Abstract
Background
The aim of this study was to identify DNA methylation sites in peripheral blood leukocytes from patients with histologically confirmed nonalcoholic fatty liver disease (NAFLD) that included simple hepatic steatosis and nonalcoholic steatohepatitis (NASH).
Material/Methods
DNA was isolated from peripheral blood leukocytes from patients with histologically diagnosed NAFLD (n=35), including simple hepatic steatosis (n=18) and NASH (n=17). Healthy controls included individuals without liver disease (n=30). DNA was hybridized, and DNA methylation was interrogated in an epigenome-wide association study (EWAS). DNA methylation levels (β-values) were correlated with serum lipid profiles, liver enzymes, and liver histology.
Results
Circulating blood leukocytes from 35 patients with NAFLD (simple steatosis and NASH) contained 65 CpG sites, which represented 60 genes that were differentially methylated when compared with healthy controls. In the simple hepatic steatosis group (n=18), 42 methylated CpG sites were found to be associated with increased levels of serum alanine aminotransferase (ALT), and 32 methylated CpG sites were associated with increased serum lipid profiles. In the NASH group (n=17), when compared with the simple hepatic steatosis group, methylated CpG sites showed significant correlations with the presence of lobular inflammation compared with hepatic steatosis and fibrosis. Six differentially methylated CpG sites were identified in the ACSL4, CRLS1, CTP1A, SIGIRR, SSBP1 and ZNF622 genes, which were associated with histologically confirmed simple hepatic steatosis and NASH.
Conclusions
The study identified some key methylated CpG sites from peripheral blood leukocytes, which might be used as serum biomarkers to stratify NAFLD patients into simple hepatic steatosis and NASH.
MeSH Keywords: DNA Methylation, Epigenomics, Fatty Liver
Background
Nonalcoholic fatty liver disease (NAFLD) is defined as the pathological accumulation of fat in liver cells and tissues in the absence of hepatotoxicity due to alcohol intake or any other identified cause. NAFLD describes a spectrum of changes in the liver associated with fat accumulation that include simple hepatic steatosis, nonalcoholic steatohepatitis (NASH), fibrosis, and liver cirrhosis [1]. The global prevalence of NAFLD has been estimated to be as high as 25.2%, with the Middle East and South America showing the highest prevalence, and Africa has the lowest prevalence [2]. In Asia, the prevalence of NAFLD has been reported to have recently increased to 27.4% [3]. This high prevalence of NAFLD in Asia might be due to changes in lifestyle and dietary patterns, including the increasing availability of highly calorific convenience foods.
Simple hepatic steatosis is a mild form of NAFLD, which may progress to NASH in some cases. Progression to NASH has been reported in 44% of patients with simple hepatic steatosis at baseline [4]. In cases of NASH, up to 2.8% of patients may develop end-stage liver cirrhosis or hepatocellular carcinoma (HCC) [5]. Therefore, because the liver changes in NASH can be progressive and irreversible, NASH has become an increasingly common reason for liver transplantation [4]. Associations between NAFLD and insulin resistance, obesity, hyperlipidemia, type 2 diabetes, and metabolic syndrome have been shown [4]. However, the pathogenesis of NAFLD and the associated phenotypes remain to be elucidated.
The progression of NAFLD to cirrhosis and HCC is variable and modified by age, gender, genetic predisposition, and epigenetic factors. Despite being an invasive procedure associated with some degree of risk and dependency on adequate, relevant liver sampling, liver biopsy remains the gold standard for assessing all form of NAFLD. Optimal non-invasive detection methods for NAFLD are still being developed. However, a new approach for understanding the pathogenesis of NAFLD might be provided by identifying epigenetic modifications [6], which are known to be involved in insulin resistance (IR) and to affect lipid metabolism, and the liver cell endoplasmic reticulum and mitochondria due to oxidative stress responses [7].
DNA methylation occurs at the cytosine base within cytosine-guanine dinucleotides, which are referred to as CpG sites. During DNA methylation, DNA methyltransferase catalyzes the transfer of a methyl group to the fifth carbon atom from 5-methylcytosine within the cytosine ring, and an increased DNA methylation level of promoters that usually correlates with low or no transcription [8]. Also, the hypermethylation of the CpG islands is usually associated with gene silencing, and global hypomethylation of genomic DNA can affect genomic stability [9].
There have been few previously published studies on the potential role of DNA methylation in NAFLD. A study using a rodent model showed that diets depleted of methyl donors could promote DNA hypomethylation of important metabolism-related genes and that this hypomethylation altered hepatic fat metabolism, resulting in simple hepatic steatosis [10]. Recent research on human liver disease has begun to apply the genome-wide association studies (GWAS) [11,12], which have shown that alterations in the pattern of DNA methylation are capable of inducing gene expression changes that are reversible and can contribute to insulin resistance and changes in lipid metabolism [13]. The use of GWAS means that there is now the potential to detect epigenetic modifiers in NAFLD, which might provide molecular tools for diagnosing, assessing the severity, and predicting disease progression [14].
Recently, studies have explored the possibility of using circulating leukocytes, derived from peripheral blood samples, for the evaluation of potential biomarkers in liver disease [15]. A recent study by Nano et al., which was the most extensive study to date using circulating leukocytes, identified CpG sites and DNA methylation levels that were associated with serum enzyme levels and simple hepatic steatosis, resulting in new insights into the epigenetic mechanisms associated with NAFLD [16].
The aims of the present study were to identify DNA methylation sites in peripheral blood leukocytes in patients with histologically confirmed NAFLD, including simple hepatic steatosis and NASH. Of particular interest was the possibility that epigenetic changes in DNA methylation were associated with clinical parameters, including serum liver enzymes, and lipid profiles, as well as the histological features of NAFLD. The use of routine blood samples was combined with findings from liver biopsy histology, with the aim of determining whether circulating leukocytes could be used to identify specific methylated CpGs that could be related to liver enzyme levels, lipid profiles, or NAFLD phenotypes, to provide novel non-invasive biomarkers for NAFLD, but mainy for NASH in this study.
Material and Methods
Study participants and study design
Between March 2012 and May 2013, 35 Chinese Han patients with NAFLD between the ages of 18–70 years were recruited to the study in Shanghai, China. Also, 30 healthy controls with normal liver function tests were recruited. Individuals were excluded from the study if they reported excessive alcohol consumption (>30 g/day for men; >20 g/day for women) or had known diseases that could cause fatty liver, such as chronic hepatitis C, autoimmune hepatitis, drug-induced liver injury or Wilson’s disease, who were being given total parenteral nutrition (TPN), exhibited other end-stage diseases or malignancy, or who had diabetes mellitus. To ensure patient confidentiality, patient anonymity was included in the study protocol, and to meet the appropriate ethical requirements, all procedures were conducted in accordance with the ethical principles of the Declaration of Helsinki.
Evaluated clinical variables
All clinical variables were evaluated according to the normal reference standards of our institution. Venous blood samples and metabolic profiles were obtained from NAFLD patients and normal controls. The body mass index (BMI) was calculated using the weight (in kilograms) divided by the square of their height (in meters) (kg/m2). The BMI classification used was the World Health Organization (WHO) classification for adults: lean individuals (BMI <18.5 kg/m2), normal individuals (BMI 18.5–24.9 kg/m2), overweight individuals (BMI 25.0–29.9 kg/m2), and obese individuals (BMI ≥30 kg/m2).
The following fasting blood biochemical tests were performed for all study participants using a conventional automated analyzer (Hitachi 7600, Tokyo, Japan): fasting blood glucose (FBG), insulin; total cholesterol (TC), triglyceride (TG), high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), very low-density lipoprotein (VLDL). Liver function tests included alanine transaminase (ALT), aspartate transaminase (AST), γ-glutamyl-transpeptidase (GGT), total bilirubin (TBil), direct bilirubin (DBil), and uric acid (UA) levels.
The homeostatic model assessment insulin resistance (HOMA-IR) index was used to assess basal glucose and insulin concentrations. HOMA-IR scores were obtained by multiplying the fasting serum insulin level (μIU/mL) by the FBG level (mmol/L), and dividing the product by 22.5.
Each study participant underwent a FibroScan® 502 test (Echosens, Paris, France) with an M-probe to measure liver stiffness (kPa) and the controlled attenuation parameter (CAP) that correlated with liver fibrosis and steatosis, respectively. All 35 patients met the histological diagnostic criteria for NAFLD. All control subjects had a CAP value <240 dB/m and a liver stiffness measurement (LSM) value <7.0 kPa, to confirm that they were free from fatty liver disease.
Liver histology
Patients with NAFLD (n=35) had undergone percutaneous liver biopsy with real-time ultrasound guidance. The liver biopsy specimens were fixed and stored in neutral buffered formalin, processed and embedded in paraffin wax blocks. The tissue sections were then cut onto glass slides. Hematoxylin and eosin (H&E) staining and Masson’s trichrome staining for collagen and reticulin were routinely performed on all biopsies, and the results were assessed under light microscopy by experienced histopathologists, who were unaware of the patient’s clinical and imaging history. NAFLD, including simple liver steatosis and NASH, was diagnosed according to the semi-quantitative histological scoring algorithm of hepatic steatosis, hepatocyte ballooning, and lobular inflammation. A SAF score was created for each case including steatosis (S), activity (A, ballooning + lobular inflammation), and liver fibrosis (F). A score that fulfilled the criteria of S≥1A≥2Fany was used to support the diagnosis of NASH [17].
DNA extraction and bisulfite conversion
DNA extraction from peripheral blood samples was performed using a nucleic acid extraction kit (Qiagen, Hilden, Germany). The quality of the DNA was determined using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific Inc., DE, USA). Bisulfite conversion of the DNA (500 ng/sample) was then performed according to the manufacturer’s protocol using the EZ DNA Methylation Kit (Zymo Research, CA, USA) and a modified thermo-cycling procedure (Illumina, San Diego, CA, USA).
DNA methylation status was analyzed with a microarray approach using the Illumina Human Methylation450 BeadChip platform (Illumina, San Diego, CA, USA) to interrogate DNA methylation in an epigenome-wide association study (EWAS). The methylation level of each cytosine was calculated as the fluorescence intensity ratio of the methylated alleles to the unmethylated alleles, based on the Infinium type I probes and the Infinium type II probes and was expressed as a β value. The β values ranged from between 0 (unmethylated) and 1 (completely methylated) according to the combination of the Cy3 and Cy5 fluorescence intensities [18]. Illumina Genome Studio® Methylation module version 1.0 (Illumina, San Diego, CA, USA) was used to calculate the β value for each CpG site. Color balance adjustment was performed to normalize the samples between the two color channels using Genome Studio Illumina software (V2010.3). GenomeStudio was used to normalize the data using different internal controls that were present on the Illumina HumanMethylation450 BeadChip platform (Illumina, San Diego, CA, USA).
All probes were subsequently filtered according to the following requirements: detection of P-values >0.05 in one or more samples; the presence of single nucleotide polymorphisms (SNPs) at the 10 bp 3′ end of the interrogating probe; and alignment with multiple locations on the X and Y chromosomes. The final number of valid CpG sites used for this study was 418,913. The global methylation level was compared between patients with NASH, patients with simple hepatic steatosis, and normal controls. The genomic distribution of the differentially methylated CpG sites was examined using the distribution of the CpG sites among all analyzed sites on the Illumina HumanMethylation450 BeadChip platform.
Pyrosequencing
The methylation of specific cytosines with CpG dinucleotides was quantified by pyrosequencing using PyroMark Q 96 MD (Qiagen, Hilden, Germany). The acyl-CoA synthetase long-chain family member 4 (ACSL4) gene was used for validation. The sequencing primers were as follows: forward (GTGATGGATTTTG-GGGTTTT), reverse (AAAACTCCCTAACCCTCAATTAC). Sequencing primers (GTATTTAGAGGGTTAG AAGTTAT) were obtained using Pyromark Assay Design software (version 2.0) (Qiagen, Hilden, Germany). Bisulfite-treated DNA was amplified via polymerase chain reaction (PCR) using the PyroMark PCR Kit (Qiagen, Hilden, Germany) and PyroMark CpG software (Qiagen, Hilden, Germany) to present the sequencing results.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database was used to analyze the effect of differentially methylated CpG sites and to identify signaling pathways that were significantly related to CpG sites (http://www.genome.jp/kegg/pathway.html). The method of detection of the false discovery rate (FDR) was used to exclude false-positive results [19,20].
Statistical analysis
Normally distributed data were expressed as the mean ± standard deviation (SD) and as numbers and percentages. The non-normally distributed data were presented as the median and interquartile range (IQR). After methylation data pre-processing [21], the Illumina Methylation Analyzer (IMA) R software package was used to perform t-tests following methylation data pre-processing. The P-values were adjusted using the Benjamini-Hochberg false discovery rate (FDR) procedure for multiple comparisons, in which an FDR <5% was considered to be statistically significant. A multivariate linear regression model was used to analyze the associations between the DNA methylation levels of the identified genes and liver enzyme levels and lipid profiles as continuous variables, adjusted by gender, age and BMI. The potential associations between the histological features of NAFLD and DNA methylation levels were evaluated using Pearson’s correlation coefficient (r). The diagnostic efficiencies of the candidate differentially methylated CpG sites were calculated through receiver-operating characteristic (ROC) curve analysis. An area under the curve (AUC) value >0.8 was interpreted as indicating very good efficiency, and an AUC value between 0.6–0.8 was interpreted as indicating good efficiency. All statistical analysis was performed using SPSS software, version 17.0 (SPSS, IBM Inc., Chicago, IL). Statistical significance was based on P-values of <0.05. Statistically significant correlations were shown using plots generated with GraphPad Prism® software, version 6.0 C (GraphPad, La Jolla, CA, USA).
Results
Demographic, clinical, and metabolic characteristics of the study participants
The demographic, clinical, and metabolic characteristics of the participants in this study are presented in Table 1. Liver biopsies from 35 patients with NAFLD showed simple hepatic steatosis (n=18) and NASH (n=17), according to the SAF score. Among the total study participants (n=65), 30.8% were women (n=20). In addition to exhibiting changes in their serum laboratory profiles, patients with NAFLD presented increased serum levels of ALT, GGT, and UA, but showed no significant differences in FBG levels. In addition to presenting high serum levels of cytokeratin (CK)18-M30 and CK18-M65, which are direct measures of liver cell damage, apoptosis, and inflammation, high LSM values were found, but only in patients with NASH.
Table 1.
Clinical characteristics of the participants.
| NASH (n=17) | Simple hepatic steatosis (n=18) | NAFLD (n=35) | Control (n=30) | NAFLD vs. CL P-value | |
|---|---|---|---|---|---|
| Sex (F) (N, %) | 12 (70.6%) | 15 (83.3%) | 8 (22.9%) | 12 (40.0%) | 0.12** |
| BMI >25 (N, %) | 12 (70.6%) | 14 (77.8%) | 26 (74.0%) | 4 (13.0%) | <0.001** |
| Metabolic syndrome (N, %) | 7 (41.2%) | 6 (33.3%) | 22 (63.0%) | 0 (0.0%) | <0.001** |
| Central obesity (N, %) | 13 (76.5%) | 13 (72.2%) | 26 (74.0%) | 9 (30.0%) | 0.01** |
| Age (y) | 37.76±12.77 | 37.72±12.48 | 37.74±12.43 | 46.63±7.24 | <0.001 |
| HT (cm) | 167.76±6.69 | 169.44±5.83 | 168.63±6.23 | 164.32±7.22 | 0.010 |
| WT (kg) | 76.88±11.15 | 79.97±13.38 | 78.47±12.27 | 62.29±9.04 | <0.001 |
| BMI | 27.29±3.4 | 27.79±3.84 | 27.55±3.59 | 23.02±2.63 | <0.001 |
| FBG (mmol/L) | 5.33±1.14 | 6.22±3.68 | 5.81±2.82 | 5.29±0.39 | 0.300 |
| TBIL (μmol/L) | 12.36±3.31 | 19.86±21.9 | 16.33±16.32 | 14.29±4.05 | 0.510 |
| DBIL (μmol/L) | 7.23±10.12 | 9.64±19.58 | 8.51±15.67 | 2.98±0.83 | 0.050 |
| GGT (u/L) | 78.25 (44.03–131.00) | 51.55 (32.50–80.70) | 65.35 (36.48–88.78) | 13.00 (11.00–17.25) | 0.020* |
| ALT (u/L) | 70.48±40.78 | 75.61±64.31 | 73.19±53.79 | 15.6±4.42 | <0.001 |
| TC (mmol/L) | 4.99±0.65 | 4.95±1.07 | 4.97±0.91 | 4.35±0.61 | <0.001 |
| TG (mmol/L) | 2.37±2.13 | 2.01±0.77 | 2.17±1.5 | 0.82±0.34 | <0.001 |
| HDL-C (mmol/L) | 1.23±0.34 | 1.19±0.28 | 1.21±0.3 | 1.43±0.25 | <0.001 |
| LDL-C (mmol/L) | 2.8±0.66 | 3.08±0.82 | 2.97±0.76 | 2.24±0.38 | <0.001 |
| UA (μmol/L) | 413.16±122.29 | 362.02±110.02 | 383.19±115.97 | 257.83±62.14 | <0.001 |
| CK18M 30 (ng/L) | 537.44±317.21 | 362.03±162.21 | <0.001*** | ||
| CK18M 65 (ng/L) | 1159.32±613.07 | 829.94±421.69 | <0.001*** | ||
| LSM (kpa) | 9.45 (6.08–14.40) | 5.70 (4.35–9.95) | 7.60 (5.40–12.00) | 4.15 (3.70–4.70) | <0.001* |
| CAP (dB/m) | 328.50 (289.25–358.75) | 310.00 (276.00–361.50) | 310.00 (277.00–359.00) | 195.00 (174.00–228.00) | <0.001* |
Mann-Whitney test;
Pearson chi-square or Fisher’s exact test;
NASH vs. Simple Hepatic Steatosis P-value.
FBG – fasting blood glucose; TC – total cholesterol; TG – triglyceride; HDL-C – high-density lipoprotein cholesterol; LDL-C – low-density lipoprotein cholesterol; ALT – alanine transaminase; AST – aspartate transaminase; GGT – γ-glutamyl-transpeptidase; TBIL – total bilirubin; DBIL – direct bilirubin; UA – uric acid; CK18 – cytokeratin-18; LSM – liver stiffness measurement; CAP – controlled attenuation parameter; N – number. Normal distribution data are expressed as the means±standard deviations and as numbers and percentages. The non-normal distribution data are presented as the median and interquartile range.
The global pattern of methylated DNA CpG sites in NAFLD
Comparisons of global methylation levels in the peripheral blood leukocytes of patients with NAFLD phenotypes compared with the normal controls are shown in Figures 1 and 2. Among a total of 418,913 probes, the average DNA methylation levels were similar in the NASH, simple hepatic steatosis, and normal control groups in terms of their correlation with either the CpG site content or the gene nearest to their functional genome. Hypomethylation was observed in the region of CpG islands, its north shore and south shore area (Figure 1). Promoter areas were defined as transcription start site (TSS)1500, TSS200, the 5′ UTR and the first exon. The DNA methylation levels were lower in promoter areas in all three groups studied (simple hepatic steatosis, NASH, and control groups) (Figure 2). The circulating blood leukocytes of the 35 patients with simple hepatic steatosis (n=18) and NASH (n=17) exhibited 65 CpG sites, which represented 60 genes that were differentially methylated, compared with normal controls. In the simple hepatic steatosis group, 42 methylated CpG sites were found to be associated with increased levels of alanine transaminase (ALT), and 32 methylated CpG sites were associated with increased serum lipid profiles, including TC, HDL-C, LDL-C, and TG levels. In the NASH group, compared with the simple hepatic steatosis group, methylated CpG sites showed more significant correlations with the presence of lobular inflammation than with hepatic steatosis and fibrosis.
Figure 1.
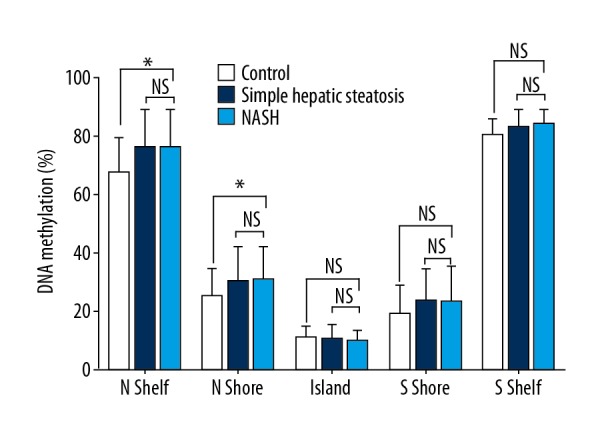
Global DNA methylation in circulating leukocytes from normal controls, patients with simple hepatic steatosis, and patients with nonalcoholic steatohepatitis (NASH), showing CpG island regions. Global DNA methylation is calculated as the average DNA methylation (%) at all CpG sites in each annotated region, according to the Illumina HumanMethylation450 BeadChip platform, which was used to interrogate DNA methylation in an epigenome-wide association study (EWAS). Shore, flanking regions of CpG islands (0–2,000 bp). Shelf, regions flanking island shores (2,000–4,000 bp from the CpG island). N – northern; S – Southern. * Significant difference (P<0.05).
Figure 2.
