Abstract
While much is known about the genes and proteins that make up the circadian clocks in vertebrates and several arthropod species, much less is known about the clock genes in many other invertebrates, including nudibranchs. The goal of this project was to identify the RNA and protein products of putative clock genes in the central nervous system of three nudibranchs, Hermissenda crassicornis, Melibe leonina, and Tritonia diomedea. Using previously published transcriptomes (Hermissenda and Tritonia) and a new transcriptome (Melibe), we identified nudibranch orthologs for the products of five canonical clock genes: brain and muscle aryl hydrocarbon receptor nuclear translocator like protein 1, circadian locomotor output cycles kaput, non-photoreceptive cryptochrome, period, and timeless. Additionally, orthologous sequences for the products of five related genes—aryl hydrocarbon receptor nuclear translocator like, photoreceptive cryptochrome, cryptochrome DASH, 6-4 photolyase, and timeout—were determined. Phylogenetic analyses confirmed that the nudibranch proteins were most closely related to known orthologs in related invertebrates, such as oysters and annelids. In general, the nudibranch clock proteins shared greater sequence similarity with Mus musculus orthologs than Drosophila melanogaster orthologs, which is consistent with the closer phylogenetic relationships recovered between lophotrochozoan and vertebrate orthologs. The suite of clock-related genes in nudibranchs includes both photoreceptive and non-photoreceptive cryptochromes, as well as timeout and possibly timeless. Therefore, the nudibranch clock may resemble the one exhibited in mammals, or possibly even in non-drosopholid insects and oysters. The latter would be evidence supporting this as the ancestral clock for bilaterians.
Introduction
Most organisms express daily rhythms of activity and physiology that persist in constant conditions and thus are referred to as circadian rhythms. These circadian rhythms are produced by molecular clocks that exhibit cyclical production of various intracellular proteins as the result of transcription-translation feedback loops. The first clock protein to be discovered was PERIOD, initially identified in Drosophila melanogaster by Konopka and Benzer (1971). Since then, homologs of PERIOD have been found in numerous other bilaterian organisms, including humans (Tei et al., 1997). Additional circadian proteins that participate in the molecular clock have been identified and sequenced in a wide array of species, ranging from cyanobacteria to mammals (reviewed in Dunlap, 1999; Zhang and Kay, 2010). Although the individual proteins can vary between widely disparate phylogenetic groups, the presence of negative and positive feedback elements regulating the transcription and translation of each other appears to be a ubiquitous component of circadian pacemakers in all organisms studied to date.
In animals, the molecular mechanisms of the circadian clock are best understood in Drosophila and mammals. While the Drosophila clock is quite derived and different from that of some other insects (Rubin et al., 2006; Zhu et al., 2008; Ingram et el., 2012), we focus on it here because it is one of the best-characterized circadian systems. Briefly, in Drosophila, the canonical core clock proteins consist of circadian locomotor output cycles kaput (CLOCK) and CYCLE, which work together to drive the production of PERIOD and TIMELESS. These latter two proteins feed back on their own transcription, forming a negative feedback loop with a circadian periodicity. A blue-light photoreceptive CRYPTOCHROME (PCRY) is important in providing light input to the clock by triggering the degradation of TIMELESS (Hardin, 2005; Rosato et al., 2006). In mammals, CLOCK and brain and muscle aryl hydrocarbon receptor nuclear translocator-like protein 1 (BMAL1; a homolog of CYCLE) heterodimerize and stimulate transcription and translation of PERIOD and a vertebrate, nonphotoreceptive CRYPTOCHROME (NPCRY). PERIOD and NPCRY then serve as the negative inhibitors of the clock (Reppert and Weaver, 2002; Lowrey and Takahashi, 2004). NPCRY is closely related to a protostome NPCRY, found in many invertebrates, but not present in Drosophila (Lin and Todo, 2005; Özturk, 2016; Michael et al., 2017). A TIMELESS protein was initially identified in mammals (Koike et al., 1998; Sangoram et al., 1998; Takumi et al., 1999), but this is now believed to actually be an ortholog of TIMEOUT (Li et al., 2016), which is a paralog of TIMELESS.
The molecular circadian clocks in mammals and fruit flies represent two major lineages of animals: deuterostome chordates and protostome ecdysozoans, respectively. As described above, these molecular clocks are quite similar operationally, but they differ in the proteins used for negative feedback and how light input is transmitted to the clock. While circadian genes have been identified in some species in the other major protostome lineage, Lophotrochozoa (Zantke et al., 2013; Bao et al., 2017; Perrigault and Tran, 2017; Schnytzer et al., 2018), we do not yet fully understand the molecular mechanisms underlying clocks in lophotrochozoans as a group. Further elucidating the molecular basis of circadian clocks in this clade may shed light on the evolution of circadian clocks.
Circadian rhythms in gastropod molluscs, which are a lophotrochozoan clade, were first discovered 50 years ago in Aplysia californica (Kupfermann, 1968; Jacklet, 1972; Block and Lickey, 1973; Lickey et al., 1977). Subsequently, circadian rhythms of locomotion, oxygen consumption, and ocular electrical activity have been identified in several other gastropods, including Bulina tropicas (Chaudhry and Morgan, 1983), Bulla gouldiana (Block and Davenport, 1982), Bursatella leachi (Block and Roberts, 1981), Helisoma trivolvis (Kavaliers, 1981), Helix aspersa (Bailey, 1981; Blanc, 1993), Hydrobia ulvae (Barnes, 1986), Limax maximus (Sokolove et al., 1977), Littorina irrorata (Shirley and Findley, 1978), Melanerita atramentosa (Zann, 1973), Melanoides tuberculata (Beeston and Morgan, 1979), and Melibe leonina (Newcomb et al., 2014). However, despite strong interest in gastropod circadian rhythms, and the advantages of this group of animals for investigating the neuronal bases of behaviors, there has been very little progress in identifying circadian genes in gastropods, with the exception of the transcript for period in Bulla gouldiana (Constance et al., 2002), the basic helix-loop-helix (bHLH)-containing proteins BMAL1 and CLOCK in Biomphalaria glabrata, Lottia gigantea, and Patella vulgate (Bao et al., 2017), and some automated annotations on GenBank at the National Center for Biotechnology Information (NCBI, Bethesda, MD). Furthermore, after submission of this paper, Schnytzer et al. (2018) published a study reporting on the transcript sequences for most of the core circadian clock genes in the limpet Cellana rota.
The goal of this study was to identify the core circadian gene products in the central nervous systems (CNS) of three nudibranchs that are common neurophysiological model systems: Hermissenda crassicornis (Eschscholtz, 1831), Melibe leonina (Gould, 1852), and Tritonia diomedea Bergh, 1894 (synonym = Tritonia tetraquetra [Pallas, 1788]), using bioinformatic analyses of recently assembled transcriptomes (Senatore et al., 2015; Tamvacakis et al., 2015). Gene products were identified in all three species for a number of canonical clock genes, as well as some associated sequences, and compared to sequences in clock proteins in other species.
