SAUR62 and SAUR75 regulate pollen tube growth through physical interaction with ribosome subunits to affect translation of cell wall and actin cytoskeleton remodeling-related transcripts.
Abstract
Successful pollen tube elongation is critical for double fertilization, but the biological functions of pollen tube genes and the regulatory machinery underlying this crucial process are largely unknown. A previous translatomic study revealed two Arabidopsis (Arabidopsis thaliana) SAUR (SMALL AUXIN UP RNA) genes, SAUR62 and SAUR75, whose expression is up-regulated by pollination. Here, we found that both SAUR62 and SAUR75 localized mainly to pollen tube nuclei. The siliques of homozygous saur62 (saur62/−), saur75 (saur75/−), and the SAUR62/75 RNA interference (RNAi) knockdown line had many aborted seeds. These lines had normal pollen viability but defective in vitro and in vivo pollen tube growth, with branching phenotypes. Immunoprecipitation with transgenic SAUR62/75-GFP flowers revealed ribosomal protein RPL12 family members as potential interacting partners, and their individual interactions were confirmed further by yeast two-hybrid and bimolecular fluorescence complementation assays. Polysome profiling showed reduced 80S ribosome abundance in homozygous saur62, saur75, ribosomal large subunit12c, and SAUR62/75 RNAi flowers, suggesting that SAUR62/75 play roles in ribosome assembly. To clarify their roles in translation, we analyzed total proteins from RNAi versus wild-type flowers by isobaric tags for relative and absolute quantitation, revealing significantly reduced expression of factors participating in pollen tube wall biogenesis and F-actin dynamics, which are critical for the elastic properties of tube elongation. Indeed, RNAi pollen tubes showed mislocalization of deesterified and esterified pectins and F-actin organization. Thus, the biological roles of SAUR62/75 and their RPL12 partners are critical in ribosomal pre-60S subunit assembly for efficient pollen tube elongation and subsequent fertilization.
Seeds or grains constitute the major source of food supply around the world, and their production depends on successful pollination, double fertilization, and subsequent embryogenesis. The pollen tube, the reduced male gametophyte of flowering plants, travels through the transmitting tract inside female tissues and delivers the male gametes (two sperm cells) to fertilize female gametes (egg and central cell) for double fertilization (Berger et al., 2008). Well-programmed microspore development, pollen germination, and tube growth are essential for successful fertilization (Malhó, 2006). Cell-cell communication and intricate signaling cascades are important steps in the pollination process (Cheung and Wu, 2008; Qin and Yang, 2011; Dresselhaus and Franklin-Tong, 2013).
The phytohormone auxin influences nearly all aspects of plant growth and development by regulating cell division, expansion, and differentiation (Chapman and Estelle, 2009). Auxin regulates these processes in part by changing the expression of a set of early auxin response genes: AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA), GRETCHEN HAGEN3 (GH3), and SMALL AUXIN UP RNA (SAUR; Hagen and Guilfoyle, 2002; Woodward and Bartel, 2005; Quint and Gray, 2006; Hayashi, 2012). AUX/IAA genes encode AUX/IAA proteins, which function as transcription repressors (Ulmasov et al., 1997), and GH3 genes encode IAA-amido synthetases, which convert active IAA to an inactive conjugated form (Staswick et al., 2005). SAURs were first identified as primary auxin-inducible genes, and their expression could be induced within 2.5 min after auxin treatment (Franco et al., 1990; Hagen and Guilfoyle, 2002). Most SAURs have no intron, cluster on chromosomes, and contain auxin-responsive elements in their promoter regions (Chen et al., 2014). Furthermore, some SAURs have an mRNA-destabilizing downstream element in the 3′ untranslated region (Gil and Green, 1996).
In addition to auxin, ethylene, brassinosteroid (BR), gibberellin (GA), abscisic acid (ABA), jasmonic acid (JA), light, and osmotic stresses regulate the expression of SAUR genes (Li et al., 2015; Ren and Gray, 2015; Sun et al., 2016), which indicates that SAURs contribute to other hormone- and environmental factor-mediated plant growth and development. Recent studies show that some SAUR genes are expressed mainly in hypocotyls or other elongating tissues, so they may play important roles in regulating cell elongation (Knauss et al., 2003; Chae et al., 2012; Spartz et al., 2012, 2014). For example, AtSAUR63 may affect auxin transport to regulate hypocotyl and stamen filament elongation (Chae et al., 2012), whereas AtSAUR19 promotes cell expansion by negatively regulating type 2C protein phosphatases belonging to the D subfamily to modulate plasma membrane H+-ATPase activity (Spartz et al., 2012, 2014). A few SAUR proteins have been found to bind to Ca2+/calmodulin proteins (Yang and Poovaiah, 2000; Reddy et al., 2002), regulate auxin synthesis and transport (Kant et al., 2009), alter apical hook development (Park et al., 2007), regulate leaf growth and senescence (Spartz et al., 2012; Hou et al., 2013), or regulate root growth and development (Kong et al., 2013; Markakis et al., 2013), but the roles of SAUR proteins during pollen tube growth remain elusive.
Plant ribosomes are composed of four rRNAs and 80 distinct ribosomal proteins (32 small and 48 large ribosomal subunit proteins) and are responsible for protein synthesis (Barakat et al., 2001). During the process of ribosome biogenesis, ribosomal proteins are synthesized in the cytoplasm and then imported into the nucleolus. Ribosome assembly begins in the nucleolus, where ribosomal proteins assemble with rRNAs to form the pre-40S and pre-60S subunits. The preribosomal subunits are then transported through the nucleus to the cytoplasm, where their assembly is completed (Byrne, 2009). Impaired ribosome biogenesis is known to cause developmental defects in yeast (Saccharomyces cerevisiae), humans, and plants (Tschochner and Hurt, 2003; Galani et al., 2004; Ruan et al., 2012). In general, ribosomal proteins are considered housekeeping proteins and required for basic architectural roles in the ribosome. However, accumulating evidence from several ribosomal protein-defective mutants highlights the extra and unique functions of ribosomal proteins. For example, mutations in genes encoding ribosomal proteins have been found to confer multiple cellular and developmental defects, including embryo lethality, late flowering, vascular pattern defects, retarded root growth, or reduced plant size (Byrne, 2009). In addition, several ribosomal protein mutants have auxin-related developmental defects. For instance, mutation of ribosomal large subunit24b (rpl24b) causes defects in the apical-basal patterning of the gynoecium resembling the auxin response factor mutants ettin/auxin response factor3 (arf3) and monopteros/arf5, and the rpl4d and rpl5a mutants show developmental defects in primary root elongation, lateral root initiation, and shoot architecture. These phenotypes are caused by abnormal auxin distribution and sensing via the translational control of auxin response factors dependent on an upstream open reading frame-dependent mechanism (Nishimura et al., 2005; Rosado et al., 2012). Some ribosomal proteins also are involved in sexual plant reproduction, and loss-of-function mutants with gametophyte defects include rpl10a, rpl18aB, rpl27aC, and rps5a (Weijers et al., 2001; Imai et al., 2008; Szakonyi and Byrne, 2011; Yan et al., 2016). All these results suggest that translational regulation as a regulatory mechanism is required for proper and complex plant growth and development.
Our previous study of translatomes of in vivo-grown Arabidopsis (Arabidopsis thaliana) pollen tubes revealed the enrichment of 41 transcripts (Lin et al., 2014), two of which belong to the SAUR family, SAUR62 and SAUR75. Like other in vivo (iv) mutants (Lin et al., 2014), both saur62 and saur75 mutants show unfertilized seeds in mature siliques. In this study, we report the exploration and detailed characterization of the biological roles of these SAURs in elongating pollen tubes. More specifically, we show that their interactions with RPL12 family proteins in the nucleus regulate ribosome assembly, composition, and the subsequent translational activities of several factors involved in wall biogenesis and dynamic F-actin organization of elongating pollen tubes.
RESULTS
SAUR62 and SAUR75 Are Expressed in Pollen Grains and Pollen Tubes
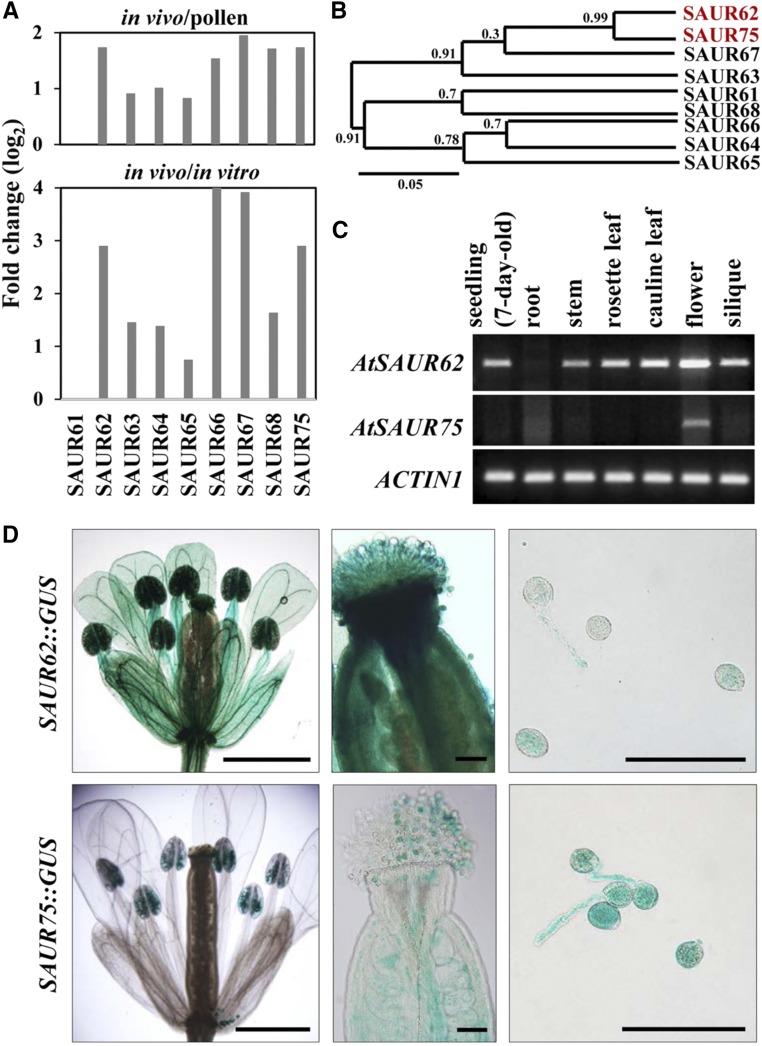
The Arabidopsis genome contains 81 SAUR genes, including two pseudogenes, and is classified into 12 clades (Ren and Gray, 2015). Our previous translatome studies (Lin et al., 2014) show that about one-third of SAURs are up-regulated after pollination and enriched in in vivo-elongating pollen tubes (Supplemental Fig. S1). Among these transcripts, nine members of clade II, SAUR61 to SAUR68 and SAUR75, drew our attention. These SAUR genes except SAUR61 showed higher expression in in vivo-grown pollen tubes, which suggested their important roles in stimulating pollen tube elongation and/or following fertilization (Fig. 1A). These nine SAUR proteins also shared high similarity (Supplemental Fig. S2), and phylogenetic analysis showed that SAUR62 and SAUR75 had the highest amino acid sequence similarity (Fig. 1B). Thus, we focused on these two closely related SAURs in this study.
