Chloroplast TOC/TIC protein translocons possess a relatively large pore size, in contrast to previous suggestions that they have strong unfoldase activity relative to other translocons.
Abstract
The degree of residual structure retained by proteins while passing through biological membranes is a fundamental mechanistic question of protein translocation. Proteins are generally thought to be unfolded while transported through canonical proteinaceous translocons, including the translocons of the outer and inner chloroplast envelope membranes (TOC and TIC). Here, we readdressed the issue and found that the TOC/TIC translocons accommodated the tightly folded dihydrofolate reductase (DHFR) protein in complex with its stabilizing ligand, methotrexate (MTX). We employed a fluorescein-conjugated methotrexate (FMTX), which has slow membrane transport rates relative to unconjugated MTX, to show that the rate of ligand accumulation inside chloroplasts is faster when bound to DHFR that is actively being imported. Stromal accumulation of FMTX is ATP dependent when DHFR is actively being imported but is otherwise ATP independent, again indicating DHFR/FMTX complex import. Furthermore, the TOC/TIC pore size was probed with fixed-diameter particles and found to be greater than 25.6 Å, large enough to support folded DHFR import and also larger than mitochondrial and bacterial protein translocons that have a requirement for protein unfolding. This unique pore size and the ability to import folded proteins have critical implications regarding the structure and mechanism of the TOC/TIC translocons.
INTRODUCTION
The import of proteins into chloroplasts is a prerequisite for photosynthesis and, therefore, plant growth and development. The protein translocons of the outer and inner chloroplast membranes (TOC and TIC, respectively) are responsible for the import of ∼95% of all chloroplast proteins from the cytoplasm and are highly conserved among all land plants (Shi and Theg, 2013a). The TOC complex is composed of Toc75, Toc34, and Toc159 at a 4:4:1 or 3:3:1 ratio (Paila et al., 2015). Toc75, a β-barrel BamA ortholog, is the major pore-forming subunit at the outer membrane. The composition of the TIC complex has been disputed, with Tic20, Tic21, and/or Tic110 proposed as major pore-forming subunits at the inner membrane (Paila et al., 2015; Bölter and Soll, 2016). High-resolution structures are available for many of the soluble domains of individual TOC/TIC components, but little is known about the assembled complex structures. The pore sizes of Toc75, Tic20, and Tic110 have been estimated to be 14 to 26 Å, 7.8 to 14.1 Å, and 15 to 34 Å, respectively, by electrophysiological measurements in proteoliposomes (Heins et al., 2002; Hinnah et al., 2002; Kovács-Bogdán et al., 2011). However, these calculations rely on many assumptions and may not reflect the full functional pore size range of the subunits within their native complex and membrane environments. The driving force for protein import derives from ATPase activity of stromal chaperones that exert a pulling force on the N terminus of precursor proteins. Stromal Hsp70 plays a major role, presumably analogous to the mechanism of mitochondrial matrix-localized mtHsp70 (Shi and Theg, 2010; Su and Li, 2010; Liu et al., 2014). Other chaperones proposed to be involved in parallel and/or in series with Hsp70 are stromal Hsp93, stromal Hsp90, and an enigmatic intermembrane space-localized Hsp70 (Flores-Pérez and Jarvis, 2013).
Having most of the TOC/TIC components identified, the mechanisms of protein translocation are still not fully understood. A fundamental question in all protein translocation systems is whether the translocon requires substrate proteins to be unfolded or allows folded proteins to cross the membrane by means of a larger pore. Proteins are generally thought to traverse membranes in an unfolded conformation, as is the case for mitochondrial membrane translocons (TOM and TIM), bacterial SecYEG, and Sec61 in the endoplasmic reticulum (Eilers and Schatz, 1986; Arkowitz et al., 1993; Matouschek, 2003; Conti et al., 2014). However, pathways that transport folded proteins also exist, such as for import into peroxisomes and Tat transport across bacterial and thylakoid membranes (Walton et al., 1995; Clark and Theg, 1997). Unlike mitochondria, chloroplasts are known to tolerate small folded proteins, for example, internally cross-linked 6.5-kD bovine pancreatic trypsin inhibitor (BPTI), but have been generally considered to unfold larger proteins (Jascur et al., 1992; Clark and Theg, 1997). The 22-kD dihydrofolate reductase (DHFR) is a model protein for folding/unfolding due to interactions with its stabilizing noncovalently bound ligand, methotrexate (MTX). MTX binding is a good indicator for DHFR folding because the kinetics of DHFR unfolding match those of DHFR/MTX complex dissociation, implying unfolded DHFR has no affinity for MTX (Touchette et al., 1986). MTX blocks DHFR translocation through the TOM/TIM and SecYEG complexes by preventing DHFR unfolding, but does not block DHFR transport through the Tat pathway (Eilers and Schatz, 1986; Arkowitz et al., 1993; Hynds et al., 1998). MTX was observed not to block DHFR import into chloroplasts, with both DHFR and MTX found in the stroma after import assays (America et al., 1994). In interpreting this experiment, the authors suggested that the TOC/TIC translocons have a strong unfoldase activity capable of stripping MTX away from DHFR. Here, we report that DHFR/MTX is in fact imported into chloroplasts as a folded complex. Additionally, we probed the TOC/TIC pore size and found it to be significantly larger than its functional cognate pore in mitochondrial TOM/TIM.
RESULTS
Utility of FMTX in Differentiating Folded and Unfolded DHFR Import
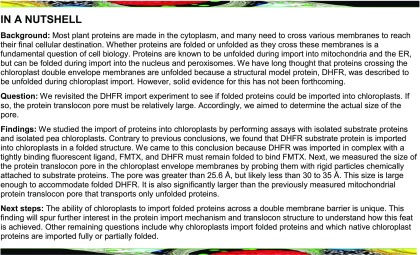
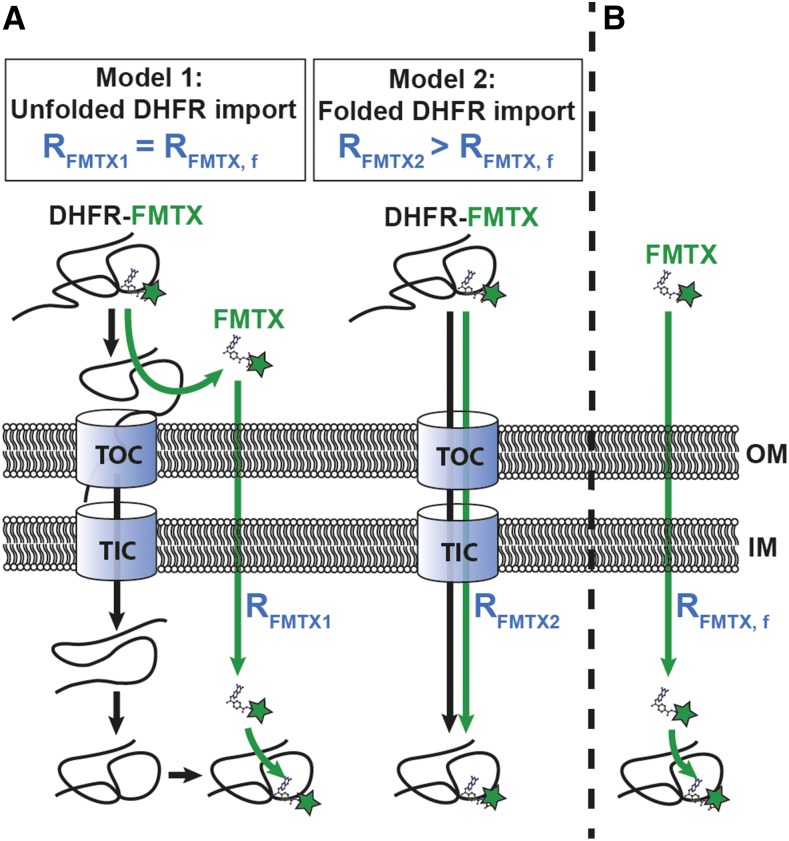
In previous studies, the mechanism of DHFR and MTX import into chloroplasts was described as the two molecules independently crossing the envelope membranes and then reassociating in the stroma (America et al., 1994). The ability of MTX to rapidly cross the chloroplast envelope membranes complicates the question of whether or not chloroplasts unfold the DHFR/MTX complex prior to import. However, fluorescein-conjugated methotrexate (FMTX) has extremely reduced passive membrane diffusion rates, unlike MTX, while retaining a high affinity for DHFR (Assaraf et al., 1989; Degan et al., 1989). These properties make FMTX an ideal substrate to test whether it is imported into chloroplasts independently or in complex with DHFR (Figure 1). If chloroplasts are able to unfold the DHFR/FMTX complex (Model 1), the rate of FMTX import (RFMTX1) should equal that of free FMTX import in the absence of DHFR import (RFMTX, f). Alternatively, if complexed DHFR/FMTX is imported (Model 2), the rate of FMTX import (RFMTX2) would not necessarily equal RFMTX, f. More specifically, RFMTX2 should be greater than RFMTX, f since FMTX displays slow membrane transport rates. Another useful property of FMTX is that it has enhanced fluorescence when bound to DHFR, which allows for quantification of the DHFR/FMTX complex (Kaufman et al., 1978; Degan et al., 1989). Such FMTX fluorescence enhancement was seen upon binding chimeric Escherichia coli DHFR fused to the Rubisco small subunit transit peptide (tp22DHFR) purified from E. coli and was reversed by addition of excess MTX to compete for tp22DHFR binding (Figure 2).
