A bZIP transcription factor enables maize to grow better under stress by promoting root growth and the expression of stress-related genes.
Abstract
In plants, bZIP (basic leucine zipper) transcription factors regulate diverse processes such as development and stress responses. However, few of these transcription factors have been functionally characterized in maize (Zea mays). In this study, we characterized the bZIP transcription factor gene ZmbZIP4 from maize. ZmbZIP4 was differentially expressed in various organs of maize and was induced by high salinity, drought, heat, cold, and abscisic acid treatment in seedlings. A transactivation assay in yeast demonstrated that ZmbZIP4 functioned as a transcriptional activator. A genome-wide screen for ZmbZIP4 targets by immunoprecipitation sequencing revealed that ZmbZIP4 could positively regulate a number of stress response genes, such as ZmLEA2, ZmRD20, ZmRD21, ZmRab18, ZmNHX3, ZmGEA6, and ZmERD, and some abscisic acid synthesis-related genes, including NCED, ABA1, AAO3, and LOS5. In addition, ZmbZIP4 targets some root development-related genes, including ZmLRP1, ZmSCR, ZmIAA8, ZmIAA14, ZmARF2, and ZmARF3, and overexpression of ZmbZIP4 resulted in an increased number of lateral roots, longer primary roots, and an improved root system. Increased abscisic acid synthesis by overexpression of ZmbZIP4 also can increase the plant’s ability to resist abiotic stress. Thus, ZmbZIP4 is a positive regulator of plant abiotic stress responses and is involved in root development in maize.
Maize (Zea mays) is one of the most important crops for livestock and humans. Its growth and development frequently are affected by various abiotic stresses, such as high salinity, drought, heat, and low temperature, which negatively affect crop productivity. Therefore, it is imperative to elucidate the mechanisms of stress responses in maize. Many previous studies have demonstrated that abscisic acid (ABA) is an important phytohormone involved in stress response and tolerance and that most stress-responsive genes are regulated by ABA (Yamaguchi-Shinozaki and Shinozaki, 2006; Wasilewska et al., 2008). Among the promoters of these stress-responsive genes, a major cis-acting element (ABRE) was characterized as being necessary for their response to ABA (Guiltinan et al., 1990; Mundy et al., 1990; Hattori et al., 2002). Subsequently, a series of basic leucine zipper (bZIP) transcription factors that bind to ABRE were identified, and they were shown to be critical for the activation of downstream gene expression (Choi et al., 2000; Uno et al., 2000).
bZIP is a large transcription factor family in plants involved in regulating stress response and hormone signal transduction (Kim et al., 2004). Based on their conserved regions, 75 bZIP genes have been identified and classified into 10 groups in Arabidopsis (Arabidopsis thaliana; Iida et al., 2005), and 89 bZIP transcription factors were divided into 11 groups in rice (Oryza sativa; Corrêa et al., 2008; Nijhawan et al., 2008). Most ABRE-binding bZIPs belong to group A, in which the expression of several members could be strongly induced by ABA and abiotic stresses (Lu et al., 2009). It has been reported that overexpression of the group A bZIP genes could improve resistance to abiotic stresses. In Arabidopsis, transgenic lines overexpressing AREB1/ABF2 showed increased resistance to multiple stresses (Kim et al., 2004; Fujita et al., 2005). ABF3/AREB2/ABF4 transgenic plants were reported to have improved drought resistance (Kang et al., 2002). In addition, areb1/areb2/abf3 triple mutants showed reduced ABA sensitivity and drought resistance, which confirmed that AREB/ABFs play key coordinating roles in ABA-mediated signaling pathways under stress (Yoshida et al., 2010). In rice, OsbZIP23 plays an essential role in ABA signaling and biosynthesis, and transgenic rice overexpressing OsbZIP23 exhibited significant drought and salt resistance (Xiang et al., 2008). OsbZIP23 directly targets a series of genes related to stress response, hormone signaling, and developmental processes; among these genes, OsPP2C49 and OsNCED4 can regulate ABA signaling and ABA levels, respectively (Zong et al., 2016).
To cope with environmental stress, plants form a unique root system and continuously optimize it to satisfy their mineral and water requirements. Under drought stress conditions, plants reduced overall lateral root initiation and elongation while elongating the primary root to reach deeper water sources in the soil and enable seedling establishment before shoot emergence under dry conditions (Xiong et al., 2006; Xu et al., 2013). In maize, the mechanisms involved in growth responses of the primary root to water stress have been studied extensively, and the role of ABA accumulation, the relationship between osmotic adjustment and root growth maintenance, and the modification of cell wall extension properties have been gradually brought to light (Yamaguchi and Sharp, 2010). Accumulation of ABA in the root apex was required for the maintenance of elongation in the apical region of the elongation zone (Saab et al., 1990, 1992). ABA may promote the elongation of water-stressed roots by regulating ion transport, osmotic potential, and cell wall extensibility in the apical region of the elongation zone (Ober and Sharp, 1994, 2003; Wu et al., 1994, 2001; Yamaguchi and Sharp, 2010). Within the maize root system, root branching is a developmentally important aspect of root system architecture enabling the plant to have a larger root surface area to absorb more water and nutrients. Root branching involves the formation of adventitious and lateral roots and their outgrowth. The adventitious roots, which include aerial nodal roots and crown roots formed at belowground nodes, are important for lodging resistance and water and mineral uptake at the mature stage in maize (Zhang et al., 2018). Lateral root development can be initiated from pericycle cells in the primary root, seminal roots, and the belowground nodes of roots, and exogenous auxin can promote pericycle cell division, which results in the production of many lateral roots (Overvoorde et al., 2010; Olatunji et al., 2017). It was reported that ABA can regulate lateral root formation. For example, the ABA biosynthesis-deficient mutants aba2-1 and aba3-1 and the ABA signaling-deficient mutant aba insensitive4 showed increases in lateral root number (Deak and Malamy, 2005; Shkolnik-Inbar and Bar-Zvi, 2010). Drought stress leads to rapid ABA accumulation (Xu et al., 2013). Mild water stress resulting in slightly elevated ABA levels could promote root growth but inhibit shoot growth, which led to an increased root-to-shoot ratio (Moriwaki et al., 2013). ABA regulates this response via the core signaling pathway (Antoni et al., 2013). In contrast, severe water stress inhibits both root and shoot growth but promotes the formation of lateral roots and drought rhizogenesis (Vartanian et al., 1994).
By RNA sequencing, global gene expression profiles involved in stress response have been monitored in different tissues of maize, including leaves, roots, and kernels (Kakumanu et al., 2012; Humbert et al., 2013; Opitz et al., 2014, 2016; Seeve et al., 2017). In addition, a number of stress-induced genes, including many transcription factors, have been identified under water deficit in maize. When maize was exposed to drought stress, root growth maintenance was important for accessing deeper water, but only a few maize transcription factor genes related to abiotic stress and root development have been characterized (Nieva et al., 2005; Zhang et al., 2008a; Ying et al., 2012). In our study, a putative bZIP transcription factor, ZmbZIP4, which showed increased transcript abundance in the transcriptome analysis of young primary root and ovary tissue in maize under water deficit (Kakumanu et al., 2012; Opitz et al., 2016; Seeve et al., 2017), was isolated, and its functional roles in stress tolerance and root development, and its expression profile under different treatments, were analyzed in maize. Transgenic maize plants overexpressing ZmbZIP4 showed enhanced tolerance to salt stress and a better root system compared with wild-type plants. Genome-wide identification of the direct target genes of ZmbZIP4 indicates that ZmbZIP4 regulates numerous processes, including stress response, hormone signaling, and root development.
RESULTS
Sequence Analysis of ZmbZIP4, a Putative bZIP Transcription Factor
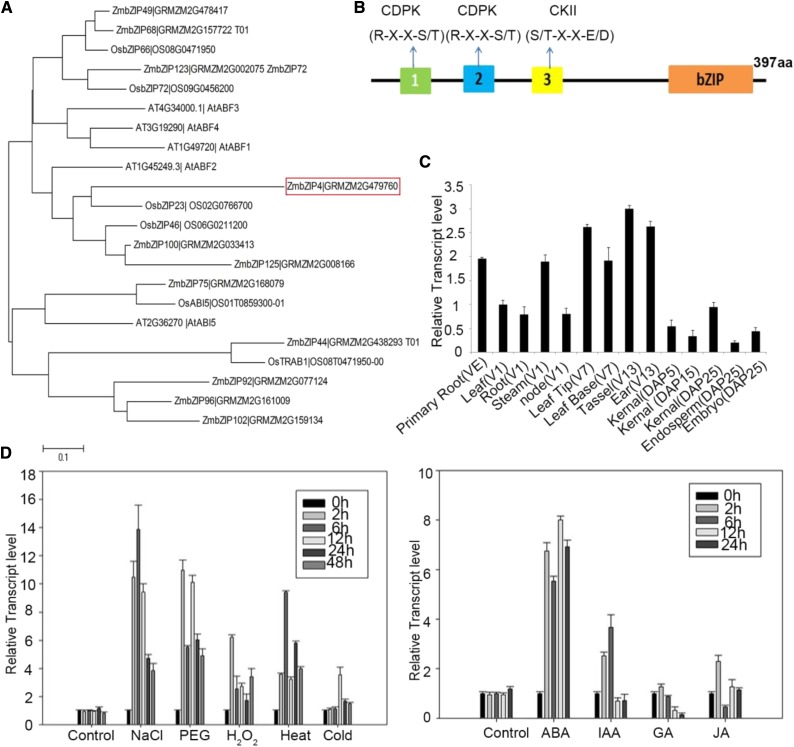
Two cDNA genes of bZIP family transcription factors were identified by microarray from a differentially expressed cDNA library of maize inbred line Q319 roots under osmotic stress (16% [w/v] PEG6000 for 3 d). These two genes were up-regulated under osmotic stress; one (GRMZM2G168079) is the homologous gene of Arabidopsis ABI5, and the other is GRMZM2G479760 (Supplemental Data Set S1), named ZmbZIP4 according to the MaizeGDB database. The phylogenetic relationship of ZmbZIP4 with the group A bZIP proteins from Arabidopsis and rice was shown with a phylogenetic tree based on the amino acid sequences of these proteins (Fig. 1A). ZmbZIP4 had the closest similarity to OsbZIP23 from rice, in terms of their amino acid sequences. The homologous gene of ZmbZIP4 in Arabidopsis is ABSCISIC ACID-RESPONSIVE ELEMENT BINDING PROTEIN1 (AREB1; i.e. ABF2), which is a basic domain/Leu zipper transcription factor that binds to the ABA-responsive element. ZmbZIP4 is a 397-amino acid protein with a typical bZIP DNA-binding domain in its C-terminal region and three conserved sequences in the N-terminal half of the protein (Fig. 1B). Several protein kinase target sites, such as R/KXXS/T for CDPK and S/TXXE/D for CKII, exist in these conserved sequences (Fig. 1B). The results show that ZmbZIP4 is a member of the group A bZIP transcription factors.
