The B-box protein BBX28 negatively regulates photomorphogenic development by functioning as a key factor in the CONSTITUTIVELY PHOTOMORPHOGENIC1-HY5 regulatory hub.
Abstract
Plants have evolved a delicate molecular system to fine-tune their growth and development in response to dynamically changing light environments. In this study, we found that BBX28, a B-box domain protein, negatively regulates photomorphogenic development in a dose-dependent manner in Arabidopsis thaliana. BBX28 interferes with the binding of transcription factor HY5 to the promoters of its target genes through physical interactions, thereby repressing its activity and negatively affecting HY5-regulated gene expression. In darkness, BBX28 associates with CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) and undergoes COP1-mediated degradation via the 26S proteasome system. Collectively, these results demonstrate that BBX28 acts as a key factor in the COP1-HY5 regulatory hub by maintaining proper HY5 activity to ensure normal photomorphogenic development in plants.
INTRODUCTION
Light is a key environmental factor that influences diverse developmental processes throughout the entire plant lifecycle. Upon germination, buried seeds develop elongated hypocotyls, closed cotyledons and curved apical hooks in order to emerge from the soil and reach the light. Subsequently, this rapid hypocotyl elongation is inhibited and the cotyledons quickly expand upon light irradiation, a process termed photomorphogenic development (Sullivan and Deng, 2003). Extensive studies have revealed that a large group of key factors work in concert to precisely regulate these distinct seedling developmental processes and their transitions (Jiao et al., 2007; Lau and Deng, 2012; Huang et al., 2014).
CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1), a central repressor of photomorphogenesis, is enriched in the nucleus in darkness (Deng et al., 1991, 1992; von Arnim and Deng, 1994). COP1 is an E3 ubiquitin ligase that targets a number of photomorphogenic-promoting factors for ubiquitination and degradation, thereby promoting skotomorphogenesis (seedling development in the dark) (Lau and Deng, 2012; Huang et al., 2014). Consistently, Arabidopsis thaliana plants with a loss of COP1 function exhibit a constitutively photomorphogenic phenotype, with expanded cotyledons and short hypocotyls, even in darkness (Deng et al., 1991, 1992). ELONGATED HYPOCOTYL5 (HY5), a bZIP transcription factor, acts as a key positive regulator of light signaling. Recessive mutations in HY5 lead to dramatically elongated hypocotyls in plants under all light conditions (Oyama et al., 1997; Ang et al., 1998). HY5 is ubiquitinated and degraded by COP1 in darkness, while it is highly abundant in the light, primarily due to the light-mediated repression of COP1 activity through multiple mechanisms (Osterlund et al., 2000; Hoecker, 2017; Podolec and Ulm, 2018). HY5 controls the expression of over 3000 genes, either positively or negatively, and regulates a large proportion of genes in a transcriptional cascade that mediates light-controlled development (Lee et al., 2007; Zhang et al., 2011). Thus, COP1 and HY5 activity is tightly under the control of light signals, and the COP1-HY5 module represents a core regulatory mechanism that operates during the dark-to-light transition. Tremendous progress has recently been made in identifying and characterizing key components in the COP1- and HY5-mediated light-signaling pathway. These factors work synergistically with COP1 and HY5 to precisely control light-mediated developmental processes in plants (Lau and Deng, 2012; Gangappa and Botto, 2014; Huang et al., 2014; Hoecker, 2017; Podolec and Ulm, 2018).
Increasing studies have shown that multiple B-box-containing (BBX) proteins are key components of the COP1-HY5 regulatory network that mediates photomorphogenesis (Khanna et al., 2009; Gangappa and Botto, 2014). BBX4, BBX21, BBX22, and BBX23 are positive regulators of light signaling (Datta et al., 2006, 2007, 2008; Fan et al., 2012; Xu et al., 2016, 2018; Zhang et al., 2017), whereas BBX19, BBX20, BBX24, BBX25, and BBX32 negatively regulate photomorphogenic development (Indorf et al., 2007; Holtan et al., 2011; Fan et al., 2012; Gangappa et al., 2013; Wang et al., 2015). In darkness, biologically active COP1 negatively regulates the abundance of these BBX proteins via its E3 ubiquitin ligase activity (Chang et al., 2011; Fan et al., 2012; Gangappa et al., 2013; Wang et al., 2015; Xu et al., 2016). In the light, the accumulated BBX proteins modulate HY5-mediated photomorphogenic development through distinct mechanisms. BBX21 directly binds to the T/G-box present in the HY5 promoter to upregulate its expression (Xu et al., 2016, 2018), while HY5 binds to the promoter of BBX22 and activates its expression (Chang et al., 2008). BBX23 synergistically works together with HY5 in the regulation of downstream target gene expression. On the other hand, both BBX24 and BBX25 associate with HY5 to repress its transcriptional activation activity (Gangappa et al., 2013). Accordingly, loss or gain of function of these individual BBX proteins leads to various photomorphogenic phenotypes in response to specific wavelengths of light (Datta et al., 2006, 2007, 2008; Holtan et al., 2011; Fan et al., 2012; Wang et al., 2015; Xu et al., 2016; Zhang et al., 2017). Therefore, BBX proteins appear to play critical roles in the COP1- and HY5-regulated signaling network to fine-tune plant growth and development.
In this study, we found that BBX28, a B-box domain protein, acts as a negative regulator of photomorphogenesis. Arabidopsis bbx28 mutant seedlings developed shortened hypocotyls, whereas transgenic plants overexpressing BBX28 displayed elongated hypocotyls in the light. BBX28 physically associates with HY5 and represses the binding of HY5 to its target sites, as well as its activation of transcriptional activation activity. Consequently, BBX28 negatively regulates HY5-mediated transcriptomic changes. BBX28 directly interacts with COP1 and undergoes COP1-mediated degradation via the 26S proteasome system in darkness. Collectively, these findings indicate that the activity of BBX28, a key component of the COP1-HY5 signaling hub, is tightly controlled by COP1 in darkness and that BBX28 represses photomorphogenesis through interacting with HY5 to inhibit its ability to bind to target sites and regulate gene expression in the light.
RESULTS
BBX28 Is a Negative Regulator of Light Signaling
Hypocotyl length, apical hook angle, and cotyledon opening angle are three extensively characterized phenotypes of seedling photomorphogenic development (Sullivan and Deng, 2003; Sentandreu et al., 2011). To investigate whether BBX28 is involved in the regulation of light signaling, we examined these phenotypes in four independent Arabidopsis bbx28 mutant alleles (bbx28-1 to bbx28-4) (Supplemental Figure 1), which were generated using the clustered regulatory interspaced short palindromic repeats (CRISPR)/Cas9 system (Feng et al., 2013). Four independent bbx28 mutants were grown under various light conditions (dark [D], white [W], blue [B], red [R], and far-red [FR]) for 4 d. In all four bbx28 mutant lines, hypocotyl length was indistinguishable from that of the wide type when grown in darkness (Figures 1A and 1B). However, all four bbx28 single mutants exhibited significantly (P < 0.0001) shortened hypocotyls compared with the wild type in all light conditions tested (W, B, R, and FR) (Figures 1C to 1J). In addition, the apical hook angle and cotyledon-opening angle of bbx28 were significantly larger than those of wild-type plants grown in darkness and in W light, respectively (Supplemental Figure 2). Together, these results demonstrate that BBX28 plays a negative role in regulating light-mediated seedling development.
