Abstract
The sterol metabolome of Acanthamoeba castellanii (Ac) yielded 25 sterols. Substrate screening of cloned AcCYP51 revealed obtusifoliol as the natural substrate which converts to Δ8,14-sterol (<95%). The combination of [2H3-methyl]methionine incubation to intact cultures showing C28-ergosterol incorporates 2-2H atoms and C29-7-dehydroporiferasterol incorporates 5 2H-atoms, the natural distribution of sterols, CYP51 and previously published sterol methyltransferase (SMT) data indicate separate Δ24(28) - and Δ25(27) -olefin pathways to C28- and C29- sterol products from the protosterol cycloartenol. In cell-based culture, we observed a marked change in sterol compositions during the growth and encystment phases monitored microscopically and by trypan blue staining; trophozoites possess C28/C29-Δ5,7-sterols, viable encysted cells (mature cyst) possess mostly C29-Δ5-sterol and non-viable encysted cells possess C28/C29-Δ5,7-sterols that turnover variably from stress to 6-methyl aromatic sterols associated with changed membrane fluidity affording lysis. An incompatible fit of steroidal aromatics in membranes was confirmed using the yeast sterol auxotroph GL7. Only viable cysts, including those treated with inhibitor, can excyst into trophozoites. 25-Azacycloartanol or voriconazole that target SMT and CYP51, respectively, are potent enzyme inhibitors in the nanomolar range against the cloned enzymes and amoeba cells. At minimum amoebicidal concentration of inhibitor amoeboid cells rapidly convert to encysted cells unable to excyst. The correlation between stage-specific sterol compositions and the physiological effects of ergosterol biosynthesis inhibitors suggests that amoeba fitness is controlled mainly by developmentally-regulated changes in the phytosterol B-ring; paired interference in the Δ5,7-sterol biosynthesis (to Δ5,7) - metabolism (to Δ5 or 6-methyl aromatic) congruence during cell proliferation and encystment could be a source of therapeutic intervention for Acanthamoeba infections
Keywords: Acanthamoeba castellanii, sterol methyltransferase, sterol evolution, aromatic sterols, trophozoite, encystment, ergosterol biosynthesis inhibitor
1. Introduction
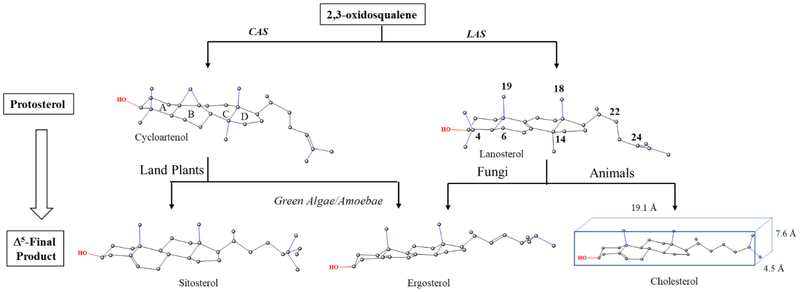
Sterol synthesis is a basic metabolic pathway of eukaryotes giving rise to essential membrane components. The biosynthesis pathway is typically divided into three routes, two of which characterize the phytosterol biosynthesis pathways in land plants and fungi yielding C29-sitosterol (24α-ethyl group) and C28-ergosterol (24β-methyl group), respectively, and in animals yielding C27-cholesterol (Fig. 1) [1–4]. These contemporary biosynthetic pathways retain ancestral features along with novelties specific to their particular lineages. Notably, the protosterol product of the 2,3-oxidosqualene synthase yielding cycloartenol or lanosterol associates phylogenetically to plant-animal divisions while the substrate acceptability and mechanisms of individual enzymes catalyzing completion of the pathway to Δ5-sterols reveal relatedness or divergence in catalytic competence across kingdoms [5, 6]. For phytosterols generating diversity, the size, location and direction (α- or β-orientation) of the side chain C24-alkyl group correlate to plant-fungal divisions or to more- or less-advanced in descent. Structural features common to sterols are a cyclopentanoperhydrophenanthrene ring system, polar C3-OH group and intact side chain of 8 to 10-carbon atoms. These characteristics contribute positively to the molecule’s overall flat shape, length and amphipathic character relevant to the architectural suitability of sterols in membranes [7]. Alternatively, there can be harmful features in intermediates that are removed during the normal course of metabolism. For example, the C4-geminal methyl groups and C14-methyl group on the protosterol, eliminated to yield Δ5-sterols, are known to affect the hydrogen bonding strength of the C3-OH group and planarity of the α-(back) face of the sterol nucleus, respectively; when intermediates that possess these features accumulate in the cell the result can be detrimental to sterol homeostasis that thereby, interferes with growth and maturation [8–10].
Figure 1.
Sterol biosynthesis diversity in eukaryote organisms
Despite the prevailing hypothesis suggesting progressive structural modifications in the protosterol formed by prokaryotes parallels a gradual cholesterol evolution in response to increases in the atmospheric concentration of oxygen [11–13] recent chemical analysis on the origin of these compounds indicate a complete sterol biosynthesis to Δ5-products may have existed in the most primitive eukaryotes, evidenced in the presence of cholestane and its C28- to C30-carbon homologs in molecular fossils found in rocks and formations between 750 Mya to 1,000 Mya [14]. Additionally, phylogenomic analysis shows the genes of 13 sterolic enzymes yielding these steranes could be codified in the genomic organization of the last eukaryotic common ancestor (LECA) approximately 2.45–2.32 Mya [15, 16] with orthologous prokaryotic enzymes likely formed during the Great Oxidation Event by horizontal gene transfers [17, 18]. Based on this new information, we surmise competing phylogenies constructed from these characters could reasonably describe the punctuated and recurrent evolution of sterol biosynthesis to amoeba ergosterol deeply rooted in primary metabolism of the LECA [19, 20]. However, relevant to the understanding of sterol biosynthesis in Acanthamoeba for the purpose of inhibition, the phyla-specific canonical routes to alternate sterol products must also be considered, because different steps in sterol metabolism – sterol side chain or nucleus revisions-could interface uniquely through stage-specific structural adjustment during the amoeba life history, as reported can occur in land plants [21, 22] to yield ergosterol convergence (Fig. 1).
Acanthamoeba spp. has a number of particularly advantageous characteristics for research on sterol function and evolution. In the first place, the differentiation process originating in the trophozoite and passing through a pseudocyst, i.e., an intermediate structure composed of a wall that may not protect against adverse conditions [23, 24], can yield multiple cyst types, including 1) viable cysts with a resistant double-layered outer wrinkled ectocyst and inner endocyst and 2) a mixture of non-viable cysts of apoptotic cells that possess a fragile cell wall and dead cells generated from autophagocytosis [25]. These studies further show azolic inhibitors-voriconazole can prevent trophozoite growth as well as activate an encystment pathway with a final trajectory ending in programed cell death [26]. Ergosterol biosynthesis inhibitors with various structures can therefore be administered and the effects on growth and differentiation measured in correlation to changes in sterol biosynthesis. Intriguingly, the unikont Acanthamoeba is distinctly different from fungi and bikont protozoa (kinetoplastids) in their sterol biosynthesis capabilities. The variant pathways for C28-ergosterol biosynthesis in parasitic protozoa also differ from the C27-cholesterol biosynthesis in the human host where sterol C24-methylation is lost [27].
The ability of Acathamoeba spp to synthesize a plant-based pathway of cycloartenol to Δ5- and Δ5,7-24β-C28- and C29-alkyl sterols was discovered in the early 1980’s by Raedorstorff and Rohmer [28, 29]. These researchers together with the Korn group demonstrated the amoeba ergosterol and 7-dehydroporiferasterol can convert to uncommon C28 - and C29-phenanthrene 6-methyl aromatic sterols [30, 31]. The Acanthamoeba system has been exploited in the collaborative laboratories of Kelly and Nes [32, 33] and more recently by Roberts and coworkers [34]. However, unlike the Rohmer and Korn groups, the more recent authors [34] have concluded that Acanthamoeba operates a fungal-based pathway of lanosterol to ergosterol and the protozoan is unable to synthesize C29-sterols. Our own preliminary work corroborated the original findings, and the purpose of the present paper is to document this in detail and to expand the study to encompass a more comprehensive evaluation of the biosynthetic enzymes and steroidal nuggets expressed variably at stage-specific times in Acanthamoeba. The approach used in our investigation has been chosen to validate the ergosterol biosynthesis pathway from cycloartenol and the existence of a new Δ25(27) -C29-sterol biosynthesis pathway in amoeba that recapitulates the green algal sterol biosynthesis pathway to C29-sterols.
Another reason for choosing Acanthamoeba was the critical factors in target deconvolution using sterolomics, bioinformatics and genetic technologies that led to enzyme-based strategies for manipulating the ergosterol biosynthesis pathway in this and related parasitic protozoa. These various investigations provide new chemical starting points against our target enzymes- the Acanthamoeba castellanii (Ac) sterol C14-demethylase (CYP51) and sterol C24/C28-methyltransferases (SMT). Inhibition of either enzyme by a rationally designed tight binding inhibitor rapidly yields trophozoite death [33]. The only real uncertainties left about the importance of phytosterol biosynthesis/metabolism in Acanthamoeba are: (i) can the sterol composition change during the amoeba life cycle, (ii) has phylogenetics influenced formation of separate routes to C28- and C29-sterols, (iii) to what extent is ergosterol biosynthesis during the Acanthamoeba encystment-excystment cycle vulnerable to inhibitors and (iv) what role is played in Acanthamoeba physiology by changing the sterol B-ring structure from Δ5,7 - to Δ5 - or 6-methyl aromatic group?
In this study, the pattern of sterol biosynthesis and accumulation in Acanthamoeba against ultrastructure and viability of the cells was determined at various stages of amoeba proliferation and encystment. In addition, we examined the physiological role of the uncommon 6-methyl aromatic sterol structures generated by the amobea through use of enzyme inhibitors and the yeast sterol auxotroph GL7. The results define the biosynthetic factors that control the changes in sterol compositions associated with amoeba differentiation, and provide the necessary foundation for further regulatory studies at the biochemical and molecular levels. Given the therapeutic importance for CYP51 and SMT as chokepoint enzymes in Acanthamoeba and other parasitic protozoa, knowledge of the diversity and function of terminal C28- and C29-sterols in the trophozoite and encysted cell can be of value in developing new inhibitor chemotypes to treat amoeba diseases.
2. Materials and methods
2.1. Strain and culture conditions.
Acanthamoeba castellanii strain ATCC 30010 was inoculated into tissue culture 25 ml T-flasks prepared with ATTC media 712. Cells were cultured axenically in 5 ml static culture at 25 °C with occasional hand shaking to keep cells in suspension. Continuous cultures were maintained by subculture of 4–5 day growth arrested cells into fresh medium. Growth experiments to establish the growth curve and for sterol analyses were typically carried out using 1 × 104 trophozoites (< 90%) or cysts derived by the methods below. The number of individual amoeba distinct from encysted cells that possess a double wall per ml was determined microscopically. For cell aggregates a best effort was made to count the number of individual encysted cells in the clump. Morphological changes and encystation ratios were calculated by counting trophozoite to cysts using a Neubauer hemocytometer under an Olympus CH-2 compound light microscope and in separate observations cell differentiation was detected with an inverted microscope monitored daily for 2 weeks. Cells were pelleted by centrifugation for 5 min at 5,000 g. Cell viability was checked by trypan blue exclusion method.
2.2. Induction of encystment.
To generate viable cysts capable of excystment, equivalent to resting or dormant cysts, three different approaches were employed. Method-1, using wild-type trophozoites, encystment was induced naturally by allowing the early growth arrested cultures (ca. 4–5 day synchronous cultures) to deplete essential growth nutrients required in metabolite biosynthesis. In a second approach encystment was forced by two replacement methods. Method-2 is salt- induced encystment and was used by Korn et al. and Mehdi and Garg [24, 30, 35]. Briefly, trophozoites were resuspended in medium containing 1 mM MgCl2 to a cell concentration of approximately 6.5 × 106 cells/ml. The culture was incubated in a 30 °C shaker at 150 rpm for 2 days. The cells were collected and treated with 1% sodium dodecyl sulfate (SDS) solution for 30 min at room temperature to remove trophozoites, fragile encysted cells and cell debris. The resulting SDS-resistant cysts were washed with PAGE’s saline solution 5 times and analyzed for sterols. Method 3 is starvation-induced encystment where the amoebae are maintained on an agar-embedment system for 2–4 weeks [23]. Encystment synchrony was evaluated morphologically using a combination of inverted microscopy, light microscopy and electron microscopy. More than 90–95% of the cells formed from these conditions yielded encysted cells with features representative of resting or dormant cysts. Resting cysts generated by the natural and replacement methods were determined viable by their Δ5-sterol profile, and in each case yielded similar excystment activities of about 100%. Typically, the viable cysts at the end of these experiments appeared as single cells in light microscopy either on the solid or in liquid medium. A third approach was used to promote encystment by administering drugs to the medium.
