Plastidial phosphoglucose isomerase determines seed yield by affecting gibberellin-mediated reproductive development and the conversion of glucose-6-phosphate to seed fatty acids and proteins.
Abstract
The plastid-localized phosphoglucose isomerase isoform PGI1 is an important determinant of growth in Arabidopsis thaliana, likely due to its involvement in the biosynthesis of plastidial isoprenoid-derived hormones. Here, we investigated whether PGI1 also influences seed yields. PGI1 is strongly expressed in maturing seed embryos and vascular tissues. PGI1-null pgi1-2 plants had ∼60% lower seed yields than wild-type plants, with reduced numbers of inflorescences and thus fewer siliques and seeds per plant. These traits were associated with low bioactive gibberellin (GA) contents. Accordingly, wild-type phenotypes were restored by exogenous GA application. pgi1-2 seeds were lighter and accumulated ∼50% less fatty acids (FAs) and ∼35% less protein than wild-type seeds. Seeds of cytokinin-deficient plants overexpressing CYTOKININ OXIDASE/DEHYDROGENASE1 (35S:AtCKX1) and GA-deficient ga20ox1 ga20ox2 mutants did not accumulate low levels of FAs, and exogenous application of the cytokinin 6-benzylaminopurine and GAs did not rescue the reduced weight and FA content of pgi1-2 seeds. Seeds from reciprocal crosses between pgi1-2 and wild-type plants accumulated wild-type levels of FAs and proteins. Therefore, PGI1 is an important determinant of Arabidopsis seed yield due to its involvement in two processes: GA-mediated reproductive development and the metabolic conversion of plastidial glucose-6-phosphate to storage reserves in the embryo.
INTRODUCTION
Oilseeds are major sources of calories for human consumption and are of a significant agricultural and industrial value. Seed number and weight are the two main components of seed yield. However, the genetic factors and the molecular and biochemical mechanisms controlling these agronomic traits are still poorly understood (Van Daele et al., 2012).
Isoprenoid hormones such as cytokinins (CKs) and gibberellins (GAs) regulate many aspects of plant growth, development, and metabolism, including shoot branching and elongation, shoot and reproductive meristem activity, the transition from vegetative growth to flowering, ovule formation, seed development, and the accumulation of seed storage compounds and, thus, seed yield (Fleet and Sun, 2005; Riefler et al., 2006; Rieu et al., 2008b; Mutasa-Göttgens and Hedden, 2009; Bartrina et al., 2011; D’Aloia et al., 2011; Chen et al., 2012a; Yamaguchi et al., 2014). In plants, these hormones are synthesized from precursors produced in the cytosol by the mevalonate pathway and in plastids by the methylerythritol 4-phosphate (MEP) pathway. The MEP pathway uses the central carbon intermediates pyruvate and glyceraldehyde 3-phosphate (GAP) to produce isopentenyl diphosphate and dimethylallyl diphosphate (DMAPP), the universal prenyl diphosphate precursors of all isoprenoids (Pulido et al., 2012; Pokhilko et al., 2015).
In oleaginous species, lipids and proteins are major contributors to seed dry weight and are thus important determinants of yield (Baud et al., 2008). Maturing oilseed embryos convert sucrose obtained from maternal tissues to lipids and proteins, which are used to support postgerminative seedling growth and establishment. De novo synthesis of fatty acids (FAs) is an ATP and NAD(P)H-dependent process that is initiated in plastids (Baud et al., 2008). Analyses of expressed sequence tags from developing seeds (White et al., 2000) and microarrays of genes expressed in seeds (Girke et al., 2000; Ruuska et al., 2002) have suggested that the major route of FA biosynthesis in the model oilseed species Arabidopsis thaliana involves the glycolytic conversion of sucrose to phosphoenolpyruvate (PEP) and pyruvate in the cytosol. PEP is then transferred to plastids by the plastidial PEP translocator (PPT) and converted to pyruvate via the action of pyruvate kinase (PK). Genetic support for this view has been obtained from characterization of mutants impaired in cytosolic GAPDH, PPT, and plastid-localized PK (Baud et al., 2007; Prabhakar et al., 2010; Guo et al., 2014). Seeds of these mutants accumulate lower levels of FAs than wild-type seeds. Recently, Lee et al. (2017) reported that seed-specific overexpression of the pyruvate transporter BASS2 increases oil content in Arabidopsis seeds, suggesting that the transport of cytosolic pyruvate into the chloroplast plays an important role in FA biosynthesis. However, metabolic flux analyses have shown that, in oilseed rape (Brassica napus) and developing Arabidopsis embryos, large proportions of FAs are synthesized from glucose-6-phosphate (G6P) in plastids via the action of phosphoglucose isomerase (PGI) coupled with pathways involving glycolysis/oxidative pentose phosphate pathway (OPPP)/Calvin-Benson enzymes (Schwender et al., 2004; Lonien and Schwender, 2009).
PGI catalyzes the reversible isomerization of fructose-6-phosphate (F6P) and G6P. This enzyme is involved in early steps of glycolysis and is thus important in the generation of ATP, reductants, and precursors (amino acids) for protein biosynthesis. PGI is also involved in the regeneration of G6P pools in the oxidative pentose phosphate OPPP, which provides metabolic intermediates for the synthesis of RNA, DNA, phenolic compounds, aromatic amino acids, and NADPH. Arabidopsis has two PGI isozymes, one in the cytosol (cytPGI) and the other in the plastid (PGI1) (Yu et al., 2000; Bahaji et al., 2015). It is widely accepted that PGI1 plays a key role in transitory starch biosynthesis in mesophyll cells of leaves, connecting the Calvin-Benson cycle with the starch biosynthetic pathway (Yu et al., 2000). We have shown that Arabidopsis mutants impaired in PGI1 accumulate low levels of CKs derived from the MEP pathway, display reduced photosynthetic capacity and slow growth phenotypes, and accumulate low levels of transitory starch in leaves, a phenotype that can be reverted to the wild type by supplementation with exogenous CK (Bahaji et al., 2015). Stimulation of photosynthesis and synthesis of active CK forms is accompanied by enhanced growth and the accumulation of exceptionally high levels of starch in the mesophyll cells of PGI1 null pgi1-2 plants (Sánchez-López et al., 2016). Thus, we have proposed that PGI1 is an important determinant of photosynthesis, transitory starch accumulation, and growth, likely due to its involvement in the synthesis of metabolic intermediates, e.g., GAP and pyruvate, required for the synthesis of plastidial isoprenoid-derived molecules (Bahaji et al., 2015).
While PGI1 appears to be involved in the production of precursors and reductants for plastidial isoprenoid-derived hormones and storage reserves such as proteins and FA and could thus potentially be a determinant of yield, its role in these processes has not been clarified through direct genetic tests. The purpose of this study was to investigate the contribution of PGI1 to seed yield via its involvement in GA metabolism and protein and FA biosynthesis. To this end, we investigated the expression pattern of PGI1 and characterized pgi1-2 plants. Our results show that PGI1 is an important determinant of seed yield in Arabidopsis due to its involvement in two processes: GA-mediated reproductive development and the metabolic conversion of plastidial G6P to embryonic FA and proteins.
RESULTS
PGI1 Is Strongly Expressed in Vascular Tissues and Developing Embryos
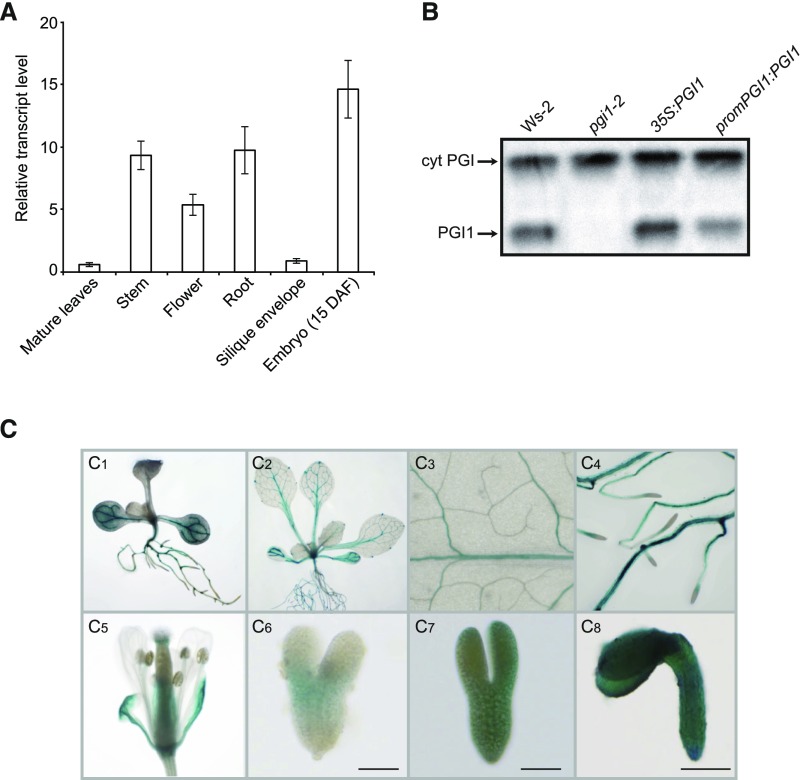
We analyzed PGI1 tissue expression profiles in wild-type (Ws-2) plants by qRT-PCR and histochemically analyzed wild-type plants transformed with constructs carrying the PGI1 promoter region (1700 bp) fused to the GUS reporter. As shown in Figure 1A, qRT-PCR analyses revealed that PGI1 is weakly expressed in mature leaves and silique envelopes but strongly expressed in roots, stems, flowers, and maturing embryos. Zymogram analyses of PGI activity further confirmed that PGI1 is expressed in maturing embryos (Figure 1B). Histochemical analyses of 10 independent promPGI1:GUS-expressing lines showed that the PGI1 promoter is strongly active in vascular tissues of roots, cotyledons, and hypocotyls of young seedlings (Figure 1C1). It is also active in vascular tissues of fully expanded mature leaves (Figures 1C2 and 1C3), roots (Figures 1C2 and 1C4), and sepals (Figure 1C5). The PGI1 promoter did not drive expression in anthers, pollen grains, or seed coat (Figure 1C5) and was weakly active in embryos at the heart developmental stage (Figure 1C6), but strongly active in maturing embryos (Figures 1C7 and 1C8). In keeping with findings of Tsai et al. (2009), compared with Figure 5, the PGI1 promoter was expressed at subdetection limits in mesophyll cells of mature leaves (Figures 1C2 and 1C3). However, this was sufficient for normal starch synthesis in leaf mesophyll cells, as mature leaves of pgi1-2 plants ectopically expressing PGI1 under the control of its own promoter (promPGI1:PGI1 plants) accumulated wild-type starch levels (Supplemental Figure 1).
