Two NAC transcription factors, ANAC087 and ANAC046, control postmortem chromatin degradation in the lateral root cap and the life span of shed columella root cap cells in the rhizosphere.
Abstract
Programmed cell death in plants occurs both during stress responses and as an integral part of regular plant development. Despite the undisputed importance of developmentally controlled cell death processes for plant growth and reproduction, we are only beginning to understand the underlying molecular genetic regulation. Exploiting the Arabidopsis thaliana root cap as a cell death model system, we identified two NAC transcription factors, the little-characterized ANAC087 and the leaf-senescence regulator ANAC046, as being sufficient to activate the expression of cell death-associated genes and to induce ectopic programmed cell death. In the root cap, these transcription factors are involved in the regulation of distinct aspects of programmed cell death. ANAC087 orchestrates postmortem chromatin degradation in the lateral root cap via the nuclease BFN1. In addition, both ANAC087 and ANAC046 redundantly control the onset of cell death execution in the columella root cap during and after its shedding from the root tip. Besides identifying two regulators of developmental programmed cell death, our analyses reveal the existence of an actively controlled cell death program in Arabidopsis columella root cap cells.
INTRODUCTION
Programmed cell death (PCD) is defined as genetically encoded, actively controlled cellular suicide. In both animals and plants, PCD processes are essential for the growth and survival of the organisms (Fuchs and Steller, 2011; Daneva et al., 2016). Plants can respond to pathogen infection with PCD processes reminiscent of inflammatory PCD types in animals (Coll et al., 2011). During periods of abiotic stress, cells, tissues, or entire organs can be sacrificed to increase the survival chances of the plant as a whole (Gadjev et al., 2008). Yet, also during undisturbed plant growth, specific tissues and cell types undergo developmental PCD (dPCD) as part of their regular differentiation program (Daneva et al., 2016; Van Durme and Nowack, 2016). Forms of dPCD are essential for plant development or reproduction. Xylem dPCD, for instance, creates a network of hollow cell corpses crucial for efficient long-distance water and solute transport in land plants (Heo et al., 2017), while during anther development, precisely timed tapetum dPCD is paramount for male fertility in angiosperms (Plackett et al., 2011).
Recently, it was shown that in Arabidopsis thaliana, differentiated root cap cells undergo dPCD to maintain root cap organ size and position at the root tip (Fendrych et al., 2014). The root cap is a structure that surrounds and protects the stem cell niche in the root tip and has important functions in root gravitropism (Kumpf and Nowack, 2015). In Arabidopsis and many other plants, the root cap consists of two tissues, the columella root cap (columella) at the very root tip and the lateral root cap (LRC) that flanks the columella and the root meristem (Figure 1A). New columella and LRC cells are continually produced by distinct stem cells, the columella initials and the epidermis/LRC initials, respectively (Dolan et al., 1993). To restrict the extent of the root cap to the meristem region, root cap cells need to be disposed of when they reach the start of the root elongation zone. In Arabidopsis, this disposal is achieved by the execution of a tightly controlled dPCD program followed by a cell-autonomous degradation of the LRC cell corpses on the root surface (Fendrych et al., 2014). In contrast, columella cells are shed from the root tip as packs of living cells to achieve root cap turnover (Vicré et al., 2005). Once PCD is initiated at the elongation zone, the distal cell death zone in the oldest LRC layer progresses toward the root tip until it reaches the columella. The most proximal dead or dying LRC cells are shed together with the living columella cells (Figure 1A) (Kumpf and Nowack, 2015). Cell death in LRC cells is characterized by a succession of cytological events (Van Hautegem et al., 2015). The first observable change in a LRC cell undergoing cell death is an abrupt acidification of the cytoplasm, which is followed by a loss of plasma membrane integrity. Subsequently, vacuolar collapse can be observed (Fendrych et al., 2014). The stepwise breakup of cellular compartments is thought to release hydrolases, which execute cell death and initiate corpse degradation during and after cellular death (Van Durme and Nowack, 2016).
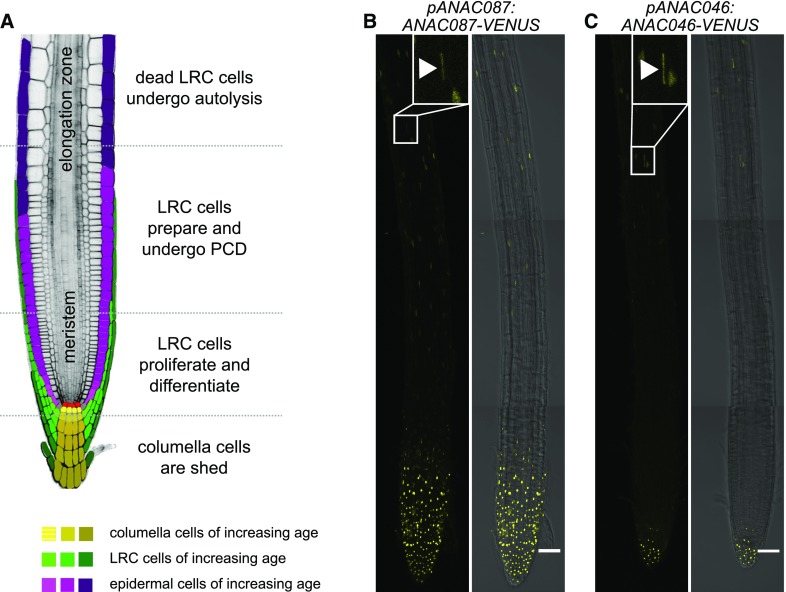
Figure 1.
ANAC087 and ANAC046 Are Expressed in the Root Cap.
(A) A schematic representation of the Arabidopsis root tip. The columella is shown in yellow, the LRC in green, the epidermis in pink, and the quiescent center in red. The different developmental stages of the root cap are indicated. (Adapted from Kumpf and Nowack, 2015.)
(B) The pANAC087:ANAC087-VENUS reporter was expressed in the entire root cap and showed weaker expression in some epidermal cells and in the differentiating TE cells (elongated nucleus indicated by arrowhead in the inset). Shown is the root of a representative 5-d-old T3 seedling. Five homozygous lines were analyzed in T3.
(C) The pANAC046:ANAC046-VENUS reporter was expressed in the columella and showed expression in some epidermal cells and in the differentiating TE cells (elongated nucleus indicated by arrowhead in the inset). Shown is the root of a representative 5-d-old T3 seedling; two homozygous lines were analyzed in T3. In (B) and (C), the VENUS signal is shown in yellow, and roots are shown without (left) and with (right) transmitted light. The VENUS signal is shown as a Z-projection, whereas the bright-field channel is shown as a single stack. Bars in (B) and (C) = 50 µm.
Despite the importance of dPCD in plants, knowledge of the molecular regulation of this process is still very limited. During leaf senescence, the NAC (no apical meristem [NAM], Arabidopsis thaliana activation factor [ATAF], cup-shaped cotyledon [CUC]) family transcription factor ORESARA1 (ORE1/ANAC092) was shown to activate transcription of BIFUNCTIONAL NUCELASE1 (BFN1) (Matallana-Ramirez et al., 2013). BFN1 was identified as one of the core dPCD-associated genes in Arabidopsis, alongside other genes, such as RIBONUCLEASE3 (RNS3), EXITUS1 (EXI1), and PUTATIVE ASPARTIC PROTEINASE A3 (PASPA3). These genes are coexpressed in several cell types or tissues undergoing dPCD, including the xylem vessels, the tapetum, the root cap, and the endosperm (Olvera-Carrillo et al., 2015).
The root cap-specific NAC transcription factor SOMBRERO (SMB/ANAC033) is not only involved in the regulation of root cap cell maturation (Bennett et al., 2010), but also in the preparation of PCD in LRC cells as the final cellular differentiation step (Fendrych et al., 2014). In the smb mutant, LRC cell death is delayed, occurring in the root elongation zone, and highly aberrant, as the expression of PCD-associated genes and the execution of postmortem cell corpse clearance is lacking (Fendrych et al., 2014). Conversely, dexamethasone-inducible overexpression lines of SMB (p35S:SMB-GR) showed tissue growth arrest and ectopic formation of tracheary element (TE)-like cells (Bennett et al., 2010). Similar to ORE1, which is not expressed in the root cap, SMB controls BFN1 expression, but it is not clear to date if this control occurs directly or indirectly. Though no bfn1 mutant phenotype has been described in senescent leaves, loss of BFN1 causes a delay in postmortem corpse clearance of LRC cells (Fendrych et al., 2014).
In this study, we investigated the transcriptional network of root cap dPCD and identified two NAC transcription factors, ANAC087 and ANAC046, as root cap cell death regulators. Ectopic overexpression of both genes was sufficient to induce a cell death process reminiscent of LRC cell death. Conversely, analyses of loss-of-function alleles revealed that ANAC087 orchestrates the postmortem nuclear degradation of LRC cells in the root elongation zone by promoting BFN1 expression. In addition, ANAC087 and ANAC046 redundantly control PCD execution in shedding and shed columella cells, revealing that columella cells execute a genetically controlled cell death program after shedding from the Arabidopsis root.
RESULTS
ANAC087 and ANAC046 Are Two PCD-Associated Transcription Factors
To identify additional regulators of root cap dPCD, we analyzed publically available transcriptome data. Applying the VisuaLRTC methodology (Parizot et al., 2010), we performed a meta-analysis to explore tissue-specific Arabidopsis mRNA microarrays. Specifically, we searched for transcription factors (TFs) that are highly expressed in two root tissues undergoing PCD, the xylem TEs and the LRC, but weakly expressed in other tissues of the root (Supplemental Data Set 1). Interestingly, among the TFs with the most preferential expression in xylem and LRC were two NAC TFs of the same clade as ORE1, ANAC087 and ANAC046 (Ooka et al., 2003). We decided to investigate these TFs in detail for their potential involvement in root cap PCD.
To analyze protein expression and localization of both TFs in the root, we generated translational reporter lines. For both TFs, we used a 4-kb promoter fragment to drive expression of the genomic sequences with introns and exons fused to the VENUS reporter gene (pANAC087:ANAC087-VENUS and pANAC046:ANAC046-VENUS). In several independent ANAC087-VENUS lines, 5-d-old seedlings showed strong nucleus-localized VENUS signals in the entire root cap and a weaker nuclear signal in the root epidermis and the TEs (Figure 1B). In several independent ANAC046 translational reporter lines, nuclear VENUS signals were found in the columella, and signals were also detected in some epidermal cells and in the differentiating TEs (Figure 1C). Interestingly, several independent transcriptional reporter lines of both TFs, which contained the same 4-kb promoter fragments driving expression of nucleus-localized tdTOMATO (pANAC087:NLS-tdTOMATO and pANAC046:NLS-tdTOMATO), did not show expression in the differentiating TE cells (Supplemental Figures 1A and 1B). These data suggest that both ANAC087 and ANAC046 expression might be fine-tuned by intronic cis-regulatory elements or that the respective fusion proteins undergo posttranscriptional modification that alters their accumulation pattern in TEs.
