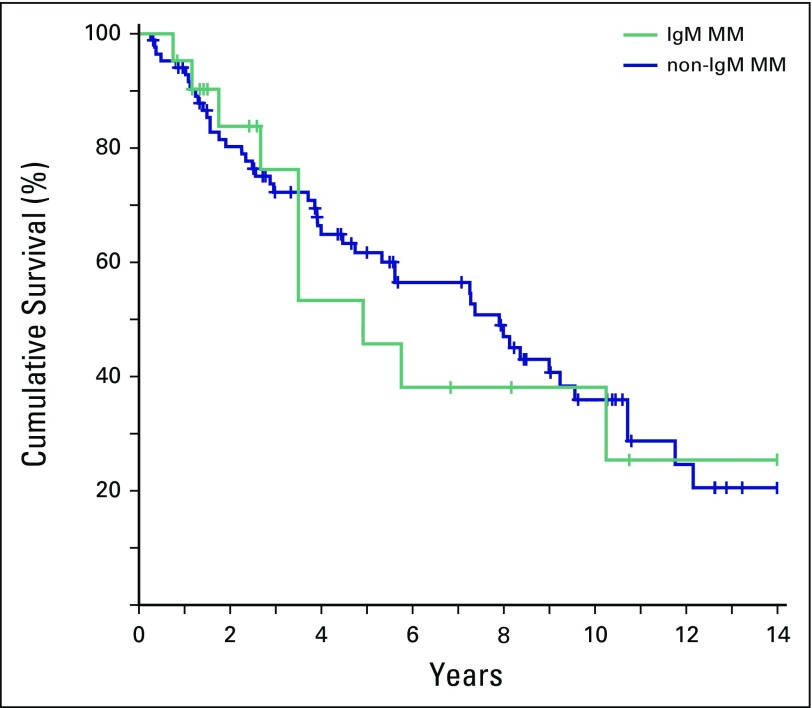
When the OS of the IgM MM group (4.9 years; 95% CI, 3.5 to ∞) was compared with a matched group of 84 patients with non-IgM MM (7.9 years; 95% CI, 5.3 to 10.75), no statistically significant difference was observed (P = .751; Fig 1). Both groups had lower OS than the median OS of the historical group of 158 symptomatic patients with WM (9.2 years).15 As previously reported, the median OS of the WM group remained largely unaffected, even when the subgroup of patients with WM requiring treatment was analyzed (9.0 years).
Fig 1.
Kaplan-Meier estimates of overall survival in immunoglobulin M (IgM) multiple myeloma (MM; n = 21, red line) and a matched cohort of non-IgM MM (n = 84, blue line). No statistical difference was found for overall survival between these two groups, with a log-rank P equal to .751.
Bone Marrow Karyotype, Fluorescence In Situ Hybridization, and DNA Flow Cytometry
In our cohort, baseline bone marrow cytogenetic data were available for 19 of 21 patients. Six of 19 patients had abnormal cytogenetics at presentation. Two of six patients had a t(11;14) translocation, one had t(14;16), and the other three had complex karyotype with hypodiploidy.
Only one patient who had interphase fluorescence in situ hybridization performed on bone marrow aspirate at baseline demonstrated t(11;14) translocation (CCND1/IgH) and a loss of chromosome 13; conventional cytogenetics for this patient was reported as normal.
Aneuploidy by DNA flow cytometry was evident in 13 of 21 patients (62%). This fact is in accordance with the already published data stating the frequency of aneuploidy in MM32 and is in striking antithesis with WM (aneuploidy was detected in 12 of 168 patients with DNA flow cytometry; unpublished data; P < .001)
Gene Expression Profiling
Of 21 patients with IgM MM, 12 had available GEP data on initial diagnosis. One patient had an upregulated CCND2.33 In six of these patients (50%), cyclin D1 gene expression was high. This is consistent with previously published data.34
To discover the features of gene expression that may be unique to IgM MM, we performed an unsupervised hierarchical cluster analyses of GEP of CD138-positive cells from the following samples: bone marrow aspirates of non-IgM MM and bone marrow aspirates of WM. Next, we added in the GEP data from bone marrow aspirates of the 21 patients with IgM MM.
A comparative genomic analysis was performed on the patients with IgM MM, non-IgM MM, and WM with available GEP data at initial diagnosis (12, 60, and 52 patients, respectively). We identified the best 1,000 probe sets (Data Supplement) that distinguish between WM and non-IgM MM. Many of these probes are for membrane proteins, such as CD19, CD20, CD22, CD24, CD138, components of the major histocompatibility complex, and adherence junctions.