Materials and Methods
Transcriptomes
Previously published CNS transcriptomes were used for Hermissenda and Tritonia (Senatore et al., 2015; Tamvacakis et al., 2015), while the Melibe transcriptome was developed using 15 specimens collected by Monterey Abalone Company (Monterey, CA). Total RNA was extracted from the CNS (the fused cerebropleural and pedal ganglia, plus the buccal ganglia), using an RNeasy Plus Universal Midi Kit (QIAGEN, Hilden, Germany). RNA integrity was confirmed by visualization of electrophoresed RNA on an ethidium bromide-stained agarose gel, as well as with a bioanalyzer, using an Agilent RNA 6000 Pico Kit (Agilent Technologies, Waldbronn, Germany). RNA was then diluted to ~100 ng μl−1 in Trisethylenediaminetetraacetic acid (EDTA) buffer (1 mmol l−1 EDTA, 10 mmol l−1 Tris, pH 7.5) and shipped frozen on dry ice to Beckman Coulter Genomics (Danvers, MA) for cDNA synthesis and Illumina sequencing. At Beckman Genomics, mRNA was isolated via polyA capture, and cDNA was synthesized and amplified using proprietary primers. Paired-end sequencing of cDNA was performed on an Illumina HiSeq2500 platform (2 × 100 bp, Illumina, San Diego, CA), followed by the removal of adaptor sequences and low-quality reads. After delivery of the data, we further trimmed paired reads with the program Sickle (Joshi and Fass, 2011) to remove 3′ and 5′ ends as well as entire reads shorter than 20 bp or with quality scores less than 20 (i.e., default parameters). A de novo transcriptome shotgun assembly (TSA) was then developed using Trinity (release 2013-08-14; Grabherr et al., 2011; Haas et al., 2013; Senatore et al., 2015; Tamvacakis et al., 2015).
Identification of homologous clock sequences
The creation of a bioinformatic pipeline (Pankey, 2018) significantly increased the efficiency with which contiguous sequences from the nudibranch transcriptomes could be identified as potential gene orthologs (Fig. 1). More importantly, this pipeline incorporated phylogenetic analyses to discriminate sequences with orthology to specific clock genes from paralogous proteins that do not serve a circadian clock function. After TSA assembly, candidate coding regions from each nudibranch transcriptome were identified via the in silico translation of the Trinity data sets using TransDecoder (Haas et al., 2013). The translated transcriptome for each nudibranch species could then be imported into the bioinformatic pipeline for the automated discovery of circadian clock gene orthologs.
Figure 1.
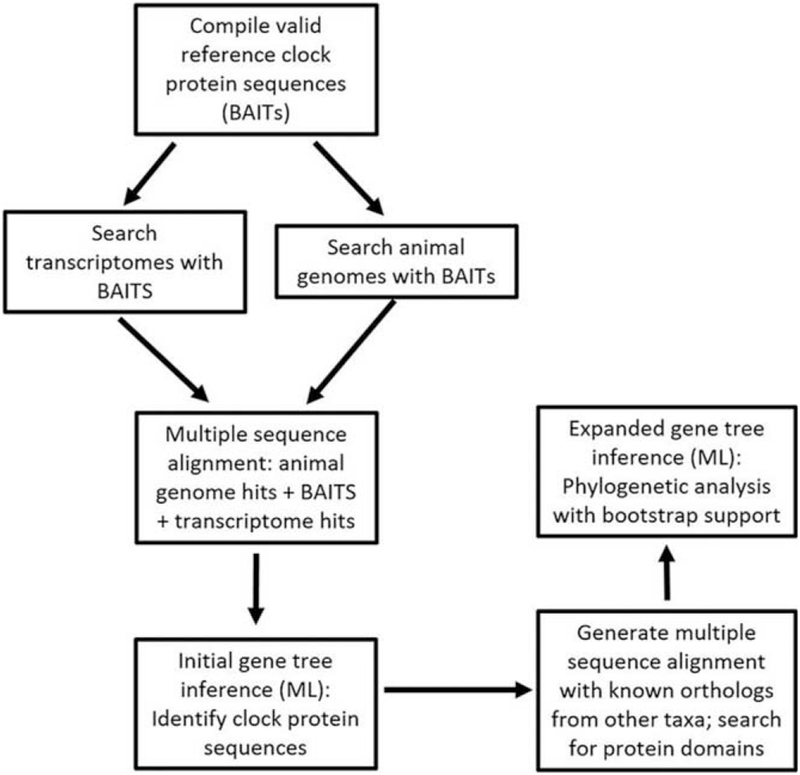
Schematic flow diagram of the steps taken to sequence and compare clock proteins. The creation of a bioinformatic pipeline significantly increased the efficiency with which contiguous sequences from the nudibranch transcriptomes could be identified as potential gene orthologs; more importantly, it accurately determined whether they were most closely associated with the products of specific clock genes or more similar to structurally related proteins that do not serve a circadian clock function. To begin, valid reference clock protein sequences (what we termed “BAIT” sequences) were compiled and used to fish out contiguous sequences from the nudibranch transcriptome assemblies. After aligning all sequences, contiguous sequences were then mapped on a phylogenetic tree with the BAIT sequences as well as closely matched protein sequences from automated BLAST searches. This enabled us to identify putative clock protein sequences from each nudibranch transcriptome. ML, maximum likelihood.
Next, various databases (e.g., GenBank, UniProt) were manually queried to locate and compile a valid reference set for each of the following protein sequences: CLOCK, PERIOD, BMAL1, CYCLE, aryl hydrocarbon receptor nuclear translocator-like (ARNTL), PCRY, NPCRY, 6-4 PHOTOLYASE (PHR), CRYPTOCHROME DASH (CRY DASH), TIMELESS, and TIMEOUT. Bait sequences are indicated in blue font in Supplementary Figures 1–5, and accession numbers for these proteins can be found in Supplementary Table 1 (Supplementary Figs. 1–9 and Supplementary Tables 1–3 are available online). These protein sequences were used as queries in BLASTp (NCBIBLAST+) searches of all curated animal genomes on NCBI, as well as the translated nudibranch transcriptomes (Altschul et al., 1990; Fig. 1). Similar to SmartBlast (NCBI), the parallel execution of these tasks efficiently identified protein sequences for targeted circadian clock genes from the nudibranch transcriptomes as well as a diverse array of metazoan taxa that aided with the subsequent rooting of phylogenetic trees and the locating of paralogous gene clades.
Last, the full-length protein sequences for each circadian gene recovered from genome searches, along with the initial query sequences, were aligned using MUSCLE, version 3.8.31 (Fig. 1; Edgar, 2004). Each nudibranch predicted peptide fragment was then iteratively aligned to the full-length alignment, using the “-profile” setting in MUSCLE. With each resulting alignment, RAxML, version 7.3.0, was used to infer an initial gene tree by maximum likelihood (ML) under PROTGAMMAWAG and to assess support for bipartitions via 1000 bootstrap replicates (see Supplementary Figs. 1–5, available online; Yang, 1996; Whelan and Goldman, 2001; Stamatakis, 2014)
Alignment, functional domains, and phylogenetic analyses of homologous clock sequences
The alignments and phylogenetic analyses described above were used as an initial step to identify candidate clock sequences in the three nudibranch transcriptomes. We then used additional alignments and phylogenetic analyses, as described below, for comparing conserved functional domains. To begin, orthologous sequences for each of the 10 circadian clock proteins targeted by this investigation were compiled from different lophotrochozoan, ecdysozoan, and chordate taxa into a reference set (see Supplementary Table 1, available online). Two multiple sequence alignments (MSAs) were sequentially performed on nudibranch protein sequences with orthology to specific clock genes, using Clustal Omega (Sievers et al., 2011). This two-step process prepared the collection of protein sequences associated with each clock gene for phylogenetic analysis by ML.
The first MSA included a circadian clock protein sequence (e.g., CLOCK) mined from each nudibranch transcriptome—when available—and the relevant orthologs from a small subset of taxa. This facilitated the efficient identification of conserved functional protein domains among lophotrochozoan, ecdysozoan, and/or chordate taxa using Normal SMART (SMART Technologies, Calgary, Alberta, Canada) (Schulz et al., 1998; Letunic et al., 2017). Once the conserved functional domains in each circadian clock protein had been identified and mapped—effectively creating a guide for their rapid identification in orthologous protein sequences from other taxa—sequences flanking these conserved domains were trimmed, and a second MSA was performed.
