Small Ubiquitin-like Modifier protein, SUMO, regulates jasmonic acid (JA) signaling by suppressing the JA receptor CORONATINE INSENSITIVE1 activity independently of JA levels.
Abstract
Plants respond rapidly to sudden environmental cues, often responding prior to changes in the hormone levels that coordinate these responses. How this is achieved is not fully understood. The integrative role of the phytohormone jasmonic acid (JA) relies upon the plant’s ability to control the levels of JASMONATE ZIM (JAZ) domain-containing repressor proteins. Here, we demonstrate that regardless of intrinsic JA levels, Small Ubiquitin-like Modifier (SUMO)-conjugated JAZ proteins inhibit the JA receptor CORONATINE INSENSITIVE1 (COI1) from mediating non-SUMOylated JAZ degradation. The SUMO-deconjugating proteases OVERLY TOLERANT TO SALT1 (OTS1) and OTS2 regulate JAZ protein SUMOylation and stability. The ots1 ots2 double mutants accumulate SUMOylated and non-SUMOylated JAZ repressor proteins but show no change in endogenous JA levels compared with wild-type plants. SUMO1-conjugated JAZ proteins bind to COI1 independently of the JA mimic coronatine. SUMO inhibits JAZ binding to COI1. We identify the SUMO interacting motif in COI1 and demonstrate that this is vital to SUMO-dependent inhibition of COI1. Necrotroph infection of Arabidopsis thaliana promotes SUMO protease degradation, and this increases JAZ SUMOylation and abundance, which in turn inhibits JA signaling. This study reveals a mechanism for rapidly regulating JA responses, allowing plants to adapt to environmental changes.
INTRODUCTION
In plants, growth must be integrated with changes in the natural environment. Modulation of hormone signaling pathways plays a key role in this process. Jasmonic acid (JA) regulates a wide spectrum of plant growth, developmental, and defense responses to pathogen attack. In this context, JA is a major coordinator of both constitutive developmental processes and in defense responses activated upon pathogen invasion. Conjugation of JA to the amino acid l-isoleucine produces the bioactive signal (3R,7S)-jasmonoyl-l-isoleucine (JA-Ile) (Fonseca et al., 2009). JA-Ile is structurally and functionally imitated by the phytotoxin coronatine produced by the bacterial pathogen Pseudomonas syringae (Feys et al., 1994). An important step in the elucidation of the jasmonate signaling pathway was made with the discovery of the JA receptor CORONATINE INSENSITIVE1 (COI1) that encodes an F-box protein acting as part of a Skip-Cullin-F-box E3 ubiquitin ligase complex, targeting proteins for proteasomal degradation (Xie et al., 1998).
The JASMONATE ZIM (JAZ) domain family of transcriptional repressors are the target substrates that associate with COI1 in a hormone-dependent manner (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008; Fonseca et al., 2009). JAZ repressors directly interact with and govern the activity of transcription factors that include the basic helix-loop-helix proteins MYC2, MYC3, and MYC4 that act redundantly to regulate a plethora of JA-mediated responses (Fernández-Calvo et al., 2011; Pauwels and Goossens, 2011). In the absence of a JA-Ile signal, JAZ proteins actively repress JA-responsive transcription factors. In response to environmental cues that upregulate JA signaling, the hormone binds to COI1 and stimulates specific binding to JAZ proteins. This leads to polyubiquitination and subsequent proteasomal degradation of JAZ proteins. JAZ degradation relieves repression of JA responsive transcriptional regulation leading to physiological changes. The integrative role of JA is heavily reliant on the plant’s ability to control JAZ protein levels; to date, this has been demonstrated to be controlled through modulating levels of JA-Ile. However, the static nature of plants dictates that they must respond rapidly to changing environments and often prior to changes in de novo JA levels. How this is achieved in plants is largely unknown.
Several ubiquitin-like proteins have been described in plants, including Small Ubiquitin-like Modifier (SUMO), which can act to stabilize proteins to which it is conjugated (Conti et al., 2008). Synthesized as an inactive precursor, SUMO proteins are processed to their mature form by SUMO proteases that cleave the C-terminal tail from the precursor. This exposes a di-glycine motif where target attachment occurs in a series of enzymatic reactions very similar to ubiquitination that includes activation, conjugation, and ligation (Jentsch and Pyrowolakis, 2000; Kerscher et al., 2006; Capili and Lima, 2007). To regulate the effects of SUMO-conjugated proteins, SUMOylation can also be reversed by SUMO-specific proteases, which release SUMO from their substrates (Hay, 2001). SUMO proteases are crucial as they function in both maturation and deconjugation. These two activities share a common catalytic mechanism, although the substrates differ in so much as maturation involves hydrolysis of an amino-linked peptide bond and deconjugation catalyzes the hydrolysis of lysine-glycine isopeptide bonds (Reverter and Lima, 2009). So far, only a few bona fide SUMO proteases have been characterized in Arabidopsis thaliana and rice (Oryza sativa) (Reeves et al., 2002; Conti et al., 2008; Srivastava et al., 2016). Previously, we identified two SUMO proteases, OVERLY TOLERANT TO SALT1 (OTS1) and OTS2 that are localized in the nucleus and act redundantly to regulate salt stress responses in Arabidopsis (Conti et al., 2008). OTS1/OTS2 regulate the abundance of SUMO conjugates in a salt stress-dependent manner and overexpressing OTS1 alone reduces salt-induced SUMO conjugate accumulation and can rescue the sensitivity of the ots1 ots2 double mutant to high salinity (Conti et al., 2008).
Once covalently conjugated, SUMO affects protein-protein interactions, subcellular localization, and stability of target proteins (Hay, 2001; Verger et al., 2003). Furthermore, SUMO may facilitate new protein-protein interactions through SUMO-interacting motifs (SIMs) and compete with other posttranslational modifications such as ubiquitination and acetylation (Kerscher et al., 2006; Hickey et al., 2012). Previously, we demonstrated that the sequestration of the GA receptor GID1 by SUMO-conjugated DELLAs leads to an accumulation of non-SUMOylated DELLAs by blocking their ubiquitination, thereby enabling beneficial growth restraint during stress (Conti et al., 2014). Here, we demonstrate a role for SUMOylation in stabilizing JAZ proteins by inhibiting COI1 from mediating JAZ repressor degradation. The SUMO protease OTS1 regulates JAZ protein stability. The ots1 ots2 double mutants accumulate SUMOylated and non-SUMOylated JAZ repressor proteins but show no change in endogenous JA levels compared with wild-type plants. SUMO1 conjugated JAZ proteins bind to COI1 independently of the JA mimic coronatine. SUMO1 inhibits JAZ binding to COI1. Botrytis cinerea infection of Arabidopsis promotes OTS1 SUMO protease degradation and consequently increases JAZ SUMOylation and abundance, inhibiting JA signaling. Our data reveal a SUMO-dependent attenuation mechanism for JA signaling in plants.
RESULTS
Mutants of the OTS SUMO Proteases Are Susceptible to the Fungal Pathogen B. cinerea and the Arthropod Herbivore Spider Mite, Tetranychus urticae
Previously, we demonstrated that the ots1 ots2 double mutant displayed enhanced resistance to virulent P. syringae pv tomato (Pst) and accumulated higher levels of salicylic acid (SA), compared with wild-type plants (Bailey et al., 2016). Furthermore, ots1 ots2 mutants exhibited upregulated expression of the SA biosynthesis gene ISOCHORISMATE SYNTHASE1 and enhanced SA-responsive PR1 expression as compared with the wild type. SA stimulates OTS1/2 degradation and promotes accumulation of SUMO1/2 conjugates. These results indicate that OTS1 and OTS2 act in a feedback loop in SA signaling and de novo OTS1/2 synthesis works antagonistically to SA-promoted degradation, thereby adjusting the abundance of the OTS1/2 to moderate SA signaling.
The SA and JA signaling pathways often act antagonistically (Glazebrook, 2005; Kazan and Manners, 2008; Koornneef and Pieterse, 2008; Pieterse, 2012). Although there are exceptions, generally it can be stated that pathogens with a predominantly biotrophic lifestyle are more sensitive to SA-induced defenses, whereas JA activates defense against necrotrophic pathogens and herbivorous insects (Glazebrook, 2005; Howe and Jander, 2008). Since, ots1 ots2 mutants were more resistant to Pst due to increased SA levels, we wanted to ascertain if they were more susceptible to a necrotrophic fungal pathogen, B. cinerea, which causes gray mold disease (Mengiste, 2012) and an arthropod herbivore (red spider mite [T. urticae]), where JA is known to play a key role.
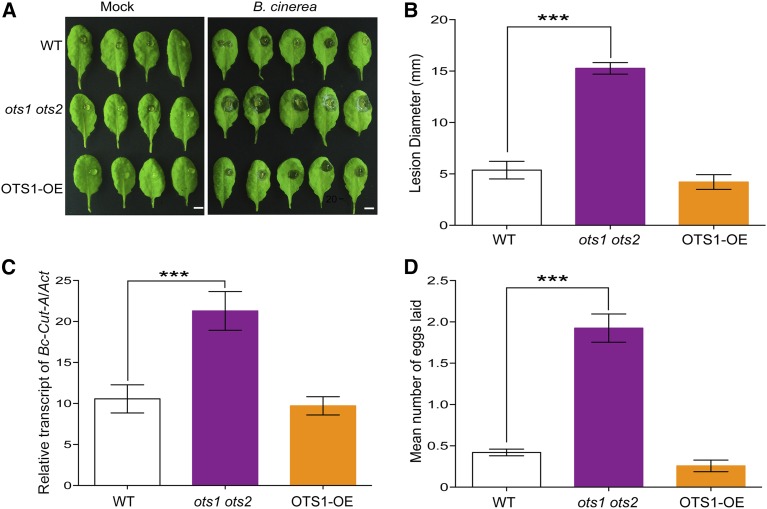
We compared the susceptibility of the wild type and ots1 ots2 double mutant and OTS1-overexpressing (OTS1-OE) (Bailey et al., 2016) plants to B. cinerea. Initially, we scored the size of the necrotic lesions on detached leaves to assess the resistance of the wild type and ots1 ots2 mutants to B. cinerea after inoculating plants with drops of fungal spore suspension onto the upper epidermis of rosette leaves. As shown in Figures 1A and 1B, disease lesions on detached leaves from the ots1 ots2 plants were significantly larger than that of the wild type, confirming that these SUMO proteases are required for resistance against B. cinerea. The previously established OTS1-OE line was also included in this analysis and showed no significant difference when compared with the wild type. The severity of symptoms seen in ots1 ots2 double mutants was also reflected in the increased fungal biomass, as indicated by RT-qPCR data that showed that ots1 ots2 mutants had ∼3-fold more fungal DNA compared with wild-type or OTS1-OE lines (Figure 1C). Spider mite fecundity assays indicated that the female mites laid significantly more eggs on ots1 ots2 mutant plants compared with the wild-type and OTS1-OE transgenic lines (Figure 1D), suggesting reduced resistance to insect herbivory in ots1 ots2 mutants. The lack of any observable phenotypic differences in disease development in the OTS1-OE lines maybe be attributed to the fact that the levels of OTS1 expression in these lines may not be significant enough to yield a tangible difference in defense phenotypes compared with wild-type plants, but they were able to complement the salt stress sensitivity phenotype in ots1 ots2 mutants (Conti et al., 2008; Bailey et al., 2016).