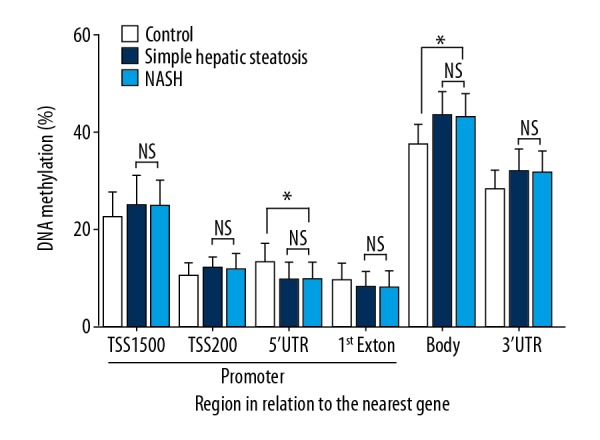
Global DNA methylation in circulating leukocytes from normal controls, patients with simple hepatic steatosis, and patients with nonalcoholic steatohepatitis (NASH) is shown for each gene region. Transcription start site (TSS) represents the proximal promoter, defined as 200 bp or 1,500 bp upstream of the TSS. * Significant difference (P<0.05).
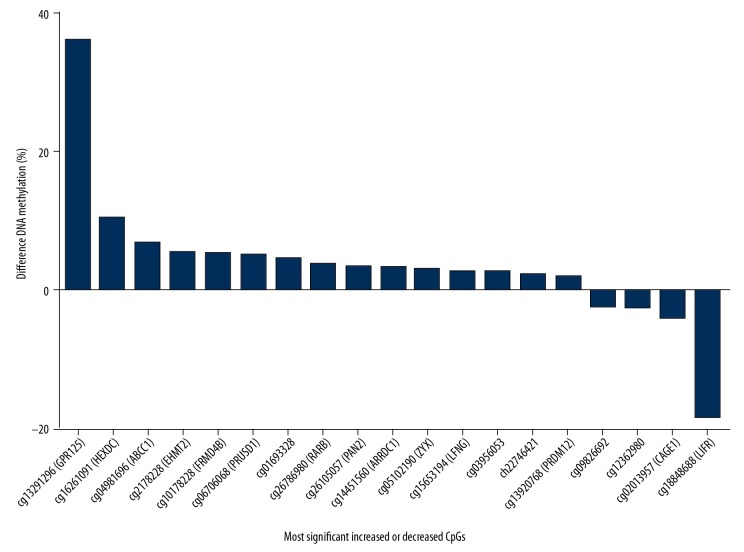
The 65 NAFLD-associated CpG sites, representing 60 genes, were found to be significantly methylated in individuals with NAFLD and normal controls based on a P-value of <0.05 and a false discovery rate (FDR) of q<0.05 (Supplementary Table 1). The methylation levels (β values) in more than half of the CpG sites (41 sites, 65.1%) were within the range of 0–0.2, suggesting that hypomethylation mostly characterized the NAFLD-associated CpG sites. Figure 3 shows the top 19 CpG sites associated with NAFLD presenting the most significant increases or decreases in methylation. Cg13291296 (coding GPR125) showed the most significant methylation level, with a 36% point change. Additionally, in an analysis of the difference in DNA methylation levels between patients with simple hepatic steatosis and normal controls, 119 CpG sites were annotated to 106 genes (Supplementary Table 2). The gender of the study participant was not found to exert a statistically significant influence on methylation levels in this study.
Figure 3.
CpG sites showing the most significant increase or decrease in DNA methylation (q<0.05) in nonalcoholic fatty liver disease (NAFLD).
Enrichment of gene-mapped CpG sites related to NAFLD in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were used to determine the enrichment of genes mapped to NAFLD-associated CpG sites before analyzing the association of clinical parameters with the NAFLD phenotypes (Supplementary Table 3). For the NAFLD-associated methylated CpG sites, the two top-ranked pathways involved ribosome biogenesis in eukaryotes (EF=8.56, q=0.010) and MAPK signaling (EF=2.59, q=0.011). Enriched genes related to metabolic pathways, cytokine-cytokine receptor interactions, and insulin signaling pathways were also found.
Because this study identified only 65 NAFLD-associated sites, all of these CpG sites were considered in the second KEGG pathway analysis for the association between DNA methylation and simple hepatic steatosis (Supplementary Table 4). This analysis identified genes that were mainly involved in vitamin digestion and absorption through enrichment factor (EF) analysis (EF=15.84, q=0.033) and in a particular pathway in cancer (EF=2.33, q=0.034). After a search of the relevant literature, genes that were involved in the endoplasmic reticulum, insulin signaling, and adipose tissue homeostasis were identified. Although these associations showed a lower EF, they are recognized as being associated with the pathogenesis of NAFLD. These simple hepatic steatosis-associated methylated CpG sites, together with the NAFLD-associated methylated CpG sites mentioned above, were then used to explore the relationship between DNA methylation and clinical parameters.
Association between DNA methylation and liver enzyme and lipid profiles
The evaluation of the relationship between DNA methylation and clinical parameters focused on the NAFLD-associated CpG sites and the other 15 selected simple hepatic steatosis-associated CpG sites (in alphabetical order by gene name: ABCC1, ATP5G1, AXIN2, CPT1A, CRLS1, DDX20, LDHB, MAPK1, NFE2L2, PMM1, PTEN, RB1, SEH1L, SGMS1, and SLC5A6) from the KEGG pathway analysis. Simple linear regression analysis showed that most of the CpG sites were closely related to lipid profiles and ALT levels. Regarding demographic and clinical parameters, these CpG sites were strongly linked to BMI and waist circumference. Multivariate linear regression analysis adjusted for age, gender, and BMI also verified the results (significant results are listed in Supplementary Table 5).
A total of 42 methylated CpG sites were found to correlate with ALT levels, and these correlations remained significant after adjustment for age, gender, and BMI. Additionally, the correlations between levels of GGT with cg03992938 (CCDC13), cg04787602 (C1orf183), and cg014451560 (ARRDC1) were statistically significant. There were 32 CpG sites associated with TG, TC, or LDL-C levels, indicating dyslipidemia. With regard to glucose metabolism in NAFLD, the CpG sites were searched, and the results showed that cg07953400 (PIGQ), cg06706068 (RPUSD1), and cg16416718 (CAMTA1) exhibited strong correlations with fasting blood glucose levels, while cg25456633, cg03992938 (CCDC13), and cg02013957 (CAGE1) were closely associated with the homeostatic model assessment insulin resistance (HOMA-IR) index.
Methylation of CpG sites associated with NAFLD and liver histology
As shown in Table 2, a total of 23 CpG sites were partially correlated with the hitological characteristics of NAFLD. Among the CpG sites associated with increased serum ALT levels, three sites were inversely correlated with hepatic steatosis: cg19634213 (PTEN) (r=−0.358, P=0.035), cg01067963 (MAPK1) (r=−0.343, P=0.044), and cg05102190 (ZYX) (r=−0.357, P=0.035). Additionally, seven CpG sites were negatively correlated with lobular inflammation: cg19634213 (PTEN) (r=−0.37, P=0.029), cg16398128 (ZNF622) (r=−0.497, P=0.002), cg22185268 (COMMD4) (r=−0.341, P=0.045), cg22721468 (SH3BP5L) (r=−0.337, P=0.048), cg15226170 (PMM1) (r=−0.369, P=0.029), cg04787602 (C1orf91) (r=−0.522, P=0.032), and cg05102190 (ZYX) (r=−0.362, P=0.033). None of the ALT-associated CpG sites were found to be correlated with liver fibrosis.
Table 2.
Correlations(r) between methylation levels and histological parameters in NAFLD.
| Illumina ID (n=35) | Steatosis | Ballooning | Inflammation | Fibrosis | ||||
|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | |
| cg15653194 (LFNG) | −0.179 | 0.491 | −0.088 | 0.737 | 0.485 | 0.049* | −0.395 | 0.117 |
| cg00150500 (HCFC1R1) | −0.381 | 0.024* | −0.337 | 0.048* | −0.346 | 0.042* | −0.179 | 0.303 |
| cg04787602 (C1orf91) | 0.286 | 0.265 | 0.382 | 0.131 | −0.522 | 0.032* | 0.355 | 0.161 |
| cg16398128 (ZNF622) | −0.119 | 0.495 | −0.031 | 0.859 | −0.497 | 0.002* | −0.054 | 0.757 |
| cg07112456 (LRWD1) | −0.526 | 0.025* | −0.439 | 0.069 | −0.132 | 0.448 | −0.095 | 0.708 |
| cg01693328 (not mapped) | 0.164 | 0.346 | 0.379 | 0.025* | 0.288 | 0.093 | 0.21 | 0.226 |
| cg15226170 (PMM1) | −0.222 | 0.199 | 0.032 | 0.854 | −0.369 | 0.029* | −0.009 | 0.961 |
| cg08013262 (INSR) | 0.145 | 0.406 | 0.337 | 0.048* | 0.047 | 0.788 | 0.083 | 0.635 |
| cg10178228 (FRMD4B) | −0.085 | 0.629 | 0.048 | 0.785 | −0.2 | 0.249 | −0.062 | 0.723 |
| cg05131957 (CRLS1) | −0.445 | 0.007* | −0.264 | 0.125 | −0.448 | 0.007* | 0.063 | 0.721 |
| cg19634213 (PTEN) | −0.358 | 0.035* | −0.047 | 0.787 | −0.370 | 0.029* | 0.132 | 0.449 |
| cg12473838 (SSBP1) | −0.704 | 0.001* | −0.542 | 0.001* | −0.531 | 0.001* | −0.219 | 0.001* |
| cg01067963 (MAPK1) | −0.343 | 0.044* | −0.165 | 0.344 | −0.177 | 0.308 | −0.002 | 0.992 |
| cg22185268 (COMMD4) | −0.04 | 0.818 | 0.187 | 0.283 | −0.341 | 0.045* | 0.066 | 0.706 |
| cg22721468 (SH3BP5L) | −0.19 | 0.275 | −0.122 | 0.484 | −0.337 | 0.048* | −0.078 | 0.654 |
| cg13463639 (SIGIRR) | 0.278 | 0.106 | 0.321 | 0.059 | 0.64 | 0.001* | 0.230 | 0.184 |
| cg13397649 (AFG3L2) | −0.375 | 0.138 | −0.543 | 0.024* | 0.093 | 0.722 | −0.286 | 0.267 |
| cg26309655 (TARS2) | 0.294 | 0.252 | 0.558 | 0.020* | −0.399 | 0.113 | 0.2 | 0.441 |
| cg04160753 (C10orf91) | −0.326 | 0.202 | 0.078 | 0.766 | −0.486 | 0.048 | −0.157 | 0.549 |
| cg05102190 (ZYX) | −0.357 | 0.035* | −0.197 | 0.256 | −0.362 | 0.033* | −0.064 | 0.716 |
| cg00574958 (CTP1A) | 0.134 | 0.441 | 0.294 | 0.087 | 0.443 | 0.008* | 0.232 | 0.180 |
| cg19878987 (LDHB) | −0.28 | 0.103 | −0.048 | 0.784 | −0.413 | 0.014* | 0.116 | 0.507 |
| cg15536552 (ACSL4) | 0.482 | 0.050* | 0.316 | 0.216 | −0.385 | 0.127 | 0.339 | 0.183 |
p<0.05.
Among the 32 CpG sites associated with lipid profiles, 11 were correlated with the histological characteristics of NAFLD: Cg00150500 (HCFC1R1) showed the strongest association with steatosis and lobular inflammation. Cg12473838 (SSBP1) showed a strong correlation with the following NAFLD histological features: steatosis (r=−0.704, P<0.001), hepatocytes ballooning (r=−0.542, P<0.001), lobular inflammation (r=−0.531, P<0.001) and fibrosis (r=−0.219, P<0.001). Additionally, cg13463639 (SIGIRR) and cg15653194 (LFNG) both showed a strong-to-moderate correlation with lobular inflammation (r=0.64, P<0.001 and r=0.485, P=0.049, respectively). No methylated CpG sites associated with FBG levels or HOMA-IR scores were correlated with the histological features of NAFLD. Pearson’s correlation coefficient (r) (Y-axis) between the liver histology parameters (X-axis) and the methylated DNA sites that were inversely or directly associated with NAFLD are plotted in Figure 4.
Figure 4.
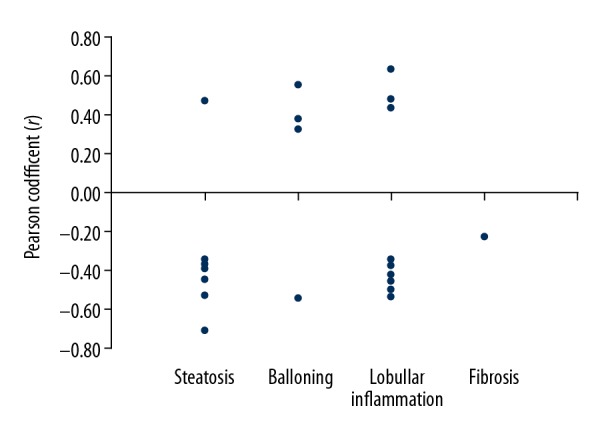
Pearson’s correlation coefficients (r) (Y-axis) between histological features (X-axis) and methylated DNA sites inversely or directly associated with nonalcoholic fatty liver disease (NAFLD).
Differentially methylated CpG (DMCpG) sites as potential biomarkers in NAFLD
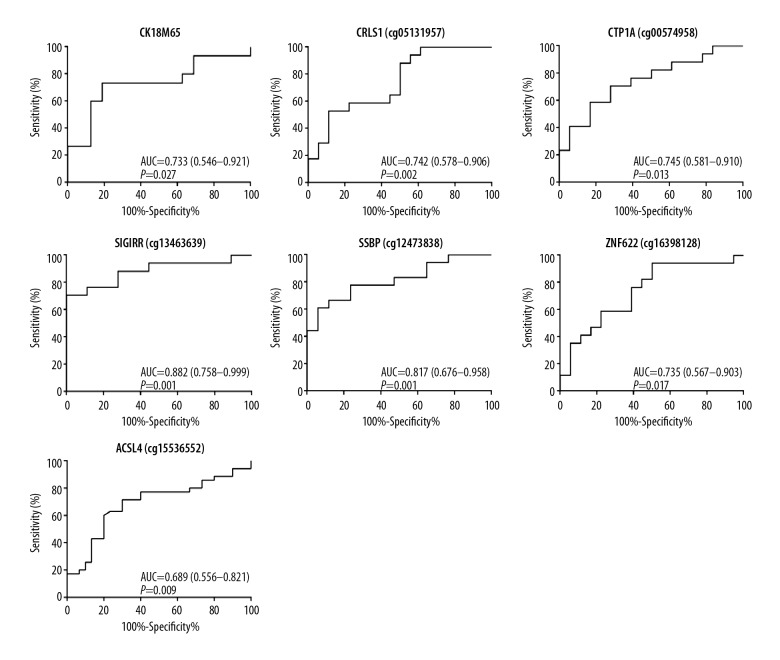
The DNA methylation levels of the CpG sites that were correlated with the histological features of NAFLD were re-tested, which showed that the methylation levels of the ACSL4, CRLS1, CTP1A, SIGIRR, SSBP1, and ZNF622 genes were significantly different in patients with NASH compared with patients with simple hepatic steatosis. The areas under the receiver-operating characteristic (ROC) curves (AUROCs) for these six sites were 0.689 (95% CI, 0.556–0.821) for ACSL4, 0.742 (95% CI, 0.578–0.906) for CRLS1, 0.745 (95% CI, 0.581–0.910) for CTP1A, 0.882 (95% CI, 0.758–0.999) for SIGIRR, 0.817 (95% CI, 0.676–0.958) for SSBP1, and 0.735 (95% CI, 0.567–0.903) for ZNF622 (Figure 5). These six sites showed significant effectiveness in discriminating between patients with NASH and patients with simple hepatic steatosis. The estimated cut-off values, sensitivities, specificities, positive predictive values (PPVs), and negative predictive values (NPVs) are listed in Table 3. The effectiveness of these six sites was compared with that of serum CK18 M65 levels as a marker of inflammation and hepatocyte injury, and no significant differences were found.
Figure 5.
Receiver-operating characteristic (ROC) curves for the methylation levels of the nonalcoholic steatohepatitis (NASH)-related differentially methylated CpG sites.
Table 3.
Comparison of the performance of each test for differentiating NASH versus simple hepatic steatosis.
| Test | AUC (95%CI) | Sig | Cut-off | Sens (%) | Spec (%) | PPV (%) | NPV (%) | Z-score | P-value |
|---|---|---|---|---|---|---|---|---|---|
| CK18M65 | 0.733 (0.546–0.921) | 0.027 | 756.35 | 73.3 | 75.0 | 78.6 | 76.5 | ||
| ACSL4 | 0.689 (0.556–0.821) | 0.009 | 0.123 | 62.9 | 57.1 | 64.9 | 78.6 | 1.258 | 0.20 |
| CTP1A | 0.745 (0.581–0.909) | 0.013 | 0.151 | 70.6 | 72.2 | 70.6 | 72.2 | 0.08 | 0.93 |
| CRLS1 | 0.742 (0.578–0.906) | 0.015 | 0.071 | 88.9 | 52.9 | 67.7 | 81.8 | 0.31 | 0.75 |
| SIGIRR | 0.882 (0.758–0.999) | 0.001 | 0.787 | 70.6 | 99 | 92.3 | 77.3 | 1.12 | 0.26 |
| SSBP1 | 0.817 (0.676–0.958) | 0.001 | 0.04 | 61.1 | 94.1 | 91.7 | 69.6 | 0.32 | 0.75 |
| ZNF622 | 0.735 (0.567–0.903) | 0.017 | 0.046 | 50.0 | 94.1 | 92.3 | 61.5 | 0.08 | 0.93 |
AUROC – area under the receiver operating characteristic curve; Sens – sensitivity; Spec – specificity; PPV – positive predictive value; NPV – negative predictive value.
Validation of ACSL4 methylation by pyrosequencing
ACSL4 (cg15536552) was selected for validation through bisulfite pyrosequencing. The pyrosequencing results indicated that the methylation level of ACSL4 (cg15536552) was consistent with that determined by the Illumina HumanMethylation450 BeadChip platform (r=0.756, P<0.0001). According to the SAF score, ACSL4 was significantly hypomethylated in patients with NASH compared with patients with simple hepatic steatosis (P=0.004).
Discussion
Epigenetic processes are now recognized to play a role in the progression of nonalcoholic fatty liver disease (NAFLD). The characterization of how these epigenetic processes result in changes associated with liver injury provides new insights related to disease diagnostics and management. DNA methylation refers to the addition of a methyl group at a CpG site, which can influence the function of DNA by activating or suppressing gene expression [22]. CpG methylation is regarded as a ‘molecular clock’ during the progression of liver phenotypes from normal liver histology to simple hepatic steatosis to inflammation and fibrosis [23].
In the present study, the application of differentially methylated CpG sites to assess NAFLD phenotypes using microarrays allowed the analysis of a large fraction of the DNA methylome and the identification of genes correlated with changes in DNA methylation in peripheral blood leukocytes. This differential methylation may directly influence normal epigenetic regulation and cause pathological changes. The main purpose of the present study was to identify differentially methylated CpG sites in peripheral blood leukocytes to determine epigenetic biomarkers that might exhibit a strong correlation with the clinical parameters and histological features of NAFLD.
First, we analyzed more than 410,000 CpG sites to demonstrate that nonalcoholic steatohepatitis (NASH) was associated with differential DNA methylation in comparison with normal livers and simple hepatic steatosis. The adoption of a statistical significance of q<0.05 enabled the identification of methylated sites that were significantly associated with NAFLD and simple hepatic steatosis. The identified NAFLD-associated CpG sites were mostly hypomethylated, a phenomenon supported by previous work by Murphy et al., who showed that the livers of individuals with advanced NAFLD exhibited more hypomethylation than those of individuals with mild NAFLD [13].
In this study, the top NAFLD-associated CpG sites, including KCNQ3, FUT11, DUSP16, CAMTA1, and GTSE1, appeared to present a close connection with NAFLD and a higher risk of developing HCC. Genes contributing to insulin resistance (EHMT2, INSR, TARS2, and BRSK2) and lipid metabolism (RXRB) were enriched. Additionally, at simple hepatic steatosis-associated CpG sites, some adipogenic and lipid metabolism genes were also enriched in adipocytokine and insulin signaling pathways (CPT1A, PTEN, LDHB, SGMS1, PMM1, ATP5G1, CRLS1, and PIGQ). These observations provide evidence that, as a condition associated with metabolic disorders, NAFLD is influenced by integrated epigenetic modifications, even at an early stage. This finding might form the basis for future identification of DNA methylation biomarkers and their use to estimate future disease risk [24].
Increased serum levels of liver enzymes such as alanine aminotransferase (ALT), aspartate transaminase (AST), and γ-glutamyl-transpeptidase (GGT) are markers of liver injury [25]. GGT affects the pro-oxidant roles played by molecular species originating during the catabolism of glutathione and promotes lipid peroxidation. Previous studies have shown a genetic effect on liver enzyme levels [26]. In this study, the majority of the NAFLD-methylated CpG sites were associated with increased ALT levels, a connection that retained its significance even after adjustments for age, gender and body mass index (BMI). The analysis focused on the correlation of these CpG sites with the histological features of NAFLD. The differential methylation of CpG sites within ZNF622, PTEN, COMMD4, SH3BP4L, PMM1, C1orf91, and ZYX was associated with lobular inflammation. Some of these genes encode key enzymes that catalyze the initial steps of lipid, acetyl-CoA, and glucose metabolism and are members of insulin-like signaling pathways. Methylation differences in ZYX and PTEN were also correlated with steatosis, although no CpG sites associated with AST were identified.