The refined protein sequences for each clock gene were then subjected to phylogenetic analysis in MEGA7 (Kumar et al., 2016), using ML with 1000 bootstrap replications. The Whelan and Goldman (WAG) model of amino acid substitution, with gamma-distributed and invariant (G+I) sites with five discrete gamma categories, was used for each analysis. The tree inference options were set to nearest-neighbor interchange, with an automatically generated (neighbor-joining [NJ] and BioNJ) initial tree. The tree with the highest log likelihood was considered the best for each phylogenetic analysis.
Results
Sequencing and TSA metrics were previously provided for Hermissenda (Tamvacakis et al., 2015) and Tritonia (Senatore et al., 2015). For Melibe, sequencing resulted in 123,087,512 paired-end reads, 94.5% of which had a Phred score greater than 30. De novo assembly with Trinity yielded 167,841 transcripts with an N50 of 1528 bp; these partitioned into 93,882 nonredundant gene clusters (unigenes). In silico translation of the transcriptome data set with TransDecoder yielded 39,641 predicted protein sequences. Paired-end reads and the TSA were uploaded to NCBI as a BioProject (accession no. PRJNA420367).
Five core clock orthologs were identified in all three nudibranch species (BMAL1, CLOCK, PERIOD, TIMELESS, and NPCRY; Table 1). In addition, we found sequences in the three species for ARNTL, TIMEOUT, and three other CRYPTOCHROMES (PCRY, PHR, and CRY DASH; Table 1). BLASTp analyses provided evidence supporting the similarity of these sequences to other molluscan clock-related proteins and a closer affinity to mammalian orthologs than those of Drosophila (Supplementary Tables 2, 3, available online). These results were further supported by the more rigorous phylogenetic analyses described below and in Supplementary Figures 1–5, available online.
Table 1.
Identified core circadian gene transcripts for the three nudibranchs
| Designation | Hermissenda crassicornis | Melibe leonina | Tritonia diomedea |
|---|---|---|---|
| clock | MG162587, C, 650 | MG189943, C, 651 | MG253827, P, 504 |
| bmal1 | MG282902, C, 698 | MG282903, P, 126 | |
| arntl | MG282904, P, 252 | MG282905, C, 631 | MG282906, C, 633 |
| period | MG282907, C, 1615 | MG282908, C, 1491 | MG427049, C, 1565 |
| npcry | MG437149, C, 561 | MG437150, C, 556 | MG437151, C, 559 |
| pcry | MG437152, C, 547 | MG470828, C, 543 | MG470829, C, 542 |
| phr | MG470830, P, 162 | MG470831, P, 236 | MG516799, C, 530 |
| cry dash | MG516800, C, 527 | MG516801, C, 561 | MG516802, C, 550 |
| timeless | MG516803, P, 608 | MG516804, P, 696 | MG516805, P, 632 |
| timeout | MG549825, C, 673 | MG549826, P, 381 | MG549827, P, * |
In the species columns, each data cell contains the accession number, the designation of a complete (C) or partial (P) sequence, and the number of amino acids in the translated protein. Sequences were determined as complete or partial on the basis of alignment with orthologous sequences and the presence of start and stop codons. Protein translations were done with the standard settings in the ExPASy Translate Tool (Artimo et al., 2012). An asterisk indicates multiple noncontiguous nucleotide sequences, which thus could not be translated.
clock
In all three nudibranchs, we identified a single RNA transcript for the gene clock (Table 1). The length of the predicted amino acid sequence was similar for Hermissenda (650 amino acids) and Melibe (651), whereas in Tritonia, CLOCK was >100 amino acids shorter (504), indicating that it was likely a partial sequence. A BLAST search indicated that Melibe CLOCK had the highest BLAST score with a predicted CLOCK sequence in the sea hare Aplysia californica (63% identity; Supplementary Table 2, available online). The nudibranch CLOCK proteins contained the conserved regions present in orthologs of other species, including a helix-loop-helix (HLH) domain, two Per-Arnt-Sim (PAS) domains, and aPAS-associated C-terminal (PAC) motif (see Fig. 2 for example of the alignment; alignments for subsequent circadian transcripts are provided in Supplementary Figs. 6–9, available online). Similar to BLASTp comparisons (Supplementary Tables 2, 3), phylogenetic analysis indicated that the nudibranch CLOCK proteins evolved from an ancestral lophotrochozoan ortholog with a closer relationship to chordate CLOCK proteins than arthropod (ecdysozoan) orthologs, suggesting a dynamic evolutionary history of duplication and losses of this homolog (Fig. 3; Supplementary Fig. 1, available online).
Figure 2.
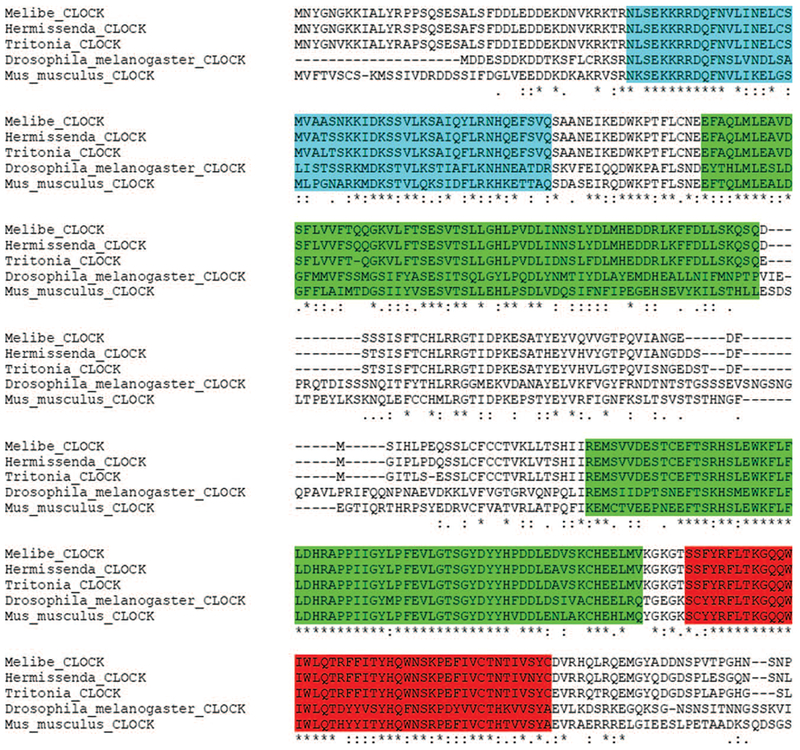
Alignment of conserved regions for CLOCK. The nudibranch CLOCK proteins contained conserved domains present in orthologs for Drosophila melanogaster and Mus musculus: blue indicates the helix-loop-helix domains, light green the Per-Arnt-Sim (PAS) domains, and red the PAS-associated C-terminal domains. Symbols immediately below the aligned sequences indicate functional similarity of residues among the CLOCK protein orthologs from the five different taxa. An asterisk indicates identical residue, a colon indicates residues with strongly similar properties, and a period indicates residues with weakly similar properties.
Figure 3.

