PUX10 is a lipid droplet-localized scaffold protein that recruits CDC48 and is involved in the degradation of proteins associated with lipid droplets.
Abstract
The number of known proteins associated with plant lipid droplets (LDs) is small compared with other organelles. Many aspects of LD biosynthesis and degradation are unknown, and identifying and characterizing candidate LD proteins could help elucidate these processes. Here, we analyzed the proteome of LD-enriched fractions isolated from tobacco (Nicotiana tabacum) pollen tubes. Proteins that were highly enriched in comparison with the total or cytosolic fraction were further tested for LD localization via transient expression in pollen tubes. One of these proteins, PLANT UBX DOMAIN-CONTAINING PROTEIN10 (PUX10), is a member of the plant UBX domain-containing (PUX) protein family. This protein localizes to LDs via a unique hydrophobic polypeptide sequence and can recruit the AAA-type ATPase CELL DIVISION CYCLE48 (CDC48) protein via its UBX domain. PUX10 is conserved in Arabidopsis thaliana and expressed in embryos, pollen tubes, and seedlings. In pux10 knockout mutants in Arabidopsis, LD size is significantly increased. Proteomic analysis of pux10 mutants revealed a delayed degradation of known LD proteins, some of which possessed ubiquitination sites. We propose that PUX10 is involved in a protein degradation pathway at LDs, mediating an interaction between polyubiquitinated proteins targeted for degradation and downstream effectors such as CDC48.
INTRODUCTION
Lipid droplets (LDs; also referred to as oleosomes and oil or lipid bodies) are specialized organelles mainly occurring in lipid-rich seeds, tapetal cells, and pollen grains (Chapman et al., 2012; Ischebeck, 2016; Huang, 2018). Additionally, all other plant cell types are considered either to contain a relatively small number of LDs or to form them only under certain environmental conditions, such as in response to abiotic stress (Gidda et al., 2016; Brocard et al., 2017; VanBuren et al., 2017). Structurally, LDs consist of a core of hydrophobic compounds, in most cases triacylglycerols (TAGs) and sterol esters, surrounded by a phospholipid monolayer (Pyc et al., 2017a). The monolayer derives from the outer layer of the endoplasmic reticulum (ER), where the LDs are initially formed, before they are released into the cytoplasm (they are distinct from plastid-derived plastoglobuli that also store neutral lipids; van Wijk and Kessler, 2017).
Several proteins are known to be involved in the proper formation of LDs (Huang, 2018). In pollen and seeds, diacylglycerol O-acyltransferase 1 and phospholipid:diacylglycerol acyltransferase 1, localized at the ER, are the main enzymes catalyzing the final step of TAG biosynthesis (Zhang et al., 2009). Situated at the ER-LD junction sites, SEIPINs are important for TAG accumulation and LD proliferation (Cai et al., 2015; Taurino et al., 2018). Other proteins are directly associated with LDs. For instance, caleosin and steroleosin (also referred to as hydroxysteroid dehydrogenases [HSDs]) are anchored to LDs by a hydrophobic hairpin sequence that contains conserved proline residues that form a so-called proline “knot” and “knob,” respectively (Shimada and Hara-Nishimura, 2010). Caleosins are considered to have a structural role in maintaining LD integrity, but some isoforms have also been reported to have peroxygenase activity (Hanano et al., 2006; Blée et al., 2014) and are thought to act after the oxidation of fatty acids by LD-localized lipoxygenases (Rudolph et al., 2011; Zienkiewicz et al., 2013). Based on genetic evidence, steroleosins are associated with brassinosteroid metabolism (Li et al., 2007; Baud et al., 2009) and have been shown to be able to act on steroid substrates as dehydrogenases, interconverting hydroxyl-groups of the steroid backbone to keto-groups (d’Andréa et al., 2007). Rather unknown on the molecular level is the function of LDAPs (LD-associated proteins; Gidda et al., 2013). These proteins, first discovered on LDs of avocado (Persea americana) mesocarp (Horn et al., 2013), are also present in Arabidopsis thaliana, where they are needed for the proper compartmentation of neutral lipids during postgerminative seedling growth and also in mature leaves, especially under drought stress conditions (Gidda et al., 2016; Kim et al., 2016). A similar role has been proposed for the LDAP interaction partner LDIP (LDAP-interacting protein; Pyc et al., 2017b).
The best studied and most prominent LD proteins, at least in seeds, tapetum, and pollen, are oleosins (Tzen et al., 1990; Lee et al., 1994; Huang, 1996; Wang et al., 1997). Similar to caleosins and steroleosins, oleosins are tightly anchored to the LD by a hydrophobic proline knot structure. Oleosins are considered LD coat proteins, which function to prevent the coalescence of LDs, especially during seed desiccation (Tzen, 2012). Oleosins might also be involved in the formation of LDs and their budding from the ER (Chapman et al., 2012). It is furthermore speculated that they might act as binding sites for other proteins (Quettier and Eastmond, 2009). In germinating seeds, when TAG, but also LD proteins are degraded, oleosins have been found to be ubiquitinated (Hsiao and Tzen, 2011; Deruyffelaere et al., 2015), which might be crucial for their breakdown.
Apart from the aforementioned proteins, only a few other LD proteins have been clearly identified in higher plants (Chapman et al., 2012; Pyc et al., 2017a; Huang, 2018), making the total number of known LD proteins relatively small in comparison to other organelles. Moreover, while hundreds of proteins have been found by proteomic screens in LD-enriched fractions from various higher plants (Jolivet et al., 2004; Liu et al., 2015), these fractions are prone to contaminations and only few of these proteins have been confirmed to be LD-associated by additional lines of evidence. Nevertheless, knowledge of LDs and their proteome has steadily increased in recent years and, in doing so, has raised a number of new and important questions about the organelle’s protein homeostasis: How is LD protein turnover regulated, and especially how are proteins removed from the LD when the organelle is being degraded, for instance, during lipolysis or in the case of protein misfolding?
Several protein degradation pathways exist in plants including autophagy (Wang et al., 2018) and degradation by the 26S proteasome (Sharma et al., 2016). In the latter case, the majority of proteins are ubiquitinated prior to their breakdown. The proteasome is localized in the cytoplasm and hence has no access to membrane proteins or proteins inside organelles (with the exception of the nucleus; Peters et al., 1994). Therefore, proteins derived, for example, from the ER lumen or membrane have to be transported to the cytoplasm before they can be degraded by the proteasome. One mechanism for the elimination of misfolded proteins from the ER lumen and membrane is the ER-associated protein degradation (ERAD) pathway. This pathway requires a unique set of proteins to recognize, ubiquitinate, retrotranslocate, and ultimately degrade misassembled proteins, and its components are mostly conserved among eukaryotes. In terms of the cytoplasmic events that take place in the ERAD pathway, a variety of proteins are known (Meusser et al., 2005). Among these proteins are the UBX (ubiquitin regulatory X domain) domain-containing proteins that can act as a scaffold to mediate contact between the ubiquitinated substrates of the ERAD pathway and downstream effector proteins like the AAA-type ATPase CELL DIVISION CYCLE48 (CDC48) protein, which is also known (in mammals) as p97/VCP (Schuberth and Buchberger, 2008).
The Arabidopsis genome encodes 16 PLANT UBX DOMAIN-CONTAINING (PUX) proteins with a diverse domain structure (Liu and Li, 2014). We identified one member of this family, PUX10, in the proteome of LD-enriched fractions isolated from tobacco (Nicotiana tabacum) pollen tubes and confirmed that the protein is localized to LDs in both pollen tubes and seeds via a hydrophobic region. Furthermore, we show that disruption of PUX10 expression influences LD size and number and that PUX10 is able to recruit the p97/VCP homolog At-CDC48A to LDs, implicating a role for PUX10 and At-CDC48A in LD protein turnover.
RESULTS
Identification of Candidate Proteins Associated with LDs from Tobacco Pollen Tubes
One strategy for improving our understanding of LDs with regard to their synthesis, degradation, and/or function is to identify proteins specifically associated with this organelle. Therefore, the first goal in this study was to investigate the proteome of LDs to extend the number of known LD-bound proteins. Tobacco pollen tubes were chosen, as LDs of this tissue have not been previously investigated by proteomics, in contrast to other studies that focused on LDs in seeds (Jolivet et al., 2004; Liu et al., 2015), seedlings (Pyc et al., 2017b), mesocarp of avocado (Persea americana) (Horn et al., 2013), and chinese tallow (Triadica sebifera; Zhi et al., 2017). In addition, tobacco pollen tubes can be easily transformed to verify the association of candidate proteins with the LDs (Müller et al., 2017), eliminating the need to switch systems from identification to verification. LD-enriched fractions were obtained together with cytosolic and total fractions, and the protein composition of all samples was analyzed by tryptic digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS). Protein composition was subsequently investigated using the label-free intensity-based absolute quantification (iBAQ) algorithm (Schwanhäusser et al., 2011) from MaxQuant (Supplemental Data Set 1), which identified a total of 1314 protein groups. The relative iBAQ values given by the software represent an estimate of the abundance of a protein in a mixture and were calculated as ‰ of all proteins in one sample (Shin et al., 2013). The enrichment was determined from the relative LD iBAQ values divided by the higher value of the total and cytosolic fractions (Supplemental Data Set 2).
The two most abundant proteins found in the tobacco pollen tube LD fraction were a caleosin (Nt-CLO1a) with an iBAQ ‰ of 158 and an enrichment factor of 1234 and an oleosin (Nt-OLE6b) with an iBAQ ‰ of 64 and an enrichment factor of 2137. The LD fraction, however, also contained contaminants from other organelles. For example, an ATP synthase subunit (Uniprot ID A0A1S4CK04) was the third most abundant protein in the LD fraction with an iBAQ ‰ of 56, but it was only enriched by a factor of 2.6 and thereby ruled out to be a LD protein, since all known LD proteins that were identified displayed much higher enrichment factors (Supplemental Data Set 3). To determine if another organelle was also strongly enriched in the LD fraction, various marker proteins that could be assigned to a specific organelle and that showed a high abundance in the total fraction were chosen (Supplemental Data Set 4). Overall, the selected marker proteins for mitochondria, plastids, peroxisomes, or the cytosol were only slightly enriched or depleted in the LD fraction (Supplemental Data Set 4). On the other hand, the three chosen marker proteins from the ER were enriched in the LD fraction by a factor of 4 to 21, indicating that the ER, to some extent, copurified with LDs, albeit the relative abundance of the ER marker proteins in the LD fraction was relatively minor compared with enrichment of homologs of known LD proteins in the LD fraction (Supplemental Data Set 3).
We also compared the isoforms of known LD proteins found in the pollen tubes on a qualitative level to the isoforms found in tobacco seeds (Supplemental Data Sets 5 and 6). This revealed that two of the oleosin and caleosin isoforms were only found in pollen tubes (Supplemental Figures 1 and 2 and Supplemental Data Sets 7 and 8). Steroleosins, however, were only detected in seeds but not in pollen tubes.For an overview of all LD-localized protein families and the protein members detected, see Supplemental Figures 1 to 10 and Supplemental Data Sets 7 to 16; all proteins were named according to their closest homolog from Arabidopsis.
Verification of LD Association by Transient Expression in Tobacco Pollen Tubes
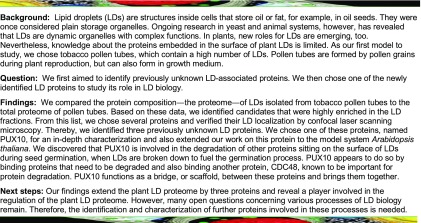
In the pollen tube data set, 18 proteins had an enrichment factor higher than 100 and an iBAQ ‰ higher than 1 (Table 1). From these proteins, at least one member of the corresponding nine gene families was cloned from tobacco pollen tube cDNA, except for Nt-OBL1, which was previously characterized (Müller and Ischebeck, 2018). All constructs were transiently expressed in tobacco pollen tubes as fusion proteins with mVenus, and Nile Red was used to stain LDs. In total, seven proteins were confirmed to be localized, at least partially, to LDs (Figure 1), including Nt-LDAP1a, Nt-LDIPa, which, notably, appeared to localize only to some LDs, the oleosin Nt-OLE6b, the caleosin Nt-CLO1b, a cycloartenol synthase Nt-CAS1b (and its Arabidopsis homolog, At-CAS1; Supplemental Figure 11), a sterol methyltransferase 1 (Nt-SMT1c), a protein formerly annotated as unknown (named here as Nt-PTLD2b for POLLEN TUBE LIPID DROPLET PROTEIN 2b), and a member of the family of PUX proteins (Nt-PUX10c), which also partially localized to the ER (Figures 2A and 2B). A homolog of Arabidopsis retinol dehydrogenase, Nt-FEY1, did not localize to LDs, but instead localized exclusively to the ER (Figure 3).
Table 1. Proteins Enriched in the LDs of Tobacco Pollen Tubes.
| Protein Type | Name | Gene ID | iBAQ ‰ | Enrichment |
|---|---|---|---|---|
| Caleosin1 | NtCLO1a | LOC107783728 | 158 ± 10 | 1234 |
| Oleosin1 | NtOLE6b | LOC107824536 | 64 ± 30 | 2137 |
| Caleosin | NtCLO1b | LOC107817909 | 40 ± 5 | 993 |
| Oleosin | NtOLE6a | LOC107780677 | 35 ± 16 | 6746 |
| LDAP1 | NtLDAP1a | LOC107765167 | 28 ± 3 | 741 |
| Cycloartenol synthase1 | NtCASb | LOC107762593 | 16 ± 4 | 1517 |
| Plant UBX domain-containing protein1 | NtPUX10c | LOC107769345 | 15 ± 3 | 341 |
| LDAP | NtLDAP1b | LOC107804268 | 7.0 ± 0.5 | n.d. in controls |
| LDAP | NtLDAP3a | LOC107827582 | 5.0 ± 0.4 | 557 |
| Cycloartenol synthase | NtCASa | LOC107826198 | 4.1 ± 1.2 | 2605 |
| Sterol methyltransferase 11 | NtSMT1c | LOC107813194 | 3.1 ± 0.3 | 219 |
| Unknown protein | NtPTLD1 | LOC107815104 | 2.8 ± 0.02 | n.d. in controls |
| Oil body lipase | NtOBL1 | LOC107788962 | 2.6 ± 0.9 | 307 |
| Plant UBX domain-containing protein | NtPUX10a | LOC107799924 | 2.2 ± 0.3 | n.d. in controls |
| LDIP1 | NtLDIPa | LOC107763626 | 1.8 ± 0.8 | n.d. in controls |
| Unknown protein | NtPTLD3 | LOC107796437 | 1.7 ± 0.8 | 249 |
| Retinol dehydrogenase2 | NtFEY1 | LOC107800303 | 1.2 ± 4 | 1087 |
| LDIP | NtLDIPb | LOC107831283 | 1.1 ± 1.6 | n.d. in controls |
| Arfaptin domain-containing protein2 | NtADCP | LOC107827250 | 3.0 ± 0.6 | 9 |
| PLAT domain-containing protein3 | NtPDCP | LOC107793109 | 2.2 ± 0.6 | 10 |
| Small G protein3 | NtRABB1b | LOC107771805 | 1.12 ± 0.16 | 36 |
| Unknown protein1 | NtPTLD4 | LOC107778489 | 0.7 ± 0.2 | n.d. in controls |
| Carotenoid cleavage dioxygenase3 | NtCCD | LOC107830214 | 0.45 ± 0.08 | 245 |
| Small G protein3 | NtRAB2c | LOC107794510 | 0.13 ± 0.11 | 15 |
LDs were isolated from tobacco pollen tubes. Their protein composition (n = 3 biological replicates) was compared to total extracts and a cytosolic fraction (both n = 4) by LC-MS/MS after a tryptic in-gel digest. iBAQ values were determined using MaxQuant software. The relative iBAQ values were calculated as ‰ of all proteins in one sample. The enrichments were each determined from the LD iBAQ ‰ value divided by the higher value of the total and cytosolic fractions. Only proteins with an enrichment factor of >100 and making up more than 1 ‰ of the total iBAQ values were considered for the upper part of the list. Further proteins analyzed in respect to their localization are depicted in the lower part. For a list of all identified proteins, see Supplemental Data Set 1. Several corresponding cDNAs were cloned and transiently expressed as mVenus fusions in tobacco pollen tubes:
protein localized to LDs (1), protein localized to the ER (2), and protein that did not localize to the LDs or the ER (3). The “±” indicates sd. n.d., not detected.