Inducible Misexpression of SMB, ANAC087, and ANAC046 Is Sufficient to Cause Ectopic Cell Death
It was shown before that ectopic overexpression of SMB leads to growth arrest and the formation of TE-like secondary cell wall thickenings (Bennett et al., 2010). To test whether this growth arrest is accompanied by cell death, we introduced a tonoplast integrity marker driven by the PASPA3 promoter (pPASPA3:ToIM) (Fendrych et al., 2014) in the p35S:SMB-GR-inducible overexpression line (Bennett et al., 2010). The pPASPA3:ToIM (tonoplast integrity marker) consists of a cytoplasmic eGFP (enhanced green fluorescent protein) marker and a vacuolar mRFP (monomeric red fluorescent protein) marker, which makes it possible to observe vacuolar collapse during PCD when both signals merge (Fendrych et al., 2014). Induction with dexamethasone resulted in ectopic expression of pPASPA3:ToIM and showed cells undergoing vacuolar collapse, indicating cell death preparation and execution outside the root cap (Figure 2A; Supplemental Movie 1). In order to test whether ANAC087 and ANAC046 would be able to induce cell death in a comparable fashion, we generated estradiol-inducible misexpression lines (Siligato et al., 2016). In these lines, expression of the estradiol-activated transcriptional activator XVE was controlled by the strong meristematic promoter pRPS5A (Weijers et al., 2001; Gao et al., 2018). To be able to follow the expression pattern of both TFs after induction, they were fused to GFP, resulting in the constructs pRPS5A:XVE>>ANAC087-GFP and pRPS5A:XVE>>ANAC046-GFP. Approximately 18 h after induction, we observed intense staining with Evans Blue (Escamez et al., 2017) in several independent misexpression lines of both ANAC087 and ANAC046, indicating wide-spread cell death in the meristem 24 h after estradiol treatment (Figure 2B). Staining with propidium iodide (PI) confirmed the occurrence of meristematic cell death in time-lapse analyses (Supplemental Figure 2A and Supplemental Movie 2), suggesting that the expression of both genes is sufficient to induce cell death outside the root cap context. Expression databases on RT-qPCR confirmed that both ANAC087 and ANAC046 were strongly upregulated 24 h after estradiol induction. However, ANAC087 overexpression did not induce ANAC046 expression and vice versa, excluding a regulatory cascade between the two TFs (Figure 2C; Supplemental Figure 2B). The ectopic cell death observed in these seedlings coincided with root growth arrest after induction by estradiol, which was observable both in microscopic and macroscopic imaging setups (Figures 2B and 2D). While in ANAC087 misexpression lines, the seedlings were able to survive longer than 5 d after induction and started to form lateral roots, the seedlings of the ANAC046 misexpression appeared entirely dead within 5 d (Figure 2D; Supplemental Figure 2C and Supplemental Movie 3). These differences between ANAC087 and ANAC046 misexpression lines might be due to different expression levels (Figure 2C; Supplemental Figure 2B) or additional differences in protein characteristics. This notwithstanding, our results indicate that inducible overexpression of both ANAC087 and ANAC046 is sufficient to trigger ectopic cell death outside the root cap context.
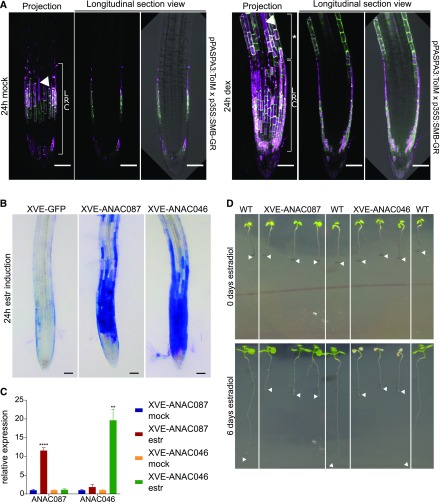
Figure 2.
Inducible Overexpression of ANAC087 and ANAC046 Leads to Ectopic Cell Death.
(A) The pPASPA3-ToIM marker showed ectopic expression and cell death in the epidermis distal of the LRC 24 h after induction of the p35S-SMB-GR (right, indicated by an asterisk). In the mock treatment (left), only regular cell death in the LRC was observed. The eGFP signal is shown in green in the cytoplasm, whereas the mRFP signal is shown in magenta in the vacuole. Merge of both signals indicates vacuolar collapse, a hallmark of cell death (arrowheads). Bars = 50 µm.
(B) Inducible overexpression of ANAC087-GFP and ANAC046-GFP (induced with estradiol [estr]) using the chimeric XVE TF and the pRPS5A promoter led to intense ectopic Evans Blue staining. Shown is the root of a representative 5-d-old T3 seedling; four homozygous lines were analyzed in T3 for ANAC087-GFP and three lines for ANAC046-GFP. In the control line, where only NLS-GFP is inducibly expressed, no ectopic Evans Blue staining was observed. Photographs show the time point of 24 h after induction. Evans Blue is shown in blue. Bars = 50 µm.
(C) As indicated by RT-qPCR analysis, 24 h induction by estradiol caused strong expression of ANAC087 and ANAC046 in the pRPS5A:XVE>>ANAC087-GFP and pRPS5A:XVE>>ANAC046-GFP line, respectively. The error bars show the se of means of three independent biological replicates (three independent treatments and three technical repeats each). t test; **P < 0.01 and ****P < 0.0001.
(D) Macroscopic images of estradiol-induced overexpression lines of ANAC087 and ANAC046 followed over time showed a different response for both TFs. Six days after estradiol treatment, XVE-ANAC087 seedlings arrested root growth, while the shoot survived, whereas the XVE-ANAC046 seedlings showed complete growth arrest and death of the seedling. In contrast, wild-type seedlings were not visually affected by the estradiol treatment. Arrowheads indicate root tips at the beginning (0 d estradiol) and the end (6 d estradiol) of the estradiol treatment. Representative full time courses are shown in Supplemental Movie 3.
SMB, ANAC087, and ANAC046 Act in Parallel to Control PCD-Associated Genes
To investigate if SMB, ANAC087, and ANAC046 are inducing a LRC-like cell death program outside the root cap, we tested whether the PCD-associated genes BFN1, RNS3, and EXI1 are downstream targets of these TFs (Olvera-Carrillo et al., 2015). First, we performed RT-qPCR-based expression analysis of smb-3, anac087-1, and anac046-1 mutant root tips. We isolated the anac087-1 and anac046-1 null-mutants from a T-DNA insertion collection (Supplemental Figures 3A and 5A), while the smb-3 mutant was described before (Willemsen et al., 2008). Based on expression patterns, root tips of anac087-1 and smb-3 were cut just above the meristem, while small root tips of the anac046-1 were cut around the quiescent center (QC). The RT-qPCR analysis showed that all three PCD-associated genes were downregulated in anac087-1 and possibly anac046-1 (Figures 3A to 3C). Downregulation of the PCD-associated genes in the anac046-1 mutant was more moderate and not statistically significant, possibly because expression of ANAC087 in the columella is sufficient to at least partly compensate for the lack of ANAC046. To gain a tissue-specific resolution of a representative PCD-associated gene, we crossed a pBFN1:NLS-tdTOMATO transcriptional reporter line with either mutant. In the wild type, the tdTOMATO signal was observed in cells of the LRC, the columella, and in differentiating xylem TEs (Supplemental Figure 4A). However, in the anac087-1 mutant background, only a very weak tdTOMATO signal could be detected in some root cap cells, while the signal in the TEs was comparable to the wild type (Supplemental Figure 4A). This observation suggests that alternative TFs might regulate BFN1 expression in the xylem. Finally, in the anac046-1 mutant background, tdTOMATO signals were not different from the wild type in the LRC, columella, and TEs (Supplemental Figure 4A). In line with the RT-qPCR data, these results indicate that ANAC087 expression is necessary and sufficient to promote BFN1 expression in the LRC and columella, while the lack of ANAC046 does not cause a significant reduction of BFN1 expression in the columella. Interestingly, ANAC087 was not downregulated in the anac046-1 mutant and vice versa (Figures 3A and 3B), and neither TF was differentially regulated in the smb mutant (Figure 3C).
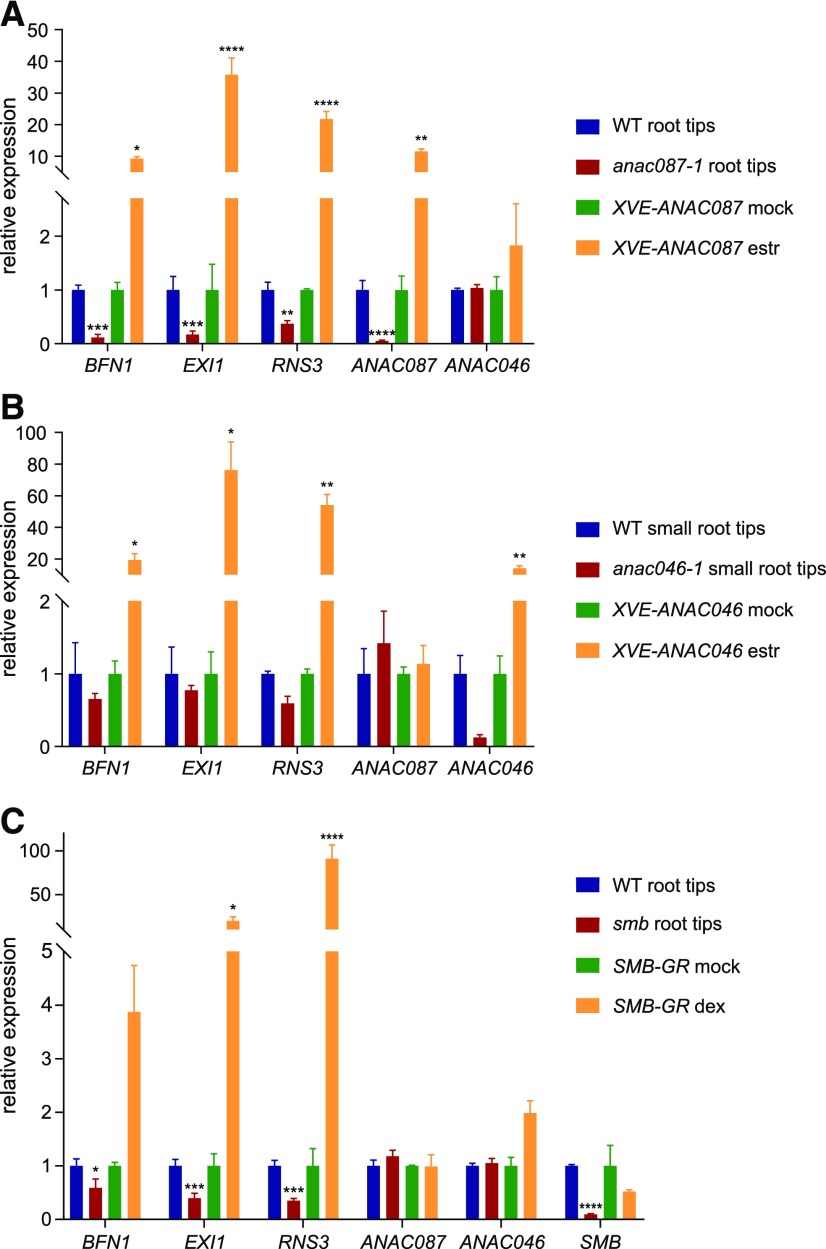
Figure 3.