Figure 1.
Expression Pattern of PGI1.
(A) qRT-PCR analysis of PGI1 expression in the indicated organs of Arabidopsis. Values represent the means ± se of four biological replicates obtained from four independent experiments, each biological replicate being a pool of the indicated organs from four plants.
(B) Zymogramic detection of PGI in proteins extracted from wild-type (Ws-2), pgi1-2, promPGI1:PGI1, and 35S:PGI1 developing seeds.
(C) Tissue-specific expression pattern of PGI1 in transgenic promPGI1:GUS plants, as manifested by GUS histochemical staining of a young seedling (C1), a 3-week-old plant (C2), a magnified expanded leaf (C3), roots (C4), a flower (C5), and three stages of embryo development ([C6] to [C8]). Heart stage embryo (C6), embryo during transition from heart to torpedo stage (C7), mature embryo (C8). DAF, days after flowering. Bars = 50 μm in (C6) and (C7) and 200 μm in (C8).
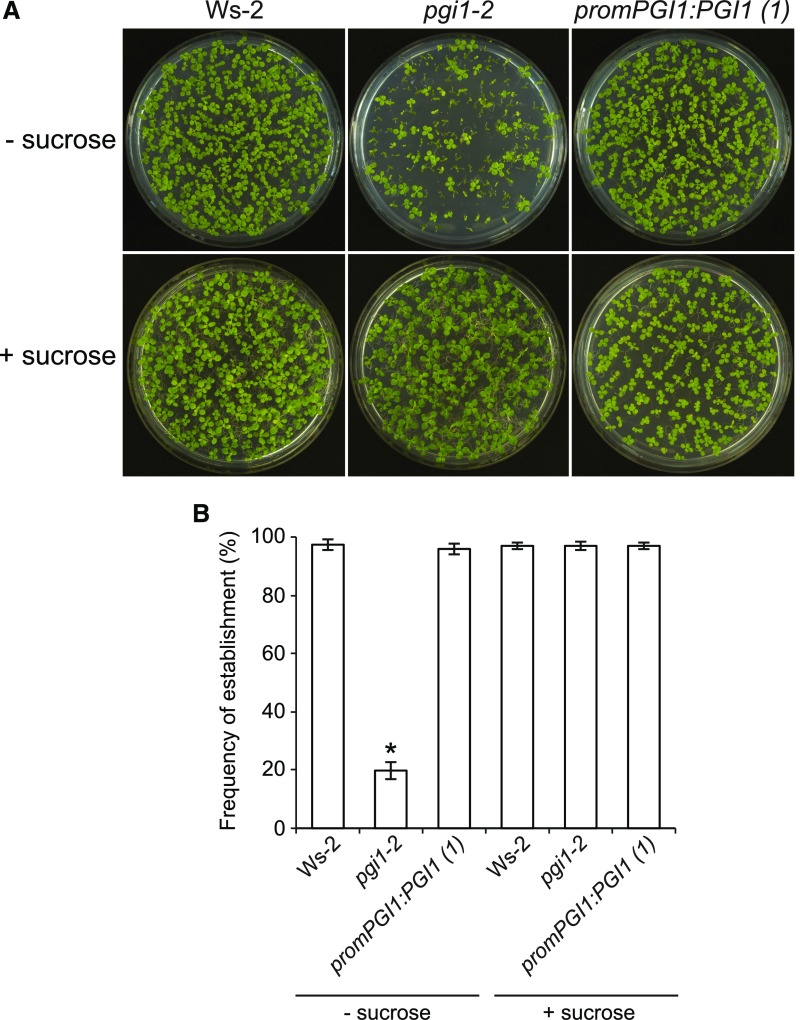
Figure 5.
pgi1-2 Seedlings Have Reduced Establishment Rates.
Photographs (A) and establishment rates (B) 12 d after sowing of wild-type (Ws-2), pgi1-2, and promPGI1:PGI1(1) plants cultured in MS with or without sucrose supplementation. Values in (B) are means ± se of four biological replicates obtained from four independent experiments, each biological replicate being ∼100 seeds. The asterisk indicates significant differences in establishment rates between wild-type and pgi1-2 plants according to Student’s t tests (P < 0.05).
Knocking out PGI1 Decreases Seed Yield
To investigate the effects that a lack of PGI1 would have on seed yield, we compared the seed weight per plant at maturity between Ws-2 and pgi1-2 plants. We also characterized aps1 plants impaired in the small subunit of ADP-glucose pyrophosphorylase (Ventriglia et al., 2008), which, like pgi1-2 plants, show a slow growth phenotype when cultured under long-day conditions and accumulate low levels of transitory starch in their leaves (Bahaji et al., 2015). As shown in Table 1, we found that seed yields of pgi1-2 plants were only ∼40% of those of wild-type plants, but wild-type yields could be restored by ectopic expression of PGI1 under its own promoter or the 35S promoter (Table 1). In clear contrast, aps1 plants had similar to wild-type seed yields (Table 1).
Table 1. Harvest Parameters for Wild-Type (Ws-2 and Col-0), pgi1-2, promPGI1:PGI1(1), 35S:PGI1, and aps1 Plants.
| Plant Line | No. of Elongated Inflorescences per Plant | No. of Siliques per Plant | No. of Seeds per Silique | Specific Seed Weight (μg Seed−1) | Seed Yield (mg Plant−1) |
|---|---|---|---|---|---|
| Ws-2 | 29.5 ± 2.5 | 684.1 ± 14.7 | 48.2 ± 0.8 | 18.1 ± 0.8 | 596.8 ± 22.1 |
| pgi1-2 | 15.3 ± 1.3* | 457.3 ± 22.4* | 47.2 ± 0.9 | 11.7 ± 0.4* | 252.5 ± 7.3* |
| promPGI1:PGI1(1) | 27.5 ± 1.9 | 682.4 ± 18.2 | 46.6 ± 1.3 | 18.0 ± 0.5 | 572.4 ± 9.8 |
| 35S:PGI1 | 28.6 ± 0.9 | 662.7 ± 21.5 | 47.3 ± 1.2 | 18.2 ± 0.6 | 570.4 ± 19.7 |
| Col-0 | 30.6 ± 2.6 | 766.1 ± 40.8 | 57.2 ± 0.9 | 17.1 ± 1.0 | 749.3 ± 11.9 |
| aps1 | 29.6 ± 3.0 | 761.6 ± 22.2 | 58.2 ± 0.9 | 17.6 ± 0.7 | 780.1 ± 19.9 |
Values are means ± se obtained from four independent experiments using 10 plants in each experiment. The asterisks indicate significant differences with respect to the wild type according to Student’s t tests (P < 0.05).
Knocking out PGI1 Reduces the Number of Seeds per Plant
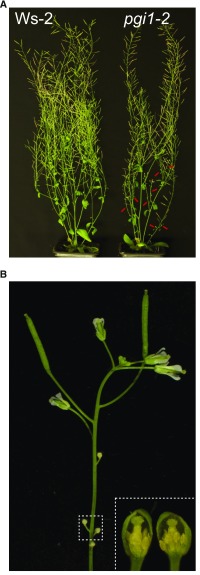
The reductions in seed yields of pgi1-2 plants could potentially be due to reductions in seed weight or in the number of either seeds per silique or siliques per plant. To differentiate between these possibilities, we first counted the numbers of inflorescences and siliques per plant and the numbers of seeds per silique in wild-type (Ws-2) and pgi1-2 plants. We also characterized aps1 plants. At first sight, we detected no major differences between the external phenotypes of Ws-2 and pgi1-2 plants at maturity (Figure 2A). Additionally, there was no significant difference in the number of seeds per silique between Ws-2 and pgi1-2 plants (Table 1). However, many inflorescences failed to elongate in pgi1-2 plants, and flowers developing first on the raceme exhibited short anthers and failed to produce siliques and seeds (Figure 2B). Consequently, the numbers of inflorescences with siliques and siliques per plant were lower in pgi1-2 plants than in Ws-2 plants (Table 1), thus causing a reduction in the number of seeds per plant. This phenotype could be rescued by the ectopic expression of PGI1 under the control of its own promoter or the 35S promoter (Table 1). Contrary to pgi1-2, aps1 plants had similar numbers of inflorescences and siliques per plant to wild-type plants (Table 1). These results demonstrate that PGI1 is an important determinant of seed yield, at least partly because of its involvement in reproductive development.
Figure 2.
Knocking out PGI1 Reduces the Number of Elongated Inflorescences and Causes Anther Development Arrest in the First Flowers of Inflorescences That Do Not Elongate.
(A) External phenotype of mature Ws-2 and pgi1-2 plants.
(B) External phenotype of a representative inflorescence that does not elongate in pgi1-2 plants and morphology of two dissected early flowers (inset) from the same inflorescence. In (A), red arrows indicate inflorescences that do not elongate in pgi1-2 plants.
The Reduced Number of Inflorescences in the pgi1-2 Mutant Is Associated with Low Bioactive GA Content
We have reported that pgi1-2 plants accumulate lower than wild-type levels of MEP-derived CKs and proposed that PGI1 may be involved in the production of precursors of MEP pathway-derived isoprenoid compounds (Bahaji et al., 2015). As GAs and CKs are important determinants of reproductive development (Rieu et al., 2008a, 2008b; Bartrina et al., 2011), we hypothesized that the reduction in the numbers of elongated inflorescences, well-developed flowers, siliques, and seeds in pgi1-2 plants could be at least partly due to reduced contents of these isoprenoid hormones. To explore this possibility further, we measured levels of different GAs in shoots (all aerial parts including rosette) at bolting time in Ws-2, pgi1-2, and promPGI1:PGI1 plants. We also counted the numbers of inflorescences and siliques in Ws-2 and pgi1-2 plants following exogenous 6-benzylaminopurine (BAP) or GA4+7 application.
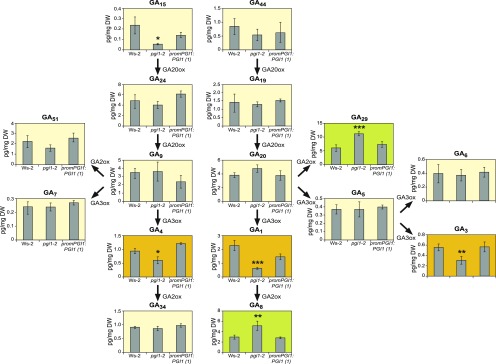
Notably, levels of GA1, GA3, and GA4—the main bioactive GAs in plants (Yamaguchi, 2008; Hedden and Thomas, 2012)—were lower in pgi1-2 shoots than in Ws-2 shoots, but wild-type levels could be restored by ectopic expression of PGI1 (Figure 3). Also, pgi1-2 plants accumulated higher levels of the inactive catabolites GA29 and GA8 than Ws-2 plants and promPGI1:PGI1-expressing pgi1-2 plants (Figure 3). Moreover, exogenous application of GA4+7 restored to wild type the numbers of elongated inflorescences and siliques of the pgi1-2 mutant (Table 2). In contrast, exogenously applied BAP did not affect the number of inflorescences and siliques (Table 2). The overall data provide strong evidence that PGI1 is an important determinant of bioactive GA content, and they indicate that the low number of inflorescences, siliques, and seeds in pgi1-2 plants is at least partly due to lower levels of active GA forms.