Figure 1.
bbx28 Is Hypersensitive to Light.
Hypocotyl length and phenotype in 4-d-old Col and four independent bbx28 single mutant seedlings grown in darkness ([A] and [B]), W (18.1 μmol/m2/s) ([C] and [D]), B (3.88 μmol/m2/s) ([E] and [F]), R (75.1 μmol/m2/s) ([G] and [H]), and FR (2.05 μmol/m2/s) ([I] and [J]) light conditions. Hypocotyl length is expressed in millimeters. Data are means ± se; n ≥ 20. Letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA with Tukey’s post-hoc analysis. The experiments were performed three times with similar results. The graphs depict the results of one of three experiments.
Dosage Dependence of BBX28 in the Repression of Photomorphogenesis
To further explore the negative role of BBX28 in light signaling, we generated constructs expressing myc-BBX28 or YFP-BBX28 under the control of the cauliflower mosaic virus 35S promoter and used them to transform wild-type plants in the Columbia (Col) background. Three myc-tagged and three YFP-tagged BBX28 transgenic lines, in which BBX28 transcript and protein levels gradually increased, were selected for further analysis (Figures 2A and 2B). The hypocotyl lengths of all transgenic lines were similar to those of the wild type in the dark (Supplemental Figure 3). However, under all light conditions tested (W, B, R, and FR), the BBX28 transgenic seedlings displayed significantly (P < 0.0001) elongated hypocotyls. More importantly, as BBX28 transcript and protein levels increased, the hypocotyls of the transgenic seedlings became longer (Figure 2). These data further support the notion that BBX28 is a negative regulator of light signaling, and they demonstrate that BBX28 levels are closely and inversely correlated with the extent of photomorphogenic development in Arabidopsis.
Figure 2.
BBX28 Transgenic Seedlings Are Hyposensitive to Light.
(A) BBX28 transcript levels in Col and various BBX28 transgenic seedlings grown in W light for 4 d, as determined by RT-qPCR. Error bars represent sd of three technical replicates.
(B) myc-BBX28 and YFP-BBX28 protein levels in each of three myc- and YFP-tagged BBX28 transgenic seedlings grown in W for 4 d, as determined by immunoblot analysis .
(C) to (J) Hypocotyl length and phenotype in 4-d-old Col and six independent BBX28 transgenic lines grown in W (33.3 μmol/m2/s) ([C] and [D]), B (3.28 μmol/m2/s) ([E] and [F]), R (75.1 μmol/m2/s) ([G] and [H]), and FR (2.05 μmol/m2/s) ([I] and [J]) conditions. Hypocotyl length is expressed in millimeters. Data are means ± se; n ≥ 20.
In (A), (D), (F), (H), and (J), letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA with Tukey’s post-hoc analysis. The experiments were performed three times with similar results. The graphs depict the results of one of three experiments.
BBX28 Genetically and Physically Interacts with HY5
HY5 is a positive regulator of photomorphogenesis, and loss of HY5 function leads to dramatically elongated hypocotyls in Arabidopsis in response to various wavelengths of light (Oyama et al., 1997; Ang et al., 1998). We thus examined the genetic link between BBX28 and HY5. The bbx28-1 single mutants developed shorter hypocotyls than the wild type, whereas hy5-215 displayed drastically elongated hypocotyls under all light conditions tested (W, B, R, and FR), as reported previously (Oyama et al., 1997). The hypocotyls of the hy5-215 bbx28-1 double mutant were longer than those of the wild type and bbx28-1 but significantly shorter than those of hy5-215 single mutant seedlings (Figure 3), suggesting that defect in BBX28 significantly compensates for the hy5 mutant phenotype.
Figure 3.
Hypocotyl Phenotype and Length in hy5-215 bbx28-1 Seedlings Grown in the Light.
Hypocotyl phenotype and length in 4-d-old Col, bbx28-1, hy5-215, and hy5-215 bbx28-1 seedlings grown in W (18.1 μmol/m2/s) ([A] and [B]), B (4.49 μmol/m2/s) ([C] and [D]), R (75.1 μmol/m2/s) ([E] and [F]), and FR (2.05 μmol/m2/s) ([G] and [H]) conditions. Hypocotyl length is expressed in millimeters. Data are means ± se; n ≥ 20. Letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA with Tukey’s post-hoc analysis. The experiments were performed three times with similar results. The graphs depict the results of one of three experiments.
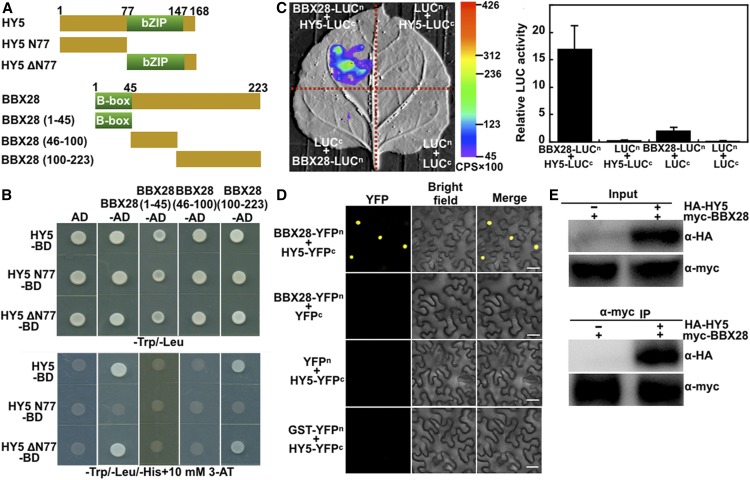
To explore the molecular nature of this genetic interaction, we performed yeast-two hybrid assays to test whether BBX28 physically interacts with HY5. BBX28 indeed interacted with HY5 in yeast cells (Figures 4A and 4B). In addition, HY5 strongly interacted with the C terminus of BBX28 (BBX28 [amino acids] 100–223) but did not associate with the N terminus of BBX28 containing a B-box domain (BBX28 1–45) or its middle portion (BBX28 46–100). Further yeast two-hybrid assays revealed that only a C-terminal truncated fragment of HY5 (HY5ΔN77), but not an N-terminal truncated fragment of HY5 (HY5 N77), interacted with full-length BBX28 or its C-terminal region (Figures 4A and 4B). Together, these data suggest that the C termini of both HY5 and BBX28 are responsible for their protein-protein interaction. To verify the BBX28-HY5 interaction in vivo, we fused BBX28 with N-terminal luciferase (LUC; BBX28-LUCn) and HY5 with C-terminal LUC (HY5-LUCc) and performed a firefly luciferase complementation imaging (LCI) assay. High luciferase activity was readily detected in Nicotiana benthamiana leaves coexpressing BBX28-LUCn and HY5-LUCc (Figure 4C); however, luciferase activity was undetectable in leaves coexpressing BBX28-LUCn and LUCc, LUCn and HY5-LUCc, or LUCn and LUCc (Figure 4C). We then performed a bimolecular fluorescence complementation (BiFC) assay and observed strong YFP signals in N. benthamiana leaf cells transiently coexpressing BBX28-YFPn (BBX28 with the N terminus of YFP) and HY5-YFPc (HY5 with the C terminus of YFP) (Figure 4D). By contrast, the negative controls, BBX28-YFPn and YFPc, YFPn and HY5-YFPc, or glutathione S-transferase (GST)-YFPn and HY5-YFPc, did not produce any detectable YFP signals in N. benthamiana leaves (Figure 4D). We performed a coimmunoprecipitation (co-IP) assay using N. benthamiana leaves transiently expressing 35S:myc-BBX28 and UBQ10:HA-HY5. As shown in Figure 4E, myc-BBX28 coimmunoprecipitated with HA-HY5, providing further evidence for the association between BBX28 and HY5 in vivo. We then performed immunoblot assays to examine whether BBX28 and HY5 are present in the same tissues in plants. Both YFP-BBX28 and HY5 were detected in the cotyledons, hypocotyls, and roots of YFP-BBX28 Col #18 transgenic seedlings (Supplemental Figure 4A). Moreover, either BBX28-CFP or YFP-HY5 alone likely resided in the nucleus when transiently expressed in N. benthamiana leaf cells. BBX28-CFP and YFP-HY5 colocalized to the nuclei of the same cells when coexpressed in N. benthamiana leaves. By contrast, the negative control, BBX28-CFP and YFP-GST, did not colocalize to the nuclei of N. benthamiana leaf cells (Supplemental Figure 4B). Together, these data suggest that BBX28 and HY5 are most likely present in the nuclei of the same cells in the same tissues.