The identification of methylation sites associated with liver enzymes and hepatic steatosis has recently been reviewed by Nano et al., who conducted an epigenome-wide association study and identified eight probes associated with serum GGT levels and one probe associated with serum ALT levels [16]. As in the present study, they identified no probe associated with serum AST levels.
Future large-scale studies may produce different results from the present study. Individuals with simple hepatic steatosis seldom progress to clinically significant liver disease and are considered to exhibit mild NAFLD. However, in patients with NASH, hepatic steatosis may trigger a fibrogenic repair process that can lead to cirrhosis or hepatocellular carcinoma (HCC). Murphy et al. found relevant differences in methylation that distinguished between patients with advanced fibrosis from those with mild fibrosis [13]. Although the present study employed the same technique for measuring DNA methylation, the approach taken for analyzing the differences between liver phenotypes was dissimilar. Also, our results supported the existence cross-talk between epigenetic features and liver enzymes at the identified CpG sites. Collectively, these observations may reflect the influential role of hepatic inflammation in simple hepatic steatosis at the epigenetic level, which contributes to progression toward NASH [27].
The second main aim of this study focused on the CpG sites correlated with lipid profiles. Dysfunctional lipid metabolism causes hepatic fat accumulation. Increased serum free fatty acids (FFA) cause increased triglyceride (TG) and very low-density lipoprotein (VLDL) levels in hepatocytes and trigger lipid peroxidation. These effects are closely related to the progression of NAFLD [28]. Also, circulating cytokine and adipokine levels as well as the associated mitochondrial dysfunction, lipotoxicity, endoplasmic reticulum damage and oxidative stress are involved in hepatic steatosis [29]. The most common pattern of dyslipidemia in NAFLD is characterized by hypertriglyceridemia, high levels of low-density lipoprotein cholesterol (LDL-C), and low levels of high-density lipoprotein cholesterol (HDL-C). In patients with NASH, there is a significant increase in the levels of oxidized LDL-C. In this study, a total of 31 CpG sites associated with LDL-C were selected after a multivariate analysis adjusted for gender, age, and BMI. Pearson’s correlation coefficient (r) was used to verify the correlations between the CpG sites and NAFLD histological features. Ten of the identified CpG sites (CTP1A, LFNG, ZNF622, HCFC1R1, CH3BP5L, COMMD4, SIGIRR, LDHB, SSBP1, and C1ofr91) were moderately correlated with lobular inflammation.
Previously published studies have shown that high serum levels of oxidized LDL can be considered a risk factor for NASH, as oxidized LDL-C interacts with immune cells and contributes to the inflammatory process, upregulates adhesion molecules, and induces inflammation by increasing reactive oxygen species (ROS) generation and apoptotic cell death, which are involved in the progression of NASH [30]. In this study, DNA methylation was measured in peripheral blood leukocytes, and the results were consistent with the serum lipid profile data. According to Pearson’s correlation coefficient (r) analysis, a close association was found between lobular inflammation and NAFLD-associated methylated CpG sites relevant to altered LDL-C levels, which might indicate that alterations in methylation levels also exert a direct or indirect influence on liver lobular inflammation that results in the onset of NASH.
The final part of this study highlighted six methylated CpG sites that differentiated NASH from simple hepatic steatosis (ACLS4, CPT1A, CRLS1, SSBP1, SIGIRR, and ZNF622). Cytokeratin (CK)18-M65 was selected as a reference standard, as it is one of the most commonly employed serum biomarkers for diagnosing NASH. A previously published meta-analysis showed that the area under the receiver-operating characteristic (AUROC) curve for CK18-M65 for the diagnosis of NASH was 0.71–0.93 (sensitivity 66%, specificity 82%) [31]. The present study showed that SSBP1 and SIGIRR presented good efficiency for discriminating between NASH and simple hepatic steatosis, with AUCs of 0.817 and 0.882, respectively.
Expression of the SSBP1 gene has been reported to be associated with the development of obesity and has been found to increase lipid accumulation in the liver. The cholesterol content of cells has been reported to be significantly increased following SSBP1 knockdown, indicating that SSBP1 could inhibit cellular cholesterol synthesis and accumulation [32]. SIGIRR (also known as TIR8) is a negative regulator of Toll-like receptor 4 (TLR4), which is upregulated in NAFLD and mediates NASH before the onset of liver fibrosis. The expression of the SIGIRR gene has also been shown to represent an important checkpoint in anti-cancer activity in natural killer (NK) cells [33].
Arachidonic acid preferred long-chain acyl-CoA synthetase (ACSL4) is a key enzyme involved in fatty acid metabolism in a variety of tissues and in hepatic steatosis, even after adjustment for BMI. Upregulation of the ACSL4 gene accelerates lipogenesis, whereas downregulation of ACSL4 prevents the accumulation of cellular cholesterol [34]. Previous study also reported leukocytic hypomethylated ACSL4 an index for borderline/definitive NASH, with odds ratio (OR) at 11.44 and 95% confidence interval (CI) from 1.04 to 125.37 (P=0.046) [35]. In that study, the NAFLD Activity Score (NAS) scoring system has been used for the stratification of clinical phenotypes of NAFLD. However, inflammation facilitates fibrosis, while the prognosis of steatosis is still controversial. NAS scoring system is no longer recommended for the diagnosis of NASH because of its low prognostic value [36]. In our study, we used SAF score instead, a system that complements the histopathological evaluation by dissociating hepatic steatosis from inflammation. This system helps to avoid some special cases from being excluded, as in some advanced cases, the liver may display ballooning and lobular inflammation with rare fat vacuoles, and provides a more accurate evaluation of NAFLD with better reproducibility [37]. In our study, ACSL4 retains an effective diagnosis value with AUROC at 0.742 (0.578–0.906) for the stratification of NASH in NAFLD.
CPT1A is a protein-encoding gene that plays an important role in the mitochondrial transport of carnitine, which results in a decrease in fatty acid beta-oxidation, and DNA methylation of the CpG sites of the CPT1A gene has been reported to be associated with increased lipid levels and metabolic syndrome [38]. The CRLS1 gene encodes cardiolipin, which is a phospholipid located in the inner mitochondrial membrane. The mitochondrial membrane is particularly susceptible to attack by ROS, resulting in damage to mitochondrial proteins, lipids and mitochondrial DNA, which can cause hepatocyte injury in NAFLD. Additionally, loss of cardiolipin leads to the generation of excessive levels of reactive oxygen species (ROS) that promote lipid peroxides and may cause oxidation of cardiolipin, catalyzed by cytochrome-c, resulting in apoptosis [39,40]. The ZNF622 gene (also known as ZPR9) encodes a multiprotein that is involved in the apoptosis signal-regulating kinase 1 (ASK1) and transforming growth factor (TGF)-β signaling pathway, and downregulation of ASK1 and TGF-β signaling activity can result in reduced intracellular lipid deposition.
Finally, this study identified a group of genes with a transcriptional status that was significantly correlated with DNA methylation levels, and this relationship differed between the NAFLD phenotypes. The findings of this study further showed that the methylated CpG sites detected in peripheral blood leukocytes were strongly associated with liver enzyme levels and lipid profiles, which are relevant to the outcomes of liver injury, impaired lipid metabolism, and the histological features of NAFLD. The findings of this study may have implications for the diagnosis of NAFLD, particularly for the diagnosis and evaluation of the severity of NASH, with differentially methylated CpG sites functioning as diagnostic or prognostic biomarkers. Epigenetic modifications might serve as malleable targets for future interventions that aim to detect or reverse advanced NAFLD.
The main challenge of this study was to assess the cause of NAFLD, as there was no evidence to explain why methylation facilitates lobular inflammation or why lobular inflammation in the progression of NASH alters methylation levels. All of these research questions require additional study. A further limitation of the study was that DNA methylation was measured in peripheral blood leukocytes, rather than in liver tissue, which may not be relevant to findings in the liver. Additionally, this study did not include gene expression analysis, and RNA samples were not available. As a result, differential DNA methylation was not studied directly from the analysis of the samples exhibiting the histological features of the NAFLD phenotypes. The whole-blood samples used in this study provided leukocytes for analysis, which were employed for quantifying DNA methylation levels, and allowed correlations with liver enzymes levels and lipid profiles to be assessed. Therefore, some CpG sites important to NAFLD might not have been detected in this study.
Conclusions
The findings of this clinical study showed that DNA methylation sites in peripheral blood leukocytes correlated with changes in serum liver enzyme levels and lipid profiles and with histologically confirmed forms of nonalcoholic fatty liver disease (NAFLD) that included simple steatosis and nonalcoholic steatohepatitis (NASH) when compared with healthy controls. The study identified six differentially methylated CpG sites in genes including ACSL4, CPT1A, SSBP1, CRLS1, ZNF622, and SIGIRR in NAFLD patients. In the simple hepatic steatosis group, 42 methylated CpG sites were found to be associated with the increased serum levels of alanine aminotransferase (ALT), and 32 methylated CpG sites were associated with the altered lipid profiles in the blood. In the NASH group, altered methylation of CpG sites showed a more close correlation with the presence of hepatic lobular inflammation, seen histologically on liver biopsy. These results suggest that changes in methylated CpG sites can be detected in peripheral blood leukocytes in patients with NAFLD, and these blood-based biomarkers may have potential value for clinical research and diagnosis of NASH.
Supplementary Tables
Supplementary Table 1.
Individual CpG sites associated with NAFLD (q<0.05) in circulating leukocytes. (N=64).
| Probe ID | p NAFLD | q NAFLD | NAFLD mean β | CL mean β | Difference (% points) | Gene symbol | UCSC RefGene Acession | Relation to gene region | Relation to CpG island |
|---|---|---|---|---|---|---|---|---|---|
| cg00150500 | 5.05E-08 | 0.003526 | 0.040611 | 0.026409 | 0.014201229 | HCFC1R1; THOC6 | NM_017885; NM_001002018; NM_001002017; NM_001142350; NM_024339 | Body; TSS1500 | Island |
| cg01693328 | 3.78E-06 | 0.037736 | 0.734933 | 0.689903 | 0.045030407 | S_Shelf | |||
| cg02013957 | 2.80E-07 | 0.008217 | 0.069828 | 0.108342 | −0.03851329 | CAGE1; RIOK1 | NM_205864; NM_031480; NM_001170693; NM_001170692 | TSS1500; Body | S_Shore |
| cg03511638 | 7.43E-06 | 0.04944 | 0.207524 | 0.192478 | 0.015045277 | FBXL15 | NM_024326; NM_024326 | 1st Exon; 5′UTR | Island |
| cg03615240 | 2.36E-06 | 0.029878 | 0.234903 | 0.217214 | 0.01768861 | Island | |||
| cg03753066 | 2.57E-06 | 0.029878 | 0.042778 | 0.033719 | 0.009058818 | PGPEP1 | NM_017712 | TSS200 | N_Shore |
| cg03956053 | 4.68E-06 | 0.040684 | 0.094807 | 0.069568 | 0.025238064 | Island | |||
| cg03992938 | 7.18E-06 | 0.04944 | 0.201834 | 0.1878 | 0.014034138 | CCDC13 | NM_144719; NM_144719 | 1st Exon; 5′UTR | Island |
| cg04160753 | 5.99E-06 | 0.046488 | 0.275213 | 0.255442 | 0.019770564 | C10orf91 | NM_173541 | TSS1500 | |
| cg04396550 | 3.68E-12 | 1.54E-06 | 0.037964 | 0.025583 | 0.012380549 | KCNQ3 | NM_004519 | TSS1500 | Island |
| cg04787602 | 2.61E-07 | 0.008217 | 0.161306 | 0.144232 | 0.017073828 | C1orf183 | NM_198926; NM_019099 | Body; TSS1500 | Island |
| cg04981696 | 5.54E-07 | 0.012898 | 0.559923 | 0.491968 | 0.067954905 | ABCC1 | NM_019862; NM_019898; NM_019899; NM_004996; NM_019900 | Body | S_Shore |
| cg05102190 | 7.38E-06 | 0.04944 | 0.292254 | 0.261536 | 0.030718122 | ZYX | NM_003461; NM_001010972 | TSS200 | Island |
| cg06706068 | 6.62E-06 | 0.04866 | 0.450319 | 0.399254 | 0.051064413 | RPUSD1 | NM_058192 | 3′UTR | N_Shore |
| cg07067744 | 6.97E-06 | 0.04944 | 0.920563 | 0.90232 | 0.018243335 | BRSK2 | NM_003957 | Body | |
| cg07112456 | 6.04E-07 | 0.013311 | 0.931972 | 0.912429 | 0.019543021 | LRWD1 | NM_152892 | Body | S_Shelf |
| cg07953400 | 1.74E-06 | 0.02696 | 0.054226 | 0.045265 | 0.008961501 | PIGQ | NM_004204; NM_148920 | TSS1500; TSS1500 | N_Shore |
| cg07956264 | 1.06E-07 | 0.006374 | 0.060192 | 0.042644 | 0.017547367 | GPATCH3 | NM_022078 | 1st Exon | Island |
| cg08013262 | 2.14E-06 | 0.028915 | 0.051125 | 0.040134 | 0.010991397 | INSR | NM_000208; NM_001079817 | Body | Island |
| cg09367046 | 6.48E-06 | 0.048457 | 0.078046 | 0.057973 | 0.020073316 | ANGEL1 | NM_015305 | TSS200 | Island |
| cg09826692 | 3.66E-07 | 0.009581 | 0.695734 | 0.717446 | −0.0217123 | Island | |||
| cg10077144 | 6.43E-06 | 0.048457 | 0.151084 | 0.136608 | 0.014476221 | Island | |||
| cg10178228 | 2.57E-06 | 0.029878 | 0.16383 | 0.11116 | 0.052670088 | FRMD4B | NM_015123 | TSS200 | S_Shore |
| cg10576516 | 2.94E-07 | 0.008217 | 0.046487 | 0.034138 | 0.01234886 | RECQL5; SAP30BP | NM_001003716; NM_013260; NM_001003715; NM_013260; NM_004259 | TSS200; 1st Exon; 5′UT | Island |
| cg12289509 | 9.38E-07 | 0.019426 | 0.12632 | 0.116298 | 0.010022888 | TUBE1 | NM_016262; NM_001033564 | Body; TSS200 | Island |
| cg12362980 | 3.73E-06 | 0.037736 | 0.890648 | 0.915371 | −0.02472296 | Island | |||
| cg13291296 | 1.94E-06 | 0.028151 | 0.928293 | 0.567811 | 0.360481701 | GPR125 | NM_145290 | Body | |
| cg13373361 | 9.74E-07 | 0.019426 | 0.972063 | 0.958036 | 0.014027139 | MYH16 | NR_002147 | Body | |
| cg13397649 | 2.39E-06 | 0.029878 | 0.184331 | 0.171045 | 0.013286234 | AFG3L2 | NM_006796 | 1st Exon | Island |
| cg13463639 | 1.14E-07 | 0.047685 | 0.795196 | 0.744952 | 0.050244062 | SIGIRR | NM_001135053; NM_021805 | TSS1500 | S_Shore |
| cg13920768 | 4.76E-06 | 0.040684 | 0.180692 | 0.16038 | 0.02031251 | PRDM12 | NM_021619 | 3′UTR | Island |
| cg14214745 | 1.61E-06 | 0.025892 | 0.030079 | 0.02396 | 0.006119444 | CRELD1 | NM_001077415; NM_001031717; NM_015513; NM_015513; NM_001077415; NM_001031717 | 1st Exon; 5′UTR | Island |
| cg14247318 | 2.91E-06 | 0.032137 | 0.06859 | 0.051255 | 0.017334521 | S_Shore | |||
| cg14451560 | 4.08E-06 | 0.038185 | 0.163369 | 0.130014 | 0.033355016 | ARRDC1 | NM_152285 | TSS1500 | N_Shore |
| cg15317837 | 4.86E-06 | 0.0407 | 0.082077 | 0.065888 | 0.016188353 | GTPBP4 | NM_012341 | TSS1500 | Island |
| cg15653194 | 1.99E-09 | 0.000271 | 0.077077 | 0.051633 | 0.025443482 | LFNG | NM_001166355; NM_002304; NM_001040168; NM_001040167 | Body; TSS200 | Island |
| cg16105594 | 1.46E-06 | 0.025002 | 0.222788 | 0.206881 | 0.015907317 | MEF2C | NM_001131005; NM_002397 | 5′UTR; TSS1500 | Island |
| cg16261091 | 5.26E-06 | 0.042403 | 0.871221 | 0.768176 | 0.103045403 | C17orf101; HEXDC | NM_175902; NM_173620; NM_024648 | Body; TSS1500 | N_Shore |
| cg16357287 | 1.96E-07 | 0.007527 | 0.033464 | 0.022286 | 0.011177644 | SEH1L | NM_001013437; NM_031216 | TSS1500 | N_Shore |
| cg16398128 | 1.66E-07 | 0.007527 | 0.038963 | 0.025503 | 0.01345955 | ZNF622 | NM_033414; NM_033414 | 5′UTR; 1st Exon | Island |
| cg16416718 | 4.04E-06 | 0.038185 | 0.168379 | 0.155538 | 0.012840944 | CAMTA1 | NM_015215 | TSS1500 | Island |
| cg16487280 | 1.11E-06 | 0.020289 | 0.163618 | 0.146336 | 0.017282797 | MYBL2 | NM_002466 | TSS1500 | Island |
| cg17162475 | 1.08E-06 | 0.020289 | 0.136101 | 0.120308 | 0.015793168 | C6orf150 | NM_138441 | TSS200 | Island |
| cg17176517 | 5.44E-06 | 0.042966 | 0.055192 | 0.043187 | 0.012004537 | SNX4 | NM_003794; NM_003794 | 5′UTR; 1st Exon | Island |
| cg18570553 | 3.67E-06 | 0.037736 | 0.144011 | 0.128948 | 0.015063351 | PRR15 | NM_175887 | TSS200 | N_Shore |
| cg18848688 | 1.42E-08 | 0.001192 | 0.143981 | 0.325489 | −0.18150742 | LIFR | NM_001127671; NM_002310 | TSS1500; 5′UTR | Island |
| cg19137662 | 1.98E-07 | 0.007527 | 0.045624 | 0.032493 | 0.013131109 | NOP56 | NM_006392; NR_031699; NR_027700; NR_003078 | Body; TSS200 | Island |
| cg19189310 | 1.49E-06 | 0.025002 | 0.201487 | 0.18607 | 0.015416665 | RPS10 | NM_001014 | 5′UTR | Island |
| cg21786114 | 2.28E-07 | 0.007946 | 0.516219 | 0.460762 | 0.055456681 | EHMT2 | NM_006709; NM_025256 | TSS1500 | N_Shore |
| cg22185268 | 3.99E-07 | 0.009831 | 0.026994 | 0.020076 | 0.006918145 | COMMD4 | NM_017828 | TSS200 | Island |
| cg22402398 | 2.50E-06 | 0.029878 | 0.939115 | 0.923689 | 0.015425828 | FGR | NM_001042747; NM_005248; NM_001042729 | TSS1500; 5′UTR | |
| cg22598233 | 3.16E-06 | 0.033997 | 0.069128 | 0.056446 | 0.012681401 | POLRMT | NM_005035 | TSS200 | Island |
| cg22721468 | 1.96E-06 | 0.028151 | 0.060214 | 0.04424 | 0.01597372 | SH3BP5L | NM_030645 | TSS1500 | Island |
| cg22746421 | 1.52E-07 | 0.007527 | 0.87434 | 0.852661 | 0.021678989 | ||||
| cg23224405 | 2.59E-09 | 0.000271 | 0.064945 | 0.045018 | 0.019927219 | DUSP16 | NM_030640; NM_030640 | 1st Exon; 5′UTR | Island |
| cg24199050 | 2.65E-06 | 0.029971 | 0.010376 | 0.006359 | 0.004017477 | GTSE1 | NM_016426; NR_024009 | TSS200 | Island |
| cg24239690 | 7.36E-06 | 0.04944 | 0.196241 | 0.181611 | 0.014630372 | FSCN1 | NM_003088 | Body | S_Shore |
| cg24689754 | 4.95E-06 | 0.0407 | 0.029538 | 0.020196 | 0.009341864 | C19orf62 | NM_001033549; NM_001033549; NM_014173; NM_014173 | 5′UTR; 1st Exon | |
| cg25295726 | 1.84E-09 | 0.000271 | 0.181557 | 0.162827 | 0.018730389 | FUT11 | NM_173540 | TSS200 | Island |
| cg25456633 | 4.20E-06 | 0.038224 | 0.897003 | 0.878185 | 0.018817636 | ||||
| cg25818813 | 4.10E-06 | 0.038185 | 0.064429 | 0.050092 | 0.014336947 | MERTK | NM_006343 | Body | Island |
| cg26105057 | 2.02E-06 | 0.028151 | 0.160883 | 0.126787 | 0.034095982 | PAN2 | NM_014871; NM_001166279; NM_001127460; NM_014871; NM_001166279 | 1st Exon; 5′UTR; TSS200 | Island |
| cg26309655 | 4.66E-06 | 0.040684 | 0.16828 | 0.148223 | 0.02005632 | TARS2 | NM_025150 | TSS200 | |
| cg12473838 | 4.88E-06 | 0.028795 | 0.040501 | 0.031202 | 0.009299747 | SSBP1 | NM_003143; NR_015392 | TSS200; Body | Island |
| cg26786980 | 6.79E-06 | 0.049042 | 0.220594 | 0.183155 | 0.037438788 | RARB | NM_016152; NM_016152; NM_000965; NM_000965 | 5′UTR; 1st Exon |
Supplementary Table 2.