Figure 1.
Proteins Localizing to LDs.
The proteins were transiently expressed in tobacco pollen tubes, as C-terminal ([D] and [E]) or N-terminal ([A] to [C] and [F] to [I]) fusions to the fluorescent protein mVenus. The tubes were cultivated for 5 to 8 h, fixed with formaldehyde, and stained with Nile Red. Then, they were monitored by confocal microscopy. Colocalization was observed in all of 7 (A), 9 (B), 11 (C), 14 ([D] and [E]), 13 (F), and 7 (G) pollen tubes. (E) is a magnified section of (D). Partial association of mVenus structures and Nile Red-stained LDs was observed in 17 of 23 ([H] and [I]) pollen tubes. Bars = 10 µm in (A) to (D) and (F) to (I) and 2 µm in (E).
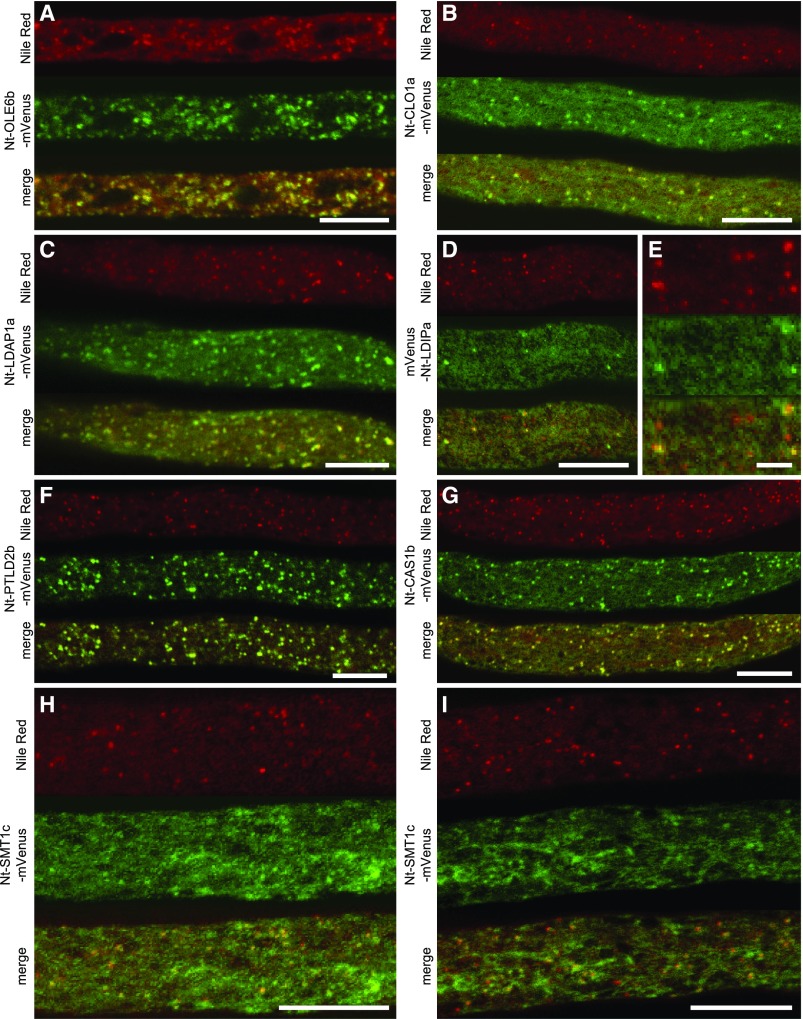
Figure 2.
The Hydrophobic Stretch Is Both Essential and Sufficient for LD Association.
(A) and (B) The full-length Nt-PUX10c protein with a C-terminal mVenus fusion localizes to LDs and to the ER highlighted by the ER marker ERD-CFP.
(C) The overall domain structure of Nt-PUX10. The protein contains a hydrophobic stretch (HS; hydrophobic region predicted with TMHMM; see Supplemental Figure 13) a UBA domain predicted to interact with ubiquitinated proteins, a UAS domain of unknown function, and a UBX domain commonly considered to interact with CDC48-type proteins. Domain prediction made with CD-Search.
(D) All variants including the region comprising amino acids 81 to 131 colocalized at least in part with LDs, while all constructs excluding this region were cytosolic (indicated by Cyt). A construct missing amino acids 1 to 88 and 120 to 465 localized to punctate structures (indicated by PS) not identical with LDs.
The proteins were transiently expressed in tobacco pollen tubes, as C- or N-terminal fusions to the fluorescent protein mVenus. The tubes were cultivated for 5 to 8 h, fixed with formaldehyde, and stained with Nile Red ([A] and [D]) or not fixed and stained (B). Similar localization was observed in n = 6 to 11 independent pollen tubes per construct. Bars = 10 µm. See Supplemental Figure 14 for magnified images.
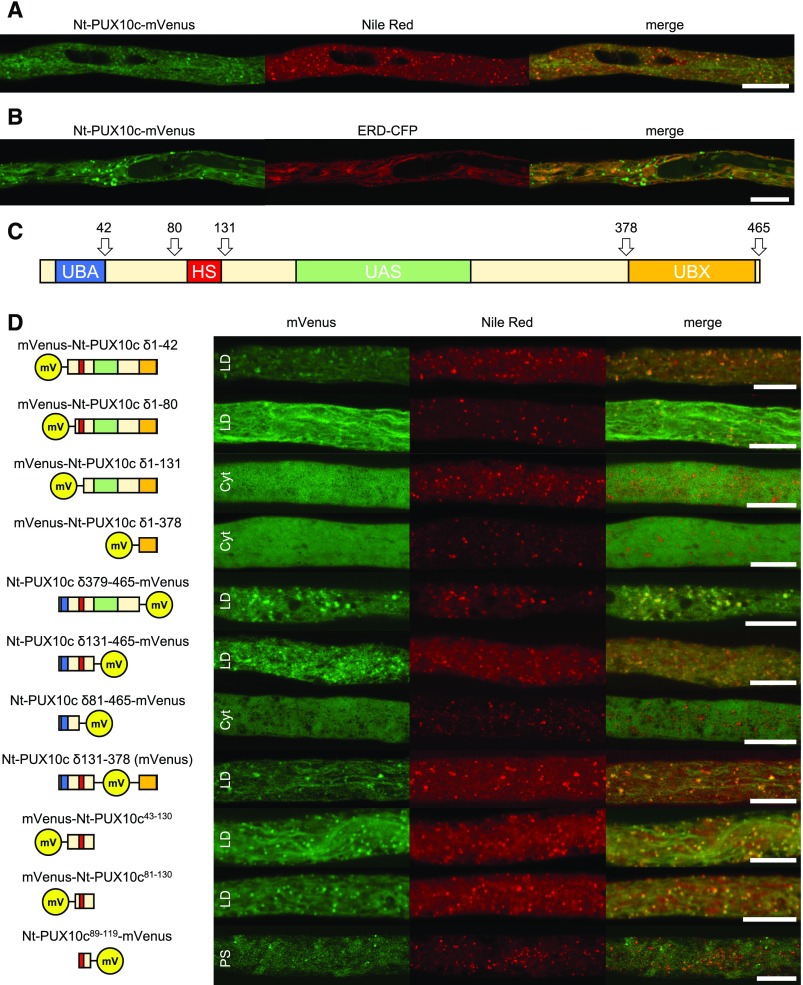
Figure 3.
Proteins Not Localizing to LDs.
The proteins were transiently expressed in tobacco pollen tubes, as N-terminal ([A] to [F] and [I]) or C-terminal ([G] and [H]) fusions to the fluorescent protein mVenus. The tubes were cultivated for 5 to 8 h, fixed with formaldehyde, and stained with Nile Red ([A], [C], [D], and [F]), only fixed ([B] and [E]), or not treated ([H] and [I]). Then, they were monitored by confocal microscopy. (C) is a magnified section of (A), showing partial association of mVenus and Nile Red fluorescence. Images are representative of 10 ([A], [C] to [E], and [H]), 14 ([B] and [I]), 7 ([C] and [F]), and 13 (G) pollen tubes. Bars = 10 µm in (A), (B), and (D) to (I) and 1 µm (C).
We also tested other selected proteins that had either high iBAQ ‰ scores or a strong enrichment and that were potentially interesting candidates based on their known cellular function(s) (Table 1, lower part). None of these proteins, however, showed any obvious localization, partial or otherwise, to LDs (Figure 3), reinforcing the premise that only the combination of a high iBAQ ‰ and a strong enrichment reflected a protein’s association with LDs. Nonetheless, one protein with unknown function termed here ARFAPTIN DOMAIN-CONTAINING PROTEIN (ADCP) localized mostly to the ER, but appeared enriched in domains interacting with LDs (Figures 3A and 3B). Other selected proteins localized to either the cytosol (the small G proteins Nt-RABB1b and Nt-RAB2c; Figures 3G and 3H), plasma membrane (carotenoid cleavage dioxygenase Nt-CCD; Figure 3I), or to unknown punctate structures (a PLAT-domain containing protein, Nt-PDCP).
Taken together, the addition of a second line of evidence for LD localization via transient expression in tobacco pollen tubes confirmed many of the positive candidates identified in our proteomics screen. One of these, Nt-PUX10c, and its homolog from Arabidopsis (Supplemental Figure 4 and Supplemental Data Set 10), were subsequently chosen for a more in-depth investigation (see below), as these proteins have previously not been described as LD proteins in plants, yet other members of the Arabidopsis family of PUX proteins are known or implied to play a central role in protein degradation (Liu and Li, 2014).
Nt-PUX10 Is Targeted to LDs by a Short, Hydrophobic Polypeptide Sequence
PUX proteins are a diverse family of proteins that share the eponymous UBX domain, but can differ in their overall domain structure (Supplemental Figure 12; Liu and Li, 2014). In Arabidopsis, 16 PUX proteins have been identified (Supplemental Figure 12) and, apart from the UBX domain, the isoforms At-PUX5, At-PUX7 to At-PUX11, and At-PUX13 to At-PUX16 also possess a putative ubiquitin-associated (UBA) domain. Additionally, At-PUX10 harbors a unique hydrophobic polypeptide sequence that is predicted to be a transmembrane domain (TMHMM Server v. 2.0; Krogh et al., 2001; Supplemental Figure 13) and is not present in any of the other members of the Arabidopsis PUX protein family (Supplemental Figure 13). Tobacco possesses six isoforms most closely related to At-PUX10 (Supplemental Figure 4 and Supplemental Data Set 10), which we named Nt-PUX10a-f, and among these, only Nt-PUX10a-d contain a hydrophobic region similar to that in At-PUX10 (Supplemental Figure 4 and Supplemental Data Set 10).
To test whether the hydrophobic region in the various PUX10 proteins that contain this sequence is necessary for LD targeting, a series of truncated constructs missing portions of the N terminus and/or the C terminus of Nt-PUX10c were generated and then transiently expressed as fusion proteins to mVenus in tobacco pollen tubes (Figure 2D; see Supplemental Figure 14 for magnified images). Overall, all the Nt-PUX10c-mVenus fusion constructs lacking the hydrophobic region (i.e., amino acids 89–119) were mislocalized to the cytoplasm, while all other fusion constructs containing this region targeted, at least partially, to LDs, including one that comprised only the 50 amino acids (amino acids 81–130; Figure 2D) spanning the hydrophobic region. An even shorter variant (amino acids 89–119) was not able to target the fluorophore to LDs. These results indicate that the novel hydrophobic region and the adjacent amino acids in Nt-PUX10 are not only necessary, but also sufficient for targeting to LDs.
Nt-PUX10 Can Recruit At-CDC48A to LDs
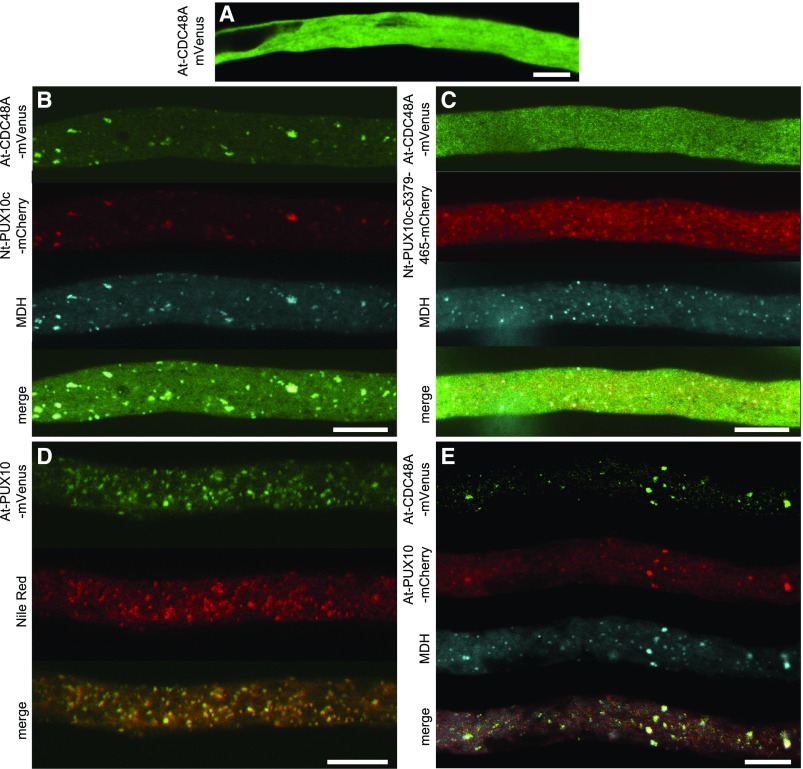
One of the described functions of UBX domain-containing proteins is to interact and recruit CDC48 (Liu and Li, 2014), an AAA-type ATPase involved in the proteome homeostasis of various cellular processes and an important player for the degradation of ubiquitinated proteins by the proteasome (Baek et al., 2013). To test if PUX10 can influence the subcellular localization of CDC48, we employed again the transient expression system in tobacco pollen tubes. We chose At-CDC48A as a potential interaction partner, as this is the best-described isoform from plants (Park et al., 2008; Copeland et al., 2016). At-CDC48A expressed alone as an mVenus fusion accumulated in the cytoplasm, with no obvious localization to any other intracellular compartment(s) (Figure 4A). However, when At-CDC48A was coexpressed with Nt-PUX10c-mCherry, both proteins clearly colocalized to LDs stained with the LD marker stain monodansylpentane (Figure 4B). Notably, this relocalization of At-CDC48A to LDs was not observed when At-CDC48A was coexpressed with Nt-PUX10c lacking its UBX domain (Figure 4C), indicating that the recruitment of At-CDC48A to LDs by Nt-PUX10c is mediated by its UBX domain. Similarly, At-CDC48A was relocalized to LDs when coexpressed with the Arabidopsis homolog of Nt-PUX10c, i.e., At-PUX10 (Figures 4D and 4E).
Figure 4.
PUX10 Can Recruit At-CDC48A to LDs in Pollen Tubes with the Help of Its UBX Domain.
The proteins were transiently expressed in tobacco pollen tubes as N-terminal fusions to the fluorescent protein mVenus or mCherry as indicated. The tubes were cultivated for 5 to 8 h, fixed with formaldehyde, and stained with the blue fluorescent LD dye monodansylpentane (MDH; [B], [C], and [E]) or Nile Red (D). Then, they were monitored by confocal microscopy. At-CDC48A expressed alone is localized in the cytosol (A) but is recruited to LDs by Nt-PUX10c (B). Nt-PUX10c missing the C-terminal UBX domain (δ379-465) still localizes to LDs but can no longer recruit At-CDC48A (C). At-PUX10 alone localizes to LDs (D) and can also recruit At-CDC48A. Images are representative of 11 (A), 9 (B), 10 (C), 12 (D), and10 (E) pollen tubes. Bars = 10 µm.