Figure 3.
Knocking out PGI1 Decreases Levels of Bioactive GAs and Increases Levels of Inactive GAs.
Contents of the indicated GAs (pg mg−1 DW) in shoots (20 d after sowing) of Ws-2 and pgi1-2 plants, and pgi1-2 plants transformed with promPGI1:PGI1(1). Plants were harvested at the end of the light phase. Active GAs (significantly less abundant in pgi1-2 plants than in Ws-2 plants) and inactive GAs (significantly more abundant in pgi1-2 plants than in Ws-2 plants) are highlighted in orange and green, respectively. Values represent the means ± se of two independent experiments, each consisting of four biological replicates corresponding to a pool of shoots from four plants. Asterisks indicate significant differences with respect to Ws-2 according to ANOVA (*P < 0.05, **P < 0.01, and ***P < 0.001).
Table 2. Number of Inflorescences and Siliques Produced by Wild-Type and pgi1-2 Plants with or without Exogenous 100 μM BAP or GA4+7 Treatment.
| Plant Line | No. of Elongated Inflorescences per Plant | No. of Siliques per Plant |
|---|---|---|
| Ws-2 | 29.5 ± 2.5 | 684.1 ± 14.7 |
| pgi1-2 | 15.3 ± 1.3* | 457.3 ± 22.4* |
| Ws-2 + GA | 27.3 ± 1.5 | 729.1 ± 34.2 |
| pgi1-2 + GA | 27.1 ± 2.2 | 715.6 ± 74.1 |
| Ws-2 + BAP | 26.4 ± 1.8 | 657.3 ± 19.3 |
| pgi1-2 + BAP | 14.4 ± 1.2* | 417.3 ± 26.4* |
Values are means ± se obtained from four independent experiments using 10 plants in each experiment. The asterisks indicate significant differences with respect to the wild type according to Student’s t tests (P < 0.05).
GA1 and GA4 are produced by the GA3-oxidase (GA3ox)-mediated oxidation of GA20 and GA9, respectively (Figure 3). These bioactive GAs can be deactivated by GA2-oxidase (GA2ox), yielding GA8 and GA34, respectively (Yamaguchi, 2008; Hedden and Thomas, 2012) (Figure 3). GA2ox can also metabolize GA20 to the inactive GA29 (Hedden and Thomas, 2012). In Arabidopsis shoots, the most strongly expressed GA2ox- and GA3ox-encoding genes are GA2ox6 and GA3ox1, respectively (Mitchum et al., 2006; Rieu et al., 2008a). Whereas GA3ox1 is upregulated by CKs, GA2ox6 is downregulated by these hormones (Kiba et al., 2005; Bhargava et al., 2013; Brenner and Schmülling, 2015). This and the finding that pgi1-2 plants accumulate low levels of CKs (Bahaji et al., 2015) would suggest that the reduced contents of bioactive GAs and enhanced levels of inactive GAs in pgi1-2 plants could be due to reductions in GA3ox1 and/or increases in GA2ox6 expression caused by the reduced CK content. To investigate this hypothesis, we conducted qRT-PCR analyses of GA2ox and GA3ox genes in shoots at bolting time in Ws-2 and pgi1-2 plants. We also analyzed the expression levels of GA20ox genes. As shown in Supplemental Figure 2, no significant differences in the expression levels of these genes were found between wild-type and pgi1-2 shoots.
Knocking out PGI1 Decreases Seed Weight and the Contents of Proteins and Fatty Acids
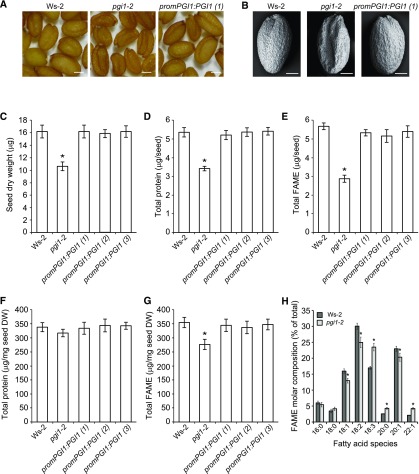
We next addressed the possibility that the low seed yields of pgi1-2 plants could also be related to reductions in seed weight. As shown in Table 1, pgi1-2 seeds were 35% lighter than Ws-2 seeds, but wild-type weights could be restored by ectopic expression of PGI1. Analyses of embryonic phenotypes in a series of developing pollinated pistils revealed no major differences between pgi1-2 seeds and Ws-2 seeds (Supplemental Figure 3), excluding the possibility that the reduced weight of pgi1-2 seeds could be due to aberrant embryo growth and development. Dry pgi1-2 seeds displayed a wrinkled phenotype (Figures 4A and 4B), accumulated 35% less dry matter than Ws-2 seeds (Figure 4C), and had lower establishment rates on MS agar with no supplemental sugar than wild-type seeds (Figure 5). These phenotypes could be restored by ectopic expression of PGI1 (Figures 4 and 5).
Figure 4.
Protein and FA Contents Are Diminished in pgi1-2 Seeds.
(A) and (B) External phenotype (A) and scanning electron micrographs (B) of seeds from wild-type (Ws-2) and pgi1-2 plants, and one line of pgi1-2 plants ectopically expressing PGI1 under the control of the PGI1 promoter [promPGI1:PGI1(1)].
(C) to (G) Seed dry weight (C), total protein content per seed (D), total FAME contents per seed (E), total protein content on DW basis (F), and total FAME content (G) on DW basis of dry seeds of the wild type, pgi1-2, and three independent lines of pgi1-2 plants ectopically expressing PGI1 under the control of the PGI1 promoter [promPGI1:PGI1(1-3)].
(H) FA profile of dry mature wild-type and pgi1-2 seeds.
Values in (C) to (H) represent the means ± se of four biological replicates obtained from four independent experiments, each biological replicate being a pool of 300 seeds (C), 200 seeds ([D] and [F]), or 20 seeds ([E], [G], and [H]). Asterisks indicate significant differences from wild-type seeds according to Student’s t tests (P < 0.05). Bars in (A) and (B) = 200 and 100 μm, respectively.
Focks and Benning (1998) suggested that the wrinkled appearance of oil-deficient wri1-1 seeds could be due to an increased sucrose content during seed development. To determine whether this might be the case in pgi1-2 seeds, we performed time-course analyses of sucrose levels in developing Ws-2 and pgi1-2 seeds. As shown in Supplemental Figure 4, there were no detectable differences between Ws-2 and pgi1-2 seeds with respect to their sucrose contents during development.
Oil and proteins are major components of dry Arabidopsis seeds (Baud et al., 2008), and seeds with reduced FA content show a wrinkled phenotype (Focks and Benning, 1998; Baud et al., 2007; Chen et al., 2015). Furthermore, seedlings of mutants with defects in the synthesis or postgerminative mobilization of seed storage lipids have reduced establishment rates in the absence of an exogenous sugar source (Hayashi et al., 1998; Cernac et al., 2006; Baud et al., 2007). Thus, these observations suggested that pgi1-2 seeds have reduced protein and lipid contents. This inference was corroborated by analysis of protein and FA levels (following methylesterification) in mature seeds. As shown in Figures 4D and 4E, pgi1-2 seeds accumulated ∼35% less protein and ∼50% less FA than Ws-2 seeds at maturity, but wild-type levels could be restored by ectopic expression of PGI1. On a dry weight (DW) basis, pgi1-2 seeds accumulated ∼8% less protein and 23% less FA than Ws-2 seeds (Figures 4F and 4G).
We also analyzed the FA composition in mature pgi1-2 seeds and, as shown in Figure 4H, found no significant differences in relative contents of the saturated FAs palmitic acid (16:0) and stearic acid (18:0) between Ws-2 and pgi1-2 seeds. In contrast, relative amounts of the desaturation intermediates, oleic (18:1), and linoleic (18:2) acids, were lower in pgi1-2 seeds than in Ws-2 seeds, while the relative amounts of the end products of desaturation and elongation, linolenic (18:3) and erucic (22:1) acids, were higher (Figure 4H). The FA composition profile of pgi1-2 seeds was similar to that of seeds of the low seed oil wri1 mutant and plants lacking the plastid-localized PK (Focks and Benning, 1998; Baud et al., 2007). These mutations impair FA biosynthesis by reducing precursor supplies. These findings indicate that PGI1 is an important determinant of seed oil accumulation through its involvement in the provision of precursors for FA synthesis.
