Screening of natural Arabidopsis accessions reveals a membrane-associated protein kinase involved in pollen aperture development and the formation of distinct membrane domains in microspores.
Abstract
Certain regions on the surfaces of developing pollen grains exhibit very limited deposition of pollen wall exine. These regions give rise to pollen apertures, which are highly diverse in their patterns and specific for individual species. Arabidopsis thaliana pollen develops three equidistant longitudinal apertures. The precision of aperture formation suggests that, to create them, pollen employs robust mechanisms that generate distinct cellular domains. To identify players involved in this mechanism, we screened natural Arabidopsis accessions and discovered one accession, Martuba, whose apertures form abnormally due to the disruption of the protein kinase D6PKL3. During pollen development, D6PKL3 accumulates at the three plasma membrane domains underlying future aperture sites. Both D6PKL3 localization and aperture formation require kinase activity. Proper D6PKL3 localization is also dependent on a polybasic motif for phosphoinositide interactions, and we identified two phosphoinositides that are specifically enriched at the future aperture sites. The other known aperture factor, INAPERTURATE POLLEN1, fails to aggregate at the aperture sites in d6pkl3 mutants, changes its localization when D6PKL3 is mislocalized, and, in turn, affects D6PKL3 localization. The discovery of aperture factors provides important insights into the mechanisms cells utilize to generate distinct membrane domains, develop cell polarity, and pattern their surfaces.
INTRODUCTION
The formation of specific cellular domains is an essential step in cell-type specification and the development of many unicellular and multicellular organisms. The formation of distinct domains within or near the plasma membrane (PM) results in symmetry breaking and the generation of cell polarity. This, in turn, contributes to such important processes as cell and tissue morphogenesis, pattern formation, cell recognition, reproduction, barrier formation, and defense against pathogens and plays a critical role in the development and survival of organisms (Nelson, 2003; Goldstein and Macara, 2007; Yang, 2008; Roppolo et al., 2011; Tepass, 2012; Nagano et al., 2016). The mechanisms involved in the formation of distinct PM domains are not well understood, particularly in plants, which lack orthologs of the classical metazoan polarity PARTITIONING DEFECTIVE proteins (Abrash and Bergmann, 2009; Geldner, 2009; Pan et al., 2015; Shao and Dong, 2016).
Pollen provides a unique and valuable model for studying polarity. The pollen surface is protected by the pollen wall exine, the specialized sporopollenin-based cell wall, which in most plants is deposited nonuniformly, leaving gaps that are either not covered by exine or have reduced exine deposition (Walker and Doyle, 1975; Furness and Rudall, 2004). These gaps are known as pollen apertures, and they play a role in plant reproduction as sites for water transport and pollen tube germination, as well as architectural details that help pollen accommodate changes in volume (Wang and Dobritsa, 2018). Apertures exhibit species-specific variations in their morphology, number, and positions and produce a variety of pollen surface patterns, with the most common pattern in eudicots consisting of three equidistant longitudinal apertures (Wodehouse, 1935; Walker and Doyle, 1975; Furness and Rudall, 2004). Aperture formation indicates that during pollen development, specific domains on the pollen surface acquire identity different from the surrounding regions and become protected from exine deposition.
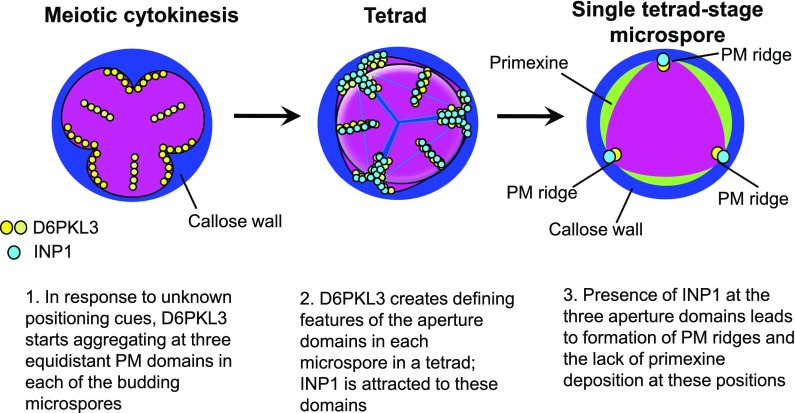
Like the pollen of many other eudicot species, the pollen of Arabidopsis thaliana forms three narrow longitudinal apertures, spaced equidistantly at the equator of the grain (Figure 1A). Aperture formation begins at the tetrad stage of pollen development. Each tetrad of microspores contains four products of male meiosis. Initially held together by the common callose wall, the microspores subsequently develop into four individual pollen grains. Previously, we demonstrated that INAPERTURATE POLLEN1 (INP1), an Arabidopsis protein of unknown biochemical function, is an essential factor for aperture formation, with apertures completely disappearing in its absence (Dobritsa and Coerper, 2012). At the PM of each of the tetrad-stage microspores, INP1 assembles into three equidistant longitudinal punctate lines that mark the positions of future apertures and indicate formation of distinct aperture domains (Dobritsa and Coerper, 2012; Reeder et al., 2016; Dobritsa et al., 2018). INP1 has a role in keeping these PM domains in close association with the overlying callose wall and protecting them from the formation of primexine, the scaffold for exine deposition (Dobritsa et al., 2018).
Figure 1.
D6PKL3 Is Required for the Formation of Normal Pollen Apertures.
(A) Wild-type (Col-0) Arabidopsis pollen has three equidistant apertures (arrows).
(B) to (H) Seven different d6pkl3 mutants (genotypes indicated) produce pollen with abnormal apertures partially covered with exine (arrowheads).
(I) and (J) Pollen grains from the F1 plants of the crosses between Mt-0 and d6pkl3-2 (I) and Mt-0 and d6pkl3-4 (J) do not show complementation of the aperture phenotype (arrowheads indicate traces of apertures).
(K) and (L) D6PKL3-YFP transgene restores aperture formation (arrows) in the Mt-0 (K) and d6pkl3-2 (L) plants. Confocal images of pollen stained with auramine O. Bars = 10 μm.
(M) Diagram of the structure of the D6PKL3 gene with two exons and an intron. The protein-coding sequence corresponds to the entire exons, starting and ending with the black boxes (untranslated regions are not shown). The sequence encoding the kinase domain is indicated in blue, and the sequences encoding the MID domain and the K/R-rich motif (K) are shown in green and yellow, respectively. Positions of the mutations in the seven d6pkl3 alleles described in this study are indicated. For point mutations, the corresponding amino acid substitutions are shown.
INP1 is unlikely to be the sole player in controlling aperture formation. In support of this notion, hoary stock (Matthiola incana), a relative of Arabidopsis that lacks pollen apertures, still possesses a functional copy of INP1, implying that the phenotype must be due to a defect in another aperture factor (Li et al., 2018). Also, although the presence of INP1 is critical for the formation of apertures in Arabidopsis, INP1 does not appear to be responsible for specifying the number and positions of aperture sites, instead recognizing and responding to aperture patterns that have already been established prior to its arrival (Reeder et al., 2016; Dobritsa et al., 2018; Li et al., 2018). The mechanisms through which INP1 localization is controlled and aperture sites are established are yet to be determined.
To uncover additional players participating in the process of pollen aperture formation, we performed a screen of natural Arabidopsis accessions and found one accession, Martuba (Mt-0), with abnormal apertures that are largely covered with exine. This defect was mapped to a gene encoding a predicted protein kinase, D6 PROTEIN KINASE-LIKE3 (D6PKL3). Here, we demonstrate that, like INP1, D6PKL3 localizes at the PM to the positions of developing aperture domains. Interestingly, D6PKL3 starts assembling at these domains earlier than INP1, before the end of meiotic cytokinesis. D6PKL3 is necessary for correct INP1 localization, suggesting that D6PKL3 acts upstream of INP1 in establishing aperture domains. Localization of D6PKL3 depends on its kinase activity and a phosphoinositide-interacting polybasic motif. We also found that PM aperture domains are enriched in two D6PKL3-cognate phosphoinositide species, phosphatidylinositol-4-phosphate [PI(4)P] and phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2], pointing to a role for phospholipids and phospholipid-D6PKL3 interactions in defining PM aperture domains. Our data provide important insights into the mechanisms contributing to the formation of apertures and pollen surface patterning and, more broadly, into the mechanisms cells use to generate distinct PM domains and control polarity.
RESULTS
The Arabidopsis Martuba Accession Develops Abnormal Apertures on the Pollen Surface
To identify genes involved in pollen aperture formation, we screened pollen from 316 natural accessions of Arabidopsis thaliana (Supplemental Data Set 1), most of which had been sequenced by the Arabidopsis 1001 Genomes Project (1001 Genomes Consortium, 2016). The screen focused on changes in pollen shape that could be detected under a dissecting microscope at low magnification, which served as a proxy for changes in pollen apertures (Dobritsa et al., 2011; Dobritsa and Coerper, 2012). One of the accessions, the Libyan accession Martuba (Mt-0), had pollen that was rounder than normal (Supplemental Figure 1). When examined by confocal microscopy, Mt-0 pollen displayed an abnormal aperture phenotype, with pollen grains either lacking apertures or, more commonly, having aperture regions that were partially covered with exine but were still recognizable (Figure 1B). In that regard, the Mt-0 phenotype was weaker than the phenotype observed in the previously discovered inp1 mutant, in which apertures disappear entirely (Dobritsa et al., 2011; Dobritsa and Coerper, 2012). The cross between Mt-0 and the reference accession Columbia (Col-0) demonstrated that this abnormal aperture phenotype behaved as a single recessive trait, with approximately one-quarter of the F2 progeny from this cross exhibiting aperture formation defects. Similar to inp1, the Mt-0 defect was sporophytic, as all pollen grains from the F1 heterozygous plants had normal apertures. In the progeny of the cross between Mt-0 and inp1, all pollen also had normal apertures (Supplemental Figure 2), indicating that the abnormal aperture phenotype of Mt-0 was due to a defect in a gene other than INP1.
The Mt-0 Pollen Aperture Defect Is Caused by a Mutation in the D6PKL3 Gene
To identify the factor responsible for the aperture defect of Mt-0, we performed positional cloning using 926 mutant F2 plants from the cross between Mt-0 and Col-0. The defect was mapped to a 125-kb region on chromosome 3 that included 38 genes in the region between At3g27330 and At3g27660. According to the available genome sequence information for Mt-0 (1001 Genomes Consortium, 2016), most of these genes contained multiple polymorphisms compared with Col-0, including insertions, deletions, and single-nucleotide polymorphisms, often leading to missense or nonsense mutations. We confirmed the enrichment of Mt-0 markers in this region by performing next-generation sequencing (NGS) on pooled genomic DNA samples from 150 F2 plants with the mutant aperture phenotype. The resolution achieved with the NGS approach was lower than the resolution of the positional cloning, yet the resequencing allowed us to verify the identity of multiple polymorphisms in Mt-0.