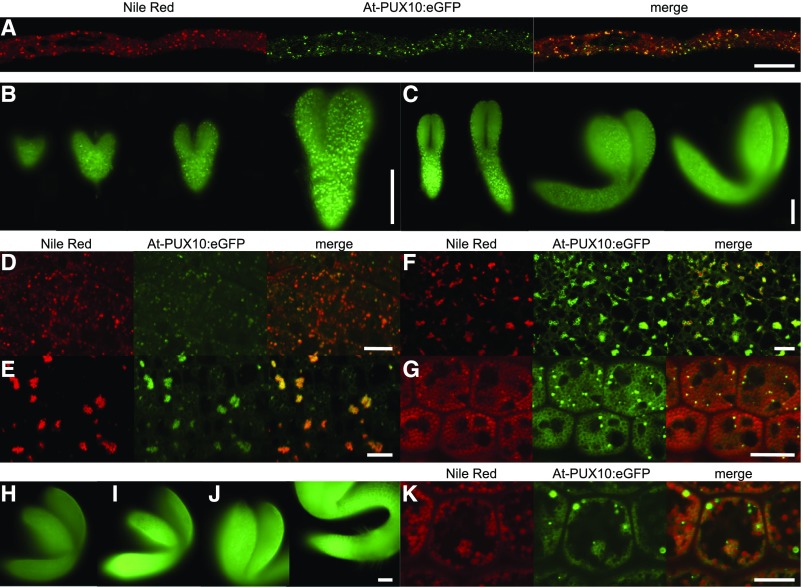
At-PUX10 Localizes to LDs in Pollen Tubes, Developing Embryos, and Seedlings
To further investigate the tissue-specific expression of At-PUX10 and its subcellular localization in a native system, the entire genomic sequence of At-PUX10, including a 642-bp promoter region in frame with an eGFP-encoding sequence (i.e., At-PUX10pro:At-PUX10-eGFP), was cloned and 10 stable transgenic lines were generated, from which three were chosen for further analysis (based on the relative strength of the fluorescence signal attributable to At-PUX10pro:At-PUX10-eGFP in transgenic seedlings). In all lines, At-PUX10-eGFP fluorescence was observed in pollen tubes grown in vitro (Figure 5) and in developing embryos starting at the heart stage through to mature embryos (Figures 5B and 5C). In the latter case, At-PUX10-eGFP fluorescence levels were somewhat weaker, but during seed germination they increased again (Figures 5H to 5J). At all stages, At-PUX10-eGFP was found predominantly on LDs (Figures 5D to 5G). Interestingly, in seedlings, At-PUX10-eGFP fluorescence was not equally distributed between all LDs, but, instead, appears to be concentrated on only a subpopulation of LDs (Figure 5K). This uneven distribution persisted in isolated LDs (Supplemental Figure 15). On the other hand, in comparison to Nt-PUX10-mVenus transiently expressed in tobacco pollen tubes (Figure 2A), no localization of At-PUX10-eGFP to ER-like structures was observed.
Figure 5.
PUX10-eGFP Is Present at LDs during Embryo Development, Seed Germination, and in Pollen Tubes When Expression Is Driven under the Native Promoter.
The construct At-PUX10pro:gAt-PUX10:eGFP was used to stably transform Arabidopsis plants. Three of originally 10 independent lines were chosen for further investigation. eGFP fluorescence colocalized with Nile Red in Arabidopsis pollen tubes. The tubes were cultivated for 2 h and then fixed with formaldehyde and stained with Nile Red. Fluorescence was also detected by epifluorescence microscopy throughout embryo development ([B] and [C]) and in germinating seedlings ([H], imbibed seed; [I], 24 h; and [J], 48 h after imbibition; both images are taken from the same seedling). All images were obtained with identical settings, under which wild-type plants did not show any significant signal. Colocalization with Nile Red-stained LDs was observed by confocal microscopy in the heart (D), torpedo (E), walking-stick (F), and mature stage (G) as well as in the seedling 24 h after imbibition (K). In the mature stage and in seedlings, fluorescence was increased around individual LDs. All images are representative of at least five embryos, seedlings per stage, or five pollen tubes for each of three independent lines. Bars = 100 µm in (B), (C), and (H) to (J) and 10 µm in (A), (D) to (G), and (K).
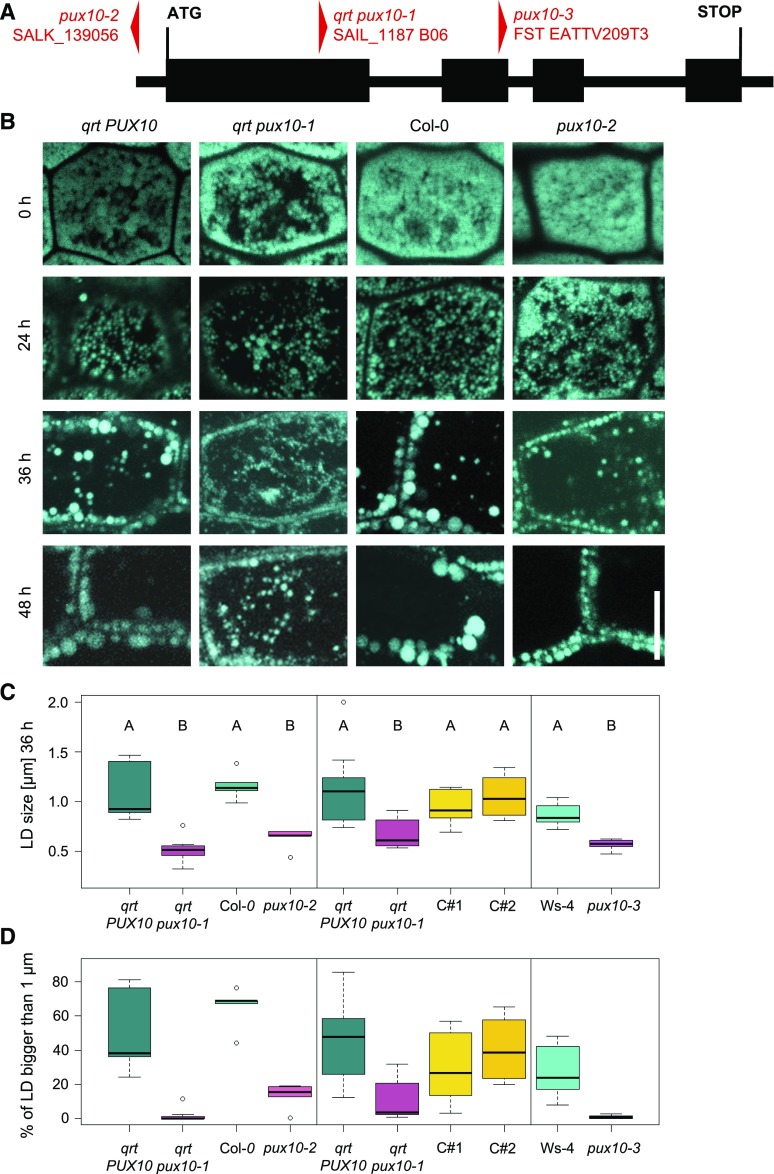
LD Size Is Altered in Arabidopsis pux10 T-DNA Insertion Mutants
For functional characterization of At-PUX10, T-DNA insertion lines of Arabidopsis were obtained from the NASC seed stock center and the insertions of the T-DNA in PUX10 were determined to be in the promoter region (SALK_139056 in the Col-0 background; referred to here as pux10-2) and the first exon (SAIL_1187 B06 in the qrt [quartet] background; qrt pux10-1). Another mutant line was obtained from INRA Versailles-Grignon Center with an insertion in the second exon of PUX10 (FST EATTV209T3 in the Ws-4 background; pux10-3). All insertions were confirmed by genotyping PCR and sequencing (Figure 6A; Supplemental Data Set 17). qrt PUX10 and qrt pux10-1 plants are offspring of the same heterozygous plant (qrt pux10-1/PUX10). Additionally, RT-qPCR was performed to assess transcript levels in each of the pux10 mutants and their respective parental backgrounds. Toward that end, RNA was isolated from dry seeds of each line and, as shown in Supplemental Figure 16, for qrt pux10-1 and pux10-2, PUX10 transcripts were reduced to 10% and 25%, respectively. For pux10-3, no PUX10 transcript was detected (Supplemental Figure 16). Despite the fact that PUX10 expression in the qrt pux10-1 mutant was not abolished completely, no PUX10 peptides were detected in all but one sample of LDs isolated from the qrt pux10-1 mutant (Supplemental Figure 17).
Figure 6.
LDs in Seedling Hypocotyls Are Smaller in the pux10 Mutants.
(A) Localization of the T-DNA insertions in PUX10. The gene consists of four exons (bold line) and three introns (thin lines). The SALK_139056 (pux10-2) insertion is localized 100 bp in 5′ direction from the start codon in the promoter region, the SAIL_1187 B06 (qrt pux10-1) insertion is 637 bp into the first exon, and the FST EATTV209T3 (pux10-3) insertion 387 bp into the second exon.
(B) to (D) Seeds were imbibed for 16 h at 4°C and subsequently grown under continuous light ([B] and [C], left panel) or long-day conditions ([C], middle and right panels).
(B) Confocal images of the hypocotyl represent single slides (0 and 24 h) or stacks (36 and 48 h). Images are excerpts from larger images and representative of 10 seedlings (from two independent experiments). Bar = 10 µm.
(C) and (D) Confocal scans from seedlings (36 h) were used for quantification and the average diameter of the LDs determined from one image of each plant (n = 4–11 independent seedlings per line, 20–188 LDs per seedling). Absolute average sizes of LDs (C) and the percentage of LDs bigger than 1 µm are presented in (C) and (D), respectively. LD size was quantified with ImageJ and statistically analyzed by one-factor ANOVA followed by Tukey’s post-hoc test. The test indicated a significant difference (P < 0.02) for all pux10 mutants in comparison to all of the respective control lines. Plots were created in R.
Results presented in (B), and in the left panels in (C) and (D), originate from the same experiment. Different panels represent independent experiments using seed material of independently grown plants.
In all three pux10 mutant lines, LD morphology during germination was altered when compared with their respective nonmutated backgrounds. For instance, in Col-0 (background to pux10-2) and qrt PUX10 (background to qrt pux10-1) seedlings, LD size increased while their number decreased during the progress of germination (Figure 6B). However, in both of the corresponding pux10 mutant lines, LDs appeared to stay smaller and were higher in number at later stages of germination (36 and 48 h). Indeed, quantification of LD sizes in five different seedlings per line (at 36 h) revealed that LDs in qrt pux10-1 and pux10-2 seedlings were significantly smaller than in the respective backgrounds (Figure 6C, left panel), and in particular the number of LDs larger than 1 µm were reduced (Figure 6D, left panel).
At a similar time point (40 h after imbibition), the LDs in the hypocotyl cells of two complemented lines (complemented with At-PUX10pro:At-PUX10-eGFP, termed complemented 1 [C#1] and complemented 2 [C#2]) were significantly larger than the LDs in the qrt pux10-1 mutant, but did not differ significantly from the qrt PUX10 background (Figure 6C, middle panel). Furthermore and similar to the results described above, the proportion of LDs with a diameter of more than 1 µm was similar in the qrt PUX10 background and the two complementation lines, but significantly reduced in the qrt pux10-1 mutant (Figure 6D, middle panel). These results were confirmed using the third independent pux10 T-DNA insertion mutant line, pux10-3. That is, LDs in pux10-3 seedlings at 40 h after imbibition were significantly smaller than those in the Ws-4 background (Figure 6C, right panel). Furthermore, almost no LDs with a diameter above 1 µm were observed in the pux10-3 mutant, whereas ∼20% of the LDs of that size were seen in Ws-4 seedlings (Figure 6D, right panel). Overall, these results confirm that the observed phenotype in LD size and number is due to a disruption of PUX10 expression.
LD Proteins Are Degraded More Slowly in the Arabidopsis pux10 Mutants
It has been shown previously that the removal of a LD proteinaffects LD morphology in plants (Pyc et al., 2017a; Huang, 2018). For instance, Siloto et al. (2006) reported that the disruption of expression of the major structural LD protein in Arabidopsis seeds, OLEOSIN1, results in a dramatic increase in LD size, indicating a link between the abundance of structural LD proteins and LD size. As such, PUX10, by recruiting CDC48 to LDs and thereby possibly regulating LD degradation, might influence the abundance of LD proteins. To test this hypothesis, we took a proteomic approach, whereby proteins isolated from homogenized seedlings of different lines (qrt PUX10, qrt pux10-1, C#1, and C#2, as well as Ws-4 and pux10-3) and different developmental stages (rehydrated seeds, and seedlings 1 to 3 d after imbibition [DAI]) were subjected to tryptic digestion and LC-MS/MS analysis (Figure 7; Supplemental Figure 16). The resulting data were processed with the MaxQuant label-free quantification (LFQ) algorithm for each time point (Supplemental Data Sets 18 to 21) and calculated as the ‰ value of total LFQ intensities (Supplemental Data Sets 22 to 27).
Figure 7.
LD Proteins Are Enriched in the pux10 Mutant Seedlings.
(A) In the qrt pux10-1 line, LD proteins are significantly more abundant at 1 and 2 DAI when compared with the qrt PUX10 line. This effect is partially rescued in the complemented lines. Proteins were isolated from total seedling homogenate of rehydrated seeds (0 DAI) and seedlings at 1, 2, and 3 DAI. The same experiment was independently conducted on seeds and seedlings of Ws-4 and pux10-3. Here, the observed difference is greatest at 0 and 1 DAI. LC-MS/MS data were processed with MaxQuant software’s LFQ algorithm. Values are given as ‰ of total LFQ intensities. Error bars correspond to sd, when comparing the summed up intensities of the 13 given proteins between the three biological replicates. The same summed up intensities for each biological replicate were used for one-factor ANOVA followed by Tukey’s post-hoc test; lowercase letters indicate statistical significance, P < 0.05.
(B) The same values as in (A) were normalized for each protein to the highest value in all compared lines and time points. Green shows high abundance and dark red not detectable. For separate graphs, see Supplemental Figure 18, and for all individual values, see Supplemental Data Sets 18 and 29.
In total, 1690 proteins were detected and quantified with the complexity of the proteome strongly increasing during germination. The sum of these relative LFQ intensities for 13 major LD proteins (i.e., the oleosins OLE1-8, HSD1, and HSD5, LDAP2, and the caleosins CLO1 and CLO2) was calculated for every time point and genetic background (Supplemental Data Set 28). While the relative abundance of the LD proteins in the total fractions strongly decreased in all lines within 3 d, at 1 and 2 DAI, significantly more of these proteins were present in the qrt pux10-1 mutant when compared with the qrt PUX10 background (Figure 7A). This effect was rescued in C#2 at 1 DAI. At 2 DAI, especially the abundances of several individual proteins (namely, OLE4, OLE5, and HSD1; Figure 7B; Supplemental Figure 18A) were higher in the qrt pux10-1 mutant than in the wild type. Similar analysis of the pux10-3 mutant line (Supplemental Data Sets 29 to 39) also showed a higher abundance of LD proteins at 0 and 1 DAI (Figure 7A). Furthermore, the degradation of LD proteins was relatively faster in comparison to that in pux10 mutant lines in the Col-0 background, presumably due to the difference in the ecotype. In the pux10-3 line, again several individual proteins showed a higher abundance, especially at 1 DAI, where the oleosins OLE1, OLE4, and OLE5 were significantly increased (Figure 7B; Supplemental Figure 18B). Overall, however, LD protein degradation was not completely abolished in any of the pux10 mutants, but merely slowed down. In contrast to protein degradation, TAG degradation and postgerminative growth were not changed as measured by the decrease of the marker fatty acid C20:1 (Supplemental Figure 19).