Figure 1.
Structural Models for DHFR/FMTX Import.
(A) Model 1: The DHFR/FMTX complex is unfolded at the chloroplast surface; both components are imported independently across the envelope membranes (OM and IM) and reassociate in the stroma. Model 2: The DHFR/FMTX complex remains intact during import across the envelope membranes.
(B) The rate of free FMTX import (RFMTX, f) in the absence of DHFR import is compared with the theoretical rates of FMTX import in Models 1 and 2 (RFMTX1 and RFMTX2) to establish a basis for differentiating the two models.
Figure 2.
Fluorescence Enhancement of FMTX upon Binding tp22DHFR.
tp22DHFR (40 nM) was added to 80 nM FMTX in import buffer followed by addition of excess MTX (800 nM) at indicated time points.
DHFR Is Imported into Chloroplasts in a Folded Complex
Separate or complexed FMTX import into the stroma was analyzed by adding FMTX to chloroplasts after or during tp22DHFR import. To test for FMTX import when complexed with DHFR, both protein and ligand were added to the initial import reaction (co-import). The reaction was stopped by dilution into cold import buffer, followed by immediate washing to remove excess FMTX. This washing effectively removed the remaining external FMTX (Supplemental Figure 1). To test for DHFR-independent FMTX import, tp22DHFR needed to be preimported so that independently imported FMTX could potentially bind to DHFR in the stroma and accumulate therein. This was achieved by importing tp22DHFR in an initial import reaction, followed by washing and thermolysin treatment to remove external protein. FMTX was introduced to the chloroplasts in a second mock import reaction with time, temperature, light, and ATP conditions identical to the initial import reaction (independent import). The only difference between the DHFR/FMTX co-import and independent FMTX import treatments was the initial location of DHFR (outside or inside the chloroplast, respectively). Thermolysin treatment of chloroplasts should not affect FMTX import since FMTX is small enough to freely pass through pores in the outer membrane and therefore would not require any outer membrane transporters (Flugge and Benz, 1984).
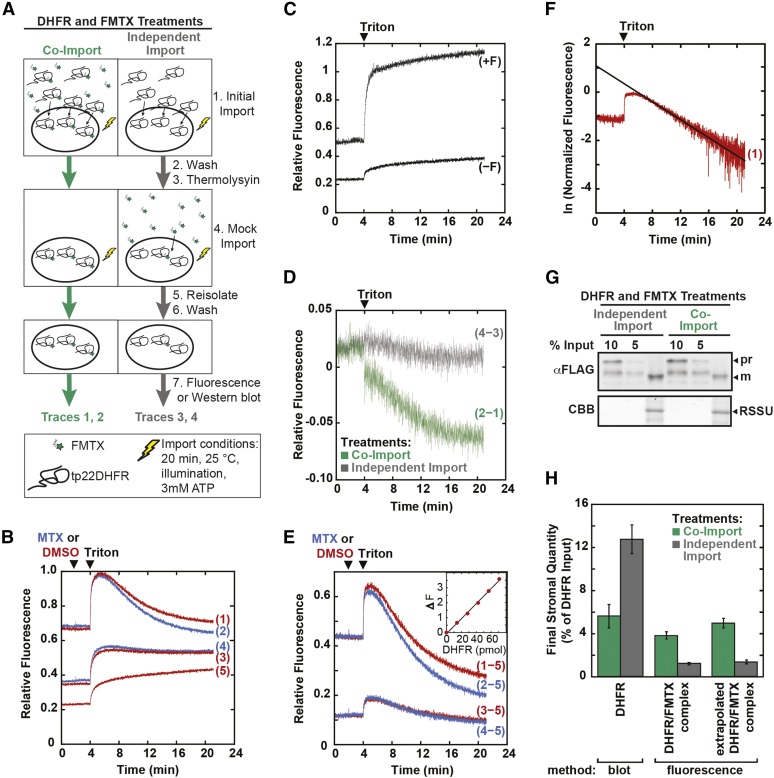
Imported chloroplast samples were reisolated, and stromal FMTX was detected by fluorescence. The initial fluorescence observed with DHFR and FMTX co-import was much higher than that of independent FMTX import, supporting the model that the folded DHFR/FMTX complex was imported (Figure 3B). Quantification of the imported tp22DHFR/FMTX complex requires debinding of FMTX with excess MTX. However, excess MTX did not efficiently replace bound FMTX in the stroma. MTX may not accumulate in the stroma at high enough concentrations to compete with FMTX binding at a detectable rate. Therefore, we solubilized the chloroplasts with 0.05% Triton after the addition of excess MTX to allow for complete FMTX debinding. Triton solubilization caused the chloroplast solution to become more transmissive to light, increasing the fluorescence signal at the 4-min time point. Solubilization also caused the effective volume surrounding the tp22DHFR/FMTX complex to increase 1000-fold from the stromal volume to the total bulk solution volume. This dilution shifted the binding equilibrium toward the unbound state, causing the fluorescence signal to decline at the 5-min time point even without the addition of excess MTX. The fluorescence decay was indeed specific to the dilution-mediated DHFR/FMTX debinding, as it was not seen upon solubilizing chloroplasts with imported precursor covalently conjugated to fluorescein (Figure 3C). In the presence of excess MTX, the fluorescence decay after solubilization was faster due to the additive effects of dilution and MTX/FMTX binding competition (Figure 3B). We observed the kinetics of FMTX replacement with MTX by subtracting the traces without MTX addition from those with MTX (Figures 3B, trace 2−1 and trace 4−3, and 3D). The fluorescence decay caused by the ligand replacement reaction was greater for DHFR/FMTX co-import than independent FMTX import. This is consistent with a larger amount of stromal DHFR/FMTX complex in the co-import treatment.
Figure 3.
FMTX Import into Chloroplasts Is Dependent upon Concurrent DHFR Import.
(A) Schematic of FMTX import experiment. tp22DHFR (930 nM) was imported into chloroplasts in 300-μL reactions with or without 8 μM FMTX as shown (initial import). Reactions were stopped with cold import buffer. Chloroplasts were washed, resuspended in 400 μg/mL thermolysin, incubated 1 h on ice, and quenched with 12.5 mM EDTA. Chloroplasts were resuspended in import buffer with ATP and with or without 8 μM FMTX as indicated and were again exposed to light for 20 min (mock import). Chloroplasts were reisolated on 40% Percoll cushions, washed with import buffer, and assayed for fluorescence in 3 mL of import buffer.
(B) Fluorescence of chloroplasts derived from DHFR and FMTX co-import (traces 1 and 2) or independent import treatments (traces 3 and 4). Trace 5 represents background chloroplast fluorescence derived from chloroplasts treated as shown in (A), except without any addition of protein or FMTX. Excess MTX (17 μM, traces 2 and 4) or DMSO (traces 1, 3, and 5) was added at t = 2 min, and 0.05% Triton X-100 (TX) was added to all traces at t = 4 min. All traces represent averages of three independent experiments.
(C) RSSUFHC precursor protein labeled with fluorescein maleimide was imported into chloroplasts, which were thermolysin treated, reisolated, and assayed for fluorescence (+F) along with a mock-import control (−F).
(D) DMSO-added traces from (B) were subtracted from MTX-added traces (traces 2−1 and 4−3) to show the MTX replacement reaction kinetics.
(E) Background fluorescence (trace 5) was subtracted from traces 1 through 4. Replicates were offset adjusted to the same initial fluorescence. tp22DHFR/FMTX complex import was determined as the change in fluorescence due to debinding of FMTX from DHFR (base fluorescence at t = 21 min in MTX added trace subtracted from peak fluorescence at t = 5 min in the DMSO added trace). Inset: Fluorescence change (ΔF) was calibrated by adding DHFR lacking tp22 fusion in 14 pmol increments to a saturating 170 nM FMTX solution in import buffer containing chloroplasts (17 μg/mL chlorophyll).
(F) The true tp22DHFR/FMTX peak at t = 4 min was determined by extrapolating linearized DMSO-added traces (1 and 3) back to t = 4 min. Decay curves were linearized by taking the natural log of normalized data. Sample extrapolation is shown.
(G) tp22DHFR import was determined by α-FLAG blotting, shown along with Rubisco small subunit (RSSU) as a loading control.
(H) tp22DHFR protein import and tp22DHFR/FMTX complex import (nonextrapolated and extrapolated) were quantified from immunoblots and fluorescence data, respectively. Error bars indicate se (n = 3). CBB, Coomassie Brilliant Blue; pr, tp22DHFR precursor; m, mature tp22DHFR.