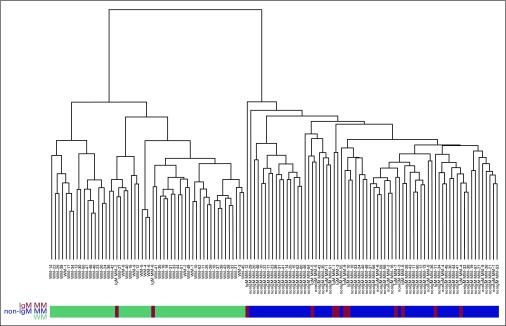
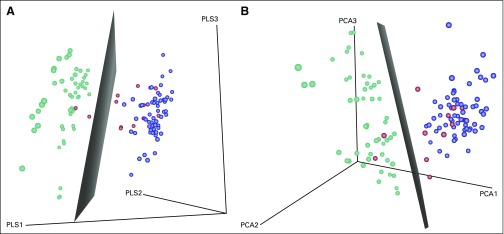
Using support vector machine analysis alone29 or together with principal component analysis27 or partial least squares,28 Figures 2 and 3 demonstrate that the majority of the IgM MM samples are on the non-IgM MM side of the boundary, indicating that the GEP of IGM MM is more closely related to non-IgM MM than it is to WM.
Fig 2.
Complete-linkage clustering analysis of Euclidean distance. Unsupervised hierarchical clustering was performed on all samples, including Waldenström macroglobulinemia (WM; green), immunoglobulin M (IgM) multiple myeloma (MM; red), and non-IgM MM (blue) gene expression profiles, using the 1,000 probesets retrieved in the filtering step.
Fig 3.
Analysis of gene expression profiles (GEPs). (A) Principal components analysis of GEPs from immunoglobulin M (IgM) multiple myeloma (MM; red), non-IgM MM (blue), and patients with Waldenström macroglobulinemia (WM; green). (B) Partial least squares analysis of GEPs from IgM MM, non-IgM MM, and WM patients. Both methods were used to reduce the dimensionality of the top 1,000 probesets in WM and non-IgM MM groups to three dimensions. The same transformation was applied to IgM MM samples. Then, a linear support vector machine model was applied to the WM non-IgM MM groups, and the boundary plane was plotted. This method reflects the same results as the hierarchical clustering analysis (Fig 2), in that the majority of the IgM MM samples (red) are on the non-IgM MM side of the boundary, indicating that the GEP of IGM MM is more closely related to non-IgM MM subtypes than it is to WM.
Clinical Practice Points
MM and WM are plasma cell–related disorders.
WM and IgM myeloma present with elevated IgM levels. However, both disorders are clearly distinct by expression of Myd88 in WM and in terms of molecular profile, clinical presentation, treatment, and prognosis.
In the setting of elevated IgM levels, the presence of bone lytic lesions, hypercalcemia, and renal failure points toward IgM myeloma, whereas the presence of splenomegaly and lymphadenopathy points toward WM.
Our study demonstrates the difference between both diseases at the gene expression profiling level and provides a useful tool when differentiating both diseases is challenging.
DISCUSSION
IgM MM is a discrete clinical entity that should be distinguished from WM because their prognoses and treatments differ greatly. Using one of the largest series of patients with IgM MM ever published, we have shown that patients with IgM MM have a clinical presentation, prognosis, and GEP similar to patients with non-IgM MM.
Bone disease is associated with approximately 79% of newly diagnosed patients with MM when observed with conventional radiologic techniques.1 In our cohort of patients with IgM MM, bone disease was evident in the majority of patients, especially when specialized radiologic techniques were incorporated into the initial work-up.
Organomegaly is one of the clinical findings that is usually associated with WM or plasma cell leukemia.35 Avet-Loiseau et al14 reported two of eight patients with IgM MM with organomegaly; however, our results were consistent with the findings of Schuster et al,36 in that none of our patients presented with organomegaly or plasma cell leukemia.
To determine whether patients with IgM MM have a different prognosis than patients with non-IgM MM, OS for these two groups was compared using the Kaplan-Meier log-rank test. Patients with IgM MM displayed a median OS of 4.9 years, similar to patients with non-IgM MM receiving similar treatments at our institution (P < .05). Patients with WM treated at our institution alternatively reported a longer median OS of 9 years.15
Previous studies16,37,38 have estimated that approximately 14% to 17% of patients with monoclonal gammopathy of undetermined significance (MGUS)–IgM type will develop a group of malignant lymphoid disorders, including non-Hodgkin lymphomas, chronic lymphocytic leukemia, and primary amyloidosis, within an average period of 4 years.
