A tightly regulated mechanism induced by blue light and brassinosteroids helps plants optimize photomorphogenesis based on the availability of external light signals and internal brassinosteroid signals.
Abstract
Cryptochromes (CRYs) are blue light photoreceptors that mediate a variety of light responses in plants and animals, including photomorphogenesis, flowering, and circadian rhythms. The signaling mechanism by which Arabidopsis thaliana cryptochromes CRY1 and CRY2 promote photomorphogenesis involves direct interactions with COP1, a RING motif-containing E3 ubiquitin ligase, and its enhancer SPA1. Brassinosteroid (BR) is a key phytohormone involved in the repression of photomorphogenesis, and here, we show that the signaling mechanism of Arabidopsis CRY1 involves the inhibition of BR signaling. CRY1 and CRY2 physically interact with BES1-INTERACTING MYC-LIKE1 (BIM1), a basic helix-loop-helix protein. BIM1, in turn, interacts with and enhances the activity of BRI1-EMS SUPPRESSOR1 (BES1), a master transcription factor in the BR signaling pathway. In addition, CRY1 and CRY2 interact specifically with dephosphorylated BES1, the physiologically active form of BES1 that is activated by BR in a blue light-dependent manner. The CRY1-BES1 interaction leads to both the inhibition of BES1 DNA binding activity and the repression of its target gene expression. Our study suggests that the blue light-dependent, BR-induced interaction of CRY1 with BES1 is a tightly regulated mechanism by which plants optimize photomorphogenesis according to the availability of external light and internal BR signals.
INTRODUCTION
Light is not only an energy source for photosynthesis but also an informational signal that profoundly influences plant growth and development throughout the plant lifecycle, from seed germination through vegetative growth to flowering and seed formation (Fankhauser and Chory, 1997; Deng and Quail, 1999). Plants have evolved several types of photoreceptors that perceive light signals and monitor dynamic changes in light quantity and quality. These include two types of blue/UV-A light receptors, the cryptochromes (CRYs) CRY1 and CRY2 and the phototropins PHOT1 and PHOT2, the red/far-red light-sensing phytochromes (PHYs) phyA to phyE, and the UV-B receptor UVR8 (Cashmore et al., 1999; Briggs and Christie, 2002; Quail, 2002; Rizzini et al., 2011). Among these, CRYs and PHYs act together to regulate a broad spectrum of physiological processes, including seedling photomorphogenesis, photoperiodic flowering, circadian rhythms, and stomatal development (Ahmad and Cashmore, 1993; Guo et al., 1998; Somers et al., 1998; Tóth et al., 2001; Mao et al., 2005; Liu et al., 2008; Kang et al., 2009). Cryptochromes are found not only in plants but also in a variety of other organisms from bacteria to humans.
In Drosophila melanogaster, cryptochrome acts as a photoreceptor to entrain the circadian clock, whereas in mammals, cryptochromes are integral components of the circadian clock (Emery et al., 1998; Kume et al., 1999). In migratory butterflies and birds, cryptochrome is responsible for sensing the Earth’s magnetic field and providing precise navigation during their long-distance migration (Gegear et al., 2010). Arabidopsis thaliana contains two homologous cryptochromes, CRY1 and CRY2. CRY1 plays a major role in mediating blue light-inhibited hypocotyl elongation, whereas CRY2 also inhibits hypocotyl growth, but primarily under low-intensity blue light (Ahmad and Cashmore, 1993; Lin et al., 1998). Consistent with these functions, CRY1, but not CRY2, is stable under high-intensity irradiation with blue light (Lin et al., 1998). CRY2 is the primary blue light photoreceptor promoting floral initiation under long-day conditions (Guo et al., 1998).
CRYs share amino acid sequence similarity to photolyases, flavoproteins that mediate light-dependent DNA repair (Sancar, 1994, 2003). However, CRYs lack photolyase activity and are characterized by distinct C-terminal domains (Lin et al., 1995). The C-terminal domains of Arabidopsis CRY1 and CRY2, known as CCT1 (CRY1 C terminus) and CCT2 (CRY2 C terminus) (Yang et al., 2000), mediate CRY signaling via direct interactions with COP1 (Wang et al., 2001a; Yang et al., 2001). COP1 is a RING-finger E3 ubiquitin ligase (Deng et al., 1992) that regulates photomorphogenesis and flowering via its interaction with and targeted degradation of specific transcription factors such as HY5 and CONSTANS (CO) (Osterlund et al., 2000; Jang et al., 2008; Liu et al., 2008). CRY1 and CRY2 also interact with the COP1 enhancer SPA1 (Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011), which in turn binds to and enhances the E3 ligase activity of COP1 (Seo et al., 2003). Interactions of CRY1 and CRY2 with COP1-SPA1 lead to the disruption of the COP1-SPA1 core complex, thus promoting the accumulation of HY5-CO. Dimerization of CRYs is mediated by CNT1 and CNT2, the N-terminal photolyase-homologous regions (PHRs) of CRY1 and CRY2 (Sang et al., 2005; Yu et al., 2007). The dimerization of CRY2 is inhibited by BIC1 (Wang et al., 2016a).
Overexpression of CNT1 in the cry1 mutant shortened hypocotyls in a blue-light-dependent manner. This phenotype was not related to HY5 protein accumulation, implying that CNT1 is involved in mediating CRY1 signaling independent of the presence of the C terminus of CRY1 (He et al., 2015). Recent studies have demonstrated that CRY1 and 2 interact with PIFs (phytochrome-interacting factors) to mediate responses to low levels of blue light or high temperature (Ma et al., 2016; Pedmale et al., 2016) and that the N terminus of CRY1 interacts with AUX/IAA proteins to stabilize these proteins and inhibit auxin signaling (Xu et al., 2018).
Brassinosteroids (BRs) are a group of polyhydroxylated plant steroid hormones that regulate numerous aspects of plant growth and development, including seed germination, stress tolerance, flowering, photomorphogenesis, and stomatal development (Vert et al., 2005; Kim et al., 2012; Zhu et al., 2013). Extensive studies of the BR signal transduction pathway have led to the discovery of key signaling components such as BRI1 (BRASSINOSTEROID INSENSITIVE1), BKI1 (BRI1 KINASE INHIBITOR1), BAK1 (BRI1-ASSOCIATED RECEPTOR KINASE1), BSU1 (BRI1-SUPPRESSOR1), BIN2 (BRASSINOSTEROID INSENSITIVE2), and BES1 (BRI1-EMS SUPPRESSOR1), also known as BZR1 (BRASSINAZOLE-RESISTANT1). When BR levels are high, BR is perceived by BRI1, a membrane-localized receptor kinase (Li and Chory, 1997; Kim et al., 2011). This releases the inhibitor BKI1 (Wang and Chory, 2006; Jaillais et al., 2011; Wang et al., 2011), activates the intracellular kinase domain of BRI1, and causes BRI1 to bind to its coreceptor BAK1 (Li et al., 2002; Nam and Li, 2002). Subsequently, the negative regulator BIN2 is dephosphorylated by BSU1 and inactivated through a multistep cascade of phosphorylation events (Tang et al., 2008; Kim et al., 2011), and the transcription factors BES1 and BZR1 are released from BIN2-induced phosphorylation or are dephosphorylated by PP2A (Wang et al., 2002; Yin et al., 2002; Zhao et al., 2002; Tang et al., 2011). Dephosphorylated BES1 and BZR1 can then bind to their target genes and regulate their expression, ultimately leading to BR responses (Yin et al., 2005; Vert and Chory, 2006; Sun et al., 2010). When BR levels are low, BIN2 is activated, and BZR1 and BES1 are phosphorylated by BIN2 and thus unable to bind to their target genes (Zhao et al., 2002; Vert and Chory, 2006). Therefore, the BR signaling mechanism involves BR-induced formation of the physiologically active, dephosphorylated forms of BES1 and BZR1.
A genetic screen for Arabidopsis photomorphogenic development mutants identified det2 (de-etiolated 2), a BR-deficient mutant with a shortened hypocotyl and expanded cotyledon phenotype in the dark (Chory et al., 1991; Li et al., 1996). Like det2, bri1, a loss-of-function mutant of a BR receptor, shows a shortened hypocotyl phenotype during photomorphogenesis (Clouse et al., 1996). BES1 and BZR1 are major transcription factors in BR signaling that play roles in regulating hypocotyl elongation. In the gain-of-function mutants of BES1 and BZR1, bes1-D and bzr1-1D, BES1 and BZR1 are highly stable. These mutants display BR-activated phenotypes including enhanced hypocotyl elongation and are insensitive to the BR biosynthesis inhibitor, brassinazole (BRZ) (Asami et al., 2000; Wang et al., 2002; Yin et al., 2002). By contrast, BES1-RNAi seedlings, in which both BES1 and BZR1 transcription is downregulated, have shortened hypocotyl and semidwarf phenotypes, similar to those of weak BR loss-of-function mutants (Yin et al., 2005). BES1 and BZR1 possess atypical basic helix-loop-helix (bHLH) DNA binding domains that have similar DNA binding specificities and transcriptional activities and can bind to E-boxes (CANNTG) and/or BR response elements (CGTGT/CG) (He et al., 2005; Yin et al., 2005; Yu et al., 2011). Many direct target genes of BZR1 and BES1 induced by BRs are involved in promoting cell elongation (Yin et al., 2005; Yu et al., 2011; Oh et al., 2012). BES1 and BZR1 can interact with other transcription factors involved in light signaling, such as PIF4 and HY5, to regulate transcription and hypocotyl elongation (Oh et al., 2012; Li and He, 2016). On the other hand, the protein stability of BES1 and BZR1 is negatively regulated by the E3 ubiquitin ligase COP1 or SINATs (SINA of Arabidopsis) through a 26S proteasome-dependent pathway (Kim et al., 2014; Yang et al., 2017). Clearly, BR plays an important role in light-regulated plant development.
In this study, we screened for proteins that interact with the N terminus of CRY1, and this led to the identification of BIM1 (BES1-INTERACTING MYC-LIKE1) as an interacting protein. BIM1 is a transcriptional regulator that interacts with BES1 to enhance its activity in BR signaling (Yin et al., 2005). We explored whether the signaling mechanism of CRY1 involved inhibition of BR signaling through direct interactions with BIM1 and/or BES1. Through a series of physiological, biochemical, genetic, and molecular studies, we demonstrate here that Arabidopsis CRY1 specifically interacts with dephosphorylated BES1 in a blue light-dependent manner to inhibit BR signaling and hypocotyl elongation. DNA-protein binding assays demonstrated that the CRY1-BES1 interaction leads to the inhibition of the DNA binding activity of BES1 and represses expression of its target genes, thereby inhibiting hypocotyl elongation. We show that BES1 is an important downstream component in the cryptochrome signaling pathway and that Arabidopsis CRY1 signaling involves the inhibition of BR signaling through its direct interaction with BES1. This interaction is thought to counteract the positive regulatory properties of the physiologically active, dephosphorylated form of BES1 in a blue light-dependent and BR-induced manner, thus inhibiting BR signaling and initiating the inhibition of hypocotyl elongation.
RESULTS
CRY1 Physically Interacts with BIM1
We previously demonstrated that the N-terminal PHR domain of CRY1 (CNT1) alone is sufficient to mediate blue light-inhibited hypocotyl elongation (He et al., 2015), prompting us to look for potential CNT1-interacting proteins. We performed GAL4 yeast two-hybrid screening using CNT1 and identified a clone encoding BIM1, a bHLH protein that interacts with BES1, the master transcription factor in the BR signaling pathway, to enhance its activity (Yin et al., 2005). To confirm the interaction between CNT1 and BIM1, we performed GAL4 yeast two-hybrid assays in darkness and blue light by cotransforming yeast cells with a bait construct expressing the GAL4 binding domain fused to CNT1 or CCT1 (C terminus of CRY1), together with a prey construct expressing BIM1 or COP1. CNT1 interacted with BIM1 in both darkness and blue light (Supplemental Figure 1A), whereas CCT1 did not, but it interacted with the COP1 control (Yang et al., 2001). We also tested whether CRY2 or its N terminus (CNT2) would interact with BIM1 and found that CRY2 and CNT2 interacted with BIM1 in a blue light-dependent manner (Supplemental Figure 1B). We performed in vitro pull-down assays using a GST-BIM1 fusion protein as bait and the MBP-CNT1, -CCT1, -CNT2, and -CCT2 (C terminus of CRY2) fusions as prey. CNT1, CNT2, and CCT2 were pulled down by BIM1 (Supplemental Figures 1C and 1D), but CCT1 and the MBP control were not. These results suggest that CNT1 mediates the interaction of CRY1 with BIM1, whereas both CNT2 and CCT2 mediate the interaction of CRY2 with BIM1.