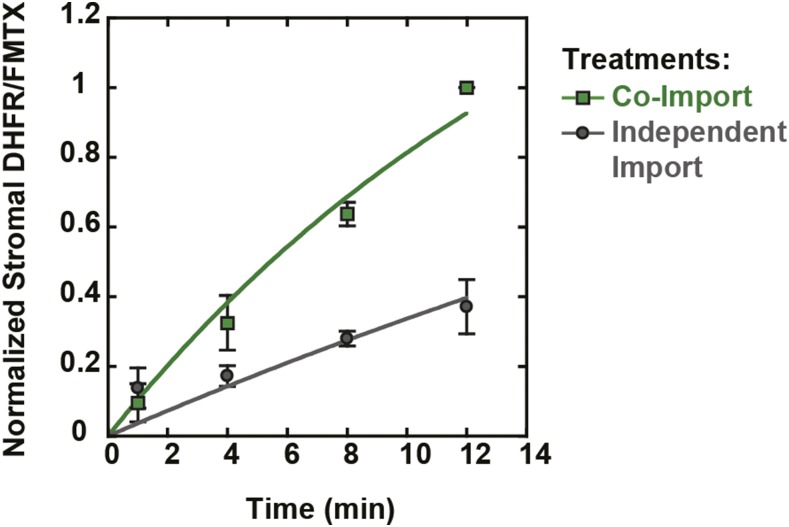
Subsequently, to quantify the DHFR/FMTX complex via the debinding fluorescence decay, we first subtracted the background chloroplast fluorescence (Figure 3B, trace 5) from the other traces (Figure 3E). We calibrated the fluorescence decay by measuring fluorescence of purified E. coli DHFR (without tp22 fusion) added stepwise to a solution of chloroplasts and saturating FMTX (Figure 3E, inset). The fluorescence decay was taken as the difference in peak fluorescence without MTX addition (at 5 min) and steady state fluorescence after complete FMTX debinding by MTX (at 21 min). However, the 5-min peak did not represent the maximum protein-bound FMTX fluorescence since the dilution effect began immediately after solubilization at 4 min and competed with the fluorescence increase caused by chloroplast solubilization between 4 and 5 min. The true peak was therefore determined by linearizing the fluorescence decays and extrapolating back to the 4-min time point when Triton was added (Figure 3F). We found that the fluorescence decay signal was specific to the DHFR/FMTX interaction and was not affected by FMTX interactions with other chloroplast components (Supplemental Figure 2). In addition to the stromal tp22DHFR/FMTX complex determined by the debinding fluorescence decay, tp22DHFR protein import was independently monitored by immunoblot analysis of replicate samples (Figure 3G). The amount of the stromal tp22DHFR/FMTX complex present was much higher when protein and ligand were co-imported than when they were independently imported, indicating that the folded tp22DHFR/FMTX complex was imported (Figure 3H). The amount of protein import measured via immunoblot matched the extrapolated import of the tp22DHFR/FMTX complex obtained from the fluorescence assay. This convergence of the data suggests an equimolar import of tp22DHFR and FMTX. Additionally, we determined the kinetics of DHFR/FMTX co-import and independent FMTX import (Figure 4). The rate of complex import was 2.9-fold greater than that of the independent FMTX import, indicating that DHFR import was rate limiting for FMTX import. We further observed a 56% inhibition of tp22DHFR import by FMTX (Figure 3H), which is similar to reported DHFR import inhibition by MTX (Kovacheva et al., 2007). This inhibition suggests that the uncomplexed tp22DHFR may in fact be somewhat unfolded and thereby imported more efficiently by chloroplasts.
Figure 4.
Kinetics of DHFR/FMTX Complex and FMTX Import.
Chloroplasts were treated with tp22DHFR and FMTX as in Figure 3A, except that import reactions were stopped at various time points. DHFR/FMTX complex import kinetics were determined by stopping the initial import reaction at the indicated time points and stopping the second mock import reaction after 12 min (co-import, green squares). FMTX import kinetics were determined by stopping the initial import reaction (with tp22DHFR) after 12 min and stopping the second mock import reaction (with FMTX) at the indicated time points (independent import, gray circles). Samples were assayed for stromal DHFR/FMTX as in Figure 3. The 12-min co-import time points were normalized to 1. Error bars indicate sd (n = 3). Lines show data fitted with a single rising exponential, both approaching the same maximum. The DHFR/FMTX co-import rate constant was 2.9-fold greater than the independent import rate constant.
Although import of the folded tp22DHFR/FMTX complex via a route other than the TOC/TIC pore is improbable, we confirmed that a native chloroplast TOC/TIC substrate competed with the complexed DHFR for import (Supplemental Figure 3). The folded tp22DHFR/FMTX complex therefore utilizes the TOC/TIC translocons.
Inhibition of DHFR Import Also Impairs FMTX Accumulation in Chloroplasts
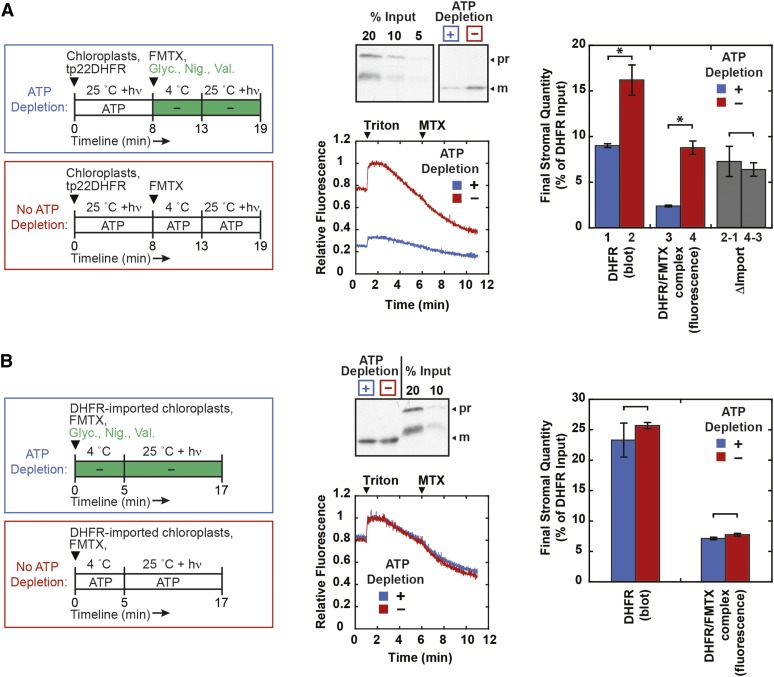
For DHFR and FMTX to be imported as a complex, FMTX ligand import should be inhibited when protein import is inhibited. To test this, protein import inhibition was achieved by depleting stromal ATP with the addition of glycerate, nigericin, and valinomycin (Scott and Theg, 1996). First, tp22DHFR was preimported with illumination to generate stromal ATP and no exogenously added ATP for 8 min, after which the reaction was stopped on ice, stromal ATP was depleted, and FMTX was added (Figure 5A). The import reaction was allowed to continue for another 6 min, during which time the FMTX could potentially cross into the stroma independently and bind to the preimported DHFR. ATP depletion did indeed inhibit protein import, since the final amount of imported DHFR was greater in control reactions that were not depleted of ATP. The final amount of imported FMTX was also greater in the control reactions, indicating that the difference between control and ATP-depleted reactions was due to DHFR/FMTX complex import. In fact, the differences between control and ATP-depleted import of both DHFR and FMTX were very similar (Figure 5A, gray bars), again suggesting a stoichiometric import of the DHFR/FMTX complex.
Figure 5.
Concurrent Inhibition of DHFR and FMTX Import into Chloroplasts.
(A) tp22DHFR (1.3 μM) was preimported into chloroplasts for 8 min at room temperature under illumination (hν) (no exogenous ATP was added), after which reactions were stopped on ice. Stromal ATP was depleted with 10 mM glycerate (Glyc.), 5 μM nigericin (Nig.), and 5 μM valinomycin (Val.) on ice for 5 min (ATP depletion). Control reactions (no ATP depletion) were mock treated with appropriate volumes of solvents (import buffer, ethanol, and DMSO). FMTX (15 μM) was added to the cold reactions at this point to allow binding to external DHFR. ATP-depleted and control reactions without ATP depletion were incubated at room temperature under illumination for an additional 6 min. Import reactions were stopped with cold import buffer. Chloroplasts were thermolysin treated and reisolated through Percoll as in Figure 3.
(B) Chloroplasts were treated essentially as in the Figure 3A independent import treatment, except that the second mock import reaction was conducted with 10 mM glycerate, 5 μM nigericin, and 5 μM valinomycin (ATP depletion) or solvent controls (no ATP depletion). Also, the initial import reaction with tp22DHFR and subsequent mock import reaction with FMTX were conducted for 16 and 12 min, respectively, at room temperature under illumination with no exogenously added ATP.
