C-lineage genes trigger nectary development in both petunia and Arabidopsis, despite their distant phylogeny, different nectary positioning, and different evolutionary trajectories.
Abstract
To attract insects, flowers produce nectar, an energy-rich substance secreted by specialized organs called nectaries. For Arabidopsis thaliana, a rosid species with stamen-associated nectaries, the floral B-, C-, and E-functions were proposed to redundantly regulate nectary development. Here, we investigated the molecular basis of carpel-associated nectary development in the asterid species petunia (Petunia hybrida). We show that its euAGAMOUS (euAG) and PLENA (PLE) C-lineage MADS box proteins are essential for nectary development, while their overexpression is sufficient to induce ectopic nectaries on sepals. Furthermore, we demonstrate that Arabidopsis nectary development also fully depends on euAG/PLE C-lineage genes. In turn, we show that petunia nectary development depends on two homologs of CRABS CLAW (CRC), a gene previously shown to be required for Arabidopsis nectary development, and demonstrate that CRC expression in both species depends on the members of both euAG/PLE C-sublineages. Therefore, petunia and Arabidopsis employ a similar molecular mechanism underlying nectary development, despite otherwise major differences in the evolutionary trajectory of their C-lineage genes, their distant phylogeny, and different nectary positioning. However, unlike in Arabidopsis, petunia nectary development is position independent within the flower. Finally, we show that the TARGET OF EAT-type BLIND ENHANCER and APETALA2-type REPRESSOR OF B-FUNCTION genes act as major regulators of nectary size.
INTRODUCTION
Despite their small size and often hidden existence inside the flower, nectaries play a major economic and ecological role in mediating insect pollination. The total economic value of insect pollination services worldwide was estimated to represent a staggering €153 billion in 2005 (Gallai et al., 2009). Unlike the highly conserved order of the major floral organs within the flower, the nectaries can be found in any position along the receptacle or in association with any of the floral organs, and more rarely on vegetative organs (Brown, 1938; Bernardello, 2007). While research in a range of different model species has greatly contributed to our understanding of the molecular mechanisms underlying the development of the major floral organs (Schwarz-Sommer et al., 1990; Coen and Meyerowitz, 1991; Krizek and Fletcher, 2005), the genetics underlying nectary development and its diversity remains poorly understood. The molecular basis of nectary development only has been studied in detail in Arabidopsis thaliana, the nectaries of which are positioned on the flower receptacle, near the base of the stamen filaments (Smyth et al., 1990). The YABBY transcription factor gene CRABS CLAW (CRC) so far is the only described example of a single gene that is essential (but not sufficient) for nectary development in Arabidopsis (Bowman and Smyth, 1999). In addition, it was shown that CRC expression is conserved in nectaries from several core eudicot species, including members of both rosids and asterids, the two major phylogenetic lineages of eudicots (Lee et al., 2005a; Fourquin et al., 2014). Furthermore, the result of a heterologous virus-induced gene silencing (VIGS) experiment suggested conservation of CRC function in Nicotiana benthamiana nectary development. In contrast, no evidence of CRC expression in nectaries of basal eudicot species was found (Lee et al., 2005a; Sun et al., 2013).
Because nectaries still develop in floral A-, B-, and C-class homeotic mutants, it was initially suggested that the Arabidopsis nectary is an ABC-independent structure (Baum et al., 2001). Later on, it was proposed that B, C, and E (SEPALLATA) functions are redundantly required for CRC activation and nectary development because nectaries were found to be absent in higher order mutants in which several homeotic functions were compromised simultaneously (Lee et al., 2005b). However, because the architecture of these mutant flowers is highly modified, it remained to be determined if the absence of nectaries in these flowers is a direct effect. Finally, it was proposed that in the absence of B- and C-class gene activities, the SHATTERPROOF1 (SHP1) and SHP2 genes (Liljegren et al., 2000) can rescue nectary development if they are ectopically expressed, as in an A-class ap2 mutant background (Lee et al., 2005b).
Here, we aimed to use the asterid model species petunia (Petunia hybrida) (Gerats and Vandenbussche, 2005; Bombarely et al., 2016; Vandenbussche et al., 2016) as an alternative entry point to better understand the molecular control of nectary development. Different from Arabidopsis in which nectaries are positioned near the base of the stamen filaments, petunia nectaries are tightly associated with the carpels, where they play a prominent role in pollination syndromes and as such have contributed to speciation in the genus Petunia (Stuurman et al., 2004; Bombarely et al., 2016). Petunia nectaries can be easily recognized as a brightly yellow/orange tissue due to carotenoid accumulation and are positioned as a ring surrounding the base of the ovary wall, with two more prominently developed regions corresponding to the junction of the two carpels. Nectary development initiates late compared with the other floral organs and starts when meiosis already occurs in the anthers (Lee et al., 2005a), defined as stage 0 flowers by Izhaki et al. (2002). A single CRC homolog in petunia has been described and its expression is associated with the development of carpels and nectaries (Lee et al., 2005a). Initially, PhCRC is expressed in carpel primordia, ovary walls, and style and stigma. In stage 0 flowers, its expression becomes uniquely localized at the base of the carpels from which the nectaries arise and persists at high levels in the nectaries throughout further development.
Here, by analyzing loss- and gain-of-function mutants of the petunia floral homeotic C-function genes PETUNIA MADS BOX GENE3 (pMADS3) and FLORAL BINDING PROTEIN6 (FBP6) (Angenent et al., 1993; Tsuchimoto et al., 1993; Kater et al., 1998; Kapoor et al., 2002; Heijmans et al., 2012), we show that petunia nectary development fully depends on the C-function and that ectopic expression of either one of its two C-function genes is sufficient to induce ectopic nectary formation at the basis of the sepals. Similar to petunia, we found that nectary development in Arabidopsis also fully depends on the C-lineage genes, redundantly encoded by AGAMOUS (AG) and SHP1/2. Furthermore, we show that CRC expression in both species fully depends on the C-lineage genes. Mutant phenotypes indicate that in petunia, as in Arabidopsis, nectary development also depends on CRC function, with the difference that the petunia CRC function is redundantly encoded by two closely related CRC genes. This indicates that petunia and Arabidopsis employ a strikingly similar molecular mechanism underlying nectary development, despite major differences in the evolutionary trajectory of their C-lineage genes, their distant phylogeny, and different nectary positioning. Finally, we show that the petunia BEN (BLIND ENHANCER) and ROB (REPRESSOR OF B-FUNCTION) genes (Morel et al., 2017), encoding members of TOE- and AP2-type AP2 transcription factors, respectively, negatively regulate the size of the nectaries by preventing their development in the apical region of the ovary. These data further expand our knowledge of the molecular mechanisms controlling nectary development in different species and offer further insight in the evolutionary trajectory of the euAP2 and MADS box transcription factor families.
RESULTS
Nectary Morphology in Wild-Type Petunia Laboratory Lines
While nectary morphology was described in detail in a commercial petunia variety (Lee et al., 2005a), we used two different petunia strains in this study, consisting of the highly active dTph1 transposon line W138 and the easily transformable W115 (Mitchell) variety (Figure 1). We found that nectary morphology in these lines and in individuals derived from crosses between these lines was similar, with the nectary positioned as a ring surrounding the base of the ovary wall, with two more prominently developed parts corresponding to the carpel junction region (Figures 1A and 1B; Supplemental Figure 1). At the cellular level, this nectariferous tissue is characterized by relatively inconspicuous cells, interspersed with modified stomata (Figures 1C and 1D), which eventually start excreting nectar.
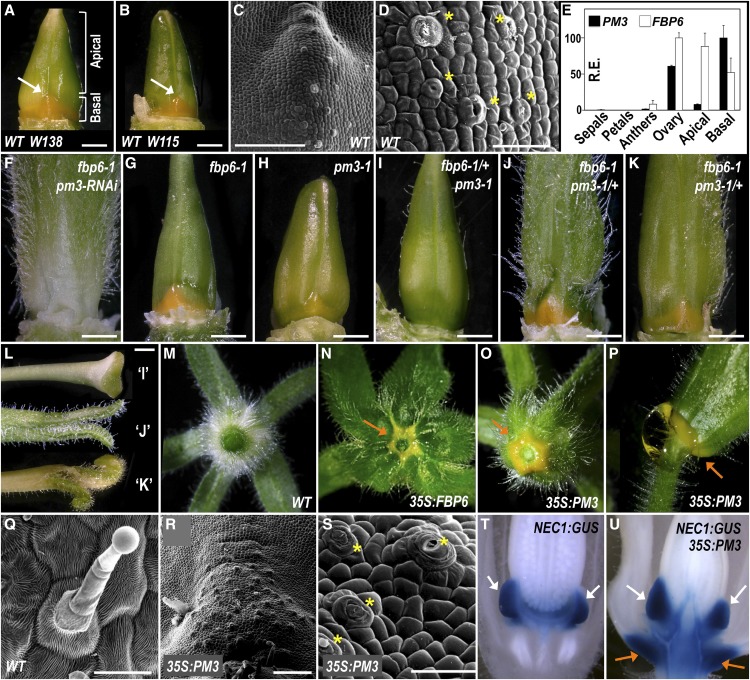
Figure 1.
Petunia Nectary Development Depends on C-Lineage Genes.
White and orange arrows indicate wild-type and ectopic nectaries, respectively. Yellow asterisks mark modified stomata for nectary secretion present on the nectary epidermis.
(A) to (D) Wild-type nectary development.
(A) and (B) Ovary with nectary at its base.
(C) Cryo-scanning electron microscopy image showing nectary at ovary base.
(D) Further magnification of (B).
(E) Expression of the C-class genes pMADS3 (PM3) and FBP6 in different floral organs and tissues just before anthesis. Bars represent the average relative expression level of three biological replicates ± se as determined by RT-qPCR analysis using ACTIN and RAN as references genes. The highest relative expression level for each gene was set to 100, and all other values were normalized to this value. Ovary: complete ovary including the nectaries. Apical: Apical part of the ovary, excluding the nectary. Basal: basal-most part of the ovary enriched in nectary tissue (as indicated in [A]). R.E., relative expression.
(F) to (K) Nectary development in C-class mutants.
(J) fbp6-1 pm3-1/+ flower with strong carpel phenotype.
(K) fbp6-1 pm3-1/+ flower with intermediate carpel phenotype.
(L) Style and stigma phenotypes corresponding to ovaries shown in (I) to (K).
(M) to (U) Ectopic nectary development in C-class gain-of-function lines compared with the wild type.
(M) to (O) Bottom views of first whorl.
(P) Big droplets of nectar are produced by the ectopic nectaries.
(Q) to (S) Cryo-scanning electron microscopy images of the abaxial surface of first-whorl organs near pedicel, showing nectary cell types in 35S:PM3 flowers ([R]and [S]), compared with the wild type (Q).
(T) NEC1:GUS reporter activity in wild-type nectaries.
(U) NEC1:GUS reporter activity in a 35S:PM3 flower showing intense staining in normal and ectopic nectaries.
Bars = 1 mm in (A), (B), and (F) to (L), 200 µm in (B) and (R), and 50 µm in (C), (Q), and (S).