The Low Fatty Acid Content of pgi1-2 Seeds Is Not Due to Reduced Active GA or CK Contents
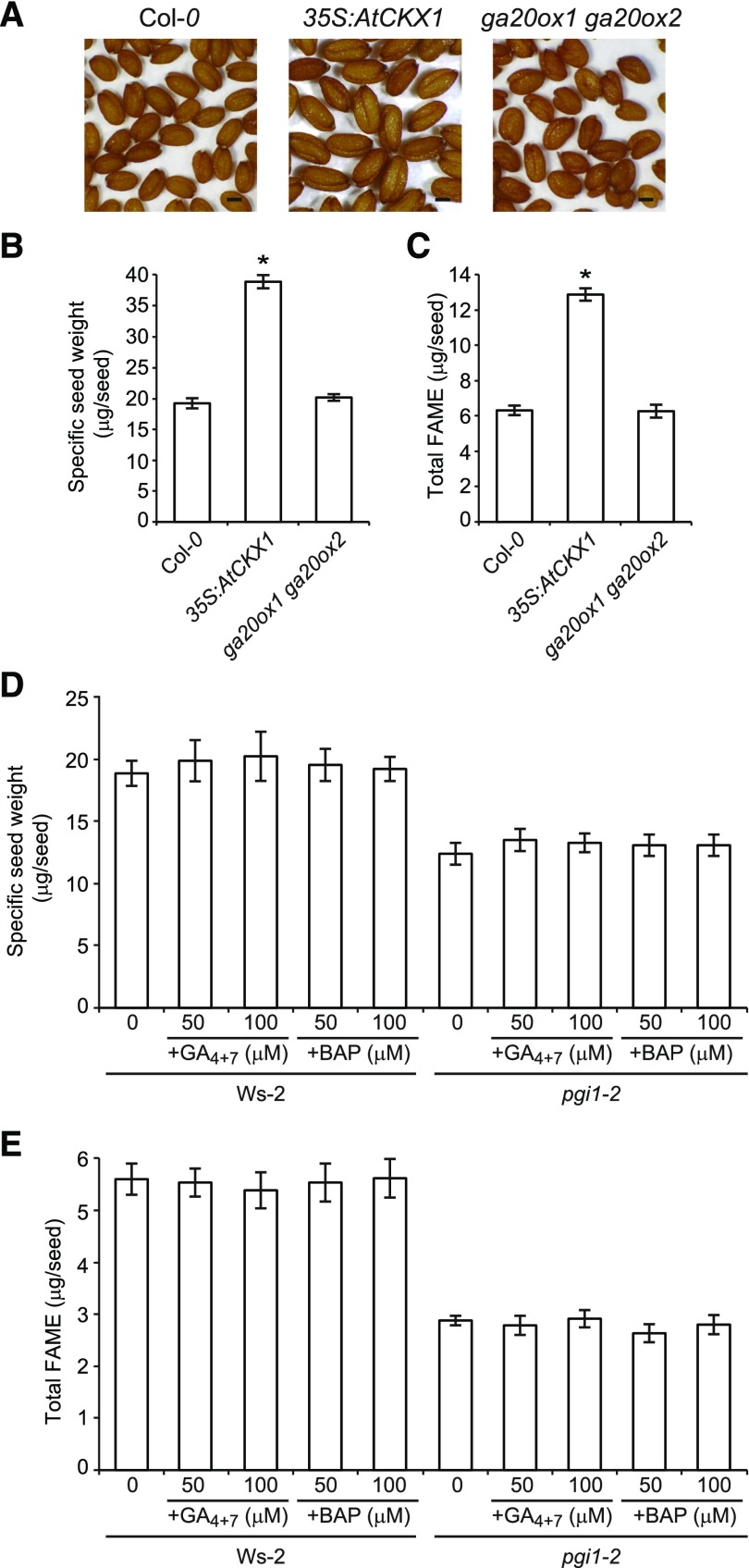
pgi1-2 plants accumulate low levels of active forms of CKs (Bahaji et al., 2015) and GAs (this work). To determine whether the reduced weight and FA content of pgi1-2 seeds could be due to low levels of active CKs or GAs, we characterized seeds of CK-deficient plants overexpressing CYTOKININ OXIDASE/DEHYDROGENASE1 (35S:AtCKX1 plants) (Werner et al., 2003) and the GA-deficient ga20ox1 ga20ox2 double mutant (Rieu et al., 2008b). Furthermore, we characterized seeds of pgi1-2 plants treated with exogenous BAP or GA4+7. In keeping with the findings of Werner et al. (2003), 35S:AtCKX1 seeds were enlarged (Figure 6A) and had almost twice the mass of wild-type seeds (Figure 6B) due to the role of CKs in controlling the cell division rate in maturing embryos (Werner et al., 2003). Additionally, the FA content of 35S:AtCKX1 seeds was twice that of wild-type seeds (Figure 6C). Conversely, the size, weight, and FA content of ga20ox1 ga20ox2 seeds were comparable to those of wild-type seeds (Figures 6A to 6C). Moreover, treatment with exogenous BAP and GA4+7 did not rescue the reduced weight and FA content phenotypes of pgi1-2 seeds (Figures 6D and 6E), showing that the low FA content of pgi1-2 seeds is not due to reduced active GA or CK contents.
Figure 6.
The Low FA Content Phenotype of pgi1-2 Seeds Is Not Due to Reduced Active CK or GA Contents.
(A) to (C) External phenotype (A), specific weight (B), and total FAME contents (C) of wild-type (Col-0), 35S:AtCKX1, and ga20ox1 ga20ox2 dry seeds.
(D) and (E) Specific weight (D) and total FAME contents (E) of dry seeds of Ws-2 and pgi1-2 plants following exogenous CK and GA4+7 application.
Bar in (A) = 200 μm. Values in (B) to (E), represent the means ± se of four biological replicates obtained from three independent experiments, each biological replicate being a pool of 300 seeds ([B] and [D]) or 20 seeds ([C] and [E]). In (B) and (C), asterisks indicate significant differences from wild-type seeds according to Student’s t tests (P < 0.05).
PGI1 Zygotically Controls FA and Protein Accumulation
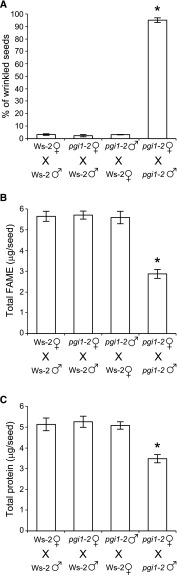
Protein and FA contents in Arabidopsis seeds can be influenced by diverse maternal factors. For example, reductions in maternal carbon supplies may cause reductions in FA accumulation in seeds (Chen et al., 2015). Proteins involved in regulating flavonoid biosynthesis in the seed coat can affect seed oil deposition by downregulating embryonic FA biosynthetic genes (Chen et al., 2012b, 2014). In addition, the homeotic regulatory gene APETALA2 can affect seed oil and protein accumulation through its action in the maternal sporophyte and endosperm genomes (Jofuku et al., 2005). pgi1-2 plants have reduced photosynthesis rates (Bahaji et al., 2015), so their low seed protein and FA contents could potentially be due to maternal factors such as limited photosynthate supplies or signals from maternal tissues. To test this possibility, we performed reciprocal crosses between pgi1-2 and Ws-2 plants and analyzed the percentages of wrinkled seeds, protein, and fatty acid methylester (FAME) content in seeds of the F1 progeny. As shown in Figure 7, there was no significant difference in the percentage of wrinkled seeds and seed FA and protein contents between the progeny of Ws-2 plants and pgi1-2 and Ws-2 crosses, showing that the low FA and protein contents of pgi1-2 seeds is dependent on the embryonic genotype but not on factors from the maternal tissues.
Figure 7.
The Phenotype of pgi1-2 Seeds Is Embryonically Controlled.
Percentage of wrinkled seeds (A), total FAME (B), and protein contents (C) in the F1 progeny of the indicated wild-type (Ws-2) and pgi1-2 crosses. Values represent the means ± se of three biological replicates obtained from three independent experiments, each biological replicate being a pool of 300 seeds (A), 20 seeds (B), or 200 seeds (C). Asterisks indicate significant differences from Ws-2 × Ws-2 cross according to Student’s t tests (P < 0.05).
Plastidial Phosphofructokinase-Mediated Metabolism of F6P Produced by PGI1 Is Not Absolutely Required for Normal Fatty Acid Biosynthesis in Arabidopsis
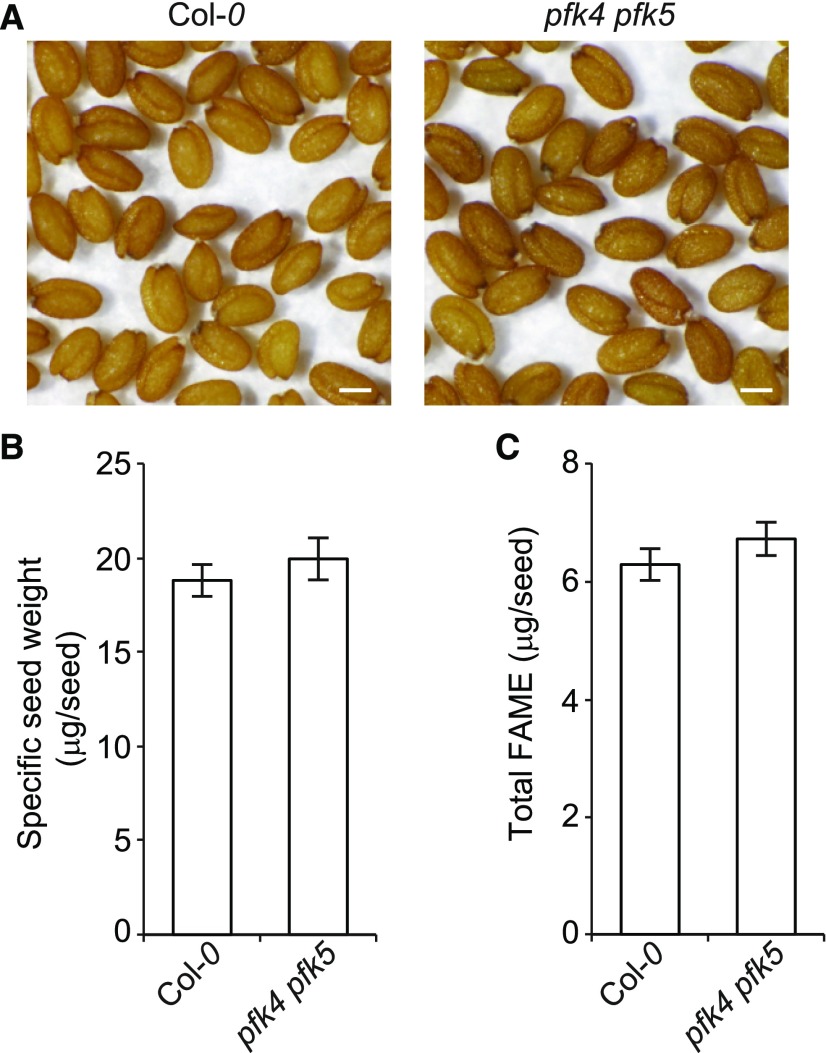
Schwender et al. (2004) comprehensively delineated every possible conversion of G6P entering the plastid into FAs in oil seed embryos using a network model consisting of glycolysis, the OPPP, the Calvin-Benson cycle, and FA synthesis enzymes. The study revealed seven flux modes that include Rubisco and that have higher carbon use efficiencies than glycolysis, two of which (modes 16 and 18) involve ATP-dependent phosphofructokinase (PFK) and bypass the glycolytic steps catalyzed by GAPDH and phosphoglycerate kinase (PGK). Two other modes (24 and 25) also have zero flux through PFK (Schwender et al., 2004). To determine whether plastidial PFK-mediated metabolism of F6P produced by PGI1 is absolutely required for normal seed FA synthesis, we measured the FA contents in seeds of the pfk4 pfk5 double knockout mutant of Arabidopsis, which has impairments in the two plastidial PFK isoforms expressed in Arabidopsis (one active, PFK4, and the other, PFK5, having weak activity when transiently expressed in wild tobacco [Nicotiana benthamiana] plants; Supplemental Figure 5). We reasoned that if the plastid-localized PFK is absolutely required for the G6P-to-FA conversion, pfk4 pfk5 seeds should accumulate low levels of FAs. Conversely, if the plastid-localized PFK-mediated metabolism of F6P produced by PGI1 is not absolutely required for normal FA biosynthesis, pfk4 pfk5 seeds should not accumulate low levels of FAs. As shown in Figure 8, the latter prediction was verified: pfk4 pfk5 seeds displayed a wild-type external phenotype and did not accumulate low levels of FA.