Individual CpG sites associated with simple hepatic steatosis (q<0.05) in circulating leukocytes. (N=119).
| Probe ID | p Simple hepatic steatosis | q Simple hepatic steatosis | Simple hepatic steatosis mean β | CL mean β | Difference (% points) | Gene symbol | UCSC RefGene acession | Relation to gene region | Relation to CpG island |
|---|---|---|---|---|---|---|---|---|---|
| cg00150500 | 0.00000 | 0.00005 | 0.04354 | 0.02641 | 0.01713 | HCFC1R1; THOC6 | NM_017885; NM_001002018; NM_001002017; NM_001142350; NM_024339 | Body; TSS1500 | Island |
| cg00574958 | 0.00000 | 0.02625 | 0.13777 | 0.19441 | −0.05664 | CPT1A; CPT1A | NM_001876; NM_001031847 | 5′UTR | N_Shore |
| cg00639286 | 0.00001 | 0.03794 | 0.11783 | 0.09329 | 0.02454 | ATOX1 | NM_004045 | Body | Island |
| cg00932007 | 0.00000 | 0.02625 | 0.07328 | 0.04803 | 0.02525 | SERINC1; PKIB | NM_020755; NM_181794; NM_020755 | 1st Exon; TSS200; 5′UTR | Island |
| cg01067963 | 0.00001 | 0.02968 | 0.06460 | 0.04919 | 0.01541 | MAPK1 | NM_138957; NM_002745 | TSS1500 | Island |
| cg01216607 | 0.00001 | 0.04888 | 0.13731 | 0.11167 | 0.02564 | ZNHIT1; PLOD3 | NM_006349; NM_001084; NM_001084 | TSS1500; 1st Exon; 5′UTR | Island |
| cg01525244 | 0.00000 | 0.00830 | 0.22203 | 0.17144 | 0.05059 | CBX7 | NM_175709 | TSS200 | N_Shore |
| cg01801101 | 0.00000 | 0.02176 | 0.31191 | 0.27460 | 0.03730 | AMZ1 | NM_133463 | 5′UTR | N_Shore |
| cg02432888 | 0.00001 | 0.03410 | 0.92622 | 0.93604 | −0.00982 | SMARCA4 | NM_001128845; NM_001128844; NM_001128848; NM_001128846; NM_003072; NM_001128849; NM_001128847 | 1st Exon; Body | N_Shelf |
| cg02581963 | 0.00000 | 0.00977 | 0.09723 | 0.06908 | 0.02816 | C10orf75 | NR_026762 | TSS200 | Island |
| cg03027241 | 0.00000 | 0.01712 | 0.35425 | 0.42982 | −0.07557 | KCNG1 | NM_002237 | 3′UTR | Island |
| cg03069383 | 0.00001 | 0.04806 | 0.22846 | 0.20088 | 0.02758 | RDH14 | NM_020905 | 1st Exon | Island |
| cg03074984 | 0.00001 | 0.04262 | 0.22030 | 0.19083 | 0.02947 | RPS6KA4 | NM_003942; NM_001006944 | TSS200 | Island |
| cg03511638 | 0.00000 | 0.02698 | 0.21013 | 0.19248 | 0.01765 | FBXL15 | NM_024326; NM_024326 | 1st Exon; 5′UTR | Island |
| cg03956053 | 0.00000 | 0.00468 | 0.10087 | 0.06957 | 0.03130 | Island | |||
| cg03992938 | 0.00000 | 0.02085 | 0.20399 | 0.18780 | 0.01619 | CCDC13 | NM_144719; NM_144719 | 1st Exon; 5′UTR | Island |
| cg04396550 | 0.00000 | 0.00000 | 0.03999 | 0.02558 | 0.01440 | KCNQ3 | NM_004519 | TSS1500 | Island |
| cg04551440 | 0.00000 | 0.02375 | 0.06975 | 0.05204 | 0.01771 | KATNAL1 | NM_001014380; NM_032116 | TSS200; 5′UTR | Island |
| cg04672769 | 0.00000 | 0.01712 | 0.10358 | 0.08340 | 0.02017 | HARS | NM_002109; NM_012208; NM_002109 | 5′UTR; TSS200; 1stExon | Island |
| cg04699668 | 0.00000 | 0.02654 | 0.78765 | 0.83452 | −0.04687 | N_Shore | |||
| cg04781764 | 0.00000 | 0.01704 | 0.86144 | 0.88530 | −0.02387 | WDR51B | NM_172240 | Body | |
| cg04787602 | 0.00000 | 0.02085 | 0.16300 | 0.14423 | 0.01876 | C1orf183; C1orf183 | NM_198926; NM_019099 | Body; TSS1500 | Island |
| cg04851352 | 0.00001 | 0.04156 | 0.32402 | 0.30766 | 0.01637 | Island | |||
| cg04981696 | 0.00000 | 0.01712 | 0.55926 | 0.49197 | 0.06730 | ABCC1 | NM_019862; NM_019898; NM_019899; NM_004996; NM_019900 | Body | S_Shore |
| cg05020775 | 0.00001 | 0.03145 | 0.11804 | 0.09411 | 0.02393 | SNPH | NM_014723 | TSS200 | Island |
| cg05102190 | 0.00000 | 0.01712 | 0.30075 | 0.26154 | 0.03922 | ZYX | NM_003461; NM_001010972 | TSS200 | Island |
| cg05131957 | 0.00000 | 0.02625 | 0.08817 | 0.06755 | 0.02062 | CRLS1; CRLS1 | NM_019095; NM_019095; NM_001127458 | 1st Exon; 5′UTR; TSS1500 | Island |
| cg05209527 | 0.00001 | 0.03018 | 0.20213 | 0.15172 | 0.05042 | SLC5A6 | NR_028323; NM_080592; NM_021095; NM_016085; NM_001170795 | TSS1500; Body; 5′UTR | S_Shore |
| cg05377161 | 0.00001 | 0.04888 | 0.11639 | 0.09404 | 0.02235 | Island | |||
| cg05629721 | 0.00000 | 0.02654 | 0.29331 | 0.24457 | 0.04875 | PSMB8 | NM_004159; NM_148919; NM_004159 | 5′UTR; TSS1500; 1st Exon | S_Shore |
| cg05977109 | 0.00000 | 0.02085 | 0.09199 | 0.06838 | 0.02361 | SGMS1 | NM_147156 | 5′UTR | Island |
| cg06276429 | 0.00001 | 0.04156 | 0.21157 | 0.17132 | 0.04025 | SLC39A7; RXRB | NM_006979; NM_021976; NM_001077516 | TSS1500; Body | N_Shore |
| cg07067659 | 0.00001 | 0.03187 | 0.03402 | 0.02441 | 0.00961 | SLC7A5 | NM_003486 | 1st Exon | Island |
| cg07112456 | 0.00001 | 0.03536 | 0.93287 | 0.91243 | 0.02044 | LRWD1 | NM_152892 | Body | S_Shelf |
| cg07953400 | 0.00001 | 0.02968 | 0.05508 | 0.04526 | 0.00982 | PIGQ | NM_004204; NM_148920 | TSS1500 | N_Shore |
| cg07956264 | 0.00000 | 0.00228 | 0.06267 | 0.04264 | 0.02003 | GPATCH3 | NM_022078 | 1st Exon | Island |
| cg08180934 | 0.00000 | 0.02625 | 0.05768 | 0.04568 | 0.01200 | STRAP | NM_007178 | TSS200 | Island |
| cg08276889 | 0.00000 | 0.02375 | 0.03778 | 0.02814 | 0.00964 | LOC100133985 | NR_024444 | Body | Island |
| cg08730728 | 0.00001 | 0.04156 | 0.10524 | 0.07919 | 0.02606 | Island | |||
| cg09090048 | 0.00001 | 0.03018 | 0.21230 | 0.18139 | 0.03091 | VPS26B; NCAPD3 | NM_052875; NM_015261 | TSS1500; Body | Island |
| cg09267087 | 0.00001 | 0.04156 | 0.67624 | 0.64183 | 0.03441 | ||||
| cg09367046 | 0.00001 | 0.04262 | 0.07982 | 0.05797 | 0.02184 | ANGEL1 | NM_015305 | TSS200 | Island |
| cg09457469 | 0.00000 | 0.01698 | 0.06248 | 0.04671 | 0.01577 | YBX2 | NM_015982 | Body | Island |
| cg09826692 | 0.00000 | 0.02375 | 0.69472 | 0.71745 | −0.02273 | Island | |||
| cg10059171 | 0.00001 | 0.03187 | 0.07502 | 0.05829 | 0.01674 | GPR89B | NM_016334; NM_016334 | 5′UTR; 1st Exon | Island |
| cg10178228 | 0.00000 | 0.00820 | 0.17082 | 0.11116 | 0.05966 | FRMD4B | NM_015123 | TSS200 | S_Shore |
| cg10369688 | 0.00001 | 0.03018 | 0.15684 | 0.13906 | 0.01778 | ZFP36 | NM_003407 | Body | Island |
| cg10381071 | 0.00000 | 0.01712 | 0.15437 | 0.20650 | −0.05213 | TLE3 | NM_020908; NM_001105192; NM_005078 | TSS1500 | Island |
| cg10576516 | 0.00000 | 0.00019 | 0.05003 | 0.03414 | 0.01589 | RECQL5; SAP30BP | NM_001003716; NM_013260; NM_001003715; NM_013260; NM_004259 | TSS200; 1st Exon; 5′UTR | Island |
| cg10965669 | 0.00001 | 0.02931 | 0.80870 | 0.82408 | −0.01538 | AXIN2 | NM_004655 | Body | Island |
| cg11319362 | 0.00001 | 0.04251 | 0.08666 | 0.06828 | 0.01838 | ARL6IP5 | NM_006407; NM_006407 | 1st Exon; 5′UTR | Island |
| cg11542063 | 0.00000 | 0.02625 | 0.05513 | 0.03884 | 0.01629 | NFE2L2 | NM_006164; NM_006164; NM_001145413; NM_001145412 | 5′UTR; 1st Exon; TSS1500 | Island |
| cg11866422 | 0.00000 | 0.02654 | 0.06337 | 0.05160 | 0.01177 | Island | |||
| cg12289509 | 0.00001 | 0.04442 | 0.12657 | 0.11630 | 0.01027 | TUBE1; C6orf225 | NM_016262; NM_001033564 | Body; TSS200 | Island |
| cg12362980 | 0.00000 | 0.00307 | 0.88730 | 0.91537 | −0.02807 | Island | |||
| cg12407666 | 0.00000 | 0.02901 | 0.12378 | 0.10250 | 0.02128 | RIC8B | NM_018157 | TSS1500 | Island |
| cg12810189 | 0.00001 | 0.03920 | 0.04387 | 0.03696 | 0.00691 | BCAP29 | NR_027830; NM_018844; NM_018844; NM_001008405 | Body; 1st Exon; 5′UTR; TSS1500 | Island |
| cg12850793 | 0.00000 | 0.02321 | 0.15979 | 0.14310 | 0.01669 | DENND5A | NM_015213 | TSS200 | Island |
| cg13110034 | 0.00001 | 0.03187 | 0.07293 | 0.06148 | 0.01145 | ANKRD34A; POLR3GL | NM_001039888; NM_032305 | TSS1500; 5′UTR | Island |
| cg13199615 | 0.00001 | 0.03018 | 0.23755 | 0.21115 | 0.02640 | KRTCAP2; TRIM46 | NM_173852; NM_025058 | TSS1500; Body | N_Shore |
| cg13398864 | 0.00000 | 0.02433 | 0.11061 | 0.08598 | 0.02463 | SEC22A | NM_012430 | TSS200 | N_Shore |
| cg13485756 | 0.00000 | 0.02698 | 0.09370 | 0.08224 | 0.01146 | RB1 | NM_000321 | TSS200 | Island |
| cg14022530 | 0.00000 | 0.02698 | 0.07371 | 0.05385 | 0.01986 | SASS6; CCDC76 | NM_194292; NM_019083 | TSS200 | Island |
| cg14214745 | 0.00000 | 0.01698 | 0.03114 | 0.02396 | 0.00718 | CRELD1 | NM_001077415; NM_001031717; NM_015513; NM_015513; NM_001077415; NM_001031717 | 1st Exon; 5′UTR | Island |
| cg14247318 | 0.00001 | 0.03018 | 0.06991 | 0.05126 | 0.01865 | S_Shore | |||
| cg15043926 | 0.00000 | 0.02698 | 0.28089 | 0.26430 | 0.01659 | STIM2 | NM_001169117; NM_020860; NM_001169118 | Body | Island |
| cg15226170 | 0.00000 | 0.02879 | 0.37491 | 0.31659 | 0.05832 | PMM1 | NM_002676 | TSS1500 | S_Shore |
| cg15324256 | 0.00000 | 0.02625 | 0.05617 | 0.04062 | 0.01555 | QTRTD1; KIAA1407 | NM_024638; NM_020817 | TSS1500; 1st Exon | Island |
| cg15324448 | 0.00000 | 0.02625 | 0.10020 | 0.08001 | 0.02019 | ZNF643 | NM_023070 | TSS200 | Island |
| cg15536552 | 0.07584 | 0.04796 | 0.20583 | 0.34740 | −0.01416 | ACSL4 | NM_022977; NM_004458 | 5′UTR; 5′UTR | N_Shore |
| cg15572086 | 0.00000 | 0.01698 | 0.06120 | 0.04806 | 0.01314 | C1orf203 | NR_027645; NR_024126; NR_024124; NR_027646; NR_024125 | TSS1500 | Island |
| cg15653194 | 0.00000 | 0.00394 | 0.07660 | 0.05163 | 0.02496 | LFNG | NM_001166355; NM_002304; NM_001040168; NM_001040167 | Body; TSS200 | Island |
| cg16007711 | 0.00000 | 0.01712 | 0.11104 | 0.08414 | 0.02689 | RHOBTB2 | NM_001160036; NM_001160037; NM_015178 | Body; 5′UTR | S_Shore |
| cg16306148 | 0.00001 | 0.03359 | 0.01234 | 0.00608 | 0.00626 | YBX1 | NM_004559 | TSS1500 | Island |
| cg16357287 | 0.00000 | 0.00228 | 0.03588 | 0.02229 | 0.01360 | SEH1L; SEH1L | NM_001013437; NM_031216 | TSS1500 | N_Shore |
| cg16398128 | 0.00000 | 0.00000 | 0.04360 | 0.02550 | 0.01810 | ZNF622 | NM_033414; NM_033414 | 5′UTR; 1st Exon | Island |
| cg16416718 | 0.00000 | 0.02879 | 0.16998 | 0.15554 | 0.01445 | CAMTA1 | NM_015215 | TSS1500 | Island |
| cg16743070 | 0.00000 | 0.01721 | 0.04222 | 0.02640 | 0.01582 | MYST3 | NM_001099413; NM_001099412; NM_006766 | 5′UTR | Island |
| cg16803522 | 0.00000 | 0.01441 | 0.03705 | 0.02603 | 0.01102 | RASGRP1 | NM_001128602; NM_005739 | TSS1500 | Island |
| cg17074213 | 0.00001 | 0.02901 | 0.20986 | 0.15861 | 0.05125 | TGFBR3 | NM_003243; NM_003243 | 1st Exon; 5′UTR | Island |
| cg17162475 | 0.00000 | 0.02375 | 0.13817 | 0.12031 | 0.01786 | C6orf150 | NM_138441 | TSS200 | Island |
| cg17168242 | 0.00000 | 0.01599 | 0.09273 | 0.08238 | 0.01035 | OTUD7B | NM_020205 | TSS200 | Island |
| cg17176517 | 0.00001 | 0.04039 | 0.05653 | 0.04319 | 0.01334 | SNX4 | NM_003794; NM_003794 | 5′UTR; 1st Exon | Island |
| cg17427781 | 0.00001 | 0.04227 | 0.08943 | 0.07059 | 0.01884 | DDX20; C1orf183 | NM_007204; NM_198926 | TSS200; Body | Island |
| cg18676790 | 0.00001 | 0.03018 | 0.27930 | 0.34935 | −0.07005 | ||||
| cg18848688 | 0.00001 | 0.02968 | 0.14296 | 0.32549 | −0.18253 | LIFR | NM_001127671; NM_002310 | TSS1500; 5′UTR | Island |
| cg19050851 | 0.00000 | 0.02690 | 0.03973 | 0.02876 | 0.01097 | ARSB | NM_000046; NM_198709 | Body; Body | Island |
| cg19137662 | 0.00000 | 0.01441 | 0.04705 | 0.03249 | 0.01455 | NOP56 | NM_006392; NR_031699; NR_027700; NR_003078 | Body; TSS200; TSS1500 | Island |
| cg19362196 | 0.00000 | 0.02625 | 0.70650 | 0.76251 | −0.05601 | STMN1 | NM_005563; NM_203399; NM_203401; NM_001145454 | TSS1500 | Island |
| cg19520115 | 0.00000 | 0.00160 | 0.12095 | 0.10488 | 0.01607 | CAPZB | NM_004930 | Body | Island |
| cg19634213 | 0.00000 | 0.01712 | 0.05716 | 0.04074 | 0.01643 | PTEN; KILLIN | NM_000314; NM_001126049 | TSS1500; 1st Exon | Island |
| cg19693031 | 0.00001 | 0.03187 | 0.80756 | 0.85146 | −0.04390 | TXNIP | NM_006472 | 3′UTR | |
| cg19878987 | 0.00000 | 0.02280 | 0.10362 | 0.08020 | 0.02342 | LDHB | NM_002300 | TSS1500 | S_Shore |
| cg20656751 | 0.00000 | 0.02625 | 0.51357 | 0.47261 | 0.04096 | NAT8L | NM_178557 | 1stExon | Island |
| cg21012729 | 0.00001 | 0.03763 | 0.08208 | 0.06527 | 0.01681 | NIN | NM_182946; NM_020921; NM_182944 | 5′UTR | Island |
| cg21373806 | 0.00001 | 0.04156 | 0.83546 | 0.86386 | −0.02840 | BAHCC1 | NM_001080519 | Body | N_Shore |
| cg21786114 | 0.00001 | 0.03187 | 0.51357 | 0.46076 | 0.05281 | EHMT2 | NM_006709; NM_025256 | TSS1500 | N_Shore |
| cg22185268 | 0.00000 | 0.00217 | 0.02871 | 0.02008 | 0.00864 | COMMD4 | NM_017828 | TSS200 | Island |
| cg22721468 | 0.00000 | 0.00228 | 0.06494 | 0.04424 | 0.02070 | SH3BP5L | NM_030645 | TSS1500 | Island |
| cg22746421 | 0.00000 | 0.01712 | 0.87645 | 0.85266 | 0.02379 | ||||
| cg22902478 | 0.00001 | 0.04156 | 0.10027 | 0.08525 | 0.01502 | LOC100130987 | NR_024469 | TSS200 | Island |
| cg23224405 | 0.00000 | 0.00011 | 0.06783 | 0.04502 | 0.02281 | DUSP16 | NM_030640; NM_030640 | 1st Exon; 5′UTR | Island |
| cg23348158 | 0.00000 | 0.01712 | 0.09342 | 0.06174 | 0.03168 | FUZ | NM_025129 | TSS200 | S_Shore |
| cg23512165 | 0.00000 | 0.02085 | 0.02886 | 0.02041 | 0.00845 | MTMR4 | NM_004687; NM_004687 | 1st Exon; 5′UTR | Island |
| cg24108286 | 0.00001 | 0.03145 | 0.25285 | 0.21178 | 0.04107 | TFDP1 | NR_026580; NM_007111 | TSS1500 | |
| cg24689754 | 0.00000 | 0.02433 | 0.03045 | 0.02020 | 0.01026 | C19orf62 | NM_001033549; NM_001033549; NM_014173; NM_014173 | 5′UTR; 1st Exon | |
| cg24728479 | 0.00001 | 0.03187 | 0.19258 | 0.17546 | 0.01712 | ZNF252; C8orf77 | NR_023392; NR_026974 | TSS1500; Body | Island |
| cg25053907 | 0.00001 | 0.02968 | 0.06377 | 0.05198 | 0.01179 | ZNF721; PIGG | NM_133474; NM_017733; NM_001127178; NM_133474 | 5′UTR; TSS200; 1st Exon | Island |
| cg25295726 | 0.00000 | 0.01101 | 0.18162 | 0.16283 | 0.01879 | FUT11 | NM_173540 | TSS200 | Island |
| cg25678532 | 0.00000 | 0.02625 | 0.05138 | 0.03889 | 0.01249 | RBBP8 | NM_203291; NM_203292; NM_002894 | TSS1500; TSS200 | Island |
| cg25818813 | 0.00000 | 0.02014 | 0.06671 | 0.05009 | 0.01661 | MERTK | NM_006343 | Body | Island |
| cg25856120 | 0.00001 | 0.04680 | 0.11157 | 0.08244 | 0.02914 | ATP5G1 | NM_005175; NM_005175; NM_001002027 | 1st Exon; 5′UTR | S_Shore |
| cg25989057 | 0.00001 | 0.02942 | 0.03813 | 0.02660 | 0.01154 | AIP | NM_003977 | TSS200 | Island |
| cg26309655 | 0.00001 | 0.04156 | 0.17136 | 0.14822 | 0.02314 | TARS2 | NM_025150 | TSS200 | |
| cg26462136 | 0.00001 | 0.03647 | 0.08411 | 0.06628 | 0.01783 | ATXN7L3 | NM_001098833; NM_020218 | TSS1500 | Island |
| cg26791384 | 0.00001 | 0.04156 | 0.25191 | 0.21622 | 0.03569 | SYCE1L | NM_001129979 | Body | Island |
| cg26824705 | 0.00001 | 0.04888 | 0.08142 | 0.06402 | 0.01740 | MUL1 | NM_024544 | TSS200 | Island |
| cg27164612 | 0.00000 | 0.02654 | 0.01342 | 0.00813 | 0.00529 | ||||
| cg27612695 | 0.00000 | 0.02625 | 0.18852 | 0.15615 | 0.03238 | SUZ12P | NR_024187 | Body | S_Shore |
Supplementary Table 3.