Figure 1.
OTS SUMO Proteases Regulate JA-Mediated Defense Responses.
(A) White light images of representative wild-type (WT), ots1 ots2, and OTS1-OE leaves showing cell death lesions at 72 h after inoculation with B. cinerea. Bars = 1 cm.
(B) Analysis of leaf lesion diameter of the wild type, ots1 ots2, and OTS1-OE at 72 h postinoculation with B. cinerea. Histograms represent the mean lesion diameter ± sd of at least 50 lesion sites from 10 plants for each genotype. Error bars represent sd from five biological replicates per experiment based on three independent experiments. Asterisks denote statistical significance of the differences between the wild type and ots1 ots2 calculated using Student’s t test (***P ≤ 0.001).
(C) Quantification of fungal growth by RT-qPCR of B. cinerea. Cutinase gene-specific primers using genomic DNA at 72 h postinoculation. Histograms represent the mean ± sd from three biological replicates. Asterisks denote statistical significance of the differences between the wild type and ots1 ots2 calculated using Student’s t test (***P ≤ 0.001).
(D) T. urticae egg counts on Arabidopsis plants 5 d after infestation with adult female mites. The average number of eggs produced per female on each genotype shown, along with standard errors. Asterisks denote statistical significance of the differences between the wild type and ots1 ots2 calculated using Student’s t test (***P ≤ 0.001).
OTS SUMO Proteases Regulate JA Responses
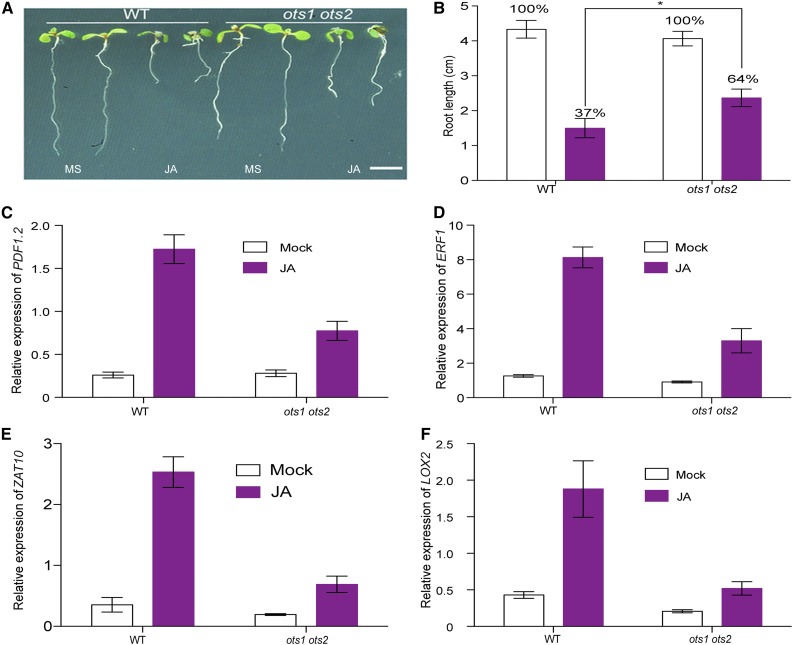
JA is well known to inhibit root growth, and this effect has been exploited in many genetic screens for plants with altered JA sensitivity. To determine the effect of JA on ots1 ots2 root growth, we grew seeds of wild-type and mutant plants in the presence of 10 μM JA and monitored root growth. Exogenous JA treatment caused significant root growth retardation in wild-type plants; however, this effect was reduced in ots1 ots2 plants (Figures 2A and 2B). ots1 ots2 mutants were at least 30% more resistant to JA mediated root growth inhibition compared with the wild type, implying that ots1 ots2 mutants are less sensitive to JA. This observation was substantiated by RT-qPCR data that demonstrated suppression of expression of downstream target genes of JA-mediated defense in ots1 ots2 mutant background (Figures 2C to 2F). Taken together, our data demonstrate that ots1 ots2 mutants have hampered JA sensitivity and/or signaling. Hence, the ots1 ots2 mutant reveals a link between SUMOylation and JA signaling.
Figure 2.
OTS SUMO Proteases Regulate JA-Mediated Growth Responses.
(A) Images of Arabidopsis seedlings of different genotypes on MS agar plates with and without JA, indicating that loss of function SUMO protease mutant ots1 ots2 shows decreased sensitivity (measured as inhibition of root growth) to exogenous JA. The wild type and ots1 ots2 mutants were grown on MS medium without or with 10 µM JA for 8 d. Bar = 1 cm.
(B) Quantification of root growth under exogenous JA treatment against that without JA treatment (designated as 100%). Values are mean ± sd of at least 20 plants of each genotype. Error bars represent sd from three biological replicates. Asterisks denote statistical significance of the differences between the wild type and ots1 ots2 calculated using Student’s t test (*P ≤ 0.05).
(C) to (F) Relative transcript levels of JA-responsive genes PDF1.2, ERF1, ZAT10, and LOX2 were measured in the wild type and ots1 ots2 mutant with and without JA treatment. Twelve-day-old seedlings were treated with 50 µM JA for 6 h, and seedlings without JA treatment were used as a mock control. Values are means ± sd of three biological replicates. At least 50 seedlings were combined into one replicate.
JAZ Proteins Are HyperSUMOylated and Stabilized in ots1 ots2 Mutants
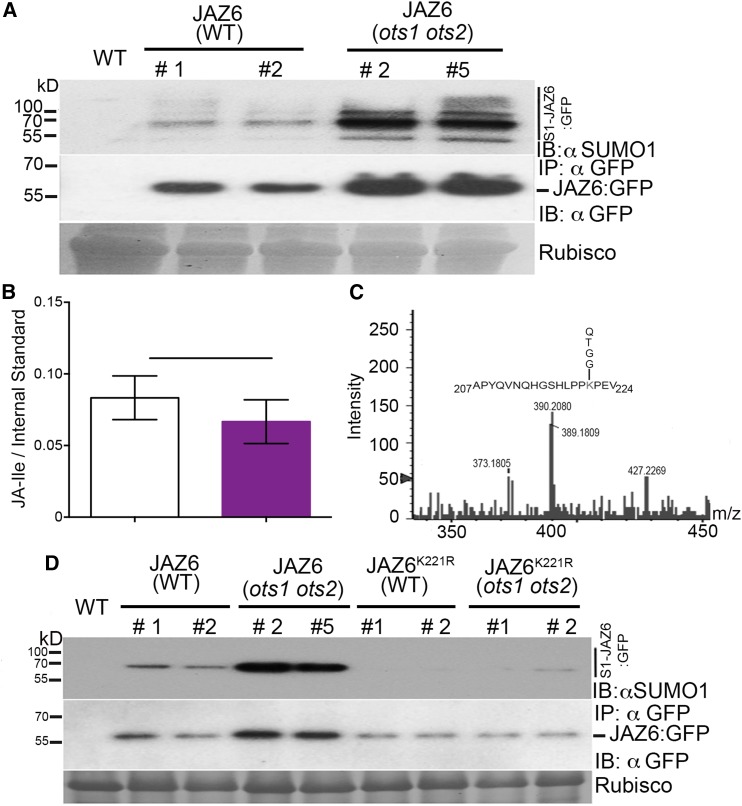
The 13 members within the JAZ repressor protein family collectively act by interacting with and regulating various transcription factors to influence diverse JA responses. This provides an overall canonical mechanism for repression of JA signaling. However, individual JAZ repressors affect specific aspects of JA signaling (Kazan and Manners, 2008). JAZ6 and 5 are known to be active in JA-mediated defense, but JAZ1 and 2 are more important for root growth (Grunewald et al., 2009; Ingle et al., 2015). Since the ots1 ots2 mutants display reduced sensitivity to JA in both defense and root growth, we wanted to ascertain the impact of the ots1 ots2 mutations on JAZ1 and JAZ6 protein abundance as proof of concept of the increased repression of the canonical mechanism for JA signaling in the ots1 ots2 mutants. Immunoblot experiments with anti-GFP antibodies revealed that 35S promoter-driven GFP-tagged JAZ1 (35S::JAZ1:GFP) and JAZ6 (35S::JAZ6:GFP) proteins were more abundant in the ots1 ots2 mutant plants compared with the wild type (Figure 3A; Supplemental Figure 1). There was no significant difference in the transcript levels of both transgenes in either genetic background (Supplemental Figure 2). OTS1 and OTS2 are SUMO proteases capable of cleaving SUMO from target proteins; therefore, we wanted to ascertain whether SUMOylation of JAZ proteins could provide a mechanism for stabilizing JAZ proteins in the ots1 ots2 background. We immunopurified the Arabidopsis JAZ6:GFP protein using GFP antibody-coated beads. Immunoblotting of GFP immunoprecipitates with Arabidopsis SUMO1-specific antibodies indicated that JAZ6:GFP was conjugated to SUMO1 (Figure 3A, upper panel). We also observed a similar pattern of SUMOylation for JAZ1:GFP (Supplemental Figure 1). This evidence indicated that the stability of JAZs as well as the SUMOylation of JAZ proteins are enhanced in the ots1 ots2 background. The increased abundance of JAZ6:GFP and JAZ1:GFP levels were not due to changes in JA levels as hormone measurements indicated that there was no significant difference in JA-Ile levels between ots1 ots2 mutants and the wild type (Figure 3B). This suggests a direct link between JAZ SUMOylation and its stability, a mechanism consistent with increased repression of the JA responses observed in these mutant plants. Intriguingly, the lack of any significant change in JA levels in the ots1 ots2 mutant plants indicate that this repression mechanism operates independently of intrinsic JA levels.