The observation of MGUS progressing to IgM MM is rarely reported. In one of those studies, Kyle et al16 reported an association between IgM MGUS and smoldering IgM MM in one patient of 213, and that patient exhibited biclonal gammopathy (IgM 386 mg/dL plus IgA λ 2840 mg/dL). Similarly, in our study, one of 21 patients presented with MGUS and progressed to IgM MM.
In general, genetic abnormalities are found in one third of patients with MM by conventional cytogenetics.39 Approximately 31% of our patients had cytogenetic abnormalities, three of whom had a t(11;14) or t(14;16) translocation, and both of these translocations are typically found in patients with MM.8
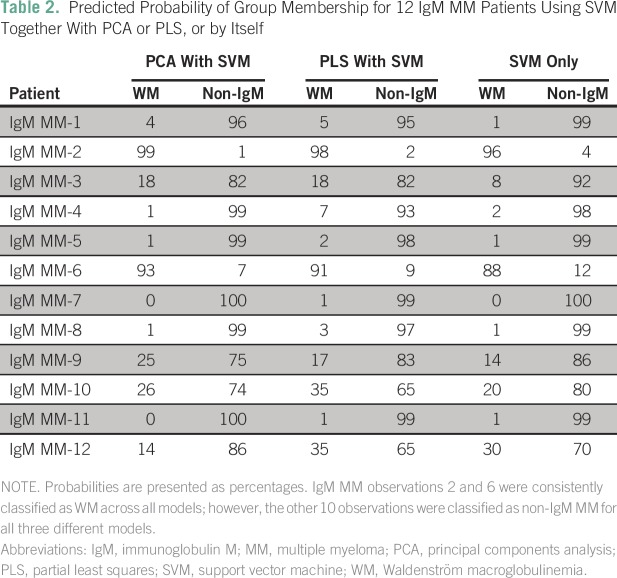
In concordance with the previous pathologic and clinical findings, GEPs for non-IgM MM and patients with WM were separated into two distinct clusters. Using these two groups as references, we added in the GEP data for patients with IgM MM. Clearly, both GEP data for IgM MM and patients with non-IgM MM clustered together, indicating that IgM MM is correctly classified as a subtype of MM, sharing the same genetic and pathologic characteristics. Only two of the 12 IgM MM samples (IgM MM-2 and IgM MM-6; Table 2) were located in the WM cluster (Fig 2), whereas the remaining 10 IgM MM samples were located in the non-IgM MM cluster.
Table 2.
Predicted Probability of Group Membership for 12 IgM MM Patients Using SVM Together With PCA or PLS, or by Itself
Although these two patients fulfilled the newest IMWG diagnostic criteria for MM,5 it should be noted that both patients did not clinically resemble typical MM. Both patients had mild to moderate plasma cell infiltration with absent or borderline immunoparesis and absent bone focal lesions by any imaging technique. Furthermore, in one of these two patients, renal insufficiency was due to light chain deposition disease without any evidence of cast nephropathy. In the other patient, bone disease was due to profound osteoporosis and associated pathologic fractures without any evidence of direct plasma cell involvement in pathology or imaging.
In conclusion, patients with IgM MM share clinical and genetic characteristics with the other subtypes of MM and have distinct differences from WM clinically, genetically, and in terms of prognosis. The IgM subtype of MM per se does not affect prognosis. All MM subtypes are affected by the established prognostic factors of MM, but the IgM subtype alone does not confer any additional prognostic indicator. In view of the remarkable differences in both treatment and prognosis between IgM MM and WM, an accurate diagnosis is essential and should be obtained with all available clinical, pathologic, and genetic assays.