Figure 1.
Phylogenetic tree and expression profiles of ZmbZIP4. A, Phylogenetic tree of ZmbZIP4 (red box) and other group A bZIP genes in maize, rice, and Arabidopsis. B, Conserved domains of the ZmbZIP4 protein. aa, Amino acids. C, Reverse transcription quantitative PCR (RT-qPCR) expression analysis of ZmbZIP4 in different tissues and organs. For each sample, the primary root was cut from seedlings that had germinated for 8 d; the root, leaf, stem, and basal node (rhizome joints) were cut from plants at the V1 stage; the leaf tip and leaf base were cut from plants at the V7 stage; and the tassel and ear were cut from plants at the V13 stage. Kernels were peeled off the ears at 10 d after planting (DAP10), DAP15, and DAP25, and the endosperm and the embryos were peeled off the ears at DAP25. D, Expression analysis of ZmbZIP4 under stress and hormone treatments. The seedlings at the three-leaf stage were treated with different abiotic stresses and phytohormones, and the expression levels of ZmbZIP4 in the root were detected by RT-qPCR. JA, Jasmonic acid. For both B and C, fold changes in RNA transcripts were calculated by the 2−ΔCt method with maize Tub (NP_001105457) as an internal control. All bars represent means ± sd (n = 3 repeats).
ZmbZIP4 Is a Stress-Responsive Transcription Factor
To examine the tissue-specific expression of ZmbZIP4 in maize seedlings and mature plants, the abundance of ZmbZIP4 transcripts was determined across different organs (Fig. 1C). Furthermore, the promoter sequence of ZmbZIP4 (Supplemental Data Set S1) contains a number of putative stress response-related cis-elements, such as an ABRE (five hits), a methyl jasmonate recognition site (three hits), and a MYBHv1 recognition site (two hits; Supplemental Table S1). To speculate on the function of ZmbZIP4, the expression profiles of ZmbZIP4 under different abiotic stresses and phytohormone treatments also were checked by RT-qPCR. The results suggested that the transcript level of ZmbZIP4 was rapidly and strongly induced by high salinity, polyethylene glycol (PEG), oxidative stress (induced by hydrogen peroxide [H2O2]), ABA treatment, indole-3-acetic acid (IAA), and heat treatment but was only slightly affected by cold, GA, and jasmonic acid (Fig. 1D).
To determine whether ZmbZIP4 has transcriptional activity, the entire coding region was fused to the GAL4 DNA-binding domain in the vector pGBKT7, and the construct was transformed into yeast YRG-2. The X-gal assay indicated that ZmbZIP4 had transcriptional activity in yeast (Supplemental Fig. S1A).
Expression Analysis of ZmbZIP4 in the Overexpression Lines and Mutants
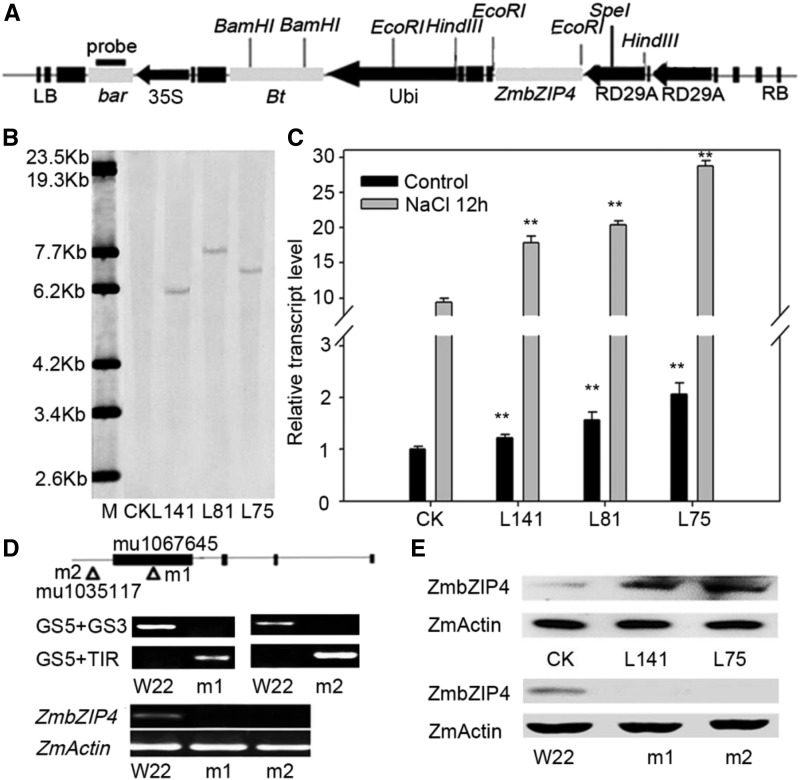
In the production of maize lines overexpressing ZmbZIP4, the stress-induced promoter RD29A was used to drive ZmbZIP4, and the herbicide resistance gene bar was used as a selection gene for transgenic screening (Fig. 2A). ZmbZIP4 was introduced into the maize inbred line DH4866, and the transgenic lines were confirmed using Southern blot. Signals of exogenous bar genes were detected with specific hybridization patterns. These results suggested that the ZmbZIP4 genes had been stably integrated into the genomes of the transgenic plants (Fig. 2B). To examine the expression level of the transgenes in those plants, total RNA was extracted from the leaves of transgenic and wild-type plants at the three-leaf stage under normal and salt-stressed treatments. RT-qPCR was used to detect the expression level of ZmbZIP4. As shown in Figure 2C, total transcripts of ZmbZIP4 were increased significantly in the transgenic lines. Western-blot analyses indicated that the protein levels of these independent lines were consistent with the transcription levels of ZmbZIP4 in these lines (Fig. 2E). In addition, two zmbzip4 UFMu mutants with different insertion sites were obtained from the Maize Genetics Cooperation Stock Center and self-fertilized to obtain homozygotes. The homozygous mutants were identified using PCR with the two primers GS5+GS3 and GS5+TIR (Fig. 2D). Both RT-qPCR and western-blot analyses revealed that the expression of ZmbZIP4 was undetectable in the mutants (Fig. 2, D and E).
Figure 2.
Molecular identification of the ZmbZIP4 transgenic lines and zmbzip4 mutants. A, T-DNA region of the plasmid of pB7WG2.0-P35S:Bar-Prd29A-Prd29A:ZmbZIP4. The black and gray boxes and black arrows indicate the T-nos, genes, and promoters, respectively. bar, The phosphinothricin acetyltransferase gene; Bt, the cryIAc gene from the Bacillus thuringiensis Ber line; LB and RB, left and right border, respectively; RD29A, the Arabidopsis RD29A promoter; 35S, the cauliflower mosaic virus 35S promoter; Ubi, the maize ubiquitin promoter. B, Southern-blot analysis of the overexpression plants (L141, L81, and L75) and the wild type (CK; DH4866). M, λ-EcoT14 I digest DNA marker. C, Real-time RT-qPCR results of the ZmbZIP4 transgenic genes in both natural and NaCl treatment conditions (100 mm NaCl treatment for 12 h). Bars represent means ± sd (n = 3 repeats). The transcript levels of the ZmbZIP4 overexpression lines were compared with their wild type (CK) under control and NaCl treatment conditions, respectively. Significant differences are indicated by asterisks (Student’s t test: **, P < 0.01). D, ZmbZIP4 UFMu mutants from the maize stock center. Triangles represent the insertion sites, and the molecular identification of the homozygous mutants (m1 and m2) was carried out by PCR and RT-qPCR. E, Western-blot assay of ZmbZIP4 in the transgenic lines and the zmbzip4 mutants.
ZmbZIP4 Is Involved in Maize Root Growth
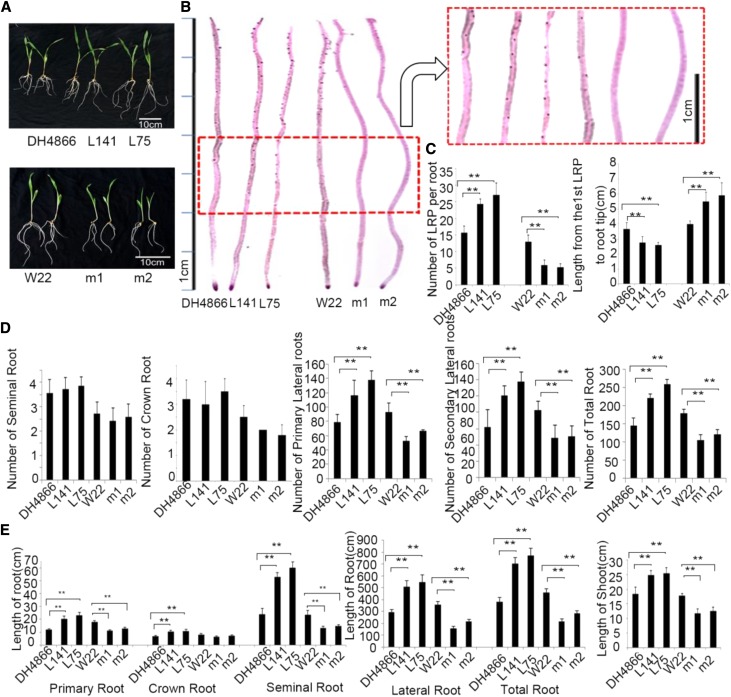
The ZmbZIP4 overexpression lines had a developed root system compared with the wild type, and the mutants were opposite (Fig. 3A). The root number, root length, and lateral root primordia (LRP) were counted on maize plants cultured in nutrient solutions. The data showed that the ZmbZIP4 overexpression lines had longer primary roots, seminal roots, and lateral roots compared with the wild type. However, the number of seminal roots and crown roots did not differ significantly between the overexpression lines and the wild type or between the mutants and the wild type (W22; Fig. 3D). When the LRP were determined by Feulgen staining, more LRP could be counted in the lateral root region of the overexpression lines and the distance from the first LRP to the root tip in the overexpression lines was shorter than that of the wild type, which meant that the LRP occurred earlier in the ZmbZIP4 overexpression lines than in the wild type. The mutant lines showed opposite alteration, with less LRP and a longer distance to the root tip compared with W22 (Fig. 3, B and C). The number of LRP of the overexpression lines was 55% to 72% more than that of the wild type, and the lateral root number was 53% to 80% more than that of the wild type. The number of LRP and the lateral root number of the zmbzip4 mutant were only 41% to 46% and 57% to 66% those of W22 (Fig. 3, C and D). Comparison of the root length of ZmbZIP4 overexpression lines, mutants, and the wild type also indicated that ZmbZIP4 positively regulated maize root growth, and the overexpression lines had much longer lateral roots and seminal roots (Fig. 3E). Therefore, the differences in the root system between the transgenic lines and the wild type were caused principally by the number and length of lateral roots.
Figure 3.