The Petunia C-Function Genes Are Required for Nectary Development
Since petunia nectaries are associated with the carpels, we hypothesized that the floral homeotic C-function, which is required for carpel and stamen identity, may also play a role in their development. Moreover, it was shown by in situ hybridization that the C-function gene pMADS3 is expressed in the nectary tissue (Kater et al., 1998). More recently, we found that the petunia C-function is encoded in a largely redundant fashion by the MADS box genes pMADS3 and FBP6, although FBP6 plays a unique role in the development of the style and stigma (Heijmans et al., 2012). To analyze their expression pattern in a quantitative manner and their possible involvement in nectary development, we compared pMADS3 and FBP6 expression levels in the basal part of the ovary that is highly enriched in nectary tissue versus the apical part of the ovary (Figure 1A) and with the other floral organs. We found that pMADS3 expression is highly enriched in the basal part compared with other floral tissues, while the mRNA level of FBP6 in the basal part is roughly similar to its expression in the remaining part of the ovary (Figure 1E). Because the two genes are well expressed in the basal part of the ovary, this suggests that both pMADS3 and FBP6 potentially may play a role in nectary development.
To further investigate this, we examined the effect of loss of function of pMADS3 and FBP6, using RNA interference (RNAi) and mutant lines, on nectary development. The fbp6-1 mutants carry a putative null fbp6 allele caused by a dTph1 insertion disrupting the first exon (Heijmans et al., 2012). In the earlier obtained pMADS3-RNAi fbp6-1 plants (Heijmans et al., 2012), we found that nectary development is completely lost (Figure 1F), suggesting that petunia nectary development depends on the C-function. However, in these flowers, the carpels are completely replaced by sepal-like organs forming the first whorl of a new flower developing in the center (Heijmans et al., 2012). Therefore, the loss of nectaries in these lines could also be an indirect effect caused by the complete loss of carpel identity. To exclude this possibility, we aimed to analyze nectary development in genetic backgrounds in which the C-function is only partially impaired, using single fbp6 and pmads3 mutants and combinations thereof. In addition, due to the RNAi approach used to silence pMADS3 expression, it could not be completely excluded that pMADS3-RNAi fbp6-1 plants lack nectaries due to off-target effects. Therefore, we generated and analyzed CRISPR-Cas9 derived null alleles of pMADS3 (Supplemental Figures 1A and 1B) as a more accurate replacement of the previously described pMADS3-RNAi knockdown line. The obtained pmads3-1 and pmads3-2 alleles contain an 8-bp deletion and 1-bp insertion, respectively, in the coding sequence of the first exon of pMADS3, encoding the MADS box DNA binding domain.
In the fbp6-1 mutants, we found that nectary development is not impaired (Figure 1G), indicating either that FBP6 is not involved in nectary development or that nectary development can be rescued by another factor in the fbp6 mutant background. Likewise, flowers of the pmads3-1 and pmads3-2 putative null mutants still develop nectaries (Figure 1H; Supplemental Figure 1D), although these systematically appeared less pigmented and are reduced in size compared with wild-type plants (Supplemental Figure 1C). Strikingly, introducing the fbp6-1 allele in a heterozygous state in the pmads3 mutant background resulted in the complete absence of nectary development (Figures 1I; Supplemental Figure 1D), despite that outer carpel wall identity, and style and stigma development was still largely intact in these lines (Figures 1L; Supplemental Figure 1D). By contrast, fbp6-1 pmads3-1/+ and fbp6-1 pmads3-2/+ flowers exhibited severe carpel identity defects, ranging from fourth-whorl organs that closely resembled sepals to less severely converted fourth-whorl organs (Figures 1J to 1L; Supplemental Figure 1D). Yet, irrespective of the severity of their carpel phenotype, all these flowers exhibited well-developed nectaries (Figures 1J and 1K; Supplemental Figure 1D). Together, this indicates that nectary development and carpel identity can be fully uncoupled and that nectary development in petunia directly depends on the C-lineage genes. In addition, the different phenotypes suggest that pMADS3 may play a more important role in nectary development compared with FBP6.
Ectopic Nectary Formation in 35S:pMADS3 and 35S:FBP6 Plants
To test whether pMADS3 and/or FBP6 is also sufficient for nectary development, we next investigated nectary development in flowers of 35S:pMADS3 and 35S:FBP6 plants. Although 35S:pMADS3 or 35S:FBP6 overexpression lines exhibit flowers with antheroid petals as expected for ectopic C-function expression in the perianth whorls, they do not develop first whorl carpels (Heijmans et al., 2012). Yet, we found that in both overexpression lines, ectopic nectary tissue develops as a ring around the pedicel, at the base of the sepals (Figures 1M to 1P). At the microscopic level, cells in this area closely resemble wild-type nectary tissue instead of wild-type sepaloid cells (Figures 1C, 1D, and 1Q to 1S). These ectopic nectaries are fully functional, since big droplets of nectar can often be observed at the base of the sepals when flowers mature (Figure 1P). Moreover, we found that the pNEC1:GUS reporter, a marker for nectaries (Ge et al., 2000) (Figure 1T), is strongly activated in these ectopic nectaries (Figure 1U), further confirming their nectary identity. Together with the loss-of-function data, this demonstrates that nectary development is redundantly regulated by both pMADS3 and FBP6 and that the potential for nectary development in petunia is not limited to the fourth floral whorl.
The Arabidopsis C-lineage Genes AG, SHP1, and SHP2 Are Redundantly Required for Nectary Development
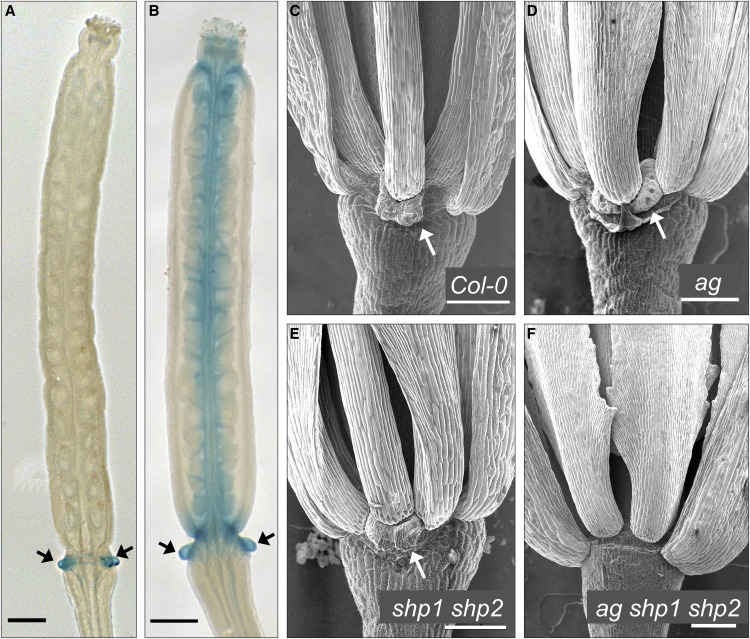
In petunia, we showed here that nectary development is redundantly regulated by pMADS3 and FBP6, which belong to the euAG and PLE C-sublineages (Kramer et al., 2004), respectively. This identified another common function for pMADS3 and FBP6, in addition to their broadly overlapping role in encoding the floral homeotic C-function (Heijmans et al., 2012). Because the current Arabidopsis model for nectary development is not clear, we decided to test if members of both the euAG and PLE C-sublineages might simply redundantly encode nectary development in Arabidopsis, as in petunia. In contrast to petunia, the euAG lineage gene AG by itself is fully required for the homeotic C-function. As such, ag mutants form flowers consisting of reiterations of sepals, petals, and petals (Yanofsky et al., 1990), but nectaries still develop at the base of the homeotically transformed third-whorl petals (Bowman and Smyth, 1999; Baum et al., 2001). This contributed to the initial idea that nectary identity in Arabidopsis would be independent of the ABC floral organ identity functions (Baum et al., 2001). Furthermore, the Arabidopsis PLE sublineage genes SHP1 and SHP2 were shown to play a specialized role in fruit dehiscence, while still retaining some common functions with AG in the development of carpels and ovules (Liljegren et al., 2000; Pinyopich et al., 2003; Colombo et al., 2010). Interestingly, a transcriptome analysis of Arabidopsis nectaries (Kram et al., 2009) showed that SHP1, SHP2, and AG all exhibit a strong nectary enriched expression profile. Moreover, GUS staining in the nectaries was shown for pSHP2:GUS (Dinneny et al., 2005) and mentioned for pAG:GUS and pSHP1:GUS transgenic lines (Sieburth and Meyerowitz, 1997; Baum et al., 2001). Confirming these reports, we observed an intense staining in the developing nectaries of pAG:GUS and pSHP1:GUS lines that we also generated in our lab (Figure 2). Their overlapping expression pattern in nectaries (Figures 2A and 2B) supported our hypothesis that AG and SHPs might have retained a common function in nectary development similar to the AG and PLE C-lineage genes in petunia. To test this genetically, we compared the phenotypes of ag, shp1 shp2, and ag shp1 shp2 mutants. We found that nectaries still develop in ag and in shp1 shp2 mutants (Figures 2D and 2E) as reported earlier (Baum et al., 2001; Lee et al., 2005b), while nectary development is completely lost in ag shp1 shp2 triple mutants (Figure 2F). This demonstrates that in Arabidopsis, nectary development is redundantly controlled by the full complement of C-lineage genes, as in petunia.
Figure 2.
Arabidopsis Nectary Development Depends on C-Lineage Genes.
(A) AG:GUS.
(B) SHP1:GUS.
(C) to (F) Scanning electron microscopy images of Arabidopsis nectaries in different genetic backgrounds. Sepals and second-whorl petals have been removed.
Bars = 200 µm in (A) and (B) and 100 µm in (C) to (F). Arrows indicate nectaries.
Activation of CRC Expression Requires C-Lineage Genes in Arabidopsis and Petunia
It has been demonstrated that CRC, which is essential for nectary development in Arabidopsis, is a direct target for activation by AG (Gómez-Mena et al., 2005; ÓMaoiléidigh et al., 2013). Yet, CRC expression is still observed in an ag background, consistent with the presence of nectaries in ag (Bowman and Smyth, 1999). This suggested that other factors can activate CRC expression in the absence of AG. Because we found that SHP1, SHP2 and AG redundantly are required for nectary development, SHP1 and SHP2 were logical candidates for also being capable of activating CRC expression. To test this, we quantified CRC expression in different mutant backgrounds by reverse transcription quantitative PCR (RT-qPCR) analysis (Figure 3). We found that ag flowers show a ∼3-fold decrease in CRC expression, consistent with earlier results (ÓMaoiléidigh et al., 2013), while the expression level in shp1 shp2 flowers was similar to that of wild-type flowers. Finally, ag shp1 shp2 flowers showed an almost complete loss of CRC expression, suggesting that CRC expression is redundantly activated by all C-lineage members (Figure 3A).