To examine whether CRY1 interacts with BIM1 in vivo, we performed protein colocalization analysis by transiently expressing full-length CRY1, CNT1, or CCT1 tagged with YFP, together with BIM1 tagged with CFP, in tobacco (Nicotiana tabacum) leaves. Both CRY1 and CNT1 localized to the same nuclear bodies as BIM1, whereas CCT1 did not (Supplemental Figure 1E). We then performed bimolecular fluorescence complementation (BiFC) assays by coexpressing CRY1 fused to the C-terminal half of YFP (CRY1-cYFP) and BIM1 fused to the N-terminal half of YFP (nYFP-BIM1) in tobacco cells. As shown in Supplemental Figure 1F, YFP fluorescence signals were clearly observed in the nuclei of cells coexpressing nYFP-BIM1 and CRY1-cYFP, as well as nYFP-BIM1 and CNT1-cYFP. By contrast, no signals were detected in the nuclei of cells coexpressing nYFP-BIM1 and CCT1-cYFP, nYFP-CCT1 and CRY1-cYFP, or nYFP-CCT1 and CNT1-cYFP, but they were observed in those coexpressing CCT1-cYFP and nYFP-SPA1. These results again suggest that CNT1 mediates the interaction of CRY1 with BIM1. Next, we performed a coimmunoprecipitation (co-IP) assay with tobacco leaves coexpressing Flag-tagged BIM1 (BIM1-Flag) and Myc-tagged CRY1 proteins (Myc-CRY1). We found that CRY1 interacted with BIM1 (Supplemental Figure 1G). Furthermore, we performed pull-down experiments with GST-BIM1 and protein extracts prepared from Arabidopsis seedlings overexpressing Myc-CRY1 (Myc-CRY1-OX) adapted in the dark and exposed to blue, red, or far-red light. GST-BIM1 pulled down Myc-CRY1 from Myc-CRY1-OX seedlings exposed to blue light, but not from dark-adapted seedlings or seedlings exposed to red or far-red light (Supplemental Figure 1H). This indicated that the CRY1-BIM1 interaction is a specific response to blue light. BIM1 consists of three domains: the N-terminal domain, bHLH domain, and C-terminal domain (Yin et al., 2005). Protein colocalization assays suggested that the N-terminal domain of BIM1 is essential for the interaction of BIM1 with CRY1 and CNT1 (Supplemental Figures 2).
CRY1 and CRY2 Interact with BES1 in a Blue Light-Dependent Manner in Arabidopsis Cells
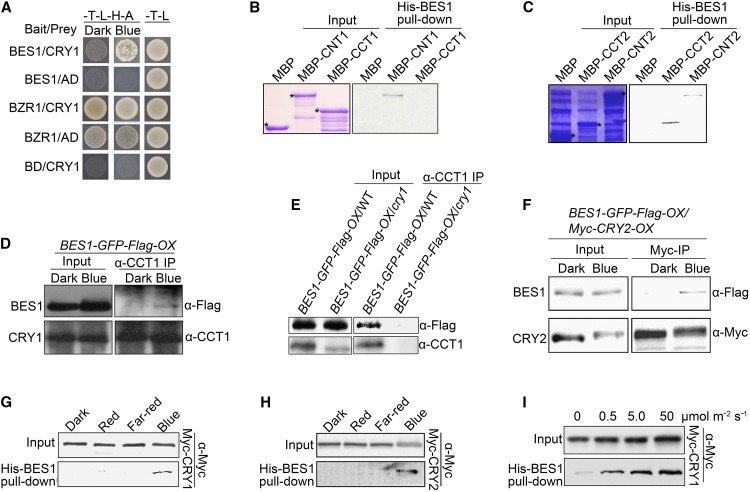
Because BIM1 is known to interact and cooperate with BES1 to regulate the expression of BR-responsive genes (Yin et al., 2005), we explored whether CRY1 might interact with BES1 and its close homolog, BZR1, in a yeast two-hybrid assay. CRY1 interacted with BES1 more strongly in blue light than in the dark, but it interacted with BZR1 strongly in both the dark and blue light (Figure 1A). It also appeared that CNT1 but not CCT1 mediated the interaction of CRY1 with BES1 (Supplemental Figure 3A). We then performed His-BES1 pull-down experiments with MBP-CNT1, -CCT1, -CNT2, and -CCT2 fusion proteins. As shown in Figures 1B and 1C, CNT1, CNT2, and CCT2 were pulled down by BES1, whereas MBP and CCT1 were not. These results indicate that CNT1, like BIM1, mediates the interaction of CRY1 with BES1, whereas both CNT2 and CCT2 mediate the interaction of CRY2 with BES1.
Figure 1.
CRY1 and CRY2 Physically Interact with BES1 in a Blue Light-Specific Manner.
(A) Yeast two-hybrid assay showing interactions of CRY1 with BES1 and BZR1. Yeast cells coexpressing the indicated combinations of constructs were grown on SD medium lacking Trp and Leu (SD-T-L) or Trp, Leu, His, and Ade (SD-T-L-H-A) medium in continuous darkness or blue light (30 μmol m−2 s−1). BD, DNA binding domain of GAL4; AD, DNA activation domain of GAL4 .
(B) and (C) His-BES1 pull-down assays showing the interactions of BES1 with CNT1 (B) or with CCT2 and CNT2 (C). His-BES1 served as bait. MBP, MBP-CNT1, -CCT1, -CCT2, and -CNT2 served as preys. Input images show Coomassie blue staining. Asterisks denote the indicated proteins. Two independent experiments were performed, with similar results.
(D) and (E) Co-IP assay showing the blue light-induced interaction of CRY1 with BES1 in Arabidopsis. BL-treated BES1-GFP-Flag-OX seedlings in the wild type ([D] and [E]) or cry1 mutant (E) background were dark-adapted or exposed to blue light (100 μmol m−2 s−1) for 1 h, followed by immunoprecipitation with an anti-CCT1 antibody. The IP (CRY1) and co-IP signals (BES1) were detected in immunoblots probed with anti-CCT1 and -Flag antibodies, respectively.
(F) Co-IP assay showing blue light-induced interaction of CRY2 with BES1 in Arabidopsis. BES1-GFP-Flag-OX/Myc-CRY2-OX seedlings treated with BL and MG132 were dark-adapted or exposed to blue light (50 μmol m−2 s−1) for 1 h, followed by immunoprecipitation with an anti-Myc antibody. The IP (CRY2) and co-IP signals (BES1) were detected in immunoblots probed with anti-Myc and -Flag antibodies, respectively.
(G) and (H) His-BES1 pull-down assays showing blue light-specific interactions of CRY1 and CRY2 with BES1. His-BES1 served as bait. Preys were protein extracts prepared from Myc-CRY1-OX (G) and MG132-treated Myc-CRY2-OX (H) seedlings that were dark-adapted and exposed to 100 μmol m−2 s−1 (G) or 50 μmol m−2 s−1 blue light (H), 20 μmol m−2 s−1 red light, or 10 μmol m−2 s−1 far-red light for 1 h, respectively. Two independent experiments were performed, with similar results.
(I) His-BES1 pull-down assay showing that CRY1-BES1 interactions are enhanced in response to increasing levels of blue light. His-BES1 served as bait. Preys were Myc-CRY1-containing protein extracts prepared from Myc-CRY1-OX seedlings that were dark-adapted or exposed to blue light for 1 h at the fluence rates noted above the image. Two independent experiments were performed, with similar results.
To investigate whether CRY1 and CRY2 interact with BES1 and BZR1 in vivo, we performed co-IP assays with protein extracts prepared from tobacco leaves expressing BES1-Flag together with Myc-CRY1 or Myc-CRY2. After we found that both CRY1 and CRY2 interacted with BES1 (Supplemental Figures 3B and 3C), we next performed BiFC assays with tobacco cells expressing nYFP-BES1 or nYFP-CCT1 together with CRY1-cYFP, CNT1-cYFP, or CCT1-cYFP and found that CRY1 and CNT1 interacted with BES1, whereas CCT1 did not (Supplemental Figure 3D). Furthermore, CRY1, CRY2, CNT1, and CNT2 localized to the same nuclear bodies as BZR1 (Supplemental Figure 3E), and both CRY1 and CRY2 interacted with BZR1 in a BiFC assay (Supplemental Figure 3F). Because it is unlikely that BES1 is phosphorylated in yeast or Escherichia coli and it interacted with CRY1 and CNT1 (Figures 1A and 1B), we reasoned that CRYs might interact with dephosphorylated BES1 in vivo.
To examine whether CRY1 interacts with BES1 in vivo, we generated transgenic seedlings overexpressing BES1 tagged with GFP and Flag in the wild-type and cry1 mutant backgrounds (BES1-GFP-Flag-OX/WT and BES1-GFP-Flag-OX/cry1) and subjected them to co-IP assays. We tagged BES1 with Flag plus GFP instead of Flag alone because the molecular weight of BES-Flag fusion protein is similar to that of the heavy chain of IgG, which might interfere with observing the co-IP signal. To ensure that the BES1-GFP-Flag fusion protein was biologically functional, we produced transgenic Arabidopsis plants overexpressing the same gain-of-function mutant form of BES1 as in the bes1-D mutant (Yin et al., 2002) tagged with GFP and Flag (mbes1-GFP-Flag-OX) and analyzed their hypocotyl elongation phenotype. The mbes1-GFP-Flag-OX seedlings exhibited an enhanced hypocotyl elongation phenotype in the dark as well as blue, red, and far-red light (Supplemental Figure 4), as reported for bes1-D (Yin et al., 2002). Moreover, treatment with brassinolide (BL) the most active form of BR (Wang et al., 2001b), induced the formation of dephosphorylated BES1-GFP-Flag (see below). These results indicate that the fusion of GFP and Flag to BES1 does not affect its function.
We performed co-IP assays with these BL-treated seedlings, which were adapted in the dark and then exposed to blue light for 1 h. As shown in Figures 1D and 1E, endogenous CRY1 coprecipitated with dephosphorylated BES1 in extracts from blue light-irradiated BES1-GFP-Flag-OX/WT seedlings, but not from blue light-irradiated BES1-GFP-Flag-OX/cry1 seedlings or dark-grown BES1-GFP-Flag-OX/WT seedlings. We also created transgenic lines overexpressing both Myc-CRY2 and BES1-GFP-Flag (BES1-GFP-Flag-OX/Myc-CRY2-OX) by performing genetic crosses between BES1-GFP-Flag-OX and Myc-CRY2-OX plants, followed by co-IP assays of seedlings treated with BL that were dark-adapted and exposed to blue light for 1 h. CRY2 interacted with dephosphorylated BES1 in blue light, but not in the dark (Figure 1F). These results demonstrate that CRY1 and CRY2 interact with BES1 in a blue light-dependent manner in Arabidopsis.
To further investigate the effects of light quality and quantity on the interactions of CRY1 and CRY2 with BES1, we performed His-BES1 pull-down experiments with Myc-CRY1- and Myc-CRY2-containing protein extracts prepared from Myc-CRY1-OX and Myc-CRY2-OX seedlings adapted in darkness and exposed to blue, red, or far-red light. BES1 pulled down CRY1 and CRY2 in extracts prepared from seedlings exposed to blue light (Figures 1G and 1H), but not from dark-adapted seedlings or seedlings exposed to red or far-red light. Moreover, the amount of CRY1 pulled down by BES1 increased as the blue light fluence rate increased (Figure 1I). Taken together, these results suggest that CRY1 and CRY2 interact with BES1 in a blue light-dependent manner in plant cells.