Figure 3.
Arabidopsis JAZ6 Protein Is SUMOylated.
(A) Immunoprecipitations (IP: αGFP) from total proteins derived from 4-week-old plant leaves of the wild type (WT) or 35S::JAZ6:GFP (wild type background) or 35S::JAZ6:GFP (ots1 ots2 background). Immunoprecipitated proteins were immunoblotted (IB) and probed with anti-GFP (αGFP) or anti-AtSUMO1/2 antibodies. S1-JAZ6:GFP indicates SUMOylated JAZ6:GFP proteins. Molecular masses are indicated on the left in kilodaltons. Ponceau staining indicating Rubisco levels was employed to determine protein loading for the immunoprecipitation. Wild-type (nontransgenic) plants served as a negative control.
(B) Estimation of JA-Ile concentrations through mass spectrometry analysis from 12-d-old seedlings of the wild type and ots1 ots2 mutant. Data presented are mean from three biological replicates. Error bars indicate sd of the means and no significant difference was observed between the genotypes after Student’s t test analysis.
(C) Relevant section of mass spectra obtained from JAZ6-SUMO1 conjugated peptide fragmentation experiments. The peak representing JAZ6 peptide sequence carrying a SUMO1 signature peptide fragment QTGG on residue Lsy-221 is indicated on the amino acid sequence.
(D) Immunoblots indicating reduced SUMOylation and protein abundance of 35S::JAZ6K221R:GFP in the wild type or 35S::JAZ6K221R:GFP in the ots1 ots2 backgrounds compared with the 35S::JAZ6:GFP in wild-type and ots1 ots2 background. Proteins were immunoblotted (IB) and probed with anti-GFP (αGFP) or anti-AtSUMO1/2 (αSUMO1) antibodies. S1-JAZ6:GFP indicates SUMOylated JAZ6:GFP proteins. Molecular masses are indicated on the left in kilodaltons. Ponceau staining indicating Rubisco levels was employed to determine protein loading for the immunoprecipitation (IP:αGFP). Wild-type (nontransgenic) plants served as a negative control.
To determine the site of SUMO conjugation on JAZ6, we exploited the bacterial SUMO conjugation system (Okada et al., 2009) to purify higher order SUMO1-JAZ6 conjugates and subjected them to mass spectrometry analysis (Supplemental Figure 3). Trypsin cleavage of SUMO-conjugated peptides leaves a four-specific amino acid (QTGG) footprint when the mass spectrometry adapted SUMO1 (Miller et al., 2010) is used to conjugate to target proteins. The peptide carrying this unique mass footprint can be manually identified from fragmented ion mass spectra of a target protein. Using this method, we successfully identified Lys-221 in JAZ6 as a SUMO1 attachment site (Figure 3C).
To test the hypothesis that SUMOylation on JAZ6K221 was responsible for the increased stability of JAZ6 we produced transgenic plants ectopically expressing via the 35S promoter, mutagenized versions of JAZ6 lacking the relevant SUMO attachment site (lysine-to-arginine mutation at position 221, K to R) (35S::JAZ6K221R:GFP) in the wild-type and ots1 ots2 backgrounds. Anti-GFP immunoblot analysis revealed that JAZ6 levels in the ots1 ots2 genetic background reverted to those levels seen in the wild-type background (Figure 3D), even though there was no significant difference in transcript levels of JAZ6 in either backgrounds (Supplemental Figure 4). We also observed a drastic reduction in the SUMOylation of JAZ6K221R:GFP (Figure 3D, upper panel). These observations, together with the finding that JAZ6 repressor accumulates in ots1 ots2 background indicate that SUMOylation of JAZ repressors modulates JA signaling.
SUMOylation of JAZ6 Modulates the Stability of JAZ Repressor after JA Treatment
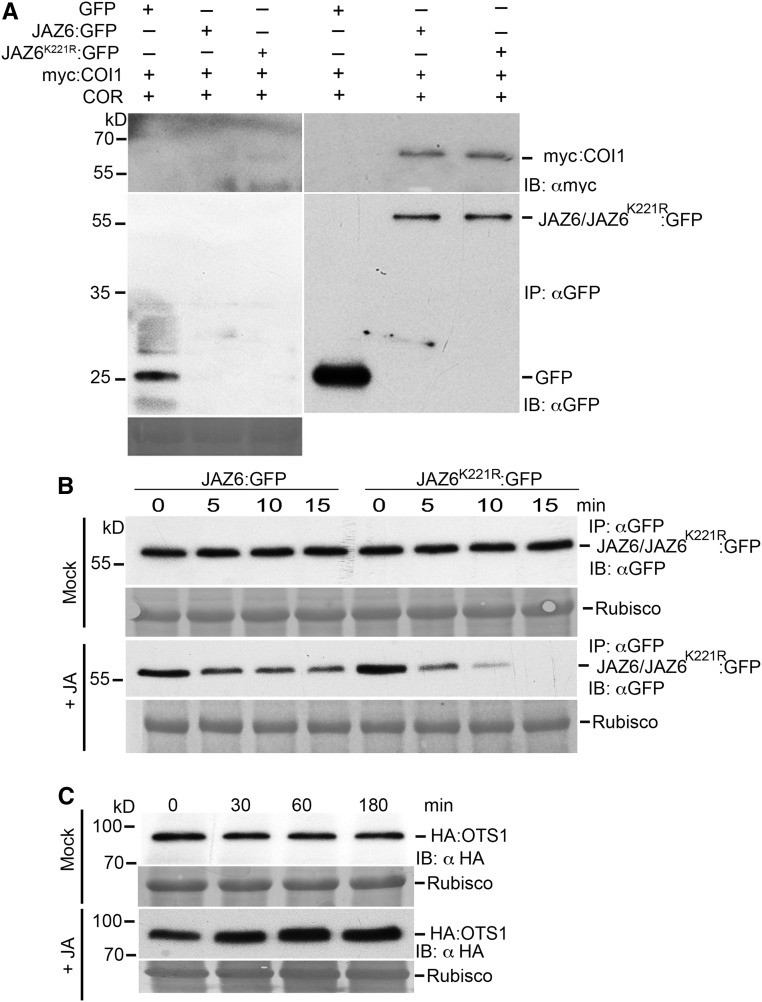
We investigated the interaction of mutated JAZ6 (JAZ6K221R) with COI1 using coimmunoprecipitation with anti-GFP beads of JAZ6:GFP and JAZ6K221R:GFP with myc tagged COI1 (myc-COI1) in the Nicotiana benthamiana transient assay system. Immunoblotting with anti-GFP and anti-myc antibodies allowed us to ascertain that the variant of JAZ6 with the mutated SUMO site actively interacted with myc-COI1 in a coronatine (JA mimic) dependent manner and thus is still functional (Figure 4A).
Figure 4.
JAZ6 SUMO Site Mutation Affects the Stability of JAZ6 Protein but Does Not Affect Its Interaction with COI1.
(A) Coimmunoprecipitation of myc:COI1 with GFP only, JAZ6:GFP, and JAZ6K221R:GFP was performed in planta using N. benthamiana transient assays to investigate the interaction of JAZ6:GFP and JAZ6K221R:GFP with myc:COI1 protein. Immunoprecipitates (IP: αGFP) were analyzed by SDS-PAGE and immunoblots were probed with αGFP to detect JAZ6:GFP and JAZ6K221R:GFP and GFP alone and with αmyc to detect myc:COI1 proteins. Ponceau staining indicating Rubisco levels was employed to determine protein loading for the immunoprecipitation (IP:αGFP).
(B) JA-mediated degradation of JAZ6:GFP and JAZ6K221R:GFP proteins. Immunoblot probed with anti-GFP antibodies showing protein levels of 35S::JAZ6:GFP and 35S::JAZ6K221R:GFP in respective seedlings treated with and without (mock treatment) JA (100 µM). Seedling samples were collected at the indicated time points. Ponceau staining indicating Rubisco levels was employed to determine protein loading for the immunoprecipitation (IP:αGFP).
(C) Immunoblots probed with αHA (IB: αHA) indicating the accumulation of HA:OTS1 protein in 12-d-old seedlings expressing 35S promoter driven HA-OTS1 transgene. Seedlings were treated with and without (mock) JA. Protein samples from seedlings were collected at the indicated time points. Ponceau red-stained Rubisco protein was used to indicate total protein levels.
JAZ6 is degraded in the presence of JA (Chini et al., 2007); therefore, we wanted to exploit this assay to determine the stability kinetics of JAZ6 and JAZ6K221R in the presence of JA. In a JAZ degradation time-course experiment, we treated JAZ6:GFP and JAZ6K221R:GFP seedlings with JA for varying periods of time, and, as indicated in Figure 4B, JAZ6K221R:GFP was more rapidly degraded and was undetectable after 15 min compared with JAZ6:GFP under the same conditions. JA treatment also promotes the accumulation of OTS1 protein (30% compared with control mock treatment as quantified by ImageJ against Rubisco), indicating that deSUMOylation of JAZ6 protein is enhanced within 30 min of JA treatment (Figure 4C). The increase in HA:OTS1 protein levels could be due to the downregulation of a potential ubiquitin E3 ligase that targets OTS1 for ubiquitin dependent proteasomal degradation. We have previously shown that OTS1 is degraded by salt and abscisic acid treatment in a proteasome-dependent manner (Conti et al., 2008; Srivastava et al., 2017). Therefore, it is likely that JA treatment triggers the downregulation of a yet undiscovered E3 ligase.