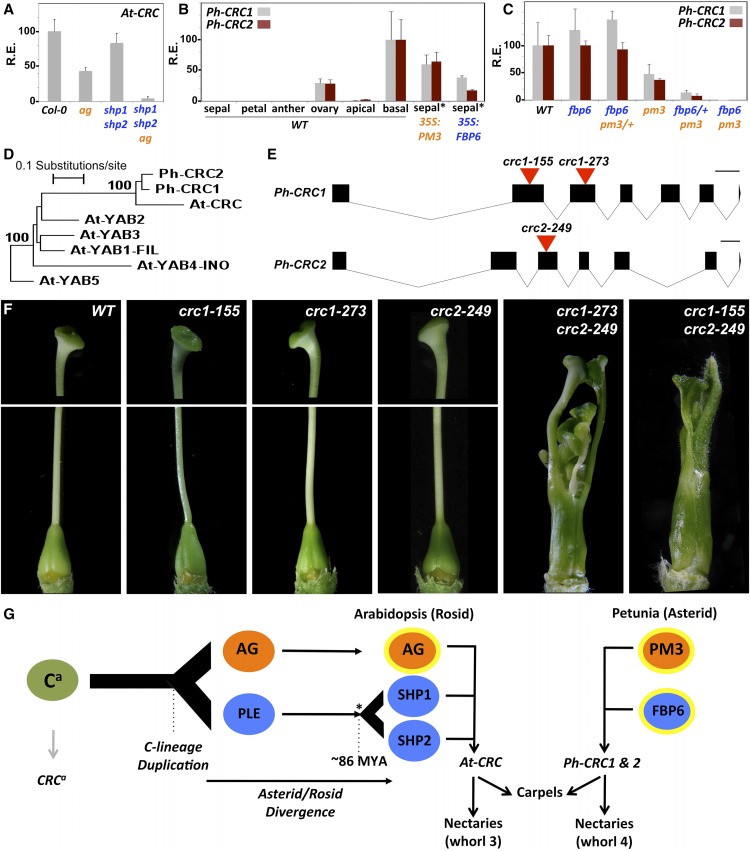
Figure 3.
C-Lineage-Dependent CRC Expression and Nectary Development in Arabidopsis and Petunia.
(A) Arabidopsis CRC RT-qPCR expression analysis in wild-type and mutant flowers. Values are relative to wild-type levels set to 100.
(B) Petunia Ph-CRC1and2 RT-qPCR expression analysis in different floral organs and tissues just before anthesis. Ovary: complete ovary including the nectaries. Apical: apical part of the ovary, excluding nectaries. Basal: basal-most part of the ovary enriched in nectary tissue (as indicated in Figure 1A). Sepal*: basal part of the sepals enriched in ectopic nectaries. The highest relative expression level for each gene was set to 100, and all other values were normalized to this value.
(C) Expression in the basal part of the fourth whorl in wild-type and C-class loss-of-function lines just before anthesis. Values are relative to wild-type levels set to 100.
Bars in (A) to (C) represent the average relative expression level of three biological replicates ± se as determined by RT-qPCR analysis using ACT8 in (A) and ACTIN and RAN in (B) and (C) as references genes. R.E., relative expression.
(D) Neighbor-joining tree of all YABBY proteins from Arabidopsis (prefix At-) and the two petunia CRC homologs (prefix Ph-) based on the alignment shown in Supplemental Figure 2B. One thousand bootstrap samples were generated to assess support for the inferred relationships. Only bootstrap percentages above 75% are shown. The tree was rooted with Arabidopsis YAB5.
(E) Genomic structure of petunia CRC1 and CRC2 genes and position of the dTph1 transposon insertions (red triangles). Black rectangles, exons; lines, introns. Bars = 100 bp.
(F) Phenotypes of crc1, crc2, and crc1 crc2 mutants compared with the wild type. All images are at the same magnification. Styles for wild type and crc single mutants are only partially shown.
(G) Evolutionary history of the C-lineage and CRC activation in Arabidopsis and petunia. Black and gray arrows represent confirmed and presumed interactions respectively. Yellow circles around C-lineage genes indicate classical C-function activity. *, Corresponds to the most recent Arabidopsis genome duplication, resulting in the duplication of the SHP gene, and of AG, but of which the second copy most likely has been lost (Causier et al., 2005). Ca and CRCa represent ancestral C-function and CRC genes. PM3, pMADS3.
To further investigate this apparently conserved link of CRC expression with both the euAG and PLE C-lineage genes in Arabidopsis, we analyzed the dependence of CRC expression on pMADS3 and FBP6 functions in petunia. Earlier, a single CRC homolog in petunia has been reported (named PhCRC) that is expressed in developing carpels and nectaries, similar to Arabidopsis CRC expression (Lee et al., 2005a). However, a transcriptome database (Zenoni et al., 2011) and the recently released petunia genome sequence (Bombarely et al., 2016) revealed that petunia contains two closely related CRC homologs, PhCRC1 (formerly PhCRC) and PhCRC2 (Figure 3D; Supplemental Figure 2). As expected based on the previously published in situ hybridization experiment (Lee et al., 2005a), we found that PhCRC1 expression was strongly enriched in the basal part of the ovary, compared with the apical part, and to the other floral organs (Figure 3B). A quasi-identical expression profile was observed for PhCRC2, suggesting that the PhCRC1/2 genes are regulated in a similar way. Furthermore, PhCRC1and PhCRC2 expression levels were dramatically upregulated in the sepals of 35S:FBP6 and 35S:pMADS3 lines exhibiting ectopic nectaries, and only fully downregulated when both fbp6 and pmads3 were homozygous mutant (Figures 3B and 3C). This demonstrates that also in petunia, CRC expression depends redundantly on the euAG and PLE C-lineage genes and that ectopic expression of the C-lineage genes in petunia is sufficient to induce ectopic CRC expression leading to ectopic nectary development.
In Arabidopsis, it has been shown by both in vitro and in vivo assays that AG activates CRC expression through binding with a specific CArG box in the CRC promoter sequence, positioned ∼3 kb upstream of the ATG (Gómez-Mena et al., 2005; ÓMaoiléidigh et al., 2013). Interestingly, this site corresponds to one of the conserved regions previously identified by phylogenetic footprinting of the CRC promoters from Arabidopsis, Lepidium africanum, and Brassica oleracea, representing three different genera within the Brassicaceae (Lee et al., 2005b). We performed a similar analysis comparing the promoter sequences of PhCRC1, PhCRC2, and two CRC genes from N. benthamiana, covering two different subfamilies within the Solanaceae. Similar to what was observed for members of the Brassicaceae, we found that their promoter sequences are conserved in modules and that two strongly conserved CarG boxes were present in all four sequences, positioned in close proximity to each other, around 3 kb upstream of the ATG start codon (Supplemental Figure 3). Together with the PhCRC1 and PhCRC2 expression profiles in wild type and loss-of-C-function mutants, this suggests that the molecular mechanism underlying C-lineage dependent CRC activation is conserved between Arabidopsis and petunia. In strong support of that, Lee et al. (2005a) found that a pAtCRC:GUS construct introduced in tobacco (Nicotiana tabacum), a species with nectaries similar to petunia, leads to GUS expression in the nectary disc and style of the gynoecium.
PhCRC1 and PhCRC2 Are Required for Nectary and Carpel Development
To our knowledge, no stable crc mutants in species other than in Arabidopsis have been described that show defects in nectary development. However, a heterologous VIGS experiment using the full-length petunia CRC1 coding sequence in N. benthamiana resulted in the loss of nectaries, while in petunia, this resulted in a loss of flower meristem determinacy, but with little visible effect on nectary development (Lee et al., 2005a). Therefore, to provide solid genetic proof for the function of the petunia CRC genes and their putative requirement for nectary development, we analyzed the effect of dTph1 transposon insertions in the PhCRC1 and PhCRC2 genes. By screening our sequence indexed dTph1 flanking sequence collection (Vandenbussche et al., 2008), we identified two independent insertions in PhCRC1 positioned in the middle of the second and third exons, at 155 and 273 bp downstream of the ATG in the coding sequence, and one insertion in the 3rd exon of PhCRC2, at 249 bp downstream of the ATG in the coding sequence (Figure 3E). Because all three insertions are located in the coding sequence, and dTph1 encodes stop codons in all six possible reading frames, these insertions are likely to disrupt gene function. However, single homozygous mutants were not visibly different from the wild type (Figure 3F). Together with their overall high sequence similarity of 90% (Supplemental Figure 2A), this suggested genetic redundancy. To test this, we created and analyzed homozygous double mutants and found that they had fully lost their nectaries (Figure 3F). These results show that CRC function in petunia is essential for nectary development, as in Arabidopsis. In addition, the internal placenta structure in the ovary of crc1 crc2 flowers was fully replaced by a variety of floral organs, including petal-, stamen-, and carpel-like organs, indicative of a loss of meristem determinacy (Supplemental Figure 2C).