Figure 8.
Plastidial PFK-Mediated Metabolism of F6P Produced by PGI1 Is Not Absolutely Required for Normal FA Biosynthesis in Arabidopsis.
External phenotype (A), specific weight (B), and total FAME contents (C) of wild-type (Col-0) and pfk4 pfk5 dry seeds. Bar in (A) = 200 μm. In (B) and (C), values represent the means ± se of three biological replicates obtained from five independent experiments, each biological replicate being a pool of 300 seeds (B) or 20 seeds (C).
DISCUSSION
PGI1 Is Expressed in Organs That Produce Active GAs and MEP Pathway-Derived CKs
We have proposed that PGI1 could be involved in the glycolytic conversion of plastidial G6P into GAP and pyruvate necessary for the synthesis of plastidial isoprenoid-derived hormones, as schematically illustrated in Figure 9A (Bahaji et al., 2015). The results presented here consistently indicate that PGI1 is expressed in organs expressing genes associated with the biosynthesis of GA and MEP pathway-derived CKs. In particular, this gene is expressed in vascular tissues (Figure 1), like genes such as GA1 (which encodes the enzyme that catalyzes the first committed step of GA biosynthesis, the ent-copalyl diphosphate synthase), GA3ox1 and GA3ox2 (which encode enzymes that catalyze the production of bioactive GAs), and several genes encoding enzymes involved in rate-limiting steps of plastidial CK biosynthesis (Silverstone et al., 1997; Miyawaki et al., 2004; Mitchum et al., 2006). The strikingly similar expression patterns of PGI1 and these genes involved in the synthesis of active GAs and CKs would suggest that primary carbohydrate metabolism and secondary isoprenoid metabolism in plastids are transcriptionally coregulated via mechanisms that harmonize GA and CK biosynthesis rates (and, thus, growth and development-related processes) with carbon supplies. Accordingly, like PGI1 and some genes involved in the synthesis of plastidial isoprenoid-derived hormones, PPT1 and PKp2 (both encoding enzymes that function in the provision of pyruvate to the plastid) are expressed in vascular tissues of adult plants and seedlings (Knappe et al., 2003; Baud et al., 2007).
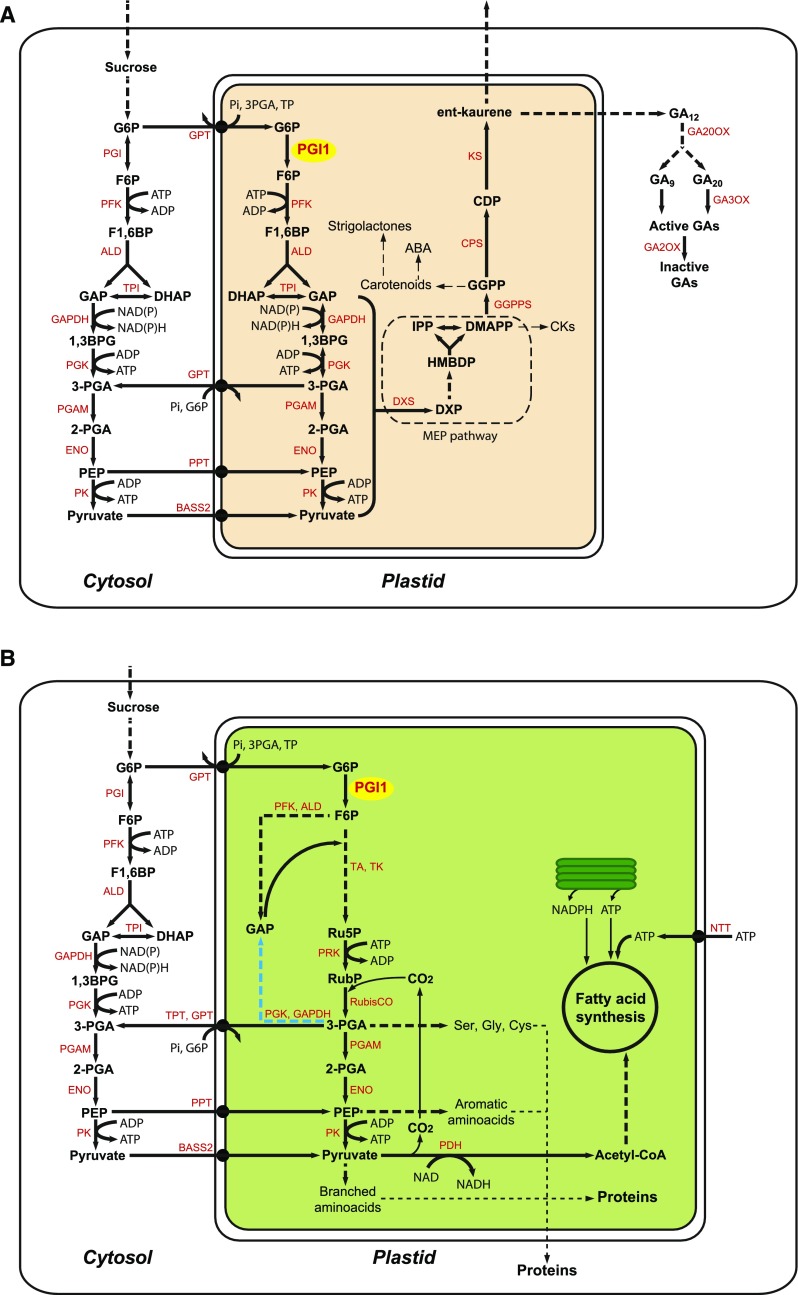
Figure 9.
Suggested Schemes for PGI1-Mediated Biosynthesis of MEP Pathway Isoprenoid-Derived CKs and GAs in Cells of Vascular Tissues, and FAs and Proteins in Mixotrophic Maturing Embryos of Arabidopsis.
In both cases, cytosolic G6P can be glycolytically converted to PEP and pyruvate, which can enter plastids via the PPT and BASS2 transporters, respectively. According to the scheme in (A), some of the cytosolic G6P can be incorporated into plastids through the hexose-P transporter (GPT) and subsequently glycolytically converted to GAP and pyruvate. Plastidial GAP and pyruvate are then metabolized by 1-deoxy-d-xylulose 5-phosphate (DXP) synthase (DXS) to products that enter the MEP pathway and fuel synthesis of isoprenoid hormones (Pulido et al., 2012; Pokhilko et al., 2015). DMAPP can be converted to ent-kaurene in a three-step process catalyzed by geranylgeranyl diphosphate (GGPP) synthase (GGPPS), ent-copalyl diphosphate synthase (CPS), and ent-kaurene synthase (KS). ent-Kaurene can be then transported to other parts of the plant or oxidized to GA12, which is converted to bioactive GA in the cytosol by GA20ox and GA3ox. DMAPP can also be converted to CKs (Spíchal, 2012). In heterotrophic cells lacking plastid-localized phosphoglyceromutase (PGAM), enolase (ENO), and the triose-P translocator (TPT), 3PGA produced in the plastid via PGI1 can be exported to the cytosol via GPT or other nonspecific transporters then glycolytically converted into PEP and pyruvate, which can then enter plastids through the PPT and the BASS2 pyruvate transporter, respectively. According to the scheme in (B), some of the cytosolic G6P can be incorporated into plastids through GPT and converted to 3PGA by the Rubisco shunt, which involves reactions of the early steps of glycolysis (PFK and aldolase), reactions of the nonoxidative pentose phosphate pathway, phosphoribulokinase (PRK), and Rubisco. In plants lacking plastidial PFK, part of the 3PGA generated can be converted to GAP necessary for Ru5P production by means of plastidial GAPDH and PGK (highlighted in blue). 3PGA can leave plastids via the TPT, the GPT, or other nonspecific transporters and glycolytically converted to PEP and pyruvate in the cytosol, which can enter plastids, as indicated above. Alternatively, and/or additionally, plastidial 3PGA can be converted to pyruvate in plastids. The ATP required for FA synthesis can be generated from photosynthesis and the PK reaction in the plastidial compartment or obtained from the cytosol via the NTT transporter (Reiser et al., 2004). NADPH and NADH can be generated in plastids from the light photosynthetic and pyruvate dehydrogenase reactions, respectively. ALD, aldolase; TA, transaldolase; TK, transketolase; TPI, triose-phosphate isomerase; PDH, pyruvate dehydrogenase; HMBDP, (E)-4-hydroxy-3-methyl-but-2-enyl diphosphate; IPP, isopentenyl diphosphate; CDP, ent-copalyl diphosphate. Enzymes are highlighted in red.
Environmental stimuli such as light, temperature, and salt stress specifically regulate the expression of GA biosynthetic enzymes, allowing GAs to translate these extrinsic signals into developmental changes according to environmental conditions (Yamaguchi, 2008). Oxidative modification of redox-sensitive proteins plays important roles in modulating rapid physiological and developmental responses of plants to changing environmental conditions (Foyer and Noctor, 2005). Notably, recent high-throughput analyses of proteins whose thiols undergo reversible oxidative modifications in Arabidopsis have revealed that PGI1 is a redox-sensitive protein that responds to agents that perturb cellular redox homeostasis and inhibit growth (Liu et al., 2014; Yin et al., 2017). It is thus tempting to speculate that PGI1 could play an important role in fine regulatory tuning of plastidial isoprenoid hormone-driven growth to a pace that is compatible with the plant’s carbon status under prevailing environmental conditions.
PGI1 Is an Important Determinant of Seed Yield Due to Its Involvement in GA-Mediated Reproductive Development
pgi1-2 plants produce fewer elongated inflorescences than Ws-2 plants, and flowers developing first on the short inflorescences fail to produce siliques and seeds (Figure 2B). Consequently, pgi1-2 plants produce fewer siliques and seeds than Ws-2 plants (Table 1). This phenotype cannot be ascribed to slow plant growth or low leaf starch content, since the number of inflorescences and the seed yield of starch-deficient aps1 plants were comparable to those of wild-type plants (Table 1).
In Arabidopsis, bioactive GAs stimulate elongation of vegetative stem internodes upon bolting and exert a positive effect on inflorescence length and flower formation, thus acting as major determinants of reproductive development and yield (Rieu et al., 2008b; Yamaguchi et al., 2014). Here, we have shown that levels of the main bioactive GAs (e.g., GA1, GA3, and GA4) are lower in pgi1-2 plants than in Ws-2 plants and pgi1-2 plants ectopically expressing PGI1 (Figure 3). These findings strongly indicate that the reduced numbers of elongated inflorescences, siliques, and seeds per pgi1-2 plant are partly due to their diminished content of active GAs. Accordingly, exogenous GA application restored to wild type the numbers of elongated inflorescences and siliques in pgi1-2 plants (Table 2). Therefore, we propose that PGI1 is a major determinant of seed yield, at least in part, due to its regulatory action on GA homeostasis and thus reproductive development.