To reduce the number of potential candidate genes, we compared the presence and absence of nonsynonymous polymorphisms between Mt-0 and 62 accessions that were scored as the wild type in our screen. This analysis helped us to narrow the pool of candidates, limiting it to five genes (see Methods) that had polymorphisms specific to Mt-0 and therefore were more likely to be responsible for the aperture defects. We then examined available T-DNA or transposon insertion lines of these remaining candidate genes for the presence of the Mt-0-like pollen phenotype. One of the genes in this region, At3g27580, known as D6PKL3 or Arabidopsis PROTEIN KINASE7, had a premature stop codon (Leu198Stop) in Mt-0 (Figure 1M), which was not found in any other accession in the 1001 Genomes database. The full-length D6PKL3 protein is composed of 578 amino acids. Thus, the stop codon at position 198, which resides within the highly conserved kinase domain (Figure 1M), is expected to generate a truncation that would fully impair the D6PKL3 protein kinase function.
The identity of a mutation in D6PKL3 as the causative defect for aperture phenotype in Mt-0 was subsequently confirmed by analysis of the D6PKL3 T-DNA insertion mutant SALK_047347 (d6pkl3-2), a previously described loss-of-function allele carrying an insertion in exon 1 (Zourelidou et al., 2009), as well as two T-DNA insertions from the Gabi-Kat collection, GK-265A02 (d6pkl3-4) and GK-169H08 (d6pkl3-5) (Figure 1M). These d6pkl3 insertion alleles all displayed the Mt-0 phenotype (Figures 1C to 1E), and, when crossed with Mt-0, these mutations failed to complement the aperture phenotype in the F1 plants (Figures 1I and 1J).
Based on the sequence information from the 1001 Genomes database, the Armenian accession Yeghegis-1 (Yeg-1) was expected to have a sizable deletion in D6PKL3. We sequenced the D6PKL3 locus from Yeg-1 and found a 607-bp deletion combined with a 16-bp insertion, a polymorphism somewhat different from the database entry (Figure 1M; Supplemental Figure 3). This mutation is expected to inactivate the protein since it causes a frameshift in the open reading frame at amino acid 193 within the kinase domain, followed by a premature stop codon (Supplemental Figure 3). Consistently, Yeg-1 pollen had the mutant aperture phenotype identical to that of Mt-0 and of the T-DNA mutants of D6PKL3 (Figure 1F).
Additionally, we discovered two point mutants with substitutions in D6PKL3 affecting conserved amino acids within the kinase domain, Glu290Lys (E290K, d6pkl3-6) and Gly506Lys (G506K, d6pkl3-7) (Figure 1M), in lines that we had recently identified in a screen for aperture mutants following EMS mutagenesis. Both mutants also had the Mt-0-like phenotype (Figures 1G and 1H) and failed to complement the aperture defect when crossed with Mt-0. Altogether, we identified seven independent mutants with defects in D6PKL3, all of which displayed identical aperture phenotypes.
For final confirmation of causality, we suppressed the pollen aperture defect of Mt-0 and d6pkl3-2 plants with a transgene expressing the D6PKL3 coding region fused to a YFP from a 2016-bp D6PKL3 promoter fragment (D6PKL3pr:D6PKL3-YFP). Apertures were restored in the T1 lines expressing this construct (n ≥ 10), indicating that D6PKL3 is sufficient to complement the aperture defects in Mt-0 and d6pkl3-2 and that the YFP fusion protein of D6PKL3 retains functionality with regard to control of aperture formation (Figures 1K and 1L).
D6PKL3 Is the Only Member of the D6 PROTEIN KINASE Family That Is Involved in Pollen Aperture Formation
D6PKL3 together with D6PK, D6PKL1, and D6PKL2 forms the D6PK subfamily within the AGCVIIIa family of Arabidopsis serine/threonine protein kinases (Zegzouti et al., 2006; Zourelidou et al., 2009; Rademacher and Offringa, 2012). D6PKL3, which shares ∼60% sequence identity with the three other proteins, is the most divergent member within this subfamily, while D6PK and D6PKL1 share 86% amino acid identity, and D6PKL2 shares 73% identity with D6PK and D6PKL1. The four kinases have overlapping functions in the control of auxin transport in vegetative tissues, and the quadruple combination of d6pk/d6pkl mutations leads to severe impairment of growth and development in roots and shoots (Zourelidou et al., 2009; Willige et al., 2013; Barbosa et al., 2014). However, no d6pkl3 single mutant phenotype had previously been reported.
Since functional redundancy with the other family members had been described, we checked pollen phenotypes of the individual d6pk/d6pkl single gene mutations alone or in combination with other mutations. Besides d6pkl3, none of the other single mutants displayed an abnormal aperture phenotype (Figure 2), and among the double, triple, and quadruple mutants, only those that contained the d6pkl3 mutation exhibited the Mt-0-like phenotype (Figure 2). Significantly, no increase in the severity of the phenotype, e.g., occurrence of completely inaperturate pollen identical to that observed in the inp1 mutant, was observed in the d6pk/d6pkl higher-order mutants, including the quadruple mutant (Figure 2), indicating that D6PKL3 is the sole member of this subfamily involved in pollen aperture formation.
Figure 2.
D6PKL3 Is the Only Member of the D6PK Subfamily of AGCVIII Kinases Involved in Pollen Aperture Formation.
Pollen phenotypes of single, double, triple and quadruple mutants with d6pk, d6pkl1, d6pkl2, and d6pkl3 mutations. Only the mutants containing the d6pkl3 mutation develop abnormal apertures. Normal apertures are indicated with arrows. Bars = 10 μm.
D6PKL3 Specifically Localizes to the Three Aperture PM Domains in Tetrad-Stage Microspores
D6PK, the founding member of the D6PK subfamily, is a polarly localized PM-associated kinase required for phosphorylation and activation of the PIN-FORMED (PIN) auxin transporters (Zourelidou et al., 2009, 2014; Willige et al., 2013; Barbosa et al., 2014, 2018; Stanislas et al., 2015). Excitingly, when we checked the localization of D6PKL3-YFP protein expressed from the D6PKL3pr:D6PKL3-YFP transgene in tetrad-stage microspores, we found that D6PKL3-YFP localizes to three equidistant plasma membrane positions in a pattern similar to the distribution of INP1, the known marker for aperture formation (Figures 3A to 3D; Supplemental Movie 1). Three pieces of evidence support the conclusion that these PM domains correspond to the aperture domains. First, the positions of the lines of D6PKL3-YFP signal coincided with the positions of membrane protrusions characteristic of aperture domains (Dobritsa et al., 2018) (Figures 3A to 3D). Second, the D6PKL3-YFP signal coincided with the signal of CFP-tagged INP1 (Figures 3M to 3O). Finally, the D6PKL3-YFP signal was concentrated directly underneath the aperture gaps in late tetrad-stage microspores, which had already initiated exine deposition and had recognizable apertures (Figures 3P to 3S). D6PKL3 is thus a polarly localized PM protein in developing pollen.
Figure 3.
D6PKL3 Accumulates at the Aperture Domains of the PM Where It Forms Punctate Lines Similar to Those of INP1.
(A) to (D) 3D reconstructions of confocal z-stacks of a tetrad of microspores showing D6PKL3-YFP (yellow) assembled into lines at three equidistant membrane domains in each microspore. The callose wall is shown in blue (A) and the PM is in magenta ([B] and [D]). (D) is a merge of (B) and (C). See also Supplemental Movie 1.
(E) to (L) YFP fluorescence in D6PKL3pr:D6PKL3-YFP sporogenic cells at different stages of development. Optical sections of microspore mother cells (MMC) and tetrads (Tds). Top images show YFP fluorescence of D6PKL3-YFP, and bottom images show the merged fluorescent signal from YFP (yellow), Calcofluor White (blue, callose wall) and CellMask Deep Red (magenta, membranous structures).
(E) and (I) MMC prior to cytokinesis has diffuse YFP signal in cytoplasm, as well as some punctate signal.
(F) and (J) MMC undergoing cytokinesis already displays peripheral D6PKL3-YFP puncta. See also Supplemental Movie 2.
(G), (H), (K), and (L) Tetrad-stage microspores develop punctate lines of D6PKL3-YFP at the PM domains.
(M) to (O) D6PKL3 and INP1 localize to the same PM domains (arrows).
(M) D6PKL3-YFP signal (yellow).
(N) INP1-CFP signal (cyan).
(O) Merged signals.
(P) to (S) D6PKL3-YFP lines are located directly underneath the apertures. Bars = 10 μm.
(P) to (R) A surface view of the late-stage tetrad shows that the positions of the D6PKL3-YFP lines (yellow) coincide with the apertures (white arrows) visible as gaps in the developing exine (blue).
(S) Signal intensity profile of the two fluorophores shown in (Q) and (R), along the red arrow drawn through the aperture in (Q) and (R). A peak of the yellow signal corresponds to the reduction in the blue signal. a.u., arbitrary units.
D6PKL3 Starts Assembling into Peripheral Punctate Lines Earlier Than INP1
INP1 exhibits diffuse cytoplasmic staining in the early stages of microsporogenesis and only starts assembling into three longitudinal punctate lines at the microspore periphery in tetrad-stage cells (Dobritsa and Coerper, 2012; Dobritsa and Reeder, 2017; Dobritsa et al., 2018). To understand the mechanism of aperture formation and the role of D6PKL3 in this process, we compared the dynamics of D6PKL3-YFP and INP1 expression and localization during pollen development.
Unlike INP1, D6PKL3 was already visible as puncta in the cytoplasm of microspore mother cells (MMCs) prior to cell division (Figures 3E and 3I). In the MMC undergoing meiotic cytokinesis, D6PKL3 punctate signal was localized at the cell periphery (Figures 3F and 3J), which was significantly earlier than the appearance of INP1 at these cellular domains (Dobritsa et al., 2018). Already during cytokinesis, D6PKL3 started forming three lines in each of the budding sister microspores, suggesting that the aperture PM domains, which will subsequently become decorated with INP1 at the tetrad stage, start developing their distinct features much earlier (Figures 3F and 3J; Supplemental Movie 2). D6PKL3-YFP completes its assembly into lines at the aperture regions during the tetrad stage (Figures 3G, 3K, 3H, and 3L). Taken together, these observations suggest that D6PKL3 already marks the future aperture regions during cytokinesis and that D6PKL3 localization to these regions precedes INP1 accumulation.