BEN and ROB Genes Restrict the Size of the Nectary Domain in the Fourth Whorl
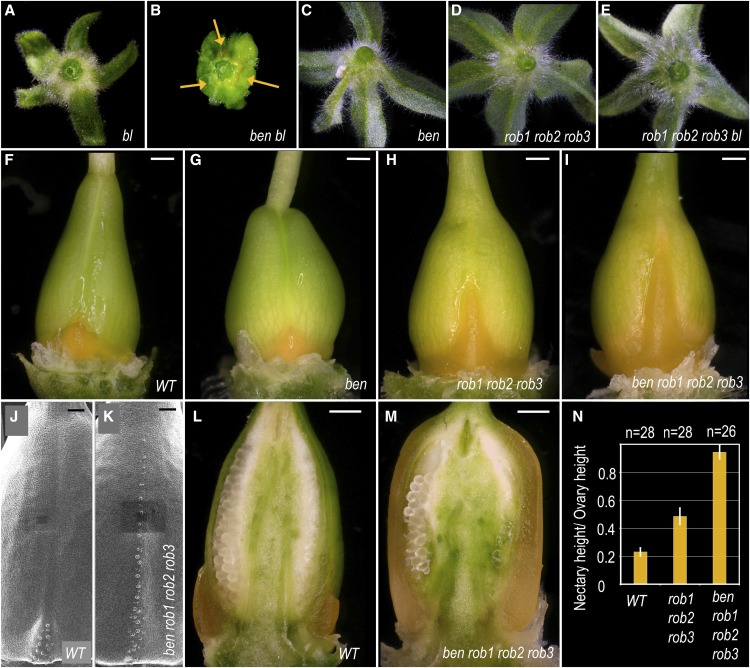
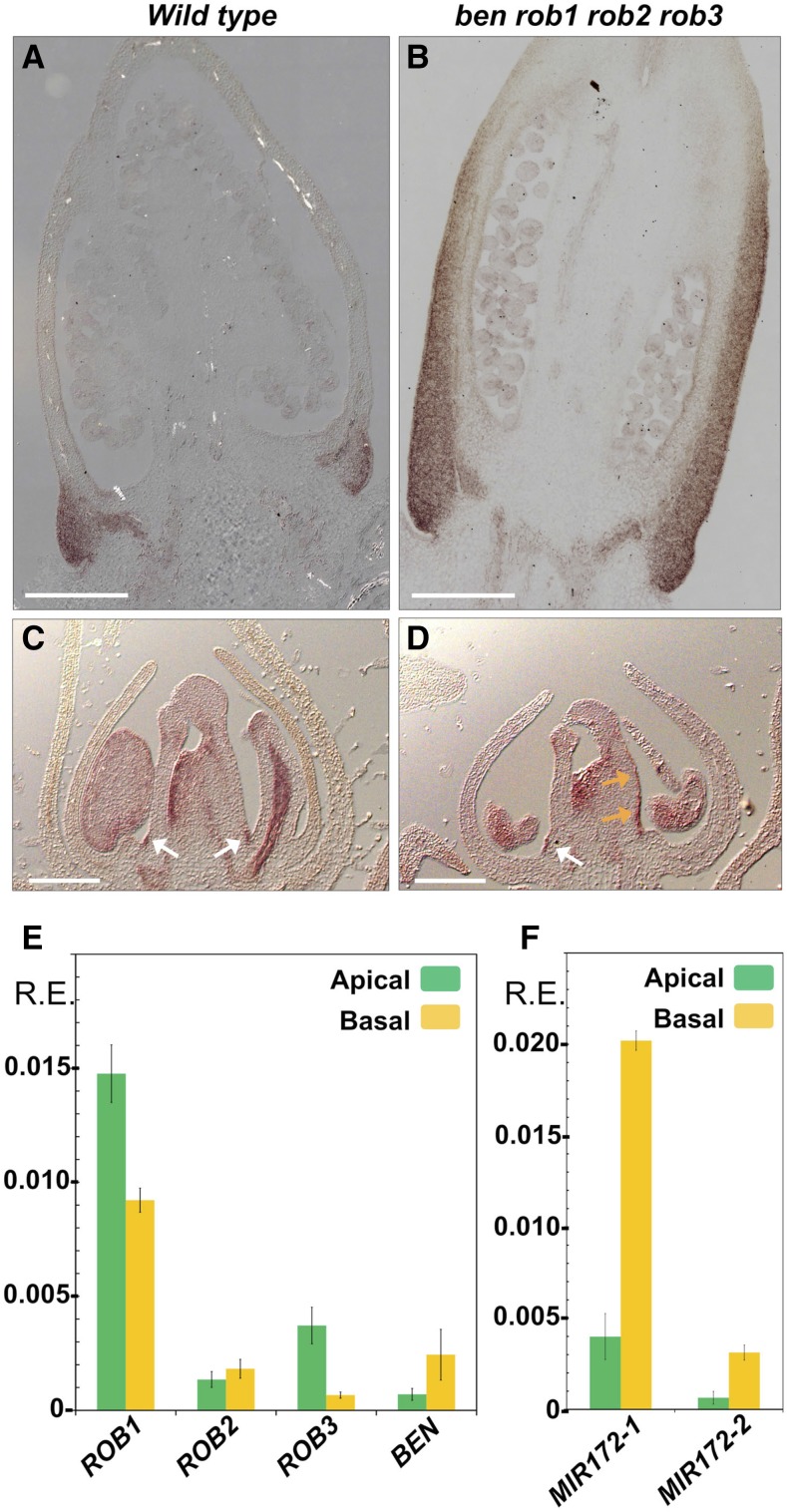
During previous research, we identified the euAP2 TOE-type transcription factor BEN as a negative regulator of the C-function in the first floral whorl, acting in parallel with the microRNA BL (Morel et al., 2017). While bl flowers are otherwise similar to 35S:pMADS3 and 35S:FBP6 flowers, the development of ectopic nectaries at the base of the 35S:pMADS3 and 35S:FBP6 sepals does not occur in bl mutants (Figure 4), although transcripts of both C-class genes are also ectopically expressed in the first whorl of bl mutants (Tsuchimoto et al., 1993; Kater et al., 1998; Rijpkema et al., 2006; Cartolano et al., 2007; Morel et al., 2017). This suggested that C-expression levels in bl sepals might not be sufficient to provoke nectary development, in contrast to the transgenic C-gene overexpression lines. To further analyze this, we looked at ben bl double mutants, which show an ∼4-fold further increase of C-gene expression levels in the first whorl compared with bl mutants (Morel et al., 2017). In agreement with our hypothesis that ectopic nectary formation requires relatively high levels of ectopic C-activity, we observed indeed that most ben bl flowers develop nectarifous tissue on the abaxial side of first-whorl organs, which are homeotically converted into carpelloid organs (Figure 4B). Note that these ectopic nectaries vary in size between flowers on the same plant and between different plants and were too small to be observed by eye (or absent) in eight of 33 flowers examined (sampled from five different plants). As expected, ben single mutants did not develop first whorl ectopic nectaries (Figure 4C), since the C-genes are not ectopically expressed in the perianth of ben mutants (Morel et al., 2017). Furthermore, we looked also at fourth-whorl nectary development in ben single mutants, but found no obvious difference with the wild type (Figures 4F and 4G). This indicates that the negative effect of BEN on nectary formation in the first whorl is either not relevant for fourth whorl nectary formation, or alternatively, that its function in the fourth whorl is masked in redundancy.
Figure 4.
BEN and ROB Genes Negatively Regulate the Size of the Nectary Domain in the Fourth Whorl.
(A) to (E) Abaxial side of first-whorl organs in various genotypes. Orange arrows indicate ectopic nectaries.
(F) to (I) Ovaries of various genotypes showing dramatic differences in nectary size (orange tissue).
(J) to (K) Scanning electron microscopy images of young ovaries, showing nectary stomata developing along the entire ovary wall in ben rob1 rob2 rob3 mutants, while restricted to the base in the wild type.
(L) and (M) Longitudinal section of wild-type and ben rob1 rob2 rob3 ovaries All bars = 0.5 mm except in (J) and (K) = 100 µm.
(N) Quantification of nectary size in different genotypes. Height of the bars represents the average ratio between nectary and ovary height ± standard variation. n = number of ovaries analyzed.
In the same study, in which we identified BEN as a repressor of the C-function, we also analyzed the function of the petunia AP2-type genes ROB1, ROB2, and ROB3, the closest homologs of Arabidopsis AP2. In contrast to BEN, and in contrast to AP2 function in Arabidopsis, there is no evidence that the ROB genes repress the C-function in the perianth (Morel et al., 2017). Indeed, rob1 rob2 rob3 mutants do not display ectopic C-function activity in the perianth, and unlike ben bl mutants, rob1 rob2 rob3 bl first-whorl organs do not become carpelloid. In line with that, we did not observe first whorl nectaries in these mutants (Figures 4D and 4E). Unexpectedly, we did find a major effect on nectary development in the fourth whorl: Nectaries in all rob1 rob2 rob3 flowers examined are considerable larger than found in the wild type (Figures 4F and 4H). In particular, the nectary domain in rob1 rob2 rob3 flowers extends further toward the apical part of the ovary, in the region where the two carpels fuse. We quantified this by determining the ratio between the height of the nectary and the total length of the ovary and found that rob1 rob2 rob3 nectaries are on average around twice as long compared with the wild type (Figure 4N). Note that rob single mutants did not exhibit this phenotype, while intermediate phenotypes could be observed in rob double mutants (Supplemental Figures 4A to 4D). Interestingly, the phenotype found in rob1 rob2 rob3 ovaries was strongly enhanced in ben rob1 rob2 rob3 quadruple mutant flowers, in which these nectary domains extended along the entire carpel fusion region of the ovary (Figures 4I and 4L to 4N). This local carpel-to-nectary conversion was already visible at relatively early developmental stages, since modified stomata could already be observed on the carpel fusion region before nectary tissue became clearly noticeable (Figures 4J and 4K). Thus, ROB and BEN genes act in a redundant but additive manner to restrict nectary development to the base of the ovary in the wild type by preventing its development in the apical regions of the ovary. Finally, we report that although BL plays a major role in repressing the C-function in the perianth in parallel with BEN, loss of BL does not seem to influence size extension of the fourth whorl nectaries along the vertical axis, since bl and ben bl nectaries are similar to the wild type, and bl rob1 rob2 rob3 nectaries comparable to rob1 rob2 rob3 nectaries. Likewise, fourth-whorl nectaries in flowers of 35S:pMADS3 plants do not show a longitudinal extension (Supplemental Figures 4E to 4H).
Molecular Analysis of the ben rob1 rob2 rob3 Fourth-Whorl Nectary Phenotype
In this manuscript, we showed that petunia nectary development depends on the C-function genes, and we demonstrated earlier that BEN negatively regulates the C-function in the first whorl, in parallel with BL (Morel et al., 2017). Furthermore, the presence of ectopic nectaries in the first whorl of 35S:PMADS3, 35S:FBP6, and ben bl flowers but not in bl mutants (Figures 1 and 4) suggest that sufficiently high levels of C-gene expression are needed to locally induce nectary development. Therefore, to explain the ben rob1 rob2 rob3 phenotype, we hypothesized that BEN and ROB genes negatively regulate nectary development in the apical part of the ovary by locally reducing C-gene expression levels under the threshold required to trigger nectary development. While in wild-type flowers FBP6 is expressed at significant levels in both apical and basal parts of the ovary, pMADS3 clearly showed much higher expression levels in the basal part of the ovary, compared with the apical part (Figure 1E). For that reason, we focused on pMADS3 to compare its expression pattern in wild-type and ben rob1 rob2 rob3 ovaries by means of in situ hybridization (Figure 5). During late wild-type ovary development (ovary diameter ∼1.5 mm), in which the nectary tissues are clearly recognizable at the base of the ovary, we found that pMADS3 expression is highly upregulated in these regions compared with the rest of the carpel tissues (Figure 5A), in agreement with earlier in situ expression data (Kater et al., 1998) and with the result of our RT-qPCR experiment (Figure 1E). By contrast, in ben rob1 rob2 rob3 flowers at a similar developmental stage, strong pMADS3 expression extends apically along the ovary wall corresponding to the ectopic nectary tissue (Figure 5B). Earlier in wild-type ovary development, when the nectaries are not yet clearly visible, pMADS3 expression can be observed in two clear foci (white arrows) at the base of the carpels, while the remaining part of the carpels show a weaker and more uniform expression (Figure 5C). In addition, strong pMADS3 expression is also observed in the placental tissues inside the ovary. In ben rob1 rob2 rob3 ovaries of a similar developmental stage, we observed sections in which pMADS3 expression extends apically along the outer carpel wall (orange arrows), while expression in other tissues was comparable to the wild type (Figure 5D). Together, these results provide support for the hypothesis that BEN and ROB genes restrict nectary development to the basis of the ovary by negatively regulating the expression levels of pMADS3 in the apical region of the ovary where the two carpels fuse. Moreover, this suggests low BEN/ROB activity at the basis of the ovary to allow high C-gene expression and associated nectary development, compared with high BEN/ROB activity in the apical region.
Figure 5.
Molecular Analysis of ben rob1 rob2 rob3 Mutants.
(A) to (D) pMADS3 in situ hybridization in the wild type and rob1 rob2 rob3 ben mutants. Ovaries ([A] and [B]) and entire flower buds ([C] and [D]). Bars = 0.5 mm in (A) and (B) and 0.25 mm in (C) and (D). White arrows in (C) indicate pMADS3 expression at the basis of the carpels where nectaries initiate. Orange arrows in (D) indicate upregulation of pMADS3 expression in the apical region of the ovary.
(E) RT-qPCR expression analysis of ROB1, ROB2, ROB3, and BEN genes.