Maximum likelihood analysis indicating the evolutionary relationships between CLOCK orthologs. The nudibranch CLOCK proteins grouped together with orthologs in other molluscs and shared a more recent common ancestor with chordate CLOCK proteins than arthropod orthologs. The tree was rooted on CLOCK from the cnidarian Nematostella vectensis. Numbers at branch points indicate bootstrap values for maximum likelihood analysis with 1000 replicates.
bmal1
A single RNA transcript for the bmal1 gene was identified in Melibe, which encoded a protein of 698 amino acids (Table 1). Only a partial bmal1 transcript could be determined in Tritonia (encoding 126 amino acids), and the bioinformatics pipeline did not identify a bmal1 sequence in Hermissenda. The predicted Melibe BMAL1 protein contained the conserved regions present in orthologs of other species, including the HLH, PAS, and PAC domains (Supplementary Fig. 6, available online). A BLAST search indicated that Melibe BMAL1 was most similar to a predicted ARNTL1 protein in the freshwater snail Biomphalaria glabrata (69% identity; Supplementary Table 2, available online).
In addition to bmal1, complete RNA transcripts of the related arntl gene were identified in all three species, and the Melibe ARNTL protein was used to root the phylogenetic analysis of BMAL1 (Fig. 4). Results of this analysis suggest that Melibe BMAL1 shares a sister grouping with the other molluscan BMAL1 (Crassostrea gigas) and that these arose from a homolog in a lophotrochozoan ancestor. As with CLOCK and BLASTp analyses (Supplementary Table 3, available online), the Melibe BMAL1 shared greater sequence similarity with chordate BMAL1 proteins than crustacean BMAL1 proteins or the related CYCLE found in insects.
Figure 4.
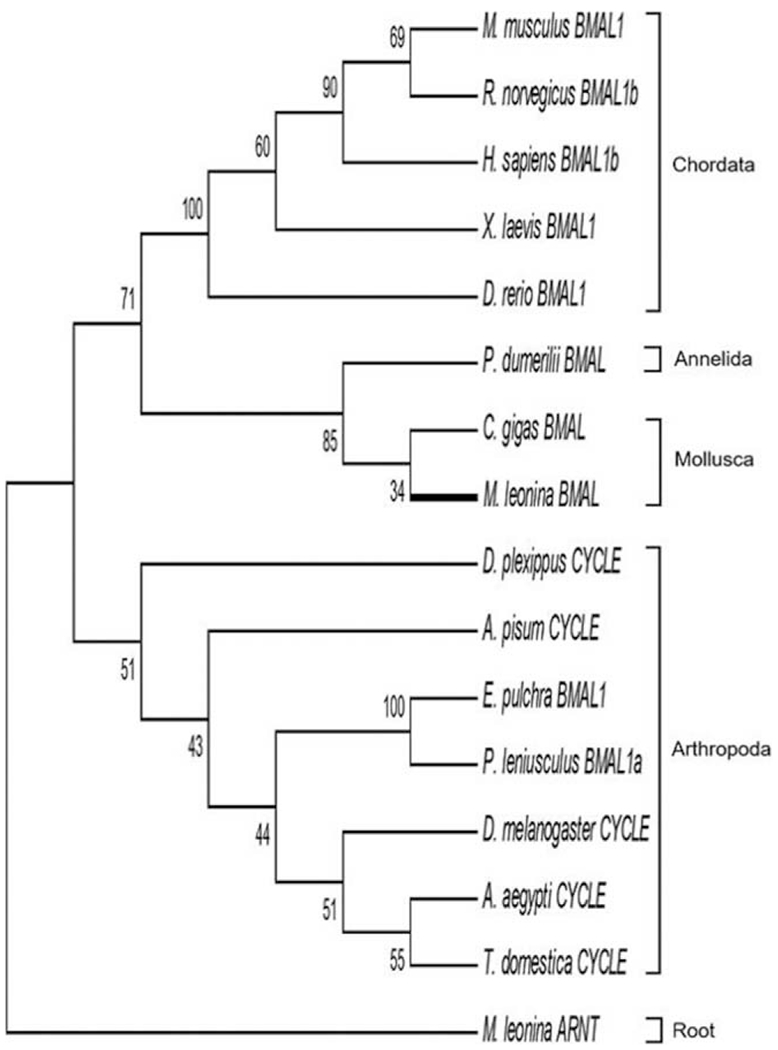
Evolutionary relationships of BMAL1/CYCLE orthologs, based on maximum likelihood analysis. In this study, a complete sequence for BMAL1 was identified only in Melibe leonina. The putative Melibe BMAL1 was closely related to orthologs in other lophotrochozoans (e.g., molluscs and annelids) and, similar to CLOCK, shared a more recent common ancestor with chordate BMAL1 proteins than arthropod orthologs. The tree was rooted on the newly identified ARNTL protein in Melibe.
period
A single transcript for the period gene was identified for all three nudibranchs (Table 1). Melibe PERIOD had the highest BLAST score to a predicted, but uncharacterized, protein in Aplysia, based on a BLASTp search (53% identity; Supplementary Table 2, available online). All three nudibranch PERIOD proteins contained the conserved HLH, PAS, and PAC domains present in orthologs of other species (Supplementary Fig. 7, available online). Phylogenetic analysis of these conserved regions suggested that, as with CLOCK and BMAL1, the nudibranch PERIOD proteins evolved from a lophotrochozoan ancestor (Fig. 5). In contrast to BLASTp comparisons that suggested that Melibe PERIOD was more similar to Mus than Drosophila (Supplementary Table 3, available online), phylogenetic analysis suggested that molluscan PERIOD proteins shared a common ancestor more recently with protostomes other than chordates (Fig. 5). However, low bootstrap support at deep nodes, together with the absence of non-bilaterian PERIOD orthologs (Supplementary Fig. 3, available online) to serve as an outgroup, hinder efforts to resolve the evolutionary history of this key player.
Figure 5.
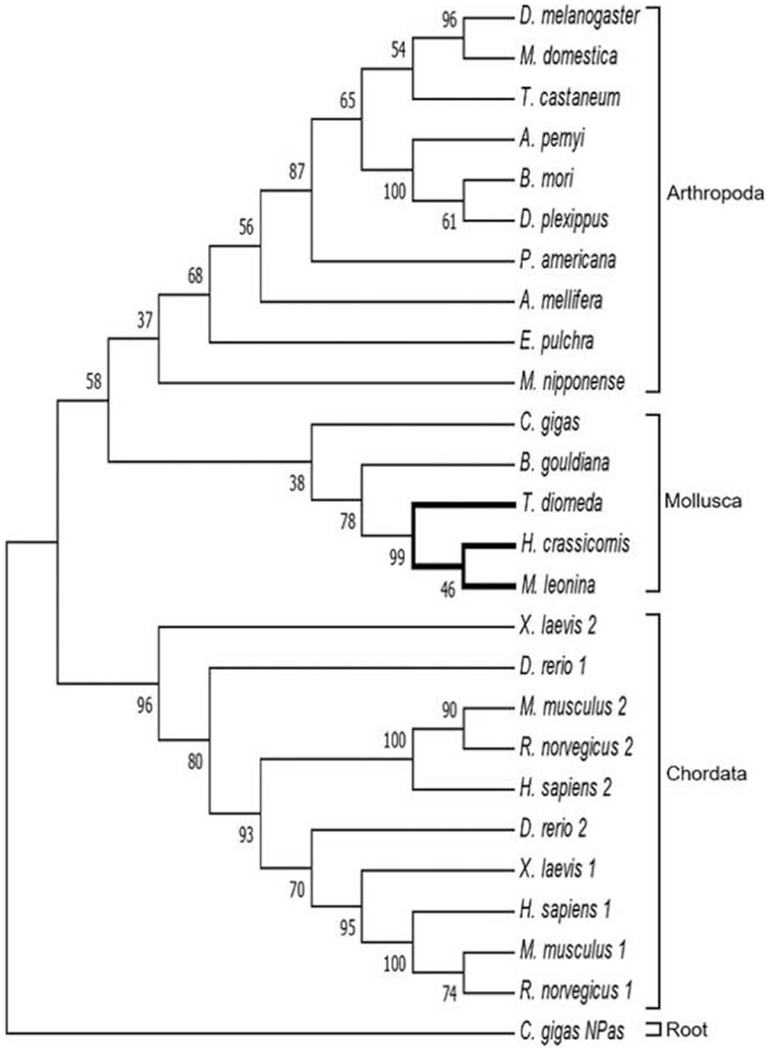
Phylogenetic tree indicating evolutionary relationships of PERIOD orthologs across taxa. The nudibranch PERIOD proteins were most closely related to other molluscs but, unlike CLOCK and BMAL1, shared more recent common ancestry with arthropods than chordates, though with low bootstrap support for this relationship. The tree was rooted on NEURONAL NPAS DOMAIN PROTEIN (NPAS) from the oyster Crassostrea gigas (Perrigault and Tran, 2017).