Collectively, our data indicate that SUMOylation at Lys-221 in JAZ6 is critical for its stability. The accumulation of OTS1 protein after JA treatment further supports the role of this SUMO protease as a regulator of JAZ-SUMOylation
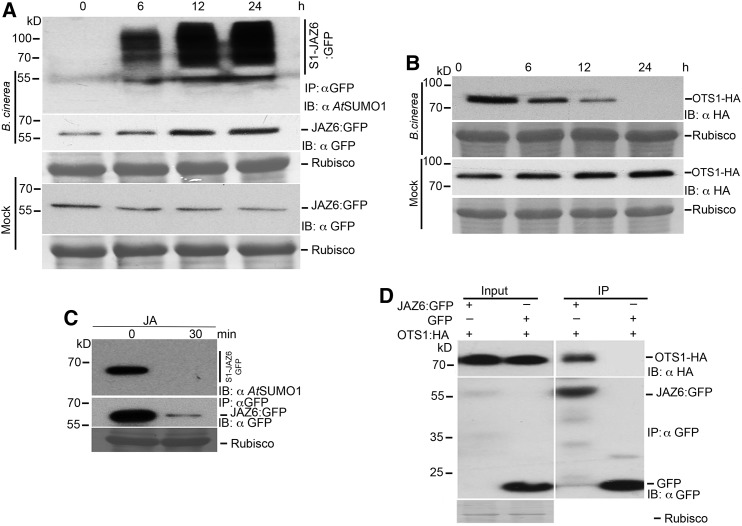
SUMOylation of JAZ6 Is Enhanced during B. cinerea Infection
Our data indicate a link between SUMOylation and JAZ protein stability through the OTS1 OTS2 SUMO proteases. We therefore hypothesized that JAZ protein SUMOylation and deSUMOylation may be a naturally occurring mechanism by which pathogens attenuate JA signaling in plants. To test this hypothesis, we challenged 35S::JAZ6:GFP transgenic plants with the virulent bacteria Pst and the fungal pathogen B. cinerea to investigate the status of JAZ6 SUMOylation. As shown in Supplemental Figure 5, JAZ6:GFP degradation begins to occur within 2 h after bacterial inoculation with a concomitant decrease in JAZ6:GFP SUMO conjugation. In contrast, B. cinerea infection leads to the accumulation of higher levels of JAZ6:GFP after 24 h and a striking increase in SUMOylated JAZ6:GFP levels (Figure 5A). This coincided with the degradation of OTS1 protein during B. cinerea infection (Figure 5B). However, JA treatment reduces SUMOylation of JAZ6:GFP with the concomitant reduction of total JAZ6:GFP protein to a similar level to that observed following Pst inoculation (Figure 5C). Coimmunoprecipitation experiments using Agrobacterium tumefaciens-mediated transient assays in N. benthamiana demonstrated that the OTS1 SUMO protease formed a protein complex with JAZ6:GFP (Figure 5D) indicating that OTS1 SUMO protease deSUMOylates JAZ repressors in the absence of pathogen infection. Taken together, our data indicate that necrotrophic pathogen attack leads to the degradation of the OTS1 SUMO protease that otherwise targets JAZ proteins for deSUMOylation. This leads to the accumulation of SUMOylated JAZ proteins, resulting in the attenuation of JA-mediated defense pathway. Since the JA pathway is vital for defense against B. cinerea, we postulate that targeted degradation of OTS1 resulting in JAZ6 protein accumulation is part of the virulence strategy of B. cinerea.
Figure 5.
SUMOylation of JAZ6 Is Enhanced during B. cinerea Infection.
(A) Immunoblots indicating significantly increased SUMOylation and protein abundance of GFP-tagged JAZ6 from 4-week-old (35S:JAZ6:GFP transgenics in wild-type background) plants infected with B. cinerea. Samples were collected at different time points after infection and mock-treated samples were used for immunoprecipitation with anti-GFP antibodies (IP: αGFP). Immunoblots (IB) were probed with GFP (IB:αGFP) or AtSUMO1/2 antibodies (IB:αSUMO1). Ponceau staining indicating Rubisco levels was employed to determine protein loading for the immunoprecipitation (IP:αGFP).
(B) Immunoblots probed with anti-HA antibodies showing HA-OTS1 levels in 35S::OTS1-HA transgenic Arabidopsis lines infected with B. cinerea. Four-week-old 35S::OTS1-HA transgenic Arabidopsis leaves were pressure infiltrated with B. cinerea and mock treated with magnesium chloride solution. Protein extracts were harvested from leaf samples collected at different time points after infection. Ponceau red stained Rubisco protein was used to indicate total protein levels.
(C) Immunoblots indicating greatly reduced SUMOylation and protein abundance of GFP-tagged JAZ6 from 15-d-old seedlings (35S:JAZ6:GFP transgenics in wild-type background) treated with 100 µM JA for 30 min. Protein samples were collected for immunoprecipitation with anti-GFP antibodies (IP: αGFP) at 0 and 30 min after treatment. Immunoblots (IB) were probed with GFP (αGFP) or AtSUMO1/2 antibodies (αSUMO1). Ponceau staining indicating Rubisco levels was employed to determine protein loading for the immunoprecipitation (IP:αGFP).
(D) Coimmunoprecipitation of HA-OTS1 with JAZ6:GFP in planta. Agrobacterium cultures containing 35S::HA-OTS1 were mixed with Agrobacterium cultures containing either 35S::GFP or 35S::JAZ6:GFP and transiently expressed in N. benthamiana. Total protein was extracted for immunoprecipitation with anti-GFP beads. Immunoprecipitates were analyzed by immunoblotting using anti-HA and anti-GFP antibodies to detect for the presence of OTS1-HA or JAZ6:GFP, respectively. Ponceau red stained Rubisco protein was used to indicate total protein levels used in the immunoprecipitation in the time points
SUMO Inhibits Binding of the JA Receptor COI1 to JAZ6 Protein
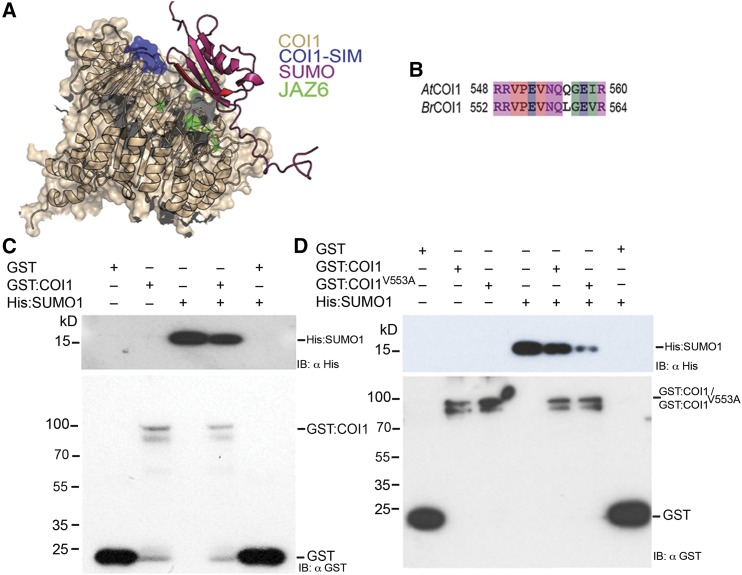
We have established that JAZ6:GFP protein is SUMOylated during B. cinerea infection and this leads to enhanced JAZ6:GFP stability. We also demonstrated that OTS SUMO proteases play a direct role in JAZ6:GFP protein stability by deSUMOylating JAZ6:GFP. We next investigated whether the SUMOylated JAZ6:GFP protein could interfere with the function of the JA receptor COI1, which is an F-box protein. Inspection of the Arabidopsis COI1 protein sequence revealed a conserved putative SIM at its C terminus (position 550–558 in Arabidopsis COI1; Figures 6A and 6B), which is also conserved in Brassica napus COI1. SIM motifs on proteins are specific consensus sequences that bind to SUMO and therefore mediate distinct protein-protein interactions (Minty et al., 2000). Depending on the ability to interact with the SIM containing proteins, SUMOylated proteins may influence cell functions (Hecker et al., 2006). These facts led us to hypothesize that SUMOylation of JAZ6 protein and the SIM in COI1 might have a significant role on COI1-JAZ interaction that results in modulating JA signaling pathway. Furthermore, a structural model of COI1 (Sheard et al., 2010) developed using PyMOL Graphics software based on the resolved structures of COI1 and JAZ suggested that free SUMO and SUMOylated JAZ proteins can occupy the same interaction face as non-SUMOylated JAZ for COI1 binding. This suggests that there may be competition between SUMOylated and non-SUMOylated JAZ proteins for COI1 binding through the SIM motif (Figure 6A).
Figure 6.
SUMO Inhibits JA Receptor COI1 Binding to JAZ Proteins.
(A) Side view of the COI1 JA receptor (beige) allows the identification of the location of the flexible loop forming the COI1 SIM (blue) residing at the interface between COI1 and the JAZ degron binding site (green). The binding of SUMO1 (pink) via its β-sheet (red) at this position can mask the COI1 domain that binds JAZ proteins. The binding of COI1 to SUMOylated JAZ through its SIM is therefore predicted to be able to disrupt binding of the non-SUMOylated JAZ to COI1.
(B) Cross-species alignment of COI1 SIM from Arabidopsis and Brassica. AtCOI1; Arabidopsis COI1; BrCOI1, B. rapa COI1. Residues are colored according to properties: red, hydrophobic; blue, acidic; magenta, basic; green, hydrophilic.
(C) GST pull-down assays between recombinant His:SUMO1 with recombinant GST:COI1 or GST only indicate that GST:COI1 binds to SUMO1.
(D) GST pull-down assays between recombinant His:SUMO1 with recombinant GST:COI1, SIM site-mutated GST:COI1V553A, or GST only. The data indicate that Val at position 553 is critical for SUMO1 binding.
To investigate the potential role of a SIM in COI1-JAZ6 interaction, we first used GST pull-down assays to examine possible interactions between COI1 and SUMO1 proteins. In the first in vitro binding experiment, COI1 was expressed in Escherichia coli as a GST fusion (GST:COI1) and immobilized on glutathione beads. SUMO1 (His:SUMO1) was expressed in E. coli and purified using nickel beads. Possible interaction between GST:COI1 and His:SUMO1 was examined by incubating the His-tagged SUMO1 protein with beads immobilized with GST:COI1 or GST only. After extensive washing of unbound molecules, the bound SUMO1 was detected by immunoblotting using a monoclonal anti-His antibody. As shown in Figure 6C, His:SUMO1 was retained on the GST:COI1 beads but not on the GST control beads, indicating that COI1 indeed possesses a bona fide SIM motif. To further validate the significance of COI1-SIM for SUMO1 binding, we mutated the core SIM amino acid residue, Val at position 553 of COI1 to Ala through site-directed mutagenesis (GST:COI1V553A) to potentially eliminate SUMO binding to COI1. GST:COI1V553A showed markedly decreased interaction with SUMO1 in comparison with its corresponding wild-type COI1, demonstrating the critical nature of the SIM motif in COI1 for SUMO1 binding (Figure 6D).