All samples in (A) and (B) were assayed for stromal DHFR/FMTX by extrapolated fluorescence or immunoblotted for stromal DHFR as in Figure 3. Representative blots and averaged fluorescence traces corrected for background chloroplast fluorescence are shown. Quantifications are shown with se (n = 3). Significant differences were determined by t tests (*P < 0.05). Quantifications with and without stromal ATP depletion in (A) were subtracted to show change in import (gray bars) for DHFR (bar 2−1) and FMTX (bar 4−3).
Although FMTX has been shown to display a passive transport mechanism in mammalian cells, we wanted to verify that ATP levels did not affect independent FMTX import into chloroplasts. To address this question, chloroplasts were preimported with DHFR, thermolysin-treated, and incubated in a second mock import reaction containing FMTX in the presence or absence of internal ATP (Figure 5B). ATP levels did not have a significant effect on FMTX uptake, confirming that the ATP-mediated differences in FMTX import observed in Figure 5A were due to complexed import of DHFR/FMTX. Although not statistically significant, there was a slight difference between independent FMTX import with or without ATP depletion. Since this difference corresponds to only 2% of the total amount of imported DHFR, it may reflect a small population of protease-protected import-intermediate tp22DHFR precursor in the intermembrane space (IMS) that was chased across the inner membrane in complex with FMTX during the second mock reaction when stromal ATP was not depleted. The ability of IMS-localized precursor to be chased into the stroma in thermolysin-treated chloroplasts has been previously reported (Scott and Theg, 1996). In summary, FMTX import into chloroplasts was ATP-dependent during concurrent DHFR import, but was ATP-independent when imported separately, indicating that FMTX was imported in complex with DHFR.
The TOC/TIC Pore Size Is Greater Than 25.6 Å
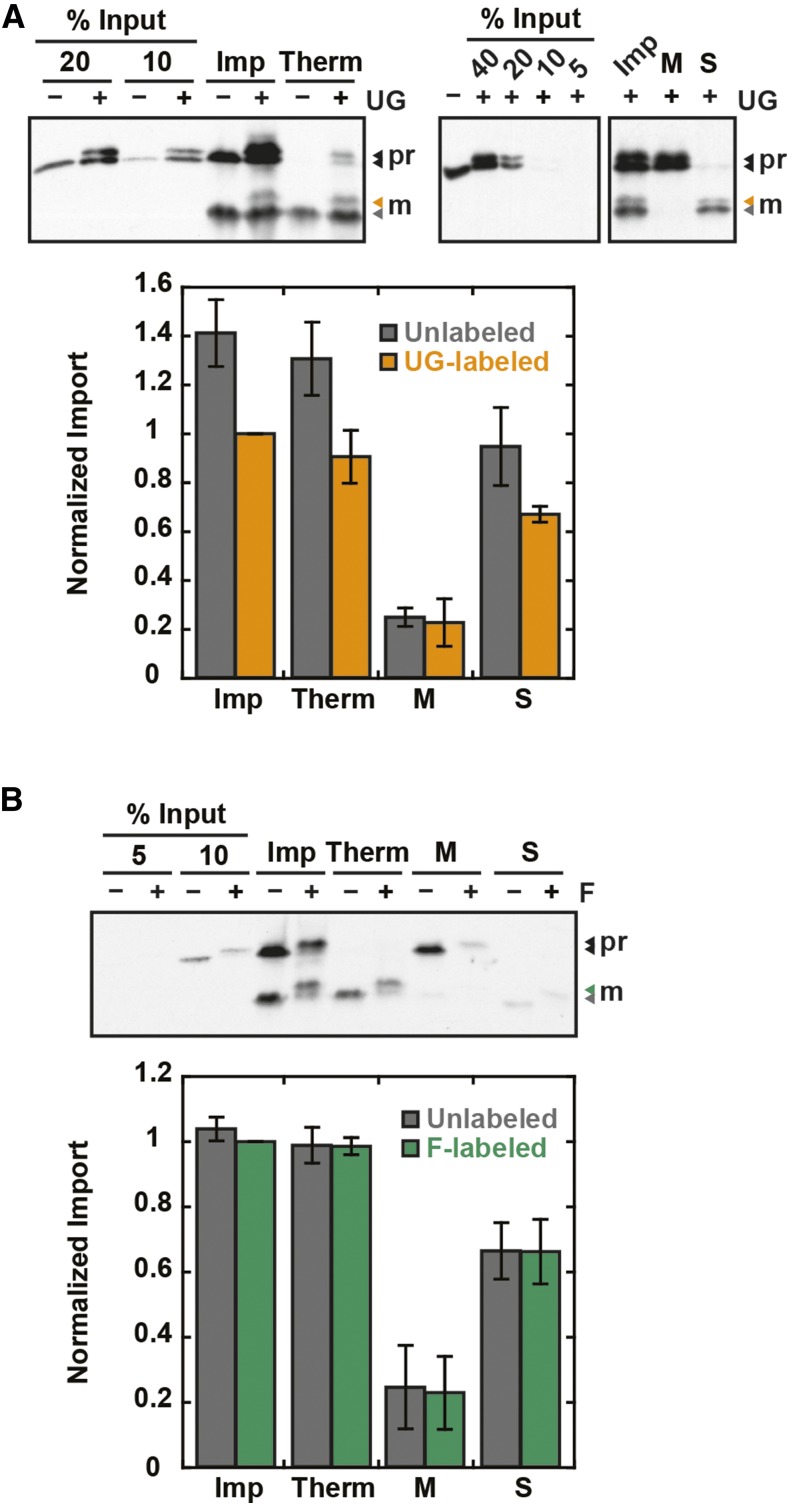
To further confirm that folded DHFR (average minor axis diameter of 27.6 Å) can pass through the TOC/TIC translocons, the functional pore size was measured by probing the translocons with particles of fixed diameter attached to precursor proteins. If the probe is imported, the TOC/TIC pore size must be larger than the probe diameter. A rigid, spherical, 20-Å monomaleimido Undecagold particle was covalently conjugated to a single C-terminal cysteine on Rubisco small subunit precursor containing FLAG and HIS tags for detection and purification (RSSUFHC). The Undecagold-labeled RSSUFHC, detected by a gel shift, was imported and localized to the chloroplast stroma (Figure 6A). The labeled mature protein was protease-protected and predominantly detected in the soluble stromal fraction. The Undecagold label did not affect the amount of soluble mature RSSUFHC relative to total mature RSSUFHC. A minor inhibitory effect of Undecagold on RSSUFHC import was observed and was further quantified in Figures 8A and 8C. To control for the effect of protein modification on import, a smaller 12- to 14-Å particle, fluorescein maleimide, was conjugated to RSSUFHC and also localized to the chloroplast stroma (Figure 6B). The smaller particle had no effect on RSSUFHC import efficiency.
Figure 6.
The TOC/TIC Pore Size Is Greater Than 20 Å.
Undecagold (UG)-labeled (A) or fluorescein (F)-labeled (B) RSSUFHC was localized to the chloroplast stroma. Import reactions were conducted for 20 min and stopped in cold import buffer (Imp). For protease treatment, chloroplasts were further resuspended in import buffer containing 400 μg/mL thermolysin (Therm), incubated 30 min on ice, and quenched with 12.5 mM EDTA. All samples were reisolated on 40% Percoll cushions and washed with import buffer. After reisolation, some non-protease-treated samples were separated into membrane (M) and soluble (S) fractions by lysis in 2 mM EDTA for 10 min on ice, mixing with 200 mM NaCl, and pelleting membranes at 16,000g. Soluble fractions were precipitated in 15% TCA on ice and washed with cold acetone. Samples were analyzed by SDS-PAGE and α-FLAG blotting along with percent input standards. Mature (M) labeled protein was quantified and, as an internal control, the unlabeled mature protein in the same lanes as the labeled protein was also quantified. Error bars indicate se (n = 3). Labeled protein Imp treatments were normalized to 1. pr, RSSUFHC precursor; m, mature RSSUFHC.
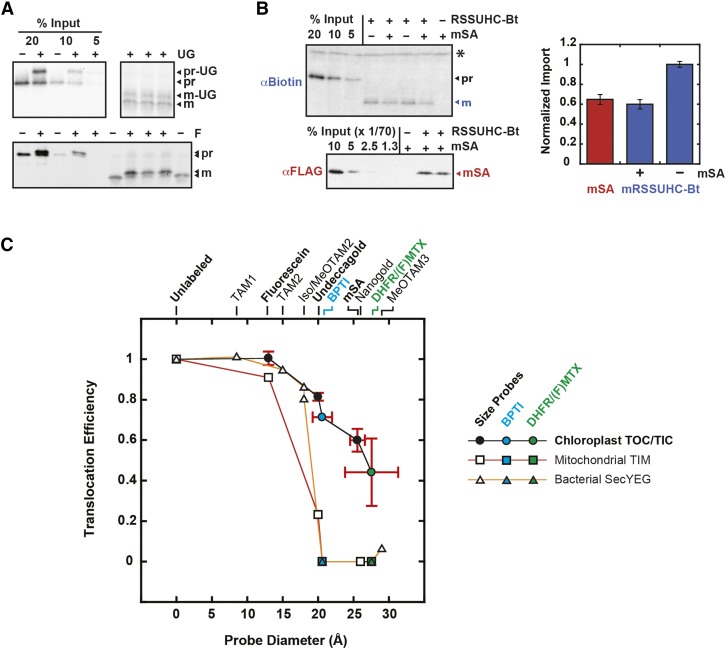
Figure 8.