Significantly enriched KEGG pathways of genes with altered DNA methylation level in NAFLD.
| Pathway ID | Description | p Value | q Value | Genes | Enrich factor | |
|---|---|---|---|---|---|---|
| hsa03008 | Ribosome biogenesis in eukaryotes | 0.004 | 0.010 | GTPBP4 | GTP binding protein4 | 8.56 |
| NOP56 | Ribonucleo protein | |||||
| hsa04010 | MAPK signaling pathway | 0.001 | 0.011 | DUSP16 | Dual specificity phosphatase 16 | 2.59 |
| MEF2C | Myocyte enhancer factor 2C |
Supplementary Table 4.
Significantly enriched KEGG pathways of genes with altered DNA methylation level in simple hepatic steatosis.
| Pathway ID | Description | p Value | q Value | Genes | Enrich factor | |
|---|---|---|---|---|---|---|
| hsa04977 | Vitamin digestion and absorption | 0.0007 | 0.033 | SLC5A6 | Solute carrier family 5 member 6 | 15.84 |
| ABCC1 | ATP binding cassette sub familyC member1 | |||||
| hsa03013 | RNAtransport | 0.0100 | 0.040 | DDX20 | DEAD-box helicase 20 | 2.50 |
| SEH1L | SEH1 like nucleoporin | |||||
| hsa05200 | Pathways in cancer | 0.0070 | 0.034 | PTEN | Phosphatase and tensin homolog | 2.33 |
| AXIN2 | Axin 2 | |||||
| MAPK1 | Mitogen-activated protein kinase 1 | |||||
| RB1 | RB transcriptional corepressor 1 |
Supplementary Table 5.
Significance of associations between biological parameters and selected methylated CpG sites.
| Illumina ID | Gene symbol | GLU | HOMAIN | TG | TC | HDL | LDL | GGT | ALT | AST | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | ||
| cg00150500 | HCFC1R1 | 0.492 | 0.736 | 0.290 | 0.433 | 0.020 | 0.545 | 0.008 | 0.027* | 0.505 | 0.516 | 0.000 | 0.005* | 0.107 | 0.132 | 0.006 | 0.057 | 0.792 | 0.668 |
| cg00574958 | CTP1A | 0.333 | 0.435 | 0.433 | 0.768 | 0.001 | 0.001* | 0.017 | 0.308 | 0.067 | 0.575 | 0.000 | 0.001* | 0.125 | 0.417 | 0.034 | 0.478 | 0.340 | 0.657 |
| cg01067963 | MAPK1 | 0.321 | 0.685 | 0.520 | 0.963 | 0.039 | 0.795 | 0.216 | 0.632 | 0.070 | 0.316 | 0.018 | 0.259 | 0.365 | 0.201 | 0.006 | 0.035* | 0.603 | 0.454 |
| cg01693328 | 0.899 | 0.875 | 0.408 | 0.665 | 0.247 | 0.816 | 0.126 | 0.170 | 0.173 | 0.441 | 0.087 | 0.165 | 0.162 | 0.174 | 0.002 | 0.009* | 0.111 | 0.185 | |
| cg01801101 | AMZ1 | 0.971 | 0.949 | 0.392 | 0.838 | 0.006 | 0.211 | 0.240 | 0.280 | 0.287 | 0.844 | 0.016 | 0.078 | 0.983 | 0.932 | 0.000 | 0.002* | 0.818 | 0.488 |
| cg02013957 | CAGE1 | 0.860 | 0.788 | 0.010 | 0.010* | 0.049 | 0.371 | 0.213 | 0.951 | 0.051 | 0.656 | 0.150 | 0.413 | 0.133 | 0.122 | 0.001 | 0.024* | 0.498 | 0.082 |
| cg03511638 | FBXL15 | 0.158 | 0.282 | 0.136 | 0.258 | 0.129 | 0.930 | 0.585 | 0.985 | 0.373 | 0.908 | 0.073 | 0.446 | 0.113 | 0.141 | 0.010 | 0.070 | 0.588 | 0.818 |
| cg03615240 | 0.147 | 0.757 | 0.286 | 0.355 | 0.101 | 0.206 | 0.036 | 0.142 | 0.816 | 0.237 | 0.012 | 0.012* | 0.179 | 0.400 | 0.000 | 0.033* | 0.061 | 0.373 | |
| cg03992938 | CCDC13 | 0.107 | 0.296 | 0.067 | 0.032* | 0.844 | 0.139 | 0.035 | 0.449 | 0.404 | 0.240 | 0.001 | 0.006* | 0.016 | 0.021* | 0.012 | 0.011* | 0.187 | 0.882 |
| cg04160753 | C10orf91 | 0.162 | 0.316 | 0.936 | 0.880 | 0.014 | 0.105 | 0.030 | 0.161 | 0.466 | 0.798 | 0.014 | 0.108 | 0.295 | 0.361 | 0.104 | 0.345 | 0.904 | 0.986 |
| cg04787602 | C1orf183 | 0.153 | 0.304 | 0.328 | 0.396 | 0.044 | 0.271 | 0.058 | 0.914 | 0.046 | 0.259 | 0.001 | 0.039* | 0.033 | 0.035* | 0.001 | 0.010* | 0.181 | 0.207 |
| cg04981696 | ABCC1 | 0.260 | 0.318 | 0.420 | 0.784 | 0.046 | 0.090 | 0.055 | 0.778 | 0.470 | 0.645 | 0.002 | 0.033* | 0.657 | 0.736 | 0.000 | 0.004* | 0.777 | 0.547 |
| cg05102190 | ZYX | 0.116 | 0.122 | 0.972 | 0.880 | 0.039 | 0.084 | 0.056 | 0.360 | 0.301 | 0.926 | 0.025 | 0.077 | 0.338 | 0.365 | 0.005 | 0.030* | 0.812 | 0.717 |
| cg05131957 | CRLS1 | 0.091 | 0.199 | 0.489 | 0.876 | 0.032 | 0.274 | 0.068 | 0.312 | 0.585 | 0.892 | 0.006 | 0.047* | 0.995 | 0.780 | 0.018 | 0.069 | 0.483 | 0.196 |
| cg05209527 | SLC5A6 | 0.143 | 0.519 | 0.254 | 0.407 | 0.141 | 0.919 | 0.040 | 0.109 | 0.563 | 0.908 | 0.005 | 0.506 | 0.923 | 0.353 | 0.000 | 0.006* | 0.478 | 0.317 |
| cg06706068 | RPUSD1 | 0.034 | 0.020* | 0.477 | 0.970 | 0.533 | 0.052 | 0.032 | 0.268 | 0.090 | 0.748 | 0.023 | 0.049* | 0.233 | 0.243 | 0.003 | 0.043* | 0.851 | 0.586 |
| cg07067744 | BRSK2 | 0.782 | 0.974 | 0.454 | 0.760 | 0.093 | 0.104 | 0.038 | 0.963 | 0.389 | 0.782 | 0.003 | 0.030* | 0.360 | 0.435 | 0.012 | 0.108 | 0.694 | 0.757 |
| cg07112456 | LRWD1 | 0.167 | 0.261 | 0.370 | 0.531 | 0.230 | 0.199 | 0.071 | 0.660 | 0.470 | 0.664 | 0.033 | 0.178 | 0.082 | 0.097 | 0.031 | 0.208 | 0.475 | 0.480 |
| cg07953400 | PIGQ | 0.001 | 0.004* | 0.625 | 0.452 | 0.233 | 0.005* | 0.000 | 0.823 | 0.916 | 0.770 | 0.000 | 0.001* | 0.250 | 0.343 | 0.013 | 0.041* | 0.939 | 0.439 |
| cg07956264 | GPATCH3 | 0.702 | 0.693 | 0.026 | 0.068 | 0.000 | 0.028* | 0.029 | 0.002* | 0.112 | 0.493 | 0.009 | 0.023* | 0.158 | 0.164 | 0.000 | 0.002* | 0.282 | 0.232 |
| cg08013262 | INSR | 0.582 | 0.678 | 0.482 | 0.695 | 0.031 | 0.046* | 0.023 | 0.242 | 0.850 | 0.569 | 0.013 | 0.061 | 0.847 | 0.952 | 0.001 | 0.005* | 0.859 | 0.992 |
| cg09367046 | ANGEL1 | 0.275 | 0.356 | 0.557 | 0.793 | 0.109 | 0.351 | 0.190 | 0.811 | 0.203 | 0.624 | 0.022 | 0.118 | 0.175 | 0.205 | 0.000 | 0.002* | 0.198 | 0.188 |
| cg10077144 | 0.722 | 0.544 | 0.467 | 0.283 | 0.006 | 0.203 | 0.029 | 0.084 | 0.645 | 0.736 | 0.007 | 0.138 | 0.950 | 0.711 | 0.002 | 0.004* | 0.776 | 0.621 | |
| cg10178228 | FRMD4B | 0.566 | 0.705 | 0.482 | 0.603 | 0.447 | 0.937 | 0.668 | 0.196 | 0.005 | 0.055 | 0.078 | 0.357 | 0.240 | 0.270 | 0.000 | 0.005* | 0.838 | 0.776 |
| cg12289509 | TUBE1 | 0.402 | 0.503 | 0.479 | 0.670 | 0.236 | 0.029* | 0.019 | 0.921 | 0.676 | 0.683 | 0.001 | 0.002* | 0.443 | 0.489 | 0.000 | 0.001* | 0.624 | 0.833 |
| cg12473838 | SSBP1 | 0.004 | 0.167 | 0.500 | 0.711 | 0.419 | 0.928 | 0.000 | 0.192 | 0.000 | 0.664 | 0.001 | 0.030* | 0.122 | 0.140 | 0.755 | 0.199 | 0.453 | 0.765 |
| cg13291296 | GPR125 | 0.652 | 0.802 | 0.697 | 0.967 | 0.034 | 0.101 | 0.064 | 0.256 | 0.063 | 0.211 | 0.010 | 0.028* | 0.242 | 0.253 | 0.012 | 0.057 | 0.752 | 0.820 |
| cg13397649 | AFG3L2 | 0.090 | 0.227 | 0.678 | 0.749 | 0.003 | 0.809 | 0.305 | 0.079 | 0.133 | 0.385 | 0.038 | 0.295 | 0.898 | 0.948 | 0.017 | 0.082 | 0.301 | 0.156 |
| cg13463639 | SIGIRR | 0.082 | 0.551 | 0.618 | 0.985 | 0.376 | 0.709 | 0.002 | 0.348 | 0.028 | 0.363 | 0.009 | 0.020* | 0.375 | 0.275 | 0.980 | 0.730 | 0.298 | 0.431 |
| cg13920768 | PRDM12 | 0.278 | 0.431 | 0.539 | 0.635 | 0.004 | 0.101 | 0.035 | 0.065 | 0.855 | 0.221 | 0.016 | 0.095 | 0.278 | 0.347 | 0.007 | 0.040* | 0.779 | 0.710 |
| cg14214745 | CRELD1 | 0.764 | 0.627 | 0.141 | 0.246 | 0.001 | 0.419 | 0.207 | 0.024* | 0.022 | 0.196 | 0.048 | 0.218 | 0.883 | 0.713 | 0.041 | 0.267 | 0.314 | 0.359 |
| cg14247318 | 0.405 | 0.309 | 0.206 | 0.361 | 0.052 | 0.009* | 0.017 | 0.146 | 0.456 | 0.718 | 0.004 | 0.004* | 0.224 | 0.215 | 0.011 | 0.031* | 0.648 | 0.821 | |
| cg14451560 | ARRDC1 | 0.132 | 0.129 | 0.282 | 0.480 | 0.075 | 0.191 | 0.117 | 0.452 | 0.044 | 0.182 | 0.022 | 0.076 | 0.040 | 0.047* | 0.014 | 0.078 | 0.796 | 0.810 |
| cg15226170 | PMM1 | 0.894 | 0.780 | 0.422 | 0.625 | 0.024 | 0.354 | 0.136 | 0.352 | 0.511 | 0.911 | 0.012 | 0.095 | 0.302 | 0.203 | 0.001 | 0.009* | 0.535 | 0.537 |
| cg15317837 | GTPBP4 | 0.920 | 0.942 | 0.194 | 0.550 | 0.005 | 0.682 | 0.507 | 0.282 | 0.012 | 0.244 | 0.166 | 0.460 | 0.650 | 0.708 | 0.016 | 0.254 | 0.127 | 0.472 |
| cg15536552 | ACSL4 | 0.001 | 0.439 | 0.001 | 0.045* | 0.001 | 0.189 | 0.050 | 0.478 | 0.670 | 0.667 | 0.001 | 0.085 | 0.119 | 0.664 | 0.001 | 0.043* | 0.071 | 0.431 |
| cg15653194 | LFNG | 0.167 | 0.052 | 0.190 | 0.843 | 0.000 | 0.001* | 0.006 | 0.010* | 0.124 | 0.320 | 0.001 | 0.001* | 0.840 | 0.955 | 0.016 | 0.453 | 0.001 | 0.111 |
| cg16105594 | MEF2C | 0.228 | 0.481 | 0.701 | 0.832 | 0.042 | 0.234 | 0.067 | 0.437 | 0.020 | 0.064 | 0.002 | 0.022* | 0.411 | 0.500 | 0.026 | 0.124 | 0.086 | 0.213 |
| cg16261091 | HEXDC | 0.722 | 0.620 | 0.073 | 0.146 | 0.005 | 0.315 | 0.242 | 0.110 | 0.093 | 0.465 | 0.044 | 0.118 | 0.097 | 0.095 | 0.004 | 0.036* | 0.561 | 0.614 |
| cg16357287 | SEH1L | 0.574 | 0.745 | 0.092 | 0.138 | 0.081 | 0.183 | 0.110 | 0.446 | 0.126 | 0.361 | 0.008 | 0.025* | 0.773 | 0.709 | 0.000 | 0.003* | 0.644 | 0.386 |
| cg16398128 | ZNF622 | 0.410 | 0.717 | 0.339 | 0.352 | 0.003 | 0.022* | 0.005 | 0.058 | 0.365 | 0.766 | 0.000 | 0.001* | 0.976 | 0.859 | 0.000 | 0.001* | 0.809 | 0.764 |
| cg16416718 | CAMTA1 | 0.001 | 0.004* | 0.739 | 0.772 | 0.200 | 0.010* | 0.001 | 0.778 | 0.387 | 0.558 | 0.000 | 0.001* | 0.053 | 0.069 | 0.031 | 0.079 | 0.984 | 0.313 |
| cg16487280 | MYBL2 | 0.082 | 0.070 | 0.708 | 0.744 | 0.183 | 0.112 | 0.064 | 0.488 | 0.989 | 0.067 | 0.006 | 0.027* | 0.671 | 0.497 | 0.081 | 0.624 | 0.004 | 0.060 |
| cg17162475 | C6orf150 | 0.317 | 0.328 | 0.201 | 0.465 | 0.052 | 0.507 | 0.341 | 0.874 | 0.052 | 0.618 | 0.032 | 0.165 | 0.562 | 0.635 | 0.000 | 0.008* | 0.611 | 0.313 |
| cg17176517 | SNX4 | 0.349 | 0.639 | 0.406 | 0.446 | 0.255 | 0.182 | 0.042 | 0.821 | 0.291 | 0.593 | 0.005 | 0.062 | 0.563 | 0.713 | 0.006 | 0.030* | 0.715 | 0.484 |
| cg18570553 | PRR15 | 0.042 | 0.098 | 0.444 | 0.310 | 0.028 | 0.043* | 0.008 | 0.387 | 0.640 | 0.712 | 0.001 | 0.009* | 0.063 | 0.084 | 0.005 | 0.035* | 0.527 | 0.705 |
| cg18848688 | LIFR | 0.885 | 0.925 | 0.125 | 0.242 | 0.003 | 0.064 | 0.051 | 0.102 | 0.023 | 0.175 | 0.005 | 0.012* | 0.224 | 0.221 | 0.002 | 0.022* | 0.651 | 0.826 |
| cg19189310 | RPS10 | 0.398 | 0.564 | 0.453 | 0.711 | 0.062 | 0.150 | 0.052 | 0.953 | 0.205 | 0.964 | 0.009 | 0.086 | 0.200 | 0.239 | 0.005 | 0.075 | 0.292 | 0.224 |
| cg19634213 | PTEN | 0.194 | 0.497 | 0.494 | 0.932 | 0.900 | 0.353 | 0.102 | 0.213 | 0.483 | 0.651 | 0.010 | 0.092 | 0.297 | 0.356 | 0.001 | 0.002* | 0.236 | 0.447 |
| cg19878987 | LDHB | 0.321 | 0.532 | 0.328 | 0.574 | 0.086 | 0.136 | 0.034 | 0.691 | 0.477 | 0.932 | 0.008 | 0.087 | 0.176 | 0.240 | 0.001 | 0.003* | 0.197 | 0.218 |
| cg21786114 | EHMT2 | 0.344 | 0.374 | 0.953 | 0.367 | 0.023 | 0.073 | 0.060 | 0.841 | 0.363 | 0.484 | 0.004 | 0.029* | 0.600 | 0.654 | 0.001 | 0.018* | 0.809 | 0.483 |
| cg22185268 | COMMD4 | 0.943 | 0.744 | 0.548 | 0.618 | 0.007 | 0.180 | 0.058 | 0.108 | 0.299 | 0.842 | 0.001 | 0.019* | 0.329 | 0.431 | 0.001 | 0.008* | 0.467 | 0.844 |
| cg22746421 | 0.798 | 0.779 | 0.030 | 0.105 | 0.130 | 0.237 | 0.205 | 0.633 | 0.011 | 0.241 | 0.021 | 0.054 | 0.302 | 0.289 | 0.002 | 0.038* | 0.163 | 0.527 | |
| cg23224405 | DUSP16 | 0.873 | 0.594 | 0.099 | 0.062 | 0.024 | 0.256 | 0.053 | 0.351 | 0.099 | 0.194 | 0.002 | 0.054 | 0.147 | 0.199 | 0.000 | 0.001* | 0.197 | 0.435 |
| cg24199050 | GTSE1 | 0.064 | 0.175 | 0.949 | 0.862 | 0.012 | 0.006* | 0.001 | 0.129 | 0.719 | 0.984 | 0.000 | 0.001* | 0.754 | 0.988 | 0.004 | 0.013* | 0.717 | 0.190 |
| cg24689754 | C19orf62 | 0.357 | 0.466 | 0.273 | 0.341 | 0.147 | 0.607 | 0.366 | 0.576 | 0.102 | 0.183 | 0.007 | 0.045* | 0.731 | 0.593 | 0.001 | 0.005* | 0.368 | 0.104 |
| cg25295726 | FUT11 | 0.685 | 0.906 | 0.175 | 0.314 | 0.056 | 0.231 | 0.094 | 0.748 | 0.030 | 0.184 | 0.007 | 0.050 | 0.112 | 0.130 | 0.003 | 0.040* | 0.699 | 0.746 |
| cg25456633 | 0.613 | 0.938 | 0.003 | 0.003* | 0.080 | 0.581 | 0.235 | 0.649 | 0.296 | 0.688 | 0.085 | 0.377 | 0.626 | 0.742 | 0.001 | 0.012* | 0.893 | 0.758 | |
| cg25818813 | MERTK | 0.131 | 0.253 | 0.968 | 0.926 | 0.087 | 0.037* | 0.015 | 0.264 | 0.848 | 0.545 | 0.000 | 0.002* | 0.478 | 0.550 | 0.000 | 0.001* | 0.179 | 0.513 |
| cg26105057 | PAN2 | 0.843 | 0.972 | 0.305 | 0.831 | 0.029 | 0.122 | 0.161 | 0.401 | 0.107 | 0.656 | 0.058 | 0.082 | 0.155 | 0.137 | 0.000 | 0.002* | 0.522 | 0.167 |
| cg26309655 | TARS2 | 0.103 | 0.299 | 0.820 | 0.667 | 0.065 | 0.172 | 0.032 | 0.301 | 0.671 | 0.670 | 0.006 | 0.077 | 0.391 | 0.514 | 0.004 | 0.008* | 0.757 | 0.542 |
| g22721468 | SH3BP5L | 0.097 | 0.225 | 0.616 | 0.649 | 0.232 | 0.106 | 0.028 | 0.816 | 0.618 | 0.787 | 0.000 | 0.009* | 0.465 | 0.569 | 0.001 | 0.008* | 0.643 | 0.882 |
1# – Uni-variate p; 2# – Multi-variate; p* p<0.05, adjusted by sex, age and BMI.