Cryptochrome
We identified four distinct cryptochrome transcripts in all of the nudibranchs: npcry, pcry, phr, and cry dash (Table 1; Supplementary Fig. 4, available online). All of the translated protein sequences were complete, with the exception of the Hermissenda and Tritonia CRY DASH proteins. BLASTp searches (Supplementary Table 2, available online) indicated that Melibe NPCRY was most similar to a predicted cryptochrome-1-like protein in Aplysia californica (85% identity). Melibe PCRY was also most similar to a predicted cryptochrome-1-like protein, but in Biomphalaria glabrata (69% identity). NPCRY is a core clock protein in mammals, and possibly many invertebrates as well, so we compared conserved regions of the nudibranch NPCRY proteins with those of the bivalve Crassostrea (Supplementary Fig. 8, available online). The nudibranch NPCRY proteins all contained the conserved DNA photolyase and flavin adenine dinucleotide (FAD) binding 7 domains, as indicated by analysis in SMART software (SMART Technologies). PCRY is a core clock protein in Drosophila and also contains the same conserved DNA photolyase and FAD binding 7 domains as NPCRY, so our ML analysis included protein sequences from both of these cryptochrome families (Fig. 6). The nudibranch NPCRY and PCRY sequences were clustered together in their respective clades and grouped with other molluscan proteins. Similar to findings in other studies, the lophotrochozoan NPCRY proteins shared a common ancestor with vertebrate NPCRY proteins. PCRY formed a clade separate from NPCRY, and molluscan PCRY proteins were homologous to insect PCRY proteins.
Figure 6.

Phylogenetic tree of NPCRY and PCRY orthologs, based on maximum likelihood analysis of conserved regions. The nudibranch NPCRY proteins were similar to other molluscs and shared more recent ancestry with orthologs in chordates than arthropods. Nudibranch PCRY proteins were also grouped with molluscs. Chordates do not have orthologs for PCRY. The tree was rooted on a CRY protein from the sponge Amphimedon queenslandica (Rivera et al., 2012).
timeless and timeout
We identified a complete RNA transcript for the timeout gene in Hermissenda, but only partial sequences for timeout in the other two species, as well as only partial transcripts for the timeless gene in all three nudibranchs (Table 1). Hermissenda TIMEOUT contained the conserved TIMELESS domain present in orthologs in other species (Supplementary Fig. 9, available online). Phylogenetic analysis with the Hermissenda TIMEOUT suggests that it shares homology with nematode and chordate orthologs (Fig. 7). Although there are no TIMEOUT sequences yet reported in other lophotrochozoans, we have recovered a clade of putative TIMEOUT orthologs that includes annelids and molluscs (Supplementary Fig. 5, available online). TIMELESS and TIMEOUT proteins fell out into respective clades, with the exception of Drosophila TIMEOUT, which was in the midst of the TIMELESS clade. However, this and numerous other branches had low bootstrap support (<50%).
Figure 7.
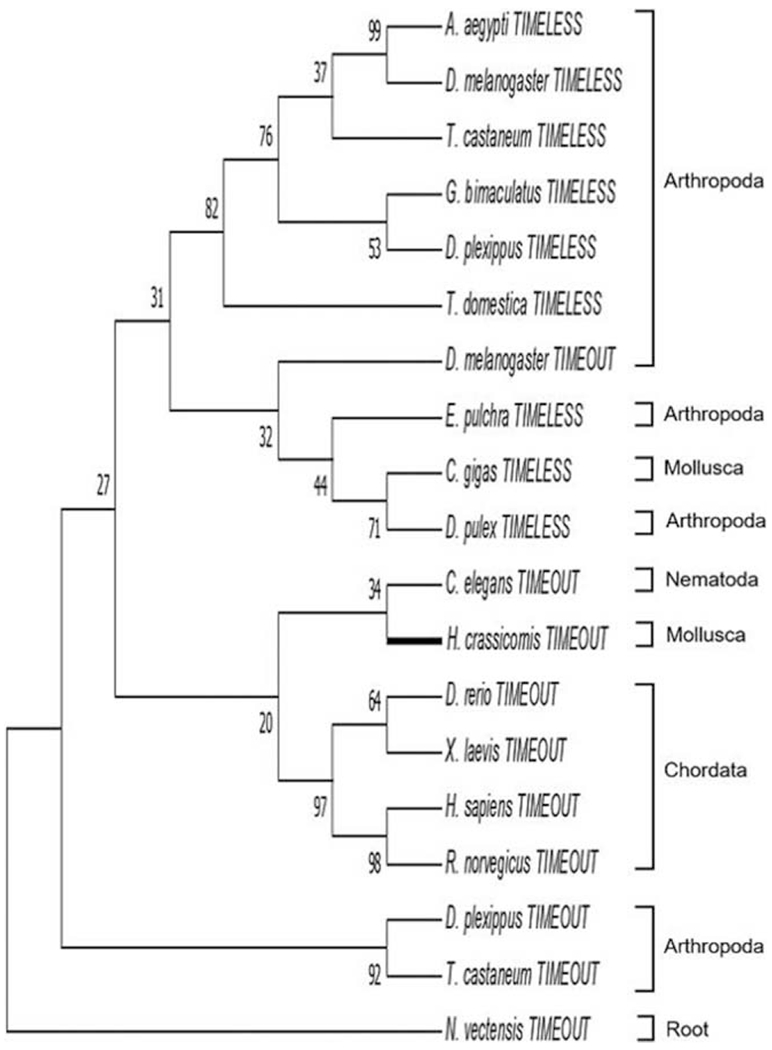
Evolutionary relationships between TIMELESS and TIMEOUT orthologs. A complete TIMEOUT sequence was identified in Hermissenda crassicornis, while only partial sequences of TIMEOUT were identified in the other two nudibranchs. Similarly, only partial sequences for TIMELESS were identified in all three nudibranchs. Therefore, only the H. crassicornis TIMEOUT was included in this maximum likelihood analysis. The putative H. crassicornis TIMEOUT shared most recent ancestry with orthologs in Caenorhabditis elegans and chordates. TIMEOUT and TIMELESS largely grouped together, with the exception of arthropod TIMEOUT proteins (see Drosophila melanogaster, Danaus plexippus, and Tribolium castaneum). However, bootstrap values for more ancestral branch points were below the typical 50%–75% threshold for high confidence. The tree was rooted on TIMEOUT in the cnidarian Nematostella vectensis.