Changes in D6PKL3 Expression May Modulate Aperture Numbers
Upon close examination of the lines formed by D6PKL3-YFP at the periphery of tetrad-stage microspores, we noticed that these lines were frequently composed of two parallel lines of D6PKL3-YFP puncta, running in close proximity to each other (Figures 3M to 3O and 4A; Supplemental Movie 3). We initially hypothesized that these double lines may serve as the borders that define the width of each aperture. However, INP1, which does not form double lines when analyzed on its own (Reeder et al., 2016; Dobritsa et al., 2018), also assembled into double lines in the presence of the D6PKL3-YFP transgene (Figures 3M to 3O), suggesting that this pattern was guided by D6PKL3 activity or distribution and that the aberrant pattern was likely the consequence of aberrant D6PKL3 transgene expression or D6PKL3-YFP function in the transgenic lines.
Figure 4.
Pollen of D6PKL3pr:D6PKL3-YFP Transgenic Lines Commonly Display Unusual Aperture Phenotypes.
(A) and (B) Tetrads in which some microspores have D6PKL3-YFP assembled either into double lines ([A], arrows) or into more than three lines ([B], arrows). See also Supplemental Movies 3 and 4.
(C) and (D) Examples of pollen grains with the double-aperture phenotype (white arrows show double apertures; red arrows point to exine in between the double apertures).
(E) and (F) Examples of pollen with four apertures (arrows; front and back views of the same pollen grain).
(G) and (H) Examples of pollen with irregular apertures (white arrows; front and back views of the same pollen grain). Red arrows point to exine inclusion in between aperture areas. Bars = 10 μm.
(I) and (J) Formation of pollen with more than three apertures correlates with the level of D6PKL3 expression.
(I) Percentage of pollen grains that developed more than three apertures in four independent homozygous transgenic D6PKL3pr:D6PKL3-YFP lines in Col-0 background (T3 generation). Pollen from five plants per each T3 line was scored (error bars indicate se; n = total number of pollen grains scored for each line).
(J) The levels of D6PKL3-YFP transcripts from the same transgenic T3 lines as in (I) were measured by qRT-PCR and normalized to the levels of the MMD1 transcripts. Error bars indicate se (n = 3 biological replicates).
The occurrence of these unusual lines formed by D6PKL3-YFP in tetrad-stage microspores of the D6PKL3pr:D6PKL3-YFP transgenic lines correlated with the development of unusual aperture patterns in mature pollen (Figures 4C and 4D). Frequently, each aperture in these pollen grains exhibited a double-aperture phenotype, defined by two narrow parallel lines separated by a thin stripe of exine (Figures 4C and 4D). Furthermore, a significant portion (5–25%) of the pollen grains from multiple D6PKL3pr:D6PKL3-YFP plants developed more than three apertures, typically four or six (Figures 4E and 4F) or had apertures that were irregularly placed on the pollen surface (Figures 4G and 4H). Although these unusual phenotypes were observed with the D6PKL3pr:D6PKL3-YFP transgene in both the d6pkl3-2 and wild-type backgrounds, they were more prevalent and noticeable in the wild-type background. Usually, in the cases when a pollen grain formed more than three apertures, the apertures did not have the double-aperture morphology (Figures 4E and 4F). The presence of supernumerary apertures in mature pollen of these transgenic plants corresponded with the formation of supernumerary equidistant punctate lines of D6PKL3-YFP in tetrad-stage microspores in these populations (Figure 4B; Supplemental Movie 4). These observations support the idea that the D6PKL3-YFP lines premark positions of aperture domains and that D6PKL3-YFP dosage may critically determine D6PKL3-YFP polar distribution and aperture number.
To clarify the origin of these unusual aperture phenotypes, we analyzed populations of pollen produced by plants from four homozygous T3 lines (5-6, 1-1, 6-5, and 4-4), containing independent insertions of the D6PKL3pr:D6PKL3-YFP transgene in the wild-type background. To test if correlation exists between the aperture phenotype and the D6PKL3 expression, we measured the transcript level of the transgenic D6PKL3 in stage-9 buds from different lines by quantitative RT-PCR and compared it with the frequency of appearance of pollen grains with higher aperture number in the same transgenic lines (Figures 4I and 4J). We noted that it was technically challenging to compare endogenous D6PKL3 expression with that of the transgenes, so instead, the expression of D6PKL3 transgenes was compared between different transgenic lines. Plants from line 5-6 produced the highest number of pollen grains with more than three apertures (∼20–25%) (Figure 4I). This line also had the highest level of transgene expression (Figure 4J), thus showing a positive correlation between the levels of D6PKL3 transgene expression and the aperture number and suggesting that elevated D6PKL3 gene expression may, at least in part, be responsible for the occurrence of the new aperture phenotype.
Taken together, these results are consistent with the notion that if D6PKL3 is expressed at higher than normal levels in transgenic plants, the excessive D6PKL3 protein either (preferentially) assembles into double lines at each of the three regular aperture domains, causing the formation of double apertures in mature pollen, or forms more than three equidistant single lines, resulting in pollen grains with more than three apertures. Thus, D6PKL3 protein dosage may be a key factor for determining the formation of polar PM domains in developing pollen.
Kinase Activity Is Required for the Role of D6PKL3 in Aperture Formation and for Its PM Localization
Like other members of the D6PK family, D6PKL3 is expected to be an active kinase (Zegzouti et al., 2006; Zourelidou et al., 2009; Willige et al., 2013; Weller et al., 2017). To evaluate whether the kinase activity of D6PKL3 is required for its role in aperture formation, we created a kinase-dead version of D6PKL3-YFP carrying a point mutation Lys211Glu (K211E) that converted the invariant lysine required for ATP binding of the kinase into a glutamic acid. Identical mutations in other members of the D6PK and AGCVIIIa kinase families had previously been shown to render them inactive (Zourelidou et al., 2009; Willige et al., 2012). The kinase-dead D6PKL3K211E was unable to rescue the aperture defect in the d6pkl3-2 mutant when expressed as a YFP-fusion from the D6PKL3 promoter fragment (7/7 lines) (Figure 5A). In addition, this mutated version failed to localize to the PM domains in microspores, instead exhibiting diffuse cytoplasmic signals (Figure 5B). These data indicate that the kinase activity of D6PKL3 is essential for both its correct protein localization and aperture formation.
Figure 5.
Kinase Activity of D6PKL3 Is Required for Its Function in Aperture Formation and for Protein Localization.
(A) The kinase-dead D6PKL3K211E-YFP fails to rescue the aperture defect in the d6pkl3-2 mutant.
(B) The kinase-dead D6PKL3K211E-YFP exhibits diffuse cytoplasmic fluorescence in tetrad-stage microspores and failed to organize into punctate lines at the aperture PM domains. Bars = 10 μm.
D6PKL3 and INP1 Depend on Each Other for Correct Peripheral Localization, but Likely Do Not Interact Directly
INP1 and D6PKL3 localize to the same PM domains (Figures 3M to 3O). To evaluate the genetic interactions between INP1 and D6PKL3, we crossed the DMC1pr:INP1-YFP line (exhibiting strong INP1-YFP expression) (Reeder et al., 2016; Dobritsa et al., 2018) into the Mt-0 and the d6pkl3-2 backgrounds. We found that, unlike in the Col-0 background (Figure 6A), INP1-YFP almost never formed punctate lines at the microspore periphery when placed in the d6pkl3 mutant background, suggesting that D6PKL3 is required for the proper localization of INP1 (Figures 6B, 6C, 6F, and 6G). The extremely rare observations of INP1-YFP peripheral puncta in the d6pkl3 mutant backgrounds are consistent with the occurrence of weak apertures in d6pkl3 pollen. The finding that D6PKL3-YFP assembles at the future aperture sites earlier than INP1 during microsporogenesis and that D6PK-YFP is able to recruit INP1 to double lines in the D6PKL3pr:D6PKL3-YFP transgenic lines is consistent with D6PKL3 acting upstream of INP1. Interestingly, when D6PKL3pr:D6PKL3-YFP was introduced into the inp1 background, D6PKL3-YFP also failed to assemble into peripheral lines (Figures 6D and 6H). We hypothesize that INP1 (e.g., its cytoplasmic pool) may be directly or indirectly necessary for targeting D6PKL3 to the PM or for its maintenance there, although the underlying mechanism is presently unclear.
Figure 6.
D6PKL3 and INP1 Require Each Other for Their Localization at the Aperture Domains.
(A) to (C) and (E) to (G) INP1-YFP assembles into punctate lines in the Col-0 background ([A] and [E]) but fails to assemble into peripheral puncta and lines in the d6pk3 mutant backgrounds ([B], [C], [F], and [G]).
(D) and (H) D6PKL3-YFP fails to display peripheral puncta and lines in the inp1 background. Top images: single optical sections of tetrads. Bottom images: 3D reconstructions from confocal z-stacks. Bars = 10 μm.
To test if INP1 and D6PKL3 may interact directly, we performed yeast two-hybrid (Y2H) interaction assays, bimolecular fluorescence complementation (BiFC) assays in leaf protoplasts, and in vitro phosphorylation assays. INP1 and D6PKL3 did not interact in yeast or protoplasts (Figures 7A and 7B). Since interactions between kinases and their substrates are often transient, we attempted to stabilize the interaction in the Y2H assay using a kinase-dead version of D6PKL3, yet the proteins still showed no interaction (Figure 7A). Similarly, the purified GST-D6PKL3 did not phosphorylate GST-INP1 in an in vitro phosphorylation assay (Figure 7C). By contrast, GST-D6PKL3 successfully phosphorylated the auxin transporter PIN1 (Figure 7C), which had previously been shown to act in vitro as a D6PKL3 phosphorylation substrate (Weller et al., 2017), therefore confirming that the recombinant GST-D6PKL3 protein retained kinase activity. We further attempted to increase the kinase activity of D6PKL3 by introducing a phosphomimic mutation (S415D) into the D6PKL3 activation loop, as similar modification has previously been shown to increase the activity of the D6PKL3-related AGCVIIIa kinase PROTEIN KINASE ASSOCIATED WITH BRX (PAX) (Marhava et al., 2018). Although compared with both the wild-type D6PKL3 and the D6PKL3S415A mutant, the activity of D6PKL3S415D toward PIN1 indeed considerably increased, it still showed no interaction with INP1 (Figure 7C).
Figure 7.
D6PKL3 and INP1 Do Not Show Interaction in Y2H, BiFC, and Kinase Phosphorylation Assays.
(A) Y2H experiments. Two separate colonies were grown for each combination of constructs. INP1/positive indicates a positive control for interaction between INP1 and the ribosomal protein S26e (At2g40510), which was found in a Hybrigenics Y2H screen as a strong interactor of INP1.