2.3. Trophozoite toxicity assay.
To determine the effect of the test compounds on the growth of Acanthamoeba, trophozoites, approximately 1 × 105 cells, were inoculated into 24 well (4 × 6) microtiter plates of 3 ml final volume taking the inoculum from cells grown in the T-flask continuous culture system. Stock inhibitor concentrations were prepared in dimethylsulfoxide (not to exceed 1% solvent) and as relevant serially diluted to final concentrations in wells that ranged from 16 nM to 30 μM (approximate to the end point for inhibitor studies administered to human epithelial kidney cells [32, 33]. Control wells containing medium and 1% DMSO solvent had no effect on the cell growth. The microliter plates were incubated at 25-°C for 48 hr and the resulting number of trophozoites to cysts counted at the end of the incubation using the hemocytometer. The percentage of viable tophozoites following to different concentrations of inhibitors was determined by the trypan blue exclusion method. The 50% effective does (IC50) value of inhibitors against tophozoite growth under the stated assay conditions was determined by linear extrapolation using GraphPad Prism (GraphPad Software Inc., CA). Cells stained blue were considered non-viable while live cells were unstained. These experiments were carried out in triplicate and at different time points. The drug concentration responsible for the minimum ameobacidal activity (MAC) was defined as the lowest concentration of the inhibitor with no visible live trophozoites, as determined by visual inspection of the treated cultures using the light microscope following trypan blue staining and inverted microscope which confirmed cell death by the absence of trophozoite cells and notable accumulation of cell debris. Observations were performed in triplicate × 2 using light microscope for counting cells. There was excellent agreement (≤10% variation for any data point) in the growth response to inhibitors in the two independent trials.
2.4. Time-dependent killing.
A log-phase culture of Trophozoites was seeded into 24-well plates as described above. Cells were then challenged with inhibitors at previously determined MAC concentrations. At 12 hr intervals, aliquots were removed and counted and cell viability determined; at 48 hr remaining cells were pelleted and resuspended into fresh medium and allowed to incubate for another 5 days and the cells counted to determine trophozoite proliferation and analyzed for sterol content. Experiments were performed with biological replicates. A metabolic marker was used to assess viable cells by means of GC-MS analysis. Viable trophozioites were associated with the production of ergosterol/7-dehydroporiferaterol and viable cysts were associated with poriferasterol production and cells in the death mode were associated with steroidal aromatics as discussed in the text.
2.5. Cysticidal assay.
Cysts, generated by the methods above, or derived from inhibitor treatment, were incubated with the anti-amoeba drug under the same assay conditions described for ameobacidal assay. Growth was defined as the presence of excysted trophozoites found growing in 24 well plates or 5 ml T-flasks after 5 days of growth.
2.6. Chemical and instrumental analysis.
The source of sterol substrates and steroidal inhibitors evaluated in this study is described in earlier papers [8, 20, 36]. Voriconazole was purchased from Sigma. All sterols were purified by HPLC to < 95% by GC analysis. S-adenosyl-L-methionine (SAM) chloride salt was purchased from Sigma, [methyl-3H3]SAM (specific activity 10–15 Ci/mMol, and diluted to 10 μCi/μMol, tetraosylate (methyl-2H3]SAM (99% atom enrichment) purchased from S/D/N isotopes (Pointe-Claire, QC), L-[methyl-2H3]methionine (Sigma). The Bradford protein assay kit was purchased from Bio-Rad and isopropyl-1-thio-β-D-galactoside (IPTG) purchased from Research Products International Corp. DNA synthesis inhibitor 5-fluor-2-deoxyuridine (FUdR) was purchased from MP Biomedicals LLC. All other reagents and chemical were from Sigma or Fisher unless otherwise noted.
Instrumental methods have been reported previously [19, 20]. Briefly, 1H-NMR spectra were recorded in CDL3 at ambient temperature using a Varian Unity Inova 500 MHz spectrometer; chemical shifts (δ, ppm) are referenced to chloroform, (δ, 7.26 ppm). Mass spectra were obtained on a Hewlett-Packard 6890 GC-HP 5973 MSD instrument (electron impact, 70eV, scan range 50–550 amu). HPLC was carried out at room temperature using a Phenomenex Luna C18-column (250 mm × 4.6. mm × 5 μM) connected to a diode array multiple wavelength diode array detector with methanol as eluent. Capillary GC (0.25 mm i.d, by 30 m fused silica column coated with Zebron ZB-5 from Phenomenex) was operated at a flow rate of He set at 1.2 ml/min, injector port at 250 0C, and temperature program of initial 170 °C, held for 1 min, and increased at 20 °C/min to 280 °C. GC analysis of sterols is reported as RRTc values referring to the retention time of sample GC peak to retention time of cholesterol peak which moved typically in the chromatogram at 13.8 min or slightly different to 14.5 min depending on whether the column tip was clipped due to age issues. In HPLC samples were separated on a Phenomenex Luna C18-column (250 mm × 4.6 mm × 5 cm) connected to a diode array multiple detector with samples eluted with methanol; the αc of the sterol is the elution time of compound relative to the elution time of cholesterol, which is 20.6 min. In sterol analysis, product distributions were determined by approximate integration of chromatographic peaks, experiments were performed in biological replicates with excellent agreement between trials (>95%). Thin layer chromatography was performed on 10 × 20 cm, 250 μM silica plates (Baker) developed twice in benzene-ether (85/15, v/v).
2.7. Cell metabolite identification.
Typically, small-scale amoeba cultures were harvested different points along the growth curve in the presence and absence of steroidal inhibitor and azole at MAC concentrations of the compound. Control or treated cultures in 5 ml or 10 ml medium dispersed in 25 ml T-flasks were inoculated with trophozoites (90%) or resting cysts (100%) and cultured for the desired time without shaking and sterol analyzed at the end of the incubation period. Independently, trophozoites were incubated in the 24-well plate system containing 1 ml total medium and after 48 h treatment the sterol composition determined by GCMS analysis. Cell pellets were split with an internal standard of 5α-cholestane added to one of the cell pellets for determination of sterol amounts in cells. Cells were saponified with 10% methanolic KOH extracted with hexanes and the neutral lipids routinely analyzed by GC-MS for structure determination and quantification. Components were identified by comparison of retention times in GC and HPLC and mass spectra with authentic standards in our own collection [19, 20, 22, 37] and were quantified by comparison of detector response with that of the internal standard. As relevant preparative scale incubation were carried to obtain a subset of amoeba sterols for 1HNMR analysis.
2.8. Scanning (SEM) and transmission (TEM) electron microscopy.
SEM: Prior to fixation, control and treated cell pellets were rinsed in 10% phosphate buffer solution, and centrifuged again to remove any salts that would give false images of crystalline-like structures in the background. After resuspension in buffer, cells were adhered onto poly-L-lysine-coated glass coverslips and subsequently fixed with a solution of 2.5% glutaraldehyde in 0.05M cacodylate buffer. Samples were postfixed for 10 min in 1.5% OsO4, dehydrated in ethanol, and critical point dried with liquid CO2. The cells were then coated with technics hummer V coater and observed under a Zeiss 540 scanning electron microscope.
TEM: Similar to SEM, prior to fixation, control and treated cell pellets, that included primarily trophozoites or cysts depending on the treatment, were rinsed in 10% phosphate buffer solution, and centrifuged again. After resuspension in buffer, cells were adhered onto poly-L-lysine-coated glass coverslips and subsequently fixed in a solution of 2.5% glutaraldehyde in 0.05M cacodylate buffer. After fixation, the cells were post-fixed with 1% (wt/vol) OsO4, dehydrated with ethanol and acetone and embedded in epoxy resin. Ultrathin sections (1 micron) were contrast stained with uranyl acetate and lead acetate and observed under a Hitahci H-8100 Transmission Electron Microscope fitted with AMT side mount digital camera.
2.9. Induction of aromatic sterol synthesis.
Following the protocol of Korn [30] enabling the rapid induction of aromatase activity through cell disruption coupled to osmotic stress, control amoeba cell pellets harboring approximate 1 × 107 cells/ml composed mostly of trophozoites (90%) were equilibrated for 20 min at zero degrees in 0.002 M CaCl2 −0.001 M Tris buffer (pH 7.0) and homogenized with three gentle strokes of a Douce homogenizer. Samples of 20 ml were incubated at 18 degrees for 6 h. At the end of the incubations, the lipids extracted with chloroform-methanol (2:1. v/v). The solvent was evaporated and the resulting organic layer analyzed by GC-MS/HPLC-UV or in separate study, preparative scale studies were carried out to isolate the major sterol products for 1HNMR.
2.10. Determination of AcCYP51 substrate affinity and product outcome
The expression of pCWori+:AcCYP51 construct in E. coli has been described previously [32]. Cytochrome P450 concentration, determined by reduced carbon monoxide difference spectra, and ligand binding studies [6, 38]. The dissociation constant of the enzyme-ligand complex (Kd) for each sterol or azole was determined by non-linear regression (Levenberg-Marquardt algorithm) using a rearrangement of the Morrison equation for tight ligand binding and the Michaelis-Menten equation when ligand binding was no longer tight [38]. Stock 2.5 mM sterol solutions were prepared in 40% (w/v) of 2-hydroxypropyl-b-cyclodextrin (HPCD). All spectral determinations were determined in triplicate. Curve-fitting of ligand binding data were performed using the computer program ProFit 6.1.12 (QuantumSoft, Zurich, Switzerland). Spectral determinations were made using quartz semi-micro cuvettes with a Hitachi-U 3310 UV/Vis spectrometer (San Jose, CA). CYP51 enzyme activity from 10 min incubation was determined in triplicate using a CYP51 reconstitution assay system (0.5 ml final volume) composed of 1.14 μM AcCYP51, 2.81 μM Aspergillus fumigatus (AfCPR1-Q4WM67) cytochrome P450 reductase, 50 mM sterol substrate, 50 mM dilauryl phosphatidylcholine, 4% HPCD, 0.4 mg ml−1 isocitrate dehydrogenase, 25 mM trisodium isocitrate, 50 mM NaCl, 5 mM MgCl2 and 40 mM MOPS (pH, 7.2). In a preliminary study to establish approximate Km and Kcat kinetic constants for obtusifoliol against pure Ac C14-demethylase, reactions were carried out at 37 °C with 1.14 μM recombinant CYP51 and 2.74 μM Homo sapiens CPR (as redox partner) at sterol concentrations of 6.25, 12.5 25, 50, 75 and 100 μM for 20 min. In a separate set of experiments, the kcat for AcCYP51 against AfCPR1 was determined under assay conditions performed with the HsCPR redox partner. Because the turnover number for obtusifoliol was distinctly low against HsCPR redox partner we chose to evaluate sterol structure-activity against the redox partner of AfCPR. Enzyme-generated products were recovered by methanolic KOH saponification followed by extraction with 2 × 3 ml n-hexane, and then evaporation to dryness using a vacuum centrifuge. Product outcome was determined by GC-MS analysis of the free sterol. Competition experiments to establish IC50 of azole against obtusifoliol were carried with the AcCYP51 and redox partner HsCPR.
2.11. AcSMT Inhibition studies
Enzymatic assay conditions with soluble (cloned) Ac24-SMT or Ac28-SMT enzyme preparation and inhibition constant (IC50) determination against natural substrate and the 25-azacycloartanol inhibitor were essentially as described in the previous report [33]. Briefly, IC50 values were determined from a dose-response curve against variable inhibitor concentrations in the range 0 nM, 3.9 nM to 1 μM in the presence of fixed-saturation- concentration of sterol acceptor (100 μM) and coenzyme (150 μM). Assays were performed in triplicate with less than 10% deviation. IC50 values were determined by standard graphical procedures for which computer-assisted linear regression analysis afforded correlation coefficients greater than 0.95 in a cases. Conversion of IC50 value to Ki as dissociation constant, based on the experimentally deduced kinetic properties of cycloartenol (24-SMT) or 24(28)-methylene lophenol (28-SMT) and 25-azacyclartanol was accomplished using the Cheng-Prussoff equation [38].
2.12. GL7 growth
Sterol structure-membrane response effects were carried out using the Saccharomyces cerevisiae strain GL7 with a defective 2,3-oxidosqualene to lanosterol synthase (LAS) auxotrophic for ergosterol, its natural membrane insert [8, 39]. GL7 was cultured on 5 mg/l and Tween 80 (15 ml/l) in yeast-peptone-dextrose medium for 72 h affording growth arrest typical of wild-type yeast of 1 × 108 cells/ml [8].