Overexpression of ZmbZIP4 promotes root system development. A, Morphology of the seedlings of ZmbZIP4 overexpression lines (L141 and L75), mutants (m1 and m2), and the wild type grown in nutrient solutions. B, Distribution of the LRP of the ZmbZIP4 overexpression lines, mutants, and the wild type. C, Number of LRP per seminal root and distance from the first lateral primordium to the root tip. D, Number of different types of roots from ZmbZIP4 overexpression lines, mutants, and the wild type grown in nutrient solutions. E, Root length and shoot length of seedlings of ZmbZIP4 overexpression lines, mutants, and the wild type grown in nutrient solutions. Bars represent means ± sd (n = 3 repeats). Significant differences are indicated by asterisks (Student’s t test: **, P < 0.01).
Performance of the Overexpressed ZmbZIP4 Plants and zmbzip4 Mutants under Stress Conditions
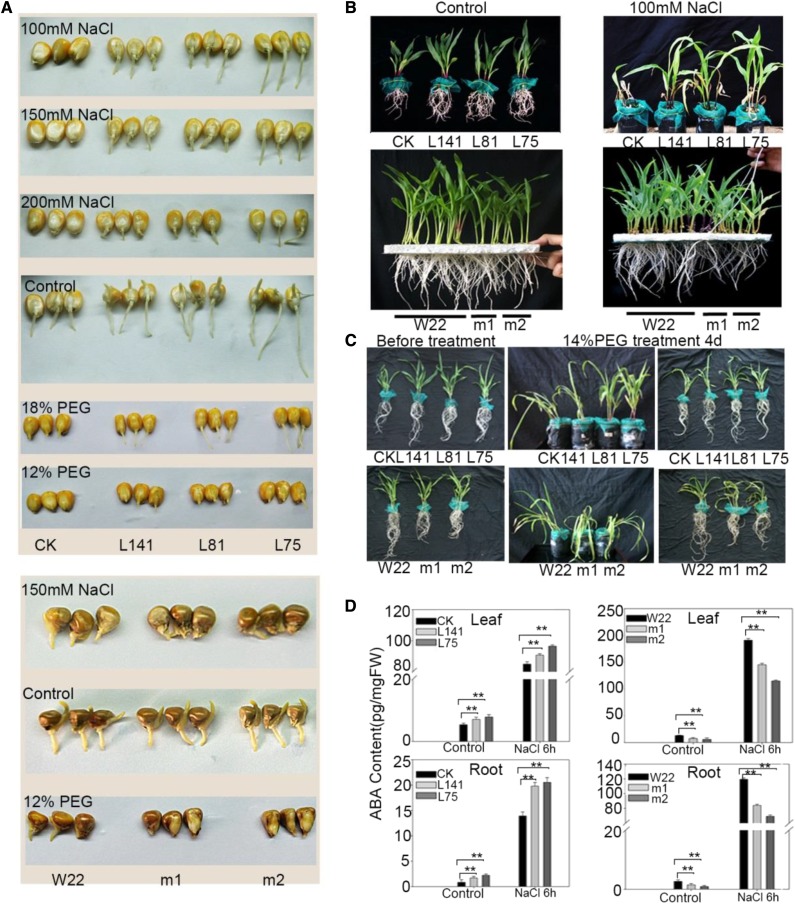
Based on the results that ZmbZIP4 expression was up-regulated by high-salinity or PEG treatment in maize, we investigated the effects of salt or drought stress on the performance of ZmbZIP4 overexpression plants and zmbzip4 mutants grown hydroponically and in soil culture. At the germination stage, the germination rates under the control condition were not significantly different between the ZmbZIP4 overexpression lines and its corresponding wild type (DH4866) or between the zmbzip4 mutants and W22. However, the mutants’ germination was delayed compared with W22 under salt or PEG treatment, and the ZmbZIP4-overexpressing lines germinated faster with higher germination rates under the stress conditions (Fig. 4A; Supplemental Fig. S2A). At the seedling stage, the ZmbZIP4-overexpressing plants exhibited not only improved salt resistance compared with the wild type (DH4866; Fig. 4B) but also a greater ability to adjust to osmotic stress (Fig. 4C; Supplemental Fig. S2B). As expected, the zmbzip4 mutants showed poor resistance to the NaCl and PEG solution treatments (Fig. 4, B and C). Moreover, the drought stress experiment in soil also showed the same results (Supplemental Fig. S3, A and B). These findings demonstrated that overexpression of ZmbZIP4 results in more drought- and salt-resistant maize.
Figure 4.
Phenotypes of the ZmbZIP4 transgenic lines and the mutant under abiotic stress. A, Germination experiment of the ZmbZIP4 overexpression lines (L141, L85, and L75 from DH4866), zmbzip4 mutants (m1 and m2 from W22), and wild-type maize (DH4866 [CK] and W22) under the NaCl and PEG treatment conditions. B, Test of seedling salt resistance in ZmbZIP4 overexpression lines, zmbzip4 mutants, and wild-type maize. C, Phenotypes of the overexpression lines, mutants, and wild-type maize under normal conditions and the 4-d drought stress treatment with a 14% PEG6000 solution. D, ABA contents in leaves and roots of the ZmbZIP4 overexpression lines, mutants, and the wild type under normal (Control) and NaCl treatment for 6 h. FW, Fresh weight. Values are means ± sd. Bars represent means ± sd (n = 3 repeats). Significant differences are indicated by asterisks (Student’s t test: **, P < 0.01).
The ABA contents in the leaves and roots of the ZmbZIP4-overexpressing lines, zmbzip4 mutants, and their wild-type controls were determined. As shown in Figure 4D, under normal conditions, the ABA content of the overexpression lines was higher in both leaves and roots than that of the wild type. When subjected to 100 mm NaCl treatment, the ABA content in the plants increased dramatically by 14.8-fold in leaves and 15.5-fold in roots compared with the control, and the ABA content in the overexpression lines also was higher than that of the wild type. However, in the zmbzip4 mutants, the ABA content was lower than that in W22 under both the normal and NaCl treatments. These results indicated that an increase of ZmbZIP4 expression level enhanced ABA accumulation in maize.
Exploration of the Target Genes of ZmbZIP4
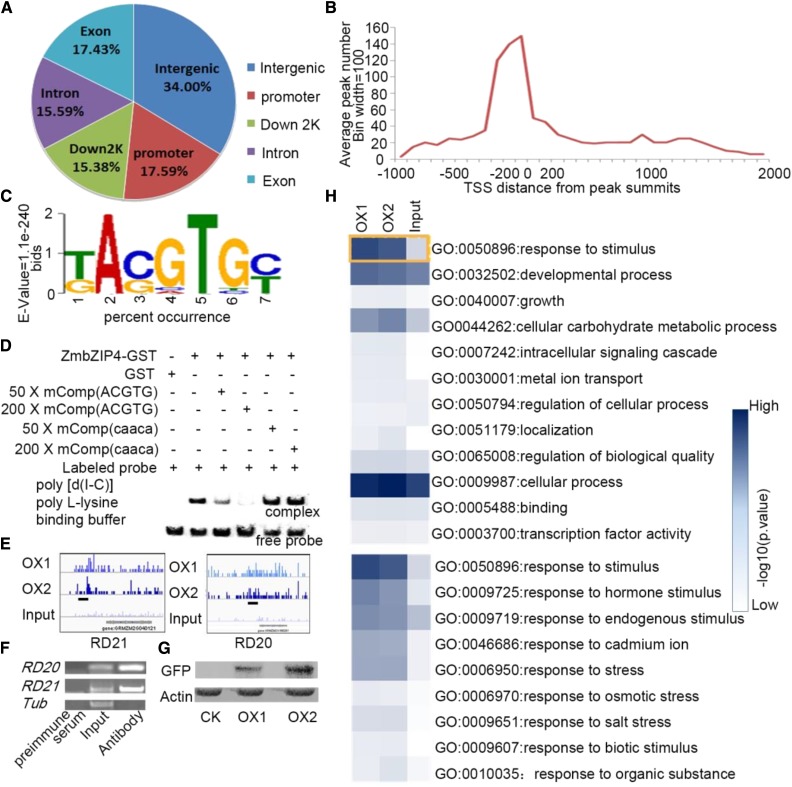
To investigate the function of ZmbZIP4 in stress resistance and plant development, the downstream target genes of ZmbZIP4 were identified by chromatin immunoprecipitation followed by sequencing (ChIP-Seq; Mundy et al., 1990; Park, 2009) with ZmbZIP4-GFP transgenic maize plants at the three-leaf stage and the monoclonal antibody of GFP. The fusion of GFP to ZmbZIP4 did not interfere with the transcription activation activity and the binding activity of ZmbZIP4 (Supplemental Fig. S1). To identify their positions, the 23 million reads in the ChIP-Seq data were mapped with the maize genome (MaizeGDB; http://www.maizegdb.org/) using the ultrafast Bowtie aligner (Langmead et al., 2009). By excluding the peaks in intergenic regions, MACS software (Zhang et al., 2008b) detected 1,868 and 2,454 genes (2,619 and 3,433 peaks) with P < 0.05 for each of the ChIP-Seq data sets. A total of 1,020 genes were found with high enrichment in both sets, and 17.59% of the reads that mapped to these genes were located in promoter regions (Fig. 5A). Additionally, except for the peaks in the intergenic regions, the distribution peak was close to the transcription start site, from −400 to +100 bp (Fig. 5B). The Gene Ontology (GO) analysis results of the 1,020 enriched genes showed significant and coincident enrichments in developmental process (GO:0032502), carbohydrate metabolic process (GO:0044262), and response to stimulus (GO:0050896) compared with the input sample data (Fig. 5H). The next hierarchical classification of this gene data involved mainly biological processes, including hormone stimulus (GO:009725), response to stress (GO:0006950), and response to cadmium ion (GO:0046686; Fig. 5H). Genes in these three GO categories that exhibited a fold enrichment greater than 10 (normalized by the input sample) and peaks located in their promoter region or first exon were selected to test their expression levels in transgenic plants. The ChIP-Seq signals of GRMZM2G04012 (ZmRD20) and GRMZM2G166281 (ZmRD21) are shown on the genome browser (Fig. 5E). The ChIP samples were quantified by ChIP-qPCR using primers specific to the promoters of ZmRD20 and ZmRD21, and the promoter region of these genes was enriched effectively and showed an apparently higher transcript level in the overexpression (OX) lines than the wild-type lines. These data demonstrated that ZmbZIP4 targeted the promoters of these genes and that these ChIP-Seq data were valid. All of them have an ACGTG motif in their promoter regions (Supplemental Table S2). To investigate the binding motifs of ZmbZIP4, ±100-bp flanking sequences around the peaks were submitted to MEME-ChIP (http://meme-suite.org/tools/meme-chip) to search for enriched motifs. The most frequently captured motifs belonged to the ABREs (Fig. 5C). An electrophoretic mobility shift assay (EMSA) with point mutations verified that ACGTG is the core sequence of ZmbZIP4 binding in the promoters of the target genes (Fig. 5D). After recombining their promoter regions into the pLacZi vector for one-hybrid yeast system assays in the YM4271 strain, the X-gal staining results proved that these regions were targeted by ZmbZIP4 in vivo (Fig. 6, A and D). The one-hybrid yeast system results using series substitutions as reporters indicated that ACGTG is the core DNA-binding motif of ZmbZIP4 (Supplemental Fig. S4). These results suggested that ZmbZIP4 could recognize ACGTG and thus regulate the downstream network.