(B) BiFC experiments. Leaf protoplasts were transformed, respectively, with an empty vector, the 35S:GFP construct (a transformation control), and the INP1-nYFP/D6PKL3-cYFP constructs.
(C) In vitro kinase activity assay using purified GST-D6PKL3 (D6PKL3), GST-D6PKL3S415A (D6PKL3S415A), or the phosphomimic variant GST-D6PKL3S415D (D6PKL3S415D). Purified GST-INP1 (INP1) and the GST-tagged cytoplasmic loop of PIN1 (PIN1) were used as substrates. CBB, Coomassie Brilliant Blue-stained gel; AR, autoradiograph.
D6PKL3 and INP1 May Localize to Different Sides of the PM at the Aperture Domains
We recently proposed that the punctate lines of INP1 might form at the outer side of the PM at the aperture domains, where INP1 may interact with both the PM and callose wall, although determining the exact subcellular locations of INP1 complexes is difficult (Dobritsa and Reeder, 2017; Dobritsa et al., 2018). To try to determine subcellular localization of the D6PKL3 puncta, we performed plasmolysis experiments on D6PKL3-YFP-expressing tetrads by incubating them in a solution of 25% sucrose. In contrast to the results of similar experiments with INP1-YFP (Dobritsa et al., 2018), we found that the D6PKL3-YFP puncta in the plasmolyzed tetrads remained associated primarily with the PM that was detached from the cell wall after plasmolysis and not with the callose wall (Figure 8A), suggesting that D6PKL3 likely localizes to the cytoplasmic side of the PM aperture domains. The differential behavior of INP1 and D6PKL3 in this experiment can be used as a further indication of the different roles of these two proteins in aperture formation.
Figure 8.
D6PKL3 Appears to Be Closely Associated with the PM and Does Not Depend on the Presence of the Callose Wall for Localization.
(A) In a plasmolyzed tetrad in which the PM has dissociated from the callose wall (arrows), the D6PKL3-YFP fluorescence remains in close contact with the PM.
(B) In the cals5-2 tetrads lacking the outer callose wall, D6PKL3-YFP signal was observed both next to the remaining callose wall in the center of the tetrad and at the tetrad periphery where no wall was present.
(C) The INP1-YFP signal preferentially clustered near the remaining callose wall in the center of the cals5-2 tetrads. Merged fluorescent signals from YFP (yellow), Calcofluor White (CW; blue, callose wall), and CellMask Deep Red (DR; magenta, membranous structures). Bars = 10 μm.
To investigate the involvement of the callose wall in D6PKL3 localization, we additionally checked the localization of D6PKL3-YFP in the callose synthase5-2 (cals5-2) mutant background (Dong et al., 2005), which lacks recognizable apertures (Dobritsa et al., 2018). In this mutant, production of the outer callose wall around the tetrad is largely abolished, but the intersporal walls, albeit weaker than in the wild type, still develop and separate individual microspores (Dong et al., 2005; Dobritsa et al., 2018). Unlike the puncta of INP1-YFP, which tended to cluster near the remaining callose walls at the center of cals5-2 tetrads (Dobritsa et al., 2018) (Figure 8C), the D6PKL3-YFP puncta were largely distributed normally, next to both the inner regions, where the callose wall still remained, and the outer regions, where the wall was missing (Figure 8B). Taken together, D6PKL3 appears to be associated with the plasma membrane, but not with the callose wall.
D6PKL3 and INP1 Bind Phosphatidylinositol Species, Some of Which Are Enriched at the Aperture PM Domains
It was recently shown that D6PK family members and related kinases can bind phospholipids and that this binding is important for the association of kinase with the PM (Stanislas et al., 2015; Barbosa et al., 2016). We used purified GST-D6PKL3 to test its ability to interact with a panel of phospholipids. In this lipid binding assay, D6PKL3 bound to all negatively charged phosphoinositides as well as to phosphatidyl serine, but not to the phospholipids with neutral head groups (phosphatidylcholine and phosphatidylethanolamine) or to the monoacidic phosphatidyl inositol; Figure 9A). Additionally, GST-INP1 showed a similar ability to bind phosphorylated PI variants (Figure 9A).
Figure 9.
D6PKL3 and INP1 Can Bind to Phospholipids, and Some Phosphoinositides Show Enrichment at the Aperture PM Domains.
(A) In vitro lipid binding assays with a panel of phospholipids and purified GST, GST-INP1, and GST-D6PKL3. LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; PI, phosphatidylinositol and its mono/bis/tris-phosphates; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine-1-phosphate; PA, phosphatidic acid; PS, phosphatidylserine.
(B) and (C) Localization of the PIP#21 sensor for PI(4)P (construct DMC1pr:mCitrine-2xPHFAPP1) in tetrad-stage (B) and in recently released free microspores (C). Positions of aperture domains and apertures are indicated with arrows. In (B), fluorescent signals are from CellMask Deep Red (DR; magenta, membranous structures), YFP (yellow), and Calcofluor White (CW; blue, callose wall on merged image). In (C), fluorescent signals are from developing exine (blue), YFP (yellow), and CellMask Deep Red (magenta, membranous structures, on merged image). YFP signals are shown both as yellow color (left image) and as color-coded heat maps of fluorescence intensity (right image).
(D) Localization of the PIP#15 sensor for PI(4,5)P2 (construct MMD1pr:mCitrine-TUBBY-C) in tetrad-stage microspores. Shown are the same types of fluorescent signals as in (B). Bars = 5 μm.
Despite their typically low abundance in cells, phosphoinositides are important players in polarity generation in many organisms (Di Paolo and De Camilli, 2006; Ischebeck et al., 2010; Tejos et al., 2014). Their asymmetric distribution can contribute to the recruitment of membrane-associated proteins to specific regions of the PM and its organization into distinct domains. The distribution of phosphoinositides in developing pollen has been unclear. To determine if any phosphoinositide species were specifically enriched at the aperture PM domains, where they could serve as molecular anchors for D6PKL3 and/or INP1, we took advantage of genetically encoded fluorescent phospholipid biosensors (PIP lines) (Simon et al., 2014, 2016). Because the UBIQUITIN10 promoter used for the original PIP lines appeared to be nonfunctional in sporogenic cells, we replaced it with the meiosis-associated DMC1 or MMD1 promoters, which are active in MMCs and microspores (Klimyuk and Jones, 1997; Yang et al., 2003; Dobritsa et al., 2018). We transformed these constructs into Arabidopsis and checked the sensor localization patterns during microspore development. In two of the PIP lines, PIP#21 [expressing mCitrine-2xPHFAPP1, a sensor for PI(4)P), and PIP#15, expressing mCitrine-Tubby-C, a sensor for PI(4,5)P2], fluorescent signal, although weak, showed some enrichment at the protrusions and ridges of the microspore PM corresponding to the characteristic sites of future apertures (Figures 9B to 9D). These results suggest that the aperture PM domains are enriched in PI(4)P and PI(4,5)P2, which could influence the distribution and aggregation of proteins required for aperture specification and formation.
A K/R-Rich Motif Contributes to D6PKL3 Localization at the Aperture Domains
In animals and plants, protein motifs with an elevated basic/hydrophobicity (BH) score are important for targeting proteins to the PM and keeping them at distinct, polarized cortex domains (Heo et al., 2006; Brzeska et al., 2010; Bailey and Prehoda, 2015; Barbosa et al., 2016; Simon et al., 2016). In D6PK and several other members of the AGCVIII family, a short, positively charged (lysine-rich) motif located in the insertion region (MID domain) separating kinase subdomains VII and VIII governs PM association (Barbosa et al., 2016) (Figures 1M and 10A). The corresponding region in D6PKL3 (amino acids 380–393) contains four lysine and two arginine residues (Figure 10A) and has a BH score > 0.6, meeting the definition of a basic/hydrophobic motif that may confer membrane association (Brzeska et al., 2010).
Figure 10.
Presence of the K/R-Rich Motif Helps D6PKL3 Associate with the Aperture PM Domains.
(A) Alignment of the protein region in the vicinity of the previously described K-rich motif in D6PK (blue line) and the corresponding regions in the other D6PK subfamily members. The K/R-rich motif of D6PKL3 is underlined (red line). Positions of amino acids replaced with Ala are indicated.
(B) The D6PKL34K2R→6A protein has lost its ability to interact with phospholipids, whereas D6PKL34K→4A still exhibits significant affinity to phospholipids. In vitro lipid binding assays with the same panel of phospholipids as in Figure 9 and with purified GST-D6PKL3, GST-D6PKL34K→4A, and GST-D6PKL34K2R→6A.
(C) to (L) Representative examples of tetrads (3D reconstructions; [C] to [G]) and pollen grains ([H] to [L]) from transgenic lines expressing the indicated K/R to A-substituted constructs. Bars = 10 μm.
To test if this lysine/arginine-rich (K/R) motif may facilitate the D6PKL3 localization to the aperture domains, we generated mutated versions of D6PKL3, replacing four, five, or all six basic residues in this region with alanine (Figure 10A). The resulting YFP-tagged D6PKL34K→4A, D6PKL34K1R→5A, and D6PKL34K2R→6A constructs, driven by the D6PKL3 promoter, were transformed into the d6pkl3-2 mutant and in Col-0, and protein localization and aperture phenotypes were analyzed in the transgenic plants (Figures 10C to 10L).
The D6PKL34K→4A construct still fully complemented the mutant aperture phenotype (3/3 lines) and exhibited easily recognizable peripheral YFP lines, indistinguishable from those of the wild-type D6PKL3 (7/8 lines) (Figures 10C and 10H). In addition, in the in vitro phospholipid binding assay, the corresponding GST-tagged version of this mutated protein also showed a significant, albeit weaker relative to the wild type, ability to interact with phospholipids. The D6PKL34K1R→5A mutation, however, had a negative effect on the formation of punctate lines at the microspore periphery (Figures 10D to 10G); 5/7 transgenic lines showed noticeable reduction in peripheral puncta, with many tetrads showing no peripheral accumulation in D6PKL3. Despite the apparently weak PM association of the D6PKL34K1R→5A protein, the aperture phenotype was rescued in the D6PKL34K1R→5A d6pkl3-2 lines (3/3 lines) (Figures 10I and 10J). However, when all six basic residues were mutated in the D6PKL34K2R→6A construct, both protein localization and aperture formation were affected most significantly, with 7/7 lines showing very few, if any, peripheral puncta and 2/4 D6PKL34K2R→6A d6pkl3-2 lines displaying a complete lack of rescue of the mutant aperture phenotype (Figures 10F, 10G, and 10L). In addition to the reduction in the in vivo localization of D6PKL34K2R→6A to aperture domains, the ability of the purified GST-D6PKL34K2R→6A protein to bind phospholipids in vitro was also compromised (Figure 10B). These data suggest that the K/R motif helps D6PKL3 to stay near aperture membrane domains: When this ability is compromised, this might affect the plasma membrane association of D6PKL3 molecules or their rate of diffusion, which may have a negative impact on aperture formation.