RESULTS
3.1. Development of novel steroidogenesis patterns with growth and differentiation.
A single cohort of viable cysts cultured on optimal growth conditions linked to extrinsic cues of salt stress or a decrease in carbon sources [23, 24] was used to observe excystment to replicative senescence-death and any associated sterol patterning. The initial experiments to observe differentiation included inoculum obtained from cysts determined to possess exlcusivelyΔ5-sterols - brassicasterol and poriferasterol. Trophozoite proliferation, from 104 resting cysts/ml, followed a logarithmic course with a doubling time of 2.8 h. There seemed to be a slight induction period related to excystment phase of approximate 8–12 h yielding trophozoites. Amoeba cell proliferation reached growth arrest at approximately 3 days affording a cell density 140 thousand cells/ml (Fig. 2, Panel A). As the amoeba continued in stationary phase, they began to progressively convert to encysted cells of mixed population of live and dead. The total cell number decreased with culture age as did the fresh weight of cultures by roughly a factor 10 at day 7 (Fig. 2, Panel B-inset). Continued incubation of the encysted cells for a month led to dead cells by trypan blue staining and to further decrease in the number of encysted cells by another factor of ten to about 104/ml cysts remaining. Because some media preparations for amoeba growth include fetal bovine serum (FBS), which we determined contains 10μM cholesterol, we added FBS as described in the American Type Culture Collection growth media or added 10μM cholesterol in DMSO to our growth media. In either case, the cholesterol supplement had only a minor stimulation effect on trophozoite growth.
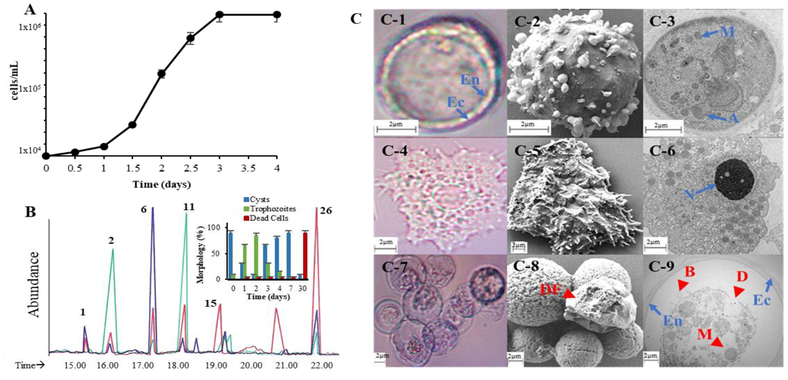
Figure 2.
Developmental changes in A. castalenii growth and sterol biosynthesis. Panel A, Proliferation of trophozoites from a true cyst inoculum. Panel B, Total ion current chromatograms of neutral lipids of trophozoites (green), true (viable) cysts (blue) and dead cells (red); GC peaks correspond to 1= brassicasterol, 2=ergosterol, 6=poriferasterol, 11=7-dehydroporiferasterol, 15= amebasterol-1 and 26 = amebasterol; inset shows quantitative changes in the population of encysted cells to trophozoite to dead cells. Panel C, C1-light micrograph (LM) of double wall viable cyst used as inoculum with rounded outer wall ectocyst (EC) and inner wall endocyst (EN), C2-scanning electron micrograph (SEM) of cyst in the process of excystment., C3- Transmission electron micrograph (TEM) of true cyst showing diagnostic mitochondria (M) and autolysosomes (A). C4-LM of trophozoite, C5-SEM of trophozoite showing acanthapodia. C6 TEM of trophozoite showing prominent central nucleolous (n), mitochondria and cytoplasmic food vacuoles. C7-LM of trypan blue stained clumped encysted cells showing many dead. C8- SEM of clumped encysted cells of control or 25-azacycloartenol treated trophozoites showing deflated (DF) cell characteristic of dead cells. L9- TEM of encysted trophozoite from L8 samples showing ectocyst-endocyst boundary and abnormal morphological features of autophagy-like structures that contain undigested organelles (arrow), blebby structures in the membranes (B, arrowheads) and disorganized membrane structures (D, arrowhead), intense vacuolization and loss of mitochondria.
To assess the conservation or change in sterol composition associated with differentiation, cells were harvested at 6 different stages in the excystment-trophozoite-encystment cycle at days 1, 2, 3, 4, 7 and 1 month post-inoculation. The analysis demonstrated developmental changes in cell morphology and viability were accompanied by remarkable changes in steroidogenesis and metabolism. First, in what may be considered the amoeba growth phase, in the early logarithmic phase of growth in days 1 to 1.5 the C30 and C31-cyclopropyl sterols -cycloartenol and 24(28)-methylene cycloartanol accumulate (Fig 2 and Suppl. Fig. 1), followed in logarithmic growth in days 2 to 3/3.5 by a shift to Δ5,7-C28- and C29-sterol end product formation in approximate 5 to 7 ratio (Fig. 2, Panel B and Suppl. Fig. 2).
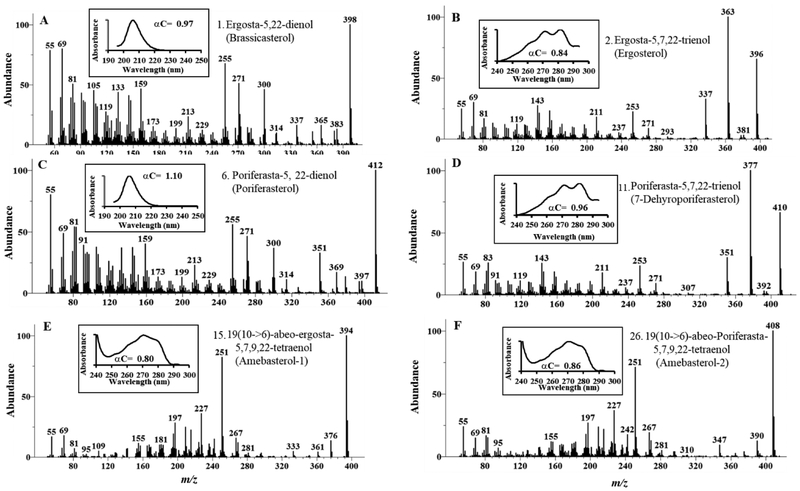
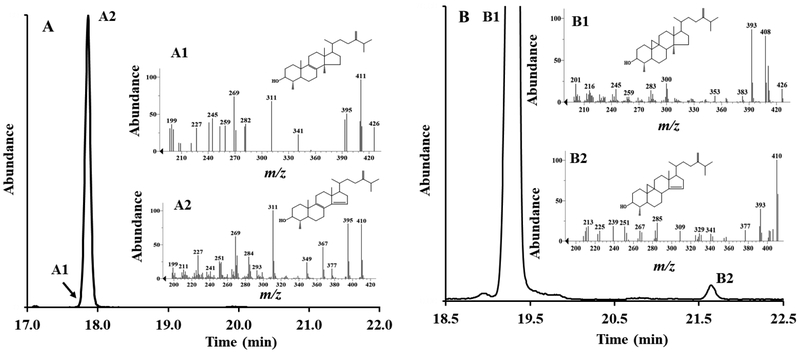
At day 4 where the total cell number was similar to that at day 3, there was a mixed population of trophozpoite and cysts and the sterol composition of the pelleted cells was a mixture of ergosterol, 7-dehydroporiferasterol, brassicasterol and poriferasterol roughly in a 50:50 ratio of Δ5,7 to Δ5-sterols. At day 7, most trophozoites had encysted (90% of total cells). The fresh weight of the cell pellet had decreased by a factor of 3 and the sterol composition of the encysted cells had reverted back to that of inoculum of Δ5- C29-sterol (90% of total sterol). (Figure 2, Panel B inset). At one-month incubation, after the remaining encysted cells had opportunity to excyst and proceed to trophozoite and then encyst again in cyclic fashion using up the carbon sources, most encysted cells were dead and contained predominantly C28- and C29- 6-methyl aromatic sterols, which we refer to as amebasterol-1 and amebasterol-2, respectively. These sterols were previously observed by Korn and Rohmer in Acanthamoeba as minor compounds (trace to 5% total sterol) [28–30, 40] from growth arrested cultures. We confirmed the structure of the major Ac sterols, as well as that of brassicasterol which is a newly characterized Ac sterol, through an examination of their chromatographic and spectral properties reported in Figure 3 (Suppl. Table 1 for 1HNMR analysis).
Figure 3.
Chromatographic and spectral characteristics of main Acanthamoeba sterols from the growth studies shown in Figure 2.
With this knowledge, the morphology and ultrastructure characteristics of trophozoites and encysted cells obtained from control (and subsequently treated cells) were determined. In light microscopy, inoculum cells generated by Method 2 against SDS (Materials and Methods) were found to possess the double-wall feature, and in transmission electron microscopy the encystment was evidenced in ultrastructure showing a nucleolus with a dense droplet in the cyst center, dense cytoplasm packed with lipid droplets, spherical mitochondria (M) and autolysosomes (A) that appear as large basophilic granules. Our observations, in agreement with Bowers and Korn [24], showed the mean diameter of rounded amoeboid cells were approximately 26.5 ± 0.17 μ whereas the mean cytoplasmic diameter of cysts was 16.2 ± 0.13 μ. The decrease in surface area during encystment of a cell of median diameter was more than 65% while the decrease in median cell volume was approximately 80%, suggesting significant loss of cell membrane from amoeba to true cyst. The sterol content in trophozoite and true cyst was 12 pg/cell and 4 pg/cell, respectively. Thus, there is both a distinct loss in total sterol and a marked metabolism of preformed Δ5,7-sterol as trophozoite converts into the true cyst form. These sterol modifications correlate to the different membrane requirements of non-encysted and encysted cells capable of excystment (Figure 2). Once true cysts differentiate into trophozoites, the resulting amoeba, observed to synthesize ergosterol and 7-dehydroporiferaterol, were examined by light and electron microscopy. These non-encysted amoebae were shown to contain typical cytoplasmic organelles, such as a nucleus with a prominent nucleolus, an abundant rough endoplasmic reticulum, mitochondria, vacuoles and lipid droplets and showed surface protrusions -acanthapodia, durable double wall of smooth ectocyst and a round endocyst (Fig. 2, Panel C).
3.2. Medium-replacement induced-encystment can lead to distinct cell types of characteristic sterol compositions.
To determine whether the unusual Δ5-sterol and 6-methyl aromatic sterol profiles observed in the stationary phase growth were associated with distinct cyst populations, we initiated encystment by Method 2. The salt stressed trophozoite cultures spontaneously encysted yielding some cell debris. GC-MS analysis of the pellet extract revealed 21 sterols (Suppl. Fig. 2, Panel E and Table 1) which is the most complex sterol composition reported for not only the amoeba but for protozoa generally. Many of these sterols represent co-metabolites and by-products. Of interest was the experimentally induced encystment of amoeba yielded a sterol mixture of Δ5,7-sterols, Δ5-sterols and 6-methyl aromatic sterols. Notably, no significant trpohozoites were detected microscopically, yet there is a significant amount of ergosterol and 7-dehydroporiferasterol in the sterol mixture suggesting that a subset of encysted cells could possess Δ5,7-sterols. As the encysted cells were processed further through SDS-treatment, the cells that remain contain predominantly brassicasterol and poriferasterol by GC-MS analysis (Supple Fig. 2, Panel F); these cells are viable and can excyst completely to trophozoites in enriched medium in 12 h. We surmise the salt step can create mixed populations of encysted cells; one population is composed of fragile double walls and membranes composed of Δ5,7-sterols or aromatic sterols and a second population is composed of resistant double-wall structures and membranes composed of Δ5- sterols.
Table 1.