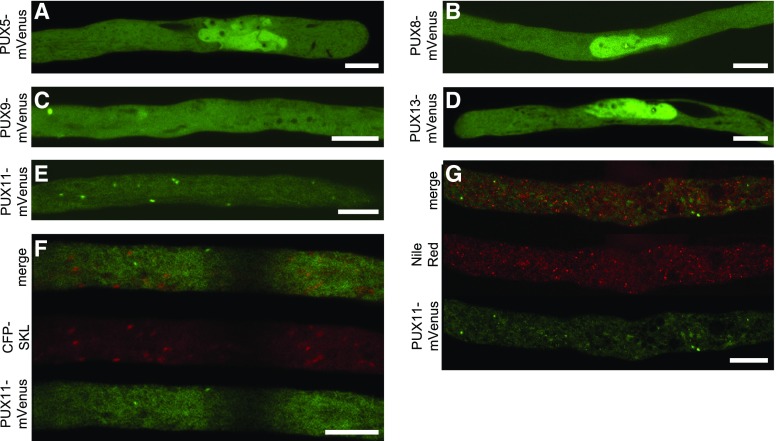
To investigate whether one of the other proteins of the PUX gene family containing the UBA and UBX domains might have redundant functions with PUX10 (Supplemental Figure 12), we cloned and analyzed the subcellular localization of the Arabidopsis proteins PUX5, PUX8, PUX9, PUX11, and PUX13 (fused to mVenus) by transient expression in tobacco pollen tubes. As shown in Figure 8, PUX5, PUX8, and PUX13 localized to the nucleus, similar to what has been described previously for PUX7 (Gallois et al., 2013), whereas PUX9 was cytosolic. PUX11 localized also to the cytoplasm, as well as to punctate structures, which were ruled out to be LDs or peroxisomes (Figure 8).
Figure 8.
Subcellular Localization of Other Members of the Arabidopsis PUX Gene Family.
To analyze whether PUX10 has functionally redundant gene family members that also localize to LD and/or the ER, the following genes were cloned, transiently expressed as N-terminal fusions to mVenus in tobacco pollen tubes, fixed, and observed by confocal microscopy. Images are representative of 6 ([A], [E], and [F]), 11 (B), 8 (C), 9 (D), and 5 (G) pollen tubes. Bars = 10 µm.
(A), (B), and (D) PUX5, PUX8, and PUX13 localize preferentially to the nucleus and the cytosol.
(C) PUX9 is localized in the cytosol.
(E) PUX11 localizes to punctuate structures in the cytosol.
(F) and (G) The structures were excluded to be LDs or peroxisomes by staining with Nile Red (G) or by coexpression with the peroxisomal marker CFP-SKL (F), respectively.
Ubiquitin Is Enriched in LD-Enriched Fractions of the qrt pux10-1 Mutant
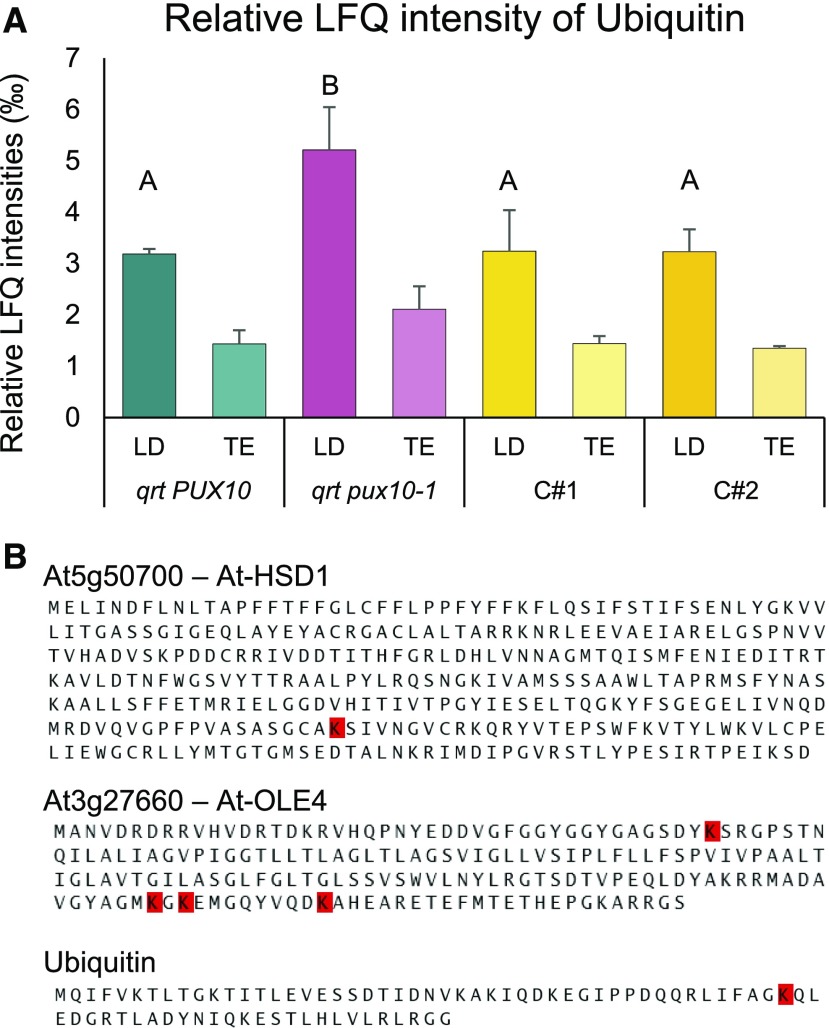
To gain more detailed insight into the protein composition of LDs of the qrt pux10-1 mutant line and to increase the chances of detecting posttranslationally modified peptides, LD-enriched fractions were analyzed in another proteomics-based approach. Here, the LD fraction from homogenized seedlings of qrt PUX10, qrt pux10-1, C#1, and C#2 at 38 h after imbibition (Supplemental Data Sets 40 to 43) was isolated, but prior to fractionation by centrifugation, aliquots of total protein were taken for comparison. Again, the different samples were tryptically digested and analyzed by LC-MS/MS. The data were processed as described above, and, overall, there was an enrichment of LD proteins in the LD fraction, as expected (Supplemental Data Set 42). In addition, the relative LFQ intensities of ubiquitin in the LD fraction of qrt pux10-1 seedlings was significantly increased compared with all control plants (Figure 9A). Ubiquitin was also 2-fold enriched in the LD fraction compared with the total fraction (P < 0.02). Moreover, an immunoblot analysis against ubiquitin indicated stronger ubiquitination of proteins in the LD fraction of the qrt pux10-1 line in comparison to the wild type and complemented lines (Supplemental Figure 20).
Figure 9.
Ubiquitin Accumulates in LD-Enriched Fractions of the qrt pux10-1 Mutant Seedlings.
(A) Proteins were isolated from LD fractions and the respective total protein fractions (TE) of seedlings at 2 DAI. Peptides derived from these fractions were analyzed via LC-MS/MS. The LFQ algorithm was applied, and values are presented as averages of three biological replicates in ‰ of total LFQ. Error bars indicate sd. Statistical analysis was performed by one-factor ANOVA followed by Tukey’s post-hoc test. Lowercase letters indicate significant differences (P < 0.05).
(B) Ubiquitination sites were detected in the LD proteins HSD1, OLE4, and ubiquitin via LC-MS/MS analysis of their tryptic peptides (see Supplemental Figures 22 to 24 for spectra). Ubiquitination sites are highlighted in red.
To detect potential ubiquitination sites on a qualitative level, the proteomic data were analyzed for peptides containing lysine residues with two attached glycines, which remain at the C terminus of the attached ubiquitin after tryptic digestion. More specifically, data obtained for qrt PUX10 and qrt pux10-1 seedlings at 1 and 2 DAI were analyzed (Supplemental Data Sets 44 to 46), and, based on this, several ubiquitination sites were identified. The ubiquitination position within the ubiquitin protein, Lys-48 (K48) was identified (Figure 9B). Ubiquitination positions were also detected for HSD1 (K269) and OLE4 (K157, K159, and K168) (Figure 9B; see Supplemental Figures 21 to 24 for fragmentation spectra). These proteins are among the proteins significantly enriched in qrt pux10-1 seedlings at 2 DAI (Supplemental Figure 18 and Supplemental Data Sets 28 and 39). All proteomic data of LDs isolated from qrt PUX10, qrt pux10-1, at 1 and 2 DAI as well as from qrt PUX10, qrt pux10-1, C#1, and C#2 at 38 h after imbibition are displayed in the supplements (Supplemental Data Sets 40, 41, 43, and 44). All ubiquitinated peptides are displayed in Supplemental Data Sets 43 and 46.
DISCUSSION
The Composition of LDs Differs from Pollen Tubes to Seeds
Tobacco pollen grains store smaller amounts of neutral lipids (2% of the dry weight; Rotsch et al., 2017) in comparison to many dicotyledon oilseeds like Arabidopsis (30% of the dry weight; Cai et al., 2017), and the LDs appear smaller (Rotsch et al., 2017). In addition, the LDs in pollen tubes and oilseeds differ in their neutral lipid composition: Sterol esters make up a much larger proportion of neutral lipids (24% on a weight basis; Rotsch et al., 2017) in pollen tubes in comparison to seeds from Arabidopsis (∼0.3% of the level of TAG; Bouvier-Navé et al., 2010). While seeds degrade their neutral lipids following germination, tobacco pollen tubes synthesize TAG (Mellema et al., 2002) and sterol esters (Müller and Ischebeck, 2018), and pollen tubes cultivated for 14 h still contain large amounts of these neutral lipids (Müller and Ischebeck, 2018). By contrast, pollen tubes from olive (Olea europaea) degrade their LDs during pollen tube growth (Zienkiewicz et al., 2013). It remains unclear, though, if LDs are synthesized de novo in tobacco pollen tubes. Evidence for the importance of LDs for pollen tube growth comes in part from recent observations of Arabidopsis pollen disrupted in all three SEIPIN genes (Taurino et al., 2018). These pollen grains contain conspicuously enlarged LDs that cannot enter the pollen tube, leading to reduced fertility. LD degradation appears to be important to some degree, as mutants disrupted in a LD-associated TAG lipase show reduced pollen tube growth (Müller and Ischebeck, 2018). Hence, while these data support the importance of LDs during pollen tube growth, it remains to be determined what specific function(s) these organelles serve. Differences in protein and/or lipid composition of LDs in different tissue types might both reflect different function(s) of the organelle. For instance, similar to other studies on pollen LDs (Kim et al., 2002; Jiang et al., 2007), we found two oleosins as two of the most abundant proteins (Table 1). Their presence could be explained by the desiccation phase that pollen grains undergo before rehydration, similar to seed desiccation, where oleosins are also present in relatively high amounts (Lee et al., 1994; Huang, 1996; Wang et al., 1997). On the other hand, we also detected steroleosins in tobacco seeds (Supplemental Data Set 6) and Arabidopsis seedlings (Supplemental Data Sets 39, 42, and 46), but not in pollen tubes (Supplemental Data Set 2), indicating that their potential function in brassinosteroid synthesis is missing in pollen tubes (Li et al., 2007). By contrast, at least three caleosin isoforms were detected in pollen tubes (Supplemental Data Set 4). Two of these caleosins, Nt-CLO1a and Nt-CLO1b, were the most and third most abundant LD proteins based on the relative iBAQ score (Table 1), indicating that caleosins are more predominant in the LDs of pollen tubes than in seeds and seedlings, where oleosins predominate (Pyc et al., 2017b; Supplemental Data Sets 42 and 46). Nt-CLO1a and Nt-CLO1b are most closely related to the isoforms CLO1 and CLO2 from Arabidopsis (Supplemental Figure 4), which display peroxygenase activity (Hanano et al., 2006; Poxleitner et al., 2006), indicating that such an activity could also be of importance in pollen tubes.
Similar to caleosins, the recently described LDAPs (Horn et al., 2013) and LDIP (Pyc et al., 2017b) showed a much higher relative abundance in tobacco pollen tubes (Supplemental Data Set 4) compared with Arabidopsis seedlings (Pyc et al., 2017b) and were not detected in tobacco seeds (Supplemental Data Set 6). Interestingly, Nt-LDIPa did not localize to all LDs, when transiently expressed in tobacco pollen tubes (Figures 1D and 1E) as its Arabidopsis homolog does in Nicotiana benthamiana leaves (Pyc et al., 2017b), suggesting the existence of different subpopulations of LDs in pollen tubes. Future studies using pollen tubes as model systems should help elucidate the function of LDIP and its sorting to distinct LDs.
Our proteomic and cell biological data indicated that sterol synthesis is a potential novel function for LDs, since we detected both a putative cycloartenol synthase (Nt-CAS1) and a cycloartenol-C-24-methyltransferase (Nt-SMT1) enriched in LD fractions (Table 1). While both Nt-CAS1 and At-CAS1 clearly localized to LDs in tobacco pollen tubes (Figure 1G; Supplemental Figure 11), the localization of Nt-SMT1 was less clear (Figured 1H and 1I), indicating that it is not exclusively localized at LDs. In contrast to At-CAS1, At-SMT1 is also less strongly enriched in Arabidopsis seedling LDs based on proteomic data (Pyc et al., 2017b; Supplemental Data Sets 42 and 46). As such, while we found on LDs two of the four enzymes required for the truncated sterol synthesis pathway from squalene to cycloeucalenol described for tobacco pollen tubes (Villette et al., 2015; Rotsch et al., 2017), it remains to be determined if sterols are synthesized on LDs in tobacco pollen tubes or on LDs in other tissues. Interestingly, some enzymes of the newly discovered alternative cholesterol synthesis pathway have been speculated to be localized at LDs (Sonawane et al., 2016), further reinforcing the growing functional repertoire of this organelle.
Another enzyme we identified enriched in the LD fraction (Table 1) is a tobacco homolog of the castor bean (Ricinus communis) oil body lipase (Rc-OBL; Eastmond, 2004). Recently, we confirmed that both Nt-OBL and its Arabidopsis homolog At-OBL1 are LD-associated and that At-OBL1 is a TAG, diacylglycerol, and 1-monoacylglycerol lipase and that the activity of At-OBL1 is important for pollen tube growth (Müller and Ischebeck, 2018). We also found a previously unknown LD protein, PTLDb (Table 1, Figure 1F), and via BLAST analysis, we identified four additional tobacco homologs (Supplemental Figure 10), all of which were enriched in LD fractions isolated from tobacco pollen tubes (Supplemental Data Set 4). PTLD proteins are evolutionarily conserved with homologs present in many species, including plants important for oil production such as olive, oil palm (Elaeis guineensis), soybean (Glycine max), and maize (Zea mays), but not in Brassicaceae. It remains to be resolved, however, if this protein is only specific for pollen tubes or if it is also found in seeds. Finally, one of the most notable findings from our proteomics analysis of the LD fraction of tobacco pollen tubes in terms of potentially novel LD proteinswere Nt-PUX10 proteins, although At-PUX10 was found to be enriched in the LD fraction of Arabidopsis seedlings (Pyc et al., 2017b; Supplemental Data Set 42). In addition, in humans, the UBX domain-containing protein UBXD8, which is normally ER localized, relocates to LDs upon treatment of cultured cells with the fatty acid oleate (Olzmann et al., 2013).
Overall, the verification of LD localization via an appropriate screening method as presented here with transient expression in tobacco pollen tubes efficiently eliminates false positive candidates determined by proteomic approaches and identified several novel LD proteins.
Determinants of LD Localization
One culprit that limits the discovery and characterization of LD proteins is the lack of a known consensus signal sequence for targeting the organelle. Targeting sequences are a conserved concept for the correct distribution of proteins to subcellular compartments and in past years, in silico analyses of sequenced genomes allowed for the reliable prediction of the subcellular localization of putative proteins (Meinken and Min, 2012). However, such a conserved sequence has not been described for known LD proteins, complicating both the discovery of additional LD proteins and the confirmation of their subcellular localization.
Nonetheless, one common feature among at least some plant LD proteins is a hydrophobic domain that presumably anchors the protein to the LD. For instance, oleosins are embedded in the TAG core of the LD through their protruding hydrophobic sequence that is predicted to form a hairpin-like structure with a proline knot motif (Jolivet et al., 2017). Similarly, caleosins and steroleosins have elongated hydrophobic sequences with a proline knot and knob, respectively, that anchors them to the LD (Tzen, 2012).