Figure 1.
Spatiotemporal expression profiles of Arabidopsis SAUR62 and SAUR75. A, Comparing the expression of SAUR62 to SAUR68 and SAUR75 in pollen grains (pollen), in vitro-cultured pollen tubes (in vitro), and in vivo-grown pollen tubes (in vivo) from previously characterized expression profiles of a translatomic study (Lin et al., 2014) where all transcripts were up-regulated significantly in in vivo-grown pollen tubes. B, Phylogenetic tree of nine members in clade II of SAURs obtained at the Website Phylogeny.fr: Tree Dyn. The numbers on the nodes of the phylogenetic tree correspond to bootstrap values. SAUR62 and SAUR75 were in the same cluster. C, RT-PCR analysis showed the ubiquitous and flower-enriched expression patterns of SAUR62 and SAUR75, respectively. ACTIN1 was used as an internal loading control. D, Promoter::GUS analyses showing pollen grains and the in vitro-cultured pollen tube expression profiles of SAUR62 and SAUR75. Bars = 1 mm in flowers and 100 μm in pistils and pollen tubes.
Tissue-specific expression profiles by reverse transcription (RT)-PCR indicated a ubiquitous expression pattern for SAUR62, including in seedlings, stems, rosette leaves, cauline leaves, flowers, and siliques, whereas SAUR75 was present mainly in flowers (Fig. 1C). To further examine the spatial expression patterns within flowers, a promoter::GUS assay was used. SAUR62::GUS was expressed in sepals, petals, stamen filament, pollen grains, pollen tubes, stigma, and style, but SAUR75::GUS was detected mainly in pollen grains, pollen tubes, and ovules (Fig. 1D; Supplemental Fig. S3). Thus, both SAUR62 and SAUR75 were expressed in mature pollen grains and pollen tubes, which suggests their potential roles in pollen development and/or pollen tube growth.
SAUR62 and SAUR75 Localize to the Nucleus of Elongating Pollen Tubes
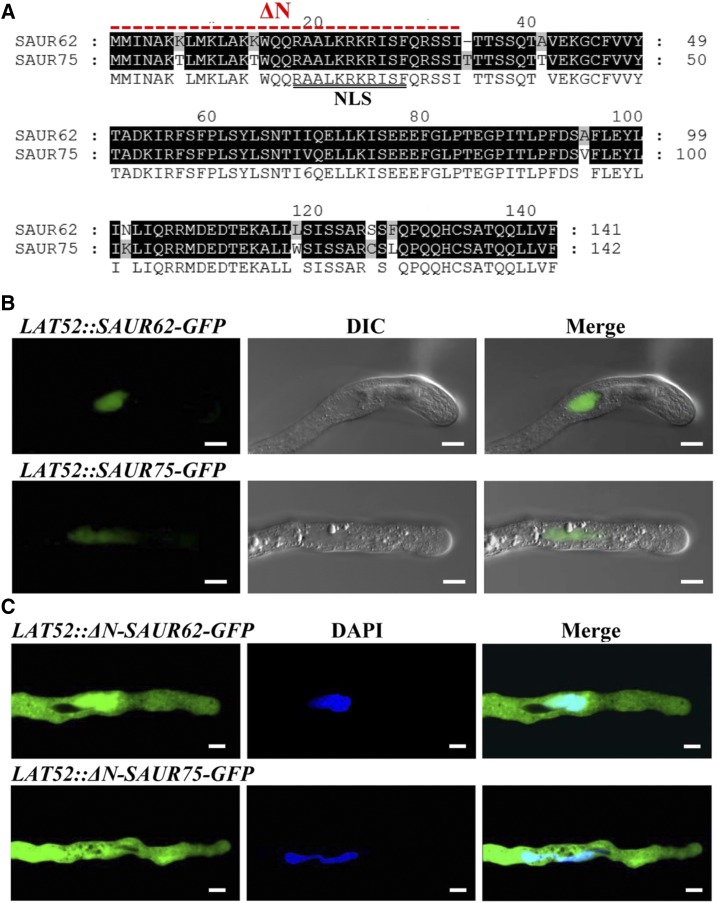
SAUR62 and SAUR75 shared 97.65% and 93.62% identity at the nucleotide and amino acid sequences, respectively, and contain a putative nuclear localization signal (NLS) at their N-terminal regions (Fig. 2A). To examine their subcellular localization, we generated transgenic Arabidopsis plants expressing SAUR62-2xGFP and SAUR75-2xGFP driven by a native SAUR62/75 promoter. SAUR62 and SAUR75 fusion proteins appeared in the nuclei of transgenic seedlings (Supplemental Fig. S4), but no GFP signal was detected in in vitro-cultured pollen tubes. We further generated transgenic Arabidopsis plants expressing SAUR62-GFP and SAUR75-GFP driven by the LAT52 promoter, a postmeiosis-specific promoter in pollen and pollen tubes. In these stable lines, no signal was detected in pollen grains and pollen tubes, but immunoblot analysis detected signals in transgenic flowers (Supplemental Fig. S5).
Figure 2.
Subcellular localization of SAUR62 and SAUR75 in elongating pollen tubes. A, Genetic Computer Group and NLS Mapper were used to obtain pairwise sequence alignment and the predicted NLS marked by the double black lines for SAUR62 and SAUR75. Black shading indicates identical sequences between SAUR62 and SAUR75. ΔN indicates the deleted amino acid sequence in constructs. B and C, Subcellular localization of LAT52 promoter-driven SAUR62/75-GFP (B) and LAT52 promoter-driven ΔN-SAUR62/75-GFP (C) fusion proteins in particle bombardment-transformed tobacco pollen tubes. The bombardment experiments for each construct were performed two independent times; each time, at least 20 fluorescent pollen tubes were observed and showed the same localization pattern. DIC, Differential interference contrast. Bars = 10 μm.
To further evaluate the subcellular localization of SAUR62 and SAUR75 and clarify the relation between their function and pollen tube growth, LAT52::SAUR62-GFP and LAT52::SAUR75-GFP plasmids were expressed transiently in tobacco (Nicotiana tabacum) pollen grains by particle bombardment. Both SAUR62-GFP and SAUR75-GFP fluorescence signals appeared in the vegetative nucleus (Fig. 2B), and their NLS was responsible for their nuclear targeting: the N-terminal domain of ΔN-SAUR62-GFP and ΔN-SAUR75-GFP lack the NLS, so the truncated SAUR62/75 started to accumulate in the cytoplasm of elongating tubes (Fig. 2C). These results suggest that SAUR62/75 are present in the nucleus of elongating tobacco pollen tubes.
SAUR62 and SAUR75 Are Required for Fertility
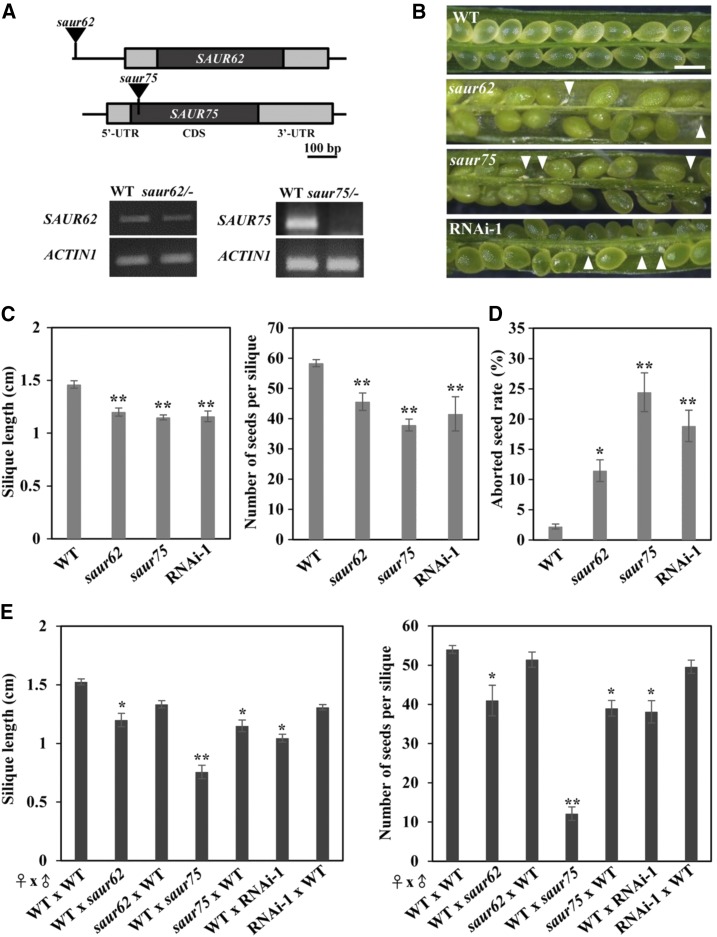
To characterize the biological function of SAUR62 and SAUR75, we analyzed their corresponding T-DNA insertion lines with individual T-DNAs inserted in the promoter and exon regions of SAUR62 and SAUR75, respectively (Fig. 3A). RT-PCR revealed that SAUR62 expression was moderately knocked down in homozygous saur62 (saur62/−) and SAUR75 expression was almost completely knocked out in homozygous saur75 (saur75/−). In addition, we generated the LAT52::SAUR62/75-RNAi mutant by targeting 231-bp fragments to the coding region for both genes under the control of the LAT52 promoter. RT-PCR confirmed reduced SAUR62 and SAUR75 transcript levels in five independent homozygous LAT52::SAUR62/75-RNAi lines as compared with the wild type (Supplemental Fig. S6). Given that SAUR62 and SAUR75 were expressed in pollen grains and pollen tubes (Fig. 1), we analyzed the self-fertilization rates in wild-type, saur62/−, saur75/−, and RNAi-1 plants. Siliques of the wild type were filled with seeds, whereas siliques of saur62, saur75, and RNAi-1 showed seed abortion (Fig. 3B), shorter siliques, and fewer seeds per silique (Fig. 3C). The seed abortion rates were higher in both mutants and RNAi-1 lines than in the wild type (Fig. 3D). These results suggest that saur62, saur75, and RNAi-1 plants had a fertilization defect.