To maintain optimum growth rates, plants have evolved homeostatic regulatory mechanisms for modulating GA levels involving both negative feedback and positive feed-forward transcriptional regulation of GA metabolism genes (Hedden and Thomas, 2012). Plants have also evolved less well-defined mechanisms for posttranscriptional regulation of GA metabolism that may involve microRNA-mediated destabilization and/or reductions in the efficiency of translation of several GA2ox, GA3ox, and GA20ox transcripts (http://sundarlab.ucdavis.edu/mirna/) (Barker, 2011) and modification of the stability of GA metabolism enzymes (Magome et al., 2004; Lee and Zeevaart, 2007; Barker, 2011). The qRT-PCR analyses conducted in this study revealed no major differences in the expression of GA metabolism genes between Ws-2 and pgi1-2 plants that could account for the observed differences in the GA contents of these genotypes (Supplemental Figure 2). This finding suggests that there are further complexities in GA homeostasis due to mechanisms in which PGI1 plays an important role in regulating the activity of GA metabolism enzymes and/or providing precursors for active GA biosynthesis.
PGI1 Is an Important Determinant of Seed Yield Due to Its Involvement in Embryonic Fatty Acid and Protein Biosynthesis
PGI1’s roles in seed FA and protein production have previously been obscure. Here, we have provided evidence that PGI1 contributes to seed yield through participation in the generation of precursors for embryonic FA and protein biosynthesis, as schematically illustrated in Figure 9B. The hypothesis is strongly supported by the following findings: pgi1-2 seeds accumulate ∼35% less protein and 50% less FAs than Ws-2 seeds, respectively (Figure 4), PGI1 is strongly expressed in the embryo (Figure 1), and PGI1 zygotically controls FA and protein accumulation (Figure 7). Two findings strongly indicate that the low FA contents of pgi1-2 seeds are not due to reduced active CK and GA contents: Seeds of CK-deficient 35S:AtCKX1 and GA-deficient ga20ox1 ga20ox2 do not accumulate low levels of FA (Figure 6C), and exogenous application of BAP and GA4+7 did not rescue the reduced weight and FA content of pgi1-2 seeds (Figures 6D and 6E). The observation that pgi1-2 seeds have elevated relative contents of linolenic and erucic acids (Figure 4H) indicates that no feedback regulation occurs in this mutant to adjust the degree of unsaturation and elongation of the FAs to the decrease of carbon flow into the FA biosynthetic pathway and that different regulators control the activities of enzymes involved in lipid metabolism during seed development.
PGI may be involved in regenerating the G6P pool in the OPPP or early steps of glycolysis. The plastidial OPPP has been regarded as one of the major sources of NADPH linked to FA biosynthesis in oilseeds (Kang and Rawsthorne, 1996; Eastmond and Rawsthorne, 1998; Hutchings et al., 2005). An objection to this hypothesis is that plastid-localized G6P dehydrogenase (which catalyzes G6P’s entry into the plastidial OPPP) is subject to redox inactivation during illumination (Née et al., 2009), so plastidial G6P dehydrogenase is likely inactive during FA accumulation in green oilseeds. Furthermore, metabolic flux analyses have indicated that the reductant produced by the plastidial OPPP accounts for only 20% of the total reductant needed for FA synthesis in oilseed embryos (Schwender et al., 2003). The glycolytic capacity in plastids of lipid-storing seeds considerably exceeds requirements for maximum rates of lipid synthesis (Eastmond and Rawsthorne, 2000). Thus, the glycolytic pathway could play an important role in the conversion of photosynthate to oil. However, previous biochemical analyses with oilseed rape and Arabidopsis embryos have shown that up to 80% of the 3PGA linked to FA biosynthesis is generated from G6P through pathways involving glycolysis/OPPP/Calvin-Benson enzymes that have higher carbon use efficiencies than glycolysis (Schwender et al., 2004; Lonien and Schwender, 2009). Some of these pathways involve plastidial PFK. Others have zero flux through PFK and derive the GAP needed for Ru5P production via 3PGA metabolism (Schwender et al., 2004; Lonien and Schwender, 2009). In this work, we found that pfk4 pfk5 seeds do not accumulate low levels of FA (Figure 8C), strongly indicating that plastidial PFK is not absolutely required for the metabolic conversion of F6P produced by PGI1 into FAs in Arabidopsis seeds. To explain why the pgi1-2 mutation reduces seed FA content but the pfk4 pfk5 mutation does not, we propose that pgi1-2 completely blocks the entry of carbon into the 3PGA-generating glycolysis/OPPP/Rubisco network, whereas pfk4 pfk5 can be compensated for by Rubisco shunt flux modes that cause some 3PGA formed in the plastids to be converted into the GAP needed for Ru5P production by plastidial GAPDH and PGK enzymes (Figure 9B). The remainder of the 3PGA produced in the plastids may be metabolized in plastids and/or exported to the cytosol, where it may be converted to PEP and pyruvate, which may move back into plastids and subsequently be converted to FAs (Figure 9B).
According to a flux map of central carbon metabolism established in steady state 13C labeling experiments by Schwender et al. (2003), PEP is preferentially transported from the cytosol to the stroma as a substrate for de novo FA synthesis in developing rapeseed embryos. The Arabidopsis genome contains two PPT-encoding genes: PPT1 and PPT2 (Knappe et al., 2003). Unlike PPT1, PPT2 is not expressed in seeds (Knappe et al., 2003) (see also http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007). Although the overall information indicates that PPT1 participates in embryonic FA biosynthesis, Prabhakar et al. (2010) showed that seeds of Arabidopsis cue1 plants with defective PPT1 can accumulate up to 85% of wild-type FA contents. This indicates that PEP may not be the major form of photosynthate that enters the plastid for subsequent conversion to FAs in Arabidopsis seeds. Moreover, the findings that pgi1-2 seeds and seeds of pkp2 plants impaired in plastidial PK accumulate 50% and 60% less FA than wild-type seeds, respectively (Baud et al., 2007; this work) strongly indicate that a sizable pool of the photosynthate linked to Arabidopsis seed FA production enters the plastid as G6P and pyruvate, as shown schematically in Figure 9B.
METHODS
Plants, Growth Conditions, and Sampling
The work was performed using Arabidopsis thaliana wild-type plants (ecotypes Col-0 and Ws-2), pgi1-2 knockout mutants (Bahaji et al., 2015), Ws-2 plants expressing GUS under the control of the 1700-bp Ws-2 PGI1 promoter (promPGI1:GUS), pgi1-2 plants expressing PGI1 under the control of the 1700-bp PGI1 promoter (promPGI1:PGI1), pgi1-2 plants expressing PGI1 under the control of the cauliflower mosaic virus 35S promoter (35S:PGI1), the ADP-glucose pyrophosphorylase null aps1 mutant (SALK_040155), CK OXIDASE/DEHYDROGENASE1-overexpressing 35S:CKX1 plants (Werner et al., 2003), the GA-deficient ga20ox1 ga20ox2 double mutant (Rieu et al., 2008b), and the pfk4 and pfk5 T-DNA insertional knockout mutants (SALK_012602 and SAIL_297_F05, respectively) obtained from the European Arabidopsis Stock Center (NASC). By crossing pfk4 with pfk5, self-pollinating the resulting heterozygous mutants, and PCR screening for homozygous progeny using the oligonucleotide primers listed in Supplemental Table 1, we produced pfk4 pfk5 double mutants. The promPGI1:PGI1, 35S:PGI1, and promPGI1:GUS plasmid constructs were produced using Gateway technology as illustrated in Supplemental Figure 6 and confirmed by sequencing. Primers used for PCR amplification of PGI1 cDNA, GUS, and the PGI1 promoter are listed in Supplemental Table 2. The plasmid constructs were transferred into Agrobacterium tumefaciens EHA105 cells by electroporation and utilized to transform Arabidopsis plants as described by Clough and Bent (1998). Transgenic promPGI1:PGI1, 35S:PGI1, and promPGI1:GUS plants were selected on medium containing antibiotics.
Plants were cultured on soil in growth chambers under a long-day photoperiod with (16 h light 22°C/8 h dark 18°C) at 90 μmol photons m−2 s–1 light intensity using a mix of Sylvania 215-W cool white fluorescent tubes and 60-W mate bulbs. For exogenous application of CKs and GAs, 1 week after bolting, pgi1-2 plants were sprayed every 2 d with the indicated concentrations of BAP or GA4+7.
Confirmation of the Knockout Status of pfk5
PFK5 contains 13 exons (Supplemental Figure 5A) and encodes a protein of 537 amino acids (Supplemental Figure 5B) with weak activity. The pfk5 allele has a T-DNA inserted in exon 12 and encodes a truncated protein (designated PFK5*) that lacks 85 amino acids at the C terminus (Supplemental Figure 5B). To explore whether PFK5* is enzymatically active, we produced PFK5, PFK5*, and PFK4 expression vectors (Supplemental Figure 7). Primers used for PCR amplification of PFK4 and PFK5 cDNA are listed in Supplemental Table 2. The plasmids were transferred to Agrobacterium GV3101, which were used for transient expression in agroinfiltrated Nicotiana benthamiana leaves. For coinfiltration of the RNA-silencing inhibitor P19, a bacterial suspension harboring pBin61-P19 (Voinnet et al., 1999) was added. The bacteria were infiltrated into the abaxial sides of leaves using a 1-mL syringe with no needle. N. benthamiana leaf samples were taken 2 d after infiltration, immediately frozen in liquid nitrogen, and subjected to ATP-PFK activity measurement analyses as described by Mustroph et al. (2007).
qRT-PCR Analyses
Total RNA was extracted from frozen organs using the TRIzol method according to Meng and Feldman (2010) and treated with RNase-free DNase (Takara). Then, 1.5 µg portions were reverse-transcribed using a mixture of oligo(dT) and random primers and an Expand Reverse Transcriptase kit (Roche) according to the manufacturer’s instructions. RT-PCR amplifications were performed using a 7900HT sequence detector system (Applied Biosystems) with Premix Ex Tag Mix (Takara RR420A) according to the manufacture’s protocol. Each amplification was performed in triplicate with 0.4 μL of the first-strand cDNA in a total volume of 20 µL. The specificity of the PCR amplicons was checked by acquiring heat dissociation curves (from 60°C to 95°C). Comparative threshold values were normalized to an EF-1α RNA internal control and compared with obtain relative expression levels. Primers used for qRT-PCR amplification of PGI1 and different GA2ox, GA3ox, and GA20ox genes are listed in Supplemental Table 3.