Both Nt-PUX10c and Rc-OBL contain N-terminal hydrophobic sequences, which are necessary for LD localization (Figure 2; Eastmond, 2004). However, the hydrophobic sequence of Nt-PUX10c alone (amino acids 94–116, Supplemental Figure 26) is not sufficient for the targeting to LDs (Nt-PUX10c89-119; Figure 2; Supplemental Figure 14). On the other hand, the inclusion of additional amino acid residues immediately adjacent to the hydrophobic sequence (as in Nt-PUX10c81-130; Figure 2D; Supplemental Figure 14) conveys proper localization to LDs. Using in silico studies, we also showed that this latter sequence, i.e., Nt-PUX10c81-130, is predicted to form an amphipathic helix that is evidently necessary for correct targeting (Supplemental Figure 26). Similarly, the PTLDs described here also contain a prominent hydrophobic region that might insert into the membrane (Supplemental Figure 27). Furthermore, Pyc et al. (2017b) recently reported that the proper targeting of At-LDIP to LDs relies on a discrete region of the protein that is predicted to form an amphipathic sequence and includes a hydrophobic sequence and adjacent residues.
How LD proteins with hydrophobic sequences are properly folded in the aqueous environment of the cytosol and finally find their correct target membrane remains to be investigated.
Other LD proteins, for example LDAPs (Gidda et al., 2016) and At-CAS1 (Supplemental Figure 25), have no obvious predicted hydrophobic regions and yet localize efficiently to the LD surface. Hence, their localization is thought to be mediated in other ways, perhaps via protein-protein or protein-lipid interactions. Evidence for protein-protein interactions mediating LD targeting comes from our experiments with PUX10 and CDC48A. The AAA-type ATPase CDC48 is a conserved ubiquitous enzyme that achieves spatial and temporal specificity through interaction with scaffold proteins. Scaffold proteins that interact with CDC48 are characterized by the presence of specific domains that mediate this interaction (Baek et al., 2013). One of those domains is the evolutionarily conserved UBX domain (Schuberth and Buchberger, 2008). Previously, it was shown for At-PUX1 and At-PUX7 via yeast two-hybrid experiments that their UBX domain can interact with CDC48 (Park et al., 2007; Gallois et al., 2013). Our transient pollen tube expression system showed that At-CDC48A relocalized from the cytoplasm when expressed on its own to LDs when coexpressed with either Nt-PUX10c or At-PUX10 (Figure 4). In a similar manner, other cytosolic factors needed to exert different LD functions at certain times may be recruited, extending the number of proteins potentially interacting with this organelle.
Differential Localization of LD Proteins within the Same Cell
While At-PUX10 could be confirmed to localize to LDs in embryos, seeds and pollen tubes (Figure 5), it does not appear to do so in a homogenous manner in all tissues. That is, following germination, PUX10 was observed to localize to specific LDs (Figure 5B). Similarly, Nt-LDIPa did not localize to all LDs when expressed transiently in tobacco pollen tubes (Figures 1D and 1E). Subpopulations of LDs were previously observed in yeast (Eisenberg-Bord et al., 2018), human cell culture (Wolins et al., 2005; Hsieh et al., 2012; Zhang et al., 2016), and Drosophila melanogaster (Wilfling et al., 2013; Thul et al., 2017). Based on these studies, different subpopulations of LDs were postulated to perform different functions. In plants, differential functionality of LD subpopulations has not been explored. Nonetheless, an attractive possibility is that LDs with different functions are established during seed germination and that PUX10 is involved in the required remodeling of the LD proteome to generate such functional subpopulations. The isolation of PUX10-enriched LDs followed by proteomic characterization might shed light on this possibility.
The Abundance of LD Proteins Appears to Influence the Size of LDs
The size of LDs is, at least partially, known to be determined by their protein-to-oil ratio. Ting et al. (1996) analyzed two different maize strains, which differ in their oil content but not in oleosin expression, and revealed differences in LD size and shape. In the high-oil strain, LDs were larger than in the strain strongly depleted of oil. Likewise, knockout mutants of OLE1 and OLE2 in Arabidopsis have enlarged LDs, especially in the case of OLE1 (Siloto et al., 2006), apparently because oleosins prevent LD fusion, or perhaps due to their role in LD synthesis.
During germination and postgerminative growth in Arabidopsis, oleosins are degraded in parallel to, or even faster than, TAG (Deruyffelaere et al., 2015; Figure 7; Supplemental Figure 19). There are hints that oleosin degradation is a prerequisite to oil breakdown as it might shield the LDs from TAG lipases (Matsui et al., 1999). Thus, the increase of LD size that is observed in Arabidopsis seedlings (Figure 6) could be caused by the degradation of LD proteins leading to the fusion of LDs if neutral lipids are not degraded at the same pace. Consistent with this conclusion, a delay in the breakdown of LD proteins in the qrt pux10-1 and pux10-3 mutant seedlings (Figure 7) could account for the reduction in the size of LDs, in comparison to the wild type (Figure 6). Alternatively, PUX10 could be involved directly or indirectly in LD fusion, but this remains speculative, as the proteins involved in LD fusion in plants, if the process even takes place, are so far not known and it is not clear how PUX10 could be involved in such a process.
Degradation of LDs and LD Proteins
The degradation of LD components has been the subject of extensive research. To date, the catabolism of the stored oil in LDs, particularly TAG, is far better understood than the breakdown of the proteins associated with the organelle. The lipases responsible for the majority of TAG degradation during postgerminative growth, SDP1 and SDP1-like, have been identified (Eastmond, 2006; Kelly et al., 2011). However, degradation of the LD proteins that “coat” the surface of the organelle is far less understood.
Recently, it was suggested that oleosins are substrates for the ubiquitin-proteasome system (UPS) of the cell. Deruyffelaere et al. (2015) proposed several ubiquitination sites in different oleosins and showed that they were polyubiquitinated. We could add more ubiquitination sites in OLE4 and HSD1, and additionally detected enhanced ubiquitination of LD proteins from qrt pux10-1 mutant seedlings (Figure 9B). However, due to the methods used, it is not possible to determine whether K48-linked ubiquitin peptides originate from oleosins. Still, our findings support the hypothesis that oleosins, and potentially other LD proteins, are substrates of the UPS and that PUX10 and CDC48 are involved in their degradation (Figure 10).
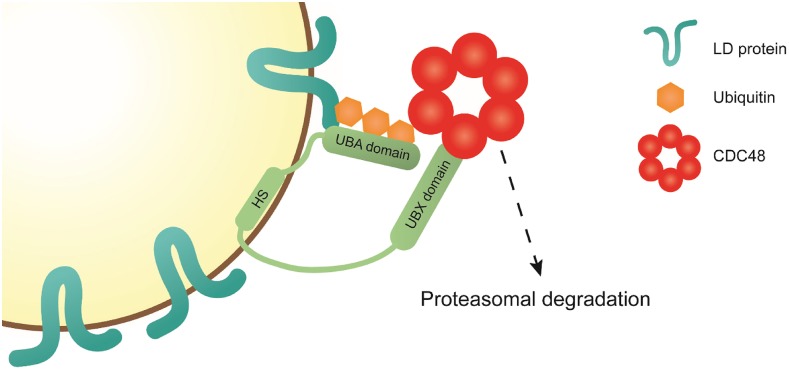
Figure 10.
Proposed Model for the Function of PUX10 on the LD.
LD proteins destined for degradation are marked with a polyubiquitin motif by K48-linked ubiquitins. This motif is recognized by the UBA domain of PUX10. PUX10 is an LD protein, attached to the organelle via its hydrophobic stretch (HS), and an amphipathic helix N-terminal of the HS. Via its UBX domain, PUX10 interacts with CDC48, recruiting this protein to the LD. CDC48 helps to channel the polyubiquitinated substrate to the proteasome. Whether CDC48 interacts directly with the proteasome, helps to extract the substrate from the LD, or recruits additional factors remains to be determined.
As PUX10 is also expressed during embryo development (Figures 5B and 5C), it might also be involved in protein turnover during this period. Furthermore, in the pux10 mutants, LD protein levels were already higher inseeds imbibed for 45 min than in those from their respective backgrounds (Figure 7; Supplemental Figure 18). While the half-life of LD proteins during embryo development is unknown, an average value of 3.5 d has been measured in leaves (Ishihara et al., 2015), a time frame much shorter than embryo development (∼3 weeks; Baud et al., 2002). Overall, the turnover of LD proteins during embryogenesis appears possible and our data supports the idea that PUX10 is involved in this process.
The characterization of the pux10 mutants also led to the conclusion that the action of PUX10 is by no means an exclusive mechanism related to LD protein turnover, since LD protein breakdown is only slowed, but was not abolished, in the pux10 mutants (Figure 7). This indicates alternative pathways or at least proteins functionally redundant to PUX10 that are able to allow normal postgerminative growth.
So far, none of the other PUX proteins tested by us and others could be found to localize to LDs (Supplemental Figure 18) or, alternatively, to the ER (Park et al., 2007; Gallois et al., 2013). In other eukaryotic systems, homologs of PUX10, such as UBX2 in yeasts and UBXD8 in mammals, can shuttle between the ER and LDs (Suzuki et al., 2012; Wang and Lee, 2012). These proteins also contain a hydrophobic domain important for their proper intracellular localization and have long been identified as key components of the ERAD in their respective systems. While the plant homologs for other ERAD components have been identified, the ERAD PUX protein remains elusive (Liu and Li, 2014). This protein would be, considering the evidence from studies with yeast and mammals, a good candidate to potentially replace the function of PUX10. As this candidate remains elusive, the possibility exists that PUX10 serves this role. When expressed in tobacco pollen tubes, NtPUX10 localizes to both LDs and the ER (Figures 2A and 2B). Due to the close structural and functional relationship between the ER and LDs, it is also possible that factors that were originally identified as components of ERAD could operate at LDs. As mentioned above, Hs-UBXD8 relocates from the ER to LDs upon LD formation (Olzmann et al., 2013). Similarly, PUX10 could be LD-localized in LD-rich tissues and remain ER-bound in others. As for the ERAD pathway, whose importance is more pronounced during stress and disease (Liu et al., 2011; Guerriero and Brodsky, 2012), a functioning protein degradation machinery on the LD might also be particularly important under stress conditions, like temperature stress or pathogen infection.
The essential role of CDC48 has been studied in plants as well as other eukaryotic systems. Park et al. (2008) demonstrated the importance of At-CDC48A during various cellular processes including cytokinesis and cellular differentiation. All T-DNA insertion lines investigated in their study displayed defects in pollen tube growth and embryo development and were seedling lethal. In another study, the same group established a functional connection between At-CDC48A and another PUX protein, At-PUX1, which regulates CDC48A’s hexametric structure and ATPase activity (Rancour et al., 2004). Loss of At-PUX1 resulted in accelerated growth of the mutant plants, an observation we could not confirm for pux10 knockout lines, suggesting that while both proteins interact with CDC48A, their physiological functions differ. Nevertheless, even if evidence increases that LD proteins are substrate of the UPS system, many questions remain. As mentioned earlier, it is unclear how much of the necessary machinery is LD-specific as opposed to “borrowed” from the ER or the cytosol. The identification of potential players, like the E3 ligase responsible for the ubiquitination of oleosins, would help shed light on the mechanism.
Alternatively, the degradation of LDs via autophagy has been shown to be pivotal in animal and fungal cells, but the importance of the process in plants remains to be determined (recently reviewed in Elander et al., 2018). Studies in algae suggest autophagy is involved in LD degradation (Zhao et al., 2014; Schwarz et al., 2017). Additionally, in Arabidopsis, the uptake of LDs into the vacuole for degradation, potentially achieved by autophagy and mediated via caleosins, has been previously proposed (Poxleitner et al., 2006). However, the focus of many studies on this topic is aimed more so at the degradation of the lipid components of LDs with the purpose of energy conversion, rather than the degradation of LD protein components. In general, the degradation of specific proteins by the UPS is a more flexible mechanism of spatial and temporal protein homeostasis.
In summary, we propose a mechanism for the degradation of LD proteins by the UPS. In our model (Figure 10), LD proteins are marked as substrate for the UPS by the addition of K48-linked polyubiquitin chains. This motif is recognized by the LD-bound PUX10 protein via its UBA domain. With its UBX domain, PUX10 interacts with the ubiquitous cytosolic AAA-type ATPase CDC48, which then channels the bound substrate to the proteasome. Whether CDC48 is responsible for removing LD proteins from the LD and/or whether it interacts directly with the proteasome or via additional factors remains to be elucidated. Furthermore, it remains to be determined whether PUX10 is involved in the degradation of ER proteins. This might be possible as PUX10 can also localize to the ER (Figure 2B), and as it has been speculated elsewhere that LDs stay connected to the ER throughout their life cycle (Goodman, 2009).
METHODS
Plant Growth
Tobacco (Nicotiana tabacum) plants were grown in the greenhouse as described (Rotsch et al., 2017). Pollen was harvested from freshly opened anther buds (six flowers of two plants per construct) when used for pollen transformation. For large-scale cultivation of pollen tubes for LD isolation, green anther buds close to dehiscence were collected and dried for 2 to 4 d.
Arabidopsis thaliana plants (ecotypes Col-0 and Ws-4) were grown on soil in a climate chamber (York) in 60% relative humidity, a constant temperature of 23°C and under 16-h-light/8-h-dark cycle with daytime lighting at 150 µmol photons m−2 s−1 (the climate chamber was equipped with LuxLine Plus F36W 830 Warm White de Luxe fluorescent tubes; Osram Silvania).
For all experiments conducted with the qrt pux10-1 and the pux10-2 mutant and their respective control lines, seeds were derived from one seed batch each originating from 10 mother plants grown side-by-side. In the case of pux10-3 and its control line, seed batches of different individual mother plants were used for each biological replicate.
For seed germination on half-strength Murashige and Skoog (MS) medium without any supplemented sucrose, seeds were first surface sterilized in sodium-chlorate solution (6% [w/v] NaClO3 and 1% [v/v] Tween 20) for 10 min, washed five times with sterile water, and resuspended in 0.1% (w/v) agar. Seeds were imbibed for a given time at 4°C in the dark and grown under 22°C 16-h-light/8-h-dark cycle with daytime lighting at 150 µmol photons m−2 s−1. In all cases, the seeds of mutant lines and their respective controls were derived from plants grown side-by-side under identical conditions.
Isolation of LD-Enriched Fractions of Tobacco Pollen Tubes
For each biological replicate, pollen was collected from ∼40 plants for a week and the experiment repeated in subsequent weeks for each biological replicate using the same plants. Two grams of pollen per replicate was resuspended in 40 mL of pollen tube medium (Read et al., 1993) and spread on a layer of 6-µm Nylon mesh in four 20-cm Petri dishes. Pollen tubes were cultivated for 2 h at 22°C. All of the following steps were performed on ice or at 4°C. The tubes were collected by centrifugation at 1000g for 1 min and the pellet manually ground with sand in 10 mL grinding buffer (50 mM Tris-HCl, pH 7.5, 10 mM KCl, and 200 µM proteinase inhibitor PMSF). An aliquot of the total extract was taken after a centrifugation at 1000g for 1 min and the rest of the extract was centrifuged at 100,000g and for 30 min in a swing-out rotor. The floating pad including the LDs was taken off with a spatula and resuspended in 1 mL grinding buffer. From the phase between the fat pad and the pellet, an aliquot was taken as a cytosolic fraction. The LD fraction was washed three times with grinding buffer using centrifugations of 20,000g for 5 min each in a tabletop centrifuge. During the isolation procedure, harsh washing steps with detergents or high salt concentrations were omitted, to avoid the detachment of loosely bound proteins.
Isolation of Total Protein of Arabidopsis Seedlings
For total extract samples, 10 to 40 mg (∼500–2000) dry seeds per biological replicate was sterilized and imbibed for 3 d at 4°C in the dark. Then, the seedlings were grown on half strength MS media for a 24-, 48-, and 72-h growth period. For 0 d after imbibition samples, seeds were soaked in water for 45 min. Seedlings were taken up in 1.6 mL grinding buffer (62.5 mM Tris-HCl, pH 7.5, 5% [w/v] SDS, 10% [v/v] glycerol, 1 mM DTT, and 200 µM PMSF) and ground with sand in a mortar for 3 min. Then, the homogenate was incubated for 5 min at 95°C, transferred to a 15-mL reaction tube and 96% ethanol was added to a total volume of 15 mL. All steps were performed on ice or at 4°C.