The apparent retention of the aperture-forming activity by D6PKL34K2R→6A (Figure 10K) in two out of the four transgenic lines suggest that protein sequence elements other than the K/R-rich motif may contribute to protein targeting to the aperture domains. Even though weakened in its interaction with the PM, D6PKL3 may still be correctly delivered to perform its function, albeit with a reduced retention time. It may also be that protein levels vary between the wild type and the mutant variant, since the K/R mutations may affect other properties of the protein, such as its stability or turnover. Previous work on the related kinase D6PK revealed that transgenic plants expressing similar K/R-mutated D6PK variants vary strongly in D6PK abundance and that this can be aligned with relevant phenotypic differences (Barbosa et al., 2016). In lines with somewhat higher overall concentrations of D6PKL3, more protein may successfully reach the aperture domains, thus resulting in aperture formation.
DISCUSSION
Pollen apertures exhibit tremendous diversity across species in regard to their numbers, positions, and morphology. They typically develop in a given species with remarkable precision and provide an excellent model for studying how cells specify and create distinct cellular and extracellular domains (Wang and Dobritsa, 2018). The large variety of aperture patterns in nature suggests that the system might be fairly responsive to genetic perturbations. Furthermore, although in some species apertures, serving as the sites for pollen tube exit, are essential for plant reproduction (Li et al., 2018), they are dispensable in Arabidopsis grown under laboratory conditions (Dobritsa et al., 2011; Dobritsa and Coerper, 2012) and, as this study shows, sometimes in nature. In this study, we discovered two natural D6PKL3-disrupting polymorphisms that exist in Arabidopsis accessions and found that D6PKL3 is involved in the formation of aperture domains. Interestingly, the screening of more than 300 accessions revealed just one accession, Mt-0, in which apertures were formed abnormally, in addition to Yeg-1, which was discovered later through a reverse-genetics approach. The apparently low frequency of pollen aperture mutants among the Arabidopsis accessions is potentially significant, as it could suggest that even though pollen grains retain fertility in the absence of apertures in this species, natural selection likely still favors pollen with normal apertures. In addition to the role of apertures in pollen tube emergence, this could be due to other roles apertures play, such as helping pollen accommodate volume changes in response to changing hydration conditions (Wodehouse, 1935; Heslop-Harrison, 1979; Katifori et al., 2010). The apparent prevalence of accessions with normal apertures among Arabidopsis populations is consistent with the results of a recent study that evaluated the effect of aperture number on pollen performance and suggested that three apertures may represent the best evolutionary tradeoff (Albert et al., 2018). It remains to be seen if the loss of normal apertures in Mt-0 and Yeg-1 represents a beneficial adaptation; for example, to specific climatic conditions to which these accessions are exposed in Libya and Armenia, respectively.
Like the other previously identified aperture factor INP1, D6PKL3 specifically accumulates at the future aperture domains in microspores. Other members of the D6PK subfamily to which D6PKL3 belongs, as well as related AGCVIII kinases, have previously been identified as critical players in the generation of polarity and the formation of distinct domains in vegetative cells. Most notably, D6PK, a paralog of D6PKL3, as well as the AGCVIII kinases PINOID, PAX, WAG1, and WAG2, phosphorylate and activate the polarly localized PIN auxin transporters (Friml et al., 2004; Michniewicz et al., 2007; Zourelidou et al., 2009; Willige et al., 2013; Barbosa et al., 2014; Weller et al., 2017; Marhava et al., 2018). In addition, both D6PK and PAX themselves exhibit polar distribution at the PM of vegetative cells (Zourelidou et al., 2009; Barbosa et al., 2014; Stanislas et al., 2015; Marhava et al., 2018). The discovery that D6PKL3, which functions in developing pollen, is also associated with very distinct aperture membrane domains suggests that members of this family have been recruited multiple times and in very diverse cell types to participate in domain formation and polarity establishment.
Importantly, D6PKL3 is the only member of the D6PK subfamily in Arabidopsis that is involved in the formation of apertures, as mutations in the three other proteins do not result in abnormal phenotypes on their own and do not enhance the d6pkl3 phenotype when tested in combination. The aperture defects are very similar in different d6pkl3 mutants, with most of the pollen grains still developing three recognizable traces of apertures placed at the normal positions. In that respect, the d6pkl3 defects are milder compared with the total lack of apertures observed in inp1 mutant pollen. It remains to be seen if this weaker phenotype is due to the involvement in aperture formation of other kinases besides D6PK, D6PKL1, and D6PKL2 (e.g., other members of the AGCVIII family) or if the role of D6PKL3 in this process, although important, is less essential than that of INP1.
As a membrane-associated kinase, D6PKL3 potentially helps create the defining features of aperture PM domains that are used to attract the downstream executor proteins, such as INP1. The results of analysis of the expression and localization of these two proteins are consistent with the model in which D6PKL3 acts upstream of INP1, since D6PKL3 starts assembling into the three characteristic punctate lines at the PM significantly earlier than INP1, with the D6PKL3 lines already becoming visible before the end of meiotic cytokinesis. This finding has important implications for understanding the mechanism through which microspores establish positions of their apertures and place them in the arrangement most prevalent among eudicots, at the sites facing each other in sister microspores. Although it has been hypothesized for a long time that the last contact points formed at the end of meiotic cytokinesis between the sister microspores may serve as the spatial cues defining aperture placement (Wodehouse, 1935; Ressayre et al., 1998, 2002; Dobritsa and Coerper, 2012), our results show that the formation of the D6PKL3-decorated domains happens prior to the formation of the last contact points and, therefore, the initial aperture positioning must utilize a different mechanism.
Interestingly, the phenotypes of transgenic D6PKL3pr:D6PKL3-YFP plants indicate that D6PKL3 is sufficient for the formation of new aperture domains and additional apertures, and thus it may be a limiting factor in aperture formation. The unusual double-aperture phenotype, commonly observed in these plants, arises due to D6PKL3 assembling into two parallel, closely positioned lines at each of the three normal aperture domains and attracting INP1 to these sites. At each aperture domain, exine is then excluded from two, rather than the typical one, linear regions and is deposited between these lines. The double-aperture phenotype, although not very common, does exist in nature. For example, pollen of several species of Passiflora (e.g., P. citrine, P. suberosa, and P. costaricensis) and Teucrium represented in PalDat (www.paldat.org), the Palynological Database of pollen morphology, displays this phenotype. Additionally, pollen of many species exhibits a feature described in the palynological literature as “ornamented aperture membrane,” referring to exine deposition within the aperture regions. This much more common morphology could also be related to the double-aperture phenotype, as apertures of at least some species with this morphology (e.g., Cretan ebony [Ebenus cretica] and sainfoin [Onobrychis viciifolia]) show similarity to double apertures (PalDat; www.paldat.org). This suggests the possibility that plant species may generate different patterns on their pollen surface through modulation of protein kinases orthologous or related to D6PKL3.
In addition to the double apertures, some pollen grains in D6PKL3-YFP transgenic lines developed more than the normal complement of apertures: These grains did not exhibit any noticeable increase in size, suggesting that this increase in aperture number was different from the previously described effect of pollen ploidy and/or size on the number of apertures (Reeder et al., 2016) and was likely the consequence of microspores containing higher than normal levels of D6PKL3. This is in contrast to the behavior of INP1, which, on its own, does not appear to be sufficient to promote aperture formation (Reeder et al., 2016).
The kinase activity of D6PKL3 is important for its membrane localization and role in aperture formation, as the kinase-dead D6PKL3 does not assemble into PM aperture lines and does not complement the d6pkl3-2 mutant aperture phenotype. In this regard, D6PKL3 behaves differently from its proposed paralog D6PK, whose kinase-dead version still exhibits polar distribution in root epidermal cells, similar to the wild-type protein (Barbosa et al., 2014). The phosphorylation targets of D6PKL3 in microspores remain to be identified, but our results indicate that INP1 may not be one of them (with the important caveat that the physical interaction between D6PKL3 and INP1 has not yet been studied in developing pollen where these proteins function). Still, INP1 depends on the presence of functional D6PKL3 for its accumulation at the aperture domains, and, in turn, the presence of INP1 appears to be necessary for the maintenance of DPKL3 at these domains, although the mechanisms through which these proteins influence each other’s localization remain to be identified.
D6PK family members have a demonstrated role in regulating the polarity of auxin transport in vegetative systems by phosphorylating PIN transporters. This invites the question of whether auxin-related mechanisms, controlled by D6PKL3, could be involved in the formation of pollen apertures. So far, significant auxin production in developing anthers, as measured by the activation of the auxin-responsive reporters DR5:GUS and DR5:GFP, was found only after the tetrad stage during which the aperture sites are specified (Cecchetti et al., 2008; Yao et al., 2018). However, the auxin biosynthesis genes YUCCA2 (YUC2) and YUC6 are expressed at earlier developmental stages in MMCs and tetrads (Cecchetti et al., 2008; Yao et al., 2018), and free indole-3-acetic acid, the most common form of auxin, is detectable during the corresponding stages of anther development (Cecchetti et al., 2013). Therefore, it cannot be completely excluded that auxin and its transport contribute to aperture formation. Yet, at this moment, there is no demonstrated role of PINs as potential substrates of D6PKL3 in pollen development, and it is thus not possible to draw any conclusions about the involvement of D6PKL3 in controlling auxin transport during the formation of aperture domains. Furthermore, a recent analysis of the yuc2 yuc6 double mutant showed that its early free-stage microspores exhibit apparently normal development of the pollen wall and apertures (Yao et al., 2018), suggesting that the role of D6PKL3 in aperture formation might differ from its role in auxin transport in vegetative tissues.