Figure 5.
ChIP-Seq assay of ZmbZIP4. A, Distribution of ZmbZIP4 transcription factor-binding sites. B, Distances of identified peaks from the transcription start sites (TSS) for ZmbZIP4. The peaks were highly enriched in the region −200 to +100 bp from the TSS. C, Binding situation of ZmbZIP4 quantified by the percentage occurrence of the ACGTG motif (x axis) against the log10 (fold enrichment in immunoprecipitation samples compared with the input sample; y axis). D, EMSA showing that ACGTG is required for ZmbZIP4 to bind to targets. GST-tagged ZmbZIP4 was used in the EMSA, and the GST tag was used as a control. E, ChIP-Seq signals of RD21 and RD20 on the genome browser. Short black lines denote regions used in ChIP-qPCR. F, ChIP-qPCR assay. Sample immunized by preimmune serum was used as the negative control. G, Immunoblotting detects the specificity of the GFP antibody; seedlings of the wild type, ZmbZIP4-GFP, were used for protein extraction. H, GO analysis results of the input sample data and two immunoprecipitation samples (OX1 and OX2) data.
Figure 6.
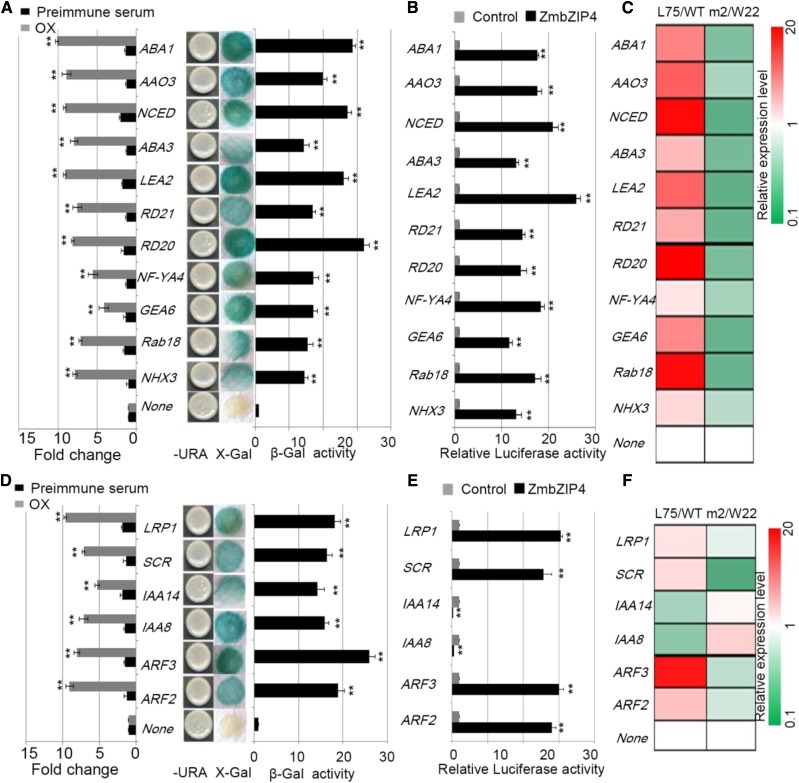
Validation of candidate downstream genes. A, ChIP-qPCR (left) and yeast one-hybrid analysis (right) of ZmbZIP4 and 11 candidate genes involved in abiotic stress response and ABA synthesis. B, Transient expression of P35S:ZmbZIP4 with promoter-Luc reporter constructs in maize protoplasts. C, Relative expression level heat map of 11 candidate genes involved in abiotic stress response and ABA synthesis in ZmbZIP4 overexpression lines and mutants compared with wild-type maize. D to F, ChIP-qPCR (left) and yeast one-hybrid analysis (right), relative luciferase activity, and relative expression level heat map of six candidate genes related to root development, respectively. All plants grew under natural conditions. All bars represent means ± sd (n = 3 repeats). Significant differences are indicated by asterisks (Student’s t test: **, P < 0.01).
Some Genes Involved in the Abiotic Stress Response and ABA Synthesis Were Regulated by ZmbZIP4
Analysis of the ZmbZIP4 ChIP-Seq data and the ChIP-qPCR identified a number of positive regulators of salt and drought stress resistance, including ZmRD20 (GRMZM2G342685), ZmRD21 (GRMZM2G166281), ZmRab18 (RESPONSIVE TO ABA 18/GRMZM2G052364), ZmNHX3 (SODIUM/HYDROGEN EXCHANGER3/GRMZM2G118019), ZmGEA6 (EM-LIKE PROTEIN GEA6/GRMZM2G162659), ZmNF-YA4 (NUCLEAR FACTOR-YA4/GRMZM2G000686), and ZmLEA2 (LATE EMBROGENESIS ABUNDANT PROTEIN/GRMZM2G704021), as ZmbZIP4 candidate target genes (Fig. 6A). The expression levels of RD20, RD21, Rab18, NHX3, GEA6, and LEA2 were all up-regulated by ZmbZIP4 (Fig. 6C). The promoters of these genes were amplified and inserted into the yeast expression vector pLacZi to detect whether ZmbZIP4 could bind to the promoters and drive the expression of the marker gene. A transactivation assay showed the ability of ZmbZIP4 to bind to these promoters (Fig. 6A). To demonstrate whether ZmbZIP4 can activate the expression of these downstream genes in vivo, the promoter regions of these genes were ligated to pGreenII0800-Luc and cotransformed with 35S:ZmbZIP4 into the protoplasts of maize, and the relative luciferase activity was measured (Fig. 6B). The results confirmed that ZmbZIP4 positively regulates the expression of RD20, RD21, Rab18, NHX3, GEA6, NF-YA4, and LEA2. ABA treatment improved the transactivation activities of ZmbZIP4 (Supplemental Fig. S5). There also are several other stress-related genes, such as some other LEA2 genes (GRMZM2G447569, GRMZM2G111679, GRMZM2G063287, and GRMZM2G042421), oxidative stress-related genes (ZmOX3/GRMZM2G031580), and K+ uptake and transporter genes (GRMZM2G455817, GRMZM2G036792, GRMZM2G118497, and GRMZM2G317728), that were up-regulated by ZmbZIP4 (Supplemental Table S2). Moreover, multiple transcription factor genes also were indicated as target genes of ZmbZIP4 by ChIP-Seq, such as the bZIP family (ZmHY5/GRMZM2G171912 and ZmbZIP63/GRMZM2G019446), the MYB family (ZmMYB12/GRMZM2G051528 and ZmMYB102/GRMZM2G166337), the NAC family (ZmNAC41/GRMZM2G179049 and ZmNAC46/GRMZM2G146380), and so on (Supplemental Table S2). Some molecular chaperones, such as heat shock proteins (ZmTMS1/GRMZM2G380889 and ZmHSFA2/GRMZM2G005815), were shown to be regulated by ZmbZIP4 (Supplemental Table S2). In the promoter regions of these genes, motifs of ACGTG exist that are the target site of ZmbZIP4. When maize plants respond to abiotic stress, ZmbZIP4 plays an important role in the regulation of transcription factors, transporters, and molecular chaperones at the transcription level and contributes to plant resistance to abiotic stress.
In the ChIP-Seq and ChIP-qPCR results, the zeaxanthin epoxidase (ZEP/ABA1), 9-cis-epoxycarotenoid dioxygenase (NCED), abscisic aldehyde oxidase (AAO3), and molybdopterin cofactor sulfurase (LOS5/ABA3) genes also were ZmbZIP4 target genes (Fig. 6A). The higher expression of these genes led to higher levels of ABA in ZmbZIP4-overexpressing lines (Fig. 4D). When maize plants suffered from salt or osmotic stress or ABA treatment, the expression of ZmbZIP4 was up-regulated (Fig. 1D), and in lines overexpressing ZmbZIP4, ABA synthesis was enhanced, which led to higher ABA levels in overexpressing lines compared with the wild type (Fig. 4D).
Multiple Root Development-Related Genes Are Targeted by ZmbZIP4
IAA8 (INDOLEACETIC ACID-INDUCED PROTEIN8), IAA14, ZmARFs (AUXIN-RESPONSIVE FACTORS), SCR (SCARECROW), and LRP1 (LATERAL ROOT PRIMORDIUM PROTEIN1) were reported to be involved in root development, especially lateral root formation (Smith and Fedoroff, 1995; Lim et al., 2005; Krichevsky et al., 2009; Arase et al., 2012; Bruno et al., 2017). The ZmbZIP4 overexpression lines had longer primary roots and seminal roots as well as more lateral roots and LRP than the wild type, while the mutants showed opposite changes (Fig. 3). The ChIP-Seq and ChIP-qPCR analyses (Fig. 6D) indicated that ZmSCR (GRMZM2G131516), ZmLRP1 (GRMZM2G450459), ZmIAA8 (GRMZM2G479834), ZmIAA14 (GRMZM2G001799), ZmARF2 (GRMZM2G338259), and ZmARF3 (GRMZM2G030710) were the ZmbZIP4 target genes. The expression of ZmSCR, ZmLRP1, ZmARF2, and ZmARF3 was up-regulated in the ZmbZIP4-overexpressing lines compared with the wild type, while the expression of ZmIAA8 and ZmIAA14 was down-regulated (Fig. 6F). The latter two are transcriptional repressors for auxin response genes. In the zmbzip4 mutant, the expression levels of these genes showed an opposite trend (Fig. 6F). At the same time, the X-gal and luciferase activity assays were used to confirm the ability of ZmbZIP4 to bind to these promoters and regulate the expression of these genes (Fig. 6, D and E). There also were several other genes related to root development, such as some d-type cyclins, ZmCYCD1;1 (GRMZM2G047637), ZmCYCD2;1 (GRMZM2G088980), and ZmCYCD6;1 (GRMZM2G050933), that were the targets of ZmbZIP4 (Supplemental Table S2). Previous research indicated that SHORT-ROOT (SHR) and SCR could directly activate a d-type cyclin to regulate formative cell divisions in root development (Sozzani et al., 2010). It could be suggested that ZmbZIP4 participates in root development in maize by regulating auxin signaling and cyclin-related genes.