Figure 3.
Mutations and knockdown expression of SAUR62 and SAUR75 significantly impair fertility. A, Schematics of the genomic fragments of SAUR62 and SAUR75 with the corresponding T-DNA insertion sites marked by black triangles. RT-PCR analysis showed that saur62/− was a knockdown mutant and saur75/− was a null mutant. ACTIN1 was used as an internal loading control. B and C, Effects of mutations of SAUR62 and SAUR75 on fertility. B, Aborted seeds (white arrowheads) in dissected siliques. Bar = 0.5 mm. C, Length and average seed number per mature silique obtained from the primary shoots of wild-type (WT), saur62, saur75, and RNAi-1 plants. Columns represent means ± se (n > 20). Asterisks indicate significant differences from the wild type (**, P < 0.01) by Student’s t test. D, Frequencies (%) of aborted seeds per silique of wild-type, saur62, saur75, and RNAi-1 plants. Columns represent means ± se (n > 30). Asterisks indicate significant differences from the wild type (*, P < 0.05 and **, P < 0.01) by Student’s t test. E, Silique length and number of seeds per silique in reciprocal pollination plants. Columns represent means ± se (n > 10). Asterisks indicate significant differences from wild-type ♀ × wild-type ♂ (*, P < 0.05 and **, P < 0.01) by Student’s t test.
To investigate whether the reduced fertility in saur62, saur75, and RNAi-1 was caused by the pistil, pollen grains, or both, we performed reciprocal crosses. With saur62/−, saur75/−, or RNAi-1 pollen grains as the male donors pollinated on wild-type stigmas, crossed siliques showed remarkably reduced silique length and seed number (Fig. 3E). When the pistils of saur75/− were pollinated with wild-type pollen grains, silique length and average number of seeds were reduced. These results indicate that the fertilization defect in saur62 and RNAi-1 was attributed to the pollen grains but not to pistils and in saur75 to both pollen grains and pistils.
Pollen Tube Growth Was Defective in saur62/75 and RNAi-1 Lines
To establish whether the reduced fertility of saur62/−, saur75/−, and RNAi-1 was due to defective pollen, we examined nuclear division by 4′,6-diamidino-2-phenylindole (DAPI) staining and pollen viability by fluorescein diacetate (FDA) and Alexander staining. DAPI staining showed that the pollen grains from saur62, saur75, and RNAi-1 were typical trinucleate pollen grains (two sperm cell nuclei and a vegetative nucleus) like wild-type pollen grains (Supplemental Fig. S7, A–D and M). FDA and Alexander staining revealed that most saur62, saur75, and RNAi-1 pollen grains showed FDA fluorescence (Supplemental Fig. S7, E–H and N) and were red in color (Supplemental Fig. S7, I–L). Therefore, the mutations of SAUR62 or SAUR75 likely do not affect pollen grain development and viability.
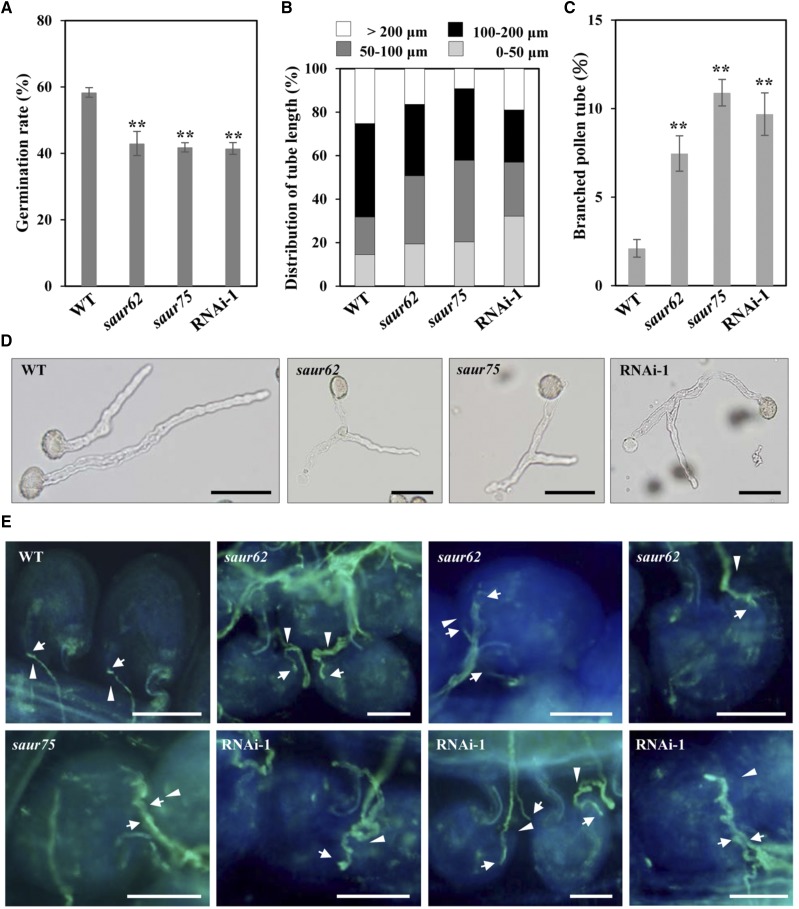
Because the single mutant and RNAi-1 lines produced normal mature pollen grains, we next examined in vitro pollen germination and tube growth of all selected plants. Pollen grains from the wild type, saur62/−, saur75/−, and RNAi-1 were germinated in vitro on Arabidopsis pollen germination medium and observed after 5 h. The pollen germination rates were lower for saur62, saur75, and RNAi-1 than for wild-type pollen: 58.35% for the wild type, 42.98% for saur62, 41.82% for saur75, and 41.48% for RNAi-1 under the same conditions (Fig. 4A). In addition, pollen tubes were markedly shorter for saur62, saur75, and RNAi-1 than for the wild type. The mean pollen tube length was 144.03 ± 42.69 μm (n > 300) for the wild type, 116.56 ± 36.97 μm (n > 300) for saur62, 102.68 ± 32.39 μm (n > 300) for saur75, and 109.18 ± 40.15 μm (n > 300) for RNAi-1 (Fig. 4B). Surprisingly, microscopic observation revealed a branching phenotype of pollen tube tips in saur62, saur75, and several RNAi lines (Fig. 4D; Supplemental Fig. S6). Even though tube branching also occurred occasionally in wild-type pollen tubes, the frequency of branched tubes in saur62, saur75, and RNAi-1 was approximately 3 times higher than in the wild type (Fig. 4C).
Figure 4.
Impaired in vitro pollen germination, tube elongation, and pollination in saur62, saur75, and RNAi-1 plants. A and B, In vitro pollen germination rate and tube length of 5-h-grown pollen tubes. Columns represent means ± se (n > 300). Asterisks indicate significant differences from the wild type (WT; **, P < 0.01) by Student’s t test. C and D, Branching phenotypes of wild-type, saur62, saur75, and RNAi-1 in vitro-cultured pollen tubes. The tip-branching phenotype (D) was confirmed by statistical analysis of the proportion of the branching phenotype (C). Columns represent means ± se (n > 300). Asterisks indicate significant differences from the wild type (**, P < 0.01) by Student’s t test. Bars = 50 μm. E, Aniline Blue staining reveals pollen tube behavior and fertilization of saur62/−, saur75/−, and RNAi-1 pollen on wild-type styles. After pollination, a wild-type ovule containing a single pollen tube (arrows) reached the micropyle (arrowheads). saur62 and RNAi-1 branched pollen tubes reached the micropyle but failed to enter it, and some pollen tubes twisted around the micropyle. Multiple saur62, saur75, and RNAi-1 pollen tubes grew toward one ovule. Bars = 50 μm.
To visualize in vivo-grown pollen tubes, pistils were stained with Aniline Blue 6 and 24 h after hand pollination (Supplemental Fig. S8). With saur62/−, saur75/−, or RNAi-1 pollen grains pollinated on wild-type gynoecia, pollen tubes showed remarkably retarded growth after 6 h of pollination. Nevertheless, they eventually reached the bottom of the transmitting tract at 24 h. The siliques from these pollination combinations showed aborted seeds dispersing randomly from the top to the base (Supplemental Fig. S9). Furthermore, cross-pollination of wild-type pollen onto the saur75/− gynoecium resulted in shorter pollen tube length and seed abortion (Supplemental Figs. S8 and S9).
To investigate the impact of pollen tubes on pollination and fertilization, Aniline Blue staining was used to investigate pollen tube growth in pistils of these mutants after self-pollination. Wild-type pollen tubes grew normally and successfully entered the micropyle; however, some pollen tubes of saur62/−, saur75/−, and RNAi-1 failed to perceive the guidance signal from the micropyle. Additionally, some branched pollen tubes of mutants could target the micropyle but failed to enter it (Supplemental Fig. S10), which explained why the ovule could not be fertilized and was consistent with the fertility defect in the homozygous saur62, saur75, and RNAi-1 plants.
To further determine whether these defects were caused by male gametophytes, wild-type pistils were pollinated with saur62/−, saur75/−, and RNAi-1 pollen and observed at 24 h after pollination. Some pollen tubes (arrows) from saur62 and RNAi-1 twisted around the micropyle (arrowheads), and some saur62 and RNAi-1 branched pollen tubes reached but failed to enter the micropyle (Fig. 4E). In addition, in wild-type ovules, only one pollen tube is allowed to move along the funiculus and penetrate the micropyle. Occasionally, multiple saur62, saur75, and RNAi-1 pollen tubes reached a single ovule, so these pollen tubes might not be able to sense or respond to the female signal, which is essential for funiculus repulsion and/or even micropylar repulsion.
In summary, saur62, saur75, and RNAi-1 showed defective in vitro and in vivo pollen tube growth. SAUR62 and SAUR75 are likely important for normal pollen germination and pollen tube growth, and the reduced fertility of the mutants results from impaired pollen tube growth. However, SAUR75 also has a subtle effect on pistil function for pollen tube growth.