Figure 4.
BBX28 Physically Interacts with HY5.
(A) Schematic diagram of various constructs used in the yeast two-hybrid assays. Numbers indicate the amino acid positions in HY5 or BBX28.
(B) Yeast two-hybrid interactions between the indicated BBX28 and HY5 proteins.
(C) Firefly LCI assay showing that BBX28 interacts with HY5 in N. benthamiana leaf cells. The BBX28-LUCn and HY5-LUCc constructs were transiently coinfiltrated into wild tobacco leaves, and luminescence intensity was detected using LB985 NightSHADE. LUCn and LUCc served as negative controls. The color scale shows the range of luminescence intensity. Error bars represent sd of three replicates.
(D) BiFC assay showing the interaction of BBX28 with HY5. Full-length BBX28 and HY5 were fused to the split N- or C-terminal (YFPn or YFPc) fragments of YFP. Unfused YFP N-terminal (YFPn), YFP C-terminal (YFPc), and GST-YFPn fragments were used as negative controls. Merge, merged images of YFP channel and bright field. Bar = 40 μm.
(E) Co-IP analysis showing that myc-BBX28 interacts with HA-HY5. Total protein was extracted from wild tobacco leaves transiently expressing 35S:myc-BBX28 alone or together with UBQ10:HA-HY5. The immunoprecipitates were detected using anti-HA and anti-myc antibodies.
BBX28 Interferes with the Binding of HY5 to Its Target Sites
Considering the genetic and physical interactions between BBX28 and HY5, we sought to assess the functional interplay between these two proteins. HY5 gene expression was not significantly altered in bbx28 and BBX28 transgenic seedlings grown in W light, and BBX28 transcript levels in hy5-215 were similar to those of Col when grown under all light conditions tested (D, W, B, R, and FR) (Supplemental Figure 5). These results suggest that BBX28 and HY5 do not affect each other at the transcriptional level. As HY5 is a b-ZIP-type transcription factor, it directly binds to the promoters of a large number of regulatory genes to mediate their expression (Lee et al., 2007; Zhang et al., 2011). HY5 tagged with histidine (HY5-His) bound to the promoters of FAR-RED ELONGATED HYPOCOTYL1 (FHY1), CHALCONE SYNTHASE (CHS), and HY5 itself in vitro electrophoretic mobility shift assay (EMSAs), which is consistent with previous studies (Figures 5A to 5C; Li et al., 2010; Abbas et al., 2014; Binkert et al., 2014; Zhang et al., 2017). Neither His-Trigger Factor (TF; a prokaryotic ribosome-associated chaperone protein) nor His-TF-BBX28 was able to bind to these DNA subfragments. As the amount of His-TF-BBX28 in the reactions increased, the binding of HY5 to FHY1, CHS, and HY5 promoter subfragments clearly decreased. However, His-TF had no effect on the binding affinity of HY5 (Figures 5A to 5C). These data suggest that BBX28 interferes with the ability of HY5 to bind to these DNA subfragments in vitro. We then performed yeast one-hybrid assays to verify these results. HY5 activated the transcription of FHY1 and CHS in yeast cells, as reported previously (Figures 5D and 5E; Li et al., 2010). BBX28 alone did not have any effect on the proFHY1:LacZ or proCHS:LacZ reporters. However, the activation of proFHY1:LacZ and proCHS:LacZ by HY5 was dramatically reduced in yeast cells cotransformed with BBX28 together with HY5, as revealed by analysis of β-galactosidase activity (Figures 5D and 5E). Moreover, HY5 alone activated the transcription of proBBX22:LUC, proCHS:LUC, or proHY5:LUC when transiently expressed in Arabidopsis protoplasts, as reported previously (Figures 5F to 5I; Gangappa et al., 2013; Zhang et al., 2017; Xu et al., 2018). BBX28 alone did not activate any of these reporters. However, in the presence of BBX28, the activation of proHY5:LUC, proCHS:LUC, and proBBX22:LUC by HY5 was significantly reduced (Figures 5F to 5I). Together, these results strongly suggest that BBX28 represses the transcriptional activation activity of HY5 through interfering with its capacity to bind to the promoters of its target genes.
Figure 5.
BBX28 Represses the Transcriptional Activation Activity of HY5.
(A) to (C) EMSA showing that the presence of increasing amounts of TF-BBX28 decreases the binding of HY5-His to the promoters of FHY1 (A), CHS (B), and HY5 (C). “−” indicates the absence of corresponding of probes or proteins. For HY5-His, “+” indicates that 5.4 pmol is present; for His-TF, “+” and “++” indicate that 2.1 and 4.2 pmol are present, respectively; for His-TF-BBX28, “+” and “++” indicate that 1.4 and 2.8 pmol are present, respectively. TF, trigger factor, a prokaryotic ribosome-associated chaperone protein. FP indicates free probe.
(D) and (E) Yeast-one hybrid assays showing that BBX28 inhibits the activation of proFHY1:LacZ (D) and proCHS:LacZ (E) by HY5. Error bars represent sd of four independent yeast cultures. Asterisks represent statistically significant differences (***P < 0.001), as determined by Student’s t test. The experiments were performed three times with similar results.