(F) Stem-loop RT-qPCR expression analysis of mature MIR172-1 and MIR172-2 microRNAs. Bars in (E) and (F) represent the average relative expression level of three biological replicates and three technical replicates ± se as determined by RT-qPCR analysis using ACTIN, GAPDH, and RAN as references genes. Apical: apical part of the ovary, excluding the nectary. Basal: basal-most part of the ovary enriched in nectary tissue (as indicated in Figure 1A). R.E., relative expression.
In Arabidopsis, it has been demonstrated that the activity of euAP2 genes (to which BEN and ROB genes belong) is regulated by miR172 microRNAs at the posttranscriptional level (Rhoades et al., 2002; Aukerman and Sakai, 2003; Kasschau et al., 2003; Chen, 2004; Schwab et al., 2005). As Arabidopsis euAP2 members, BEN and ROB genes all contain a highly conserved miR172 recognition site (Morel et al., 2017), while in the Petunia axillaris genome sequence, nine different miR172 loci were identified (Bombarely et al., 2016). Two of these loci encode an identical mature microRNA (miR172-1), while the remaining seven loci also encode an identical mature microRNA (miR172-2) differing only at two nucleotide positions with miR172-1 located at both extremities (Supplemental Figure 5). To further understand how the activity of BEN and ROB genes restricts wild-type nectary development to the base of the ovary, we compared ROB1, ROB2, ROB3, BEN, miR172-1, and miR172-2 expression levels in apical and basal parts of the ovary. We found that ROB1 and ROB3 were expressed more strongly in the apical part compared with the basal part, but no clear differences could be found for ROB2, while BEN appeared to display the inverse pattern (Figure 5E). Interestingly, however, stem-loop RT-qPCR analysis (Chen et al., 2005) showed that both mature miR172-1 and miR172-2 microRNAs were expressed at ∼5-fold higher levels in the basal part compared with the apical part of the ovary (Figure 5F), suggesting lower BEN/ROB activity in the basal part compared with the apical part of the ovary.
DISCUSSION
C-Lineage-Dependent CRC Expression and Nectary Development in Arabidopsis and Petunia
The molecular basis of nectary development and its connection with the floral ABC gene network only has been studied in detail in Arabidopsis, a rosid species with stamen-associated nectaries (Smyth et al., 1990; Bowman and Smyth, 1999; Baum et al., 2001; Lee et al., 2005b; Kram et al., 2009). In this study, we investigated the molecular basis of nectary development in petunia, an asterid species with nectaries associated with its carpels. By analyzing various loss and gain-of-function mutants of the petunia C-function genes, we found that nectary development fully depends on the activity of these genes. We showed that the petunia C-function MADS box genes pMADS3 and FBP6 redundantly are required for nectary development, with pMADS3 playing a more important role compared with FBP6, and that ectopic expression of either pMADS3 or FBP6 is sufficient to induce the development of fully functional nectaries at the base of the sepals.
pMADS3 and FBP6 are C-lineage genes belonging to the AG subfamily of MADS box genes, which has undergone two major duplications. One occurred before the split between monocots and dicots, giving rise to the C- and D-lineages (Kramer et al., 2004; Zahn et al., 2006), and a second duplication within the C-lineage occurred in the eudicots, before the split between asterids and rosids (Figure 3G), giving rise to the euAG and PLENA sublineages (Kramer et al., 2004; Zahn et al., 2006). While petunia contains one copy of each type, the most recent genome duplication in Arabidopsis (Figure 3G) has yielded two copies in the PLE sublineage (SHP1 and SHP2), while a second AG copy most likely has been lost after duplication (Causier et al., 2005). Here, we showed that AG and SHPs have retained a common function in nectary development similar to the AG and PLE C-lineage genes in petunia. Note that it was suggested earlier that in the absence of B- and C-class gene activities, SHP1/2 might rescue nectary development if they are ectopically expressed, as in an A-class ap2 mutant background (Lee et al., 2005b). However, the observation that nectary development is completely lost in ag shp1 shp2 triple mutants while nectaries still develop in ag and in shp1 shp2 mutants (Figure 2) clearly demonstrates that SHP1 and SHP2 do not need to be in an ap2 genetic context to play a role in nectary formation. This is also fully supported by a transcriptome analysis of Arabidopsis nectaries (Kram et al., 2009) showing that SHP1, SHP2, and AG all show a strong nectary enriched expression profile in the wild type. Interestingly, the strong staining in the nectaries of pAG:GUS and pSHP:GUS lines suggest that the cis-element(s) required for nectary expression reside(s) in the SHP and AG promoter regions, and thus for AG not in its second regulatory intron that otherwise contains all required elements to provide a normal C-function expression pattern (Sieburth and Meyerowitz, 1997). This is in line with the idea that SHPs have subfunctionalized mainly through loss of elements in the second intron regulatory region compared with AG (Hong et al., 2003; Causier et al., 2005).
Next, we showed that in both petunia and Arabidopsis, CRC expression depends on members of both PLE and AG sublineage genes, explaining why CRC expression and nectary development is still observed in ag mutants. Finally, using dTph1 transposon mutants, we provided solid genetic proof that petunia nectary development requires CRC activity, redundantly encoded by PhCRC1 and PhCRC2 (Figure 3; Supplemental Figure 2). This redundancy possibly may explain why a previous VIGS-based silencing experiment using the full-length PhCRC1 had little effect on petunia nectary development (Lee et al., 2005a). On the other hand, a strong floral indeterminacy was observed in the VIGS plants, a phenotype that we only observed in homozygous crc1 crc2 double mutants, indicating that the PhCRC1-VIGS construct was not gene specific. Compared with the relatively mild phenotype of Arabidopsis crc mutants, the strong floral indeterminacy phenotype in petunia crc1 crc2 mutants suggests that the CRC genes in petunia play a more critical role during gynoecium development, as proposed also for, e.g., the CRC orthologs DL in rice (Oryza sativa; Yamaguchi et al., 2004) and EcCRC in California poppy (Eschscholzia californica; Orashakova et al., 2009). In line with that, it was recently shown in Arabidopsis that CRC and KNUCKLES (KNU) (Payne et al., 2004) synergistically regulate floral meristem termination (Breuil-Broyer et al., 2016; Yamaguchi et al., 2017), as illustrated by the highly indeterminate phenotype of crc knu double mutants. This shows that Arabidopsis CRC also plays a major role in floral meristem termination but that parallel pathways in Arabidopsis are more capable to rescue floral meristem termination in a crc mutant background compared with other species.
In Arabidopsis, it was proposed that the B-function proteins PI and AP3 also promote nectary development, based on the absence of nectary tissue in pi ag and ap3 ag flowers (Lee et al., 2005b). However, both pi and ap3 mutants produce nectaries (Baum et al., 2001), and the lack of nectary tissue in pi ag and ap3 ag flowers may, as also proposed by the authors, be due to the completely different organization of the floral whorls in these mutants, consisting of the reiteration of the first two whorls, rather than due to a direct effect of B-function genes on nectary formation. The B-function MADS box transcription factors AP3 and PI were shown to function as obligate heterodimeric partners and are required to be coexpressed for their nuclear localization (Goto and Meyerowitz, 1994; Jack et al., 1994; Krizek and Meyerowitz, 1996; McGonigle et al., 1996; Riechmann et al., 1996; Yang et al., 2003). Since only AP3 is expressed in developing nectaries (Baum et al., 2001; Kram et al., 2009), it seems unlikely that B-class genes would directly promote nectary formation, unless PI protein produced in adjacent cell layers would be invading the nectary tissues or, alternatively, if nectary formation would not require heterodimerization. More recently, it was shown that AP3 and PI directly suppress the expression of CRC (Wuest et al., 2012), which seems also difficult to unite with a model in which the B-function genes would promote nectary formation. Nevertheless, the nectaries in ap3 and pi mutants exhibit changes in size and morphology (Baum et al., 2001). Therefore, the (indirect) effect of the B-function on nectary development remains not fully understood.
Finally, the Arabidopsis SEP1-3 proteins (Pelaz et al., 2000) also have been proposed to be required for nectary development, based on the absence of nectaries in triple sep1 sep2 sep3 mutants (Lee et al., 2005b). Since the C-class proteins require complex formation with SEP proteins for their function (Honma and Goto, 2001), this is fully compatible with our model in which nectary development depends on the C-lineage MADS box proteins.
Antiquity of the C-Lineage Gene/CRC Module and Nectary Development?
Altogether, the now available data demonstrate that in both Arabidopsis and petunia, a strikingly similar mechanism underlies nectary development, consisting of euAG and PLE C-lineage members that are redundantly required for CRC expression, which on its turn is required for nectary development (Figure 3G). Because all C-lineage genes from petunia (pMADS3 and FBP6) and Arabidopsis (AG, SHP1, and SHP2) are able to activate CRC expression, this suggests that C-lineage gene dependent CRC activation already existed before the split between asterids and rosids, and before the duplication event leading to the euAG and PLE sublineages (Kramer et al., 2004; Causier et al., 2005; Zahn et al., 2006). This could suggest a common evolutionary origin for nectary development in the two major core eudicot lineages, at least for species in which the nectaries are associated with the reproductive organs, and thus residing within the classical C-function expression domain. However, it cannot be excluded that the recruitment of the C-lineage gene/CRC module for nectary development has occurred independently in the asterid and rosid lineages, since the antiquity of the C-lineage gene/CRC module may simply reflect the ancestry of CRC function in carpel development (Fourquin et al., 2005).
BEN and ROB Negatively Regulate the Size of the Petunia Nectaries in the Fourth Whorl
In this study, we identified BEN and ROB genes as major negative regulators of the nectary domain, as illustrated by the dramatic increase in nectary size in ben rob1 rob2 rob3 mutants (Figure 4). The clear upregulation of pMADS3 in the apical region of ben rob1 rob2 rob3 ovaries (Figures 5A to 5D) indicate that BEN/ROB genes restrict nectary development to the base of the ovary by negatively regulating the C-function in the apical part during later stages of ovary development, when nectaries initiate. For BEN, this novel function did not come as a big surprise, since we showed earlier that BEN negatively regulates C-class gene expression levels in the perianth, in parallel with BL (Morel et al., 2017). By contrast, the involvement of the ROB genes in this process was rather unexpected, since we previously could not observe any indications that they play a role in repressing the C-function in the perianth, in contrast to the function of AP2, their closest homolog in Arabidopsis. Instead, we previously found that the ROB genes repress the B-function in the first whorl, together with BEN (Morel et al., 2017). The results presented in this study thus suggest that the ROB genes do encode a C-class repressor function as their Arabidopsis AP2 counterpart, but in a highly subfunctionalized way restricted to fourth whorl development and associated nectary development.