Photoexcited CRY1 and CRY2 Interact Specifically with Dephosphorylated BES1
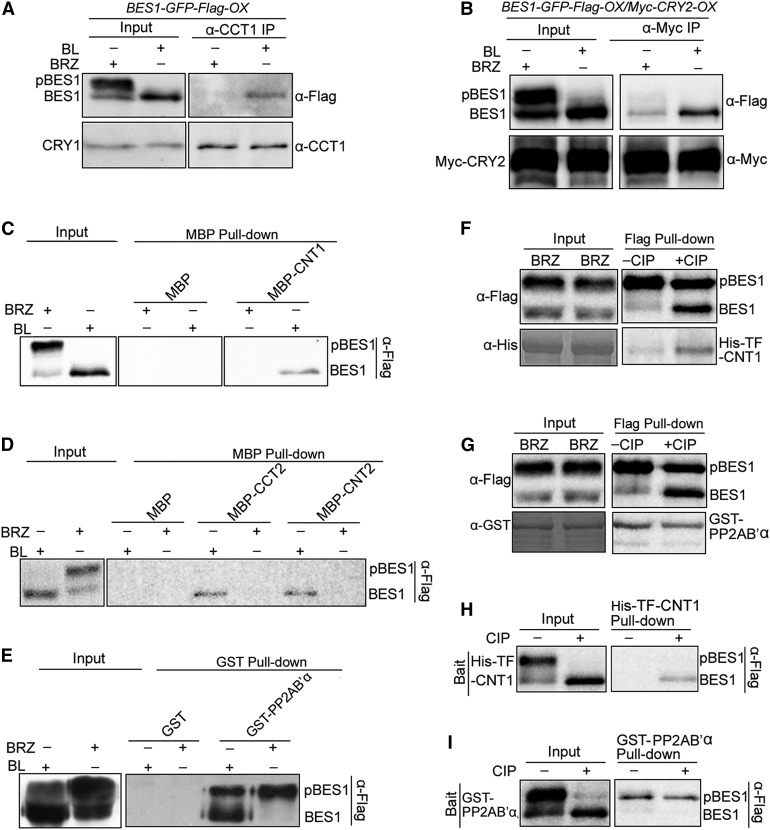
Arabidopsis contains two forms of BES1: physiologically active dephosphorylated BES1 and physiologically inactive phosphorylated BES1, which are induced by BL and the BR biosynthesis inhibitor BRZ, respectively (Asami et al., 2000; Yin et al., 2002; Vert and Chory, 2006). To determine whether phosphorylation affects the interactions of BES1 with CRYs, we performed co-IP assays with blue light-irradiated BES1-GFP-Flag-OX/WT and (BES1-GFP-Flag-OX/Myc-CRY2-OX) seedlings that had been treated with BL or BRZ. Intriguingly, endogenous CRY1 coprecipitated with dephosphorylated BES1, but not with phosphorylated BES1 (Figure 2A). Similarly, CRY2 specifically coprecipitated with dephosphorylated BES1 (Figure 2B), indicating that photoexcited CRY1 and CRY2 interact specifically with dephosphorylated BES1.
Figure 2.
CRY1 and CRY2 Interact Specifically with Dephosphorylated BES1.
(A) Co-IP assay showing the specific interaction of CRY1 with the dephosphorylated form of BES1 in Arabidopsis. Dark-adapted BES1-GFP-Flag-OX seedlings (in the wild type background) were treated with BL or BRZ and exposed to blue light (100 μmol m−2 s−1) for 1 h, followed by immunoprecipitation with an anti-CCT1 antibody. The IP (CRY1) and co-IP signals (BES1) were detected in immunoblots probed with anti-CCT1 and -Flag antibodies, respectively. pBES1 and BES1 represent phosphorylated BES1 and dephosphorylated BES1, respectively.
(B) Co-IP assay showing the specific interaction of CRY2 with the dephosphorylated form of BES1 in Arabidopsis. Dark-adapted (BES1-GFP-Flag-OX/Myc-CRY2-OX) seedlings were treated with BL or BRZ and MG132 and exposed to blue light (50 μmol m−2 s−1) for 1 h, followed by immunoprecipitation with an anti-Myc antibody. The IP (CRY2) and co-IP signals (BES1) were detected in immunoblots probed with anti-Myc and -Flag antibodies, respectively.
(C) to (E) Pull-down assays showing specific interactions of CNT1, CNT2, and CCT2 with the dephosphorylated form of BES1. MBP, MBP-CNT1, -CNT2, -CCT2, and GST-PP2AB’α served as baits. BES1-Flag-containing protein extracts prepared from BES1-Flag-OX/WT seedlings treated with BL or BRZ served as preys. Two or three independent experiments were performed, with similar results.
(F) to (I) Pull-down assays showing the interaction of CNT1 with purified dephosphorylated BES1. BES1 and pBES1 were prepared by immunoprecipitation of protein extracts from BRZ-treated BES1-Flag-OX/WT seedlings using anti-Flag beads, followed by treatment with or without calf intestine phosphatase. Flag bead-bound BES1 and pBES1 served as baits ([F] and [G]), and BES1 and pBES1 eluted from the Flag beads served as preys ([H] and [I]). PP2AB’α interacted with purified phosphorylated BES1, which served as a control. Two independent experiments were performed, with similar results.
To confirm the specific interactions of CRY1 and CRY2 with dephosphorylated BES1, we generated transgenic Arabidopsis plants overexpressing BES1-Flag (BES1-Flag-OX/WT) and performed pull-down assays using MBP-CNT1, -CNT2, -CCT2, and MBP as baits and protein extracts prepared from BES1-Flag-OX/WT seedlings treated with BL and BRZ as preys. The GST-PP2AB’α fusion protein, which preferably binds to phosphorylated BZR1, served as a control (Tang et al., 2011). As shown in Figures 2C and 2D, CNT1, CNT2, and CCT2 pulled down dephosphorylated BES1 but not phosphorylated BES1. By contrast, PP2AB’α preferentially pulled down phosphorylated BES1 (Figure 2E). Because treatment with calf intestine phosphatase increases the formation of dephosphorylated BES1 (Fankhauser et al., 1999; Yin et al., 2002), we conducted a pull-down assay using purified phosphorylated and dephosphorylated BES1 proteins as bait or prey, which were obtained through immunoprecipitation of extracts prepared from BRZ-treated BES1-Flag-OX/WT seedlings using anti-Flag beads and subsequent treatment with or without CIP. When BES1 was used as bait, CNT1 was primarily pulled down by dephosphorylated BES1 (Figure 2F), whereas PP2AB’α was pulled down by phosphorylated BES1 (Figure 2G). When BES1 was used as prey, only dephosphorylated BES1 was pulled down by CNT1 (Figure 2H), whereas phosphorylated BES1 was pulled down by PP2AB’α (Figure 2I). Taken together, these results demonstrate that CRY1 and CRY2 interact specifically with dephosphorylated BES1.
CRY1 and CRY2 Mediate Blue Light Inhibition of BR Responses
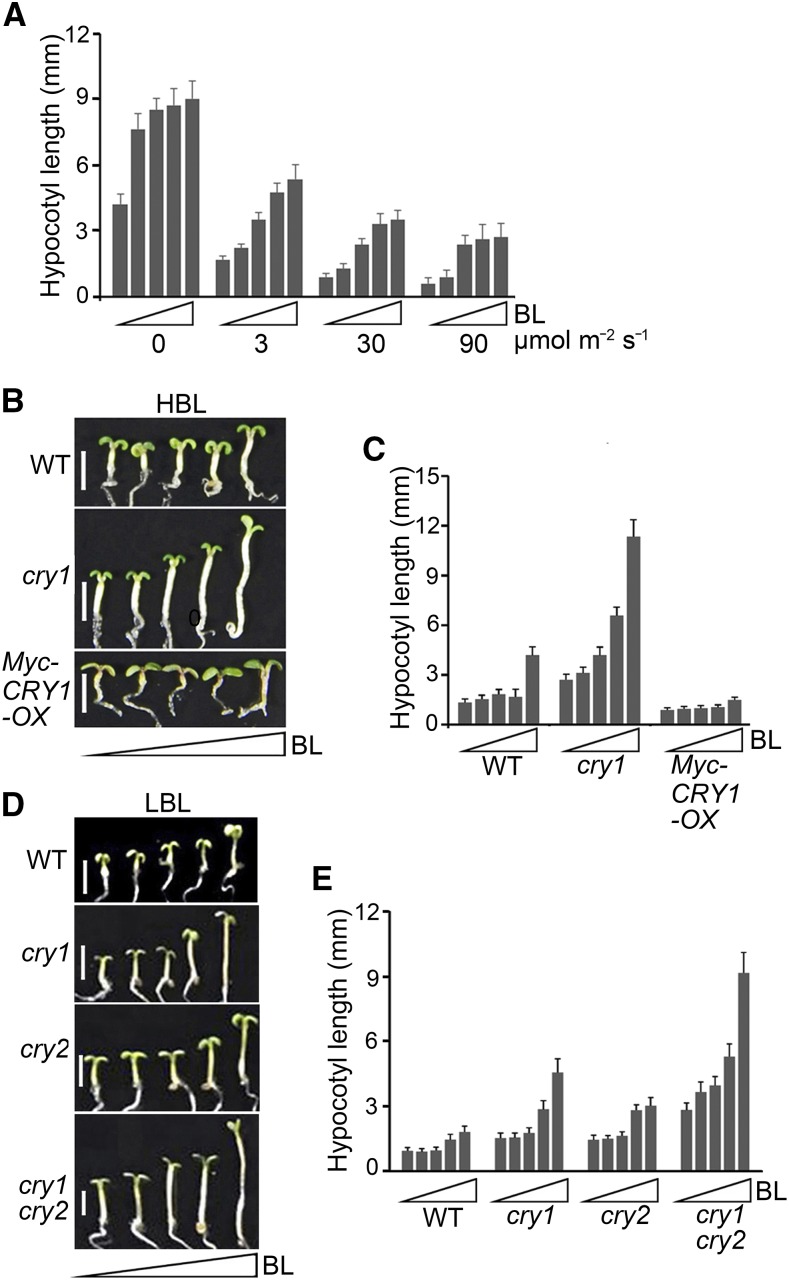
To examine the physiological significance of the interactions of CRY1 and CRY2 with BES1, we analyzed the responsiveness of dark- and blue light-grown wild-type seedlings to BRZ. Dark-grown seedlings showed significantly greater inhibition of hypocotyl elongation in response to BRZ than blue light-grown seedlings (Supplemental Figures 5A and 5B). We then analyzed the responsiveness of blue light-grown wild-type, cry1, and Myc-CRY1-OX seedlings to BRZ. The cry1 mutant was significantly more responsive to BRZ than the wild type, whereas Myc-CRY1-OX seedlings were significantly less responsive to BRZ than the wild type (Supplemental Figures 5C and 5D). These results suggest that CRY1-mediated blue light signaling inhibits BR biosynthesis under continuous blue light. To minimize the potential differences in BR contents in wild-type seedlings grown in darkness versus blue light or in cry1, cry2, cry1 cry2, and Myc-CRY1-OX seedlings grown in blue light in subsequent studies, we examined the responsiveness of these seedlings to gradually increasing concentrations of BL in the presence of 2 μM BRZ, the concentration we found to have the greatest inhibitory effect on BR biosynthesis. Blue light-grown wild-type seedlings exhibited much less sensitivity to BL than dark-grown seedlings (Figure 3A; Supplemental Figure 6A). Moreover, as the blue light fluence rate increased, the sensitivity of wild-type seedlings to BL decreased. These results suggest that blue light suppresses BR signaling. Because CRY1 inhibits hypocotyl elongation under both high and low levels of blue light (HBL and LBL), whereas CRY2 represses hypocotyl elongation under LBL (Lin et al., 1998), we investigated the involvement of CRY1 and CRY2 in regulating BR responses in HBL and LBL, respectively. Under HBL, as the BL concentration increased, the cry1 mutant showed significantly more responsiveness to BL than the wild type, whereas Myc-CRY1-OX seedlings responded very little to BL (Figures 3B and 3C; Supplemental Figure 6B) This suggested that CRY1 mediates the inhibition of BR responses by HBL. Under LBL, both the cry1 and cry2 single mutants were more sensitive to BL than the wild type, but the cry1 cry2 double mutant was much more sensitive to BL than the cry1 and cry2 single mutants (Figures 3D and 3E; Supplemental Figure 6C). These results indicate that CRY1 and CRY2 act together to mediate LBL inhibition of BR responses.
Figure 3.
CRYs Are Involved in Mediating the Inhibition of BR Responses in Arabidopsis Seedlings in Response to Blue Light.
(A) Analysis of blue light inhibition of BR responses. Wild type seedlings were grown for 6 d on 0.5× MS plates supplemented with 2 μM BRZ plus a gradient of concentrations of BL (small triangles represent 0, 1, 10, 100, and 1000 nM) in the dark and increasing intensities of blue light (0, 3, 30, and 90 μmol m−2 s−1), and then hypocotyl length was measured. Three independent experiments were performed, with similar results.