Footnotes
Source of support: This study was supported by the State Key Development Program for Basic Research of China (Ref: 2012CB517501), the National Natural Science Foundation of China (Refs: 81070322, 81270491, 81470840), and the 100 Talents Program of the Shanghai Board of Health (Ref: XBR2011007)
References
- 1.Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67(1):328–57. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 2.Younossi ZM, Koenig AB, Abdelatif D, et al. Global epidemiology of nonalcoholic fatty liver disease. Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 3.Fan JG, Kim SU, Wong VW. New trends on obesity and NAFLD in Asia. J Hepatol. 2017;67(4):862–73. doi: 10.1016/j.jhep.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 4.McPherson S, Hardy T, Henderson E, et al. Evidence of NAFLD progression from steatosis to fibrosing-steatohepatitis using paired biopsies: Implications for prognosis and clinical management. J Hepatol. 2015;62(5):1148–55. doi: 10.1016/j.jhep.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 5.Tian JB, Zhong R, Liu C, et al. Association between bilirubin and risk of non-alcoholic fatty liver disease based on a prospective cohort study. Sci Rep. 2016;6:310006. doi: 10.1038/srep31006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmer V, Lammert F. Genetics and epigenetics in the fibrogenic evolution of chronic liver diseases. Best Pract Res Clin Gastroenterol. 2011;25(2):269–80. doi: 10.1016/j.bpg.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 7.Podrini C, Borghesan M, Greco A, et al. Redox homeostasis and epigenetics in non-alcoholic fatty liver disease (NAFLD) Curr Pharm Design. 2013;19(15):2737–46. doi: 10.2174/1381612811319150009. [DOI] [PubMed] [Google Scholar]
- 8.Illingworth RS, Bird AP. CpG islands – ‘A rough guide’. Febs Lett. 2009;583(11):1713–20. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Neuschwander-Tetri BA, Loomba R, Sanyal AJ, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): A multicentre, randomised, placebo-controlled trial. Lancet. 2015;385(9972):956–65. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordero P, Campion J, Milagro FI, Martinez JA. Transcriptomic and epigenetic changes in early liver steatosis associated to obesity: Effect of dietary methyl donor supplementation. Mol Genet Metab. 2013;110(3):388–95. doi: 10.1016/j.ymgme.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 11.Zeybel M, Hardy T, Robinson SM, et al. Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin Epigenetics. 2015;7:25. doi: 10.1186/s13148-015-0056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahrens M, Ammerpohl O, von Schonfels W, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18(2):296–302. doi: 10.1016/j.cmet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Murphy SK, Yang HN, Moylan CA, et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145(5):1076–87. doi: 10.1053/j.gastro.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallego-Duran R, Romero-Gomez M. Epigenetic mechanisms in non-alcoholic fatty liver disease: An emerging field. World J Hepatol. 2015;7(24):2497–502. doi: 10.4254/wjh.v7.i24.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardy T, Zeybel M, Day CP, et al. Plasma DNA methylation: A potential biomarker for stratification of liver fibrosis in non-alcoholic fatty liver disease. Gut. 2017;66(7):1321–28. doi: 10.1136/gutjnl-2016-311526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nano J, Ghanbari M, Wang W, et al. Epigenome-wide association study identifies methylation sites associated with liver enzymes and hepatic steatosis. Gastroenterology. 2017;153(4):1096–106e2. doi: 10.1053/j.gastro.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Bedossa P, Poitou C, Veyrie N, et al. Histopathological algorithm and scoring system for evaluation of liver lesions in morbidly obese patients. Hepatology. 2012;56(5):1751–59. doi: 10.1002/hep.25889. [DOI] [PubMed] [Google Scholar]
- 18.Du P, Kibbe WA, Lin SM. Lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–48. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 19.Dupuy D, Bertin N, Hidalgo CA, et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol. 2007;25(6):663–68. doi: 10.1038/nbt1305. [DOI] [PubMed] [Google Scholar]
- 20.Draghici S, Khatri P, Tarca AL, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17(10):1537–45. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilhelm-Benartzi CS, Koestler DC, Karagas MR, et al. Review of processing and analysis methods for DNA methylation array data. Br J Cancer. 2013;109(6):1394–402. doi: 10.1038/bjc.2013.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusminski CM, Holland WL, Sun K, et al. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat Med. 2012;18(10):1539–49. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikeska T, Craig JM. DNA methylation biomarkers: Cancer and beyond. Genes. 2014;5(3):821–64. doi: 10.3390/genes5030821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pratt DS, Kaplan MM. Primary care: Evaluation of abnormal liver-enzyme results in asymptomatic patients. N Engl J Med. 2000;342(17):1266–71. doi: 10.1056/NEJM200004273421707. [DOI] [PubMed] [Google Scholar]
- 26.van Beek JHDA, Lubke GH, de Moor MHM, et al. Heritability of liver enzyme levels estimated from genome-wide SNP data. Eur J Hum Genet. 2015;23(9):1223–28. doi: 10.1038/ejhg.2014.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu WS, Baker RD, Bhatia T, et al. Pathogenesis of nonalcoholic steatohepatitis. Cell Mol Life Sci. 2016;73(10):1969–87. doi: 10.1007/s00018-016-2161-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alkhouri N, Eng K, Lopez R, Nobili V. Non-high-density lipoprotein cholesterol (non-HDL-C) levels in children with nonalcoholic fatty liver disease (NAFLD) Springerplus. 2014;3:407. doi: 10.1186/2193-1801-3-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koppe SWP. Obesity and the liver: Nonalcoholic fatty liver disease. Transl Res. 2014;164(4):312–22. doi: 10.1016/j.trsl.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Stroka KM, Levitan I, Aranda-Espinoza H. OxLDL and substrate stiffness promote neutrophil transmigration by enhanced endothelial cell contractility and ICAM-1. J Biomech. 2012;45(10):1828–34. doi: 10.1016/j.jbiomech.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwok R, Tse YK, Wong GL, et al. Systematic review with meta-analysis: Non-invasive assessment of non-alcoholic fatty liver disease – the role of transient elastography and plasma cytokeratin-18 fragments. Aliment Pharmacol Ther. 2014;39(3):254–69. doi: 10.1111/apt.12569. [DOI] [PubMed] [Google Scholar]
- 32.Krishnan J, Danzer C, Simka T, et al. Dietary obesity-associated Hif1a activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the Sirt2-NAD+ system. Genes Dev. 2012;26(3):259–70. doi: 10.1101/gad.180406.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song CY, Zeng X, Wang Y, et al. Sophocarpine attenuates toll-like receptor 4 in steatotic hepatocytes to suppress pro-inflammatory cytokines synthesis. J Gastroen Hepatol. 2015;30(2):405–12. doi: 10.1111/jgh.12691. [DOI] [PubMed] [Google Scholar]
- 34.Cui M, Xiao ZL, Sun BD, et al. Involvement of cholesterol in hepatitis B virus X protein-induced abnormal lipid metabolism of hepatoma cells via up-regulating miR-205-targeted ACSL4. Biochem Bioph Res Commun. 2014;445(3):651–55. doi: 10.1016/j.bbrc.2014.02.068. [DOI] [PubMed] [Google Scholar]
- 35.Zhang RN, Pan Q, Zheng RD, et al. Genome-wide analysis of DNA methylation in human peripheral leukocytes identifies potential biomarkers of nonalcoholic fatty liver disease. Int J Mol Med. 2018;42(1):443–52. doi: 10.3892/ijmm.2018.3583. [DOI] [PubMed] [Google Scholar]
- 36.European Association for the Study of the Liver (EASL); European Association for the Study of Diabetes (EASD); European Association for the Study of Obesity (EASO) EASL-EASD-EASO Clinical Practice Guidelines for the management of oholic fatty liver disease. J Hepatol. 2016;64(6):1388–402. doi: 10.1016/j.jhep.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 37.Bedossa P. Utility and appropriateness of the fatty liver inhibition of progression (FLIP) algorithm and steatosis, activity, and fibrosis (SAF) score in the evaluation of biopsies of nonalcoholic atty liver disease. Hepatology. 2014;60:565–75. doi: 10.1002/hep.27173. [DOI] [PubMed] [Google Scholar]
- 38.Das M, Sha J, Hidalgo B, et al. Association of DNA methylation at CPT1A locus with metabolic syndrome in the genetics of lipid lowering drugs and diet network (GOLDN) study. PLoS One. 2016;11(1):e0145789. doi: 10.1371/journal.pone.0145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paradies G, Paradies V, Ruggiero FM, Petrosillo G. Oxidative stress, cardiolipin and mitochondrial dysfunction in nonalcoholic fatty liver disease. World J Gastroenterol. 2014;20(39):14205–18. doi: 10.3748/wjg.v20.i39.14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dolinsky VW, Cole LK, Sparagna GC, Hatch GM. Cardiac mitochondrial energy metabolism in heart failure: Role of cardiolipin and sirtuins. Biochim Biophys Acta. 2016;1861(10):1544–54. doi: 10.1016/j.bbalip.2016.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Individual CpG sites associated with NAFLD (q<0.05) in circulating leukocytes. (N=64).
| Probe ID | p NAFLD | q NAFLD | NAFLD mean β | CL mean β | Difference (% points) | Gene symbol | UCSC RefGene Acession | Relation to gene region | Relation to CpG island |
|---|---|---|---|---|---|---|---|---|---|
| cg00150500 | 5.05E-08 | 0.003526 | 0.040611 | 0.026409 | 0.014201229 | HCFC1R1; THOC6 | NM_017885; NM_001002018; NM_001002017; NM_001142350; NM_024339 | Body; TSS1500 | Island |
| cg01693328 | 3.78E-06 | 0.037736 | 0.734933 | 0.689903 | 0.045030407 | S_Shelf | |||
| cg02013957 | 2.80E-07 | 0.008217 | 0.069828 | 0.108342 | −0.03851329 | CAGE1; RIOK1 | NM_205864; NM_031480; NM_001170693; NM_001170692 | TSS1500; Body | S_Shore |
| cg03511638 | 7.43E-06 | 0.04944 | 0.207524 | 0.192478 | 0.015045277 | FBXL15 | NM_024326; NM_024326 | 1st Exon; 5′UTR | Island |
| cg03615240 | 2.36E-06 | 0.029878 | 0.234903 | 0.217214 | 0.01768861 | Island | |||
| cg03753066 | 2.57E-06 | 0.029878 | 0.042778 | 0.033719 | 0.009058818 | PGPEP1 | NM_017712 | TSS200 | N_Shore |
| cg03956053 | 4.68E-06 | 0.040684 | 0.094807 | 0.069568 | 0.025238064 | Island | |||
| cg03992938 | 7.18E-06 | 0.04944 | 0.201834 | 0.1878 | 0.014034138 | CCDC13 | NM_144719; NM_144719 | 1st Exon; 5′UTR | Island |
| cg04160753 | 5.99E-06 | 0.046488 | 0.275213 | 0.255442 | 0.019770564 | C10orf91 | NM_173541 | TSS1500 | |
| cg04396550 | 3.68E-12 | 1.54E-06 | 0.037964 | 0.025583 | 0.012380549 | KCNQ3 | NM_004519 | TSS1500 | Island |
| cg04787602 | 2.61E-07 | 0.008217 | 0.161306 | 0.144232 | 0.017073828 | C1orf183 | NM_198926; NM_019099 | Body; TSS1500 | Island |
| cg04981696 | 5.54E-07 | 0.012898 | 0.559923 | 0.491968 | 0.067954905 | ABCC1 | NM_019862; NM_019898; NM_019899; NM_004996; NM_019900 | Body | S_Shore |
| cg05102190 | 7.38E-06 | 0.04944 | 0.292254 | 0.261536 | 0.030718122 | ZYX | NM_003461; NM_001010972 | TSS200 | Island |
| cg06706068 | 6.62E-06 | 0.04866 | 0.450319 | 0.399254 | 0.051064413 | RPUSD1 | NM_058192 | 3′UTR | N_Shore |
| cg07067744 | 6.97E-06 | 0.04944 | 0.920563 | 0.90232 | 0.018243335 | BRSK2 | NM_003957 | Body | |
| cg07112456 | 6.04E-07 | 0.013311 | 0.931972 | 0.912429 | 0.019543021 | LRWD1 | NM_152892 | Body | S_Shelf |
| cg07953400 | 1.74E-06 | 0.02696 | 0.054226 | 0.045265 | 0.008961501 | PIGQ | NM_004204; NM_148920 | TSS1500; TSS1500 | N_Shore |
| cg07956264 | 1.06E-07 | 0.006374 | 0.060192 | 0.042644 | 0.017547367 | GPATCH3 | NM_022078 | 1st Exon | Island |
| cg08013262 | 2.14E-06 | 0.028915 | 0.051125 | 0.040134 | 0.010991397 | INSR | NM_000208; NM_001079817 | Body | Island |
| cg09367046 | 6.48E-06 | 0.048457 | 0.078046 | 0.057973 | 0.020073316 | ANGEL1 | NM_015305 | TSS200 | Island |
| cg09826692 | 3.66E-07 | 0.009581 | 0.695734 | 0.717446 | −0.0217123 | Island | |||
| cg10077144 | 6.43E-06 | 0.048457 | 0.151084 | 0.136608 | 0.014476221 | Island | |||
| cg10178228 | 2.57E-06 | 0.029878 | 0.16383 | 0.11116 | 0.052670088 | FRMD4B | NM_015123 | TSS200 | S_Shore |
| cg10576516 | 2.94E-07 | 0.008217 | 0.046487 | 0.034138 | 0.01234886 | RECQL5; SAP30BP | NM_001003716; NM_013260; NM_001003715; NM_013260; NM_004259 | TSS200; 1st Exon; 5′UT | Island |
| cg12289509 | 9.38E-07 | 0.019426 | 0.12632 | 0.116298 | 0.010022888 | TUBE1 | NM_016262; NM_001033564 | Body; TSS200 | Island |
| cg12362980 | 3.73E-06 | 0.037736 | 0.890648 | 0.915371 | −0.02472296 | Island | |||
| cg13291296 | 1.94E-06 | 0.028151 | 0.928293 | 0.567811 | 0.360481701 | GPR125 | NM_145290 | Body | |
| cg13373361 | 9.74E-07 | 0.019426 | 0.972063 | 0.958036 | 0.014027139 | MYH16 | NR_002147 | Body | |
| cg13397649 | 2.39E-06 | 0.029878 | 0.184331 | 0.171045 | 0.013286234 | AFG3L2 | NM_006796 | 1st Exon | Island |
| cg13463639 | 1.14E-07 | 0.047685 | 0.795196 | 0.744952 | 0.050244062 | SIGIRR | NM_001135053; NM_021805 | TSS1500 | S_Shore |
| cg13920768 | 4.76E-06 | 0.040684 | 0.180692 | 0.16038 | 0.02031251 | PRDM12 | NM_021619 | 3′UTR | Island |
| cg14214745 | 1.61E-06 | 0.025892 | 0.030079 | 0.02396 | 0.006119444 | CRELD1 | NM_001077415; NM_001031717; NM_015513; NM_015513; NM_001077415; NM_001031717 | 1st Exon; 5′UTR | Island |
| cg14247318 | 2.91E-06 | 0.032137 | 0.06859 | 0.051255 | 0.017334521 | S_Shore | |||
| cg14451560 | 4.08E-06 | 0.038185 | 0.163369 | 0.130014 | 0.033355016 | ARRDC1 | NM_152285 | TSS1500 | N_Shore |
| cg15317837 | 4.86E-06 | 0.0407 | 0.082077 | 0.065888 | 0.016188353 | GTPBP4 | NM_012341 | TSS1500 | Island |
| cg15653194 | 1.99E-09 | 0.000271 | 0.077077 | 0.051633 | 0.025443482 | LFNG | NM_001166355; NM_002304; NM_001040168; NM_001040167 | Body; TSS200 | Island |
| cg16105594 | 1.46E-06 | 0.025002 | 0.222788 | 0.206881 | 0.015907317 | MEF2C | NM_001131005; NM_002397 | 5′UTR; TSS1500 | Island |
| cg16261091 | 5.26E-06 | 0.042403 | 0.871221 | 0.768176 | 0.103045403 | C17orf101; HEXDC | NM_175902; NM_173620; NM_024648 | Body; TSS1500 | N_Shore |
| cg16357287 | 1.96E-07 | 0.007527 | 0.033464 | 0.022286 | 0.011177644 | SEH1L | NM_001013437; NM_031216 | TSS1500 | N_Shore |
| cg16398128 | 1.66E-07 | 0.007527 | 0.038963 | 0.025503 | 0.01345955 | ZNF622 | NM_033414; NM_033414 | 5′UTR; 1st Exon | Island |
| cg16416718 | 4.04E-06 | 0.038185 | 0.168379 | 0.155538 | 0.012840944 | CAMTA1 | NM_015215 | TSS1500 | Island |
| cg16487280 | 1.11E-06 | 0.020289 | 0.163618 | 0.146336 | 0.017282797 | MYBL2 | NM_002466 | TSS1500 | Island |
| cg17162475 | 1.08E-06 | 0.020289 | 0.136101 | 0.120308 | 0.015793168 | C6orf150 | NM_138441 | TSS200 | Island |
| cg17176517 | 5.44E-06 | 0.042966 | 0.055192 | 0.043187 | 0.012004537 | SNX4 | NM_003794; NM_003794 | 5′UTR; 1st Exon | Island |
| cg18570553 | 3.67E-06 | 0.037736 | 0.144011 | 0.128948 | 0.015063351 | PRR15 | NM_175887 | TSS200 | N_Shore |
| cg18848688 | 1.42E-08 | 0.001192 | 0.143981 | 0.325489 | −0.18150742 | LIFR | NM_001127671; NM_002310 | TSS1500; 5′UTR | Island |
| cg19137662 | 1.98E-07 | 0.007527 | 0.045624 | 0.032493 | 0.013131109 | NOP56 | NM_006392; NR_031699; NR_027700; NR_003078 | Body; TSS200 | Island |
| cg19189310 | 1.49E-06 | 0.025002 | 0.201487 | 0.18607 | 0.015416665 | RPS10 | NM_001014 | 5′UTR | Island |
| cg21786114 | 2.28E-07 | 0.007946 | 0.516219 | 0.460762 | 0.055456681 | EHMT2 | NM_006709; NM_025256 | TSS1500 | N_Shore |
| cg22185268 | 3.99E-07 | 0.009831 | 0.026994 | 0.020076 | 0.006918145 | COMMD4 | NM_017828 | TSS200 | Island |
| cg22402398 | 2.50E-06 | 0.029878 | 0.939115 | 0.923689 | 0.015425828 | FGR | NM_001042747; NM_005248; NM_001042729 | TSS1500; 5′UTR | |
| cg22598233 | 3.16E-06 | 0.033997 | 0.069128 | 0.056446 | 0.012681401 | POLRMT | NM_005035 | TSS200 | Island |
| cg22721468 | 1.96E-06 | 0.028151 | 0.060214 | 0.04424 | 0.01597372 | SH3BP5L | NM_030645 | TSS1500 | Island |
| cg22746421 | 1.52E-07 | 0.007527 | 0.87434 | 0.852661 | 0.021678989 | ||||
| cg23224405 | 2.59E-09 | 0.000271 | 0.064945 | 0.045018 | 0.019927219 | DUSP16 | NM_030640; NM_030640 | 1st Exon; 5′UTR | Island |
| cg24199050 | 2.65E-06 | 0.029971 | 0.010376 | 0.006359 | 0.004017477 | GTSE1 | NM_016426; NR_024009 | TSS200 | Island |
| cg24239690 | 7.36E-06 | 0.04944 | 0.196241 | 0.181611 | 0.014630372 | FSCN1 | NM_003088 | Body | S_Shore |
| cg24689754 | 4.95E-06 | 0.0407 | 0.029538 | 0.020196 | 0.009341864 | C19orf62 | NM_001033549; NM_001033549; NM_014173; NM_014173 | 5′UTR; 1st Exon | |
| cg25295726 | 1.84E-09 | 0.000271 | 0.181557 | 0.162827 | 0.018730389 | FUT11 | NM_173540 | TSS200 | Island |
| cg25456633 | 4.20E-06 | 0.038224 | 0.897003 | 0.878185 | 0.018817636 | ||||
| cg25818813 | 4.10E-06 | 0.038185 | 0.064429 | 0.050092 | 0.014336947 | MERTK | NM_006343 | Body | Island |
| cg26105057 | 2.02E-06 | 0.028151 | 0.160883 | 0.126787 | 0.034095982 | PAN2 | NM_014871; NM_001166279; NM_001127460; NM_014871; NM_001166279 | 1st Exon; 5′UTR; TSS200 | Island |
| cg26309655 | 4.66E-06 | 0.040684 | 0.16828 | 0.148223 | 0.02005632 | TARS2 | NM_025150 | TSS200 | |
| cg12473838 | 4.88E-06 | 0.028795 | 0.040501 | 0.031202 | 0.009299747 | SSBP1 | NM_003143; NR_015392 | TSS200; Body | Island |
| cg26786980 | 6.79E-06 | 0.049042 | 0.220594 | 0.183155 | 0.037438788 | RARB | NM_016152; NM_016152; NM_000965; NM_000965 | 5′UTR; 1st Exon |
Supplementary Table 2.