The restriction of nectary development to the base of the carpels in the wild type assumes high BEN/ROB repressive activity in the apical part of the ovary compared with low activity in the region where the nectaries form. In line with that, we found that both ROB1 and ROB3 displayed higher transcript levels in the apical versus the basal part of the ovary (Figure 5E), but this was not the case for ROB2 and BEN. However, both MIR172 variants were expressed at 5-fold higher levels in the basal part compared with the apical part of the ovary. This supports a model in which high miR172 activity leads to a low BEN/ROB activity at the basis of the ovary, allowing high C-gene expression and nectary development. In the more apical region of the ovary, the inverse pattern then leads to a repression of nectary development. In Arabidopsis, it has been shown that both translational inhibition and cleavage of transcripts contribute to miR172-dependent AP2 regulation, but the relative contribution of each process is obscured by a feedback loop in which AP2 negatively regulates its own transcription (Aukerman and Sakai, 2003; Kasschau et al., 2003; Chen, 2004; Schwab et al., 2005). In addition, the differential effect of MIR172a overexpression on the steady state transcript levels of its different targets in Arabidopsis indicate that individual members of the euAP2 family exhibit differences in the efficiency of their feedback regulation as well as differences in the extent of MIR-dependent cleavage versus translational inhibition (Schwab et al., 2005). This may explain why BEN and the three ROB genes do not all show similar apical/basal expression level ratios.
Although the repression of the C-function in the apical part of the ovary is sufficient to explain the absence of apical nectary development, it is likely that BEN and ROB genes in addition repress (an) other factors(s) required for nectary development. In support of that is the observation that the ovaries of 35S:pMADS3 flowers (Supplemental Figure 4H) do not show the strong apical nectary extension as found in ben rob1 rob2 rob3 mutants. Possible candidates to represent these other factors are one or more members of the SEPALLATA subfamily, since SEP3 was also shown to be repressed by AP2 in Arabidopsis, at least in the first floral whorl (Krogan et al., 2012). However, the regulatory interactions between BEN/ROB function and the seven members of the petunia SEP/AGL6 clade (Rijpkema et al., 2009) remains to be elucidated.
While a dramatic and persistent overproliferation of the nectaries as observed in petunia ben rob1 rob2 rob3 mutants remains to be described in Arabidopsis, there are indications that AP2, and possible other members of the euAP2 lineage may also negatively regulate the size of the nectary domain. For example, it has been reported that in ap2 ag double mutants, more nectary glands per reiteration are present compared single ag mutants and that the shape of these nectaries is variable (Baum et al., 2001). On the other hand, the strong ap2-2 mutant flowers appear to have less nectary glands compared with the wild type, since nectaries normally only develop at lateral positions, possibly because ap2 flowers display severe defects in organ primordia initiation. Interestingly, however, it was mentioned that in these mutants sometimes nectary glands develop along the margin of the first-whorl medial organs instead of between them (Baum et al., 2001). In petunia, a strong overproliferation of nectary tissue is only observed when the function of several euAP2 members was compromised simultaneously. So far, a nectary phenotype only has been described for Arabidopsis ap2. It is therefore conceivable that a clear phenotype will only be obtained in plants combining mutations in different members of the euAP2 lineage, although it is not clear to what extent the expected organ primordia initiation defects will possibly obscure the interpretation of the nectary phenotypes. Besides AP2, Arabidopsis contains five additional euAP2 genes, consisting of TARGET OF EAT1 (TOE1), TOE2, TOE3, SCHLAFMÜTZE, and SCHNARCHZAPFEN, which redundantly act as floral repressors (Aukerman and Sakai, 2003; Jung et al., 2007; Mathieu et al., 2009; Yant et al., 2010). Among these genes, TOE1 and TOE3 exhibit highest similarity with petunia BEN and ROB genes, respectively (Morel et al., 2017). Furthermore, it was found that TOE3, in addition to its role as a flowering time gene, also represses AG expression during flower development (Jung et al., 2014). TOE1 and TOE3 could be therefore interesting candidates to investigate their role in nectary development, in combination with AP2.
Other Factors Required for Nectary Identity and Position-(In)dependent Nectary Development
Even though the C-lineage gene/CRC module is required to confer nectary identity in petunia and Arabidopsis flowers, the function (and expression domain) of C-lineage and CRC genes is obviously much broader than nectary development. Therefore, the restriction of nectaries at the base of the carpels in wild-type petunia, and near the base of the stamen, filaments in Arabidopsis must depend on the local presence of one or more additional genetic factors that in combination with the C-lineage gene/CRC module locally specify nectary identity. In Arabidopsis, nectaries are strongly linked to the third floral whorl, independently from the identity of the different types of floral organs that may develop in the third whorl in various homeotic mutant backgrounds (Baum et al., 2001). Likewise, in flowers in which stamens develop in ectopic positions, the position of the nectaries remains restricted to the third-whorl stamens (Baum et al., 2001). In petunia, it appears that nectaries are less position dependent compared with Arabidopsis because fully functional ectopic nectaries develop at the basis of the sepals when either one of the C-lineage genes is ectopically expressed. This could suggest that at least one of these unknown additional nectary identity factors is more broadly expressed in petunia compared with Arabidopsis: Besides being present at the base of the carpels where nectaries normally develop, it must be expressed at least also at the base of sepals, allowing nectary development when the C-lineage gene/CRC module becomes locally active. Identification of this factor and possibly other unknown players will be crucial to further unravel the combinatorial code underlying nectary identity in petunia and Arabidopsis. Nevertheless, with the current knowledge we can already explain/speculate why the two species develop nectaries on different positions. First of all, the restriction of this unknown nectary factor to the basis of the third whorl in Arabidopsis is sufficient to explain why in contrast to petunia, no nectary development occurs at the basis of its ovary, despite local expression of CRC. In turn, petunia CRC expression is not detected at the basis of its stamens (Lee et al., 2005a), unlike in Arabidopsis. Likewise, while we observed clear pMADS3 expression at the basis of the carpels where nectary development initiates (Figure 5), no comparable pMADS3 expression was found near the basis of the stamen filaments. Since the C-lineage/CRC module is essential for nectary development, a local difference in C-lineage-dependent CRC activation must therefore be a major contributing factor in their different nectary positioning.
Diversity of Nectary Positioning and the C-Lineage/CRC Module
Most of the rosid and asterid species have their nectaries associated either with the stamens or carpels (Bernardello, 2007). Because (1) both positions reside within the classical C-function expression domain and (2) CRC expression in nectaries has shown to be conserved in a number of higher eudicot species (Lee et al., 2005a), it is likely that reproductive organ associated nectary development in rosids and asterids might more generally occur via a C-lineage/CRC module as identified in petunia and Arabidopsis.
However, nectary position within the plant kingdom is much more diverse: A number of species develop nectaries in the perianth, or even outside the flower, such as in leaf axils or on leaves and bracts (Brown, 1938; Bernardello, 2007), tissues in which C-lineage genes would expected to be lowly expressed if at all. Interestingly, Lee and colleagues (Fourquin et al., 2005) detected CRC expression in extrafloral nectaries, e.g., in the leaf axil positioned nectaries of Capparis flexuosa, and in the nectaries that develop from the midvein of leaves and cotyledons and on the involucral bracts in cotton (Gossypium hirsutum). On the other hand, CRC expression could not be detected in the nectaries of the basal eudicot species Aquilegia formosa (Fourquin et al., 2005) or Epimedium sagittatum (Sun et al., 2013). In summary, nectary development may involve CRC function, but its activation may not necessarily depend on C-lineage genes, or nectary development can even occur independently from CRC. Nectary development in these species may therefore depend on a different mechanism(s) as identified for petunia and Arabidopsis, in line with the hypothesis that nectaries evolved multiple times independently (Brown, 1938).
METHODS
Plant Material and Genotyping
Petunia (Petunia hybrida) plants were grown in a greenhouse (16 h day/8 h night: natural light supplemented with Philips Sodium HPS 400W SON-T AGRO light bulbs; 55,000 lumens) with conditions that were further influenced by local seasonal changes (45.72°N 4.82°E). Petunia crc1 and crc2 mutants were identified by searching our sequence-indexed dTph1 transposon database (Vandenbussche et al., 2008), which has been considerably expanded in recent years. Exact insert positions were determined by aligning the transposon flanking sequences with the corresponding genomic and coding sequences. The insertion alleles were named after their exact insert position, expressed in base pairs downstream of the ATG in the coding sequence (Figure 3E). Offspring of candidate insertion lines were grown and genotyped by PCR using gene-specific primer pairs flanking the insertion site (Supplemental Table 1). The following thermal profile was used for segregation analysis PCR: 11 cycles (94°C for 15 s, 71°C for 20 s minus 1°C/cycle, 72°C for 30 s), followed by 40 cycles (94°C for 15 s, 60°C for 20 s, and 72°C for 30 s). The different crc1 crc2 double mutants were obtained by crossing homozygous crc1-155 and crc1-273 plants with a homozygous crc2-249 individual. Double mutants were obtained in the F2 generation that was fully genotyped. 35S:pMADS3 NEC1:GUS plants were obtained in the offspring of a cross between a 35S:pMADS3 line (Heijmans et al., 2012) and a NEC1:GUS line (Ge et al., 2000). The two different CRISPR-Cas9-derived pmads3 alleles (Supplemental Figure 1) were generated using the protocol and vectors described by Schiml et al. (2016). Briefly, the 5′-GAGGAAAGATTGAGATCAAG-3′ PAM sequence was cloned by restriction into the pEN-Chimera vector (Schiml et al., 2016), giving the pEN-Chimera-pMADS3 vector. Gateway LR recombination was performed using pEN-Chimera- pMADS3 and ProDE-Cas9, leading to the production of the plant transformation vector pDE-Cas9-pMADS3. This construct was transformed in the Mitchell background by leaf-disc transformation (Horsch et al., 1985). T-DNA-positive lines were then screened for the presence of gene editing by amplifying and sequencing pMADS3 genomic DNA. Plants carrying the 8-bp deletion allele (pmads3-1) were genotyped by PCR using a primer pair (Supplemental Table 1) flanking the mutation site, and PCR products were analyzed by 4% agarose gel electrophoresis. Genotyping of the 1-bp insertion allele (pmads3-2) was done by direct sequencing of PCR products. The pmads3 alleles were combined with the fbp6-1 mutant by crossing. All other petunia transgenic lines and transposon mutants used in this study have been previously described. These include fbp6-1; fbp6-1 pMADS3-RNAi and 35S:FBP6 (Heijmans et al., 2012); bl-2 (Cartolano et al., 2007); ben-724, ben-724 bl-2; rob1-61, rob2-915, rob3-935, rob1-61 rob2-915, rob1-61 rob2-915 rob3-935, rob1-61 rob2-915 rob3-935 bl-2, rob1-61 rob2-915 rob3-935 ben-724 (Morel et al., 2017). Arabidopsis thaliana plants were grown under long days (16 h day/8 h night, 21°C). ag-6+/− (Prunet et al., 2008) plants were crossed with shp1 shp2 (Liljegren et al., 2000) to obtain triple mutants and genotypes were confirmed on a phenotypic basis for ag-6 and by PCR for shp1 and shp2 as previously described (Liljegren et al., 2000).
Oligonucleotides
Primers used in this study are listed in Supplemental Table 1.