Expression of BFN1, EXI1, and RNS3 Is Regulated by ANAC087, ANAC046, and SMB.
(A) RT-qPCR analysis showed reduced expression of PCD-associated genes (BFN1, EXI1, and RNS3) in root tips of the anac087-1 mutant (root tips were cut just above the LRC), whereas these genes were upregulated in the ANAC087-inducible overexpression line. t test; *P < 0.05, **P < 0.01, and ****P < 0.0001.
(B) RT-qPCR analysis for small root tips of the anac046-1 mutant (root tips were cut just above the columella) and the ANAC046-inducible overexpression line. Expression of ANAC046 is not altered in the anac087-1 mutant or ANAC087 overexpression line and vice versa. The error bars in (A) and (B) show se of three independent biological replicates (three independent treatments and three technical repeats each). t test; *P < 0.05 and **P < 0.01.
(C) PCD-associated genes showed increased expression upon induction of the p35S:SMB-GR (shown as SMB-GR) and decreased expression in the smb mutant (root tips were cut just above the LRC), as indicated by RT-qPCR analysis. As application of dexamethasone (dex) only leads to activation of the highly expressed SMB-GR fusion protein, SMB RNA expression is not increased. The error bars show se of three independent biological replicates (three independent treatments and three technical repeats each). t test; *P < 0.05, ***P < 0.001, and ****P < 0.0001.
In a complementary approach, we showed that 24 h after estradiol induction, SMB, ANAC087, and ANAC046 misexpression led to significant (P < 0.0001) upregulation of BFN1, RNS3, and EXI1 (Figures 3A to C). However, none of the TF genes were differentially expressed in any of the inducible misexpression lines. Together, these results indicate that the three core PCD-associated genes BFN1, EXI1, and RNS3 are regulated by SMB, ANAC087, and ANAC046 and that these TFs do not work in a regulatory cascade to control PCD-associated gene expression.
To investigate whether BFN1, EXI1, and RNS3 are direct or indirect targets of ANAC087 and ANAC046, we performed an electrophoretic mobility shift assay (EMSA) testing the binding of HIS6MBP (hexahistidine-maltose binding protein)-tagged TF proteins to a labeled 40-bp promoter fragment. As the DNA binding sites of ANAC087 and ANAC046 are unknown, we analyzed a protein binding microarray together with coexpression data to predict putative binding sites for ANAC087 and ANAC046 (Lindemose et al., 2014; Gao et al., 2018). Interestingly, for each PCD-associated gene, both TFs shared predicted binding sites (Supplemental Figure 4B). The clear band shifts in the EMSA suggested that both ANAC087 and ANAC046 proteins were able to physically bind to promoter fragments of the tested PCD-associated gene in vitro (Figure 4A). To confirm this result in vivo, we performed a chromatin immunoprecipitation experiment followed by quantitative PCR (ChIP-qPCR; Supplemental Figure 4C). For ANAC087, no significant enrichment of any tested downstream target could be detected (Figure 4B), indicating that the in vitro binding of ANAC087 to the three promoter fragments might not be occurring in vivo. However, for ANAC046, ChIP-qPCR showed enrichment of the promoter fragments of BFN1, EXI1, and RNS3 (Figure 4C). This enrichment was statistically significant (P < 0.01) for BFN1 and RNS3, but not for EXI1. These results indicate that, in contrast to ANAC087, ANAC046 is interacting with the promoters of BFN1, RNS3, and possibly EXI1 in vivo.
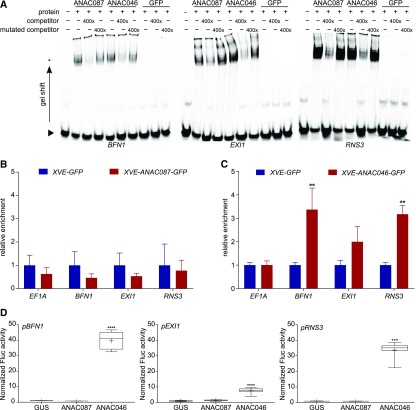
Figure 4.
BFN1, EXI1, and RNS3 Are Indirect Targets of ANAC087 and Direct Targets of ANAC046.
(A) EMSA for binding of ANAC087 and ANAC046 HIS6MBP-tagged proteins to labeled 40-bp promoter fragments of BFN1, EXI1, and RNS3. Both proteins caused a slower migration of the protein-probe complex (band shift indicated by an asterisk) compared with the free probe (indicated by arrowhead). When a 400× concentrated unlabeled competitor probe was added, the signal of the protein-probe complex decreased, whereas when a mutated, unlabeled competitor probe was added, the signal intensity was restored, indicating the specificity of the interaction. When the negative control HIS6MBP-labeled GFP or no protein was added, no band shift was observed.
(B) and (C) ChIP-qPCR analysis showed no enrichment of BFN1, EXI1, or RNS3 promoter fragments when ANAC087-GFP was immunoprecipitated. However, when ANAC046-GFP was pulled down, promoter fragments of BFN1, EXI1, and RNS3 were enriched, as detected by RT-qPCR. The graph shows relative enrichment compared with estradiol induced pRPS5A:XVE>>GFP seedlings. As a negative control, the housekeeping gene EF1A was used, which did not show enrichment. Error bars indicate se of four biological replicates (protein extractions from four independently grown samples). t test; **P < 0.01.
(D) TEAs show increased activity of firefly luciferase (Fluc) from the 2-kb promoters of pBFN1, pEXI1, and pRNS3 when ANAC046 was present, indicating that ANAC046 was able to activate transcription from these promoters. ANAC087 was not able to induce promoter activity of pBFN1, pEXI1, or pRNS3. As a negative control, GUS was used, which is not able to induce transcription. Activation is shown relative to GUS. Box plot whiskers indicate maximum and minimum values; + indicates mean of eight biological replicates (eight independent protoplast transformations). Kruskal-Wallis nonparametric test ***P < 0.001 and ****P < 0.0001.
To test whether the binding of ANAC046 to the promoters of BFN1, EXI1, and RNS3 results in gene expression activation, we performed a transient expression assay (TEA) in tobacco (Nicotiana tabacum) protoplasts (Vanden Bossche et al., 2013). In the TEA, ANAC046 was able to strongly activate the three tested 2-kb gene promoters, as indicated by the elevated luciferase signals observed. In accordance with the ChIP-qPCR results, the promoters of BFN1 and RNS3 were more efficiently activated than the promoter of EXI1 (Figure 4D). Unlike ANAC046, ANAC087 was not able to activate any of the tested promoters (Figure 4D), which was in line with the ChIP-qPCR results. In addition, the TEA did not show a more efficient reporter gene expression when both TFs were expressed simultaneously, indicating that ANAC087 and ANAC046 do not act synergistically in activating these PCD-associated genes (Supplemental Figure 4D). In conclusion, our results show that BFN1, EXI1, and RNS3 are most likely direct targets of ANAC046, whereas they are probably indirect targets of ANAC087.
ANAC087 Regulates Postmortem Nuclear Degradation and PCD Onset in the LRC
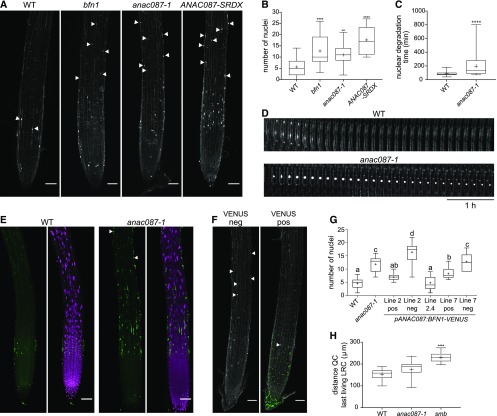
To investigate a possible role of ANAC087 in root cap PCD, the anac087-1 null mutant was analyzed for a root cap phenotype. Staining with PI revealed that the root surface of the anac087-1 mutant was covered with significantly (P < 0.01) more stained nuclear remnants compared with wild-type roots (Figures 5A and 5B). This phenotype is reminiscent of the bfn1 mutant (Fendrych et al., 2014), indicating that nuclear DNA degradation—a hallmark of cell-autonomous cell corpse clearance—is impaired in anac087-1. This phenotype was confirmed in an independent anac087-2 mutant allele, and it could be rescued by introducing the pANAC087:ANAC087-VENUS construct in the anac087-1 mutant background (Supplemental Figures 3A to 3E). Additionally, a ANAC087 dominant-negative repressor controlled by the root cap-specific SMB promoter (pSMB:ANAC087-SRDX) copied the root cap phenotype of the anac087-1 mutant in a single-insertion line (Figures 5A and 5B). Interestingly, when a strong, multiple-locus line of ANAC087-SRDX was analyzed, a smb-like phenotype was observed (Supplemental Figure 3F), suggesting that ANAC087-SRDX is able to also repress SMB target genes when strongly expressed. NAC TFs are known to interact with similar binding sites (Bennett et al., 2010; Lindemose et al., 2014), and overexpression of dominant-repressive protein varieties might lead to off-target repression.