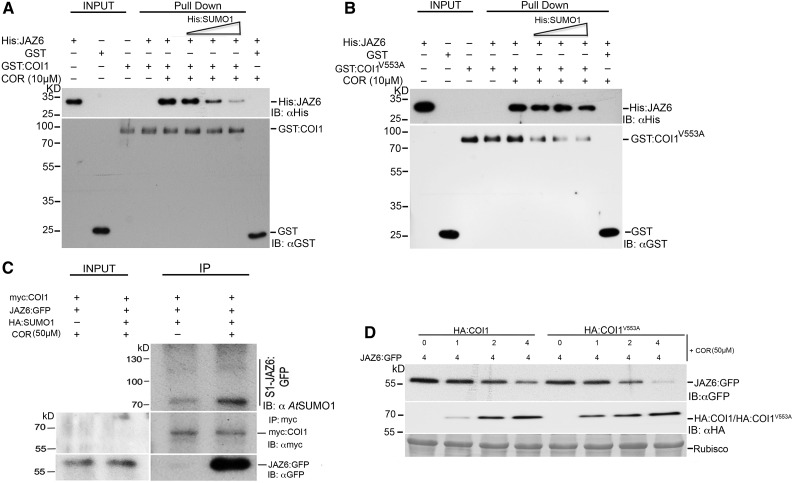
SUMOylated JAZ6 Inhibits Non-SUMOylated JAZ and COI1 Interaction
Coronatine is a high-affinity analog of JA-Ile (Katsir et al., 2008; Fonseca et al., 2009; Sheard et al., 2010) produced by pathogens to overcome SA-induced resistance (Brooks et al., 2004, 2005). Coronatine can mimic JA-Ile to relieve transcriptional repression of JA-responsive genes by promoting the interaction of the COI1 F-box protein with the JAZ transcriptional repressors. To test whether SUMO1 affected the interaction between COI1 and JAZ6, we performed in vitro GST pull-down assays between GST:COI1 and His:JAZ6 with increasing amounts of recombinant His:SUMO1. This experiment demonstrated that COR-dependent interaction of GST-COI1 with His:JAZ6 can be inhibited by His:SUMO1 (Figure 7A). However, the inhibitory effect of His:SUMO1 was significantly less efficient when GST-tagged COI1V553A SIM mutant was used instead of the wild-type COI1 with His:JAZ6 (Figure 7B). We also examined the possible in vivo interaction of SUMOylated JAZ6 and COI1 in planta via coimmunoprecipitation assays. SUMOylated JAZ6:GF interacts with myc-COI1 in planta independently of coronatine (Figure 7C), suggesting that this mechanism operates regardless of endogenous JA levels.
Figure 7.
SUMOylated JAZ6 Negatively Regulates COI1-JAZ6 Interaction.
(A) GST pull-down assays indicate that interaction between His:JAZ6 and GST:COI1 is weakened by the addition of increasing amounts of His:SUMO1 protein. His:JAZ6 protein mixed with different amounts of His:SUMO1 and pulled down with either GST:COI1 or GST alone in the presence or absence of coronatine (10 µM). The eluates were then probed with anti-His tag (αHis) or anti-GST (αGST) antibodies to detect His:JAZ6 or GST-tagged proteins, respectively.
(B) GST pull-down assays performed as above but with GST:COI1 replaced by the COI1 SIM mutant GST:COI1V553 indicate that mutation of Val to Ala rescues the interaction between GST:COI1V553A and His:JAZ6 even in the presence of His:SUMO1 protein. The eluates were probed with anti-His tag (αHis) or anti-GST (αGST) antibodies to detect His-JAZ6 or GST-tagged proteins, respectively.
(C) Coimmunoprecipitation of JAZ6:GFP with myc:COI1 in planta indicates that SUMOylated JAZ6:GFP binds to myc:COI1 even in the absence of JA mimic coronatine. Agrobacterium culture containing 35S::JAZ6:GFP was mixed with Agrobacterium cultures containing both 35S::myc:COI1 and 35S::HA:SUMO1 and transiently expressed in N. benthamiana. Total protein was extracted for immunoprecipitation with anti-myc antibodies (IP; αmyc) to pull down myc-COI1 and the immunoprecipitates were probed with anti-SUMO, anti-myc (IB: αmyc), and anti-GFP(IB: αGFP) antibodies to detect for the presence of SUMOylated and non-SUMOylated JAZ6:GFP and myc:COI1. Ponceau staining indicating Rubisco levels was employed to determine protein loading for the immunoprecipitation (IP:αGFP).
(D) In vivo degradation of JAZ6 was observed in coinfiltration experiments with increasing amounts of HA:COI1 or HA:COI1V553A in presence of 50 µM coronatine. The ratio of the relative concentration of agrobacteria used in the different coinfiltrations is indicated by numbers (top). Cell extracts were analyzed by immunoblot analysis with anti-GFP and anti-HA antibodies. Immunoblot analysis indicated that JAZ6:GFP was more unstable in plants transiently expressing HA:COI1V553A when compared with plants expressing HA:COI1. Ponceau red-stained Rubisco protein was used as a loading control.
The enhanced interaction between the GST-tagged SIM mutant of COI1 (COI1V553A) and His:JAZ6, even in the presence of His:SUMO1, raises the possibility that JAZ proteins may be degraded more rapidly in the presence of the COI1V553A SIM mutant. We therefore tested HA:COI1 and HA:COI1V553A mediated degradation of JAZ6:GFP in N. benthamiana transient assays. The results showed that plants expressing HA:COI1V553A degrade JAZ6:GFP more rapidly than plants expressing wild-type HA:COI1 in the presence of coronatine (Figure 7D; Supplemental Figure 6). These data demonstrate that HA:COI1V553 is not only active as a JA receptor but, since it is not under the repression of SUMO1, HA:COI1V553A is more potent in mediating JAZ6:GFP degradation. These observations provide a mechanism for SUMOylated JAZ to disrupt the interaction of COI1 with non-SUMOylated JAZ allowing the accumulation of the repressor. Since SUMOylated JAZ6:GFP interacted with myc-COI1 even in the absence of the JA mimic coronatine, it is highly likely that this COI1 inhibition by SUMO1 is JA independent.
COI1 SIM Mutant Suppresses JA Insensitivity of SUMO Protease (ots1 ots2) Mutants
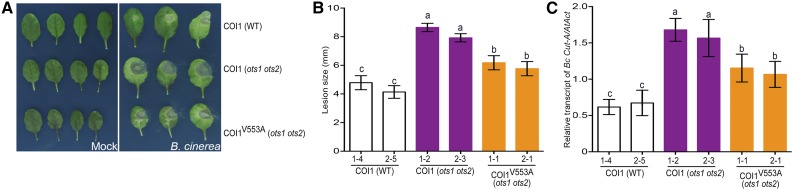
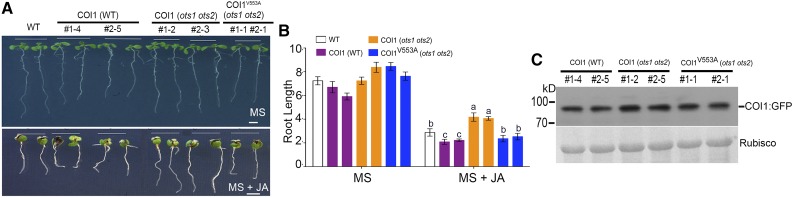
COI1 forms a functional E3 ubiquitin ligase SCFCOI1 and acts as an essential component of JA perception machinery by stimulating the degradation of JAZ proteins (Chini et al., 2007; Pauwels and Goossens, 2011). Results from Figure 7 indicate that SUMO inhibits COI binding to JAZ proteins via the SIM motif therefore we postulated that disrupting the SIM motif in COI1 should lead to increased JA signaling by promoting faster degradation of JAZ repressors as seen in the N. benthamiana transient assays (Figure 7D). To test the significance of the SIM motif of COI1 in JA signaling, we generated Arabidopsis transgenics overexpressing (under the 35S promoter) wild-type COI1 (35S::COI1:GFP) or the SIM variant COI1V553A (35S::COI1V553A:GFP) in the ots1 ots2 double mutant background where there are higher levels of both SUMOylated JAZ and non-SUMOylated JAZs. We anticipated that by overexpressing the SIM disrupted COI1V553A:GFP, we should overcome JA insensitivity mediated by increased JAZ levels in the ots1 ots2 double mutant. As controls, we also expressed wild-type COI1 (35S::COI1:GFP) in the wild-type Col-0 background. Both in B. cinerea infection assays (Figures 8A to 8C) and in root growth inhibition assays (Figures 9A to 9C), we observed that the transgenic plants expressing COI1V553A:GFP (in the ots1 ots2 background) were more sensitive to JA than the corresponding plants expressing wild-type COI1:GFP (Supplemental Figure 7). The comparable protein levels of the respective transgenes (Figure 9C) demonstrated that SUMOylated JAZ proteins suppress JA signaling by inhibiting COI1 from targeting non-SUMOylated JAZ proteins for ubiquitin-dependent degradation.
Figure 8.
COI1 SIM Mutant Suppresses B. cinerea Susceptibility.
(A) White light images of representative leaves from 4-week-old transgenic plants expressing 35S::COI1:GFP in the wild type (wild type), ots1 ots2, and 35S::COI1V553A:GFP in the ots1 ots2 background at 72 h after infection with mock (left panel) or B. cinerea spores (right panel).
(B) Quantification of lesion sizes on rosette leaves at 72 h after infection with B. cinerea spores. Values represent the means ± sd of three biological replicates of 4-week-old transgenic plants. The letters indicate averages that are statistically significantly different from each other (*P < 0.05 and **P< 0.01).
(C) Quantification of fungal growth by real-time PCR on B. cinerea genomic DNA with B. cinerea cutinase gene-specific primers at 72 h after infection. Histograms represent the means ± sd of three biological replicates of 4-week-old transgenic plants. The letters indicate averages that are statistically significantly different (P < 0.05) from each other. n = 15 to 20, 4-week-old plants in each replicate.
Figure 9.
COI1 SIM Mutant Restores JA Sensitivity in the ots1 ots2 Mutant Background.