SAUR62 and SAUR75 Are Involved in Ribosome Assembly
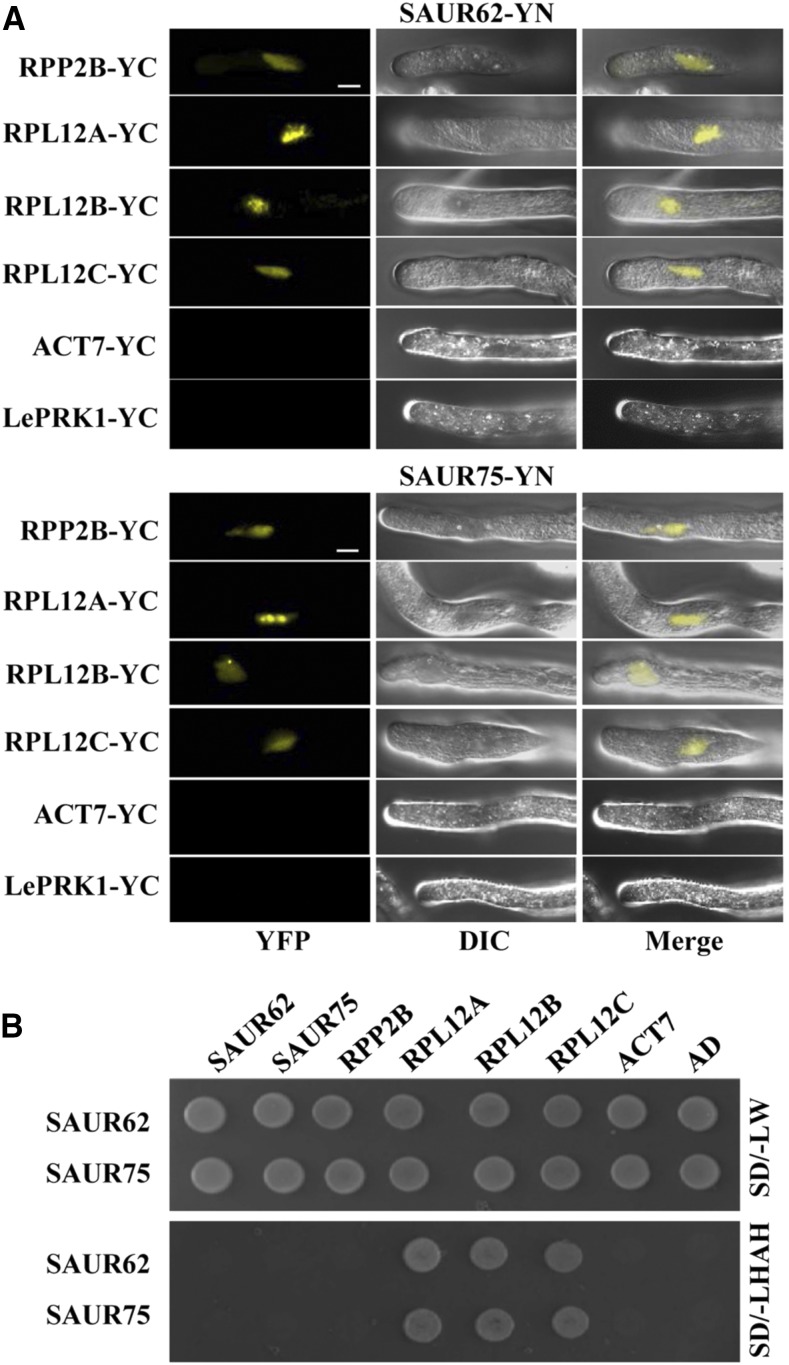
Although many SAUR genes have been studied in Arabidopsis, the molecular mechanisms of their roles in pollen tube growth remain to be elucidated. The possible roles of SAUR62 and SAUR75 during pollination could be elucidated by examining the potential interacting partners of SAUR62 and SAUR75. We performed immunoprecipitation with anti-GFP antibody followed by mass spectrometry (MS) analysis with total proteins extracted from LAT52::SAUR62-GFP and LAT52::SAUR75-GFP open flowers and identified several candidates, including RPL12 family proteins (RPL12A/B/C; Supplemental Table S1). According to previously published microarray data, all three RPL12 genes are expressed in mature pollen grains, in vitro pollen tubes, and in vivo-grown pollen tubes (Supplemental Fig. S11A), which suggests that the RPL12 family members are putative interaction candidates for SAUR62/75. Unlike SAUR62 and SAUR75, which are localized predominantly in the nucleus (Fig. 2B), RPL12A and RPL12C were found in both the nucleus and the cytoplasm (Supplemental Fig. S11B). To explore the roles of RPL12 in pollen tube elongation and pollination/fertilization, we investigated the phenotypes of available RPL12A and RPL12C mutants. Homozygous rpl12a (rpl12a/−) and rpl12c (rpl12c/−) mutants showed similar defects in fertility, pollen germination, and tube elongation (Supplemental Fig. S12), as was found in saur62/75 and RNAi-1 (Figs. 3, B–D, and 4, A and B). We further confirmed that SAUR62/75 interacted with RPL12A and RPL12C by bimolecular fluorescence complementation (BiFC; Fig. 5A) and yeast two-hybrid (Y2H) assays (Fig. 5B).
Figure 5.
RPL12A/B/C are SAUR62/75-interacting proteins. A, BiFC assays of SAUR62-YN (containing the N terminus of YFP) and/or SAUR75-YN with RPP2B-YC (containing the C terminus of YFP), RPL12A-YC, RPL12B-YC, RPL12C-YC, ACT7-YC, or LePRK1-YC in particle bombardment-transformed tobacco pollen tubes confirmed the interaction between SAUR62/75 and individual RPL12s. LePRK1-YC was used as a negative control. Bars = 10 μm. B, Y2H assays of SAUR62 and SAUR75 with RPP2B, RPL12A, RPL12B, RPL12C, ACT7, or AD on SD/-LW (synthetic dropout-Leu/-Trp) and SD/-LWAH (synthetic dropout-Leu/-Trp/-Ade/-His) medium were used to verify the interactions between SAUR62/75 and their interaction partners. Empty vector (AD, pGADT7) was used as a negative control.
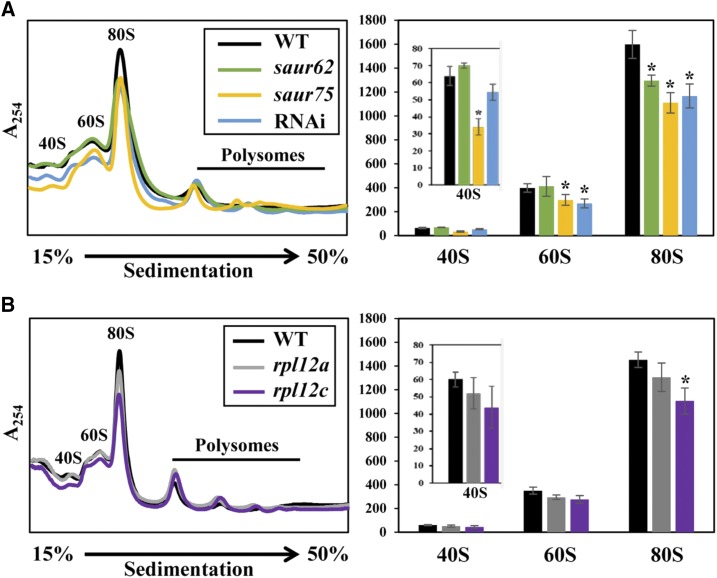
The interactions between individual SAUR62/75 and RPL12A/C combinations occurred exclusively in the vegetative nuclei of pollen tubes. Because RPL12A/C are components of the ribosomal pre-60S subunit, their interactions in the nucleus might be important for pre-60S subunit composition before being transported to the cytosol for maturation and 80S monosome assembly. To investigate a potential role for SAUR62 and SAUR75 in ribosomal presubunit biogenesis, 80S monosome assembly, and subsequent polysome complex formation, we performed polysome profiling. As compared with the wild type, saur62, saur75, RNAi, and rpl12c flowers showed significantly reduced abundance of 80S monosomes but no significant difference at the level of polysomes. In addition, the abundance of 40S and 60S ribosomal subunits was reduced in saur75 and the abundance of 60S ribosomal subunits was reduced in RNAi lines (Fig. 6). These results show that SAUR62/75 associate with the RPL12 family proteins, as evidenced by immunoprecipitation and mass spectrometry analysis (IP-MS), BiFC, and Y2H assays, and reveal an important role for SAUR62 and SAUR75 in ribosomal pre-60S subunit assembly.
Figure 6.
Polysome profiling of wild-type (WT), saur62, saur75, RNAi, rpl12a, and rpl12c flowers. Polysomes extracted from open flowers from the wild type, saur62, saur75, and RNAi (A) and the wild type, rpl12a, and rpl12c (B) were fractionated on a 15% to 50% Suc density gradient, followed by UV254 absorbance profiling. Absorbance peaks with the corresponding positions of 40S ribosomal subunits, 60S ribosomal subunits, and 80S ribosomes are indicated. The quantified abundance of ribosome subunits was derived from three independent biological replicates. Columns represent means ± se. Asterisks indicate significant differences from the wild type (*, P < 0.05) by Student’s t test.
Dynamics of Cell Wall Pectin and F-Actin Are Disturbed in RNAi Pollen Tubes
Polysome profiling showed that SAUR62 and SAUR75 are required for the proper assembly of ribosomes, the major components of the translational machinery. It is ideal but technically not feasible to compare the global pollen tube protein profiles in wild-type and RNAi flowers. With limited working materials, we could use only wild-type and RNAi flowers for total protein extraction and subsequent isobaric tags for relative and absolute quantitation (iTRAQ) analysis to compare their total protein profiles. By using a pollen-specific promoter to knock down the expression of SAUR62/75 in RNAi lines, we expected that individual proteins contributed by the defective pollen tubes would show a very minor fold change in levels, because proteins of in vivo-grown pollen tubes contribute only a very small portion of the total proteins obtained from a whole flower. iTRAQ analysis identified 15 proteins involved in ribosome biogenesis with differential expression changes in RNAi (Supplemental Table S2), which suggests that the loss of SAUR62/75 might affect the homeostatic levels of proteins involved in ribosome biogenesis. Nevertheless, two major protein groups with unique functions in pectin wall biogenesis and F-actin organization were down-regulated significantly in RNAi (Supplemental Table S3). We found reduced levels of several pectin methylesterases (PMEs), PME inhibitors, and pectin lyases as well as F-actin profiling, actin-depolymerizing factors, and LIM domain-containing proteins (LIMs), for example.