Chromatographic and spectral properties of sterols from Acanthamoeba cells or substrats/products of incubations with SMT and CYP51 enzyme preparations.1
| Structure2 | Sterol (systematic name) | Trivia Name | RRT c3 | M+(amu) | UV (λmax, nm) | TLC (Rf) |
|---|---|---|---|---|---|---|
| 1 | Ergosta-5,22-dienol | Brassicasterol | 1.06 | 398 | 205 | 0.42 |
| 2 | Ergosta-5,7,22-trienol | Ergosterol | 1.12 | 396 | 282 | 0.42 |
| 3 | Ergosta-7,22-dienol | 22-Dehydrofungisterol | 1.14 | 398 | 205 | 0.42 |
| 4 | Ergost-5-enol | Ergost-5-enol | 1.15 | 400 | 205 | 0.42 |
| 5 | Ergosta-7, 24(28)-dienol | Ergosta-7, 24(28)- dienol | 1.17 | 398 | 205 | 0.42 |
| 6 | Poriferasta-5,22 -dienol | Poriferasterol | 1.21 | 412 | 205 | 0.42 |
| 7 | Ergosta-5,7-dienol | Ergosta-5,7-dienol | 1.22 | 398 | 282 | 0.42 |
| 8 | Ergost-7-enol | Fungisterol | 1.24 | 400 | 205 | 0.42 |
| 9 | 4α, 14α-Dimethylergosta-8,24(28)- dienol | Obtusifoliol | 1.26 | 426 | 205 | 0.53 |
| 10 | 4α-Methylergosta-8, 14, 24(28)-trienol | 4α-Methylergosta- 8, 14, 24(28)- trienol | 1.27 | 426 | 242 | 0.53 |
| 11 | Poriferasta-5,7,22-trienol | 7-Dehydroporiferast erol (7-DHP) | 1.28 | 410 | 282 | 0.42 |
| 12 | Poriferast-5-enol | Clionasterol | 1.30 | 414 | 205 | 0.42 |
| 13 | Poriferasta-7,25(27)-dienol | Poriferasta-7,25(27)-dienol | 1.32 | 412 | 205 | 0.42 |
| 14 | 4α-Methylergosta-7, 24(28)-dienol | 24-Methylenelophenol | 1.34 | 412 | 205 | 0.53 |
| 15 | 19(10->6)-abeo-Ergosta-5,7,9,22-tetraenol | Amebasterol-1 | 1.36 | 394 | 270 | 0.47 |
| 16 | 4α- 14α-Dimethylcycloart-24(28)-enol | Cycloeucalenol | 1.36 | 426 | 205 | 0.53 |
| 17 | Poriferasta-5,7 -dienol | 7-Dehydroclionasterol | 1.38 | 412 | 205 | 0.42 |
| 18 | Poriferast-7-enol | Dihydroclionasterol | 1.41 | 414 | 205 | 0.42 |
| 19 | Lanosta-9(11),24-dienol | Parkeol | 1.42 | 426 | 205 | 0.65 |
| 20 | Poriferasta-5,7,25(27)-trienol | Poriferasta-5,7,25(27)-trienol | 1.43 | 410 | 282 | 0.42 |
| 21 | Cycloart-24(25)-enol | Cycloartenol | 1.45 | 426 | 205 | 0.65 |
| 22 | 19(10->6)-abeo-Ergosta-5,7,9-trienol | Amebasterol-3 | 1.49 | 396 | 270 | 0.47 |
| 23 | 4α-Methylporiferasta-7,25-dienol | 4α-Methylporiferasta-7,25-dienol | 1.51 | 426 | 205 | 0.53 |
| 24 | 4α-Methylcycloart-14, 24(28)-dienol | 4α-Methylcycloart- 14, 24(28)-dienol | 1.52 | 410 | 205 | 0.53 |
| 25 | 4α-Methylporiferasta-7, 24(28)£-dienol | 4α-Methylporiferasta- 7, 24(28)E-dienol | 1.54 | 426 | 205 | 0.53 |
| 26 | 19(10->6)-abeo-Poriferasta-5,7,9,22-tetraenol | Amebasterol-2 | 1.58 | 408 | 270 | 0.47 |
| 27 | Cycloart-24(28)-enol | 24-Methylenecycloart anol |
1.58 | 440 | 205 | 0.65 |
| 28 | 4α-Methylporiferasta-7,24(28)Z-dienol | Citostradienol | 1.59 | 426 | 205 | 0.53 |
| 29 | 19(10->6)-abeo-Poriferasta-5,7,9,22,25-pentaenol | Amebasterol-4 | 1.63 | 406 | 270 | 0.47 |
| 30 | 19(>6)-abeo-Poriferasta-5,7,9,25(27)-tetraenol | Amebasterol-5 | 1.66 | 408 | 270 | 0.47 |
| 31 | 19(10->6)-abeo-Poriferasta-5,7,9-trienol | Amebasterol-6 | 1.71 | 410 | 270 | 0.47 |
| 32 | Lanosta-8,24(28)-dienol | Eburicol | 1.43 | 440 | 205 | 0.65 |
| 33 | Lanosta-8,24(25)-dienol | Lanosterol | 1.32 | 426 | 205 | 0.65 |
| 34 | Lanost-8-enol | 24,25-Dihydrolanosterol | 1.24 | 428 | 205 | 0.65 |
| 35 | 14α-Methylcholest-8,24(25)-dienol | 14α-Methylzymosterol | 1.08 | 398 | 205 | 0.53 |
| 36 | Lanosta-7,24(25)-dienol | Δ7-lanosterol | 1.44 | 426 | 205 | 0.65 |
| 37 | 4α, 14α-Dimethylcholesta-8,24(25)-dienol | 31-Norlanosterol | 1.16 | 412 | 205 | 0.53 |
Analytical procedures are described in Materials and Methods;
See supplemental Figure 3 for a key to structures;
Relative retention time to Cholesterol.
A crucial observation made by the Korn group to prepare an abundance of 6-methyl aromatic sterols of the phenanthrene skeleton was to generate a cell-free system of Ac in high salt solution yielding a microsomal (membranes) subcellular fraction. Under these conditions, the ergosterol and 7-dehydroporiferasterol rapidly convert in stoichiometric fashion to amebasterol-1 and amebasterol-2; the aromatic compounds remain membrane-bound [30]. The relevance of the Korn study is to show both that the phenanthrene steroids can be easily obtained in amounts necessary for structure determination and that these aromatic compounds can associate with the endoplasmic-plasma-membrane fraction of microsomes in intact cells. Notably, this approach used by Rohmer on the related Acanthamoeba species-polyphaga yielded sufficient compound to be characterized by 13C/1H-NMR to prove the structure and stereochemistry of the metabolites as new class of steroids [31]. We repeated these studies and observed Ac homogenates generate 6-methyl aromatic sterols from Δ5,7 -sterols in amounts reported by Korn over the 6 h incubation period. The 6-methyl aromatic sterols can remain stable for at least 2 months (Suppl. Figure 2, Panels G and H).
3.3. Catalytic competence of AcCYP51relative to Ac24-SMT and Ac28-SMT.
The functional expression in E. coli of pC Wori+:AcCYP51 construct and wild-type cDNA of 24-SMT and 28-SMT have been described previously [32, 33]. To elucidate the diversity of substrate preference that could influence the order of intermediates in C28- and C29- sterol synthesis, eleven 14-methyl sterols were evaluated for substrate recognition by AcCYP51 (Table 2, Suppl. Fig. 3 to 6). When the redox partner for AcCYP51 activity was AfCPR1 the rate of formation of product was greater by a factor of 10 compared to using the HsCPR redox partner affording approximately 7.6 ± 0.05 min−1. Consequently, we used the AfCPR1 redox partner to determine structure-activity.
Table 2.
Comparison of binding and catalytic properties of AcCYP51 and AcSMTs for sterol.
| Substrate1 | AcCYP51 | SMT5 | |||
|---|---|---|---|---|---|
| Kd (μM)2 | AfCPR kcat (min−1)3 | 24-SMT kcat(min−1) | 28-SMT kcat(min−1) | ||
| 10 | 4α, 14α-Dimethylergosta-8,24(28)-dienol | 41 ±3 | 7.6 ±0.05 | 0 | 0 |
| 32 | Lanosta-8,24(28)-dienol | 51 ±10 | 3.2 ±0.04 | nd | nd |
| 33 | Lanosta-8,24(25)-dienol | 46 ±3 | 1.7 ±0.01 | 1.50 ±0.01 | 0.23 ±0.01 |
| 34 | Lanost-8-enol | 60±12 | 2.0 ±0.002 | nd | nd |
| 35 | 14α-Methylcholest-8,24(25)-dienol | 12 ±2 | 1.6 ±0.08 | 1.50 ±0.05 | 0.39 ±0.02 |
| 24 | 4α-Methylcycloart-24(28)-enol | No fit | 0.04 ±0.01 | nd | nd |
| 21 | Cycloart-24(25)-enol | 18 ±5 | 0 | 1.50 ±0.07 | 0.45 ± 0.03 |
| 27 | Cycloart-24(28)-enol | No fit | 0 | 0 | 0.23 ±0.05 |
| 36 | Lanosta-7,24(25)-dienol | Nd4 | 0 | nd | nd |
| 37 | 4α, 14α-Dimethyl cholesta-8,24(25)-dienol | nd | 0 | 1.50 ±0.04 | 0.74 ±0.02 |
| 14 | 4α-Methylergosta-7,24(28)-dienol | nd | 0 | 0 | 0.80 ± 0.04 |
See supplemental Figure 3 for a key to structures;
Kd value was determined by progressive titration of 4 M AcCYP51 with sterol in 40% (W/V) HPCP;
kcat was determined in the CYP51 reconstitution assays for 50 μM sterol using AfCPR redox partners;
nd, not determined;
Kinetic constants for SMT were determined previously for these sterols assayed in the range 5 μM to 150 μM against 150 μM SAM and 80 μg SMT as described in reference 33.
After incubation in the usual way, no metabolism occurred with cycloartenol, whereas obtusifoliol was fully metabolized by AcCYP51 under the same conditions as evidenced in GCMS analysis showing the Δ8,14 –product in 95% yield (Suppl. Fig. 4). Because the rate of product formation against the AfCPR1 partner was much higher than when using the HsCPR redox partner, we could now detect some small conversion of cycloeucalenol. This was evident in two ways; in catalytic assay to establish competence afforded a rate of 0.04 ± 0.01 min−1 and in GCMS analysis showing a new cyclosteroid product (M+410, ~1% yield,) that contained the Δ8-structure (Suppl. Fig. 4, structure 24 in Table 1). While doubtful the new metabolite converts to Δ5,7 -sterol under physiological conditions, it does shows mechanistically that the Δ8-bond is not essential for CYP51 activity. In contrast, a Δ8-bond in the CYP51 substrate is required in plant and animal sterol biosynthesis [41–43]. Except for cycloeucalenol as substrate, there was general agreement between affinity and catalytic competence determined by the sterol difference spectra and binding constant (Kd) and efficiency for substrate to be converted to 14-desmethyl product (kcat). Those substrates having a 9,19-cyclopropyl group, typical of intermediates in the plant sterol biosynthesis pathway, poorly bind to the enzyme (cycloeucalenol) or not at all (cycloartenol and 24(28)-methylene cycloartanol). Structural isomers of the cyclopropyl sterol that possess a Δ7-bond do not bind while those that possess a Δ8-bond bind well with obtusifoliol (Suppl. Fig 3–10) the apparent favored substrate for AcCYP51. While not readily evident from the affinity constant (Kd), but apparent from the rate data (kcat) through comparison of pairs of substrates that differ in side chain construction for a Δ24(28) –group of lanosterol (Suppl. Fig. 3 – 33) versus eburicol (Suppl. Fig. 3 – 32), obtusifoliol (Suppl. Fig. 3– 10) versus 31-norlanosterol (Suppl. Fig. 3– 37), it appears that AcCYP51 prefers a substrate that possesses the 24(28)-bond (Table 2) consistent with its phylogenetic association to land plant enzymes [6] and provides rationale for the biosynthetic ordering of intermediates.
Thus, in good agreement with results of the titration experiments and catalytic parameters of the AcCYP51 the enzyme clearly prefers obtusifoliol (Table 2) yielding an optimal rate for C14-demethylation of 7.6 min−1. The value is close to the Vmax reported for kinetoplastid protozoan and land plant CYP51and is approximately 6 fold lower than those for the fungal and human isoforms [6]. On the basis of the previously determined catalytic competence of cloned AcSMTs (referenced in Table 2), it has been shown cycloartenol [kcat 1.5 min−1 /Km 44 μM] is the favored substrate for 24-SMT and the enzyme generated product is 24(28)-methylene cycloartanol while 24(28)-methylene lophenol [kcat 0.8 min−1/ Km 25 μM] is the favored substrate for 28-SMT which converts to multiple 24-ethyl sterol Δ25(27) - and Δ24(28) - products [33]. Comparative analysis of the sterol C14-demethylase and C24-methylase specificity reveal neither of the AcSMT recognizes obtusifoliol as substrate and that AcCYP51 fails to productively accept either of the AcSMT favored substrate.
3.4. Identification of a new sterol biosynthesis in protozoa; evidence for separate intermediates to C28 - and C29-sterols.