In addition to the requirement for the functional kinase active site, the K/R-rich region of D6PKL3 helps keep the protein at the PM, with the mutations in these basic residues leading to a significant reduction in the ability of D6PKL3 to form peripheral puncta and lines. In many organisms, phosphoinositides, although found in very small quantities, serve as important players in the formation of distinct PM domains and the generation of polarity. In multiple systems and biological processes, these phosphorylated versions of phosphatidyl inositol are distributed nonuniformly in the PM and are often associated with non-uniformly localized proteins. The creation of specific domain identities and the breaking of local symmetry, mediated by the distribution of phosphoinositide variants, contribute to the acquisition of different fates by daughter cells after division, the formation of cellular outgrowths (including pollen tubes), the generation of apico-basal and planar polarity, and many other developmental and physiological processes (Kost et al., 1999; Pinal et al., 2006; Comer and Parent, 2007; Martin-Belmonte and Mostov, 2008; Tejos et al., 2014; Sekereš et al., 2015). Like several other members of the D6PK and AGCVIII families (Stanislas et al., 2015; Barbosa et al., 2016; Simon et al., 2016), D6PKL3 interacts with several phosphoinositides, with the interaction dependent on the presence of the K/R-rich motif in D6PKL3. Two D6PKL3-cognate phosphoinositides, PI(4)P and PI(4,5)P2, exhibited some enrichment in the PM aperture domains, suggesting that these domains may have specific lipid signatures that can facilitate protein recruitment to these domains.
Based on the results presented here, we propose the following working model for aperture formation (Figure 11). We propose that D6PKL3 reads positional cues that appear during male meiotic cytokinesis and accumulates at the three specific equidistant membrane domains in each of the four budding microspores. A combination of kinase activity and the presence of the K/R-rich motif-phospholipid interactions at the PM are necessary for keeping D6PKL3 at these domains. The presence of D6PKL3 and other factors, e.g., its phosphorylation substrates, creates the distinguishing features of these domains, with both sides of the PM possibly undergoing changes. During the tetrad stage, INP1 becomes attracted to the D6PKL3-decorated membrane domains and aggregates there, potentially assembling in complexes on their outer surfaces. The accumulation of INP1 at these domains contributes to the formation of membrane ridges that keep aperture domains tethered to the overlying callose wall, preventing the deposition of primexine and, eventually, exine at these sites. In addition to explaining a functional role of D6PKL3 in pollen development, our study may also contribute to the understanding of the cell biological processes that control polarity in other cellular systems in plants.
Figure 11.
Working Model for the Roles of D6PKL3 and INP1 in Pollen Aperture Formation.
Callose wall is indicated in blue, MMC and microspores are in magenta, and primexine is in green. D6PKL3 and INP1 proteins are represented by yellow/beige and blue circles, respectively.
METHODS
Plant Materials and Growth Conditions
Plants were grown at 20 to 22°C under a 16-h-light/8-h-dark cycle in growth chambers or in a greenhouse at the Biotechnology facility at OSU. White light (120–150 μmol m−2 s−1) from F96T12 fluorescent lamps supplemented with 60-W incandescent bulbs was used in the growth chambers. In the greenhouse, the ambient light was supplemented with the light from 400-W high-intensity discharge metal halide lamps. The list of Arabidopsis (Arabidopsis thaliana) accessions that were screened is provided in Supplemental Data Set 1. The following genotypes were used in this study: Columbia (Col-0), inp1-1 (Dobritsa and Coerper, 2012), cals5-2 (Dong et al., 2005), Martuba (Mt-0, CS77112), Yeghegis-1 (Yeg-1, CS76394), d6pkl3-2 (SALK_047347), d6pkl3-4 (GK-265A02, CS308942), d6pkl3-5 (GK-169H08, CS317999), d6pk-1 (SALK_061847), d6pkl1-1 (SALK_056618), d6pkl2-2 (SALK_086127), and the previously described combinations of mutations in the D6PK family of genes (Zourelidou et al., 2009). The point mutations d6pkl3E290K (d6pkl3-6) and d6pkl3G506K (d6pkl3-7) were identified in our recent forward genetic screen performed on EMS-mutagenized M2 plants of the Landsberg erecta (Ler) background. The aperture phenotype of Ler pollen is indistinguishable from that of Col-0.
Screen of Arabidopsis Natural Accessions
For the pollen aperture screen, flowers of 316 natural accessions (Supplemental Data Set 1) donated to the ABRC by Magnus Nordberg (Gregor Mendel Institute, Vienna, Austria) and grown by the ABRC at OSU were collected into Eppendorf tubes and stored at −80°C until ready to be processed. For screening, pollen from these flowers was brushed onto a glass slide and allowed to dry for ∼10 min. The pollen was then examined for abnormalities under a dissecting microscope (Nikon SMZ745; magnification 75×) as previously described (Dobritsa et al., 2011). Particular attention was paid to such characteristics as pollen shape, size, light reflection (glossiness), and tendency to form clumps.
Mapping the Source of the Mt-0 Aperture Defect
Mt-0 was crossed with Col-0, and the resulting F2 population was screened under a dissecting microscope for the presence of the round-pollen mutant phenotype. DNA was isolated from 926 mutants, and we first performed bulked segregant analysis to identify the general genomic interval in which the Mt-0 mutation was located, followed by map-based positional cloning (Lukowitz et al., 2000). The INDEL-based PCR markers for this analysis were generated using combined information from the 1001 Genomes Project database (http://signal.salk.edu/atg1001/index.php; 1001 Genomes Consortium, 2016) and the Arabidopsis Mapping Platform (http://amp.genomics.org.cn/; Hou et al., 2010). These markers are listed in Supplemental Data Set 2. The mutation was first mapped to a 198-kb interval on chromosome 3 between markers Mt0-3-10Mb-K (10,086,975 bp) and Mt0-3-10Mb-H (10,285,600 bp). We then further reduced this interval to a 125-kb region between 10,118,890 and 10,244,584 bp by Sanger-sequencing selected sites predicted to be polymorphic between Col-0 and Mt-0 in the two plants that still contained heterozygous markers at the borders of the previous 198-kb interval. In parallel with the map-based cloning, we also used NGS on a pooled sample containing DNA from 150 mutant F2 plants. SHORE v0.9.3 (http://shore.sourceforge.net/wiki/index.php/SHORE_Overview) (Ossowski et al., 2008) was used to map reads and identify polymorphisms against the TAIR10 genome. SHOREmap v3.0 (http://bioinfo.mpipz.mpg.de/shoremap/) was then used to identify Mt-0 marker enrichment and annotate the effects of polymorphisms on genes. Although the NGS analysis confirmed the enrichment of Mt-0 markers around the 10-Mb region of chromosome 3 and verified the presence of specific polymorphisms predicted by the 1001 Genomes database, its resolution was lower than that of the map-based cloning. To determine which of the 38 genes in this 125-kb interval was responsible for the aperture defect in Mt-0, we looked at the polymorphisms predicted to affect protein sequences, first concentrating on polymorphisms between Mt-0 and Col-0 (most of the genes had them), and then comparing the Mt-0 sequences with those from 62 additional accessions that were scored as wild-type for their pollen phenotype. This approach allowed us to narrow the list of the most likely candidates to just five genes (At3g27400, At3g27420, At3g27530, At3g27580, and At3g27590). We then evaluated mutants with the T-DNA insertions in these genes available from ABRC for the presence of the Mt-0-like phenotypes. Only the mutants of At3g27580, which had a premature stop codon in Mt-0, displayed these phenotypes.
Transgenic Constructs
All primers used in this study are described in Supplemental Table 1. To create the D6PKL3pr:D6PKL3-YFP construct, the D6PKL3 promoter and open reading frame were amplified, respectively, with primer pairs AD229/230 and AD228/221 and placed into pGR111 (Dobritsa et al., 2010) upstream of the YFP gene with the In-Fusion HD Cloning Kit (Clontech). To create DMC1pr:mCitrine-2xPHFAPP1 [PIP#21 for PI(4)P], mCitrine-2xPHFAPP1 was amplified using the original PIP#21 line (Simon et al., 2014) with PIPline-AgeI-F and PIPline-SpeI-R primers, digested with AgeI/SpeI, and replaced the INP1 gene in the DMC1pr:INP1-YFP-pGR111 construct (Dobritsa et al., 2018). To create MMD1pr:mCitrine-TUBBY-C [PIP#15 for PI(4,5)P2], mCitrine-TUBBY-C was amplified using the original PIP#15 line with PIPline-AgeI-F and PIPline-XbaI-R primers, digested with AgeI/XbaI, and replaced the INP1 gene in the MMD1pr:INP1-YFP-pGR111 construct (Dobritsa et al., 2018). Other PIP constructs were made in a similar manner using DNA from PIP lines created by Yvon Jaillais’ group (Simon et al., 2014). All constructs were verified by sequencing and transformed into the Agrobacterium tumefaciens strain GV3101. Wild-type Col-0 or d6pkl3-2 plants were then transformed by floral dip (Clough and Bent, 1998), transgenic plants were selected with BASTA, and the presence of transgenes was confirmed with specific primers.
Site-Directed Mutagenesis
D6PKL3pr:D6PKL3K211E-YFP, D6PKL3pr:D6PKL34K→4A-YFP, D6PKL3pr:D6PKL34K1R→5A-YFP, D6PKL3pr:D6PKL34K2R→6A-YFP, GST-D6PKL3S415D, GST-D6PKL3S415A, GST-D6PKL34K→4A, and GST-D6PKL34K2R→6A were created by PCR-based site-directed mutagenesis (Sawano and Miyawaki, 2000; Zourelidou et al., 2009), using D6PKL3pr:D6PKL3-YFP and GST-D6PKL3 with AD325 primer for kinase-dead D6PKL3K211E constructs, AD345 for D6PKL34K→4A, AD346 for D6PKL34K2R→6A, AD400 for GST-D6PKL3S415D, and AD401 for D6PKL3S415A. D6PKL34K1R→5A was fortuitously created in the reaction with AD346 primer and confirmed by sequencing.
Confocal Microscopy
Preparation and imaging of mature pollen and tetrads, as well as the plasmolysis experiments, were performed as previously described (Dobritsa et al., 2018). Imaging was done on a Nikon A1+ confocal microscope with a 100× oil-immersion objective (NA = 1.4), using the 3× confocal zoom for pollen grains and the 5× zoom for tetrads. To image pollen grains, pollen was placed into a 5-μL drop of auramine O solution (0.001%; diluted in water from the 0.1% stock prepared in 50 mM Tris-HCl), allowed to hydrate, covered with a #1.5 cover slip, and sealed with nail polish. Exine was excited with a 488-nm laser and fluorescence was collected at 500 to 550 nm. To visualize apertures on the entire surface, images from the front and back of the pollen grains were taken.
To image tetrads, anthers were dissected out of stage-9 flower buds (Smyth et al., 1990) and placed into the Vectashield antifade solution (Vector Labs) supplemented with membrane stain CellMask Deep Red (Molecular Probes) (5 μg/mL) and calcofluor white (0.02%). Anthers were covered with a cover slip, to which gentle pressure was applied to release tetrads. CFP and YFP were excited, respectively, with 457- and 514-nm lasers, and their emissions were collected, respectively, at 465 to 500 nm and at 522 to 555 nm. Calcofluor white was excited with 405-nm laser and collected at 424 to 475 nm, and CellMask Deep Red dye was excited with 640-nm laser and collected at 663 to 738 nm. Z-stacks of tetrads were obtained with a step size of 500 nm and 3D reconstructed using NIS Elements v.4.20 (Nikon).