Discussion
We report the sequences of putative core clock gene products in three nudibranchs: Hermissenda crassicornis, Melibe leonina, and Tritonia diomedea. The majority of the predicted proteins have higher percent similarity with orthologs in Mus than those in Drosophila (Supplementary Table 3, available online). This similarity to chordate sequences was supported by most phylogenetic analyses, with the inclusion of numerous orthologs in other species (Figs. 3–7). The group of putative clock proteins identified in the nudibranchs suggests that the gastropod clocks are likely to function more like the mammalian clock than the Drosophila clock.
Bioinformatics pipeline
The nudibranch transcripts were identified using an automated bioinformatics pipeline that used manually determined bait sequences (usually from canonical species such as Drosophila and Mus) to fish out contiguous sequences from the transcriptome assemblies. These were then mapped on a phylogenetic tree with the bait sequences, as well as additional closely matched protein sequences from automated BLAST searches. Compared to simple BLAST-style searches of a transcriptome, this pipeline significantly facilitated the ease with which contiguous sequences could be identified as potential orthologs, and, more importantly, it accurately determined whether they were most closely associated with the products of specific clock genes or whether they were more similar to paralogs that likely do not serve a circadian clock function. For example, in the early stages of our transcriptome searches, we identified a gene product in Melibe using BLAST-style queries with BMAL1 and CYCLE bait sequences. However, it was not until we used the automated pipeline that we realized that the Melibe sequence that we had identified as a putative BMAL1 or CYCLE ortholog actually fell out with non-circadian ARNTL proteins. We were then further able to identify a transcript that more closely matched BMAL1 from related species (Fig. 4). The pipeline is a LINUX-based sequence of bioinformatics modules, using freely available online databases and the transcriptome of a selected organism. We have made this pipeline available for download (Pankey, 2018).
Nudibranch clock sequences
In mammals, CLOCK and BMAL1 work together to drive the transcription of other clock genes, including period and npcry (Gekakis et al., 1998), whereas in flies, CLOCK works with CYCLE, a homolog of mammalian BMAL1, to perform a similar function (Rutila et al., 1998). Even though a complete sequence for BMAL1 could only be found in Melibe, a partial sequence was identified in Tritonia. Bioinformatics analysis indicated that these proteins shared common ancestry with other BMAL1 proteins, as opposed to the homologous insect CYCLE (Fig. 4). This mirrors the situation in other lophotrochozoans investigated to date (Arendt et al., 2004; Zantke et al., 2013; Bao et al., 2017; Perrigault and Tran, 2017), as well as in crustaceans (Zhang et al., 2013) and some insects (Rubin et al., 2006).
Another key difference between the mammalian clock and the dipteran clock is the molecular partner for PERIOD. In Drosophila, PERIOD heterodimerizes with TIMELESS, and the pair act as a brake on their own transcription, as well as that of other clock-associated genes (Gekakis et al., 1995). Although it is hypothesized that TIMELESS and its paralog TIMEOUT were both present early in animals, TIMELESS has been lost in lineages leading to Caenorhabditis elegans and deuterostomes (Li et al., 2016). Thus, chordate proteins initially named TIMELESS have now proven to actually be orthologs of TIMEOUT. In regard to the mammalian circadian clock, PERIOD works with NPCRY, instead of TIMELESS, to perform a similar negative feedback function (Kume et al., 1999). Drosophila has lost the gene npcry, so TIMELESS may have been co-opted to serve a similar function. All three nudibranchs in this study had both TIMELESS and TIMEOUT, as well as NPCRY. While there is no clear evidence for a role for TIMEOUT in circadian clocks, it remains to be determined whether TIMELESS and/or NPCRY are involved in the negative feedback loop of the nudibranch circadian clock.
Animal cryptochromes are thought to have evolved from ancestral photolyases associated with light-dependent DNA repair (Mei and Dvornyk, 2015) and now include regulators of circadian rhythms (Özturk et al., 2007; Michael et al., 2017). Animal cryptochromes can be photoreceptive (PCRY), providing direct light input into the circadian clock, or non-photoreceptive (NPCRY), possibly acting as transcriptional repressors of clock genes. In this study, in addition to all three nudibranchs expressing related DNA repair genes phr and cry dash, they also exhibited gene products for both pcry and npcry. The presence of both pcry and npcry is similar to many invertebrates, although Drosophila has only pcry (Emery et al., 1998; Stanewsky et al., 1998); and vertebrates (Hsu et al., 1996; Todo et al., 1996) and some insects (Rubin et al., 2006; Ingram et al., 2012) have only npcry. It has been hypothesized that in animals that have both pcry and npcry, both of these genes may be important in circadian clock function, with PCRY acting as a blue-light photoreceptor to provide light input to the clock and NPCRY serving a transcriptional repressor function for the clock (Yuan et al., 2007). Further research will be necessary to determine whether both of these cryptochromes are involved with the circadian clock of nudibranchs.
While many animals appear to have single orthologs of the canonical core clock genes seen in Drosophila and Mus, there have also been instances during animal evolution where these clock genes have duplicated one or more times. For example, mammals have three period genes (period1, period2, and period3) (Shearman et al., 1997; Zylka et al., 1998), although only period1 and period2 play a significant role in the central clock mechanism (Bae et al., 2001); and they have two paralogs for npcry (referred to as cryptochrome1 and cryptochrome2) (Hsu et al., 1996). In invertebrates, the horseshoe crab Limulus polyphemus has two paralogs for cycle and three gene copies for period (Chesmore et al., 2016); and the silk moth Antheraea pernyi has two paralogs for period, located on different sex chromosomes (Gotter et al., 1999). In our study, we found only single copies of transcripts for all of the canonical clock genes. Therefore, we did not find any evidence of clock gene duplication, such as occurred in ecdysozoans (arthropods) and deuterostomes (mammals).
Evolution of circadian clocks
The two best-studied clock gene networks in the animal kingdom are those in Drosophila and mammals. Therefore, we compared the Melibe proteins with orthologs in Drosophila and Mus (Supplementary Table 3, available online). In almost all cases, the Melibe proteins had a higher similarity with those found in the mouse. The lone exception was TIMELESS, but that is likely due to the fact that TIMELESS has been lost in mammals, and the timeless gene in Mus is actually timeout (Li et al., 2016). Based on evolutionary relationships, this similarity of Melibe clock sequences to those of mammals may be due to recent divergence and/or rapid gene evolution of Drosophila. While the overall feedback loop of circadian clocks in Drosophila and mammals is similar, and there are numerous homologous genes in these clocks, there are distinct differences as well. For example, TIMELESS is the transcriptional repressor partner for PERIOD in Drosophila (Gekakis et al., 1995), whereas NPCRY serves that function in mammals (Kume et al., 1999). In addition, PCRY provides light input to the clock in Drosophila (Emery et al., 1998; Stanewsky et al., 1998), whereas MELANOPSIN plays this role in mammals (Hattar et al., 2002; Panda et al., 2002; Ruby et al., 2002). These differences raise the question of what the ancestral clock was like before the divergence of protostomes and deuterostomes. Lophotrochozoans, such as molluscs, represent an animal clade that can potentially shed some light on this question. Recent research on the oyster Crassostrea gigas found that the bivalve circadian clock may be intermediate to that seen in mammals and Drosophila, with both TIMELESS and NPCRY possibly acting as transcriptional repressors and PCRY providing light input to the clock (Perrigault and Tran, 2017). This is actually similar to the proposed clock mechanism in some non-drosopholid insects, such as butterflies (Zhu et al., 2008), and the chelicerate Limulus (horseshoe crab) (Chesmore et al., 2016). Considering that the nudibranchs in this study expressed a suite of clock genes most similar to butterflies, horseshoe crabs, and oysters, this potentially increases the number of clades with this type of circadian clock.