(B) and (C) Analysis of the involvement of CRY1 in the inhibition of BR responses by high-intensity blue light (HBL). Wild-type, cry1, and Myc-CRY1-OX seedlings were grown on the same medium as in (A) in blue light (30 μmol m−2 s−1) for 6 d (B), and hypocotyl length was measured (C). Triangles represent concentration gradients of BL from 0 to 1000 nM. Bars in (B) = 2.5 mm. Two independent experiments were performed, with similar results.
(D) and (E) Analysis of the involvement of CRY1 and CRY2 in the inhibition of BR responses by low-intensity blue light (LBL). Wild-type, cry1, cry2, and cry1 cry2 seedlings were grown on 0.5× MS plates supplemented with 2 μM BRZ plus gradient concentrations of BL (0, 1, 10, 100, and 1000 nM) under low-intensity blue light (5 μmol m−2 s−1) for 5 d (D), and hypocotyl length was measured (E). Triangles represent concentration gradients of BL from 0 to 1000 nM. Bars in (D) = 2.5 mm.
Data in (A), (C), and (E) are means ± sd (n > 20 seedlings per treatment and genotype).
BES1 Acts Genetically Downstream of CRY1 and CRY2 to Regulate Hypocotyl Elongation in Blue Light
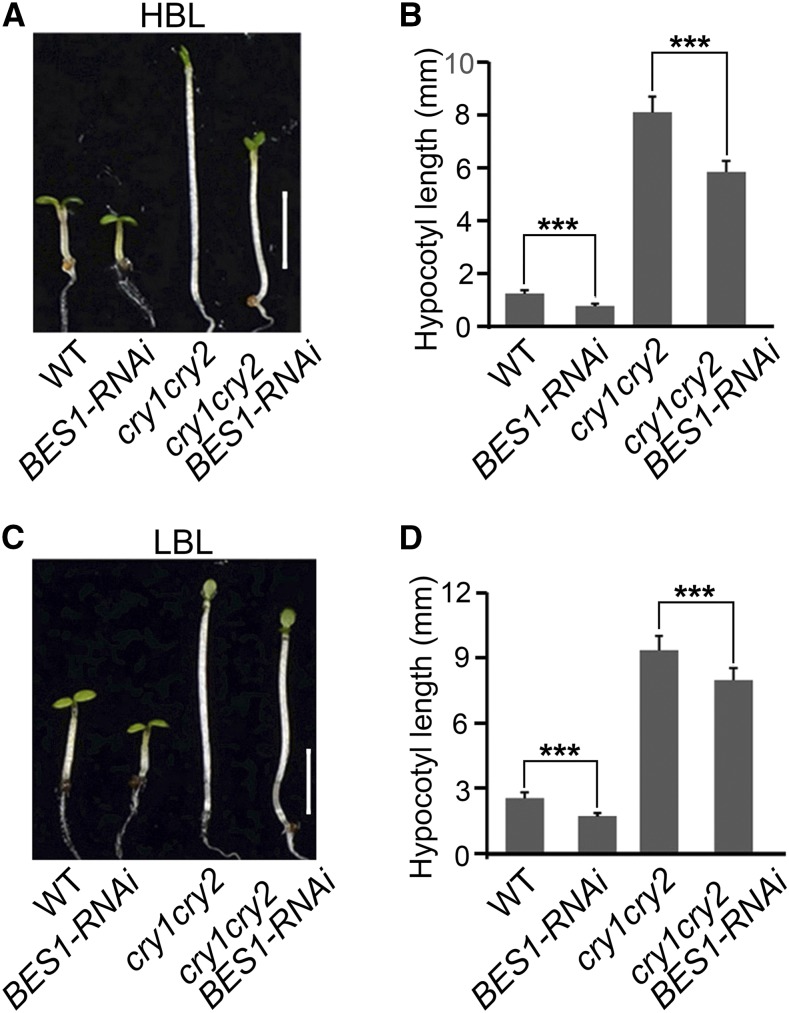
To evaluate the role of BES1 in regulating photomorphogenic development, we examined hypocotyl elongation in BES1-RNAi seedlings in which both BES1 and BZR1 were downregulated (Yin et al., 2005) in blue, red, and far-red light. As expected, BES1-RNAi seedlings developed significantly shorter hypocotyls than the wild type in darkness, blue, red, and far-red light (Supplemental Figure 7). We then constructed transgenic Arabidopsis plants overexpressing the same gain-of-function mutant form of BES1 found in the bes1-D mutant (Yin et al., 2002) tagged with Flag (mbes1-Flag-OX) (Supplemental Figure 8A). We found that the mbes1-Flag-OX seedlings exhibited an enhanced hypocotyl elongation phenotype in blue light (Supplemental Figures 8B and 8C). These results indicate that BES1 promotes hypocotyl elongation in darkness, blue, red, and far-red light. Next, we introgressed BES1-RNAi into the cry1 cry2 double mutant background by genetic crossing and generated the cry1 cry2 BES1-RNAi line. The cry1 cry2 BES1-RNAi seedlings developed significantly shorter hypocotyls than cry1 cry2 in both LBL and HBL (Figure 4; Supplemental Figures 6D and 6E), especially in HBL. Given the observations that CRY1 and CRY2 interact with BES1 and BZR1 (Figures 1 and 2; Supplemental Figure 3) and that CRY1 and CRY2 have other downstream components such as COP1 and SPAs (Wang et al., 2001a; Yang et al., 2001; Lian et al., 2011; Liu et al., 2011), these results suggest that BES1 and BZR1 act downstream of CRY1 and CRY2 to regulate hypocotyl elongation in blue light.
Figure 4.
BES1 Acts Genetically Downstream of CRY1 and CRY2 to Regulate Hypocotyl Elongation in Blue Light.
Genetic interaction analysis showing that BES1 and BZR1 act downstream of CRY1 and CRY2 in regulating hypocotyl elongation under high ([A] and [B]) and low ([C] and [D]) intensities of blue light. Wild-type, BES1-RNAi, cry1 cry2, and cry1 cry2 BES1-RNAi seedlings were grown in high-intensity blue light (HBL; 30 μmol m−2 s−1) (A) or low-intensity blue light (LBL; 5 μmol m−2 s−1) (C) for 5 d, and hypocotyl length was then measured ([B] and [D]). Bars in (A) and (C) = 2.5 mm. Two independent experiments were performed, with similar results. Data in (B) and (D) are means ± sd (n > 20 seedlings per treatment and genotype); asterisks indicate significant difference at P < 0.001.
CRY1 Negatively Regulates the Expression of BES1 Target Genes
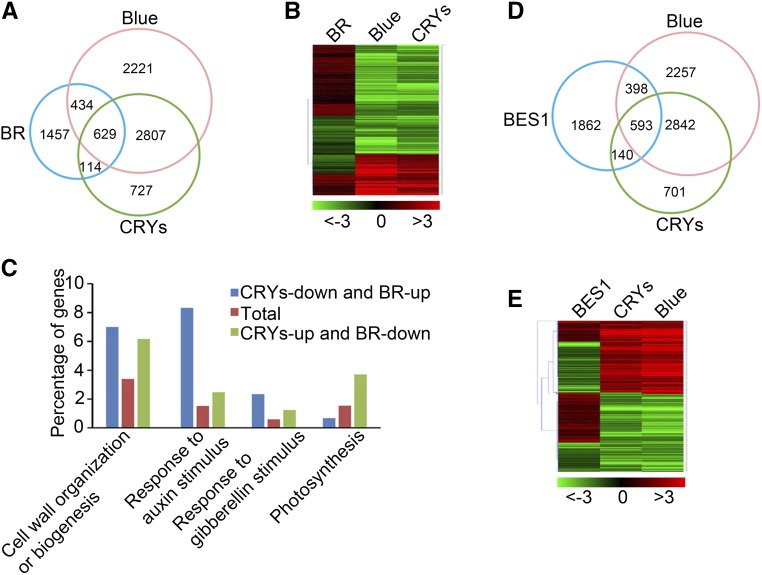
Our findings that CRY1 and CRY2 interact with BES1 and that BES1 acts downstream of CRY1 and CRY2 to regulate hypocotyl elongation in blue light suggest that CRYs might regulate the expression of BES1 target genes. To test this possibility, we compared genes regulated by blue light and CRYs (He et al., 2015) with those regulated by BRs (Oh et al., 2014) and identified 629 genes that are coregulated by CRY-mediated blue light signaling and BR signaling (Figure 5A; Supplemental Data Set 1), including 381 genes (60%) with opposite expression patterns (Figure 5B). Gene Ontology (GO) analysis indicated that the GO terms involving responses to growth-promoting hormones including auxin and gibberellins, as well as those involved in cell wall organization or biogenesis, were highly enriched among genes that are positively regulated by BL but negatively regulated by CRY-mediated blue light signaling. By contrast, the genes that were repressed by BL but activated by CRY-mediated blue light signaling are mainly involved in cell wall organization or biogenesis and photosynthesis (Figure 5C). We then compared the genes regulated by blue light and CRYs with those regulated by BES1, which were identified in BL-treated bes1-D seedlings (Yu et al., 2011), and identified 593 genes that were coregulated by CRY-mediated blue light signaling and BES1 (Figure 5D; Supplemental Data Set 2), including 368 genes (65%) that exhibited opposite expression patterns (Figure 5E).
Figure 5.
CRYs Regulate Numerous BR- and BES1-Regulated Genes.
(A) Venn diagram showing the sets of genes regulated by BR, blue light, and/or CRYs. BR, genes regulated by brassinosteroid (WT+BL versus WT-BL); Blue, genes regulated by blue light (WT-blue versus WT-dark); CRYs, genes regulated by CRY1 and CRY2 (WT versus cry1 cry2), respectively.
(B) Heat map of the 629 overlapping genes shown in (A) regulated by BR, blue light, and CRYs. Red and green represent induced and repressed genes, respectively. The colored bar beneath the map denotes the log2 value of fold change.
(C) GO analysis of genes shown in (B) that are downregulated by CRYs and upregulated by BR or upregulated by CRYs and downregulated by BR. The numbers on each column denote the percentage of genes in each GO category.
(D) Venn diagram showing the overlapping genes among genes regulated by blue light, CRYs, and BES1. BES1, blue, and CRYs indicate genes regulated by BES1, blue, and CRYs, respectively. BES1, genes regulated by BES1 (bes1-D+BL versus WT+BL); Blue, genes regulated by blue light (WT-blue versus WT-dark); CRYs, genes regulated by CRY1 and CRY2 (WT versus cry1 cry2).
(E) Heat map of the 593 overlapping genes shown in (D) regulated by blue light, CRYs, and BES1. Red and green represent induced and repressed genes, respectively. The colored bar beneath the map denotes the log2 value of fold change.
Next, we chose six BR-activated genes, ACS5, PRE5, PRE6, IAA19, SAUR15 (also known as SAUR-AC1), and SAUR68 (also known as SAUR-AC1-LIKE), which are direct target genes of BES1/BZR1 and promote hypocotyl elongation (Yin et al., 2002; Yu et al., 2011; Oh et al., 2012; Wang et al., 2016b), and subjected them to RT-qPCR analysis in wild-type and cry1 seedlings. The seedlings were grown in darkness for 5 d and exposed to blue light for 1 h with and without BL treatment. These genes were more strongly upregulated by BL in cry1 than in the wild type (Supplemental Figure 9A). Moreover, we examined the dynamic changes in the expression of BR biosynthesis genes DWF4 and CPD, which are also direct target genes of BES1 and/or BZR1 and are known to be repressed by BR (Vert and Chory, 2006; Yu et al., 2011), in wild-type and cry1 seedlings treated with BL under blue light conditions. As shown in Supplemental Figure 9B, the expression levels of DWF4 and CPD were significantly lower in cry1 than in the wild type at 0.5 to 2 h and 0.5 h after BL treatment, respectively. These results suggest that CRY1 regulates the expression of BES1/BZR1 target genes in response to blue light.