DISCUSSION
ZmbZIP4 Participates in the Multiple Abiotic Stress Responses of Maize
In our study, ZmbZIP4 had the highest similarity to the OsbZIP23 gene of rice at the amino acid sequence level. Previous research has shown that OsbZIP23 plays an essential role in drought resistance in rice (Xiang et al., 2008; Zong et al., 2016). Transgenic maize plants overexpressing ZmbZIP4 showed enhanced tolerance to salt and drought stress compared with wild-type plants, very similar to the OsbZIP23-overexpressing plants in rice. A study in Arabidopsis explored AREB1/ABF2, which is the homolog of ZmbZIP4, and found that it could participate in the regulation of sugar signaling and multiple abiotic stresses (Kim et al., 2004; Fujita et al., 2005). We also observed that the expression of some key glycometabolism enzyme genes was regulated by overexpressing ZmbZIP4 (Supplemental Table S2). More importantly, the expression of ZmbZIP4 was induced not only by drought and ABA treatments but also by salt stress, high temperature, and reactive oxygen species stress (Fig. 1D). In the promoter sequences of ZmbZIP4, it has light-responsive, methyl jasmonate-responsive, and ABA-responsive cis-acting regulatory elements, anaerobic induction, and some dehydration response elements such as TC-rich repeats (Supplemental Table S1), which is consistent with the expression pattern. Overexpression lines of ZmbZIP4 showed better survivability after suffering from high salt stress in the germination and seedling stages, and the mutants were sensitive to abiotic stresses (Fig. 4, A and B). Several regulator genes involved in the response to abiotic stress were positively regulated by ZmbZIP4, such as ZmRD20, ZmRD21, ZmRab18, ZmNHX3, ZmLEAs, and ZmGEA6. Osmotic regulation is important for plant drought resistance. Among them, LEA proteins are highly hydrophilic proteins that function in plant abiotic stress and protect the cell by stabilizing cellular components in response to water loss (Chakrabortee et al., 2007). It has been reported that the transformation of the LEA gene into a number of plant species can confer tolerance to drought stress (Park et al., 2011; Bao et al., 2017). The RD20 gene can be induced by various abiotic stresses, such as drought, salt stress, cold, and wounding, and now often is used as a stress marker gene in Arabidopsis (Magnan et al., 2008; Alexandre et al., 2009). RD20 played an important role in drought tolerance through stomatal control under water deficit conditions. The rd20 knockout plants present a higher transpiration rate that correlates with enhanced stomatal opening and a reduced tolerance to drought compared with the wild type (Aubert et al., 2010). RAB18 encodes a hydrophilic Gly-rich protein that accumulates specifically in Arabidopsis exposed to low temperature or desiccation and in response to exogenous ABA application (Lång and Palva, 1992; Ghelis et al., 2000; Nylander et al., 2001). Salinity causes ion-specific stresses that result from the altered K+/Na+ ratios, leading to a buildup of Na+ and Cl−, which is detrimental to plants. Plants can remove Na+ ions out of the cytosol by transporting them into the vacuolar lumen or out of the cell using Na+/H+ exchangers localized in the vacuolar and plasma membranes, respectively. NHX3 is a vacuolar Na+/H+ antiporter, and constitutive expression of AtNHX3 in sugar beet (Beta vulgaris) conferred augmented resistance to high salinity in transgenic plants (Liu et al., 2008, 2010). It was implied that abiotic stress or ABA could induce the expression of ZmbZIP4 and that ZmbZIP4 positively regulates the resistance of maize to multiple abiotic stresses (Fig. 7).
Figure 7.
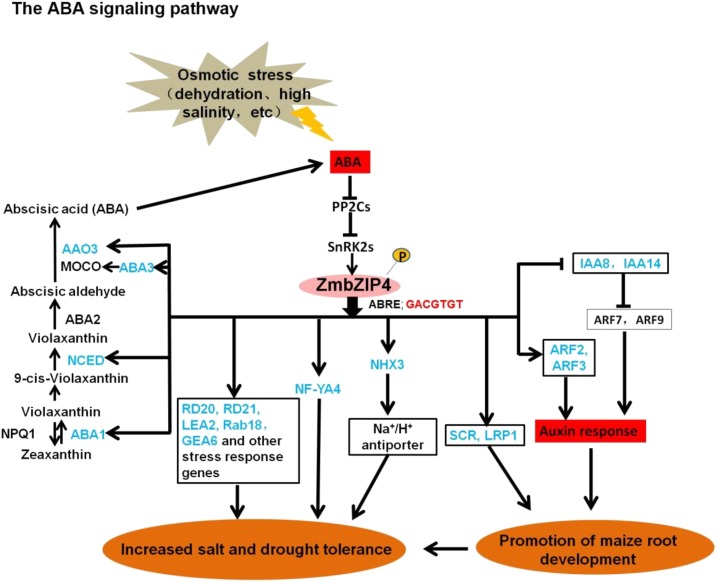
Proposed model for the role of ZmbZIP4 in maize root development and stress response. ABA could be induced by abiotic stress and then regulate the stress response via the core signaling pathway, which includes the PYR/PYL/RCAR receptor, PP2C proteins, SnRK2 family members, AREB/ABF transcription factors, and downstream regulated genes; ZmbZIP4 belonged to the transcription factor of the AREB/ABF subfamily. ZmbZIP4 could bind directly to the promoters of several genes that participated in ABA biosynthesis, which led to the ABA accumulation. Furthermore, ZmbZIP4 also could directly regulate some stress-related genes, transcription factors, and the Na+/H+ antiporter NHX3 to increase stress resistance. In addition, ZmbZIP4 also could target to the promoters of SCR, LRP1, and several auxin response genes to promote maize root development. Blue font indicates the genes targeted directly by ZmbZIP4.
ZmbZIP4 Regulates ABA Accumulation
ABA plays an important role in the adaptive responses of plants to environmental stresses. In ABA biosynthesis, the first step that is more specific to the ABA biosynthesis pathway is the epoxidation of zeaxanthin and antheraxanthin to violaxanthin catalyzed by a ZEP (Thompson et al., 2000). Overexpression of ZEP in Arabidopsis conferred greater abiotic tolerance, indicating that this enzyme might be limiting with regard to some stress response (Park et al., 2008). After a series of structural modifications, violaxanthin is converted to 9-cis-epoxycarotenoid. The rate-limiting step of ABA biosynthesis is the formation of xanthoxin via oxidative cleavage from either of these precursors by the enzyme NCED (Tan et al., 1997). Overexpression of NCED leads to higher levels of ABA, a reduction of the transpiration rate in leaves, and an enhanced level of drought tolerance (Iuchi et al., 2001). The xanthoxin is converted to ABA through a two-step reaction via ABA aldehyde. A short-chain alcohol dehydrogenase/reductase, encoded by the AtABA2 gene (González-Guzmán et al., 2002), catalyzes the first step of this reaction and generates ABA aldehyde. The last step in the synthesis of ABA is catalyzed by abscisic aldehyde oxidase (AAO). This AAO requires a sulfurase form of a molybdenum cofactor (MoCo) for its activity (Seo et al., 2000, 2004; Bittner et al., 2001). The MoCo sulfurase is encoded by the osmotically responsive gene LOS5/ABA3 (Bittner et al., 2001; Xiong et al., 2001). Overexpressing AtLOS5 in transgenic maize increased ABA levels and increased salt stress tolerance-mediated root ion fluxes and leaf water status under salt stress (Zhang et al., 2016). Analysis of the ZmbZIP4 ChIP-Seq data identified a number of genes related to ABA biosynthesis as candidate ZmbZIP4 target genes, such as ZmABA1, ZmNCED, ZmAAO3, and ZmLOS5 (Fig. 6, A–C). ZmbZIP4 overexpression led to higher levels of ABA in leaves and roots under both normal and salt stress conditions (Fig. 4C). It was clear that an early reaction of plants under drought conditions is an increase in ABA level, which triggers the expression of ABA-responsive genes and induces stomatal closure. ABA regulates the stress response via the core signaling pathway (Cutler et al., 2010), which includes the PYR/PYL/RCAR receptor, PP2C proteins, SnRK2 family members, AREB/ABF transcription factors and downstream regulated genes, and the ABA-activated signaling pathway. In our study, we found that ZmbZIP4 belongs to the AREB/ABF subfamily of transcription factors. The ZmbZIP4 overexpression lines had increased ABA levels, while the ABA levels of zmbzip4 mutants were lower compared with the wild type. ZmbZIP4 also targets ZmNCED, ZmABA1, ZmAAO3, and ZmLOS5, which are involved in ABA biosynthesis, and higher levels of ZmbZIP4 promote ABA accumulation by regulating ABA synthesis. Together, our data suggested that, in maize, ABA biosynthesis and signaling are regulated by ZmbZIP4 via a complex pathway (Fig. 7).
ZmbZIP4 Is Involved in the Regulation of Maize Root Development
The root system of a plant is a crucial factor for plant survival under stress. Changes in root architecture are correlated tightly with perturbations in environmental conditions. Plants could adapt their root system architecture by modulating primary, lateral, or adventitious root growth as well as by modulating root hair length and distribution (Malamy, 2005). In this study, the overexpression of ZmbZIP4 enhanced maize root development and growth compared with the wild type (DH4866), and the mutants showed the opposite pattern. Because of the differences in development conditions between the DH4866 and W22 genotypes, we used two controls. The increased lateral root number and length allow the plant to have a larger root surface area to absorb more water and nutrients. The growth and elongation of the seminal roots and primary roots were promoted in the ZmbZIP4 transgenic lines under optimal and stress conditions, which led the plant to absorb the deeper water in the soil during osmotic stress. Combining the root phenotypes of the zmbzip4 mutant, ZmbZIP4 was found to act as a positive regulatory factor in root development. Multiple root development-related genes, including ZmSCR, ZmLRP1, ZmARF2, and ZmARF3, were targeted by ZmbZIP4, and their expression was up-regulated in the overexpression lines and reduced in the mutants. Another two genes, ZmIAA8 and ZmIAA14, transcriptional repressors, also were targets of ZmbZIP4, and their expression levels were down-regulated in the overexpression line and up-regulated in the mutants. These genes are involved in auxin signaling. Auxin is important for lateral root initiation and subsequent LRP development (Laskowski et al., 1995; Himanen et al., 2002). Central regulators of auxin signaling include the TRANSPORT INHIBITOR RESPONSE1 (TIR1) protein, auxin/indole acetic acid (Aux/IAA) proteins, and ARF proteins (Mockaitis and Estelle, 2008). IAA8 encodes an Aux/IAA protein and has been reported to be involved in the auxin-activated signaling pathway in Arabidopsis (Dreher et al., 2006) and lateral root formation, and the process is regulated through the interaction with the TIR1 auxin receptor and ARF transcription factors in the nucleus (Arase et al., 2012). The slr mutant, which carries a gain-of-function mutation in domain II of IAA14, has no lateral roots and exhibits other auxin-related phenotypes, including limited root hair formation and reduced root gravitropism (Fukaki et al., 2002). IAA14 can interact with ARF7 and ARF19. The slr mutation blocks auxin-induced pericycle cell divisions for lateral root initiation, indicating that auxin-responsive transcription mediated by SLR/IAA14 is important for lateral root formation. At low intracellular auxin concentrations, Aux/IAA proteins act as transcriptional repressors that interact with ARF proteins via their domains III and IV. The ARF proteins of these complexes interact with auxin-responsive elements in the promoters of downstream genes, thereby repressing their transcription (Woodward and Bartel, 2005). Thus, down-regulated expression levels of ZmIAA8 and ZmIAA14 in the ZmbZIP4 overexpression lines may be beneficial to the derepression of ARF proteins and, thereby, promote the expression of downstream genes related to root development.