Going further back in evolutionary time, cnidarians also express orthologs of clock, bmal1 and cycle, and both pcry and npcry (reviewed in Reitzel et al., 2013). However, to date there is no evidence of period or timeless homologs existing in cnidarians. Therefore, the metazoan ancestral clock may resemble that present in non-drosopholid insects, horseshoe crabs, oysters, and nudibranchs, with the exception that NPCRY may have been the sole transcriptional repressor of clock and bmal1 and cycle. Additional studies in other lophotrochozoans and cnidarians, as well as other phylogenetically informative clades near the base of Metazoa (e.g., placozoans), should shed additional light on the evolution of these animal circadian clocks.
Supplementary Material
Acknowledgments
Funding to JMN and WHW was provided by New Hampshire Institutional Development Award (IDeA) Network of Biomedical Research Excellence (NH-INBRE), P20GM103506, from the National Institute of General Medical Sciences of the National Institutes of Health. Funding to PSK and AS was provided by National Science Foundation–Division of Integrative Organismal Systems (IOS)–1455527. We thank W. Kelley Thomas, Jordan Ramsdell, and Joseph Sevigny, at the University of New Hampshire, for assistance with the bioinformatics analyses and uploading of sequences to the National Center for Biotechnology Information, and Jonathan Boykin, at Georgia State University, for facilitating access to transcriptome data.
Abbreviations:
- ARNTL
aryl hydrocarbon receptor nuclear translocator like
- bHLH
basic helix-loop-helix
- BMAL1
brain and muscle aryl hydrocarbon receptor nuclear translocator like protein 1
- CLOCK
circadian locomotor output cycles kaput
- CNS
central nervous system
- CRY DASH
cryptochrome DASH
- FAD
flavin adenine dinucleotide
- G+I
gamma-distributed andinvariant
- ML
maximum likelihood
- MSA
multiple sequence alignments
- NCBI
National Center for Biotechnology Information
- NPCRY
non-photoreceptive cryptochrome
- PAC
Per-Arnt-Sim-associated C-terminal
- PAS
Per-Arnt-Sim
- PCRY
photoreceptive cryptochrome
- PHR
6-4 photolyase
- TSA
transcriptome shotgun assembly
Footnotes
Online enhancement: supplementary appendix.
Literature Cited
- Altschul SF, Gish W, Miller W, Meyers EW, and Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Arendt D, Tessmar-Raible K, Snyman H, Dorresteijn AW, and Wittbrodt J. 2004. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science 306: 869–871. [DOI] [PubMed] [Google Scholar]
- Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E et al. 2012. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40: W597–W603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, and Weaver DR. 2001. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30: 525–536. [DOI] [PubMed] [Google Scholar]
- Bailey SER 1981. Circannual and circadian rhythms in the snail Helix aspersa (Müller) and the photoperiodic control of annual activity and reproduction. J. Comp. Physiol 142: 89–94. [Google Scholar]
- Bao Y, Xu F, and Shimeld SM. 2017. Phylogenetics of lophotrochozoan bHLH genes and the evolution of lineage-specific gene duplicates. Genome Biol. Evol 9: 869–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RSK 1986. Daily activity rhythms in the intertidal gastropod Hydrobia ulvae (Pennant). Estuar. Coast. Shelf Sci. 22: 325–334. [Google Scholar]
- Beeston DC, and Morgan E. 1979. A crepuscular rhythm of locomotor activity in the freshwater prosobranch, Melanoides tuberculata (Müller). Anim. Behav 27: 284–291. [Google Scholar]
- Blanc A 1993. Ultradian and circadian rhythmicity of behavioral activities in the young snail Helix aspersa maxima (Gastropoda, Helicidae). Can. J. Zool 71: 1506–1510. [Google Scholar]
- Block GD, and Davenport PA. 1982. Circadian rhythmicity in Bulla gouldiana: role of the eyes in controlling locomotor behavior. J. Exp. Zool. 224: 57–63. [Google Scholar]
- Block GD, and Lickey ME. 1973. Extraocular photoreceptors and oscillators can control the circadian rhythm of behavioral activity in Aplysia. J. Comp. Physiol. 84: 367–374. [Google Scholar]
- Block GD, and Roberts MH. 1981. Circadian pacemaker in the Bursatella eye: properties of the rhythm and its effect on locomotor behavior. J. Comp. Physiol. 142: 403–410. [Google Scholar]
- Chaudhry MA, and Morgan E. 1983. Circadian variation in the behavior and physiology of Bulinus tropicas (Gastropoda: Pulmonata). Can. J. Zool. 61: 909–914. [Google Scholar]
- Chesmore KN, Watson III WH, and Chabot CC. 2016. Identification of putative circadian clock genes in the American horseshoe crab, Limulus polyphemus. Comp. Biochem. Physiol. D Genom. Proteom. 19: 45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constance CM, Green CB, Tei H, and Block GD. 2002. Bulla gouldiana period exhibits unique regulation at the mRNA and protein levels. J. Biol. Rhythm. 17: 413–427. [DOI] [PubMed] [Google Scholar]
- Dunlap JC 1999. Molecular bases for circadian clocks. Cell 96: 271–290. [DOI] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, and Rosbash M. 1998. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95: 669–679. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Saez L, Delahaye-Brown A-M, Myers MP, Sehgal A, Young MW, and Weitz CJ. 1995. Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science 270: 811–815. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, and Weitz CJ. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280: 1564–1569. [DOI] [PubMed] [Google Scholar]
- Gotter AL, Levine JD, and Reppert SM. 1999. Sex-linked period genes in the silkmoth, Antheraea pernyi: implications for circadian clock regulation and the evolution of sex chromosomes. Neuron 24: 953–965. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q et al. 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE 2005. The circadian timekeeping system of Drosophila. Curr. Biol. 15: R714–R722. [DOI] [PubMed] [Google Scholar]
- Hattar S, Liao H-W, Takao M, Berson DM, and Yau K-W. 2002. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science 295: 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang R-P, Todo T, Wei Y-F, and Sancar A. 1996. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry 35: 13871–13877. [DOI] [PubMed] [Google Scholar]
- Ingram KK, Kutowoi A, Wurm Y, Shoemaker D, Meier R, and Block GD. 2012. The molecular clockwork of the fire ant Solenopsis invicta. PLoS One 7: e45715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacklet JW 1972. Circadian locomotor activity in Aplysia. J. Comp. Physiol. 79: 325–341. [Google Scholar]
- Joshi NA, and Fass JN. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files, version 1.33. [Online]. Available: https://github.com/najoshi/sickle [2018, May 2].