CRY1 Inhibits the DNA Binding Activity of BES1
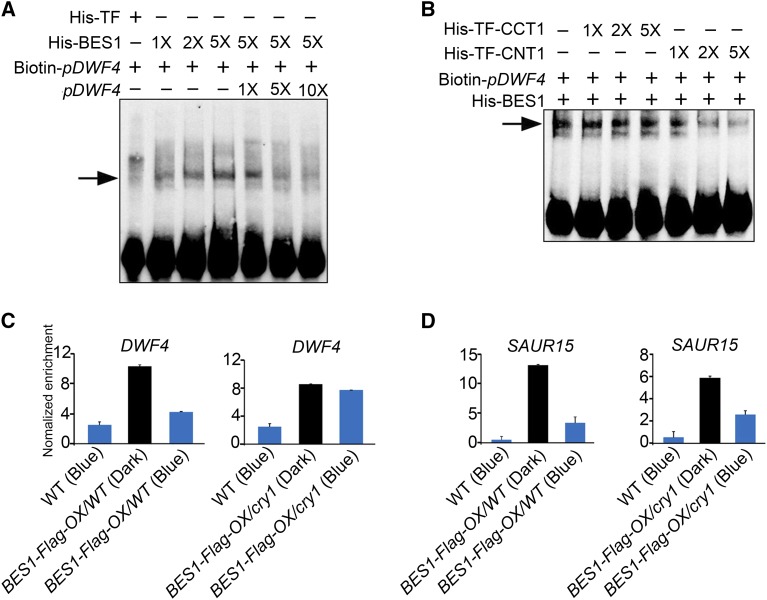
The finding that CRY1 directly interacts with dephosphorylated BES1 to regulate the expression of its target genes and BR responses prompted us to investigate whether CRY1 regulates the DNA binding activity of BES1. To test this possibility, we performed electrophoretic mobility shift assays (EMSAs) to investigate the binding of BES1 to the promoter sequence of the BR signaling marker gene, DWF4, which is also a BES1 target gene (Yu et al., 2011). Control experiments showed that BES1 bound to the promoter sequence of DWF4, which contains an E-box sequence (CANNTG), in a protein concentration-dependent manner and that the binding capacity was reduced by competition from cold competitor (Figure 6A). By contrast, this binding was not detected using His-tagged trigger factor (His-TF). As expected, the intensity of DNA bands whose size shifted in response to BES1 decreased with increasing amounts of CNT1 (Figure 6B), but did not with increasing amounts of CCT1. These results indicate that CNT1, but not CCT1, inhibits the DNA binding activity of BES1 in vitro, consistent with the finding that CNT1 but not CCT1 interacts with BES1 (Figures 1B and 2C; Supplemental Figures 3A and 3D).
Figure 6.
CRY1 Inhibits the DNA Binding Activity of BES1.
(A) and (B) An EMSA showing that CNT1 inhibits the DNA binding activity of BES1. EMSA was performed with His-BES1 and His-TF proteins using Biotin-pDWF4 as probes. 1×, 2×, 5× and 1×, 5×, 10× in (A) indicate the amount of His-BES1 and cold competitor DNA relative to the initial concentration of His-BES1 and Biotin-pDWF4 probes, respectively. 1×, 2×, and 5× in (B) indicate the amount of His-TF-CCT1 and His-TF-CNT1 relative to that of His-BES1, respectively. Arrows indicate the BES1-DNA complex. Signals at the bottom indicate free probes.
(C) and (D) ChIP-qPCR assay showing that CRY1 inhibits the DNA binding activity of BES1 in Arabidopsis. ChIP-qPCR was performed with BES1-Flag-OX seedlings in the wild type and cry1 mutant background treated with BL for 3 h and dark-adapted or exposed to blue light (100 μmol m−2 s−1) for 1 h. The promoter fragments of DWF4 (C), SAUR15 (D), and TA3 (negative control) were immunoprecipitated with an anti-Flag antibody. IP/input was calculated via a comparison with the Ct values between the immunoprecipitate and input, and relative enrichment was normalized to the negative control TA3. Data are mean values of three biological replicates ± sd.
To further explore whether CRY1 regulates the DNA binding activity of BES1 in planta, we crossed BES1-Flag-OX/WT with cry1 mutant plants and obtained cry1 mutant plants overexpressing BES1-Flag (BES1-Flag-OX/cry1). Unexpectedly, the BES1-Flag protein level was much higher in the wild type than in the cry1 mutant background due to a certain degree of gene silencing (Supplemental Figures 10A and 10B). We performed ChIP-qPCR (chromatin immunoprecipitation followed by quantitative PCR) assays with BL-treated BES1-Flag-OX/WT and BES1-Flag-OX/cry1 plants that were first dark-adapted and then exposed to blue light for 1 h. Immunoblot analysis indicated that 1 h of blue light irradiation did not affect the level of dephosphorylated BES1 protein in plants in the wild type and cry1 mutant backgrounds (Supplemental Figure 10C). We then evaluated the effects of 1 h blue light irradiation on the binding of BES1 to the promoters of BR signaling marker genes DWF4 and SAUR15/SAUR-AC1 (Yu et al., 2011) in the wild-type and cry1 backgrounds. Compared with darkness, 1 h of blue light irradiation led to ∼60% and 10% decreases in the binding of BES1 to DWF4 in the wild type and cry1, respectively (Figure 6C). Similarly, 1 h of blue light exposure resulted in ∼74 and 56% decreases in the binding of BES1 to SAUR15 in the wild type and cry1, respectively (Figure 6D). These results indicate that the binding of BES1 to both DWF4 and SAUR15 is repressed by photoactivated CRY1, especially its binding to DWF4. These results, together with the finding that CRY1 physically interacts with BES1 in a blue light-dependent manner (Figures 1D to 1G), suggest that the blue light-triggered interaction of CRY1 with BES1 results in the inhibited DNA binding activity of BES1 in planta.
DISCUSSION
The C terminus of CRY1 mediates CRY1 signaling through direct interactions with COP1 and SPA1 (Wang et al., 2001a; Yang et al., 2001; Lian et al., 2011; Liu et al., 2011). The N terminus of CRY1 serves as a FAD/FMN chromophore binding domain and mediates the homodimerization of CRY1 (Lin et al., 1995; Sang et al., 2005). We recently showed that the N terminus of CRY1 is involved in mediating CRY1 signaling independently of its C terminus (He et al., 2015). In this study, our screen for CRY1 N terminus-interacting proteins led to the identification of BIM1 as a new CRY1-interacting protein. This transcriptional regulator interacts with the master transcription factor BES1 in the BR signaling pathway to enhance BES1 activity. This finding prompted us to investigate a possible role for cryptochrome in mediating blue light repression of BR signaling. We successfully identified the molecular link between cryptochrome signaling and BR signaling, which is mediated through a blue light-dependent interaction of CRY1 with BES1. Because BES1 plays a major role in regulating BR signaling and BIM1 appears to enhance BES1 activity (Yin et al., 2005), we focused on the mechanism by which CRY1 mediates blue light-regulated inhibition of BR signaling through the CRY1-BES1 interaction.
We performed a series of biochemical assays to evaluate the direct interactions of CRY1 and CRY2 with BES1, including yeast two-hybrid, pull-down, protein colocalization, BiFC, and co-IP assays. The combined results from these assays clearly demonstrated that CRY1 and CRY2 interacted specifically with dephosphorylated BES1 in a blue light-dependent manner and that the capacity for the CRY1-BES1 interaction positively correlated with the intensity of blue light. Given the previous finding that the formation of the physiologically active (dephosphorylated) form of BES1 is induced by BR, our results indicate that both blue light and BR signals are required for the interactions of CRY1 and CRY2 with BES1. Both CRY1 and CRY2 interact with COP1 via their C termini (Yang et al., 2000; Wang et al., 2001a), and CRY1 interacts with SPA1 via its C terminus (Lian et al., 2011; Liu et al., 2011), whereas CRY2 interacts with SPA1 via its N terminus (Zuo et al., 2011). This study revealed differences in the domains that mediate the interactions of CRY1 and CRY2 with BIM1 and BES1. It appears that the N terminus of CRY1 mediates the interactions of CRY1 with BIM1 and BES1, whereas both the N and C termini of CRY2 mediate the interactions of CRY2 with BIM1 and BES1.
Our analysis of the responsiveness of wild type, cry1, cry2, cry1 cry2, and CRY1-overexpressing seedlings to BL suggested that blue light inhibits BR responses and that CRY1 and CRY2 are responsible for mediating this process. Based on previous studies (Lin et al., 1998) and these results, we conclude that CRY1 plays a role in inhibiting BR signaling under both high and low intensities of blue light, whereas CRY2 mainly mediates the repression of BR signaling by low intensities of blue light. Analysis of hypocotyl elongation phenotype of BES1-RNAi and mbes1-overexpressing seedlings under monochromatic light conditions demonstrated that BES1 promotes hypocotyl elongation in blue, red, and far-red light. Genetic interaction analysis of CRY1 and CRY2 with BES1 and BZR1 supported our conclusion that BES1 acts downstream of CRY1 and CRY2 to regulate hypocotyl elongation under blue light conditions. Consistent with the results of biochemical, physiological, and genetic analyses, genome-wide gene expression analysis indicated that CRY1 and CRY2 mediate blue light regulation of numerous BR-responsive genes and that a large proportion of BES1-regulated genes are regulated oppositely by CRY1 and CRY2. Furthermore, protein-DNA binding analyses, such as EMSA and ChIP-qPCR assays, in conjunction with protein-protein interaction analysis, indicated that the blue light-triggered CRY1-BES1 interaction results, consecutively, in the inhibition of the DNA binding activity of BES1, the repression of BES1 target gene expression, and the suppression of hypocotyl elongation. Taken together, we conclude that the signaling mechanism of Arabidopsis cryptochromes involves the inhibition of BR signaling through blue light-dependent and BR-induced interactions of cryptochromes with BES1. Interestingly, analysis of the genes coregulated by CRY1, CRY2, and BES1 (Supplemental Data Set 1) suggested that the expression of auxin-responsive genes is repressed by CRY1 and CRY2 but induced by BES1 (Supplemental Table 1), consistent with recent studies showing that CRY1 mediates blue light-inhibited auxin signaling (Xu et al., 2018), whereas BES1-mediated BR signaling promotes auxin signaling (Tian et al., 2018).
A recent study demonstrated that the UV-B receptor UVR8 interacts with both BIM1 and BES1 to regulate transcription and BR signaling (Liang et al., 2018). Notably, there are differences between the mechanisms underlying CRY1, CRY2, and UVR8 signaling. Specifically, CRY1 and CRY2 interact with SPA1 (Lian et al., 2011; Liu et al., 2011) and BIM1 and BES1 (this report) in a blue light-dependent manner, whereas UVR8 interacts with BIM1 and BES1 regardless of the presence of UV-B light (Liang et al., 2018). It is likely that blue light irradiation triggers a redox reaction and activates CRY1 and CRY2 through blue light-induced conformational changes and dimerization (Cashmore et al., 1999; Wang et al., 2016a), whereas UV-B light activates UVR8 by promoting its monomerization and nuclear localization (Kaiserli and Jenkins, 2007; Rizzini et al., 2011). We speculate that phytochrome interacts with BIM1 and BES1 to regulate BR signaling based on the following findings: (1) PhyB interacts with the known CRY1-interacting proteins such as COP1 and SPAs (Seo et al., 2004; Jang et al., 2010; Lu et al., 2015; Sheerin et al., 2015); (2) CRY1 and phyB directly interact with AUX/IAA proteins in a blue and red light-dependent manner, respectively, to stabilize these proteins and inhibit auxin signaling (Xu et al., 2018); (3) CRY1, CRY2, and UVR8 interact with BIM1 and BES1 to inhibit BR signaling (Liang et al., 2018; this report); and (4) BES1 regulates photomorphogenesis in blue, red, and far-red light (this report).
BES1 and BZR1, the major transcription factors involved in BR signaling, play essential roles in regulating BR-responsive gene expression. These proteins can be directly phosphorylated by BIN2 kinase or dephosphorylated by PP2A (Wang et al., 2002; Yin et al., 2002; Zhao et al., 2002; Tang et al., 2011). In this study, we found that BES1 promotes hypocotyl elongation in blue, red, and far-red light. Given the findings that BL treatment rapidly induces the dephosphorylation of BES1 and BZR1 in planta (Wang et al., 2002; Yin et al., 2002) and that BRZ treatment or defects in BR biosynthesis or BR signaling result in the phosphorylation of BES1 and /BZR1 (Asami et al., 2000; Wang et al., 2002; Yin et al., 2002), we conclude that the activation of BR signaling leads to the formation of physiologically active dephosphorylated BES1 and BZR1. In this study, we showed how BR signaling is involved in light-regulated plant development by demonstrating that the DNA binding activity of BES1 is inhibited by the direct interaction of photoactivated cryptochrome with dephosphorylated BES1. Because BIM1 cooperates with and enhances BES1 activity (Yin et al., 2005), it is possible that the CRY1-BIM1 interaction results in the inhibition of BES1 activity.