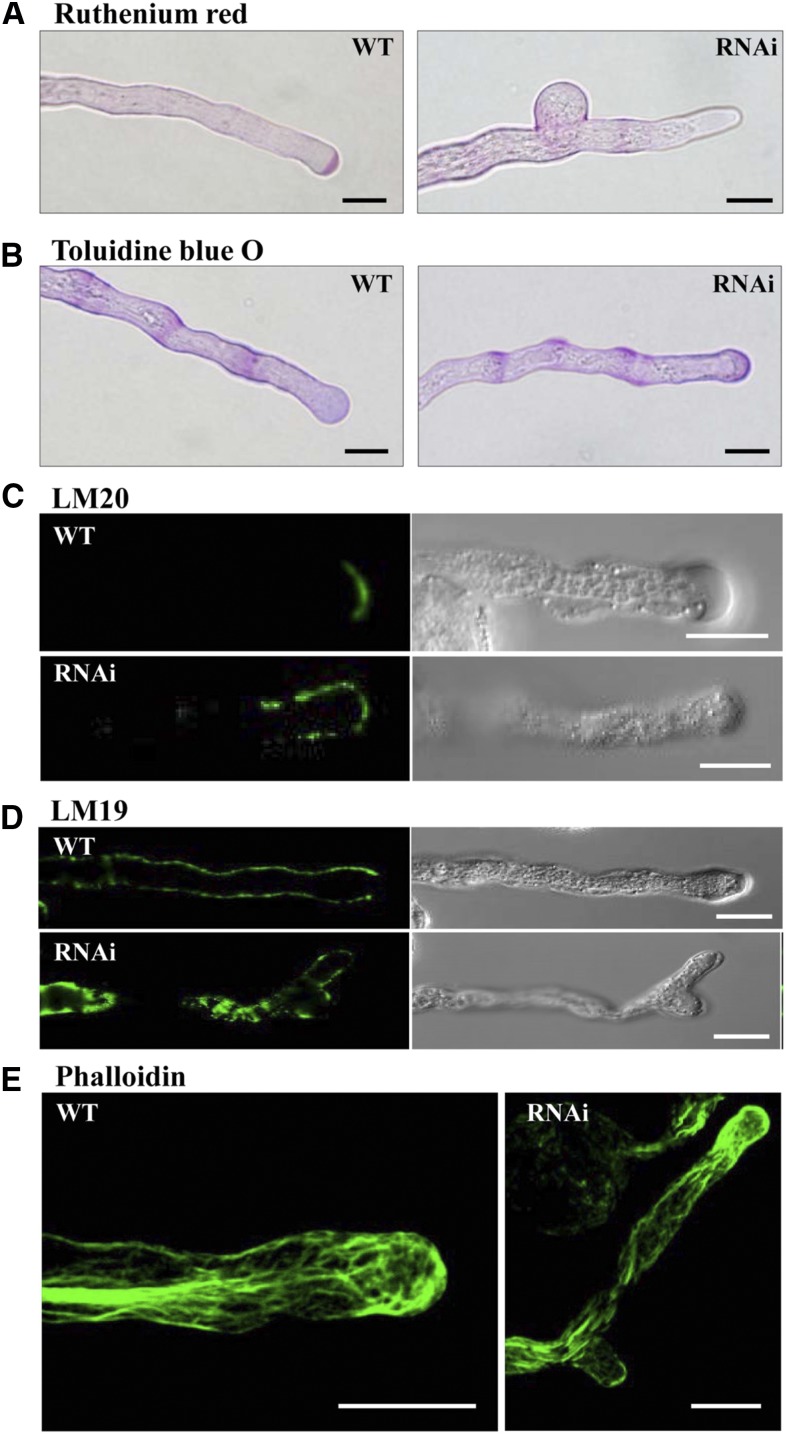
The saur62/75 and RNAi plants exhibited impaired pollen germination and pollen tube growth and many down-regulated proteins involved in the remodeling of pectins and actin cytoskeleton, which suggested altered deposition of cell wall components and distribution of actin filaments in RNAi pollen tubes. Pectins are the major component of the pollen tube cell wall and are deposited as a high-methylesterified form at the pollen tube tip by exocytosis. At the subapical region, methylesterified pectins are deesterified by PMEs (Bosch et al., 2005; Bosch and Hepler, 2005). To investigate the distribution of the major pectic polysaccharide domains in the pollen tube wall, we used Ruthenium Red and Toluidine Blue O for esterified and deesterified pectin staining, respectively. Ruthenium Red staining revealed the typical tip-localized esterified pectins in wild-type pollen tubes. However, esterified pectins were restricted to the tube tip and also started to expand to the subapical region in RNAi pollen tubes (Fig. 7A). For deesterified pectin distribution, Toluidine Blue O staining revealed a more regular banding pattern at the shank region of wild-type pollen tubes, but RNAi pollen tubes exhibited highly deesterified pectins at the tip region (Fig. 7B).
Figure 7.
Altered distribution of cell wall materials and F-actin cytoskeleton in RNAi pollen tubes. Wild-type (WT) and RNAi pollen tubes cultured 5 h in vitro were collected and stained with Ruthenium Red (A; esterified pectins), Toluidine Blue O (B; deesterified pectins), and Alexa Fluor 488 phalloidin (E; actins) and labeled with LM20 (C; esterified pectins) and LM19 (D; deesterified pectins). Bars = 10 μm.
To further validate the distribution of homogalacturonan with high and low degrees of methylesterification, we used the monoclonal antibodies LM20 and LM19, respectively (Verhertbruggen et al., 2009). In wild-type pollen tubes, the LM20 antibody specifically localized esterified pectins restricted to the apical region (Fig. 7C), which was consistent with Ruthenium Red staining (Fig. 7A). In RNAi pollen tubes, LM20 signals exhibited a broader staining pattern extending from the apical region to the distal subapical/shank region (Fig. 7C). The LM19 antibody defined deesterified pectins along the shank of the tube but was excluded from the apex of wild-type pollen tubes. However, in RNAi pollen tubes, LM19-labeled deesterified pectins were present in the shank region and also extended into the apical regions (Fig. 7D).
F-actin filament and its binding proteins serve as key contributors to pollen tube morphology (Vidali and Hepler, 2001). We visualized F-actin organization in pollen tubes by fluorescent phalloidins. In wild-type pollen tubes, F-actin filaments arranged into longitudinal cables in the shank region of the pollen tube, with some shorter actin filaments in the apical region, whereas RNAi pollen tubes exhibited fewer longitudinal cables in the shank region and the accumulation of F-actin filaments in the tip region (Fig. 7E).
The combined data indicated that the loss function of SAUR62 and SAUR75 resulted in the down-regulation of factors critical for the elastic properties of elongating pollen tubes. The increased apical deposition of deesterified pectin and the abnormal distribution of actin filament in pollen tubes might alter the elastic properties of the cell wall and underlie the pollen tube branching phenotype.
DISCUSSION
The Arabidopsis genome contains 79 SAUR family genes that encode proteins shown to participate in the regulation of diverse cellular and developmental processes in response to various hormones and environmental cues (Ren and Gray, 2015). However, the biological functions of SAURs in regulating pollen tube growth and the subsequent pollination are largely unknown. Here, we show the unique pollen tube roles of two SAUR proteins, SAUR62 and SAUR75, that regulate 80S monosome assembly via their interactions with the ribosomal protein RPL12 family. The 80S monosome is the fundamental core of the translational machinery and is responsible for several key factors involved in dynamic pectin wall biogenesis and F-actin organization, which are critical for sufficient plasticity at the apical dome of the elongating pollen tube. Accordingly, we suggest that SAUR62/75 and their partner RPL12 proteins may play important roles in the ribosome assembly and translation of certain transcripts for proper pollen tube elongation.
SAUR62 and SAUR75 were highly expressed in in vivo-grown elongating pollen tubes (Fig. 1; Supplemental Fig. S1), which suggests that these genes are induced by communication between pollen tubes and female tissues during pollination. This finding might explain why abolished and/or knocked down expression of SAUR62/75 significantly retarded pollen tube elongation and also affected fertility (Figs. 3 and 4). When saur62/−, saur75/−, and RNAi pollen grains were cross-pollinated with wild-type or corresponding self pistils, both fertility (silique lengths and total seed number per silique) and in vivo pollen tube length were reduced significantly (Fig. 3E; Supplemental Figs. S8 and S9). In addition, after 24 h of pollination, saur62/−, saur75/−, and RNAi-1 pollen tubes within wild-type pistils showed pollen tubes twisting around the micropyle; some branched pollen tubes reached but failed to enter the micropyle, or multiple pollen tubes reached a single ovule (Fig. 4E). In most flowering plants, only one pollen tube is allowed to move along the funiculus and penetrate the micropyle during pollination (Berger et al., 2008). Occasionally, multiple saur62, saur75, and RNAi-1 pollen tubes reached a single ovule, which suggests that SAUR62 and SAUR75 might be important for the pollen tube perceiving or responding to the fertilized micropylar signal associated with the repulsion of additional pollen tubes. Several studies have shown that failure to fertilize the egg cell or the central cell results in the attraction of a second pollen tube targeting the same ovule (Beale et al., 2012; Kasahara et al., 2012; Maruyama et al., 2013). Further study could investigate the identity of the missing pollen tube-expressing factor in saur62, saur75, and RNAi-1 pollen tubes and its biological role in funiculus and/or micropylar guidance during pollination.
The promoter::GUS data showed that both SAUR62 and SAUR75 were expressed in pollen grains, elongating pollen tubes, gynoecia, and ovules (Fig. 1D; Supplemental Fig. S3), which suggests their functions in both male and female gametophytes. Silique development and in vivo pollen tube length were not impaired with wild-type pollen grains pollinated on saur62/− and RNAi pistils but were impaired with wild-type pollination on saur75/− pistils (Fig. 3E; Supplemental Figs. S8 and S9). Because saur62/− is a knockdown mutant and saur75/− is a knockout mutant (Fig. 3A), genetic differences in these mutants might explain the impaired seed development (both average silique length and seed number per silique) observed only with wild-type pollen grains pollinated on saur75/− but not saur62/− and RNAi pistils.