(A) Image of representative 10-d-old seedlings grown in MS and MS + JA (10 μM) and the effect of JA on root length of different transgenic plants. Bar = 1 cm.
(B) Mean root length of 10-d-old seedlings in the presence of 10 μM JA relative to the controls. Values represent the means ± sd of three biological replicates. The letters indicate significant differences between the wild type and the transgenic lines of COI1 (wild type), COI1 (ots1 ots2), and COI1V553A (ots1 ots2) in presence of JA. n = 35 to 40 seedlings each replicate.
(C) Immunoblots probed with αGFP indicating COI1:GFP and COI1V553A:GFP protein levels in the wild type and ots1 ots2 background.
DISCUSSION
Given the importance of jasmonates as endogenous developmental regulators in plants, and as primary responders against pathogen attack, improving our understanding of their mechanisms of recognition and signaling has far-reaching importance for plant biology. This study reveals a control feature of the JA pathway with demonstrable implications for developmental processes and adaptive responses in plants.
The SUMO-SIM interaction is emerging as a key theme in molecular signaling in a wide range of organisms (Geiss-Friedlander and Melchior, 2007). This study describes how the SUMO-SIM “molecular glue” paradigm operates within plants to block ubiquitination of target proteins (sequestering COI1 needed for ubiquitinating JAZ repressors). Through this study, we unravel a mechanism for attenuating JA signaling through SUMOylation of the JAZ repressor proteins. Three clear lines of evidence support this conclusion. First, hormone analysis indicates that there is no significant change in JA levels in the ots1 ots2 double mutants, although these mutants accumulate JAZ1 and 6 repressor proteins as well as their SUMOylated forms. Second, in Figure 7D, we provide data on JAZ6 degradation kinetics demonstrating that, at the same level of JA, the COI1 SIM mutant (that is no longer under SUMO mediated repression) is more efficient in causing the degradation of JAZ6:GFP compared with the wild type. Third, Figure 7C shows that only SUMOylated JAZ6:GFP, but not JAZ, is able to interact with myc-COI1 in the absence of the JA mimic (coronatine). This study identifies a mechanism that can operate independently of JA to suppress COI1 activity. This provides direct evidence that intrinsic hormonal levels in planta do not affect SUMOylated JAZ from inhibiting COI1 and degrading non-SUMOylated JAZ and therefore attenuating JA signaling. This mechanism may allow plants to develop a rapid adaptive response prior to changes in JA levels. Attenuation of JA signaling has been reported to occur by an increase in JAZ repressor gene expression (Chico et al., 2008) and by the degradation of Je-Ile (Aubert et al., 2015; Smirnova et al., 2017). Here, we identify a posttranslational mechanism for repressing JA signaling that operates within hours of B. cinerea infection that does not require changes in JA levels. This process affords a new layer of regulation in hormone signaling, allowing plants to rapidly apply “brakes” on JA responses without the need for changes in hormone levels.
The JA mimic coronatine is an important component of the armory of phytopathogenic P. syringae used to infect and cause disease in Arabidopsis. Mutants deficient in JA signaling were found to be more resistant to virulent Pst as in ots1 ots2 double mutants that also have elevated SA levels (Bailey et al., 2016; de Torres Zabala et al., 2016). Coronatine-mediated activation of JA signaling contributes to disease development and Arabidopsis challenged with coronatine deficient Pst also have enhanced levels of SA (Laurie-Berry et al., 2006; Geng et al., 2012). A JA signaling repression mechanism that operates independently of coronatine will allow plants to counter Pst infection and evidence that de novo JA levels accumulated very late in Pst infected Arabidopsis argues for the existence of such a mechanism (de Torres Zabala et al., 2016). Here, we postulate that plants have exploited the SUMO system to attenuate JA signaling to enhance defense against biotrophic and hemibiotrophic phytopathogens.
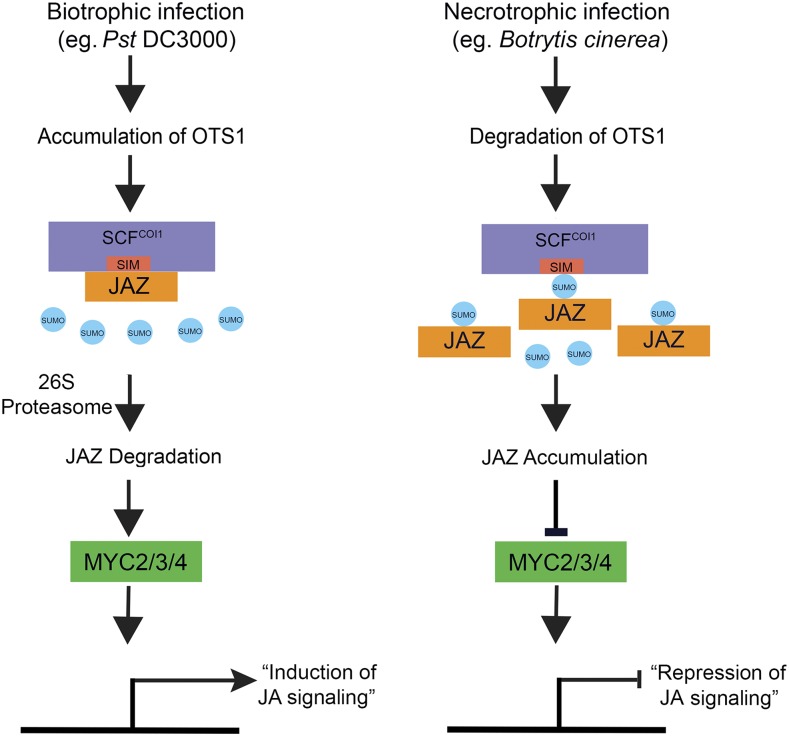
In our model (Figure 10), we suggest that virulent Pst infection promotes deSUMOylation of JAZ repressors, which promotes COI1-JAZ interaction to activate JA signaling. Activation of JA signaling pathways results in the suppression of SA signaling. By contrast, during necrotrophic infection such as B. cinerea, OTS SUMO protease is degraded, and this upregulates SUMOylation of JAZ6:GFP protein possibly by the change in equilibrium in favor of SUMO E2s, which have been known to SUMOylate targets directly, and/or SIZ1 SUMO E3, which have been implicated in pathogen responses in Arabidopsis (Lee et al., 2007). The consequent reSUMOyation of JAZ proteins would inhibit JAZ6-COI1 interaction preventing JAZ6 degradation thus repressing JA signaling.
Figure 10.
A Model for Repression of JA Signaling by SUMOylated JAZ Proteins.
During biotroph infection, such as by Pst, bacterial coronatine promotes JA signaling by activating the 26S proteasome-mediated degradation of JAZ repressors by the JA receptor COI1. Part of this process involves JA-mediated accumulation of the SUMO deconjugating protease OTS1 that rapidly deSUMOylates JAZ repressors and facilitates COI1 access to JAZ for degradation. JAZ repressor turnover activates JA-responsive gene expression through the transcriptional regulators, such as MYC2/MYC3/MYC4. On the other hand, infection by necrotrophs, such as the fungal pathogen B. cinerea, stimulates degradation of the SUMO-deconjugating protease OTS1. This leads to the accumulation of SUMOylated JAZ proteins (this does not preclude increased SUMO conjugating via hitherto unknown mechanisms) that inhibit COI1 mediated degradation of non-SUMOylated JAZ repressors, consequently, suppressing JA signaling.
Interestingly, the SUMO site in JAZ6 is located in the C-terminal JAS motif that has been shown to interact with, not only COI1, but also a range of transcription factors (Yan et al., 2007; Melotto et al., 2008; Staswick, 2008) whose activity is repressed by JAZ proteins. The impact of SUMOylation on JAZ repressor interaction with cognate transcription factors is not known and requires further investigation.
DELLA growth regulators restrain plant growth, whereas gibberellic acid (GA) promotes growth by targeting DELLAs for destruction. Different studies have demonstrated that DELLA restraint is a crucial mechanism for plants to modulate growth according to environmental cues (Achard et al., 2008; de Lucas et al., 2008; Achard and Genschik, 2009). We previously demonstrated that a proportion of DELLAs are conjugated to the SUMO protein and the extent of conjugation increases during stress, similar to JAZ1 and JAZ6. We identified a SIM in the GA receptor GID1 and demonstrated that SUMO-conjugated DELLA binds to this motif in a GA-independent manner (Conti et al., 2008). The consequent sequestration of GID1 by SUMO-conjugated DELLAs leads to an accumulation of non-SUMOylated DELLAs resulting in beneficial growth restraint during stress. For example, DELLAs sequester light responsive and phytochrome interacting transcription factors, such as PIF3 and PIF4, and inhibit hypocotyl elongation in the light (de Lucas et al., 2008; Feng et al., 2008). In this context, the JAZ proteins appear to play an analogous role in inhibiting transcription factor activity. Primary root growth of ots1 ots2 mutants is less hindered by exogenous JA treatment, and this is likely to be due to the suppression of the inhibitory effect of MYC2 on root development due to the accumulation of JAZ proteins in the ots1 ots2 mutants.
In all of these cases, the common central thread is the relative abundance of DELLAs and JAZ repressors, which is modulated by changes in GA and JA levels, respectively. We have demonstrated that dwarfism can be reversed independently of GA levels by modifying the SUMOylation status of DELLAs and that this mechanism is particularly important for plant growth under stress (Conti et al., 2014). Recent evidence indicates that DELLA and JAZ proteins directly interact to mediate crosstalk between GA and JA. The discovery that both DELLA and JAZ proteins are SUMOylated leads to the possibility that SUMO may provide a new facet to this crosstalk. Thus, this study provides an important insight into the integrative role of hormones in controlling plant growth and defense.
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana lines used in this study are in Col-0 (wild type) background. Seeds were plated on 0.5× Murashige and Skoog (MS) medium, and 0.8% agar (w/v) on vertical plates, stratified for 2 d at 4°C, and then transferred to growth chambers at 21°C under a long-day cycle (16 h light/8 h dark, 300 µmol m−2 s−1, provided by a combination of cool-white fluorescent bulbs, supplemented by incandescent lighting). The ots1 ots2 double mutants were used as described previously (Bailey et al., 2016).