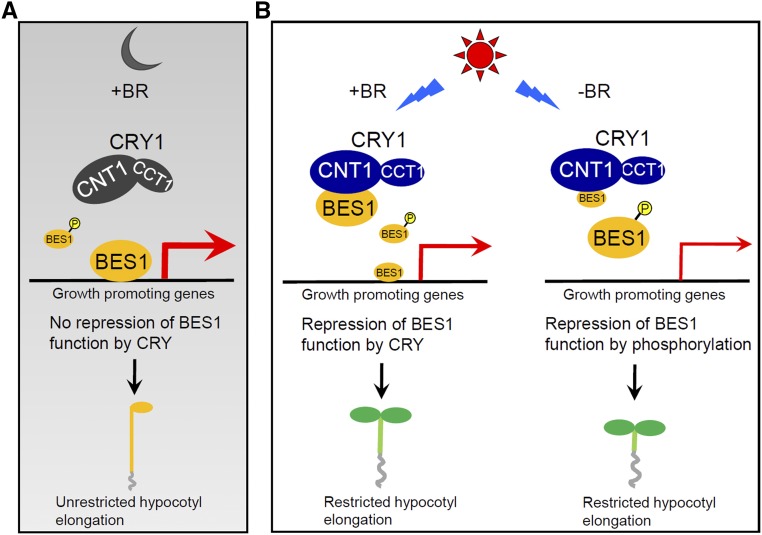
Based on previous and current findings, we propose a model describing how CRY1 inhibits BR signaling to repress hypocotyl elongation. According to this model, when BR is available, BES1 is largely dephosphorylated. In darkness, CRY1 is inactive and unable to interact with dephosphorylated BES1. BES1 is thus able to induce the expression of growth-promoting genes, thereby enhancing hypocotyl elongation (Figure 7A). Upon blue light irradiation, CRY1 undergoes conformational changes and becomes activated and is therefore able to interact with dephosphorylated BES1 through its N terminus to inhibit its DNA binding activity. As a result, the expression of growth-promoting genes is repressed and hypocotyl elongation is inhibited (Figure 7B, left panel). However, when BR is not available, BES1 is largely phosphorylated, and photoactivated CRY1 is not able to interact with phosphorylated BES1 (Figure 7B, right panel). We interpret the finding that CRY1 interacts specifically with dephosphorylated BES1 in a blue light-dependent manner to mean that the interaction of CRY1 with BES1 is dependent on the availability of both light and BR signals in plants. This is biologically meaningful because only photoexcited CRY1 and BR-induced dephosphorylated BES1 are physiologically active. Such a strictly regulated interaction mechanism would give plants the ability to optimize growth and development according to the availability of external light signals and internal BR signals in nature. Given our findings that dark-grown wild-type Arabidopsis seedlings were much more sensitive to BRZ than light-grown seedlings (Supplemental Figure 5A) and that cry1 seedlings were significantly more responsive to BRZ than wild type in blue light (Supplemental Figure 5B), it is possible that CRY1 is involved in repressing BR biosynthesis under constant blue light conditions. Interestingly, expression analysis of the BR biosynthesis genes DWF4 and CPD suggested that, in response to short periods of blue light irradiation, CRY1 inhibits the repression of BR biosynthesis gene expression by BR (Supplemental Figure 9B). This suggests that CRY1 represses the feedback inhibition loop of BR biosynthesis under transient blue light conditions. Additional studies will be needed to confirm these possibilities and to evaluate their biological significance.
Figure 7.
Model Describing How CRY1 Regulates BR Signaling.
When BR is available, BES1 is largely dephosphorylated. In the dark (A), CRY1 is inactive and unable to inhibit BR signaling because it is not capable of interacting with dephosphorylated BES1. In the light (B), CRY1 is activated and interacts with dephosphorylated BES1 via the CRY1 N terminus (CNT1) to inhibit its DNA binding activity (left side). When BR is unavailable, BES1 is largely phosphorylated. Photoactivated CRY1 is not able to interact with phosphorylated BES1 (right side). Thicker and thinner red arrows indicate higher and lower levels of gene expression, respectively. +BR and −BR indicate the availability and unavailability of BR in plants, respectively. Larger and smaller font sizes for BES1 represent higher and lower levels of dephosphorylated or phosphorylated BES1, respectively.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana plants used in this study were of the Columbia ecotype. The cry1, cry2, and cry1 cry2 mutants, the transgenic lines overexpressing Myc-tagged full-length CRY1 and CRY2 (Myc-CRY1-OX and Myc-CRY2-OX), and the BES1-RNAi line were described previously (Yang et al., 2001; Mao et al., 2005; Sang et al., 2005; Yin et al., 2005). The MCS (multiple cloning site)-3×Flag from the pBS-3×Flag (Liu et al., 2008) fragment was cloned into the plant expression vector pHB (Mao et al., 2005) to generate pHB-3×Flag. The full-length cDNA sequences encoding BES1 and its gain-of-function mutant, mbes1 (Yin et al., 2005), were constructed into pHB-3×Flag to generate constructs overexpressing BES1-Flag and mbes1-Flag (Pro35S:BES1-Flag and Pro35S:mbes1-Flag), respectively. A fragment encoding the BES1-GFP-Flag/mbes1-GFP-Flag fusion protein was cloned into plant expression vector pKYL71 (Schardl et al., 1987) to generate the constructs Pro35S:BES1-GFP-Flag and Pro35S:mbes1-GFP-Flag. All constructs were confirmed by DNA sequencing. All primers used for vector construction in this study are listed in Supplemental Data Set 3.
All constructs were transferred into Agrobacterium tumefaciens strain GV3101. Transgenic lines overexpressing BES1-GFP-Flag, mbes1-GFP-Flag, BES1-Flag, and mbes1-Flag in the wild-type background (BES1-GFP-Flag-OX, mbes1-GFP-Flag-OX, BES1-Flag-OX, and mbes1-Flag-OX) were screened on MS plates containing either 100 mg/mL kanamycin or 50 mg/mL hygromycin. The Pro35S:BES1-GFP-Flag, Pro35S:BES1-Flag, and BES1-RNAi transgenes in the wild-type background were introgressed into the cry1 or cry1 cry2 mutant background by genetic crossing, respectively, to generate BES1-GFP-Flag-OX/cry1, BES1-Flag-OX/cry1, and cry1 cry2 BES1-RNAi plants. These were confirmed by phenotypic and/or immunoblot analysis. A transgenic line overexpressing both BES1-GFP-Flag and Myc-CRY2 (BES1-GFP-Flag-OX/Myc-CRY2-OX) was obtained by genetic crossing between BES1-GFP-Flag-OX and Myc-CRY2-OX plants and confirmed by immunoblot analysis.
Imbibed seeds were stored at 4°C for 3 d and grown on half-strength MS nutrient medium (Murashige and Skoog, 1962) plus 1% sucrose with 0.8% agar at 22°C under white light (100 μmol m−2 s−1). Monochromatic blue, red, and far-red lights were generated from blue LEDs (λ max = 469 nm), red LEDs (λ max = 680 nm), and far-red LEDs (λ max = 730 nm) of an LED screen (Qiding), as described previously (Kang et al., 2009). Light spectra and intensity were measured with a HandHeld spectroradiometer (ASD) and a Li250 quantum photometer (Li-Cor).
Yeast Two-Hybrid Assays
GAL4 yeast two-hybrid screening and yeast two-hybrid assays were performed according to the manufacturer’s instructions (Matchmaker; Clontech). The Arabidopsis cDNA library cloned in the prey vector pGAD-T7 (AD) was produced by OE BioTech. The bait plasmid pGBKT7 (BD) and the prey library DNA were cotransformed into yeast strain AH109 (Saccharomyces cerevisiae). Approximately 1 × 107 transformants were screened each time to select colonies grown in blue light with 10 mM 3-amino-1,2,4-triazole.
For the yeast two-hybrid assay, the cDNAs of CNT1, CCT1, CRY2, CNT2, CCT2, BES1, and BZR1 were cloned into the BD vector, and the cDNAs of CRY1, COP1, BES1, and BIM1 were cloned into the AD vector. Combinations of BD and AD vectors were cotransformed into AH109 cells via polyethylene glycol/lithium acetate transformation (Gietz et al., 1992). Transformed yeast cells were spread onto synthetic dropout nutrient medium (SD)-Trp/Leu/His/Ade (SD-T-L-H-A) plates supplemented with 10 mM or 5 mM 3-amino-1,2,4-triazole. Half of the plates were exposed to blue light (30 μmol m−2 s−1), and the other half were kept in the dark for the interaction test (Luo et al., 2014; Xu et al., 2016). At least three independent experiments were performed, and the results of one representative experiment are shown.
Protein Colocalization Analyses
Construction of the Pro35S:CFP, 35S:YFP, Pro35S:CRY1-YFP, Pro35S:CRY2-YFP, and Pro35S:CFP-SPA1 cassettes was described previously (Lian et al., 2011; Luo et al., 2014; Lu et al., 2015). The Pro35S:GUS-NLS-YFP, Pro35S:BIM1-CFP, Pro35S:BIM1ΔNT-CFP, Pro35S:BIM1ΔCT-CFP, Pro35S:BIM1ΔbHLH-CFP, Pro35S:BZR1-CFP, Pro35S:YFP-NLS-GUS-CCT1, and Pro35S:YFP-NLS-CNT1 cassettes were cloned into the pHB vector. GV3101 cultures harboring constructs expressing CFP fusion proteins, YFP fusion proteins, and the gene silencing repressor p19 were mixed at a ratio of 1:1:1 and introduced into tobacco (Nicotiana tabacum) leaves by infiltration. Protein colocalization was detected by confocal microscopy (Leica TCSSP5II confocal laser scanning microscope).
BiFC Assays
The vectors pXY104 and pXY106 were used to produce constructs for the BiFC assays, and they carry fragments encoding the C- and N-terminal halves of YFP (cYFP and nYFP), respectively (Luo et al., 2014; Lu et al., 2015). Construction of the Pro35S:nYFP-SPA1 cassettes was described previously (Lu et al., 2015). The cDNA fragments encoding CRY1, CNT1, CCT1, BES1, and BZR1 were fused to the fragment encoding the C terminus of YFP, and the fragments encoding CRY1, CRY2, CCT1, and BES1 were fused to the fragment encoding the N terminus of YFP. All vectors were transformed into Agrobacterium strain GV3101. GV3101 cultures harboring constructs expressing nYFP fusion proteins and cYFP fusion proteins were mixed at a ratio of 1:1 and introduced into tobacco leaves through infiltration. BiFC signals were detected by confocal microscopy as described above.
Pull-Down Assays Using Purified Proteins
Pull-down assays were performed as described previously with minor modifications (Xu et al., 2016). The CNT1, CCT1, CNT2, CCT2, BES1, and BIM1 cDNA fragments were individually cloned into the pMAL-c2X, pET21a, or pGEX-4T-1 vector. MBP-CNT1 (residues 1–489), MBP-CCT1 (residues 490–681), MBP-CNT2 (residues 1–485), MBP-CCT2 (residues 486–612), GST-BIM1, and His-BES1 proteins were expressed in Escherichia coli (BL21). MBP-CNT1, MBP-CCT1, MBP-CNT2, and MBP-CCT2 fusion proteins were purified according to the protocol from the manufacturer (NEB). Bait proteins GST-BIM1 and His-BES1 were incubated with GST beads (GE) or Ni-NTA Agarose (Qiagen) in non-EDTA buffer (20 mM Tris-HCl, pH 7.5, and 100 mM NaCl) at 4°C for 2 h and then washed three times with the same buffer. The beads were resuspended in 1 mL non-EDTA buffer, and 50 to 100 ng of MBP, MBP-CNT1, MBP-CCT1, MBP-CNT2, or MBP-CCT2 prey protein was added to the solution. The mixture was incubated at 4°C for 1 h and rinsed three times. Proteins were eluted into SDS loading buffer and examined by immunoblot analysis. Prey proteins were detected using an anti-MBP antibody (NEB catalog no. E8032S; 1:1000), and bait proteins were visualized by Coomassie Brilliant Blue staining.