Import Efficiencies of Fixed-Diameter Probes.
(A) and (B) Eight-minute import reactions of fluorescein (F)-labeled and Undecagold (UG)-labeled RSSUFHC (A) and mSA-labeled RSSUHC-Bt (B) are shown. RSSUHC-Bt (8.3 μg/mL in import reaction) was incubated with a 70-fold molar excess of mSA in import buffer prior to import. All reactions were stopped after 8 min with cold 400 μg/mL thermolysin and incubated 1 h on ice. Samples were subsequently treated as in Figures 6 and 7. Mature RSSU (mRSSUHC-Bt) and mSA were quantified to illustrate their equimolar import ratio.
(C) Import efficiencies of all three labeled proteins (defined as the labeled:unlabeled mature protein ratio) were plotted as a function of probe size (black circles). The internal control unlabeled protein was used for fluorescein and Undecagold quantifications. Error bars (sd) represent multiple replicates from two independent experiments. For comparison, tp22DHFR/FMTX import efficiency was plotted as the ratio of tp22DHFR import in the presence and absence of FMTX derived from Figure 3H (green circle, error bar = sd, n = 3). Protein dimensions were plotted as minor axis diameters with error bars representing sd away from a perfectly circular cross section in the minor axis plane. Mitoplast TIM complex import rates for fluorescein (13 Å), Undecagold (20 Å), and Nanogold (26 Å) size probes were taken from Schwartz and Matouschek (1999). The Undecagold import efficiency was plotted as the labeled:unlabeled ratio of preprotein extent of import after 4 min since the authors noted that the initial import rates overestimate the import efficiency. Bacterial SecYEG transport rates for tetraarylmethane derivative size probes (TAMs) were replotted from Bonardi et al. (2011) with the unlabeled substrate transport rate normalized to 1. Internally cross-linked BPTI translocation efficiency through TOC/TIC, TIM, and SecYEG complexes (cyan circle/square/triangle) and DHFR/MTX translocation efficiency through TIM and SecYEG complexes (green square/triangle) also derive from previous publications (Eilers and Schatz, 1986; Schiebel et al., 1991; Jascur et al., 1992; Arkowitz et al., 1993; Clark and Theg, 1997). Asterisk indicates nonspecific chloroplast protein.
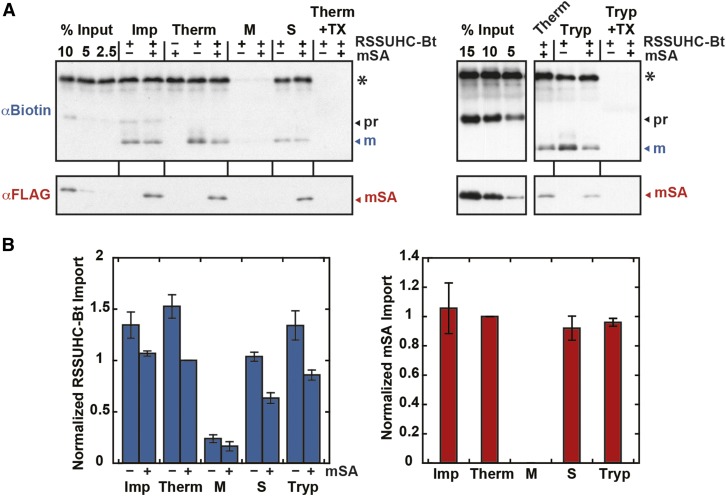
The TOC/TIC translocons were additionally probed with cylindrical monomeric streptavidin (mSA) with a 25.6-Å diameter and a 2.8 nM Kd for biotin (Lim et al., 2013). His-tagged Rubisco small subunit precursor was biotinylated at a single C-terminal cysteine (RSSUHC-Bt), to which His- and FLAG-tagged mSA was noncovalently bound. Although mSA is not a rigid particle like Undecagold, it can still be used as a size probe since it must remain folded to retain affinity for RSSUHC-Bt during import. Without RSSUHC-Bt present, mSA was not imported into chloroplasts [Figure 7A, (−)RSSUHC-Bt]. When imported with RSSUHC-Bt, mSA was localized to the chloroplast stroma, indicating that the maximum TOC/TIC pore size is greater than 25.6 Å (Figure 7). Imported mSA and RSSUHC-Bt were found in the soluble stromal fraction. They were membrane-protected from thermolysin and trypsin proteases but were protease-sensitive upon incubation with Triton to solubilize the membranes. mSA did not affect the relative amount of mature RSSUHC-Bt in the soluble fraction. Import of mSA into chloroplasts was saturated above a 35-fold molar excess of mSA over RSSUHC-Bt in the import reaction (Supplemental Figure 4). Under these saturating conditions, mSA was imported into chloroplasts at an equimolar ratio with RSSUHC-Bt (Figure 8B), ruling out the possibility that the RSSUHC-Bt/mSA complex was dissociated by a strong unfoldase activity applied on mSA at the chloroplast membrane surface.
Figure 7.
The TOC/TIC Pore Size Is Greater Than 25.6 Å.
Biotinylated RSSUHC (RSSUHC-Bt, 8.3 μg/mL in import reaction) was incubated with 17.5- to 70-fold molar excess mSA in import buffer prior to import reactions. Thermolysin treatments were conducted as in Figure 6. Stromal localization was determined as in Figure 6, except that the membrane/stroma fractionation was done on thermolysin-treated chloroplasts. For trypsin (Tryp) treatments, postimport chloroplasts were resuspended in import buffer containing 240 μg/mL trypsin, incubated 30 min at 25°C, resuspended again in 1 mg/mL soybean trypsin inhibitor, reisolated on 40% Percoll, and washed with import buffer. Additionally, protease treatments were conducted in the presence of 1% Triton X-100 (TX). RSSUHC-Bt and mSA were detected on α-biotin and α-FLAG blots, respectively, along with standards that represent a molar percentage of RSSUHC-Bt input. Error bars indicate se (n = 3). mSA-labeled mature RSSUHC or mSA from the Therm treatment were normalized to 1. pr, RSSUHC-Bt precursor; m, mature RSSUHC-Bt. Asterisk indicates nonspecific chloroplast protein.
The TOC/TIC Pore Size Is Larger Than That of TOM/TIM and SecYEG
Efficiencies of fixed-diameter probe import were determined as the ratio of probe-labeled to -unlabeled precursor imported after 8-min reactions (Figure 8AB). The 8-min time point falls at the end of the linear portion of the reaction, and therefore correlates with the initial import rate (Supplemental Figure 5). As is to be expected, the probe import efficiency decreased with increasing probe size (Figure 8C). For our analysis, we plotted the protein dimensions based on the minor axis diameters since the longest major axis could, in principle, be imported perpendicular to the TOC/TIC pore diameter. Thus, we consider these minor axis values to represent the minimum pore size dimensions. The import efficiency of mSA supports the model of folded DHFR import since they have roughly similar minor axis diameters. The probes were imported more efficiently into chloroplasts than identical (20 Å Undecagold) or similarly sized probes (26 Å Nanogold) were imported into mitoplasts through the TIM complex as determined by Schwartz and Matouschek (1999) (Figure 8C). In our hands, the Nanogold-precursor conjugate was unstable in the presence of chloroplasts and could not be used as a probe. The TOC/TIC pore is also larger than the bacterial SecYEG pore (Figure 8C).
DISCUSSION
Readdressing the tolerance of the TOC/TIC translocons for folded proteins, we show that their functional pore size is larger than previously realized. We provide three lines of evidence to show that the DHFR/FMTX complex can be imported in a folded conformation: (1) FMTX import was significantly faster during concurrent DHFR import than independent import. (2) Inhibition of DHFR import yielded a proportional decrease in FMTX import. (3) The TOC/TIC pore size was greater than 25.6 Å, which is larger than the TOM/TIM and SecYEG pores and also large enough to accommodate folded DHFR. The comparatively larger pore in chloroplasts is consistent with previous observations that the small internally cross-linked BPTI protein can be imported into chloroplasts, but cannot be transported through mitochondrial or Sec translocons (Figure 8C) (Schiebel et al., 1991; Jascur et al., 1992; Clark and Theg, 1997). Tetrameric avidin has previously been shown to block chloroplast import, suggesting the TOC/TIC pore size has an upper limit of 50 Å (Froehlich and Keegstra, 1997). However, based on the reduced import efficiencies of mSA and DHFR/FMTX (Figure 8C), the maximum pore size appears likely to be no more than 30 to 35 Å. In mitochondria, the TIM complex pore size is only slightly larger than 20 Å (Schwartz and Matouschek, 1999). The TOM pore is larger than the TIM pore, but less than 26 Å. Here, the TOC pore size was not determined independently of TIC, but it is tempting to speculate that the TOC pore may be larger since the inner membrane is solute selective, while the outer membrane is not (Heldt and Sauer, 1971). Since FMTX does have some inhibitory effect on DHFR import efficiency, DHFR is likely imported in a somewhat unfolded state in the absence of FMTX. The foldedness of proteins with diameters roughly 20 to 30 Å during import is probably determined by both the stability and size of the protein. Proteins below 20 Å are likely to remain folded during their import, while those above 30 to 35 Å would at least be partially unfolded. However, given the pore size that we measured here, even large proteins may retain significant amounts of residual structure based on their particular mechanical unfolding pathways.