Isolation of PhCRC2 and Sequence Analyses
The full-length PhCRC2 coding sequence was amplified using oligos PhCRC2-fw and PhCRC2-rv, and sequenced. We refer to the originally identified PhCRC (AY854801) as PhCRC1. For the phylogenetic analysis (Figure 3D), the conserved region indicated in Supplemental Figure 2B was used to generate the neighbor-joining tree, which on itself was computed with Treecon software (Van de Peer and De Wachter, 1994) using (1) distance estimation options: Tajima and Nei Distance Calculation; insertions and deletion not taken into account; alignment positions: all; bootstrap analysis: yes, 1000 samples. (2) Infer tree topology options: neighbor-joining; bootstrap analysis: yes. (3) Root unrooted trees options: outgroup option: single sequence (forced); bootstrap analysis: yes. Select root: At-YAB5. Sequence identifiers for the Arabidopsis YABBY genes are indicated in the legend of Supplemental Figure 2. The alignment is shown in Supplemental File 1. For the CRC promoter analysis (Supplemental Figure 3), genomic fragments from Petunia axillaris (Bombarely et al., 2016) and Nicotiana benthamiana (http://benthgenome.qut.edu.au/) containing 5 kb of sequence upstream of the putative ATG start codon followed by the full coding sequence were analyzed via the mVISTA website (http://genome.lbl.gov/vista/mvista/submit.shtml) (Frazer et al., 2004) using the MLAGAN alignment program (Brudno et al., 2003). The alignment is shown in Supplemental File 2. Conservation parameters for plotting Vista graphs were set at 70% minimum conservation identity (Min_Id) and 50-bp minimum length for a conserved noncoding sequence. The 5-kb promoter regions were scanned for the presence of CarG boxes via JASPAR (http://jaspar.genereg.net/) (Khan et al., 2018) using the MA0005.1 and MA0005.2 matrix profiles for AG and applying a relative profile score threshold of 85%. Predicted pre-miR172 sequences (Bombarely et al., 2016) were aligned using ClustalW (Thompson et al., 1994) and visualized in Bioedit (Hall, 1999) (Supplemental Figure 5A). Minimal free energy RNA secondary structure prediction of the P. axillaris pre-miR172 sequences (Supplemental Figure 5B) was generated via the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) (Gruber et al., 2008).
GUS Staining, Imaging, and Microscopy
pAG:GUS and pSHP1:GUS constructs were made using the Gateway system with oligo pairs pAG-fw/pAG-rev and pSHP1-fw/pSHP1-rev to amplify the AG and SHP1 promoter sequences, respectively. DNA sequences were cloned into the pENTR/D-topo vector (Life Technologies) and then transferred into the pKGWFS7 vector (Karimi et al., 2002) for plant transformation. GUS activity for petunia and Arabidopsis lines was analyzed by incubating tissues overnight at 37°C in staining solution (0.1% Triton X-100, 2 mM Fe2+CN, 2 mM Fe3+CN, and 1 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid in 50 mM phosphate buffer, pH 7.0). Samples were cleared using 70% ethanol and photographed with a standard digital camera. Petunia samples for cryo-scanning electron microscopy shown in Figure 1 were analyzed as previously described (Vandenbussche et al., 2009). Arabidopsis scanning electron microscopy images shown in Figure 2 and petunia scanning electron microscopy images shown in Figure 4 were obtained with a HIROX SH-1500 benchtop environmental electron microscope equipped with a cooled stage. Macroscopic floral phenotypes were imaged by conventional digital photography using a glass plate as a support and black velvet tissue around 10 cm below the glass plate in order to generate a clean black background. When needed, backgrounds were further equalized by removing dust particles and light reflections with Photoshop. Images shown in Figures 1A, 1B, 1F to 1L, 4F to 4I, and 4L to 4M and Supplemental Figures 1D and 4A to 4H were obtained using a Keyence VHX-900F digital microscope. Nectary surface measurements (Supplemental Figure 1C) were done using the standard software package of the Keyence VHX-900F digital microscope. Ovaries observed to determine nectary height/ovary height ratio’s (Figure 4N) were analyzed under a standard binocular microscope equipped with a PC-connected camera. Due to the large size of the ovaries shown in Figures 5A and 5B, images were obtained based on an assembly of smaller pictures acquired by a Zeiss AxioImager Z.1 microscope with motorized platform and reconstructed using MetaMorph software version 7.8.13.0 (Molecular Devices). Images in Figures 5C and 5D were photographed using a Zeiss Imager M2 microscope equipped with an AxioCam MRc camera (Zeiss).
RT-qPCR Gene Expression Analysis and in Situ Hybridization
For RT-qPCR experiments shown in Figures 1E and 3B, total RNA was isolated and purified from different plant tissues using either TRIzol reagent (Invitrogen) as described by the manufacturer and an additional sodium acetate/ethanol precipitation step, or by using the RNeasy Plant Mini Kit (Qiagen), also as recommended by the manufacturer. cDNA synthesis was performed using the iScript cDNA synthesis kit (Bio-Rad), using 1 µg RNA per reaction. cDNA-specific primer pairs for RT-qPCR (Supplemental Table 1) were designed using Beacon Designer 4 software (Premier Biosoft International). RT-qPCR was performed using SybrGreen mix (Bio-Rad), according to the manufacturer’s protocol, in a total volume of 25 µL. All reactions took place in a Bio-Rad MyIQ iCycler, using a two-step protocol: 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 58°C for 1 min. Afterwards, the reaction mixtures were subjected to the machine’s standard melting curve analysis. The average PCR amplification efficiency for each primer pair was calculated using LinRegPCR (Ruijter et al., 2009). Then, using the efficiency and Ct-values, relative normalized expression levels were calculated using PhACTIN and PhRAN as reference genes (Rieu and Powers, 2009; Mallona et al., 2010). Three biological replicates (material harvested from three different plants) were used for each experiment. The expression levels of the genes of interest were normalized to the highest value in the series (set to 100).
For RT-qPCR experiments shown in Figures 3A, 3C, 5E, and 5F, total RNA was extracted using the Spectrum Plant Total RNA kit (Sigma-Aldrich). Note that for the RT-qPCR experiment in Figure 5F, an increased amount of binding solution was used to enrich for small RNAs, as recommended in the protocol (Sigma-Aldrich). The RNAs were then treated with Turbo DNA-free DNase I (Ambion). RNA was reverse transcribed using RevertAid M-MuLV reverse transcriptase (Fermentas) according to the manufacturer’s protocol. For the analysis of miRNA expression (Figure 5F), stem-loop primers were added to the cDNA synthesis mix according to the protocol developed by Chen et al. (2005). RT-qPCR was performed in an optical 384-well plate in the QuantStudio 6 Flex real-time PCR system (Applied Biosystems) using FastStart Universal SYBR Green Master (Rox; Roche), in a final volume of 10 µL, according to the manufacturer’s instructions. The following standard thermal profile was used: 95°C for 10 min, 40 cycles of 95°C for 10 s, and 60°C for 30 s. Data were analyzed using QuantStudio 6 and 7 Flex Real-Time PCR System Software v1.0 (Applied Biosystems). cDNA-specific primer pairs for RT-qPCR (Supplemental Table 1) were designed using the online PrimerQuest tool (Integrated DNA Technologies). PCR efficiency (E = 10−1/slope) was calculated from the data obtained from the standard curve amplification. Relative expression values on the y axes are the average of nine data points resulting from the technical triplicates of three biological replicates ± sd and normalized to the geometrical average of three E−ΔCt, where ΔCt = CtGOI − CtACTIN, GAPDH and RAN (petunia) or ACT8 (Arabidopsis). The expression levels of the genes of interest in Figures 3A and 3C were further normalized relative to the wild type in the series (set to 100). For the petunia expression studies, RNA was extracted from indicated tissues harvested from flowers just before anthesis. For CRC expression analysis in Arabidopsis wild type and mutants (Figure 3A), stage 14 flowers (Smyth et al., 1990) were sampled for the wild type and shp1 shp2, and of equivalent stages (based on position in the inflorescence) for ag and ag shp1 shp2 flowers. For pMADS3 in situ hybridization, a gene-specific fragment was generated by PCR using a primer pair containing the T7 transcription site in the 5′ region of the reverse primer (Supplemental Table 1). Probe synthesis and in situ hybridization were performed as described previously (Ferrandiz and Sessions, 2008a, 2008b).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession number KJ739883.
Supplemental Data
Supplemental Figure 1. Creation and analysis of CRISPR-Cas9-derived pmads3 alleles.
Supplemental Figure 2. Petunia has two CRC homologs.
Supplemental Figure 3. Comparison of the 5′ upstream regions of Petunia axillaris (Pax) and Nicotiana benthamiana (Nb) CRC genes.
Supplemental Figure 4. Additional 4th-whorl nectary phenotypes.
Supplemental Figure 5. The Petunia axillaris miR172 family.
Supplemental Table 1. Oligo sequences used in this study.
Supplemental Data File 1. Alignment of the conserved region shown in Supplemental Figure 2B and used to generate the NJ tree in Figure 3D.
Supplemental Data File 2. Alignment (MLAGAN) of Pax and Nb CRC genomic sequences for mVISTA analysis.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank G. Angenent for sharing the NEC1 reporter line, and the greenhouse, logistics, and secretarial teams at the Laboratoire Reproduction et Développement des Plantes (ENS de Lyon) for their support. K.H. was supported by NWO Grant 818.02.012. M.V. and K.A. were supported by a CNRS ATIP-AVENIR grant.
AUTHOR CONTRIBUTIONS
M.V., K.H., K.A., M.C., and P.M. conceived and designed the experiments. P.M., K.H., K.A., M.C., A.B., S.R.B., P.C., C.T., and M.V. performed the experiments. P.M., K.H., K.A., M.C., C.T., and M.V. analyzed the data. M.V., K.H., and P.M. wrote the article with feedback from C.T.
Footnotes
Articles can be viewed without a subscription.