Pollen tubes of saur62, saur75, and RNAi lines showed shorter and branching phenotypes, which might account for male sterility in these lines (Figs. 3 and 4; Supplemental Fig. S6). iTRAQ analysis suggested that down-regulated proteins with unique functions in pectin wall biogenesis and F-actin organization were down-regulated in RNAi flowers (Supplemental Table S3). Pectins are the major component of the pollen tube primary wall, are polymerized in the Golgi, and then are secreted into the apical wall in a highly methylesterified state (Bosch et al., 2005; Bosch and Hepler, 2005). Incorrect deposition of cell wall components could cause various pollen tube defects, including short and swollen, short and burst, or branched pollen tubes (Jiang et al., 2005; Iwai et al., 2006; Wang et al., 2013; Leroux et al., 2015). Methylesterified pectins recognized by Ruthenium Red or LM20 localize predominantly at the tip region of elongating pollen tubes (Verhertbruggen et al., 2009; Boisson-Dernier et al., 2013), but deesterified pectins recognized by Toluidine Blue O or LM19 are absent from the tip and localize along the shank wall (Verhertbruggen et al., 2009; Hoedemaekers et al., 2015). In the elongating RNAi pollen tubes, esterified pectins misdistribute along the shank wall (Fig. 7, A and C), and the rigidity of the cell wall decreases to result in pollen tube rupture at random sites. Nevertheless, overaccumulation of deesterified pectins at the pollen tube tip (Fig. 7, B and D) will thicken the apical cell wall, which thus loses flexibility and retards pollen tube elongation. Therefore, altered deposition of both esterified and deesterified pectin in RNAi pollen tubes clearly explains the observed phenotypes. Proper organization of the actin cytoskeleton is important for pollen tube growth. In addition, the tube cell wall architecture is controlled and organized by the fine apical F-actin cytoskeleton, where the secreted vesicles driving the intracellular transport of the Golgi contribute the wall and membrane components essential for pollen tube growth (Qu et al., 2015). We have shown that disturbing the apical dynamic F-actin organization significantly retards pollen tube elongation and results in branched pollen tubes (Wang et al., 2008). The down-regulation of several F-actin-binding factors affects the dynamic F-actin organization at the tip region and results in growth retardation and tube tip branching (Fig. 7E).
Diverse subcellular locations have been reported for several SAUR fusion proteins in the nucleus (Knauss et al., 2003; Park et al., 2007), cytosol (Kant et al., 2009; Kong et al., 2013), and plasma membrane (Chae et al., 2012; Spartz et al., 2012). We found SAUR62/75-GFP localized predominantly in the vegetative nucleus of the pollen tube by particle bombardment-mediated transient assay in tobacco pollen tubes (Fig. 2). However, we did not observe the GFP signal in pollen tubes of LAT52::SAUR62-GFP and LAT52::SAUR75-GFP transgenic plants, but we observed GFP signals in the nucleus but not the nucleolus of hypocotyls (SAUR62-2xGFP) or petioles (SAUR75-2xGFP) in corresponding SAUR::SAUR-2xGFP seedlings (Supplemental Fig. S5). ZmSAUR2, the Arabidopsis SAUR62/75 homolog protein in maize (Zea mays), is a short-lived (about 7 min) nuclear protein (Knauss et al., 2003). Other studies showed that AtSAUR19 and AtSAUR63 are almost completely degraded at 30 min and may be regulated by the 26S proteasome (Chae et al., 2012; Spartz et al., 2012). In immunoblot assays, we detected the presence of SAUR62-GFP and SAU752-GFP in corresponding transgenic but not wild-type flowers (Supplemental Fig. S4), which suggests that SAUR62 and SAUR75 are nuclear proteins with very short half-lives in pollen tubes. Therefore, SAUR62/75 might be degraded rapidly before their detection in elongating pollen tubes.
There are 249 ribosomal genes in the Arabidopsis genome (Barakat et al., 2001); our previous translatome data suggested that 213 were highly expressed in in vivo-grown pollen tubes during pollination (Lin et al., 2014). In addition, defective 80S monosome assembly is compromised by the significantly up-regulated biosynthesis of many ribosome proteins (Supplemental Table S2) to support the extensive translation activity in rapidly elongating pollen tubes. SAUR62/75 associate with the RPL12 family, as evidenced by IP-MS (Supplemental Table S1), BiFC, and Y2H assays (Fig. 5). SAUR62/75 interact with the RPL12 family in the nucleus (Fig. 5A), and polysome profiling experiments showed reduced 80S monosome fractions in saur62, saur75, RNAi, and rpl12c flowers, but polysome production was not affected (Fig. 6). From their exclusively nuclear localization, SAUR62/75 may modulate RPL12 protein activity and thereby affect pre-60S subunit assembly or their composition before export to the cytoplasm, where they mature and assemble into functional ribosomes for translation. In addition, proteins involved in pollen tube wall biogenesis and F-actin dynamic organization were predominantly down-regulated in RNAi flowers (Fig. 7; Supplemental Table S3), which implies that certain transcripts might be recruited selectively to the translational machinery to fulfill the rapid pollen tube elongation during pollination. Horiguchi et al. (2012) proposed three possible models of ribosome biogenesis (ribosome insufficiency, heterogeneity, and aberrancy) in regulating plant development. We speculate that the amount of fully functional ribosomes used for translating certain proteins required for proper pollen tube elongation might be impaired and result in the retarding and/or branching phenotypes in RNAi lines. We need further research into the underlying mechanism of the interaction of SAUR62/75 and RPL12 in regulating the assembly of functional 80S ribosomes to regulate certain groups of mRNAs essential for rapid pollen tube growth. These results highlight the unique roles of SAUR62/75 in associating with the RPL12 family in ribosome assembly and, thereafter, possibly selective translation of certain groups of proteins to coordinate prompt pollen tube elongation during pollination.
SAUR61 to SAUR68 and SAUR75 were induced by auxin in seedlings and flowers (Paponov et al., 2008), and auxin has been found to be important for stamen development, including anther dehiscence, pollen maturation, and filament elongation (Cecchetti et al., 2008). Although the precise roles of auxin mediating pollen tube growth during pollination remain unclear, many studies suggest that auxin participates in regulating pollen tube elongation and function. For instance, some studies show an increase in free IAA content in the pistil after pollination (Kovaleva and Zakharova, 2003; Aloni et al., 2006), and exogenous IAA promotes in vitro pollen tube growth in tobacco and Torenia fournieri (Chen and Zhao, 2008; Wu et al., 2008). The mutant pin-formed8 (pin8), an auxin efflux carrier mutant, showed reduced pollen germination rate as compared with the wild type, but overexpression of PIN8 (LAT52::PIN8) increased pollen tube elongation (Ding et al., 2012). In addition, our previous translatomic studies showed up-regulated transcripts of PIN3, PIN4, the auxin receptor TRANSPORT INHIBITOR RESPONSE1, ARFs, and many auxin response genes during pollination (Lin et al., 2014).
Here, we functionally studied two closely related SAURs, SAUR62 and SAUR75, which might be involved in the regulation of translational machinery via the interaction between SAUR62/75 and RPL12 to impair ribosome assembly. These functionally and/or structurally distinct ribosomes selectively recruit some transcripts for translation to support rapid pollen tube elongation during pollination. The loss of function of SAUR62/75 quantitatively reduced the levels of proteins participating in pollen tube wall formation or F-actin organization, eventually disturbing the organization of the tube wall and cytoskeleton and causing a branched pollen tube phenotype. These findings provide important insights into the molecular mechanism of SAUR proteins in translational regulation during pollen tube elongation.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used in this study. Two T-DNA insertion lines, SALK_024165 (saur62) and SALK_076536 (saur75), were obtained from the Arabidopsis Biological Resource Center. Wild-type, saur62, saur75, and transgenic plants were grown in a growth chamber at 22°C with a photoperiod of 16 h of light/8 h of dark.
Phylogenetic Analysis
Amino acid sequences of SAUR61 to SAUR68 and SAUR75 were obtained from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) and used for phylogenetic tree analysis. The phylogenetic tree of SAUR61 to SAUR68 and SAUR75 was obtained at the Web site Phylogeny.fr: Tree Dyn. (http://www.phylogeny.fr/). The numbers on the nodes of the phylogenetic tree correspond to bootstrap values.
RT-PCR Analysis
Total RNA was extracted from different tissues by using a plant total RNA purification kit (GeneMark), then treated with the TURBO DNA-free Kit (Ambion) to remove contaminating genomic DNA. First-strand cDNA was synthesized by using the M-MLV reverse transcriptase kit (Invitrogen) according to the supplier’s instructions. ACTIN1 was used as an internal control for RT-PCR. The primer pairs used in the assays are listed in Supplemental Table S4.
Molecular Cloning and Generation of Transgenic Plants
To construct the SAUR62::GUS and SAUR75::GUS expression vectors, ∼1.2- and ∼1-kb DNA fragments containing the promoters and 5′ untranslated regions of SAUR62 and SAUR75, respectively, were cloned into the pBX-2 vector (Ho et al., 2000). For the RNAi construct, a 231-bp similar sequence of SAUR62/75 was amplified and cloned into the pKANNIBAL vector (Wesley et al., 2001) carrying the LAT52 promoter. All SAUR62::GUS, SAUR75::GUS, and LAT52::SAUR62/75-RNAi constructs were subcloned into the pSMY1H binary vector (Ho et al., 2000) and then transformed into wild-type plants by the floral dip method (Clough and Bent, 1998). For transient expression assays in pollen, SAUR62 and SAUR75 coding regions and their deletion fragments were cloned into the G39 vector (Gui et al., 2014), and the constructs were transformed into tobacco (Nicotiana tabacum) pollen by particle bombardment.
GUS Histochemical Assay
Plant materials were fixed in 90% (v/v) acetone at −20°C for 1 h, rinsed twice with 100 mm sodium phosphate buffer (pH 7), and then placed into X-Gluc reaction solution (100 mm sodium phosphate, pH 7, 10 mm EDTA, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, 0.1% Triton X-100, 10% (v/v) methanol, and 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-d-GlcA) at 37°C overnight. The stained tissues were decolorized in 70% ethanol at 37°C for 24 h and observed with an Olympus BX51 microscope.
Pollen Development and Viability Assays
For nucleus staining, mature pollen grains were stained with 1 μL of DAPI solution (1 mg mL−1) in 1 mL of mounting buffer (0.1 m Tris-HCl, pH 9, and 50% [v/v] glycerol), and pollen grains were observed with a laser scanning confocal microscope (Carl Zeiss; LSM510) after 20 min. For Alexander staining, mature anthers before anthesis were cut and fixed in an acetic acid:ethanol solution (1:3 [v/v]) overnight, then incubated in Alexander’s solution (Alexander, 1969). Stained anthers were observed by bright-field microscopy (Olympus; BX51). For staining with FDA, mature pollen grains were collected from plants and stained with 1 μL of 0.1% (w/v) FDA in 20 μL of liquid pollen germination medium for 5 min. Images were captured by laser scanning confocal microscopy (Carl Zeiss; LSM510).
Pollen Germination, Observation, and Cytological Observation
Mature pollen grains from 20 open flowers were collected in liquid pollen germination medium (1 mm KCl, 5 mm CaCl2, 0.8 mm MgSO4, 1.5 mm H3BO3, 15% (w/v) Suc, 10 μm myoinositol, and 5 mm MES, pH 5.8) with vigorous shaking for 5 min. The mixture was centrifuged at 11,000 rpm for 5 min at room temperature. After removing flower tissues and supernatant, the pollen pellet was resuspended in 100 μL of liquid pollen germination medium and transferred onto solid pollen germination medium (1 mm KCl, 5 mm CaCl2, 0.8 mm MgSO4, 1.5 mm H3BO3, 19.8% (w/v) Suc, 10 μm myoinositol, 5 mm MES, pH 5.8, and 1% [w/v] agarose) for 5 h at room temperature in the dark. The pollen germination rates and elongating pollen tubes were observed by light microscopy (Olympus; BX51). The pollen tubes were measured by using ImageJ.