Isolation of LD-Enriched Fractions from Arabidopsis Seedlings
Two hundred milligrams (∼10,000) of Arabidopsis seeds per replicate were sterilized and stratified for 3 d in the dark at 4°C. Then, seedlings were grown for 38 h (Supplemental Data Sets 40 to 43) or 1 and 2 d (Supplemental Data Sets 44 to 46) in 16-h-light/8-h-dark cycles at 22°C on half-strength MS media plates without sucrose. Seedlings were taken up in 3 mL grinding buffer (10 mM sodium phosphate buffer, pH 7.4, 0.5 mM Lohman’s reagent, and 10 mM N-ethylmaleimide), ground with sand for 3 min with a mortar and pestle. The resulting homogenate was spun for 10 s at 100g and a 200-μL aliquot of the supernatant (“total extract” sample) was taken. The remaining homogenate was centrifuged at 20,000g for 20 min at 4°C and the resulting fat pad was washed twice with grinding buffer, using the same centrifugation step. All steps were performed on ice or at 4°C.
Proteomics Sample Preparation and LC/MS Analysis of Peptides
All protein fractions were precipitated and defatted with a final concentration of 80% ethanol and washed with 96% ethanol. The protein pellets were redissolved in 6 M urea and 5% (w/v) SDS. Protein concentrations were determined with a Pierce BCA protein assay kit (Thermo Fisher Scientific). Ten to twenty micrograms of protein was run on a SDS-PAGE gel until they entered the separation gel. A single gel piece per sample containing all proteins was excised, subjected to tryptic digestion, and derivatized as described (Shevchenko et al., 2006). Peptides were desalted over an Empore Octadecyl C18 47-mm extraction disks 2215 (Supelco) according to Rappsilber et al. (2007), dried, and dissolved in a 20 μL 0.1% formic acid. Then, the peptides were subjected to LC-MS/MS (Schmitt et al., 2017). In detail, nano-flow liquid chromatography was done with the RSLCnano Ultimate 3000 (Thermo Fisher Scientific) system. Peptides of 1 to 3 μL sample solution were loaded with 0.07% trifluoroacetic acid on an Acclaim PepMap 100 precolumn (100 µm × 2 cm, C18, 3 µm, 100 Å; Thermo Fisher Scientific) at a flow rate of 20 µL/min for 3 to 6 min. Analytical peptide separation by reverse phase chromatography was performed at a flow rate of 300 nL/min on an Acclaim PepMap RSLC column (75 µm × 50 cm, C18, 3 µm, 100 Å; Thermo Fisher Scientific). A gradient from 98% solvent A (0.1% formic acid) and 2% solvent B (80% acetonitrile, 0.1% formic acid) to 32% B was applied within 94 min and to 65% B within the next 26 min. Optima LC-MS solvents and acids were purchased from Thermo Fisher Scientific.
Nano-electrospray ionization mass spectrometry was done with the Orbitrap Velos Pro hybrid mass spectrometer (Thermo Fisher Scientific). Chromatographically eluting peptides were online ionized by nano-electrospray ionization using the Nanospray Flex Ion Source (Thermo Fisher Scientific) at 1.5 kV (liquid junction) and continuously transferred into the mass spectrometer. Full scans within the mass range of 300 to 1850 m/z were taken with the Orbitrap-FT analyzer at a resolution of 30,000 followed by data-dependent top 15 CID fragmentation (dynamic exclusion enabled) within the ion trap Velos Pro analyzer. LC-MS method programming and data acquisition was performed with XCalibur 2.2 software (Thermo Fisher Scientific).
Computation Analysis of MS/MS2 Raw Data
MS/MS2 raw data processing for peptide analysis, protein identification, and quantification were performed with MaxQuant 1.5.6.0 or MaxQuant 1.6.1.0 (the ladder was used to analyze proteomic data related to the pux10-3 mutant line; Cox and Mann, 2008).
Protein abundance was quantified using an LFQ label-free quantification algorithm implemented in MaxQuant software (Schaab et al., 2012) using the settings as described in the meta data files (Supplemental Data Sets 47 to 52). Default settings were used. Additionally, in group-specific parameter setting, label-free quantification was enabled, and the intensity determination was set to total sum. Within the global parameters, the PSM false discovery rate and protein false discovery rate were set to 0.02. Match between runs, dependent peptides, and iBAQ were enabled. After processing, the values were calculated as per mille of all values in one sample.
The iBAQ algorithm rather than the LFQ algorithm was used to calculate enrichment of proteins in subcellular fractions, as it is better suited to detect small abundance levels, and as the LFQ algorithm gave unrealistically high enrichment levels.
For tobacco, the N. tabacum Uniprot database was used (as of August 16, 2017) and for Arabidopsis the TAIR10 database.
Molecular Cloning
Molecular cloning into the vectors pUC-LAT52-mVenusC, pUC-LAT52-mVenusN, and pUC-LAT52-mCherryC (Mähs et al., 2013; Steinhorst et al., 2015) as well as pLatMVC-GW, pLatMVN-GW (Müller et al., 2017), and pGWB604 (Nakamura et al., 2010) was performed using classical restriction sites or Gateway cloning as described (Müller et al., 2017). For a complete list of primers and strategies used, see Supplemental Data Set 17. All constructs used in this study were subject to full sequencing of the gene and its attachment sites to the promoter and the fluorophore. Sanger sequencing was performed by GATC Biotech (now Eurofins). The ERD-CFP construct for pollen tube expression has been previously described (Müller et al., 2017).
Particle Bombardment and Microscopy of Tobacco Pollen Tubes
The transformation of pollen grains by particle bombardment, the cultivation of the pollen tubes on microscope slides, Nile Red and Bodipy 505/515 staining as well as confocal microscopy were performed as described (Müller et al., 2017). Monodansylpentane (Abgent) was excited at 405 nm using a HFT 405/514/633 as major beam splitter and detected at a wavelength of 422 to 486 nm.
Identification of T-DNA Insertion Lines and Generation of Transgenic Lines
The Arabidopsis mutant line described here as qrt pux10-1 was obtained as SAIL_1187B06 from the SAIL collection (Sessions et al., 2002) and is in the qrt background (Preuss et al., 1994). This line and the control line used (qrt PUX10) derive from the same line heterozygous for the pux10-1 mutant allele. A second mutant line, pux10-2, was obtained as SALK_139056 from the SALK collection (Alonso et al., 2003). A third independent mutant line, pux10-3, was obtained as FST EATTV209T3 from INRA in the Ws-4 background (Samson et al., 2002).
Homozygous plants were identified by performing PCR on genomic DNA using REDTaq ReadyMix (Sigma-Aldrich) according to the manufacturer’s instructions.
Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998). Arabidopsis Col-0 plants were transformed with Agrobacterium tumefaciens strain EHA105 containing pGWB604-At-PUX10pro:gAt-PUX10-eGFP construct. For complementation, those lines were crossed with qrt pux10-1 plants and the F2-progeny was selected by PCR for lines homozygous for pux10-1.
PUX10 Expression Analysis via RT-qPCR
Ten milligrams (∼500) of dry seeds per biological replicate and line was frozen in liquid nitrogen and homogenized with the help of a retch mill. RNA was isolated with the Spectrum Plant Total RNA Kit (Sigma-Aldrich). The RNA was treated with DNaseI (Thermo Fisher Scientific) according to the manufacturer’s instructions. For cDNA synthesis, 1 µg of DNaseI-treated RNA was reverse transcribed (Revert Aid Minus Reverse Transcriptase; Thermo Fisher Scientific). The reaction product was diluted 1:10 in double-distilled water before the RT-qPCR, and 2.5 μL of diluted cDNA per sample and Takyon No Rox SYBR MasterMix dTTP Blue (Eurogentec) were used for the RT-qPCR reaction. The following PCR program was applied in an iQ5 qPCR cycler (Bio-Rad Laboratories): 95°C for 1 min 20 s (95°C for 20 s, 58°C for 20 s, 72°C for 40 s) × 39, 72°C 4 min (for primers, see Supplemental Data Set 17). At4g12590 was used as a reference gene (Dekkers et al., 2012).
Microscopy of Arabidopsis Embryos, Seeds, Seedlings, and Pollen Tubes
Embryos were obtained from plants grown on soil and seeds were germinated on half-strength MS medium. Arabidopsis embryos and seedlings were gently squeezed out of the seed coat by pressure between a microscope slide and a cover slip. They were then stained with either Nile Red or Bodipy 505/515 (Thermo Fisher Scientific) for at least 5 min in a reaction cup before they were retransferred to a microscope slide.
Confocal microscopy was performed with a Zeiss LSM 510 confocal microscope (Carl Zeiss). eGFP was excited at 488 nm using a 488-nm major beam splitter and detected at a wavelength of 496 to 530 nm; Bodipy 505/515 was excited at 514 nm using a HFT 405/514/633 as major beam splitter and detected at a wavelength of 530 to 600 nm. Diameters of the LDs contained in those cells were quantified with ImageJ (Schneider et al., 2012). Epifluorescence was observed on a epifluorescence microscope (BX51; Olympus) supplied with an U-MF2 filter cube for mVenus fluorescence and recorded with an ORCA-Flash 4.0 V2 camera (Hamamatsu Photonics).
Pollen of Arabidopsis Col-0 At-PUX10pro:At-PUX10-eGFP plants were grown in vitro by gently brushing open 10 flowers over a microscope slide and covering them with Arabidopsis pollen tube medium (0.01% [w/v] boric acid, 5 mM CaCl2, 5 mM KCl, 1 mM MgSO4, and 10% [w/v] sucrose). The microscope slides were incubated for 3 h in the dark. Pollen tubes were stained with 0.5% (w/v) Nile Red and fixed with 5% (v/v) formaldehyde both dissolved in Arabidopsis pollen tube medium. Pollen tubes were imaged by confocal microscopy as described above.
Quantification of Fatty Acids
Arabidopsis seeds were sterilized and stratified at 4°C in the dark for 24 h and subsequently germinated on half-strength MS medium in a long-day chamber (16 h light/8 h dark). Seeds and seedlings (10 per biological replicate) were collected for analysis every 24 h and used directly for the generation of fatty acid methyl esters. C15:0 TAG (Sigma-Aldrich) was added as an internal standard. Fatty acid methyl esters were quantified by gas chromatography analysis with flame ionization detection as described (Hornung et al., 2002).
Immunoblotting
Twenty micrograms of protein, derived from proteins extracted from 200 mg seedlings per biological replicate, was dissolved in 6 M urea and 5% (w/v) SDS. Then, the samples were run on a 12% or 13% SDS gel (LD and total samples, respectively). Proteins were transferred from the gel on a Nitrocellulose membrane (Roti-NC, 0.2 µm; Carl Roth) in 1× transfer buffer (25 mM Tris, 192 mM glycine, and 20% ethanol). Before blocking, the membrane was stained with Ponceau S (5% [v/v] acetic acid and 0.1% [w/v] Ponceau S; Serva) for 30 min, destained with double-distilled water, and documented. Then, the membrane was blocked in 1× TBS-T buffer (50 mM Tris, 0.15 M NaCl, and 0.05% [v/v] Tween 20, pH to 7.6 with HCl) supplemented with 3% (w/v) milk powder (Carl Roth) for 1 h at room temperature under agitation. The membrane was incubated with the first antibody, a rabbit antibody recognizing an epitope of Arabidopsis UBIQUITIN 11 (At4g05050, AS08 307 A by Agrisera), overnight at 4°C under agitation. The antibody was diluted 1:5000 in 1× TBS-T supplemented with 3% milk powder. After that incubation, the membrane was washed four times for 10 min per wash with 1× TBS-T supplemented with 3% milk powder and incubated with the secondary antibody for 1 h 30 min under agitation at room temperature. The goat anti rabbit IgG alkaline phosphates antibody (catalog no. A3678, lot no. 036K6031; Sigma-Aldrich) was diluted 1:30,000 in TBS-T supplemented with 3% milk powder. Then, the membrane was washed twice 10 min with 1× TBS-T and twice for 5 min with 1× TBS (50 mM Tris, and 0.15 M NaCl, pH to 7.6 with HCl). For revelation, the membrane was first equilibrated for 10 min with AP buffer (100 mM Tris, 100 mM NaCl, and 5 mM MgCl2, pH 9.5) and then incubated in 10 mL AP buffer supplemented with 33 μL BCIP (50 mg/mL in DMF) and 66 μL NBT (50 mg/mL in DMF) in the dark until bands were visible. The reaction was stopped with double-distilled water and the membrane documented.
Bioinformatics
Phylogenetic trees were created with MEGA version X software using MUSCLE alignment with gap penalties set to −9 for gap open and to −3 for gap extension (Edgar, 2004; Kumar et al., 2018). The aligned protein sequences were used for phylogenetic tree construction using the maximum likelihood method based on the JTT matrix-based model (Jones et al., 1992). The phylogeny was tested with the Bootstrap method set for 1000 replicates (Felsenstein, 1985). No phylogeny testing was performed for the LDIP phylogenetic tree.
Hydropathy analyses of proteins were performed on the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al., 2001). All statistical analysis was performed in and all box plots created with RStudio 0.98.1060. Tobacco proteins were blasted against the Arabidopsis Uniprot database (8/3/2018) using the BLAST 2.8.0 algorithm (Altschul et al., 1997). Helical wheel projection of amino acids was performed with HeliQuest (Gautier et al., 2008).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: At-PUX10, AT4G10790; At-OLE1, AT4G25140; At-OLE2, AT5G40420; At-OLE3, AT5G51210; At-OLE4, AT3G27660; At-OLE5, AT3G01570; At-OLE6, AT1G48990; At-OLE7, AT2G25890; At-OLE8, AT3G18570; At-HSD1, AT5G50600/AT5G50700; At-HSD5, AT4G10020, At-CLO1, At4G26740; At-CLO2, AT5G55240; At-LDAP1, AT1G67360; At-LDAP2, AT2G47780; At-LDAP3, AT3G05500; At-UBQ5, AT3G62250; At-PUX5, AT4G15410; At-PUX8, AT4G11740; At-PUX9, AT4G00752; At-PUX11, AT2G43210; At-PUX13, AT4G23040; At-CAS1, At2G07050; At-CDC48A, At3G09840; Nt-CLO1a, LOC107783728; Nt-OLE6b, LOC107824536; Nt-CLO1b, LOC107817909; Nt-OLE6a, LOC107780677; Nt-LDAP1a, LOC107765167; Nt-CASb, LOC107762593; Nt-PUX10c, LOC107769345; Nt-LDAP1b, LOC107804268; Nt-LDAP3a, LOC107827582; Nt-CASa, LOC107826198; Nt-SMT1c, LOC107813194; Nt-PTLD1b, LOC107815104; Nt-OBL1, LOC107788962; Nt-PUX10a, LOC107799924; Nt-LDIPa, LOC107763626; Nt-PTLD2a, LOC107796437; Nt-FEY1, LOC107800303; Nt-LDIPb, LOC107831283; Nt-ADCP, LOC107827250; Nt-PDCP, LOC107793109; Nt-RAB18, LOC107762524; Nt-PTLD2b, LOC107778489; Nt-CCD, LOC107830214; and Nt-RAB2c, LOC107794510. All proteomic data, including raw data files, MaxQuant search files, as well as protein group and peptide search results created by MaxQuant are available on ProteomeXchange/PRIDE (Vizcaíno et al., 2014) under the identifiers PXD009184, PXD009186, PXD009207, PXD009397, PXD009248, and PXD009247.
Supplemental Data
Supplemental Figure 1. Phylogenetic tree of Arabidopsis thaliana and Nicotiana tabacum oleosin proteins.
Supplemental Figure 2. Phylogenetic tree of Arabidopsis thaliana and Nicotiana tabacum caleosin proteins.
Supplemental Figure 3. Phylogenetic tree of Arabidopsis thaliana and Nicotiana tabacum steroleosin proteins.
Supplemental Figure 4. Phylogenetic tree of Arabidopsis thaliana and Nicotiana tabacum plant UBX domain-containing proteins 10 (PUX10).
Supplemental Figure 5. Phylogenetic tree of Arabidopsis thaliana and Nicotiana tabacum oil body lipases.