Quantitative RT-PCR
Total RNA was extracted from homozygous D6PKL3pr:D6PKL3-YFP T3 plants as previously described (Reeder et al., 2016). The primer pair AD347/348 was used to amplify the YFP-containing transgene, and the primer pair AD349/350 was used for the control gene MMD1 expressed in MMCs and tetrads (Yang et al., 2003; Dobritsa et al., 2018). Three biological replicates (each consisting of cDNA prepared from nonoverlapping pools of ∼150 stage-9 flower buds from at least 40 individual plants) and two technical replicates (repeated measurements of the biological samples) were used for each of the four transgenic lines. After qRT-PCR, the amounts of the transcripts were calculated relative to standard curves obtained from serial dilutions of the YFP transgene-carrying genomic DNA. For each sample, the amount of the D6PKL3-YFP transgenic transcripts was then normalized to the amount of the MMD1 transcripts used as an endogenous control.
Protein Purification
To create GST-INP1 and GST-D6PKL3 constructs, the ORFs of INP1 and D6PKL3 were amplified with BamHI-INP1-EF/NotI-INP1-HR and AD283/284 primers, respectively. The PCR products were digested with BamHI/NotI and BamHI/SalI, respectively, and placed into the pGEX-6p-1 vector (GE Healthcare). Constructs expressing the GST-fused proteins were transformed into BL21(DE3) cells. Cultures (250 mL), grown at 37°C for 5 h until OD600 0.5 to 1.0, were induced with 0.5 mM IPTG and grown at 30°C for an additional 2 h. The cells were pelleted, sonicated in PBS with protease inhibitor (Complete, EDTA-free; Roche), and GST fusions were purified from supernatants with Glutathione Agarose 4B (Macherey-Nagel) per the manufacturer’s instructions.
Protein Phosphorylation Assay
In vitro phosphorylation experiments were performed using purified recombinant GST-D6PKL3, GST-D6PKL3S415A, and GST-D6PKL3S415D in combination with purified recombinant GST-INP1 and the GST-tagged cytoplasmic loop of PIN1 (Zourelidou et al., 2014). The recombinant proteins were incubated for 30 min at 28°C in phosphorylation buffer (25 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 2.0 mM EDTA, and 1 μL protease inhibitor [Complete, EDTA-free; Roche]), supplemented with 10 μCi [γ-32P]ATP (370 MBq, specific activity 185 TBq; Hartmann Analytic). Each 20-μL reaction was stopped by adding 5× Laemmli buffer and loaded on a 10% SDS-PAGE. The protein extracts were separated on a 10% SDS-PAGE. The gel was exposed to an x-ray film and then rehydrated and stained with Coomassie Brilliant Blue to serve as a loading control.
Lipid Binding Assays
Lipid binding assays were performed on PIP-strip membranes (Echelon Biosciences; P-6001) as previously described (Barbosa et al., 2016). Binding of GST-fused proteins was detected with primary rabbit anti-GST (Sigma-Aldrich; G7781; 1:2000 in TBS-T [0.1% Tween] buffer with 4% BSA) and secondary anti-rabbit IgG-peroxidase conjugated antibodies (KPL; 5220-0283; 1:8000 in TBS-T buffer with 4% BSA). Free GST was used as a negative control. After the final washes, the membranes were processed using SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific; no. 34577) and imaged with a MyECL imager (Thermo Scientific).
Yeast Two-Hybrid Assay
The NMY51 yeast strain (a gift from Marisa Otegui) was used for Y2H assays. The bait construct pB29-INP1 was constructed by Hybrigenics. The open reading frame regions of INP1 and D6PKL3 were cloned into the pP6 prey vector using restriction enzymes NcoI-BamHI for INP1 and BamHI-XbaI for D6PKL3. Positive bait-prey cotransformants were selected on synthetic dropout medium lacking Leu and Trp (-LW). To test for interaction, cotransformed yeast cells were grown either on the medium that, in addition to Leu and Trp, lacked His and contained 3 mM 3-amino-1,2,4-triazole (-LWH+3-AT) or on the medium lacking Leu/Trp/His/adenine (-LWHA).
BiFC Assay
To create BiFC constructs, the INP1 and D6PKL3 open reading frames were amplified with AD277/278 and AD279/280 primers, respectively, digested with SalI/BamHI, and cloned into the pA7-nYFP and pA7-cYFP split-YFP vectors under the control of the CaMV 35S promoter (Chen et al., 2006; Kang et al., 2010). The constructs were cotransformed into protoplasts isolated from wild-type Col-0 leaves as described (Yoo et al., 2007). A 35Spr:GFP construct (Kang et al., 2010) was used as a control for transformation efficiency. Protoplasts cotransformed with INP1-nYFP and D6PKL3-cYFP were excited with a 514-nm laser, and YFP emission was collected at 522 to 555 nm.
Accession Numbers
The Arabidopsis Genome Initiative accession numbers for the genes used in this study are the following: At5g55910 (D6PK), At4g26610 (D6PKL1), At5g47750 (D6PKL2), At3g27580 (D6PKL3), At4g22600 (INP1), and At2g13680 (CALS5). The NGS data were deposited into the Sequence Read Archive under accession number SRP158504.
Supplemental Data
Supplemental Figure 1. Shape of the Mt-0 pollen is distinguishable from the shape of the wild-type Col-0 pollen at the dissecting-microscope level.
Supplemental Figure 2. Pollen of F1 plants from a cross between Mt-0 and inp1 develops normal apertures.
Supplemental Figure 3. Alignment of the predicted D6PKL3 proteins from Col-0 and Yeg-1.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1. Natural accessions of Arabidopsis screened for pollen morphological phenotypes.
Supplemental Data Set 2. Molecular markers differentiating between Mt-0 and Col-0 that were used to map the Mt-0 aperture locus.
Supplemental Movie 1. Volume reconstruction of a z-stack of confocal sections through a tetrad of microspores expressing D6PKL3pr:D6PKL3-YFP.
Supplemental Movie 2. Volume reconstruction of a z-stack of confocal sections through a dividing microspore mother cell expressing D6PKL3pr:D6PKL3-YFP.
Supplemental Movie 3. D6PKL3-YFP can form double lines at the aperture domains.
Supplemental Movie 4. D6PKL3-YFP can assemble into more than three lines per microspore.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
Funding for this project was provided to A.A.D. by the U.S. National Science Foundation (MCB-1517511 and MCB-1817835). B.T.H. was funded by the National Institute of General Medical Sciences of the National Institute of Health (Award T32GM007103). C.S. was funded by the Deutsche Forschungsgemeinschaft (SCHW751/12-2). The undergraduate research of Z.T.W. was supported through the NSF-REU supplement mechanism. We are grateful to members of the Dobritsa lab for discussions and to Sarah Reeder for critical reading of the manuscript. We thank the ABRC at the Ohio State University for providing seed stocks, Keith Slotkin (OSU) for providing mutagenized seeds, Peng Li (OSU and Tsinghua University) for creating the DMC1pr:INP1-CFP construct, undergraduate students Prativa Amom and Michelle Tan (OSU) for finding two EMS mutants of D6PKL3, Marisa Otegui (University of Wisconsin, Madison) for the yeast strain, and Yvon Jaillais (ENS, Lyon, France) for PIP constructs.