(F) Schematic representation of various constructs used in the transient transfection assay in Arabidopsis protoplasts. Arrow after the 35S promoter indicates the transcriptional start site. −994, −1000, and −752 indicate the length of the BBX22, CHS, and HY5 promoter sequence that was fused to the firefly luciferase gene to create the reporter construct, respectively.
(G) to (I) Bar graphs showing that BBX28 represses the activation of the proBBX22:LUC (G), proCHS:LUC (H), and proHY5:LUC (I) reporters by HY5. Error bars represent sd of three independent transient transfections in Arabidopsis protoplasts. Asterisks represent statistically significant differences (***P < 0.001), as determined by Student’s t test. The experiments were performed three times with similar results.
To support the conclusion that BBX28 negatively interferes with HY5-regulated gene expression, we examined the expression of HY5-regulated genes in bbx28 mutant and BBX28 transgenic plants via RT-qPCR. As expected, the transcript levels of all HY5-activated genes tested [ALTERNATIVE NAD(P)H DEHYDROGENASE1, SECRETION-ASSOCIATED RAS1, and BLUE-LIGHT INHIBITOR OF CRYPTOCHROMES1, At2g15020; Lee et al., 2007; Zhang et al., 2011; Wang et al., 2016, 2017] were upregulated in bbx28 but downregulated in BBX28-overexpressing seedlings compared with the wild type. By contrast, the expression levels of all HY5-repressed genes tested (AGAMOUS-LIKE58 and ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE2; Lee et al., 2007; Zhang et al., 2011) were reduced in bbx28 but upregulated in BBX28 transgenic seedlings compared with the wild type (Supplemental Figure 6). These data are in agreement with the finding (from biochemical analysis) that BBX28 interacts with HY5, consequently repressing the ability of HY5 to modulate downstream target gene expression.
BBX28 Genetically and Physically Interacts with COP1
COP1 is a central repressor of photomorphogenesis, and multiple BBX proteins function in various COP1-regulated processes (Datta et al., 2006, 2007, 2008; Xu et al., 2016). We therefore investigated the genetic interaction between BBX28 and COP1. Arabidopsis cop1-6, a weak allele of cop1, displayed a constitutively photomorphogenic phenotype in darkness and dramatically shortened hypocotyls under various light conditions, as reported previously (McNellis et al., 1994). However, the bbx28-4 cop1-6 double mutant developed significantly shorter hypocotyls than the cop1-6 single mutant in darkness. By contrast, the hypocotyl length of YFP-BBX28 cop1-6 #18 was indistinguishable from that of Col and bbx28-4 in darkness, suggesting that overexpressing BBX28 largely suppresses the constitutively photomorphogenic phenotype of cop1-6 (Figures 6A and 6B). Under all light conditions tested (W, B, R, and FR), the hypocotyls of bbx28-4 cop1-6 were significantly shorter than those of cop1-6, while YFP-BBX28 cop1-6 #18 hypocotyls were longer than those of cop1-6 but shorter than those of Col and YFP-BBX28 Col #18 (Supplemental Figure 7). These genetic observations demonstrate that the loss of function of BBX28 enhances the mutant phenotype of cop1 in both the dark and light, whereas the gain of function of BBX28 strongly suppresses this phenotype in the dark, but only partially suppresses it in the light.
Figure 6.
BBX28 Genetically and Physically Interacts with COP1.
(A) and (B) Hypocotyl phenotype (A) and length (B) in 4-d-old Col, bbx28-4, YFP-BBX28 Col #18, cop1-6, bbx28-4 cop1-6, and YFP-BBX28 cop1-6 #18 seedlings grown in darkness. Hypocotyl length is expressed in millimeters. Data are means ± se; n ≥ 20. Letters above the bars indicate significant differences (P < 0.05), as determined by one-way ANOVA with Tukey’s post-hoc analysis. The experiments were performed three times with similar results. The graphs depict the results of one of three experiments.
(C) Schematic diagram of the domain structure of COP1 and the truncated COP1 proteins. Numbers indicate the amino acid positions in COP1.
(D) Yeast two-hybrid interactions between the indicated COP1 protein and BBX28.
(E) BiFC assay showing the interaction of BBX28 with COP1. Full-length BBX28 and COP1 were fused to the split N- or C-terminal (YFPn or YFPc) fragments of YFP. Unfused YFPn, YFPc, and GST-YFPn fragments were used as negative controls. Merge, merged images of YFP channel and bright field. Bar = 80 μm.
(F) Co-IP analysis showing that myc-BBX28 interacts with COP1 in Arabidopsis seedlings. Four-day-old W light-grown Col and myc-BBX28 Col #44 seedlings were transferred to darkness for 48 h and subjected to a co-IP assay using anti-myc antibodies, with the immunoprecipitates were detected using anti-COP1 and anti-myc antibodies, respectively. Actin served as a negative control.
Next, we performed yeast two-hybrid assays to examine whether BBX28 interacts with COP1. As shown in Figures 6C and 6D, BBX28 indeed interacted with COP1 in yeast cells. Moreover, domain-mapping assays demonstrated that COP1 strongly interacted with the C terminus of BBX28 (BBX28 100–223) but did not interact with the N terminus of BBX28 containing a B-box domain (BBX28 1–45) and its middle portion (BBX28 46–100). These results suggest that the C terminus of BBX28 mediates its interaction with COP1 in yeast cells. To further map the domain in COP1 responsible for its interaction with BBX28, we performed yeast two-hybrid assays using various COP1 truncated fragments (Figure 6C). None of the COP1 truncated fragments interacted with full-length BBX28 or with any BBX28 truncated fragment (Figure 6D), suggesting that the overall structure of COP1 is required for its interaction with BBX28. To verify the interaction between BBX28 and COP1, we performed a BiFC assay. When N. benthamiana leaves were transiently cotransfected with constructs expressing BBX28-YFPn and COP1 fused with the C terminus of YFP (COP1-YFPc), strong YFP fluorescent signals were clearly observed (Figure 6E). However, leaves coexpressing BBX28-YFPn and YFPc, YFPn and COP1-YFPc, or GST-YFPn and COP1-YFPc did not show any detectable YFP signals (Figure 6E). We then performed co-IP assays using myc-BBX28 Col #44 transgenic plants and monoclonal myc antibodies. myc-BBX28 strongly coimmunoprecipitated endogenous COP1 in myc-BBX28 Col #44 transgenic Arabidopsis seedlings (Figure 6F). Together, these data demonstrate that BBX28 directly interacts with COP1 in planta.