Supplemental Figure 6. Phylogenetic tree of Arabidopsis thaliana and Nicotiana tabacum lipid droplet-associated proteins.
Supplemental Figure 7. Phylogenetic tree of Arabidopsis thaliana and Nicotiana tabacum lipid droplet-associated protein-interacting proteins.
Supplemental Figure 8. Phylogenetic tree of Arabidopsis thaliana and Nicotiana tabacum cycloartenol synthase proteins.
Supplemental Figure 9. Phylogenetic tree of Arabidopsis thaliana and Nicotiana tabacum sterol methyltransferase proteins.
Supplemental Figure 10. Phylogenetic tree of “pollen tube lipid droplet protein” from Nicotiana tabacum.
Supplemental Figure 11. The cycloartenol synthase At-CAS1, Arabidopsis homolog to the LD-localized Nt-CAS1b, localizes to LDs.
Supplemental Figure 12. Arabidopsis thaliana plant UBX domain-containing (PUX) protein gene family.
Supplemental Figure 13. Hydropathy profiles of the Arabidopsis PUX proteins and NtPUX10c.
Supplemental Figure 14. Localization of Nt-PUX10c and truncated variants (magnified images).
Supplemental Figure 15. Uneven distribution of At-PUX10-eGFP on LD in germinating seeds remains after isolation.
Supplemental Figure 16. Relative expression of PUX10 in mutants is strongly reduced compared with the mutant backgrounds.
Supplemental Figure 17. Relative LFQ intensity of PUX10 protein on LDs of the qrt pux10-1 mutant and controls.
Supplemental Figure 18. LD proteins accumulate in the pux10 mutant seedlings.
Supplemental Figure 19. Postgerminative fatty acid degradation in qrt pux10-1 and the nonmutated background qrt PUX10.
Supplemental Figure 20. Antiubiquitin immunoblots.
Supplemental Figure 21. Annotated fragmentation spectrum of identified ubiquitination site of ubiquitin presented in Figure 9B.
Supplemental Figure 22. Annotated fragmentation spectrum of identified ubiquitination site of At-OLE4 presented in Figure 9B.
Supplemental Figure 23. Annotated fragmentation spectra of identified ubiquitination site of At-OLE4 presented in Figure 9B.
Supplemental Figure 24. Annotated fragmentation spectrum of identified ubiquitination site of At-HSD1 presented in Figure 9B.
Supplemental Figure 25. Hydropathy profiles of At-CAS1 and Nt-LDAP1a.
Supplemental Figure 26. Helical wheel projection of amino acids 93 to 111 of At-PUX10.
Supplemental Figure 27. Hydropathy profiles of the N. tabacum PTLD proteins.
Supplemental Data Set 1. Proteins found in tobacco pollen tubes (raw iBAQ and LFQ values).
Supplemental Data Set 2. Proteins found in tobacco pollen tubes (normalized and sorted data).
Supplemental Data Set 3. Lipid droplet proteins found in tobacco pollen tubes.
Supplemental Data Set 4. Organellar marker proteins in lipid droplet fractions of tobacco pollen tubes.
Supplemental Data Set 5. Proteins found in tobacco seeds (raw iBAQ and LFQ values).
Supplemental Data Set 6. Proteins found in tobacco seeds (normalized and sorted data).
Supplemental Data Set 7. Sequence alignment of Arabidopsis thaliana and Nicotiana tabacum oleosin proteins.
Supplemental Data Set 8. Sequence alignment of Arabidopsis thaliana and Nicotiana tabacum caleosin proteins.
Supplemental Data Set 9. Sequence alignment of Arabidopsis thaliana and Nicotiana tabacum steroleosin proteins.
Supplemental Data Set 10. Sequence alignment of Arabidopsis thaliana and Nicotiana tabacum plant UBX domain-containing proteins 10 (PUX10).
Supplemental Data Set 11. Sequence alignment of Arabidopsis thaliana and Nicotiana tabacum oil body lipases.
Supplemental Data Set 12. Sequence alignment of Arabidopsis thaliana and Nicotiana tabacum lipid droplet-associated proteins.
Supplemental Data Set 13. Sequence alignment of Arabidopsis thaliana and Nicotiana tabacum lipid droplet-associated protein-interacting proteins.
Supplemental Data Set 14. Sequence alignment of Arabidopsis thaliana and Nicotiana tabacum cycloartenol synthase proteins.
Supplemental Data Set 15. Sequence alignment of Arabidopsis thaliana and Nicotiana tabacum sterol methyltransferase proteins.
Supplemental Data Set 16. Sequence alignment of “pollen tube lipid droplet protein” from Nicotiana tabacum.
Supplemental Data Set 17. Cloning strategies used and list of primers.
Supplemental Data Set 18. Protein composition of rehydrated seeds of pux10-1 and controls (raw iBAQ and LFQ values of total extracts).
Supplemental Data Set 19. Protein composition of seedlings 1 d after imbibition of pux10-1 and controls (raw iBAQ and LFQ values of total extracts).
Supplemental Data Set 20. Protein composition of seedlings 2 d after imbibition of pux10-1 and controls (raw iBAQ and LFQ values of total extracts).
Supplemental Data Set 21. Protein composition of seedlings 3 d after imbibition of pux10-1 and controls (raw iBAQ and LFQ values of total extracts).
Supplemental Data Set 22. Protein composition of rehydrated seeds of pux10-1 and controls (normalized iBAQ and LFQ values of total extracts).
Supplemental Data Set 23. Protein composition of seedlings 1 d after imbibition of pux10-1 and controls (normalized iBAQ and LFQ values of total extracts).
Supplemental Data Set 24. Protein composition of seedlings 2 d after imbibition of pux10-1 and controls (normalized iBAQ and LFQ values of total extracts).
Supplemental Data Set 25. Protein composition of seedlings 3 d after imbibition of pux10-1 and controls (normalized iBAQ and LFQ values of total extracts).
Supplemental Data Set 26. Protein composition of rehydrated seeds and seedlings 1 to 3 d after imbibition of pux10-1 and controls (averages of LFQ values of total extracts).
Supplemental Data Set 27. Protein composition of rehydrated seeds and seedlings 1 to 3 d after imbibition of pux10-1 and controls (averages of iBAQ values of total extracts).
Supplemental Data Set 28. Abundance of lipid droplet proteins of rehydrated seeds and seedlings 1 to 3 d after imbibition of pux10-1 and controls (averages of LFQ values of total extracts).
Supplemental Data Set 29. Protein composition of rehydrated seeds of Ws-4 and pux10-3 - raw iBAQ and LFQ values of total extracts.
Supplemental Data Set 30. Protein composition of seedlings 1 d after imbibition of Ws-4 and pux10-3 (raw iBAQ and LFQ values of total extracts).
Supplemental Data Set 31. Protein composition of seedlings 2 d after imbibition of Ws-4 and pux10-3 (raw iBAQ and LFQ values of total extracts).
Supplemental Data Set 32. Protein composition of seedlings 3 d after imbibition of Ws-4 and pux10-3 (raw iBAQ and LFQ values of total extracts).
Supplemental Data Set 33. Protein composition of rehydrated seeds of Ws-4 and pux10-3 (normalized iBAQ and LFQ values of total extracts).
Supplemental Data Set 34. Protein composition of seedlings 1 d after imbibition of Ws-4 and pux10-3 (normalized iBAQ and LFQ values of total extracts).
Supplemental Data Set 35. Protein composition of seedlings 2 d after imbibition of Ws-4 and pux10-3 (normalized iBAQ and LFQ values of total extracts).
Supplemental Data Set 36. Protein composition of seedlings 3 d after imbibition of Ws-4 and pux10-3 (normalized iBAQ and LFQ values of total extracts).
Supplemental Data Set 37. Protein composition of rehydrated seeds and seedlings 1 to 3 d after imbibition of Ws-4 and pux10-3 (averages of LFQ values of total extracts).
Supplemental Data Set 38. Protein composition of rehydrated seeds and seedlings 1 to 3 d after imbibition of Ws-4 and pux10-3 (averages of iBAQ values of total extracts).
Supplemental Data Set 39. Abundance of lipid droplet proteins of rehydrated seeds and seedlings 1 to 3 d after imbibition of Ws-4 and pux10-3 (averages of LFQ intensity values of total extracts).
Supplemental Data Set 40. Proteins found in seedlings 38 h after imbibition of pux10-1 and controls (raw iBAQ and LFQ values).
Supplemental Data Set 42. Proteins found in seedlings 38 h after imbibition of pux10-1 and controls (normalized iBAQ and LFQ values).
Supplemental Data Set 42. Marker proteins found in seedlings 38 h after imbibition of pux10-1 and controls (normalized iBAQ and LFQ values).
Supplemental Data Set 43. Ubiquitinated peptides found in seedlings 38 h after imbibition of pux10-1 and controls.
Supplemental Data Set 44. Proteins found in seedlings 1 and 2 d after imbibition of pux10-1 and controls (raw iBAQ and LFQ values).
Supplemental Data Set 45. Proteins found in seedlings 1 and 2 d after imbibition of pux10-1 and controls (normalized iBAQ and LFQ values).
Supplemental Data Set 46. Ubiquitinated peptides found in seedlings 1 and 2 d after imbibition of pux10-1 and controls.
Supplemental Data Set 47. Metadata: Nicotiana tabacum pollen tube lipid droplet proteome.
Supplemental Data Set 48. Metadata: Nicotiana tabacum seed lipid droplet proteome.
Supplemental Data Set 49. Metadata: Seedling proteome of Arabidopsis thaliana qrt pux10-1 mutant versus the wild type and complemented lines.
Supplemental Data Set 50. Metadata: Seedling proteome of Arabidopsis thaliana pux10-3 mutant versus the wild type.
Supplemental Data Set 51. Metadata: Proteome of Arabidopsis thaliana qrt pux10-1 mutant versus wild type and complemented line lipid droplet and total fractions 38 h after imbibition.
Supplemental Data Set 52. Metadata: Proteome of Arabidopsis thaliana qrt pux10-1 mutant lipid droplets versus the wild type 24 and 48 h after imbibition.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Ivo Feussner for support and many helpful discussions and Christiane Gatz and Alexander Stein for valuable advice. We thank Kent Chapman, John Dyer, and Robert Mullen for discussing our work with us and for critically reading the manuscript. We also thank Jörg Großhans and Steven A. Johnson for granting access to their confocal microscopes as well as Florian Wegwitz and Johannes Sattmann for their assistance. We thank Tsuyoshi Nakagawa (Shimane University) for providing the vector pGWB604 that contains the bar gene, which was identified by Meiji Seika Kaisha; and Leonie Steinhorst and Jörg Kudla (University of Münster) for generating and providing plasmids. We thank Kaneschka Yaqubi, Anna-Lena Gippert, Jörg Heinz, Maurice Hädrich, Lara Baumblüth, and Milena Lewandowska for help in the lab and Benedikt Ni for proofreading. This work was supported by funding from the Deutsche Forschungsgemeinschaft (IS 273/2-2 to T.I. and GR1945/3-1, SFB937/TP10, and INST1525/16-1 FUGG to Jörg Großhans) and the Studienstiftung des Deutschen Volkes (to F.K.K.).
AUTHOR CONTRIBUTIONS
F.K.K., G.H.B., and T.I. designed the work. F.K.K., L.A.M., A.O.M., K.S., K.F.B., O.V., and T.I. performed research. F.K.K., L.A.M., K.S., O.V., G.H.B., and T.I. analyzed data. F.K.K. and T.I. wrote the manuscript. All authors critically read and revised the manuscript and approved the final version.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek G.H., Cheng H., Choe V., Bao X., Shao J., Luo S., Rao H. (2013). Cdc48: a swiss army knife of cell biology. J. Amino Acids 2013: 183421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S., et al. (2009). Regulation of HSD1 in seeds of Arabidopsis thaliana. Plant Cell Physiol. 50: 1463–1478. [DOI] [PubMed] [Google Scholar]
- Baud S., Boutin J.-P., Miquel M., Lepiniec L., Rochat C. (2002). An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 40: 151–160. [Google Scholar]
- Blée E., Boachon B., Burcklen M., Le Guédard M., Hanano A., Heintz D., Ehlting J., Herrfurth C., Feussner I., Bessoule J.-J. (2014). The reductase activity of the Arabidopsis caleosin RESPONSIVE TO DESSICATION20 mediates gibberellin-dependent flowering time, abscisic acid sensitivity, and tolerance to oxidative stress. Plant Physiol. 166: 109–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier-Navé P., Berna A., Noiriel A., Compagnon V., Carlsson A.S., Banas A., Stymne S., Schaller H. (2010). Involvement of the phospholipid sterol acyltransferase1 in plant sterol homeostasis and leaf senescence. Plant Physiol. 152: 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard L., Immel F., Coulon D., Esnay N., Tuphile K., Pascal S., Claverol S., Fouillen L., Bessoule J.J., Bréhélin C. (2017). Proteomic analysis of lipid droplets from Arabidopsis aging leaves brings new insight into their biogenesis and functions. Front. Plant Sci. 8: 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Goodman J.M., Pyc M., Mullen R.T., Dyer J.M., Chapman K.D. (2015). Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 27: 2616–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., McClinchie E., Price A., Nguyen T.N., Gidda S.K., Watt S.C., Yurchenko O., Park S., Sturtevant D., Mullen R.T. (2017). Mouse fat storage‐inducing transmembrane protein 2 (FIT2) promotes lipid droplet accumulation in plants. Plant Biotechnol. J. 15: 824– 836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K.D., Dyer J.M., Mullen R.T. (2012). Biogenesis and functions of lipid droplets in plants. J. Lipid Res. 53: 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Copeland C., Woloshen V., Huang Y., Li X. (2016). AtCDC48A is involved in the turnover of an NLR immune receptor. Plant J. 88: 294–305. [DOI] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- d’Andréa S., Canonge M., Beopoulos A., Jolivet P., Hartmann M.A., Miquel M., Lepiniec L., Chardot T. (2007). At5g50600 encodes a member of the short-chain dehydrogenase reductase superfamily with 11beta- and 17beta-hydroxysteroid dehydrogenase activities associated with Arabidopsis thaliana seed oil bodies. Biochimie 89: 222–229. [DOI] [PubMed] [Google Scholar]
- Dekkers B.J., Willems L., Bassel G.W., van Bolderen-Veldkamp R.P., Ligterink W., Hilhorst H.W., Bentsink L. (2012). Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol. 53: 28–37. [DOI] [PubMed] [Google Scholar]
- Deruyffelaere C., Bouchez I., Morin H., Guillot A., Miquel M., Froissard M., Chardot T., D’Andrea S. (2015). Ubiquitin-mediated proteasomal degradation of oleosins is involved in oil body mobilization during post-germinative seedling growth in Arabidopsis. Plant Cell Physiol. 56: 1374–1387. [DOI] [PubMed] [Google Scholar]
- Eastmond P.J. (2004). Cloning and characterization of the acid lipase from castor beans. J. Biol. Chem. 279: 45540–45545. [DOI] [PubMed] [Google Scholar]
- Eastmond P.J. (2006). SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Bord M., et al. (2018). Identification of seipin-linked factors that act as determinants of a lipid droplet subpopulation. J. Cell Biol. 217: 269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elander P.H., Minina E.A., Bozhkov P.V. (2018). Autophagy in turnover of lipid stores: trans-kingdom comparison. J. Exp. Bot. 69: 1301–1311. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Gallois J.L., Drouaud J., Lécureuil A., Guyon-Debast A., Bonhomme S., Guerche P. (2013). Functional characterization of the plant ubiquitin regulatory X (UBX) domain-containing protein AtPUX7 in Arabidopsis thaliana. Gene 526: 299–308. [DOI] [PubMed] [Google Scholar]
- Gautier R., Douguet D., Antonny B., Drin G. (2008). HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics 24: 2101–2102. [DOI] [PubMed] [Google Scholar]
- Gidda S.K., et al. (2013). Lipid droplet-associated proteins (LDAPs) are involved in the compartmentalization of lipophilic compounds in plant cells. Plant Signal. Behav. 8: e27141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda S.K., Park S., Pyc M., Yurchenko O., Cai Y., Wu P., Andrews D.W., Chapman K.D., Dyer J.M., Mullen R.T. (2016). Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol. 170: 2052–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman J.M. (2009). Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J. Lipid Res. 50: 2148–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero C.J., Brodsky J.L. (2012). The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol. Rev. 92: 537–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanano A., Burcklen M., Flenet M., Ivancich A., Louwagie M., Garin J., Blée E. (2006). Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J. Biol. Chem. 281: 33140–33151. [DOI] [PubMed] [Google Scholar]