Pore size should affect ion leakage through the active TOC/TIC translocons. Because the chloroplast envelope membranes are not energy transducing like the bacterial plasma and mitochondrial inner membranes, it may be that the larger TOC/TIC pore diameter can be tolerated even if it leaks some ions. It should be noted that the Tat pathway transports folded proteins across energy-transducing membranes without leaking ions (Teter and Theg, 1998). However, Tat transport seems to employ unique mechanisms that would not allow for direct comparison in this regard to more typical proteinaceous pores (Hou et al., 2018). It remains to be determined how leaky the active TIC complex is to ions.
Our prior understanding of protein foldedness during chloroplast import has mostly been limited to conclusions drawn from protease sensitivity assays of tightly folded precursors bound to the chloroplast surface. These studies with different precursors yielded conflicting results. Purified ferredoxin reductase was found to be protease-resistant when bound to intact chloroplasts, but a purified chimeric OE33-RicinA protein was observed to be protease-sensitive (Walker et al., 1996; Ottado and Ceccarelli, 1998). An in vitro-translated ferredoxin-DHFR fusion protein complexed with MTX was found to be protease-sensitive when bound to purified envelope membranes (Guéra et al., 1993). In vitro-translated plastocyanin-DHFR fusion protein complexed with MTX was found to be protease-sensitive when bound to intact chloroplasts (America et al., 1994). In this case, DHFR/MTX was bound to chloroplasts, which were reisolated and washed before protease treatment. The MTX was likely diluted away due to equilibrium debinding during the reisolation and washing steps, just as FMTX was debound by dilution in Figure 3B. The same study reported a lack of [3H]-MTX associated with chloroplast-bound DHFR; however, the [3H]-MTX was again likely lost during chloroplast reisolation. It seems improbable that these tightly folded proteins could be unfolded by a passive mechanism at the chloroplast surface, yet an energy dependent unfoldase at the outer membrane has not been found. The quantitative import of mSA with RSSUHC-Bt also indicates that there is no global unfoldase activity at the chloroplast surface (Figure 8B).
Revisiting previous studies on transit peptide length requirements, the TOC/TIC pore size measured here seems to clarify inconsistent conclusions. Tightly folded substrates with long loosely structured N-terminal extensions (including targeting sequences) are able to be imported efficiently into both mitochondria and chloroplasts as the transit peptide/presequence must reach internal chaperones that provide the energy for unfolding and/or translocation. Mitochondrial preproteins require an ∼80-residue N-terminal extension to span two membranes and reach mtHsp70 in the matrix (Schleiff and Becker, 2011). Here also, the chloroplast data are conflicting, with ferredoxin reductase (FNR) requiring roughly 80 residues for efficient import and titin requiring only 60 (Rial et al., 2002; Bionda et al., 2010). It was suggested that the unfoldase activity for titin resided in the IMS such that the transit peptide only had to cross one membrane to reach the chaperone. However, the average minor axis diameters of FNR and titin are 40 and 22 Å, respectively, which suggests that titin does not necessarily need to be unfolded prior to chloroplast import, unlike FNR. The force required for mechanical unfolding of titin by atomic force microscopy is roughly twice the DHFR/MTX unfolding force, which strongly suggests titin would also remain folded during chloroplast import since it is both smaller and more stable than DHFR/MTX (Best et al., 2003; Ainavarapu et al., 2005). IMS ATPase activity has been implicated in precursor translocation through TOC when uncoupled from TIC, but no IMS chaperone has been directly implicated in the coupled TOC/TIC import process to date (Scott and Theg, 1996; Flores-Pérez and Jarvis, 2013; Bionda et al., 2016). While it remains possible for IMS ATPase activity to play a role in precursor unfolding, the simpler explanation for the FNR/titin discrepancy would be related to the respective sizes of the proteins. Accordingly, folded proteins too large to pass through the TOC/TIC pore likely require the 80 residue N-terminal extension for efficient import, as in mitochondria.
Extensive atomic force microscopy data on the mechanical unfolding pathway of titin make it an ideal translocon-mediated protein-unfolding model. With mechanical force applied to the N and C termini of titin, the N-terminal β-strand A unravels first at ∼100 pN, followed by the next β-strand A’ at 200 pN, after which the rest of the protein is easily unfolded at lower force (Best et al., 2003). Stabilizing mutations in the A strand inhibit import efficiency of titin into mitochondria, while destabilizing mutations enhance import (Sato et al., 2005). The localized N-terminal stability of proteins is important for mitochondrial import because transient N-terminal unfolding is rate limiting. In chloroplasts, destabilizing strand A’ mutations, but not strand A mutations, affected import efficiency, and it was concluded that chloroplasts have a fundamentally different unfoldase mechanism, possibly involving an IMS chaperone (Ruprecht et al., 2010). An alternate explanation is that strand A mutations did not affect chloroplast import because titin is small and stable enough to be imported as a folded protein and unfolding of the A strand does not significantly reduce the minor axis diameter of the protein. Unlike strand A, destabilizing mutations in strand A’ and the core of titin would allow the entire protein to unfold, thereby increasing chloroplast import efficiency. Strand A’ mutations probably did not affect mitochondrial import because mtHsp70 turnover and not protein unfolding was rate limiting (Sato et al., 2005). Due to the complication of folded titin import into chloroplasts, the mechanism of TOC/TIC-mediated protein unfolding remains unclear.
Yet further consequences of folded protein import into chloroplasts include structural implications for the TOC and TIC pores. The transport pathways of substrates utilizing β-barrel translocases such as mitochondrial Tom40 and bacterial FhaC pass through the central channel of the β-barrels, as determined by extensive translocon to substrate cross-linking studies (Baud et al., 2014; Shiota et al., 2015). By analogy, the transport pathway of Toc75 substrates should also be through the central channel, but this has not been tested. Although the central channel is too small to accommodate folded DHFR, it has been speculated that the weakly interacting first and last β-strands of Toc75 could open (Paila et al., 2015). These Toc75 β-strands are structurally similar to those of its bacterial ortholog, BamA, and dissimilar from those of other β-barrel translocases (i.e., Tom40), which have more tightly interacting terminal β-strands. The emerging model of BamA/Sam50 function in OMP biogenesis involves the terminal β-strands opening and pairing with newly inserted β-strands of substrate OMPs (Höhr et al., 2018). In this model, once the substrate OMP is fully inserted into the membrane, its terminal β-strands would be paired with the terminal β-strands of BamA forming one large β-barrel. The substrate OMP would subsequently bud off from BamA and close its β-barrel structure. This mechanism requires BamA to be somewhat flexible and lends credibility to the prospect of the Toc75 β-barrel opening and potentially forming a large pore in cooperation with other TOC components. Pore flexibility is certainly not uncommon and is important even for SecYEG (Hamman et al., 1998), where restricting the opening of the lateral gate caused transport inhibition of soluble proteins (Bonardi et al., 2011). The Toc159 membrane domain has been cross-linked to precursor proteins and may therefore cooperate with Toc75 to form part of the functional pore (Kouranov and Schnell, 1997). Multiple Toc75 subunits within the TOC complex may also cooperate to form the pore. A low-resolution EM structure of the TOC complex reveals four pores (possibly four Toc75 subunits) surrounding a central “finger” domain (possibly Toc159) (Schleiff et al., 2003). The four pores seem to be connected in the center near the “finger” domain, which suggests that the Toc75 terminal β-strands may already be open in the resting (nontransporting) state and may then expand to accommodate larger substrates. It is much harder to speculate on the nature of the TIC pore since there is very little structural information and continued controversy over the identity of the complex components. If multiple TIC complexes exist, they may also have different maximum pore sizes. Both TOC and TIC pores are likely to be somewhat flexible and expandable to accommodate folded proteins.
Overall, we have shown that the 22-kD DHFR is imported into chloroplasts in a folded complex with its ligand and that the TOC/TIC pore size is large enough to accommodate this structure. The pore, measured to be greater than 25.6 Å, is significantly larger than that of the bacterial SecYEG and cognate mitochondrial TOM/TIM translocons, both of which only translocate unfolded proteins.