There is no clear information about the C29-sterol synthesis pathway. In the first round of trophozoite sterol analyses, we identified 15 neutral sterols (other than the novel aromatic sterols) from the MgCl2 treated cultures (compounds 1, 2, 3, 4, 5, 6, 7, 8, 9, 11, 12, 13, 17, 18 and 20, Suppl. Fig. 2 and 3) and from logarithmic phase cultures 4 more sterols were detected (Suppl. Fig 1 16, 19, 21 and 27 and illustrated in Suppl. Fig. 3) while additional sterols were obtained from treated cultures corresponding to many of the compounds previously isolated from Acanthamoeba polyphaga, including cycloartenol, 24(28)-methylene cycloartanol, obtusifoliol, 24β-ethyl cholesterol and 24β-methyl cholesterol [28, 29]. Our chemical identification of Ac cycloartenol by GC-MS against a standard available to us and of parkeol synthesized from cycloartenol by acid-induced rearrangement of the cyclopropane ring to the 9(11)-structure [44] agrees with the careful biochemical and enzymological studies of Rohmer showing as well that cycloartenol is the sole product of 2,3-oxidosqualene cyclization to protosterol in A. polyphaga and 9(11)-metabolites occur as byproducts in trace amounts [28, 29]. Our observations combined with that of Rohmer for cycloartenol rather than lanosterol as the true protosterol for Acanthamoeba species is supported by Ac genome mining for sterolic enzymes where a single 2,3-oxidosqualene to protosterol synthase (cyclase) is detected corresponding to the cycloartenol synthase (CAS1). Recent phylogenetic studies have shown that CAS1 can be distinguished from the gene for lanosterol synthase (LAS1) through a signature peptide sequence with a conserved residue at amino acid position-442 (Ac numbering system) for isoleucine rather than valine characteristic of the lanosterol synthase (LAS1), as shown in supplemental Figure 7 [13, 45 and references cited therein]. Testing effects of substitution of the conserved residues in and around position 442 (Ac numbering) on CAS1 and LAS1 have led to the notion that high conservation of the amino acid composition in these regions represents an important structural basis for evolutionary conservation of protosterol formation [13], and indicate that cycloartenol evolved before lanosterol.
For elucidation of new biochemical transformations and biosynthetic pathways mass spectra of sterols identified in GC-MS analysis are extremely powerful tools in providing information about multiple structural features of the molecule. In many cases authentic compounds are recorded in the recently released NIST library of chemical standards. A molecular ion is normally observed in the mass spectrum of free sterol which is reason we have studied this sterol form in GC-MS analysis. It is preferable to consider fragmentation patterns and subtleties in ion intensity associated with the ring and side chain in formulating sterol structure [46]. For those that rely on the NIST library to assign structure to sterols caution should be exercised since partial information is obtained from observation of molecular ion and base peak of target sterol as match indicators and thus can lead to erroneous structure assignments for several key compounds as reported in reference 34. Importantly, many reference sterols that occur naturally are not entered into the NIST library. For example, the aromatic sterols identified in this study, e.g., amebasterol-1 of M+ 394 amu, is matched to 9-methyl anthrasteroid (anthraergostatraenol) in the NIST library with 81.1% match of 86.7 % probability. This seemingly excellent match in the database generates an incorrect structure for the Ac compound while the correct structure has yet to be deposited into the NIST library. Indeed, Rohmer proved through NMR studies, and confirmed here, the structure of this Ac metabolite as the phenanthrene skeleton harboring an aromatic B-ring with the C19 methyl rearranged to the C6-position [31]. Another relevant example for potential inaccuracy in structure determination as it relates to Ac sterol biosynthesis pathways comes from the supposed mass spectrum match of ergosterol which has the Δ5,7,22 -triene system. Its Δ25(27) -ergostene isomer, like that of its Δ24(28) - and Δ24(25) -ergostene isomers, have identical mass spectra and one of more of these isomers can co-elute with ergosterol in GC unless appropriate chromatographic conditions are met [19, 46]. Referencing the NIST library is further limited in the identification of the set of B-ring structural isomers of Δ8 versus Δ7-ergostenes, for the set of conjugated B/C-ring dienes of Δ5,7(8), Δ8,14(15), Δ5,8(9), Δ6,8(14) -ergostene isomers and for the C24-alkyl epimers (for example) poriferasterol (24β-ethyl) and stigmasterol (24α-ethyl) [46–49]; in the usual practice, identification of the individual isomers is confirmed through a comparison of chromatographic and spectral constants of the unknown as the free alcohol or as a C3-derivatized compound to assist in chromatographic resolution against equivalent data obtained on authentic standards or for determination of stereochemistry at C24 through NMR of the pure compound or as necessary to synthesize the applicable standard [48].
Previously isolated sterols from Acanthamoeba are variably positioned as intermediates in biosynthetic schemes for ergosterol and 7-dehydroporiferasterol synthesis, including a hypothetical terminal step whereby Δ5- C29-monoene sterols (24-ethyl cholesterol), formed from Δ5,7-sterol, convert to final Δ5,22-diene sterol, as can occur in land plants. Curiously, the Δ25(27) -C29-sterols that we observed in trace amounts in A. castellanii were detected also in trace amounts in A. polyphaga and A. culbertsoni by research groups with expertise in natural product characterizations [28, 50]. However, the earlier investigators failed to consider the biosynthetic importance of Δ25(27) -sterols to ergosterol and 7-dehydroporiferasterol synthesis, relegating them to by-products in the sterol metabolome. The accumulation of ergsost-5-enol and poriferast-5-enol [structures 4 and 14 in Suppl. Fig. 3] which arise from the salt treatments is problematic and may not relate to their biosynthetic intermediate role in Δ5,7-sterol formation of ergosterol and 7-dehydroporiferasterol as proposed [28]. Rather, these sterols may be considered metabolically inert end products formed along with brassicasterol and poriferasterol from a Δ7-reductase of broad substrate specificity during encystment and not from trophozoite growth (Fig. 6). As our studies suggest, the Δ7-reductase appears only to be active during the encystment process.
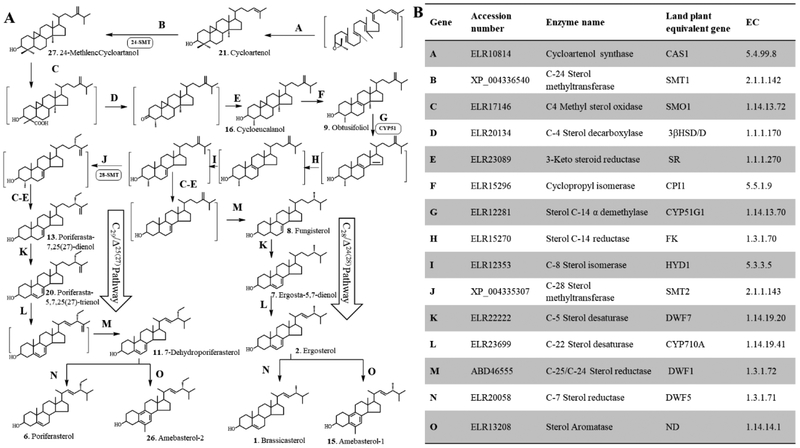
Figure 6.
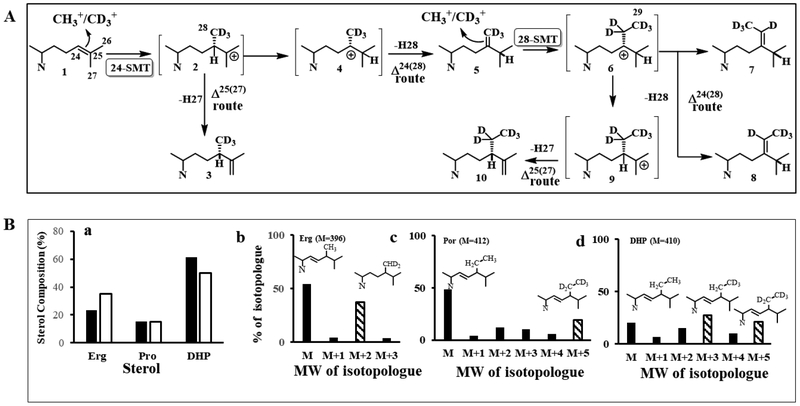
Biosynthetic model for the formation of phytosterols in Acanthamoeba. (A) Proposed separate Δ24(28) - and Δ25(27) -olefin intermediates to C28- and C29-sterol products. (B). Sterolic genes identified in the Ac Genebank compared to known sterolic genes from land plants and humans (aromatase).
Incubation of [methyl-2H3]methionine with trophozoites revealed growth arrested cultures possessed three major isotopically labeled sterols corresponding to ergosterol, poriferasterol and 7-dehydroporiferasterol; for ergosterol the mass spectrum showed the dideuterated species (M+ 396 → M+ 398) while the mass spectrum of 7- dehydroporiferasterol showed the pentadeuterated species (M+ 410 → M+ 415) and the mass spectrum of poriferasterol showed the pentadeteuterated species (M+ 412 → M+ 417) (Figure 2, Panels E and F). The fragment containing the high end molecular cluster at m/z 396 for ergosterol and m/z 410 for 7-dehydroporiferasterol derived from the methyl cation on SAM was shifted by 2 Da or by 5 Da, respectively, in the mass spectra of the labeled sterol (Suppl. Fig. 3) These labeling results show a mixed biosynthetic pathway in which ergosterol is formed by an Δ24(28) - sterol methylation pathway similar to land plants which incorporate 2 D-2H3-SAM atoms at C28 while the 7-dehydroporiferasterol and poriferasterol sterol C -methylation pathways are similar to the Δ25(27)29 –C29-sterol methylation pathway in green algae which incorporate 5-D 2H3-SAM atoms into the C29-sterol [19, 45]. In contrast, land plant C29-sterol biosynthesis 4D 2H3-SAM atoms are incorporated into the 24-ethyl group consistent with the involvement of a Δ24(28) -C29--sterol intermediate [51]. The enzyme-catalyzed conversion of Δ24(25) -sterol substrate to C28- and C29-sterol products were considerably altered from the sterol profile of control. As shown (Figure 2, Panel B), the composition of the sterol mixtures obtained from cultures grown on normal and deuterium-labeled methionine showed that the proportion of C28-sterol was markedly increased while the C29 sterols decreased in the cultures grown in the presence of [methyl-2H3]methionine. These labeling experiments reveal the operation of a pronounced deuterium isotope effect and indicates that the proton loss from a C28 intermediate (Figure 5, Panel A) is the rate-limiting step in formation of C29-sterol products. Thus, in the presence of substrate 28-SMT affording C29-sterols can in some circumstances significantly alter the course and equilibrium of metabolic sequences affecting the balance of C28 to C29-sterols during growth and differentiation. For C29-sterols there is switch to an alternate route utilizing Δ25(27)-intermediates in contrast to the previously reported route of Δ24(28) -intermediates [28]. It should be remembered the cell pellet used for our sterol analysis was from a culture composed of two populations of cells—trophozoites that synthesize Δ5,7 -sterols and encysted cells that contain Δ5- sterols. Given the differentiation processes discussed in section 3.1, it is more likely the poriferasterol is derived from the 7-dehydroporiferasterol associated with trophozoite to encysted cell formation. It is also notable the phytosterols synthesized in land plants possess side chain C24/methyl/ethyl structures that are α-oriented while our 1HNMR analysis of the A. castellanii ergosterol and 7-dehydroporiferasterol indicated the C24-methyl/ethyl structures are β-oriented similar to the side chain structures of C28-and C29- sterols in the green alga Chlamydomona reinhardtii [19].
Figure 5.
Sterol C24-methylation activity in Acanthamoeba. (A) Proposed sterol C24-methylation pathway in Acanthamoeba showing incorporation of SAM and [methyl-2H3]SAM = D3-SAM at C24 and C28 of the sterol side chain. (B) Mass spectral analysis of sterols (ergosterol, erg, poriferasterol, por, and 7-dhydroporiferasterol, DHP) recovered from trophozoites incubated in the presence and absence of [methyl-2H3]methionine.
Conventional co-metabolite ordering in Ac sterol biosynthesis suggests the amoeba operates a land plant phytosterol biosynthesis pathway via the cycloartenol to 24(28)-methylenecycloartanol Δ24(28) -pathway to C28- and C29-sterol final products. However, through a comparison of the Ac genomic database with land plant (Arabidopsis) genomic database (and for aromatase the human genome database) together we our chemical analysis of trophozoites and encysted cells, we reconstructed the cycloartenol to 6-methyl aromatic sterol biosynthetic pathways, taking into account phylogenetic differences in the sterol methylation of Δ25(27) – (green algae) and Δ24(28) -olefin (land plant) routes and in the formation of Δ5,7- sterols versus Δ5-sterols (Fig. 6). The reconstruction of the A. castellanii sterol pathway underscores the metabolic divergence of amoeba and recurrent evolution. The results show: (i) completion to a Δ5-monoene sterol atypical of green algae, (ii) selective sterol methylation for the 24β- orientation typical of green algae (iii) in vitro AcSMT assays show obtusifoliol is not a substrate and that Δ25(27) –sterol is a product of the enzyme catalyzed reaction consistent with ordering of intermediates based on biosynthetic reasonableness, and (iv) 2H-methinone fed to Ac show the C28 - and C29- sterol products are formed by separate Δ24(28) – C24-methylene and Δ25(27) -olefin C24-ethyl intermediates, respectively, and that any Δ24(28) −24-ethyl sterols formed enzymatically are redundant not incorporated into 7-dehydroporiferaterol under physiological conditions.