BBX28 Undergoes COP1-Mediated Degradation in Darkness
Considering that BBX28 plays a critical role in light signaling, we investigated whether BBX28 itself is regulated by light signals. BBX28 gene expression was significantly reduced in seedlings grown in various wavelengths of light compared with dark-grown seedlings (Figure 7A). By contrast, myc-BBX28 protein levels were higher in myc-BBX28 Col #44 transgenic seedlings grown under various light conditions compared with dark-grown seedlings (Figure 7B). These results indicate that light signals negatively regulate BBX28 at the transcriptional level, whereas they promote the accumulation of BBX28 at the protein level. Next, we examined whether BBX28 is degraded via the 26S proteasome system. YFP-BBX28 protein levels markedly increased in dark-grown YFP-BBX28 Col #10 plants in response to treatment with 50 μM MG132 (a proteasome inhibitor); this increase became more obvious when the concentration of MG132 increased to 100 μM (Figure 7C). These observations suggest that BBX28 undergoes 26S proteasome-mediated degradation in etiolated seedlings. Given that the BBX28-COP1 interaction and BBX28 are subjected to 26S proteasome-mediated degradation in the dark (Figures 6 and 7A to 7C), we investigated whether COP1 controls BBX28 levels. To this end, the cop1-6 mutation was introduced into YFP-BBX28 Col #18 transgenic plants by genetic crossing. YFP-BBX28 cop1-6 #18 accumulated much higher levels of YFP-BBX28 compared with YFP-BBX28 Col #18 seedlings grown in darkness (Figure 7D), suggesting that COP1 promotes the degradation of BBX28 in dark-grown Arabidopsis seedlings.
Figure 7.
BBX28 Undergoes COP1-Mediated Degradation in Darkness.
(A) BBX28 transcript levels in 4-d-old Col grown in various light conditions (D, W, B, R, and FR), as determined by RT-qPCR. Error bars represent sd of three technical replicates. Asterisks represent statistically significant differences (***P < 0.01), as determined by Student’s t test.
(B) myc-BBX28 protein levels in 4-d-old myc-BBX28 Col #44 transgenic seedlings grown in various light conditions (D, W, B, R, and FR), as determined by immunoblot analysis. Col served as a negative control.
(C) YFP-BBX28 protein levels in 4-d-old dark-grown YFP-BBX28 Col #10 transgenic seedlings treated with various concentrations (0, 50, and 100 μm) of MG132. Col treated with DMSO served as a negative control.
(D) YFP-BBX28 protein levels in YFP-BBX28 Col #18 and YFP-BBX28 cop1-6 #18 grown in darkness for 4 d. cop1-6 served as a negative control.
In (B) to (D), actin was used as a loading control.
DISCUSSION
Skotomorphogenic and photomorphogenic growth are two key steps that enable a germinated, buried seed to emerge from the soil and give rise to a healthy seedling. A variety of wavelength-specific photoreceptors, E3 ubiquitin ligases, transcription factors, and so on function synergistically in regulating these two critical developmental processes. Photoreceptors transduce light signals to downstream factors that in turn initiate the reprogramming of gene expression in Arabidopsis (Ma et al., 2001; Tepperman et al., 2004). COP1 is a central repressor of light signaling, and it represses photomorphogenesis through ubiquitinating and degrading downstream substrates including HY5 in darkness (Lau and Deng, 2012; Huang et al., 2014). Upon light irradiation, COP1 is inactivated through multiple regulatory mechanisms, allowing HY5 to accumulate (Hoecker, 2017; Podolec and Ulm, 2018). This HY5 rapidly upregulates or downregulates the expression of over 3000 genes to promote photomorphogenesis (Lee et al., 2007; Zhang et al., 2011). Therefore, the COP1-HY5 regulatory module functions at the center of a transcriptional and posttranslational network hub involved in the light signal transduction pathway.
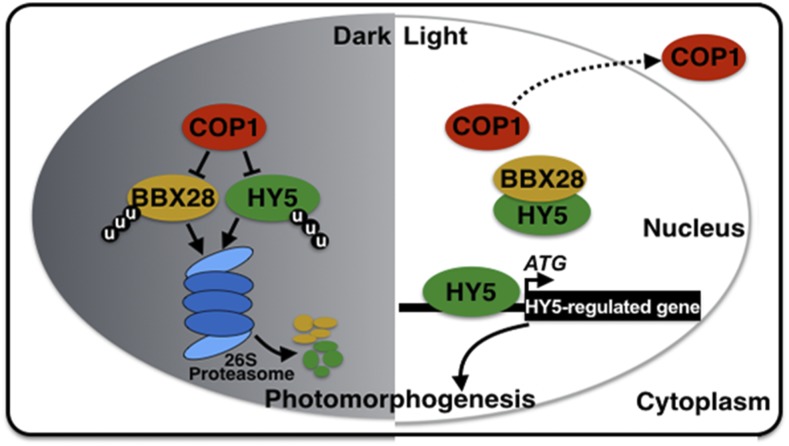
Our results indicate that BBX28, a repressor of light signaling, participates in the COP1-HY5-mediated developmental transition in Arabidopsis seedlings in response to light. BBX28 genetically and physically interacted with COP1 (Figure 6). Degradation of BBX28 occurred via the 26S proteasome pathway in darkness; this molecular event was dependent on COP1 (Figures 6 and 7). These findings suggest that BBX28 is a downstream substrate for COP1 and that etiolated seedlings maintain very low levels of BBX28. Light triggered the accumulation of BBX28 (Figure 7B), most likely due to the light-mediated inactivation of COP1 (Hoecker, 2017; Podolec and Ulm, 2018). Like BBX28, HY5 accumulates in the light. These two proteins associated with each other (Osterlund et al., 2000; Figure 4), suggesting that BBX28 and HY5 might function together in the control of seedling development in response to light. BBX28 was able to interfere with the binding of HY5 to the promoters of its target genes, which in turn negatively affected its ability to control target gene expression (Figure 5; Supplemental Figure 6). All of these findings support the notion that BBX28 modulates the activity of the COP1-HY5 module and the process regulated by this module. Loss or gain of function of BBX28 enhanced or suppressed the mutant phenotype of cop1-6, respectively, implying that BBX28 has a positive effect on COP1 activity. BBX28 represses photomorphogenesis (Figures 1 and 2; Supplemental Figure 2), whereas HY5 is a positive regulator of light signaling (Oyama et al., 1997; Ang et al., 1998). Both BBX28 and HY5 regulate photomorphogenic development in a dose-dependent manner (Osterlund et al., 2000; Figure 2). These findings suggest that BBX28 quantitatively modulates the activity of the COP1-HY5 module, particularly the output conferred by HY5. Collectively, our results indicate that COP1 targets both BBX28 and HY5 for ubiquitination and degradation via the 26S proteasome system in darkness. Upon light illumination, both BBX28 and HY5 accumulate due to the light-mediated inactivation of COP1. BBX28 forms heterodimers with HY5 through their respective C-terminal regions, which in turn interferes with the binding of HY5 to its target sites and represses its transcriptional activation activity. This mechanism may serve to precisely modulate the biochemical ability of HY5 to fine-tune the expression of thousands of genes, as well as photomorphogenic growth (Figure 8).
Figure 8.
A Proposed Working Model Depicting How the COP1-BBX28-HY5 Regulatory Module Mediates Light Signaling.