GUS Expression Analysis
Expression of the GUS reporter gene was monitored using the histochemical staining assay described by Jefferson et al. (1987).
Morphological Analysis of Seeds
Seeds were observed and photographed using an Olympus MVX10 stereomicroscope. Scanning electron microscopy analyses of gold-coated dry seeds were performed using a JEOL JSM-5610LV scanning electron microscope.
Metabolite Analysis
Seed Fatty Acid Content Analysis
Sets of 20 dry seeds were prepared in Teflon-lined screw-capped glass tubes and incubated at 90°C for 90 min in 1 mL of 1 M HCl in methanol, with 20 µg of heptadecanoic acid (C17:0) as an internal standard, and 300 μL of toluene. After cooling to room temperature, FAMEs were extracted in 1 mL hexane following the addition of 1.5 mL of 0.9% (w/v) NaCl. The hexane phase was transferred to a gas chromatography vial after vigorous vortex mixing and centrifuged for 3 min at 10,000g. Portions (1 μL) of the samples were analyzed using a gas chromatography-mass spectrometry system consisting of a 7890A GC equipped with an Agilent J&W DB-WAX column (diameter, 0.25 mm; film thickness, 0.25 μm; length, 30 m) coupled to a 5975C Inert XL MSD mass selective detector (Agilent Technologies). The GC settings were as follows: carrier gas, helium (1 mL/min); injection mode, split (1:50); injector temperature, 250°C; GC temperature program, 1 min at 50°C then 25°C/min rise to 200°C followed by 3°C/min rise to 230°C and finally 18 min hold at 230°C. The MSD settings were: solvent delay, 5 min; ion source temperature, 230°C; fragmentation energy, 70 eV; scanning rate, 20 scans/min (40 to 500 m/z). Data were acquired with ChemStation software (Agilent).
Seed Protein Content Analysis
Sets of 200 dry seeds were ground in a mortar and pestle. The powder was resuspended in 60 μL of extraction buffer (50 mM HEPES, 5 mM MgCL2, 5 mM DTT, 1 mM PSMF, 10% [v/v] ethylene glycol [pH 7.5], 1 mM EDTA, and 1% Polyclar), sonicated, and centrifuged at 10,000g. The supernatant thus obtained was kept on ice and the pellet was treated with 60 μL of 1 M NaOH and centrifuged at 10,000g. The supernatants were used to quantify protein levels using a Bio-Rad DC protein assay kit.
Seed Sucrose Content Analysis
For sucrose extraction, sets of 50 seeds were homogenized in 500 μL of 90% (v/v) ethanol, left at 70°C for 90 min, and centrifuged at 13,000g for 10 min. Sucrose content from the supernatants was determined by HPLC with pulsed amperometric detection on a DX-500 Dionex system by gradient separation with a CarboPac 10 column according to the application method suggested by the supplier (100 mM NaOH/100 mM sodium acetate to 100 mM NaOH/500 mM sodium acetate in 40 min).
Leaf Starch Content Analysis
Fully expanded source leaves of plants were harvested at the end of the light period, freeze-clamped, and ground to a fine powder in liquid nitrogen with a pestle and mortar. Starch was measured using an amyloglucosidase-based test kit (Boehringer Mannheim).
Native Gel PGI Activity Assays
PGI zymograms were acquired as described by Bahaji et al. (2015). Briefly, protein extracts of wild-type, pgi1-2, promPGI1:PGI1, and 35S:PGI1 seeds collected 15 d after flowering were loaded onto a 7.5% (w/v) polyacrylamide gel. After electrophoresis, the gels were stained by incubating in darkness at room temperature with 0.1 M Tris-HCl (pH 8.0), 5 mM F6P, 1 mM NAD+, 4 mM MgCl2, 0.2 mM methylthiazolyldiphenyl-tetrazolium bromide (Sigma-Aldrich M5655), 0.25 mM phenazine methosulfate (Sigma-Aldrich P9625), and 1 unit/mL of G6P dehydrogenase from Leuconostoc mesenteroides (Sigma-Aldrich G8404).
Quantitative Analysis of Endogenous GAs
GA samples were prepared and analyzed as described by Urbanová et al. (2013), with some modifications. Briefly, freeze-dried plant tissue samples (5–10 mg) were ground to a fine consistency using 3-mm zirconium oxide beads (Retsch) and a MM 301 mill (Retsch) vibrating at 30 Hz for 3 min, with 1 mL of ice-cold 80% acetonitrile containing 5% formic acid as extraction solution. The samples were then extracted overnight at 4°C using a Stuart SB3 benchtop laboratory rotator (Bibby Scientific) after adding 17 internal GAs standards ([2H2]GA1, [2H2]GA3, [2H2]GA4, [2H2]GA5, [2H2]GA6, [2H2]GA7, [2H2]GA8, [2H2]GA9, [2H2]GA15, [2H2]GA19, [2H2]GA20, [2H2]GA24, [2H2]GA29, [2H2]GA34, [2H2]GA44, [2H2]GA51, and [2H2]GA53; purchased from OlChemIm). The homogenates were centrifuged at 36,670g and 4°C for 10 min, and the corresponding supernatants were further purified using reversed-phase and mixed mode SPE cartridges (Waters) and analyzed by ultra-high-performance chromatography-tandem mass spectrometry (UHPLC-MS/MS; Micromass). GAs were detected using the multiple-reaction monitoring mode of the transition of the ion [M–H]− to the appropriate product ion. Masslynx 4.1 software (Waters) was used to analyze the data, and the standard isotope dilution method (Rittenberg and Foster, 1940) was used to quantify the GAs.
Statistical Analysis
The data presented are the means (±se) from at least three independent experiments, with three to five biological replicates for each experiment. The significance of differences between control and transgenic lines was statistically evaluated with Student’s t test using SPSS software. Differences were considered significant if P < 0.05. In GA analyses, significance was determined by one-way univariate ANOVA for parametric data and Kruskal Wallis tests for nonparametric data, using open source R 2.15.1 software (http://cran.r-project.org/). Tukey’s HSD post-hoc tests were applied for multiple comparisons after ANOVA.
Accession Numbers
Sequence data from the article can be found in the GenBank/EMBL libraries under the following accession numbers: PGI1 (At4g24620), PKK4 (At5g61580), PFK5 (At2g22480), GA2ox1 (At1g78440), GA2ox2 (At1g30040), GA2ox3 (At2g34555), GA2ox4 (At1g47990), GA2ox6 (At1g02400), GA2ox7 (At1g50960), GA2ox8 (At4g21200), GA3ox1 (At1g15550), GA3ox2 (At1g80340), GA3ox3 (At4g21690), GA3ox4 (At1g80330), GA20ox1 (At4g25420), GA20ox2 (At5g51810), GA20ox3 (At5g07200), GA20ox4 (At1g60980), and GA20ox5 (At1g44090).
Supplemental Data
Supplemental Figure 1. Starch content in mature leaves of Ws-2, pgi1-2 and promPGI1:PGI1 plants.
Supplemental Figure 2. Expression levels of different GA2ox, GA3ox, and GA20ox genes in wild-type (Ws-2) and pgi1-2 shoots.
Supplemental Figure 3. Embryo development in wild-type (Ws-2) and pgi1-2 embryos at different developmental stages.
Supplemental Figure 4. Time course of sucrose content in developing Ws-2 and pgi1-2 seeds.
Supplemental Figure 5. Confirmation of the knockout status of the pfk4 pfk5 mutant.
Supplemental Figure 6. Stages in construction of the promPGI1:GUS, 35S:PGI1, and promPGI1:PGI1 plasmids used to produce promPGI1:GUS-, 35S:PGI1-, and promPGI1:PGI1-expressing plants.
Supplemental Figure 7. Stages in construction of the 35S:PFK4, 35S:PFK5, and 35S:PFK5* plasmids used for transient expression in N. benthamiana leaves.
Supplemental Table 1. Primers used for PCR screening of the pfk4 pfk5 mutant.
Supplemental Table 2. Primers used to produce the promPGI1:GUS, promPGI1:PGI1 35S:PGI1, 35S:PFK4, 35S:PFK5, and 35S:PFK5* plasmids.
Supplemental Table 3. Primers used in qRT-PCR analyses.
Supplemental Table 4. ANOVA table.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Francisco Carreto-Cano (Institute of Agrobiotechnology of Navarra) for technical support. We also thank T. Werner (Freie Universität Berlin, Germany), R.E. Häusler (University of Cologne, Germany), and Peter Hedden (Rothhamsted Research, UK) who kindly provided the 35S:AtCKX1, pgi1-2, and ga20ox1 ga20ox2 mutants, respectively. This work was partially supported by the Comisión Interministerial de Ciencia y Tecnología and Fondo Europeo de Desarrollo Regional (Spain) (Grants BIO2013-49125-C2-1-P and BIO2016-78747-P), the Ministry of Education, Youth, and Sports of the Czech Republic (Grant LO1204 from the National Program of Sustainability I), the Government of Navarra (ref. P1004 PROMEBIO), the Università degli Studi di Milano (UNIMI-RTD-A, Linea2-DBS 2017-2018), and the H2020-MSCA-RISE Project (ExpoSeed GA-691109).
AUTHOR CONTRIBUTIONS
A.B., G.A., I.E., A.M.S.-L., F.J.M., N.D.D., L.S., K.D., and J.P.-R. designed the experiments and analyzed the data. A.B., G.A., I.E., S.G.-A., R.J.B., M.C.S., D.T., E.C., M.A.M., and E.B.-F. performed most of the experiments. A.B., I.E., K.D., and J.P.-R. supervised the experiments. A.B., F.J.M., and J.P.-R. wrote the article with contributions from all the authors. J.P.-R. conceived the project and research plans.