- Kavaliers M 1981. Circadian and ultradian activity rhythms of a freshwater gastropod, Helisoma trivolvis: the effects of social factors and eye removal. Behav. Neural Biol. 32: 350–363. [DOI] [PubMed] [Google Scholar]
- Koike N, Hida A, Numano R, Hirose M, Sakaki Y, and Tei H. 1998. Identification of the mammalian homologues of the Drosophila timeless gene, Timeless1. FEBS Lett. 441: 427–431. [DOI] [PubMed] [Google Scholar]
- Konopka RJ, and Benzer S. 1971. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68: 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, and Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, and Reppert SM.1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205. [DOI] [PubMed] [Google Scholar]
- Kupfermann I 1968. A circadian locomotor rhythm in Aplysia californica. Physiol. Behav. 3: 179–181. [Google Scholar]
- Letunic I, and Bork P. 2017. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46: D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Su Y, Tu J, Wei R, Fan Z, Yin H, Hu Y, Xu H, Yao Y, Yang D et al. 2016. Evolutionary conservation of the circadian gene timeout in Metazoa. Anim. Biol. 66: 1–11. [Google Scholar]
- Lickey ME, Wozniak JA, Block GD, Hudson DJ, and Augter GK. 1977. The consequences of eye removal for the circadian rhythm of behavioral activity in Aplysia. J. Comp. Physiol. A Sens. Neural Behav. Physiol. 118: 121–143. [Google Scholar]
- Lin C, and Todo T. 2005. The cryptochromes. Genome Biol. 6: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, and Takahashi JS. 2004. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. Genom. Hum. Genet. 5: 407–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Q, and Dvornyk V. 2015. Evolutionary history of the photolyase/cryptochrome superfamily in eukaryotes. PLoS One 10: e0135940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AK, Fribourgh JL, Van Gelder RN, and Partch CL. 2017. Animal cryptochromes: divergent roles in light perception, circadian timekeeping and beyond. Photochem. Photobiol. 93: 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb JM, Kirouac LE, Naimie AA, Bixby KA, Lee C, Malanga S, Raubach M, and Watson WH III. 2014. Circadian rhythms of crawling and swimming in the nudibranch mollusc Melibe leonina. Biol. Bull. 227: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özturk N 2016. Phylogenetic and functional classification of the photolyase/cryptochrome family. Photochem. Photobiol. 93: 104–111. [DOI] [PubMed] [Google Scholar]
- Özturk N, Song S-H, Özgür S, Selby CP, Morrison L, Partch C, Zhong D, and Sancar A. 2007. Structure and function of animal cryptochromes. Cold Spring Harb. Symp. Quant. Biol. 72: 119–131. [DOI] [PubMed] [Google Scholar]
- Panda S, Sato TK, Castrucci AM, Rollag MD, DeGrip WJ, Hogenesch JB, Provencio I, and Kay SA. 2002. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science 298: 2213–2216. [DOI] [PubMed] [Google Scholar]
- Pankey MS 2018. Melibe transcriptome miner. [Online]. Available: https://github.com/scriptomika/MelibeTxMiner [2018, April 18].
- Perrigault M, and Tran D. 2017. Identification of the molecular clockwork of the oyster Crassostrea gigas. PLoS One 12: e0169790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Tarrant AM, and Levy O. 2013. Circadian clocks in the Cnidaria: environmental entrainment, molecular regulation, and organismal outputs. Integr. Comp. Biol. 53: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, and Weaver DR. 2002. Coordination of circadian timing in mammals. Nature 418: 935–941. [DOI] [PubMed] [Google Scholar]
- Rivera AS, Ozturk N, Fahey B, Plachetzki DC, Degnan BM, Sancar A, and Oakley TH. 2012. Blue-light-receptive cryptochrome is expressed in a sponge eye lacking neurons and opsin. J. Exp. Biol. 215: 1278–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato E, Tauber E, and Kyriacou CP. 2006. Molecular genetics of the fruit-fly circadian clock. Eur. J. Hum. Genet. 14: 729–738. [DOI] [PubMed] [Google Scholar]
- Rubin EB, Shemesh Y, Cohen M, Elgavish S, Robertson HM, and Block GD. 2006. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 16: 1352–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby NF, Brennan TJ, Xie X, Cao V, Franken P, Heller HC, and O’Hara BF. 2002. Role of melanopsin in circadian responses to light. Science 298: 2213–2216. [DOI] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, and Hall JC. 1998. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93: 805–814. [DOI] [PubMed] [Google Scholar]
- Sangorum AM, Saez L, Antoch MP, Gekakis N, Staknis D, Whiteley A, Fruechte EM, Vitaterna MH, Kazuhiro S, King DP et al. 1998. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron 21: 1101–1113. [DOI] [PubMed] [Google Scholar]
- Schnytzer Y, Simon-Blecher N, Li J, Ben-Asher HW, Salmon-Divon M, Achituv Y, Hughes ME, and Levy O. 2018. Tidal and diel orchestration of behavior and gene expression in an intertidal mollusc. Sci. Rep. 8: 4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, and Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95: 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senatore A, Edirisinghe N, and Katz. PS 2015. Deep mRNA sequencing of the Tritonia diomedea brain transcriptome provides access to gene homologues for neuronal excitability, synaptic transmission and peptidergic signaling. PLoS One 10: e0118321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF Jr., and Reppert SM. 1997. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19: 1261–1269. [DOI] [PubMed] [Google Scholar]
- Shirley TC, and Findley AM. 1978. Circadian rhythm of oxygen consumption in the marsh periwinkle, Littorina irrorata (Say, 1822). Comp. Biochem. Physiol. A Comp. Physiol. 59: 339–342. [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolove PG, Beiswanger CM, Prior DJ, and Gelperin A. 1977. A circadian rhythm in the locomotor behavior of the giant garden slug Limax maximus. J. Exp. Biol. 66: 47–64. [DOI] [PubMed] [Google Scholar]
- Stamatakis A 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, and Hall JC. 1998. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95: 681–692. [DOI] [PubMed] [Google Scholar]
- Takumi T, Nagamine Y, Miyake S, Matsubara C, Taguchi K, Takekida S, Sakakida Y, Nishikawa K, Kishimoto T, Niwa S et al. 1999. A mammalian ortholog of Drosophila timeless, highly expressed in SCN and retina, forms a complex with mPER1. Genes Cells 4: 67–75. [DOI] [PubMed] [Google Scholar]
- Tamvacakis AN, Senatore A, and Katz PS. 2015. Identification of genes related to learning and memory in the brain transcriptome of the mollusk, Hermissenda crassicornis. Learn. Mem. 22: 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, and Sakaki Y. 1997. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 389: 512–516. [DOI] [PubMed] [Google Scholar]
- Todo T, Ryo H, Yamamoto K, Toh H, Inui T, Ayaki H, Nomura T, and Ikenaga M. 1996. Similarity among the Drosophila (6–4)photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science 272: 109–112. [DOI] [PubMed] [Google Scholar]
- Whelan S, and Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18: 691–699. [DOI] [PubMed] [Google Scholar]
- Yang Z 1996. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol. Evol. 11: 367–372. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Metterville D, Briscoe AD, and Reppert SM. 2007. Insect cryptochromes: gene duplication and loss define diverse ways to construct circadian clocks. Mol. Biol. Evol. 24: 948–955. [DOI] [PubMed] [Google Scholar]
- Zann LP 1973. Interactions of the circadian and circatidal rhythms of the littoral gastropod Melanerita atramentosa (Reeve). J. Exp. Mar. Biol. Ecol. 11: 249–261. [Google Scholar]
- Zantke J, Ishikawa-Fujiwara T, Arboleda E, Lohs C, Schipany K, Hallay N, Straw AD, Todo T, and Tessmar-Raible K. 2013. Circadian and circalunar clock interactions in a marine annelid. Cell Rep. 5: 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, and Kay SA. 2010. Clocks not winding down: unraveling circadian networks. Nat. Rev. 11: 764–776. [DOI] [PubMed] [Google Scholar]
- Zhang L, Hastings MH, Green EW, Tauber E, Sladek M, Webster SG, Kyriacou CP, and Wilcockson DC. 2013. Dissociation of circadian and circatidal timekeeping in the marine crustacean Eurydice pulchra. Curr. Biol. 23: 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Sauman I, Yuan Q, Casselman A, Emery-Le M, Emery P, and Reppert SM. 2008. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS One 6: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Weaver DR, and Reppert SM. 1998. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 20: 1103–1110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