In darkness, COP1 mediates the degradation of BBX28 and HY5 via the 26S proteasome system to promote skotomorphogenesis. Upon light illumination, COP1 activity is inhibited in a light-dependent manner, allowing BBX28 and HY5 to accumulate. HY5 regulates the expression of numerous downstream target genes to promote photomorphogenesis. BBX28 interacts with HY5 to interfere with its binding to target sites and repress its transcriptional activity, which in turn negatively regulates photomorphogenesis. u, ubiquitin.
Light initiates a complicated but delicate transcriptional cascade to promote light-mediated developmental processes in plants. Previous studies suggest that HY5 is a high hierarchical transcription factor that initiates and controls the light-mediated transcriptional cascade. HY5 constitutively binds to its target sites in both the dark and light, supporting the notion that the binding of HY5 alone might be insufficient for regulating the expression of target genes and that additional factors are required for its proper transcriptional activation activity (Lee et al., 2007; Zhang et al., 2011). Previous work and this study have identified and characterized a set of HY5-interacting factors including HY5-HOMOLOG, G-BOX BINDING FACTOR1, CALMODULIN7, BBX21, BBX22, BBX23, BBX24, BBX25, BBX28, and BBX32 (Holm et al., 2002; Datta et al., 2007, 2008; Holtan et al., 2011; Singh et al., 2012; Gangappa et al., 2013; Abbas et al., 2014; Zhang et al., 2017; this study). These regulators interact with HY5 to enhance or repress its transcriptional activation activity, suggesting that various factors converge on HY5 to orchestrate a transcriptional cascade during light-mediated seedling development. Notably, the finding that multiple BBXs act as coactivators or corepressors of HY5 activity suggests that BBXs play critical roles in the HY5-mediated transcriptional network and that a transcriptional cascade controlled by BBXs-HY5 may be a central theme of seedling development.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana cop1-6 (McNellis et al., 1994), hy5-215 (Ang and Deng, 1994), and bbx28 (bbx28-1 to bbx28-4) (this study) mutants are in the Col-0 ecotype. Seeds were surface sterilized with 30% commercial Clorox bleach and 0.02% Triton X-100 for 5 min, washed three times with sterile water, and sown on 1× Murashige and Skoog (MS) medium supplemented with 0.8% Agar (Difco) and 1% sucrose. The seeds were stratified in darkness for 3 d at 4°C and transferred to light chambers (Percival Scientific) maintained at 22°C. The fluence rates of the light growth chambers were 18.1 μmol/m2/s for W light, 3.88 μmol/m2/s for B light, 75.1 μmol/m2/s for R light, and 2.05 μmol/m2/s for FR light.
Plasmid Construction
The full-length BBX28, COP1, or HY5 open reading frame sequence was cloned into the pDONR-221 or pDONR-223 vector (Invitrogen) and introduced into the plant binary vector pEarley Gateway 203, pEarley Gateway 104, pSPYNE, or pSPYCE (Earley et al., 2006) under the control of the 35S promoter using Gateway LR Clonase enzyme mix (Invitrogen). pEarley Gateway-myc-BBX28, pEarley Gateway-YFP-BBX28, pSPYNE-35S-BBX28, pSPYCE-35S-COP1, and pSPYCE-35S-HY5 constructs were generated.
To generate pGADT7-BBX28, pGADT7-BBX28 (1–45), pGADT7-BBX28 (46–100), and pGADT7-BBX28 (100–223) constructs, full-length BBX28, BBX28 (1–45), BBX28 (46–100), and BBX28 (100–223) fragments were amplified by PCR with the respective pairs of primers and then cloned into the NdeI/XhoI sites of the pGADT7 vector (BD Clontech). To generate the pGBKT7-COP1, pGBKT7-COP1 N282, pGBKT7-COP1 coil, and pGBKT7-COP1 WD40 constructs, full-length COP1, COP1 N282, COP1 coil, and COP1 WD40 fragments were amplified by PCR with the respective pairs of primers and cloned into the EcoRI/PstI sites of the pGBKT7 vector (BD Clontech). To generate the pGBKT7-HY5, pGBKT7-HY5 N77, and pGBKT7-HY5ΔN77 constructs, full-length HY5, HY5N77, and HY5ΔN77 fragments were amplified by PCR with the respective pairs of primers and cloned into the NdeI/PstI sites of the pGBKT7 vector (BD Clontech).
To produce the constructs for the LCI assays, the full-length BBX28 and HY5 sequences were amplified by PCR with the respective pairs of primers and cloned into the KpnI/SalI or BamHI/SalI sites of pCambia1300-nLUC or pCambia1300-cLUC, respectively (Chen et al., 2008). For the protoplast assays, the 994-bp BBX22, 1000-bp CHS, or 752-bp HY5 promoter sequences upstream of ATG were amplified by PCR with the respective pairs of primers and cloned into the KpnI/PstI sites of the pGreen0800II-LUC vector. The full-length BBX28 sequence was amplified by PCR with the respective pairs of primers and cloned into the NdeI/XhoI sites of the pCold-His-TF vector (Takara). UBQ10:HA-HY5 was amplified by PCR and cloned into the EcoRI/KpnI sites of the pCambia1300 vector. The 752-bp CHS promoter sequence upstream of ATG was amplified by PCR with the respective pairs of primers and cloned into the KpnI/XhoI sites of the pLacZ-2u vector (Lin et al., 2007). The pCambia1300-35S:P19 (for suppressing posttranscriptional gene silencing; Sparkes et al., 2006; Liu et al., 2010), pCambia1300-Lucc-HY5 (Li et al., 2010), pLacZ-2u-FHY1 (Li et al., 2010), and pET28a-His-HY5 (Xu et al., 2016) constructs were described previously. The primers used for plasmid construction are listed in Supplemental Table 1.
Transgenic Plants
The pEarley Gateway-myc-BBX28 and pEarley Gateway-YFP-BBX28 constructs were transformed into Agrobacterium tumefaciens GV3101 by the freeze-thaw method and introduced into Col plants via the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on MS medium containing 20 mg/L Basta.
Measurement of Hypocotyl Length
To measure the hypocotyl length of seedlings, seeds were sown on plates and stratified at 4°C in darkness for 3 d, followed by incubation in continuous white light for 8 h to induce uniform germination. The seeds were transferred to D, W, B, R, and FR light conditions and incubated at 22°C for 4 d (Xu et al., 2016). The hypocotyl length of seedlings was measured using ImageJ software.
Co-IP Assays and Immunoblot Analysis
For co-IP assays using Nicotiana benthamiana leaves, Agrobacterium strain GV3101 cells carrying the 35S:myc-BBX28 or UBQ10:HA-HY5 constructs were transiently infiltrated into N. benthamiana leaves. The plants were grown under long-day conditions for 3 d and lysed. The extracts were incubated with 4 μL of anti-myc antibodies (1:250 v/v) (Sigma-Aldrich; catalog no. M4439) coupled with 25 μL of Protein-A Sepharose (GE Healthcare) for 3 h at 4°C. The Sepharose was washed three times with protein extraction buffer. The precipitates were eluted into 100 mM glycine (pH 2.5) and 100 mM NaCl, immediately neutralized by 2 M Tris-HCl, pH 9.0, and 100 mM NaCl, and concentrated using StrataClean Resin (Stratagene) prior to immunoblot analysis.