Figure 5.
ANAC087 Is Involved in Regulation of Postmortem Nuclear Degradation.
(A) Numerous nuclear remnants were found on the PI-stained root surface of bfn1, anac087-1, and ANAC087-SRDX, as indicated by arrowheads. Fusion of the SRDX domain to a TF turns this TF into a dominant repressor of its target genes (Hiratsu et al., 2003).
(B) Quantification of the number of nuclear remnants shown in (A). Box plots show data from at least 14 roots. Whiskers indicate minimum and maximum values; + indicates average. ANOVA; **P < 0.01, ***P < 0.001, and ****P < 0.0001.
(C) Nuclei of the anac087-1 took longer to be degraded after cell death compared with nuclei from the wild type. Box plots show data from at least 34 nuclei from at least 4 different roots. Whiskers indicate minimum and maximum values; + indicates average. t test; ****P < 0.0001.
(D) Kymograph showing the delay of nuclear degradation in the anac087-1 mutant compared with the wild type. Cells were imaged every 10 min for 5 h.
(E) Multiple TUNEL-positive nuclei were found on the anac087-1 mutant root surface compared with the wild type (indicated by arrowheads). Signal of the fluorescein is shown in green, and DAPI signal is shown in magenta.
(F) Introducing a pANAC087:BFN1-VENUS construct into the anac087-1 mutant resulted in rescue of the mutant phenotype (VENUS pos). In contrast, in segregating VENUS-negative sister plants (VENUS neg), the anac087-1 phenotype was observed. Green signal indicates VENUS; the white signal shows PI staining. Bars in (A), (E), and (F) = 50 µm.
(G) Quantification of the number of nuclear remnants observed in segregating and homozygous pANAC087:BFN1-VENUS lines in the anac087-1 mutant background. Line 2 positive and line 7 positive show a partial rescue of the anac087-1 phenotype. Negative sister plants of the same segregating lines (line 2 neg and line 7 neg) do not show a rescue of the anac087-1 mutant phenotype. Line 2.4 is a homozygous T3 line and shows complete rescue of the anac087-1 mutant phenotype. The experiment was performed on at least 10 roots per genotype. Box plot whiskers indicate minimal and maximal values; + indicates mean. ANOVA; different letters indicate statistically significant differences.
(H) The distance from the QC to the beginning of the last living LRC cell (closest to the transition zone) was altered in the smb mutant, but not in the anac087-1 mutant compared with the wild type, which indicates that timing of LRC PCD is not changed in the anac087-1 mutant. The experiment was performed on at least 13 roots per genotype. Box plot whiskers indicate minimal and maximal values; + indicates mean. Kruskal-Wallis nonparametric test; **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Time-lapse imaging confirmed a bfn1-like delayed nuclear degradation phenotype in anac087-1 and ANAC087-SRDX LRC cells (Figures 5C and 5D; Supplemental Figures 3G and 3H). Additionally, we used terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) to follow DNA fragmentation in the anac087-1 mutant in situ. In a TUNEL assay, a deoxynucleotide tagged with a fluorochrome is attached to a free 3′-hydroxyl group of the DNA, indicating DNA fragmentation by an increase in cellular fluorescence. As in the bfn1 mutant, a delay in DNA degradation was indicated by the numerous TUNEL positive nuclei on the anac087-1 mutant root surface (Figure 5E; Supplemental Figure 3I). These data indicate that ANAC087 is necessary to efficiently complete nuclear fragmentation rather than initiating it, as was already suggested before for BFN1 (Fendrych et al., 2014). Intriguingly, when a functional BFN1-VENUS fusion protein (Fendrych et al., 2014) was expressed under the control of the ANAC087 promoter and transformed into the anac087-1 mutant, we observed a partial rescue of the anac087-1 mutant phenotype (Figures 5F and 5G). These results indicate that the detected bfn1-like phenotype in the anac087-1 mutant is partly caused by the lack of BFN1 expression.
The role of BFN1 is restricted to postmortem chromatin degradation, and in contrast to the smb mutant, the bfn1 mutant does not show a delay of cell death at the edge of the LRC (Fendrych et al., 2014). Likewise, when we measured the distance from the QC to the last living LRC cells in anac087-1 seedlings, we observed that, in contrast to the smb mutant, this distance was not significantly increased (Figure 5H). These data suggest that, unlike SMB, ANAC087 does not regulate the timing of LRC PCD in the distal LRC. Instead, ANAC087 plays a central role in organizing postmortem nuclear degradation by indirectly activating the expression of BFN1 and likely other hydrolytic enzymes.
ANAC046 and ANAC087 Redundantly Promote Cell Death in the Columella Root Cap
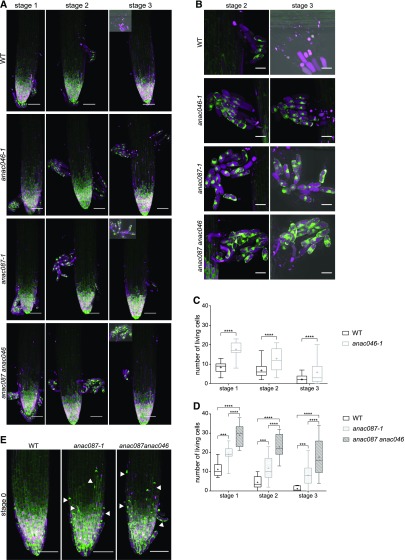
To test the putative involvement of ANAC046 in root cap PCD, we isolated and analyzed an anac046 mutant allele, anac046-1. As expected based on the lack of ANAC046 expression in the LRC, no PCD-related phenotype could be detected in this part of the root cap (Supplemental Figures 5B and 5C). Prompted by ANAC046 expression in the columella, we investigated the behavior of anac046-1 mutant columella cells. To this end, we used a ToIM driven by the POLYUBIQUITIN10 promoter (pUBQ10:ToIM), which is strongly expressed in the columella. To be able to analyze shedding root cap cells, we analyzed ToIM-expressing anac046-1 mutants growing in imaging chambers at 12-h intervals for several days. We did not observe any aberration in the shedding process of anac046-1 columella cells, but surprisingly, we discovered that shedding and shed anac046-1 columella cells remained alive significantly (P < 0.0001) longer than the wild-type columella cells (Figures 6A to 6C). Already in stage 1 (defined by the visual onset of columella cell separation, which occurs in ∼5.5-d-old seedlings), more living columella cells were found in the mutant compared with the wild type, indicating that in the wild type, PCD of some columella cells is already executed before shedding. Also in stage 2 and 3 (12 h and 24 h after onset of shedding respectively), significantly (P < 0.0001) more columella cells were found alive in the anac046-1 mutant than in the wild type. Even in stage 4 (36 h after shedding), often living, shed columella cells could still be detected in the anac046-1 mutant, while this was rarely observed in wild-type plants.
Figure 6.
ANAC046 and ANAC087 Redundantly Control Columella PCD.
(A) pUBQ:ToIM expression in the wild type, anac046-1, anac087-1, and anac087 anac046 was analyzed every 12 h (stages 1–3), with the visual onset of root cap shedding defined as stage 1. All three mutant lines show longer living shed columella cells compared with the wild type. EGFP signal is shown in green in the cytoplasm, and mRFP signal is shown in magenta in the vacuole. Vacuolar collapse indicating imminent cell death is visualized by merging of the green and magenta signals.
(B) Higher magnification of shed root cap cells from stage 2 and 3 shown in (A). Bars = 20 µm.
(C) and (D) Quantification of living, shed cells from the wild type and the anac046-1 mutant (C) and the wild type, anac087-1, and anac087 anac046 mutants (D) at three stages after shedding shows that columella cells of the mutants lived longer compared with the wild type. The columella cells of the anac087 anac046 double mutant also lived longer compared with the anac087-1 single mutant. Box plot whiskers show minimum and maximum values; + indicates mean of at least 10 roots. Poisson regression; ***P < 0.001 and ****P < 0.0001.
(E) At stage 0 (defined as 12 h before onset of shedding), anac087-1 and anac087 anac046 mutants with the pUBQ:ToIM show very elongated cells close to the root tips (indicated by arrowheads), which sometimes initiate shedding before the columella. eGFP is shown in green, and mRFP is shown in magenta. Bars in (A) and (E) = 50 µm.
To confirm the anac046-1 mutant phenotype, we created a bona fide null mutant using CRISPR/Cas9 technology (anac046-2). This allele has a 7-bp insertion in the C-terminal half of the ANAC046 gene (Supplemental Figure 5A). The resulting premature stop codon should lead to the formation of a truncated protein lacking the molecular recognition motif necessary for transcriptional activation and protein interactions (O’Shea et al., 2015). The anac046-2 mutant showed a comparable columella phenotype to anac046-1 (Supplemental Figures 5D and 5E). Additionally, a dominant-negative version of ANAC046 (pSMB:ANAC046-SRDX) was created. Because no ToIM marker was present in this line, seedlings were stained with fluorescein diacetate (FDA) and PI to identify living and dead cells, respectively. Similar to the anac046-1 mutant, 6-d-old pSMB:ANAC046-SRDX seedlings (approximately corresponding to stage 2) showed longer-living columella cells after shedding (Supplemental Figures 5F and 5G). In summary, our results demonstrate that shedding and shed columella cells undergo an actively controlled cell death process, the timing of which is controlled by ANAC046.
As ANAC046 and ANAC087 are coexpressed in the columella, we also analyzed the columella in anac087-1 and in an anac087-1 anac046-1 double mutant. Similar to anac046-1, anac087-1 columella cells stayed alive significantly (P < 0.001) longer than the wild type in different stages after shedding (Figures 6A, 6B, and 6D). Interestingly, the proximal LRC cells adjacent to the columella also stayed alive longer, which was indicated by the presence of living elongated cells close to the root tip at stage 0 (12 h before the onset of shedding; Figure 5D). In some cases, these elongated proximal LRC cells were shed alive as single cells or together with the columella (Figures 6A and 6E). In the anac087-1 anac046-1 double mutant, the shed columella cells were even longer lived than in either single mutant, creating an additive intensification of the phenotype (Figures 6A, 6B, and 6D). At all stages investigated, significantly (P < 0.0001) more root cap cells were alive in the double mutant compared with either single mutant (Supplemental Figure 5D), and often cells of anac087-1 anac046-1 were still alive in stage 4 and even in stage 5, which was never observed in the wild type and only very rarely in either single mutant (Supplemental Figure 5H). As in the anac087-1 mutant, proximal LRC cells died late in the double mutant (Figure 6E; Supplemental Movie 4). However, the cell death time point of the distal LRC cells at the end of the root meristem was not distinguishable from the wild type, indicating that unlike in the smb mutant, PCD of the distal LRC is not delayed in the anac087-1 anac046-1 double mutant (Supplemental Figure 5C). Taken together, ANAC087 promotes postmortem degradation in the distal LRC, as well as controlling cell death timing in the proximal LRC and the columella. In the shedding columella cells, ANA046 and ANAC087 jointly control the timely onset of PCD, demonstrating that a dedicated cell death program restricts the life span of columella cells in the rhizosphere.