- Horn P.J., James C.N., Gidda S.K., Kilaru A., Dyer J.M., Mullen R.T., Ohlrogge J.B., Chapman K.D. (2013). Identification of a new class of lipid droplet-associated proteins in plants. Plant Physiol. 162: 1926–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung E., Pernstich C., Feussner I. (2002). Formation of conjugated Delta11Delta13-double bonds by Delta12-linoleic acid (1,4)-acyl-lipid-desaturase in pomegranate seeds. Eur. J. Biochem. 269: 4852–4859. [DOI] [PubMed] [Google Scholar]
- Hsiao E.S.L., Tzen J.T.C. (2011). Ubiquitination of oleosin-H and caleosin in sesame oil bodies after seed germination. Plant Physiol. Biochem. 49: 77–81. [DOI] [PubMed] [Google Scholar]
- Hsieh K., Lee Y.K., Londos C., Raaka B.M., Dalen K.T., Kimmel A.R. (2012). Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 125: 4067–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.H.C. (1996). Oleosins and oil bodies in seeds and other organs. Plant Physiol. 110: 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.H.C. (2018). Plant lipid droplets and their associated oleosin and other proteins: potential for rapid advances. Plant Physiol. 176: 1894–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T. (2016). Lipids in pollen—They are different. Biochim. Biophys. Acta 1861: 1315–1328. [DOI] [PubMed] [Google Scholar]
- Ishihara H., Obata T., Sulpice R., Fernie A.R., Stitt M. (2015). Quantifying protein synthesis and degradation in Arabidopsis by dynamic 13CO2 labeling and analysis of enrichment in individual amino acids in their free pools and in protein. Plant Physiol. 168: 74–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P.-L., Wang C.-S., Hsu C.-M., Jauh G.-Y., Tzen J.T.C. (2007). Stable oil bodies sheltered by a unique oleosin in lily pollen. Plant Cell Physiol. 48: 812–821. [DOI] [PubMed] [Google Scholar]
- Jolivet P., Roux E., D’Andrea S., Davanture M., Negroni L., Zivy M., Chardot T. (2004). Protein composition of oil bodies in Arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 42: 501–509. [DOI] [PubMed] [Google Scholar]
- Jolivet P., Aymé L., Giuliani A., Wien F., Chardot T., Gohon Y. (2017). Structural proteomics: Topology and relative accessibility of plant lipid droplet associated proteins. J. Proteomics 169: 87–98. [DOI] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kelly A.A., Quettier A.-L., Shaw E., Eastmond P.J. (2011). Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol. 157: 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.Y., Park K.Y., Seo Y.S., Kim W.T. (2016). Arabidopsis small rubber particle protein homolog SRPs play dual roles as positive factors for tissue growth and development and in drought stress responses. Plant Physiol. 170: 2494–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.U., Hsieh K., Ratnayake C., Huang A.H. (2002). A novel group of oleosins is present inside the pollen of Arabidopsis. J. Biol. Chem. 277: 22677–22684. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Bih F.Y., Learn G.H., Ting J.T.L., Sellers C., Huang A.H.C. (1994). Oleosins in the gametophytes of Pinus and Brassica and their phylogenetic relationship with those in the sporophytes of various species. Planta 193: 461–469. [DOI] [PubMed] [Google Scholar]
- Li F., Asami T., Wu X., Tsang E.W., Cutler A.J. (2007). A putative hydroxysteroid dehydrogenase involved in regulating plant growth and development. Plant Physiol. 145: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li J. (2014). Endoplasmic reticulum-mediated protein quality control in Arabidopsis. Front. Plant Sci. 5: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wang C., Chen F., Shen S. (2015). Proteomic analysis of oil bodies in mature Jatropha curcas seeds with different lipid content. J. Proteomics 113: 403–414. [DOI] [PubMed] [Google Scholar]
- Liu L., Cui F., Li Q., Yin B., Zhang H., Lin B., Wu Y., Xia R., Tang S., Xie Q. (2011). The endoplasmic reticulum-associated degradation is necessary for plant salt tolerance. Cell Res. 21: 957–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähs A., Steinhorst L., Han J.P., Shen L.K., Wang Y., Kudla J. (2013). The calcineurin B-like Ca2+ sensors CBL1 and CBL9 function in pollen germination and pollen tube growth in Arabidopsis. Mol. Plant 6: 1149–1162. [DOI] [PubMed] [Google Scholar]
- Matsui K., Hijiya K., Tabuchi Y., Kajiwara T. (1999). Cucumber cotyledon lipoxygenase during postgerminative growth. Its expression and action on lipid bodies. Plant Physiol. 119: 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinken J., Min J. (2012). Computational prediction of protein subcellular locations in eukaryotes: an experience report. Computational Molecular Biology 2: 1–7. [Google Scholar]
- Mellema S., Eichenberger W., Rawyler A., Suter M., Tadege M., Kuhlemeier C. (2002). The ethanolic fermentation pathway supports respiration and lipid biosynthesis in tobacco pollen. Plant J. 30: 329–336. [DOI] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. (2005). ERAD: the long road to destruction. Nat. Cell Biol. 7: 766–772. [DOI] [PubMed] [Google Scholar]
- Müller A.O., Ischebeck T. (2018). Characterization of the enzymatic activity and physiological function of the lipid droplet-associated triacylglycerol lipase AtOBL1. New Phytol. 217: 1062–1076. [DOI] [PubMed] [Google Scholar]
- Müller A.O., Blersch K.F., Gippert A.L., Ischebeck T. (2017). Tobacco pollen tubes - a fast and easy tool for studying lipid droplet association of plant proteins. Plant J. 89: 1055–1064. [DOI] [PubMed] [Google Scholar]
- Nakamura S., Mano S., Tanaka Y., Ohnishi M., Nakamori C., Araki M., Niwa T., Nishimura M., Kaminaka H., Nakagawa T., Sato Y., Ishiguro S. (2010). Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci. Biotechnol. Biochem. 74: 1315–1319. [DOI] [PubMed] [Google Scholar]
- Olzmann J.A., Richter C.M., Kopito R.R. (2013). Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc. Natl. Acad. Sci. USA 110: 1345–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Rancour D.M., Bednarek S.Y. (2007). Protein domain-domain interactions and requirements for the negative regulation of Arabidopsis CDC48/p97 by the plant ubiquitin regulatory X (UBX) domain-containing protein, PUX1. J. Biol. Chem. 282: 5217–5224. [DOI] [PubMed] [Google Scholar]
- Park S., Rancour D.M., Bednarek S.Y. (2008). In planta analysis of the cell cycle-dependent localization of AtCDC48A and its critical roles in cell division, expansion, and differentiation. Plant Physiol. 148: 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.-M., Franke W.W., Kleinschmidt J.A. (1994). Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J. Biol. Chem. 269: 7709–7718. [PubMed] [Google Scholar]
- Poxleitner M., Rogers S.W., Lacey Samuels A., Browse J., Rogers J.C. (2006). A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J. 47: 917–933. [DOI] [PubMed] [Google Scholar]
- Preuss D., Rhee S.Y., Davis R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460. [DOI] [PubMed] [Google Scholar]
- Pyc M., Cai Y., Greer M.S., Yurchenko O., Chapman K.D., Dyer J.M., Mullen R.T. (2017a). Turning over a new leaf in lipid droplet biology. Trends Plant Sci. 22: 596–609. [DOI] [PubMed] [Google Scholar]
- Pyc M., Cai Y., Gidda S.K., Yurchenko O., Park S., Kretzschmar F.K., Ischebeck T., Valerius O., Braus G.H., Chapman K.D., Dyer J.M., Mullen R.T. (2017b). Arabidopsis lipid droplet-associated protein (LDAP) - interacting protein (LDIP) influences lipid droplet size and neutral lipid homeostasis in both leaves and seeds. Plant J. 92: 1182–1201. [DOI] [PubMed] [Google Scholar]
- Quettier A.-L., Eastmond P.J. (2009). Storage oil hydrolysis during early seedling growth. Plant Physiol. Biochem. 47: 485–490. [DOI] [PubMed] [Google Scholar]
- Rancour D.M., Park S., Knight S.D., Bednarek S.Y. (2004). Plant UBX domain-containing protein 1, PUX1, regulates the oligomeric structure and activity of arabidopsis CDC48. J. Biol. Chem. 279: 54264–54274. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., Ishihama Y. (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2: 1896–1906. [DOI] [PubMed] [Google Scholar]
- Read S., Clarke A., Bacic A. (1993). Stimulation of growth of cultured Nicotiana tabacum W 38 pollen tubes by poly (ethylene glycol) and Cu (II) salts. Protoplasma 177: 1–14. [Google Scholar]
- Rotsch A.H., Kopka J., Feussner I., Ischebeck T. (2017). Central metabolite and sterol profiling divides tobacco male gametophyte development and pollen tube growth into eight metabolic phases. Plant J. 92: 129–146. [DOI] [PubMed] [Google Scholar]
- Rudolph M., Schlereth A., Körner M., Feussner K., Berndt E., Melzer M., Hornung E., Feussner I. (2011). The lipoxygenase-dependent oxygenation of lipid body membranes is promoted by a patatin-type phospholipase in cucumber cotyledons. J. Exp. Bot. 62: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson F., Brunaud V., Balzergue S., Dubreucq B., Lepiniec L., Pelletier G., Caboche M., Lecharny A. (2002). FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaab C., Geiger T., Stoehr G., Cox J., Mann M. (2012). Analysis of high accuracy, quantitative proteomics data in the MaxQB database. Mol. Cell Proteomics 11: M111.014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt K., Smolinski N., Neumann P., Schmaul S., Hofer-Pretz V., Braus G.H., Valerius O. (2017). Asc1p/RACK1 connects ribosomes to eukaryotic phosphosignaling. Mol. Cell. Biol. 37: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuberth C., Buchberger A. (2008). UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell. Mol. Life Sci. 65: 2360–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011). Global quantification of mammalian gene expression control. Nature 473: 337–342. [DOI] [PubMed] [Google Scholar]
- Schwarz V., Andosch A., Geretschläger A., Affenzeller M., Lütz-Meindl U. (2017). Carbon starvation induces lipid degradation via autophagy in the model alga Micrasterias. J. Plant Physiol. 208: 115–127. [DOI] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B., Joshi D., Yadav P.K., Gupta A.K., Bhatt T.K. (2016). Role of ubiquitin-mediated degradation system in plant biology. Front. Plant Sci. 7: 806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1: 2856–2860. [DOI] [PubMed] [Google Scholar]
- Shimada T.L., Hara-Nishimura I. (2010). Oil-body-membrane proteins and their physiological functions in plants. Biol. Pharm. Bull. 33: 360–363. [DOI] [PubMed] [Google Scholar]
- Shin J.-B., et al. (2013). Molecular architecture of the chick vestibular hair bundle. Nat. Neurosci. 16: 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siloto R.M.P., Findlay K., Lopez-Villalobos A., Yeung E.C., Nykiforuk C.L., Moloney M.M. (2006). The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane P.D., et al. (2016). Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat. Plants 3: 16205. [DOI] [PubMed] [Google Scholar]
- Steinhorst L., Mähs A., Ischebeck T., Zhang C., Zhang X., Arendt S., Schültke S., Heilmann I., Kudla J. (2015). Vacuolar CBL-CIPK12 Ca(2+)-sensor-kinase complexes are required for polarized pollen tube growth. Curr. Biol. 25: 1475–1482. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Otsuka T., Ohsaki Y., Cheng J., Taniguchi T., Hashimoto H., Taniguchi H., Fujimoto T. (2012). Derlin-1 and UBXD8 are engaged in dislocation and degradation of lipidated ApoB-100 at lipid droplets. Mol. Biol. Cell 23: 800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taurino M., Costantini S., De Domenico S., Stefanelli F., Ruano G., Delgadillo M.O., Sánchez-Serrano J.J., Sanmartín M., Santino A., Rojo E. (2018). SEIPIN proteins mediate lipid droplet biogenesis to promote pollen transmission and reduce seed dormancy. Plant Physiol. 176: 1531–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul P.J., Tschapalda K., Kolkhof P., Thiam A.R., Oberer M., Beller M. (2017). Targeting of the Drosophila protein CG2254/Ldsdh1 to a subset of lipid droplets. J. Cell Sci. 130: 3141–3157. [DOI] [PubMed] [Google Scholar]
- Ting J.T.L., Lee K., Ratnayake C., Platt K.A., Balsamo R.A., Huang A.H.C. (1996). Oleosin genes in maize kernels having diverse oil contents are constitutively expressed independent of oil contents. Size and shape of intracellular oil bodies are determined by the oleosins/oils ratio. Planta 199: 158–165. [DOI] [PubMed] [Google Scholar]
- Tzen J.T.C. (2012). Integral proteins in plant oil bodies. Int. Sch. Res. Notices Botany 2012: 1–16. [Google Scholar]
- Tzen J.T.C., Lai Y.K., Chan K.L., Huang A.H.C. (1990). Oleosin isoforms of high and low molecular weights are present in the oil bodies of diverse seed species. Plant Physiol. 94: 1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R., Wai C.M., Zhang Q., Song X., Edger P.P., Bryant D., Michael T.P., Mockler T.C., Bartels D. (2017). Seed desiccation mechanisms co-opted for vegetative desiccation in the resurrection grass Oropetium thomaeum. Plant Cell Environ. 40: 2292–2306. [DOI] [PubMed] [Google Scholar]
- van Wijk K.J., Kessler F. (2017). Plastoglobuli: plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu. Rev. Plant Biol. 68: 253–289. [DOI] [PubMed] [Google Scholar]
- Villette C., Berna A., Compagnon V., Schaller H. (2015). Plant sterol diversity in pollen from angiosperms. Lipids 50: 749–760. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J.A., et al. (2014). ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-W., Lee S.-C. (2012). The ubiquitin-like (UBX)-domain-containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J. Cell Sci. 125: 2930–2939. [DOI] [PubMed] [Google Scholar]
- Wang P., Mugume Y., Bassham D.C. (2018). New advances in autophagy in plants: Regulation, selectivity and function. Semin. Cell Dev. Biol. 80: 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.W., Balsamo R.A., Ratnayake C., Platt K.A., Ting J.T.L., Huang A.H.C. (1997). Identification, subcellular localization, and developmental studies of oleosins in the anther of Brassica napus. Plant J. 11: 475–487. [DOI] [PubMed] [Google Scholar]
- Wilfling F., et al. (2013). Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 24: 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins N.E., Quaynor B.K., Skinner J.R., Schoenfish M.J., Tzekov A., Bickel P.E. (2005). S3-12, Adipophilin, and TIP47 package lipid in adipocytes. J. Biol. Chem. 280: 19146–19155. [DOI] [PubMed] [Google Scholar]
- Zhang M., Fan J., Taylor D.C., Ohlrogge J.B. (2009). DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Wang Y., Cui L., Deng Y., Xu S., Yu J., Cichello S., Serrero G., Ying Y., Liu P. (2016). Morphologically and functionally distinct lipid droplet subpopulations. Sci. Rep. 6: 29539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Dai J., Wu Q. (2014). Autophagy-like processes are involved in lipid droplet degradation in Auxenochlorella protothecoides during the heterotrophy-autotrophy transition. Front. Plant Sci. 5: 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi Y., et al. (2017). Comparative lipidomics and proteomics of lipid droplets in the mesocarp and seed tissues of Chinese tallow (Triadica sebifera). Front. Plant Sci. 8: 1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienkiewicz A., Zienkiewicz K., Rejón J.D., Rodríguez-García M.I., Castro A.J. (2013). New insights into the early steps of oil body mobilization during pollen germination. J. Exp. Bot. 64: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]