For the phenotypic analysis of pollen tubes, pistils were harvested after pollination and fixed in an acetic acid:ethanol solution (1:3 [v/v]) for 2 h, then softened in an 8 m NaOH solution overnight, and stained with Aniline Blue for 5 h in the dark. The Aniline Blue buffer contained 0.1% (w/v) Aniline Blue in 0.1 m K2HPO4-KOH buffer, pH 11. The stained pollen tubes in the pistils were observed with an Olympus BX51 microscope with UV light emission.
For actin cytoskeleton staining with Alexa Fluor 488 phalloidin (Molecular Probes), pollen tubes were fixed with 300 μm 3-maleimidobenzoic acid N-hydroxysuccinimide ester in liquid pollen germination medium for 1 h, then washed with TBS-T (50 mm Tris-HCl, 200 mm NaCl, and 0.05% [v/v] Nonidet P-40, pH 7.4), and stained with 1 μL of 300 units per 1.5 mL of Alexa Fluor 488 phalloidin in 40 μL of TBS-T buffer for 5 min. Images were captured by laser scanning confocal microscopy (Carl Zeiss; LSM510).
For pectin staining, pollen tubes were stained with 0.01% (w/v) Ruthenium Red (Sigma) in liquid pollen germination medium and stained with Toluidine Blue O (Sigma) with 15 μL of 0.05% (w/v) Toluidine Blue O in 0.1 m phosphate buffer at pH 6.8 in 20 μL of liquid pollen germination medium for 20 min, then images were captured with a light microscope (Olympus; BX51). For the immunolocalization of methylesterified and unesterified homogalacturonan, pollen tubes were fixed in 4% formaldehyde in PIPES buffer (50 mm PIPES, 1 mm EGTA, 5 mm MgSO4, 0.5 mm CaCl2, and 0.1% [v/v] Triton X-100, pH 7) for 1 h. The fixed pollen tubes were washed three times with PBS (100 mm potassium phosphate, 138 mm NaCl, and 2.7 mm KCl, pH 7.3) and then incubated in blocking buffer (0.8% [w/v] BSA, 0.1% [w/v] gelatin, and 2 mm NaN3 in PBS) for 30 min. After removing the blocking buffer, samples were incubated for 1 h at room temperature with primary antibodies for LM19 and LM20 (Verhertbruggen et al., 2009; 1:100 dilution in blocking buffer). Samples were rinsed three times with PBS and incubated for 30 min at room temperature with Alexa Fluor 488-conjugated secondary antibody (1:100 dilution in blocking buffer). Samples were washed five times with PBS before analysis by laser scanning confocal microscopy (Carl Zeiss; LSM510).
Y2H Assay
The cDNAs were cloned into the pGBKT7 and pGADT7 vectors. Pairs of pGBKT7 and pGADT7 plasmids were cotransformed into the yeast (Saccharomyces cerevisiae) strain AH109 following the Match Maker GAL4 Two-Hybrid System according to the supplier’s instructions (Clontech). Primary transformants were selected on synthetic dropout medium lacking Trp and Leu and confirmed again by colony PCR before growing on synthetic dropout medium lacking Ade, His, Trp, and Leu.
Pollen Bombardment and BiFC Assays
cDNAs were cloned into the K307 (YN) and the K309 (YC) vectors (Gui et al., 2014). Plasmid pairs were cotransformed into tobacco pollen by particle bombardment. For particle bombardment, mature pollen grains from 10 open flowers were collected in liquid pollen germination medium with vigorous shaking for 5 min. The mixture was centrifuged at 11,000 rpm for 5 min at room temperature. After removing flower tissues and supernatant, the pollen pellet was resuspended in 1 mL of liquid pollen germination medium and transferred onto germination medium-immersed filter paper in a 90-mm petri dish. The YC- and YN-containing plasmids (3.5 μg each) were incubated with 1.875 mg of gold particles (1 μm) and bombarded into pollen grains by using the PDS-1000/He Biolistic Particle Delivery System (Bio-Rad) at 1,100 p.s.i., 27 mm Hg vacuum, 1-cm gap distance, and 9-cm particle flight distance. Bombarded pollen grains were washed immediately from filter paper with 1 mL of liquid pollen germination medium and cultured in solid pollen germination medium (10% [w/v] Suc, 1.27 mm CaCl2, 0.162 mm H3BO3, 0.99 mm KNO3, pH 5.2, and 1% agarose) for 3 to 5 h. Images were captured by Zeiss LSM 510 confocal microscopy.
Polysome Profiling
Open flowers were collected and ground with liquid nitrogen. Approximately 1,000 μL of frozen tissue powder was extracted with 500 μL of polysome extraction buffer (200 mm Tris-HCl, pH 8, 50 mm KCl, 25 mm MgCl2, 50 μg mL−1 cycloheximide, 400 units mL−1 RNasin [Promega], 2% [v/v] polyoxyethylene 10 tridecyl ether, and 1% [w/v] deoxycholic acid). The resuspended mixture was incubated on ice for 5 min and then centrifuged at 13,000 rpm for 10 min at 4°C. The supernatant was transferred to a new prechilled 1.5-mL tube, and OD260 was measured using NanoDrop. The same OD amount of supernatant was loaded onto the 11-mL Suc gradient (15% [w/v]–50% [w/v]) and spun at 35,000 rpm for 3.5 h at 4°C. The distribution of the nucleic acids was examined by a UV254 absorbance profile (model #UA-6; ISCO).
IP-MS
Open flowers (∼2 g) of the wild type, LAT52::SAUR62-GFP, and LAT52::SAUR75-GFP were ground under liquid nitrogen before the addition of 2 mL of lysis buffer (150 mm NaCl, 100 mm Tris-HCl, pH 8, 5 mm EDTA, 10 mm DTT, and 0.1% [v/v] Nonidet P-40) supplemented with protease inhibitor (Roche). Homogenates were centrifuged at 13,000 rpm for 15 min at 4°C to remove cellular debris. The supernatants were coimmunoprecipitated using GFP-trap beads (Chromotek) and then incubated with gentle end-over-end mixing (15 rpm) for 1.5 h at 4°C. The mixtures were centrifuged at 2,700g for 2 min at 4°C. After washing the pellet three times with wash buffer (150 mm NaCl, 100 mm Tris-HCl, pH 8, 5 mm EDTA, 10 mm DTT, and 0.1% [v/v] Nonidet P-40), immunoaffinity complexes were eluted with elution buffer (0.1 m Gly, pH 2.5) followed by neutralization with 1 m Tris base. The protein components of the immunoprecipitates were determined by MS analysis as described previously (Tamura et al., 2010). The LC-nESI-Q Exactive mass spectrometer (Thermo Fisher Scientific) coupled with an online nanoUHPLC system (Dionex UltiMate 3000 Binary RSLCnano) was used. Peptide and protein identification were performed using Proteome Discoverer software (version 1.4; Thermo Fisher Scientific) with the Mascot version 2.4 search engine against a TAIR10 database with 27,416 protein sequence entries.
iTRAQ Analysis
Open flowers (∼1 g) from wild-type and RNAi plants were ground in liquid nitrogen in a prechilled mortar with use of a pestle. Proteins were extracted with extraction buffer (8 m urea and 50 mm Tris-HCl, pH 8.5) supplemented with protease inhibitor (Roche). Protein sample preparation, iTRAQ labeling, protein determination by liquid chromatography-MS/MS, and data analysis were performed as described previously (Lan et al., 2011). Differentially regulated proteins were identified by change in expression of 1.3-fold or greater for up-regulated proteins and 0.7-fold or less for down-regulated proteins and at least two unique peptides identified.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SAUR62 (AT1G29430) and SAUR75 (AT5G27780).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression profiles of 79 Arabidopsis SAUR family genes in flower buds (pollen grains) and in vivo- and in vitro-cultured pollen tubes.
Supplemental Figure S2. Amino acid sequence alignment of Arabidopsis SAUR61 to SAUR68 and SAUR75.
Supplemental Figure S3. Promoter::GUS assay of seedlings and ovules.
Supplemental Figure S4. Subcellular localization of SAUR62 and SAUR75 in stably transformed plants.
Supplemental Figure S5. Immunoblot analysis of lysates from wild-type and transgenic (SAUR62/75-GFP) plants.
Supplemental Figure S6. Expression of SAUR62 and SAUR75 in RNAi flowers and phenotyping of selected RNAi pollen tubes.
Supplemental Figure S7. Pollen viability of the wild type, saur62, saur75, and RNAi-1.
Supplemental Figure S8. SAUR62 and SAUR75 are required for normal pollen tube growth in vivo.
Supplemental Figure S9. Silique phenotypes after reciprocal cross-pollination of saur62/−, saur75/−, RNAi-1, and wild-type plants.
Supplemental Figure S10. Aniline Blue-stained gynoecia reveal impaired pollen tube growth in self-pollinated saur62/−, saur75/−, and RNAi-1 flowers.
Supplemental Figure S11. Expression levels and subcellular localization of RPL12s in pollen grains and elongating pollen tubes.
Supplemental Figure S12. Phenotyping of the rpl12a/− and rpl12c/− mutants.
Supplemental Table S1. Potential SAUR62/75-interacting candidates obtained from the in vivo immunoprecipitation experiment.
Supplemental Table S2. iTRAQ analysis showing differentially regulated proteins involved in ribosome biogenesis in flowers of RNAi lines.
Supplemental Table S3. iTRAQ analysis showing down-regulation of two groups of proteins involved in cell wall biogenesis and dynamic F-actin organization in RNAi flowers.
Supplemental Table S4. Primer sequences used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Yun Xiang (MOE Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University) for providing the pKANNIBAL plasmid. We thank Dr. Wei-Hua Tang (National Key Laboratory of Plant Molecular Genetics, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for providing the BiFC plasmid. We also thank colleagues at the Institute of Plant and Microbial Biology, Academia Sinica: Dr. Shu-Hsing Wu for assistance in polysome profiling; Ms. Mei-Jane Fang (DNA Analysis Core Laboratory) and Dr. Tuan-Nan Wen (Proteomics Core Laboratory) for assistance with DNA sequencing and iTRAQ, respectively.
Footnotes
This work was supported by research grants from Academia Sinica (Taiwan; AS-105-TP-B03) and the Ministry of Science and Technology (Taiwan; MOST 104-2321-B-001-008 and MOST 105-2311-B-001-074-MY3) to G.-Y.J.