Individual CpG sites associated with simple hepatic steatosis (q<0.05) in circulating leukocytes. (N=119).
| Probe ID | p Simple hepatic steatosis | q Simple hepatic steatosis | Simple hepatic steatosis mean β | CL mean β | Difference (% points) | Gene symbol | UCSC RefGene acession | Relation to gene region | Relation to CpG island |
|---|---|---|---|---|---|---|---|---|---|
| cg00150500 | 0.00000 | 0.00005 | 0.04354 | 0.02641 | 0.01713 | HCFC1R1; THOC6 | NM_017885; NM_001002018; NM_001002017; NM_001142350; NM_024339 | Body; TSS1500 | Island |
| cg00574958 | 0.00000 | 0.02625 | 0.13777 | 0.19441 | −0.05664 | CPT1A; CPT1A | NM_001876; NM_001031847 | 5′UTR | N_Shore |
| cg00639286 | 0.00001 | 0.03794 | 0.11783 | 0.09329 | 0.02454 | ATOX1 | NM_004045 | Body | Island |
| cg00932007 | 0.00000 | 0.02625 | 0.07328 | 0.04803 | 0.02525 | SERINC1; PKIB | NM_020755; NM_181794; NM_020755 | 1st Exon; TSS200; 5′UTR | Island |
| cg01067963 | 0.00001 | 0.02968 | 0.06460 | 0.04919 | 0.01541 | MAPK1 | NM_138957; NM_002745 | TSS1500 | Island |
| cg01216607 | 0.00001 | 0.04888 | 0.13731 | 0.11167 | 0.02564 | ZNHIT1; PLOD3 | NM_006349; NM_001084; NM_001084 | TSS1500; 1st Exon; 5′UTR | Island |
| cg01525244 | 0.00000 | 0.00830 | 0.22203 | 0.17144 | 0.05059 | CBX7 | NM_175709 | TSS200 | N_Shore |
| cg01801101 | 0.00000 | 0.02176 | 0.31191 | 0.27460 | 0.03730 | AMZ1 | NM_133463 | 5′UTR | N_Shore |
| cg02432888 | 0.00001 | 0.03410 | 0.92622 | 0.93604 | −0.00982 | SMARCA4 | NM_001128845; NM_001128844; NM_001128848; NM_001128846; NM_003072; NM_001128849; NM_001128847 | 1st Exon; Body | N_Shelf |
| cg02581963 | 0.00000 | 0.00977 | 0.09723 | 0.06908 | 0.02816 | C10orf75 | NR_026762 | TSS200 | Island |
| cg03027241 | 0.00000 | 0.01712 | 0.35425 | 0.42982 | −0.07557 | KCNG1 | NM_002237 | 3′UTR | Island |
| cg03069383 | 0.00001 | 0.04806 | 0.22846 | 0.20088 | 0.02758 | RDH14 | NM_020905 | 1st Exon | Island |
| cg03074984 | 0.00001 | 0.04262 | 0.22030 | 0.19083 | 0.02947 | RPS6KA4 | NM_003942; NM_001006944 | TSS200 | Island |
| cg03511638 | 0.00000 | 0.02698 | 0.21013 | 0.19248 | 0.01765 | FBXL15 | NM_024326; NM_024326 | 1st Exon; 5′UTR | Island |
| cg03956053 | 0.00000 | 0.00468 | 0.10087 | 0.06957 | 0.03130 | Island | |||
| cg03992938 | 0.00000 | 0.02085 | 0.20399 | 0.18780 | 0.01619 | CCDC13 | NM_144719; NM_144719 | 1st Exon; 5′UTR | Island |
| cg04396550 | 0.00000 | 0.00000 | 0.03999 | 0.02558 | 0.01440 | KCNQ3 | NM_004519 | TSS1500 | Island |
| cg04551440 | 0.00000 | 0.02375 | 0.06975 | 0.05204 | 0.01771 | KATNAL1 | NM_001014380; NM_032116 | TSS200; 5′UTR | Island |
| cg04672769 | 0.00000 | 0.01712 | 0.10358 | 0.08340 | 0.02017 | HARS | NM_002109; NM_012208; NM_002109 | 5′UTR; TSS200; 1stExon | Island |
| cg04699668 | 0.00000 | 0.02654 | 0.78765 | 0.83452 | −0.04687 | N_Shore | |||
| cg04781764 | 0.00000 | 0.01704 | 0.86144 | 0.88530 | −0.02387 | WDR51B | NM_172240 | Body | |
| cg04787602 | 0.00000 | 0.02085 | 0.16300 | 0.14423 | 0.01876 | C1orf183; C1orf183 | NM_198926; NM_019099 | Body; TSS1500 | Island |
| cg04851352 | 0.00001 | 0.04156 | 0.32402 | 0.30766 | 0.01637 | Island | |||
| cg04981696 | 0.00000 | 0.01712 | 0.55926 | 0.49197 | 0.06730 | ABCC1 | NM_019862; NM_019898; NM_019899; NM_004996; NM_019900 | Body | S_Shore |
| cg05020775 | 0.00001 | 0.03145 | 0.11804 | 0.09411 | 0.02393 | SNPH | NM_014723 | TSS200 | Island |
| cg05102190 | 0.00000 | 0.01712 | 0.30075 | 0.26154 | 0.03922 | ZYX | NM_003461; NM_001010972 | TSS200 | Island |
| cg05131957 | 0.00000 | 0.02625 | 0.08817 | 0.06755 | 0.02062 | CRLS1; CRLS1 | NM_019095; NM_019095; NM_001127458 | 1st Exon; 5′UTR; TSS1500 | Island |
| cg05209527 | 0.00001 | 0.03018 | 0.20213 | 0.15172 | 0.05042 | SLC5A6 | NR_028323; NM_080592; NM_021095; NM_016085; NM_001170795 | TSS1500; Body; 5′UTR | S_Shore |
| cg05377161 | 0.00001 | 0.04888 | 0.11639 | 0.09404 | 0.02235 | Island | |||
| cg05629721 | 0.00000 | 0.02654 | 0.29331 | 0.24457 | 0.04875 | PSMB8 | NM_004159; NM_148919; NM_004159 | 5′UTR; TSS1500; 1st Exon | S_Shore |
| cg05977109 | 0.00000 | 0.02085 | 0.09199 | 0.06838 | 0.02361 | SGMS1 | NM_147156 | 5′UTR | Island |
| cg06276429 | 0.00001 | 0.04156 | 0.21157 | 0.17132 | 0.04025 | SLC39A7; RXRB | NM_006979; NM_021976; NM_001077516 | TSS1500; Body | N_Shore |
| cg07067659 | 0.00001 | 0.03187 | 0.03402 | 0.02441 | 0.00961 | SLC7A5 | NM_003486 | 1st Exon | Island |
| cg07112456 | 0.00001 | 0.03536 | 0.93287 | 0.91243 | 0.02044 | LRWD1 | NM_152892 | Body | S_Shelf |
| cg07953400 | 0.00001 | 0.02968 | 0.05508 | 0.04526 | 0.00982 | PIGQ | NM_004204; NM_148920 | TSS1500 | N_Shore |
| cg07956264 | 0.00000 | 0.00228 | 0.06267 | 0.04264 | 0.02003 | GPATCH3 | NM_022078 | 1st Exon | Island |
| cg08180934 | 0.00000 | 0.02625 | 0.05768 | 0.04568 | 0.01200 | STRAP | NM_007178 | TSS200 | Island |
| cg08276889 | 0.00000 | 0.02375 | 0.03778 | 0.02814 | 0.00964 | LOC100133985 | NR_024444 | Body | Island |
| cg08730728 | 0.00001 | 0.04156 | 0.10524 | 0.07919 | 0.02606 | Island | |||
| cg09090048 | 0.00001 | 0.03018 | 0.21230 | 0.18139 | 0.03091 | VPS26B; NCAPD3 | NM_052875; NM_015261 | TSS1500; Body | Island |
| cg09267087 | 0.00001 | 0.04156 | 0.67624 | 0.64183 | 0.03441 | ||||
| cg09367046 | 0.00001 | 0.04262 | 0.07982 | 0.05797 | 0.02184 | ANGEL1 | NM_015305 | TSS200 | Island |
| cg09457469 | 0.00000 | 0.01698 | 0.06248 | 0.04671 | 0.01577 | YBX2 | NM_015982 | Body | Island |
| cg09826692 | 0.00000 | 0.02375 | 0.69472 | 0.71745 | −0.02273 | Island | |||
| cg10059171 | 0.00001 | 0.03187 | 0.07502 | 0.05829 | 0.01674 | GPR89B | NM_016334; NM_016334 | 5′UTR; 1st Exon | Island |
| cg10178228 | 0.00000 | 0.00820 | 0.17082 | 0.11116 | 0.05966 | FRMD4B | NM_015123 | TSS200 | S_Shore |
| cg10369688 | 0.00001 | 0.03018 | 0.15684 | 0.13906 | 0.01778 | ZFP36 | NM_003407 | Body | Island |
| cg10381071 | 0.00000 | 0.01712 | 0.15437 | 0.20650 | −0.05213 | TLE3 | NM_020908; NM_001105192; NM_005078 | TSS1500 | Island |
| cg10576516 | 0.00000 | 0.00019 | 0.05003 | 0.03414 | 0.01589 | RECQL5; SAP30BP | NM_001003716; NM_013260; NM_001003715; NM_013260; NM_004259 | TSS200; 1st Exon; 5′UTR | Island |
| cg10965669 | 0.00001 | 0.02931 | 0.80870 | 0.82408 | −0.01538 | AXIN2 | NM_004655 | Body | Island |
| cg11319362 | 0.00001 | 0.04251 | 0.08666 | 0.06828 | 0.01838 | ARL6IP5 | NM_006407; NM_006407 | 1st Exon; 5′UTR | Island |
| cg11542063 | 0.00000 | 0.02625 | 0.05513 | 0.03884 | 0.01629 | NFE2L2 | NM_006164; NM_006164; NM_001145413; NM_001145412 | 5′UTR; 1st Exon; TSS1500 | Island |
| cg11866422 | 0.00000 | 0.02654 | 0.06337 | 0.05160 | 0.01177 | Island | |||
| cg12289509 | 0.00001 | 0.04442 | 0.12657 | 0.11630 | 0.01027 | TUBE1; C6orf225 | NM_016262; NM_001033564 | Body; TSS200 | Island |
| cg12362980 | 0.00000 | 0.00307 | 0.88730 | 0.91537 | −0.02807 | Island | |||
| cg12407666 | 0.00000 | 0.02901 | 0.12378 | 0.10250 | 0.02128 | RIC8B | NM_018157 | TSS1500 | Island |
| cg12810189 | 0.00001 | 0.03920 | 0.04387 | 0.03696 | 0.00691 | BCAP29 | NR_027830; NM_018844; NM_018844; NM_001008405 | Body; 1st Exon; 5′UTR; TSS1500 | Island |
| cg12850793 | 0.00000 | 0.02321 | 0.15979 | 0.14310 | 0.01669 | DENND5A | NM_015213 | TSS200 | Island |
| cg13110034 | 0.00001 | 0.03187 | 0.07293 | 0.06148 | 0.01145 | ANKRD34A; POLR3GL | NM_001039888; NM_032305 | TSS1500; 5′UTR | Island |
| cg13199615 | 0.00001 | 0.03018 | 0.23755 | 0.21115 | 0.02640 | KRTCAP2; TRIM46 | NM_173852; NM_025058 | TSS1500; Body | N_Shore |
| cg13398864 | 0.00000 | 0.02433 | 0.11061 | 0.08598 | 0.02463 | SEC22A | NM_012430 | TSS200 | N_Shore |
| cg13485756 | 0.00000 | 0.02698 | 0.09370 | 0.08224 | 0.01146 | RB1 | NM_000321 | TSS200 | Island |
| cg14022530 | 0.00000 | 0.02698 | 0.07371 | 0.05385 | 0.01986 | SASS6; CCDC76 | NM_194292; NM_019083 | TSS200 | Island |
| cg14214745 | 0.00000 | 0.01698 | 0.03114 | 0.02396 | 0.00718 | CRELD1 | NM_001077415; NM_001031717; NM_015513; NM_015513; NM_001077415; NM_001031717 | 1st Exon; 5′UTR | Island |
| cg14247318 | 0.00001 | 0.03018 | 0.06991 | 0.05126 | 0.01865 | S_Shore | |||
| cg15043926 | 0.00000 | 0.02698 | 0.28089 | 0.26430 | 0.01659 | STIM2 | NM_001169117; NM_020860; NM_001169118 | Body | Island |
| cg15226170 | 0.00000 | 0.02879 | 0.37491 | 0.31659 | 0.05832 | PMM1 | NM_002676 | TSS1500 | S_Shore |
| cg15324256 | 0.00000 | 0.02625 | 0.05617 | 0.04062 | 0.01555 | QTRTD1; KIAA1407 | NM_024638; NM_020817 | TSS1500; 1st Exon | Island |
| cg15324448 | 0.00000 | 0.02625 | 0.10020 | 0.08001 | 0.02019 | ZNF643 | NM_023070 | TSS200 | Island |
| cg15536552 | 0.07584 | 0.04796 | 0.20583 | 0.34740 | −0.01416 | ACSL4 | NM_022977; NM_004458 | 5′UTR; 5′UTR | N_Shore |
| cg15572086 | 0.00000 | 0.01698 | 0.06120 | 0.04806 | 0.01314 | C1orf203 | NR_027645; NR_024126; NR_024124; NR_027646; NR_024125 | TSS1500 | Island |
| cg15653194 | 0.00000 | 0.00394 | 0.07660 | 0.05163 | 0.02496 | LFNG | NM_001166355; NM_002304; NM_001040168; NM_001040167 | Body; TSS200 | Island |
| cg16007711 | 0.00000 | 0.01712 | 0.11104 | 0.08414 | 0.02689 | RHOBTB2 | NM_001160036; NM_001160037; NM_015178 | Body; 5′UTR | S_Shore |
| cg16306148 | 0.00001 | 0.03359 | 0.01234 | 0.00608 | 0.00626 | YBX1 | NM_004559 | TSS1500 | Island |
| cg16357287 | 0.00000 | 0.00228 | 0.03588 | 0.02229 | 0.01360 | SEH1L; SEH1L | NM_001013437; NM_031216 | TSS1500 | N_Shore |
| cg16398128 | 0.00000 | 0.00000 | 0.04360 | 0.02550 | 0.01810 | ZNF622 | NM_033414; NM_033414 | 5′UTR; 1st Exon | Island |
| cg16416718 | 0.00000 | 0.02879 | 0.16998 | 0.15554 | 0.01445 | CAMTA1 | NM_015215 | TSS1500 | Island |
| cg16743070 | 0.00000 | 0.01721 | 0.04222 | 0.02640 | 0.01582 | MYST3 | NM_001099413; NM_001099412; NM_006766 | 5′UTR | Island |
| cg16803522 | 0.00000 | 0.01441 | 0.03705 | 0.02603 | 0.01102 | RASGRP1 | NM_001128602; NM_005739 | TSS1500 | Island |
| cg17074213 | 0.00001 | 0.02901 | 0.20986 | 0.15861 | 0.05125 | TGFBR3 | NM_003243; NM_003243 | 1st Exon; 5′UTR | Island |
| cg17162475 | 0.00000 | 0.02375 | 0.13817 | 0.12031 | 0.01786 | C6orf150 | NM_138441 | TSS200 | Island |
| cg17168242 | 0.00000 | 0.01599 | 0.09273 | 0.08238 | 0.01035 | OTUD7B | NM_020205 | TSS200 | Island |
| cg17176517 | 0.00001 | 0.04039 | 0.05653 | 0.04319 | 0.01334 | SNX4 | NM_003794; NM_003794 | 5′UTR; 1st Exon | Island |
| cg17427781 | 0.00001 | 0.04227 | 0.08943 | 0.07059 | 0.01884 | DDX20; C1orf183 | NM_007204; NM_198926 | TSS200; Body | Island |
| cg18676790 | 0.00001 | 0.03018 | 0.27930 | 0.34935 | −0.07005 | ||||
| cg18848688 | 0.00001 | 0.02968 | 0.14296 | 0.32549 | −0.18253 | LIFR | NM_001127671; NM_002310 | TSS1500; 5′UTR | Island |
| cg19050851 | 0.00000 | 0.02690 | 0.03973 | 0.02876 | 0.01097 | ARSB | NM_000046; NM_198709 | Body; Body | Island |
| cg19137662 | 0.00000 | 0.01441 | 0.04705 | 0.03249 | 0.01455 | NOP56 | NM_006392; NR_031699; NR_027700; NR_003078 | Body; TSS200; TSS1500 | Island |
| cg19362196 | 0.00000 | 0.02625 | 0.70650 | 0.76251 | −0.05601 | STMN1 | NM_005563; NM_203399; NM_203401; NM_001145454 | TSS1500 | Island |
| cg19520115 | 0.00000 | 0.00160 | 0.12095 | 0.10488 | 0.01607 | CAPZB | NM_004930 | Body | Island |
| cg19634213 | 0.00000 | 0.01712 | 0.05716 | 0.04074 | 0.01643 | PTEN; KILLIN | NM_000314; NM_001126049 | TSS1500; 1st Exon | Island |
| cg19693031 | 0.00001 | 0.03187 | 0.80756 | 0.85146 | −0.04390 | TXNIP | NM_006472 | 3′UTR | |
| cg19878987 | 0.00000 | 0.02280 | 0.10362 | 0.08020 | 0.02342 | LDHB | NM_002300 | TSS1500 | S_Shore |
| cg20656751 | 0.00000 | 0.02625 | 0.51357 | 0.47261 | 0.04096 | NAT8L | NM_178557 | 1stExon | Island |
| cg21012729 | 0.00001 | 0.03763 | 0.08208 | 0.06527 | 0.01681 | NIN | NM_182946; NM_020921; NM_182944 | 5′UTR | Island |
| cg21373806 | 0.00001 | 0.04156 | 0.83546 | 0.86386 | −0.02840 | BAHCC1 | NM_001080519 | Body | N_Shore |
| cg21786114 | 0.00001 | 0.03187 | 0.51357 | 0.46076 | 0.05281 | EHMT2 | NM_006709; NM_025256 | TSS1500 | N_Shore |
| cg22185268 | 0.00000 | 0.00217 | 0.02871 | 0.02008 | 0.00864 | COMMD4 | NM_017828 | TSS200 | Island |
| cg22721468 | 0.00000 | 0.00228 | 0.06494 | 0.04424 | 0.02070 | SH3BP5L | NM_030645 | TSS1500 | Island |
| cg22746421 | 0.00000 | 0.01712 | 0.87645 | 0.85266 | 0.02379 | ||||
| cg22902478 | 0.00001 | 0.04156 | 0.10027 | 0.08525 | 0.01502 | LOC100130987 | NR_024469 | TSS200 | Island |
| cg23224405 | 0.00000 | 0.00011 | 0.06783 | 0.04502 | 0.02281 | DUSP16 | NM_030640; NM_030640 | 1st Exon; 5′UTR | Island |
| cg23348158 | 0.00000 | 0.01712 | 0.09342 | 0.06174 | 0.03168 | FUZ | NM_025129 | TSS200 | S_Shore |
| cg23512165 | 0.00000 | 0.02085 | 0.02886 | 0.02041 | 0.00845 | MTMR4 | NM_004687; NM_004687 | 1st Exon; 5′UTR | Island |
| cg24108286 | 0.00001 | 0.03145 | 0.25285 | 0.21178 | 0.04107 | TFDP1 | NR_026580; NM_007111 | TSS1500 | |
| cg24689754 | 0.00000 | 0.02433 | 0.03045 | 0.02020 | 0.01026 | C19orf62 | NM_001033549; NM_001033549; NM_014173; NM_014173 | 5′UTR; 1st Exon | |
| cg24728479 | 0.00001 | 0.03187 | 0.19258 | 0.17546 | 0.01712 | ZNF252; C8orf77 | NR_023392; NR_026974 | TSS1500; Body | Island |
| cg25053907 | 0.00001 | 0.02968 | 0.06377 | 0.05198 | 0.01179 | ZNF721; PIGG | NM_133474; NM_017733; NM_001127178; NM_133474 | 5′UTR; TSS200; 1st Exon | Island |
| cg25295726 | 0.00000 | 0.01101 | 0.18162 | 0.16283 | 0.01879 | FUT11 | NM_173540 | TSS200 | Island |
| cg25678532 | 0.00000 | 0.02625 | 0.05138 | 0.03889 | 0.01249 | RBBP8 | NM_203291; NM_203292; NM_002894 | TSS1500; TSS200 | Island |
| cg25818813 | 0.00000 | 0.02014 | 0.06671 | 0.05009 | 0.01661 | MERTK | NM_006343 | Body | Island |
| cg25856120 | 0.00001 | 0.04680 | 0.11157 | 0.08244 | 0.02914 | ATP5G1 | NM_005175; NM_005175; NM_001002027 | 1st Exon; 5′UTR | S_Shore |
| cg25989057 | 0.00001 | 0.02942 | 0.03813 | 0.02660 | 0.01154 | AIP | NM_003977 | TSS200 | Island |
| cg26309655 | 0.00001 | 0.04156 | 0.17136 | 0.14822 | 0.02314 | TARS2 | NM_025150 | TSS200 | |
| cg26462136 | 0.00001 | 0.03647 | 0.08411 | 0.06628 | 0.01783 | ATXN7L3 | NM_001098833; NM_020218 | TSS1500 | Island |
| cg26791384 | 0.00001 | 0.04156 | 0.25191 | 0.21622 | 0.03569 | SYCE1L | NM_001129979 | Body | Island |
| cg26824705 | 0.00001 | 0.04888 | 0.08142 | 0.06402 | 0.01740 | MUL1 | NM_024544 | TSS200 | Island |
| cg27164612 | 0.00000 | 0.02654 | 0.01342 | 0.00813 | 0.00529 | ||||
| cg27612695 | 0.00000 | 0.02625 | 0.18852 | 0.15615 | 0.03238 | SUZ12P | NR_024187 | Body | S_Shore |
Supplementary Table 3.