AUTHOR CONTRIBUTIONS
B.H.L., R.J.S., C.S., and A.A.D. conceived and designed the experiments. B.H.L., Z.T.W., M.Z., B.T.H., and A.A.D. performed the experiments. B.H.L. and A.A.D. analyzed the data. B.H.L., C.S., and A.A.D. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- 1001 Genomes Consortium (2016). 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrash E.B., Bergmann D.C. (2009). Asymmetric cell divisions: a view from plant development. Dev. Cell 16: 783–796. [DOI] [PubMed] [Google Scholar]
- Albert B., Ressayre A., Dillmann C., Carlson A.L., Swanson R.J., Gouyon P.-H., Dobritsa A.A. (2018). Effect of aperture number on pollen germination, survival and reproductive success in Arabidopsis thaliana. Ann. Bot. 121: 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M.J., Prehoda K.E. (2015). Establishment of Par-polarized cortical domains via phosphoregulated membrane motifs. Dev. Cell 35: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa I.C.R., Zourelidou M., Willige B.C., Weller B., Schwechheimer C. (2014). D6 PROTEIN KINASE activates auxin transport-dependent growth and PIN-FORMED phosphorylation at the plasma membrane. Dev. Cell 29: 674–685. [DOI] [PubMed] [Google Scholar]
- Barbosa I.C.R., Shikata H., Zourelidou M., Heilmann M., Heilmann I., Schwechheimer C. (2016). Phospholipid composition and a polybasic motif determine D6 PROTEIN KINASE polar association with the plasma membrane and tropic responses. Development 143: 4687–4700. [DOI] [PubMed] [Google Scholar]
- Barbosa I.C.R., Hammes U.Z., Schwechheimer C. (2018). Activation and polarity control of PIN-FORMED auxin transporters by phosphorylation. Trends Plant Sci. 23: 523–538. [DOI] [PubMed] [Google Scholar]
- Brzeska H., Guag J., Remmert K., Chacko S., Korn E.D. (2010). An experimentally based computer search identifies unstructured membrane-binding sites in proteins: application to class I myosins, PAKS, and CARMIL. J. Biol. Chem. 285: 5738–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchetti V., Altamura M.M., Falasca G., Costantino P., Cardarelli M. (2008). Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchetti V., Altamura M.M., Brunetti P., Petrocelli V., Falasca G., Ljung K., Costantino P., Cardarelli M. (2013). Auxin controls Arabidopsis anther dehiscence by regulating endothecium lignification and jasmonic acid biosynthesis. Plant J. 74: 411–422. [DOI] [PubMed] [Google Scholar]
- Chen S., Tao L., Zeng L., Vega-Sanchez M.E., Umemura K., Wang G.-L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7: 417–427. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Comer F.I., Parent C.A. (2007). Phosphoinositides specify polarity during epithelial organ development. Cell 128: 239–240. [DOI] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P. (2006). Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657. [DOI] [PubMed] [Google Scholar]
- Dobritsa A.A., Coerper D. (2012). The novel plant protein INAPERTURATE POLLEN1 marks distinct cellular domains and controls formation of apertures in the Arabidopsis pollen exine. Plant Cell 24: 4452–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Reeder S.H. (2017). Formation of pollen apertures in Arabidopsis requires an interplay between male meiosis, development of INP1-decorated plasma membrane domains, and the callose wall. Plant Signal. Behav. 12: e1393136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Lei Z., Nishikawa S., Urbanczyk-Wochniak E., Huhman D.V., Preuss D., Sumner L.W. (2010). LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol. 153: 937–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Geanconteri A., Shrestha J., Carlson A., Kooyers N., Coerper D., Urbanczyk-Wochniak E., Bench B.J., Sumner L.W., Swanson R., Preuss D. (2011). A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiol. 157: 947–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Kirkpatrick A.B., Reeder S.H., Li P., Owen H.A. (2018). Pollen aperture factor INP1 acts late in aperture formation by excluding specific membrane domains from exine deposition. Plant Physiol. 176: 326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Hong Z., Sivaramakrishnan M., Mahfouz M., Verma D.P.S. (2005). Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 42: 315–328. [DOI] [PubMed] [Google Scholar]
- Friml J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865. [DOI] [PubMed] [Google Scholar]
- Furness C.A., Rudall P.J. (2004). Pollen aperture evolution--a crucial factor for eudicot success? Trends Plant Sci. 9: 154–158. [DOI] [PubMed] [Google Scholar]
- Geldner N. (2009). Cell polarity in plants: a PARspective on PINs. Curr. Opin. Plant Biol. 12: 42–48. [DOI] [PubMed] [Google Scholar]
- Goldstein B., Macara I.G. (2007). The PAR proteins: fundamental players in animal cell polarization. Dev. Cell 13: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo W.D., Inoue T., Park W.S., Kim M.L., Park B.O., Wandless T.J., Meyer T. (2006). PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 314: 1458–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1979). An interpretation of the hydrodynamics of pollen. Am. J. Bot. 66: 737–743. [Google Scholar]
- Hou X., Li L., Peng Z., Wei B., Tang S., Ding M., Liu J., Zhang F., Zhao Y., Gu H., Qu L.-J. (2010). A platform of high-density INDEL/CAPS markers for map-based cloning in Arabidopsis. Plant J. 63: 880–888. [DOI] [PubMed] [Google Scholar]
- Ischebeck T., Seiler S., Heilmann I. (2010). At the poles across kingdoms: phosphoinositides and polar tip growth. Protoplasma 240: 13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.G., Price J., Lin P.-C., Hong J.C., Jang J.-C. (2010). The arabidopsis bZIP1 transcription factor is involved in sugar signaling, protein networking, and DNA binding. Mol. Plant 3: 361–373. [DOI] [PubMed] [Google Scholar]
- Katifori E., Alben S., Cerda E., Nelson D.R., Dumais J. (2010). Foldable structures and the natural design of pollen grains. Proc. Natl. Acad. Sci. USA 107: 7635–7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk V.I., Jones J.D.G. (1997). AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 11: 1–14. [DOI] [PubMed] [Google Scholar]
- Kost B., Lemichez E., Spielhofer P., Hong Y., Tolias K., Carpenter C., Chua N.-H. (1999). Rac homologues and compartmentalized phosphatidylinositol 4, 5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Ben-Menni Schuler S., Reeder S.H., Wang R., Suárez Santiago V.N., Dobritsa A.A. (2018). INP1 involvement in pollen aperture formation is evolutionarily conserved and may require species-specific partners. J. Exp. Bot. 69: 983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Gillmor C.S., Scheible W.-R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123: 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhava P., Bassukas A.E.L., Zourelidou M., Kolb M., Moret B., Fastner A., Schulze W.X., Cattaneo P., Hammes U.Z., Schwechheimer C., Hardtke C.S. (2018). A molecular rheostat adjusts auxin flux to promote root protophloem differentiation. Nature 558: 297–300. [DOI] [PubMed] [Google Scholar]
- Martin-Belmonte F., Mostov K. (2008). Regulation of cell polarity during epithelial morphogenesis. Curr. Opin. Cell Biol. 20: 227–234. [DOI] [PubMed] [Google Scholar]
- Michniewicz M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130: 1044–1056. [DOI] [PubMed] [Google Scholar]
- Nagano M., Ishikawa T., Fujiwara M., Fukao Y., Kawano Y., Kawai-Yamada M., Shimamoto K. (2016). Plasma membrane microdomains are essential for Rac1-RbohB/H-mediated immunity in rice. Plant Cell 28: 1966–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W.J. (2003). Adaptation of core mechanisms to generate cell polarity. Nature 422: 766–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R.M., Lanz C., Warthmann N., Weigel D. (2008). Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Chen J., Yang Z. (2015). Auxin regulation of cell polarity in plants. Curr. Opin. Plant Biol. 28: 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinal N., Goberdhan D.C.I., Collinson L., Fujita Y., Cox I.M., Wilson C., Pichaud F. (2006). Regulated and polarized PtdIns(3,4,5)P3 accumulation is essential for apical membrane morphogenesis in photoreceptor epithelial cells. Curr. Biol. 16: 140–149. [DOI] [PubMed] [Google Scholar]
- Rademacher E.H., Offringa R. (2012). Evolutionary adaptations of plant AGC kinases: From light signaling to cell polarity regulation. Front. Plant Sci. 3: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder S.H., Lee B.H., Fox R., Dobritsa A.A. (2016). A ploidy-sensitive mechanism regulates aperture formation on the Arabidopsis pollen surface and guides localization of the aperture factor INP1. PLoS Genet. 12: e1006060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressayre A., Godelle B., Mignot A., Gouyon P.H. (1998). A morphogenetic model accounting for pollen aperture pattern in flowering plants. J. Theor. Biol. 193: 321–334. [DOI] [PubMed] [Google Scholar]
- Ressayre A., Godelle B., Raquin C., Gouyon P.H. (2002). Aperture pattern ontogeny in Angiosperms. J. Exp. Zool. 294: 122–135. [DOI] [PubMed] [Google Scholar]
- Roppolo D., De Rybel B., Dénervaud Tendon V., Pfister A., Alassimone J., Vermeer J.E.M., Yamazaki M., Stierhof Y.-D., Beeckman T., Geldner N. (2011). A novel protein family mediates Casparian strip formation in the endodermis. Nature 473: 380–383. [DOI] [PubMed] [Google Scholar]
- Sawano A., Miyawaki A. (2000). Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28: E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekereš J., Pleskot R., Pejchar P., Žárský V., Potocký M. (2015). The song of lipids and proteins: dynamic lipid-protein interfaces in the regulation of plant cell polarity at different scales. J. Exp. Bot. 66: 1587–1598. [DOI] [PubMed] [Google Scholar]
- Shao W., Dong J. (2016). Polarity in plant asymmetric cell division: Division orientation and cell fate differentiation. Dev. Biol. 419: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M.L.A., Platre M.P., Assil S., van Wijk R., Chen W.Y., Chory J., Dreux M., Munnik T., Jaillais Y. (2014). A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J. 77: 322–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M.L.A., Platre M.P., Marquès-Bueno M.M., Armengot L., Stanislas T., Bayle V., Caillaud M.-C., Jaillais Y. (2016). A PtdIns(4)P-driven electrostatic field controls cell membrane identity and signalling in plants. Nat. Plants 2: 16089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislas T., Hüser A., Barbosa I.C.R., Kiefer C.S., Brackmann K., Pietra S., Gustavsson A., Zourelidou M., Schwechheimer C., Grebe M. (2015). Arabidopsis D6PK is a lipid domain-dependent mediator of root epidermal planar polarity. Nat. Plants 1: 15162. [DOI] [PubMed] [Google Scholar]
- Tejos R., Sauer M., Vanneste S., Palacios-Gomez M., Li H., Heilmann M., van Wijk R., Vermeer J.E.M., Heilmann I., Munnik T., Friml J. (2014). Bipolar plasma membrane distribution of phosphoinositides and their requirement for auxin-mediated cell polarity and patterning in Arabidopsis. Plant Cell 26: 2114–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U. (2012). The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28: 655–685. [DOI] [PubMed] [Google Scholar]
- Walker J.A., Doyle J.A. (1975). The bases of angiosperm phylogeny: Palynology. Ann. Miss. Bot. Gard. 62: 664–723. [Google Scholar]
- Wang R., Dobritsa A.A. (2018). Exine and aperture patterns on the pollen surface: Their formation and roles in plant reproduction. In Annual Plant Reviews, Roberts J.A., ed (John Wiley & Sons; ), pp. 1–40. [Google Scholar]
- Weller B., Zourelidou M., Frank L., Barbosa I.C.R., Fastner A., Richter S., Jürgens G., Hammes U.Z., Schwechheimer C. (2017). Dynamic PIN-FORMED auxin efflux carrier phosphorylation at the plasma membrane controls auxin efflux-dependent growth. Proc. Natl. Acad. Sci. USA 114: E887–E896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige B.C., Ogiso-Tanaka E., Zourelidou M., Schwechheimer C. (2012). WAG2 represses apical hook opening downstream from gibberellin and PHYTOCHROME INTERACTING FACTOR 5. Development 139: 4020–4028. [DOI] [PubMed] [Google Scholar]
- Willige B.C., Ahlers S., Zourelidou M., Barbosa I.C.R., Demarsy E., Trevisan M., Davis P.A., Roelfsema M.R.G., Hangarter R., Fankhauser C., Schwechheimer C. (2013). D6PK AGCVIII kinases are required for auxin transport and phototropic hypocotyl bending in Arabidopsis. Plant Cell 25: 1674–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodehouse R.P. (1935). Pollen Grains: Their Structure, Identification and Significance in Science and Medicine. (New York, London: McGraw-Hill; ). [Google Scholar]
- Yang Z. (2008). Cell polarity signaling in Arabidopsis. Annu. Rev. Cell Dev. Biol. 24: 551–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Makaroff C.A., Ma H. (2003). The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15: 1281–1295. [PMC free article] [PubMed] [Google Scholar]
- Yao X., Tian L., Yang J., Zhao Y.-N., Zhu Y.-X., Dai X., Zhao Y., Yang Z.-N. (2018). Auxin production in diploid microsporocytes is necessary and sufficient for early stages of pollen development. PLoS Genet. 14: e1007397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zegzouti H., Li W., Lorenz T.C., Xie M., Payne C.T., Smith K., Glenny S., Payne G.S., Christensen S.K. (2006). Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. J. Biol. Chem. 281: 35520–35530. [DOI] [PubMed] [Google Scholar]
- Zourelidou M., et al. (2014). Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases, D6 PROTEIN KINASE and PINOID. eLife 3: e02860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zourelidou M., Müller I., Willige B.C., Nill C., Jikumaru Y., Li H., Schwechheimer C. (2009). The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development 136: 627–636. [DOI] [PubMed] [Google Scholar]