The up-regulated expression of ZmARF2 and ZmARF3 in the ZmbZIP4 overexpression lines also could promote the expression of some auxin-induced genes. Their combined role could greatly enhance root development. Previous results showed that the GRAS family transcription factors SHR and SCR are required for the establishment and maintenance of separate cortical and endodermal cell layers (Di Laurenzio et al., 1996; Sabatini et al., 2003; Koizumi et al., 2012). The transcription factor SCR is a key regulator of primary root stem cell differentiation/maintenance and radial patterning (Helariutta et al., 2000) and is expressed in the cortex/endodermis initial cell and the endodermis. SCR binds to its own promoter in the presence of SHR (Cui et al., 2007) and jointly regulates quiescent center markers (WUSCHEL-RELATED HOMEOBOX; Sarkar et al., 2007). Both SHR and SCR directly affect the expression of CYCD6;1 in cortical endodermal daughter cells, triggering asymmetric periclinal division that produces the endodermis and cortex (Sozzani et al., 2010). SCR regulation of asymmetric cell division is essential for generating the radial organization of the Arabidopsis root (Di Laurenzio et al., 1996). The scr-1 mutant lacks an appropriately specified quiescent center, and stem cell activity is ultimately lost, thereby leading to the loss of meristem maintenance and root growth (Sabatini et al., 2003). The ChIP-Seq results indicate that ZmbZIP4 also can regulate the expression of some cyclin genes, such as CYCD1;1, CYCD2;1, and CYCD6;1 (Supplemental Table S2). Among these genes, CYCD6;1 has been reported to be involved in cortex/endodermis asymmetric stem cell division to regulate root development in Arabidopsis (Cruz-Ramírez et al., 2012; Lee et al., 2016). In this study, to better reveal the function of ZmbZIP4 in maize, we used two different ecotypes, DH4866 and W22, as the background of the overexpressed and mutant maize, respectively. The expression levels of ZmbZIP4 in both ecotypes showed a positive correlation with the development of the root system. Above all, ZmbZIP4 may act as a positive regulatory factor in the root development of maize (Fig. 7).
ABA and Auxin Signaling Interact to Modulate Root Growth in ZmbZIP4 Overexpression Lines
In our study, ZmbZIP4 overexpression increased root development and stress resistance in maize. The ChIP-Seq results indicate that ZmbZIP4 regulates a number of ABA and auxin signaling-related genes. ZmbZIP4 also can promote ABA accumulation by regulating ABA synthesis. It was postulated that ZmbZIP4 participates in the cross talk of ABA and auxin signaling.
In our study, multiple auxin signaling-related genes, ZmSCR, ZmIAA8, ZmIAA14, ZmARF2, and ZmARF3, were targeted by ZmbZIP4. In Arabidopsis, the expression of ARF2 was reported to be ABA inducible, and ARF2 coordinated with PLTs and PINs to orchestrate the ABA-mediated regulation of root meristem activity. The arf2 mutant was more sensitive to the ABA-mediated inhibition of primary root elongation (Wang et al., 2011; Promchuea et al., 2017). During drought stress, ABA has been found to transcriptionally enhance the expression of the auxin transporter genes AUX1 and PIN2 in the root tip to activate proton secretion. This increase in proton secretion is needed to promote the elongation of primary root and root hair development under conditions of moderate drought stress (Xu et al., 2013). Moreover, ZmbZIP4 overexpression led to an accumulation of ABA (Fig. 4C). ABA also can promote lateral root growth recovery through a pathway mediated by the action of the ABA receptor PYL8. PYL8 interacts directly with a group of transcription factors, MYB77, MYB44, and MYB73, leading to the enhancement of auxin-dependent transcription. MYB77 forms a heterodimer with ARF7 and increases the transcription of ARF7 target genes. This is a synergistic action of ABA and auxin in controlling the growth of plant lateral roots (Zhao et al., 2014). Although there are still gaps in our understanding of the molecular mechanisms, it is clear that drought stress, ABA, and auxin are closely interlinked within a general framework in which extrinsic environmental signals impinge on intrinsic auxin signaling (Hong et al., 2013).
Above all, in ZmbZIP4 overexpression maize, ABA and auxin participate together in root development under normal and stress conditions.
CONCLUSION
ZmbZIP4 is involved in root system development and resistance to abiotic stress in maize. Overexpression lines of ZmbZIP4 developed a much better root system under normal conditions and showed a higher germination rate and survivability when suffering from serious abiotic stresses. The ChIP-Seq data offered evidence that ZmbZIP4 targets genes with an ACGTG consensus sequence in their promoters. The overexpression of ZmbZIP4 up-regulated some crucial genes related to abiotic stress responses and root development. ZmbZIP4 is a crucial upstream regulator of abiotic stress resistance in maize.
MATERIALS AND METHODS
Phylogenetic Analysis of the ZmbZIP4 Gene Family
Sequences of ZmbZIP4 and homologous genes in maize (Zea mays), rice (Oryza sativa), and Arabidopsis (Arabidopsis thaliana) were obtained from NCBI (https://www.ncbi.nlm.nih.gov/). Amino acid sequences coded by these genes were aligned by ClustalW (Thompson et al., 1997) using default parameters. The neighbor-joining method was used with bootstrap values from 1,000 replicates at each branch. A phylogenetic tree was constructed with the neighbor-joining method in MEGA (Saitou and Nei, 1987; Kumar et al., 2004) and presented by using TreeView (Page, 1996).
Expression Analysis of ZmbZIP4
The seeds of the inbred line DH4866 (provided by Shandong Denghai Seeds) were surface sterilized with 70% (v/v) ethanol for 5 min, followed by several rinses with tap water, and then sown in pots (35 cm in diameter and 22 cm in height) containing a mixture of vermiculite and organic fertilizer (2:1, v/v) with total N 1.2 g kg−1, Pi 90 mg kg−1, and K+ 160 mg kg−1. The seedlings were grown in a greenhouse under natural conditions and watered once every 3 d. The primary root from seedlings germinated for 8 d (VE stage); roots, stems, basal nodes, and leaves of seedlings at the three-leaf stage (V1 stage); leaf tips and leaf bases of the ear internode (V7 stage); immature tassels and ears from plants in the V13 stage; kernels from the ears at DAP5, DAP15, and DAP25; and endosperms and embryos from the ears at DAP25 were used for organ-specific expression analysis.
The seeds of the maize inbred line DH4866 were surface sterilized and germinated in filter paper for 4 d (28°C, dark); then, seedlings with a primary root length of 2 cm were transferred to Hoagland nutrient solution and grown at a photon flux density of 250 μmol m−2 s−1 (14 h/10 h of light/dark) at 32°C/25°C (day/night) in a greenhouse until the plants reached the three-leaf stage. The nutrient solution was aerated with a mini air pump and supplemented with fresh solution to ensure that the seedlings received a sufficient amount each day. The seedlings at the three-leaf stage were either watered with 18% (w/v) PEG6000 (Mr 5,000–7,000; Merck Schuchardt; −0.77 MPa), watered with 200 mm NaCl, exposed to heat shock stress (plants exposed to 41°C), exposed to cold (plants transferred to a 4°C culturing room), or exposed to oxidative stress (plants soaked with 1% [v/v] H2O2 solution); the seedlings in each treatment were sampled at the designated times (0, 2, 6, 12, 24, and 48 h). For phytohormone treatment, ABA, IAA, GA, and jasmonic acid at 0.1 mm concentration were added to the culture solution for the maize plants, and the roots were taken in each treatment at 0, 2, 6, 12, and 24 h. Expression levels of ZmbZIP4 in roots under the stress treatments, including NaCl, PEG, H2O2 (0, 2, 6, 12, 24, and 48 h), and heat (0, 2, 6, 12, and 24 h), were determined. The roots of treated seedlings were harvested at the given time points, frozen immediately in liquid nitrogen, and stored at −80°C. Total RNA was extracted with TRIzol Reagent and treated with RNase-free DNase. cDNA synthesis was performed with an RT reagent kit (Takara) according to the manufacturer’s protocol. Real-time RT-qPCR was performed on an ABI 7500 RT-PCR instrument. The primers used for RT-qPCR are listed in Supplemental Table S3.
Cloning of the ZmbZIP4 Gene and Construction of a Transformation Vector
The full-length cDNA sequence of ZmbZIP4 was amplified from the cDNA library of the maize inbred line Qi319 (seeds stored in our laboratory) by PCR using a SMART RACE cDNA Amplification Kit (Clontech). The amplified products were purified and cloned into the pEASY-Blunt Cloning vector (Takara) for sequencing. Then, ZmbZIP4 was inserted into an EcoRI site of the donor vector of a Gateway system including a bar gene; the donor fragment was recombined into a descendant vector of pB7WG2.0 using the LR Gateway reaction, with an LR Clonase enzyme kit (Invitrogen).
Maize Transformation and Confirmation of Transgenic Lines
The transformation vector was introduced into Agrobacterium tumefaciens strain LBA4404 with the freeze-thaw method. The A. tumefaciens-induced maize shoot-tip transformation was performed as described by Li et al. (2011). The maize inbred line DH4866 was used as the plant receptor. T1 transgenic plants were detected using Basta herbicide (0.4% [v/v] effective concentration). The plants showing Basta resistance were selected for self-pollination. T2 and T3 transgenic plants were self-pollinated to produce progeny. In addition, two zmbzip4 UFMu mutants with different insertion sites were obtained from the Maize Genetics Cooperation Stock Center and self-pollinated to obtain homozygotes. Total RNA was extracted from leaves of the transgenic plants at the three-leaf stage. The transcription level was detected using RT-qPCR as described previously.
Polyclonal Antibody of ZmbZIP4
Full-length ZmbZIP4 was cloned by PCR, and the products were digested by BamHI/EcoRI and connected into the pGEX-4T-2; then, the plasmid was transformed into the BL21 strain. The prokaryotic expression strain was cultivated, and the expression of fusion protein ZmbZIP4-GST was induced as described on pages 1221 to 1231 in Molecular Cloning: A Laboratory Manual (III)(Sambrock and Russel, 2001). The purification of the soluble ZmbZIP4-GST protein was performed with Chelating Sepharose Fast Flow (GE Healthcare). The preparation of the polyclonal antibody was performed by Abmart.