Significantly enriched KEGG pathways of genes with altered DNA methylation level in NAFLD.
| Pathway ID | Description | p Value | q Value | Genes | Enrich factor | |
|---|---|---|---|---|---|---|
| hsa03008 | Ribosome biogenesis in eukaryotes | 0.004 | 0.010 | GTPBP4 | GTP binding protein4 | 8.56 |
| NOP56 | Ribonucleo protein | |||||
| hsa04010 | MAPK signaling pathway | 0.001 | 0.011 | DUSP16 | Dual specificity phosphatase 16 | 2.59 |
| MEF2C | Myocyte enhancer factor 2C |
Supplementary Table 4.
Significantly enriched KEGG pathways of genes with altered DNA methylation level in simple hepatic steatosis.
| Pathway ID | Description | p Value | q Value | Genes | Enrich factor | |
|---|---|---|---|---|---|---|
| hsa04977 | Vitamin digestion and absorption | 0.0007 | 0.033 | SLC5A6 | Solute carrier family 5 member 6 | 15.84 |
| ABCC1 | ATP binding cassette sub familyC member1 | |||||
| hsa03013 | RNAtransport | 0.0100 | 0.040 | DDX20 | DEAD-box helicase 20 | 2.50 |
| SEH1L | SEH1 like nucleoporin | |||||
| hsa05200 | Pathways in cancer | 0.0070 | 0.034 | PTEN | Phosphatase and tensin homolog | 2.33 |
| AXIN2 | Axin 2 | |||||
| MAPK1 | Mitogen-activated protein kinase 1 | |||||
| RB1 | RB transcriptional corepressor 1 |
Supplementary Table 5.
Significance of associations between biological parameters and selected methylated CpG sites.
| Illumina ID | Gene symbol | GLU | HOMAIN | TG | TC | HDL | LDL | GGT | ALT | AST | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | 1# | 2# | ||
| cg00150500 | HCFC1R1 | 0.492 | 0.736 | 0.290 | 0.433 | 0.020 | 0.545 | 0.008 | 0.027* | 0.505 | 0.516 | 0.000 | 0.005* | 0.107 | 0.132 | 0.006 | 0.057 | 0.792 | 0.668 |
| cg00574958 | CTP1A | 0.333 | 0.435 | 0.433 | 0.768 | 0.001 | 0.001* | 0.017 | 0.308 | 0.067 | 0.575 | 0.000 | 0.001* | 0.125 | 0.417 | 0.034 | 0.478 | 0.340 | 0.657 |
| cg01067963 | MAPK1 | 0.321 | 0.685 | 0.520 | 0.963 | 0.039 | 0.795 | 0.216 | 0.632 | 0.070 | 0.316 | 0.018 | 0.259 | 0.365 | 0.201 | 0.006 | 0.035* | 0.603 | 0.454 |
| cg01693328 | 0.899 | 0.875 | 0.408 | 0.665 | 0.247 | 0.816 | 0.126 | 0.170 | 0.173 | 0.441 | 0.087 | 0.165 | 0.162 | 0.174 | 0.002 | 0.009* | 0.111 | 0.185 | |
| cg01801101 | AMZ1 | 0.971 | 0.949 | 0.392 | 0.838 | 0.006 | 0.211 | 0.240 | 0.280 | 0.287 | 0.844 | 0.016 | 0.078 | 0.983 | 0.932 | 0.000 | 0.002* | 0.818 | 0.488 |
| cg02013957 | CAGE1 | 0.860 | 0.788 | 0.010 | 0.010* | 0.049 | 0.371 | 0.213 | 0.951 | 0.051 | 0.656 | 0.150 | 0.413 | 0.133 | 0.122 | 0.001 | 0.024* | 0.498 | 0.082 |
| cg03511638 | FBXL15 | 0.158 | 0.282 | 0.136 | 0.258 | 0.129 | 0.930 | 0.585 | 0.985 | 0.373 | 0.908 | 0.073 | 0.446 | 0.113 | 0.141 | 0.010 | 0.070 | 0.588 | 0.818 |
| cg03615240 | 0.147 | 0.757 | 0.286 | 0.355 | 0.101 | 0.206 | 0.036 | 0.142 | 0.816 | 0.237 | 0.012 | 0.012* | 0.179 | 0.400 | 0.000 | 0.033* | 0.061 | 0.373 | |
| cg03992938 | CCDC13 | 0.107 | 0.296 | 0.067 | 0.032* | 0.844 | 0.139 | 0.035 | 0.449 | 0.404 | 0.240 | 0.001 | 0.006* | 0.016 | 0.021* | 0.012 | 0.011* | 0.187 | 0.882 |
| cg04160753 | C10orf91 | 0.162 | 0.316 | 0.936 | 0.880 | 0.014 | 0.105 | 0.030 | 0.161 | 0.466 | 0.798 | 0.014 | 0.108 | 0.295 | 0.361 | 0.104 | 0.345 | 0.904 | 0.986 |
| cg04787602 | C1orf183 | 0.153 | 0.304 | 0.328 | 0.396 | 0.044 | 0.271 | 0.058 | 0.914 | 0.046 | 0.259 | 0.001 | 0.039* | 0.033 | 0.035* | 0.001 | 0.010* | 0.181 | 0.207 |
| cg04981696 | ABCC1 | 0.260 | 0.318 | 0.420 | 0.784 | 0.046 | 0.090 | 0.055 | 0.778 | 0.470 | 0.645 | 0.002 | 0.033* | 0.657 | 0.736 | 0.000 | 0.004* | 0.777 | 0.547 |
| cg05102190 | ZYX | 0.116 | 0.122 | 0.972 | 0.880 | 0.039 | 0.084 | 0.056 | 0.360 | 0.301 | 0.926 | 0.025 | 0.077 | 0.338 | 0.365 | 0.005 | 0.030* | 0.812 | 0.717 |
| cg05131957 | CRLS1 | 0.091 | 0.199 | 0.489 | 0.876 | 0.032 | 0.274 | 0.068 | 0.312 | 0.585 | 0.892 | 0.006 | 0.047* | 0.995 | 0.780 | 0.018 | 0.069 | 0.483 | 0.196 |
| cg05209527 | SLC5A6 | 0.143 | 0.519 | 0.254 | 0.407 | 0.141 | 0.919 | 0.040 | 0.109 | 0.563 | 0.908 | 0.005 | 0.506 | 0.923 | 0.353 | 0.000 | 0.006* | 0.478 | 0.317 |
| cg06706068 | RPUSD1 | 0.034 | 0.020* | 0.477 | 0.970 | 0.533 | 0.052 | 0.032 | 0.268 | 0.090 | 0.748 | 0.023 | 0.049* | 0.233 | 0.243 | 0.003 | 0.043* | 0.851 | 0.586 |
| cg07067744 | BRSK2 | 0.782 | 0.974 | 0.454 | 0.760 | 0.093 | 0.104 | 0.038 | 0.963 | 0.389 | 0.782 | 0.003 | 0.030* | 0.360 | 0.435 | 0.012 | 0.108 | 0.694 | 0.757 |
| cg07112456 | LRWD1 | 0.167 | 0.261 | 0.370 | 0.531 | 0.230 | 0.199 | 0.071 | 0.660 | 0.470 | 0.664 | 0.033 | 0.178 | 0.082 | 0.097 | 0.031 | 0.208 | 0.475 | 0.480 |
| cg07953400 | PIGQ | 0.001 | 0.004* | 0.625 | 0.452 | 0.233 | 0.005* | 0.000 | 0.823 | 0.916 | 0.770 | 0.000 | 0.001* | 0.250 | 0.343 | 0.013 | 0.041* | 0.939 | 0.439 |
| cg07956264 | GPATCH3 | 0.702 | 0.693 | 0.026 | 0.068 | 0.000 | 0.028* | 0.029 | 0.002* | 0.112 | 0.493 | 0.009 | 0.023* | 0.158 | 0.164 | 0.000 | 0.002* | 0.282 | 0.232 |
| cg08013262 | INSR | 0.582 | 0.678 | 0.482 | 0.695 | 0.031 | 0.046* | 0.023 | 0.242 | 0.850 | 0.569 | 0.013 | 0.061 | 0.847 | 0.952 | 0.001 | 0.005* | 0.859 | 0.992 |
| cg09367046 | ANGEL1 | 0.275 | 0.356 | 0.557 | 0.793 | 0.109 | 0.351 | 0.190 | 0.811 | 0.203 | 0.624 | 0.022 | 0.118 | 0.175 | 0.205 | 0.000 | 0.002* | 0.198 | 0.188 |
| cg10077144 | 0.722 | 0.544 | 0.467 | 0.283 | 0.006 | 0.203 | 0.029 | 0.084 | 0.645 | 0.736 | 0.007 | 0.138 | 0.950 | 0.711 | 0.002 | 0.004* | 0.776 | 0.621 | |
| cg10178228 | FRMD4B | 0.566 | 0.705 | 0.482 | 0.603 | 0.447 | 0.937 | 0.668 | 0.196 | 0.005 | 0.055 | 0.078 | 0.357 | 0.240 | 0.270 | 0.000 | 0.005* | 0.838 | 0.776 |
| cg12289509 | TUBE1 | 0.402 | 0.503 | 0.479 | 0.670 | 0.236 | 0.029* | 0.019 | 0.921 | 0.676 | 0.683 | 0.001 | 0.002* | 0.443 | 0.489 | 0.000 | 0.001* | 0.624 | 0.833 |
| cg12473838 | SSBP1 | 0.004 | 0.167 | 0.500 | 0.711 | 0.419 | 0.928 | 0.000 | 0.192 | 0.000 | 0.664 | 0.001 | 0.030* | 0.122 | 0.140 | 0.755 | 0.199 | 0.453 | 0.765 |
| cg13291296 | GPR125 | 0.652 | 0.802 | 0.697 | 0.967 | 0.034 | 0.101 | 0.064 | 0.256 | 0.063 | 0.211 | 0.010 | 0.028* | 0.242 | 0.253 | 0.012 | 0.057 | 0.752 | 0.820 |
| cg13397649 | AFG3L2 | 0.090 | 0.227 | 0.678 | 0.749 | 0.003 | 0.809 | 0.305 | 0.079 | 0.133 | 0.385 | 0.038 | 0.295 | 0.898 | 0.948 | 0.017 | 0.082 | 0.301 | 0.156 |
| cg13463639 | SIGIRR | 0.082 | 0.551 | 0.618 | 0.985 | 0.376 | 0.709 | 0.002 | 0.348 | 0.028 | 0.363 | 0.009 | 0.020* | 0.375 | 0.275 | 0.980 | 0.730 | 0.298 | 0.431 |
| cg13920768 | PRDM12 | 0.278 | 0.431 | 0.539 | 0.635 | 0.004 | 0.101 | 0.035 | 0.065 | 0.855 | 0.221 | 0.016 | 0.095 | 0.278 | 0.347 | 0.007 | 0.040* | 0.779 | 0.710 |
| cg14214745 | CRELD1 | 0.764 | 0.627 | 0.141 | 0.246 | 0.001 | 0.419 | 0.207 | 0.024* | 0.022 | 0.196 | 0.048 | 0.218 | 0.883 | 0.713 | 0.041 | 0.267 | 0.314 | 0.359 |
| cg14247318 | 0.405 | 0.309 | 0.206 | 0.361 | 0.052 | 0.009* | 0.017 | 0.146 | 0.456 | 0.718 | 0.004 | 0.004* | 0.224 | 0.215 | 0.011 | 0.031* | 0.648 | 0.821 | |
| cg14451560 | ARRDC1 | 0.132 | 0.129 | 0.282 | 0.480 | 0.075 | 0.191 | 0.117 | 0.452 | 0.044 | 0.182 | 0.022 | 0.076 | 0.040 | 0.047* | 0.014 | 0.078 | 0.796 | 0.810 |
| cg15226170 | PMM1 | 0.894 | 0.780 | 0.422 | 0.625 | 0.024 | 0.354 | 0.136 | 0.352 | 0.511 | 0.911 | 0.012 | 0.095 | 0.302 | 0.203 | 0.001 | 0.009* | 0.535 | 0.537 |
| cg15317837 | GTPBP4 | 0.920 | 0.942 | 0.194 | 0.550 | 0.005 | 0.682 | 0.507 | 0.282 | 0.012 | 0.244 | 0.166 | 0.460 | 0.650 | 0.708 | 0.016 | 0.254 | 0.127 | 0.472 |
| cg15536552 | ACSL4 | 0.001 | 0.439 | 0.001 | 0.045* | 0.001 | 0.189 | 0.050 | 0.478 | 0.670 | 0.667 | 0.001 | 0.085 | 0.119 | 0.664 | 0.001 | 0.043* | 0.071 | 0.431 |
| cg15653194 | LFNG | 0.167 | 0.052 | 0.190 | 0.843 | 0.000 | 0.001* | 0.006 | 0.010* | 0.124 | 0.320 | 0.001 | 0.001* | 0.840 | 0.955 | 0.016 | 0.453 | 0.001 | 0.111 |
| cg16105594 | MEF2C | 0.228 | 0.481 | 0.701 | 0.832 | 0.042 | 0.234 | 0.067 | 0.437 | 0.020 | 0.064 | 0.002 | 0.022* | 0.411 | 0.500 | 0.026 | 0.124 | 0.086 | 0.213 |
| cg16261091 | HEXDC | 0.722 | 0.620 | 0.073 | 0.146 | 0.005 | 0.315 | 0.242 | 0.110 | 0.093 | 0.465 | 0.044 | 0.118 | 0.097 | 0.095 | 0.004 | 0.036* | 0.561 | 0.614 |
| cg16357287 | SEH1L | 0.574 | 0.745 | 0.092 | 0.138 | 0.081 | 0.183 | 0.110 | 0.446 | 0.126 | 0.361 | 0.008 | 0.025* | 0.773 | 0.709 | 0.000 | 0.003* | 0.644 | 0.386 |
| cg16398128 | ZNF622 | 0.410 | 0.717 | 0.339 | 0.352 | 0.003 | 0.022* | 0.005 | 0.058 | 0.365 | 0.766 | 0.000 | 0.001* | 0.976 | 0.859 | 0.000 | 0.001* | 0.809 | 0.764 |
| cg16416718 | CAMTA1 | 0.001 | 0.004* | 0.739 | 0.772 | 0.200 | 0.010* | 0.001 | 0.778 | 0.387 | 0.558 | 0.000 | 0.001* | 0.053 | 0.069 | 0.031 | 0.079 | 0.984 | 0.313 |
| cg16487280 | MYBL2 | 0.082 | 0.070 | 0.708 | 0.744 | 0.183 | 0.112 | 0.064 | 0.488 | 0.989 | 0.067 | 0.006 | 0.027* | 0.671 | 0.497 | 0.081 | 0.624 | 0.004 | 0.060 |
| cg17162475 | C6orf150 | 0.317 | 0.328 | 0.201 | 0.465 | 0.052 | 0.507 | 0.341 | 0.874 | 0.052 | 0.618 | 0.032 | 0.165 | 0.562 | 0.635 | 0.000 | 0.008* | 0.611 | 0.313 |
| cg17176517 | SNX4 | 0.349 | 0.639 | 0.406 | 0.446 | 0.255 | 0.182 | 0.042 | 0.821 | 0.291 | 0.593 | 0.005 | 0.062 | 0.563 | 0.713 | 0.006 | 0.030* | 0.715 | 0.484 |
| cg18570553 | PRR15 | 0.042 | 0.098 | 0.444 | 0.310 | 0.028 | 0.043* | 0.008 | 0.387 | 0.640 | 0.712 | 0.001 | 0.009* | 0.063 | 0.084 | 0.005 | 0.035* | 0.527 | 0.705 |
| cg18848688 | LIFR | 0.885 | 0.925 | 0.125 | 0.242 | 0.003 | 0.064 | 0.051 | 0.102 | 0.023 | 0.175 | 0.005 | 0.012* | 0.224 | 0.221 | 0.002 | 0.022* | 0.651 | 0.826 |
| cg19189310 | RPS10 | 0.398 | 0.564 | 0.453 | 0.711 | 0.062 | 0.150 | 0.052 | 0.953 | 0.205 | 0.964 | 0.009 | 0.086 | 0.200 | 0.239 | 0.005 | 0.075 | 0.292 | 0.224 |
| cg19634213 | PTEN | 0.194 | 0.497 | 0.494 | 0.932 | 0.900 | 0.353 | 0.102 | 0.213 | 0.483 | 0.651 | 0.010 | 0.092 | 0.297 | 0.356 | 0.001 | 0.002* | 0.236 | 0.447 |
| cg19878987 | LDHB | 0.321 | 0.532 | 0.328 | 0.574 | 0.086 | 0.136 | 0.034 | 0.691 | 0.477 | 0.932 | 0.008 | 0.087 | 0.176 | 0.240 | 0.001 | 0.003* | 0.197 | 0.218 |
| cg21786114 | EHMT2 | 0.344 | 0.374 | 0.953 | 0.367 | 0.023 | 0.073 | 0.060 | 0.841 | 0.363 | 0.484 | 0.004 | 0.029* | 0.600 | 0.654 | 0.001 | 0.018* | 0.809 | 0.483 |
| cg22185268 | COMMD4 | 0.943 | 0.744 | 0.548 | 0.618 | 0.007 | 0.180 | 0.058 | 0.108 | 0.299 | 0.842 | 0.001 | 0.019* | 0.329 | 0.431 | 0.001 | 0.008* | 0.467 | 0.844 |
| cg22746421 | 0.798 | 0.779 | 0.030 | 0.105 | 0.130 | 0.237 | 0.205 | 0.633 | 0.011 | 0.241 | 0.021 | 0.054 | 0.302 | 0.289 | 0.002 | 0.038* | 0.163 | 0.527 | |
| cg23224405 | DUSP16 | 0.873 | 0.594 | 0.099 | 0.062 | 0.024 | 0.256 | 0.053 | 0.351 | 0.099 | 0.194 | 0.002 | 0.054 | 0.147 | 0.199 | 0.000 | 0.001* | 0.197 | 0.435 |
| cg24199050 | GTSE1 | 0.064 | 0.175 | 0.949 | 0.862 | 0.012 | 0.006* | 0.001 | 0.129 | 0.719 | 0.984 | 0.000 | 0.001* | 0.754 | 0.988 | 0.004 | 0.013* | 0.717 | 0.190 |
| cg24689754 | C19orf62 | 0.357 | 0.466 | 0.273 | 0.341 | 0.147 | 0.607 | 0.366 | 0.576 | 0.102 | 0.183 | 0.007 | 0.045* | 0.731 | 0.593 | 0.001 | 0.005* | 0.368 | 0.104 |
| cg25295726 | FUT11 | 0.685 | 0.906 | 0.175 | 0.314 | 0.056 | 0.231 | 0.094 | 0.748 | 0.030 | 0.184 | 0.007 | 0.050 | 0.112 | 0.130 | 0.003 | 0.040* | 0.699 | 0.746 |
| cg25456633 | 0.613 | 0.938 | 0.003 | 0.003* | 0.080 | 0.581 | 0.235 | 0.649 | 0.296 | 0.688 | 0.085 | 0.377 | 0.626 | 0.742 | 0.001 | 0.012* | 0.893 | 0.758 | |
| cg25818813 | MERTK | 0.131 | 0.253 | 0.968 | 0.926 | 0.087 | 0.037* | 0.015 | 0.264 | 0.848 | 0.545 | 0.000 | 0.002* | 0.478 | 0.550 | 0.000 | 0.001* | 0.179 | 0.513 |
| cg26105057 | PAN2 | 0.843 | 0.972 | 0.305 | 0.831 | 0.029 | 0.122 | 0.161 | 0.401 | 0.107 | 0.656 | 0.058 | 0.082 | 0.155 | 0.137 | 0.000 | 0.002* | 0.522 | 0.167 |
| cg26309655 | TARS2 | 0.103 | 0.299 | 0.820 | 0.667 | 0.065 | 0.172 | 0.032 | 0.301 | 0.671 | 0.670 | 0.006 | 0.077 | 0.391 | 0.514 | 0.004 | 0.008* | 0.757 | 0.542 |
| g22721468 | SH3BP5L | 0.097 | 0.225 | 0.616 | 0.649 | 0.232 | 0.106 | 0.028 | 0.816 | 0.618 | 0.787 | 0.000 | 0.009* | 0.465 | 0.569 | 0.001 | 0.008* | 0.643 | 0.882 |
1# – Uni-variate p; 2# – Multi-variate; p* p<0.05, adjusted by sex, age and BMI.