For co-IP assays using Arabidopsis seedlings, 1 mg of total proteins was extracted from 4-d-old Arabidopsis seedlings in protein extraction buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% (v/v) glycerol, 0.1% Tween 20, 1 mM PMSF, and 1× complete Protease Inhibitor Mixture (Roche). The extracts were incubated with 4 μL of anti-myc antibodies (Sigma-Aldrich) coupled with 25 μL of Protein-A Sepharose (GE Healthcare) for 3 h at 4°C. The Sepharose was washed three times with protein extraction buffer. The precipitates were eluted into 100 mM glycine (pH 2.5) and 100 mM NaCl, immediately neutralized by 2 M Tris-HCl, pH 9.0, and 100 mM NaCl, and concentrated using StrataClean Resin (Stratagene) prior to immunoblot analysis.
For immunoblot analysis, wild-type or mutant Arabidopsis seedlings were homogenized in protein extraction buffer containing 100 mM NaH2PO4, 10 mM Tris-HCl, 200 mM NaCl, 8 M urea, pH 8.0, 1 mM PMSF, and 1× complete protease inhibitor cocktail (Roche). Primary antibodies used in this study were anti-COP1 (1:1000 v/v) (McNellis et al., 1994), anti-cMyc (1:1000 v/v) (Sigma-Aldrich; catalog no. M4439), anti-HA (1:1000 v/v) (Sigma-Aldrich; catalog no. H3663), anti-GFP (1:5000 v/v) (Abmart; catalog no. M20004M), and anti-Actin (1:2000 v/v) (Sigma-Aldrich; catalog no. A0480).
EMSA
EMSA was performed using biotin-labeled probes and a Light Shift Chemiluminescent EMSA kit (Thermo Scientific) as described previously (Xu et al., 2014). The promoter subfragments of FHY1 (143 bp, −280 to −138 bp), CHS (150 bp, −547 to −398 bp), and HY5 (163 bp, −414 to −252 bp) upstream of ATG were PCR-amplified, mixed with biotin, and incubated in UV light for 30 min for biotin labeling. Next, purified His-TF-BBX28, HY5-His, or His-TF proteins as indicated were incubated together with 40 fmol biotin-labeled probes in a 20-μL reaction mixture containing 10 mM Tris-HCl, pH 7.5, 0.05% Nonidet P-40, 10 mM MgCl2, 5% (v/v) glycerol, and 0.1 μg/mL poly (dI∙dC). The reactions were incubated at 25°C for 20 min, followed by separation on 6% native polyacrylamide gels in 0.5× TBE buffer. The gels were electroblotted to Hybond N+ (Millipore) nylon membranes in 0.5× TBE for 40 min, and the labeled probes were detected according to the manufacturer’s protocols provided with the EMSA kit. The probes used in this study are listed in Supplemental Table 1.
Yeast One-Hybrid and Two-Hybrid Assays
For the yeast one-hybrid assay, the respective combinations of AD-fusion effectors and LacZ reporters were cotransformed into yeast strain EGY48, and transformants were selected and grown on SD/-Trp-Ura dropout medium. Yeast transformation and liquid assay were conducted as described in the Yeast Protocols Handbook (BD Clontech). Yeast two-hybrid assays were performed using the Matchmaker GAL4 Two-Hybrid System (BD Clontech). The respective combinations of pGAD-T7 and pGBKT7 fusion plasmids were cotransformed into yeast strain AH109. The empty pGAD-T7 and pGBKT7 vectors were cotransformed in parallel as negative controls. To assess protein interactions, the transformed yeast cells were grown on SD/-3 (-His/-Leu/-Trp) dropout medium. The interactions were observed after 3 d of incubation at 30°C. Yeast transformation was performed as described in the Yeast Protocols Handbook (BD Clontech).
BiFC Assays
YFPn- and YFPc-fused plasmids were transformed into Agrobacterium strain GV3101, and the indicated transformant pairs were infiltrated into N. benthamiana leaves. A Carl Zeiss confocal laser scanning microscope (LSM510 Meta) was used to detect YFP fluorescence signals. YFP fluorescence was excited by a 514 nm laser and detected between 517 and 589 nm (Kudla and Bock, 2016).
Firefly LCI Assays
LCI assays were performed as described previously (Chen et al., 2008). The LUCn- and LUCc-fused plasmids were transformed into Agrobacterium strain GV3101, and the indicated transformants pairs were infiltrated into N. benthamiana leaves. The plants were grown in the light for 3 d, and luciferase activity was measured using LB985 NightShade (Berthold Technologies). An exposure time of 2 min with 3×3 binning was used for all images taken. Relative LUC activity is equivalent to luminescence intensity/10 mm2 leaf area. The experiments were performed three times, and each data point was obtained from the results of three replicate experiments.
Protoplast Assay
Arabidopsis mesophyll cell protoplasts were prepared and transfected as described previously (Yoo et al., 2007). The promoter-reporter used were the 994-bp BBX22, 1000-bp CHS, or 752 -bp HY5 promoters driving the firefly luciferase gene (pGreen0800II-proBBX22:LUC, pGreen0800II-proCHS:LUC, and pGreen0800II-proHY5-LUC). pCambia1300-UBQ10:HA-HY5 and pEarley Gateway-35S:myc-BBX28 were used as the effectors. A Dual Luciferase kit (Promega) was used to detect reporter activity. The Renilla luciferase gene driven by the cauliflower mosaic virus 35S promoter was used as an internal control.
Quantitative RT-PCR
Total RNA was extracted from 4-d-old Arabidopsis seedlings grown under white light using an RNeasy Plant Mini Kit (Qiagen). cDNA was synthesized from 2 μg of total RNA using the 5X All-In-One RT Master Mix cDNA synthesis system (Applied Biological Materials) according to the manufacturer’s instructions. The cDNA was subjected to RT-qPCR using the StepOnePlus Real-time PCR detection system (Applied Biosystems) and SYBR Green PCR Master Mix (Takara). The experiments were performed three times. PCR was performed in triplicate for each sample, and the expression levels were normalized to that of the housekeeping gene PP2A. The primers used in this study are listed in Supplemental Table 1.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 5.0 (GraphPad Software). To determine statistical significance, one-way ANOVA with Tukey’s post-hoc test was employed (Supplemental Table 2).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: BBX28 (At4g27310), HY5 (At5g11260), and COP1 (At2g32950).
Supplemental Data
Supplemental Figure 1. Mutations in the four independent bbx28 alleles created by the CRISPER/Cas9 method.
Supplemental Figure 2. Apical hook and cotyledon phenotypes of bbx28 seedlings grown in darkness and white light.