METHODS
Plasmid Constructs
Pea (Pisum sativum) RSSU cDNA was cloned into pET23a with 5ʹ NdeI and 3ʹ XhoI restriction sites. The three native cysteines were mutated (C-1S, C41V, and C112V) and a single C-terminal cysteine was inserted after the 6xHis tag by site-directed mutagenesis, yielding the plasmid pET23a-RSSUHC. A C-terminal FLAG tag was added 5ʹ of the His tag to yield pET23a-RSSUFHC. The pET23a-tp22mDHFR plasmid was cloned by fusing the sequence encoding the first 79 residues of pea RSSU from the plasmid pET23a-tp22GFP (Shi and Theg, 2013b) with mouse cytosolic DHFR DNA from the plasmid AtPC-DHFR (Hageman et al., 1990) flanked by 5ʹ PstI and 3ʹ XhoI sites. The pET23a-tp22DHFR plasmid was made by replacing the coding sequence for RSSU residues 80 to 180 in RSSUHC-pET23a with that for Escherichia coli DHFR residues 2 to 159 flanked by two FLAG tags and 5ʹ BamHI/3ʹ XhoI sites. tp22DHFR (from E. coli) was made because full-length tp22mDHFR (from mouse) could not be expressed in E. coli. The coding sequence for E. coli DHFR without the tp22 transit peptide fusion was cloned into pET23a with 5ʹ NdeI and 3ʹ XhoI sites. pRSET-mSA was a gift from Sheldon Park (Addgene plasmid no. 39860) (Lim et al., 2013). It encodes mSA with 6xHis and FLAG tags for purification and detection.
Protein Production and Labeling
Proteins were expressed in E. coli BL21 cells and purified under native (DHFR lacking tp22 fusion) or denaturing (other purified proteins) conditions with Ni-NTA Agarose according to the manufacturer (Qiagen). mSA was refolded by rapid dilution into a stirred PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.2) solution overnight at 4°C. Soluble mSA was concentrated and buffer exchanged into 50 mM NaCl and 50 mM HEPES, pH 7.2, with Ultra-15 10K centrifugal filters (Amicon). DHFR lacking tp22 fusion and all other purified proteins were respectively buffer exchanged from elution buffer into buffer A (50 mM HEPES, pH 7.2) or buffer B (8 M urea and 50 mM HEPES, pH 7.2). Proteins were quantified on Coomassie-stained SDS-PAGE gels against BSA standards. [3H]-tp22mDHFR was in vitro-transcribed with T7 RNA polymerase and translated with wheat germ extract according to the manufacturer (Promega).
Maleimide reactions were performed in buffer A or B, both containing 7 mM TCEP, for 2 h at 4°C or 25°C, respectively. Fluorescein maleimide (Vector Labs), monomaleimido Undecagold (Nanoprobes), and biotin maleimide (Sigma-Aldrich) stocks (10, 1, and 100 mM, respectively) were made in DMSO. RSSUFHC (1 mg/mL) was reacted with 0.2 mM fluorescein maleimide or 0.05 mM Undecagold and added directly to import reactions. RSSUHC was labeled with 5 mM biotin maleimide, precipitated with 15% TCA on ice, washed with cold acetone, and resuspended in Buffer B.
Growth Conditions
Pea (P. sativum var Little Marvel) seeds (Harris Seeds) were soaked in water for several hours and grown in a tray of potting soil topped with a thin layer of vermiculite in a controlled environment chamber (20°C, 12/12-h light/dark cycle, 50 μE/m2s PAR).
Chloroplast Import Assays
Chloroplasts were isolated from 8- to 12-d-old peas and quantified by chlorophyll absorbance as described previously (Lo and Theg, 2011). Import reactions were conducted under 100 μE/m2s light with precursor protein (33 μg/mL unless otherwise specified) and isolated chloroplasts (0.33 mg/mL chlorophyll) in import buffer (330 mM sorbitol, 3 mM MgCl2, and 50 mM Tricine-KOH, pH 8.0). Except for those in Figure 5, reactions included exogenously added ATP (3 mM). All protease treatments included 5 mM CaCl2. Those that included 1% Triton X-100 were subsequently precipitated in 15% TCA on ice and washed with cold acetone. All samples, including percent input standards, were finally solubilized in 2× Laemmli sample buffer and analyzed by SDS-PAGE followed by fluorography or immunoblotting. Input standards for mSA import experiments were diluted with cold 2× sample buffer containing solubilized chloroplasts. Fluorography and fluorograph quantification were performed as previously described (Shi and Theg, 2013b). Independent experiments (n) were conducted on separate days with different chloroplast preparations. Replicates indicate individual import reactions conducted with the same chloroplast preparation.
Immunoblotting
SDS-PAGE resolved proteins were transferred to PVDF membranes. α-FLAG blots were blocked with 5% fat-free milk in TBST (150 mM NaCl, 0.1% Tween 20, and 50 mM Tris, pH 7.5) and probed with monoclonal α-FLAG (Invitrogen; MA1-91878) diluted 1:1000 in blocking buffer. Rabbit α-mouse IgG-HRP secondary antibody (SCBT; sc-358920) was diluted 1:10,000 in blocking buffer. α-Biotin blots were blocked with 1% gelatin in TBST and probed with Avidin-HRP (Sigma-Aldrich; A3151) diluted 1:10,000 in blocking buffer. Blots were incubated with ECL substrate (GE Amersham) and developed on film or with a ChemiDoc imager (Bio-Rad).
Fluorescence Spectroscopy
Fluorescence measurements were conducted in a Fluorolog 3-22 spectrofluorometer (Horiba) set to λex = 494 nm, λem = 518 nm, and 5-nm slit widths. Kinetics were measured in stirred 3-mL cuvettes.
Accession Numbers
Protein sequences from this article can be found under UniProt accession numbers P0ABQ4 (E. coli DHFR), P00375 (mouse DHFR), P07689 (RSSU), and P14226 (OE33).
Supplemental Data
Supplemental Figure 1. Wash steps effectively removed residual FMTX.
Supplemental Figure 2. DHFR/FMTX complex quantification is not confounded by other FMTX interactions.
Supplemental Figure 3. Import competition of tp22mDHFR/MTX with prOE33.
Supplemental Figure 4. Saturation curve of mSA import.
Supplemental Figure 5. Time-course import of RSSUFHC.
Supplemental Table 1. List of primers.
Acknowledgments
We thank Jonathan Ho and Johnathan Keilman for cloning and purifying RSSUHC and tp22DHFR, respectively. We thank Kentaro Inoue, Hsou-min Li, and Enoch Baldwin for very helpful discussions and Anthony Ho for assistance with fluorimetry. This work was supported in part by National Science Foundation Grant MCB-1330321 and Department of Energy Grant DE-SC0017035 to S.M.T.
AUTHOR CONTRIBUTIONS
I.G., L.-X.S., M.L., and S.M.T. designed research. I.G., L.-X.S., and M.L. performed research and contributed new reagents/analytical tools. I.G. and S.M.T. wrote the manuscript.