DISCUSSION
In this study, we showed that ANAC087 and ANAC046 have partially redundant tasks in regulating two distinct aspects of PCD in different root cap tissues. Both ANAC087 and ANAC046 were already shown to be associated with plant PCD, but their roles in dPCD had been obscure. ANAC087 was found to be expressed downstream of the master regulator ORE1 during leaf senescence (Kim et al., 2014) and was recently shown to directly bind to the promoters of some chlorophyll catabolic genes (Oda-Yamamizo et al., 2016). More recently still, an oilseed rape (Brassica napus) homolog of ANAC087 has been shown to modulate reactive oxygen species accumulation followed by cell death in the context of stress, hormone treatments, and senescent leaves. Interestingly, this TF directly promotes the expression of several cell death-associated genes when overexpressed in wild tobacco (Nicotiana benthamiana), including the tobacco ZINNIA ENDONUCLEASE1 (NbZEN1), a tobacco homolog of BFN1 (Yan et al., 2018).
Similarly, Arabidopsis ANAC046 was shown to interact with RADICAL-INDUCED CELL DEATH, a protein involved in oxidative stress responses (Jaspers et al., 2009). Furthermore, ANAC046 was previously examined for a function in chlorophyll degradation during leaf senescence (Oda-Yamamizo et al., 2016). Recently, mutant anac046 and ANAC046-SRDX plants were found to show enhanced growth and delayed leaf senescence, whereas constitutive overexpression of ANAC046 resulted in slower germination, smaller plants, and accelerated leaf senescence (Oda-Yamamizo et al., 2016). However, in our hands, inducible overexpression lines of ANAC046 in 5-d-old seedlings resulted in complete growth arrest followed by the death of the entire seedling (Figures 2B and 2D). Conceivably, the use of inducible overexpression enabled us to select lines with high expression levels, which would not have been recovered via a constitutive overexpression approach. Furthermore, it was shown that BFN1, EXI1, and RNS3 are downstream targets of ANAC046 in senescing leaves (Oda-Yamamizo et al., 2016). A microarray experiment revealed that all three PCD-associated genes were downregulated in the anac046 and ANAC046-SRDX mutants and upregulated in the ANAC046 overexpression line, which was confirmed in our experiments. Furthermore, several other PCD-associated genes, including C-TERMINALLY ENCODED PEPTIDE1 (CEP1), METACASPASE9 (MC9), PUTATIVE ASPARTIC PROTEINASE A3 (PASPA3), and DUF679 DOMAIN MEMBRANE PROTEIN4 (DMP4), were shown to be downstream targets of ANAC046 (Olvera-Carrillo et al., 2015; Oda-Yamamizo et al., 2016). It is tempting to speculate that, besides promoting chlorophyll degradation, ANAC046 might be involved in regulating PCD processes that terminate the senescence process in aging leaves.
Our data show that ANAC046 and ANAC087 are involved in regulating dPCD of the root cap, indicating that both TFs might be PCD regulators in several tissues activated under different circumstances, including cellular differentiation, senescence, and stresses. Interestingly, our transcriptional and translational reporters revealed that while ANAC087 and ANAC046 are preferentially expressed in cells preparing for PCD, there is also some degree of expression in tissues not undergoing cell death. This suggests that these TFs might have other functions besides regulating PCD. Either expression levels determine the functions of these TFs with respect to their role in cell death, or specifically expressed interaction partners fine-tune PCD versus non-PCD roles in different tissues.
Our results indicate that, on the one hand, ANAC087 regulates postmortem degradation of dying cells in the distal part of the LRC, whereas on the other hand, it regulates the timing of PCD onset in the proximal part of the LRC and in the columella. These results suggest that the cell death programs in the distal and proximal regions of the LRC are differentially regulated (Figure 7). In the distal LRC, the timing of PCD execution is controlled by SMB, while ANAC087 seems to merely coordinate one aspect of PCD execution, postmortem corpse clearance. However, the smb-like phenotype of the strong pSMB:ANAC087-SRDX line might suggest a partially redundant role of ANAC087 in controlling the timing of PCD onset in the distal LRC. In the proximal LRC and the columella, however, ANAC087 takes on a more SMB-like function by regulating PCD timing in the LRC and columella cells before, during, and after columella sloughing. ANAC046 is specifically expressed in the columella, and redundantly with ANAC087, promotes the timely execution of PCD in this tissue (Figure 7). Whether ANAC046 and ANAC087 also control postmortem degradation in the columella is difficult to determine. Analyses of single time points are not able to discriminate between a delay in cell death execution and a delay in postmortem degradation. Time courses to track the degradation of individual columella cells are problematic due to the sloughing of these cells and potential adverse effects of the PI stain over time. Nevertheless, our data suggest that ANAC087 indirectly controls the expression of downstream PCD-associated genes, whereas ANAC046 directly binds to and activates their promoters. These results indicate that additional TFs downstream of ANAC087 might be involved in the control of dPCD-associated genes, but so far, the identity of these downstream TFs is still unknown.
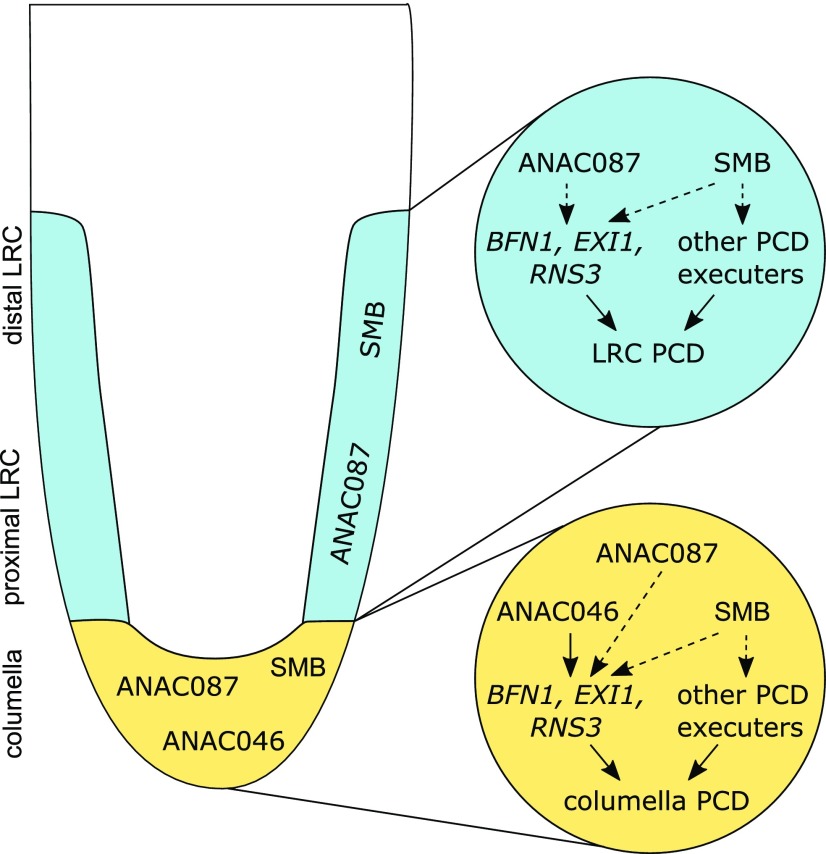
Figure 7.
Model: SMB, ANAC087, and ANAC046 Control Different Aspects of Root Cap Cell Death.
In Arabidopsis, PCD is the final step of differentiation of both the columella and the LRC. In the columella (shown in yellow), ANAC087 and ANAC046 redundantly control the timing of PCD onset by regulating the expression of at least three PCD-associated genes: BFN1, EXI1, and RNS3. Furthermore, SMB is expressed in the columella (Bennett et al., 2010), and SMB was shown to function not only upstream of BFN1, EXI1, and RNS3, but also of some additional PCD-associated genes (Fendrych et al., 2014). In the LRC (shown in blue), so far only ANAC087 and SMB have been shown to be involved in PCD regulation. Again, both ANAC087 and SMB orchestrate the expression of BFN1, EXI1, and RNS3, whereas SMB has some additional PCD-associated targets (Fendrych et al., 2014). However, even though both ANAC087 and SMB are expressed in the entire LRC, it appears that PCD is differentially regulated at the distal part compared with the proximal zone.
Similar to ANAC087 and ANAC046, SMB also causes growth arrest (Bennett et al., 2010) and ectopic cell death (Figure 2A) when inducibly overexpressed. This indicates that several PCD-related NAC TFs are sufficient to induce PCD when ectopically overexpressed. Furthermore, our RT-qPCR data on smb mutant root tips and SMB inducible overexpression lines showed that BFN1, EXI1, and RNS3 are downstream targets of SMB (Figure 3C). These results indicate that SMB, ANAC087, and ANAC046 share a similar spectrum of target genes and are likely components of an integral transcriptional PCD network, even though there is no evidence that they form a transcriptional cascade. Our data suggest that distinct, but likely interconnected pathways exist to fine-tune PCD execution and to ensure proper timing of PCD in tissues that are destined to die. A similar fine-tuning of PCD regulation was observed in senescing leaves, where independent pathways regulate senescence, ultimately resulting in terminal senescence and cell death (Kim et al., 2014). In senescent leaves, the TF EIN2 was found to control the entire senescence regulatory network on top of a transcriptional cascade. It is possible that a similar regulatory cascade also exists in root cap cell death, but additional TFs will have to be identified to test this hypothesis.