The Drought and Salt Stress Experiment
Maize seeds (the transgenic lines, the zmbzip4 mutants, and their origin lines) were surface sterilized and germinated on moist filter paper in sterile culture flasks (28°C, dark). The filter papers were soaked with NaCl solutions of different concentrations (0, 100, 150, and 200 mm) and PEG solutions of different concentrations (0% [m/v], 12%/−0.36 MPa, and 18%/−0.77 MPa). The PEG solutions were measured with an osmometer (Fiske Micro-Osmometer model 210) to quantify osmolyte concentrations (mol L−1). Water potential was calculated using the following equation: water potential [MPa] = − (concentration [mol L−1]) * gas constant [8.314 Pa * L/(mol * K)] * temperature [298.15 K] (Opitz et al., 2014). Fifty seeds were used as one sample, and each treatment was repeated three times.
For the PEG and NaCl treatments, seedlings of DH4866 at the three-leaf stage were cultured with hydroponics for 4 d with Hoagland nutrient solution supplemented with a 14% (m/v) PEG6000 (−0.49 MPa) or a 100 mm NaCl (−0.4 MPa) solution. The nutrient solutions were aerated with a mini air pump and supplemented with fresh solution to maintain the volume. Then, the phenotypes of the plants were measured, and the leaf water potentials of the plants in the PEG treatments were measured.
For drought stress in soil, seeds were sown in a soil box (25 × 18 × 16 cm) and seedlings at the three-leaf stage were exposed to a drought stress treatment by stopping watering and sheltering them from rain. The leaf water potentials of the ZmbZIP4 transgenic lines and mutants were measured when the plants were treated for 6 d (soil water content decreased to 7.7%). After drought for 8 d and rewatering for 1 h, the recovered conditions of the overexpression lines and mutants were recorded and compared with those of their wild types. At last, the dry weights of the plants under normal and drought conditions were measured.
Determination of ABA Content in Maize
For each replicate, approximately 200 mg (fresh weight) of maize leaves or roots was homogenized under liquid nitrogen, weighed, and extracted with cold methanol and [2H6]ABA (internal standard; OlChemIm) for 24 h. Endogenous ABA was purified and measured as described previously (Fu et al., 2012) with some modifications in detection conditions. Liquid chromatography-tandem mass spectrometry analysis was performed on an ultra-performance liquid chromatography system (Waters) coupled to the 5500 Qtrap system (AB Sciex). Liquid chromatography separation used a BEH C18 column (1.7 mm, 2.1 × 150 mm; Waters) with mobile phase 0.05% HAc (A) and 0.05% HAc in ACN (B), and the gradient was set with initial 20% B and increased to 70% B within 6 min. ABA was detected in MRM mode with transition 263/153. Quantitation was done using the isotope dilution method (Fu et al., 2012).
Transactivation and One-Hybrid Assays in Yeast
For the transactivation assay, full-length ZmbZIP4 was inserted into the EcoRI and BamHI sites of pGBKT7-BD. The plasmid was transformed into the yeast YRG-2 strain (Agilent Stratagene) following the Yeast Protocols Handbook (Clontech). In addition, the β-galactosidase activities were examined by X-gal staining.
For the one-hybrid assays, the coding sequence of ZmbZIP4 was obtained by PCR, and the products were digested by EcoRI and BamHI and then inserted into the pGADT7-AD vector containing the GAL4 active domain. The promoters of the candidate target genes were cloned into the KpnI and XhoI sites of pLacZi. Plasmids were transformed in pairs in the yeast strain YM4271 (Invitrogen). The β-galactosidase activities were examined by X-gal staining and measured by o-nitrophenyl-β-d-galactopyranoside assay as described in the Yeast Protocols Handbook (Clontech).
To verify the effects of fusion protein ZmbZIP4-GFP on the transcript activation or the binding activity ability of ZmbZIP4, full-length ZmbZIP4-GFP, GFP, and ZmbZIP4 were inserted into the EcoRI and BamHI sites of pGBKT7-BD or pGADT7-AD for the one-hybrid yeast assay and transformed into YRG-2 or YM4271, respectively.
ChIP-Seq and ChIP-qPCR
The construct pCAMBIA1302-35S::ZmbZIP4:GFP was used for transformation of the young embryos of maize inbred line Qi319 via a gene gun (Bio-Rad). Stable transgenic maize and the anti-GFP antibody (ChIP-grade ab290 from Abcam) were used for the ChIP-Seq assay. ChIP was performed with the method described originally by Kaufmann et al. (2010) with minor modifications. In brief, 1.2 g of roots (V1 stage) of ZmbZIP4-GFP transgenic plants was collected for the ChIP assay. The nuclei suspensions were sheared to 200 to 500 bp by ultrasonic treatment (Bioruptor; Picoruptor; 15 cycles of 30 s on and 30 s off). Sequencing was performed with the Illumina HiSeq 4000 platform using aligner and parameter (Clean Parameter soap_mm_gz -p 4 -v 5 -s 35 -m 0 -x 600) and peak caller and parameter (Peak Calling Parameter macs14 -g 411831487 -p 1e-5 -w-space 50 -m 10, 30). The ChIP-Seq raw data have been uploaded to a public database: https://doi.org/10.6084/m9.figshare.6225806.v2.
The independent overexpression lines OX1 and OX2 and the input samples were used in the ChIP-qPCR assay, and all the samples were diluted to 10 ng µL−1 and reacted with 5 μL of SYBR Premix Ex Taq (2×), 0.2 μL of PCR forward primer (10 μm), 0.2 μL of PCR reverse primer (10 μm), and 1 μL of DNA template. The primers used to amplify the enriched region of the target genes are listed in Supplemental Table S3.
EMSA
EMSA was performed referenced to the DIG Gel Shift Kit, 2nd Generation (Roche). The labeled 50-bp key fragments or excess unlabeled fragments of the promoter interacted with ZmbZIP4-GST and poly(dI-dC) at 25°C for 1 h. The samples were subjected to electrophoresis under 80 V on 8% PAGE gels running with 0.5× TEA buffer at 4°C in the dark for 1 h. Then, the generated chemiluminescent signals were recorded on an imaging device according to the methods of the DIG Gel Shift Kit, 2nd Generation (Roche).
Luciferase Assay
Promoter fragments (sequences were referenced to the maize genome [https://www.maizegdb.org/], and the primers are listed in Supplemental Table S3) in the pGreenII 0800:Luc vector were cotransformed with 35S:ZmbZIP4 into the maize protoplasts. The isolation of protoplasts and transformation were performed according to the protocol described previously (Yoo et al., 2007; Cao et al., 2014). Protoplasts were harvested by centrifugation at 100g for 1 to 2 min after incubation for 18 h. For the ABA treatment, after incubation of the transfected protoplasts for 14 h, half of the protoplasts were treated with 100 μm ABA and then incubated for another 4 h. The luciferase activity (FLuc/Rluc) was measured after cell lysis using the Double-Luciferase Reporter Assay Kit (TransDetect).
Accession Numbers
Sequence data from this article can be found in the MaizeGDB data libraries (https://www.maizegdb.org/) under the accession numbers of the genes listed in Supplemental Table S3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Assay of the transcription factor activity in yeast.
Supplemental Figure S2. Germination rates and solute potentials of the ZmbZIP4 overexpression lines, zmbzip4 mutants, and wild-type maize under stress conditions.
Supplemental Figure S3. Phenotypes of the ZmbZIP4 transgenic lines and the mutant under drought stress conditions.
Supplemental Figure S4. Determination of the core binding motif by yeast one-hybrid analysis of ZmbZIP4.
Supplemental Figure S5. Improvement of transactivation activities of ZmbZIP4 by ABA treatment.
Supplemental Table S1. Different cis-acting elements in the promoters of ZmbZIP4, OsbZIP23, and ABF2.
Supplemental Table S2. Some target genes of ZmbZIP4 in maize.
Supplemental Table S3. Primers used in this study.
Supplement Data Set S1. Peptide and promoter sequences of ZmbZIP4.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jinfang Chu (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for help with the determination of ABA content. We thank the Maize Genetics Cooperation Stock Center for the maize mutants.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant no. 31571674 to J.Z.) and National Major Projects for Genetically Modified Organisms Breeding in China (grant no. 2016ZX08003004-003 to J.Z.).
Articles can be viewed without a subscription.