AUTHOR CONTRIBUTIONS
Conception and design: Shebli Atrash, Xenofon Papanikolaou, Bart Barlogie
Administrative support: Bart Barlogie
Provision of study materials or patients: Xenofon Papanikolaou
Collection and assembly of data: Shebli Atrash, Al-Ola Abdallah
Data analysis and interpretation: Shebli Atrash, Qing Zhang, Caleb Stein
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Shebli Atrash
No relationship to disclose
Qing Zhang
Patents, Royalties, Other Intellectual Property: Patent applications related to use of gene expression profiles in cancer medicine
Xenofon Papanikolaou
No relationship to disclose
Caleb Stein
No relationship to disclose
Al-Ola Abdallah
No relationship to disclose
Bart Barlogie
Consulting or Advisory Role: Celgene, Genzyme
Speakers' Bureau: Celgene, Millennium
Research Funding: Celgene, Novartis
Patents, Royalties, Other Intellectual Property: Patent applications related to use of gene expression profiles in cancer medicine
REFERENCES
- 1.Kyle RA, Gertz MA, Witzig TE, et al. Review of 1027 patients with newly diagnosed multiple myeloma. Mayo Clin Proc. 2003;78:21–33. doi: 10.4065/78.1.21. [DOI] [PubMed] [Google Scholar]
- 2.Dimopoulos MA, Panayiotidis P, Moulopoulos LA, et al. Waldenström’s macroglobulinemia: Clinical features, complications, and management. J Clin Oncol. 2000;18:214–226. doi: 10.1200/JCO.2000.18.1.214. [DOI] [PubMed] [Google Scholar]
- 3.Konoplev S, Medeiros LJ, Bueso-Ramos CE, et al. Immunophenotypic profile of lymphoplasmacytic lymphoma/Waldenström macroglobulinemia. Am J Clin Pathol. 2005;124:414–420. doi: 10.1309/3G1X-DX0D-VHBN-VKB4. [DOI] [PubMed] [Google Scholar]
- 4.Owen RG, Treon SP, Al-Katib A, et al. Clinicopathological definition of Waldenstrom’s macroglobulinemia: Consensus panel recommendations from the Second International Workshop on Waldenstrom’s macroglobulinemia. Semin Oncol. 2003;30:110–115. doi: 10.1053/sonc.2003.50082. [DOI] [PubMed] [Google Scholar]
- 5. Swerdlow SH, Campo E, Harris NL, et al (eds): WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (ed 4). Geneva, Switzerland, WHO-OMS, 2008. [Google Scholar]
- 6.King RL, Howard MT, Hodnefield JM, et al. IgM multiple myeloma: Pathologic evaluation of a rare entity. Am J Clin Pathol. 2013;140:519–524. doi: 10.1309/AJCP0N7IELYUNJGZ. [DOI] [PubMed] [Google Scholar]
- 7.Bakkus MH, Heirman C, Van Riet I, et al. Evidence that multiple myeloma Ig heavy chain VDJ genes contain somatic mutations but show no intraclonal variation. Blood. 1992;80:2326–2335. [PubMed] [Google Scholar]
- 8.Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001;20:5611–5622. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- 9.Bergsagel PL, Kuehl WM, Zhan F, et al. Cyclin D dysregulation: An early and unifying pathogenic event in multiple myeloma. Blood. 2005;106:296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaughnessy J, Jr, Gabrea A, Qi Y, et al. Cyclin D3 at 6p21 is dysregulated by recurrent chromosomal translocations to immunoglobulin loci in multiple myeloma. Blood. 2001;98:217–223. doi: 10.1182/blood.v98.1.217. [DOI] [PubMed] [Google Scholar]
- 11.Chesi M, Bergsagel PL, Brents LA, et al. Dysregulation of cyclin D1 by translocation into an IgH gamma switch region in two multiple myeloma cell lines. Blood. 1996;88:674–681. [PubMed] [Google Scholar]
- 12.Hurt EM, Wiestner A, Rosenwald A, et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell. 2004;5:191–199. doi: 10.1016/s1535-6108(04)00019-4. [DOI] [PubMed] [Google Scholar]
- 13.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 14.Avet-Loiseau H, Garand R, Lodé L, et al. 14q32 Translocations discriminate IgM multiple myeloma from Waldenstrom’s macroglobulinemia. Semin Oncol. 2003;30:153–155. doi: 10.1053/sonc.2003.50053. [DOI] [PubMed] [Google Scholar]
- 15.Usmani S, Sexton R, Crowley J, et al. Autologous stem cell transplantation as a care option in Waldenström’s macroglobulinemia. Clin Lymphoma Myeloma Leuk. 2011;11:139–142. doi: 10.3816/CLML.2011.n.032. [DOI] [PubMed] [Google Scholar]
- 16.Kyle RA, Therneau TM, Rajkumar SV, et al. Long-term follow-up of IgM monoclonal gammopathy of undetermined significance. Blood. 2003;102:3759–3764. doi: 10.1182/blood-2003-03-0801. [DOI] [PubMed] [Google Scholar]
- 17.Greipp PR, San Miguel J, Durie BG, et al. International staging system for multiple myeloma. J Clin Oncol. 2005;23:3412–3420. doi: 10.1200/JCO.2005.04.242. [DOI] [PubMed] [Google Scholar]