3.5. Dependence of inhibitor effect on the physiological state of the amoeba cells
The primary mechanism of the azole and steroidal inhibitor effect- inhibition of ergosterol biosynthesis in amoeba cells- has as a prerequisite an active cellular metabolism for the biosynthetic renewal of ergosterol with proliferation and growth. It is understandable that dormant cysts could only be affected at high concentrations of these drugs (>100 μM), if at all. However, unclear is the extent to which the encysted cell can alter its sterol profile in response to an ergosterol biosynthesis inhibitor. The aza analog of the C25-carbocation intermediate of the C24-methylation reaction, a steroidal inhibitor, against Ac24SMT and Ac28-SMT and the tight binding inhibitor voriconazole against AcCYP51 were utilized to determine the relative importance of active site interactions of select sterol biosynthesis enzymes responsive to these drugs in vitro as well as in cell-based studies
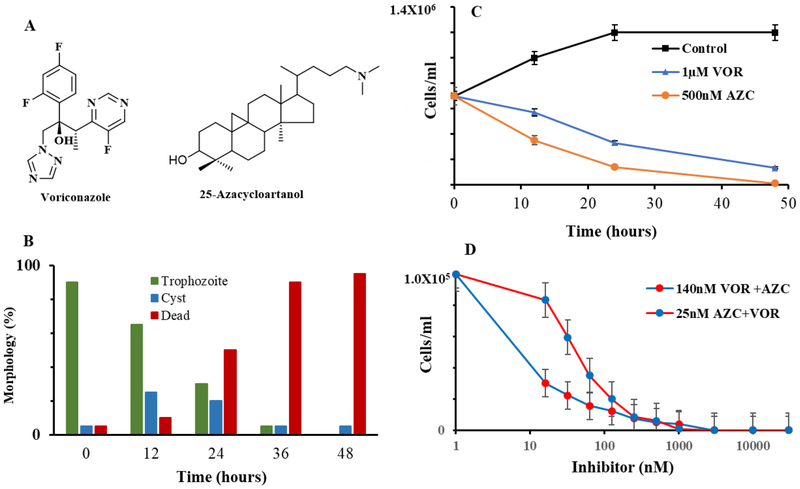
The effect of azole and 25-azacycloartanol (Fig. 7, Panel A) on rapidly proliferating trophozoites was much the same—both compounds prevented amoeba growth at low inhibitor concentrations of IC50 values 390 nM and 25 nM, respectively and MAC values between 1–3 μM (Fig. 7, Panel B). Sterol analysis of mostly encysted cells revealed mark changes to the C28- and C29- sterol balance associated with encystment (Suppl. Fig. 2, Panel C and Suppl. Fig. 9) and the total sterol content was less than in trophozoite by approximately 50% on per cell or pellet basis. Although steroidal inhibitor treatment resulted in reduced C29-sterol levels, showing disruption in substrate pools, there was not much accumulation of intermediates cycloartenol or obtusifoliol, suggesting a down-regulation of de novo sterol synthesis as trophozoite converts to encysted cell as reported elsewhere [35]. In neither treatment was there significant accumulation of 6-methyl aromatic sterol. However, after the treated cells were incubated for another 8 days all the Δ5,7-sterol had converted to 6-methyl aromatic sterol accompanied by a ten- fold decline in fresh weight of the pellet. When the concentration of inhibitor is increased 10 to 50 fold, i.e., increasing stress, the resulting 48 cell pellet is composed exclusively of dead cells and the sterol profile consists mostly of 6-methyl aromatic sterol.
Figure 7.
Effects of ergosterol biosynthesis inhibitors as single or combination treatment on Acanthamoeba growth and encystment prepared by Method-1 (Materials and Methods). Panel A, structures of growth inhibitors. Panel B, Time-kill studies of 25-azacycloartanol (AZC) and voriconazole (VOR). Panel C, Quantification of AZC treated cysts (viable and clumped non-viable) and dead cells determined microscopically and by trypan blue staining. Panel D. A. castellanii treated with VOR and AZC at the previously determined IC50 concentration and then using the pair of compounds at varied concentration of the inhibitor from 16 nM to 3 μM. Data represent mean ±sd. P < 0.05, determined by T-test. In each case data are from a single experiment which is representative of three independent experiments.
To determine whether FUdR, a DNA synthesis inhibitor [52] that should prevent ergosterol biosynthesis globally, had an effect on differentiation, spontaneous encystment was evaluated after the cultures containing inhibitor reached stationary phase of control. The resulting cells had a population of cysts equal to the parent trophozoite inoculum of approximately 2–3 ×105 cells/ml. Effectively all of these encysted cells (>95%) were alive according to trypan blue staining. The FUdR treated cells showed a significant increase in Δ5-sterol of brassicasterol and poriferasterol (40 % total sterol) which associates with true cyst formation (Suppl. Fig. 10). In contrast, for treatments with voriconazole or 25-azacyloartanol the number of cells at 48 h was reduced by a factor of 10 to 2–3 × 104 cells/ml, most were dead by trypan blue staining (Fig. 7, Panel B) and they (encysted cells) possessed mostly ergosterol and 7-dehydroporiferasterol (Suppl. Fig. 9) with no accompanying 6-methyl aromatic sterol.
Time-killing experiments were carried out by the addition of inhibitor at approximate MAC levels to the culture medium for 48 incubations. Figure 7 (Panel C) shows that medium inoculated with a population of 1 × 105 cells/ml trophozoites (90%) affords continued trophozoite proliferation, without significant encystment (data not shown), to a stationary phase population of 1 × 106 cell/ml. Alternatively, in the presence of the test inhibitors, at about the MAC, the trophozoite population drops to zero. This stressing of amoeba stimulates trophozoite to encyst, which as inhibitor pressure is maintained yields a decrease in the total number of encysted cells from approximate 3 × 104 encysted cells/ml at 12 h to an approximate 5 ×102 to 5 ×103 cells/ml at 48 h growth. Trypan blue staining of the encysted cells revealed two dominant populations characterized by those that are viable or non-viable and dead. When the 48 h treated pellets were transferred to fresh medium and incubated for a week, 25-azacycloartanol treatments showed no growth while from the voriconazole treatment a few thousand trophozoites were evident, suggesting the predominant population of encysted cells viewed as dead by trypan blue staining were indeed dead in terms of excystment.
Because the clumped cells were in much greater density in treated cells than in control, it was straightforward to examine them by scanning electron microscopy (SEM) and transmission election microscopy (TEM). Thus, SEM and TEM images of 25-azacycloartanol-generated cysts (VOR looked much the same) from the 48 h harvest were virtually the same as those of control cysts originating in T-flasks showing one or more flaccid cells clumped together with rounded cells (cf. Figure 2, Panel C). Fine structure of the treated cells also showed a deformation in cell wall associated with an aberrant thickening or partially formed ectocyst and endocyst, and the cell interior looked severely damaged with (i) formation of blebby structures in the cell membrane, (ii) broken membrane structures and (iii) intense vacuolization and loss of mitochondria typically associated with autophagy-like structures in the center of the cell that contained undigested organelle.
To determine whether inhibitors designed to block two essential enzymes in the post-squalene segment of the ergosterol biosynthesis pathway are more potent in combination than when added to the medium singly, we monitored the effects of combining subinhibitory concentrations of one drug against varied concentrations of the other drug on trophozoite multiplication. Complete eradication of the amoeba parasite is obtained when using the drugs in combination at concentrations as low as equal to their IC50 values which in our studies for voriconazole was 140 nM ± 10 nM compared to our previously reported 390 nm [32] and for 25-azacycloartanol is 25 nM ± 2 nM (Fig. 7, Panel D). The number of dead cells at 48 h in the combination drug experiment of 25 nM 25-azacycloartanol (25 nM) and voriconazole (100 nM) was similar to the number of dead cells when the drugs were administered individually to the medium (Fig. 7, Panel B), and no trophozoites were detected after 5 days following resuspension of treated pellet into fresh medium. These results suggest that combined treatment of two or more rationally designed inhibitors at sub-MAC concentration can successfully interfere with sterol biosynthesis to prevent leakiness at the target enzyme and in so doing limits production of the downstream C28- and C29-sterols, supporting a role for AcCYP51 and AcSMTs in the control of metabolic flux through distinct intermediates.
3.6. Excystment in response to inhibitor treatment.
In a separate experiment the anti-amoeba effect of the drugs on excystment was determined with cysts derived from the SDS-treated process. Using MAC concentrations of the ergosterol biosynthesis inhibitors, all the resting cysts convert to trophozoites by 5 days, indicating that excystment is insensitive to these treatments. In similar fashion, the FUdR treated resting cells easily excysted into trophozoites by 3 days. During these experiments, increase numbers of empty cyst walls were evident in cell mixture, consistent with excystment.
3.7. Enzymatic basis of action and selectivity.
All the available experimental evidence indicates that AcCYP51 and 24-/28-AcSMT are the primary target of the medical azole and steroidal N-inhibitor, respectively, since we observed small accumulations of obtusifoliol and cycloartenol in the presence of voriconazole and 24(R,S),25-epiminolanosterol [32, 33]. The target enzymes were characterized from cloned genes introduced into E. coli and solubilized for activity assay. In our earlier study, we observed that voriconazole is a specific and reversible inhibitor of AcCYP51 exhibiting an IC50 of 390 nM and its MAC is approximately 3 μM [32, 33]. 25-Azacycloartanol tested as a true substrate analog inhibitor is approximately as effective against 24-AcSMT and 28-AcSMT as voriconazole against AcCYP51. The inhibitor against trophozoite proliferation generates an IC50 of 25 nM ± 0.25 nM and MAC of 1 μM ± 0.05 μM (data not shown) and against 24-SMT incubated with cycloartenol as substrate or 28-SMT incubated with 24(28)-methylene lophenol as substrate yields IC50 values of 15 nm (Ki = 13 nM) and 22 nM (Ki = 8 nM) (Suppl. Fig. 11). The measured inhibition of the AcCYP51 and SMT activities is quantitatively sufficient to account for the inhibition of ergosterol and 7-dehydroporifersterol biosynthesis in amoeba.
3.8. Effect of Acanthamoeba sterols on growth of GL7
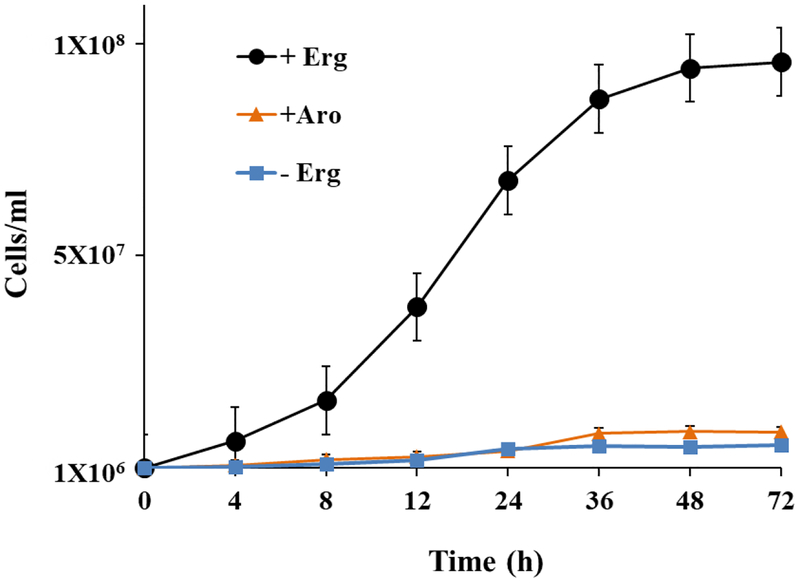
To assess the structural specificity for sterols as membrane inserts directly, we used the yeast GL7 auxotrophic for sterol. Close interactions between sterol supplementation and the growth response was detected. Ergosterol supplementation to GL7 afforded growth arrest in 72 h affording approximate 1 × 108 cells/ml, typical of wild-type yeast, while cultures without added ergosterol yielded a no growth-response, as reported previously (Fig. 8) [8, 39, 53]. Ergosterol was recovered in unchanged form from the cells at approximately 20 fg/cell. Microscopic examination showed the GL7 yeast at growth arrest revealed mostly single cells or those that were undergoing budding. Alternate sterol supplements fed to GL7, 7-dehydroporiferasterol, brassicasterol, poriferasterol reached a stationary phase of 1 × 108 cells/ml similar to the ergosterol fed cells and each of the sterols was recovered from cells in unchanged form at 20 fg/cell. Alternatively, cells fed the 6-methyl aromatic sterol supplement failed to proliferate by 72 and microscopically many cells were clumped or dead by trypan blue staining typical of yeast cultured on a steroidal inhibitor [53]. Past interpretation using the GL7 system showing a no growth response following administration of nutritionally adequate amounts of sterol is the supplement failed to adequately fit the membrane structure [7, 8].