Articles can be viewed without a subscription.
References
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Aloni R, Aloni E, Langhans M, Ullrich CI (2006) Role of auxin in regulating Arabidopsis flower development. Planta 223: 315–328 [DOI] [PubMed] [Google Scholar]
- Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J (2001) The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 127: 398–415 [PMC free article] [PubMed] [Google Scholar]
- Beale KM, Leydon AR, Johnson MA (2012) Gamete fusion is required to block multiple pollen tubes from entering an Arabidopsis ovule. Curr Biol 22: 1090–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Hamamura Y, Ingouff M, Higashiyama T (2008) Double fertilization: caught in the act. Trends Plant Sci 13: 437–443 [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Lituiev DS, Nestorova A, Franck CM, Thirugnanarajah S, Grossniklaus U (2013) ANXUR receptor-like kinases coordinate cell wall integrity with growth at the pollen tube tip via NADPH oxidases. PLoS Biol 11: e1001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Hepler PK (2005) Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 17: 3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M, Cheung AY, Hepler PK (2005) Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol 138: 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME. (2009) A role for the ribosome in development. Trends Plant Sci 14: 512–519 [DOI] [PubMed] [Google Scholar]
- Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M (2008) Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. Plant Cell 20: 1760–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71: 684–697 [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M (2009) Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43: 265–285 [DOI] [PubMed] [Google Scholar]
- Chen D, Zhao J (2008) Free IAA in stigmas and styles during pollen germination and pollen tube growth of Nicotiana tabacum. Physiol Plant 134: 202–215 [DOI] [PubMed] [Google Scholar]
- Chen Y, Hao X, Cao J (2014) Small auxin upregulated RNA (SAUR) gene family in maize: identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. J Integr Plant Biol 56: 133–150 [DOI] [PubMed] [Google Scholar]
- Cheung AY, Wu HM (2008) Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu Rev Plant Biol 59: 547–572 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Ding Z, Wang B, Moreno I, Dupláková N, Simon S, Carraro N, Reemmer J, Pěnčík A, Chen X, Tejos R, et al. (2012) ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat Commun 3: 941. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Franklin-Tong N (2013) Male-female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol Plant 6: 1018–1036 [DOI] [PubMed] [Google Scholar]
- Franco AR, Gee MA, Guilfoyle TJ (1990) Induction and superinduction of auxin-responsive mRNAs with auxin and protein synthesis inhibitors. J Biol Chem 265: 15845–15849 [PubMed] [Google Scholar]
- Galani K, Nissan TA, Petfalski E, Tollervey D, Hurt E (2004) Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J Biol Chem 279: 55411–55418 [DOI] [PubMed] [Google Scholar]
- Gil P, Green PJ (1996) Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: the 3′ untranslated region functions as an mRNA instability determinant. EMBO J 15: 1678–1686 [PMC free article] [PubMed] [Google Scholar]
- Gui CP, Dong X, Liu HK, Huang WJ, Zhang D, Wang SJ, Barberini ML, Gao XY, Muschietti J, McCormick S, et al. (2014) Overexpression of the tomato pollen receptor kinase LePRK1 rewires pollen tube growth to a blebbing mode. Plant Cell 26: 3538–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Hayashi K. (2012) The interaction and integration of auxin signaling components. Plant Cell Physiol 53: 965–975 [DOI] [PubMed] [Google Scholar]
- Ho SL, Tong WF, Yu SM (2000) Multiple mode regulation of a cysteine proteinase gene expression in rice. Plant Physiol 122: 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoedemaekers K, Derksen J, Hoogstrate SW, Wolters-Arts M, Oh SA, Twell D, Mariani C, Rieu I (2015) BURSTING POLLEN is required to organize the pollen germination plaque and pollen tube tip in Arabidopsis thaliana. New Phytol 206: 255–267 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Van Lijsebettens M, Candela H, Micol JL, Tsukaya H (2012) Ribosomes and translation in plant developmental control. Plant Sci 191-192: 24–34 [DOI] [PubMed] [Google Scholar]
- Hou K, Wu W, Gan SS (2013) SAUR36, a small auxin up RNA gene, is involved in the promotion of leaf senescence in Arabidopsis. Plant Physiol 161: 1002–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai A, Komura M, Kawano E, Kuwashiro Y, Takahashi T (2008) A semi-dominant mutation in the ribosomal protein L10 gene suppresses the dwarf phenotype of the acl5 mutant in Arabidopsis thaliana. Plant J 56: 881–890 [DOI] [PubMed] [Google Scholar]
- Iwai H, Hokura A, Oishi M, Chida H, Ishii T, Sakai S, Satoh S (2006) The gene responsible for borate cross-linking of pectin rhamnogalacturonan-II is required for plant reproductive tissue development and fertilization. Proc Natl Acad Sci USA 103: 16592–16597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Yang SL, Xie LF, Puah CS, Zhang XQ, Yang WC, Sundaresan V, Ye D (2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Bi YM, Zhu T, Rothstein SJ (2009) SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol 151: 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara RD, Maruyama D, Hamamura Y, Sakakibara T, Twell D, Higashiyama T (2012) Fertilization recovery after defective sperm cell release in Arabidopsis. Curr Biol 22: 1084–1089 [DOI] [PubMed] [Google Scholar]
- Knauss S, Rohrmeier T, Lehle L (2003) The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J Biol Chem 278: 23936–23943 [DOI] [PubMed] [Google Scholar]
- Kong Y, Zhu Y, Gao C, She W, Lin W, Chen Y, Han N, Bian H, Zhu M, Wang J (2013) Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol 54: 609–621 [DOI] [PubMed] [Google Scholar]
- Kovaleva L, Zakharova E (2003) Hormonal status of the pollen-pistil system at the progamic phase of fertilization after compatible and incompatible pollination in Petunia hybrida L. Sex Plant Reprod 16: 191–196 [Google Scholar]
- Lan P, Li W, Wen TN, Shiau JY, Wu YC, Lin W, Schmidt W (2011) iTRAQ protein profile analysis of Arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol 155: 821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux C, Bouton S, Kiefer-Meyer MC, Fabrice TN, Mareck A, Guénin S, Fournet F, Ringli C, Pelloux J, Driouich A, et al. (2015) PECTIN METHYLESTERASE48 is involved in Arabidopsis pollen grain germination. Plant Physiol 167: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZG, Chen HW, Li QT, Tao JJ, Bian XH, Ma B, Zhang WK, Chen SY, Zhang JS (2015) Three SAUR proteins SAUR76, SAUR77 and SAUR78 promote plant growth in Arabidopsis. Sci Rep 5: 12477. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lin SY, Chen PW, Chuang MH, Juntawong P, Bailey-Serres J, Jauh GY (2014) Profiling of translatomes of in vivo-grown pollen tubes reveals genes with roles in micropylar guidance during pollination in Arabidopsis. Plant Cell 26: 602–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhó R. (2006) The pollen tube: a model system for cell and molecular biology studies. Plant Cell Monogr 3: 1–13 [Google Scholar]
- Markakis MN, Boron AK, Van Loock B, Saini K, Cirera S, Verbelen JP, Vissenberg K (2013) Characterization of a small auxin-up RNA (SAUR)-like gene involved in Arabidopsis thaliana development. PLoS ONE 8: e82596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Hamamura Y, Takeuchi H, Susaki D, Nishimaki M, Kurihara D, Kasahara RD, Higashiyama T (2013) Independent control by each female gamete prevents the attraction of multiple pollen tubes. Dev Cell 25: 317–323 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Wada T, Yamamoto KT, Okada K (2005) The Arabidopsis STV1 protein, responsible for translation reinitiation, is required for auxin-mediated gynoecium patterning. Plant Cell 17: 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JA, Palme K (2008) Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant 1: 321–337 [DOI] [PubMed] [Google Scholar]
- Park JE, Kim YS, Yoon HK, Park CM (2007) Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci 172: 150–157 [Google Scholar]
- Qin Y, Yang Z (2011) Rapid tip growth: insights from pollen tubes. Semin Cell Dev Biol 22: 816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Jiang Y, Chang M, Liu X, Zhang R, Huang S (2015) Organization and regulation of the actin cytoskeleton in the pollen tube. Front Plant Sci 5: 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Gray WM (2006) Auxin signaling. Curr Opin Plant Biol 9: 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VS, Ali GS, Reddy AS (2002) Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J Biol Chem 277: 9840–9852 [DOI] [PubMed] [Google Scholar]
- Ren H, Gray WM (2015) SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant 8: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado A, Li R, van de Ven W, Hsu E, Raikhel NV (2012) Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc Natl Acad Sci USA 109: 19537–19544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Sun L, Hao Y, Wang L, Xu J, Zhang W, Xie J, Guo L, Zhou L, Yun X, et al. (2012) Ribosomal RACK1 promotes chemoresistance and growth in human hepatocellular carcinoma. J Clin Invest 122: 2554–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70: 978–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM (2014) SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Wang J, Gao Z, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen H (2016) Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci USA 113: 6071–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakonyi D, Byrne ME (2011) Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J 65: 269–281 [DOI] [PubMed] [Google Scholar]
- Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I (2010) Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22: 4084–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H, Hurt E (2003) Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhertbruggen Y, Marcus SE, Haeger A, Ordaz-Ortiz JJ, Knox JP (2009) An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr Res 344: 1858–1862 [DOI] [PubMed] [Google Scholar]
- Vidali L, Hepler PK (2001) Actin and pollen tube growth. Protoplasma 215: 64–76 [DOI] [PubMed] [Google Scholar]
- Wang HJ, Wan AR, Jauh GY (2008) An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol 147: 1619–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang W, Wang YQ, Liu YY, Wang JX, Zhang XQ, Ye D, Chen LQ (2013) Arabidopsis galacturonosyltransferase (GAUT) 13 and GAUT14 have redundant functions in pollen tube growth. Mol Plant 6: 1131–1148 [DOI] [PubMed] [Google Scholar]
- Weijers D, Franke-van Dijk M, Vencken RJ, Quint A, Hooykaas P, Offringa R (2001) An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128: 4289–4299 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JZ, Lin Y, Zhang XL, Pang DW, Zhao J (2008) IAA stimulates pollen tube growth and mediates the modification of its wall composition and structure in Torenia fournieri. J Exp Bot 59: 2529–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Chen D, Wang Y, Sun Y, Zhao J, Sun M, Peng X (2016) Ribosomal protein L18aB is required for both male gametophyte function and embryo development in Arabidopsis. Sci Rep 6: 31195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW (2000) Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J Biol Chem 275: 3137–3143 [DOI] [PubMed] [Google Scholar]