Pull-Down Assays Using Protein Extracts from Seedlings
To investigate the blue light specificity of the CRY1-BIM1 and CRY1/CRY2-BES1 interactions, His-BES1 and GST-BIM1 bait proteins were incubated with 10 μL Ni-NTA agarose beads and 10 μL GST agarose beads for 1 h and washed three times with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.2% Triton X-100). The prey was protein extracts from Myc-CRY1-OX and MG132-treated Myc-CRY2-OX seedlings that were dark-adapted for 4 d and then either kept in darkness for 1 h or exposed to 100 μmol m−2 s−1 (Myc-CRY1-OX) or 50 μmol m−2 s−1 blue light (Myc-CRY2-OX), 20 μmol m−2 s−1 red light, or 10 μmol m−2 s−1 far-red light for 1 h. (Note that MG132 inhibits the 26S proteasome, which is used to inhibit the degradation of Myc-CRY2 protein triggered by blue light.) To investigate the dependence of the CRY1-BES1 interaction on the blue light fluence rate, protein extracts from Myc-CRY1-OX seedlings that were dark-adapted for 4 d and then either kept in darkness for 1 h or exposed to different fluence rates of blue light for 1 h were used as prey. All seedlings were homogenized in lysis buffer plus 1 mM Pefabloc (Roche Diagnostics) and Cocktail (Roche Diagnostics) and 50 μM MG132 (Merck). After centrifugation, the supernatant was incubated with BES1 protein-bound Ni-NTA agarose beads or BIM1 protein-bound GST beads for 30 min and washed four to five times with 1 mL lysis buffer for each time. The precipitates were eluted into 20 μL SDS 1× loading buffer and subjected to immunoblot analysis with an anti-Myc antibody (Millipore; catalog no. 05-724; 1:2000).
To analyze the effects of phosphorylation of BES1 on its interactions with CNT1/CNT2/CCT2, MBP, MBP-CNT1, MBP-CNT2, MBP-CCT2, GST, and GST-PP2AB’α proteins were used as baits, with MBP, GST, and GST-PP2AB’α serving as controls. The proteins were incubated with 10 μL MBP agarose beads (NEB) and GST agarose beads (GE) for 1 h and washed three times. Protein extracts from BES1-Flag-OX/WT seedlings, which were grown on half-strength MS plates supplemented with or without 2 μM BRZ in white light (100 μmol m−2 s−1) for 5 d, dark-adapted for 3 d, treated with or without BL, and incubated in darkness for 3 h before exposure to 100 μmol m−2 s−1 blue light for 1 h, served as prey. All seedlings were harvested in dim green safe light and homogenized in lysis buffer. After centrifugation, the supernatant was incubated with the baits for 30 min and washed four to five times with 1 mL lysis buffer each time. The precipitates were eluted into 20 μL SDS 1× loading buffer and subjected to immunoblot analysis with an anti-Flag antibody (Sigma-Aldrich; catalog no. F3165; 1:4000).
To analyze the interactions of the immunoprecipitated phosphorylated BES1 (pBES1) and dephosphorylated BES1 (BES1) with CNT1, pBES1 and BES1 were prepared as follows: BES1-Flag-OX/WT seedlings were grown on half-strength MS plates supplemented with 2 μM BRZ in white light (100 μmol m−2 s−1) for 5 d, dark-adapted for 3 d, and then exposed to 100 μmol m−2 s−1 blue light for 1 h. The protein extracts from these BRZ-treated seedlings were immunoprecipitated with anti-Flag M2 Affinity Gel and treated with or without calf intestine phosphatase (NEB) (Yin et al., 2002). At this point, pBES1 and BES1 were bound to the Flag beads, which served as baits. His-TF-CNT1 and GST-PP2AB’α were used as preys and were incubated with these baits for 30 min and washed four to five times with 1 mL lysis buffer each time. The precipitates were eluted into 20 μL SDS 1× loading buffer and subjected to immunoblot analysis with anti-His (mouse monoclonal; catalog no. 28004; 1:2000) and -GST antibodies (JiangLai Biological; catalog no.AbM59001-2H5-PU; 1:1000). When pBES1 and BES1 were used as preys, these proteins were prepared by washing the Flag bead-bound pBES1 and BES1 three times with lysis buffer and eluting the protein from the beads with 0.5 μg/μL 3×Flag peptide (Sigma-Aldrich) in 20 μL lysis buffer. The eluted pBES1 and BES1 proteins, which were attached to Ni-NTA agarose beads and GST beads, respectively, were incubated with bait proteins His-TF-CNT1 and GST-PP2AB’α for 30 min and washed four to five times with 1 mL lysis buffer each time. The precipitates were eluted into 20 μL SDS 1× loading buffer and subjected to immunoblot analysis with an anti-Flag antibody (Sigma-Aldrich; catalog no. F3165; 1:4000).
Co-IP Assays
Co-IP assays in Arabidopsis were performed as described previously (Lian et al., 2011), with minor modifications. For the co-IP assay of the blue light-dependent CRY1-BES1 interaction, BES1-GFP-Flag-OX/WT or BES1-GFP-Flag-OX/cry1 seedlings were grown in white light (100 μmol m−2 s−1) for 4 to 5 d and dark-adapted for 3 d. Half of the dark-adapted BES1-GFP-Flag-OX/WT and BES1-GFP-Flag-OX/cry1 seedlings treated with 1 μM BL were incubated in darkness for 3 h before being exposed to 100 μmol m−2 s−1 blue light for 1 h, and the other half were incubated in darkness for 4 h. For the co-IP assay of specific interactions of CRY1 and CRY2 with dephosphorylated BES1, BES1-GFP-Flag-OX/WT and BES1-GFP-Flag-OX/Myc-CRY2-OX seedlings were grown on half-strength MS plates supplemented with or without 2 μM BRZ in white light (100 μmol m−2 s−1) for 5 d, dark-adapted for 3 d, treated with or without BL, and then kept in darkness for 3 h before being exposed to 100 or 50 μmol m−2 s−1 blue light for 1 h. All seedlings were harvested under a dim green safe light (bulb type: QD-058G54-220, fluence rate: 25 μmol m−2 s−1, spectral quality: 520–525 nm) and homogenized in lysis buffer (Luo et al., 2014). Total protein concentrations were determined by the Bradford assay (Bio-Rad), and equal amounts of total protein in 1 mL of lysis buffer were incubated with anti-CCT1 antiserum (Sang et al., 2005) attached to Dynabeads protein G beads (GE Healthcare) or anti-Myc agarose beads (Sigma-Aldrich) at 4°C for 2 h. The immunoprecipitates were washed two to three times with wash buffer (Luo et al., 2014) and the eluted into 20 μL SDS 1× loading buffer and subjected to immunoblot analysis with anti-CCT1, anti-Flag (Sigma-Aldrich), or anti-Myc (Millipore) antibodies.
For the co-IP assays in tobacco cells, the Pro35S:Myc-CRY1, Pro35S:Myc-CRY2, and Pro35S:cLUC (residues 322–550), NLS-Flag cassettes were constructed as described previously (Sang et al., 2005; Xu et al., 2016). The Pro35S:BES1-Flag and Pro35S:BIM1-Flag cassettes were cloned into pHB. The fragment encoding 6×Myc was cloned into pHB to generate pHB-6×Myc. For anti-Flag IP, Flag-tagged BIM1/BES1 proteins were coexpressed with Myc-CRY1 or Myc-CRY2 in tobacco leaves. Equal amounts of total protein in 1 mL of lysis buffer from tobacco cells coexpressing BIM1-Flag and Myc-CRY1 or BES1-Flag and Myc-CRY1/Myc-CRY2 were incubated with 10 μL anti-Flag M2 Affinity Gel (Sigma-Aldrich) at 4°C for 2 h. The precipitates were eluted into 20 μL SDS 1× loading buffer and subjected to immunoblot analysis with anti-Myc (Millipore) and anti-Flag (Sigma-Aldrich) antibodies.
BL and BRZ Treatments
BL and BRZ (Sigma-Aldrich) stocks were prepared in 80% ethanol or DMSO. Wild-type, cry1, cry2, cry1 cry2, and Myc-CRY1-OX seedlings were grown on 0.5× MS plates supplemented with a gradient of concentrations of BRZ (0, 0.1, 0.2, 0.5, 1.0, and 2.0 μM) or a gradient of concentrations of BL (0, 1, 10, 100, and 1000 nM) plus BRZ (2 μM) at 4°C for 3 d, incubated in white light overnight, and transferred to blue, red, or far-red light for an additional 5 to 6 d. The seedlings were photographed, and hypocotyl lengths were measured with Image J software.
RT-qPCR
Total RNA was isolated with an RNAprep Plant kit (Tiangen) and reverse-transcribed to cDNA using an iScript cDNA synthesis kit (Bio-Rad). SYBR Premix Ex Taq II (TaKaRa) was used for qPCR on a CFX96 real-time system (Bio-Rad). The reaction conditions included 40 to 50 amplification cycles (30 s at 94°C; 5 s at 94°C; 30 s at 60°C). Three technical replicates were performed for each cDNA sample. The relative quantification was calculated with the 2−ΔΔCt method using ACT2 and ACT7 as the reference genes(Zhang et al., 2014). The primers used are listed in Supplemental Table 2.
EMSAs
EMSA was performed with 100 ng of His-BES1, His-TF, His-TF-CCT1, or His-TF-CNT1, which was expressed in E. coli, concentrated, and purified with Ni-NTA Agarose (Qiagen) and Amicon Ultra centrifugal filter units (Ultra-4; MWCO:10 kD) (Merck Millipore; catalog no. UFC801024). Biotin-labeled probes were obtained by synthesizing, annealing, and labeling the probes with biotin, followed by annealing. The binding reaction was performed in 20 μL binding buffer [10× binding buffer, 100 mM MgCl2, 1 µg/μL poly (dI·dC), and 1% Nonidet P-40] with 200 fmol probe and 100 ng His-BES1 protein according to the manufacturer’s instructions (Thermo Scientific) (Zhang et al., 2014). After 20 min incubation at room temperature, the reactions were resolved by 6% native polyacrylamide gel electrophoresis at 4°C, and the DNA-protein complexes were transferred to a nylon membrane (GE Healthcare). Biotin-labeled probes were detected with HRP-conjugated streptavidin and visualized with an ECL detection kit.
ChIP-qPCR
ChIP was performed as described previously (Zhang et al., 2014) with minor modifications. Briefly, wild-type, BES1-Flag-OX/WT, and BES1-Flag-OX/cry1 seedlings were grown in white light (100 μmol m−2 s−1) for 4 to 5 d and placed in darkness for an additional 2 to 3 d. Half of the BES1-Flag-OX/WT and BES1-Flag-OX/cry1 seedlings were treated with 1 μM BL in darkness for 3 h before being exposed to 100 μmol m−2 s−1 blue light for an additional 1 h, and the other half of these seedlings were treated with 1 μM BL and kept in darkness for 4 h. Wild-type seedlings were treated with 1 μM BL and incubated in darkness for 3 h before being exposed to 100 μmol m−2 s−1 blue light for 1 h. All seedlings were harvested in the dim green safe light described above or in blue light (100 μmol m−2 s−1) and fixed in fixation buffer by vacuum infiltration at 4°C. Glycine was added to the samples to a final concentration of 0.125 M under a vacuum to stop the reaction. The sonicated protein-DNA complexes were immunoprecipitated with an anti-Flag antibody (Sigma-Aldrich). The resulting DNA was used for qPCR. SYBR Premix Ex Taq II (TaKaRa) was used for qPCR on a CFX96 real-time system (Bio-Rad). The reaction conditions included 40 to 50 amplification cycles (30 s at 94°C; 5 s at 94°C; 30 s at 60°C). Three technical replicates were performed for each cDNA sample. The relative quantification was calculated with the 2−ΔΔCt method.
IP/input was calculated by comparing the Ct values between immunoprecipitates and input. The TA3 fragment was used as a control (Yin et al., 2005).
RNA-Seq Data Processing
Venn diagrams were generated using Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html). Heat maps were generated by hierarchical clustering analysis using MeV 4.7 software. GO enrichment analysis was performed using agriGO (http://bioinfo.cau.edu.cn/agriGO/). The significantly enriched GO terms of the DEGs were identified by FDR (q-value) < 0.05 (Wang et al., 2016b).
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank database or the Arabidopsis Genome Initiative database under the following accession numbers: CRY1 (AT4G08920), CRY2 (AT1G04400), BIM1 (AT5G08130), BES1 (AT1G19350), BZR1 (AT1G75080), ACS5 (AT5G65800), PRE5 (AT3G28857), PRE6 (AT1G26945), IAA19 (AT3G15540), SAUR15 (AT4G38850), SAUR68 (AT1G29490), DWF4 (AT3G50660), and CPD (AT5G05690). The accession numbers of the previously produced microarray or RNA-seq data sets used in this study are GSE58552 [blue (WT-blue versus WT-dark), CRYs (WT versus cry1 cry2)] (He et al., 2015), and GSE51772 [(BL (WT+BL versus WT-BL)] (Oh et al., 2014). The RNA-seq data showing BES1-regulated genes (bes1-D+BL versus WT+BL) were from Yu et al. (2011).