Figure 8.
Saccharomyces cerevisiae strain GL7 defective in ergosterol biosynthesis incubated with nutritional sterols supplements as described in Materials and Methods; ergosterol (black circle), 6-methyl sterol aromatics from A. castellanii (orange circle) or no sterol (blue circle). Note, 7-dehydroporiferasterol, brassicasterol and poriferasterol supplements supported growth in similar fashion to ergosterol-grown cells.
DISCUSSION
In spite of the therapeutic importance for an exacting sterol composition in parasitic protozoa [27], relatively little is known about the regulation and function of phytosterols in amoeba. This lack of information on metabolic controls is a serious impediment to the development of strategies for targeted inhibition of phyla-specific enzymes, such as the sterol C24-methylase, that can occur in the protozoan pathogen while absent from the human host. Previous functional and phylogenetic analysis of amoeba steroidogenesis has left the role and origins of C28- and C29- sterols somewhat opaque. In particular, while it seems clear the Acanthamoeba can synthesize ergosterol and 7-dehydroporiferasterol, it has been assumed there is one set of terminal compounds – Δ5,7-sterols - that act unaltered as membrane inserts and for the sterol biosynthetic genes to be descended from plants. However, the possibility for evolutionary accommodation of stage-specific differences in Δ5-, Δ5,7- and 6-methyl aromatic compounds in Acanthamoeba was indicated by GC-MS analysis of mixed populations of trophozoites and encysted cells, although it was ignored, leading to the view that Δ5- sterols are the precursor of Δ5,7-sterols, that the C24-methylation Δ24(28) -route typical of land plants yield intermediates for the synthesis of C28- and C29-sterols and for the sterolic enzymes to lack substrate specificity affording criss-crossing routes of major and minor biosynthesis pathways to final Δ5,7-sterol products [2, 29, 54]. Subsequent co-metabolite and phylogenetic analysis based on an erroneous characterization of many of the reported amoeba sterols, incorrect bioinformatic analyses for identity of the 2,3-oxidosqualene cyclase as lanosterol synthase and failure to report a SMT2-type isoform required in the synthesis of 24-ethyl sterols [34] developed an enigmatic history for Acanthamoeba ergosterol biosynthesis pathway as originating with the fungi.
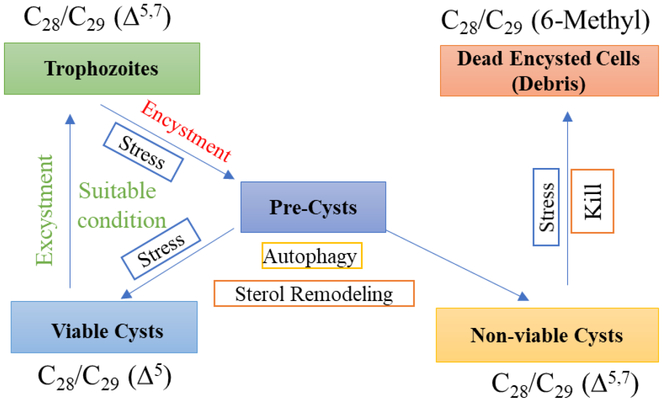
Here and in our previous paper [33] we provide evidence for the first time that indicates the sterol biosynthetic enzymes in Acanthamoeba were composed of catalytically constrained participants of strict substrate specificity, for sterol methylases to be responsible for generating distinct Δ24(28) - and Δ25(27) -olefin intermediates that convert to functional C28- and C29-sterol products of varied nucleus structures and that the conventional sterols typical of protozoa ergosterol and 7-dehydroporiferaterol can be replaced with the uncommon brassicasterolporiferasterol and amebasterol-1 and amebasterol-2 steroidal pairs during the encystment phase of the amoeba life history. Because methylation patterns in large part determine product outcome, subtle alterations in methyltransferase substrate selectivity have a profound impact on the balance of C28- and C29-sterols, which was noted following inhibitor treatment. The step-wise alteration in the sterol compositions observed during a normal excystment-encystment cycle yielding Δ5-sterols and 6-methyl aromatic sterols from a Δ5,7-sterol parent compound is not consistent with the limited sterol diversity in protozoa, rather it supports a simple model of cryptic unikont evolution not necessarily observed in the bikont amoeba Naegleria wherein a Δ5,7–sterol biosynthetic renewal is crucial to trophozoite proliferation, Δ5-sterol synthesis is essential for the production of a resting cyst capable of excystment and 6-methyl aromatic sterols, derived from Δ5,7-sterols in encysted cells considered non-viable for excystment, promote cell lysis and death as outlined in the proposed model of Acanthamoeba sterol biosynthesis and function shown in Figure 9.
Figure 9.
Proposed model for stage-specific sterols in growth and encystment leading to alternate trajectories for the production of viable or non-viable cysts.
Comparing the ontogenetically regulated sterol compositions in trophozoites and encysted cells and catalytic analysis of relevant chokepoint enzymes AcCYP51 and AcSMT is phylogenetically interesting yet connects to the functional importance of sterol products through modified structural features that occur in the stage-specific furnished molecule. Thus, proliferating trophozoites synthesize C28- and C -Δ5,7 29 -sterol products typical of kinetoplastids, such as Trypanosma cruzi, and the green algae synthesize ergosterol and 7-dehydroporiferasterol [19, 20, 57] but these organisms synthesize the C28- and C29-sterols through different biosynthesis routes. It appears AcCYP51 possesses substrate preference and catalytic competence more like that of plant, and hence protozoa, than of fungal or human CYP51 [6]. The substrate specificity of AcCYP51 places C14-demethylation after the first sterol methylation step (Fig. 6). However, while T. cruzi has a single SMT1 which is Δ24(25) - and Δ24(28) -substrate bifunctional capable of generating a single Δ24(28)-Z ethylidene C29 – product [55–57] and green alga SMT is substrate bifunctional capable of generating a single Δ25(27)-C29-sterol product [8], A. castellanii has two SMTs to carry out these sterol methylations at C24 with the second methylation step yielding the 24β-stereochemistry. As we discovered in this investigation through isotopically labeling studies to amoeba, the AcSMT2 (or 28-SMT) can, unlike the T. cruzi SMT1, synthesize the C -Δ25(27)29 -olefin outcome and the resulting intermediate is precursor to the 7-dehydroporiferasterol andporiferasterol. Fungi, on the other hand, synthesize a single SMT1 operating the Δ24(28) -route specific for zymosterol and ergosterol while land plants possess two SMTs and both operate the Δ24(28) –route such that separate intermediates are involved in the formation of C28α-campesterol and C29α-sitosterol, respectively [20]. The lineage-specific recurrence of an ancient Δ25(27) -sterol metabolic pathway affording primitive 24β-ethyl sterols in the unikonts suggest divergence with the ancestral SMT occurred independently multiple times during the course of eukaryote evolution and within the protozoa product-specific families of SMTs evolved unique to the Acanthamoeba. The notion of divergent evolution rather than convergent evolution for the amoeba SMTs is supported by the conservation of sequence similarity and gene structure among the diverse SMTs isolated throughout the eukaryotic domain to date [8, 59, 60].
The results of studies carried out so far on stage-specific sterol profiling in Acanthamoeba and for GL7 growth responses to sterol supplementation lead to the important conclusion that the membranes of trophozoites and resting cysts have certain structural requirements for sterol that are remarkably similar to yeast. They are clearly associated with the three-dimensional character of the molecule since C28- and/or C29-sterols are consistently found in the cells. In the trophozoite and resting cyst, the sterols possess a double bond in ring B of Δ5,7-bond (trophozoite) or Δ5-bond (resting cyst). The terminal formation of Δ5- sterols could represent a fine tuning in nucleus planarity but the structural change does not qualify for a significant effect on membrane fluidity. We surmise the conserved metabolism of Δ5,7-sterols to form a Δ5-sterol relates to its homoallylic effect on the C-O bond at C3. For the Δ5-sterol, donation of electrons through an inductive effect, analogously to the well-studied 3,5-cyclosteroid rearrangement, should strengthen the H-O bond and therefore weaken somewhat and fine-tune any hydrogen bonding of the sterol to phospholipid in the lipid leaflet of the resting cyst. It should be remembered that steric interference from C4-methyl groups on the hydrogen bonding ability of C3- group can weaken sterol-lipid interactions and therefore is reason for removal of C4 to generate an acceptable sterol membrane insert.
In depending upon Δ5,7-sterols to act as the encysted cell sterols, it is important not to overlook these cells are on a trajectory for lysis and death, which associate with rapid turnover of the Δ5,7-sterol, now as intermediates, to a 6-methyl aromatic sterol; cell death from these sterols is observed in GL7 growth experiments as well. The B-ring aromatic sterols effectively replace Δ5,7-sterols as the membrane component. It appears the architectural unsuitability of the phenanthrene sterols stems mainly from their diminished hydrogen-bonding strength of the C3-group resulting from the altered electronegativity of the polar group affected by the double bond conjugations in the nearby aromatic ring. This is consistent with their less polar chromatographic character in TLC observed too for 4-methyl sterols relative to 4-desmethyl sterols. Alternatively, contribution from the π-lobe negativity in the B-ring could minimize Van der Walls’ forces, disrupting lipid flexibility in the plasmamembrane.
Apart from these considerations and relevant to our research program in rational drug design [27, 33, 58], a major question brought forward by the developmentally regulated sterol biosynthetic enzymes of Acanthamoeba is to what extent they are exploitable by chemotherapy as the relevant sterol nucleus demethylase enzyme in Acanthamobea differs critically from those of its human host and the substrate requirements for sterol side chain methylases are unlike any of those for enzymes in the cholesterol biosynthesis pathway. Thus, the CYP51 and SMT enzymes in amoeba ergosterol biosynthesis represent potential species-specific drug targets. These enzymes at MAC concentrations of approximate 1 μM generate trophozoite kill, with the remaining viable cysts following excystment lysing as the inhibitor pressure continues. Although trophozoite kill is foremost the focus of our research, prevention of viable cysts that contain brassicaterol and poriferasterol is also relevant. In the latter case, the possibility for a transcriptional up-regulation of active Δ7-reductase affecting the conversion of Δ5,7 - to Δ5 -sterol during encystment and its impairment by an appropriate sterol biosynthesis inhibitor would, in combination therapy with azole or steroidal inhibitor, enable prevention or cure of diseases from Acanthamoeba. It could be as well the dramatic change in sterol profiles are a combination of anabolic and catabolic processes since the current study focuses on providing measurements of steady levels of particular sterols and are not measuring dynamic changes associated with biosynthesis. Further work is warranted to determine whether additional sterol metabolites or derivatives contribute to the changes noted in sterol-controlled encystment events and whether their formation are differentially sensitive to sterol biosynthesis inhibitors.
Conclusions.
To summarize the evidence, our findings revealed a new C29 Δ25(27) -sterol pathway in protozoa showing a phylogenetic closeness of Acanthamoeba to green alga and provide a powerful strategy to enhance anti-amoeba efficacy using combinations of azole and steroidal inhibitor to render resistant amoeba pathogens responsive to ergosterol biosynthesis inhibitor treatment. The capacity of steroidal inhibitors yet to be discovered (e.g., Δ7-reductase inhibitor) to modulate core encystation pathway yielding non-viable cells destined to die overcomes a major challenge for small molecule efficacy against the cysticidal effects of drugs necessary to prevent recurrence of the disease. The potency of tight binding inhibitors against A. castellanii target enzymes was confirmed by time-kill curves and through encystment-excystment assays. Finally, strict structural specificities for Δ5,7 - and Δ5-sterols in the autophagic stage of encystment correlated to altered membrane structures of encysted cells yielding non-viable or viable cysts; the 6-methyl aromatic sterols represent chemical signatures for amoeba death accordingly these compounds can be medical diagnostics.
Supplementary Material
Figure 4.
GC-MS analysis of the AcCYP51-generated products. (A) The total ion current chromatogram (insert- mass spectra of relevant GC peaks) of incubation with obtusifoliol. (B) The total ion current chromatogram (insert-mass spectra of relevant GC peaks) of incubation with cycloeucalenol.
Highlights:
Acanthamoeba castellanii cells generate 25 sterols, a mixture of 4-methyl intermediates and Δ5-final products, affording a biosynthetic pathway recurrent from an ancient algal predecessor; separate intermediates via the Δ24(28) -methyl olefin route and Δ25(27) - ethyl olefin route determine the C28- and C29-sterol balance in the amoeba.
Cloned AcCYP51 favors obtusifoliol as substrate, but can convert cycloeucalenol weakly to Δ14-product.
Δ5,7 - and Δ5-sterols are differentially correlated to trophozoite growth and mature cyst viability and excystment, respectively, while 6-methyl aromatic sterols are a chemical signature indicative of encysted cells on the path to death.