References
- Bahaji A., et al. (2015). Plastidic phosphoglucose isomerase is an important determinant of starch accumulation in mesophyll cells, growth, photosynthetic capacity, and biosynthesis of plastidic cytokinins in Arabidopsis. PLoS One 10: e0119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker R. (2011). Gibberellin Biosynthesis and Signalling in Arabidopsis Root Growth. PhD dissertation (Nottingham, UK: University of Nottingham). [Google Scholar]
- Bartrina I., Otto E., Strnad M., Werner T., Schmülling T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S., Wuillème S., Dubreucq B., de Almeida A., Vuagnat C., Lepiniec L., Miquel M., Rochat C. (2007). Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J. 52: 405–419. [DOI] [PubMed] [Google Scholar]
- Baud S., Dubreucq B., Miquel M., Rochat C., Lepiniec L. (2008). Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. The Arabidopsis Book 6: e0113, doi/10.1199/tab.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A., Clabaugh I., To J.P., Maxwell B.B., Chiang Y.-H., Schaller G.E., Loraine A., Kieber J.J. (2013). Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol. 162: 272–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner W.G., Schmülling T. (2015). Summarizing and exploring data of a decade of cytokinin-related transcriptomics. Front. Plant Sci. 6: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A., Andre C., Hoffmann-Benning S., Benning C. (2006). WRI1 is required for seed germination and seedling establishment. Plant Physiol. 141: 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.-Q., Lin I.W., Qu X.-Q., Sosso D., McFarlane H.E., Londoño A., Samuels A.L., Frommer W.B. (2015). A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27: 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Du X., Zhu Y., Wang Z., Hua S., Li Z., Guo W., Zhang G., Peng J., Jiang L. (2012a). Seed Fatty Acid Reducer acts downstream of gibberellin signalling pathway to lower seed fatty acid storage in Arabidopsis. Plant Cell Environ. 35: 2155–2169. [DOI] [PubMed] [Google Scholar]
- Chen M., Wang Z., Zhu Y., Li Z., Hussain N., Xuan L., Guo W., Zhang G., Jiang L. (2012b). The effect of transparent TESTA2 on seed fatty acid biosynthesis and tolerance to environmental stresses during young seedling establishment in Arabidopsis. Plant Physiol. 160: 1023–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Xuan L., Wang Z., Zhou L., Li Z., Du X., Ali E., Zhang G., Jiang L. (2014). TRANSPARENT TESTA8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis. Plant Physiol. 165: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- D’Aloia M., Bonhomme D., Bouché F., Tamseddak K., Ormenese S., Torti S., Coupland G., Périlleux C. (2011). Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J. 65: 972–979. [DOI] [PubMed] [Google Scholar]
- Eastmond P.J., Rawsthorne S. (1998). Comparison of the metabolic properties of plastids isolated from developing leaves or embryos of Brassica napus L. J. Exp. Bot. 49: 1105–1111. [Google Scholar]
- Eastmond P.J., Rawsthorne S. (2000). Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos. Plant Physiol. 122: 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet C.M., Sun T.-P. (2005). A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8: 77–85. [DOI] [PubMed] [Google Scholar]
- Focks N., Benning C. (1998). wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118: 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke T., Todd J., Ruuska S., White J., Benning C., Ohlrogge J. (2000). Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 124: 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ma F., Wei F., Fanella B., Allen D.K., Wang X. (2014). Cytosolic phosphorylating glyceraldehyde-3-phosphate dehydrogenases affect Arabidopsis cellular metabolism and promote seed oil accumulation. Plant Cell 26: 3023–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Toriyama K., Kondo M., Nishimura M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Thomas S.G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444: 11–25. [DOI] [PubMed] [Google Scholar]
- Hutchings D., Rawsthorne S., Emes M.J. (2005). Fatty acid synthesis and the oxidative pentose phosphate pathway in developing embryos of oilseed rape (Brassica napus L.). J. Exp. Bot. 56: 577–585. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku K.D., Omidyar P.K., Gee Z., Okamuro J.K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. USA 102: 3117–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang F., Rawsthorne S. (1996). Metabolism of glucose-6-phosphate and utilization of multiple metabolites for fatty acid synthesis by plastids from developing oilseed rape embryos. Planta 199: 321–327. [Google Scholar]
- Kiba T., Naitou T., Koizumi N., Yamashino T., Sakakibara H., Mizuno T. (2005). Combinatorial microarray analysis revealing arabidopsis genes implicated in cytokinin responses through the His->Asp Phosphorelay circuitry. Plant Cell Physiol. 46: 339–355. [DOI] [PubMed] [Google Scholar]
- Knappe S., Löttgert T., Schneider A., Voll L., Flügge U.-I., Fischer K. (2003). Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis--AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J. 36: 411–420. [DOI] [PubMed] [Google Scholar]
- Lee D.J., Zeevaart J.A.D. (2007). Regulation of gibberellin 20-oxidase1 expression in spinach by photoperiod. Planta 226: 35–44. [DOI] [PubMed] [Google Scholar]
- Lee E.-J., Oh M., Hwang J.-U., Li-Beisson Y., Nishida I., Lee Y. (2017). Seed-specific overexpression of the pyruvate transporter BASS2 increases oil content in Arabidopsis seeds. Front. Plant Sci. 8: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Zhang H., Wang H., Xia Y. (2014). Identification of redox-sensitive cysteines in the Arabidopsis proteome using OxiTRAQ, a quantitative redox proteomics method. Proteomics 14: 750–762. [DOI] [PubMed] [Google Scholar]
- Lonien J., Schwender J. (2009). Analysis of metabolic flux phenotypes for two Arabidopsis mutants with severe impairment in seed storage lipid synthesis. Plant Physiol. 151: 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome H., Yamaguchi S., Hanada A., Kamiya Y., Oda K. (2004). dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 37: 720–729. [DOI] [PubMed] [Google Scholar]
- Meng L., Feldman L. (2010). A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnol. J. 5: 183–186. [DOI] [PubMed] [Google Scholar]
- Mitchum M.G., Yamaguchi S., Hanada A., Kuwahara A., Yoshioka Y., Kato T., Tabata S., Kamiya Y., Sun T.P. (2006). Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 45: 804–818. [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Matsumoto-Kitano M., Kakimoto T. (2004). Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37: 128–138. [DOI] [PubMed] [Google Scholar]
- Mustroph A., Sonnewald U., Biemelt S. (2007). Characterisation of the ATP-dependent phosphofructokinase gene family from Arabidopsis thaliana. FEBS Lett. 581: 2401–2410. [DOI] [PubMed] [Google Scholar]
- Mutasa-Göttgens E., Hedden P. (2009). Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 60: 1979–1989. [DOI] [PubMed] [Google Scholar]
- Née G., Zaffagnini M., Trost P., Issakidis-Bourguet E. (2009). Redox regulation of chloroplastic glucose-6-phosphate dehydrogenase: a new role for f-type thioredoxin. FEBS Lett. 583: 2827–2832. [DOI] [PubMed] [Google Scholar]
- Pokhilko A., Bou-Torrent J., Pulido P., Rodríguez-Concepción M., Ebenhöh O. (2015). Mathematical modelling of the diurnal regulation of the MEP pathway in Arabidopsis. New Phytol. 206: 1075–1085. [DOI] [PubMed] [Google Scholar]
- Prabhakar V., et al. (2010). Phosphoenolpyruvate provision to plastids is essential for gametophyte and sporophyte development in Arabidopsis thaliana. Plant Cell 22: 2594–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P., Perello C., Rodríguez-Concepción M. (2012). New insights into plant isoprenoid metabolism. Mol. Plant 5: 964–967. [DOI] [PubMed] [Google Scholar]
- Reiser J., Linka N., Lemke L., Jeblick W., Neuhaus H.E. (2004). Molecular physiological analysis of the two plastidic ATP/ADP transporters from Arabidopsis. Plant Physiol. 136: 3524–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riefler M., Novak O., Strnad M., Schmülling T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I., Eriksson S., Powers S.J., Gong F., Griffiths J., Woolley L., Benlloch R., Nilsson O., Thomas S.G., Hedden P., Phillips A.L. (2008a). Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20: 2420–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I., Ruiz-Rivero O., Fernández-García N., Griffiths J., Powers S.J., Gong F., Linhartova T., Eriksson S., Nilsson O., Thomas S.G., Phillips A.L., Hedden P. (2008b). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53: 488–504. [DOI] [PubMed] [Google Scholar]
- Rittenberg D., Foster G.L. (1940). A new procedure for quantitative analysis by isotope dilution, with application to the determination of amino acids and fatty acids. J. Biol. Chem. 133: 737–744. [Google Scholar]
- Ruuska S.A., Girke T., Benning C., Ohlrogge J.B. (2002). Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-López Á.M., et al. (2016). Arabidopsis responds to Alternaria alternata volatiles by triggering plastid phosphoglucose isomerase-independent mechanisms. Plant Physiol. 172: 1989–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J., Ohlrogge J.B., Shachar-Hill Y. (2003). A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J. Biol. Chem. 278: 29442–29453. [DOI] [PubMed] [Google Scholar]
- Schwender J., Goffman F., Ohlrogge J.B., Shachar-Hill Y. (2004). Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432: 779–782. [DOI] [PubMed] [Google Scholar]
- Silverstone A.L., Chang C., Krol E., Sun T.P. (1997). Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 12: 9–19. [DOI] [PubMed] [Google Scholar]
- Spíchal L. (2012). Cytokinins-recent news and views of evolutionally old molecules. Funct. Plant Biol. 39: 267–284. [DOI] [PubMed] [Google Scholar]
- Tsai H.L., Lue W.L., Lu K.J., Hsieh M.H., Wang S.M., Chen J. (2009). Starch synthesis in Arabidopsis is achieved by spatial cotranscription of core starch metabolism genes. Plant Physiol. 151: 1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanová T., Tarkowská D., Novák O., Hedden P., Strnad M. (2013). Analysis of gibberellins as free acids by ultra performance liquid chromatography-tandem mass spectrometry. Talanta 112: 85–94. [DOI] [PubMed] [Google Scholar]
- Van Daele I., Gonzalez N., Vercauteren I., de Smet L., Inzé D., Roldán-Ruiz I., Vuylsteke M. (2012). A comparative study of seed yield parameters in Arabidopsis thaliana mutants and transgenics. Plant Biotechnol. J. 10: 488–500. [DOI] [PubMed] [Google Scholar]
- Ventriglia T., Kuhn M.L., Ruiz M.T., Ribeiro-Pedro M., Valverde F., Ballicora M.A., Preiss J., Romero J.M. (2008). Two Arabidopsis ADP-glucose pyrophosphorylase large subunits (APL1 and APL2) are catalytic. Plant Physiol. 148: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Pinto Y.M., Baulcombe D.C. (1999). Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96: 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.A., Todd J., Newman T., Focks N., Girke T., de Ilárduya O.M., Jaworski J.G., Ohlrogge J.B., Benning C. (2000). A new set of Arabidopsis expressed sequence tags from developing seeds. The metabolic pathway from carbohydrates to seed oil. Plant Physiol. 124: 1582–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N., Winter C.M., Wu M.-F., Kanno Y., Yamaguchi A., Seo M., Wagner D. (2014). Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science 344: 638–641. [DOI] [PubMed] [Google Scholar]
- Yin Z., Balmant K., Geng S., Zhu N., Zhang T., Dufresne C., Dai S., Chen S. (2017). Bicarbonate induced redox proteome changes in Arabidopsis suspension cells. Front. Plant Sci. 8: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T.S., Lue W.L., Wang S.M., Chen J. (2000). Mutation of Arabidopsis plastid phosphoglucose isomerase affects leaf starch synthesis and floral initiation. Plant Physiol. 123: 319–326. [DOI] [PMC free article] [PubMed] [Google Scholar]