The root cap is known to be important for protecting the stem cells and receiving environmental stimuli to direct root growth (Kumpf and Nowack, 2015). Additionally, in many species, including pea (Pisum sativum) and maize (Zea mays), living border cells that are shed from the root cap into the rhizosphere have been shown to function in protecting the plant against pathogens (Cannesan et al., 2011; Hawes et al., 2012). Also, the living, sloughed border-like cells in Arabidopsis have been implicated in pathogen defense (Driouich et al., 2013). Here, we show that ANAC087 and ANAC046 control a cell death program that restricts the life span of shed columella cells. Previous reports indicate that shed Arabidopsis columella cells can stay alive up to 48 h in liquid medium and then gradually die (Plancot et al., 2013). However, we observed that in wild-type seedlings, most columella cells had died ∼24 h after shedding. This difference might be due to the use of slightly older seedlings (10 d old) or different growth conditions in the previously published study (Plancot et al., 2013).
To date, we can only speculate about the adaptive advantage of columella cells undergoing PCD after shedding. Perhaps columella PCD is merely a vestigial process that does not play any crucial role in root growth and development in Arabidopsis. However, it is possible that, under natural conditions, a certain life span of columella cells terminated by controlled PCD is advantageous for the plant’s interaction with microbiota in the rhizosphere. With the rapidly growing interest in the workings and functions of the plant rhizosphere (Mauchline and Malone, 2017), it would be very interesting to follow this hypothesis in the future.
METHODS
Meta-Analysis of Publically Available Transcriptome Data
An Excel spreadsheet was created using the VisuaLRTC methodology (Parizot et al., 2010) to identify genes that are highly expressed in the LRC and TEs compared with other root tissues. Microarray hybridization files for the following marker lines were retrieved from the Gene Expression Omnibus database: S17, S32, COBL9, J0121, S4, SUC2, J2501, RM1000, PET111, AGL42, LRC, GL2, SCR, J0571, WOL, CORTEX, APL, and S18. J2661 hybridization files were kindly provided by Mitchell Levesque from Philip Benfey’s lab at Duke University, Durham, NC (Levesque et al., 2006). Normalization was performed using the robust multiarray average method (Irizarry et al., 2003b) and “present,” “marginal,” and “absent” calls were generated using the MAS5 (Irizarry et al., 2003a) algorithm in R. Affymetrix probe sets to AGI ID assignment was done using the affy_ATH1_array_elements-2010-12-20.txt file downloaded from TAIR (www.arabidopsis.org) for 21,107 probe sets corresponding to unique AGI IDs. In order to find genes more highly expressed in the lateral root cap and the xylem TEs (LRC and S18, respectively) compared with other tissues (all tissues previously described, excluding LRC, S18, and the columella PET111, which can share homologies with the LRC), we selected genes that are systematically “present” and more highly expressed in both LRC and S18 compared with the other tissues. Additionally, we limited this list to TFs only according to the supplemental data from Mitsuda and Ohme-Takagi (2009). The result of this query is provided in Supplemental Data Set 1 with genes ranked according to the enrichment of expression in the LRC and TEs compared with the other tissues. From the 15 resulting TFs, we selected two NAC TFs for further analysis.
Plant Material and Growth Conditions
All Arabidopsis thaliana seedlings were grown vertically on 0.5× Murashige and Skoog (MS) medium (0.1 g/L MES, pH 5.8 [KOH], and 0.8% plant agar) (Murashige and Skoog, 1962) for 5 d in day/night conditions (16 h light, 8 h dark, 22°C) under white light emitted by fluorescent lamps (intensity 120 µmol m2 s−1) before analysis, unless stated otherwise. Mutant lines of anac087 and anac046 were ordered from the NASC stock center in Nottingham, UK. For verification of homozygous mutants, primers P11-P12, P13-P14, and P24-P25 were used together with GABI-Kat (GK), SALK, and Saskatoon (SK) specific left border primers. The pRPS5A:XVE>>NLS-GUS-GFP line was used by Gao et al. (2018) and the ToIM marker was published by Fendrych et al. (2014) and here cloned under the control of the pUBIQUITIN10 promoter (kindly provided by Matthias Van Durme). ANAC046 CRISPR lines were created using sgRNA pairs listed in Supplemental Table 1. Primers for these sgRNAs were annealed and ligated into pMR217 and pMR218 (Ritter et al., 2017). Entry clones were then recombined into pDE-Cas9 (Fauser et al., 2014), and the obtained expression clones were transformed into Col-0 plants using the Agrobacterium tumefaciens floral dip method (Clough and Bent, 1998). Homozygous T2 lines were selected for the production of T3 seeds and selected for the absence of the Cas9 gene. To induce overexpression in the inducible overexpression lines, 5-d-old seedlings were transferred to plates with 0.5× MS medium supplemented with 10 µM estradiol (dissolved in ethanol) or ethanol (mock) before analysis. For all transgenic lines generated, several independent transformation events were generated, and at least two representative independent single-insertion lines were analyzed in detail.
Cloning
All fragments were cloned into Gateway-compatible entry clones (Invitrogen). To create the entry clones, DNA fragments were generated from template-DNA using specific primers and high-fidelity Phusion DNA polymerase (New England Biolabs) in a standard PCR. All cloning primers are listed in Supplemental Table 2.
The 4-kb promoter fragments of ANAC087 and ANAC046 were amplified from genomic DNA using primers P1-P2 and P15-P16, respectively, and cloned into pDONRP4P1r by restriction enzyme digestion and ligation. The ANAC087 and ANAC046 coding sequences without stop codon were amplified from cDNA using primers P3-P4 and P17-P18, respectively, and both PCR products were recombined into pDONR221 using BP Clonase (Invitrogen) to obtain pEN-L1-ANAC087-L2 and pEN-L1-ANAC046-L2. For the translational reporters of both TFs, a fusion protein with VENUS was created. ANAC087 was amplified from genomic DNA using primers P3-P5, VENUS was amplified from a VENUS containing plasmid (PSB Gateway Vector collection) using primers P6-P7, and both fragments were fused in a PCR using primers P3-P7. Similar to ANAC087, ANAC046 was amplified from genomic DNA using primers P17-P19, VENUS was amplified from a VENUS containing plasmid (PSB Gateway Vector collection) using primers P20-P7, and both fragments were fused in a PCR using primers P17-P7. The obtained ANAC087-VENUS and ANAC046-VENUS fragments were recombined into pDONR221 using BP Clonase (Invitrogen) to create pEN-L1-ANAC087-VENUS-L2 and pEN-L1-ANAC046-VENUS-L2. The 3′ untranslated regions (UTRs) from ANAC087 and ANAC046 were amplified from genomic DNA using primers P8-P9 and P21-P22, respectively. The PCR fragments were then recombined into pDONRP2R-P3 using BP-Clonase (Invitrogen) to obtain pEN-R2-ANAC087-3UTR-L3 and pEN-R2-ANAC046-3UTR-L3. Finally, the SRDX fusions were created by a two-step PCR. For ANAC087, first, a DNA fragment was amplified from cDNA using primers P3-P10, and the obtained DNA fragment was used as a template in a new PCR using primers P3-P26. Similar to ANAC087, for ANAC046, first a DNA fragment was amplified from cDNA using primers P17-P23, and the obtained DNA fragment was used as a template in a new PCR using primers P17-P26. The obtained DNA fragments were recombined into pDONR221 using BP Clonase (Invitrogen) to create pEN-L1-ANAC087-SRDX-L2 and pEN-L1-ANAC046-SRDX-L2. Other entry clones that were used are pEN-L4-pSMB-L1 from Fendrych et al. (2014), pEN-L1-NLS-tdTOMATO-L2, pEN-R2-VENUSstopL3, and pEN-R2-GFP-L3, which were obtained from the PSB Gateway Vector collection, and pEN-L4-pRPS5A-XVE-R1 (Gao et al., 2018).
The obtained entry clones were recombined into Gateway destination vectors pB7m34GW or pB7m24GW using LR Clonase II plus enzyme mix (Invitrogen) to create the expression clones pANAC087:tdTOMATO, pANAC046:tdTOMATO, pANAC087:ANAC087-VENUS-3′UTR, pANAC046:ANAC046-VENUS-3′UTR, pSMB:ANAC087-SRDX, pSMB:ANAC046-SRDX, pRPS5A:XVE>>ANAC087-GFP, and pRPS5A:XVE>>ANAC046-GFP. These expression clones were transformed into competent Agrobacterium cells (C58C1 pMP90) using electroporation, and these bacteria were used for a modified floral dip (Clough and Bent, 1998) into Col-0 plants. Homozygous, single-locus T3 lines were used for analysis, unless stated otherwise.
Confocal Imaging
Confocal imaging was done on the LSM710 (Zeiss) using the Plan Apochromat 20× objective (numerical aperture 0.8), unless stated otherwise. All images were in 8 bit with at least 2× line averaging.
For the reporter lines, 5-d-old seedlings were mounted on a glass slide in 1/10 MS medium. tdTOMATO was excited by the 561-nm laser and emission detected between 570 and 564 nm. The VENUS fusion proteins were excited by the 514-nm line of the argon laser and detected between 525 and 620 nm. Static images of 5-d-old mutants and SRDX lines stained with PI (1/10 MS supplemented with 10 µg/mL PI) were obtained with the Plan Apochromat 10× objective (numerical aperture 0.45) using the 561-nm laser and detection between 570 and 645 nm in channel 1 and between 650 and 755 nm in channel 2. To create kymographs, seedlings were transferred to a LabTekII chamber (Nunc) and covered by an agar slab (0.5× MS) supplemented with 10 µg/mL PI and imaged every 10 min (anac087-1 and wild type) or 12 min (SRDX and wild type). PI was imaged as for the static images. Rescue lines were imaged sequentially for VENUS and PI in track 1 and track 2, respectively. Excitation and emission of VENUS and PI were as described before. Imaging of the TUNEL samples was done using the 488-nm laser line to excite the fluorescein, and emission was detected between 495 and 550 nm in the first track. In the second track, 4′,6-diamidino-2-phenylindole (DAPI) was excited by the 405-nm laser and detected between 420 and 480 nm.
In the inducible overexpression lines, seedlings treated for 24 h with estradiol or ethanol (mock) were mounted on a glass slide in 1/10 MS supplemented with 10 µg/mL PI. GFP and PI were detected simultaneously in the same track. GFP was excited by the 488-nm line of the argon laser and detected between 495 and 550 nm in channel 1, whereas PI was detected between 580 and 735 nm in channel 2. For the overnight time course, 6 h induced seedlings were transferred to a LabTekII chamber (Nunc) and covered with an agar slab (0.5× MS) supplemented with 10 µg/mL PI and 10 µM estradiol. Seedlings were imaged every 30 min for 16 h on the LSM5 Exciter (Zeiss), which was flipped for vertical imaging. Imaging of GFP and PI was performed as described before.