Voriconazole inhibits AcCYP51 as other azoles and 25-azacycloartanol inhibits AcSMT in the low nanmolar range; these drugs are potent inhibitors of trophozoite growth and impair encystment toward viable cyst production in the range of 500 nM to 3 μM.
Acknowledgements
This work was supported by the National Institutes of Health Grant R21(R33) AI119782 (WDN). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Abbreviations:
- Ac
Acanthamoeba castellanii
- SMT
sterol methyl transferase
- GC-MS
gas-chromatography-mass spectroscopy
- VOR
voriconazole
- AZC
25-azacycloartanol
- RRTc
retention time of unknown to retention time of cholesterol in GC
- Mya
million years ago
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Nes WD Biosynthesis of cholesterol and other sterols. Chem. Rev 111 (2011) 6423–6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Benveniste P Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol 55 (2004) 429–457. [DOI] [PubMed] [Google Scholar]
- [3].Nes WR & McKean ML Biochemistry of Steroids and Other Isoprenoids University Park Press, Baltimore: (1977) pp 325–555. [Google Scholar]
- [4].Kodner RB Sterols in a unicellular relative of the metazoans. Proc. Natl. Acad. Sci. USA 105 (2008) 9897–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nes WR and Nes WD Lipids in Evolution Plenum Press, New York: (1980). [Google Scholar]
- [6].Hargrove TY, et al. Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14α-demethylase CYP51 from Leishmania infantum. J. Biol. Chem 286 (2011) 26838–26848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bloch KE Sterols structure and membrane function. CRC Crit. Rev. Biochem 14 (1983) 47–82. [DOI] [PubMed] [Google Scholar]
- [8].Nes WD et al. The structural requirements of sterols for membrane function in Saccharomyces cerevisiae. Arch. Biochem. Biophys 300 (1993) 724–733. [DOI] [PubMed] [Google Scholar]
- [9].Lepesheva GI et al. Sterol 14α-demethylase inhibitors as potential target for antitrypanosomal therapy: Enzyme inhibition and parasite growth. Chem. & Biol 14 (2007) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].De Souza W & Fernandes Rodrigues JC Sterol biosynthesis pathway as target for anti-trypanosomal drugs. Interdisciplin. Perspect. Infect. Dis 2009 (2009) 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bloch KE Blondes in Venetian Paintings, the Nine-Banded Armadillo, and other Essays in Biochemistry, Yale University Press, New Haven: (1994). [Google Scholar]
- [12].Ourisson G and Nakatani. The terpenoid theory of the origin of cellular life: the evolution of terpenoids to cholesterol. Chem. Biol 1 (1994) 11–23. [DOI] [PubMed] [Google Scholar]
- [13].Summons RE et al. Steroids and triterpenoids and molecular oxygen. Phil. Trans. R. Soc 361 (2006) 951–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Knoll AH Paleobiological perspectives on early eukaryotic evolution In The Origin and Evolution of Eukaryotes, (Keeling PJ and Koonin EV, eds). Cold Spring Harbor Laboratory Press; (2014) pp 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Desmond E and Gribaldo S Phylogenomics of sterol synthesis: Insights into the origin, evolution and diversity of a key eukaryote feature. Genome Biol. Evol 1 (2011) 364–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gold DA et al. Paleoproterozoic sterol biosynthesis and the rise of oxygen. Nature 543 (2017) 420–423. [DOI] [PubMed] [Google Scholar]
- [17].Rezen T et al. New aspects on lanosterol 14alpha-demethylase and cytochrome P450 evolution: Lanosterol/cycloartenol diversification and lateral transfer. J. Mol. Evol 59 (2004) 51–58 (2004). [DOI] [PubMed] [Google Scholar]
- [18].Wei JH, Yin X, Welander PV Sterol synthesis in diverse bacteria. Front. Microbiol 7 (2016) 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Miller MB et al. Evolutionarily conserved Δ25(27)-ergosterol biosynthesis pathway in the alga Chlamydomonas reinhardtii is distinct from the Δ24(28)-pathway to fungal ergosterol. J. Lipid Res 53 (2012) 1636–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Haubrich BA et al. Characterization, mutagenesis and mechanistic analysis of an ancient algal sterol C24-methyltransferase: Implications for understanding sterol evolution in the green lineage. Phytochemistry 113 (2015) 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Garg VK Nes WR Changes in Δ5 – and Δ7- sterols during germination and seedling development of C. maxima. Lipids 20 (1985) 876–884. [Google Scholar]
- [22].Guo D, Venkatramesh M Nes WD Developmental regulation of sterol biosynthesis in Zea mays. Lipids 30 (1996) 203–219. [DOI] [PubMed] [Google Scholar]
- [23].Neff RJ, Ray SA, Benton WF, Wilborn M Chapter 4 Induction of synchronous encystment (differentiation) in Acanthamoeba sp., In: Prescott DM, Editor(s), Methods in Cell Biology, Academic Press, Vol. 1 (1964) pp 55–83. [Google Scholar]
- [24].Bowers B, Korn ED The fine structure of Acanthamoeba castellanii (Neff strain). J Cell. Biol 41 (1969) 786–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Moon E-K, Kim S-E, Hong Y, Chung D-II, Goo Y-K, Kong H-H. Autophagy inhibitors as a potential antiamoebic treatment for Acanthamoeba keratitis. Antimicrob. Agents Chemotherap 59 (2015) 4020–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Martin-Navarro CM, et al. Statins and voriconazole induce programmed cell death in Acanathamoeba castellanii. Antimicrob. Agents Chemotherap 59 (2015) 2817–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Haubrich BA, et al. Discovery of an ergosterol-signaling factor that regulates Trypanosoma brucei growth. J. Lipid Res 56 (2015) 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Raederstorff D, Rohmer M Sterol biosynthesis de novo via cycloartenol by the soil amoeba Acanthamoeba polyphaga. Biochem. J 231 (1985) 609–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Raederstorff D, Rohmer M The action of the systemic fungicides tridemorph and fenpropimorh on sterol biosynthesis by the soil amoeba Acanthamoeba polyphaga. Eur. J. Biochem 164 (1987) 421–426. [DOI] [PubMed] [Google Scholar]
- [30].Korn ED, et al. The enzymatic aromatization of the B-ring of Δ5,7-sterol. Biochim. Biophys. Acta 187 (1969) 553–563. [DOI] [PubMed] [Google Scholar]
- [31].Bisseret P, Adam H, Rohmer M Structural elucidation of ring-B aromatic sterols of the soil amoeba Acanthamoeba polyphaga. J. Chem. Soc. Chem. Commun (1987) 693–695. [Google Scholar]
- [32].Lamb DC, et al. Azole antifungal agents to treat the human pathogens Acathanomoeba castellanii and Acanthamoeba polyphagy through inhibition of sterol 14-demethylase (CYP51). Antimicrob. Agents Chemother 59 (2015) 4707–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kildane ME, et al. Sterol methyltransferase a target for anti-amoeba therapy: Towards transition state analog and suicide substrate design. J. Lipid Res 58 (2017) 2310–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Thomson S, et al. Characterisation of sterol biosynthesis and validation of 14α-demethylase as a drug target in Acanthamoeba. Sci. Rep 7 (2017) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mehdi H, Garg NK. Changes in the lipid composition and activities of isocitrate dehydrogenase and isocitrate lyase during encystation of Acanthamoeba culbertsoni strain A1. Trans. Soc R. Tropical Med. Hygen 81 (1987) 633–636 [DOI] [PubMed] [Google Scholar]
- [36].Zhou W, Lepesheva GI, Waterman MR Nes WD Mechanistic analysis of a multiple product sterol methyltransferase from Trypanosoma brucei implicated in ergosterol biosynthesis.. J. Biol. Chem 281 (2006) 6290–6296. [DOI] [PubMed] [Google Scholar]
- [37].Xu S, Norton RA, Crumley FG Nes WD Comparison of the chromatographic properties of sterols, select additional steroids and triterpenoids: Gravity-flow liquid chromatography, thin-layer chromatography, gas-liquid chromatography, and high performance liquid chromatography. J. Chromatogr 452 (1988) 377–398. [DOI] [PubMed] [Google Scholar]
- [38].Copeland RA Enzymes 2nd Edition Wiley-VCH; New York: (2000). [Google Scholar]
- [39].Ramgopal M, Bloch K Sterol syngerism in yeast. Proc. Natl. Acad 80 (1983) 712–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Smith FR Korn ED 7-Dehydrostigmasterol and ergosterol: The major sterols of an amoeba. J. Lipid Res 9 (1968) 405–408. [PubMed] [Google Scholar]
- [41].Nes WD, et al. Concerning the role of 24,25-dihydrolanosterol and lanostanol in sterol biosynthesis by cultured cells. Steroids 53 (1988) 461–475. [DOI] [PubMed] [Google Scholar]
- [42].Taton M and Rahier A Properties and structural requirements for substrate specificity of cytochrome P450-dependent obtusifoliol 14-alpha demethylase from maize (Zea mays) seedlings. Biochem. J 277 (1990) 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bellamine A, et al. Structural requirements for substrate recognition of Mycobacterium tuberculosis 14a-demethylase: implications for sterol biosynthesis. J. Lipid Res 42 (2001) 128–136. [PubMed] [Google Scholar]
- [44].Nes WD, et al. 9β,19-Cyclosterol analysis by 1H- and 13C-NMR, crystallographic observations and molecular mechanics calculations. J. Amer. Chem. Soc 120 (1998) 5970–5980. [Google Scholar]
- [45].Gas-Pascual E et al. Plant oxidosqualene metabolism: Cycloartenol synthase-dependent sterol biosynthesis in Nicotiana benthamiana. PlosOne 9 (2014) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Goad LJ and Akihisa T Analysis of Sterols Blackie Academic, London: 1997. 437pp. [Google Scholar]
- [47].Zhou W, Cross GAM, Nes WD J. Lipid Res 48 (2007) 665–673. [DOI] [PubMed] [Google Scholar]
- [48].Nes WD, Heupel RC Le PH Biosynthesis of ergosta-6(7),8(14),22(23)-trien-3b-ol by Gibberella fujikuori: Its importance to ergosterol’s biosynthesis pathway. Chem. Commun (1985) 1431–1433. [Google Scholar]
- [49].Muller C, Binder U, Bracher F Giera M Antifungal drug testing by combining minimal inhibitory concentration testing with target identification by gas chromatography-mass spectroscopy. Nat. Protocols 12 (2017) 947–963. [DOI] [PubMed] [Google Scholar]
- [50].Mehdi H Sterols of Acanthamoeba culbertsoni strain A-1. Steroids 551 (1988) 551–558. [DOI] [PubMed] [Google Scholar]
- [51].Nes WR Biochemistry of plant sterols. Adv. Lipid Res 15 (1977) 233–359. [Google Scholar]
- [52].Roti LW Stevens AR Effect of 5-bromodeoxyuridine on growth, encystment, and excystment of Acanthamoeba castelanii. J. Cell Biol 61 (1974) 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nes WR Dhanuka IC Inhibition of sterol synthesis by Δ5-sterols in a sterol auxotroph of yeast defective in oxidosqualene cyclase and cytochrome P-450. J. Biol. Chem 263 (1988) 11844–11850. [PubMed] [Google Scholar]
- [54].Roberts CWR et al. Fatty acid and sterol metabolism: potential antimicrobial targets in apicomplexan and trypanosomatid parasitic protozoa. Mol. Biochem. Parasitol 126 (2003) 129–142. [DOI] [PubMed] [Google Scholar]
- [55].Urbina JA et al. Modification of the sterol composition of Trypanosoma (Schizotrypanum) cruzi epimastigotes by D24(25)-sterol methyltransferase inhibitors and their combinations with ketoconazole. Mol. Biochem. Para 34 (1995) 199–210. [DOI] [PubMed] [Google Scholar]
- [56].Sowa MA Characterization and inhibition of C24-sterol methyltransferase of Trypanosoma cruzi Master of Science, Texas Tech University; (2016) 1–65. [Google Scholar]
- [57].Sterol methylltransferse enzyme and its chemotherapeutic implications for Chagas disease Doctoral Dissertation, Texas Tech University; (2014) 1–76. [Google Scholar]
- [58].Kanagasabai R, et al. Disruption of ergosterol biosynthesis, growth and the morphological transition in Candida albicans by sterol methyltransferase inhibitors containing sulfur at C-25 in the sterol side chain. Lipids, 39 (2004) 737–746. [DOI] [PubMed] [Google Scholar]
- [59].Gold DA et al. Sterol and genomic analyses validate the sponge biomarker hypothesis. Proc. Natl. Acad. Sci. USA 113 (2016) 2684–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Neelakandan AK et al. Cloning, functional expression, and phylogenetic analysis of plant 24C-methyltransferases involved in sitosterol biosynthesis. Phytochemistry, 70 (2009) 1982–1998. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.