References
- Ainavarapu S.R., Li L., Badilla C.L., Fernandez J.M. (2005). Ligand binding modulates the mechanical stability of dihydrofolate reductase. Biophys. J. 89: 3337–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- America T., Hageman J., Guéra A., Rook F., Archer K., Keegstra K., Weisbeek P. (1994). Methotrexate does not block import of a DHFR fusion protein into chloroplasts. Plant Mol. Biol. 24: 283–294. [DOI] [PubMed] [Google Scholar]
- Arkowitz R.A., Joly J.C., Wickner W. (1993). Translocation can drive the unfolding of a preprotein domain. EMBO J. 12: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaraf Y.G., Seamer L.C., Schimke R.T. (1989). Characterization by flow cytometry of fluorescein-methotrexate transport in Chinese hamster ovary cells. Cytometry 10: 50–55. [DOI] [PubMed] [Google Scholar]
- Baud C., Guérin J., Petit E., Lesne E., Dupré E., Locht C., Jacob-Dubuisson F. (2014). Translocation path of a substrate protein through its Omp85 transporter. Nat. Commun. 5: 5271. [DOI] [PubMed] [Google Scholar]
- Best R.B., Fowler S.B., Herrera J.L., Steward A., Paci E., Clarke J. (2003). Mechanical unfolding of a titin Ig domain: structure of transition state revealed by combining atomic force microscopy, protein engineering and molecular dynamics simulations. J. Mol. Biol. 330: 867–877. [DOI] [PubMed] [Google Scholar]
- Bionda T., Tillmann B., Simm S., Beilstein K., Ruprecht M., Schleiff E. (2010). Chloroplast import signals: the length requirement for translocation in vitro and in vivo. J. Mol. Biol. 402: 510–523. [DOI] [PubMed] [Google Scholar]
- Bionda T., Gross L.E., Becker T., Papasotiriou D.G., Leisegang M.S., Karas M., Schleiff E. (2016). Eukaryotic Hsp70 chaperones in the intermembrane space of chloroplasts. Planta 243: 733–747. [DOI] [PubMed] [Google Scholar]
- Bölter B., Soll J. (2016). Once upon a time: Chloroplast protein import research from infancy to future challenges. Mol. Plant 9: 798–812. [DOI] [PubMed] [Google Scholar]
- Bonardi F., Halza E., Walko M., Du Plessis F., Nouwen N., Feringa B.L., Driessen A.J. (2011). Probing the SecYEG translocation pore size with preproteins conjugated with sizable rigid spherical molecules. Proc. Natl. Acad. Sci. USA 108: 7775–7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.A., Theg S.M. (1997). A folded protein can be transported across the chloroplast envelope and thylakoid membranes. Mol. Biol. Cell 8: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti B.J., Elferich J., Yang Z., Shinde U., Skach W.R. (2014). Cotranslational folding inhibits translocation from within the ribosome-Sec61 translocon complex. Nat. Struct. Mol. Biol. 21: 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degan P., Carpano P., Cercignani G., Montagnoli G. (1989). A fluorescence study of substrate and inhibitor binding to bovine liver dihydrofolate reductase. Int. J. Biochem. 21: 291–295. [DOI] [PubMed] [Google Scholar]
- Eilers M., Schatz G. (1986). Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature 322: 228–232. [DOI] [PubMed] [Google Scholar]
- Flores-Pérez Ú., Jarvis P. (2013). Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta 1833: 332–340. [DOI] [PubMed] [Google Scholar]
- Flugge U.I., Benz R. (1984). Pore-forming activity in the outer-membrane of the chloroplast envelope. FEBS Lett. 169: 85–89. [Google Scholar]
- Froehlich J.E., Keegstra K. (1997). Identification of a translocation intermediate occupying functional protein import sites in the chloroplastic envelope membrane. J. Biol. Chem. 272: 8077–8082. [DOI] [PubMed] [Google Scholar]
- Guéra A., America T., van Waas M., Weisbeek P.J. (1993). A strong protein unfolding activity is associated with the binding of precursor chloroplast proteins to chloroplast envelopes. Plant Mol. Biol. 23: 309–324. [DOI] [PubMed] [Google Scholar]
- Hageman J., Baecke C., Ebskamp M., Pilon R., Smeekens S., Weisbeek P. (1990). Protein import into and sorting inside the chloroplast are independent processes. Plant Cell 2: 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman B.D., Hendershot L.M., Johnson A.E. (1998). BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell 92: 747–758. [DOI] [PubMed] [Google Scholar]
- Heins L., Mehrle A., Hemmler R., Wagner R., Küchler M., Hörmann F., Sveshnikov D., Soll J. (2002). The preprotein conducting channel at the inner envelope membrane of plastids. EMBO J. 21: 2616–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldt H.W., Sauer F. (1971). The inner membrane of the chloroplast envelope as the site of specific metabolite transport. Biochim. Biophys. Acta 234: 83–91. [DOI] [PubMed] [Google Scholar]
- Hinnah S.C., Wagner R., Sveshnikova N., Harrer R., Soll J. (2002). The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides. Biophys. J. 83: 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhr A.I.C., Lindau C., Wirth C., Qiu J., Stroud D.A., Kutik S., Guiard B., Hunte C., Becker T., Pfanner N., Wiedemann N. (2018). Membrane protein insertion through a mitochondrial β-barrel gate. Science 359: eaah6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B., Heidrich E.S., Mehner-Breitfeld D., Brüser T. (2018). The TatA component of the twin-arginine translocation system locally weakens the cytoplasmic membrane of Escherichia coli upon protein substrate binding. J. Biol. Chem. 293: 7592–7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynds P.J., Robinson D., Robinson C. (1998). The sec-independent twin-arginine translocation system can transport both tightly folded and malfolded proteins across the thylakoid membrane. J. Biol. Chem. 273: 34868–34874. [DOI] [PubMed] [Google Scholar]
- Jascur T., Goldenberg D.P., Vestweber D., Schatz G. (1992). Sequential translocation of an artificial precursor protein across the two mitochondrial membranes. J. Biol. Chem. 267: 13636–13641. [PubMed] [Google Scholar]
- Kaufman R.J., Bertino J.R., Schimke R.T. (1978). Quantitation of dihydrofolate reductase in individual parental and methotrexate-resistant murine cells. Use of a fluorescence activated cell sorter. J. Biol. Chem. 253: 5852–5860. [PubMed] [Google Scholar]
- Kouranov A., Schnell D.J. (1997). Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 139: 1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacheva S., Bedard J., Wardle A., Patel R., Jarvis P. (2007). Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J. 50: 364–379. [DOI] [PubMed] [Google Scholar]
- Kovács-Bogdán E., Benz J.P., Soll J., Bölter B. (2011). Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol. 11: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K.H., Huang H., Pralle A., Park S. (2013). Stable, high-affinity streptavidin monomer for protein labeling and monovalent biotin detection. Biotechnol. Bioeng. 110: 57–67. [DOI] [PubMed] [Google Scholar]
- Liu L., McNeilage R.T., Shi L.X., Theg S.M. (2014). ATP requirement for chloroplast protein import is set by the Km for ATP hydrolysis of stromal Hsp70 in Physcomitrella patens. Plant Cell 26: 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S.M., Theg S.M. (2011). Protein targeting across and into chloroplast membranes. Methods Mol. Biol. 684: 139–157. [DOI] [PubMed] [Google Scholar]
- Matouschek A. (2003). Protein unfolding--an important process in vivo? Curr. Opin. Struct. Biol. 13: 98–109. [DOI] [PubMed] [Google Scholar]
- Ottado J., Ceccarelli E.A. (1998). A fully active FAD-containing precursor remains folded up to its translocation across the chloroplast membranes. Eur. J. Biochem. 253: 132–138. [DOI] [PubMed] [Google Scholar]
- Paila Y.D., Richardson L.G., Schnell D.J. (2015). New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J. Mol. Biol. 427: 1038–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rial D.V., Lombardo V.A., Ceccarelli E.A., Ottado J. (2002). The import of ferredoxin-NADP+ reductase precursor into chloroplasts is modulated by the region between the transit peptide and the mature core of the protein. Eur. J. Biochem. 269: 5431–5439. [DOI] [PubMed] [Google Scholar]
- Ruprecht M., Bionda T., Sato T., Sommer M.S., Endo T., Schleiff E. (2010). On the impact of precursor unfolding during protein import into chloroplasts. Mol. Plant 3: 499–508. [DOI] [PubMed] [Google Scholar]
- Sato T., Esaki M., Fernandez J.M., Endo T. (2005). Comparison of the protein-unfolding pathways between mitochondrial protein import and atomic-force microscopy measurements. Proc. Natl. Acad. Sci. USA 102: 17999–18004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel E., Driessen A.J., Hartl F.U., Wickner W. (1991). Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell 64: 927–939. [DOI] [PubMed] [Google Scholar]
- Schleiff E., Becker T. (2011). Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 12: 48–59. [DOI] [PubMed] [Google Scholar]
- Schleiff E., Soll J., Küchler M., Kühlbrandt W., Harrer R. (2003). Characterization of the translocon of the outer envelope of chloroplasts. J. Cell Biol. 160: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M.P., Matouschek A. (1999). The dimensions of the protein import channels in the outer and inner mitochondrial membranes. Proc. Natl. Acad. Sci. USA 96: 13086–13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.V., Theg S.M. (1996). A new chloroplast protein import intermediate reveals distinct translocation machineries in the two envelope membranes: energetics and mechanistic implications. J. Cell Biol. 132: 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. (2010). A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. (2013a). The chloroplast protein import system: from algae to trees. Biochim. Biophys. Acta 1833: 314–331. [DOI] [PubMed] [Google Scholar]
- Shi L.X., Theg S.M. (2013b). Energetic cost of protein import across the envelope membranes of chloroplasts. Proc. Natl. Acad. Sci. USA 110: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T., et al. (2015). Molecular architecture of the active mitochondrial protein gate. Science 349: 1544–1548. [DOI] [PubMed] [Google Scholar]
- Su P.H., Li H.M. (2010). Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter S.A., Theg S.M. (1998). Energy-transducing thylakoid membranes remain highly impermeable to ions during protein translocation. Proc. Natl. Acad. Sci. USA 95: 1590–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchette N.A., Perry K.M., Matthews C.R. (1986). Folding of dihydrofolate reductase from Escherichia coli. Biochemistry 25: 5445–5452. [DOI] [PubMed] [Google Scholar]
- Walker D., Chaddock A.M., Chaddock J.A., Roberts L.M., Lord J.M., Robinson C. (1996). Ricin A chain fused to a chloroplast-targeting signal is unfolded on the chloroplast surface prior to import across the envelope membranes. J. Biol. Chem. 271: 4082–4085. [DOI] [PubMed] [Google Scholar]
- Walton P.A., Hill P.E., Subramani S. (1995). Import of stably folded proteins into peroxisomes. Mol. Biol. Cell 6: 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]