- 18. doi: 10.1038/leu.2015.15. Khan R, Apewokin S, Grazziutti M, et al: Renal insufficiency retains adverse prognostic implications despite renal function improvement following Total Therapy for newly diagnosed multiple myeloma. Leukemia 29:1195-1201, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terpos E, Katodritou E, Roussou M, et al. High serum lactate dehydrogenase adds prognostic value to the international myeloma staging system even in the era of novel agents. Eur J Haematol. 2010;85:114–119. doi: 10.1111/j.1600-0609.2010.01466.x. [DOI] [PubMed] [Google Scholar]
- 20.Avet-Loiseau H. Role of genetics in prognostication in myeloma. Best Pract Res Clin Haematol. 2007;20:625–635. doi: 10.1016/j.beha.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Krausz Y, Zlotnick A. Macroglobulinemia of Waldenström associated with severe osteolytic lesions. Acta Haematol. 1977;58:307–311. doi: 10.1159/000207842. [DOI] [PubMed] [Google Scholar]
- 22.Berman HH. Waldenstroöm’s macroglobulinemia with lytic osseous lesions and plasma-cell morphology. Report of a case. Am J Clin Pathol. 1975;63:397–402. doi: 10.1093/ajcp/63.3.397. [DOI] [PubMed] [Google Scholar]
- 23.Mehmood K, Naqvi IH, Shah SR, et al. Waldenstroms macroglobulinemia patient presenting with rare ‘lytic’ lesions and hypercalcemia: A diagnostic dilemma. J Clin Diagn Res. 2014;8:FD10–FD11. doi: 10.7860/JCDR/2014/8184.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. The R Foundation: The R Project for Statistical Computing. http://www.R-project.org/
- 25.Ho DE, Imai K, King G, et al. MatchIt : Nonparametric preprocessing for parametric causal inference. J Stat Softw. 2011;42:1–28. [Google Scholar]
- 26.Dudoit S, Fridlyand J, Speed TP. Comparison of discrimination methods for the classification of tumors using gene expression data. J Am Stat Assoc. 2002;97:77–87. [Google Scholar]
- 27.
- 28.ter Braak CJF, de Jong S. The objective function of partial least squares regression. J Chemometr. 1998;12:41–54. [Google Scholar]
- 29.10.1145/1961189.1961199. [DOI]
- 30.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Durie BG, Salmon SE. A clinical staging system for multiple myeloma. Correlation of measured myeloma cell mass with presenting clinical features, response to treatment, and survival. Cancer. 1975;36:842–854. doi: 10.1002/1097-0142(197509)36:3<842::aid-cncr2820360303>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 32.Barlogie B, Alexanian R, Pershouse M, et al. Cytoplasmic immunoglobulin content in multiple myeloma. J Clin Invest. 1985;76:765–769. doi: 10.1172/JCI112033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian E, Sawyer JR, Heuck CJ, et al. In multiple myeloma, 14q32 translocations are nonrandom chromosomal fusions driving high expression levels of the respective partner genes. Genes Chromosomes Cancer. 2014;53:549–557. doi: 10.1002/gcc.22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Troussard X, Avet-Loiseau H, Macro M, et al. Cyclin D1 expression in patients with multiple myeloma. Hematol J. 2000;1:181–185. doi: 10.1038/sj.thj.6200025. [DOI] [PubMed] [Google Scholar]
- 35.Fernández de Larrea C, Kyle RA, Durie BGM, et al. Plasma cell leukemia: Consensus statement on diagnostic requirements, response criteria and treatment recommendations by the International Myeloma Working Group. Leukemia. 2013;27:780–791. doi: 10.1038/leu.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuster SR, Rajkumar SV, Dispenzieri A, et al. IgM multiple myeloma: Disease definition, prognosis, and differentiation from Waldenstrom’s macroglobulinemia. Am J Hematol. 2010;85:853–855. doi: 10.1002/ajh.21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kyle RA, Garton JP. The spectrum of IgM monoclonal gammopathy in 430 cases. Mayo Clin Proc. 1987;62:719–731. doi: 10.1016/s0025-6196(12)65225-2. [DOI] [PubMed] [Google Scholar]
- 38.Tsai H-T, Caporaso NE, Kyle RA, et al. Evidence of serum immunoglobulin abnormalities up to 9.8 years before diagnosis of chronic lymphocytic leukemia: A prospective study. Blood. 2009;114:4928–4932. doi: 10.1182/blood-2009-08-237651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawyer JR, Waldron JA, Jagannath S, et al. Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genet Cytogenet. 1995;82:41–49. doi: 10.1016/0165-4608(94)00284-i. [DOI] [PubMed] [Google Scholar]