Agrobacterium-Mediated Transformation of Arabidopsis for the Generation of Transgenic Plants
The constructs were transformed into Agrobacterium tumefaciens GV1301 and transferred into Arabidopsis using the floral dip method. Agrobacterium cells containing the appropriate construct were collected by centrifugation and resuspended using 5% (w/v) sucrose solution until the OD600 of the cell suspension was 0.8. Silwet L-77, as a strong surfactant, was added to the sucrose solution to obtain a final concentration of 0.05% (v/v). Developing Arabidopsis inflorescences were dipped into Agrobacterium cell suspensions for ∼15 s, and transformed plants were subsequently grown in darkness horizontally for 16 to 24 h. The seeds of treated plants were harvested after Agrobacterium-mediated transformation and seedlings of transformants were obtained by glufosinate screening. Transgenic lines expressing JAZ6-GFP and COI1-GFP and their mutant variants in the ots1 ots2 double mutant and wild-type Col-0 were generated by floral dips of the respective constructs in Agrobacterium as described above. Two independent lines containing single insertions in T3 generation homozygous transgenic plants with comparable level of transcripts (Supplemental Figures 2 and 4) were used for further experiments.
Analysis of Disease and Herbivore Resistance
Pseudomonas syringae pv tomato DC3000 (Pst) was grown on King’s B medium plates with appropriate antibiotics and incubated for 2 d at 28°C. Pst infection was performed as previously described (Bailey et al., 2016). Briefly, bacterial cells were collected by centrifugation (2500g) and resuspended in 10 mM MgCl2. Pressure infiltration of Pst (cfu 1 × 106; OD600 = 0.002) was performed using a needleless syringe. Whole leaves were harvested at the indicated time point after bacterial challenge, frozen immediately in liquid nitrogen, and used for immunoblotting. Three independent experiments were performed for the protein accumulation analysis. Each replicate consisted of rosette leaves of at least three plants grown in individual pots. Collection of B. cinerea spores and plant inoculation was performed as described previously (Bailey et al., 2016). In short, B. cinerea was subcultured on sterile Petri dishes with potato dextrose agar medium 2 weeks prior to use of the spores. Subcultures were incubated in the dark at 25°C. Spores were harvested in water, and inoculums were filtered to remove hyphae and then resuspended in potato dextrose broth to a concentration of 105 spores/mL. Leaf 7 from each of the plants was detached and placed on a bed of 0.8% (w/v) agar in three plates. Half of the leaves were inoculated with 5-μL droplets of B. cinerea inoculum and the other half were mock inoculated with 5 μL of sterile potato dextrose broth. Each plate contains 24 infected and 24 uninfected randomly arranged leaves. Trays were covered with lids and kept under the same conditions as for plant growth, except that the relative humidity was raised to 90%. Lesion perimeters were determined from photographs taken 48 and 72 h after inoculation using image analysis software ImageJ (http://rsb.info.nih.gov/ij/). Mean lesion perimeters of 20 leaves from 20 plants of different genotypes were compared. Cultures of red spider mite (Tetranychus urticae) were maintained on French bean plants. Adult female mites were collected from stock plants and released onto leaves (five mites per plant) of Arabidopsis plants grown in controlled environments under standard conditions. After 5 d, eggs were counted using a binocular microscope.
Site-Directed Mutagenesis
Wild-type sequences of JAZ6 and COI1 were amplified by PCR from Arabidopsis and cloned into pENTR/D-TOPO (Invitrogen). Mutated versions of JAZ6 and COI1 were generated by site-directed mutagenesis using the pENTR/D-TOPO clones as template. Oligonucleotide primers used to introduce the mutations are listed in Supplemental Table 1. The introduction of mutations was confirmed by sequencing, performed both before and after introduction of the mutated JAZ6 and COI1 coding sequences into pEarlyGate103/201/203 destination vectors using LR Clonase (Invitrogen). The pEarlyGate 103/201/203 vector drives expression with the cauliflower mosaic virus 35S promoter with GFP, HA, and myc tags. The GFP-tagged constructs were introduced into Columbia-0 (wild type) and ots1 ots2 double mutant background plants via Agrobacterium-mediated transformation.
Generation of JAZ Expression Constructs and Transgenic Lines
To generate the 35S::JAZ6 construct, the JAZ6 cDNA was cloned into the pENTR/D-TOPO (Invitrogen) vector and recombining the plasmid pENTR/D-TOPO with the binary vector pEarlygate-103 vectors to generate overexpression constructs. The JAZ6K221R allele was generated according to the quick-change site-directed mutagenesis kit with mutagenic oligos (JAZ6K221R FP/RP). The resulting plasmid was recombined with the pEarlygate 103 vector to obtain the 35S::JAZ6K221R:GFP fusion. The COI1 ORF was amplified by PCR from whole cDNAs from seedlings with COI1-specific oligos and cloned into pENTR/D-TOPO to yield the entry clone. The 35S::COI1:GFP construct was generated by recombining the plasmid entry vector with the binary vector pEarlygate 101 vector. The COI1V553A allele was generated according to the quick-change site-directed mutagenesis kit with mutagenic oligos (COI1V553AFP/RP). For GST pull-down assays, fusion constructs GST:COI1, His:JAZ6, and His:SUMO1 were generated by recombining entry vector plasmids with destination vectors pDEST15 (GST tag) and pDEST17 (His tag) vectors. Transgenic plants were generated and analyzed as described above.
Total RNA Extraction and RT-qPCR
Twelve-day-old wild-type ots1 ots2 plant leaves were frozen in liquid nitrogen and ground to a fine powder in pestle and mortar. RNA was extracted using the Spectrum Plant Total RNA kit (Sigma-Aldrich) following the manufacturer’s recommendations. RNA was quantified by measuring absorbance at wavelengths of 260 and 280 nm using a NanoDrop 1000 spectrophotometer (Thermo Scientific). RNA was DNase treated with Promega DNase I and cDNA synthesis conducted using Invitrogen SuperScript-II reverse transcriptase following the manufacturer’s guidelines. Seedlings were exposed to mock (MS) or 50 μM MeJA before being harvested and RNA extracted. One microgram of total RNA was used for cDNA synthesis and RT-qPCR analysis was performed.
The RT-qPCR assay was conducted as described previously (Conti et al., 2008), using SYBR green master mix (Applied Biosystems) and used for qPCR with a Rotor-Gene-Q (Qiagen). Amplification was followed by a melt curve analysis. The 2−∆∆Ct method was used for relative quantification (Livak and Schmittgen, 2001). To detect transcript levels, oligos for specific genes were used (Supplemental Table 1). Oligonucleotides amplifying ACTIN were used for normalization.
Quantification of JA-Ile from Arabidopsis Tissues
JA-IIe was quantified essentially as previously described (Forcat et al., 2008) with slight modification. Twelve-day-old seedlings grown on 0.5× MS plates were harvested into liquid nitrogen. Samples were ground using a mortar and pestle, and 10 mg powdered tissue aliquots were weighed into microcentrifuge tubes and extracted with 400 μL of 10% (v/v) methanol containing 1% (v/v) acetic acid to which internal standards (10 ng of JA) had been added. Following removal of the supernatant, the pellet was reextracted (400 μL of 10% methanol; 1% acetic acid). Following a 30-min incubation on ice, the extract was centrifuged and the supernatants pooled. Samples were then analyzed by mass spectrometry using a Sciex Q TRAP 6500 hybrid triple-quadrupole analyzer linked to Shimadzu Nexera UHPLC system. Samples were separated on a Phenomenex Luna Omega Polar column (1.6 µm, 100 × 2.1 mm) using mobile phases of 0.1% (v/v) formic acid (A) and 0.1% formic acid in methanol (B) at a flow rate of 200 µL min−1, starting at 5% B, held for 2 min, with a linear gradient to 95% B at 9 min, held for 2.9 min, with a total run time of 12.2 min. The column was equilibrated at 5% B for 5 min between runs.
Bioinformatical Analysis of Protein Structures
The structural model of the COI1 SIM site interaction was developed using PyMOL software. The coordinates for each structure were downloaded from the PDB (files 3OGK and 1A5R) and the binding sites of SUMO1 and COI1 were mapped onto the protein.
Recombinant Protein and GST Pull-Down Assay
Recombinant protein expression and production in Escherichia coli were as previously described (Srivastava et al., 2016) with slight modifications. COI1 (GST:COI1), SUMO1 (His:SUMO1), and JAZ6 (His:JAZ6) were expressed in BL21 (DE3) cells. GST-COI1 protein was overexpressed and purified from E. coli using Glutathione sepharose 4B beads (GE). His:SUMO1 and His:JAZ6 protein was overexpressed and purified from E. coli (BL21) cells using Ni-NTA agarose beads (Qiagen). For in vitro binding experiments, GST and GST-COI1 (2.0 μg) protein was bound to a GST column by incubating with in vitro pull-down buffer for 2 h at 4°C. Excess unbound protein was washed off and His:SUMO1 proteins were added in equimolar ratio and incubated in 500 μL in vitro pull-down buffer (50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 0.2% [v/v] glycerol, 1 mM EDTA, 0.1% Nonidet P-40 [v/v], 1 mM PMSF, and 1× protease inhibitor cocktail [Sigma-Aldrich]) at 4°C. The GST beads were collected by brief centrifugation and (input was collected separately) washed three times with 1 mL of in vitro pull-down buffer. Pellets were resuspended in 1× SDS loading buffer, boiled for 5 min, and analyzed by SDS-PAGE for protein binding. Both input (2%) and pull-down samples were probed with anti-GST and anti-His antibodies.
Reconstitution of SUMOylation in E. coli
In order to perform SUMOylation reactions in E. coli, we transformed the His:JAZ6 plasmid in two different strains containing SUMO conjugation machinery with SUMO1 modified to expose the C-terminal Gly-Gly (GG) sequence and as a negative control, SUMO1 with the C-terminal Gly-Gly mutated to Ala-Ala (AA) (Okada et al., 2009). For SUMOylation reactions, proteins were purified from freshly transformed E. coli using 1 mL His-Trap nickel affinity columns (GE Healthcare) and probed with anti-MBP and anti-AtSUMO1 antibodies to investigate the SUMOylation of JAZ6 in vitro.
Mass Spectrometry Analysis
The reaction was performed in a single cell system and the protein purified using His-Trap columns and samples were loaded with 4× SDS loading buffer. Five individual reactions were combined and separated by 10% acrylamide SDS-PAGE gel. Gels were stained for total proteins with Coomassie Brilliant Blue and subsequently destained with 10% acetic acid, 40% methanol, and 50% water and washed with double-distilled water. Protein bands were sliced for mass spectrometry analysis.