References
- Angenent G.C., Franken J., Busscher M., Colombo L., van Tunen A.J. (1993). Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J. 4: 101–112. [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum S.F., Eshed Y., Bowman J.L. (2001). The Arabidopsis nectary is an ABC-independent floral structure. Development 128: 4657–4667. [DOI] [PubMed] [Google Scholar]
- Bernardello G. (2007). A systematic survey of floral nectaries. In Nectaries and Nectar, Nicolson S.W., Nepi M., Pacini E., eds (Springer; ), pp. 19–128. [Google Scholar]
- Bombarely A., et al. (2016). Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2: 16074. [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126: 2387–2396. [DOI] [PubMed] [Google Scholar]
- Breuil-Broyer S., Trehin C., Morel P., Boltz V., Sun B., Chambrier P., Ito T., Negrutiu I. (2016). Analysis of the Arabidopsis superman allelic series and the interactions with other genes demonstrate developmental robustness and joint specification of male-female boundary, flower meristem termination and carpel compartmentalization. Ann. Bot. 117: 905–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W.H. (1938). The bearing of nectaries on the phylogeny of flowering plants. Proc. Am. Philos. Soc. 79: 549–595. [Google Scholar]
- Brudno M., Do C.B., Cooper G.M., Kim M.F., Davydov E., Green E.D., Sidow A., Batzoglou S.; NISC Comparative Sequencing Program (2003). LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartolano M., Castillo R., Efremova N., Kuckenberg M., Zethof J., Gerats T., Schwarz-Sommer Z., Vandenbussche M. (2007). A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat. Genet. 39: 901–905. [DOI] [PubMed] [Google Scholar]
- Causier B., Castillo R., Zhou J., Ingram R., Xue Y., Schwarz-Sommer Z., Davies B. (2005). Evolution in action: following function in duplicated floral homeotic genes. Curr. Biol. 15: 1508–1512. [DOI] [PubMed] [Google Scholar]
- Chen C., et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E.S., Meyerowitz E.M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37. [DOI] [PubMed] [Google Scholar]
- Colombo M., Brambilla V., Marcheselli R., Caporali E., Kater M.M., Colombo L. (2010). A new role for the SHATTERPROOF genes during Arabidopsis gynoecium development. Dev. Biol. 337: 294–302. [DOI] [PubMed] [Google Scholar]
- Dinneny J.R., Weigel D., Yanofsky M.F. (2005). A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132: 4687–4696. [DOI] [PubMed] [Google Scholar]
- Ferrandiz C., Sessions A. (2008a). Nonradioactive in situ hybridization of RNA probes to sections of plant tissues. Cold Spring Harb. Protoc. 2008: pdb.prot4943. [DOI] [PubMed] [Google Scholar]
- Ferrandiz C., Sessions A. (2008b). Preparation and hydrolysis of digoxygenin-labeled probes for in situ hybridization of plant tissues. Cold Spring Harb. Protoc. 2008: pdb.prot4942. [DOI] [PubMed] [Google Scholar]
- Fourquin C., Vinauger-Douard M., Fogliani B., Dumas C., Scutt C.P. (2005). Evidence that CRABS CLAW and TOUSLED have conserved their roles in carpel development since the ancestor of the extant angiosperms. Proc. Natl. Acad. Sci. USA 102: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourquin C., Primo A., Martínez-Fernández I., Huet-Trujillo E., Ferrándiz C. (2014). The CRC orthologue from Pisum sativum shows conserved functions in carpel morphogenesis and vascular development. Ann. Bot. 114: 1535–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I. (2004). VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32: W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallai N., Salles J., Settele J., Vaissiere B.E. (2009). Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ. 68: 810–821. [Google Scholar]
- Ge Y.X., Angenent G.C., Wittich P.E., Peters J., Franken J., Busscher M., Zhang L.M., Dahlhaus E., Kater M.M., Wullems G.J., Creemers-Molenaar T. (2000). NEC1, a novel gene, highly expressed in nectary tissue of Petunia hybrida. Plant J. 24: 725–734. [DOI] [PubMed] [Google Scholar]
- Gerats T., Vandenbussche M. (2005). A model system for comparative research: Petunia. Trends Plant Sci. 10: 251–256. [DOI] [PubMed] [Google Scholar]
- Gómez-Mena C., de Folter S., Costa M.M., Angenent G.C., Sablowski R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132: 429–438. [DOI] [PubMed] [Google Scholar]
- Goto K., Meyerowitz E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8: 1548–1560. [DOI] [PubMed] [Google Scholar]
- Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. (2008). The Vienna RNA websuite. Nucleic Acids Res. 36: W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Heijmans K., Ament K., Rijpkema A.S., Zethof J., Wolters-Arts M., Gerats T., Vandenbussche M. (2012). Redefining C and D in the petunia ABC. Plant Cell 24: 2305–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong R.L., Hamaguchi L., Busch M.A., Weigel D. (2003). Regulatory elements of the floral homeotic gene AGAMOUS identified by phylogenetic footprinting and shadowing. Plant Cell 15: 1296–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529. [DOI] [PubMed] [Google Scholar]
- Horsch R.B., Fry J.E., Hoffmann N.L., Eichholtz D., Rogers S.G., Fraley R.T. (1985). A simple and general method for transferring genes into plants. Science 227: 1229–1231. [DOI] [PubMed] [Google Scholar]
- Izhaki A., Borochov A., Zamski E., Weiss D. (2002). Gibberellin regulates post-microsporogenesis processes in petunia anthers. Physiol. Plant. 115: 442–447. [DOI] [PubMed] [Google Scholar]
- Jack T., Fox G.L., Meyerowitz E.M. (1994). Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Seo Y.H., Seo P.J., Reyes J.L., Yun J., Chua N.H., Park C.M. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Lee S., Yun J., Lee M., Park C.M. (2014). The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Sci. 215-216: 29–38. [DOI] [PubMed] [Google Scholar]
- Kapoor M., Tsuda S., Tanaka Y., Mayama T., Okuyama Y., Tsuchimoto S., Takatsuji H. (2002). Role of petunia pMADS3 in determination of floral organ and meristem identity, as revealed by its loss of function. Plant J. 32: 115–127. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kasschau K.D., Xie Z., Allen E., Llave C., Chapman E.J., Krizan K.A., Carrington J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell 4: 205–217. [DOI] [PubMed] [Google Scholar]
- Kater M.M., Colombo L., Franken J., Busscher M., Masiero S., Van Lookeren Campagne M.M., Angenent G.C. (1998). Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 10: 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., et al. (2018). JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46: D260–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kram B.W., Xu W.W., Carter C.J. (2009). Uncovering the Arabidopsis thaliana nectary transcriptome: investigation of differential gene expression in floral nectariferous tissues. BMC Plant Biol. 9: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E.M., Jaramillo M.A., Di Stilio V.S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166: 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B.A., Fletcher J.C. (2005). Molecular mechanisms of flower development: an armchair guide. Nat. Rev. Genet. 6: 688–698. [DOI] [PubMed] [Google Scholar]
- Krizek B.A., Meyerowitz E.M. (1996). The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122: 11–22. [DOI] [PubMed] [Google Scholar]
- Krogan N.T., Hogan K., Long J.A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139: 4180–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Baum S.F., Oh S.H., Jiang C.Z., Chen J.C., Bowman J.L. (2005a). Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development 132: 5021–5032. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Baum S.F., Alvarez J., Patel A., Chitwood D.H., Bowman J.L. (2005b). Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell 17: 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren S.J., Ditta G.S., Eshed Y., Savidge B., Bowman J.L., Yanofsky M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770. [DOI] [PubMed] [Google Scholar]
- Mallona I., Lischewski S., Weiss J., Hause B., Egea-Cortines M. (2010). Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle B., Bouhidel K., Irish V.F. (1996). Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 10: 1812–1821. [DOI] [PubMed] [Google Scholar]
- Morel P., Heijmans K., Rozier F., Zethof J., Chamot S., Bento S.R., Vialette-Guiraud A., Chambrier P., Trehin C., Vandenbussche M. (2017). Divergence of the floral A-function between an asterid and a rosid species. Plant Cell 29: 1605–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ÓMaoiléidigh D.S., Wuest S.E., Rae L., Raganelli A., Ryan P.T., Kwasniewska K., Das P., Lohan A.J., Loftus B., Graciet E., Wellmer F. (2013). Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25: 2482–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orashakova S., Lange M., Lange S., Wege S., Becker A. (2009). The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant J. 58: 682–693. [DOI] [PubMed] [Google Scholar]
- Payne T., Johnson S.D., Koltunow A.M. (2004). KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131: 3737–3749. [DOI] [PubMed] [Google Scholar]
- Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203. [DOI] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G.S., Savidge B., Liljegren S.J., Baumann E., Wisman E., Yanofsky M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88. [DOI] [PubMed] [Google Scholar]
- Prunet N., Morel P., Thierry A.M., Eshed Y., Bowman J.L., Negrutiu I., Trehin C. (2008). REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 20: 901–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., Krizek B.A., Meyerowitz E.M. (1996). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93: 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I., Powers S.J. (2009). Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21: 1031–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema A.S., Royaert S., Zethof J., van der Weerden G., Gerats T., Vandenbussche M. (2006). Analysis of the Petunia TM6 MADS box gene reveals functional divergence within the DEF/AP3 lineage. Plant Cell 18: 1819–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijpkema A.S., Zethof J., Gerats T., Vandenbussche M. (2009). The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J. 60: 1–9. [DOI] [PubMed] [Google Scholar]
- Ruijter J.M., Ramakers C., Hoogaars W.M., Karlen Y., Bakker O., van den Hoff M.J., Moorman A.F. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiml S., Fauser F., Puchta H. (2016). CRISPR/Cas-mediated site-specific mutagenesis in Arabidopsis thaliana using Cas9 nucleases and paired nickases. Methods Mol. Biol. 1469: 111–122. [DOI] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Huijser P., Nacken W., Saedler H., Sommer H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250: 931–936. [DOI] [PubMed] [Google Scholar]
- Sieburth L.E., Meyerowitz E.M. (1997). Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9: 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman J., Hoballah M.E., Broger L., Moore J., Basten C., Kuhlemeier C. (2004). Dissection of floral pollination syndromes in Petunia. Genetics 168: 1585–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Huang W., Li Z., Lv H., Huang H., Wang Y. (2013). Characterization of a Crabs Claw gene in basal eudicot species Epimedium sagittatum (Berberidaceae). Int. J. Mol. Sci. 14: 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto S., van der Krol A.R., Chua N.H. (1993). Ectopic expression of pMADS3 in transgenic petunia phenocopies the petunia blind mutant. Plant Cell 5: 843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M., Janssen A., Zethof J., van Orsouw N., Peters J., van Eijk M.J., Rijpkema A.S., Schneiders H., Santhanam P., de Been M., van Tunen A., Gerats T. (2008). Generation of a 3D indexed Petunia insertion database for reverse genetics. Plant J. 54: 1105–1114. [DOI] [PubMed] [Google Scholar]
- Vandenbussche M., Horstman A., Zethof J., Koes R., Rijpkema A.S., Gerats T. (2009). Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell 21: 2269–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche M., Chambrier P., Rodrigues Bento S., Morel P. (2016). Petunia, your next supermodel? Front. Plant Sci. 7: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y., De Wachter R. (1994). TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10: 569–570. [DOI] [PubMed] [Google Scholar]
- Wuest S.E., O’Maoileidigh D.S., Rae L., Kwasniewska K., Raganelli A., Hanczaryk K., Lohan A.J., Loftus B., Graciet E., Wellmer F. (2012). Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc. Natl. Acad. Sci. USA 109: 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N., Huang J., Xu Y., Tanoi K., Ito T. (2017). Fine-tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nat. Commun. 8: 1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Nagasawa N., Kawasaki S., Matsuoka M., Nagato Y., Hirano H.Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Fanning L., Jack T. (2003). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33: 47–59. [DOI] [PubMed] [Google Scholar]
- Yanofsky M.F., Ma H., Bowman J.L., Drews G.N., Feldmann K.A., Meyerowitz E.M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39. [DOI] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T.T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn L.M., Leebens-Mack J.H., Arrington J.M., Hu Y., Landherr L.L., dePamphilis C.W., Becker A., Theissen G., Ma H. (2006). Conservation and divergence in the AGAMOUS subfamily of MADS-box genes: evidence of independent sub- and neofunctionalization events. Evol. Dev. 8: 30–45. [DOI] [PubMed] [Google Scholar]
- Zenoni S., D’Agostino N., Tornielli G.B., Quattrocchio F., Chiusano M.L., Koes R., Zethof J., Guzzo F., Delledonne M., Frusciante L., Gerats T., Pezzotti M. (2011). Revealing impaired pathways in the an11 mutant by high-throughput characterization of Petunia axillaris and Petunia inflata transcriptomes. Plant J. 68: 11–27. [DOI] [PubMed] [Google Scholar]