Supplemental Figure 3. Hypocotyl phenotype and length in various BBX28 transgenic seedlings grown in darkness.
Supplemental Figure 4. Colocalization of BBX28 and HY5.
Supplemental Figure 5. HY5 and BBX28 transcript levels in bbx28-1, BBX28 transgenic seedlings, and the hy5-215 single mutant, as revealed by RT-qPCR.
Supplemental Figure 6. HY5-regulated gene expression in bbx28-1 and BBX28 transgenic seedlings, as revealed by RT-qPCR.
Supplemental Figure 7. Hypocotyl phenotype and length in various mutant seedlings grown under various light conditions.
Supplemental Table 1. Primers used in this study.
Supplemental Table 2. ANOVA table.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jian-Kang Zhu for CRISPR/Cas9 plasmids and Jigang Li for pLacZ-2u-FHY1 plasmids. This work was supported by grants from National Key R&D Program of China (2017YFA0503800), by the National Natural Science Foundation of China (31330048 and 31621001), by the Peking-Tsinghua Center for Life Sciences, by NIH Grants GM-47850 and GM-109076, by the China postdoctoral Science Foundation, and by Southern University of Science and Technology.
AUTHOR CONTRIBUTIONS
D.X. and X.W.D. designed the project. F.L., Y.J., D.X., J.L., and T.Y. performed the research. D.X., L.F, J.S.L., Z.J.C., and X.W.D. analyzed the data. D.X. and X.W.D. wrote the article. Z.J.C. revised the article.
References
- Abbas N., Maurya J.P., Senapati D., Gangappa S.N., Chattopadhyay S. (2014). Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 26: 1036–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L.H., Deng X.W. (1994). Regulatory hierarchy of photomorphogenic loci: allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6: 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222. [DOI] [PubMed] [Google Scholar]
- Binkert M., Kozma-Bognár L., Terecskei K., De Veylder L., Nagy F., Ulm R. (2014). UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell 26: 4200–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.S., Li Y.H., Chen L.T., Chen W.C., Hsieh W.P., Shin J., Jane W.N., Chou S.J., Choi G., Hu J.M., Somerville S., Wu S.H. (2008). LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 54: 205–219. [DOI] [PubMed] [Google Scholar]
- Chang C.S., Maloof J.N., Wu S.H. (2011). COP1-mediated degradation of BBX22/LZF1 optimizes seedling development in Arabidopsis. Plant Physiol. 156: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi G.H., Deng X.W., Holm M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi C., Johansson H., Holm M. (2007). SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Johansson H., Hettiarachchi C., Irigoyen M.L., Desai M., Rubio V., Holm M. (2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.W., Caspar T., Quail P.H. (1991). cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5: 1172–1182. [DOI] [PubMed] [Google Scholar]
- Deng X.W., Matsui M., Wei N., Wagner D., Chu A.M., Feldmann K.A., Quail P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801. [DOI] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Fan X.Y., Sun Y., Cao D.M., Bai M.Y., Luo X.M., Yang H.J., Wei C.Q., Zhu S.W., Sun Y., Chong K., Wang Z.Y. (2012). BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 5: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Zhang B., Ding W., Liu X., Yang D.L., Wei P., Cao F., Zhu S., Zhang F., Mao Y., Zhu J.K. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23: 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa S.N., Botto J.F. (2014). The BBX family of plant transcription factors. Trends Plant Sci. 19: 460–470. [DOI] [PubMed] [Google Scholar]
- Gangappa S.N., Crocco C.D., Johansson H., Datta S., Hettiarachchi C., Holm M., Botto J.F. (2013). The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25: 1243–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U. (2017). The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol. 37: 63–69. [DOI] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtan H.E., et al. (2011). BBX32, an Arabidopsis B-Box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 156: 2109–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Deng X.W. (2014). Beyond repression of photomorphogenesis: role switching of COP/DET/FUS in light signaling. Curr. Opin. Plant Biol. 21: 96–103. [DOI] [PubMed] [Google Scholar]
- Indorf M., Cordero J., Neuhaus G., Rodríguez-Franco M. (2007). Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J. 51: 563–574. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230. [DOI] [PubMed] [Google Scholar]
- Khanna R., Kronmiller B., Maszle D.R., Coupland G., Holm M., Mizuno T., Wu S.H. (2009). The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J., Bock R. (2016). Lighting the way to protein-protein interactions: recommendations on best practices for bimolecular fluorescence complementation analyses. Plant Cell 28: 1002–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17: 584–593. [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., Tongprasit W., Zhao H., Lee I., Deng X.W. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Gao S., Martinez C., He G., Zhou Z., Huang X., Lee J.H., Zhang H., Shen Y., Wang H., Deng X.W. (2010). Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Ma L., Li J., Qu L., Hager J., Chen Z., Zhao H., Deng X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolec R., Ulm R. (2018). Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 45: 18–25. [DOI] [PubMed] [Google Scholar]
- Sentandreu M., Martín G., González-Schain N., Leivar P., Soy J., Tepperman J.M., Quail P.H., Monte E. (2011). Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis. Plant Cell 23: 3974–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Ram H., Abbas N., Chattopadhyay S. (2012). Molecular interactions of GBF1 with HY5 and HYH proteins during light-mediated seedling development in Arabidopsis thaliana. J. Biol. Chem. 287: 25995–26009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025. [DOI] [PubMed] [Google Scholar]
- Sullivan J.A., Deng X.W. (2003). From seed to seed: the role of photoreceptors in Arabidopsis development. Dev. Biol. 260: 289–297. [DOI] [PubMed] [Google Scholar]
- Tepperman J.M., Hudson M.E., Khanna R., Zhu T., Chang S.H., Wang X., Quail P.H. (2004). Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 38: 725–739. [DOI] [PubMed] [Google Scholar]
- von Arnim A.G., Deng X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045. [DOI] [PubMed] [Google Scholar]
- Wang Q., et al. (2016). Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354: 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. (2017). A CRY-BIC negative-feedback circuitry regulating blue light sensitivity of Arabidopsis. Plant J. 92: 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.Q., Sarmast M.K., Jiang J., Dehesh K. (2015). The transcriptional regulator BBX19 promotes hypocotyl growth by facilitating COP1-mediated EARLY FLOWERING3 degradation in Arabidbopsis. Plant Cell 27: 1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Li J., Gangappa S.N., Hettiarachchi C., Lin F., Andersson M.X., Jiang Y., Deng X.W., Holm M. (2014). Convergence of Light and ABA signaling on the ABI5 promoter. PLoS Genet. 10: e1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Jiang Y., Li J., Lin F., Holm M., Deng X.W. (2016). BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 113: 7655–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Jiang Y., Li J., Holm M., Deng X.W. (2018). B-box domain protein BBX21 promotes photomorphogenesis. Plant Physiol. 176: 2365–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., Deng X.W. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358. [DOI] [PubMed] [Google Scholar]
- Zhang X., Huai J., Shang F., Xu G., Tang W., Jing Y., Lin R. (2017). A PIF1/PIF3–HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol. 174: 2487–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]