In Vivo Protein Degradation Assays
Protein degradation assays were performed as described previously with slight modifications (Bueso et al., 2014). For in vivo protein degradation experiments, Agrobacterium cultures containing constructs that express JAZ6:GFP, HA:COI1/ HA:COI1V553A, or HA:SUMO1 and the silencing suppressor p19 were coinfiltrated at different ratios in Nicotiana benthamiana leaves. Three days after infiltration, samples were collected, ground in liquid nitrogen, and immediately placed on ice in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Nonidet P-40, 10 mM MgCl2, and protease inhibitor tablets) for protein extraction. Homogenates were cleared by centrifugation at 13,000 rpm at 4°C for 15 min, and supernatants were used for protein immunoblot analysis.
Protein Extraction and Immunoblot Analysis
Frozen plant tissue was ground to a fine powder with a chilled pestle and mortar. Protein extraction buffer (50 mM Tris/HCl, pH 8.5, 4% SDS [w/v], 2% β-mercaptoethanol [v/v], and 10 mM EDTA) and protease inhibitor tablet were added at a ratio of 1:1 (w/v). The mixture was centrifuged at 12,000g at 4°C for 10 min. The protein concentration was determined using a Direct Detect Infra-red Spectrometer (EMD Millipore), and samples were equalized with the addition of extraction buffer. Protein loading dye (4×) was added and the samples were separated on SDS-PAGE gels. Proteins were transferred to PVDF membranes and blocked with 5% (w/v) semiskimmed milk powder at room temperature and probed with the respective antibodies. Secondary horseradish peroxidase-conjugated antibodies were applied before developing the blots with x-ray film using an automated developer.
Statistical Analysis
All statistical analysis was performed using GraphPad Prism 6 software. One-way or two-way ANOVAs were performed at a significance level of P < 0.05, P < 0.01, or P < 0.001. All root phenotype experiments had at least an n of 25 to 30 seedlings in each biological replication. Data represent an average of three individual biological replicates.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AT1G19180, AT1G72450, AT2G39940, AT1G60220, and AT1G10570.
Supplemental Data
Supplemental Figure 1. JAZ1 protein accumulation and SUMOylation.
Supplemental Figure 2. Relative transcript levels of JAZ1 and JAZ6 in different transgenic plants.
Supplemental Figure 3. Reconstituted in vitro SUMOylation assay of JAZ6:MBP fusion protein.
Supplemental Figure 4. Relative transcript levels of JAZ6 in different transgenic plants.
Supplemental Figure 5. Pst DC3000 infection negatively regulates JAZ6:GFP SUMOylation and accumulation.
Supplemental Figure 6. COI1V553A:GFP SIM mutant plants show significantly increased resistance to B. cinerea infection and spider mite infestation in the ots1 ots2 genetic background.
Supplemental Table 1. List of DNA oligonucleotides used in this study.
Supplemental File 1. ANOVA tables.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
A.S. and A.K.S. were supported by the Biotechnology and Biological Sciences Research Council (Grant BB/M022048/1) and European Research Council for research funding.
AUTHOR CONTRIBUTIONS
A.K.S. and A.S. designed the research and analyzed the data. A.K.S. performed most of the experiments assisted by B.O., C.W., P.S., G.S.A., and C.Z. A.K.S., M.G., M.R.R., E.F., and A.S. wrote the article. All authors read and commented on the manuscript.
Footnotes
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Achard P., Genschik P. (2009). Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J. Exp. Bot. 60: 1085–1092. [DOI] [PubMed] [Google Scholar]
- Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert Y., Widemann E., Miesch L., Pinot F., Heitz T. (2015). CYP94-mediated jasmonoyl-isoleucine hormone oxidation shapes jasmonate profiles and attenuates defence responses to Botrytis cinerea infection. J. Exp. Bot. 66: 3879–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M., Srivastava A., Conti L., Nelis S., Zhang C., Florance H., Love A., Milner J., Napier R., Grant M., Sadanandom A. (2016). Stability of small ubiquitin-like modifier (SUMO) proteases OVERLY TOLERANT TO SALT1 and -2 modulates salicylic acid signalling and SUMO1/2 conjugation in Arabidopsis thaliana. J. Exp. Bot. 67: 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks D.M., Hernández-Guzmán G., Kloek A.P., Alarcón-Chaidez F., Sreedharan A., Rangaswamy V., Peñaloza-Vázquez A., Bender C.L., Kunkel B.N. (2004). Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol. Plant Microbe Interact. 17: 162–174. [DOI] [PubMed] [Google Scholar]
- Brooks D.M., Bender C.L., Kunkel B.N. (2005). The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 6: 629–639. [DOI] [PubMed] [Google Scholar]
- Bueso E., Rodriguez L., Lorenzo-Orts L., Gonzalez-Guzman M., Sayas E., Muñoz-Bertomeu J., Ibañez C., Serrano R., Rodriguez P.L. (2014). The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 80: 1057–1071. [DOI] [PubMed] [Google Scholar]
- Capili A.D., Lima C.D. (2007). Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J. Mol. Biol. 369: 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico J.M., Chini A., Fonseca S., Solano R. (2008). JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 11: 486–494. [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Conti L., Price G., O’Donnell E., Schwessinger B., Dominy P., Sadanandom A. (2008). Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell 20: 2894–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L., Nelis S., Zhang C., Woodcock A., Swarup R., Galbiati M., Tonelli C., Napier R., Hedden P., Bennett M., Sadanandom A. (2014). Small Ubiquitin-like Modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev. Cell 28: 102–110. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M., Zhai B., Jayaraman S., Eleftheriadou G., Winsbury R., Yang R., Truman W., Tang S., Smirnoff N., Grant M. (2016). Novel JAZ co-operativity and unexpected JA dynamics underpin Arabidopsis defence responses to Pseudomonas syringae infection. New Phytol. 209: 1120–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chico J.M., Solano R. (2009). The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12: 539–547. [DOI] [PubMed] [Google Scholar]
- Forcat S., Bennett M.H., Mansfield J.W., Grant M.R. (2008). A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8: 947–956. [DOI] [PubMed] [Google Scholar]
- Geng X., Cheng J., Gangadharan A., Mackey D. (2012). The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. Plant Cell 24: 4763–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Grunewald W., Vanholme B., Pauwels L., Plovie E., Inzé D., Gheysen G., Goossens A. (2009). Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep. 10: 923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay R.T. (2001). Protein modification by SUMO. Trends Biochem. Sci. 26: 332–333. [DOI] [PubMed] [Google Scholar]
- Hecker C.M., Rabiller M., Haglund K., Bayer P., Dikic I. (2006). Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281: 16117–16127. [DOI] [PubMed] [Google Scholar]
- Hickey C.M., Wilson N.R., Hochstrasser M. (2012). Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13: 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66. [DOI] [PubMed] [Google Scholar]
- Ingle R.A., Stoker C., Stone W., Adams N., Smith R., Grant M., Carré I., Roden L.C., Denby K.J. (2015). Jasmonate signalling drives time-of-day differences in susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea. Plant J. 84: 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., Pyrowolakis G. (2000). Ubiquitin and its kin: how close are the family ties? Trends Cell Biol. 10: 335–342. [DOI] [PubMed] [Google Scholar]
- Katsir L., Chung H.S., Koo A.J., Howe G.A. (2008). Jasmonate signaling: a conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11: 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2008). Jasmonate signaling: toward an integrated view. Plant Physiol. 146: 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. (2006). Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22: 159–180. [DOI] [PubMed] [Google Scholar]
- Koornneef A., Pieterse C.M. (2008). Cross talk in defense signaling. Plant Physiol. 146: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie-Berry N., Joardar V., Street I.H., Kunkel B.N. (2006). The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol. Plant Microbe Interact. 19: 789–800. [DOI] [PubMed] [Google Scholar]
- Lee J., et al. (2007). Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 49: 79–90. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Melotto M., Mecey C., Niu Y., Chung H.S., Katsir L., Yao J., Zeng W., Thines B., Staswick P., Browse J., Howe G.A., He S.Y. (2008). A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 55: 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50: 267–294. [DOI] [PubMed] [Google Scholar]
- Miller M.J., Barrett-Wilt G.A., Hua Z., Vierstra R.D. (2010). Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 16512–16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty A., Dumont X., Kaghad M., Caput D. (2000). Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 275: 36316–36323. [DOI] [PubMed] [Google Scholar]
- Okada S., Nagabuchi M., Takamura Y., Nakagawa T., Shinmyozu K., Nakayama J., Tanaka K. (2009). Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol. 50: 1049–1061. [DOI] [PubMed] [Google Scholar]
- Pauwels L., Goossens A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C.M. (2012). Prime time for transgenerational defense. Plant Physiol. 158: 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P.H., Murtas G., Dash S., Coupland G. (2002). early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129: 5349–5361. [DOI] [PubMed] [Google Scholar]
- Reverter D., Lima C.D. (2009). Preparation of SUMO proteases and kinetic analysis using endogenous substrates. Methods Mol. Biol. 497: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E., Marquis V., Poirier L., Aubert Y., Zumsteg J., Ménard R., Miesch L., Heitz T. (2017). Jasmonic acid oxidase 2 hydroxylates jasmonic acid and represses basal defense and resistance responses against Botrytis cinerea infection. Mol. Plant 10: 1159–1173. [DOI] [PubMed] [Google Scholar]
- Srivastava A.K., Zhang C., Yates G., Bailey M., Brown A., Sadanandom A. (2016). SUMO is a critical regulator of salt stress responses in rice. Plant Physiol. 170: 2378–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A.K., Zhang C., Caine R.S., Gray J., Sadanandom A. (2017). Rice SUMO protease Overly Tolerant to Salt 1 targets the transcription factor, OsbZIP23 to promote drought tolerance in rice. Plant J. 92: 1031–1043. [DOI] [PubMed] [Google Scholar]
- Staswick P.E. (2008). JAZing up jasmonate signaling. Trends Plant Sci. 13: 66–71. [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Verger A., Perdomo J., Crossley M. (2003). Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 4: 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yan Y., Stolz S., Chételat A., Reymond P., Pagni M., Dubugnon L., Farmer E.E. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19: 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]