Supplemental Data
Supplemental Figure 1. CRY1 Physically Interacts with BIM1 through Its N Terminus.
Supplemental Figure 2. The N-Terminal Domain of BIM1 Might Mediate the Interaction with CRY1.
Supplemental Figure 3. CRY1 and CRY2 Physically Interact with BES1 and BZR1.
Supplemental Figure 4. Analysis of the Photomorphogenic Phenotypes of mbes1-GFP-Flag-OX Seedlings Grown under Monochromatic Light Conditions.
Supplemental Figure 5. CRY1 Is Involved in Mediating Blue Light Inhibition of Arabidopsis Seedlings’ Responsiveness to BRZ.
Supplemental Figure 6. An Additional Biological Replicate Showing That CRYs Are Involved in the Inhibition of the Responses of Arabidopsis Seedlings to BR by Blue Light and Act Upstream of BES1 to Regulate Hypocotyl Elongation in Blue Light.
Supplemental Figure 7. BES1 Promotes Hypocotyl Elongation under Monochromatic Light Conditions.
Supplemental Figure 8. Analysis of the Photomorphogenic Phenotypes of mbes1-Flag-OX Grown in Blue Light.
Supplemental Figure 9. CRY1 Regulates the Expression of BES1 Target Genes.
Supplemental Figure 10. Analysis of BES1-Flag Fusion Protein and BES1-Flag Transcript Levels in Transgenic Arabidopsis Overexpressing BES1-Flag.
Supplemental Table 1. Differential Expression of Auxin-Responsive Genes Coregulated by CRYs and BES1.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Genes Regulated by Blue Light, CRYs, and BR.
Supplemental Data Set 2. Genes Regulated by Blue Light, CRYs, and BES1.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Yan-Hai Yin and Xue-Lu Wang for kindly providing the BES1-RNAi line and BiFC vectors. This work was supported by The National Key Research and Development Program of China, Ministry of Science and Technology of the People’s Republic of China (Grant 2017YFA0503800) and National Natural Science Foundation of China grants to H.Q.Y. (31530085, 91217307, and 90917014) and H.L.L. (31570282 and 31170266).
AUTHOR CONTRIBUTIONS
W.W. and H.-Q.Y. designed the experiments. W.W., X.L, L.L., H.L., Z.M., P.X., T.G., F.X., S.D., X.C., S.W., and H.S. carried out the experiments. W.W. collected the data. W.W. and H.-Q.Y. wrote the manuscript with input from all other authors.
References
- Ahmad M., Cashmore A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166. [DOI] [PubMed] [Google Scholar]
- Asami T., Min Y.K., Nagata N., Yamagishi K., Takatsuto S., Fujioka S., Murofushi N., Yamaguchi I., Yoshida S. (2000). Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol. 123: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs W.R., Christie J.M. (2002). Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 7: 204–210. [DOI] [PubMed] [Google Scholar]
- Cashmore A.R., Jarillo J.A., Wu Y.J., Liu D. (1999). Cryptochromes: blue light receptors for plants and animals. Science 284: 760–765. [DOI] [PubMed] [Google Scholar]
- Chory J., Nagpal P., Peto C.A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3: 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S.D., Langford M., McMorris T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.W., Quail P.H. (1999). Signalling in light-controlled development. Semin. Cell Dev. Biol. 10: 121–129. [DOI] [PubMed] [Google Scholar]
- Deng X.W., Matsui M., Wei N., Wagner D., Chu A.M., Feldmann K.A., Quail P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801. [DOI] [PubMed] [Google Scholar]
- Emery P., So W.V., Kaneko M., Hall J.C., Rosbash M. (1998). CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95: 669–679. [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Chory J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13: 203–229. [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Yeh K.C., Lagarias J.C., Zhang H., Elich T.D., Chory J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284: 1539–1541. [DOI] [PubMed] [Google Scholar]
- Gegear R.J., Foley L.E., Casselman A., Reppert S.M. (2010). Animal cryptochromes mediate magnetoreception by an unconventional photochemical mechanism. Nature 463: 804–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St Jean A., Woods R.A., Schiestl R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Yang H., Mockler T.C., Lin C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363. [DOI] [PubMed] [Google Scholar]
- He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S.B., Wang W.X., Zhang J.Y., Xu F., Lian H.L., Li L., Yang H.Q. (2015). The CNT1 domain of Arabidopsis CRY1 alone is sufficient to mediate blue light inhibition of hypocotyl elongation. Mol. Plant 8: 822–825. [DOI] [PubMed] [Google Scholar]
- Jaillais Y., Hothorn M., Belkhadir Y., Dabi T., Nimchuk Z.L., Meyerowitz E.M., Chory J. (2011). Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 25: 232–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Henriques R., Seo H.S., Nagatani A., Chua N.H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserli E., Jenkins G.I. (2007). UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C.Y., Lian H.L., Wang F.F., Huang J.R., Yang H.Q. (2009). Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21: 2624–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Jeong Y.J., Corvalán C., Fujioka S., Cho S., Park T., Choe S. (2014). Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis. Plant J. 77: 737–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Burlingame A.L., Wang Z.Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Michniewicz M., Bergmann D.C., Wang Z.Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482: 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K., Zylka M.J., Sriram S., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert S.M. (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205. [DOI] [PubMed] [Google Scholar]
- Li J., Chory J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938. [DOI] [PubMed] [Google Scholar]
- Li Q.F., He J.X. (2016). BZR1 interacts with HY5 to mediate brassinosteroid- and light-regulated cotyledon opening in Arabidopsis in darkness. Mol. Plant 9: 113–125. [DOI] [PubMed] [Google Scholar]
- Li J., Nagpal P., Vitart V., McMorris T.C., Chory J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401. [DOI] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222. [DOI] [PubMed] [Google Scholar]
- Lian H.L., He S.B., Zhang Y.C., Zhu D.M., Zhang J.Y., Jia K.P., Sun S.X., Li L., Yang H.Q. (2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25: 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T., Mei S., Shi C., Yang Y., Peng Y., Ma L., Wang F., Li X., Huang X., Yin Y., Liu H. (2018). UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev. Cell 44: 512–523. [DOI] [PubMed] [Google Scholar]
- Lin C., Robertson D.E., Ahmad M., Raibekas A.A., Jorns M.S., Dutton P.L., Cashmore A.R. (1995). Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269: 968–970. [DOI] [PubMed] [Google Scholar]
- Lin C., Yang H., Guo H., Mockler T., Chen J., Cashmore A.R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95: 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zuo Z., Liu H., Liu X., Lin C. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25: 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.J., Zhang Y.C., Li Q.H., Sang Y., Mao J., Lian H.L., Wang L., Yang H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.D., Zhou C.M., Xu P.B., Luo Q., Lian H.L., Yang H.Q. (2015). Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant 8: 467–478. [DOI] [PubMed] [Google Scholar]
- Luo Q., Lian H.L., He S.B., Li L., Jia K.P., Yang H.Q. (2014). COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26: 2441–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Li X., Guo Y., Chu J., Fang S., Yan C., Noel J.P., Liu H. (2016). Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 113: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J., Zhang Y.C., Sang Y., Li Q.H., Yang H.Q. (2005). From The Cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl. Acad. Sci. USA 102: 12270–12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nam K.H., Li J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212. [DOI] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Bai M.Y., Arenhart R.A., Sun Y., Wang Z.Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: 03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Pedmale U.V., Huang S.C., Zander M., Cole B.J., Hetzel J., Ljung K., Reis P.A.B., Sridevi P., Nito K., Nery J.R., Ecker J.R., Chory J. (2016). Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164: 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3: 85–93. [DOI] [PubMed] [Google Scholar]
- Rizzini L., Favory J.J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G.I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106. [DOI] [PubMed] [Google Scholar]
- Sancar A. (1994). Structure and function of DNA photolyase. Biochemistry 33: 2–9. [DOI] [PubMed] [Google Scholar]
- Sancar A. (2003). Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103: 2203–2237. [DOI] [PubMed] [Google Scholar]
- Sang Y., Li Q.H., Rubio V., Zhang Y.C., Mao J., Deng X.W., Yang H.Q. (2005). N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17: 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl C.L., Byrd A.D., Benzion G., Altschuler M.A., Hildebrand D.F., Hunt A.G. (1987). Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61: 1–11. [DOI] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999. [DOI] [PubMed] [Google Scholar]
- Seo H.S., Watanabe E., Tokutomi S., Nagatani A., Chua N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18: 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin D.J., Menon C., zur Oven-Krockhaus S., Enderle B., Zhu L., Johnen P., Schleifenbaum F., Stierhof Y.D., Huq E., Hiltbrunner A. (2015). Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Devlin P.F., Kay S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490. [DOI] [PubMed] [Google Scholar]
- Sun Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kim T.W., Oses-Prieto J.A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A.L., Wang Z.Y. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Lv B., Ding T., Bai M., Ding Z. (2018). Auxin-BR interaction regulates plant growth and development. Front. Plant Sci. 8: 2256–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R., Kevei E., Hall A., Millar A.J., Nagy F., Kozma-Bognár L. (2001). Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 127: 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Chory J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100. [DOI] [PubMed] [Google Scholar]
- Vert G., Nemhauser J.L., Geldner N., Hong F., Chory J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21: 177–201. [DOI] [PubMed] [Google Scholar]
- Wang X., Chory J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313: 1118–1122. [DOI] [PubMed] [Google Scholar]
- Wang H., Ma L.G., Li J.M., Zhao H.Y., Deng X.W. (2001a). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158. [DOI] [PubMed] [Google Scholar]
- Wang H., Yang C., Zhang C., Wang N., Lu D., Wang J., Zhang S., Wang Z.X., Ma H., Wang X. (2011). Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 21: 825–834. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zuo Z., Wang X., Gu L., Yoshizumi T., Yang Z., Yang L., Liu Q., Liu W., Han Y.J., Kim J.I., Liu B., et al. (2016a). Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354: 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.X., Lian H.L., Zhang L.D., Mao Z.L., Li X.M., Xu F., Li L., Yang H.Q. (2016b). Transcriptome analyses reveal the involvement of both C and N termini of Cryptochrome 1 in its regulation of phytohormone-responsive gene expression in Arabidopsis. Front. Plant Sci. 7: 294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Seto H., Fujioka S., Yoshida S., Chory J. (2001b). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383. [DOI] [PubMed] [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Xu F., Li T., Xu P.B., Li L., Du S.S., Lian H.L., Yang H.Q. (2016). DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett. 590: 541–549. [DOI] [PubMed] [Google Scholar]
- Xu F., He S., Zhang J., Mao Z., Wang W., Li T., Hua J., Du S., Xu P., Li L., Lian H., Yang H.-Q. (2018). Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol. Plant 11: 523–541. [DOI] [PubMed] [Google Scholar]
- Yang H.Q., Wu Y.J., Tang R.H., Liu D., Liu Y., Cashmore A.R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827. [DOI] [PubMed] [Google Scholar]
- Yang H.Q., Tang R.H., Cashmore A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Li C., Cai Z., Hu Y., Nolan T., Yu F., Yin Y., Xie Q., Tang G., Wang X. (2017). SINAT E3 ligases control the light-mediated stability of the brassinosteroid-activated transcription factor BES1 in Arabidopsis. Dev. Cell 41: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005). A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259. [DOI] [PubMed] [Google Scholar]
- Yu X., Shalitin D., Liu X., Maymon M., Klejnot J., Yang H., Lopez J., Zhao X., Bendehakkalu K.T., Lin C. (2007). Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc. Natl. Acad. Sci. USA 104: 7289–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646. [DOI] [PubMed] [Google Scholar]
- Zhang J.Y., He S.B., Li L., Yang H.Q. (2014). Auxin inhibits stomatal development through MONOPTEROS repression of a mobile peptide gene STOMAGEN in mesophyll. Proc. Natl. Acad. Sci. USA 111: E3015–E3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Peng P., Schmitz R.J., Decker A.D., Tax F.E., Li J. (2002). Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 130: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.Y., Sae-Seaw J., Wang Z.Y. (2013). Brassinosteroid signalling. Development 140: 1615–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21: 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]