The seedlings of the pUBQ-ToIM line were grown for 4 d on 0.5× MS medium and then transferred to a LabTekII chamber (Nunc) covered by an agar slab (0.5× MS). After transfer, the seedlings were returned to the growth room for 24 h and then imaged every 12 h for 3 d. In between two imaging moments, chambers with seedlings were transferred back to the growth room. GFP and mRFP were imaged simultaneously using the 488-nm laser line to excite GFP and the 561-nm laser line to excite mRFP. Emission was detected for GFP in channel 1 between 495 and 550 nm and for mRFP in channel 2 between 595 and 715 nm. The long time courses following roots of pUBQ-ToIM in different backgrounds for 24 h were performed on the LSM5 Exciter (Zeiss) using the 20× objective. GFP was excited using the 488-nm laser line of the argon laser and was detected using the band-pass 505-530 filter. MRFP was excited using the 543-nm laser and detected using the long-pass 650 filter. To follow the root, the root TipTracker software was used (von Wangenheim et al., 2017). For the FDA-PI viability staining, 6-d-old seedlings were mounted on a glass slide in FDA solution (eight droplets of dissolved FDA [2 mg in 1 mL acetone] in 10 mL 0.5M sucrose) supplemented with 10 µg/μL PI. FDA and PI were imaged sequentially using the 488-nm laser line in track 1 to excite FDA and the 561-nm laser line in track 2 to excite PI. Emission was detected between 495 and 550 nm and between 580 and 715 nm for FDA and PI, respectively. Image analysis was performed in FIJI (Schneider et al., 2012).
CANON Imaging and Image Processing
To image the seedlings macroscopically over time, a Canon camera (EOS 650D) was used. Seedlings were transferred to a 0.5× MS plate with 10 µM estradiol and imaged every 30 min for 6 d using the EOS utility software (Canon). Images were imported into FIJI (Schneider et al., 2012) to create movies and time stamp.
Evans Blue Staining
For Evans Blue staining, 5-d-old seedlings were transferred to a 0.5× MS plate with 10 µM estradiol and incubated for ∼24 h. Seedlings were then stained for 30 min in a 2% solution of Evans Blue (Sigma-Aldrich) in water. Seedlings were then washed in distilled water and mounted on a glass slide. Samples were imaged on an Olympus BX51 microscope with a 10× objective (numerical aperture 0.25).
TUNEL
For the TUNEL staining, the in situ cell death detection kit, Fluorescein (Roche) was used. Fixation and all wash steps were performed in a 12-well plate, and only the TUNEL reaction was performed in a 1.5-mL Eppendorf tube. Approximately 10 seedlings per condition were used.
Seedlings were fixed for 1 h in 4% paraformaldehyde (pH 7.4) under vacuum at room temperature and subsequently washed five times with 1 mL PBS. Then, seedlings were permeabilized for 2 min on ice with 1 mL 0.1% Triton X-100 in 0.1% sodium citrate and subsequently washed again five times with 1 mL of PBS. The positive control was treated with 1 mL DNaseI solution (1 mL PBS with 0.025 μL DNaseI [Sigma-Aldrich] and 2% BSA) for 15 min at room temperature, and washed three times with 1 mL PBS. Label solutions for the negative and positive control and for the test seedlings were prepared in a tube according to the manual. Seedlings were added to the tubes and incubated for 60 min at 37°C in the dark. Afterwards, the seedlings were transferred again to a 12-well plate and washed three times with 1 mL of PBS. Finally, seedlings were mounted in Citifluor (antifading agent) with 0.25 µg/mL DAPI.
RT-qPCR
For RT-qPCR of mutant lines, mutant and wild-type seedlings were grown for 5 d on 0.5× MS medium. Using a binocular microscope (Leica), ∼200 big root tips (cut just above the meristem) were harvested for anac087-1 and 500 small root tips (cut around the QC) for anac046-1. For the inducible overexpression lines, whole seedlings induced for 24 h by estradiol or mock treatment were harvested. For each line, three biological replicates (independently repeated treatments) were tested. After RNA extraction using the Spectrum Plant Total RNA Kit (Sigma-Aldrich), 1 µg of RNA was used for DNA synthesis with the iScript cDNA synthesis kit (Bio-Rad). The RT-qPCR was performed with the LightCycler 480 (Roche) using SYBR green for detection of double-stranded DNA. Analysis of the RT-qPCR data was done using qBase+ software (Biogazelle), with PEX4 (AT5G25760), UNK (AT4G16100), EF1A (AT5G60390), and UBL5 (AT5G42300) as housekeeping genes, and later the data were normalized against wild-type or mock samples. Primers used in the RT-qPCR are listed in Supplemental Table 3.
EMSA
To create recombinant tagged proteins, ANAC087, ANAC046 (entry clones from TAIR), and GFP (control) were fused to HIS6MBP by cloning into the pDEST-HisMBP destination vector (Nallamsetty et al., 2005) and transformed into competent BL21 DE3 Escherichia coli cells. When bacterial growth reached its mid-exponential phase, the culture was induced by 1 M IPTG for 16 h at 18°C. After centrifugation, the pellet was resuspended in 50 mL lysis buffer (50 mM Tris, pH 8, 5 mM imidazole, pH 8, 500 mM NaCl, 0.1% Triton X-100, and protease inhibitors). The samples were sonicated using a Digital Sonifier (Branson Ultrasonics) and then centrifuged for 20 min at 4°C. After washing the column containing Ni2+-NTA resin (Qiagen) five times with water and five times with binding buffer (50 mM Tris, pH 8, 5 mM imidazole, pH 8, and 250 mM NaCl), the supernatant was added to bind to the column. Next, the column was washed 10 times with washing buffer (50 mM Tris, pH 8, 60 mM imidazole, pH 8, and 250 mM NaCl) and then eluted with elution buffer (50 mM Tris, pH 8, 400 mM imidazole, pH 8, and 250 mM NaCl). Finally, a PD-10 desalting columns (GE Healthcare) were used to remove the imidazole and dissolve proteins in Buffer1 (20 mM Tris, pH 7.5, and 200 mM NaCl). Fractions of 0.5 mL were collected and protein concentrations were determined using a Pierce BCA protein assay kit (Life Technologies).
For the EMSA, TF binding sites were predicted using protein binding microarray data combined with coexpression data of possible target genes (Lindemose et al., 2014). The 8-bp binding sites obtained from the protein binding microarray were located in the promoters of target genes and extended to the left and right to obtain a 40-bp promoter fragment. For BFN1, EXI1, and RNS3, these 40-bp promoter fragments were ordered as 40-bp primers labeled at the 5′ end with IRDye 700. Primer sequences can be found in Supplemental Table 4. Primer sequences for BFN1 were the same as in Matallana-Ramirez et al. (2013). Primers for the unlabeled competitor and unlabeled mutant competitor probes can also be found in Supplemental Table 4. After annealing of the primers, EMSA was performed using the Li-Cor Odyssey EMSA buffer kit, and imaging was done on the Odyssey imaging system (Li-Cor).
ChIP-qPCR
For the ChIP, ∼200 mg of seeds of the inducible overexpression lines (pRPS5A:XVE>>ANAC087-GFP or pRPS5A:XVE>>ANAC046-GFP) or the control (pRPS5A:XVE>>GFP) were grown on plates with a nylon mesh. Five-day-old seedlings were sprayed with liquid 0.5× MS medium supplemented with 50 µM estradiol and 0.01% Triton. Seedlings were harvested in liquid nitrogen 24 h (for the pRPS5A:XVE>>ANAC046-GFP line) or 30 h (for the pRPS5A:XVE>>ANAC087-GFP line) after estradiol induction. Seedlings were ground in liquid nitrogen and 2 g of plant powder was used per ChIP. The chromatin cross-linking, sonication, immunoprecipitation, reverse cross-linking, and DNA extraction were performed as described (Ritter et al., 2017). The Chip-qPCR was performed as described before. Primers for the ChIP-qPCR can be found in Supplemental Table 3. Normalized percentage of input was calculated using this formula: 
TEA
The transfection of BY2 tobacco protoplasts and the luciferase assay were performed as described (Vanden Bossche et al., 2013).
Statistical Analysis
For the statistical analysis, GraphPad Prism (GraphPad Software) was used, unless stated otherwise. For the counting of nuclear remnants, and FDA-PI staining, a one-way ANOVA with Dunn’s multiple testing correction was used. See Supplemental Table 5 for an overview on ANOVA testing. For the nuclear counting in the pANAC087:BFN1-VENUS rescue experiment, ANOVA was followed by Tukey. For the distance to QC en TEA’s, a Kruskal-Wallis nonparametric test was used. For the RT-qPCRs, nuclear degradation time, and ChIP-qPCR, a Student’s t test was used with a Holm-Sidak correction for multiple testing.
The statistical analysis for counting of living, detached columella cells in different pUBQ-ToIM lines was performed using a Poisson regression. Since the data did not look Poisson distributed, a linear mixed model was fitted to the count data. Based on a saturated mean model, including the main effects of line and stage (as categorical predictor variable) and their interaction term, several covariances were tested to take into account the correlation between measures performed on the same root. Based on the BIC (for anac087-1, anac046-2, and anac087 anac046) or AIC (for anac046-1) values, an unstructured covariance was used. Since the experiment was repeated three times, an additional random effect for repeat was added to the model. For the fixed effects part of the model, a likelihood ratio test indicated that the model with stage as numerical covariate was better than the model with stage as categorical predictor variable (P = 0.07). The Kenward-Roger approximation for computing the denominator degrees of freedom for the tests of fixed effects was applied. Differences of interest were estimated using Wald tests. Appropriate adjustment of P values was done with Tukey’s method. Assumptions were verified with residual diagnostics. The analysis was performed with the mixed procedure of SAS (version 9.4 of the SAS System for windows 7 64 bit; copyright 2002-2012, SAS Institute).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ANAC087 (AT5G18270), ANAC046 (AT3G04060), SMB (AT1G79580), BFN1 (AT1G11190), PASPA3 (AT4G04460), RNS3 (AT1G26820), EXI1 (At2g14095), CEP1 (AT5G50260), MC9 (AT5G04200), DMP4 (AT4G18425), UBQ10 (AT4G05320), and RPS5A (AT3G11940). Mutants used in this study had the following accession numbers: GK622H06 (anac087-1), SALK_011205c (anac087-2), and SK2690 (nac046-1). The following microarray hybridization files were retrieved from the Gene Expression Omnibus database: GEO Series GSE8934 (S17, S32, COBL9, J0121, S4, SUC2, J2501, and RM1000), GEO Series GSE7641 (PET111 and AGL42), GEO Series GSE5749 (LRC, GL2, SCR, J0571, and WOL), and GEO Series GSE16468 (CORTEX, APL, and S18).