References
- Alexandre C, Möller-Steinbach Y, Schönrock N, Gruissem W, Hennig L (2009) Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol Plant 2: 675–687 [DOI] [PubMed] [Google Scholar]
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Peirats-Llobet M, Pizzio GA, Fernandez MA, De Winne N, De Jaeger G, Dietrich D, Bennett MJ, et al. (2013) PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol 161: 931–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arase F, Nishitani H, Egusa M, Nishimoto N, Sakurai S, Sakamoto N, Kaminaka H (2012) IAA8 involved in lateral root formation interacts with the TIR1 auxin receptor and ARF transcription factors in Arabidopsis. PLoS ONE 7: e43414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert Y, Vile D, Pervent M, Aldon D, Ranty B, Simonneau T, Vavasseur A, Galaud JP (2010) RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol 51: 1975–1987 [DOI] [PubMed] [Google Scholar]
- Bao F, Du D, An Y, Yang W, Wang J, Cheng T, Zhang Q (2017) Overexpression of Prunus mume dehydrin genes in tobacco enhances tolerance to cold and drought. Front Plant Sci 8: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner F, Oreb M, Mendel RR (2001) ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J Biol Chem 276: 40381–40384 [DOI] [PubMed] [Google Scholar]
- Bruno L, Pacenza M, Forgione I, Lamerton LR, Greco M, Chiappetta A, Bitonti MB (2017) In Arabidopsis thaliana cadmium impact on the growth of primary root by altering SCR expression and auxin-cytokinin cross-talk. Front Plant Sci 8: 1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Yao D, Lin F, Jiang M (2014) PEG-mediated transient gene expression and silencing system in maize mesophyll protoplasts: a valuable tool for signal transduction study in maize. Acta Physiol Plant 36: 1271–1281 [Google Scholar]
- Chakrabortee S, Boschetti C, Walton LJ, Sarkar S, Rubinsztein DC, Tunnacliffe A (2007) Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc Natl Acad Sci USA 104: 18073–18078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Corrêa LG, Riaño-Pachón DM, Schrago CG, dos Santos RV, Mueller-Roeber B, Vincentz M (2008) The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS ONE 3: e2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Ramírez A, Díaz-Triviño S, Blilou I, Grieneisen VA, Sozzani R, Zamioudis C, Miskolczi P, Nieuwland J, Benjamins R, Dhonukshe P, et al. (2012) A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150: 1002–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Deak KI, Malamy J (2005) Osmotic regulation of root system architecture. Plant J 43: 17–28 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433 [DOI] [PubMed] [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18: 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Chu J, Sun X, Wang J, Yan C (2012) Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Anal Sci 28: 1081–1087 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Ghelis T, Dellis O, Jeannette E, Bardat F, Cornel D, Miginiac E, Rona JP, Sotta B (2000) Abscissic acid specific expression of RAB18 involves activation of anion channels in Arabidopsis thaliana suspension cells. FEBS Lett 474: 43–47 [DOI] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte WR Jr, Quatrano RS (1990) A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250: 267–271 [DOI] [PubMed] [Google Scholar]
- Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A (2002) Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol 43: 136–140 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T (2002) Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14: 2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JH, Seah SW, Xu J (2013) The root of ABA action in environmental stress response. Plant Cell Rep 32: 971–983 [DOI] [PubMed] [Google Scholar]
- Humbert S, Subedi S, Cohn J, Zeng B, Bi YM, Chen X, Zhu T, McNicholas PD, Rothstein SJ (2013) Genome-wide expression profiling of maize in response to individual and combined water and nitrogen stresses. BMC Genomics 14: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K (2005) RARTF: database and tools for complete sets of Arabidopsis transcription factors. DNA Res 12: 247–256 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333 [DOI] [PubMed] [Google Scholar]
- Kakumanu A, Ambavaram MM, Klumas C, Krishnan A, Batlang U, Myers E, Grene R, Pereira A (2012) Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol 160: 846–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Østerås M, Farinelli L, Krajewski P, Angenet GC (2010) Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat Protoc 5: 457–472 [DOI] [PubMed] [Google Scholar]
- Kim S, Kang JY, Cho DI, Park JH, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40: 75–87 [DOI] [PubMed] [Google Scholar]
- Koizumi K, Hayashi T, Gallagher KL (2012) SCARECROW reinforces SHORT-ROOT signaling and inhibits periclinal cell divisions in the ground tissue by maintaining SHR at high levels in the endodermis. Plant Signal Behav 7: 1573–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A, Zaltsman A, Kozlovsky SV, Tian GW, Citovsky V (2009) Regulation of root elongation by histone acetylation in Arabidopsis. J Mol Biol 385: 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163 [DOI] [PubMed] [Google Scholar]
- Lång V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20: 951–962 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM (1995) Formation of lateral root meristems is a two-stage process. Development 121: 3303–3310 [DOI] [PubMed] [Google Scholar]
- Lee SA, Jang S, Yoon EK, Heo JO, Chang KS, Choi JW, Dhar S, Kim G, Choe JE, Heo JB, et al. (2016) Interplay between ABA and GA modulates the timing of asymmetric cell divisions in the Arabidopsis root ground tissue. Mol Plant 9: 870–884 [DOI] [PubMed] [Google Scholar]
- Li Z, Gao Q, Liu Y, He C, Zhang X, Zhang J (2011) Overexpression of transcription factor ZmPTF1 improves low phosphate tolerance of maize by regulating carbon metabolism and root growth. Planta 233: 1129–1143 [DOI] [PubMed] [Google Scholar]
- Lim J, Jung JW, Lim CE, Lee MH, Kim BJ, Kim M, Bruce WB, Benfey PN (2005) Conservation and diversification of SCARECROW in maize. Plant Mol Biol 59: 619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang Q, Yu M, Zhang Y, Wu Y, Zhang H (2008) Transgenic salt-tolerant sugar beet (Beta vulgaris L.) constitutively expressing an Arabidopsis thaliana vacuolar Na/H antiporter gene, AtNHX3, accumulates more soluble sugar but less salt in storage roots. Plant Cell Environ 31: 1325–1334 [DOI] [PubMed] [Google Scholar]
- Liu H, Tang R, Zhang Y, Wang C, Lv Q, Gao X, Li W, Zhang H (2010) AtNHX3 is a vacuolar K+/H+ antiporter required for low-potassium tolerance in Arabidopsis thaliana. Plant Cell Environ 33: 1989–1999 [DOI] [PubMed] [Google Scholar]
- Lu G, Gao C, Zheng X, Han B (2009) Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229: 605–615 [DOI] [PubMed] [Google Scholar]
- Magnan F, Ranty B, Charpenteau M, Sotta B, Galaud JP, Aldon D (2008) Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J 56: 575–589 [DOI] [PubMed] [Google Scholar]
- Malamy JE. (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ 28: 67–77 [DOI] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Moriwaki T, Miyazawa Y, Kobayashi A, Takahashi H (2013) Molecular mechanisms of hydrotropism in seedling roots of Arabidopsis thaliana (Brassicaceae). Am J Bot 100: 25–34 [DOI] [PubMed] [Google Scholar]
- Mundy J, Yamaguchi-Shinozaki K, Chua NH (1990) Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc Natl Acad Sci USA 87: 1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva C, Busk PK, Domínguez-Puigjaner E, Lumbreras V, Testillano PS, Risueño MC, Pagès M (2005) Isolation and functional characterisation of two new bZIP maize regulators of the ABA responsive gene rab28. Plant Mol Biol 58: 899–914 [DOI] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV (2001) Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol 45: 263–279 [DOI] [PubMed] [Google Scholar]
- Ober ES, Sharp RE (1994) Proline accumulation in maize (Zea mays L.) primary roots at low water potentials. I. Requirement for increased levels of abscisic acid. Plant Physiol 105: 981–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober ES, Sharp RE (2003) Electrophysiological responses of maize roots to low water potentials: relationship to growth and ABA accumulation. J Exp Bot 54: 813–824 [DOI] [PubMed] [Google Scholar]
- Olatunji D, Geelen D, Verstraeten I (2017) Control of endogenous auxin levels in plant root development. Int J Mol Sci 18: E2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz N, Paschold A, Marcon C, Malik WA, Lanz C, Piepho HP, Hochholdinger F (2014) Transcriptomic complexity in young maize primary roots in response to low water potentials. BMC Genomics 15: 741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz N, Marcon C, Paschold A, Malik WA, Lithio A, Brandt R, Piepho HP, Nettleton D, Hochholdinger F (2016) Extensive tissue-specific transcriptomic plasticity in maize primary roots upon water deficit. J Exp Bot 67: 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P, Fukaki H, Beeckman T (2010) Auxin control of root development. Cold Spring Harb Perspect Biol 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Park PJ. (2009) ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet 10: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Seok HY, Park BK, Kim SH, Goh CH, Lee BH, Lee CH, Moon YH (2008) Overexpression of Arabidopsis ZEP enhances tolerance to osmotic stress. Biochem Biophys Res Commun 375: 80–85 [DOI] [PubMed] [Google Scholar]
- Park SC, Kim YH, Jeong JC, Kim CY, Lee HS, Bang JW, Kwak SS (2011) Sweetpotato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic- and salt stress-tolerance of transgenic calli. Planta 233: 621–634 [DOI] [PubMed] [Google Scholar]
- Promchuea S, Zhu Y, Chen Z, Zhang J, Gong Z (2017) ARF2 coordinates with PLETHORAs and PINs to orchestrate ABA-mediated root meristem activity in Arabidopsis. J Integr Plant Biol 59: 30–43 [DOI] [PubMed] [Google Scholar]
- Saab IN, Sharp RE, Pritchard J, Voetberg GS (1990) Increased endogenous abscisic acid maintains primary root growth and inhibits shoot growth of maize seedlings at low water potentials. Plant Physiol 93: 1329–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saab IN, Sharp RE, Pritchard J (1992) Effect of inhibition of abscisic acid accumulation on the spatial distribution of elongation in the primary root and mesocotyl of maize at low water potentials. Plant Physiol 99: 26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S, Heidstra R, Wildwater M, Scheres B (2003) SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev 17: 354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sambrock J, Russel DW (2001) Molecular Cloning: A Laboratory Manual (3rd edition). Immunology 49: 895–909
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T (2007) Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814 [DOI] [PubMed] [Google Scholar]
- Seeve CM, Cho IJ, Hearne LB, Srivastava GP, Joshi T, Smith DO, Sharp RE, Oliver MJ (2017) Water-deficit-induced changes in transcription factor expression in maize seedlings. Plant Cell Environ 40: 686–701 [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJ, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JA, Koornneef M, Kamiya Y, Koshiba T (2000) The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA 97: 12908–12913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Aoki H, Koiwai H, Kamiya Y, Nambara E, Koshiba T (2004) Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol 45: 1694–1703 [DOI] [PubMed] [Google Scholar]
- Shkolnik-Inbar D, Bar-Zvi D (2010) ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 22: 3560–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DL, Fedoroff NV (1995) LRP1, a gene expressed in lateral and adventitious root primordia of Arabidopsis. Plant Cell 7: 735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN (2010) Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JA, McCarty DR (1997) Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA 94: 12235–12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Parker RA, Morpeth DR, Burbidge A, Taylor IB (2000) Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid. Plant Mol Biol 42: 833–845 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian N, Marcotte L, Giraudat J (1994) Drought rhizogenesis in Arabidopsis thaliana: differential responses of hormonal mutants. Plant Physiol 104: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Hua D, He J, Duan Y, Chen Z, Hong X, Gong Z (2011) Auxin Response Factor2 (ARF2) and its regulated homeodomain gene HB33 mediate abscisic acid response in Arabidopsis. PLoS Genet 7: e1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilewska A, Vlad F, Sirichandra C, Redko Y, Jammes F, Valon C, Frei dit Frey N, Leung J (2008) An update on abscisic acid signaling in plants and more.... Mol Plant 1: 198–217 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Spollen WG, Sharp RE, Hetherington PR, Fry SC (1994) Root growth maintenance at low water potentials: increased activity of xyloglucan endotransglycosylase and its possible regulation by abscisic acid. Plant Physiol 106: 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Thorne ET, Sharp RE, Cosgrove DJ (2001) Modification of expansin transcript levels in the maize primary root at low water potentials. Plant Physiol 126: 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Ishitani M, Lee H, Zhu JK (2001) The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Wang RG, Mao G, Koczan JM (2006) Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiol 142: 1065–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Jia L, Shi W, Liang J, Zhou F, Li Q, Zhang J (2013) Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytol 197: 139–150 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Sharp RE (2010) Complexity and coordination of root growth at low water potentials: recent advances from transcriptomic and proteomic analyses. Plant Cell Environ 33: 590–603 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Ying S, Zhang DF, Fu J, Shi YS, Song YC, Wang TY, Li Y (2012) Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 235: 253–266 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Zhang J, Yu H, Zhang Y, Wang Y, Li M, Zhang J, Duan L, Zhang M, Li Z (2016) Increased abscisic acid levels in transgenic maize overexpressing AtLOS5 mediated root ion fluxes and leaf water status under salt stress. J Exp Bot 67: 1339–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wollenweber B, Jiang D, Liu F, Zhao J (2008a) Water deficits and heat shock effects on photosynthesis of a transgenic Arabidopsis thaliana constitutively expressing ABP9, a bZIP transcription factor. J Exp Bot 59: 839–848 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008b) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang X, Lin Z, Wang J, Xu M, Lai J, Yu J, Lin Z (2018) The genetic architecture of nodal root number in maize. Plant J 93: 1032–1044 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Xing L, Wang X, Hou YJ, Gao J, Wang P, Duan CG, Zhu X, Zhu JK (2014) The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci Signal 7: ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W, Tang N, Yang J, Peng L, Ma S, Xu Y, Li G, Xiong L (2016) Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol 171: 2810–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]