Supplemental Data
Supplemental Figure 1. Transcriptional reporter lines of ANAC087 and ANAC046.
Supplemental Figure 2. Independent overexpression lines of ANAC087 and ANAC046.
Supplemental Figure 3. Independent mutant lines and rescue lines of anac087-1.
Supplemental Figure 4. BFN1-VENUS rescue of anac087-1, EMSA binding sites, and TEA for ANAC087 and ANAC046 binding to the promoters pBFN1, pEXI1, and pRNS3.
Supplemental Figure 5. Independent mutant lines of anac046-1.
Supplemental Table 1. Primers used for CRISPR.
Supplemental Table 2. Primers used for cloning.
Supplemental Table 3. Primers used for qPCR.
Supplemental Table 4. Primers used for EMSA.
Supplemental Table 5. ANOVA table.
Supplemental Data Set 1. Identification of transcription factor genes more highly expressed in the lateral root cap and the tracheary elements compared with all other root tissue layers.
Supplemental Movie 1. pPASPA3-ToIM in inducible overexpression line of SMB.
Supplemental Movie 2. Confocal time course of inducible overexpression lines.
Supplemental Movie 3. Macroscopic time course of inducible overexpression lines.
Supplemental Movie 4. Long time course pUBQ-ToIM in the wild type and in anac087 anac046.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the members of the PCD lab for discussions and critical feedback on the manuscript. We also thank Veronique Storme for assistance with statistical analysis, Annick Bleys for help with the manuscript, and Robert Hauschild for helping to implement the TipTracker software in our imaging equipment. We thank Matthias Van Durme for cloning of the pUBQ promoter. We gratefully acknowledge funding by the ERC StG PROCELLDEATH (Project 639234) to M.K.N.
AUTHOR CONTRIBUTIONS
M.H. designed the research, performed experiments, analyzed data, and wrote the article. R.A.B., N.S., M.C.R., F.D.W., J.M., and M.F. designed and performed experiments, and analyzed data. B.P. produced the meta-analyses tool and analyzed data. M.K. cloned the promoters of ANAC087 and ANAC046. M.N. designed and directed the research and wrote the manuscript.
Footnotes
Articles can be viewed without a subscription.
References
- Bennett T., van den Toorn A., Sanchez-Perez G.F., Campilho A., Willemsen V., Snel B., Scheres B. (2010). SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 22: 640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannesan M.A., Gangneux C., Lanoue A., Giron D., Laval K., Hawes M., Driouich A., Vicré-Gibouin M. (2011). Association between border cell responses and localized root infection by pathogenic Aphanomyces euteiches. Ann. Bot. 108: 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Coll N.S., Epple P., Dangl J.L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneva A., Gao Z., Van Durme M., Nowack M.K. (2016). Functions and regulation of programmed cell death in plant development. Annu. Rev. Cell Dev. Biol. 32: 441–468. [DOI] [PubMed] [Google Scholar]
- Dolan L., Janmaat K., Willemsen V., Linstead P., Poethig S., Roberts K., Scheres B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84. [DOI] [PubMed] [Google Scholar]
- Driouich A., Follet-Gueye M.L., Vicré-Gibouin M., Hawes M. (2013). Root border cells and secretions as critical elements in plant host defense. Curr. Opin. Plant Biol. 16: 489–495. [DOI] [PubMed] [Google Scholar]
- Escamez S., Bollhöner B., Tuominen H. (2017). Quick histochemical staining methods to detect cell death in xylem elements of plant tissues. Methods Mol. Biol. 1544: 27–36. [DOI] [PubMed] [Google Scholar]
- Fauser F., Schiml S., Puchta H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79: 348–359. [DOI] [PubMed] [Google Scholar]
- Fendrych M., Van Hautegem T., Van Durme M., Olvera-Carrillo Y., Huysmans M., Karimi M., Lippens S., Guérin C.J., Krebs M., Schumacher K., Nowack M.K. (2014). Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr. Biol. 24: 931–940. [DOI] [PubMed] [Google Scholar]
- Fuchs Y., Steller H. (2011). Programmed cell death in animal development and disease. Cell 147: 742–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I., Stone J.M., Gechev T.S. (2008). Programmed cell death in plants: new insights into redox regulation and the role of hydrogen peroxide. Int. Rev. Cell Mol. Biol. 270: 87–144. [DOI] [PubMed] [Google Scholar]
- Gao Z., et al. (2018). KIRA1 and ORESARA1 terminate flower receptivity by promoting cell death in the stigma of Arabidopsis. Nat. Plants 4: 365–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes M.C., Curlango-Rivera G., Xiong Z.G., Kessler J.O. (2012). Roles of root border cells in plant defense and regulation of rhizosphere microbial populations by extracellular DNA ‘trapping’. Plant Soil 355: 1–16. [Google Scholar]
- Heo J.O., Blob B., Helariutta Y. (2017). Differentiation of conductive cells: a matter of life and death. Curr. Opin. Plant Biol. 35: 23–29. [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739. [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. (2003a). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003b). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Jaspers P., Blomster T., Brosche M., Salojarvi J., Ahlfors R., Vainonen J.P., Reddy R.A., Immink R., Angenent G., Turck F., Overmyer K., Kangasjarvi J. (2009). Unequally redundant RCD1 and SRO1 mediate stress and developmental responses and interact with transcription factors. Plant J. 60: 268–279. [DOI] [PubMed] [Google Scholar]
- Kim H.J., et al. (2014). Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65: 4023–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumpf R.P., Nowack M.K. (2015). The root cap: a short story of life and death. J. Exp. Bot. 66: 5651–5662. [DOI] [PubMed] [Google Scholar]
- Levesque M.P., Vernoux T., Busch W., Cui H., Wang J.Y., Blilou I., Hassan H., Nakajima K., Matsumoto N., Lohmann J.U., Scheres B., Benfey P.N. (2006). Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemose S., Jensen M.K., Van de Velde J., O’Shea C., Heyndrickx K.S., Workman C.T., Vandepoele K., Skriver K., De Masi F. (2014). A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res. 42: 7681–7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallana-Ramirez L.P., Rauf M., Farage-Barhom S., Dortay H., Xue G.-P., Dröge-Laser W., Lers A., Balazadeh S., Mueller-Roeber B. (2013). NAC transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol. Plant 6: 1438–1452. [DOI] [PubMed] [Google Scholar]
- Mauchline T.H., Malone J.G. (2017). Life in earth - the root microbiome to the rescue? Curr. Opin. Microbiol. 37: 23–28. [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Ohme-Takagi M. (2009). Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 50: 1232–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nallamsetty S., Austin B.P., Penrose K.J., Waugh D.S. (2005). Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci. 14: 2964–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea C., Kryger M., Stender E.G., Kragelund B.B., Willemoës M., Skriver K. (2015). Protein intrinsic disorder in Arabidopsis NAC transcription factors: transcriptional activation by ANAC013 and ANAC046 and their interactions with RCD1. Biochem. J. 465: 281–294. [DOI] [PubMed] [Google Scholar]
- Oda-Yamamizo C., Mitsuda N., Sakamoto S., Ogawa D., Ohme-Takagi M., Ohmiya A. (2016). The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci. Rep. 6: 23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvera-Carrillo Y., et al. (2015). A conserved core or programmed cell death indicator genes discriminates developmentally and environmentally induced programmed cell death in plants. Plant Physiol. 169: 2684–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10: 239–247. [DOI] [PubMed] [Google Scholar]
- Parizot B., De Rybel B., Beeckman T. (2010). VisuaLRTC: a new view on lateral root initiation by combining specific transcriptome data sets. Plant Physiol. 153: 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett A.R.G., Thomas S.G., Wilson Z.A., Hedden P. (2011). Gibberellin control of stamen development: a fertile field. Trends Plant Sci. 16: 568–578. [DOI] [PubMed] [Google Scholar]
- Plancot B., Santaella C., Jaber R., Kiefer-Meyer M.C., Follet-Gueye M.L., Leprince J., Gattin I., Souc C., Driouich A., Vicré-Gibouin M. (2013). Deciphering the responses of root border-like cells of Arabidopsis and flax to pathogen-derived elicitors. Plant Physiol. 163: 1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter A., et al. (2017). The transcriptional repressor complex FRS7-FRS12 regulates flowering time and growth in Arabidopsis. Nat. Commun. 8: 15235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siligato R., et al. (2016). Multisite Gateway-compatible cell type-specific gene-inducible system for plants. Plant Physiol. 170: 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Bossche R., Demedts B., Vanderhaeghen R., Goossens A. (2013). Transient expression assays in tobacco protoplasts. Methods Mol. Biol. 1011: 227–239. [DOI] [PubMed] [Google Scholar]
- Van Durme M., Nowack M.K. (2016). Mechanisms of developmentally controlled cell death in plants. Curr. Opin. Plant Biol. 29: 29–37. [DOI] [PubMed] [Google Scholar]
- Van Hautegem T., Waters A.J., Goodrich J., Nowack M.K. (2015). Only in dying, life: programmed cell death during plant development. Trends Plant Sci. 20: 102–113. [DOI] [PubMed] [Google Scholar]
- Vicré M., Santaella C., Blanchet S., Gateau A., Driouich A. (2005). Root border-like cells of Arabidopsis. Microscopical characterization and role in the interaction with rhizobacteria. Plant Physiol. 138: 998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wangenheim D., Hauschild R., Fendrych M., Barone V., Benková E., Friml J. (2017). Live tracking of moving samples in confocal microscopy for vertically grown roots. eLife 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Franke-van Dijk M., Vencken R.J., Quint A., Hooykaas P., Offringa R. (2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128: 4289–4299. [DOI] [PubMed] [Google Scholar]
- Willemsen V., Bauch M., Bennett T., Campilho A., Wolkenfelt H., Xu J., Haseloff J., Scheres B. (2008). The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell 15: 913–922. [DOI] [PubMed] [Google Scholar]
- Yan J., Tong T., Li X., Chen Q., Dai M., Niu F., Yang M., Deyholos M.K., Yang B., Jiang Y.Q. (2018). A novel NAC-type transcription factor, NAC87, from oilseed rape modulates reactive oxygen species accumulation and cell death. Plant Cell Physiol. 59: 290–303. [DOI] [PubMed] [Google Scholar]